Abstract
Loss-of-function mutations in the X-linked endosomal Na+/H+ exchanger 6 (NHE6) cause Christianson syndrome in males. Christianson syndrome involves endosome dysfunction leading to early cerebellar degeneration, as well as later-onset cortical and subcortical neurodegeneration, potentially including tau deposition as reported in post-mortem studies. In addition, there is reported evidence of modulation of amyloid-β levels in experimental models wherein NHE6 expression was targeted. We have recently shown that loss of NHE6 causes defects in endosome maturation and trafficking underlying lysosome deficiency in primary mouse neurons in vitro. For in vivo studies, rat models may have an advantage over mouse models for the study of neurodegeneration, as rat brain can demonstrate robust deposition of endogenously-expressed amyloid-β and tau in certain pathological states. Mouse models generally do not show the accumulation of insoluble, endogenously-expressed (non-transgenic) tau or amyloid-β. Therefore, to study neurodegeneration in Christianson syndrome and the possibility of amyloid-β and tau pathology, we generated an NHE6-null rat model of Christianson syndrome using CRISPR-Cas9 genome-editing. Here, we present the sequence of pathogenic events in neurodegenerating NHE6-null male rat brains across the lifespan. NHE6-null rats demonstrated an early and rapid loss of Purkinje cells in the cerebellum, as well as a more protracted neurodegenerative course in the cerebrum. In both the cerebellum and cerebrum, lysosome deficiency is an early pathogenic event, preceding autophagic dysfunction. Microglial and astrocyte activation also occur early. In the hippocampus and cortex, lysosome defects precede loss of pyramidal cells. Importantly, we subsequently observed biochemical and in situ evidence of both amyloid-β and tau aggregation in the aged NHE6-null hippocampus and cortex (but not in the cerebellum). Tau deposition is widely distributed, including cortical and subcortical distributions. Interestingly, we observed tau deposition in both neurons and glia, as has been reported in Christianson syndrome post-mortem studies previously. In summary, this experimental model is among very few examples of a genetically modified animal that exhibits neurodegeneration with deposition of endogenously-expressed amyloid-β and tau. This NHE6-null rat will serve as a new robust model for Christianson syndrome. Furthermore, these studies provide evidence for linkages between endolysosome dysfunction and neurodegeneration involving protein aggregations, including amyloid-β and tau. Therefore these studies may provide insight into mechanisms of more common neurodegenerative disorders, including Alzheimer’s disease and related dementias.
Keywords: rat model, lysosomes, neurodegeneration, tau, amyloid beta
Mutations in the endosomal Na+/H+ exchanger 6 (NHE6) cause Christianson syndrome. Lee et al. use CRISPR-Cas9 genome editing to generate a rat model of Christianson syndrome. This model displays early endo-lysosomal defects followed by neurodegeneration including aggregation of endogenous tau and amyloid beta.
Introduction
Defects in the endolysosomal system have been linked with neurodegenerative disorders as well as neurodevelopmental disorders.1–3 Christianson syndrome is a monogenic endosomal disorder that exhibits neurodevelopmental and neurodegenerative pathologies. Christianson syndrome is caused by loss-of-function mutations in the X-linked, endosomal Na+/H+ exchanger 6 (NHE6, encoded by SLC9A6). Male patients with Christianson syndrome exhibit postnatal microcephaly, developmental delay, lack of speech, epilepsy and progressive cerebellar ataxia.4–6 In addition to neurodevelopmental pathologies, Christianson syndrome involves neurodegenerative pathologies, including cerebellar degeneration, which may lead to inability to walk.5 Further, neurodegenerative disease with diffuse tau aggregation has been identified in Christianson syndrome patients, although post-mortem studies to date have been limited.7 Garbern et al.7 reported widespread cortical and subcortical neuronal loss, gliosis and neuronal and glial tau deposition, reminiscent of corticobasal degeneration. Additionally, we and others have found that a subset of females who are NHE6 mutation carriers may be diagnosed clinically with disorders that are associated with tau deposition, such as corticobasal degeneration, progressive supranuclear palsy and atypical parkinsonism.8,9 Notably, in post-mortem human brains from normal and pathological ageing studies, decreased NHE6 expression was correlated with greater tau deposition.8
NHE6 is an endosomal transmembrane protein that functions in regulating the pH of the endosomal lumen as well as in endosomal trafficking and signalling.10,11 The vacuolar ATPase acidifies endosomes by actively pumping protons into the endosomes, while NHE6 permits proton leak from the lumen to tightly regulate endosomal acidification and maturation of the lysosome. NHE6 is abundantly expressed in axons and dendrites and regulates synaptic development and plasticity.10,12 Loss of NHE6 leads to the over-acidification of endosomes and decreases in neuronal arborization.10 Our recent study of NHE6 function in primary neurons in vitro demonstrates that loss of NHE6 leads to defects in endosome maturation and trafficking, contributing to lysosome deficiency.11 NHE6-null mouse models exhibit reduced brain volume, increased gliosis, cerebellar degeneration and accumulation of lysosomal substrates in neurons.13–15 Mouse models of neurodegenerative disorders, including of Alzheimer’s disease, do not generally display deposition of endogenously expressed tau,16–18 and mouse models of Christianson syndrome also do not exhibit tau deposition.13 One study reports increases in amyloid-β by ELISA in the NHE6-null mouse brain,19 although an increase in amyloid-β plaques are not reported in human post-mortem studies of Christianson syndrome.7 Further still, in a recent study by Pohlkamp et al.,20 decreases in amyloid-β plaque load are observed in a transgenic mouse model of early-onset Alzheimer’s disease with loss of NHE6. Overall, additional in vivo study of amyloid-β deposition in NHE6-null brain is warranted, particularly in systems with endogenously-expressed (non-transgenic or mutant) amyloid-β species.
In this study, we genetically-engineered a rat model to study neurodegenerative pathologies associated with loss of NHE6 in the ageing brain, including tau deposition. Rats are more genetically and physiologically similar to humans,21–23 and exhibit deposition of endogenously expressed tau and amyloid-β in some pathological situations.24–27 Here, we define the sequence of pathogenic events leading to neurodegeneration in the NHE6-null brain (Supplementary Fig. 1). We observe early and rapid degeneration in the cerebellum, and a more protracted course of neurodegeneration in the cerebrum. In both the cerebellum and in the cerebrum, we observe early evidence of lysosome dysfunction which precedes neuronal loss. In the hippocampus and cortex, lysosome defects occur early, which precede subsequent autophagic defects and evidence of tau deposition, as well as amyloid-β aggregates in aged mutant rat brains. Interestingly, tau deposits were found in cortical and subcortical neurons and glial, consistent with prior post-mortem studies. In summary, we demonstrate that this new NHE6-null rat will serve as a robust model for Christianson syndrome. In addition, these studies have broad impact for investigating the potential linkages between endolysosome dysfunction and protein aggregation in other neurodegenerative disorders including amyloid-β and tau-related disorders such as Alzheimer’s disease.
Materials and methods
Animal procedures and genotyping
The NHE6-null rat model was generated on a Sprague-Dawley background using CRISPR/Cas9 (GenOway). Guide RNA sequence is the following: 5′-CGGCTGTGTAACCCTGATGA-3′. Cas9-mediated cleavage at exon 7 in the Slc9a6 locus resulted in the insertion of 2 bp, generating a frameshifts and premature stop codon. The Slc9a6 rat gene located on the X chromosome is composed of 18 exons. ATG initiation codons are located in exons 1, 3 and 4. We targeted exon 7, which is the most upstream exon among the other possible exons to disrupt the expression of all isoforms. Two male founders were generated and used for breeding. The wild-type and knock-out alleles were sequenced for genotyping and the mRNA level was confirmed by quantitative real-time PCR. Loss of NHE6 protein expression was validated by western blot. As Christianson syndrome is an X-linked condition affecting males, only male rats were used for this study. We do not observe any phenotypic differences in offspring from each of the different founders. NHE6-null mice generation has been previously reported.10 Only male mice at 24 months were used. All animal work was conducted under the guidelines of the Center for Animal Resources and Education (CARE) with a protocol (IACUC 18-11-0002) approved by the Brown University and UC Davis Institutional Animal Care and Use Committee. For the cohort for behavioural studies at UC Davis, male NHE6-null rats were shipped from Brown University, and then litters were bred at UC Davis. All experimental procedures were consistent with the US National Institutes of Health Guide for the Care and Use of Laboratory Animals (National Research Council, 8th edition). Rat tails were clipped and externally genotyped (Transnetyx).
For the UC Davis Cohort, NHE6-null males were bred with wild-type Sprague-Dawley females purchased from Envigo (East Millstone) in a conventional rat vivarium at UC Davis. From the resulting nine litters, male null and male wild-type littermates were selected for behavioural testing. To identify rats, pups were given paw tattoos on postnatal Day 2 using non-toxic animal tattoo ink (Ketchum Manufacturing Inc). Rats were given additional identifying marks on the tail at weaning using non-toxic permanent marker. Tattoos and tail marks were coded to allow investigators to carry out and score behaviours blind to genotype. At postnatal Day 2, a small tissue sample from the tail was collected for genotyping, which was carried out by Transnetyx.
Tissue preparation for sequential tau and amyloid-β extraction
Wild-type and NHE6-null rats were euthanized with CO2 and brains were removed, dissected and cut in half. Each hemisphere was weighed, snap frozen and homogenized for amyloid-β and tau extraction. More specifically, the right hemisphere was processed for sequential tau extraction, while the left hemisphere was processed for amyloid-β ELISA assays. Male wild-type and NHE6-null mice10 at 24 months were processed in the same procedures.
Sequential tau extraction
Sequential tau extraction method was modified from previously published study.28 More details are described in the Supplementary material.
Amyloid-β extraction and ELISA assay
This method is described in the previously published study.29 More details are described in the Supplementary material.
ThioflavinS staining
ThioflavinS staining is previously described.30 Briefly, brain sections were mounted on slides and air dried at room temperature for overnight. The slides were incubated in 0.05% ThioflavinS (Sigma #T1892) in 50% ethanol for 8 min in dark. ThioflavinS solution was always freshly prepared and filtered before the procedure. The slides were transferred to 80% ethanol for 15 s and washed in tap water for 1 min. The sections were washed with PBS before 2 h blocking with 10% normal goat serum (NGS) in PBS with 0.25% Triton X-100 (PBS-X; Sigma #T8787) at room temperature. The sections were incubated with NeuN primary antibody (1:500) for overnight at 4°C. After washing with PBS, the sections were incubated with Alexa Fluor 633 secondary antibodies (1:500, Invitrogen) diluted in 10% NGS in PBS-X for 2 h. The sections were washed with Tris-buffered saline (TBS) then coverslipped with Flouromount-G mounting solution.
Statistical analysis
Statistical tests for each experiment are provided in the main text as well as the figure legends. Two-tailed unpaired t-test with Welch’s correction was performed for comparison between two groups. To determine time-dependent effect on more than two groups, two-way ANOVA was performed. Detailed information about statistical analysis including post hoc analysis and F-values is described in the Supplementary material.
Data availability
All results in the main figures and supplementary figures are available upon request. Detailed methods are also described in the Supplementary material.
Results
Generation of NHE6-null rats using CRISPR/Cas9 genome editing
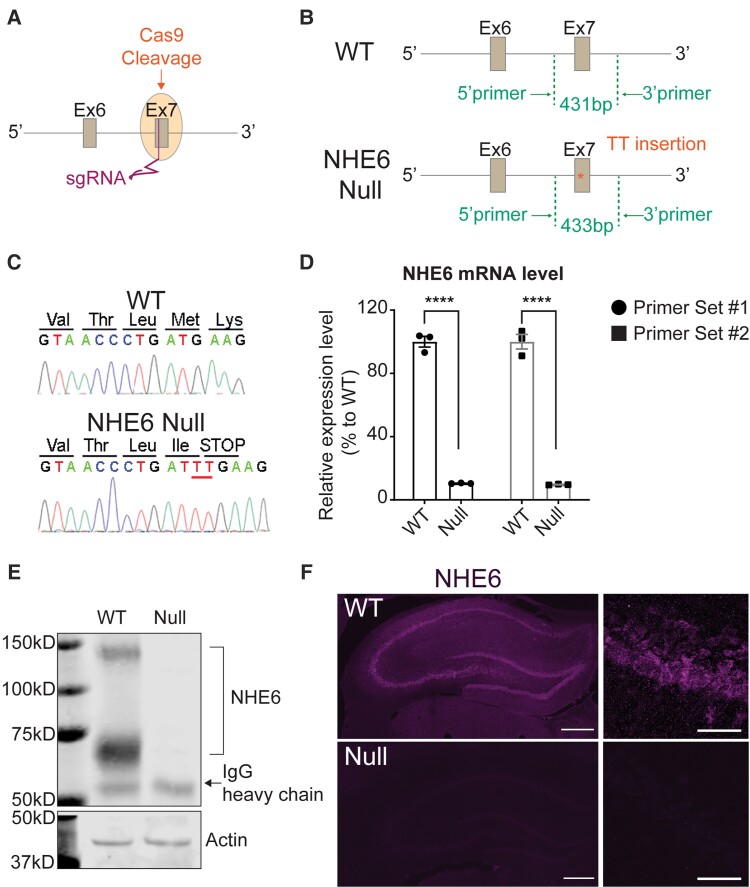
We generated NHE6-null mutant rats on a Sprague-Dawley background by CRISPR/Cas9 genome editing. Guide RNAs (sgRNA) targeting exon 7 of the rat Slc9a6 gene, which encodes NHE6, were microinjected into the pronucleus (Fig. 1A). Cas9-mediated cleavage at exon 7 led to a 2 base-pair (bp) TT insertion to generate a premature stop codon (Fig. 1B). Two independent mosaic male founders were recovered with the same TT insertion. Genomic DNA sequence, isolated from tail biopsy, shows the sequence in the wild-type rat and the 2-bp insertion predicted to cause a premature nonsense mutation in the Slc9a6 gene (Fig. 1C). This 2-bp insertion was successfully transmitted via the germ line. The edited mutant mRNA is predicted to be vulnerable to nonsense mediated mRNA decay, as has been observed consistently in Slc9a6 previously.31 We confirmed this by measuring the mRNA level in wild-type and NHE6-null rat brains (Fig. 1D). We also validated the absence of NHE6 protein in NHE6 null brains by western blot (Fig. 1E), as well as immunohistochemistry (Fig. 1F).
Figure 1.
Generation and validation of NHE6-null rats. (A) Two single guide RNAs (sgRNA) were inserted into exon 7 of the endogenous Slc9a6 gene. (B) Schematic representation of the targeted Slc9a6 locus harbouring the 2-bp (TT) insertion. The insertion generated a premature stop codon. (C) Sequences of wild-type (WT) and NHE6-null rats. Genomic DNA sequence, isolated from rat tail biopsy, shows the sequence in the wild-type rat and the 2-bp insertion causing a premature stop codon in NHE6. (D) The mRNA level of NHE6 in wild-type and NHE6-null rat brain was measured by quantitative real-time PCR. Two different sets of primers were used to validate the mRNA level. The mRNA level was normalized against the reference gene. Two-tailed unpaired t-test with Welch’s correction was used (P < 0.0001 for both primer sets, n = 3 for each genotype). (E) Absence of NHE6 protein in NHE6-null rat brain. Immunoprecipitation (IP) of NHE6 in the whole brain lysates from wild-type and NHE6-null rats validates the absence of NHE6 protein. Actin was used as a loading control. IgG Heavy chain was detected at ∼50 kDa after IP with NHE6 antibody. (F) Validation of NHE6 protein absence by immunofluorescence. Brain sections from wild-type and NHE6-null rats at 2 months were stained with NHE6 antibody. Scale bar = 500 µm.
Progressive neurodegeneration of NHE6-null rat brains
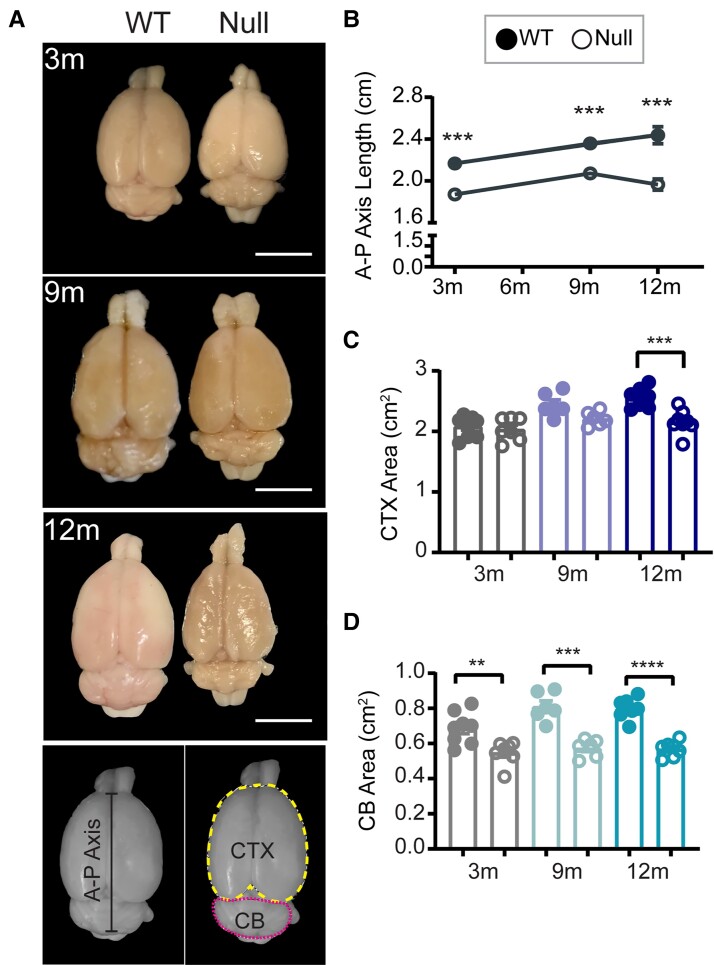
Data from ageing Christianson syndrome patients and NHE6-null mice demonstrate mixed neurodevelopmental and neurodegenerative pathology.5,8,13,14 To understand the trajectories of brain morphology changes in male NHE6-null rats, we measured the length of the anterior-posterior (A-P) axis of brains from wild-type and their NHE6-null littermates at 3, 9 and 12 months (Fig. 2A and B). The A-P length of 3-month-old NHE6-null rats was shorter than wild-type even though the brain size of NHE6-null rats continued to increase (Fig. 2B). After 9 months, the A-P length of NHE6-null continuously decreased compared to wild-type. To closely examine the overall brain size differences, we also measured the area of the cerebral cortex (CTX) and the cerebellum. The CTX area of NHE6-null rats was significantly reduced at 12 months compared to wild-type (Fig. 2C). By comparison, the cerebellum area of NHE6-null rats was smaller than wild-type as early as 3 months (Fig. 2D). Overall, these data are consistent with early neurodegeneration of the cerebellum and a more protracted timewise degeneration of structures within the cerebrum.
Figure 2.
Brain length and size analysis of NHE6-null rats across the lifespan. (A) Representative images of wild-type (WT) and NHE6-null male rats at 3, 6, 12 months (m). Scale bar = 500 µm. The length of the anterior-posterior (A-P) axis was measured at the interhemisphere divided from the tip of cerebrum to the end of cerebellum (CB) as indicated in the panel. The dotted lines indicate the measured area of the CTX (yellow) and cerebellum (magenta). (B) The A-P length of NHE6 was shorter than wild-type at 3 months (P < 0.0001). The length of wild-type and NHE6-null rat brain continued to increase until 9 months (P < 0.0001). At 12 months, the A-P length of NHE6-null rat brain was significantly decreased compared to that of 12-month-old wild-type (P = 0.0006; two-tailed unpaired t-test for each time point). (C) The CTX area of NHE6-null decreased at 12 months compared to that of wild-type (two-tailed unpaired t-test with Welch’s correction, P = 0.0009). (D) The cerebellum area of NHE6-null was smaller than that of wild-type across the lifespan (P = 0.0044 for 3 months, P = 0.0002 for 9 months, P < 0.0001 for 12 months, two-tailed unpaired t-test with Welch’s correction). The number of animals was for B–D: 3-month-old wild-type = 8, 3-month-old Null = 7, 9-month-old wild-type = 6, 9-month-old Null = 6, 12-month-old wild-type = 7, 12-month-old Null = 8. Data are presented as mean ± SEM. Asterisks represent P-values as follows: *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001, ****P < 0.0001.
Early cerebellar degeneration in NHE6-null rats
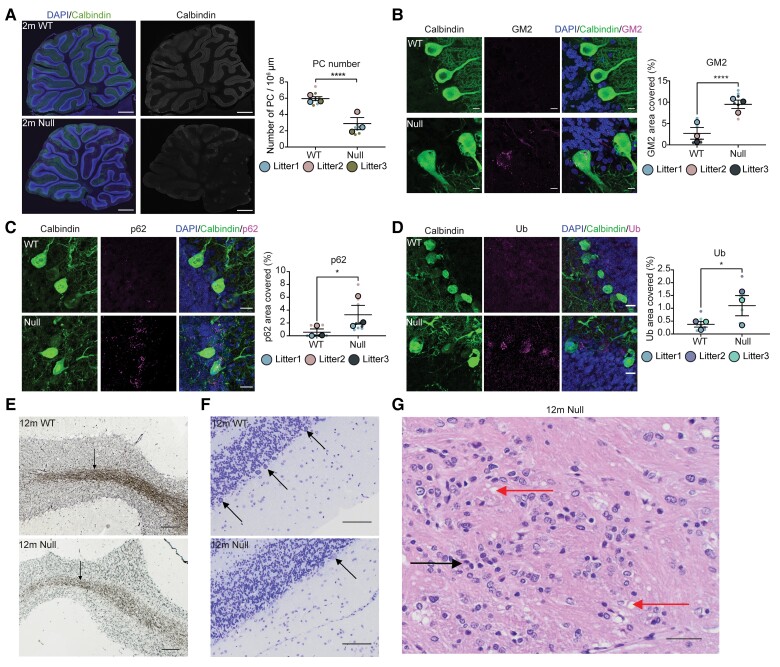
Cerebellar degeneration is a prominent feature in patients with Christianson syndrome.5 On gross examination in the NHE6-null rat, we observed reductions in cerebellar size as early as 3 months (Fig. 2D). To investigate cerebellar pathology, we stained sagittal cerebellar sections with calbindin, which labels Purkinje cells from wild-type and NHE6-null rats. At 2 months, we observed a significant reduction of calbindin staining in NHE6-null rats compared to wild-type (Fig. 3A). In terms of pathophysiological mechanisms, prior studies have demonstrated neuropathological hallmarks of lysosomal dysfunction, such as accumulation of GM2 ganglioside in NHE6-null mouse brains.14,15 To investigate GM2 ganglioside accumulation in the cerebellum in the NHE6-null rat, the cerebellar sections from wild-type and NHE6-null rats at 2 months were co-stained with calbindin and GM2 (Fig. 3B). Strikingly, GM2 was detected in most Purkinje cell of NHE6-null rats, while it was not observed in those of wild-types. In addition to defects in lysosome function, indicators of autophagy dysfunction were evident, including the accumulation of p62 and ubiquitin (Ub), which was observed in Purkinje cells at 2 months (Fig. 3C and D).
Figure 3.
Early cerebellar degeneration. (A) Sagittal cerebellar sections and quantification from 2-month-old wild-type (WT) and NHE6-null rats were stained with a Purkinje cell (PC) marker, calbindin along with DAPI. The staining of calbindin reduced in NHE6-null rats. Scale bar = 500 µm. The number of Purkinje cell cells was manually counted and divided by area (µm2). Two-tailed unpaired t-test with Welch’s correction was performed (P < 0.0001). (B) Cerebellar sections and its quantification from wild-type and NHE6-null rats at 2 months were stained with calbindin and GM2. GM2/calbindin-positive cells were detected only in NHE6-null rats. Scale bar = 10 µm. The quantification shows the per cent of area covered by GM2 (P < 0.0001). (C) Cerebellar sections were stained with p62 and calbindin. NHE6-null rats at 2 months showed increases in p62 staining (P = 0.02). (D) Cerebellar sections were stained with ubiquitin (Ub) and calbindin. The staining of Ub was increased in NHE6-null rats at 2 months (P = 0.0113). Two-tailed unpaired t-test with Welch’s correction was performed. Values from each brain section (three sections/each animal) are clustered in different colour codes according to each animal and plotted as a small dot. Means from each biological replicate are overlaid on the top of the full dataset as a bigger dot. P-value and standard error of the mean (SEM) were calculated using values from all sections from all animals (wild-type = 3, Null = 3) used in biological replicates. All data are presented as mean ± SEM. (E) Bielschowsky’s silver staining of cerebellum at 12 months. The staining in the paramedian lobule of cerebellum from NHE6-null rats prominently decreased compared to wild-type. Scale bar = 100 µm. (F) Nissl staining of paramedian lobule of cerebellum at 12 months. Arrows indicate Purkinje cell cells. The loss of Purkinje cell cells was observed in NHE6-null rats. Insets showed diffusing Nissl staining of Purkinje cell cells in NHE6-null rats. Scale bar = 100 µm. (G) Haematoxylin and eosin staining of wild-type and NHE6-null rats at 12 months. Multifocal vacuolization (red arrow) was observed in the paramedian lobule of cerebellum along with gliosis (black arrow) in NHE6-null rats. Scale bar = 100 µm.
While the neurodegeneration in the NHE6-null cerebellum is relatively rapid, we set out to sequence the pathophysiological events (Supplementary Fig. 1). Notably, we observed the reduction of Purkinje cell number in NHE6-null cerebellum and the increase in GM2 staining as early as 1 month (Supplementary Fig. 2A and B). However, no significant differences in p62 and Ub staining were observed (Supplementary Fig. 2C and D). These results suggest that the lysosomal pathology is primary in the NHE6-null cerebellum, occurring earliest and at the same time as the first Purkinje cell loss, and that defects in autophagy follow. To define the progress of cerebellar pathology and axonal loss, we performed Bielschowsky’s silver staining in cerebellum of NHE6-null rats at 12 months. Silver staining was profoundly decreased in NHE6-null cerebellum compared to wild-type reflecting axonal loss (Fig. 3E), along with reduced Nissl staining for Purkinje cell bodies (Fig. 3F). In addition, using haematoxylin and eosin staining in cerebellar sections from wild-type and NHE6-null rats at 12 months, we observed multifocal vacuolization in the molecular layer from the paramedian lobule of cerebellum in NHE6-null rats, suggesting endolysosomal pathology (Fig. 3G). In conclusion, we observed early lysosome dysfunction and Purkinje cell degeneration in the NHE6-null rat cerebellum.
Lysosome defects precede autophagy dysfunction in NHE6-null rat cerebrum
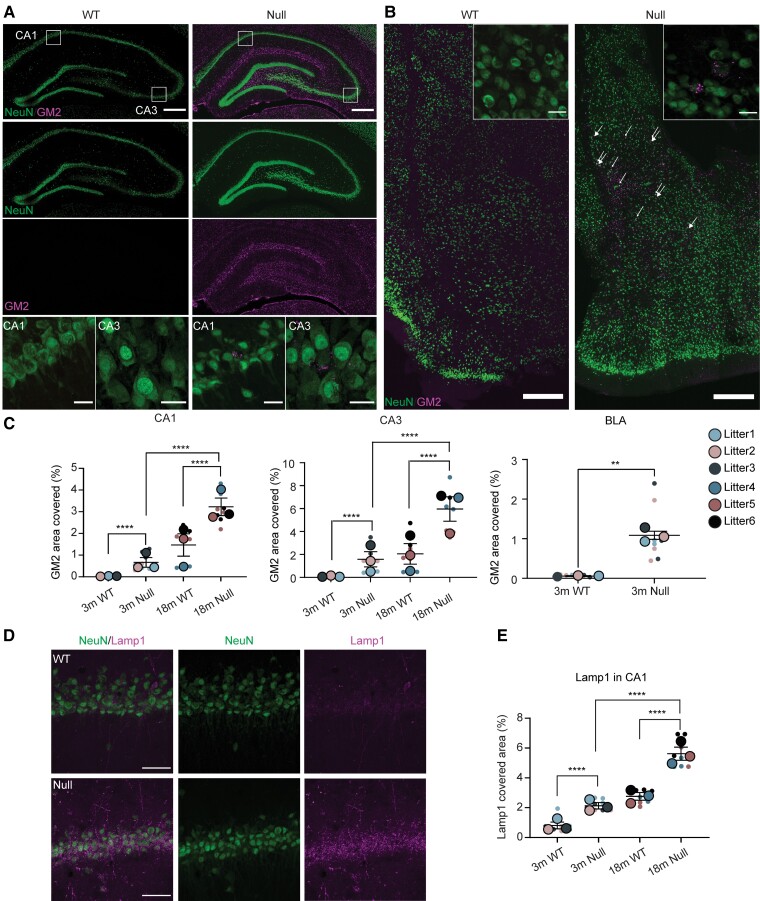
While the cerebellum demonstrates rapid neurodegeneration, neurodegeneration in the NHE6-null rat cerebrum demonstrated a more protracted time course. This longer time course permitted a clearer sequencing of neuropathological events (Supplementary Fig. 1). As in the cerebellum, endolysosomal dysfunction has been observed across the CNS in the Christianson syndrome mouse model, including accumulation of GM2.3,14,32 Christianson syndrome mice displayed abnormal GM2 ganglioside accumulation in the hippocampus (HP) and basolateral amygdala (BLA).14 To investigate the extent to which there is GM2 accumulation, and the timing of this accumulation in the hippocampus and BLA from NHE6-null rat, we co-stained the brain sections from wild-type and NHE6-null rats with GM2 and a neuronal marker, NeuN (Fig. 4A and B). GM2 prominently accumulates in neurons of the CA1, CA3 and BLA region of NHE6-null rats compared to wild-types (Fig. 4A and B) as early as 3 months. GM2 ganglioside was largely not detected in wild-types (Fig. 4A and B). GM2 accumulation increased prominently in the CA1 and CA3 at 18 months in the NHE6-null rat brain. To examine if this aberrant GM2 accumulation is associated with other features of lysosomal dysfunction, we stained the brain sections of NHE6-null and wild-type at 3 months and 18 months with a lysosomal marker, Lamp1, along with a neuronal marker, NeuN (Fig. 4D and E). NHE6-null rats showed the increase in Lamp1 staining at 3 months compared to wild-type. Age-dependent accumulation of Lamp1 was observed at 18-month-old NHE6-null rats.
Figure 4.
Lysosomal defects in NHE6-null rats. (A) GM2 ganglioside staining in the CA1 and CA3 of wild-type (WT) and NHE6-null rats at 3 months. Coronal brain sections were stained with GM2 (magenta) along with NeuN (green). In the CA1 and CA3 region, GM2 prominently accumulates in neurons of NHE6-null rats compared to wild-type. White boxes in the merged images indicate the location of magnified GM2 images. Scale bar = 500 µm; scale bar for magnified images = 50 µm. (B) GM2 staining in basolateral amygdala (BLA) region of wild-type and NHE6-null rats at 3 months. Brain sections were stained with GM2 (magenta) and NeuN (green). GM2 was detected in neurons in NHE6-null rats. The GM2 staining was prominently detected in neurons in NHE6-null rats. Arrows indicates GM2-accumulating neurons. Insets show GM2-positive neuron cells. Scale bar = 500 µm; scale bar for insets = 50 µm. (C) Quantification of GM2 covered area (%) in CA1 (P < 0.0001), CA3 (P < 0.0001) and BLA (P = 0.0006) regions of wild-type and NHE6-null rats from 3 and 18 months. Two-way ANOVA was performed followed by Tukey’s honestly significant difference (HSD) (wild-type = 3, Null = 3 for each time point). For BLA, unpaired t-test with Welch’s correction was conducted. (D) Lamp1 (lysosomal marker) staining with NeuN in wild-type and NHE6-null rats at 3 months and 18 months. The Lamp1 staining in the CA1 region and other regions increases in neurons of NHE6-null rats compared to wild-type. Scale bar= 50 µm. (E) Quantification of Lamp1 covered area (%) at 3 and 18 months (P < 0.0001). Two-way ANOVA was conducted followed by Tukey’s HSD (wild-type = 3, Null = 3 for each time point). Values from each brain section (three sections/each animal) are clustered in different colour codes according to each animal and plotted as a small dot. Means from each biological replicate are overlaid on the top of the full dataset as a bigger dot. P-value and SEM were calculated using values from all sections from all animals used in biological replicates. All data are presented as mean ± SEM.
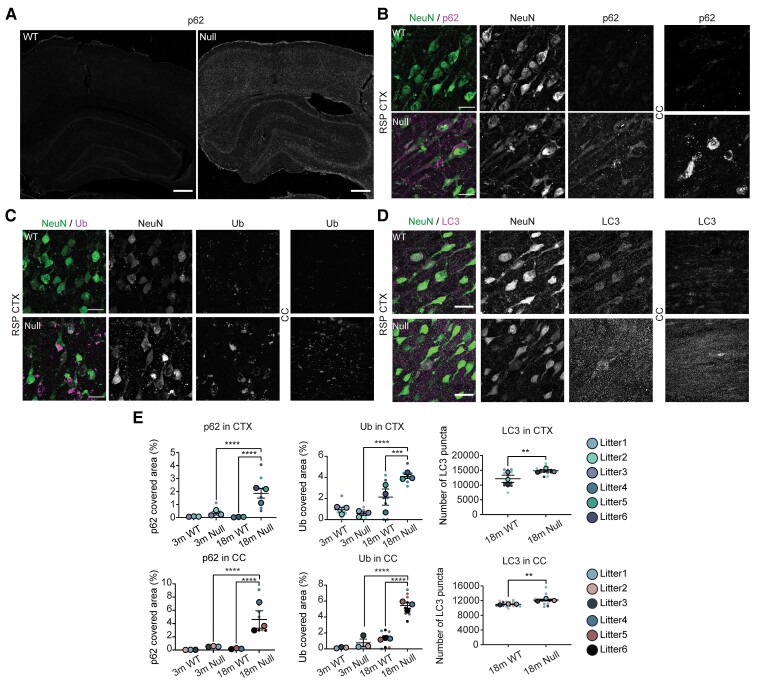
We additionally examined autophagic dysfunction in NHE6-null and wild-type in the HP and neocortex. Brain sections were stained with markers reflecting autophagic dysfunction p62, Ub and LC3. We did not observe any significant differences in p62 and Ub staining at 3 months between control and NHE6-null brains. Strong staining of p62 in the hippocampus and CTX from NHE6-null rats at 18 months was detected while that of wild-types was only weakly observed (Fig. 5A, B and E). The Ub and LC3 staining was also prominently detected in the hippocampus and CTX from NHE6-null rats at 18 months (Fig. 5C–E). Overall, these data indicate that lysosomal dysfunction occurs at an early stage in NHE6-null brain preceding later yet prominent autophagic defects.
Figure 5.
Autophagy dysfunction occurs after lysosome defects in NHE6-null rats. (A) p62 staining in wild-type (WT) and NHE6-null rats at 18 months. The p62 staining was detected in the hippocampus (HP), CTX and corpus callosum (CC) region of NHE6-null rats. Scale bar = 500 µm. (B) Magnified images of p62 staining in the retrosplenial (RSP) CTX and CC in wild-type and NHE6-null rats at 18 months. Scale bar = 20 µm. (C) Ub staining in wild-type and NHE6-null rats at 18 months. Neuronal Ub inclusion bodies were detected in the RSP CTX and CC of NHE6-null rats. The Ub staining was observed in the CC region. Scale bar = 20 µm. (D) LC3 staining in wild-type and NHE6-null rats at 18 months. Numbers of LC3 puncta were increased in the RSP CTX (P = 0.0011) and CC (P = 0.0055) of NHE6-null rats. (E) Quantification of p62 and Ub covered area (%) in the CC and CTX regions at 3 and 18 months (P < 0.0001). Two-way ANOVA was conducted followed by Tukey’s HSD (wild-type = 3, Null = 3 for each time point). The number of LC3 puncta was counted at 18 months (P = 0.0011 for CTX, P = 0.0055 for CC). Two-tailed unpaired t-test with Welch’s correction was performed (wild-type = 3, Null = 3). All data are presented as mean ± SEM. Asterisks represent P-values as follows: *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001, ****P < 0.0001.
Neuronal loss and axonal pathology in NHE6-null cortex and hippocampus
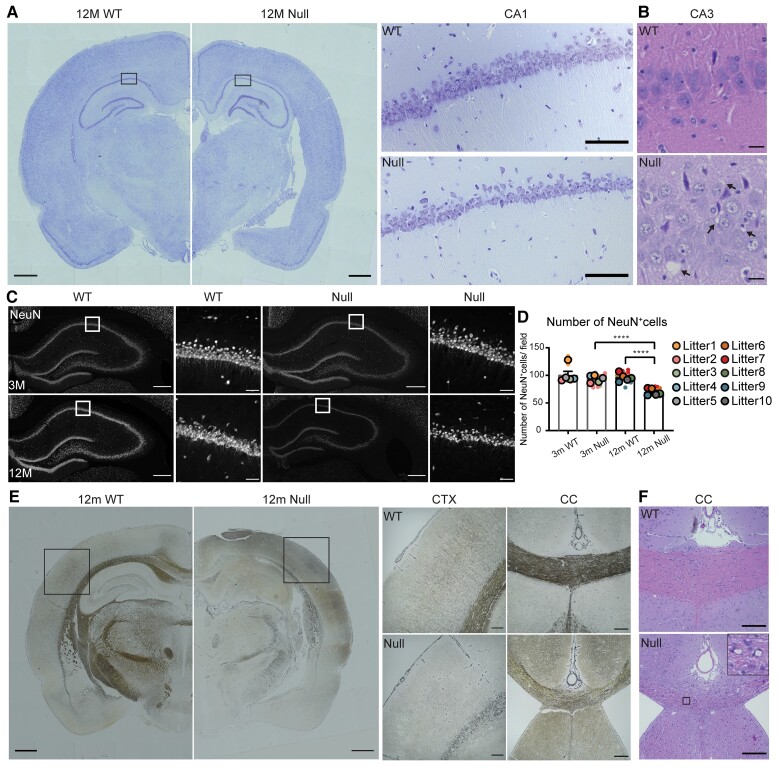
We next set out to define the extent and timing of neuronal loss in the NHE6-null rat cerebrum. While we previously noted lysosome pathology in the hippocampus as early as 3 months, we did not observe neuronal loss until approximately 1 year of age. Brain atrophy and loss of cells was observed in the hippocampus, piriform/entorhinal CTX and amygdala along with enlarged ventricles (Fig. 6A). The hippocampus volume, piriform/entorhinal CTX volume, lateral ventricle volume, cortical layer and dentate gyrus thickness were measured in wild-type and NHE6-null rats (Supplementary Fig. 3). Multifocal vacuolization was also observed in the CA3 regions of NHE6-null rat brains at 12 months with haematoxylin and eosin staining (Fig. 6B). Notably, we observed fewer neurons in the hippocampus of NHE6-null rats. To determine whether this reduction in neuronal number is a developmental phenotype or if it represents progressive neuronal loss, we stained brain sections from NHE6-null rats and their wild-type littermates at 3 and 12 months with NeuN (Fig. 6C). The number of NeuN-positive cells in the CA1 was similar between wild-type and NHE6-null rats at 3 months. At 12 months, NHE6-null rats displayed a significant reduction in the number of NeuN-positive cells in the CA1 area (Fig. 6D). Overall, these data indicate that while lysosome pathology is prominently noted in the NHE6-null hippocampus and CTX by 3 months, overt neuronal loss follows indicators of lysosome dysfunction.
Figure 6.
Neuronal loss and axonal pathology in NHE6-null rats. (A) Representative images of 12-month-old wild-type (WT) and NHE6-null rat brain sections stained with Nissl. The overall brain size of NHE6-null rat decreased. Enlarged ventricles and loss of piriform/entorhinal CTX were observed. Boxes indicate the location of the magnified hippocampal images. In the CA1 region, the Nissl staining decreased in NHE6-null rats. Scale bar for the whole brain image = 500 µm; magnified image = 200 µm. Quantification for volumes of the hippocampus (HP), the piriform/entorhinal CTX, and the lateral ventricle, cortical layer thickness and dentate gyrus thickness is presented in Supplementary Fig. 3. (B) Haematoxylin and eosin staining in the CA3 region of HP from 12-month-old wild-type and NHE6-null rats. Multifocal vacuolization was observed in NHE6-null rats. Scale bar = 20 µm. Arrows indicate vacuoles. (C) Representative images of NeuN staining. Brain sections from wild-type and NHE6-null rats at 3 and 12 months were stained with a neuronal marker, NeuN. White boxes indicate the location of magnified images. Scale bar for the whole hippocampal image = 200 µm; magnified images = 50 µm. (D) Quantification of number of NeuN-positive cells in the CA1 region of wild-type and NHE6-null rats at 3 and 12 months. At 12 months, the number of NeuN-positive cells decreased in NHE6-null rats compared to 12-month-old wild-type and 3-month-old NHE6-null rats. Two-way ANOVA was performed followed by Tukey’s HSD (wild-type = 5, Null = 5 for each time point). Tukey’s adjusted P = 0.0062 (Null at 3 months versus Null at 14 months), P = 0.0028 (wild-type at 12 months versus Null at 12 months). Values from each brain section (four sections/each animal) were clustered in different colour codes according to each animal and plotted as a small dot. Means from each biological replicate are overlaid on the top of the full dataset as a bigger dot. P-value and SEM were calculated using values from all sections from all animals used in biological replicates. All data are presented as mean ± SEM. Asterisks represent P-values as follows: *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001, ****P < 0.0001. (E) Bielschowsky’s silver staining of 12-month-old wild-type and NHE6-null rats. The staining of cortical region and major axonal tracks including corpus callosum (CC) are reduced in NHE6-null rats. Boxes indicate the location of the magnified cortical images. In CTX, the staining was barely detected in NHE6-null rats. Thinning of CC was observed in NHE6-null rats. Scale bar for the whole brain image = 500 µm; magnified image= 200 µm. (F) Haematoxylin and eosin staining of 12-month-old wild-type and NHE6-null rats. Multifocal vacuolization of the CC and adjacent grey matter was observed in NHE6-null rats. Insets represent the magnified image of vacuolization. Scale bar = 200 µm.
To investigate the possibility of axonal pathology, neurofibrils and senile plaques, we performed Bielschowsky’s silver staining in ageing NHE6-null rats and their wild-type littermates at 12 months. The silver staining was markedly reduced in the CTX and corpus callosum (CC), reflecting loss of axonal tracks of NHE6-null rats (Fig. 6E). However, we did not observe any evidence of neurofibrils and senile plaques in NHE6-null rats at 12 months. Also, we examined brain sections from wild-type and null rats at 12 months with haematoxylin and eosin. We observed multifocal vacuolization in the white matter, such as in the CC, and the grey matter adjacent to major fibre tracks, suggestive of endosomal dysfunction underlying axonal degeneration (Fig. 6F).
Increased astrogliosis and microgliosis in NHE6-null rats
Glial activation is a common process in neurodegeneration and has been previously reported in NHE6-null mice.13,33 We evaluated gliosis and staged the timing of glial activation relative to other events, i.e. lysosome and autophagy dysfunction and neuronal loss. The cerebellum of NHE6-null rats showed increased IBA1 staining and GFAP covered area at 2 months (Supplementary Fig. 4A). No glial activation was noted at 1 month in cerebellum. In the cerebrum of NHE6-null rats, the covered area of GFAP- and IBA1-positive cells significantly increased in the CA1, CA3, CC and piriform CTX of NHE6-null rats compared to wild-type at 3 months (Supplementary Fig. 4B–E). Overall, therefore, we observed glial activation relatively early in the neurodegenerative process in NHE6-null brain. This is particularly discernible in the cerebrum, where the timeline of neurodegeneration is more protracted and glial activation appears early concurrently with lysosome dysfunction, preceding autophagic dysfunction and neuronal loss.
NHE6-null rats exhibit early motor behavioural deficits
Christianson syndrome patients have worsening ataxia with age.5 To investigate gross motor exploration in a novel arena in NHE6-null rats, we conducted an open field test at postnatal Day 20 (P20) and P54. We measured animal movement and time spent in the arena using beam breaks for 30 min. The number of beam breaks between horizontal and vertical infrared beams was counted as metrics of horizontal and vertical activities, respectively. We analysed horizontal/vertical activities (Supplementary Fig. 5A and B), total distance travelled (Supplementary Fig. 5C) and duration of time spent in the centre (centre time; Supplementary Fig. 5D). At P20, no significant differences were observed between wild-type and NHE6-null rats. However, at P54, NHE6-null rats showed reduced horizontal and vertical activities compared to wild-type. The total distance NHE6-null rats travelled was shorter than wild-types. In addition, NHE6-null rats spent less time in the centre compared to wild-types. We also performed repeated measures two-way ANOVA with Sidak’s post hoc for each parameter to ensure the effect of time on each genotype. All parameters yielded significant interaction, time and genotype effects. All statistical analyses are summarized in Supplementary Table 1 and Supplementary Fig. 5.
We also performed the accelerating rotarod test from 1 month to 2 months (Supplementary Fig. 6A). At 1 month, NHE6-null rats presented a trend for worse performance. However, there was no significant difference in latency to fall. However, at 2 months, the latency to fall was significantly reduced in NHE6-null rats compared to wild-types. After 3 months, NHE6-null rats cannot perform the test because they are unable to stand on a rod. We additionally pursued the rotarod test at 2 months of age to measure motor learning and coordination (Supplementary Fig. 6B). NHE6-null rats and their littermates were tested for three successive days. The latency to fall in NHE6-null rats was significantly decreased compared to wild-types on all three test days. Also, the performance of wild-types significantly improved every day, while that of NHE6-null rats only increased from Day 1 to Day 3. This indicates the motor learning of NHE6-null rats is slower than that of wild-types.
To investigate gait abnormalities and movement of NHE6-null rats further, we conducted DigiGait analysis at 2 months to 4 months (Supplementary Fig. 6C). Rats were placed on a transparent treadmill and their ventral side was recorded to capture their strides and gait. The hindlimb ataxia coefficient was calculated from 2 months to 4 months in wild-type and NHE6-null rats. This index, which approximates stride variability, gradually increased in ageing NHE6-null rats while not changing in wild-types. We measured the body weight of wild-type and NHE6-null rats across the lifespan, monthly for 12 months (Supplementary Fig. 7) and determined that NHE6-null rats weigh less than age-sex matched littermates after 6 weeks. Reduced body mass index is a common feature of Christianson syndrome.5 However, with regard to motor strength, we do not see significant differences in forelimb/hindlimb grip strength (Supplementary Fig. 6D and E), suggesting that defective motor abilities were not a result of differences in strength. Overall, the early timing of motor defects in the NHE6-null rat are concurrent with degeneration of the cerebellum.
Late tau-associated pathologies in NHE6-null rats
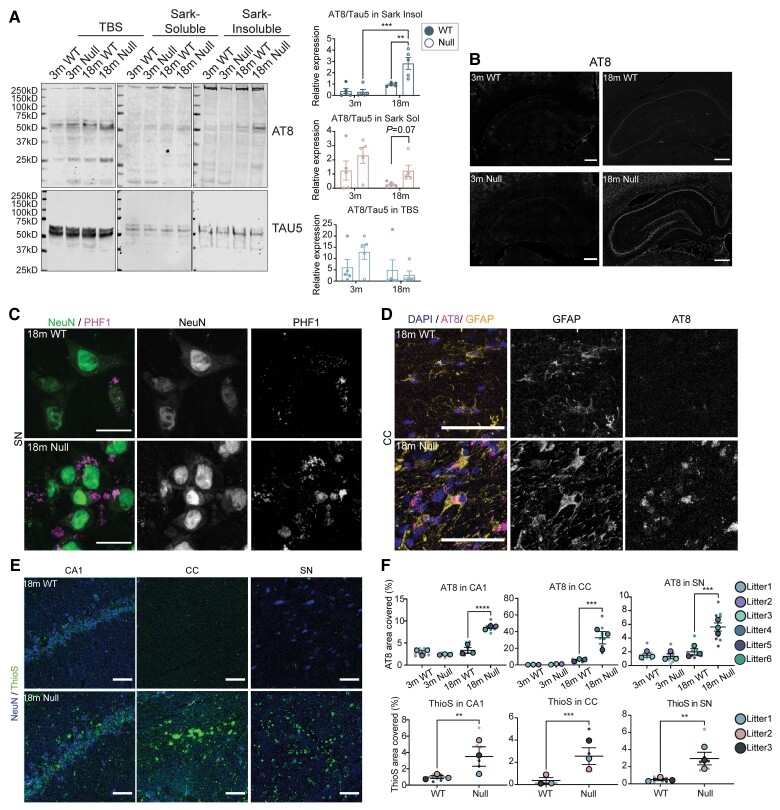
A previous post-mortem study in Christianson syndrome patient brains reported diffuse tau deposition.7 To investigate the potential for aberrant tau accumulation in NHE6-null rats, we performed sequential tau extraction from the brain at 3 months and 18 months to examine the solubility of tau (Fig. 7A). The sequentially extracted brain samples from wild-type and NHE6-null rats were immunoblotted with AT8, which recognizes tau phosphorylated at serine 202 and 205, along with TAU5, which recognizes both non-phosphorylated and phosphorylated tau.24,34 Sequential extraction involved collecting TBS-soluble, TBS-insoluble Sarkosyl-soluble and Sarkosyl-insoluble fractions. Interestingly, significant differences between NHE6-null and wild-type was observed in the Sarkosyl-insoluble fraction at 18 months. NHE6 null brain tissue displayed an elevated level of phosphorylated tau relative to total tau [Fig. 7A; d = 0.79 (medium-large effect size) for TBS fraction, d = 0.71 (small medium effect size) for Sarkosyl-soluble fraction, d = 1.595 (large effect size) for Sarkosyl-insoluble fraction].
Figure 7.
Tau-associated pathologies in the aged brains of NHE6-null rats. (A) Brain tissue from NHE6 wild-type (WT) and null rats at 3 months and 18 months were sequentially extracted in TBS, sarkosyl-containing buffer and urea-containing buffer. The whole lanes of AT8 and TAU5 were used for quantification. Representative western blot images of tau fractionation are shown. For quantification of tau fractionation, densitometry of AT8 and TAU5 was measured, using the signal present in the entire lane. Data are expressed as AT8 signal relative to TAU5 signal. The insoluble AT8 fraction increased in NHE6-null rats compared to wild-type at 18 months (P = 0.0027 for wild-type versus Null at 18 months, P = 0.00034 for Null at 3 versus 18 months). Two-way ANOVA with Tukey’s HSD was performed (wild-type = 5, Null = 5 for each time point). (B) AT8 immunostaining in the HP and CTX region at 18 months. The overall increase in AT8 staining was exhibited in NHE6-null rats. Scale bar = 500 µm. (C) PHF1 and NeuN staining in the SN region of wild-type and NHE6-null rats at 18 months. PHF1-positive NeuN staining was observed in NHE6-null rats. Scale bar = 20 µm. (D) AT8 and GFAP staining in the CC region of wild-type and NHE6-null rats at 18 months. AT8 and GFAP-positive staining was profoundly detected in NHE6-null rats. Arrow indicates AT8/GFAP-positive stained cells. Scale bar = 20 µm. (E) ThioflavinS (ThioS) staining from wild-type and NHE6-null rats at 18 months. Prominent ThioflavinS staining was detected in the CA1, CC and SN regions of NHE6-null rats. (F) Quantification of AT8 covered area (%) in the CC (P = 0.0009), SN (P = 0.0008) and CA1 (P < 0.0001) regions of wild-type and NHE6-null rats at 3 and 18 months was calculated. Two-way ANOVA followed by Tukey’s HSD was conducted. Also, ThioflavinS covered area (%) n the CC (P = 0.0007), SN (P = 0.0011) and CA1 (P = 0.0072) at 18 months was quantified. Two-tailed unpaired t-test with Welch’s correction was performed. Values from each brain section (three sections/each animal) are clustered in different colour codes according to each animal and plotted as a small dot. Means from each biological replicate are overlaid on the top of the full dataset as a bigger dot. P-value and SEM were calculated using values from all sections from all animals (wild-type = 3, Null = 3) used in biological replicates. All data are presented as mean ± SEM. Data are presented as mean ± SEM. Asterisks represent P-values as follows: *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001, ****P ≤ 0.0001.
The previous report from Christianson syndrome post-mortem brain indicates diffuse neuronal and glial tau deposits, including cortically and subcortically, such as in white matter and substantia nigra.7 In order to visualize tau inclusions in different regions of the brain in our Christianson syndrome rat model, we stained Christianson syndrome rats at 3, 12 and 18 months using PHF1 and AT8, antibodies that recognize phosphorylated tau. Increased phosphorlyated tau staining using AT8 antibodies was first evident in the hippocampus and CTX regions of NHE6-null brains at 18 months (Fig. 7B). PHF1-positive inclusions were detected in NeuN-positive cells in the brain including in the substantia nigra of NHE6-null rats at 18 months with minimal signal observed in wild-type littermates (Fig. 7C). Notably, we also observed AT8-positive inclusions in glia (GFAP-positive astrocytes) in the CC region of NHE6-null rats compared to controls (Fig. 7D). The AT8-covered area was measured in the CC, substantia nigra and CA1 regions (Fig. 7F). We did not observe the increase of AT8 staining in the cerebellum region of NHE6-null rats (Supplementary Fig. 8). We also observed strong ThioflavinS staining in NHE6-null brain sections at 18 months (Fig. 7E and F). ThioflavinS-positivity indicates the presence of β-pleated sheets within aggregating proteins such as tau or amyloid.
Aggregation of amyloid-β in aged brains of NHE6-null rats without an increase in overall amyloid-β levels
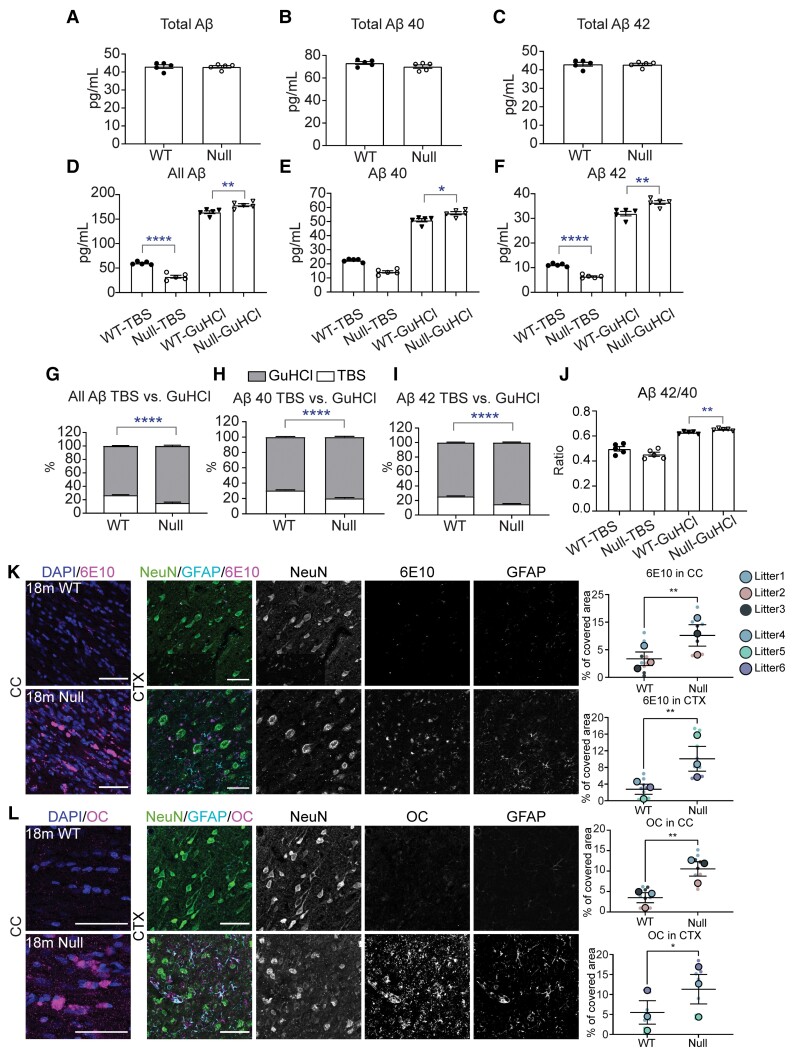
Loss of NHE6 results in acidification of endosomes and reduction of pH in endosomes has been hypothesized to increase β-secretase activity, an activity central to amyloid-β generation.10,19 However, increased amyloid-β plaque deposition was not reported in Christianson syndrome patients’ brain tissue and it is unclear if NHE6 loss will affect amyloid-β homeostasis in vivo.7 We conducted a side-by-side comparison of amyloid-β accumulation in ageing mouse and rat brains using ELISA assays of amyloid-β species (Fig. 8, Supplementary Fig. 9 and Supplementary Tables 2 and 3). In mice, we find a potential albeit modest increase (6%) in total amyloid-β (the sum of amyloid-β42, amyloid-β40 and amyloid-β38 species) at 24 months (P = 0.0385), wherein this increase appears to be largely composed of amyloid-β40 (P = 0.0292) (Supplementary Fig. 8 and Supplementary Table 2). By contrast, in rats, we do not see an increase in total amyloid-β or in amyloid-β40; however, we observe an increase in amyloid-β species in the GuHCl soluble fraction and a corresponding decrease in the TBS soluble fraction at 18 months (Fig. 8A–J and Supplementary Table 3). In NHE6-null rat brains, we were able to visualize extracellular amyloid-β deposition using immunofluorescence with anti-amyloid-β antibodies (clone: 6E10) (Fig. 8K). Also, strong immunofluorescence using an amyloid-β antibody (clone: OC) against oligomeric amyloid fibrils was prominently detected in NHE6-null rats (Fig. 8L). Strong ThioflavinS staining provides supporting evidence for the aggregation of amyloid-β in β-pleated sheets (Fig. 7E and F). Statistical analyses for the amyloid-β ELISA studies in rats and mice, including P-values, % change of NHE6-null over controls and Cohen’s d calculations, are summarized in Supplementary Tables 2 and Table 3. Effect sizes of amyloid-β aggregation were stronger in rats as compared to mice. Notably, ratios of the more pathogenic GuHCl-soluble amyloid-β42 to amyloid-β40 also increased in NHE6-null rats (Fig. 8J) yet was unchanged in mice (Supplementary Fig. 9J).
Figure 8.
Aggregation of amyloid-β in NHE6-null rats without an increase in overall amyloid-β level. (A–C) The level of amyloid-βs including amyloid-β38, amyloid-β40 and amyloid-β42 did not change from wild-type (WT) and NHE6-null rats at 18 months. (D–F) GuHCl-soluble fractions in all amyloid-β, amyloid-β40 and amyloid-β42 from NHE6-null rats increased while TBS-soluble fractions decreased. (G–I) A proportion of GuHCl-soluble fraction increases while TBS-soluble fraction decreases in all amyloid-β, amyloid-β40 and amyloid-β42. (J) Ratio of amyloid-β42/40 increase in GuHCl-soluble fraction (wild-type = 5, Null = 5). All statistical analysis details described in detail in the main text and Supplementary Table 3. (K and L) APP/amyloid-β staining in the CC and CTX region of wild-type and NHE6-null rats at 18 months. Increase in 6 ×1010 and fibrillar amyloid-β oligomer (clone: OC) was detected in NHE6-null rats. Two-tailed unpaired t-test with Welch’s correction was performed (wild-type = 3, Null = 3). Values from each brain section (three sections/each animal) are clustered in different colour codes according to each animal and plotted as a small dot. Means from each biological replicate are overlaid on the top of the full dataset as a bigger dot. P-value and SEM were calculated using values from all sections from all animals used in biological replicates. All data are presented as mean ± SEM. Data are presented as mean ± SEM. Asterisks represent P-values as follows: *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001, ****P ≤ 0.0001.
Discussion
Mutations in the endosomal NHE6 cause Christianson syndrome, an X-linked disorder associated with intellectual disability with developmental delay, postnatal microcephaly, absent speech, progressive ataxia and epilepsy.4–6 A prior post-mortem study of two males with NHE6 mutations by Garbern et al.7 revealed widespread neuronal loss, with neuronal and glial tau deposition in a pattern reported to be similar to corticobasal degeneration. Also, recent studies from our group and other colleagues reported that females who are heterozygous for loss-of-function NHE6 mutations may be diagnosed with clinical disorders that are associated with tau deposition.7–9 Further, we have also shown that post-mortem human brains, from the large Religious Order Study and Rush Memory and Aging Project, demonstrate that decreased NHE6 expression is correlated with greater tau deposition.8 In this study, we utilized a new NHE6-null rat model to identify and sequence the pathogenic events across the lifespan during neurodegeneration associated with loss of NHE6, including the possibility of tau accumulation.
In this study, we have developed an initial sequence of neuropathological events (Supplementary Fig. 1). This sequencing of events provides support for the idea that lysosomal defects are early and primary. In both the cerebellum and the cerebrum, lysosomal dysfunction is among the earliest pathological events. In a recent study, we demonstrated mechanistically that loss of NHE6 leads to defects in endosome maturation and subsequently to lysosome dysfunction in primary NHE6-null mouse neurons.11 In the current study, lysosome dysfunction occurs earlier than other pathological events such as autophagic defects; thereby these data suggest that autophagic defects may result from these lysosomal defects. In the cerebrum, where neurodegeneration is more protracted, we observed early evidence of lysosome dysfunction and glial activation by 3 months, followed by neuronal loss.
One of our key findings in this study is endogenous tau defects in NHE6-null rat brains. We utilized two different tau antibodies recognizing phosphorylated forms of tau which have been used in human brain samples.7 An increase in insoluble tau aggregates was detected in NHE6-null rats (Fig. 7). Also, we demonstrated tau accumulation in both neurons and glia (Fig. 7B–D). NHE6-null rats also exhibited glial tau accumulation in the CC and neuronal accumulation in the substantia nigra as well as in the hippocampus. This pattern is similar to what has been described previously in human post-mortem studies in Christianson syndrome.7 However, this tau phenotype has not been recapitulated in the Christianson syndrome mouse models as reported previously, likely due to limitations of the mouse model to demonstrate tau pathology.13–15
We also investigated amyloid-β accumulation in both NHE6-null rats and NHE6-null mice. Amyloid-β is produced by the cleavage of amyloid precursor protein (APP) by β-secretase (BACE1) and γ-secretase in the amyloidogenic pathway.35 BACE1 shows its maximal activity in an acidic environment.36 Thereby, acidification of endosomes as seen in NHE6-null neurons,10 may be hypothesized to enhance APP processing to amyloid-β production. This has been studied previously in in vitro cell models involving over-expression of APP in HEK293T cells with knock-down of NHE6.19 Also, a previous study in NHE6-null mice reported an increase in amyloid-β peptide levels in brain.37 Of note, the prior post-mortem studies in Christianson syndrome have reported that there is not increased amyloid-β plaques in human Christianson syndrome brain from men who died in their forties.7 In the current study, we evaluated the level of amyloid-β40 and amyloid-β42 for both Christianson syndrome mouse and rat models (Supplementary Figs 8 and 9). In rats, we do not see increased overall amyloid-β levels. Notably however, NHE6-null rats do present a shift to more insoluble amyloid-β after 18 months, while the total level was not changed. Also, increases in ThioflavinS-staining (Fig. 7E) and an oligomeric amyloid-β staining (clone: OC; Fig. 8L) in NHE6-null brains also support the presence of amyloid aggregates. In contrast, NHE6-null mice did not demonstrate strong evidence of amyloid-β aggregation even at 24 months. While we do not see increased aggregation of amyloid-β in mice, total amyloid-β may have been modestly elevated in mice in agreement with Prasad and Rao.37
Interestingly, recent studies from Pohlkamp et al.20 have reported a decrease in amyloid-β plaque load in a mouse model with humanized APP, also with the early-onset Alzheimer’s disease Swedish mutation and the 227 Beyreuther/Iberian mutation. The experimental systems that we study here and that of Pohlkamp et al.20 are quite different. For example, their system involves the early-onset mutant APP. Also, they knockdown NHE6 later in adulthood using a conditional system, as they argue that reduction of NHE6 in adulthood may be a viable treatment paradigm for reducing amyloid-β plaques in aged Alzheimer’s disease brain. However, they were not able to study changes in tau deposition or rescue of neuronal death. Importantly, we are also able to examine tau deposition in the rat and we see elevations in tau deposition in aged rat NHE6-null rat brain. Further work will be necessary to evaluate the treatment strategy proposed by Pohlkamp et al.,20 and we believe that the rat model presented here will have some advantages as a complementary in vivo system to dissect the relevant mechanisms.
Differences between rat and mouse models with regard to tau and amyloid-β phenotypes have been previously reported.24,25 The Alzheimer’s disease rat model by Cohen et al.24 overexpresses mutant human Swedish APP (APPswe) and presenilin 1 (PS1ΔE9) genes, and manifested tau pathology, based on endogenously expressed tau, along with neuronal loss. This Alzheimer’s disease rat model is considered as a complement to existing amyloid-β-overexpressing transgenic Alzheimer’s disease mice since these transgenic mice do not present tau deposition.38–41 Also, a transgenic rat model of tauopathy expressing a full length human tau with the P301S mutation has been proposed to be phenotypically closer to human patients than transgenic tau mice models.34 This tau rat model exhibited aggregates of both human tau and endogenous rat tau along with key degenerative features such as brain atrophy and ventricular dilation. However, the transgenic tau mice models only showed the human tau filamentous aggregates. There are various possibilities for why rats may present tau-related pathology while mice do not. One possibility is that rats have the full set of six tau isoforms found in humans, while mice only have three of the human isoforms.42 Tau pathology and amyloid-β aggregates are indeed notable findings of broad significance to neurodegenerative disease, corroborated here by both biochemical and microscopic evidence. Importantly, these events are relative late in the pathogenesis occurring in aged animals at and after 18 months (Supplementary Fig. 1). Therefore, tau and amyloid-β pathology may contribute to Christianson syndrome progression; however, these events may not be a part of the primary pathogenesis.
Defective endo-lysosomal function is one of the earliest features in Christianson syndrome-related neurodegeneration and potentially in other more common neurodegenerative diseases, including as proposed in Alzheimer’s disease.43–49 Here, we provide additional support that endo-lysosomal dysfunction is associated with some features found in Alzheimer’s disease. NHE6 localizes in endolysosomes to regulate luminal pH as well as endosomal trafficking and signalling.10,11 NHE6-null rats presented lysosomal dysfunction in neurons at 3 months prior to neuronal loss, and this phenotype worsens over time (Figs 3 and 4). We further observe that this lysosomal dysfunction may later disrupt autophagic clearance of toxic materials such as ubiquitin-positive inclusion bodies and tau aggregates (Fig. 5).50–52 Notably, a recent study reported transgenic mice over-expressing Rab5 in neurons also exhibited Alzheimer’s disease-like features such as cholinergic neuronal loss and elevated phosphorylated tau in the absence of amyloid-β accumulation.53 This might indicate a convergent endo-lysosomal pathway mediated by Rab5 or NHE6 to cause Alzheimer’s disease like phenotypes.
Increased astrogliosis and microgliosis are pathological hallmarks in brain regions affected by neurodegeneration.54 In the NHE6-null rat brain, glial activation appears fairly early in the timeline of pathogenesis. In NHE6-null cerebrum at 3 months (Supplementary Fig. 4B–E), we observed elevated gliosis in the regions, coincident with lysosome dysfunction, preceding autophagic dysfunction, neuronal loss and tau pathology. However, future research will endeavour to determine the precise molecular events that lead to glial activation and the role of activated glia in the causes of neuronal loss and other neuropathology.
Our current study indicates that rat is a strong model to study neurodegeneration resulting from a rare genetic disorder such as in Christianson syndrome, as well as to study more general neurological disorders, such as those involving amyloid-β and tau pathology. Our study demonstrates tau-associated pathology in NHE6-null rats, which appears to align strongly with Christianson syndrome post-mortem studies. Defects in tau and amyloid-β accumulation in this genetically mutated rat also provide strong evidence that rats are valuable animal models to supplement the existing mouse models in neurodegeneration research. Since rats are more genetically and physiologically close to humans than mice,21–23 experiments in rats may provide important insights into the neurodegenerative mechanisms of Christianson syndrome and potentially in more common neurological disorders. This experimental rat model is also an important complement to the currently limited human post-mortem studies in Christianson syndrome,7 and provides a strong experimental system to interrogate linkages between endolysosomal dysfunction and tau and amyloid-β aggregations.
Supplementary Material
Acknowledgements
We are grateful to Dr Konstantin Dobrenis for the anti-GM2 antibody.
Abbreviation
- CC =
corpus callosum
- CTX
cortex
- NHE6
Na+/H+ exchanger 6
Contributor Information
YouJin Lee, Department of Molecular Biology, Cell Biology and Biochemistry, Brown University, Providence, RI 02912, USA; Center for Translational Neuroscience, Carney Institute for Brain Science and Brown Institute for Translational Science (BITS), Brown University, Providence, RI 02912, USA.
Morgan R Miller, Department of Molecular Biology, Cell Biology and Biochemistry, Brown University, Providence, RI 02912, USA; Center for Translational Neuroscience, Carney Institute for Brain Science and Brown Institute for Translational Science (BITS), Brown University, Providence, RI 02912, USA.
Marty A Fernandez, Ann Romney Center for Neurologic Diseases, Brigham and Women’s Hospital and Harvard Medical School, Boston, MA, USA.
Elizabeth L Berg, MIND Institute and Department of Psychiatry and Behavioural Sciences, University of California Davis School of Medicine, Sacramento, CA, USA.
Adriana M Prada, Department of Molecular Biology, Cell Biology and Biochemistry, Brown University, Providence, RI 02912, USA; Center for Translational Neuroscience, Carney Institute for Brain Science and Brown Institute for Translational Science (BITS), Brown University, Providence, RI 02912, USA.
Qing Ouyang, Department of Molecular Biology, Cell Biology and Biochemistry, Brown University, Providence, RI 02912, USA; Center for Translational Neuroscience, Carney Institute for Brain Science and Brown Institute for Translational Science (BITS), Brown University, Providence, RI 02912, USA.
Michael Schmidt, Department of Molecular Biology, Cell Biology and Biochemistry, Brown University, Providence, RI 02912, USA; Center for Translational Neuroscience, Carney Institute for Brain Science and Brown Institute for Translational Science (BITS), Brown University, Providence, RI 02912, USA.
Jill L Silverman, MIND Institute and Department of Psychiatry and Behavioural Sciences, University of California Davis School of Medicine, Sacramento, CA, USA.
Tracy L Young-Pearse, Ann Romney Center for Neurologic Diseases, Brigham and Women’s Hospital and Harvard Medical School, Boston, MA, USA.
Eric M Morrow, Department of Molecular Biology, Cell Biology and Biochemistry, Brown University, Providence, RI 02912, USA; Center for Translational Neuroscience, Carney Institute for Brain Science and Brown Institute for Translational Science (BITS), Brown University, Providence, RI 02912, USA.
Funding
NIH/NINDS/NIA R01NS113141, NIH/NIMH R01MH105442, NIH/NIMH R01MH102418, NIH/NIA F32AG066372. This research was supported in part by the following: NIH/NIMH grants R01MH105442, R01MH102418 (to E.M.M.), NIH/NINDS/NIA grant R01NS113141 (to E.M.M.), and NIH/NIA grant F32AG066372 (to Y.L.). Other grants are MIND Institute’s Intellectual and Developmental Disabilities Resource Center HD079125 (PI, Abedutto). J.L.S. and E.L.B. are supported by NIH/NINDS R01NS097808 and the Foundation for Angelman Syndrome Therapeutics.
Competing interests
The authors report no competing interests.
Supplementary material
Supplementary material is available at Brain online.
References
- 1. Yap CC, Winckler B. Adapting for endocytosis: roles for endocytic sorting adaptors in directing neural development. Front Cell Neurosci. 2015;9:119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Nixon RA. Endosome function and dysfunction in Alzheimer’s disease and other neurodegenerative diseases. Neurobiol Aging. 2005;26(3):373–382. [DOI] [PubMed] [Google Scholar]
- 3. Small SA, Petsko GA. Endosomal recycling reconciles the Alzheimer’s disease paradox. Sci Transl Med. 2020;12(572):eabb1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Christianson AL, Stevenson RE, van der Meyden CH, et al. X linked severe mental retardation, craniofacial dysmorphology, epilepsy, ophthalmoplegia, and cerebellar atrophy in a large South African kindred is localised to Xq24-q27. J Med Genet. 1999;36(10):759–766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Pescosolido MF, Stein DM, Schmidt M, et al. Genetic and phenotypic diversity of NHE6 mutations in Christianson syndrome. Ann Neurol. 2014;76(4):581–593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Gilfillan GD, Selmer KK, Roxrud I, et al. SLC9A6 mutations cause X-linked mental retardation, microcephaly, epilepsy, and ataxia, a phenotype mimicking Angelman syndrome. Am J Hum Genet. 2008;82(4):1003–1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Garbern JY, Neumann M, Trojanowski JQ, et al. A mutation affecting the sodium/proton exchanger, SLC9A6, causes mental retardation with tau deposition. Brain. 2010;133(Pt 5):1391–1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Pescosolido MF, Kavanaugh BC, Pochet N, et al. Complex neurological phenotype in female carriers of NHE6 mutations. Mol Neuropsychiatry. Apr 2019;5(2):98–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Sinajon P, Verbaan D, So J. The expanding phenotypic spectrum of female SLC9A6 mutation carriers: a case series and review of the literature. Hum Genet. 2016;135(8):841–850. [DOI] [PubMed] [Google Scholar]
- 10. Ouyang Q, Lizarraga SB, Schmidt M, et al. Christianson syndrome protein NHE6 modulates TrkB endosomal signaling required for neuronal circuit development. Neuron. 2013;80(1):97–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Pescosolido MF, Ouyang Q, Liu JS, Morrow EM. Loss of Christianson syndrome Na+/H+ exchanger 6 (NHE6) causes abnormal endosome maturation and trafficking underlying lysosome dysfunction in neurons. J Neurosci. 2021;41(44):9235–9256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Deane EC, Ilie AE, Sizdahkhani S, Das Gupta M, Orlowski J, McKinney RA. Enhanced recruitment of endosomal Na+/H+ exchanger NHE6 into Dendritic spines of hippocampal pyramidal neurons during NMDA receptor-dependent long-term potentiation. J Neurosci. 2013;33(2):595–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Xu M, Ouyang Q, Gong J, et al. Mixed neurodevelopmental and neurodegenerative pathology in Nhe6-Null mouse model of Christianson syndrome. eNeuro. 2018;4(6):ENEURO.0388-17.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Stromme P, Dobrenis K, Sillitoe RV, et al. X-linked Angelman-like syndrome caused by Slc9a6 knockout in mice exhibits evidence of endosomal-lysosomal dysfunction. Brain. 2011;134(Pt 11):3369–3383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Sikora J, Leddy J, Gulinello M, Walkley SU. X-linked Christianson syndrome: heterozygous female Slc9a6 knockout mice develop mosaic neuropathological changes and related behavioural abnormalities. Dis Model Mech. 2016;9(1):13–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Spires TL, Hyman BT. Transgenic models of Alzheimer’s disease: learning from animals. NeuroRx. 2005;2(3):423–437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Sasaguri H, Nilsson P, Hashimoto S, et al. APP mouse models for Alzheimer’s disease preclinical studies. EMBO J. 2017;36(17):2473–2487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Drummond E, Wisniewski T. Alzheimer’s disease: experimental models and reality. Acta Neuropathol. 2017;133(2):155–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Prasad H, Rao R. The Na+/H+ exchanger NHE6 modulates endosomal pH to control processing of amyloid precursor protein in a cell culture model of Alzheimer disease. J Biol Chem. 2015;290(9):5311–5327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Pohlkamp T, Xian X, Wong CH, et al. NHE6-depletion corrects ApoE4-mediated synaptic impairments and reduces amyloid plaque load. Elife. 2021;10:e72034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Gibbs RA, Weinstock GM, Metzker ML, et al. Genome sequence of the Brown Norway rat yields insights into mammalian evolution. Nature. 2004;428(6982):493–521. [DOI] [PubMed] [Google Scholar]
- 22. Jacob HJ, Kwitek AE. Rat genetics: attaching physiology and pharmacology to the genome. Nat Rev Genet. 2002;3(1):33–42. [DOI] [PubMed] [Google Scholar]
- 23. Francis C, Natarajan S, Lee MT, et al. Divergence of RNA localization between rat and mouse neurons reveals the potential for rapid brain evolution. BMC Genomics. 2014;15:883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Cohen RM, Rezai-Zadeh K, Weitz TM, et al. A transgenic Alzheimer rat with plaques, tau pathology, behavioural impairment, oligomeric aβ, and frank neuronal loss. J Neurosci. 2013;33(15):6245–6256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Do Carmo S, Cuello AC. Modeling Alzheimer’s disease in transgenic rats. Mol Neurodegener. 2013;8:37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Iwata A, Chen X-H, McIntosh TK, Browne KD, Smith DH. Long-term accumulation of amyloid-beta in axons following brain trauma without persistent upregulation of amyloid precursor protein genes. J Neuropathol Exp Neurol. 2002;61(12):1056–1068. [DOI] [PubMed] [Google Scholar]
- 27. Purushothuman S, Marotte L, Stowe S, Johnstone DM, Stone J. The response of cerebral cortex to haemorrhagic damage: experimental evidence from a penetrating injury model. PLoS One. 2013;8(3):e59740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. de Calignon A, Polydoro M, Suarez-Calvet M, et al. Propagation of tau pathology in a model of early Alzheimer’s disease. Neuron. 2012;73(4):685–697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lagomarsino VN, Pearse RV II, Liu L, et al. Stem cell-derived neurons reflect features of protein networks, neuropathology, and cognitive outcome of their aged human donors. Neuron. 2021;109(21):3402.e9–3420.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. DeVos SL, Miller RL, Schoch KM, et al. Tau reduction prevents neuronal loss and reverses pathological tau deposition and seeding in mice with tauopathy. Sci Transl Med. 2017;9(374):eaag0481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lizarraga SB, Ma L, Maguire AM, et al. Human neurons from Christianson syndrome iPSCs reveal mutation-specific responses to rescue strategies. Sci Transl Med. 2021;13(580):eaaw0682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Nixon RA. The aging lysosome: An essential catalyst for late-onset neurodegenerative diseases. Biochim Biophys Acta Proteins Proteom. 2020;1868(9):140443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Hammond TR, Marsh SE, Stevens B. Immune signaling in neurodegeneration. Immunity. 2019;50(4):955–974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Malcolm JC, Breuillaud L, Do Carmo S, et al. Neuropathological changes and cognitive deficits in rats transgenic for human mutant tau recapitulate human tauopathy. Neurobiol Dis. 2019;127:323–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Selkoe DJ, Hardy J. The amyloid hypothesis of Alzheimer’s disease at 25 years. EMBO Mol Med. 2016;8(6):595–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Vassar R, Bennett BD, Babu-Khan S, et al. Beta-secretase cleavage of Alzheimer’s amyloid precursor protein by the transmembrane aspartic protease BACE. Science. 1999;286(5440):735–741. [DOI] [PubMed] [Google Scholar]
- 37. Prasad H, Rao R. Amyloid clearance defect in ApoE4 astrocytes is reversed by epigenetic correction of endosomal pH. Proc Natl Acad Sci USA. 2018;115(28):E6640–E6649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Wilcock DM, Lewis MR, Van Nostrand WE, et al. Progression of amyloid pathology to Alzheimer’s disease pathology in an amyloid precursor protein transgenic mouse model by removal of nitric oxide synthase 2. J Neurosci. 2008;28(7):1537–1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Oddo S, Caccamo A, Shepherd JD, et al. Triple-transgenic model of Alzheimer’s disease with plaques and tangles: intracellular Abeta and synaptic dysfunction. Neuron. 2003;39(3):409–421. [DOI] [PubMed] [Google Scholar]
- 40. Padmanabhan J, Levy M, Dickson DW, Potter H. Alpha1-antichymotrypsin, an inflammatory protein overexpressed in Alzheimer’s disease brain, induces tau phosphorylation in neurons. Brain. 2006;129(Pt 11):3020–3034. [DOI] [PubMed] [Google Scholar]
- 41. Colton CA, Wilcock DM, Wink DA, Davis J, Van Nostrand WE, Vitek MP. The effects of NOS2 gene deletion on mice expressing mutated human AbetaPP. J Alzheimers Dis. 2008;15(4):571–587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Hanes J, Zilka N, Bartkova M, Caletkova M, Dobrota D, Novak M. Rat tau proteome consists of six tau isoforms: implication for animal models of human tauopathies. J Neurochem. 2009;108(5):1167–1176. [DOI] [PubMed] [Google Scholar]
- 43. Small SA, Simoes-Spassov S, Mayeux R, Petsko GA. Endosomal traffic jams represent a pathogenic hub and therapeutic target in Alzheimer’s disease. Trends Neurosci. 2017;40(10):592–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Karch CM, Goate AM. Alzheimer’s disease risk genes and mechanisms of disease pathogenesis. Biol Psychiatry. 2015;77(1):43–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Nixon RA. Amyloid precursor protein and endosomal-lysosomal dysfunction in Alzheimer’s disease: inseparable partners in a multifactorial disease. FASEB J. 2017;31(7):2729–2743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Kwart D, Gregg A, Scheckel C, et al. A large panel of isogenic APP and PSEN1 mutant human iPSC neurons reveals shared endosomal abnormalities mediated by APP β-CTFs, Not Aβ. Neuron. 2019;104(5):1022. [DOI] [PubMed] [Google Scholar]
- 47. Knupp A, Mishra S, Martinez R, et al. Depletion of the AD risk gene SORL1 selectively impairs neuronal endosomal traffic independent of amyloidogenic APP processing. Cell Rep. 2020;31(9):107719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Evans LD, Wassmer T, Fraser G, et al. Extracellular monomeric and aggregated tau efficiently enter human neurons through overlapping but distinct pathways. Cell Rep. 2018;22(13):3612–3624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Young JE, Fong LK, Frankowski H, Petsko GA, Small SA, Goldstein LSB. Stabilizing the retromer complex in a human stem cell model of Alzheimer’s disease reduces TAU phosphorylation independently of amyloid precursor protein. Stem Cell Reports. 2018;10(3):1046–1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Nixon RA, Yang D-S. Autophagy and neuronal cell death in neurological disorders. Cold Spring Harb Perspect Biol. 2012;4(10):a008839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Komatsu M, Waguri S, Chiba T, et al. Loss of autophagy in the central nervous system causes neurodegeneration in mice. Nature. 2006;441(7095):880–884. [DOI] [PubMed] [Google Scholar]
- 52. Wang Y, Kruger U, Mandelkow E, Mandelkow E-M. Generation of tau aggregates and clearance by autophagy in an inducible cell model of tauopathy. Neurodegener Dis. 2010;7(1–3):103–107. [DOI] [PubMed] [Google Scholar]
- 53. Pensalfini A, Kim S, Subbanna S, et al. Endosomal dysfunction induced by directly overactivating Rab5 recapitulates prodromal and neurodegenerative features of Alzheimer’s disease. Cell Rep. 2020;33(8):108420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Ransohoff RM. How neuroinflammation contributes to neurodegeneration. Science. 2016;353(6301):777–783. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All results in the main figures and supplementary figures are available upon request. Detailed methods are also described in the Supplementary material.