Abstract
The development of a vaccine to prevent congenital human cytomegalovirus (HCMV) disease is a public health priority. We tested rhesus CMV (RhCMV) prototypes of HCMV vaccine candidates in a seronegative macaque oral challenge model. Immunogens included a recombinant pentameric complex (PC; gH/gL/pUL128/pUL130/pUL131A), a postfusion gB ectodomain, and a DNA plasmid that encodes pp65-2. Immunization with QS21-adjuvanted PC alone or with the other immunogens elicited neutralizing titers comparable to those elicited by RhCMV infection. Similarly, immunization with all 3 immunogens elicited pp65-specific cytotoxic T-cell responses comparable to those elicited by RhCMV infection. RhCMV readily infected immunized animals and was detected in saliva, blood, and urine after challenge in quantities similar to those in placebo-immunized animals. If HCMV evades vaccine-elicited immunity in humans as RhCMV evaded immunity in macaques, a HCMV vaccine must elicit immunity superior to, or different from, that elicited by the prototype RhCMV vaccine to block horizontal transmission.
Keywords: cytomegalovirus, vaccine, neutralizing antibody, protective efficacy, rhesus macaque, RhCMV
Rhesus CMV prototypes of HCMV vaccine candidates were tested in a seronegative macaque oral challenge model. Adjuvanted pentameric complex and postfusion gB subunits with pp65-2–encoding DNA elicited strong serum neutralizing and T-cell responses but were insufficient to prevent infection.
Human cytomegalovirus (HCMV) infection of immune-competent individuals generally manifests with an acute phase followed by a life-long latent phase and is often asymptomatic. However, new infection or reactivation from latent HCMV infection in immunosuppressed patients, such as transplant recipients or those with AIDS, can cause serious disease and even death. Primary HCMV infection of a seronegative pregnant woman results in transmission to her fetus in approximately 30% of cases [1, 2]. Approximately 10% of congenitally infected infants will have symptoms at birth, with approximately 30%–40% of those exhibiting severe disease, which can include hearing loss, vision loss, microcephaly, and developmental delay. Nonprimary infection of a seropositive pregnant woman or reactivation of her latent HCMV also can result in transplacental transmission. The probability of transmission to the fetus and severe sequelae may be substantially lower after nonprimary maternal infection, though studies of the outcomes of nonprimary maternal infection are inconsistent [1, 2]. A prophylactic HCMV vaccine is urgently needed to prevent congenital HCMV disease, as well as HCMV disease in immunosuppressed patients.
At least 3 viral glycoproteins or glycoprotein complexes mediate HCMV entry into cells: glycoprotein B (gB), which is required for CMV entry into all target cells [3]; the gH/gL/pUL128/pUL130/pUL131 heteropentamer complex (PC) [3], which is important for entry into epithelial and endothelial cells; and gH/gL/gO, which is important for entry into fibroblasts [4–7]. The viral fusion glycoprotein, gB, transitions from a metastable prefusion conformation to a stable postfusion conformation as it fuses membranes during cell entry [8, 9]. Postfusion gB has been isolated in a well-characterized form stable enough to test its immunogenicity. Postfusion gB elicits strong nonneutralizing antibody responses, strong complement-dependent neutralizing responses, and much weaker complement-independent neutralizing responses [10, 11]. A subunit vaccine candidate containing an adjuvanted, recombinant, postfusion CMV gB antigen has demonstrated statistically significant protection from horizontal transmission of CMV to seronegative new mothers [12], and a trend toward protection of seronegative adolescent girls [13]. Although the observed level of protection was insufficient to justify further development of a postfusion gB-based vaccine, the findings suggest that mechanisms other than virus neutralization may have some protective capacity. Prefusion HCMV gB has been stabilized for structure determination [14], but there are as yet no reports of the immunogenicity of a preparation of gB that has been convincingly demonstrated to be in the prefusion conformation.
The PC is the dominant target for neutralizing antibody (nAb) responses in natural HCMV infections [9, 15], and sera from mice and monkeys immunized with recombinant PC neutralize HCMV infection of epithelial and endothelial cells and fibroblasts to high titers [16–19]. Directly testing the protective efficacy of HCMV antigens in animal models is not feasible because HCMV only infects humans. Infection of specific-pathogen–free (SPF) rhesus macaques by RhCMV is a rigorous animal model for CMV pathogenesis and vaccine efficacy [20]. Vaccination of rhesus macaques with modified vaccinia virus Ankara (MVA) vectors expressing soluble or membrane-associated RhCMV PC did not efficiently prevent RhCMV infection after subcutaneous challenge, but did reduce viral loads [21]. The nAb response elicited by MVA-vectored PC immunization was modest and far below responses elicited by recombinant HCMV PC immunization of mice or natural HCMV infection of humans. A nucleoside modified RNA-based vaccine candidate that encodes the human CMV PC and postfusion gB is in clinical testing [22].
Here, we report the immunogenicity of RhCMV PC and its protective efficacy against oral challenge of SPF rhesus macaques with UCD52, a virulent strain of RhCMV that has a functional PC [23]. In addition, we tested the protective efficacy of immunization with RhCMV PC together with 2 other RhCMV immunogens—postfusion gB and DNA that encodes RhCMV pp65-2, an orthologue of the HCMV pp65 tegument protein.
METHODS
Vaccine Immunogens
Expression and Purification of Recombinant RhCMV UCD52 PC
The PC was expressed in SF9 insect cells from a single baculovirus with the RedVax GmbH platform design, which is based on the Multibac system [24, 25]. The 5 coding sequences (accession numbers: HQ667932-gH; HQ667933-gL; GU552456-UL128, -UL130, -UL131a) were synthesized with codon optimization for insect cell expression. Glycoprotein H is the only protein in the PC with a transmembrane domain, and its coding sequence was truncated to express a soluble ectodomain with a C-terminus at amino acid 695. The recombinant RhCMV strain UCD52 [26] PC was isolated from 12 L of SF9 culture medium using affinity chromatography with Ni Sepharose (GE Healthcare) followed by cation exchange chromatography with Toyopearl-SP 650M (Tosoh Bioscience). The final preparation of PC was quantified by micro-BCA (Invitrogen) and characterized by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE).
Production of RhCMV gB674
The N-terminal 674 residues of UCD52 gB (GenBank accession ADD92468), named gB674, with a C-terminal 6×His tag were expressed as a soluble ectodomain in Expi293F cells (Invitrogen) by transient transfection. Five liters of HEK293 conditioned culture medium was concentrated 14× and 3× diafiltered into binding buffer (20 mM NaPO4, 300 mM NaCl, pH 7.4) by 10 kDa molecular weight cutoff tangential flow filtration. The diafiltrate was applied to a 5-cm × 17-cm Ni Sepharose 6 FF column (GE Healthcare) and washed with 20 mM NaPO4, 150 mM NaCl, 0.14% Triton X-100, pH 6.8–7, followed by 20 mM NaPO4, 300 mM NaCl, 100 mM imidazole, pH 6.8–7. gB674 was eluted in 20 mM NaPO4, 150 mM NaCl, 500 mM imidazole, pH 6.8–7. The pooled fractions were concentrated to 45 mL by tangential flow filtration and applied to a Superdex 200 pg (Cytiva) size-exclusion column (5 cm × 55 cm) in 20 mM NaPO4, 150 mM NaCl, pH 6.8–7. The purification yielded 300 mg of pure gB674 protein.
UCD52 pp65-2 DNA
The coding sequence of RhCMV UCD52 pp65-2 (GenBank accession QQL11309) was codon-optimized for mammalian cell expression and cloned into an expression cassette pJV7563 as previously described [27]. The plasmid was expanded in BD1175 host cells (a noncommercial product of Pfizer, Inc) and purified by anion exchange chromatography (Fractogel TMAE [M] resin; EMD Millipore). Isoforms of the purified plasmid were analyzed by anion exchange high-performance liquid chromatography using a CIMac pDNA-0.3 analytical column. The final product contained 89% supercoiled DNA and was passed through a 0.22-µm filter.
RhCMV Strain and Monkey Cells
RhCMV strain UCD52, primary monkey kidney epithelial cells (MKE) and telomerized rhesus fibroblast cells (Telo-RF) were prepared as described [28–30]. UCD52 viral stock for animal challenge was passaged 4 times on MKE cells and expanded once on Telo-RF cells. Virus stocks were titrated on MKE or Telo-RF cells using a single-round entry assay in which virus-infected cells were enumerated by immunostaining for immediate-early-1 (IE1) 48 hours after virus inoculation.
Vaccination and Challenge Studies in SPF Rhesus Macaques
SPF rhesus macaques (Macaca mulatta) were bred and maintained at the California National Primate Research Center as reported [31]. All animal protocols were approved in advance by the Institutional Animal Care and Use Committees of the University of California, Davis (UC Davis) and Pfizer. UC Davis is fully accredited by the Association for Assessment and Accreditation of Laboratory Animal Care. All animals were seronegative for RhCMV prior to immunization or inoculation. The animals were 12–24 months of age. About half were male, and half were female. The protein antigens and pp65-2 DNA used for vaccination tested negative for adventitious agents and had endotoxin levels below the maximum permissible.
Groups of 6 rhesus macaques were injected intramuscularly (IM) with the protein immunogens adjuvanted with QS-21 3 times at 4-week intervals. One group was immunized with pp65-2 DNA by electroporation in 1 limb and with the protein immunogens in the contralateral limb. For DNA vaccination, 2 mg of plasmid was formulated in 0.5 mL phosphate-buffered saline and delivered into the quadriceps muscle using the TriGrid Delivery System by Ichor Medical Systems following the manufacturer’s protocol (Ichor Medical Systems). A group of 6 animals was mock vaccinated by electroporation as control.
Two weeks after the third vaccination, the vaccine- or placebo-immunized animals were challenged by 5 weekly inoculations of 8 × 105 plaque-forming units (pfu) of RhCMV UCD52 (GenBank accession MT157339) into the buccal pouch, as previously reported [32]. Samples of blood (EDTA-treated plasma and peripheral blood mononuclear cells [PBMCs]), saliva (oral swab), and urine (cystocentesis complemented by pan catch when needed) were collected by conventional methods and used for serological and virological testing using previously described methods with some modifications [26, 33–36].
RESULTS
Expression and Characterization of Recombinant UCD52 PC
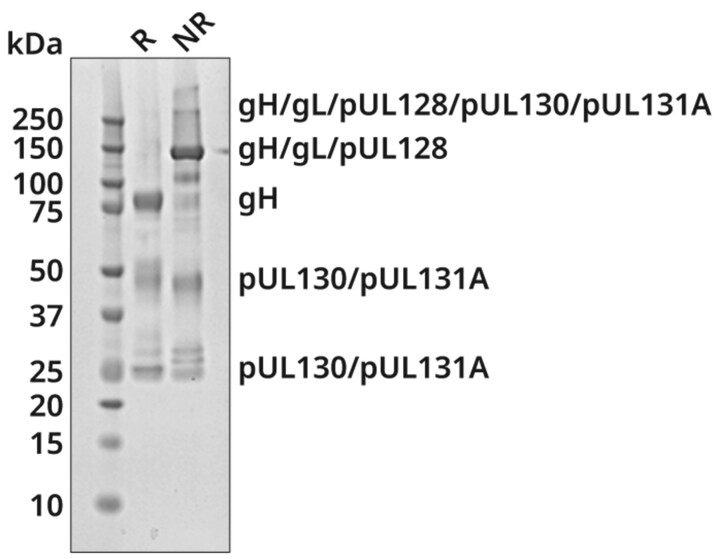
The PC was purified by affinity chromatography from recombinant baculovirus-infected insect cells by nickel affinity chromatography using the C-terminal histidine tag appended to gH, followed by cation exchange chromatography. SDS-PAGE under reducing and nonreducing conditions produced a pattern of bands similar to that of the HCMV PC published by others (Figure 1) [37]. We confirmed the identities of the major resolved bands to be gH, gL, pUL128, pUL130, and pUL131A by liquid chromatography with tandem mass spectrometry (LC-MS/MS; Supplementary Table 1).
Figure 1.
Expression and characterization of recombinant UCD52 PC. The presence of the 5 components of the PC (gH, gL, pUL128, pUL130, and pUL131A) was confirmed by SDS-PAGE analysis using 4%–12% bis-tris polyacrylamide gel. The PC samples were analyzed at nonreducing (NR) and reducing (R) conditions. Protein bands were visualized with Coomassie blue staining, and the proteins corresponding to individual bands were identified by LC-MS/MS (see Supplementary Table 1). Abbreviations: LC-MS/MS, liquid chromatography with tandem mass spectrometry; PC, pentameric complex; SDS-PAGE, sodium dodecyl sulfate polyacrylamide gel electrophoresis.
Expression and Characterization of Recombinant, Postfusion gB674
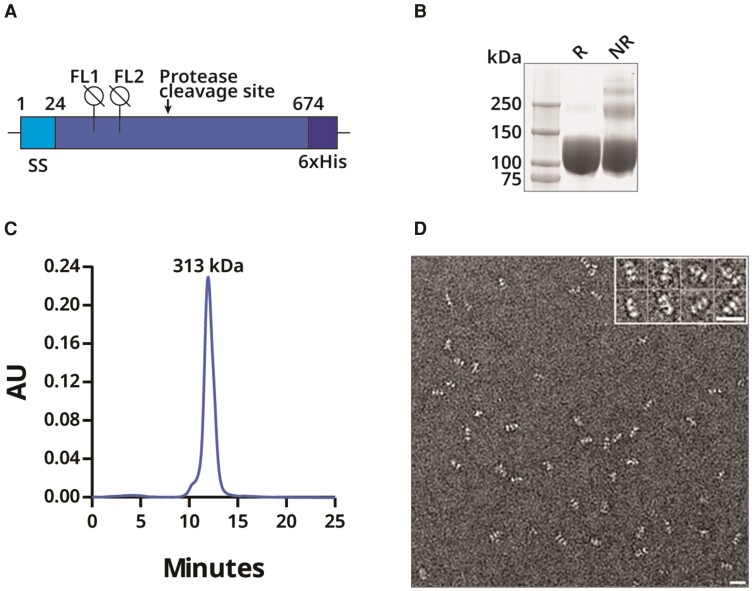
Guided by X-ray crystal structures of HCMV postfusion gB [8, 9], we truncated RhCMV gB (GenBank Accession GU552457) at residue 674 to produce a soluble, recombinant, trimeric ectodomain. The resulting construct, gB674, includes the signal peptide, mutation of the furin recognition motif (from 429RRKR432 to ATKA), 3 mutations (F128G, I129H, and W213A) in the 2 fusion loops to prevent aggregation, and a GSG linker to a C-terminal 6×His purification tag (Figure 2A). After expression in mammalian cells and purification, reducing SDS-PAGE analysis of gB674 produced a single, uniform monomer band with a migration around 100–130 kDa, which is higher than the approximately 76 kDa of the predicted molecular weight of the polypeptide, possibly due to glycosylation (Figure 2B). By nonreducing SDS-PAGE, more slowly migrating species were seen, consistent with trimer formation and a higher-order association. By size exclusion chromatography in the presence of 1 mM EDTA, a single, homogeneous peak with an apparent molecular weight of 313 kDa was observed, consistent with a gB674 trimer (Figure 2C). The gB674 preparation had a purity of >98% by SDS-PAGE. Transmission electron microscopy after negative staining revealed monodispersed gB674 trimers with rod-like shapes approximately 17 nm in length and with 3 globular densities, consistent with postfusion HCMV gB crystal structures (Figure 2D) [8].
Figure 2.
Expression and characterization of recombinant gB674 from UCD52 RhCMV. A, Design of gB674 [23, 24]. B, Coomassie-stained SDS-PAGE of purified gB674 separated under reducing (R) or nonreducing (NR) conditions. C, Size exclusion chromatography analysis of gB674. D, Electron microscopy of negatively stained gB674 at a magnification of 1:50 000. The white bar represents 20 nm, and the inset shows a subset of the particles. Abbreviations: 6×His, 6-histidine affinity tag; FL, fusion loop; gB, glycoprotein B; RhCMV, rhesus cytomegalovirus; SDS-PAGE, sodium dodecyl sulfate polyacrylamide gel electrophoresis; SS, signal sequence.
Design of a DNA Immunogen Encoding UCD52 pp65-2
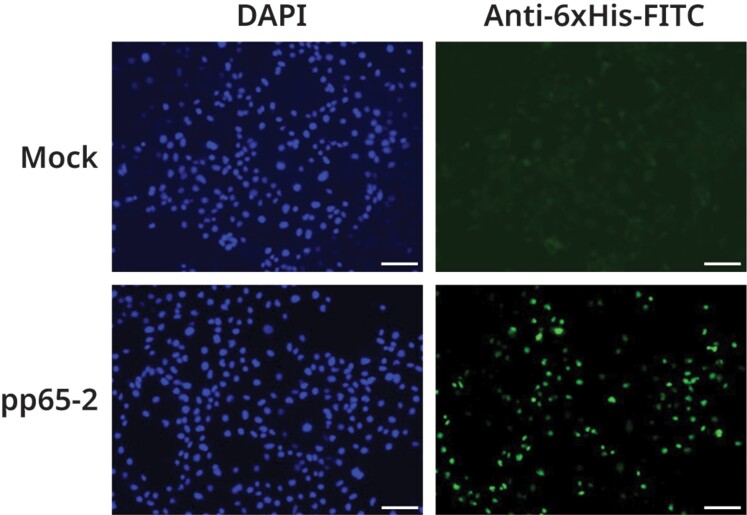
HCMV pp65 is a tegument phosphoprotein and a major target for cytotoxic T-cell responses during natural infection [38, 39]. RhCMV encodes 2 homologues of HCMV pp65: pp65-1 and pp65-2, the latter being the more important cytotoxic T-cell target in chronically RhCMV-infected monkeys [40]. In transiently transfected COS-7 cells, a plasmid encoding the UCD52 RhCMV pp65-2 open reading frame under control of the CMV promoter directed high-level expression of pp65-2, as detected by immunostaining of nuclei with an antibody that recognized the 6×His tag appended to the pp65-2 C-terminus (Figure 3).
Figure 3.
Fluorescence microscopy of RhCMV UCD52 pp65-2 expressed from a transfected DNA vaccine vector. An anti-6×His antibody conjugated with FITC (green) was used to assay the presence of histidine-tagged UCD52 pp65-2 in mock transfected (right upper panel) or DNA vaccine vector transfected (right lower panel) COS-7 cells. Nuclei of the same cells were stained with DAPI (blue in the left panels). Scale bar = 100 µm. Abbreviations: DAPI, 4′,6-diamidino-2-phenylindole; FITC, fluorescein isothiocyanate; RhCMV, rhesus cytomegalovirus.
Immunogenicity of PC, gB674, and pp65 in Rhesus Macaques
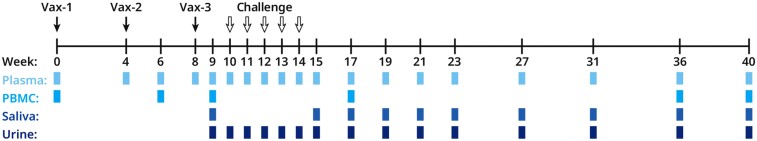
We immunized 4 groups of 6 SPF rhesus macaques at weeks 0, 4, and 8: (1) buffer only (20 mM Tris, 150 mM NaCl, 6 mM EDTA, 0.1% PS80, pH 7.7); (2) PC; (3) PC and gB674; or (4) PC, gB674, and pp65-2 DNA. QS-21 adjuvant (50 µg/0.5 mL) was included in all groups. Protein antigens were delivered by IM injection in 1 arm, and the pp65-encoding DNA was delivered to the contralateral arm by electroporation (Figure 4). Prior to immunization, all animals were confirmed to be seronegative for RhCMV gB-binding IgG. We collected plasma, PMBCs, saliva, and urine at the indicated timepoints (Figure 4).
Figure 4.
Vaccination, challenge, and sample collection timeline. Vaccinations denoted with solid black arrows, oral virus challenges denoted with hollow black arrows, the indicated sample collections denoted with colored boxes. Abbreviations: PBMC, peripheral blood mononuclear cell; Vax, vaccination.
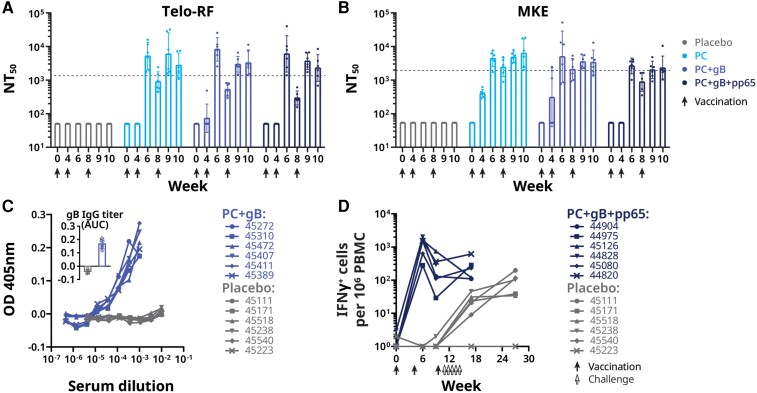
Plasma neutralizing titers against UCD52 were measured without complement on Telo-RF (fibroblast) and MKE (epithelial) target cells (Figure 5A and 5B). Neutralizing titers in animals immunized with PC alone or with PC and gB674 were undetectable 4 weeks after the first dose, and were modestly higher than the geometric mean of the neutralizing titers from 20 naturally infected adult rhesus macaques 2 weeks after the second and third doses, respectively, when the animals were challenged with RhCMV. Neutralizing titers were similar 2 weeks following the second and third doses, as were neutralizing titers measured on fibroblasts or on epithelial cells. Notably, none of the 6 control animals had detectable RhCMV neutralizing titers. All 6 animals immunized with PC and gB674 developed antibodies that bound a tagless version of the gB antigen, detectable in week 10 (2 weeks after dose 3) plasma diluted 1:100 000 (Figure 5C). The control animals remained negative for gB-binding antibodies prior to virus challenge.
Figure 5.
Immune responses elicited by immunization of rhesus macaques with QS-21 and placebo, PC, gB674, and pp65-encoding DNA. A and B, RhCMV neutralizing responses in plasma samples drawn at the indicated time points as measure on Telo-RF (A) and MKE (B). Geometric mean of NT50 from 20 naturally RhCMV-infected rhesus macaques are indicated by the dotted lines. C, RhCMV gB-binding IgG responses in immunized rhesus macaques measured by ELISA (OD 405 nm; inset, AUC). D, IFN-γ ELISPOT responses to pp65-2 DNA vaccination in PBMCs obtained at the indicated times points. Frequency of RhCMV pp65-2–specific IFN-γ–responding cells was enumerated to IFN-γ+ cells per 1 × 106 PBMCs. Each symbol represents the mean of technical triplicates from individual animals. Each animal is represented by its ID number. Abbreviations: AUC, area under the curve; ELISA, enzyme-linked immunosorbent assay; ELISPOT, enzyme-linked immunospot; gB, glycoprotein B; IFN-γ, interferon-γ; IgG, immunoglobulin G; MKE, primary monkey kidney epithelial cell; NT50, mean neutralizing titer; OD, optical density; PBMCs, peripheral blood mononuclear cells; RhCMV, rhesus cytomegalovirus; Telo-RF, transformed fibroblast.
By week 6 (2 weeks after dose 2), PBMCs from all 6 animals immunized with PC, gB674, and pp65-encoding DNA responded to stimulation with a pp65 peptide pool by ELISPOT, with approximately 1000 cells secreting interferon-γ (IFN-γ) per 1 × 106 PBMC (Figure 5D). This level of response is similar to that of PBMCs from naturally infected rhesus macaques (data not shown). In the placebo group, approximately 2 or fewer cells per 1 × 106 PBMC obtained before RhCMV challenge secreted IFN-γ after stimulation with the peptides (Figure 5D).
Protection from RhCMV Challenge
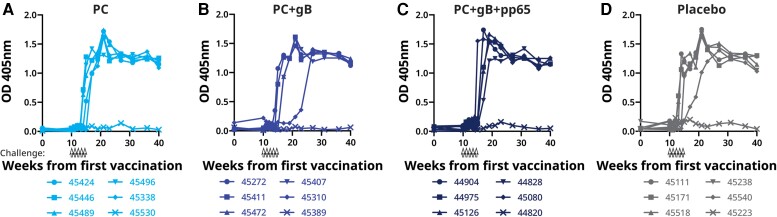
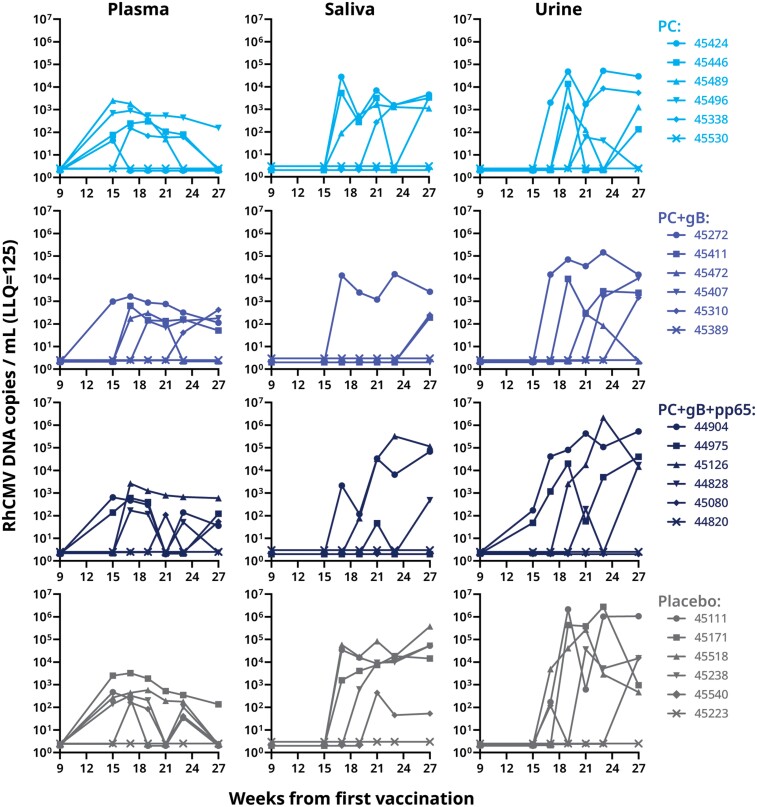
The macaques were orally challenged with 8 × 105 pfu of RhCMV UCD52 weekly for 5 weeks (weeks 10–14) starting 2 weeks after the third vaccine dose (Figure 4). Infection was detected by seroconversion to UCD52 viral interleukin 10 (vIL-10; a RhCMV antigen not used for immunization) and by detection of UCD52 DNA by quantitative polymerase chain reaction (qPCR). Following oral UCD52 challenge, 5 of 6 animals in the control-immunized group and in each of the 3 vaccinated groups developed vIL-10–binding antibody responses and had detectable viral DNA in plasma, saliva, or urine (Figure 6 and Figure 7). We did not observe significant differences in the gB-binding IgG levels and neutralization titers between the nonseroconverted and seroconverted animals within the same group. Detection of viral DNA was intermittent in some animals. Overall, similar quantities of viral DNA were detected after challenge in animals immunized with the RhCMV antigens and in those immunized with the control preparation.
Figure 6.
RhCMV vIL-10–binding IgG antibody in immunized and RhCMV strain UCD52-challenged rhesus macaques. A–D, ELISA results from the groups of animals immunized with PC, PC+gB, PC+gB+pp65 and placebo at various time points. Oral inoculations with RhCMV are indicated by hollow black arrows. The presence of anti–vIL-10 antibody was determined by ELISA. Because RhCMV vIL-10 is not in the vaccine preparations, acquisition of vIL-10–binding antibody is a biomarker of virus acquisition. Absorbance values >0.3 at OD 405 nm indicate seroconversion. Each animal is represented by its ID number. Abbreviations: ELISA, enzyme-linked immunosorbent assay; gB, glycoprotein B; IgG, immunoglobulin G; OD, optical density; PC, pentameric complex; RhCMV, rhesus cytomegalovirus; vIL-10, viral interleukin 10.
Figure 7.
RhCMV DNA detection by qPCR in plasma, saliva, and urine samples from immunized and RhCMV-challenged rhesus macaques. RhCMV was inoculated weekly during weeks 10–14. Each animal is represented by its ID number. Abbreviations: gB, glycoprotein B; LLQ, lower limit of quantification; PC, pentameric complex; qPCR, quantitative polymerase chain reaction; RhCMV, rhesus cytomegalovirus.
DISCUSSION
Immunization of women and adolescent girls with an adjuvanted postfusion gB subunit vaccine provides partial, transient protection from horizontal transmission of HCMV, despite the modest neutralizing titers elicited by this immunogen [12, 13]. Because the PC elicits much higher neutralizing titers than does postfusion gB, and pp65 elicits cytotoxic T cells, it is possible that adding a PC antigen (and potentially a pp65 antigen) to a gB-containing vaccine could provide a much greater level and duration of protection. On the other hand, the ability of HCMV to cause repeated infections—even in those who are already chronically infected and have both neutralizing antibodies and T cells that recognize HCMV [1]—raises questions about whether such immunization can prevent HCMV disease. Therefore, we sought a preclinical proof-of-concept before advancing an HCMV vaccine to clinical testing.
We tested whether a prototype vaccine containing RhCMV postfusion gB and PC with QS-21 adjuvant and pp65-encoding DNA could protect rhesus macaques from oral challenge with RhCMV. QS-21 is a potent vaccine adjuvant that enhances both antibody and cell-mediated responses and is in the formulation of a widely used licensed vaccine [41]. We delivered the RhCMV challenge orally rather than by injection because mucosal exposure is arguably a more relevant model for the horizontal transmission of HCMV in human populations [23]. We chose UCD52 as the challenge strain because, unlike the related strain UCD59, UCD52 contains a functional PC [29, 42]. Immunization of the animals 3 times with preparations that contained PC elicited neutralizing titers as high, or higher than, those elicited by natural RhCMV infection. Moreover, adding postfusion gB elicited strong gB-binding antibodies, and adding electroporated DNA encoding RhCMV pp65 elicited strong T-cell responses that recognized RhCMV pp65 peptides. However, these immune responses did not prevent RhCMV infection after oral inoculation, did not prevent spread to the kidney or the genitourinary tract (as evidenced by viral DNA in the urine), and did not consistently reduce the amount or duration of viral shedding. This failure of protection is reproducible, as we had obtained similar results in a previous study in the same UCD52 challenge model after immunization with a heterologous UCD59 PC antigen, which elicited similar RhCMV neutralizing titers to those observed in the current study (Supplementary Figures 1 and 2). These findings demonstrate the ability of RhCMV to evade vaccine-elicited immunity.
CMV has several immune evasion mechanisms, and infection at mucosal surfaces is difficult to prevent. Once CMV infects at a mucosal surface, it may spread cell to cell and be carried in the bloodstream inside infected cells [43]. More than 40 gene products of the HCMV genome are believed to have immune evasion functions [44]. To protect against CMV, it may be necessary not only to elicit high titers of neutralizing antibodies and strong CD8+ T-cell responses, but also to elicit immune responses that can counter viral immune evasion and augment mucosal immunity. In addition, analysis of binding of cell surface and soluble gB by poorly neutralizing serum IgG from participants in human trials suggests that binding to prefusion gB is potentially associated with protection from horizontal transmission [45]. We recently determined the structure of prefusion gB [14] and are working to produce conformationally homogenous prefusion and postfusion preparations of CMV gB to study the properties of each conformer as antigen and immunogen. Another consideration is that the basis for protection against vertical transmission could be different, with different vaccination outcomes, as suggested by passive transfer experiments to prevent vertical transmission in CD4+ T-cell–depleted pregnant rhesus macaques [46].
One limitation of this study is the small number of immunized macaques. Protection against experimental horizontal transmission was not observed with 6 seronegative macaques receiving each vaccine or placebo preparation. In a human trial of protection against natural horizontal transmission in which 234 seronegative women were immunized with MF59-adjuvanted postfusion gB and 230 received placebo, 50% protection was observed (P = .02) [12]; in a second trial in which 195 seronegative adolescent girls received the same vaccine and 207 received placebo, the level of protection did not reach statistical significance [13]. Thus, if the experimental RhCMV vaccine preparations prevented horizontal transmission in macaques at the modest level observed in human trials of a MF59-adjuvanted postfusion gB subunit vaccine candidate, the number of macaques would be insufficient to detect statistically significant protection.
Another limitation of this study is the need to challenge the rhesus macaques with a homologue of HCMV rather than with HCMV itself. It is possible that rhesus macaques are less amenable to vaccine-induced protection against RhCMV than humans are against HCMV. The UCD52 challenge virus contains genetic changes in UL128 and RL13 [29, 42]. Loss of the gene encoding RL13 has been associated with increased HCMV replication in cell culture in the presence of neutralizing antiserum, as well as with viral resistance to antibody-mediated cytotoxic cells [47, 48]. Although it is possible that our viral challenge regimen of 5 doses of 8 × 105 pfu per oral inoculum is greater than typical human exposures, the challenge dose does not seem excessive, with only 4 of 5 placebo-immunized macaques showing signs of RhCMV infection after challenge in this study. We previously reported that, after a single oral inoculum of 8 × 105 pfu of RhCMV strain UCD52, 3 of 4 unimmunized macaques were infected; with 8 × 104 pfu, 1 of 4 was infected; and with 8 × 103 pfu, zero of 4 were infected [23]. Moreover, 8 × 105 pfu is comparable to the range of RhCMV genome copies (103–106/mL by qPCR) persistently shed in the saliva of animals naturally exposed to endemic strains of RhCMV circulating in group-housed macaques [49]. Human children also shed large quantities of CMV in saliva [50], and multiple daily household exposures of caregivers are possible.
Because transmission of HCMV to the fetus during the first trimester is more likely to cause severe congenital disease than transmission later in pregnancy, in many cases it may be too late to start immunization with a future CMV vaccine by the time pregnancy is recognized and prenatal care is begun. Therefore, for maximal public health impact, it may be necessary to immunize before the childbearing years and achieve a duration of robust prevention lasting through the childbearing years. For these reasons, there is great value in a stringent animal model of protection from CMV infection. Partial, transient protection from CMV is unlikely to be sufficient to prevent congenital disease with real-world use. A prototype vaccine that robustly protects rhesus macaques from RhCMV challenge would provide a strong guide to the composition of a human CMV vaccine that merits clinical testing. Future studies may include further refinement of the RhCMV challenge model, and may include additional CMV vaccine antigens, such as antigens that could elicit immunity that can block HCMV immune evasion.
Supplementary Data
Supplementary materials are available at The Journal of Infectious Diseases online (http://jid.oxfordjournals.org/). Supplementary materials consist of data provided by the author that are published to benefit the reader. The posted materials are not copyedited. The contents of all supplementary data are the sole responsibility of the authors. Questions or messages regarding errors should be addressed to the author.
Supplementary Material
Contributor Information
Julia Li, Vaccine Research and Development, Pfizer, Inc, Pearl River, New York, USA.
Sabine Wellnitz, Vaccine Research and Development, Pfizer, Inc, Pearl River, New York, USA.
Xiaoyuan S Chi, Vaccine Research and Development, Pfizer, Inc, Pearl River, New York, USA.
Yujuan Yue, Center for Comparative Medicine, University of California Davis, Davis, California, USA.
Kimberli A Schmidt, Groton Center for Chemistry, Pfizer, Inc, Groton, Connecticut, USA.
Nancy Nguyen, Groton Center for Chemistry, Pfizer, Inc, Groton, Connecticut, USA.
Wei Chen, Vaccine Research and Development, Pfizer, Inc, Pearl River, New York, USA.
Irina Yurgelonis, Vaccine Research and Development, Pfizer, Inc, Pearl River, New York, USA.
Eduardo Rojas, Vaccine Research and Development, Pfizer, Inc, Pearl River, New York, USA.
Yuhang Liu, Groton Center for Chemistry, Pfizer, Inc, Groton, Connecticut, USA.
Jakob Loschko, Vaccine Research and Development, Pfizer, Inc, Pearl River, New York, USA.
Eneida Pollozi, Vaccine Research and Development, Pfizer, Inc, Pearl River, New York, USA.
Yury V Matsuka, Vaccine Research and Development, Pfizer, Inc, Pearl River, New York, USA.
Elie Needle, Vaccine Research and Development, Pfizer, Inc, Pearl River, New York, USA.
Eugene Vidunas, Vaccine Research and Development, Pfizer, Inc, Pearl River, New York, USA.
Robert G K Donald, Vaccine Research and Development, Pfizer, Inc, Pearl River, New York, USA.
Justin Moran, Vaccine Research and Development, Pfizer, Inc, Pearl River, New York, USA.
Kathrin U Jansen, Vaccine Research and Development, Pfizer, Inc, Pearl River, New York, USA.
Philip R Dormitzer, Vaccine Research and Development, Pfizer, Inc, Pearl River, New York, USA.
Peter A Barry, Center for Comparative Medicine, University of California Davis, Davis, California, USA; Department of Pathology and Laboratory Medicine, University of California Davis, Davis, California, USA; California National Primate Research Center, University of California Davis, Davis, California, USA.
Xinzhen Yang, Vaccine Research and Development, Pfizer, Inc, Pearl River, New York, USA.
Notes
Acknowledgments. The authors acknowledge useful scientific discussion from Drs Christian Schaub, John Johnson, David Keeney, Stanley Mullen, Luke Handke, Paul Liberator, Seungil Han, Xiayang Qiu, and William Gruber; and technical support from Vidia Roopchand, David Keeney, Stan Mullen, Kwok Lee, Willie Sun, Rupal Majmudar, Oleg Jouravlev, Alexey Gribenko, Kunal Bakshi, and Zhiqing He. The authors thank Aaron G. Wexler (Pfizer) for writing, editorial, and graphic design support.
Financial support. This work was supported by the Division of Worldwide Research and Development, Pfizer, Inc.
Potential conflicts of interest. Y. Y., K. A. S., N. N., and P. A. B. were employees of the University of California, Davis when working on the studies under a Research Collaboration Contract with Pfizer, Inc; J. L., S. W., X. C., I. Y., W. C., E. R., Y. L., J. L., E. P., Y. V. M., E. N., E. V., R. G. K. D., J. M., K. J., P. R. D., and X. Y. are or were employees of Pfizer, Inc or its subsidiary and may have been Pfizer shareholders while working on the studies. P. R. D. is currently a GlaxoSmithKline employee and shareholder.
All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
Presented in part: Workshop of Cytomegalovirus Infection: Advancing Strategies for Prevention and Treatment, sponsored by Maternal and Pediatric Infectious Disease Branch and National Institute of Allergy and Infectious Diseases, 4–6 September 2018, Rockville, MD; and 7th International Congenital CMV Conference and 17th International CMV Workshop, 7–11 April 2019, Birmingham, Alabama. Data in this paper were summarized in Plotkin SA, Boppana SB. Vaccination against the human cytomegalovirus. Vaccine 2019 37:7437–42
References
- 1. Kenneson A, Cannon MJ. Review and meta-analysis of the epidemiology of congenital cytomegalovirus (CMV) infection. Rev Med Virol 2007; 17:253–76. [DOI] [PubMed] [Google Scholar]
- 2. Britt WJ. Maternal immunity and the natural history of congenital human cytomegalovirus infection. Viruses 2018; 10:405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Revello MG, Gerna G. Human cytomegalovirus tropism for endothelial/epithelial cells: scientific background and clinical implications. Rev Med Virol 2010; 20:136–55. [DOI] [PubMed] [Google Scholar]
- 4. Wang D, Shenk T. Human cytomegalovirus virion protein complex required for epithelial and endothelial cell tropism. Proc Natl Acad Sci U S A 2005; 102:18153–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Vanarsdall AL, Ryckman BJ, Chase MC, Johnson DC. Human cytomegalovirus glycoproteins gB and gH/gL mediate epithelial cell-cell fusion when expressed either in cis or in trans. J Virol 2008; 82:11837–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Stegmann C, Hochdorfer D, Lieber D, et al. A derivative of platelet-derived growth factor receptor alpha binds to the trimer of human cytomegalovirus and inhibits entry into fibroblasts and endothelial cells. PLoS Pathog 2017; 13:e1006273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Wu Y, Prager A, Boos S, et al. Human cytomegalovirus glycoprotein complex gH/gL/gO uses PDGFR-α as a key for entry. PLoS Pathog 2017; 13:e1006281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Burke HG, Heldwein EE. Crystal structure of the human cytomegalovirus glycoprotein B. PLoS Pathog 2015; 11:e1005227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Chandramouli S, Ciferri C, Nikitin PA, et al. Structure of HCMV glycoprotein B in the postfusion conformation bound to a neutralizing human antibody. Nat Commun 2015; 6:8176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Loomis RJ, Lilja AE, Monroe J, et al. Vectored co-delivery of human cytomegalovirus gH and gL proteins elicits potent complement-independent neutralizing antibodies. Vaccine 2013; 31:919–26. [DOI] [PubMed] [Google Scholar]
- 11. Spaete RR. A recombinant subunit vaccine approach to HCMV vaccine development. Transplant Proc 1991; 23(Suppl 3):90–6. [PubMed] [Google Scholar]
- 12. Pass RF, Zhang C, Evans A, et al. Vaccine prevention of maternal cytomegalovirus infection. N Engl J Med 2009; 360:1191–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Bernstein DI, Munoz FM, Callahan ST, et al. Safety and efficacy of a cytomegalovirus glycoprotein B (gB) vaccine in adolescent girls: a randomized clinical trial. Vaccine 2016; 34:313–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Liu Y, Heim KP, Che Y, et al. Prefusion structure of human cytomegalovirus glycoprotein B and structural basis for membrane fusion. Sci Adv 2021; 7:eabf3178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Macagno A, Bernasconi NL, Vanzetta F, et al. Isolation of human monoclonal antibodies that potently neutralize human cytomegalovirus infection by targeting different epitopes on the gH/gL/UL128-131A complex. J Virol 2010; 84:1005–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wen Y, Monroe J, Linton C, et al. Human cytomegalovirus gH/gL/UL128/UL130/UL131A complex elicits potently neutralizing antibodies in mice. Vaccine 2014; 32:3796–804. [DOI] [PubMed] [Google Scholar]
- 17. Kabanova A, Perez L, Lilleri D, et al. Antibody-driven design of a human cytomegalovirus gHgLpUL128L subunit vaccine that selectively elicits potent neutralizing antibodies. Proc Natl Acad Sci U S A 2014; 111:17965–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Chiuppesi F, Wussow F, Johnson E, et al. Vaccine-derived neutralizing antibodies to the human cytomegalovirus gH/gL pentamer potently block primary cytotrophoblast infection. J Virol 2015; 89:11884–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Wang D, Freed DC, He X, et al. A replication-defective human cytomegalovirus vaccine for prevention of congenital infection. Sci Transl Med 2016; 8:362ra145. [DOI] [PubMed] [Google Scholar]
- 20. Itell HL, Kaur A, Deere JD, Barry PA, Permar SR. Rhesus monkeys for a nonhuman primate model of cytomegalovirus infections. Curr Opin Virol 2017; 25:126–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Wussow F, Yue Y, Martinez J, et al. A vaccine based on the rhesus cytomegalovirus UL128 complex induces broadly neutralizing antibodies in rhesus macaques. J Virol 2013; 87:1322–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Plotkin SA, Wang D, Oualim A, et al. The status of vaccine development against the human cytomegalovirus. J Infect Dis 2020; 221(Suppl 1):S113–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Yue Y, Chang WLW, Li J, et al. Pathogenesis of wild-type-like rhesus cytomegalovirus strains following oral exposure of immune-competent rhesus macaques. J Virol 2022; 96:e0165321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Bieniossek C, Richmond TJ, Berger I. MultiBac: multigene baculovirus-based eukaryotic protein complex production. Curr Protoc Protein Sci 2008; Chapter 5:Unit 5.20. [DOI] [PubMed] [Google Scholar]
- 25. Fitzgerald DJ, Berger P, Schaffitzel C, Yamada K, Richmond TJ, Berger I. Protein complex expression by using multigene baculoviral vectors. Nat Methods 2006; 3:1021–32. [DOI] [PubMed] [Google Scholar]
- 26. Abel K, Martinez J, Yue Y, et al. Vaccine-induced control of viral shedding following rhesus cytomegalovirus challenge in rhesus macaques. J Virol 2011; 85:2878–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Loudon PT, Yager EJ, Lynch DT, et al. GM-CSF increases mucosal and systemic immunogenicity of an H1N1 influenza DNA vaccine administered into the epidermis of non-human primates. PLoS One 2010; 5:e11021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Yue Y, Kaur A, Lilja A, Diamond DJ, Walter MR, Barry PA. The susceptibility of primary cultured rhesus macaque kidney epithelial cells to rhesus cytomegalovirus strains. J Gen Virol 2016; 97:1426–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Oxford KL, Strelow L, Yue Y, et al. Open reading frames carried on UL/b′ are implicated in shedding and horizontal transmission of rhesus cytomegalovirus in rhesus monkeys. J Virol 2011; 85:5105–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Chang WL, Kirchoff V, Pari GS, Barry PA. Replication of rhesus cytomegalovirus in life-expanded rhesus fibroblasts expressing human telomerase. J Virol Methods 2002; 104:135–46. [DOI] [PubMed] [Google Scholar]
- 31. Barry PA, Strelow L. Development of breeding populations of rhesus macaques (Macaca mulatta) that are specific pathogen-free for rhesus cytomegalovirus. Comp Med 2008; 58:43–6. [PMC free article] [PubMed] [Google Scholar]
- 32. dela Pena MG, Strelow L, Barry PA, Abel K. Use of specific-pathogen-free (SPF) rhesus macaques to better model oral pediatric cytomegalovirus infection. J Med Primatol 2012; 41:225–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Yue Y, Kaur A, Eberhardt MK, et al. Immunogenicity and protective efficacy of DNA vaccines expressing rhesus cytomegalovirus glycoprotein B, phosphoprotein 65-2, and viral interleukin-10 in rhesus macaques. J Virol 2007; 81:1095–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Abel K, Strelow L, Yue Y, Eberhardt MK, Schmidt KA, Barry PA. A heterologous DNA prime/protein boost immunization strategy for rhesus cytomegalovirus. Vaccine 2008; 26:6013–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Chang WL, Barry PA. Attenuation of innate immunity by cytomegalovirus IL-10 establishes a long-term deficit of adaptive antiviral immunity. Proc Natl Acad Sci U S A 2010; 107:22647–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Eberhardt MK, Deshpande A, Chang WL, Barthold SW, Walter MR, Barry PA. Vaccination against a virus-encoded cytokine significantly restricts viral challenge. J Virol 2013; 87:11323–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Ciferri C, Chandramouli S, Donnarumma D, et al. Structural and biochemical studies of HCMV gH/gL/gO and pentamer reveal mutually exclusive cell entry complexes. Proc Natl Acad Sci U S A 2015; 112:1767–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. McLaughlin-Taylor E, Pande H, Forman SJ, et al. Identification of the major late human cytomegalovirus matrix protein pp65 as a target antigen for CD8+ virus-specific cytotoxic T lymphocytes. J Med Virol 1994; 43:103–10. [DOI] [PubMed] [Google Scholar]
- 39. Wills MR, Carmichael AJ, Mynard K, et al. The human cytotoxic T-lymphocyte (CTL) response to cytomegalovirus is dominated by structural protein pp65: frequency, specificity, and T-cell receptor usage of pp65-specific CTL. J Virol 1996; 70:7569–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Yue Y, Kaur A, Zhou SS, Barry PA. Characterization and immunological analysis of the rhesus cytomegalovirus homologue (Rh112) of the human cytomegalovirus UL83 lower matrix phosphoprotein (pp65). J Gen Virol 2006; 87:777–87. [DOI] [PubMed] [Google Scholar]
- 41. Pulendran B, Arunachalam PS, O'Hagan DT. Emerging concepts in the science of vaccine adjuvants. Nat Rev Drug Discov 2021; 20:454–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Taher H, Mahyari E, Kreklywich C, et al. In vitro and in vivo characterization of a recombinant rhesus cytomegalovirus containing a complete genome. PLoS Pathog 2020; 16:e1008666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Landolfo S, Gariglio M, Gribaudo G, Lembo D. The human cytomegalovirus. Pharmacol Ther 2003; 98:269–97. [DOI] [PubMed] [Google Scholar]
- 44. Patro ARK. Subversion of immune response by human cytomegalovirus. Front Immunol 2019; 10:1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Jenks JA, Nelson CS, Roark HK, et al. Antibody binding to native cytomegalovirus glycoprotein B predicts efficacy of the gB/MF59 vaccine in humans. Sci Transl Med 2020; 12:eabb3611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Nelson CS, Cruz DV, Tran D, et al. Preexisting antibodies can protect against congenital cytomegalovirus infection in monkeys. JCI Insight 2017; 2:e94002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Stanton RJ, Baluchova K, Dargan DJ, et al. Reconstruction of the complete human cytomegalovirus genome in a BAC reveals RL13 to be a potent inhibitor of replication. J Clin Invest 2010; 120:3191–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Murrell I, Bedford C, Ladell K, et al. The pentameric complex drives immunologically covert cell-cell transmission of wild-type human cytomegalovirus. Proc Natl Acad Sci U S A 2017; 114:6104–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Eberhardt MK, Deshpande A, Fike J, et al. Exploitation of interleukin-10 (IL-10) signaling pathways: alternate roles of viral and cellular IL-10 in rhesus cytomegalovirus infection. J Virol 2016; 90:9920–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Stowell JD, Mask K, Amin M, et al. Cross-sectional study of cytomegalovirus shedding and immunological markers among seropositive children and their mothers. BMC Infect Dis 2014; 14:568. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.