Abstract
Background
Following widespread use of the Haemophilus influenzae serotype b (Hib) vaccine, H. influenzae serotype a (Hia) has emerged as an important pathogen in children in some regions. We describe the clinical features and molecular epidemiology of invasive Hia disease in children in Utah over an 11-year period.
Methods
We identified cases of invasive Hia disease, defined as detection of Hia from a normally sterile site, in children aged <18 years from Utah between 2007 and 2017. Medical records were reviewed to determine demographic characteristics and clinical outcomes. Available Hia isolates were genotyped using multilocus sequence typing, and phylogenetic division was determined using sodC polymerase chain reaction. Presence of the putative virulence-associated IS1016-bexA duplication-deletion was evaluated.
Results
We identified 51 children with invasive Hia. The average annual incidence was 1.7 cases per 100 000 children aged <5 years; 4.8 cases per 100 000 children aged <1 year. The median age was 11.3 months. The most common clinical presentation was meningitis (53%), followed by pneumonia (14%) and septic arthritis (14%). Twenty-two children (43%) required admission to an intensive care unit; 1 died. Sequence type (ST) 62, phylogenetic division II isolates caused 75% (21/28) of disease. No isolates contained the virulence-associated IS1016-bexA duplication-deletion.
Conclusions
Hia is a significant cause of severe invasive bacterial infection in Utah. The majority of infections were caused by ST62 isolates, a phylogenetic division II Hia type that lacks the IS1016-bexA duplication-deletion. Hia ST62 has not been commonly reported elsewhere, suggesting a unique molecular epidemiology in our population.
Keywords: children, Haemophilus influenza, Hia, meningitis, ST62
Haemophilus influenzae is an important cause of serious invasive disease in infants, young children, and older adults. Haemophilus influenzae strains are classified by the presence (serotypes a–f) or absence (nontypeable) of a polysaccharide capsule [1]. The presence of a polysaccharide capsule is associated with increased disease severity [2–4]. Encapsulated H. influenzae can cause meningitis, septicemia, pneumonia, septic arthritis, cellulitis, and epiglottitis and is associated with significant morbidity and mortality in children [2, 5–7].
Historically, H. influenzae serotype b (Hib) was the primary cause of invasive H. influenzae disease in children in the United States and throughout the world [8, 9]. Following introduction of the Hib-specific conjugate vaccine in the late 1980s, the incidence of invasive Hib disease decreased dramatically [2, 6]. However, since widespread use of the Hib vaccine, there has been an increase in invasive H. influenzae disease caused by non-b serotypes, particularly H. influenzae serotype a (Hia) in children [6, 7, 10–12]. Clinical presentation of children with invasive Hia resembles that of Hib in both syndromes (meningitis, pneumonia, septic arthritis, and cellulitis) and severity [7, 10, 13, 14].
Determinants of H. influenzae virulence are not fully understood. The presence of the type b capsular locus is associated with increased pathogenicity and is a major virulence determinant [15, 16]. Studies have also reported an association between more severe disease and Hia strains with a Hib-like genetic variant, the IS1016-bexA duplication-deletion, which results in a tandem duplication of the capsule region and is hypothesized to increase capsule production and thus pathogenicity [12, 13, 17–19]. Differences in pathogenicity have also been associated with specific electrophoretic types, for example, some phylogenetic division I isolates are associated with increased virulence when compared with isolates of the same capsular type in phylogenetic division II [20]. Variations in lipopolysaccharide type and immunoglobulin A protease have also been implicated in H. influenzae pathogenicity [21, 22].
Here, we report on the epidemiology and clinical presentation of invasive Hia infection in Utah children over an 11-year period. We examine the molecular epidemiology and prevalence of the IS1016-bexA duplication-deletion.
METHODS
Setting and Study Population
We identified children aged <18 years residing in Utah with invasive Hia infection treated at Intermountain Healthcare facilities between 1 January 2007 and 31 December 2017 via both prospective surveillance from 2009–2017 as well as a retrospective review from 2007 to 2009. Intermountain Healthcare is a Utah-based not-for-profit healthcare system with 22 hospitals, including Primary Children’s Hospital (PCH), a 289-bed children’s hospital that serves as both a pediatric community hospital for Salt Lake County, Utah, and as the only tertiary care pediatric referral center in the state of Utah. Approximately 90% of pediatric admissions in the state of Utah occur in hospitals within the Intermountain Healthcare system.
Invasive disease was defined according to the following Centers for Disease Control and Prevention (CDC) definition: a clinically compatible case confirmed by isolation of Hia from a normally sterile site, such as blood, cerebrospinal fluid, joint fluid, pleural fluid, or pericardial fluid. Demographic characteristics and clinical information were obtained from the Intermountain Healthcare Enterprise Data Warehouse (IHC EDW). Manual review of the medical records was performed to confirm diagnosis and validate electronic data for all patients.
The University of Utah and the PCH institutional review boards approved this study with a waiver of informed consent.
Bacterial Isolates and Molecular Typing
The microbiology laboratory at PCH collects all H. influenzae isolates recovered from a normally sterile site. Beta-lactamase testing was performed in individual microbiology laboratories; results were extracted from the IHC EDW. Isolates from outlying facilities were transferred to PCH when available. Haemophilus influenzae were identified by morphology (Gram stain and growth on chocolate agar) followed by confirmation with mass spectrometry (matrix-assisted laser desorption/ionization time-of-flight). Capsular serotyping was performed by slide agglutination with H. influenzae a–f typing antisera at the Utah Department of Health laboratory. Bacterial isolates were stored frozen at –80°C until further analysis.
Hia isolates were inoculated on chocolate agar and incubated at 37°C with 5% carbon dioxide for 22–24 hours prior to DNA extraction. Bacterial DNA was extracted using the DNeasy Blood and Tissue Kit (Qiagen, Venlo, the Netherlands). Polymerase chain reaction (PCR) amplification of the intergenic region between the bexA and sodC genes was performed as previously described and was used to infer the phylogenetic group of each isolate; sodC-negative strains belong to phylogenetic division I and sodC-positive isolates belong to phylogenetic division II [23, 24]. Multilocus sequence typing (MLST) was performed as previously described utilizing the H. influenzae MLST website (https://pubmlst.org/hinfluenzae) [25, 26]. Isolate sequence type (ST) was determined using total genome sequencing data as previously described [27]. Evaluation for the presence or absence of the IS1016-bexA duplication-deletion was performed using PCR as previously described [28]. A phylogenetic division I IS1016-bexA duplication-deletion–positive Hib strain was used as a positive control. PCR products, when present, were confirmed by agarose gel electrophoresis and Sanger sequencing.
Statistical Analyses
All children who met the clinical case definition for invasive Hia disease were included in the description of clinical syndromes. Incidence rates were based on Utah residents. Rates were calculated using the mean of annual age-specific estimates from 2007–2017 from the Utah Department of Health [29]. Population by race and ethnicity was estimated from US census bureau estimates of race and ethnicity composition of the Utah population from 2017. Exact binomial confidence intervals (Clopper-Pearson) were calculated using STATA 15 (StataCorp, College Station, TX).
RESULTS
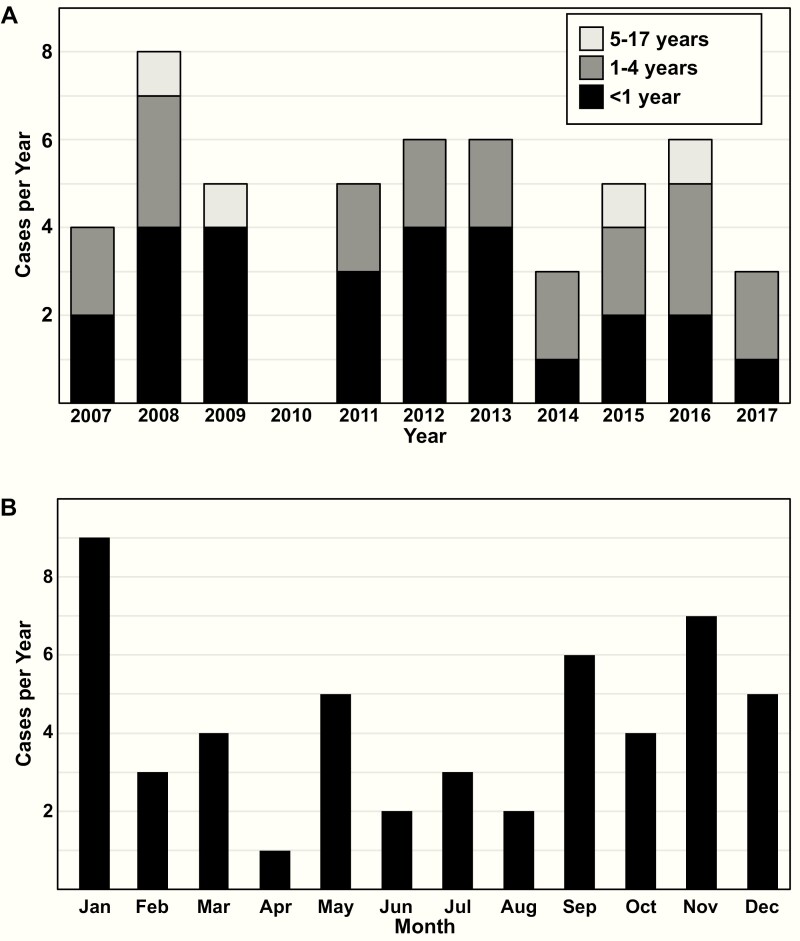
We identified 51 children with invasive Hia infections treated at IHC facilities between 2007 and 2017. Twenty-seven cases (52.9%) occurred in children aged <1 year and 47 (92%) cases occurred in children aged <5 years (Table 1). The incidence was highest among children aged <1year (4.8/100 000; 95% confidence interval, 3.1 to 6.9). Incidence rates were similar by race and ethnicity, although the confidence intervals are wide (Table 2). The annual number of cases was similar over the 11 years of the study (Figure 1A), with only modest seasonality (Figure 1B).
Table 1.
Demographic and Clinical Characteristics of Patients With Invasive Haemophilus influenzae Serotype a Disease
| Demographic Characteristic | Value (n = 51) |
|---|---|
| Age, median (IQR), months | 11.3 (6.2–41) |
| Age group, n (%), years | |
| <1 | 27 (52.9) |
| 1–4 | 20 (39.2) |
| 5–17 | 4 (7.9) |
| Male sex, n (%) | 27 (53) |
| Race/ethnicity, n (%) | |
| White | 39 (76.5) |
| Black | 2 (3.9) |
| Asian/Native, Hawaiian/Pacific Islander | 2 (3.9) |
| Hispanic | 3 (5.9) |
| American Indian/Alaskan Native | 1 (2) |
| Unknown | 4 (7.8) |
| Clinical characteristics | |
| Disease classification, n (%) | |
| Meningitisa | 27 (52.9) |
| Pneumonia with bacteremia | 7 (13.7) |
| Septic arthritisa | 7 (13.7) |
| Bacteremia (all other sources)b | 10 (19.6) |
| Outcomes | |
| Hospital admission, n (%) | 49 (96) |
| Hospital LOS, days, median (IQR) | 4.8 (2–10) |
| ICU admission, n (%) | 22 (43.1) |
| ICU LOS, median (IQR), days | 2 (1.4–5.6) |
| Death, n (%) | 1 (2) |
Abbreviations: ICU, intensive care unit; IQR, interquartile range; LOS, length of stay.
aOne patient was classified as having both arthritis and meningitis.
bBacteremia was in association with central line associated blood stream infection (1), peritonitis (1), acute otitis media (3), or no identifiable source (4).
Table 2.
Incidence of Invasive Haemophilus influenzae Serotype a Disease by Age and Race/Ethnicity
| Demographic Characteristics | Incidence/100 000a | 95% Confidence Interval |
|---|---|---|
| Age group, y | ||
| <1 | 4.8 | 3.1–6.9 |
| 1–4 | 0.89 | 0.5–1.4 |
| <5 | 1.7 | 1.2–2.2 |
| 5–17 | 0.03 | 0.004–0.1 |
| All ages | 0.5 | 0.4–0.7 |
| Race/ethnicity (aged <5 years) | ||
| White | 1.5 | 0.1–2 |
| Black | 5.1 | 0.6–18.3 |
| Asian/Native, Hawaiian/Pacific Islander | 7.1 | 0.86–25.6 |
| Hispanic | 0.3 | 0.06–14.1 |
| American Indian/Alaskan Native | 2.4 | 0.6–13.1 |
aAge- or race/ethnicity-specific incidence.
Figure 1.
Number of invasive Haemophilus influenzae serotype a infections, Utah, 2007 to 2017. (A) Number of cases per year by age group; (B) Number of cases by month of occurrence.
The most common clinical manifestation was meningitis (27 children; 54%), followed by bacteremic pneumonia (7 children; 14%) and septic arthritis (7 children; 14%; Table 1). Hia was detected in the blood of 43 children, including 85% (23/27) of children with meningitis and 57% (4/7) of children with septic arthritis. Of the 27 children with meningitis, 7 (25.9%) developed seizures, 14 (51.8%) had intracranial complications that led to prolonged antibiotic therapy, and 7 (25.9%) suffered hearing loss. Infection with a beta-lactamase–producing isolate was relatively uncommon, occurring in only 5 of 48 children (10.4%).
All but 2 children with invasive disease were hospitalized for treatment of their infection. For those hospitalized, the median hospital length of stay was 4.8 days, ranging from 1 to 48 days (Table 1). Twenty-two children (43.1%) were admitted to the intensive care unit (ICU). One 5-month-old with meningitis and septic shock died. No child developed purpura fulminans.
Twenty-eight Hia isolates (56%) were available for molecular analysis (Table 3). There were no significant differences in patient demographics, clinical characteristics, or outcomes between those with and without an available isolate (Supplementary Table 1). MLST demonstrated that the most common ST was ST62, which accounted for 75% (21/28) of isolates, followed by ST576 (4/28, 14.2%) and ST56 (3/28, 10.7%). Based on available molecular data, there did not appear to be differences in circulating strains over time (Supplementary Figure 1). All ST576 and ST56 isolates lacked sodC and were classified into phylogenetic division I, while all ST62 contained sodC and were classified into phylogenetic division II.
Table 3.
Genetic Characterization of Available Haemophilus influenzae Serotype a Isolates
| Multilocus Sequence Typing | na(%) | Phylogenetic Division | IS1016-bexA Duplication-Deletion |
|---|---|---|---|
| 56 | 3 (10.7) | I | Not present |
| 62 | 21 (75) | II | Not present |
| 576 | 4 (14.2) | I | Not present |
aAvailable isolates, n=28.
The IS1016-bexA duplication-deletion was not detected in any Hia isolates in our collection. There were no significant associations between the clinical characteristics of patients, including disease classifications and outcomes, and any specific ST. Meningitis occurred in 33.3% (1/3) of children with ST56, 61.9% (13/21) of children with ST62, and 25% (1/4) of children with ST576 isolates. ICU admission occurred in 66% (2/3) of children with ST56, 57% (12/21) of children with ST62, and 50% (2/4) of children with ST 576 isolates. The only death occurred in an infant infected with a ST62 isolate.
Discussion
In this study, we describe the clinical and molecular epidemiology of a large series of cases of invasive Hia disease in children over a 11-year period in Utah. The incidence of invasive Hia disease in our population was 4.8/100 000 among infants aged <1 year and 1.7/100 000 among children aged <5 years, which is substantially higher than estimates from the 10 surveillance sites in the CDC Active Bacterial Core (ABC) surveillance system but lower than rates reported from Alaska [6, 10, 30]. Hia caused severe disease; roughly half of the children in our study had meningitis and a similar proportion required ICU care. Molecular analysis showed a predominance of phylogenetic division II ST62 strains, which has not been reported elsewhere. Despite the severe disease, no Hia isolates contained the IS1016-bexA duplication-deletion, a putative virulence factor. Our data suggest a unique epidemiology of disease in our population.
Prior to the introduction of Hib vaccine in the United States, the incidence of invasive Hib disease was approximately 54 cases per 100 000 children aged <5 years [2]. Vaccination has dramatically decreased the incidence of Hib more than 200-fold to 0.13 cases/100 000 in children aged < 5 years [6]. Over time, non-b H. influenzae serotypes as well as nontypeable H. influenzae have emerged as important causes of illness. In particular, Hia disease has emerged among children [6, 7, 31]. The incidence of Hia in children may be increasing, with a number of reports of invasive Hia disease coming from Alaska, northern Canada as well as New Mexico and Utah [7, 10, 13, 14, 30, 32–35]. The incidence of Hia disease has been found to be particularly high among indigenous populations, such as American Indians and Alaska Natives, ranging from 15.2 per 100 000 children aged <5 years to 34.4 per 100 000 for infants aged <1 year [6, 10, 14, 35].
Two earlier studies examined Hia disease in Utah. One study included a cluster of 5 cases over a 10-month period in 1998–1999, including 1 case of purpura fulminans [13]. A second epidemiologic study from 1998–2008 demonstrated an increasing incidence of invasive Hia disease, from 0.8/100 000 among children aged <5 years in 1998 to 2.6/100 000 in 2008 [7]. A recent report on invasive H. influenzae incidence conducted through ABC surveillance sites reported an incidence for Hia of 1.52/100 000 among children aged <1 year and 0.4/100 000 among children aged 1 to 4 years [6]. These rates are substantially lower than in the earlier series from Utah and in our current study. The incidence of Hia in Utah is lower than that of reports in Alaska and Canada, where disease occurs predominantly in indigenous populations. The majority of cases (76.5%) occurred among white children, reflecting the racial makeup of the state where 78% of people are non-Hispanic, non-Latino white. When we calculated race- and ethnicity-specific rates, we did not observe any meaningful difference in incidence of invasive Hia disease.
Clinical syndromes associated with invasive Hia disease in this study were severe and resembled those for Hib. More than 50% of children had meningitis and nearly half required ICU care. The severity of disease presentation among patients in this study is similar to previously published reports of both Hia and Hib [2, 5, 6, 10, 14, 30, 34, 35]. Mortality was low in our cohort compared with other studies, particularly those reporting on Hia disease among indigenous populations [10, 30]. Plumb et al reported a case fatality rate of 11% (4/36), while Millar et al reported 2.6% (2/76). Affected children in these studies frequently lived in rural communities with challenging access to tertiary care [10, 14, 30, 35]. Despite the large geographic area of Utah, there is a well-organized network of rural care with relatively efficient emergency transportation to the tertiary care pediatric center in Salt Lake City. This difference in mortality could also be due to circulation of a less virulent type of Hia (ST62) as the predominant cause of disease in our population. However, the only death in our population was associated with ST62, and disease severity measured by clinical syndrome and ICU admission was not different for ST62 isolates compared with clonal complex (CC) 23 isolates.
The molecular epidemiology of invasive Hia in Utah appears to be unique from other sites reporting significant invasive Hia disease. The majority (21/28; 75%) of isolates that were typed belonged to phylogenetic division II ST62. Although characterization by ST and/or phylogenetic division is not always reported in epidemiologic studies, ST62 has not, to our knowledge, previously been described as a predominant ST causing invasive disease in a population or geographic area. In Canada and Alaska, the majority of invasive Hia disease has been caused by isolates from phylogenetic division I, particularly CC23 isolates (ST23, ST56, and ST576). In a large surveillance study in Brazil, all Hia isolates belonged to CC23 [14, 17, 24, 30, 36, 37]. Musser et al analyzed a large cohort of colonizing and invasive H. influenzae isolates and reported a strong association between phylogenetic division I Hia isolates and invasive disease [20]. Interestingly, we identified several ST62 or related isolates in the MLST Database reported from New Mexico. It is possible that this represents the prevalence of ST62 across a larger region [26]. Unfortunately, there is a paucity of longitudinal and geographically dispersed data on the molecular epidemiology of invasive Hia disease. While several studies have reported an association between more severe disease and phylogenetic division I ST4 or CC23 strains of Hia, the severe disease associated with ST62 in our region suggests that the molecular determinants of virulence are not yet fully understood [18, 20].
Based on the earlier study of Hia in Utah that showed severe disease caused by isolates that contained the IS1016-bexA duplication-deletion, we had hypothesized that currently circulating strains may also have this genotype [13]. Surprisingly, all 28 isolates that we evaluated were negative for this putative virulence factor. In other published studies, only ST4 Hia isolates contained the IS1016-bexA duplication-deletion [18, 24, 38]. Our data would thus suggest that isolates involved in the 1998–1999 cluster of cases did not persist in our population and that the IS1016-bexA duplication-deletion is not required for H. influenzae virulence.
Our study has several limitations. Cases of Hia in our study outside of PCH were identified retrospectively, and thus we may have missed cases that were treated at other sites in our hospital system, sites outside of our system, or in which serotyping was not performed. This may have resulted in an underestimate of the incidence of invasive Hia. While more than half of our cases had isolates available for molecular analysis, it is possible that the pattern of Hia ST distribution or molecular characteristics would be different if we had a more complete collection. Both the clinical and molecular epidemiology of Hia appears to be different in Utah when compared with Alaska and Canada, but we do not know how it compares with other US populations. Isolates from before 2006, including those from the previously reported cluster, were not available to us at the time of molecular analysis and comparisons to the more contemporary cases.
Conclusions
Invasive Hia infections are an important cause of severe disease in Utah, primarily affecting children aged <5 years. The clinical presentation of disease is dominated by meningitis and is similar to prior reports of both Hib and Hia. Two-thirds of Hia isolates available for typing belonged to ST62, a phylogenetic division II Hia type that lacks the IS1016-bexA duplication-deletion. ST62 has not been widely reported or studied, suggesting a unique molecular epidemiology in our population. Further research is needed to understand the epidemiology of Hia disease in children in other regions and to explore the molecular mechanisms of Hia virulence.
Supplementary Material
Note
Acknowledgments. We thank the Primary Children’s Hospital Microbiology Laboratory for their assistance with isolate identification and collection as well as the Utah Public Health Laboratory for their assistance with serotyping of isolates. This study made use of the Haemophilus influenzae multilocus sequence typing website (https://pubmlst.org/hinfluenzae/), which is sited at the University of Oxford (Jolley KA, Bray JE, Maiden MCJ. Open-access bacterial population genomics: BIGSdb software, the PubMLST.org website and their applications. Wellcome Open Res 2018; 3:124). The development of this site was funded by the Wellcome Trust.
Potential conflicts of interest. All authors: No reported conflicts of interest. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
Contributor Information
Hillary Crandall, Department of Pediatrics, University of Utah School of Medicine, Salt Lake City, Utah, USA.
Jennifer Christiansen, Department of Pediatrics, University of Utah School of Medicine, Salt Lake City, Utah, USA.
Alyssa A Varghese, Department of Pediatrics, University of Utah School of Medicine, Salt Lake City, Utah, USA.
Adam Russon, Department of Pediatrics, University of Utah School of Medicine, Salt Lake City, Utah, USA.
E Kent Korgenski, Department of Pediatrics, University of Utah School of Medicine, Salt Lake City, Utah, USA; Pediatric Clinical Program, Intermountain Health Care, Salt Lake City, Utah, USA.
Erika K Bengtson, Department of Pediatrics, University of Utah School of Medicine, Salt Lake City, Utah, USA.
Mandy Dickey, Department of Microbiology, Primary Children’s Hospital, Salt Lake City, Utah, USA.
Jarrett Killpack, Department of Pediatrics, University of Utah School of Medicine, Salt Lake City, Utah, USA.
Elizabeth D Knackstedt, Department of Pediatrics, University of Utah School of Medicine, Salt Lake City, Utah, USA.
Judy A Daly, Department of Microbiology, Primary Children’s Hospital, Salt Lake City, Utah, USA; Department of Pathology, University of Utah School of Medicine, Salt Lake City, Utah, USA.
Krow Ampofo, Department of Pediatrics, University of Utah School of Medicine, Salt Lake City, Utah, USA.
Andrew T Pavia, Department of Pediatrics, University of Utah School of Medicine, Salt Lake City, Utah, USA.
Anne J Blaschke, Department of Pediatrics, University of Utah School of Medicine, Salt Lake City, Utah, USA.
References
- 1. Pittman M. Variation and type specificity in the bacterial species Haemophilus influenzae. J Exp Med 1931; 53:471–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Peltola H. Worldwide Haemophilus influenzae type b disease at the beginning of the 21st century: global analysis of the disease burden 25 years after the use of the polysaccharide vaccine and a decade after the advent of conjugates. Clin Microbiol Rev 2000; 13:302–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Kroll JS, Moxon ER. Capsulation and gene copy number at the cap locus of Haemophilus influenzae type b. J Bacteriol 1988; 170:859–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Weller PF, Smith AL, Anderson P, Smith DH. The role of encapsulation and host age in the clearance of Haemophilus influenzae bacteremia. J Infect Dis 1977; 135:34–41. [DOI] [PubMed] [Google Scholar]
- 5. Monge S, Mollema L, de Melker H, Sanders E, van der Ende A, Knol M. Clinical characterization of invasive disease caused by Haemophilus influenzae Serotype b in a high vaccination coverage setting. J Pediatric Infect Dis Soc 2018; 8(3):261–4. [DOI] [PubMed] [Google Scholar]
- 6. Soeters HM, Blain A, Pondo T, et al. Current epidemiology and trends in invasive Haemophilus influenzae disease— United States, 2009–2015. Clin Infect Dis 2018; 67(6):881–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bender JM, Cox CM, Mottice S, et al. Invasive Haemophilus influenzae disease in Utah children: an 11-year population-based study in the era of conjugate vaccine. Clin Infect Dis 2010; 50:e41–6. [DOI] [PubMed] [Google Scholar]
- 8. Broome CV. Epidemiology of Haemophilus influenzae type b infections in the United States. Pediatr Infect Dis J 1987; 6:779–82. [DOI] [PubMed] [Google Scholar]
- 9. Watt JP, Wolfson LJ, O’Brien KL, et al. Burden of disease caused by Haemophilus influenzae type b in children younger than 5 years: global estimates. Lancet 2009; 374:903–11. [DOI] [PubMed] [Google Scholar]
- 10. Plumb ID, Lecy KD, Singleton R, et al. Invasive Haemophilus influenzae serotype a infection in children: clinical description of an emerging pathogen— Alaska, 2002–2014. Pediatr Infect Dis J 2018; 37:298–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Tsang RSW, Proulx JF, Hayden K, et al. Characteristics of invasive Haemophilus influenzae serotype a (Hia) from Nunavik, Canada and comparison with Hia strains in other North American Arctic regions. Int J Infect Dis 2017; 57:104–7. [DOI] [PubMed] [Google Scholar]
- 12. Ulanova M, Tsang RSW. Haemophilus influenzae serotype a as a cause of serious invasive infections. Lancet Infect Dis 2014; 14:70–82. [DOI] [PubMed] [Google Scholar]
- 13. Adderson EE, Byington CL, Spencer L, et al. Invasive serotype a Haemophilus influenzae infections with a virulence genotype resembling Haemophilus influenzae type b: emerging pathogen in the vaccine era? Pediatrics 2001; 108:E18. [DOI] [PubMed] [Google Scholar]
- 14. Kelly L, Tsang RS, Morgan A, Jamieson FB, Ulanova M. Invasive disease caused by Haemophilus influenzae type a in Northern Ontario First Nations communities. J Med Microbiol 2011; 60:384–90. [DOI] [PubMed] [Google Scholar]
- 15. Moxon ER, Vaughn KA. The type b capsular polysaccharide as a virulence determinant of Haemophilus influenzae: studies using clinical isolates and laboratory transformants. J Infect Dis 1981; 143:517–24. [DOI] [PubMed] [Google Scholar]
- 16. Zwahlen A, Kroll JS, Rubin LG, Moxon ER. The molecular basis of pathogenicity in Haemophilus influenzae: comparative virulence of genetically-related capsular transformants and correlation with changes at the capsulation locus cap. Microb Pathog 1989;7: 225–35. [DOI] [PubMed] [Google Scholar]
- 17. Kapogiannis BG, Satola S, Keyserling HL, Farley MM. Invasive infections with Haemophilus influenzae serotype a containing an IS1016-bexA partial deletion: possible association with virulence. Clin Infect Dis 2005; 41:e97–103. [DOI] [PubMed] [Google Scholar]
- 18. Lima JB, Ribeiro GS, Cordeiro SM, et al. Poor clinical outcome for meningitis caused by Haemophilus influenzae serotype A strains containing the IS1016-bexA deletion. J Infect Dis 2010; 202:1577–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kroll JS, Moxon ER, Loynds BM. An ancestral mutation enhancing the fitness and increasing the virulence of Haemophilus influenzae type b. J Infect Dis 1993; 168:172–6. [DOI] [PubMed] [Google Scholar]
- 20. Musser JM, Kroll JS, Moxon ER, Selander RK. Evolutionary genetics of the encapsulated strains of Haemophilus influenzae. Proc Natl Acad Sci U S A 1988; 85:7758–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Resman F, Manat G, Lindh V, Murphy TF, Riesbeck K. Differential distribution of IgA-protease genotypes in mucosal and invasive isolates of Haemophilus influenzae in Sweden. BMC Infect Dis 2018; 18:592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Zwahlen A, Rubin LG, Moxon ER. Contribution of lipopolysaccharide to pathogenicity of Haemophilus influenzae: comparative virulence of genetically-related strains in rats. Microb Pathog 1986; 1:465–73. [DOI] [PubMed] [Google Scholar]
- 23. Satola SW, Schirmer PL, Farley MM. Genetic analysis of the capsule locus of Haemophilus influenzae serotype f. Infect Immun 2003; 71:7202–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Sill ML, Zhou J, Law DK, et al. Molecular characterization of four Haemophilus influenzae serotype a strains isolated from patients in Quebec, Canada. Can J Microbiol 2007; 53:1191–4. [DOI] [PubMed] [Google Scholar]
- 25. Meats E, Feil EJ, Stringer S, et al. Characterization of encapsulated and noncapsulated Haemophilus influenzae and determination of phylogenetic relationships by multilocus sequence typing. J Clin Microbiol 2003; 41:1623–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Haemophilus influenzae MLST Databases. Available at: https://pubmlst.org/hinfluenzae/. Accessed 15 January 2019.
- 27. Larsen MV, Cosentino S, Rasmussen S, et al. Multilocus sequence typing of total-genome-sequenced bacteria. J Clin Microbiol 2012; 50:1355–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Leaves NI, Falla TJ, Crook DW. The elucidation of novel capsular genotypes of Haemophilus influenzae type b with the polymerase chain reaction. J Med Microbiol 1995; 43:120–4. [DOI] [PubMed] [Google Scholar]
- 29. Public Health Indicator Based Information System. Available at: https://ibis.health.utah.gov/query/. Accessed 15 January 2019.
- 30. Bruce MG, Zulz T, DeByle C, et al. Haemophilus influenzae serotype a invasive disease, Alaska, USA, 1983–2011. Emerg Infect Dis 2013; 19:932–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Rubach MP, Bender JM, Mottice S, et al. Increasing incidence of invasive Haemophilus influenzae disease in adults, Utah, USA. Emerg Infect Dis 2011; 17:1645–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Brown VM, Madden S, Kelly L, Jamieson FB, Tsang RS, Ulanova M. Invasive Haemophilus influenzae disease caused by non-type b strains in Northwestern Ontario, Canada, 2002–2008. Clin Infect Dis 2009; 49:1240–3. [DOI] [PubMed] [Google Scholar]
- 33. McConnell A, Tan B, Scheifele D, et al. Invasive infections caused by Haemophilus influenzae serotypes in twelve Canadian IMPACT centers, 1996–2001. Pediatr Infect Dis J 2007; 26:1025–31. [DOI] [PubMed] [Google Scholar]
- 34. Rotondo JL, Sherrard L, Helferty M, Tsang R, Desai S. The epidemiology of invasive disease due to Haemophilus influenzae serotype a in the Canadian North from 2000 to 2010. Int J Circumpolar Health 2013; 72: 1, 21142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Millar EV, O’Brien KL, Watt JP, et al. Epidemiology of invasive Haemophilus influenzae type A disease among Navajo and White Mountain Apache children, 1988–2003. Clin Infect Dis 2005; 40:823–30. [DOI] [PubMed] [Google Scholar]
- 36. Tsang RSW, Ulanova M. The changing epidemiology of invasive Haemophilus influenzae disease: emergence and global presence of serotype a strains that may require a new vaccine for control. Vaccine 2017; 35:4270–5. [DOI] [PubMed] [Google Scholar]
- 37. Tuyama M, Correa-Antonio J, Schlackman J, et al. Invasive Haemophilus influenzae disease in the vaccine era in Rio de Janeiro, Brazil. Mem Inst Oswaldo Cruz 2017; 112:196–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Tsang RS, Shuel M, Wylie J, Lefebvre B, Hoang L, Law DK. Population genetics of Haemophilus influenzae serotype a in three Canadian provinces. Can J Microbiol 2013; 59:362–4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.