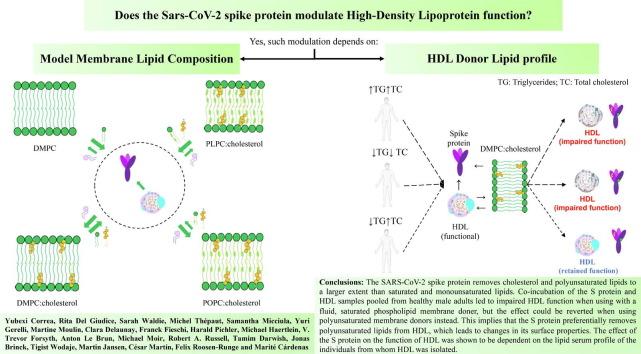
Graphical abstract
Keywords: COVID-19, SARS-CoV-2 spike protein, Deuterated cholesterol, Lipids, HDL, Neutron reflection, Cholesterol efflux capacity, Lipid metabolism
Abstract
There is a close relationship between the SARS-CoV-2 virus and lipoproteins, in particular high-density lipoprotein (HDL). The severity of the coronavirus disease 2019 (COVID-19) is inversely correlated with HDL plasma levels. It is known that the SARS-CoV-2 spike (S) protein binds the HDL particle, probably depleting it of lipids and altering HDL function. Based on neutron reflectometry (NR) and the ability of HDL to efflux cholesterol from macrophages, we confirm these observations and further identify the preference of the S protein for specific lipids and the consequent effects on HDL function on lipid exchange ability. Moreover, the effect of the S protein on HDL function differs depending on the individuals lipid serum profile. Contrasting trends were observed for individuals presenting low triglycerides/high cholesterol serum levels (LTHC) compared to high triglycerides/high cholesterol (HTHC) or low triglycerides/low cholesterol serum levels (LTLC). Collectively, these results suggest that the S protein interacts with the HDL particle and, depending on the lipid profile of the infected individual, it impairs its function during COVID-19 infection, causing an imbalance in lipid metabolism.
1. Introduction
SARS-CoV-2 causes COVID-19, which emerged from China in December 2019, and was declared a global pandemic in March 2020. The lethality of COVID-19 seems to have a strong dependency on pre-existing comorbid conditions, including cardiovascular diseases, diabetes, and hypertension [1]. There is a correlation between the plasma lipid profile and the severity of COVID-19 infection: decreased levels of low-density lipoprotein-cholesterol (LDL-C), high-density lipoprotein-cholesterol (HDL-C), and total cholesterol, as well as variable changes in triglyceride levels in patients, have been observed in patients affected by the disease [2], [3]. These phenomenological findings suggest an interaction of the SARS-CoV-2 virus with lipid metabolism, and in particular, HDL, whose mechanistic molecular underpinnings so far remain unclear.
HDL is responsible for the mechanism known as Reverse Cholesterol Transport (RCT) in which excess cholesterol is removed from peripheral tissue and carried to the liver where it is recycled or removed from the body by the gallbladder [4]. For this process to be carried out, Apolipoprotein A-I (ApoA-I), the major protein component of HDL, serves as a ligand for different cell membrane receptors, including the Scavenger receptor class B type 1 (SR-B1). SR-B1 also facilitates the entry of SARS-CoV-2 into the cells, in addition to improving its attachment to the cell membrane [5]. The SARS-CoV-2 spike protein (S) exposed on the virus surface is proposed to bind HDL for the virus to hijack the interaction pathway that HDL uses to interact with SR-B1 [5]. The widely used COVID-19 vaccines from Pfizer-BioNTech (Comirnaty®) and Moderna (Spikevax™) use messenger ribonucleic acid (m-RNA) to generate an immune response to the S protein by mediating the production of S protein into the body [6]. Thus, it might be expected that the S protein derived from the vaccine would be able to interact with HDL via the same pathways as it does for the S protein attached to the virus particle. This mechanism may underlie some of the adverse effects observed upon the administration of m-RNA vaccines [7]. For instance, it is reported that a 60-year-old man with heterozygous familial hypercholesterolemia (HeFH) and high triglycerides (TG) presented unusually high TG values (1300 mg/dL) one day after receiving his second Pfizer-BioNTech vaccine injection [8]. This suggests that the S protein could affect lipid metabolism in patients with elevated cholesterol and TG levels, and such an effect should be independent of the antibody response to the vaccine.
We [9] and other scientists [10] have previously shown that the S protein removes lipids from model membranes. We have also shown that the S protein interferes with lipid removal and exchange processes carried out by HDL in model membranes composed of saturated phospholipids (specifically 1,2-dimyristoyl-sn-glycerophosphatidylcholine, DMPC) and cholesterol (in an 80:20 M ratio) [9].
In this work, we further investigate the S protein capacity to modulate HDL function, including Reverse Cholesterol Transport (RCT). In particular, we focus on the potential role of the lipid donor type, i.e. the presence of cholesterol or the use of unsaturated lipids containing oleic versus linoleic acid [11]. This is important since it is well known that several viruses [12], [13] including SARS-CoV-2 [14] require and recognize the presence of cholesterol in the membranes to facilitate their entry into the host [15]. This interaction is mediated by the presence of cholesterol recognition amino acid consensus (CRAC) and inverted cholesterol recognition (CARC) motifs in the S protein that could be responsible for facilitating binding to cholesterol-rich membrane regions [16]. Moreover, the S protein was reported to contain a pocket that binds linoleic acid [17] and our previous work suggests that HDL preferentially uptakes saturated over unsaturated lipids [18].
Noting the potential preference of the S protein for specific lipid species, we further investigate whether the function of HDL samples purified from donors with different lipid serum profiles is distinctly affected by the S protein.
This work uses a combination of state-of-the-art biophysical techniques such as neutron reflectometry (NR) together with traditional biochemical experiments including cholesterol efflux capacity (CEC) which is part of the RCT. NR enables monitoring the integrity and composition of supported lipid bilayers (SLBs) upon incubation with S protein and/or HDL. Lipid donors include saturated, monounsaturated, and polyunsaturated phospholipids containing cholesterol, and experiments are performed under physiological-like conditions. Through CEC experiments, the ability of HDL to promote cholesterol efflux was determined from J774A.1 macrophage cells in the presence and absence of the S protein.
2. Materials and methods
2.1. Materials
D2O (99,9 %) and Tris buffer saline (50 mM Tris buffer, 150 mM NaCl, pH 7.4) in the form of TBS tablets were purchased from Sigma Aldrich. 1,2-dimyristoyl-D54-sn-glycerophosphatidylcholine (d-DMPC, 1,2-dimyristoyl-sn-glycerophosphatidylcholine (DMPC), hydrogenated cholesterol (h-cholesterol, >99%) and 23–(dipyrrometheneboron difluoride)-24-nor-cholesterol (TopFluor® Cholesterol, >99%) were bought from Avanti Polar Lipids. 1-Palmitoyl-d31-2-linoleoyl-d31-glycerol-3-phosphocholine (d-PLPC) was produced using previously reported methods for the synthesis of mixed acyl glycerophospholipids [19] from palmitic acid-d31 and linoleic acid-d31. 1-Palmitoyl-d31- 2-oleoyl-d33-glycerol-3-phosphocholine (d-POPC) was produced using previously reported methods for the synthesis of mixed acyl glycerophospholipids [19] from palmitic acid-d31 and oleic acid-d33 [20] . Deuterated cholesterol (average 80% D as determined by mass spectrometry, d-cholesterol) was produced using a previously reported method (ca. 90% D2O in the growth medium, for experiments at ANSTO) [21] or perdeuterated cholesterol produced in the Deuteration Laboratory (d-Lab) of the Life Sciences Group at Institut Laue-Langevin (ILL) as described in ref [22], [23] (for experiments at the ILL). Centrifugal devices, MicrosepTM Advance for sample buffer exchange, Fetal bovine serum (FBS) and PD10 columns were purchased from VWR. Bovine Serum Albumin fatty acid-free (BSA), Acyl-CoA: Cholesterol Acyltransferase (ACAT) inhibitor and Methyl-β-cyclodextrin were purchased from Sandoz.
2.2. Expression and purification of the spike protein 6P
Expression of SARS-CoV-2 (S-trimer 6P [24]) was performed in EXPI293 cells according to a previous protocol and purified by HisTrap HP and Superose 6 size exclusion chromatography (SEC) using superose 6 increase 10/300 GL column operated in a running buffer composed of 25 mM Tris HCl pH 8, 150 mM Sodium Chloride (NaCl) [25].
2.3. HDL isolation from human plasma
HDL isolated from human plasma was provided by Karolinska University Hospital (Stockholm, Sweden) and University Medical Center Freiburg (Freiburg, Germany). For this study, no personal information was used, and the samples were anonymized (Sweden) and used with ethical permission from the Stockholm and Freiburg Ethics Committee. Sequential ultracentrifugation was used to isolate HDL by first separating VLDL at a density of 1.019 g/mL for 24 hours under 40000 rpm (Beckman Coulter Optima XPN-80, rotor 70Ti) at 4 °C, followed by separating LDL at a density of 1.065 g/mL for another 24 hours under 40000 rpm at 4 °C. Finally, the total HDL fraction was collected at a density of 1.1 g/mL for 48 hours under 40000 rpm at 4 °C. Then, the HDL samples were stored at −80 °C in 50% sucrose, 150 mM NaCl, 24 mM ethylenediaminetetraacetic acid (EDTA), and pH 7.4. Further subfractionation to obtain HDL3, HDL2a and HDL2b was done by using a density of 1.11 g/mL for 48 hours at 50000 rpm at 4 °C. Upon thawing, the sucrose was removed by using PD10 columns or size exclusion chromatography (Superose 6 Increase 10/300 GL column, Cytiva) in 50 mM Tris-HCl 150 mM NaCl, pH 7.4. After purification, the samples were purged with Nitrogen and stored at 4 °C. Total protein concentration was determined by bicinchoninic acid assay (BCA assay). Pooling of samples took place after purification with ultracentrifugation. The samples were used within a maximum of 3 days of being thawed. The integrity of the HDL samples was monitored via dynamic light scattering (DLS) and SEC.
2.4. Model membranes
Lipid films were prepared by dissolving the deuterated phospholipids and d-cholesterol in chloroform and mixing at the desired molar ratio (i.e., 80:20 mol% phospholipid: cholesterol), followed by evaporation of the organic solvent under nitrogen gas. Once the film was formed any residual chloroform was removed by leaving the films under a vacuum overnight and then stored at −20 °C until needed.
SLBs were prepared via a small unilamellar vesicle fusion method following previously reported protocols [26]. First, multilamellar vesicles were formed by hydrating lipid films with Milli-Q water and the mixture was kept in a bath sonicator at a temperature between 40 and 50 °C for 1 h. Second, the suspension was tip sonicated (5 min on and 5 min off) for 15 min at 20% power (400 Watts, Branson 450 Digital Sonifier w/ Probe). Before injection, 0.2 mg/mL lipid vesicles were mixed with an equal volume of 4 mM Calcium Chloride (CaCl2). The vesicles were introduced into the cells by a pump at 1 mL/min and incubated for 20 min. This was followed by extensive rinsing with Milli-Q water and h-TBS buffer (TBS in H2O).
2.5. Neutron reflectometry experiments
NR experiments were performed on FIGARO [27] and D17 [28] at the ILL (Grenoble, France), and on SPATZ [29] at ANSTO (Sydney, Australia) using a setup for solid/liquid interfaces.
The silicon blocks (cut along the 111 planes) were cleaned in the following series of solvents: Chloroform, ethanol, acetone, and Milli-Q water for 20 min each in a bath sonicator. The O-ring and polyether ether ketone (PEEK) components were cleaned in Hellmanex 2% (v/v) solution and Milli-Q water twice with bath sonication and rinsed with Milli-Q water between each sonication. Before assembling the solid–fluid cells, the blocks were extensively rinsed with water, dried with a flow of Nitrogen gas, and exposed for 30 min to ozone. The solid–fluid cells were finally assembled in wet conditions. During the assembly, the substrate was sealed with the polished surface in contact with the liquid, and the cells were connected to an HPLC pump to allow the exchange of solvent with D2O, H2O, and contrast-matched silicon (CmSi = 38:62 v/v D2O: H2O).
The bespoke solid/liquid cells were pre-equilibrated at 37 °C and the surfaces were characterized in H2O and D2O to determine the roughness and the thickness of the silicon oxide layer. Then, the SLBs were formed and characterized in three isotropic contrasts composed of h-TBS (50 mM Tris buffer, 150 mM NaCl, pH 7.4 in H2O), d-TBS (50 mM Tris buffer, 150 mM NaCl, pH 7.4 in D2O), and CmSi-Tris (50 mM Tris buffer, 150 mM NaCl, pH 7.4 in CmSi).
For the experiments carried out at the ILL, both instruments operate in time-of-flight mode, with a horizontal scattering plane for D17 and a vertical scattering plane for FIGARO. Two incoming angles were selected on each instrument, 0.8° and 3° on D17 (http://dx.doi.org/10.5291/ILL-DATA.9-13-1000)h, and 0.8° and 3.2° on FIGARO (http://dx.doi.org/10.5291/ILL-DATA.8-05-466), to cover a scattering angle (Q) range between 0.003 and 0.25 A-1 with a wavelength, λ, range of (2 < λ < 20 A) on FIGARO and (2 < λ < 30 A) on D17, the upper Q limit being set by the sample background signal. The background was measured on the left and right sides of the reflectivity signal and subtracted from the measured reflected intensity. The data were normalized to the direct beam measured at the same instrumental configurations; the slit settings were selected to provide a constant surface under-illumination at the different angles of incidence (about 70% of the total substrate surface). The instrumental resolution ΔQ/Q was between 2 and 4 %.
Multidimensional TOF data were converted into reflectivity curves R(Q), by using the Cosmos software available at the ILL [30].
For SPATZ, the momentum transfer range and Q-resolution used were 0.011 Å−1 < Q < 0.273 Å−1 and ΔQ/Q = 5.8%, respectively. The instrument uses the time-of-flight principle and a disc chopper pairing of discs 1 and 2 sets 480 mm apart and running at 25 Hz was used (2.5 Å < λ < 20 Å). Collimation slits used were set to 2.66 mm and 0.75 mm for an angle of incidence of 0.85° and 10.94 mm and 3.09 mm for 3.5°, providing an illuminated footprint of 55 mm along the beam. Data were reduced with the refnx software package [31], where the time-of-flight is converted to wavelength which is used to calculate Q, accounts for detector efficiency, re-bins the data to instrument resolution, subtracts the background, stitches the data from the two angles of incidence together at the appropriate overlap region, and normalise the reflected intensity to the incident intensity (so that for samples showing total reflection this is equal to 1.
The SLBs were fitted using a 5-layer model as previously described [18] constituted by the oxide layer, a solvent layer, and three layers that correspond to the heads-tails-heads of the lipid membrane. During data fitting, the lipid bilayer was constrained to be symmetric: i.e., the coverage, the thickness, and the scattering length density (SLD) of the heads were the same for both leaflets (the tail region of both leaflets was considered as a single layer). Additionally, the roughness was constrained to be the same across the whole bilayer. The proteins, i.e., HDL and/or S, were introduced via a syringe pump at a rate of 1 mL/min at concentrations of 0.132 mg/mL and 0.05 mg/mL (0.1 mM), respectively. After 5 h of incubation, the samples were rinsed with h-TBS and re-characterized in all three contrasts.
To fit the data in the three isotropic contrasts after protein incubation and rinsing, the head-group thickness was kept constant whereas the thickness and SLD of the tails were allowed to vary though constrained to be the same across all three contrasts. A layer of HDL and/or S protein was added on top of the SLB. For HDL, we have previously shown that this model was sufficient to represent the data [32] since any protein penetration into the SLB core can with confidence be disregarded due to the lack of SLB core change when performing experiments on hydrogenated SLB [32]. Therefore, we can assign the change on SLD to lipid exchange only. For the S protein, on the other hand, no lasting adsorbed layer was detected at all with NR.
The percentage of lipid deposition is then calculated from the difference in SLD before and after exposure to lipoproteins and/or S protein. The kinetic for the pooled and individual HDL samples were fitted keeping the SLB structure constant due to a lack of information to resolve both the SLD core change and the SLB structure when using only one isotropic contrast. For this reason, the kinetics data (Supporting Information Figure S5) was analysed using the Q-values between 0.02 and 0.1 Å−1 for which the changes in the scattering length of the core are most prominent in H2O.
The experiments were performed only once due to the restricted availability of beam time. All reflectivity profiles, best fits and scattering length density profiles as well as parameters used for the best fits are provided in the Supporting Information. Data fitting was performed using the MOTOFIT package [33], and to determine the errors of the fits, a Monte Carlo error analysis (MCMC) was performed using genetic optimization. Error propagation was used in the calculation of % Lipids removed and deposited, see Supporting Information for details on modeling and estimation of these values.
2.6. Macrophages cell culture and cholesterol efflux capacity determination
Macrophages were cultured as previously described [34]. J774A.1 macrophage cells were kindly provided by Dr. César Martín (Basque Country University). Cells were maintained in Dulbecco’s modified Eagles Medium (DMEM, low glucose) supplemented with 100 µg/mL streptomycin, 100 U/mL penicillin, MycoZapTM Prophylactic, 10% fetal bovine serum (FBS, v/v) and plated in 96 well plates at a density of 2 × 104 cells per well. After 72 h in the culture medium, the macrophages were loaded with TopFluor-cholesterol in RPMI-1640 containing 4 μg/mL ACAT inhibitor, 4% BSA, and 2% FBS [34]. Upon 45 min loading time, the cells were washed and incubated at 37 °C for 15 h in resting media composed of RPMI-1640 containing 2 μg/mL ACAT inhibitor and 2% BSA. Then, either HDL, the S protein, or HDL/S protein mixtures were added to the macrophage culture in 25 mM MEM-HEPES, pH 7.4, containing 2 μg/ml ACAT inhibitor. The concentrations used for the samples were 10 and 5 μg/mL for HDL and the S protein, respectively. Such concentrations were chosen to keep the same molar ratio as for NR experiments and to keep the range of the acceptor concentration to what is required by this method. A 10 μg/mL BSA solution was used as an internal control in this experiment to get non-specific efflux. Protein concentration was determined by Detergent Compatible (DC) assay.
The samples were incubated for 6 h with either of the protein samples. At the end of the incubation time, the supernatant was collected, and the cell monolayers lysed with a buffer composed of 5.3 mM sodium fluoride (NaF), 1.5 mM sodium phosphide (NaP), 0.1% deoxycholic acid, 0.1% sodium dodecyl sulfate (SDS), 1% NP-40, 0.1 mM egtazic acid (EGTA), and 0.1 mM EDTA in 50 mM Tris-HCl at pH 7.5 for 30 min at room temperature, under agitation.
Finally, the fluorescence of the tracer of both cell lysates and the supernatant was measured using a multi-mode microplate reader (SynergyTM HTX). TopFluor-cholesterol displays excitation/emission maxima of 480/508 nm, respectively.
2.7. Quantification and Statistical analysis
To determine the specific cholesterol efflux capacity of the samples, the fluorescence intensity was used as follows:
The specific cholesterol efflux capacity is calculated as a difference between the percentage of cholesterol efflux and non-specific efflux:
Data were represented as mean ± SE. Measurements were made in quadruplicate per sample. Statistical analysis was assessed by unpaired two-tailed Student's t-test using OriginLab version 2021 (OriginLab Corporation, Northampton, MA, USA). Significant differences were considered at ∗ p < 0.05, ∗∗p < 0.01, and ***p < 0.001 compared to HDL.
3. Results and discussions
3.1. HDL's ability to remove lipids is the highest for unsaturated lipids in the absence of cholesterol and the lowest for polyunsaturated lipids in the presence of cholesterol
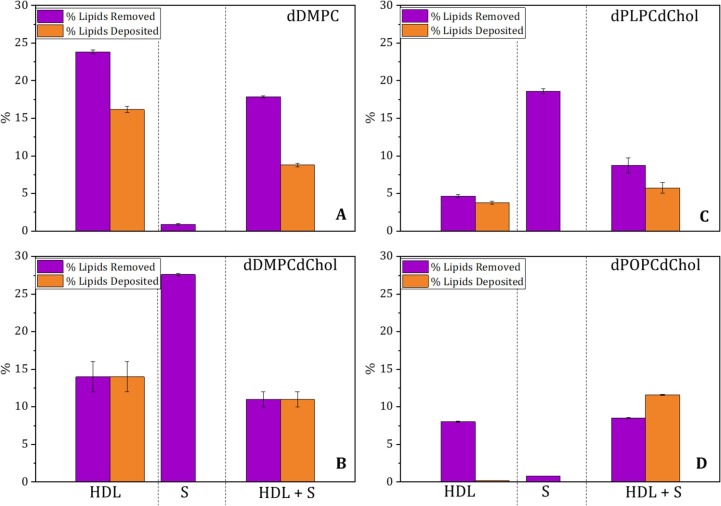
In our previous report [9], we showed that the S protein affected HDL's ability to exchange fats on model lipid membranes composed of a fluid, saturated phospholipid (DMPC) containing cholesterol. However, mammalian membranes are rich in unsaturated fats, and cholesterol is expected to modulate the S protein binding to membranes [35]. We have since focused on HDL lipid exchange ability using donor membranes comprised of saturated (DMPC), monounsaturated (POPC), and polyunsaturated (PLPC) phospholipids in the presence and absence of cholesterol. We collected steady-state NR data for donor lipid membranes that were exposed for 5 h to 0.05 mg/mL S protein and 0.132 mg/mL HDL (pooled from three healthy adults) measured in three isotropic contrasts. From the simultaneous NR data analysis of the three isotropic contrasts, the effect of membrane incubation with HDL, the S protein, and a mixture of HDL and S protein can be measured with great accuracy [32] (Fig. 1 , Supporting Information Figure S1-S3).
Fig. 1.
HDL's ability to remove and deposit lipids in fluid-saturated membranes in the absence (A) and presence of cholesterol (B), as well as in polyunsaturated (C) and monounsaturated (D) membranes in the presence of cholesterol. Data were analyzed after 5 h of incubation of the SLBs with either HDL (0.132 mg/mL), S protein (0.05 mg/mL), or a mixture of both HDL and S protein (0.132 and 0.05 mg/mL, respectively) in h-TBS at 37 °C. The NR profiles and best fits are given in Supporting Information Figures S1, S2, and S3. Errors are derived from Bayesian MCMC analysis.
The NR data analysis on HDL in the absence of the S protein (Fig. 1A) shows that HDL removed roughly 24% of lipids from membranes composed of saturated, fluid phospholipids (d-DMPC). In the presence of cholesterol, HDL removed about 14% of lipids in saturated, fluid membranes composed of d-DMPC: d-cholesterol 80:20 mol% (Fig. 1B and ref. [9]). When considering cholesterol-containing monounsaturated (d-POPC) or polyunsaturated (d-PLPC) membranes (Fig. 1D, C), HDL lipid removal was significantly lower (roughly 8 and 5 %, respectively) than for saturated lipids in the presence or absence of cholesterol. Thus, the capacity of HDL lipid removal decreases when cholesterol is present in saturated fluid membranes, and unsaturated lipids (regardless of the unsaturation level) are removed to a lesser extent than saturated lipids, in agreement with previous results [18]. However, HDL removes more monounsaturated than polyunsaturated lipids (Fig. 1D and C). Net differences in lipid removal between previous and current experiments might be due to different lipid profiles in the HDL samples, which were unknown in previous publications [36] due to blind pooling.
3.2. The S protein affects the HDL function, and this depends on the lipid donor type in the model membrane
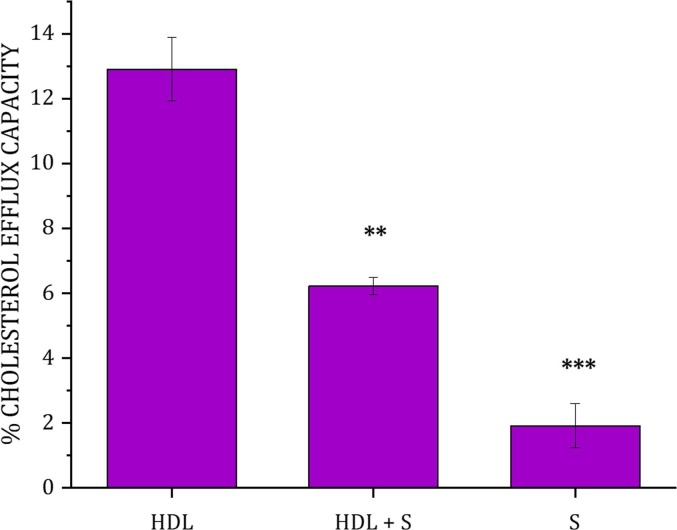
Since the presence of cholesterol in the membrane influences the extent of lipid removal, and the main HDL function is the removal of cholesterol from the body, the effect of the S protein on the HDL-mediated CEC in J774A.1 macrophage was assessed (Fig. 2 ). The data show that, while the S protein is responsible for only a low amount of cholesterol efflux (2% of the total cholesterol), the incubation of cells with the mixture of HDL and S protein decreased the cholesterol efflux capacity of HDL by a factor of 2 (from 12 to 6%). Our results are in agreement with recent cholesterol efflux capacity experiments of HDL that were found to significantly decrease in COVID-19 patients [37]. Together, these results strongly suggest that the presence of the S protein interferes with the efflux process mediated by HDL.
Fig. 2.
Cholesterol efflux Capacity of HDL in the presence and absence of the S protein from J774A.1 macrophages. The data represent the means ± SE. of at least three independent measurements. Levels of significance were determined by unpaired two-tailed Student's t-test using Origin version 2021 (OriginLab Corporation, Northampton, MA, USA). Significant differences were considered at ∗ p < 0.05, ∗∗p < 0.01, and ***p < 0.001 compared to HDL.
Next, we studied the extent to which the HDL function is affected by co-incubation with the S protein as a function of the lipid type used in the model membrane (Fig. 1). The presence of S proteins reduced the ability to remove and deposit lipids by HDL with the most reduction taking place for saturated, fluid membranes, and this being followed by cholesterol-containing saturated membranes. For cholesterol-containing monounsaturated and polyunsaturated membranes, the presence of S protein increased HDL's ability to remove lipids with the strongest effect observed for polyunsaturated membranes. Indeed, for monounsaturated membranes, there was a net deposition of lipids by HDL in the presence of the S protein. Thus, the S protein exerts an effect on the interaction process between HDL and the lipid membrane that prevents HDL from fulfilling its function: i.e., removal of saturated lipids and cholesterol. However, this is not the case when polyunsaturated lipids are used as lipid donors (in the model membrane), where an improved function was seen when coincubation with the S protein took place.
It is known that the S protein has a certain affinity for HDL and the presence of cholesterol [38]. Studies have shown that the S protein can bind to both cholesterol and HDL and this association can improve the entry of SARS-CoV-2 and its replication [39]. Therefore, it is possible that because of this association, lipid metabolism is affected. Indeed, numerous studies indicate that there is a correlation between HDL levels and COVID-19 disease severity [40], [41].
To rationalize the results, control experiments were performed with the S protein in the absence of HDL (Fig. 1 and Supporting Information Figure S2). Here we observed that the S protein ability to remove lipids was the highest for cholesterol-containing unsaturated membranes, followed by cholesterol-containing polyunsaturated membranes. We obtain minimal removal for monounsaturated and saturated membranes without cholesterol (Fig. 1). This result points towards the S protein preference for cholesterol and polyunsaturated lipids. In addition, the ability of the S protein to remove cholesterol from donor membranes [9] mirrors its cholesterol efflux capacity (Fig. 2). Structurally, this ability is probably due to the presence of cholesterol recognition amino acid consensus (CRAC) and inverted cholesterol recognition motifs (CARC) present in the S protein [42]. These motifs were previously shown to mediate the S protein binding to cholesterol [5]. Additional Attenuated Total Reflection –Fourier Transform Infrared Spectroscopy (ATR-FTIR) studies were carried out for the S protein against model membranes composed of d-DMPC [9], d-DMPC: h-cholesterol [9], d-PLPC: h-cholesterol and d-POPC: h-cholesterol under identical conditions as for NR (Supporting Information Figure S4). The results show qualitatively the same trend as for NR (Fig. 1) with a clear decrease in the C-D2 peak symmetric upon incubation with the S protein (which suggests lipid removal by the S protein) for membranes composed of d-DMPC: h-cholesterol (Supporting Information, Figure S4C) and d-PLPC: h-cholesterol (Supporting Information, Figure S4B) but minimal effects on membranes composed of d-POPC: h-cholesterol or d-DMPC.
We hypothesize that cholesterol and polyunsaturated lipids are specifically removed from HDL by the S protein since these are the ones preferentially removed by the S protein from donor model membranes. Such targeted removal could lead to the remodeling of the HDL surface, which in turn might affect its functionality. For example, the cholesterol content in membranes regulates the membrane fluidity and membrane packing, depending on the acyl chain composition of the lipids present [43]. A change in the HDL lipid composition might translate to a shift in the HDL-apolipoprotein/associated protein composition, which affects HDL functionality [44].
3.3. The plasma lipid profile strongly affects the capacity for lipid removal and the ability to efflux cholesterol of individual HDL samples and determines whether the S protein has a supporting or detrimental effect
Our previous studies have focused on lipoprotein fractions pooled from three healthy patients [32] for which no information on the lipid serum profile from the donors was available. This could explain the slight variability observed in the net lipid removal and deposition between the HDL samples used here and those used previously [9] under the same experimental conditions. For testing the hypothesis about the role of HDL biochemical composition on the extent to which S regulates HDL function, we purified HDL samples from individuals with (1) low serum triglycerides and low total serum cholesterol levels (LTLC), (2) high serum triglycerides and high total serum cholesterol levels (HTHC), and (3) low serum triglyceride and high serum total cholesterol levels (LTHC). TG and total serum cholesterol (TC) values below 150 and 200 mg/dL respectively, are considered optimal; while optimal HDL-C, levels should be greater than or equal to 60 mg/dL [45]. For the specific lipid plasma values in the samples used in this study see Table 1 .
Table 1.
Plasma lipid profile values for normolipidemic and hypertriglyceridemic individuals were used, in terms of TG, TC, HDL-C, and ApoA-I. The table shows the values for the HDL samples used for NR and CEC experiments.
| Sample | Experiment | TG (mg/dL) | TC (mg/dL) | HDL-C (mg/dL) | ApoA-I (g/L) |
|---|---|---|---|---|---|
| LTLC | NR | 63.8 (Low) | 177.9 (Low) | 77.3 (High) | 1.75 |
| CEC | 97.6 (Low) | 115 (Low) | 37 (Low) | 0.98 | |
| LTHC | NR | 50.4 (Low) | 297.8 (High) | 96.7 (High) | 2.05 |
| CEC | 140 (Low) | 290 (High) | 73.7 (High) | 1.68 | |
| HTHC | NR | 283.4 (High) | 274.5 (High) | 50.3 (Intermediate) | 1.47 |
| CEC | 636 (High) | 229 (High) | 24.90 (Low) | 0.75 |
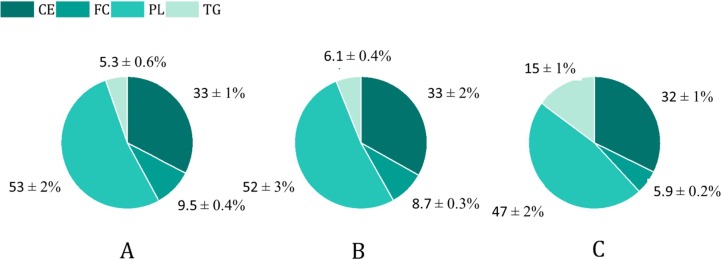
The lipid composition of the total HDL fraction indicates that the HDL particles from the LTHC individual contain higher free cholesterol/free cholesterol + phospholipids ratio ((FC)/(FC + PL)) than the other two samples (Fig. 3 ). This difference may suggest that the surface of the HDL particles from the LTHC individual is denser due to the closer packing of the PL monolayer, which should translate in lower lipid fluidity [46]. Moreover, the ratio of the surface lipids (PL + FC) to the core lipids (CE + TG) was higher for the LTHC HDL sample. This may in turn indicate that the HDL particles are larger or have a shape other than spherical. These differences in the surface composition of HDL might be key to determining how the S protein interacts with the HDL and remodeling its function.
Fig. 3.
Lipid composition represented in the molar percentage of total HDL fractions from individuals presenting (A) LTHC, (B) LTLC, and (C) HTHC lipid serum profile. CE: Cholesteryl Ester, FC: Free Cholesterol, PL: Phospholipids, and TG: Triglycerides. Lipid values were normalized to ApoA-I. The (PL + FC)/(CE + TG) ratio for total HDL fractions from LTHC, LTLC, and HTHC was 2.0 ± 0.2, 1.5 ± 0.2, and 1.1 ± 0.1 respectively. The FC/(FC + PL) ratio for the total HDL fractions from LTHC, LTLC, and HTHC was 0.15 ± 0.02, 0.14 ± 0.02, and 0.11 ± 0.02 respectively. N = 1. The coefficient of variation for the within-run imprecision was 2.1%, 1.6%, 1.0%, and 1.4% for TG, CE, FC, and PL respectively.
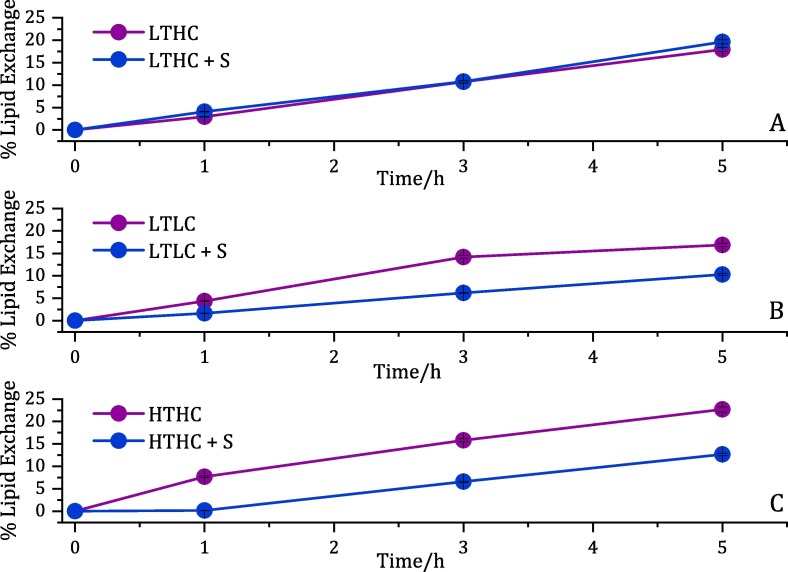
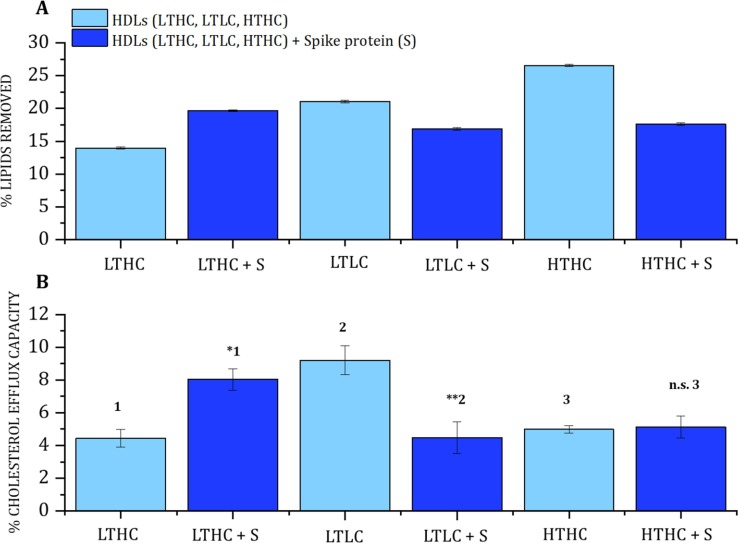
These HDL fractions were then used for CEC and NR experiments against d-DMPC:d-cholesterol 80:20 mol% lipids as model donor membranes. The kinetics for lipid exchange during the first 5 h (including a combination of both lipid removal and deposition) was followed by NR for each HDL sample (Table 1) in the presence and absence of the S protein in h-TBS at 37 °C (Fig. 4 and Supporting Information Figure S5). From the kinetics, it is clear that the HDL ability to exchange lipids was compromised only for the LTLC and HDL HTHC samples, while no noticeable effect was observed for the LTHC samples. The net lipid removal and deposition for each type of individual HDL were determined through NR data analysis for three isotropic contrasts (Fig. 5 A and Supporting Information Fig. S6-S8). A different set of samples collected from other individuals with similar lipid serum profiles (Table 1, CEC lines) was chosen for CEC determination (Fig. 5B).
Fig. 4.
Kinetics of individual HDL lipid exchange capacity upon 5 h of incubation in h-TBS at 37 °C. The extent of lipids removed by the S protein in model membranes composed of d-DMPC: d-cholesterol at a molar ratio of 80:20 mol% in the presence of (A) LTHC, (B) LTLC, and (C) HTHC HDL.
Fig. 5.
Effect of the S protein on HDL capacity to remove lipids and mediate the efflux of cholesterol on HDL samples from individuals with low triglycerides/high cholesterol (LTHC), low triglycerides/low cholesterol (LTLC), and high triglycerides/high cholesterol (HTHC) plasma levels. (A) lipid removal by NR. (B) cholesterol efflux capacity. The lipid donor for NR was d-DMPC: d-cholesterol 80:20 mol%. The data represent the means ± SE. of at least three independent measurements, errors are derived from Bayesian MCMC analysis. Levels of significance were determined by unpaired two-tailed Student's t-test. Significant differences were considered at ∗ p < 0.05, ∗∗p < 0.01, and ***p < 0.001 compared to HDL LTHC1, HDL LTLC2, and HDL HTHC3. The NR profiles and best fits are given in Supporting Information Figure S6.
The extent of lipid removal by HDL (Fig. 5A) followed the trend: LTLC > HTHC > LTHC, which is the same trend observed for the cholesterol efflux (LTLC > HTHC > LTHC), see Fig. 5B. Cholesterol efflux capacity is affected by different factors such as size and composition of HDL, the concentration of HDL-C, the concentration of ApoA-I, gender, etc [47]. Interestingly, the LTHC sample with the lowest cholesterol efflux capacity also presented the lowest ability to remove saturated lipids in the presence of cholesterol. This sample has both the highest (PL + FC)/(CE + TG) ratio and the highest FC/(FC + PC) ratio (Fig. 3), which suggests a distinctive HDL structure - at least on the surface structure - in LTHC as compared to LTLC and HTHC. A denser HDL surface could result in a higher enthalpy penalty for further lipid/cholesterol incorporation from a donor membrane.
Co-incubation with the S protein led to worsened HDL function for the LTLC HDL samples both in terms of lipid removal and cholesterol efflux capacity (Fig. 5), in agreement with the results for the pooled HDL sample (Fig. 1). For HTHC HDL, there was significant lipid removal but no measurable effect on cholesterol efflux. Surprisingly, a significant increase in HDL's ability to remove lipids and mediate the efflux of cholesterol was detected for the LTHC HDL sample.
The data hereby presented suggests that individuals with a lipid serum profile characterised by low triglycerides and high total cholesterol present some particularity that affects the S protein interaction with HDL enhancing lipid removal and cholesterol efflux capacity.
Since both HDL and S protein remove lipids and mediate the efflux of cholesterol, and assuming these two do not interact and affect each other’s function, the expected net effect upon co-incubation should simply be the sum of the individual effects. In contrast, for HTHC and LTLC HDL, there is a clear decrease in both the capacity to remove lipids and to mediate the efflux of cholesterol for HDL in the presence of S protein. This suggests a strong interaction between the S protein and HDL that remodels HDL composition, probably affecting its structure. For LTHC HDL, on the other hand, lipid removal increased from 13,9% to 19,7% in the absence or presence of the S protein, respectively. The effect of co-incubation is an additional 5,8% lipid removal, which is smaller than the sum of the removal by LTHC HDL and S protein alone (Fig. 1C) (roughly 41,6% due to the additional 27,6% lipid removal by the S protein alone). Similarly, the cholesterol efflux ability is 4.4%, 1.9%, and 8% (Fig. 2) for LTHC HDL, the S protein, and their mixtures respectively. The result for the mixture is closer to the expected outcome if there is no net interaction, i.e., 6.3%. Thus, LTHC HDL particles present some key, so far unknown properties (of biochemical or structural nature) that seem to block the interaction with the S protein. From the biochemical data (Fig. 3), we could postulate that the higher surface density (packing) for HDL lipid monolayer creates a stronger enthalpic barrier for lipid removal by the S protein.
3.4. Biochemical parameters of HDL subfractions as a potential origin for divergent virus response
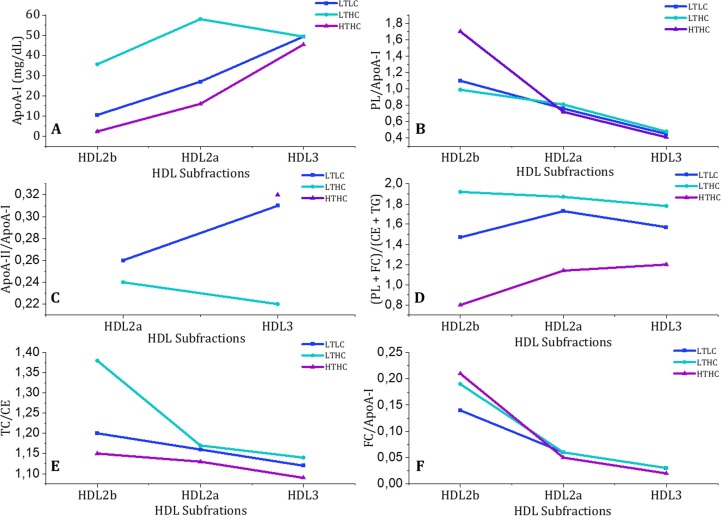
A closer look into the plasma biochemical values of the HDL subfractions for each HDL sample is given in Fig. 6 , to identify further biochemical parameters that make LTHC HDL different from LTLC and LTLC HDL. HDL can be divided into different subpopulations or subfractions for which each HDL particle varies in protein and lipid content, size, and density [48]. HDL3 are the smallest protein-enriched particles that are subsequently converted into the large HDL2, which have other subdivisions like HDL2b and HDL2a [49]. Fig. 6A shows that the amount of HDL2a and b present in LTHC HDL differs compared to that of HTHC and LTLC HDL. Assuming that the PL (Fig. 6B) and FC (Fig. 6F) determine the available surface area of the particle and that they all have a spherical shape, then these numbers should indicate the HDL particle size. The LTHC and LTLC HDL subfractions appear to be of similar size, except for the HDL2b subfraction in HTHC HDL, based on the PL and FC numbers. ApoA-II content, although not detectable in all HDL2b and in the HDL2a of HDL HTHC, is lower in the LTHC HDL fraction compared to the other two samples (Fig. 6C). The ratio of the surface lipids (PL + FC) to the core lipids (CE + TG) (Fig. 6D) for the subfractions shows that the LTHC does not follow the same trend in the LTHC HDL as compared to the other two ones with respect to HDL2b. Finally, LTHC HDL has a higher ratio of TC/CE due to the contribution of FC as well as the previous parameters (see Fig. 6E) and the HDL subfractions had similar FC/ApoA-I ratios among the samples (See Fig. 6F). In summary, Fig. 6 suggests that not only the content of FC but also the ApoA-I abundance make LTHC HDL distinguishable from the particles from the other serum lipid profiles studied.
Fig. 6.
Biochemical values of HDL subfractions for LTHC, LTLC, and HTHC samples separated by density. (1) ApoA-I concentration, (B) Phospholipid/ApoA-I, (C) ApoA-II/ApoA-I, (D) (Phospholipids + Free Cholesterol)/ (Cholesteryl ester + Triglycerides) ratios, (E) Total Cholesterol/Cholesteryl Ester, and (F) Free Cholesterol/ApoA-I ratios.
It is well known that the different HDL subpopulations have different lipid compositions including the ratio of phospholipids, sphingomyelin, and ceramides [50]. Such lipid composition can be a determinant of cholesterol efflux capacity since this aspect influences the surface lipid packing [51]. Membrane fluidity and curvature represent an important factor in the fusion process between the enveloped virus and the host membrane. The presence of cholesterol, phosphatidylethanolamine, and ceramides affects membrane fluidity and promotes the negative curvature of the membrane which is critical for viral fusion [35]. Retroviruses such as HIV and Rhinoviruses can exploit cholesterol and ceramide-enriched membranes to facilitate entry into the cells [52], [53].
In summary, this work suggests that HDL particles interact differently with the S protein due to their lipid composition and relative amount of a given subfraction (HDL2a). The ratio of surface lipids (PL + FC) and core lipids (CE + TG) differed for the LTHC HDL sample which suggests that HDL surface properties might be key to regulating how S affects its composition. These findings could indicate that individuals with an LTHC profile are at lower risk to have the lipid metabolism highjacked by the SARS-CoV-2 virus and could present a weaker interaction with the S protein upon vaccination. Given that the S protein preferentially removes polyunsaturated lipids and cholesterol, the S protein could potentially remove these lipids specifically from HDL particles. This change in lipid composition could potentially lead to remodeling on the HDL surface composition which in turn inhibits the ability of HDL to remove lipids and efflux cholesterol (since the HDL surface becomes even denser) [54]. Detailed compositional, structural, and functional studies on a substantially higher number of samples are needed to understand whether these results can be extended to more patients with different lipid profiles. Further studies are required to establish to which extent the HDL surface is remodeled by the S protein.
4. Conclusions
In this work, the S protein capacity to modulate HDL function was studied, by NR, ATR-FTIR and RCT experiments. In our previous research [9], we focus on the effect of co-incubation of S protein and HDL pooled from three healthy adult male donors in model membranes composed of fluid saturated lipids and cholesterol. Now, we attempted to decipher the role of the presence of unsaturated lipids containing oleic versus linoleic acid in combination with cholesterol and the effect of the lipid profile of the HDL donor. Linoleic acid was included in the model membranes because the S protein was reported to contain a pocket that binds linoleic acid specifically [17]. The hypothesis was that the HDL capacity to remove and exchange lipids is impaired in presence of the S protein and this depends on the lipid saturation of the model membrane, cholesterol, and the lipid profile of the donor. Our research suggests that the S protein from the SARS-CoV-2 virus has a stronger ability to remove cholesterol and polyunsaturated lipids compared to saturated and monounsaturated ones. Co-incubation of the S protein and HDL samples pooled from healthy adults with a fluid, saturated phospholipid model led to impaired HDL function, but the effect could be reverted when polyunsaturated lipids were used instead. This implies that the S protein preferentially removes polyunsaturated lipids from HDL, which leads to changes in its surface properties or remodelling. Previously, it was shown that COVID-19 induces a remodelling of HDL proteome and increases immune response and inflammation [55]. Here we show that the S protein changes HDL function in a manner that depends on the lipid serum profile. HDL samples with low triglycerides and high cholesterol serum levels (LTHC) presented an inverse trend induced by the Spike protein to that of low triglycerides/low cholesterol (LTLC) or high triglycerides/high cholesterol (HTHC) serum levels. This could indicate that individuals with an LTHC profile have a lower tendency to have the lipid metabolism highjacked by the SARS-CoV-2 virus and could present a weaker interaction with the S protein upon COVID-19 illness or vaccination against COVID-19. A larger study with samples taken from individuals having and not having had COVID-19 or received vaccination against it during the last 6 months should be performed to confirm the current findings. Further studies on reconstituted HDL samples with controlled lipid cargo are planned to demonstrate the importance of the lipid cargo on the interaction with the S protein.
CRediT authorship contribution statement
Yubexi Correa: Data curation, Formal analysis, Writing – original draft, Investigation, Methodology, Visualization. Rita Del Giudice: Formal analysis, Writing – review & editing. Sarah Waldie: Investigation, Writing – review & editing. Michel Thépaut: Resources, Writing – review & editing. Samantha Micciula: Data curation, Writing – review & editing. Yuri Gerelli: Investigation, Writing – review & editing. Martine Moulin: Resources, Writing – review & editing. Clara Delaunay: Resources, Writing – review & editing. Franck Fieschi: Resources, Writing – review & editing. Harald Pichler: Resources, Writing – review & editing. Michael Haertlein: Resources, Writing – review & editing. V. Trevor Forsyth: Resources, Writing – review & editing. Anton Le Brun: Data curation, Writing – review & editing. Michael Moir: Resources, Writing – review & editing. Robert A. Russell: Resources, Writing – review & editing. Tamim Darwish: Resources, Writing – review & editing. Jonas Brinck: Resources, Writing – review & editing. Tigist Wodaje: Resources. Martin Jansen: Formal analysis, Resources, Writing – review & editing. César Martín: Investigation, Resources, Formal analysis, Writing – review & editing. Felix Roosen - Runge: Formal analysis, Writing – review & editing. Marité Cárdenas: Conceptualization, Formal analysis, Writing – original draft, Writing – review & editing, Supervision, Methodology, Funding acquisition.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
MC, YC, and SW acknowledge economic support from the Swedish Research Council (grants number 2018-03990 and 2018-04833). MC thanks the Wennergren Foundation for funding (grant number SSh2020-0005). IBS acknowledges integration into the Interdisciplinary Research Institute of Grenoble (IRIG, CEA). VTF acknowledges support from the UK Engineering and Physical Sciences Council (EPSRC) which funded the creation of the Deuteration Laboratory within the Life Sciences Group at the ILL [GR/R99393/01 and EP/C015452/1]. This work used the platforms of the Grenoble Instruct-ERIC center (ISBG; UAR 3518 CNRS-CEA-UGA-EMBL) within the Grenoble Partnership for Structural Biology (PSB), supported by FRISBI (ANR-10-INBS-0005-02) and GRAL, financed within the University Grenoble Alpes graduate school (Ecoles Universitaires de Recherche) CBH-EUR-GS (ANR-17-EURE-0003). Operations for the Spatz neutron reflectometerand the National Deuteration Facility at ANSTO are supported by the National Characterisation Research Infrastructure Strategy (NCRIS), an Australian Government initiative.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jcis.2023.04.137.
Appendix A. Supplementary material
The following are the Supplementary data to this article:
Data availability
Data will be made available on request.
References
- 1.Wu Z., McGoogan J.M. Characteristics of and Important Lessons from the Coronavirus Disease 2019 (COVID-19) Outbreak in China: Summary of a Report of 72314 Cases from the Chinese Center for Disease Control and Prevention. J. Am. Med. Assoc. 2020;323:1239–1242. doi: 10.1001/jama.2020.2648. [DOI] [PubMed] [Google Scholar]
- 2.Fan J., Wang H., Ye G., Cao X., Xu X., Tan W., Zhang Y. Letter to the Editor: Low-density lipoprotein is a potential predictor of poor prognosis in patients with coronavirus disease 2019. Metabolism. 2020;107 doi: 10.1016/j.metabol.2020.154243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Feingold K.R. The bidirectional link between HDL and COVID-19 infections. J. Lipid Res. 2021;62 doi: 10.1016/J.JLR.2021.100067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Adorni M.P., Ronda N., Bernini F., Zimetti F. High density lipoprotein cholesterol efflux capacity and atherosclerosis in cardiovascular disease: Pathophysiological aspects and pharmacological perspectives. Cells. 2021;10:1–37. doi: 10.3390/cells10030574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wei C., Wan L., Yan Q., Wang X., Zhang J., Yang X., Zhang Y., Fan C., Li D., Deng Y., Sun J., Gong J., Yang X., Wang Y., Wang X., Li J., Yang H., Li H., Zhang Z., Wang R., Du P., Zong Y., Yin F., Zhang W., Wang N., Peng Y., Lin H., Feng J., Qin C., Chen W., Gao Q., Zhang R., Cao Y., Zhong H. HDL-scavenger receptor B type 1 facilitates SARS-CoV-2 entry. Nat. Metab. 2020;2:1391–1400. doi: 10.1038/s42255-020-00324-0. [DOI] [PubMed] [Google Scholar]
- 6.Hahn W.O., Wiley Z. COVID-19 Vaccines. Infect. Dis. Clin. North Am. 2022;36:481–494. doi: 10.1016/j.idc.2022.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Trougakos I.P., Terpos E., Alexopoulos H., Politou M., Paraskevis D., Scorilas A., Kastritis E., Andreakos E., Dimopoulos M.A. Adverse effects of COVID-19 mRNA vaccines: the spike hypothesis. Trends Mol. Med. 2022;28:542–554. doi: 10.1016/j.molmed.2022.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cheung B., Hwang J., Stolarczyk A., Mahlof E.N., Block R.C. Case study of hypertriglyceridemia from COVID-19 Pfizer-BioNTech vaccination in a patient with familial hypercholesteremia. Eur. Rev. Med. Pharmacol. Sci. 2021;25:5525–5528. doi: 10.26355/eurrev_202109_26664. [DOI] [PubMed] [Google Scholar]
- 9.Correa Y., Waldie S., Thépaut M., Micciula S., Moulin M., Fieschi F., Pichler H., Trevor Forsyth V., Haertlein M., Cárdenas M. SARS-CoV-2 spike protein removes lipids from model membranes and interferes with the capacity of high density lipoprotein to exchange lipids. J. Colloid Interface Sci. 2021;602:732–739. doi: 10.1016/j.jcis.2021.06.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Luchini A., Micciulla S., Corucci G., Batchu K.C., Santamaria A., Laux V., Darwish T., Russell R.A., Thepaut M., Bally I., Fieschi F., Fragneto G. Lipid bilayer degradation induced by SARS-CoV-2 spike protein as revealed by neutron reflectometry. Sci. Rep. 2021;11:14867. doi: 10.1038/s41598-021-93996-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang Y., Zhang J., Li Q., Wu Y., Wang D., Xu L., Zhang Y., Wang S., Wang T., Liu F., Zaky M.Y., Hou S., Liu S., Zou K., Lei H., Zou L., Liu H. Cholesterol content in cell membrane maintains surface levels of ErbB2 and confers a therapeutic vulnerability in ErbB2-positive breast cancer, Cell Commun. Signal. 2019;17:1–12. doi: 10.1186/s12964-019-0328-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bajimaya S., Frankl T., Hayashi T., Takimoto T. Cholesterol is required for stability and infectivity of influenza A and respiratory syncytial viruses. Virology. 2017;510:234–241. doi: 10.1016/j.virol.2017.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Osuna-Ramos J.F., Reyes-Ruiz J.M., del Ángel R.M. The Role of Host Cholesterol During Flavivirus Infection. Front. Cell. Infect. Microbiol. 2018;8:1–12. doi: 10.3389/fcimb.2018.00388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li X., Zhu W., Fan M., Zhang J., Peng Y., Huang F., Wang N., He L., Zhang L., Holmdahl R., Meng L., Lu S. Dependence of SARS-CoV-2 infection on cholesterol-rich lipid raft and endosomal acidification, Comput. Struct. Biotechnol. J. 2021;19:1933–1943. doi: 10.1016/j.csbj.2021.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ono A., Freed E.O. In: Adv. Roy P., editor. Virus Res., Preface; Maryland: 2005. Role of Lipid Rafts in Virus Replication; pp. 311–358. [DOI] [PubMed] [Google Scholar]
- 16.A. Dunina-barkovskaya, Cholesterol Recognition Motifs (CRAC) in the S Protein of Coronavirus : A Possible Target for Antiviral Therapy ?, in: W.S. Aronow (Ed.), Dyslipidemia, London, 2021. https://doi.org/10.5772/intechopen.95977.
- 17.Toelzer C., Gupta K., Yadav S.K.N., Borucu U., Davidson A.D., Williamson M.K., Shoemark D.K., Garzoni F., Staufer O., Milligan R., Capin J., Mulholland A.J., Spatz J., Fitzgerald D., Berger I., Schaffitzel C. Free fatty acid binding pocket in the locked structure of SARS-CoV-2 spike protein. Science. 2020;(80-.). 370:725–730. doi: 10.1126/science.abd3255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Waldie S., Sebastiani F., Browning K., Maric S., Lind T.K., Yepuri N., Darwish T.A., Moulin M., Strohmeier G., Pichler H., Skoda M.W.A., Maestro A., Haertlein M., Forsyth V.T., Bengtsson E., Malmsten M., Cárdenas M. Lipoprotein ability to exchange and remove lipids from model membranes as a function of fatty acid saturation and presence of cholesterol. Biochim. Biophys. Acta - Mol. Cell Biol. Lipids. 2020;1865 doi: 10.1016/j.bbalip.2020.158769. [DOI] [PubMed] [Google Scholar]
- 19.Moir M., Yepuri N.R., Marshall D.L., Blanksby S.J., Darwish T.A. Synthesis of Perdeuterated Linoleic Acid-d31 and Chain Deuterated 1-Palmitoyl-2-linoleoyl-sn-glycero-3-phosphocholine-d62. Adv. Synth. Catal. 2022 doi: 10.1002/adsc.202200616. [DOI] [Google Scholar]
- 20.Darwish T.A., Luks E., Moraes G., Yepuri N.R., Holden P.J., Jamesa M. Synthesis of deuterated [D32]oleic acid and its phospholipid derivative [D64]dioleoylsn- glycero-3-phosphocholine. J. Label. Compd. Radiopharm. 2013;56:520–529. doi: 10.1002/jlcr.3088. [DOI] [PubMed] [Google Scholar]
- 21.Sebastiani F., Yanez Arteta M., Lerche M., Porcar L., Lang C., Bragg R.A., Elmore C.S., Krishnamurthy V.R., Russell R.A., Darwish T., Pichler H., Waldie S., Moulin M., Haertlein M., Forsyth V.T., Lindfors L., Cárdenas M. Apolipoprotein E Binding Drives Structural and Compositional Rearrangement of mRNA-Containing Lipid Nanoparticles. ACS Nano. 2021;15:6709–6722. doi: 10.1021/acsnano.0c10064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Moulin M., Strohmeier G.A., Hirz M., Thompson K.C., Rennie A.R., Campbell R.A., Pichler H., Maric S., Forsyth V.T., Haertlein M. Perdeuteration of cholesterol for neutron scattering applications using recombinant Pichia pastoris. Chem. Phys. Lipids. 2018;212:80–87. doi: 10.1016/j.chemphyslip.2018.01.006. [DOI] [PubMed] [Google Scholar]
- 23.Haertlein M., Moulin M., Devos J.M., Laux V., Dunne O., Trevor Forsyth V. Biomolecular Deuteration for Neutron Structural Biology and Dynamics. Methods Enzymol. 2016;566:113–157. doi: 10.1016/BS.MIE.2015.11.001. [DOI] [PubMed] [Google Scholar]
- 24.Schaub J.M., Chou C.W., Kuo H.C., Javanmardi K., Hsieh C.L., Goldsmith J., DiVenere A.M., Le K.C., Wrapp D., Byrne P.O., Hjorth C.K., Johnson N.V., Ludes-Meyers J., Nguyen A.W., Wang N., Lavinder J.J., Ippolito G.C., Maynard J.A., McLellan J.S., Finkelstein I.J. Expression and characterization of SARS-CoV-2 spike proteins. Nat. Protoc. 2021;16:5339–5356. doi: 10.1038/s41596-021-00623-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Thépaut M., Luczkowiak J., Vivès C., Labiod N., Bally I., Lasala F., Grimoire Y., Fenel D., Sattin S., Thielens N., Schoehn G., Bernardi A., Delgado R., Fieschi F. DC/L-SIGN recognition of spike glycoprotein promotes SARS-CoV-2 trans-infection and can be inhibited by a glycomimetic antagonist. PLoS Pathog. 2021;17:1–34. doi: 10.1371/journal.ppat.1009576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lind T.K., Cárdenas M., Wacklin H.P. Formation of supported lipid bilayers by vesicle fusion: Effect of deposition temperature. Langmuir. 2014;30:7259–7263. doi: 10.1021/la500897x. [DOI] [PubMed] [Google Scholar]
- 27.Campbell R.A., Wacklin H.P., Sutton I., Cubitt R., Fragneto G. FIGARO: The new horizontal neutron reflectometer at the ILL. Eur. Phys. J. Plus. 2011;126:1–22. doi: 10.1140/epjp/i2011-11107-8. [DOI] [Google Scholar]
- 28.Cubitt R., Fragneto G. D17: the new reflectometer at the ILL. 2002;331:329–331. doi: 10.1007/s003390201611. [DOI] [Google Scholar]
- 29.Le Brun A., Huang T.-Y., Pullen S., Nelson A., Spedding J., Holt S. Spatz: the time-of-flight neutron reflectometer with vertical sample geometry at the OPAL research reactor. J. Appl. Crystallogr. 2023;56 doi: 10.1107/S160057672201086X. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gutfreund P., Saerbeck T., Gonzalez M.A., Pellegrini E., Laver M., Dewhurst C., Cubitt R. Towards generalized data reduction on a chopperbased time-of-flight neutron reflectometer. J. Appl. Crystallogr. 2018;51:606–615. doi: 10.1107/S160057671800448X. [DOI] [Google Scholar]
- 31.Nelson A.R.J., Prescott S.W. Refnx: Neutron and X-ray reflectometry analysis in python. J. Appl. Crystallogr. 2019;52:193–200. doi: 10.1107/S1600576718017296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Browning K.L., Lind T.K., Maric S., Fredrikson G.N. Human Lipoproteins at Model Cell Membranes: Effect of Lipoprotein Class on Lipid Exchange. Sci. Rep. 2017;7:1–11. doi: 10.1038/s41598-017-07505-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nelson A. Co-refinement of multiple-contrast neutron/X-ray reflectivity data using MOTOFIT. J. Appl. Crystallogr. 2006;39:273–276. doi: 10.1107/S0021889806005073. [DOI] [Google Scholar]
- 34.Jebari-benslaiman S., Uribe K.B., Benito-vicente A., Galicia-garcia U. Cholesterol Efflux E fficiency of Reconstituted HDL Is Affected by Nanoparticle Lipid Composition. Biomedicines. 2020;8:373. doi: 10.3390/biomedicines8100373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Theken K.N., Tang S.Y., Sengupta S., FitzGerald G.A. The roles of lipids in SARS-CoV-2 viral replication and the host immune response. J. Lipid Res. 2021;62 doi: 10.1016/J.JLR.2021.100129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Waldie S., Del Giudice R., Cárdenas M. Structure of Lipoproteins and Their Capacity for Lipid Exchange: Relevance for Development of Atherosclerosis and Its Treatment by HDL Therapy. Dyslipidemia. 2021 doi: 10.5772/intechopen.95868. [DOI] [Google Scholar]
- 37.Stadler J.T., Mangge H., Rani A., Curcic P., Herrmann M., Prüller F., Marsche G. Low HDL Cholesterol Efflux Capacity Indicates a Fatal Course of COVID-19. Antioxidants. 2022;11:1–14. doi: 10.3390/antiox11101858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kluck G.E.G., Yoo J.A., Sakarya E.H., Trigatti B.L. Good cholesterol gone bad? HDL and COVID-19. Int. J. Mol. Sci. 2021;22 doi: 10.3390/ijms221910182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wei C., Wan L., Yan Q., Wang X., Zhang J., Zhang Y., Sun J., Yang X., Gong J., Fan C., Yang X., Wang Y., Wang X., Li J., Yang H., Li H., Zhang Z., Wang R., Du P., Zong Y., Yin F., Zhang W., Peng Y., Lin H., Zhang R., Chen W., Gao Q., Cao Y., Zhong H. SARS-CoV-2 manipulates the SR-B1-mediated HDL uptake pathway for its entry. Nat. Metab. 2020;2:1391–1400. doi: 10.1038/s42255-020-00324-0. [DOI] [PubMed] [Google Scholar]
- 40.Huang S., Zhou C., Yuan Z., Xiao H., Wu X. The clinical value of high-density lipoprotein in the evaluation of new coronavirus pneumonia. Adv. Clin. Exp. Med. 2021;30:153–156. doi: 10.17219/ACEM/130606. [DOI] [PubMed] [Google Scholar]
- 41.Wang G., Zhang Q., Zhao X., Dong H., Wu C., Wu F., Yu B., Lv J., Zhang S., Wu G., Wu S., Wang X., Wu Y., Zhong Y. Low high-density lipoprotein level is correlated with the severity of COVID-19 patients: An observational study. Lipids Health Dis. 2020;19:1–7. doi: 10.1186/s12944-020-01382-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Y. Peng, L. Wan, C. Fan, P. Zhang, X. Wang, Cholesterol Metabolism — Impacts on SARS-CoV-2 Infection Prognosis Abstract :, MedRxiv. (2020) 1–15. https://doi.org/10.1101/2020.04.16.20068528.
- 43.Dawaliby R., Trubbia C., Delporte C., Noyon C., Ruysschaert J.M., Van Antwerpen P., Govaerts C. Phosphatidylethanolamine is a key regulator of membrane fluidity in eukaryotic cells. J. Biol. Chem. 2016;291:3658–3667. doi: 10.1074/jbc.M115.706523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Waldie S., Sebastiani F., Moulin M., Del Giudice R., Paracini N., Roosen-Runge F., Gerelli Y., Prevost S., Voss J.C., Darwish T.A., Yepuri N., Pichler H., Maric S., Forsyth V.T., Haertlein M., Cárdenas M. ApoE and ApoE Nascent-Like HDL Particles at Model Cellular Membranes: Effect of Protein Isoform and Membrane Composition. Front. Chem. 2021;9:1–15. doi: 10.3389/fchem.2021.630152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lee Y., Siddiqui W. Cholesterol levels. NCBI Bookshelf. 2019 doi: 10.1097/01.NURSE.0000383897.65472.6b. [DOI] [Google Scholar]
- 46.Subczynski W.K., Pasenkiewicz-Gierula M., Widomska J., Mainali L., Raguz M. High Cholesterol/Low Cholesterol: Effects in Biological Membranes: A Review. Cell Biochem. Biophys. 2017;75:369–385. doi: 10.1007/s12013-017-0792-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Talbot C.P.J., Plat J., Ritsch A., Mensink R.P. Determinants of cholesterol efflux capacity in humans. Prog. Lipid Res. 2018;69:21–32. doi: 10.1016/j.plipres.2017.12.001. [DOI] [PubMed] [Google Scholar]
- 48.Wieczorek E., Ćwiklińska A., Kuchta A., Kortas-Stempak B., Gliwińska A., Jankowski M. The differential effects of HDL subpopulations on lipoprotein lipase (LPL)-mediated VLDL catabolism. Biomedicines. 2021;9 doi: 10.3390/biomedicines9121839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Camont L., Chapman M.J., Kontush A. Biological activities of HDL subpopulations and their relevance to cardiovascular disease. Trends Mol. Med. 2011;17:594–603. doi: 10.1016/j.molmed.2011.05.013. [DOI] [PubMed] [Google Scholar]
- 50.Camont L., Lhomme M., Rached F., Le Goff W., Nègre-Salvayre A., Salvayre R., Calzada C., Lagarde M., Chapman M.J., Kontush A. Small, dense high-density lipoprotein-3 particles are enriched in negatively charged phospholipids: Relevance to cellular cholesterol efflux, antioxidative, antithrombotic, anti-inflammatory, and antiapoptotic functionalities. Arterioscler. Thromb. Vasc. Biol. 2013;33:2715–2723. doi: 10.1161/ATVBAHA.113.301468. [DOI] [PubMed] [Google Scholar]
- 51.Gulshan K., Brubaker G., Wang S., Hazen S.L., Smith J.D. Sphingomyelin depletion impairs anionic phospholipid inward translocation and induces cholesterol efflux. J. Biol. Chem. 2013;288:37166–37179. doi: 10.1074/jbc.M113.512244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Waheed A.A., Freed E.O. The role of lipids in retrovirus replication. Viruses. 2010;2:1146–1180. doi: 10.3390/v2051146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dreschers S., Franz P., Dumitru C.A., Wilker B., Jahnke K., Gulbins E. Cellular Physiology and Biochemistr y Biochemistry Infections with Human Rhinovirus Induce the Formation of Distinct Functional Membrane Domains. Cell Physiol Biochem. 2007;20:241–254. doi: 10.1159/000104170. [DOI] [PubMed] [Google Scholar]
- 54.Fidalgo Rodríguez J.L., Dynarowicz-Latka P., Miñones Conde J. Effects of saturated and polyunsaturated fatty acids on interactions with cholesterol versus 7-ketocholesterol in Langmuir monolayers and their potential biological implications. Colloids Surf. B Biointerfaces. 2019;174:189–198. doi: 10.1016/j.colsurfb.2018.08.052. [DOI] [PubMed] [Google Scholar]
- 55.D. Souza, A. Martins, L. Rosa-Fernandes, L. Rocha, G. Alexandria, S. Bhosale, F. de Rose Ghilardi, T. Falcão, A. Justi, J. carlos Nicolau, C. Marinho, C. Wrenger, M. Larsen, R. Focaccia, P. Di Mascio, G. Palmisano, G. Ronsein, HDL proteome remodeling associates with COVID-19 severity, J. Clin. Lipidol. 15 (2021) 796–804. https://doi.org/https://doi.org/10.1016/j.jacl.2021.10.005. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data will be made available on request.