Summary
Embryonic stem cells (ESCs) are superior to differentiated cells to maintain genome stability, but the underlying mechanisms remain largely elusive. R-loops are constantly formed during transcription and are inducers of DNA damage if not resolved. Here we report that mouse ESCs (mESCs) can efficiently prevent unscheduled R-loop formation, and a long noncoding RNA Lnc530 plays regulatory role. Lnc530 is expressed in mESCs and localizes on R-loops. Depletion of Lnc530 in mESCs causes R-loop accumulation and DNA damage, whereas forced expression of Lnc530 in differentiated cells suppresses the R-loop formation. Mechanistically, Lnc530 associates with DDX5 and TDP-43 in an inter-dependent manner on R-loops. Formation of Lnc530-DDX5-TDP-43 complex substantially increases the local protein levels of DDX5 and TDP-43, both of which play critical roles in R-loop regulation. This study uncovers an efficient strategy to prevent R-loop accumulation and preserve genomic stability in mESCs and possibly other stem cell types.
Keywords: lncRNA, R-loops, mouse embryonic stem cells, genomic stability
Highlights
-
•
Lnc530 is expressed in mouse ESCs and localizes on R-loops
-
•
Lnc530 constrains R-loop levels and preserves genomic stability
-
•
Lnc530-TDP-43-DDX5 form a complex on R-loops
In this article, Zheng and colleagues identify a long noncoding RNA Lnc530, which regulates R-loop homeostasis. Lnc530 localizes on R-loops where it physically interacts with DDX5 and TDP-43 and increases their accumulation on R-loops. This study uncovers an efficient strategy to prevent R-loop formation and preserve genomic stability in mouse embryonic stem cells.
Introduction
Embryonic stem cells (ESCs) are derived from the inner cell mass of blastocysts. Due to the characteristics of pluripotency and self-renewal, ESCs are widely utilized in basic research and have promising applications in cell-based regenerative medicine (Kimbrel and Lanza, 2015; Takahashi and Yamanaka, 2016). Aside from the property of pluripotency, ESCs have super-stable genome, which is crucial for the maintenance of stem cell identity (Maynard et al., 2008). Previous studies compared the genome mutation rates of mouse ESCs (mESCs) with their isogenic mouse embryonic fibroblasts (MEFs), and found that the mutation rate of mESCs was 100 times lower than that of MEFs (Cervantes et al., 2002; Tichy and Stambrook, 2008). Although some pioneer works revealed that the mESC-specific mechanisms play essential roles to ensure efficient genome stability regulation (Wang et al., 2021; Xiong et al., 2015; Zhang et al., 2019; Zhao et al., 2015, 2018), how mESCs maintain the genome stability during the rapid self-renewal remains largely unclear. Investigating the underlying mechanisms would expand the knowledge and provide clues to develop strategies to improve the safety of stem cell applications (Andrews et al., 2022).
R-loops are transient structures consisting of a DNA-RNA hybrid and a displaced single-stranded DNA (ssDNA) (Li and Manley, 2005; Sanz et al., 2016). R-loops (regulatory or unscheduled) can form co-transcriptionally when the nascent RNA hybridizes with the template DNA (Aguilera and Garcia-Muse, 2012). Distinct from the short form of RNA-DNA hybrid formed during transcription and lagging-strand DNA replication, R-loops generally span for 100–2,000 base pairs and play double-edged roles in many cellular activities (Costantino and Koshland, 2015). Regulatory R-loops are essential in a number of physiological processes, including transcription regulation (Skourti-Stathaki and Proudfoot, 2014), immunoglobulin class switching (Bhatia et al., 2017), homologous recombination (HR)-mediated DNA damage repair (Ohle et al., 2016; Yasuhara et al., 2018), telomere lengthening (Arora et al., 2014; Balk et al., 2013), and epigenetic modification of chromatin (Castellano-Pozo et al., 2013; Skourti-Stathaki et al., 2014). However, unscheduled or aberrant R-loops represent a major threat to genome stability. They can induce transcription stress and generate transcription-associated genomic instability (Aguilera and Gomez-Gonzalez, 2017; Hamperl and Cimprich, 2016). Moreover, R-loops can also be recognized by several nucleases and converted into DNA breaks (Sollier et al., 2014). In mitotic cells, R-loops could be a roadblock to DNA replication (Garcia-Muse and Aguilera, 2019). When the DNA replication fork encounters transcription machinery in the head-on orientation, the transcription-replication collisions (TRCs) stall transcription and induce persistent R-loop formation. R-loops conversely block replication and transcription, cause potential replication stress-associated DNA damage (Hamperl et al., 2017; Kim et al., 2020; Sankar et al., 2016), and impair self-renewal (Abakir et al., 2020; Pappa et al., 2019). Thus, R-loop formation and resolution must be carefully regulated to avoid aberrant R-loop accumulation.
To protect the genome from excessive R-loop accumulation, cells deploy different mechanisms to prevent R-loop formation and to resolve existing R-loops. By utilizing DNA-RNA immunoprecipitation (DRIP) or proximity labeling combined with mass spectrometry (MS) analyses, previous studies have identified the interactome of R-loops (Wang et al., 2018; Yan et al., 2022). Several hundreds of proteins have been identified, and the largest category is the RNA-DNA helicases, particularly DEAD (Asp-Glu-Ala-Asp) or DEAH (Asp-Glu-Ala-His)-box members, whose mutations affect the formation and resolution of R-loops (Bjorkman et al., 2020; Frame and North, 2021; Grunseich et al., 2018) and are associated with human diseases including amyotrophic lateral sclerosis (ALS) and frontotemporal dementia (FTD) (Skourti-Stathaki and Proudfoot, 2014; Walker et al., 2017; Wang et al., 2018). RNase H is also key to R-loop resolution in vivo (Cerritelli and Crouch, 2009), and deficiency of the enzyme causes a severe neuroinflammatory disease, Aicardi Goutières syndrome (AGS) (Cristini et al., 2022). In addition, other factors that facilitate the RNA processing or alter the DNA topology can affect R-loop formation and resolution (Luo et al., 2022; Stolz et al., 2019). Telomeric-repeat-containing RNA (TERRA) preferentially associates with short telomeres through the formation of telomeric R-loop structures to regulate telomeric chromatin structure and suppress DNA damage (Feretzaki et al., 2020; Vohhodina et al., 2021).
ESCs proliferate rapidly (e.g., 10–12 h per cell cycle in mESCs). The frequent DNA replication increases the chance of TRCs and enhances the opportunity to form undesirable R-loops. However, the genome of mESCs remains super-stable, raising the hypothesis that mESCs might be able to efficiently prevent undesirable R-loop formation or accumulation. In this study, we reported that an mESC-expressed long noncoding RNA (lncRNA), Lnc530, played essential roles in preventing R-loop accumulation. Lnc530 associated with DDX5 and TDP-43, two R-loop regulators, on R-loops and increased their local protein levels to enhance R-loop regulation and genomic stability.
Results
mESCs are able to efficiently prevent R-loop formation
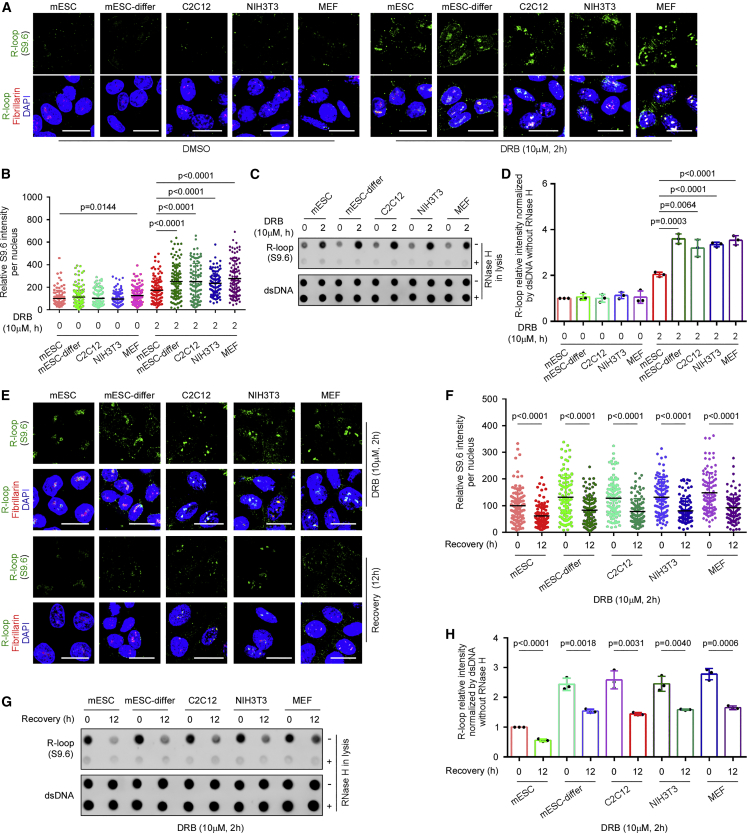
To investigate whether mESCs can prevent R-loop accumulation with higher efficiency, we utilized the reversible transcription inhibitor 5,6-dichloro-1-β-d-ribofuranosyl-benzimidazole (DRB) to induce R-loop formation (Scalera et al., 2021) in mESCs and several types of differentiated cells, including MEFs, NIH3T3, mouse myoblasts C2C12, and isogenic mESC-differentiated cells (mESC-differ) cultured in medium depleted of LIF (Figure S1A). R-loop formation was detected by immunofluorescent staining with S9.6 antibody, which specifically recognizes the DNA-RNA hybrids (Boguslawski et al., 1986; Bou-Nader et al., 2022). Treatment with DRB for 2 h induced a lower level of R-loops in mESCs than in all types of differentiated cells (Figures 1A and 1B). R-loop levels were also assessed by dot blotting of nuclear DNA with S9.6 antibody, and a consistent result was obtained (Figures 1C and 1D). To rule out the possibility that mESCs display lower transcriptional activity that contributes to less R-loop formation, we compared the transcriptional activity among these cell types. Nascent RNAs were labeled with uridine analog 5-ethynyluridine (EU) for 30 min (Kim et al., 2020) and were detected by immunofluorescent staining. As shown (Figure S1B), the transcriptional activity in mESCs was comparable with or even higher than that in all the differentiated cells. Thus, mESCs were more efficient than differentiated cells to prevent aberrant R-loop formation. We then washed away the DRB and investigated the resolution of existing R-loops in mESCs and differentiated cells. The immunofluorescence and dot-blot analyses consistently showed that the resolution rates of existing R-loops did not differ between mESCs and differentiated cells (Figures 1E–1H). Taken together, these observations suggested that mESCs, compared with the differentiated cells, could efficiently prevent R-loop formation.
Figure 1.
mESCs have high efficiency to prevent R-loop formation
(A) Immunostaining detection of R-loops in mESC, mESC-differentiated cells (mESC-differ), C2C12, NIH3T3, and MEFs. R-loop was labeled by S9.6 (green), and nucleolus was labeled by fibrillarin (red). Scale bars, 20 μm. See also Figure S1.
(B) Quantification of S9.6 staining intensity in (A). At least 20 visual fields containing 500 cells were analyzed in three independent experiments.
(C and D) Detection (C) and quantification (D) of R-loops by dot blotting of genomic DNA from the mESCs and differentiated cells. Double-stranded DNA (dsDNA) was used as internal reference.
(E and F) Dynamics of R-loop resolution. Representative images (E) and quantification (F) of S9.6 staining intensity per nucleus in mESCs and differentiated cells. At least 20 visual fields containing 500 cells were analyzed in three independent experiments. Scale bars, 20 μm.
(G and H) (G) Dot-blot analysis showed the similar kinetics of R-loop resolution in mESCs and differentiated cells. (H) The quantification of dot blot intensity. dsDNA was internal reference. All experiments were repeated three times with similar results. Data are shown as mean ± SD, two-tailed Student’s t test.
mESC-expressed lncRNA Lnc530 prevents R-loop accumulation and DNA damage
We previously identified an uncharacterized lncRNA NONMMUT019530 (NONCODE ID, Lnc530 for short) in studying the regulation of genomic stability in mESCs. Lnc530 was expressed in mESCs at a higher level compared with several differentiated cell types, including isogenic mESC-differentiated cells (Figure S2A). The expression of Lnc530 also decreased in the process of mESC differentiation via embryoid body (EB) formation (Figure S2B). Intriguingly, some regions of mouse brain expressed higher levels of Lnc530 than mESCs (Figure S2C). These expression patterns indicated that Lnc530 might function in mESCs and brain. Based on mouse reference genome assembly version mm10, the Lnc530 gene has two genomic copies and is located on the plus strand of chromosome 14 (Figure S2D). Full-length (FL) analysis by 3′- and 5′-end rapid amplification of cDNA ends (RACE) showed that it contained two exons and had two isoforms (long and short isoforms) (Figure S2E). Because no suitable primer was available to distinguish the expression of two isoforms of Lnc530 by quantitative RT-PCR, we were unable to know their relative abundance. Although Lnc530 transcripts contain one potential coding frame, in vitro translation assay failed to detect any translation of protein or peptide (Figure S2F). Thus, Lnc530 is a potential noncoding RNA, and the sequences of two isoforms are listed in Figures S3A and S3B.
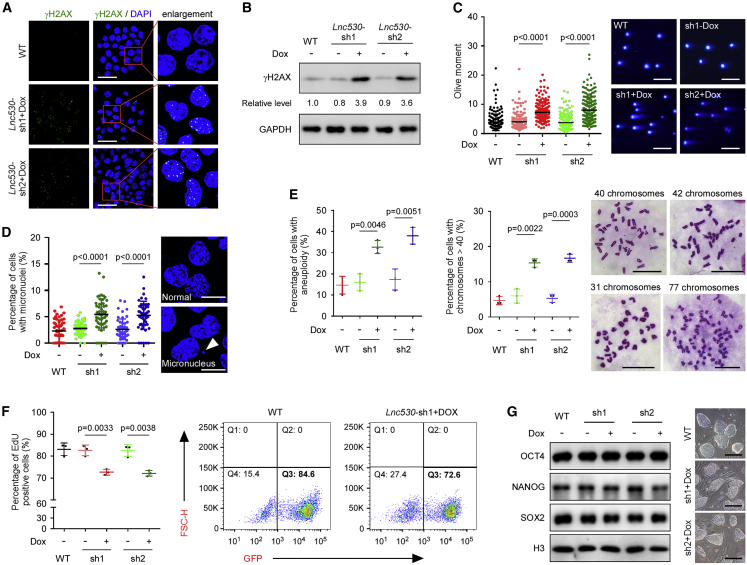
To investigate whether Lnc530 plays role in regulating genomic stability of mESCs, we designed two independent doxycycline (Dox)-inducible short hairpin RNAs (shRNAs), which targeted both isoforms (Figure S3C). Lnc530 was efficiently knocked down by two shRNAs in mESCs (Figure S3C). Depletion of Lnc530 induced DNA double-strand breaks (DSBs), monitored by γH2AX accumulation (Figures 2A and 2B) and comet tail lengthening in neutral comet assay (Figure 2C). DSB generation is often associated with other forms of genomic instability. Concordantly, Lnc530 knockdown (KD) mESCs displayed higher rates of micronuclei (Figure 2D) and aneuploidy (Figure 2E). Because genomic instability can impair mESC self-renewal, we examined the influence of Lnc530 KD on ESC proliferation as well as pluripotency marker expressions. Mitotic cells were labeled by thymidine analog 5-Ethynyl-2'-deoxyuridine (EdU), and flow cytometry analysis showed that Lnc530 KD impaired mESC proliferation (Figure 2F). However, the pluripotency marker expressions were not affected, and Lnc530 KD mESCs displayed normal colony morphology (Figure 2G).
Figure 2.
Loss of Lnc530 causes genomic instability in mESCs
(A and B) Immunostaining (A) and immunoblotting (B) showed that depletion of Lnc530 increased DSB formation as monitored by increased γH2AX level. Scale bars, 40 μm.
(C) Neutral comet assay validated the increased DSB level in Lnc530 KD mESCs. Left panel shows the quantification of comet tail length. Right panel shows representative images. At least 200 tails were analyzed in each group. Scale bars, 400 μm.
(D) More Lnc530 KD mESCs contained micronuclei. Left panel shows the quantification of micronuclei formation. Right panel shows representative images (arrow). At least 50 visual fields containing 1,000 cells were analyzed in each group. Scale bars, 10 μm.
(E) More Lnc530 KD mESCs displayed aneuploidy. Left panel shows the quantification of aneuploidy. Middle panel shows the quantification of aneuploidy with chromosome numbers >40. Right panel shows representative images. At least 150 metaphase spreads were examined in three replications in each group. Scale bars, 20 μm.
(F) Flow cytometry assay showed that the proliferation of Lnc530 KD mESCs was impaired. Left panel shows the quantification result. Right panel shows representative images of flow cytometry.
(G) Immunoblotting showed that the protein expression of core pluripotency regulators SOX2, NANOG, and OCT4 was not changed in Lnc530 KD mESCs (left). Moreover, Lnc530 KD mESCs had normal colony morphology (right). Scale bars, 200 μm. All experiments were independently repeated three times with similar results. The relative protein levels in (B) were normalized by GAPDH. Data are shown as mean ± SD, two-tailed Student’s t test.
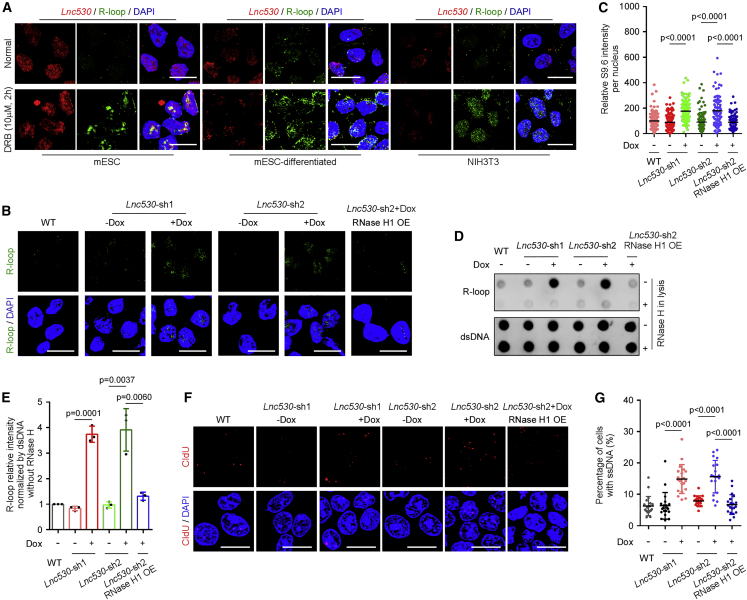
To understand how Lnc530 regulates the genomic stability in mESCs, we first examined its subcellular localization by fluorescence in situ hybridization (FISH). Intriguingly, Lnc530 localized on R-loops after DRB treatment in mESCs, whereas its distribution on R-loops was not obviously detected in differentiated cells (Figure 3A). This observation implicated a potential role of Lnc530 in regulating R-loops to ensure genomic stability in mESCs. We thus investigated the influence of Lnc530 KD on R-loop accumulation. Immunostaining with S9.6 antibody revealed that Lnc530 KD mESCs contained a higher level of R-loops than KD control ESCs under normal culture conditions (Figures 3B and 3C). The increase in R-loop accumulation after Lnc530 KD was further validated by dot-blot analysis (Figures 3D and 3E). R-loops consist of displaced ssDNA. Concordantly, thymidine analog 5-Chloro-2'-deoxyuridine (CIdU) labeling under the natural condition detected an increased level of ssDNA in Lnc530 KD mESCs (Figures 3F and 3G). We also over-expressed FLAG-tagged RNase H1, which resolves R-loops, in Lnc530 KD cells (Figure S3D). The increases in R-loop accumulation and ssDNA formation were counteracted by over-expression of RNase H1 in Lnc530 KD mESCs (Figures 3B–3G), again supporting the regulation of Lnc530 on R-loops. Because aberrant R-loop accumulation induces DNA damage and genomic instability, we proposed that Lnc530 localized on R-loops to prevent R-loop accumulation and preserve genomic stability in mESCs.
Figure 3.
Loss of Lnc530 perturbs R-loop homeostasis
(A) Immunostaining and RNA-FISH show that Lnc530 was localized on R-loops in mESCs rather than in differentiated cells after DRB treatment. Scale bars, 20 μm.
(B and C) Representative images (B) and quantification (C) of S9.6 staining in mESCs. Lnc530 KD increased the R-loop level in mESCs. At least 20 visual field containing 500 cells were analyzed in three independent experiments. Scale bars, 20 μm.
(D and E) Dot-blot analysis (D) showed that R-loop level were increased in Lnc530 KD mESCs. The quantification of dot blot intensity is shown in (E). dsDNA was internal reference.
(F and G) Immunostaining (F) and quantification (G) of ssDNA signal in mESCs. ssDNA was monitored by CIdU (red). Lnc530 KD increased the level of ssDNA in mESCs. At least 20 visual fields containing 1,000 cells were analyzed in each group. Scale bars, 20 μm. All experiments were independently repeated three times with similar results. Data are shown as mean ± SD, two-tailed Student’s t test.
Lnc530 associates with DDX5 and TDP-43 in an inter-dependent manner
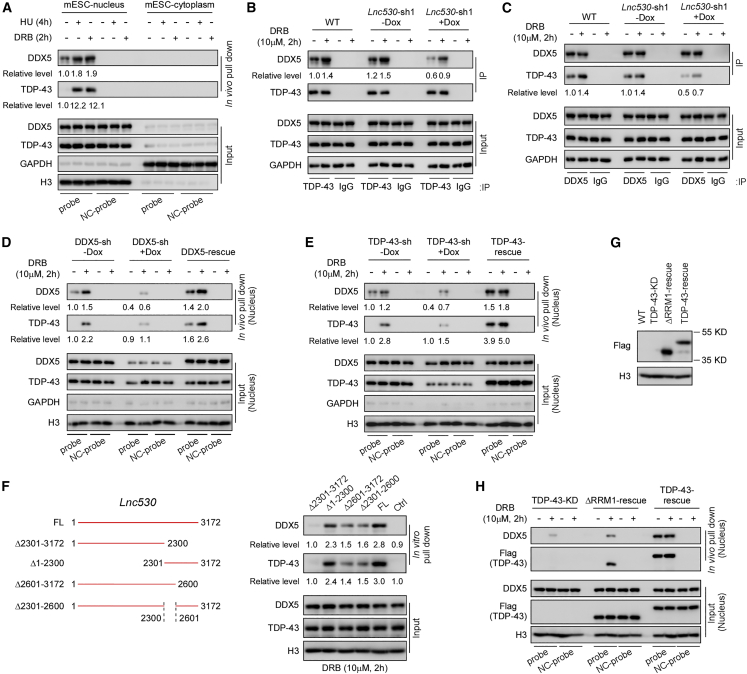
To understand how Lnc530 regulates R-loops, we performed in vivo RNA pull-down combined with MS analysis to identify the potential interaction proteins of Lnc530. MS analysis of in vivo pull-down samples reproducibly identified a list of binding candidates in two replicates (Table S1). Among these candidates, DDX5 and TDP-43 (also known as TARDBP) were previously reported to associate with and regulate R-loops (Giannini et al., 2020; Le et al., 2021; Mersaoui et al., 2019; Wood et al., 2020). We then focused on the two proteins and validated their interaction with Lnc530. In vivo cross-linking followed by nucleus-cytoplasm fractionation and RNA pull-down showed that Lnc530 was associated with DDX5 and TDP-43 in the nucleus. Induction of R-loops by DRB or hydroxylurea (HU) treatment drastically enhanced their interaction. However, no visible interaction was detected in cytoplasm under the normal culture or drug treatment conditions (Figure 4A). Thus, Lnc530 can interact with DDX5 and TDP-43 in the nucleus of mESCs.
Figure 4.
Lnc530-TDP-43-DDX5 form a complex under transcription stress condition
(A) mESCs were fractionated into compartments of cytoplasm and nucleus. In vivo RNA pull-down assay detected the association of Lnc530 with TDP-43 and DDX5 in nucleus. NC-probe, negative control probe.
(B and C) Reciprocal immunoprecipitation (IP) showed that DDX5 and TDP-43 associated with each other and the interaction was impaired in Lnc530 KD mESCs.
(D–F) (D) In vivo RNA pull-down assay showed that Lnc530-TDP-43 association was impaired in DDX5 KD mESCs. Re-expression of DDX5 (DDX5-rescue) restored the interaction. NC-probe, negative control probe. See also Figure S3 (E) In vivo RNA pull-down showed that Lnc530-DDX5 association was impaired in TDP-43 KD mESCs. Re-expression of TDP-43 (TDP-43-rescue) restored the interaction. NC-probe, negative control probe. See also Figure S3 (F) In vitro RNA pull-down using full-length (FL) and variable truncated Lnc530 revealed the fragments in Lnc530 responsible for binding to TDP-43/DDX5. Anti-sense Lnc530 was used as control (Ctrl).
(G) Immunoblotting confirmed the expression of TDP-43-ΔRRM1 (ΔRRM1-rescue) and WT TDP-43 (TDP-43-rescue) in TDP-43 KD mESCs.
(H) In vivo RNA pull-down assay revealed that Lnc530 could not interact with TDP-43-ΔRRM1 mutant protein. NC-probe, negative control probe. All experiments were independently repeated three times with similar results. The relative protein levels in (A)–(F)) were normalized by input.
We next asked whether Lnc530-DDX5-TDP-43 associated with each other. To this end, we performed reciprocal immunoprecipitation (IP). DDX5 physically interacted with TDP-43, and their association was enhanced by R-loop induction (Figures 4B and 4C). Notably, DDX5-TDP-43 interaction was significantly decreased by Lnc530 KD (Figures 4B and 4C), suggesting that the DDX5-TDP-43 association requires Lnc530. We also depleted DDX5 by Dox-inducible shRNA (Figure S3E) and examined its influence on TDP-43 and Lnc530. DDX5 KD did not affect the expression of TDP-43 or Lnc530 (Figures S3F and S3G). However, it attenuated Lnc530-TDP-43 interaction, which was successfully restored by re-expression of FLAG-tagged DDX5 in KD cells (Figures 4D and S3E). Similarly, depletion of TDP-43 reduced the Lnc530-DDX5 interaction, and re-expression of FLAG-tagged TDP-43 rescued the defect (Figures 4E and S3H). Altogether, these data demonstrate that Lnc530-DDX5-TDP-43 form a complex in an inter-dependent manner.
To provide more details on their interaction, we went on to map the TDP-43-DDX5 binding sites on Lnc530. The CatRAPID online tool (Armaos et al., 2021; Ribeiro et al., 2018) predicted that the two proteins had same putative binding sites on Lnc530, which locate at the fragments of 2,395–2,522 base pair (bp) and 2,899–3,026 bp. We then generated a series of Lnc530 truncates including Δ2,301–3,172 (deletion of 2,301–3,172), Δ1–2,300 (deletion of 1–2,300), Δ2,301–2,600 (deletion of 2,301–2,600), and Δ2,601–3,172 (deletion of 2,601–3,172) (Figure 4F). In vitro RNA pull-down assay revealed that, compared with the FL Lnc530 (long isoform), Δ2,301–3,172 failed to pull down TDP-43 and DDX5, whereas Δ1–2,300 displayed normal pull-down efficiency. Notably, two truncates, Δ2,301–2,600 and Δ2,601–3,172, which lack either one of the two predicted binding sites, showed reduced ability to interact with TDP-43 and DDX5 (Figure 4F). These results suggested that the DDX5-TDP-43 protein complex bound to the sites located within 2,301–2,600 and 2,601–3,172 in Lnc530.
TDP-43 is an RNA-binding protein containing two RNA recognition motifs (RRMs), RRM1 and RRM2 (Buratti and Baralle, 2001; D'Ambrogio et al., 2009). The online tool catRAPID predicted that Lnc530 might interact with RRM1 of TDP-43. To test this hypothesis, we generated FLAG-tagged ΔRRM1 mutation (deletion of 106–175 amino acids) and re-expressed the mutant protein in TDP-43 KD mESCs (Figure 4G). In vivo RNA pull-down assay revealed that ΔRRM1 compromised the interaction of Lnc530 with DDX5 and TDP-43 (Figure 4H), indicating that RRM1 motif in TDP-43 was involved in mediating the interaction of Lnc530 with the DDX5-TDP-43 protein complex.
Lnc530-DDX5-TDP-43 complex localizes on R-loops as a whole
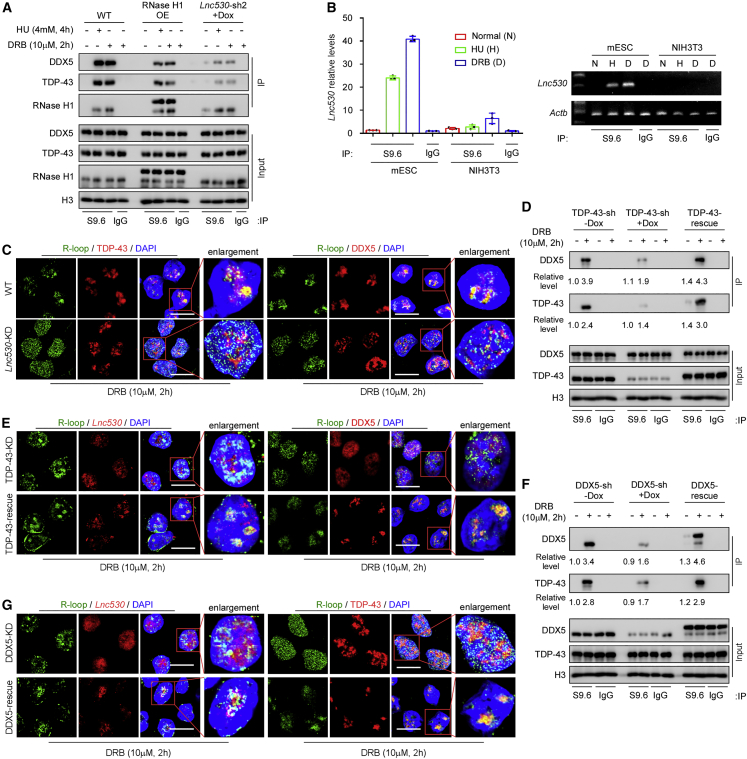
Next, we examined whether the Lnc530-DDX5-TDP-43 complex localized on R-loops in mESCs. R-loops were enriched by IP with S9.6 antibody. After treatment with DRB or HU to stimulate R-loop formation in mESCs, we detected the robust accumulation of DDX5, TDP-43, and Lnc530 on R-loops (Figures 5A and 5B). Forced expression of FLAG-tagged RNase H1 in wild-type (WT) mESCs (Figure S4A) concordantly decreased the levels of DDX5 and TDP-43 in precipitates (Figure 5A).
Figure 5.
Lnc530-TDP-43-DDX5 complex is localized on R-loop under transcription stress condition
(A) S9.6 IP showed that TDP-43 and DDX5 were accumulated on R-loops in mESCs under transcription stress condition. RNase H1 over-expression (OE) or Lnc530 KD reduced the accumulation of TDP-43 and DDX5 on R-loops.
(B) S9.6 IP combined with RT-qPCR analysis for detection of Lnc530 on R-loops in mESCs and NIH3T3. Left panel shows the relative levels of Lnc530. Right panel shows the RT-PCR bands. Actb was set as internal reference.
(C) Immunostaining showed that Lnc530 KD impaired the accumulation of TDP-43 (left) or DDX5 (right) on R-loops. Scale bars, 20 μm.
(D) S9.6 IP showed that TDP-43 KD impaired the allocation of DDX5 on R-loops. Re-expression of TDP-43 (TDP-43-rescue) rescued the defect.
(E) Immunostaining showed that accumulation of Lnc530 (left) or DDX5 (right) on R-loops was impaired in TDP-43 KD mESCs. Re-expression of TDP-43 (TDP-43-rescue) rescued the defect. Scale bars, 20 μm.
(F) S9.6 IP showed that DDX5 KD impaired the association of TDP-43 with R-loops, which was restored by re-expression of DDX5 (DDX5-rescue).
(G) Immunostaining revealed that accumulation of Lnc530 (left) and TDP-43 (right) on R-loops was impaired in DDX5 KD mESCs, and the defects were rescued by re-expression of DDX5 (DDX5-rescue). Scale bars, 20 μm. All experiments were independently repeated three times with similar results. The relative protein levels in (D) and (F) were normalized by input.
Because Lnc530-DDX5-TDP-43 form a complex in an inter-dependent manner, we speculated that depletion of each component influenced the recruitment of other components to R-loops. Under DRB treatment conditions, Lnc530 KD mESCs contained more R-loops (Figures S4B and S4C). However, significantly less DDX5 and TDP-43 were co-immunoprecipitated with R-loops in Lnc530 KD mESCs under HU or DRB treatment conditions (Figure 5A). Because Lnc530 KD did not affect the protein expression of DDX5 and TDP-43 (Figure S4D), this result suggested that the allocation of DDX5 and TDP-43 on R-loops was impaired by Lnc530 KD. Consistently, immunostaining validated the reduced localization of the two proteins on R-loops in Lnc530 KD mESCs (Figure 5C). Similar to Lnc530 KD, depletion of TDP-43 compromised the allocation of DDX5 and Lnc530 on R-loops, and re-expression of TDP-43 rescued the phenotypes (Figures 5D and 5E). The accumulation of TDP-43 and Lnc530 on R-loops was also reduced or recovered when DDX5 was depleted or re-expressed, respectively (Figures 5F and 5G). Thus, these data altogether support that the Lnc530-DDX5-TDP-43 complex is associated with R-loops.
DDX5 and TDP-43 mediate the regulation of Lnc530 on R-loops
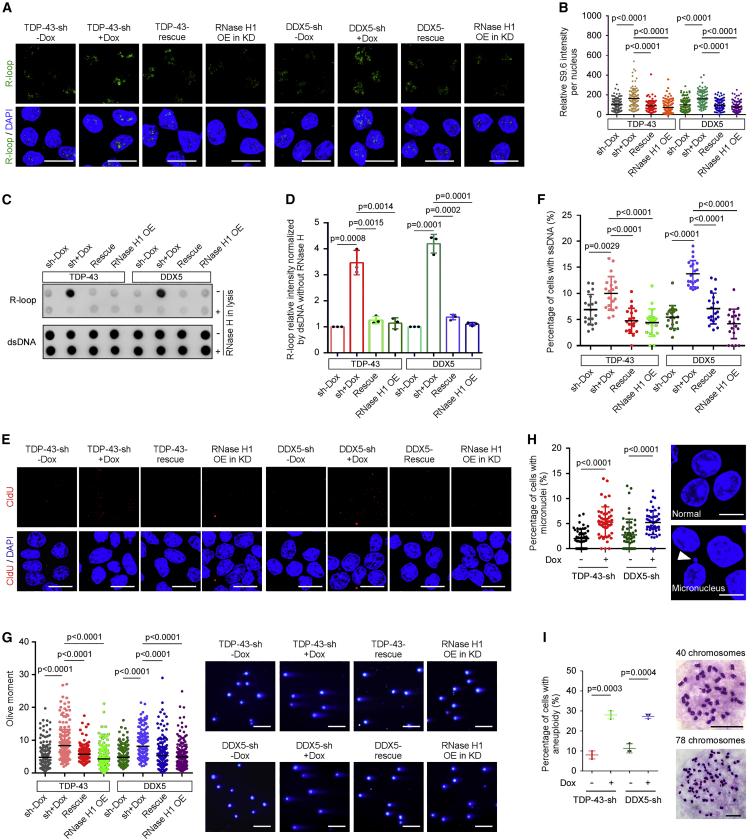
DDX5 and TDP-43 can regulate R-loops in differentiated cells (Giannini et al., 2020; Le et al., 2021; Mersaoui et al., 2019; Wood et al., 2020). We wondered whether DDX5 and TDP-43 played conserved roles in the context of mESCs. Immunostaining showed that, under normal culture conditions, KD of DDX5 or TDP-43 induced R-loop accumulation, which was alleviated by over-expression of FLAG-tagged RNase H1 (Figure S5A) or re-introduction of respective protein (Figures 6A and 6B). Consistent results were obtained by dot-blot analysis of chromatin (Figures 6C and 6D). We also examined the amount of displaced ssDNA in R-loops. Accompanying the induction of DNA-RNA hybrids, a large amount of ssDNA appeared in DDX5 KD and TDP-43 KD mESCs, respectively. Over-expression of RNase H1 or re-expression of respective protein decreased the ssDNA level (Figures 6E and 6F). Concordantly, DDX5 KD or TDP-43 KD mESCs contained higher levels of DNA DSBs (Figure 6G), micronuclei (Figure 6H), and aneuploidy (Figure 6I). The DNA DSB defect was rescued by over-expression of RNase H1 or respective protein (Figure 6G). These results suggest that DDX5 and TDP-43 prevent R-loop accumulation and mediate the regulatory function of Lnc530 on R-loops in mESCs.
Figure 6.
Loss of TDP-43 or DDX5 induces R-loop accumulation and DNA damage
(A) Immunostaining showed that TDP-43 KD or DDX5 KD induced the R-loop formation in mESCs. Re-expression of TDP-43 (TDP-43-rescue), DDX5 (DDX5-rescue), or RNase H1 OE in KD cells rescued the defect. Scale bar, 20 μm.
(B) Quantification of R-loop immunostaining intensity in (A). At least 20 visual fields containing 500 cells were analyzed in three independent experiments.
(C) Dot-blot analysis on genomic DNA confirmed that KD of TDP-43 or DDX5 increased R-loop levels in mESCs. Re-expression of TDP-43 or DDX5, or RNase H1 OE in KD cells rescued the defect.
(D) Quantification of dot-blot intensity in (C), with dsDNA as internal reference.
(E) CIdU incorporation assay showed that KD of TDP-43 or DDX5 increased the level of ssDNA monitored by CIdU (red). Re-expression of TDP-43 or DDX5, or RNase H1 OE in KD cells rescued the defect. Scale bars, 20 μm.
(F) Quantification of immunostaining intensity in (E). At least 20 visual fields containing 1,000 cells were analyzed in each group.
(G) Neutral comet assay revealed that KD of TDP-43 or DDX5 increased DNA DSBs level. Re-expression of TDP-43 or DDX5, or RNase H1 OE in KD cells rescued the defect. Left panel shows the quantification of comet tail length. Right panel shows representative images. At least 200 tails were analyzed in each group. Scale bar, 400 μm.
(H) More DDX5 KD and TDP-43 KD mESCs contained micronuclei. Left panel shows quantification result. Right panel shows the representative image of micronucleus (arrow). At least 50 visual fields containing 1,000 cells were analyzed in each group. Scale bars, 10 μm.
(I) DDX5 KD and TDP-43 KD mESCs displayed higher rate of aneuploidy. Left panel shows quantification result. Right panel shows representative images. At least 150 metaphase spreads were examined in three replications in each group. Scale bars, 20 μm. All experiments were independently repeated three times with similar results. Data are shown as mean ± SD, two-tailed Student’s t test.
To further verify the conclusion that DDX5 and TDP-43 mediate the function of Lnc530, we over-expressed the FLAG-tagged TDP-43 and DDX5 proteins in Lnc530 KD mESCs (Figure S5B), and examined whether it rescued the defects caused by Lnc530 KD. Over-expression of TDP-43 and DDX5 suppressed the R-loop induction caused by Lnc530 KD under normal culture or DRB treatment conditions, as assessed by immunostaining (Figures S5C and S5D) as well as dot-blot analysis of chromatin (Figure S5E). Similarly, the defects of DNA DSBs (Figure S5F) and micronuclei formation (Figure S5G) in Lnc530 KD mESCs were rescued by over-expression of TDP-43 and DDX5. However, aneuploidy could not be rescued (Figure S5H). Taken together, these data support that DDX5 and TDP-43 mediate the regulation of Lnc530 on R-loops.
Ectopic expression of Lnc530 in differentiated cells reduces the aberrant R-loop formation by increasing the recruitment of DDX5-TDP-43 to R-loops
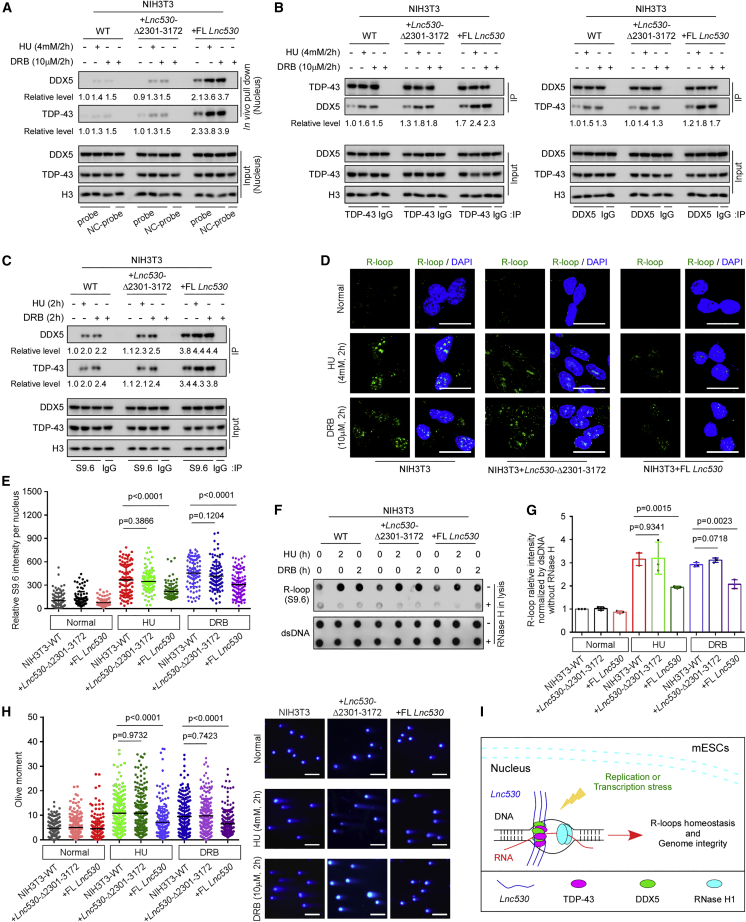
mESCs express Lnc530 at higher levels than many types of differentiated cells. Lnc530 in mESCs promotes the association of Lnc530-DDX5-TDP-43 complex with R-loops to prevent the aberrant R-loop accumulation. We wondered whether forced expression of Lnc530 in differentiated cells could be sufficient to induce Lnc530-DDX5-TDP-43 interaction and to increase the recruitment of DDX5-TDP-43 to R-loops, leading to enhanced R-loop regulation. To this end, we ectopically expressed FL Lnc530 in NIH3T3 cells. As a negative control, truncated Lnc530 Δ2,301–3,172, which could not interact with DDX5-TDP-43, was over-expressed in NIH3T3 cells (Figure S6A). In vivo RNA pull-down assay validated the interaction of DDX5-TDP-43 with exogenous FL Lnc530 under normal culture and treatment conditions (Figure 7A). Reciprocal IP also showed that DDX5-TDP-43 association was stimulated by expression of FL Lnc530 rather than truncated Lnc530 Δ2,301–3,172 (Figure 7B). Consequently, more DDX5 and TDP-43 were recruited to R-loops in NIH3T3 cells expressing FL but not truncate Lnc530 (Figure 7C). These results suggested that ectopic expression of Lnc530 was sufficient to evoke the assembly of Lnc530-DDX5-TDP-43 complex on R-loops in differentiated cells. In line with the increased accumulation of DDX5 and TDP-43 on R-loops, ectopic expression of FL Lnc530, but not truncated Lnc530, in NIH3T3 cells increased the competence to prevent R-loop formation under HU or DRB treatment conditions, as shown by immunostaining (Figures 7D and 7E) as well as dot-blot analysis (Figures 7F and 7G). Concordantly, neutral comet assay revealed that FL Lnc530-expressing NIH3T3 cells contained lower levels of DNA DSBs than WT NIH3T3 cells or NIH3T3 cells expressing truncated Lnc530 after DRB or HU treatment (Figure 7H).
Figure 7.
Ectopic expression of Lnc530 in NIH3T3 cells decreases R-loop formation and increases genome stability
(A) In vivo RNA pull-down assay confirmed that under HU or DRB treatment conditions, TDP-43 and DDX5 interacted with FL Lnc530 but not Lnc530 Δ2,301–3,172 ectopically expressed in NIH3T3 cells. NC-probe, negative control probe.
(B) Reciprocal IP showed that ectopic expression of FL Lnc530, but not Lnc530 Δ2,301–3,172, increased TDP-43-DDX5 association in NIH3T3 cells. See also Figure S6.
(C) S9.6 IP showed that ectopic expression of FL Lnc530, but not Lnc530 Δ2,301–3,172, increased the accumulation of TDP-43 and DDX5 on R-loops in NIH3T3 cells.
(D and E) Immunostaining (D) and quantification (E) revealed that OE of FL Lnc530, but not Lnc530 Δ2,301–3,172, reduced the R-loop formation after HU or DRB treatment. At least 20 visual fields containing 500 cells were analyzed in three independent experiments. Scale bars, 20 μm.
(F and G) Dot-blot analysis (F) and quantification (G) confirmed that ectopic expression of FL Lnc530, but not Lnc530 Δ2,301–3,172, decreased the R-loop formation after HU or DRB treatment. dsDNA was used as internal reference.
(H) Neutral comet assay showed that forced expression of FL Lnc530, but not Lnc530 Δ2,301–3,172, alleviated DNA DSB formation after HU or DRB treatment. Left panel shows the quantification of comet tail length. Right panel shows representative images. At least 200 tails were analyzed in each group. Scale bar, 400 μm.
(I) Working model. LncRNA Lnc530 recruited more TDP-43 and DDX5 proteins to R-loops to efficiently prevent undesirable R-loop formation. All experiments were independently repeated three times with similar results. The relative protein levels in (A)–(C) were normalized by input. Data are shown as mean ± SD, two-tailed Student’s t test.
To further validate the observations, we repeated the experiments in isogenic mESC-differentiated cells. mESCs were differentiated for 14 days as described above. FL Lnc530 was over-expressed in mESC-differentiated cells (Figure S6B). Consistently, over-expression of Lnc530 suppressed the R-loop accumulation in isogenic differentiated cells as detected by immunostaining (Figures S6C and S6D) and dot-blot analyses (Figures S6E and S6F). The DNA DSBs level in mESC-differentiated cells was also decreased after over-expression of Lnc530 (Figure S6G). Taken together, we proposed a working model in which Lnc530 increases the cellular ability to prevent undesirable R-loop formation by driving the Lnc530-DDX5-TDP-43 complex assembly and elevating the local concentrations of DDX5 and TDP-43 to regulate R-loops (Figure 7J).
Discussion
Unscheduled R-loop accumulation can promote DNA damage and chromosome instability. R-loop homeostasis must be tightly regulated to maintain genomic stability. In this study, we reported that mESCs, which possess superior stable genome over differentiated cells, had high capacity to prevent aberrant R-loop formation to avoid R-loop-induced DNA damage. Intriguingly, we found that an mESC-expressed lncRNA Lnc530 attributed to this capacity. Lnc530 localizes on R-loops, where it physically interacts with DDX5 and TDP-43, thereby increasing the recruitment of two proteins to R-loops. DDX5 and TDP-43 are two essential R-loop regulators and their increased recruitment to R-loops enhanced the homeostasis regulation of R-loops. Lnc530-DDX5-TDP-43 association on R-loops occurred in NIH3T3 cells as well as in mESC-differentiated cells when Lnc530 was ectopically expressed. It seemed that the interaction did not require other mESC-specific factors and might be autonomous. Thus, we identified an lncRNA that can localize on R-loops to regulate R-loop homeostasis. To our knowledge, this is the first work to report the R-loop-resided regulatory lncRNA. In the future, systemic identification of R-loop-associated lncRNAs by utilizing synthetic DNA-RNA hybrids would provide more insights into the regulation of R-loop homeostasis.
How Lnc530 stimulates the assembly of Lnc530-DDX5-TDP-43 complex to increase the local concentrations of DDX5 and TDP-43 is currently unknown. However, RNA-dependent condensate formation via phase separation could be an intriguing hypothesis. RNA and interacting RNA-binding proteins can form condensates that specifically concentrate biomolecules to efficiently regulate key biological processes (Banani et al., 2017; Shin and Brangwynne, 2017). Both DDX5 and TDP-43 contain intrinsically disordered regions (IDRs) essential for induction of phase separation and formation of cellular condensates. Moreover, TDP-43 can undergo phase separation. In the future, whether Lnc530-DDX5-TDP-43 form condensates via phase separation warrants investigation.
Lnc530 interacted with TDP-43 and modulated its role. Dysfunction of TDP-43 was linked to neurodegenerative disorders, including ALS and frontotemporal dementia (FTD) (Hill et al., 2016; Mann et al., 2019; Neumann et al., 2006; Yin et al., 2012). Notably, we detected abundant expression of Lnc530 in different brain regions of mice, with the levels higher than that in mESCs. These observations suggested that Lnc530 could modulate the functions of TDP-43 in brain and therefore be involved in the pathogenesis of ALS and FTD. Lnc530 is partially conserved between mouse and human. The functions of human Lnc530 and its involvement in brain diseases warrant further study.
Experimental procedures
Resource availability
Corresponding author
Further information and requests for resources and reagents should be directed to the corresponding author, Ping Zheng (zhengp@mail.kiz.ac.cn).
Materials availability
All unique/stable reagents generated in this study are available from the corresponding author, Ping Zheng, with a completed Materials Transfer Agreement.
Cell lines and culture
MEFs were obtained from embryonic day 13.5 (E13.5) embryos of CD1 mice. MEFs, C2C12, and NIH3T3 cells were cultured in DMEM supplemented with 10% fetal bovine serum (Gibco, 10099141). WT mESCs were derived previously in our laboratory (Zhao et al., 2015) and cultured in DMEM/F12 medium supplemented with 20% Knockout serum replacement (Gibco, 10828028), 2 mM L-glutamine, 1 mM sodium pyruvate, 0.1 mM β-mercaptoethanol, 1% non-essential amino acids, and 1,000 units/mL mouse leukemia inhibitory factor (mLIF, Millipore, ESG1107).
qRT-PCR
Total RNA was extracted using Trizol (Tiangen, DP424), and 1.0 μg of total RNA was reverse transcribed using PrimeScript RT Reagent Kit (Takara, RR037A). qPCR was performed on CFX96 Real-Time System (Bio-Rad) using the TB Green Premix Ex Taq II Kit (Takara, RR820A). The primers for qPCR are listed in Table S2.
Lentivirus-mediated gene manipulation
The primers for gene cloning are listed in Table S3 and for gene knockdown by shRNAs in Table S4. All the shRNAs were constructed into pTRIPZ plasmid. FL and truncated Lnc530 were amplified using overlap PCR and inserted into pEasy-Blunt plasmid (Transgene, CB101-01). pTOMO-EGFP-1×FLAG-TDP-43 was acquired from Prof. Baowei Jiao at the Kunming Institute of Zoology, Chinese Academy of Sciences. FLAG-tagged Tdp-43-ΔRRM1 coding region (CDS) was amplified using overlap PCR and inserted into modified pTOMO-EGFP plasmid. Rnase H1 and Ddx5 coding regions (CDS) were inserted into modified pTOMO-EGFP-3×FLAG plasmid. Lnc530 ORF and Floped CDS containing FLAG tag were inserted into pcDNA3.1(+) plasmid. For expression of Lnc530, FL and truncated Lnc530-Δ2,301–3,172 were inserted into pCDH-CMV-GFP-Puro plasmid.
For the virus package, the plasmids were mixed with packaging plasmids psPAX2 and pMD2.G, and co-transfected into 293T cells. For gene knockdown and Lnc530 expression, the puromycin (0.5 μg/mL) was added into culture medium for 48-h selection after lentivirus infection. For the pTOMO-mediated gene expression, GFP-positive cells were sorted by flow cytometry.
Immunofluorescent staining
Cells were fixed in 4% paraformaldehyde (PFA) for 10 min, followed by treatment with 0.2% Triton X-100, and blocked with 3% BSA at room temperature. The blocked cells were incubated with primary and secondary antibodies diluted in blocking solution, and the nuclei were counterstained with DAPI. Slides were sealed with glycerin. Images were captured using a confocal microscope (Olympus, FV1000).
For R-loop immunostaining, cells were first permeabilized with CSKT buffer (10 mM PIPES [pH = 6.8], 100 mM NaCl, 3 mM MgCl2, 0.3 M sucrose, and 0.5% Triton X-100) at 4°C for 10 min. After that, procedures were carried out as in immunofluorescence staining.
RNA transcription assay was performed using EU-Click Kit (RiboBio, C10316-1). Cells were labeled with 500 μM 5-ethynyl uridine (EU) for 30 min, and immunostaining was carried out according to the manufacturer’s protocol. Fluorescence intensity was measured by ImageJ (Version 1.52), and statistical analyses were performed using GraphPad Prism (Version 6.01). Antibody information was listed in Table S5.
Dot-blot analysis
Nucleic acids in nuclei were extracted by SDS/proteinase K buffer at 37°C overnight, followed by phenol-chloroform extraction and ethanol precipitation. DNA samples were treated with or without RNase H (New England Biolabs, M0297L), and blotted onto NC membrane. The membranes were air-dried, and blocked with 5% non-fat milk, followed by immunoblotting with primary and secondary antibodies. Images were collected from the Protein Simple FluorChem system (Protein Simple, FluorChem M FM0-561) and quantified using ImageJ (Version 1.52).
RNA-FISH
The probes and hybridization kit (C10910) were ordered from Guangzhou RiboBio. RNA-FISH was performed according to the manufacturer’s protocol. Briefly, cells were permeabilized with CSKT buffer and fixed in 4% PFA. Hybridization was performed at 37°C overnight. Glass covers were washed three times with 4× saline sodium citrate buffer (SSC), once with 2× SSC, and once with 1× SSC at 42°C. Immunostaining was performed as described in section of immunofluorescent staining.
Immunoblotting
Total protein was extracted using RIPA buffer (Beyotime, P0013B) and subjected to SDS-PAGE gel separation. Proteins were then transferred to polyvinylidene fluoride (PVDF) membrane. Membranes were blocked and incubated with antibodies. Images were collected from the Protein Simple FluorChem system (Protein Simple, FluorChem M FM0-561) and quantified using ImageJ (Version 1.52). Antibody information is listed in Table S5.
IP
For IP analysis, cells were collected and lysed in RIPA buffer (Beyotime, P0013). The extracts were incubated with 2 μg of antibody or immunoglobulin G (IgG) and 30 μL of protein A/G-MagPoly beads (Abmart, A10002M) at 4°C overnight. Beads were washed and boiled with 2× SDS loading buffer for downstream immunoblotting analysis.
S9.6 IP
S9.6 IP analysis was performed as described previously (Cristini et al., 2018). Briefly, cells were re-suspended in RSB buffer (10 mM Tris-HCl [pH = 7.5], 200 mM NaCl, 2.5 mM MgCl2, 0.5% Triton X-100) and sonicated for 10 min. The extracts were incubated with S9.6 antibody or IgG and beads, supplemented with 2 μL of RNase A (New England Biolabs, T3018L) at 4°C overnight. Beads were washed and eluted for downstream immunoblotting analysis. For detection of Lnc530 on R-loops, the assay was performed as described previously (Gagliardi and Matarazzo, 2016).
In vivo RNA pull-down and MS analysis
The probes labeled with biotin at 3ʹ end were ordered from Guangzhou RiboBio. In vivo RNA pull-down assay was performed as previously described (Chu et al., 2017). Briefly, cells were cross-linked with 265-nm UV light at 400 mJ of energy, and treated with CSKT buffer. After spinning, the pellet was re-suspended in DNase I buffer (50 mM Tris [pH = 7.5], 0.5% NP-40, 0.1% sodium lauroyl sarcosine, protease inhibitors, SUPERase-In, 600 U RNase free DNase I, 10 mM vanadyl ribonucleoside complex) and incubated at 37°C for 1 h. Supernatants were incubated with probes and M-280 streptavidin Dynabeads (Thermo, 00781251) at 65°C for 15 min. The proteins were eluted for downstream MS or immunoblotting analysis. Detailed methods for MS are available in the supplemental information.
Statistical analyses
Statistical analyses were performed by two-tailed Student’s t test, two-way ANOVA with Bonferroni post using GraphPad Prism (Version 6.01). p < 0.05 was considered significant. All experiments were repeated three times using independent biological samples.
Author contributions
D.G. performed most of the experiments, analyzed the data, and prepared the figures. L.W. obtained the FL Lnc530. H.Z. and J.G. performed the MS analysis. W.Z. supervised the figure preparation. P.Z. supervised the study and wrote the manuscript.
Acknowledgments
We thank Professor Baowei Jiao at Kunming Institute of Zoology, Chinese Academy of Sciences, for providing the pTOMO-IRES-EGFP-FLAG-TDP-43 plasmid, and Professor Hongjie Yao at Guangzhou Institute of Biomedicine and Health, Chinese Academy of Sciences, for providing S9.6 antibody. This work was supported by grants from the National Natural Science Foundation of China (31930027).
Conflict of interests
The authors declare no competing interests.
Published: March 16, 2023
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.stemcr.2023.02.003.
Supplemental information
Data and code availability
All data needed to evaluate the conclusions in this paper are present in the paper and/or the supplemental information. The MS data for in vivo RNA pull-down have been deposited to the ProteomeXchange Consortium via the iProX repository with the dataset identifier PXD040075.
References
- Abakir A., Giles T.C., Cristini A., Foster J.M., Dai N., Starczak M., Rubio-Roldan A., Li M., Eleftheriou M., Crutchley J., et al. N(6)-methyladenosine regulates the stability of RNA:DNA hybrids in human cells. Nat. Genet. 2020;52:48–55. doi: 10.1038/s41588-019-0549-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aguilera A., Garcia-Muse T. R loops: from transcription byproducts to threats to genome stability. Mol. Cell. 2012;46:115–124. doi: 10.1016/j.molcel.2012.04.009. [DOI] [PubMed] [Google Scholar]
- Aguilera A., Gómez-González B. DNA-RNA hybrids: the risks of DNA breakage during transcription. Nat. Struct. Mol. Biol. 2017;24:439–443. doi: 10.1038/nsmb.3395. [DOI] [PubMed] [Google Scholar]
- Andrews P.W., Barbaric I., Benvenisty N., Draper J.S., Ludwig T., Merkle F.T., Sato Y., Spits C., Stacey G.N., Wang H., Pera M.F. The consequences of recurrent genetic and epigenetic variants in human pluripotent stem cells. Cell Stem Cell. 2022;29:1624–1636. doi: 10.1016/j.stem.2022.11.006. [DOI] [PubMed] [Google Scholar]
- Armaos A., Colantoni A., Proietti G., Rupert J., Tartaglia G.G. catRAPID omics v2.0: going deeper and wider in the prediction of protein-RNA interactions. Nucleic Acids Res. 2021;49:W72–W79. doi: 10.1093/nar/gkab393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arora R., Lee Y., Wischnewski H., Brun C.M., Schwarz T., Azzalin C.M. RNaseH1 regulates TERRA-telomeric DNA hybrids and telomere maintenance in ALT tumour cells. Nat. Commun. 2014;5:5220. doi: 10.1038/ncomms6220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balk B., Maicher A., Dees M., Klermund J., Luke-Glaser S., Bender K., Luke B. Telomeric RNA-DNA hybrids affect telomere-length dynamics and senescence. Nat. Struct. Mol. Biol. 2013;20:1199–1205. doi: 10.1038/nsmb.2662. [DOI] [PubMed] [Google Scholar]
- Banani S.F., Lee H.O., Hyman A.A., Rosen M.K. Biomolecular condensates: organizers of cellular biochemistry. Nat. Rev. Mol. Cell Biol. 2017;18:285–298. doi: 10.1038/nrm.2017.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhatia V., Herrera-Moyano E., Aguilera A., Gómez-González B. The role of replication-associated repair factors on R-loops. Genes. 2017;8:171. doi: 10.3390/genes8070171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Björkman A., Johansen S.L., Lin L., Schertzer M., Kanellis D.C., Katsori A.M., Christensen S.T., Luo Y., Andersen J.S., Elsässer S.J., et al. Human RTEL1 associates with Poldip3 to facilitate responses to replication stress and R-loop resolution. Genes Dev. 2020;34:1065–1074. doi: 10.1101/gad.330050.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boguslawski S.J., Smith D.E., Michalak M.A., Mickelson K.E., Yehle C.O., Patterson W.L., Carrico R.J. Characterization of monoclonal antibody to DNA.RNA and its application to immunodetection of hybrids. J. Immunol. Methods. 1986;89:123–130. doi: 10.1016/0022-1759(86)90040-2. [DOI] [PubMed] [Google Scholar]
- Bou-Nader C., Bothra A., Garboczi D.N., Leppla S.H., Zhang J. Structural basis of R-loop recognition by the S9.6 monoclonal antibody. Nat. Commun. 2022;13:1641. doi: 10.1038/s41467-022-29187-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buratti E., Baralle F.E. Characterization and functional implications of the RNA binding properties of nuclear factor TDP-43, a novel splicing regulator of CFTR exon 9. J. Biol. Chem. 2001;276:36337–36343. doi: 10.1074/jbc.M104236200. [DOI] [PubMed] [Google Scholar]
- Castellano-Pozo M., Santos-Pereira J.M., Rondón A.G., Barroso S., Andújar E., Pérez-Alegre M., García-Muse T., Aguilera A. R loops are linked to histone H3 S10 phosphorylation and chromatin condensation. Mol. Cell. 2013;52:583–590. doi: 10.1016/j.molcel.2013.10.006. [DOI] [PubMed] [Google Scholar]
- Cerritelli S.M., Crouch R.J. Ribonuclease H: the enzymes in eukaryotes. FEBS J. 2009;276:1494–1505. doi: 10.1111/j.1742-4658.2009.06908.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cervantes R.B., Stringer J.R., Shao C., Tischfield J.A., Stambrook P.J. Embryonic stem cells and somatic cells differ in mutation frequency and type. Proc. Natl. Acad. Sci. USA. 2002;99:3586–3590. doi: 10.1073/pnas.062527199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu H.P., Cifuentes-Rojas C., Kesner B., Aeby E., Lee H.G., Wei C., Oh H.J., Boukhali M., Haas W., Lee J.T. TERRA RNA antagonizes ATRX and protects telomeres. Cell. 2017;170:86–101.e16. doi: 10.1016/j.cell.2017.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costantino L., Koshland D. The Yin and yang of R-loop biology. Curr. Opin. Cell Biol. 2015;34:39–45. doi: 10.1016/j.ceb.2015.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cristini A., Groh M., Kristiansen M.S., Gromak N. RNA/DNA hybrid interactome identifies DXH9 as a molecular player in transcriptional termination and R-loop-associated DNA damage. Cell Rep. 2018;23:1891–1905. doi: 10.1016/j.celrep.2018.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cristini A., Tellier M., Constantinescu F., Accalai C., Albulescu L.O., Heiringhoff R., Bery N., Sordet O., Murphy S., Gromak N. RNase H2, mutated in Aicardi-Goutieres syndrome, resolves co-transcriptional R-loops to prevent DNA breaks and inflammation. Nat. Commun. 2022;13:2961. doi: 10.1038/s41467-022-30604-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Ambrogio A., Buratti E., Stuani C., Guarnaccia C., Romano M., Ayala Y.M., Baralle F.E. Functional mapping of the interaction between TDP-43 and hnRNP A2 in vivo. Nucleic Acids Res. 2009;37:4116–4126. doi: 10.1093/nar/gkp342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feretzaki M., Pospisilova M., Valador Fernandes R., Lunardi T., Krejci L., Lingner J. RAD51-dependent recruitment of TERRA lncRNA to telomeres through R-loops. Nature. 2020;587:303–308. doi: 10.1038/s41586-020-2815-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frame J.M., North T.E. Ddx41 loss R-loops in cGAS to fuel inflammatory HSPC production. Dev. Cell. 2021;56:571–572. doi: 10.1016/j.devcel.2021.02.014. [DOI] [PubMed] [Google Scholar]
- Gagliardi M., Matarazzo M.R. RIP: RNA immunoprecipitation. Methods Mol. Biol. 2016;1480:73–86. doi: 10.1007/978-1-4939-6380-5_7. [DOI] [PubMed] [Google Scholar]
- García-Muse T., Aguilera A. R loops: from physiological to pathological roles. Cell. 2019;179:604–618. doi: 10.1016/j.cell.2019.08.055. [DOI] [PubMed] [Google Scholar]
- Giannini M., Bayona-Feliu A., Sproviero D., Barroso S.I., Cereda C., Aguilera A. TDP-43 mutations link Amyotrophic Lateral Sclerosis with R-loop homeostasis and R loop-mediated DNA damage. PLoS Genet. 2020;16:e1009260. doi: 10.1371/journal.pgen.1009260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grunseich C., Wang I.X., Watts J.A., Burdick J.T., Guber R.D., Zhu Z., Bruzel A., Lanman T., Chen K., Schindler A.B., et al. Senataxin mutation reveals how R-loops promote transcription by blocking DNA methylation at gene promoters. Mol. Cell. 2018;69:426–437.e7. doi: 10.1016/j.molcel.2017.12.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamperl S., Bocek M.J., Saldivar J.C., Swigut T., Cimprich K.A. Transcription-replication conflict orientation modulates R-loop levels and activates distinct DNA damage responses. Cell. 2017;170:774–786.e19. doi: 10.1016/j.cell.2017.07.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamperl S., Cimprich K.A. Conflict resolution in the genome: how transcription and replication make it work. Cell. 2016;167:1455–1467. doi: 10.1016/j.cell.2016.09.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill S.J., Mordes D.A., Cameron L.A., Neuberg D.S., Landini S., Eggan K., Livingston D.M. Two familial ALS proteins function in prevention/repair of transcription-associated DNA damage. Proc. Natl. Acad. Sci. USA. 2016;113:E7701–E7709. doi: 10.1073/pnas.1611673113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S., Kang N., Park S.H., Wells J., Hwang T., Ryu E., Kim B.G., Hwang S., Kim S.J., Kang S., et al. ATAD5 restricts R-loop formation through PCNA unloading and RNA helicase maintenance at the replication fork. Nucleic Acids Res. 2020;48:7218–7238. doi: 10.1093/nar/gkaa501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimbrel E.A., Lanza R. Current status of pluripotent stem cells: moving the first therapies to the clinic. Nat. Rev. Drug Discov. 2015;14:681–692. doi: 10.1038/nrd4738. [DOI] [PubMed] [Google Scholar]
- Le R., Huang Y., Zhang Y., Wang H., Lin J., Dong Y., Li Z., Guo M., Kou X., Zhao Y., et al. Dcaf11 activates Zscan4-mediated alternative telomere lengthening in early embryos and embryonic stem cells. Cell Stem Cell. 2021;28:732–747.e9. doi: 10.1016/j.stem.2020.11.018. [DOI] [PubMed] [Google Scholar]
- Li X., Manley J.L. Inactivation of the SR protein splicing factor ASF/SF2 results in genomic instability. Cell. 2005;122:365–378. doi: 10.1016/j.cell.2005.06.008. [DOI] [PubMed] [Google Scholar]
- Luo H., Zhu G., Eshelman M.A., Fung T.K., Lai Q., Wang F., Zeisig B.B., Lesperance J., Ma X., Chen S., et al. HOTTIP-dependent R-loop formation regulates CTCF boundary activity and TAD integrity in leukemia. Mol. Cell. 2022;82:833–851.e11. doi: 10.1016/j.molcel.2022.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann J.R., Gleixner A.M., Mauna J.C., Gomes E., DeChellis-Marks M.R., Needham P.G., Copley K.E., Hurtle B., Portz B., Pyles N.J., et al. RNA binding antagonizes neurotoxic phase transitions of TDP-43. Neuron. 2019;102:321–338.e8. doi: 10.1016/j.neuron.2019.01.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maynard S., Swistowska A.M., Lee J.W., Liu Y., Liu S.T., Da Cruz A.B., Rao M., de Souza-Pinto N.C., Zeng X., Bohr V.A. Human embryonic stem cells have enhanced repair of multiple forms of DNA damage. Stem Cell. 2008;26:2266–2274. doi: 10.1634/stemcells.2007-1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mersaoui S.Y., Yu Z., Coulombe Y., Karam M., Busatto F.F., Masson J.Y., Richard S. Arginine methylation of the DDX5 helicase RGG/RG motif by PRMT5 regulates resolution of RNA:DNA hybrids. EMBO J. 2019;38:e100986. doi: 10.15252/embj.2018100986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann M., Sampathu D.M., Kwong L.K., Truax A.C., Micsenyi M.C., Chou T.T., Bruce J., Schuck T., Grossman M., Clark C.M., et al. Ubiquitinated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Science. 2006;314:130–133. doi: 10.1126/science.1134108. [DOI] [PubMed] [Google Scholar]
- Ohle C., Tesorero R., Schermann G., Dobrev N., Sinning I., Fischer T. Transient RNA-DNA hybrids are required for efficient double-strand break repair. Cell. 2016;167:1001–1013.e7. doi: 10.1016/j.cell.2016.10.001. [DOI] [PubMed] [Google Scholar]
- Pappa S., Padilla N., Iacobucci S., Vicioso M., Álvarez de la Campa E., Navarro C., Marcos E., de la Cruz X., Martínez-Balbás M.A. PHF2 histone demethylase prevents DNA damage and genome instability by controlling cell cycle progression of neural progenitors. Proc. Natl. Acad. Sci. USA. 2019;116:19464–19473. doi: 10.1073/pnas.1903188116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribeiro D.M., Zanzoni A., Cipriano A., Delli Ponti R., Spinelli L., Ballarino M., Bozzoni I., Tartaglia G.G., Brun C. Protein complex scaffolding predicted as a prevalent function of long non-coding RNAs. Nucleic Acids Res. 2018;46:917–928. doi: 10.1093/nar/gkx1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sankar T.S., Wastuwidyaningtyas B.D., Dong Y., Lewis S.A., Wang J.D. The nature of mutations induced by replication-transcription collisions. Nature. 2016;535:178–181. doi: 10.1038/nature18316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanz L.A., Hartono S.R., Lim Y.W., Steyaert S., Rajpurkar A., Ginno P.A., Xu X., Chédin F. Prevalent, dynamic, and conserved R-loop structures associate with specific epigenomic signatures in mammals. Mol. Cell. 2016;63:167–178. doi: 10.1016/j.molcel.2016.05.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scalera C., Ticli G., Dutto I., Cazzalini O., Stivala L.A., Prosperi E. Transcriptional stress induces chromatin relocation of the nucleotide excision repair factor XPG. Int. J. Mol. Sci. 2021;22:6589. doi: 10.3390/ijms22126589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin Y., Brangwynne C.P. Liquid phase condensation in cell physiology and disease. Science. 2017;357:eaaf4382. doi: 10.1126/science.aaf4382. [DOI] [PubMed] [Google Scholar]
- Skourti-Stathaki K., Kamieniarz-Gdula K., Proudfoot N.J. R-loops induce repressive chromatin marks over mammalian gene terminators. Nature. 2014;516:436–439. doi: 10.1038/nature13787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skourti-Stathaki K., Proudfoot N.J. A double-edged sword: R loops as threats to genome integrity and powerful regulators of gene expression. Genes Dev. 2014;28:1384–1396. doi: 10.1101/gad.242990.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sollier J., Stork C.T., García-Rubio M.L., Paulsen R.D., Aguilera A., Cimprich K.A. Transcription-coupled nucleotide excision repair factors promote R-loop-induced genome instability. Mol. Cell. 2014;56:777–785. doi: 10.1016/j.molcel.2014.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stolz R., Sulthana S., Hartono S.R., Malig M., Benham C.J., Chedin F. Interplay between DNA sequence and negative superhelicity drives R-loop structures. Proc. Natl. Acad. Sci. USA. 2019;116:6260–6269. doi: 10.1073/pnas.1819476116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K., Yamanaka S. A decade of transcription factor-mediated reprogramming to pluripotency. Nat. Rev. Mol. Cell Biol. 2016;17:183–193. doi: 10.1038/nrm.2016.8. [DOI] [PubMed] [Google Scholar]
- Tichy E.D., Stambrook P.J. DNA repair in murine embryonic stem cells and differentiated cells. Exp. Cell Res. 2008;314:1929–1936. doi: 10.1016/j.yexcr.2008.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vohhodina J., Goehring L.J., Liu B., Kong Q., Botchkarev V.V., Jr., Huynh M., Liu Z., Abderazzaq F.O., Clark A.P., Ficarro S.B., et al. BRCA1 binds TERRA RNA and suppresses R-Loop-based telomeric DNA damage. Nat. Commun. 2021;12:3542. doi: 10.1038/s41467-021-23716-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker C., Herranz-Martin S., Karyka E., Liao C., Lewis K., Elsayed W., Lukashchuk V., Chiang S.C., Ray S., Mulcahy P.J., et al. C9orf72 expansion disrupts ATM-mediated chromosomal break repair. Nat. Neurosci. 2017;20:1225–1235. doi: 10.1038/nn.4604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang I.X., Grunseich C., Fox J., Burdick J., Zhu Z., Ravazian N., Hafner M., Cheung V.G. Human proteins that interact with RNA/DNA hybrids. Genome Res. 2018;28:1405–1414. doi: 10.1101/gr.237362.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Li J., Zhou H., Zhang W., Gao J., Zheng P. A novel lncRNA Discn fine-tunes replication protein A (RPA) availability to promote genomic stability. Nat. Commun. 2021;12:5572. doi: 10.1038/s41467-021-25827-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood M., Quinet A., Lin Y.L., Davis A.A., Pasero P., Ayala Y.M., Vindigni A. TDP-43 dysfunction results in R-loop accumulation and DNA replication defects. J. Cell Sci. 2020;133:jcs244129. doi: 10.1242/jcs.244129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong J., Todorova D., Su N.Y., Kim J., Lee P.J., Shen Z., Briggs S.P., Xu Y. Stemness factor Sall4 is required for DNA damage response in embryonic stem cells. J. Cell Biol. 2015;208:513–520. doi: 10.1083/jcb.201408106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Q., Wulfridge P., Doherty J., Fernandez-Luna J.L., Real P.J., Tang H.Y., Sarma K. Proximity labeling identifies a repertoire of site-specific R-loop modulators. Nat. Commun. 2022;13:53. doi: 10.1038/s41467-021-27722-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasuhara T., Kato R., Hagiwara Y., Shiotani B., Yamauchi M., Nakada S., Shibata A., Miyagawa K. Human Rad52 promotes XPG-mediated R-loop processing to initiate transcription-associated homologous recombination repair. Cell. 2018;175:558–570.e11. doi: 10.1016/j.cell.2018.08.056. [DOI] [PubMed] [Google Scholar]
- Yin H.Z., Nalbandian A., Hsu C.I., Li S., Llewellyn K.J., Mozaffar T., Kimonis V.E., Weiss J.H. Slow development of ALS-like spinal cord pathology in mutant valosin-containing protein gene knock-in mice. Cell Death Dis. 2012;3:e374. doi: 10.1038/cddis.2012.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W., Chen Z., Zhang D., Zhao B., Liu L., Xie Z., Yao Y., Zheng P. KHDC3L mutation causes recurrent pregnancy loss by inducing genomic instability of human early embryonic cells. PLoS Biol. 2019;17:e3000468. doi: 10.1371/journal.pbio.3000468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao B., Zhang W., Cun Y., Li J., Liu Y., Gao J., Zhu H., Zhou H., Zhang R., Zheng P. Mouse embryonic stem cells have increased capacity for replication fork restart driven by the specific Filia-Floped protein complex. Cell Res. 2018;28:69–89. doi: 10.1038/cr.2017.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao B., Zhang W.D., Duan Y.L., Lu Y.Q., Cun Y.X., Li C.H., Guo K., Nie W.H., Li L., Zhang R., Zheng P. Filia is an ESC-specific regulator of DNA damage response and safeguards genomic stability. Cell Stem Cell. 2015;16:684–698. doi: 10.1016/j.stem.2015.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data needed to evaluate the conclusions in this paper are present in the paper and/or the supplemental information. The MS data for in vivo RNA pull-down have been deposited to the ProteomeXchange Consortium via the iProX repository with the dataset identifier PXD040075.