Abstract
Background and Methods:
The Melanoma Institute of Australia (MIA) and Memorial Sloan Kettering Cancer Center (MSKCC) nomograms were developed to help guide sentinel lymph node biopsy (SLNB) decisions. Although statistically validated, whether these prediction models provide clinical benefit at NCCN guideline-endorsed thresholds is unknown. We conducted a net benefit analysis to quantify the clinical utility of these nomograms at risk thresholds of 5-10% compared to the alternative strategy of biopsying all patients. External validation data for MIA and MSKCC nomograms were extracted from respective published studies.
Results:
The MIA nomogram provided added net benefit at a risk threshold of 9% but net harm at 5-8% and 10%. The MSKCC nomogram provided added net benefit at risk thresholds of 5% and 9-10% but net harm at 6-8%. When present, the magnitude of net benefit was small (1-3 net avoidable biopsies per 100 patients).
Conclusion:
Neither model consistently provided added net benefit compared to performing SLNB for all patients.
Discussion:
Based on published data, use of the MIA or MSKCC nomograms as decision making tools for SLNB at risk thresholds of 5-10% does not clearly provide clinical benefit to patients.
Keywords: Sentinel lymph node biopsy, melanoma, clinical utility
Introduction:
Information provided by sentinel lymph node biopsy (SLNB) in patients with cutaneous melanoma can provide prognostic information and guide future therapeutic decisions [1 2]. Prior studies have identified associations between SLNB positivity and clinicopathologic factors (Breslow thickness, age, ulceration), such that these predictors are incorporated into the National Comprehensive Cancer Network (NCCN) guidelines regarding indication for the procedure. [3–5]. SLNB should be considered in patients with a ≥5% individual risk of positivity (that is, T1b or T1a with adverse features) and offered to those with a >10% risk of positivity (that is, T2a and above) [5]. As these risk thresholds are relatively low, 80% or more of patients who undergo SLNB will not have nodal metastasis [6]. Although the surgical complications directly associated with SLNB are relatively minor, there may be significant economic and clinical implications associated with doing these procedures unnecessarily [7].
Prediction models using clinicopathological factors have been developed to further assist clinicians when deciding whether a patient should undergo SLNB, namely the Melanoma Institute of Australia (MIA) and Memorial Sloan Kettering Cancer Center (MSKCC) nomograms. These models were created using large melanoma databases and each has been externally validated in separate populations [8 9]. NCCN guidelines specifically state that “…the likelihood of a positive SLNB may be informed by the use of optimized multivariable nomograms/risk calculators…” [5]. These tools are available to clinicians and patients online and provide a percent likelihood of SLN metastasis. Ideally, these models would have high sensitivity (that is, only recommend SLNB in patients who truly have nodal metastasis) and high specificity (that is, discourage SLNB in patients who do not have nodal metastasis). In practice however, prediction models vary in terms of their sensitivity and specificity. A more sensitive and less specific model may lead to unnecessary SLNB. A less sensitive and more specific model may lead to missing nodal metastasis. This forces clinicians to consider the trade-offs between different endpoints for these models as well as other clinical strategies, like biopsy all or biopsy no patients.
Net benefit is a relatively novel measure that can help quantify which prediction model best balances potential harms and benefits [10]. This is done by converting the benefits and harms to the same scale by way of a risk threshold [11]. The net benefit of competing models at several clinically appropriate risk thresholds (5-10% risk of nodal metastasis) can then be compared to the net benefit of the two default strategies, SLNB for all patients or SLNB for no patients. The strategy (biopsy based on results of prediction model(s) vs. biopsy all patients vs. biopsy no patients) that has the highest net benefit across the range of clinically appropriate risk thresholds should be chosen. Use of net benefit analyses have been recommended in editorials published in leading medical journals [10 12–16]. Of note, the net benefit calculation can identify the most optimal strategy for selecting patients for SLNB, but it does not address controversies of whether the SLNB procedure itself is beneficial or the circumstances in which it should be recommended.
In this study we ascertained whether the net benefit of using the MIA or MSKCC nomograms was greater than the net benefit of treating all patients with SLNB at the guideline-endorsed risk thresholds. We extracted data from previous studies that externally validated each nomogram independently and conducted a net benefit analysis to determine if either nomogram provided additional clinical utility.
Methods:
Institutional Review Board review was not required because data from validation studies are publicly available.
MIA nomogram and validation data:
The MIA nomogram was developed in 2014 with deidentified data from the MIA database. Patients who had undergone SLNB between 2003 and 2014 and had complete clinicopathologic information were used to develop the nomogram (n= 3477). Only clinicopathologic variables that were found to be predictive of SLN metastasis, based on multivariable logistic regression, were used to develop the MIA nomogram. The predictive variables included patient age, tumor thickness, melanoma subtype, mitoses, ulceration, and lymphovascular invasion [9].
We extracted model classification measures from an MIA validation study conducted on a population-based Dutch pathology database. Dutch patients with invasive melanoma who had undergone SLNB and who met the same inclusion criteria as the MIA nomogram development study were included in this validation study. Patient-specific probabilities were calculated based on five of the six MIA nomogram covariates. Because the Dutch pathology database does not track exact mitotic rate count, and this is an optional parameter in the MIA nomogram, this variable was omitted in this validation study and predictions were made without it [17].
MSKCC nomogram and validation data:
The MSKCC nomogram was developed in 2005 using a prospective database of all melanoma patients undergoing SLNB biopsy at MSKCC between February 1991 and November 2003. Patients who had complete clinicopathologic information available were used to develop the nomogram (n=979). Only clinicopathologic variables that were found to be predictive of SLN metastasis, based on logistic regression, were used to develop the MSKCC nomogram. The predictive variables included patient age, Breslow thickness, Clark level, tumor location, and presence of ulceration [8].
We extracted model classification measures from two separate validation studies. The first study used an Italian melanoma database where patients were prospectively selected between 1993 and 2008. Patients were included in the validation study if SLNB was performed with the same selection criteria used at MSKCC, if all clinicopathological variables were available, and if the SLNB technique was similar to how SLNB is performed at MSKCC. Patient-specific probabilities were calculated based on the five MSKCC nomogram covariates. [18].
The second MSKCC validation study was performed on an Irish melanoma database where all newly diagnosed patients were selected between 2006 and 2012. Inclusion criteria were identical to the Italian validation study inclusion criteria. Patient-specific probabilities were calculated based on the five MSKCC nomogram covariates [19].
Net benefit calculation:
We calculated net benefit as the number of patients correctly predicted to have nodal metastasis (true positives) minus the adjusted number of patients incorrectly predicted to have nodal metastasis (false positives), at each risk threshold (Table 1). These values are converted to the same scale by way of the risk threshold (Equation 1, see below). The exchange rate is derived from the threshold probability that defines a positive result. For example, a 10% risk threshold implies that the harm of not biopsying one positive SLN is equivalent to the harm of unnecessarily performing SLNB on nine patients without SLN metastasis.
| Equation 1: |
Table 1:
Definitions of true positives, false positives, false negatives, and true negatives
| SLNB positive | SLNB negative | |
|---|---|---|
|
Nomogram predicted probability ≥ p |
Correctly performed SLNB on a patient with a positive node (True positive) |
Performed SLNB on a patient with a negative node (False positive) |
|
Nomogram predicted probability < p |
Failed to perform SLNB on a patient with a positive node (False negative) |
Correctly avoided SLNB on a patient with a negative node (True negative) |
p = probability (risk threshold) that defines a positive result
The net benefit of the MSKCC and MIA nomograms and a SLNB for all strategy (100% sensitivity; 0% specificity) were calculated at 5-10% risk thresholds [5]. The unit of net benefit is true positives.
True/False positivity is defined for a given risk threshold. For example, at the 5% risk threshold, a patient predicted to have 6% risk of a positive SLNB and who ultimately had a negative SLNB would be considered a false positive. If the threshold for performing the procedure were 10%, however, that same patient with a predicted 6% risk would not undergo SLNB and would be classified as a true negative (Table 1).
Results:
MIA nomogram:
A total of 3049 Dutch patients met inclusion criteria in the MIA validation study. The median Breslow thickness was 1.8mm. There were 370 (12.1%) T1, 1427 (46.8%) T2, 922 (30.2%) T3, and 330 (10.8%) T4 patients. The SLNB positivity rate was 22.7%. The negative predictive values (NPV) at 5% and 10% risk thresholds were 91.3% and 89.9%, respectively (Table 2).
Table 2:
Classification and utility measures of MIA nomogram at guideline-endorsed thresholds
| Risk Threshold | Sensitivity (%) | Specificity (%) | TP | FP | PPV (%) | NPV (%) | Overall SLNB reduction ratea (%) | Net Benefit of the modelb | Net Benefit SLNB for all | Net benefit compared to SLNB for allc | Net avoidable interventions per 100 patients compared to SLNB for all d |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 5% | 99.7 | 0.9 | 689 | 2337 | 22.8 | 91.3 | 0.8 | 0.1856 | 0.1859 | −0.0003 | None |
| 6% | 99.1 | 2.1 | 685 | 2308 | 22.9 | 89.3 | 1.8 | 0.1763 | 0.1773 | −0.0009 | None |
| 7% | 98.8 | 4.1 | 683 | 2261 | 23.2 | 92.4 | 3.4 | 0.1682 | 0.1684 | −0.0002 | None |
| 8% | 97.4 | 7.8 | 673 | 2174 | 23.6 | 91.1 | 6.6 | 0.1587 | 0.1594 | −0.0007 | None |
| 9% | 95.7 | 13.7 | 661 | 2035 | 24.5 | 91.5 | 11.6 | 0.1508 | 0.1501 | 0.0006 | 0.65 |
| 10% | 92.8 | 18.9 | 641 | 1912 | 25.1 | 89.9 | 16.3 | 0.1406 | 0.1407 | −0.0001 | None |
The MIA nomogram provided added net benefit at a risk threshold of 9% but net harm at 5-8% and 10% (Table 2 and Figure 1). For example, at a 5% risk threshold, the MIA model accurately predicted nodal metastasis in 689 patients (true positives) and incorrectly predicted nodal metastasis in 2337 patients (false positives). Using the model would have avoided SLNB in 23 patients (0.8%), and two patients with a positive node would have been missed. This results in a net benefit of 0.1856 at a 5% risk threshold. Alternatively, simply offering SLNB for all patients would have resulted in 691 true positives and 2358 false positives. By definition, no patients would have avoided SLNB, so no positive nodes would have been missed. SLNB for all at a 5% risk threshold results in a net benefit of 0.1859. At a 10% risk threshold, the MIA model predicted 641 true positives and 1912 false positives. Using the model would have avoided SLNB in 496 patients (16.3%), and 50 patients with a positive node would have been missed. This results in a net benefit of 0.1405 at a 10% risk threshold. SLNB for all patients would have resulted in 691 true positives and 2358 false positives, with a calculated net benefit of 0.1407 at a risk threshold of 10%.
Figure 1:
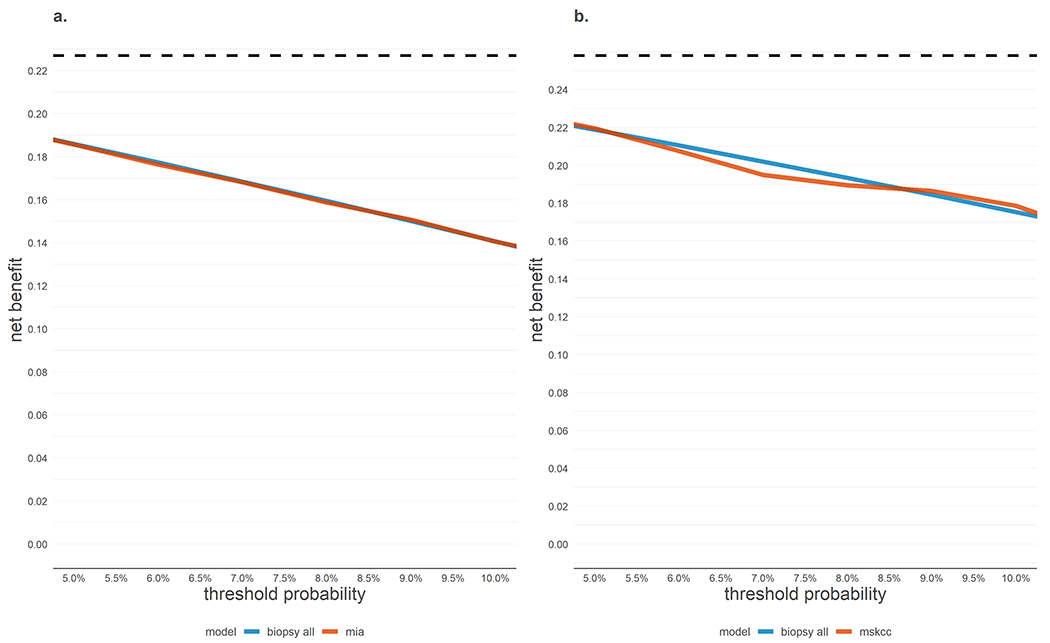
Net benefit decision curves for the MIA and MSKCC SNLB prediction models on respective external validation datasets.
Net benefit by threshold probability for (a) MIA model and (b) MSKCC model. Neither model showed a higher net benefit compared to performing SLNB for all patients across 5-10% risk thresholds. Net true positives are calculated by subtracting the harms of unnecessary SLNB (false positives) from the benefits of appropriate treatment (true positives) using the threshold to determine the exchange rate. The maximum achievable net benefit (perfect prediction, dashed lines) is the disease prevalence (0.227 in Figure 1a and 0.258 in Figure 1b).
Only at the risk threshold of 9% does the MIA nomogram have added net benefit compared to SLNB for all (+0.0006). To appreciate the magnitude of clinical benefit this provides, we can express net benefit in terms of its ability to reduce unnecessary interventions when the most common strategy is to biopsy all patients (that is, expressing net benefit as true negatives rather than true positives). Thus, at a risk threshold of 9%, use of the MIA model would be the equivalent of a strategy that reduced the number of unnecessary SLNB by about 1 per 154 patients without missing biopsy for any patients with nodal metastasis.
MSKCC nomogram:
A combined 667 patients met inclusion criteria in the MSKCC validation studies. This included 543 Italian patients (median age 56 years with 71 (13%) T1, 238 (43.8%) T2, 135 (24.8%) T3, 99 (18.2%) T4 patients) and 124 Irish patients (mean age 47 years and mean Breslow thickness 2.6mm). The combined SLNB positivity rate was 25.8%. The NPVs at 5% and 10% risk thresholds were 95.7% and 90.8%, respectively (Table 3).
Table 3:
Classification and utility measures of MSKCC nomogram at guideline-endorsed thresholds
| Risk Threshold | Sensitivity (%) | Specificity (%) | TP | FP | PPV (%) | NPV (%) | Overall SLNB reduction ratea (%) | Net Benefit of the modelb | Net Benefit SLNB for all | Net benefit compared to SLNB for allc | Net avoidable interventions per 100 patients compared to SLNB for all d |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 5% | 98.3 | 13.5 | 67 | 428 | 28.3 | 95.7 | 10.5 | 0.2196 | 0.2188 | 0.0008 | 1.50 |
| 6% | 95.3 | 19 | 94 | 401 | 29 | 92.2 | 15.3 | 0.2075 | 0.2105 | −0.003 | None |
| 7% | 91.9 | 25.1 | 124 | 371 | 29.9 | 89.9 | 20.7 | 0.195 | 0.202 | −0.007 | None |
| 8% | 90.7 | 31.3 | 155 | 340 | 31.5 | 90.6 | 25.6 | 0.1896 | 0.1933 | −0.0038 | None |
| 9% | 90.1 | 37.6 | 186 | 309 | 33.4 | 91.6 | 30.4 | 0.1866 | 0.1845 | 0.0021 | 2.12 |
| 10% | 87.8 | 42 | 208 | 287 | 34.5 | 90.8 | 34.3 | 0.1786 | 0.1754 | 0.0032 | 2.85 |
The overall SLN biopsy reduction rate is the proportion of total cases predicted to have a risk of SLN positivity lower than the risk threshold = (true negatives + false negatives) ÷ total population.
Maximum net benefit is equal to SLNB positivity rate in each validation cohorts, distinctly (22.7% for the Dutch cohort and 25.8% for the Italian-Irish cohort).
The units of net benefit are true positives. The clinical impact can be assessed using the reciprocal of the difference in net benefit (1÷difference in net benefit). For example, if the net benefit of model A is 0.01 greater than model B, use of model A will lead to 1 more net true positive treated compared to model B for every 100 patients.
The units of net avoidable interventions are true negatives. Expressing net benefit in terms of avoided unnecessary SLN biopsies may be preferred if the default reference strategy is biopsy all. , where exchange rate = (.
SLNB = sentinel lymph node biopsy.
The MSKCC nomogram provided added net benefit at risk thresholds of 5% and 9-10% but net harm at 6-8% (Table 3 and Figure 1). For example, at a 5% risk threshold, the MSKCC model predicted 169 true positives and 428 false positives. Using the model would have avoided SLNB in 70 patients (16.3%), and three patients with a positive node would have been missed. This results in a net benefit of 0.2196 at a 5% risk threshold. Alternatively, simply offering SLNB for all patients would have resulted in 172 true positives and 495 false positives. SLNB for all at a 5% risk threshold results in a net benefit of 0.2188. At a 10% risk threshold, the MSKCC model predicted 151 true positives and 287 false positives. Using the model would have avoided SLNB in 229 patients (34.3%), and 21 patients with a positive node would have been missed. This results in a net benefit of 0.1785 at a 10% risk threshold. The default SLNB for all patients with a risk threshold of 10% would have resulted in 172 true positives and 495 false positives, with a calculated net benefit of 0.1754.
Only at risk thresholds of 5% and 9-10% does the MSKCC nomogram have added net benefit compared to SLNB for all. To appreciate the magnitude of clinical benefit, we can express net benefit in terms of its ability to reduce unnecessary interventions when the most common strategy is to biopsy all patients (that is, expressing net benefit as true negatives rather than true positives). Use of the MSKCC model would be the equivalent of a strategy that reduced the number of unnecessary SLNB without missing biopsy for any patients with metastasis by 1 per 67 patients at 5%, 1 per 47 patients at 9%, and 1 per 35 patients at 10% risk thresholds.
Discussion:
Prediction models are traditionally assessed based on measures like their sensitivity, specificity, calibration, or discrimination [20]. While these measures provide insight on the statistical performance of a model, they fail to elucidate if the use of a model is of clinical value [10]. By weighting the benefits and harms on the same scale, we have determined that neither the MIA nor the MSKCC nomograms consistently provide added clinical benefit across risk thresholds of 5-10% compared to a SLNB for all strategy.
The calculation of clinical utility (net benefit) is dependent on the exchange rate between harm and benefit. Statistically, the exchange rate is derived from the risk threshold, but in practice can be thought about in relation to pathological staging. A 5% probability of a positive SLNB is the minimum probability at which the NCCN recommends considering SLNB. This inherently means that when performing SLNB on 20 patients with T1b melanoma, the NCCN deems it appropriate to biopsy 19 patients who do not have nodal metastasis to find 1 patient who does. At this risk threshold the exchange rate becomes (1/19), meaning the value of finding one true positive SLNB outweighs reducing unnecessary biopsies by a factor of 19 [21]. If the threshold for intervention were higher, as is often the case where there is higher risk of morbidity associated with an intervention, then greater value is placed on reducing unnecessary interventions. The 10% threshold for example, at which the procedure is recommended and not just considered, means that when performing SLNB on 10 patients the surgeon is willing to biopsy 9 patients who do not have nodal metastasis to find 1 patient who does.
To help illustrate the use of a net benefit analysis, we can use an insurance reimbursement plan as a practical analogy. Here net benefit is measured in dollars and a clinician is refunded for every true positive node identified but fined for every unnecessary biopsy. At a risk threshold of 5%, an insurance provider would agree to pay a clinician $100 for every correctly identified melanoma patient with nodal metastasis, and fine the clinician $5.26 ($100/19) for every unnecessary biopsy in a patient without nodal metastasis. In a cohort of 100 patients suspected to have nodal metastasis with prevalence of 22.7%, if a clinician were to use the MIA model to predict nodal metastasis, they would be reimbursed $1856, less than the $1859 if they had simply biopsied all patients.
The analogy is dependent on how much an individual weighs the benefits and the harms of SLNB. If SLNB proved to have significantly higher complication rates and proved to be a poor prognostic indicator, an insurance plan may place more value in reducing the number of SLNB performed rather than in finding every truly positive SLN. In general, higher risk thresholds value specificity more than sensitivity whereas lower risk thresholds value sensitivity more than specificity. It is possible that these models may provide net benefit at risk thresholds not currently endorsed by NCCN guidelines, such as 15-20%.
There are several limitations to this study. First, mitotic rate was omitted in the MIA validation study on Dutch patients since it is not adequately tracked in that database. This may hinder the MIA nomogram’s ability to accurately predict nodal metastasis and skew the net benefit analysis. Additionally, we did not directly compare the performance of the MIA and MSKCC nomograms. A net benefit analysis comparing these models can only be performed if the models were used on the same group of patients, which to our knowledge has not yet been reported.
We acknowledge that our findings are related to the very low risk threshold at which SLNB is currently recommended. However, these are the risk thresholds routinely endorsed to guide selection for SLNB. At these low-risk thresholds (5-10%), it is comparatively more difficult to create a model that provides net benefit over a treatment for all strategy. Further studies of relative clinical utility of additional variables that may be predictive of nodal metastasis, such as gene expression models, may help better define which model(s) should be used [22].
What do these findings mean? By using either nomogram as an additional selection step in an attempt to reduce unnecessary biopsies, clinicians would not be capitalizing on the benefits of finding every patient with nodal metastasis. This analysis indicates that use of either model as an additional selection step for patients with 5-10% risk of nodal metastasis may be unnecessary.
In summary, based on published data, use of the MIA or MSKCC nomograms as decision making tools for SLNB at risk thresholds of 5-10% does not clearly and consistently provide added clinical benefit to patients. This analysis indicates that use of either model is not currently justified.
Synopsis:
Using the MIA or MSKCC nomograms to guide sentinel lymph node biopsy decisions does not provide added net benefit compared to performing sentinel lymph node biopsy on all patients at risk thresholds of 5-10%.
Funding and Support:
This research was funded in part through the MSKCC institutional NIH/NCI Cancer Center Support Grant P30 CA008748. The funders had no role in the design and conduct of the study; collection, management, analysis, and interpretation of the data; preparation, review, or approval of the manuscript; and decision to submit the manuscript for publication.
Disclosures:
Hosein, Drebin, Kurtansky, Coit, and Marchetti report no conflicts of interest. Bartlett reports receiving institutional research support from Skyline Dx (Rotterdam, Netherlands and San Diego, United States) pertaining to the Merlin Assay. Olofsson Bagge has received institutional research grants from Bristol-Myers Squibb (BMS) and SkyLineDx, speaker honorarium from Roche and Pfizer and has served on advisory boards for Amgen, BD/BARD, Bristol-Myers Squibb (BMS), Merck Sharp & Dohme (MSD), Novartis, Roche and Sanofi Genzyme.
Footnotes
Prior Presentation: None
References:
- 1.Faries MB, Thompson JF, Cochran AJ, et al. Completion Dissection or Observation for Sentinel-Node Metastasis in Melanoma. N Engl J Med 2017;376(23):2211–22 doi: 10.1056/NEJMoa1613210 [published Online First: Epub Date]|. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Leiter U, Stadler R, Mauch C, et al. Complete lymph node dissection versus no dissection in patients with sentinel lymph node biopsy positive melanoma (DeCOG-SLT): a multicentre, randomised, phase 3 trial. Lancet Oncol 2016;17(6):757–67 doi: 10.1016/s1470-2045(16)00141-8 [published Online First: Epub Date]|. [DOI] [PubMed] [Google Scholar]
- 3.Breslow A Tumor thickness, level of invasion and node dissection in stage I cutaneous melanoma. Ann Surg 1975;182(5):572–5 doi: 10.1097/00000658-197511000-00007 [published Online First: Epub Date]|. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.In ‘t Hout FE, Haydu LE, Murali R, Bonenkamp JJ, Thompson JF, Scolyer RA. Prognostic importance of the extent of ulceration in patients with clinically localized cutaneous melanoma. Ann Surg 2012;255(6):1165–70 doi: 10.1097/SLA.0b013e31824c4b0b [published Online First: Epub Date]|. [DOI] [PubMed] [Google Scholar]
- 5.NCCN Clinical Practice Guidelines in Oncology - Cutaneous Melanoma. Secondary NCCN Clinical Practice Guidelines in Oncology - Cutaneous Melanoma 2022. https://www.nccn.org/professionals/physician_gls/pdf/cutaneous_melanoma.pdf.
- 6.Morton DL, Thompson JF, Cochran AJ, et al. Sentinel-node biopsy or nodal observation in melanoma. N Engl J Med 2006;355(13):1307–17 doi: 10.1056/NEJMoa060992 [published Online First: Epub Date]|. [DOI] [PubMed] [Google Scholar]
- 7.Wrightson WR, Wong SL, Edwards MJ, et al. Complications associated with sentinel lymph node biopsy for melanoma. Ann Surg Oncol 2003;10(6):676–80 doi: 10.1245/aso.2003.10.001 [published Online First: Epub Date]|. [DOI] [PubMed] [Google Scholar]
- 8.Wong SL, Kattan MW, McMasters KM, Coit DG. A nomogram that predicts the presence of sentinel node metastasis in melanoma with better discrimination than the American Joint Committee on Cancer staging system. Ann Surg Oncol 2005;12(4):282–8 doi: 10.1245/aso.2005.05.016 [published Online First: Epub Date]|. [DOI] [PubMed] [Google Scholar]
- 9.Lo SN, Ma J, Scolyer RA, et al. Improved Risk Prediction Calculator for Sentinel Node Positivity in Patients With Melanoma: The Melanoma Institute Australia Nomogram. Journal of Clinical Oncology 2020;38(24):2719–27 doi: 10.1200/jco.19.02362 [published Online First: Epub Date]|. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vickers AJ, Van Calster B, Steyerberg EW. Net benefit approaches to the evaluation of prediction models, molecular markers, and diagnostic tests. Bmj 2016;352:i6 doi: 10.1136/bmj.i6 [published Online First: Epub Date]|. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vickers AJ, van Calster B, Steyerberg EW. A simple, step-by-step guide to interpreting decision curve analysis. Diagnostic and Prognostic Research 2019;3(1):18 doi: 10.1186/s41512-019-0064-7 [published Online First: Epub Date]|. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kerr KF, Brown MD, Zhu K, Janes H. Assessing the Clinical Impact of Risk Prediction Models With Decision Curves: Guidance for Correct Interpretation and Appropriate Use. J Clin Oncol 2016;34(21):2534–40 doi: 10.1200/jco.2015.65.5654 [published Online First: Epub Date]|. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fitzgerald M, Saville BR, Lewis RJ. Decision curve analysis. Jama 2015;313(4):409–10 doi: 10.1001/jama.2015.37 [published Online First: Epub Date]|. [DOI] [PubMed] [Google Scholar]
- 14.Localio AR, Goodman S. Beyond the usual prediction accuracy metrics: reporting results for clinical decision making. Ann Intern Med 2012;157(4):294–5 doi: 10.7326/0003-4819-157-4-201208210-00014 [published Online First: Epub Date]|. [DOI] [PubMed] [Google Scholar]
- 15.McLernon DJ, Giardiello D, Van Calster B, et al. Assessing Performance and Clinical Usefulness in Prediction Models With Survival Outcomes: Practical Guidance for Cox Proportional Hazards Models. Ann Intern Med 2023;176(1):105–14 doi: 10.7326/m22-0844 [published Online First: Epub Date]|. [DOI] [PubMed] [Google Scholar]
- 16.de Hond AAH, Leeuwenberg AM, Hooft L, et al. Guidelines and quality criteria for artificial intelligence-based prediction models in healthcare: a scoping review. npj Digital Medicine 2022;5(1):2 doi: 10.1038/s41746-021-00549-7 [published Online First: Epub Date]|. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.El Sharouni MA, Varey AHR, Witkamp AJ, et al. Predicting sentinel node positivity in patients with melanoma: external validation of a risk-prediction calculator (the Melanoma Institute Australia nomogram) using a large European population-based patient cohort. Br J Dermatol 2021;185(2):412–18 doi: 10.1111/bjd.19895 [published Online First: Epub Date]|. [DOI] [PubMed] [Google Scholar]
- 18.Pasquali S, Mocellin S, Campana LG, et al. Maximizing the clinical usefulness of a nomogram to select patients candidate to sentinel node biopsy for cutaneous melanoma. Eur J Surg Oncol 2011;37(8):675–80 doi: 10.1016/j.ejso.2011.05.007 [published Online First: Epub Date]|. [DOI] [PubMed] [Google Scholar]
- 19.Woods JF, De Marchi JA, Lowery AJ, Hill AD. Validation of a nomogram predicting sentinel lymph node status in melanoma in an Irish population. Ir J Med Sci 2015;184(4):769–73 doi: 10.1007/s11845-014-1166-4 [published Online First: Epub Date]|. [DOI] [PubMed] [Google Scholar]
- 20.Ramspek CL, Jager KJ, Dekker FW, Zoccali C, van Diepen M. External validation of prognostic models: what, why, how, when and where? Clinical Kidney Journal 2020;14(1):49–58 doi: 10.1093/ckj/sfaa188 [published Online First: Epub Date]|. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Van Calster B, Wynants L, Verbeek JFM, et al. Reporting and Interpreting Decision Curve Analysis: A Guide for Investigators. Eur Urol 2018;74(6):796–804 doi: 10.1016/j.eururo.2018.08.038 [published Online First: Epub Date]|. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Grossman D, Okwundu N, Bartlett EK, et al. Prognostic Gene Expression Profiling in Cutaneous Melanoma: Identifying the Knowledge Gaps and Assessing the Clinical Benefit. JAMA dermatology 2020;156(9):1004–11 doi: 10.1001/jamadermatol.2020.1729 [published Online First: Epub Date]|. [DOI] [PMC free article] [PubMed] [Google Scholar]


