Abstract
Background
Systemic inflammatory scores may aid prognostication and patient selection for trials. We compared five scores in advanced pancreatic adenocarcinoma (PDAC).
Methods
Unresectable/metastatic PDAC patients enrolled in the Comprehensive Molecular Characterisation of Advanced Pancreatic Ductal Adenocarcinoma for Better Treatment Selection trial (NCT02750657) were included. Patients had pre-treatment biopsies for whole genome and RNA sequencing. CD8 immunohistochemistry was available in a subset. The neutrophil-to-lymphocyte ratio (NLR), platelet-to-lymphocyte ratio, Prognostic Nutritional Index, Gustave Roussy Immune Score (GRIm-S), and Memorial Sloan Kettering Prognostic Score (MPS) were calculated. Overall survival (OS) was estimated using Kaplan–Meier methods. Associations between inflammatory scores, clinical/genomic characteristics, and OS were analysed.
Results
We analysed 263 patients. High-risk NLR, GRIm-S and MPS were poorly prognostic. The GRIm-S had the highest predictive ability: median OS 6.4 vs. 10 months for high risk vs. low-risk (P < 0.001); HR 2.26 (P < 0.001). ECOG ≥ 1, the basal-like subtype, and low-HRDetect were additional poor prognostic factors (P < 0.01). Inflammatory scores did not associate with RNA-based classifiers or homologous recombination repair deficiency genotypes. High-risk MPS (P = 0.04) and GRIm-S (P = 0.02) patients had lower median CD8 + tumour-infiltrating lymphocytes.
Conclusions
Inflammatory scores incorporating NLR have prognostic value in advanced PDAC. Understanding immunophenotypes of poor-risk patients and using these scores in trials will advance the field.
Subject terms: Pancreatic cancer, Cancer genomics
Introduction
Despite the use of combination cytotoxic chemotherapy regimens, median overall survival (OS) for patients with advanced pancreatic adenocarcinoma (PDAC) is less than 1 year [1, 2]. Novel systemic and biological therapies are needed to improve outcomes for this patient population. While the option of clinical trial participation should be considered for all patients with PDAC, it can be difficult for clinicians to determine in a single consultation whether a patient will remain well for long enough to derive benefit from clinical trial participation. Strategies to improve patient selection for clinical trials are needed.
Several systemic inflammatory scores have been developed to aid in prognostication and patient selection for clinical trials in oncology [3–9]. These scores use a composite of variable laboratory values to categorise patients into different prognostic risk groups. There has been no study comprehensively comparing the performance of these prognostic scores in patients with advanced PDAC receiving contemporary systemic therapy.
The objectives of our study were to: (1) measure and compare the ability of each prognostic score to predict overall survival in patients with advanced PDAC and, (2) characterise associations between clinical and genomic characteristics and prognostic risk scores within a clinical trial setting.
Methods
Patient cohort
We used data from the Comprehensive Molecular Characterisation of Advanced Pancreatic Ductal Adenocarcinoma for Better Treatment Selection (COMPASS) trial (NCT02750657) for this study [10]. COMPASS was a multicenter prospective cohort study of patients with advanced (defined as locally advanced unresectable or metastatic) PDAC who had not yet started first-line palliative-intent chemotherapy. Patients had to have an ECOG performance status of 0–1 and be suitable to receive either modified FOLFIRINOX (mFOLFIRINOX; 5-fluorouracil, leucovorin, irinotecan, and oxaliplatin) or gemcitabine with nab-paclitaxel as per physicians choice. Following local institutional research ethics board approval and informed consent, patients were enrolled at five Canadian cancer centres between December 2015 and August 2020. Patient demographics, treatment details, and radiographic response to treatment according to the response evaluation criteria in solid tumours (RECIST) version 1.1 were prospectively collected using an electronic database [11]. For this present study, patients from COMPASS with baseline pre-treatment laboratory results (within 14 days of enrolment) sufficient to calculate each of the systemic inflammatory scores described below were included. Patients with missing baseline bloodwork such that the scores could not all be calculated were excluded from our study.
Genomic analyses
As part of the COMPASS trial, all patients underwent fresh tumour biopsy prior to treatment initiation for whole genome and RNA sequencing (Supplementary Appendix). The modified Moffitt classification based on gene expression signatures was used to categorise tumours into a classical versus basal-like subtype as previously described (Supplementary Fig. 1) [10, 12, 13]. The HRDetect score represents a weighted algorithmic score developed by Davies et al. incorporating whole genomic features of homologous recombination repair deficiency (HRD). This includes signature 3, the burden of indels with microhomology, characteristic rearrangement signatures and the genomic score used in the Myriad MyChoice HRD assay. This score was applied to each sample [14]. Patients with HRDetect >0.7 were classified as high, i.e., Homologous recombination repair deficient (HRD) in accordance with the previously published threshold.
Calculation of prognostic risk scores
Laboratory values collected within 14 days of enrolment were used to calculate the following prognostic scores for each patient: neutrophil-to-lymphocyte ratio (NLR; absolute neutrophil count divided by absolute lymphocyte count. NLR > 5 = high), platelet-to-lymphocyte ratio (PLR; platelet count divided by absolute lymphocyte count. PLR > 150 = high), prognostic nutritional index (PNI = serum albumin + 5 × lymphocyte count [109 cells/L]. PNI < 45 = high risk), Gustave Roussy Immune Score (GRIm-S; comprised of the NLR, serum albumin, and serum lactate dehydrogenase [LDH]. Values of NLR > 6 were assigned 1 point, albumin <35 = 1 point, LDH > upper limit of normal [ULN] = 1 point. The sum of points from these three components represents the GRIm-S, where a score of ≥2 = high risk), Memorial Sloan Kettering Prognostic Score (MPS; NLR > 4 and albumin <40 = high risk) (Supplementary Table 1) [3, 7–9].
CD8+tumour-infiltrating lymphocyte (TIL) staining using immunohistochemistry
Formalin-fixed, paraffin-embedded (FFPE) derived slides were available in 136 patients included. Immunohistochemical analysis of CD8 + tumour-infiltrating lymphocytes was performed using the CD8-4B11-L-CE antibody. Tumoural regions were reviewed and annotated by a pathologist and digital image analysis (QuPath) enumerated the number of positive cells per mm2. Samples were classified as CD8hi vs CD8lo using the median number/mm2.
Statistical analyses
OS was estimated using the Kaplan–Meier method and compared between risk groups for each prognostic score using the log-rank test. Cox proportional hazards models were used to analyse the association between each prognostic score and OS, adjusting for baseline clinical and genomic factors. Harrell’s c-index was used to compare the predictive discrimination for each prognostic score [15]. C-index values range between 0.5 (no predictive discrimination) to 1 (perfect predictive ability). Logistic regression models were used to analyse the association between clinical/genomic characteristics and prognostic scores. The differences in median CD8 + TIL according to score was calculated using the Mann–Whitney test. Analyses were performed using R version 4.1.2 and Prism version 9.4.1.
Results
Clinical cohort
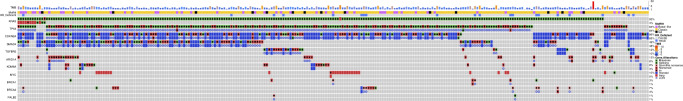
In total, 268 patients with advanced pancreatic ductal adenocarcinoma from the COMPASS study were screened, of whom 263 were included, with median follow-up of 32.9 months (95% CI 15.9–64.2). Baseline characteristics at enrolment are displayed in Table 1. The median age of patients was 64 years (range 19–84), 60% (N = 157) were male, 86% (N = 226) had metastatic disease, and 37% (N = 98) had an Eastern Cooperative Oncology Group (ECOG) clinical performance status of 0 versus 62% (N = 165) ECOG ≥ 1. The frequency of common driver mutations was as follows: KRAS mutation 93%, TP53 84%, CDKN2A 85%, SMAD4 48% (Fig. 1). The modified Moffitt tumour subtype was classical in 80% (N = 198), and the HRDetect score was high (>0.7) in 13% (N = 31) of patients. First-line chemotherapy was mFOLFIRINOX in 54% (N = 143) and gemcitabine and nab-paclitaxel in 37% (N = 98). The proportion of high-risk patients identified by each prognostic score was: NLR 32% (N = 85), PLR 63% (N = 167), PNI 22% (N = 58), GRIm-S 18% (N = 47), MPS 17% (N = 46) (Table 2). Of the patients who clinically deteriorated and were unable to have chemotherapy, half had high GRIm-S (N = 7/15; 47%).
Table 1.
Baseline clinical and genomic characteristics of advanced PDAC patients.
| Variable | All (N = 263) |
|---|---|
| Age, median (range) | 64 (19–84) |
| Sex | |
| Male | 157 (60%) |
| Female | 106 (40%) |
| ECOG | |
| 0 | 98 (37%) |
| ≥1 | 165 (63%) |
| Stage | |
| Locally advanced | 37 (14%) |
| Metastatic | 226 (86%) |
| Chemotherapy | |
| FOLFIRINOX | 143 (54%) |
| Gemcitabine and nab-paclitaxel | 98 (37%) |
| Other | 7 (3%) |
| None | 15 (6%) |
| Moffitt subtype | |
| Classical | 198 (80%) |
| Basal-like | 49 (20%) |
| HRDetect | |
| >0.7 | 31 (13%) |
Fig. 1. Oncoprint showing genomic features of cases enrolled on the COMPASS trial.
Rows at top display the tumour mutational burden (TMB), Moffitt classifer, and Homologous Recombination Repair (HR) status for each case. The common driver alterations are thereafter depicted. Each column represents a single case.
Table 2.
Categorisation of advanced PDAC patients into risk groups using systemic inflammatory scores.
| Score | Risk category | N = 263 (%) |
|---|---|---|
| Neutrophil-to-lymphocyte ratio (NLR) | Low | 178 (68%) |
| High | 85 (32%) | |
| Platelet-to-lymphocyte ratio (PLR) | Low | 96 (37%) |
| High | 167 (63%) | |
| Prognostic Nutritional Index (PNI) | Low | 205 (78%) |
| High | 58 (22%) | |
| Gustave Roussy Immune (GRIm-S) | Low | 216 (82%) |
| High | 47 (18%) | |
| Memorial Sloan Kettering Prognostic Score (MPS) | Not high | 217 (83%) |
| High | 46 (17%) |
Association of prognostic scores with OS
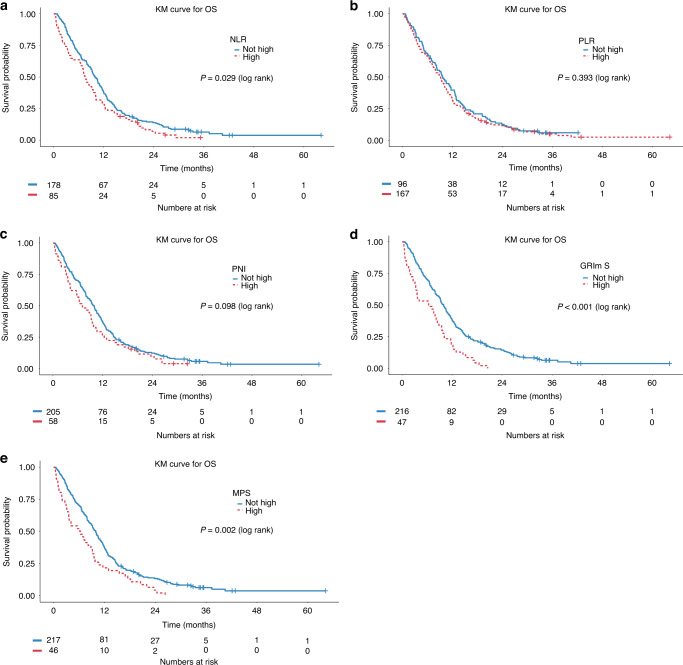
Median OS for in the intention-to-treat population was 9.3 months (95% CI 8–10.2). Kaplan–Meier estimates of OS stratified by prognostic risk category for each score are presented in Fig. 2. Patients with high-risk NLR, GRIm-S and MPS had shorter OS (Table 3). There was no difference in OS between high- and low-risk patients using PLR and PNI. A number of associations were found on univariable models (Supplementary Table 2). In multivariable analyses, the high-risk NLR, GRIm-S and MPS were associated with poor prognosis (Table 4). PLR and PNI were not prognostic. In all models, ECOG 0, the classical subtype and high HRDetect score were significantly associated with improved OS (Supplementary Tables 3–7). Predictive discrimination was similar for all models (c-index range 0.608–0.629; Table 4).
Fig. 2. Kaplan–Meier survival curves.
Kaplan–Meier survival curves for OS stratified by a Neutrophil-to-lymphocyte ratio (NLR), b Platelet-tolymphocyte ratio (PLR), c Prognostic Nutritional Index (PNI), d Gustave Roussy Immune Score (GRIm-S), and e Memorial Sloan Kettering Prognostic Score (MPS).
Table 3.
Median overall survival for risk groups according to prognostic scores.
| Prognostic score | Median overall survival (months) | Log-rank P value | |
|---|---|---|---|
| High risk | Not high risk | ||
| NLR | 7.6 | 10.0 | 0.029 |
| PLR | 9.2 | 9.6 | 0.393 |
| PNI | 7.0 | 10.0 | 0.098 |
| GRIm-S | 6.4 | 10.0 | <0.001 |
| MPS | 6.3 | 10.0 | 0.002 |
Bold values denote statistically significant P values.
Table 4.
Predictive accuracy of prognostic scores.
| Score | Adjusted hazard ratioa (95% CI; P value) | Predictive discrimination for OS (c-index) |
|---|---|---|
| NLR | 1.70 (1.27, 2.28; P < 0.001) | 0.627 (0.585, 0.668) |
| PLR | 1.17 (0.87, 1.56; P = 0.3) | 0.610 (0.569, 0.652) |
| PNI | 1.27 (0.92, 1.77; P = 0.15) | 0.608 (0.567, 0.649) |
| GRIm-S | 2.26 (1.57, 3.25; P < 0.001) | 0.629 (0.588, 0.671) |
| MPS | 1.51 (1.07, 2.15; P = 0.02) | 0.618 (0.577, 0.658) |
aMultivariable model for each prognostic score adjusted for age, sex, ECOG, Moffitt subtype and HRDetect score.
Bold values denote statistically significant P values.
Association of clinical/genomic and immune features with prognostic scores
Univariable analyses found a number of associations between with high GRIm-S (Supplementary Table 8); however, only male sex remained significantly associated with high GRIm-S on multivariable analysis (HR 5.52, 95% CI 1.94–15.71, P = 0.0014) (Table 5). There was no significant association between GRIm-S and age, ECOG status, baseline tumour burden (RECIST sum of diameters), or RNA or DNA-based classifiers on multivariable analysis (Table 5). There was no association between the MPS and male sex (HR 1.55, 95% CI 0.68–3.55, P = 0.3) or any other clinical or genomic features (Supplementary Tables 9 and 10). The median number of CD8 + TIL in the cohort was 136/mm2. There were no differences in median CD8 stratifying by the NLR, PNI and PLR scores. However, we saw higher median CD8 + TILs in patients with low MPS scores (N = 13, 152/mm2) vs high MPS (N = 118, 63.2/mm2), P = 0.04. In the high GRIm-S cohort (n = 16), the median CD8 was 59/mm2 compared to 152/ mm2 in low GRIm-S (n = 115), P = 0.02.
Table 5.
Multivariable analysis for association between clinicopathologic factors and high GRIm-S.
| Covariate | OR (95% CI) | P value | Global P value |
|---|---|---|---|
| Age | 0.99 (0.95–1.04) | 0.75 | |
| Gender | 0.0014 | ||
| Female | Reference | ||
| Male | 5.52 (1.94, 15.71) | ||
| ECOG | 0.28 | ||
| 0 | Reference | ||
| 1–2 | 1.62 (0.67–3.92) | ||
| Moffitt subtype | 0.8 | ||
| Basal-like | Reference | ||
| Classical | 1.14 (0.40–3.26) | ||
| HRDetect | 0.23 | ||
| Low | Reference | ||
| High | 1.91 (0.66–5.53) | ||
| Response | 0.17 | ||
| PR/CR | Reference | ||
| PD | 0.69 (0.21, 2.29) | 0.54 | |
| SD | 0.48 (0.17, 1.34) | 0.16 | |
| NE | 1.79 (0.55, 5.80) | 0.33 | |
| Baseline tumour burden | 1.01 (1.00, 1.01) | 0.12 |
Bold values denote statistically significant P values.
Discussion
In this large prospective cohort study of patients with advanced PDAC receiving first-line systemic therapy, we found that systemic inflammatory scores incorporating NLR were highly prognostic. These are tools that could be easily used in the clinical setting, and may help in patient selection for clinical trials.
Despite extensive efforts to improve clinical outcomes for patients with advanced PDAC, treatment resistance remains a challenge and novel strategies in systemic and other therapies are urgently needed. Although studies to date of single or dual immune checkpoint inhibition have not been successful in PDAC, ongoing trials of combination strategies targeting immune checkpoints and other components of the tumour microenvironment are being conducted [16–18]. While clinical trial participation should be considered for all patients, the aggressive behaviour of PDAC can make it difficult to predict which patients will survive long enough to derive benefit from a clinical trial, especially after failure of first-line therapy. It would be helpful to clinicians to have a readily available prognostic tool to complement clinical judgement.
Since chronic inflammation is known to contribute to cancer development and progression, several clinical scoring systems have incorporated laboratory markers associated with inflammatory response as variables in prognostic risk calculation [19]. The Glasgow Prognostic Score (GPS) and the subsequent modified GPS (mGPS) use serum albumin and C-reactive protein (CRP) to risk-stratify patients [20]. Imaoka et al. showed that the mGPS was prognostic in patients with PDAC [21]. However, CRP is not usually measured in routine clinical practice, including within our COMPASS trial, which may decrease the practicality of this tool.
The NLR is another measure of inflammation that has been associated with survival in solid tumours, and can be calculated from a complete blood count which is measured in all patients undergoing systemic therapy [7, 22]. Other scores have built upon the NLR by taking into consideration other clinical or biochemical variables, such as the GRIm-S, which was developed as a tool to aid in patient selection for Phase I immune checkpoint therapy trials [4]. In the initial discovery and validation cohorts, patients with a high GRIm-Score had significantly shorter overall survival. However, these patients predominantly had solid tumours where immune checkpoint inhibitors are more commonly used, and PDAC patients were underrepresented. The applicability of this score in PDAC was unknown prior to our study. More recently, Lebenthal et al. found that the Memorial Sloan Kettering Prognostic Score (MPS), a composite of albumin and NLR, was prognostic in PDAC [9]. Notably, the NLR cut-off used, whether alone or as part of a composite score, has varied.
Despite the existence of these different systemic inflammatory prognostic scores, none are currently used widely. To our knowledge, no previous study comprehensively compared the performance of these scores in advanced PDAC patients. Of the prognostic scores we studied, the ones that included NLR were all prognostic, which may reflect the inflammatory drive promoting progression in PDAC. The GRIm-S was superior to NLR alone, as it incorporates other measures of aggressive biology such as LDH which may predict for higher disease burden, and albumin which may reflect worse nutritional or clinical performance status.
In our study, genomic classifiers such as the classical Moffitt tumour RNA subtype and a high HRDetect DNA score were significant predictors of more favourable OS, consistent with previous work [13, 23]. While we anticipate that molecular prognostic risk stratification will become increasingly relevant in clinical practice, these tests may not always be accessible in a timely manner outside of an academic cancer centre. The systemic inflammatory scores we studied are comprised of laboratory values routinely measured in patients undergoing systemic therapy, and could be used in settings where genomic analyses are not readily available. These scores did not associate with RNA-based classifiers or HRD scores and can therefore provide additional prognostic information and highlight heterogeneity within the subtypes.
The objectivity and simplicity of these tools makes them an attractive option for use in the clinical setting. In our study, 6% of patients deteriorated and died without receiving chemotherapy despite having an ECOG performance status of 0 or 1 at enrolment. Of these patients, half had a high GRIm-S. With ongoing trials exploring different immune strategies in PDAC, this score can be helpful in guiding patient selection for clinical trials, and should be considered for integration into clinical trial design. Recently, one prospective clinical trial for patients with metastatic PDAC has made use of the GRIm-S as part of its eligibility criteria, highlighting the direct clinical applicability of this score (NCT04999969). In addition, we hypothesise that patients with a high GRIm-S or MPS have immune-excluded tumours and therefore may represent those unlikely to derive benefit from immune checkpoint inhibitors. These scores may also help clinicians frame discussions about prognosis and therapeutic decision-making with patients and family members.
We acknowledge the limitations of our study. This was a retrospective analysis of the COMPASS trial data, so our findings require prospective validation. The patients included in our study all had performance status of ECOG 0 or 1, so the generalisability to patients with worse clinical status is unknown. Notwithstanding these limitations, our study demonstrates that systemic inflammatory scores such as the GRIm-S can be used as simple prognostic tools to aid treatment decisions and patient selection for clinical trials.
Conclusion
Systemic inflammatory scores incorporating NLR have prognostic value in previously untreated ECOG 0–1 patients with advanced PDAC and can easily be used in the clinical setting. The GRIm-S had the highest predictive ability for OS and those with high scores appear particularly immune-excluded. Future clinical trials should consider these scores for use as a prognostic factor in patient selection.
Supplementary information
Acknowledgements
This study was conducted with the support of the Ontario Institute for Cancer Research (PanCuRx Translational Research Initiative) through funding provided by the Government of Ontario, the Wallace McCain Centre for Pancreatic Cancer supported by the Princess Margaret Cancer Foundation, Pancreatic Cancer Canada, the Terry Fox Research Institute, and the Canadian Cancer Society Research Institute.
Author contributions
LXM: conceptualisation, data curation, investigation and writing (original draft, review and editing). YW: data curation, investigation and writing (original draft, review and editing). OE-G: formal analysis and writing (review and editing). MJA: data curation, investigation and writing (original draft, review and editing). GHJ: data curation, investigation and writing (original draft, review and editing). AZ: data curation, investigation and writing (original draft, review and editing). AD: data curation, investigation and writing (original draft, review and editing). SR: data curation, investigation and writing (original draft, review and editing). SH: data curation, investigation and writing (original draft, review and editing). MT: investigation and writing (original draft, review and editing). RR: investigation and writing (original draft, review and editing). JB: investigation and writing (original draft, review and editing). JW: data curation, investigation and writing (original draft, review and editing). FN: data curation, investigation and writing (original draft, review and editing). SEF: data curation, investigation and writing (original draft, review and editing). GZ: investigation and writing (original draft, review and editing). SG: data curation, investigation, resources and funding acquisition and writing (original draft, review and editing). RCG: data curation, investigation and writing (original draft, review and editing). RK: data curation, investigation and writing (review and editing). NC: data curation, investigation and writing (review and editing). BTG: data curation, investigation and writing (review and editing). JJK: data curation, investigation, resources and funding acquisition and writing (original draft, review and editing). GMO’K: conceptualisation, data curation, investigation, resources and funding acquisition and writing (original draft, review and editing). The final manuscript was approved by all authors.
Funding
This study was funded by the Ontario Institute for Cancer Research (PanCuRx Translational Research Initiative), the Princess Margaret Cancer Foundation, Pancreatic Cancer Canada, a Canadian Cancer Society Research Institute grant (702316), and charitable donations from the Canadian Friends of the Hebrew University (Alex U. Soyka) and the Lebovic Chair in Hepatobiliary/Pancreatic Surgical Oncology.
Data availability
All RNA sequencing data have been deposited in the European Genome-Phenome Archive (EGA) at EGAD00001009409.
Competing interests
The authors declare no competing interests.
Ethics approval and consent to participate
This study was performed in accordance with the Declaration of Helsinki. The study was approved by the University Health Network Research Ethics Board.
Consent for publication
Not applicable.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version contains supplementary material available at 10.1038/s41416-023-02214-0.
References
- 1.Conroy T, Desseigne F, Ychou M, Bouche O, Guimbaud R, Becouarn Y, et al. FOLFIRINOX versus gemcitabine for metastatic pancreatic cancer. N Engl J Med. 2011;364:1817–25. doi: 10.1056/NEJMoa1011923. [DOI] [PubMed] [Google Scholar]
- 2.Von Hoff DD, Ervin T, Arena FP, Chiorean EG, Infante J, Moore M, et al. Increased survival in pancreatic cancer with nab-paclitaxel plus gemcitabine. N Engl J Med. 2013;369:1691–703. doi: 10.1056/NEJMoa1304369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Buzby GP, Mullen JL, Matthews DC, Hobbs CL, Rosato EF. Prognostic nutritional index in gastrointestinal surgery. Am J Surg. 1980;139:160–7. doi: 10.1016/0002-9610(80)90246-9. [DOI] [PubMed] [Google Scholar]
- 4.Onodera T, Goseki N, Kosaki G. Prognostic nutritional index in gastrointestinal surgery of malnourished cancer patients. Nihon Geka Gakkai Zasshi. 1984;85:1001–5. [PubMed] [Google Scholar]
- 5.Forrest LM, McMillan DC, McArdle CS, Angerson WJ, Dunlop DJ. Evaluation of cumulative prognostic scores based on the systemic inflammatory response in patients with inoperable non-small-cell lung cancer. Br J Cancer. 2003;89:1028–30. doi: 10.1038/sj.bjc.6601242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Templeton AJ, Ace O, McNamara MG, Al-Mubarak M, Vera-Badillo FE, Hermanns T, et al. Prognostic role of platelet to lymphocyte ratio in solid tumors: a systematic review and meta-analysis. Cancer Epidemiol Biomark Prev. 2014;23:1204–12. doi: 10.1158/1055-9965.EPI-14-0146. [DOI] [PubMed] [Google Scholar]
- 7.Templeton AJ, McNamara MG, Seruga B, Vera-Badillo FE, Aneja P, Ocana A, et al. Prognostic role of neutrophil-to-lymphocyte ratio in solid tumors: a systematic review and meta-analysis. J Natl Cancer Inst. 2014;106:dju124. doi: 10.1093/jnci/dju124. [DOI] [PubMed] [Google Scholar]
- 8.Bigot F, Castanon E, Baldini C, Hollebecque A, Carmona A, Postel-Vinay S, et al. Prospective validation of a prognostic score for patients in immunotherapy phase I trials: the Gustave Roussy Immune Score (GRIm-Score) Eur J Cancer. 2017;84:212–8. doi: 10.1016/j.ejca.2017.07.027. [DOI] [PubMed] [Google Scholar]
- 9.Lebenthal JM, Zheng J, Glare PA, O’Reilly EM, Yang AC, Epstein AS. Prognostic value of the Memorial Sloan Kettering Prognostic Score in metastatic pancreatic adenocarcinoma. Cancer. 2021;127:1568–75. doi: 10.1002/cncr.33420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Aung KL, Fischer SE, Denroche RE, Jang GH, Dodd A, Creighton S, et al. Genomics-driven precision medicine for advanced pancreatic cancer: early results from the COMPASS trial. Clin Cancer Res. 2018;24:1344–54. doi: 10.1158/1078-0432.CCR-17-2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Eisenhauer EA, Therasse P, Bogaerts J, Schwartz LH, Sargent D, Ford R, et al. New response evaluation criteria in solid tumours: revised RECIST guideline (version 1.1) Eur J Cancer. 2009;45:228–47. doi: 10.1016/j.ejca.2008.10.026. [DOI] [PubMed] [Google Scholar]
- 12.Moffitt RA, Marayati R, Flate EL, Volmar KE, Loeza SG, Hoadley KA, et al. Virtual microdissection identifies distinct tumor- and stroma-specific subtypes of pancreatic ductal adenocarcinoma. Nat Genet. 2015;47:1168–78. doi: 10.1038/ng.3398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.O’Kane GM, Grunwald BT, Jang GH, Masoomian M, Picardo S, Grant RC, et al. GATA6 expression distinguishes classical and basal-like subtypes in advanced pancreatic cancer. Clin Cancer Res. 2020;26:4901–10. doi: 10.1158/1078-0432.CCR-19-3724. [DOI] [PubMed] [Google Scholar]
- 14.Davies H, Glodzik D, Morganella S, Yates LR, Staaf J, Zou X, et al. HRDetect is a predictor of BRCA1 and BRCA2 deficiency based on mutational signatures. Nat Med. 2017;23:517–25. doi: 10.1038/nm.4292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Harrell FE, Jr, Lee KL, Mark DB. Multivariable prognostic models: issues in developing models, evaluating assumptions and adequacy, and measuring and reducing errors. Stat Med. 1996;15:361–87. doi: 10.1002/(SICI)1097-0258(19960229)15:4<361::AID-SIM168>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 16.Royal RE, Levy C, Turner K, Mathur A, Hughes M, Kammula US, et al. Phase 2 trial of single agent Ipilimumab (anti-CTLA-4) for locally advanced or metastatic pancreatic adenocarcinoma. J Immunother. 2010;33:828–33. doi: 10.1097/CJI.0b013e3181eec14c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.O’Reilly EM, Oh DY, Dhani N, Renouf DJ, Lee MA, Sun W, et al. Durvalumab with or without tremelimumab for patients with metastatic pancreatic ductal adenocarcinoma: a phase 2 randomized clinical trial. JAMA Oncol. 2019;5:1431–8. doi: 10.1001/jamaoncol.2019.1588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kamath SD, Kalyan A, Kircher S, Nimeiri H, Fought AJ, Benson A, 3rd, et al. Ipilimumab and gemcitabine for advanced pancreatic cancer: a phase Ib study. Oncologist. 2020;25:e808–e15. doi: 10.1634/theoncologist.2019-0473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Grivennikov SI, Greten FR, Karin M. Immunity, inflammation, and cancer. Cell. 2010;140:883–99. doi: 10.1016/j.cell.2010.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McMillan DC. An inflammation-based prognostic score and its role in the nutrition-based management of patients with cancer. Proc Nutr Soc. 2008;67:257–62. doi: 10.1017/S0029665108007131. [DOI] [PubMed] [Google Scholar]
- 21.Imaoka H, Mizuno N, Hara K, Hijioka S, Tajika M, Tanaka T, et al. Evaluation of modified Glasgow prognostic score for pancreatic cancer: a retrospective cohort study. Pancreas. 2016;45:211–7. doi: 10.1097/MPA.0000000000000446. [DOI] [PubMed] [Google Scholar]
- 22.Yang JJ, Hu ZG, Shi WX, Deng T, He SQ, Yuan SG. Prognostic significance of neutrophil to lymphocyte ratio in pancreatic cancer: a meta-analysis. World J Gastroenterol. 2015;21:2807–15. doi: 10.3748/wjg.v21.i9.2807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.O’Kane GM, Denroche R, Picardo SL, Zhang A, Holter S, Grant RC, et al. Homologous recombination deficiency (HRD) scoring in pancreatic ductal adenocarcinoma (PDAC) and response to chemotherapy. J Clin Oncol. 2020;38:741. doi: 10.1200/JCO.2020.38.4_suppl.741. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All RNA sequencing data have been deposited in the European Genome-Phenome Archive (EGA) at EGAD00001009409.