Abstract
Genetic and phylogenetic characterization of Cryptosporidium isolates at two loci (18S rRNA gene and heat shock gene) from both Australian and United States dogs demonstrated that dog-derived Cryptosporidium isolates had a distinct genotype which is conserved across geographic areas. Phylogenetic analysis provided support for the idea that the “dog” genotype is, in fact, a valid species.
Protozoan parasites of the genus Cryptosporidium are found worldwide in over 170 different host species (19). Cryptosporidium parvum causes the majority of mammalian infections. In the immunocompetent host, infection is self-limiting, lasting from a few days to 3 weeks, with possible morbidity in young animals (6). In the immunocompromised host, infection may result in chronic debilitating diarrhea with dehydration, malabsorption, wasting, and death (6).
There have been few reports of Cryptosporidium infection in dogs, with the majority of cases involving pups less than 6 months of age. The first evidence of cryptosporidiosis in dogs was reported in 1981 by Tzipori and Campbell, who detected Cryptosporidium antibodies in 16 of 20 canine serum samples (27). Wilson et al. reported the first clinical case of canine cryptosporidiosis 2 years later, when life cycle stages characteristic of Cryptosporidium were identified in a 1-week-old pup suffering from acute diarrhea (28).
Simpson et al. examined 101 fecal samples collected from dogs at boarding kennels in Scotland and found that all were negative for Cryptosporidium (22). Similarly, Bugg et al. screened 421 fecal samples from dogs in Western Australia, using both microscopy and PCR, none of which were positive for Cryptosporidium oocysts (1). In contrast, 2% of stray dogs sampled in California (4), 1% of specimens from seven public parks in Scotland (9), and 1.8% of samples from urban dogs and 9.2% of samples collected from public parks in Hobart, Tasmania, were positive for Cryptosporidium oocysts (13). Johnston and Gasser screened fecal samples from dogs in the Geelong and Melbourne in Australia and reported Cryptosporidium prevalences ranging from 0.7 to 19.6% (10).
There are up to 10 valid named Cryptosporidium species. These include C. parvum from humans and many mammals, C. muris from rodents, C. andersoni from ruminants, C. felis from cats, C. wrairi from guinea pigs, C. meleagridis and C. baileyi from birds, C. serpentis and C. saurophilim from reptiles, and C. nasorum from fish (6, 11, 29). There is strong evidence that the most widely studied species, C. parvum, is not a uniform species but consists of numerous distinct genotypes (14, 15, 16, 17, 29).
Cryptosporidium oocysts excreted by dogs are morphologically similar to those of C. parvum and have therefore been assumed to be zoonotic. However, recent genetic evidence suggests that dogs harbor a Cryptosporidium type genetically distinct from that detected in immunologically competent humans (29). The aim of this study was to compare dog-derived Cryptosporidium isolates from different geographic locations at two genetic loci (18S rRNA gene [rDNA] and heat shock gene Hsp-70) and to conduct a full phylogenetic analysis of dog-derived isolates at the 18S rDNA locus, as well as a wide range of other Cryptosporidium isolates or species at the same locus, in order to more clearly understand the species status of the “dog” genotype.
The sources of the dog-derived Cryptosporidium isolates studied are listed in Table 1. Cryptosporidium oocysts were initially identified using a malachite green negative strain (5) and then concentrated from intestinal scrapings and purified using phosphate-buffered saline–ether sedimentation and Ficoll gradient centrifugation (12). Dog-derived oocysts from the United States were identified by screening of fecal specimens with an immunofluorescence kit (Merifluor, Cincinnati, Ohio). Oocysts were purified by sucrose-percoll centrifugation.
TABLE 1.
Dog-derived Cryptosporidium isolates used in this study
| Isolate code | Host age | Host sex, breed | Geographic origin | Sourcea |
|---|---|---|---|---|
| AusDog1 | 9 wk | Male, bullmastiff | Queensland, Australia | MU |
| AusDog2 | 11 yr | Female, Akita | Western Australia | MU |
| AusDog3 | 22 wk | Male, schnauzer | Western Australia | MU |
| USADog1 | 4 mo | Female, NDb | Georgia | CDC |
| USADog2 | 4 mo | Female, ND | Georgia | CDC |
| USADog3 | 4 mo | Female, ND | Georgia | CDC |
| USADog4 | 2 yr | Female, ND | Georgia | CDC |
| USADog5 | >1 yr | Female, mixed | United States | USDA |
MU, Murdoch University, Western Australia; CDC, Centers for Disease Control and Prevention, Atlanta, Ga.; USDA, U.S. Department of Agriculture, Beltsville, Md.
ND, not determined.
DNA was extracted from purified oocysts as previously described (14), and a 713-bp region of the 18S rDNA was amplified and sequenced using the primers 18S-SF (forward primer; 5′ AGTCATAGTCTTGTCTCAAAGATT 3′) and 18SiR (reverse primer; 5′ CCTGCTTTAAGCACTCTAATTTTC 3′) as previously described (18). This region encompasses the 5′ end of the 18S rDNA and includes a variable region which has previously been shown to be capable of discriminating among all Cryptosporidium species, as well as multiple genotypes within C. parvum (14, 15, 17, 29). A 311-bp region of the heat shock gene Hsp-70 was also amplified using the forward primer HSPF1 (5′-ATG TCT GAA GGT CCA GCT ATT GGT ATT GA-3′) and the reverse primer HSPR3 (5′ GAT TGG CTT RTC CTT YGG RCC 3′). DNA was amplified using 2 mM MgCl2 and 45 rounds of amplification at an annealing temperature of 52°C.
Nucleotide sequences were aligned using Clustal X (25). Phylogenetic trees were constructed by three methods. Distance-based neighbor-joining (NJ) analysis and parsimony analysis were performed using PAUP, version 4.0b2 (Sinauer Associates, Sunderland, Mass.), and maximum-likelihood analyses were performed using PUZZLE, version 4.1 (24). NJ analyses were conducted using Tamura-Nei distance estimates. Parsimony analyses were conducted using either the branch and bound or heuristic search option of PAUP*. Bootstrap analyses were conducted using 1,000 replicates. Phylograms were drawn using the TreeView program (20).
Sequence analysis of a short region of the Hsp-70 gene revealed that dog-derived Cryptosporidium isolates were identical to each other but genetically distinct from other genotypes of C. parvum. Representative sequences are listed in Fig. 1.
FIG. 1.
Alignments of Hsp-70 sequence data from various Cryptosporidium isolates. Genotypes: KSU1 and C1, cattle; H1, human; K1, marsupial; AusDog1 and USADog2, dog. Asterisks indicate identical bases.
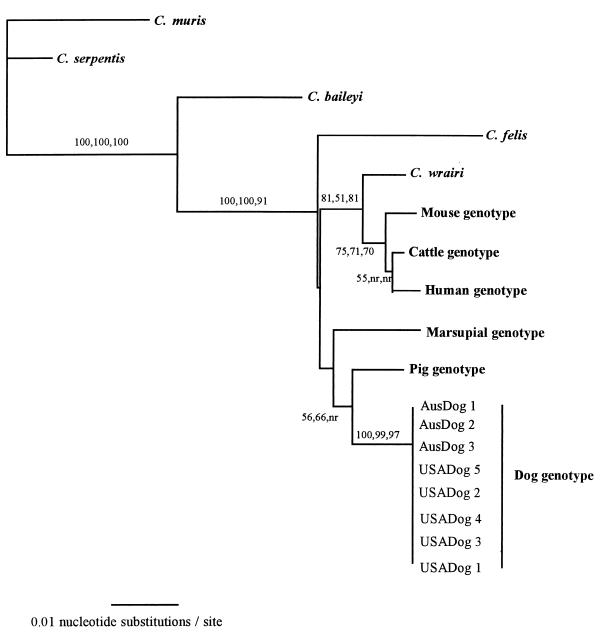
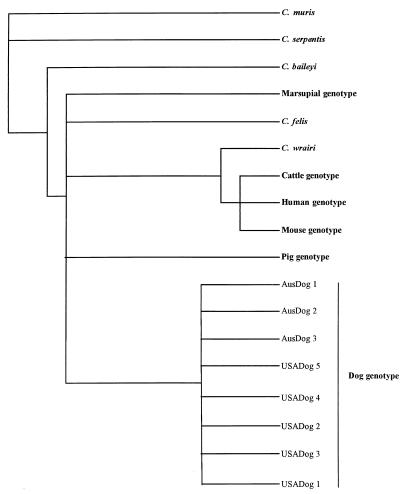
Similarly, all dog-derived Cryptosporidium isolates from the United States and Australia also possessed identical 18S rDNA sequences. The DNA sequences from these isolates were 98.3 to 97% similar to the other C. parvum sequences and 95.9 to 91.9% similar to those of other Cryptosporidium species. Phylogenetic analysis of the sequence data (Fig. 2, NJ tree illustrated) placed all of the C. parvum isolates, as well as C. felis and C. wrairi, into the same cluster. This grouping was strongly supported by distance-based NJ, parsimony, and maximum-likelihood analyses. As previously reported (17, 29), C. wrairi grouped with the mouse, human, and cattle genotypes. The isolates of C. parvum from dogs formed a broad cluster with the pig and marsupial genotypes. However, this grouping was weakly supported by the distance-based NJ and parsimony methods and not recovered by maximum-likelihood analysis. A consensus tree (Fig. 3) is presented to illustrate the groupings that are well supported by the analyses.
FIG. 2.
Evolutionary relationships of Cryptosporidium isolates inferred by NJ analysis of Tamura-Nei distances calculated from pairwise comparisons of 18S rDNA sequences. Percentages of bootstrap support (>50%) from 1,000 replicate samples (analyzed by the NJ, parsimony, and maximum-likelihood methods, respectively) are indicated at the left of the supported node. nr, node not recovered by method. Additional Cryptosporidium 18S rDNA sequences obtained from GenBank were bovine C. parvum isolate C1 (AF108864), human C. parvum isolate H7 (AF108865), mouse genotype C. parvum isolate M24 (AF108863), a pig genotype isolate (AF108861), a marsupial genotype isolate (AF108860), C. wrairi (AF115378), C. felis (AF108862), C. baileyi (AF093495), and C. serpentis (AF108866).
FIG. 3.
Parsimony consensus tree.
In this study, Cryptosporidium was detected in two immunologically stressed Australian domestic dogs. No clinical data were available for the third Australian dog. The first Australian dog-derived isolate (AusDog1), was from a 9-week-old male bullmastiff admitted to the Murdoch University Veterinary Hospital with vomiting, lethargy, and diarrhea. Serum analysis indicated a recent canine parvovirus infection (3). The dog had recently arrived in Western Australia from Queensland. The second Australian dog (AusDog2) was an 11-year-old female spayed Akita. This dog was receiving treatment for immune-mediated thrombocytopenia. The dog developed hemorrhagic diarrhea, anemia, and severe hydronephrosis with acute pyronephrosis and was euthanatized. With both dogs, low numbers of Cryptosporidium were detected in the feces. The U.S. dogs were asymptomatic.
Previous documented cases of canine cryptosporidiosis have involved symptomatic and asymptomatic animals, often with other concurrent infections. Sisk et al. (23) reported histological evidence of cryptosporidiosis in two 6-week-old pups, with one pup's death attributed to the pesticide toxophene and that of the other attributed to unspecified causes. Fukushima and Helman (7) reported an incidental finding of Cryptosporidium in a 3-month-old pup with canine distemper virus. Similarly, Turnwald et al. (26) detected Cryptosporidium oocysts in the feces of a 6-month-old female pup with canine distemper virus. In both cases, it was speculated that canine distemper caused immunosuppression, leading to increased susceptibility to cryptosporidial infection. The first case of chronic cryptosporidiosis was reported in an adult dog which had a 2-year history of intermittent diarrhea and weight loss (8). Immunosuppression, the cause of which could not be determined, was implicated as a factor increasing the susceptibility of the animal to infection. In the present study, the fact that AusDog1 and -2 were immunosuppressed appeared to predispose them to the occurrence of cryptosporidiosis. The immune status of the remaining dogs is unknown.
To date, seven distinct genotypes have been identified within what is currently recognized as C. parvum: a human genotype found only in humans; a cattle genotype found in livestock (cattle, sheep, and goats), which appears to be zoonotic, as it has been found in humans; as well as pig, marsupial, mouse, ferret, and, more recently, dog genotypes (14–17, 29).
Genetic analysis of both the 18S rDNA and Hsp-70 loci indicated that both Australian dogs were infected with a distinct Cryptosporidium genotype that appears to be conserved across geographic areas, as dog-derived Cryptosporidium isolates from the United States were identical to the Australian dog isolates at both loci.
Phylogenetic analysis of 18S rDNA sequence data obtained from dog-derived Cryptosporidium isolates from Australia and the United States, as well as other Cryptosporidium genotypes and species, revealed the dog genotype to be very distinct. The genetic similarity between the dog and the human and cattle genotypes was 97%. This is on a par with the genetic similarity between two accepted Cryptosporidium species, namely C. muris and C. serpentis (97%), and less than the similarity among C. parvum (cattle and human), C. wrairi, and C. meleagridis (99 to 98.4%). The level of genetic divergence between the dog genotype and other Cryptosporidium genotypes, combined with its apparently host-adapted nature, provides support for the idea that the dog genotype is, in fact, a separate species.
The infectivity of canine isolates for immunocompetent humans has not been demonstrated, although it has been shown that dogs can be experimentally infected with oocysts of human origin (2). It is speculated that humans may acquire infection from naturally infected dogs. Zoonotic transmission from a dog was suspected in one case when a veterinary student working in a ward where an infected dog was being cared for developed acute self-limiting diarrhea and Cryptosporidium oocysts were identified in her feces (8). In the present study, in the case of AusDog1, neither the dog's littermate nor the owners or veterinary personnel were Cryptosporidium positive, although opportunity for cross-infection existed. The dog genotype has, however, recently been identified in human immunodeficiency virus-infected human patient in the United States (21), and clearly, further studies are required in order to understand the full public health significance of the Cryptosporidium dog genotype, as well as other genotypes and species, for immunocompromised hosts.
In conclusion, the results of this study indicate that dogs harbor a genetically distinct dog-adapted Cryptosporidium strain which is conserved across geographic areas and which may well be a valid species. Further genetic and biological studies are required to fully determine the prevalence, transmission dynamics, and species status of the Cryptosporidium dog genotype.
Acknowledgments
We thank Aileen Elliot for microscopy assistance.
This study was supported by the Public Health Research and Development Committee (PHRDC) of the National Health and Medical Research Council of Australia. U. Morgan is a PHRDC Research Fellow.
REFERENCES
- 1.Bugg R J, Robertson I D, Elliot A D, Thompson R C A. Gastrointestinal parasites of urban dogs in Perth, Western Australia. Vet J. 1999;157:295–301. doi: 10.1053/tvjl.1998.0327. [DOI] [PubMed] [Google Scholar]
- 2.Current W L, Reese N C, Ernst J V, Bailey W S, Heyman M B, Weinstein W M. Human cryptosporidiosis in immunocompetent and immunodeficient persons. N Engl J Med. 1983;308:1252–1257. doi: 10.1056/NEJM198305263082102. [DOI] [PubMed] [Google Scholar]
- 3.Denholm, K. M., H. Haitjema, B. Gwynne, U. Morgan, and P. J. Irwin. Parvovirus and concurrent Cryptosporidium infection in a dog. Aust. Vet. J., in press. [DOI] [PubMed]
- 4.El-Ahraf A, Tacal J V, Sobih M, Amin M, Lawrence W, Wilcke B W. Prevalence of cryptosporidiosis in dogs and human beings in San Bernardino County California. J Am Vet Med Assoc. 1991;198:631–634. [PubMed] [Google Scholar]
- 5.Elliot A, Morgan U M, Thompson R C A. Improved staining method for detecting Cryptosporidium oocysts in stools using malachite green. J Gen Appl Microbiol. 1999;45:139–142. doi: 10.2323/jgam.45.139. [DOI] [PubMed] [Google Scholar]
- 6.Fayer R, Speer C A, Dubey J P. The general biology of Cryptosporidium. In: Fayer R, editor. Cryptosporidium and cryptosporidiosis. Boca Raton, Fla: CRC Press, Inc.; 1997. pp. 1–42. [Google Scholar]
- 7.Fukushima K, Helman R G. Cryptosporidiosis in a pup with distemper. Vet Pathol. 1984;21:247–248. doi: 10.1177/030098588402100218. [DOI] [PubMed] [Google Scholar]
- 8.Greene C E, Jacobs G J, Prickett D. Intestinal malabsorption and cryptosporidiosis in an adult dog. J Am Vet Med Assoc. 1999;197:365–367. [PubMed] [Google Scholar]
- 9.Grimason A M, Smith H V, Parker J F W, Jackson M H, Smith P G, Girdwood R W A. Occurrence of Giardia sp. cysts and Cryptosporidium sp. oocysts in faeces from public parks in the west of Scotland. Epidemiol Infect. 1993;110:641–645. doi: 10.1017/s0950268800051062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Johnston J, Gasser R B. Copro-parasitological survey of dogs in southern Victoria. Aust Vet Pract. 1993;23:127–131. [Google Scholar]
- 11.Lindsay D S, Upton S J, Owens D S, Morgan U M, Mead J R, Blagburn B L. Cryptosporidium andersoni n. sp. (Apicomplexa: Cryptosporiidae) from Cattle, Bos taurus. J Eukaryot Microbiol. 2000;47:91–95. doi: 10.1111/j.1550-7408.2000.tb00016.x. [DOI] [PubMed] [Google Scholar]
- 12.Meloni B P, Thompson R C A. Simplified methods for obtaining purified oocysts from mice and for growing Cryptosporidium parvum in vitro. J Parasitol. 1996;82:757–762. [PubMed] [Google Scholar]
- 13.Milstein T C, Goldsmid J M. The presence of Giardia and other zoonotic parasites of urban dogs in Hobart, Tasmania. Aust Vet J. 1995;72:154–155. doi: 10.1111/j.1751-0813.1995.tb15042.x. [DOI] [PubMed] [Google Scholar]
- 14.Morgan U M, Constantine C C, Forbes D A, Thompson R C A. Differentiation between human and animal isolates of Cryptosporidium parvum using rDNA sequencing and direct PCR analysis. J Parasitol. 1997;83:825–830. [PubMed] [Google Scholar]
- 15.Morgan U M, Sargent K D, Deplazes P, Forbes D A, Spano F, Hertzberg H, Elliot A, Thompson R C A. Molecular characterisation of Cryptosporidium from various hosts. Parasitology. 1998;117:31–37. doi: 10.1017/s0031182098002765. [DOI] [PubMed] [Google Scholar]
- 16.Morgan U M, Xiao L, Fayer R, Lal A A, Thompson R C A. Variation in Cryptosporidium: towards a taxonomic revision of the genus. Int J Parasitol. 1999;29:1733–1751. doi: 10.1016/s0020-7519(99)00109-5. [DOI] [PubMed] [Google Scholar]
- 17.Morgan U M, Monis P T, Fayer R, Deplazes P, Thompson R C A. Phylogenetic relationships amongst isolates of Cryptosporidium: evidence for several new species. J Parasitol. 1999;85:1126–1133. [PubMed] [Google Scholar]
- 18.Morgan U M, Xiao L, Fayer R, Graczyk R T K, Lal A A, Deplazes P, Thompson R C A. Phylogenetic analysis of 18S rDNA sequence data and RAPD analysis of Cryptosporidium isolates from captive reptiles. J Parasitol. 1999;83:525–530. [PubMed] [Google Scholar]
- 19.O'Donoghue P J. Cryptosporidium and cryptosporidiosis in man and animals. Int J Parasitol. 1995;25:139–195. doi: 10.1016/0020-7519(94)e0059-v. [DOI] [PubMed] [Google Scholar]
- 20.Page R D M. TREEVIEW: an application to display phylogenetic trees on personal computers. Comp Appl Biosci. 1996;12:357–358. doi: 10.1093/bioinformatics/12.4.357. [DOI] [PubMed] [Google Scholar]
- 21.Pieniazek N J, Bornay-Llinares F J, Slemenda S B, da Silva A J, Moura I N S, Arrowood M J, Ditrich O, Addiss D G. Cryptosporidium genotypes in HIV-infected persons. Emerg Infect Dis. 1999;3:444–449. doi: 10.3201/eid0503.990318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Simpson J W, Burnie A G, Miles R S, Scott J L, Lindsay D I. Prevalence of Giardia and Cryptosporidium infection in dogs in Edinburgh. Vet Rec. 1988;123:445. doi: 10.1136/vr.123.17.445. [DOI] [PubMed] [Google Scholar]
- 23.Sisk D B, Gosser H S, Styer E L. Intestinal cryptosporidiosis in two pups. J Am Vet Med Assoc. 1984;184:835–836. [PubMed] [Google Scholar]
- 24.Strimmer K, Von Haeseler A. Quartet puzzling: a quartet maximum likelihood method for reconstructing tree topologies. Mol Biol Evol. 1996;13:964–969. [Google Scholar]
- 25.Thompson J D, Gibson T J, Plewniak F, Jeanmougin F, Higgins D G. The Clustal X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Turnwald G H, Barta O, Taylor W, Kreeger J, Coleman S U, Pourciau S S. Cryptosporidiosis associated with immunosuppression attributable to distemper in a pup. J Am Vet Med Assoc. 1988;192:79–81. [PubMed] [Google Scholar]
- 27.Tzipori S, Campbell I. Prevalence of Cryptosporidium antibodies in 10 animal species. J Clin Microbiol. 1981;14:455–456. doi: 10.1128/jcm.14.4.455-456.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wilson R B, Holscher M A, Lyle S J. Cryptosporidiosis in a pup. J Am Vet Med Assoc. 1983;183:1005–1006. [PubMed] [Google Scholar]
- 29.Xiao L, Morgan U M, Limor J, Escalante A A, Arrowood M, Shulaw W, Thompson R C A, Fayer R, Lal A A. Genetic diversity within Cryptosporidium parvum and related Cryptosporidium species. Appl Environ Microbiol. 1999;65:3386–3391. doi: 10.1128/aem.65.8.3386-3391.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]