Abstract
Disturbed gut microbiota is a potential factor in the pathogenesis of major depressive disorder (MDD), yet whether gut microbiota dysbiosis is associated with the severity of MDD remains unclear. Here, we performed shotgun metagenomic profiling of cross-sectional stool samples from MDD (n = 138) and healthy controls (n = 155). The patients with MDD were divided into three groups according to Hamilton Depression Rating Scale 17 (HAMD-17), including mild (n = 24), moderate (n = 72) and severe (n = 42) individuals, respectively. We found that microbial diversity was closely related to the severity of MDD. Compared to HCs, the abundance of Bacteroides was significantly increased in both moderate and severe MDD, while Ruminococcus and Eubacterium depleted mainly in severe group. In addition, we identified 99 bacteria species specific to severity of depression. Furthermore, a panel of microbiota marker comprising of 37 bacteria species enabled to effectively distinguish MDD patients with different severity. Together, we identified different perturbation patterns of gut microbiota in mild-to-severe depression, and identified potential diagnostic and therapeutic targets.
Subject terms: Depression, Diagnostic markers
Introduction
Major depressive disorder (MDD) is the most common form of psychiatric and emotional disorder, affecting >350 million people [1]. Meanwhile, MDD has a high relapse rate [2] and causes an enormous social cost [3]. The baseline symptom severity of depression is one of the important factors influencing the treatment outcome [4, 5]. Mild to moderate depression can be treated conservatively without aggressive psychopharmacology [6], while severe depression may require antipsychotic, electroconvulsive, or other forms of therapy [4]. Clinically, the misdiagnosis of MDD will lead to ineffective antidepressant treatment, and even aggravate the disease. In addition, there is already a high risk of suicide or self-injury in severe cases of MDD. Therefore, it is of great clinical significance to identify new biomarkers for MDD patients with different severity, and this is crucial for early intervention.
Recently, growing evidence indicates that gut microbiota plays an essential role in the development and progression of mental disorders [7]. Alteration of gut microbiota was speculated to be the potential etiology of MDD, as it can affect the host’s brain function and behavior through the “gut-brain axis” [8]. Microbial biomarkers have been shown to be novel diagnostic and differential diagnostic tools, and help to identify new molecular therapeutic targets for diseases [9–13]. Generally, 16 S rRNA sequencing is mainly used to characterize the bacterial microbial composition, and explore the association between altered gut microbiota and various diseases [12, 13]. Previous studies have found that Bacteroides is the hub of perturbed gut microbiota in unipolar depression, while enriched Prevotella is the characteristic of bipolar depression [14]. In addition, patients with current active MDD (a-MDD) showed significantly increased in Alistipes and Anaerostipes as well as completely depleted Dialister, while mild symptoms (r-MDD) had higher abundance of Bilophila [15]. However, another study found Bacteroidetes, Proteobacteria, and Actinobacteria were strongly increased in a-MDD and r-MDD, whereas Firmicutes was significantly reduced [16]. These studies suggest that the gut microbiota may be different in MDD patients with different severity. However, due to the relative limited resolution of 16 S rRNA analysis, the identification level of bacteria can only be accurate to the genus level, and yield a small amount of information on species diversity. To make up for the gap of knowledge in this field, here, metagenome sequencing was used to characterize the gut microbial composition and function of MDD (n = 138), including mild (n = 24), moderate (n = 72) and severe patients (n = 42), and healthy controls (HCs, n = 155). Firstly, we sought to explore whether the whole microbial signature of MDD patients with different severity was significantly different from that in HCs. Next, we integrated the microbiota and related functional information through network analysis and correlated it with disease severity to further reveal how the signature of these disturbances changed as the disease worsened. Finally, we identified potential microbial markers related to severity of depression, and further tested their discriminative performance, thus making our findings a useful resource for the study of microbiome perturbations in depression.
Methods
Subject recruitment
The subjects included in this study were derived from our previous clinical cohort [17]. The study protocol was reviewed and approved by the Human Research and Ethics Committee of Beijing Anding Hospital (no. 2017-24), Capital Medical University (China). All recruited subjects signed a written informed consent. Each patient satisfied the MDD diagnostic criteria of the Diagnostic and Statistical Manual of Mental Disorders, 4th Edition (DSM-IV). The severity of MDD was staged with the HAMD-17 scale [18]. Depression severity stratification ranges as follows:[19] mild depression (score, 8–16); moderate depression (score, 17–23); and severe depression (score, ≥24). A total of 155 healthy controls (HCs; age, 29.13 ± 8.03; BMI, 22.38 ± 3.34) and 138 untreated MDD patients were recruited (age, 29.28 ± 7.10; BMI, 22.44 ± 3.41), including mild group (n = 24; age, 29.34 ± 7.64; BMI, 22.56 ± 3.99), moderate group (n = 72; age, 29.99 ± 7.31; BMI, 22.69 ± 3.21), severe group (n = 42; age, 28.08 ± 6.36; BMI, 21.93 ± 3.40). All patients provided written informed consent to participate. Patients that were excluded from this study were those who (1) had bipolar disorders, schizophrenic, schizoaffective, or other Axis I psychiatric disorders; (2) had the serious chronic somatic disease (diabetes, cardiovascular disease, thyroid disease, cancer, etc.); (3) alcohol and substance abuse, acute intoxication; (4) were pregnant or breastfeeding; (5) changed diet habit or used antibiotic within one month before sampling.
Metagenomic analysis of fecal samples
Fecal DNA extraction
All samples were collected from the clinical center. Briefly, fresh stool samples were collected and contained in sterile tubes in the morning (7–10 am) and stored at 4 °C, then transferred to a −80 °C refrigerator for subsequent processing within 6 h. According to the manufacturer’s instruction, we extracted the whole genomic DNA from fecal samples with the E.Z.N.A. Soil DNA Kit (Omega Bio-Tek, Norcross, GA, USA). Determination of the extracted DNA’s concentration and purity was performed on the TBS-380 and NanoDrop2000 separately. Then, we checked the quality on 1% agarose gel. DNA was fragmented randomly to an average size of about 300 bp by Covaris M220. Construction of paired-end library was accomplished by NEXTFLEX Rapid DNA-Seq (Bio-Scientific, Austin, TX, USA). Library was subjected to paired-end sequencing on Illumina NovaSeq (Illumina Inc., San Diego, CA, USA). To avoid batch effect, all samples were assayed in the same batch.
Quality control of raw sequences and data analysis
Low-quality sequences (sequences that were shorter than 50 bp or homopolymers that were longer than 10 bp or contained ambiguous base calls) in raw FASTQ files were filtered by Sickle (https://github.com/najoshi/sickle). Metagenomic data were aligned to the human genome using Burrows-Wheeler Aligner (http://bio-bwa.sourceforge.net), and the host genes were removed. Clean data were assembled to contigs by MEGAHIT, and the contigs with a minimum length of 300 bp were kept. Metagene was used to predict open reading frames from each assembly contigs [20]. All genes predicted to have 95% sequence identity were clustered using CD-HIT [21]. After completing the above procedures, reads were mapped to the representative sequences using SOAPaligner.
Metagenome data analysis
The gene set was annotated for bacteria based on the NCBI database using Diamond (version 0.8.35). For assessing the gut microbiota species of MDD patients, each gene was assigned to the highest-scoring taxonomy based on a unified database. Non-redundant gene set was aligned against the KEGG database with an e value cutoff of 1 × 10−5 [22], and abundance of the KO was calculated from the sum of the abundances of the genes corresponding to the KO. The gene expression value of gene set which used for species and function annotation were all based on the Reads Per Kilobase Million (RPKM). The α-diversity indexes were calculated by past 4.0. The α-diversity analysis was performed based on 4 indexes (Dominance, Simpson, Shannon and Evenness). Principal coordinate analysis (PCoA) based on Bray-Curtis distance was used to evaluate the overall difference of bacterial communities among HCs and MDD subgroups [23]. Permutational multivariate analysis of variance (PERMANOVA) was used to test the overall and pairwise group differences. PCoA analysis and PERMANOVA were based on the relative abundance of species. Samples were clustering into enterotypes in genus level by Dirichlet multinomial mixtures (DMM) approach as previous study described [24]. Optimal number of clusters was determined by Calinski-Harabasz index. Enterotypes analysis was based on the abundance of genus.
Combined biomarker for MDD subgroups
High relative abundance bacteria were selected for following analysis at species level (prevalence >20%, average relative abundance >0.01%), then unclassified species was excluded. Linear discriminant analysis effect size (LEfSe) analysis was used to identify the differentially enriched bacteria and KOs among HCs and 3 MDD subgroups (LDA score >2.5, https://huttenhower.sph.harvard.edu/galaxy/). The diagnostic performance was quantified by Random Forest classifier and tested by 5-fold cross-validation. Receiver operating characteristic (ROC) curve was plotted to estimate the diagnostic efficacy.
Construction of co-occurrence network of gut bacteria
Based on abundance data of metagenome, Sparse correlations for compositional data (SparCC) algorithm was used to calculate the correlations between all the differentially enriched bacteria and KOs which defined by LEfSe analysis (p < 0.05, http://mem.rcees.ac.cn:8081) [25]. The result was visualized by Cytoscape (version 3.9.0) and Graphpad Prime 8.
Statistical analysis
Statistical analysis was performed using SPSS (version 22.0). Continuous variables were analyzed by one-way ANOVA test (mean±SD) followed by LSD′s multiple comparison or non-parametric factorial Kruskal–Wallis sum-rank test (mean±SEM), p values of pairwise comparisons of the Kruskal–Wallis sum-rank test were corrected by Holm-Bonferroni method. Categorical variables were performed by chi-square test. Statistical significance level was set at p < 0.05.
Results
Clinical characteristics of the subjects
In this study, we included in a total of 155 HCs and 138 MDD patients. All MDD patients were treatment-naive, and there was no significant difference in gender (p = 0.71; Chi-square test), age (p = 0.62; one-way ANOVA), and body mass index (BMI; p = 0.71; one-way ANOVA) among HCs and 3 MDD subgroups (Table S1).
Altered gut microbiota among HCs and MDD subgroups
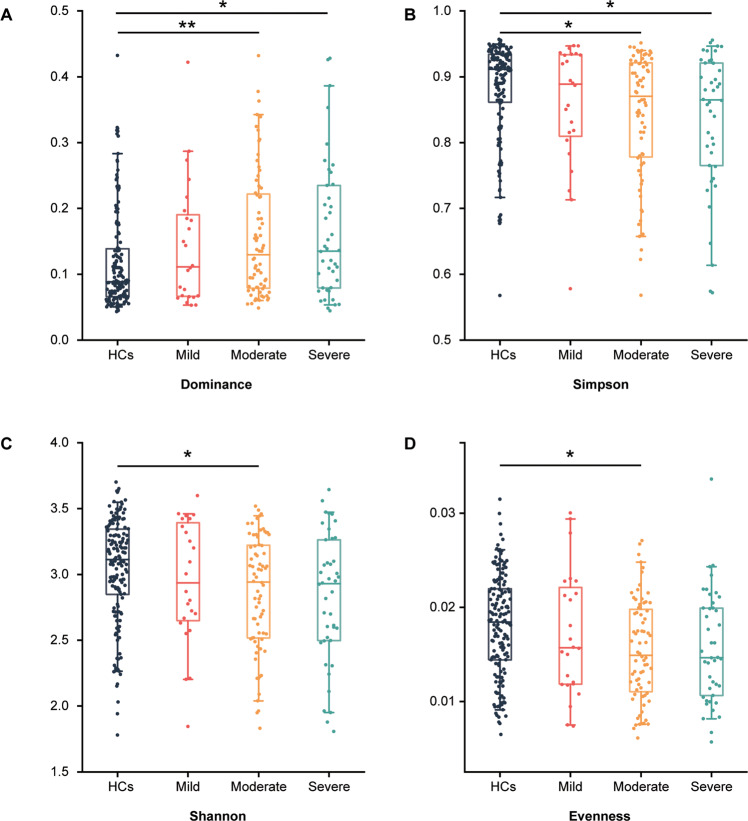
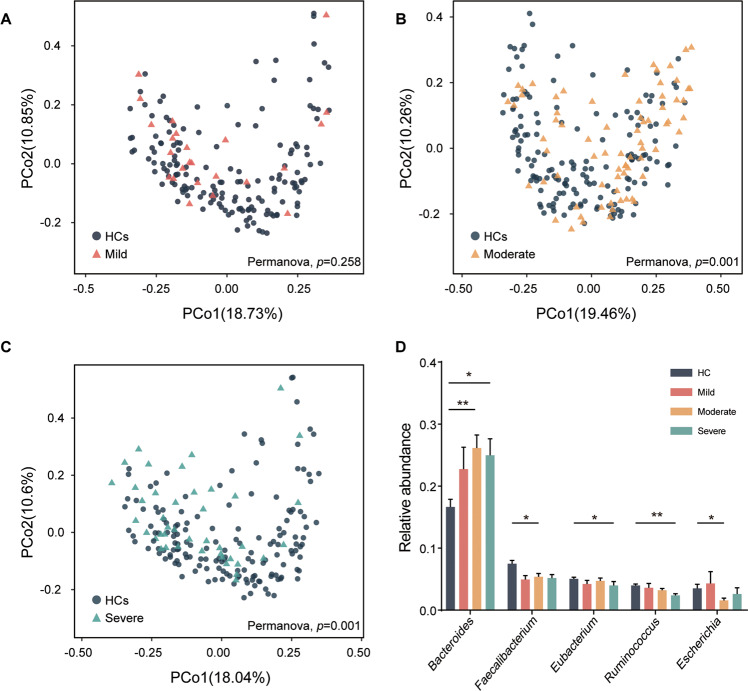
Initially, the α-diversity was compared among HCs and 3 MDD subgroups. Consequently, we found that the simpson index of moderate and severe groups was significantly decreased, while there was no significant difference between the mild group and the others (Fig. 1A, B). In addition, the shannon and evenness indexes were decreased in moderate group (Fig. 1C, D). These findings suggested that the community richness and diversity were associated with MDD severity. To explore the difference in microbial signatures among HCs and 3 subgroups, PCoA was performed. We found that the whole microbial compositions of the moderate and severe groups were different from those of the HCs; while the compositions of 3 MDD subgroups were not different from each other (Fig. 2A–C, Fig. S2).
Fig. 1. The α-diversity index analysis among HCs and MDD subgroups.
A–D The box plots showing α-phylogenetic diversity analysis results, the dominance index increased in moderate and severe groups relative to HCs while simpson was decreased, while there was no significant difference between mild and HCs. In moderate group, the shannon and evenness indexes were significantly lower than that in HCs. (HCs, n = 155; mild, n = 24; moderate, n = 72; severe, n = 42; *p < 0.05; **p < 0.01; Kruskal–Wallis rank sum test).
Fig. 2. Gut microbial characteristics among HCs and MDD subgroups.
A–C Principal co-ordinates analysis (PCoA) was conducted based on species level and Bray-Curtis distance. PERMANOVA test showed that the general characteristics of microbiota in moderate and severe groups were significantly different from HCs, while there was no significant difference between mild group and HCs. In addition, we did not find the separated characteristics of microbiota by comparing disease subgroups (see Fig. S2). D The bar plot showed the differentially enriched genus in the most top 10 high relative abundance genus. Bacteroides was significantly increased in moderate and severe group, while the reduction of 2 genera (Faecalibacterium and Escherichia) in moderate group and 2 genera (Eubacterium and Ruminococcus) in severe group, respectively. (*p < 0.05; **p < 0.01; Kruskal–Wallis rank sum test).
Next, we investigated the high relative abundance (top 10) bacteria of HCs and the 3 MDD subgroups at family and genus levels. Overall, at the family level, Bacteroidaceae, Lachnospiraceae, Ruminococcaceae and Prevotellaceae were the major high abundance bacterial taxa (Fig. S1A). At genus level, Bacteroides, Faecalibacterium, Blautia, Prevotellaceae were the major high abundance bacterial taxa (Fig. S1B). Here, we found that, compared to HCs, Bacteroides were remarkably enriched in moderate and severe groups, Faecalibacterium and Escherichia were decreased in moderate group, while Ruminococcus and Eubacterium were decreased only in severe group (Fig. 2D). Next, we explored the distribution of different enterotype in HCs and 3 MDD subgroups. Based on Dirichlet multinomial mixtures (DMM) approach, we observed 5 enterotypes. Except for the classical enterotypes (Bacteroides; Prevotella) [26–28], we distinguished 2 new enterotype which dominated by Blautia and Faecalibacterium. Faecalibacterium (31.6%) were significantly more abundant in HCs, whereas 2 types of Bacteroides (Bac, Bac2) were more abundant in 3 MDD subgroups (mild: 33.3%, moderate: 36.1%, severe: 31.0%). 3 MDD subgroups shared a similar trend that Bac and Bac 2 were the major enterotypes in microbiota (Fig. S1C).
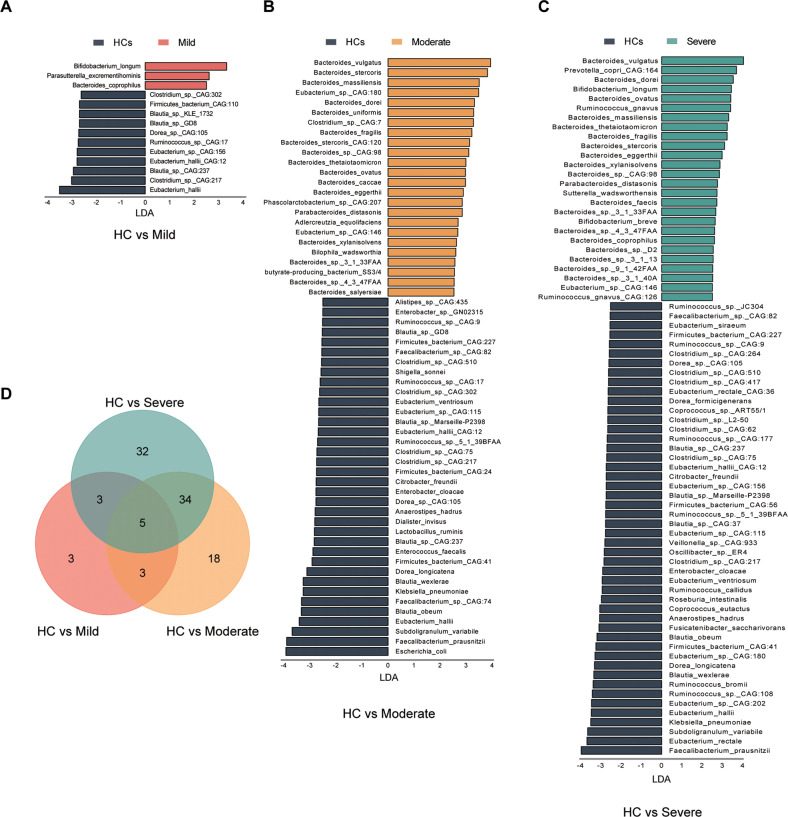
To further gain insight into the alterations of gut microbiota and the consequence of the profound bacteria derangement on metabolic function, LDA Effect Size (LEfSe) analysis was performed pairwise on bacteria between HCs and MDD subgroups (LDA > 2.5, p < 0.05). Three groups of bacteria were identified through three pairwise comparisons of LEfSe analysis (Table S2–4). Compared with HCs, mild group had less differentially enriched bacteria at the set threshold. We found 14 differentially enriched bacteria in mild group, 60 in moderate group, and 74 in severe group (Fig. 3A–C). Those decreased gut bacteria of the mild group mainly belong to Blautia (3 species) and Eubacterium (3 species). Blautia (5 species), Clostridium (4 species) and Eubacterium (4 species) were decreased in moderate group, while Bacteroides (16 species) were remarkable increased in moderate group. Consistently, Blautia (5 species), Clostridium (7 species) and Eubacterium (8 species) were also decreased in severe group, Ruminococcus (7 species) were decreased too. The increased bacteria remained Bacteroides (18 species) in severe group (Table S2–4). Venn diagram showed that there were 99 differentially enriched bacteria among HCs and 3 MDD subgroups. Three bacteria were exclusive in mild group, 18 in moderate group and 32 in severe group (Fig. 3D). Interestingly, we found that 4 of the 18 unique bacteria of moderate group belonged to Bacteroides. Five of the 32 unique bacteria of severe belonged to Bacteroides, 4 belonged to Clostridium and 6 belonged to Ruminococcus. Likewise, we found 3 differentially enriched KOs in moderate group and 5 in severe group (K03088, K21572, K07133, Table S5-6). There were 2 KOs that were exclusive in severe group (K01190, K12373, Table S6). K01190 encodes lactase to uptake and hydrolyze lactose [29]. K12373 encodes hexosaminidase involve in glycosphingolipid biosynthesis, and the lack of hexosaminidase could cause gangliosidosis [30].
Fig. 3. Differentially enriched bacteria species of HCs and the 3 MDD subgroups.
LDA Effect Size (LEfSe) analysis was performed to identify the bacteria that were differentially enriched in HCs and the 3 MDD subgroups in pairs (LDA > 2.5, p < 0.05). A–C Bar plot displayed the differentially enriched species between HC and MDD subgroups. D Venn diagram showed that moderate and severe group shared 39 differentially enriched bacteria, while only 5 bacteria were shared in the 3 MDD subgroups.
Concordance of microbial variation in patients with moderate and severe MDD
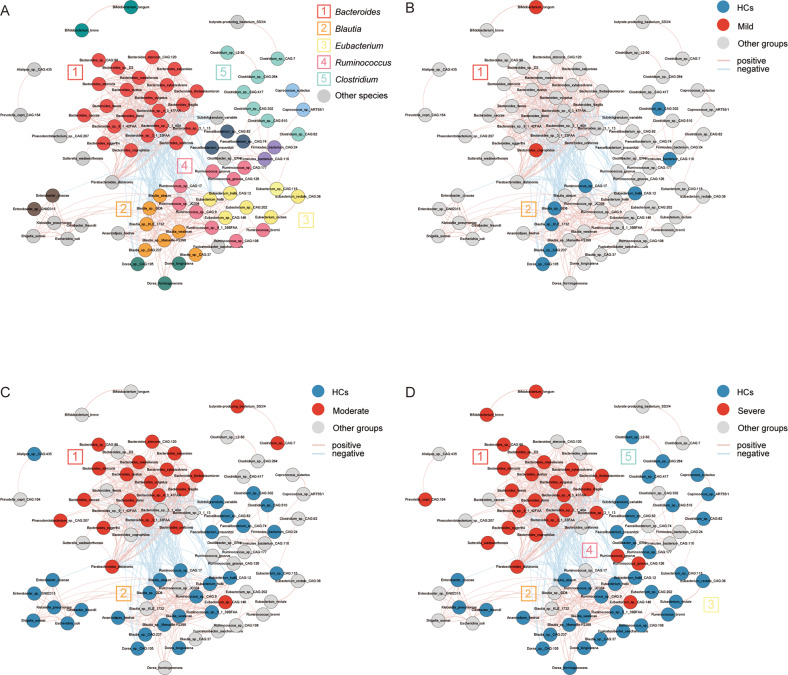
In order to explore the potential correlations among the differentially enriched microbiota, SparCC correlation analysis was performed to assess the co-occurrence relationship between the differential enriched bacteria. Based on these results, we constructed a co-occurrence network (p < 0.05, r2 > 0.25). In genus level, Bacteroides enriched markedly in every MDD subgroup (Fig. 4A–D). Notably, we found that the dominant depleted bacteria varied in MDD subgroups. The characteristic microbiota of the severe group were Ruminococcus (6/9), Eubacterium (6/7), Blutia (5/7) and Clostridium (5/7) (Fig. 4D), while that of moderate group were Blutia (5/7) (Fig. 4C). The enriched bacteria Bacteroides showed a strong negative correlation with the depleted bacteria, particularly the Blautia, Ruminococcus, Eubacterium (Fig. 4A). This phenomenon was indicative of a competitive antagonistic relationship among those bacteria. In addition, we also explored the correlation among those changed bacteria and KOs. The differentially enriched KOs and species that highly correlated with either KOs were reserved to construct the correlation-based heatmap (r2 > 0.25). Overall, Bacteroides were positively correlated with KOs and Blautia, Eubacterium, Ruminococcus were negatively correlated with KOs (Fig. S3A). Obviously, K12373 and K21572 were highly positively correlated with 21 Bacteroides while negatively correlated with Blautia and Ruminococcus (Fig. S3B). Compared with K12373, there were 3 more species of Blautia associated with K21572 (Fig. S3B).
Fig. 4. The co-occurrence network constructed from the relative abundance of differentially enriched bacteria species among HCs and MDD subgroups.
The network was mapped based on the result (p < 0.05, r2 > 0.25). A Correlation among abundances of all differentially enriched bacteria were analyzed with SparCC algorithm, clusters were assigned a particular color and main clusters had a corresponding number. B, D Species enriched in MDD subgroups were highlighted in red and depleted ones were in blue. Pink lines represented positive correlations and blue lines means negative correlations. B Bacteria altered little in mild group, some scattered species were depleted in mild groups associated with each other weakly and had no correlation with the only enriched Bacteroides in cluster 1. C Bacteria enriched in moderate groups were almost all belong to Bacteroides in cluster 1, the major depleted genus was Blutia in cluster 2, which have a significantly negative association with Bacteroides. D Gut microbiota of severe group appeared fierce disturbance featured with markable increase of Bacteroides in cluster 1. Ruminococcus, Blautia, Eubacterium and Clostridium (cluster 2, 3, 4, 5) were obviously decreased as a group. There were complex positive correlations within cluster 2, 3, 4 and a significantly negative correlation between the decreased group and Bacteroides. It revealed that there was a potential antagonistic relationship between the decreased group and Bacteroides.
Identification of faecal bacteria species as potential biomarker for different severity of MDD
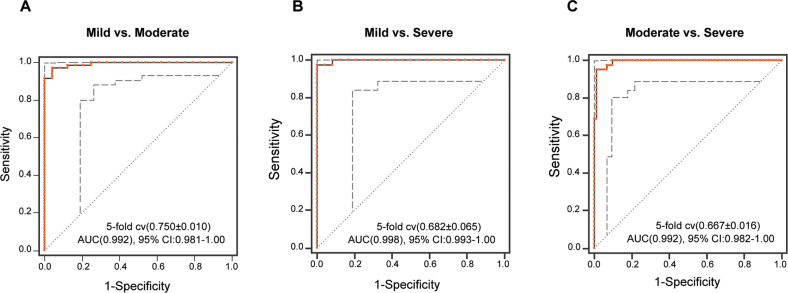
The differentially enriched bacteria of HCs and 3 subgroups were trained separately by random forest classifier (RF) for binary classification. According to Gini importance, 37 bacteria were screened out from 3 pairs of differentially enriched bacteria to construct the biomarker panel for discriminating different groups (Gini importance>0.02, Table S7). The diagnostic performance of paired identification was evaluated by receiver operating characteristic (ROC) curves and quantified by the area under the curve (AUC). AUC of the bacterial biomarker ranged from 0.992 to 0.998 (mild versus moderate, AUC = 0.992; mild versus severe, AUC = 0.998; moderate versus severe, AUC = 0.992; Fig. 5A–C). It reveals that the diagnostic performance of biomarkers was excellent.
Fig. 5. Diagnosing MDD subgroups from gut microbiome features.
A–C Based on the importance value of random forest analysis (>2%) between MDD subgroups and HCs, 37 species was identified (see Table S2–4 for importance value). This microbial panel enabled the differentiation between any 2 subgroups with high diagnostic accuracy (AUC, 0.992–0.998).
Sex-specific of altered gut microbiota
Considering of the gender differences in the prevalence and clinical manifestations of depression, we explored the gender bias of gut microbiota characteristic. In both female and male groups, species diversity declined significantly in MDD subgroups relative to HCs (Fig. S4A, B). Shannon and simpson indexes were decreased in MDD subgroups. MDD subgroups were remarkably separated from HCs in both female and male groups (Fig. S5A, B). The changes in composition were also similar in two genders. Compared the percentage of top 10 bacteria in samples, we found the Bacteroidaceae increased while Ruminococcaceae decreased at family level in MDD subgroups, Bacteroides increased while Faecalibacterium decreased at genus level in MDD subgroups (Fig. S6).
LEfSe analysis was performed to screen out the differentially enriched bacteria (LDA > 2.5, p < 0.05). Ultimately, we identified 38 species differentially enriched bacteria in female group (Fig. S7A, Table S8), most of them belonged to Bacteroides (11 species), Eubacterium (7 species), Clostridium (4 species) and Blautia (4 species); and 68 species differential enriched bacteria in male group (Fig.S7B, Table S9), most of them belonged to Bacteroides (17 species), Eubacterium (6 species), Clostridium (4 species), Blautia (6 species) and Ruminococcus (4 species).
Identically, we constructed co-occurrence networks based on SparCC analysis of differentially enriched bacteria (p < 0.05, r2 > 0.25). In female group, there were 3 hub clusters, the Bacteroides showed a negative correlation with Eubacterium and Blautia (Fig. S8A). In the male group, a positively correlated network was found among Blautia, Ruminococcus, Eubacterium and Coprococcus, while they showed negative correlations with Bacteroides (Fig. S8B).
Discussion
The link between changes in the gut microbiome and MDD has been supported by several studies [14, 17]. Here, we showed that these changes could reflect the severity of MDD. In this study, we found that the gut microbiota of moderate and severe MDD patients were characterized by the enrichment of Bacteroidetes, while Ruminococcus and Eubacterium were depleted in the severe patients. Consistently, the major enterotype of HCs was Faecalibacterium while which of MDD subgroups were Bac and Bac 2. In addition, we also identified a microbial marker panel which is capable of distinguishing MDD patients with different severity.
It is generally believed that high diversity of gut microbiome is a sign of healthy status [31, 32]. Some studies have found that the gut microbial diversity reduced in depression, bipolar disorder and schizophrenia [33]. In addition, reduced diversity has been associated with disease severity and higher risk of death in patients with diseases such as bronchiectasis [34], cystic fibrosis [35] and ulcerative colitis [36]. Consistently, we found that the simpson index decreased only in the moderate and severe MDD groups at the genus level. In addition, the Venn diagram showed that the number of shared species between moderate and severe is higher than that of other groups. Bar plot showed that Bacteroides were significantly enriched in moderate and severe subgroups, while there was no change in the mild group. These findings suggested that the gut microbial composition remains relatively stable during the early stages of MDD; but with increase in disease severity, the disturbances of gut microbiota become inevitable. Furthermore, based on the LDA effect size analysis, the differentially expressed microbiota in the MDD subgroups were identified. We constructed the co-occurrence network of perturbed microbiota of MDD with mild to severe. We found that the depletion of Blautia and Eubacterium were common features of MDD patients; enriched Bacteroides were characteristic of moderate and severe MDD. Consistently, our previous study proved that increase of Bacteroides was a signature of MDD [17]. Bacteroides was reported involved in immune system maturation, tumor formation and activation of autoimmune disease by affecting T cell’s function and promote cognitive impairment disease pathologies through activating microglia [37–40]. Bacteroides faecis helps maintaining the epithelial barrier integrity and increasing the gut IgA level to reduce inflammatory bowel disease [41, 42]. Because of related closely, Bacteroides dorei and Bacteroides vulgatus are often researched as a group, they were been reported playing an important role in brown adipose tissue metabolism and suppressing proinflammatory immune responses [43, 44]. Similarly, researched revealed that Bacteroides uniformis and Bacteroides eggerthii were involved in pathological mechanism of obesity, metabolism, and colitis [45–47]. In some studies, Blautia have been reported as potential probiotics that can induce anti-inflammatory peripheral immune response, alleviate obesity-related disease and regulate metabolism through cross-feeding with other bacteria [48–50]. Also, it has been reported that Eubacterium can produce short-chain fatty acids, which play an important role in regulating cell metabolism, immune and endocrine response [51, 52]. In addition, unlike mild and moderate patients, the highly concentrated clusters in severe MDD were dominated by decreased 7 Ruminococcus, 8 Eubacterium, 5 Clostridium and 7 Clostridium. Depletion of these potential probiotics may contribute to the development of depression. Compared with patients with mild and moderate depression, the severe individuals need more active physical and drug therapy. Therefore, combining a probiotic intervention strategy with conventional treatment will be helpful in promoting the improvement of both disease recovery and quality of life. In addition, we found that K21572 (susD) and K12373 (HEXA) may be key node connecting Blautia and Bacteroides, showing completely opposite correlation with these two microbial clusters. susD is an outer membrane protein. It is the main starch binding protein on the surface of Bacteroides, and can effectively use polysaccharides as a source of carbon and energy [53]. Further work characterizing the interaction between Blautia and Bacteroides could elucidate its role in MDD severity. K12373 (HEXA) is a hexose kinas, play an important role in sugar metabolism in Bacteroides fragilis [54]. Likewise, K03088 (rpoE) is an important part of transcription activation factor that binds to RNA polymerase complex to regulate gene expression in bacteria [55]. In sight of the positive association of K03088 and Bacteroides, the increased expression of K03088 might indicate the function enhancement of Bacteroides.
Motivated by the results that showed that there were alterations in the gut microbiota in moderate and severe MDD, we constructed a random forest model with 37 bacteria species. The AUC value of the classification of MDD subgroups were 0.992 to 0.998; suggesting a high diagnostic value. Overall, this finding provided evidence that gut microbiota-targeted biomarkers may become potential non-invasive tools for MDD stratification.
Additionally, we initially explored the gender bias in the structure of gut microbiota. The prevalence of MDD in women was twice of in men [56, 57], we wondered if there is a potential relationship between gender preference of disease and gut microbiota. Generally, we found that the changes of microbiota were similar in female and male. Previous study declared that Bacteroides and Prevotella had higher abundance in male [58]. In our study, male group owned more differentially enriched bacteria and most of them were belonged Bacteroides. Further research was required to figure out if this was related to gender differences in MDD. In order to obtain consolidation evidence, it is necessary to further expand the study cohort to determine the gender bias of gut microbiota in MDD.
The limitations of this study are: (i) The sample size of the three MDD subgroups is relatively small, and the samples were collected from a clinical center, therefore regional variations cannot be ruled out; (ii) All patients were not under medication; therefore, follow-up studies are required to explore whether this biomarker panel can be used to monitor treatment response; (iii) Given that fecal bacteria transplantation could transfer depressive phenotypes from humans to mice, it will be important to determine the correlation between the disturbance of gut microbiota and disease severity in animal models, and uncover the underlying mechanisms.
Taken together, we analyzed and found the unique and common alterations in gut microbiota across different disease severities. Microbiota may affect physiological functions through mutual synergy or antagonism, and the status may shift from balance to imbalance as symptoms get worse. Furthermore, we identified a novel combined biomarker that could discriminate different severity subgroups with high accuracy. In conclusion, our study provides a new direction for understanding the progression of MDD, and a potential promising strategy for developing a novel method for objectively assessing the severity of MDD.
Supplementary information
Acknowledgements
This work was supported by the Projects of International Cooperation and Exchanges NSFC (81820108015), The Natural Science Foundation Project of China (81971296, 82171523, 82201688, 82171526), China Postdoctoral Science Foundation (2021MD703928, 2022MD713717), CQMU Program for Youth Innovation in Future Medicine, Chongqing Science &Technology Commission (cstc2019 jcyjjqX0009, cstc2021jscx-msxm0026, cstc2021jscx-msxm0025), Beijing Talents Project, Beijing Scholar 2021 Program.
Author contributions
Designed the experiments: PX and PZ. Performed the metagenomic analysis: XH, YFL, JW, YH, XYZ and HPZ. Analyzed the metagenomic data: XH, YFL, OO, PJX, JJZ, ZLS, ML, GFZ and JY. Drafted the paper: PX, PZ, HX, XMT and LW. Revised the paper for intellectual content: PX and PZ.
Data availability
The metagenomic sequencing data were deposited in the China National GeneBank DataBase (CNGBdb) (https://db.cngb.org/; project ID: CNP0001162).
Code availability
All the source codes are available at https://github.com/mokmoksunshine/changes_of_gut_microbiota_severity.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Xi Hu, Yifan Li.
These authors jointly supervised this work: Jian Yang, Peng Zheng, Peng Xie.
Contributor Information
Jian Yang, Email: yangjian@ccmu.edu.cn.
Peng Zheng, Email: peng-zheng@foxmail.com.
Peng Xie, Email: xiepeng@cqmu.edu.cn.
Supplementary information
The online version contains supplementary material available at 10.1038/s41398-023-02436-z.
References
- 1.Uchida S, Yamagata H, Seki T, Watanabe Y. Epigenetic mechanisms of major depression: Targeting neuronal plasticity. Psychiatry Clin Neurosci. 2018;72:212–27. doi: 10.1111/pcn.12621. [DOI] [PubMed] [Google Scholar]
- 2.Kato M, Hori H, Inoue T, Iga J, Iwata M, Inagaki T, et al. Discontinuation of antidepressants after remission with antidepressant medication in major depressive disorder: a systematic review and meta-analysis. Mol Psychiatry. 2021;26:118–33. doi: 10.1038/s41380-020-0843-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Puzhko S, Schuster T, Barnett TA, Renoux C, Rosenberg E, Barber D, et al. Evaluating Prevalence and Patterns of Prescribing Medications for Depression for Patients With Obesity Using Large Primary Care Data (Canadian Primary Care Sentinel Surveillance Network) Front Nutr. 2020;7:24. doi: 10.3389/fnut.2020.00024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Davidson JRT. Major depressive disorder treatment guidelines in America and Europe. J Clin Psychiatry. 2010;71:e04. doi: 10.4088/JCP.9058se1c.04gry. [DOI] [PubMed] [Google Scholar]
- 5.Kirsch I, Deacon BJ, Huedo-Medina TB, Scoboria A, Moore TJ, Johnson BT. Initial severity and antidepressant benefits: a meta-analysis of data submitted to the Food and Drug Administration. PLoS Med. 2008;5:e45. doi: 10.1371/journal.pmed.0050045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Paris J. The mistreatment of major depressive disorder. Can J Psychiatry. 2014;59:148–51. doi: 10.1177/070674371405900306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Järbrink-Sehgal E, Andreasson A. The gut microbiota and mental health in adults. Curr Opin Neurobiol. 2020;62:102–14. doi: 10.1016/j.conb.2020.01.016. [DOI] [PubMed] [Google Scholar]
- 8.Zheng P, Zeng B, Zhou C, Liu M, Fang Z, Xu X, et al. Gut microbiome remodeling induces depressive-like behaviors through a pathway mediated by the host’s metabolism. Mol psychiatry. 2016;21:786–96. doi: 10.1038/mp.2016.44. [DOI] [PubMed] [Google Scholar]
- 9.Aron-Wisnewsky J, Vigliotti C, Witjes J, Le P, Holleboom AG, Verheij J, et al. Gut microbiota and human NAFLD: disentangling microbial signatures from metabolic disorders. Nat Rev Gastroenterol Hepatol. 2020;17:279–97. doi: 10.1038/s41575-020-0269-9. [DOI] [PubMed] [Google Scholar]
- 10.Thomas AM, Manghi P, Asnicar F, Pasolli E, Armanini F, Zolfo M, et al. Metagenomic analysis of colorectal cancer datasets identifies cross-cohort microbial diagnostic signatures and a link with choline degradation. Nat Med. 2019;25:667–78. doi: 10.1038/s41591-019-0405-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fuentes-Chust C, Parolo C, Rosati G, Rivas L, Perez-Toralla K, Simon S, et al. The Microbiome Meets Nanotechnology: Opportunities and Challenges in Developing New Diagnostic Devices. Adv Mater. 2021;33:e2006104. doi: 10.1002/adma.202006104. [DOI] [PubMed] [Google Scholar]
- 12.Shang J, Zhang Y, Guo R, Liu W, Zhang J, Yan G, et al. Gut Microbiome Analysis Can Be Used as a Noninvasive Diagnostic Tool and Plays an Essential Role in the Onset of Membranous Nephropathy. Adv Sci. (Weinh) 2022;9:e2201581. doi: 10.1002/advs.202201581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jin J, Gao L, Zou X, Zhang Y, Zheng Z, Zhang X, et al. Gut Dysbiosis Promotes Preeclampsia by Regulating Macrophages and Trophoblasts. Circ Res. 2022;131:492–506. doi: 10.1161/CIRCRESAHA.122.320771. [DOI] [PubMed] [Google Scholar]
- 14.Zheng P, Yang J, Li Y, Wu J, Liang W, Yin B, et al. Gut Microbial Signatures Can Discriminate Unipolar from Bipolar Depression. Adv Sci (Weinh) 2020;7:1902862. doi: 10.1002/advs.201902862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Caso JR, MacDowell KS, González-Pinto A, García S, de Diego-Adeliño J, Carceller-Sindreu M, et al. Gut microbiota, innate immune pathways, and inflammatory control mechanisms in patients with major depressive disorder. Transl Psychiatry. 2021;11:645. doi: 10.1038/s41398-021-01755-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jiang H, Ling Z, Zhang Y, Mao H, Ma Z, Yin Y, et al. Altered fecal microbiota composition in patients with major depressive disorder. Brain, Behav, Immun. 2015;48:186–94. doi: 10.1016/j.bbi.2015.03.016. [DOI] [PubMed] [Google Scholar]
- 17.Yang J, Zheng P, Li Y, Wu J, Tan X, Zhou J, et al. Landscapes of bacterial and metabolic signatures and their interaction in major depressive disorders. Science advances. 2020;6:eaba8555. doi: 10.1126/sciadv.aba8555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hamilton M. A rating scale for depression. J Neurol Neurosurg Psychiatry. 1960;23:56–62. doi: 10.1136/jnnp.23.1.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zimmerman M, Martinez JH, Young D, Chelminski I, Dalrymple K. Severity classification on the Hamilton Depression Rating Scale. J Affect Disord. 2013;150:384–8. doi: 10.1016/j.jad.2013.04.028. [DOI] [PubMed] [Google Scholar]
- 20.Noguchi H, Park J, Takagi T. MetaGene: prokaryotic gene finding from environmental genome shotgun sequences. Nucl Acids Res. 2006;34:5623–30. doi: 10.1093/nar/gkl723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fu L, Niu B, Zhu Z, Wu S, Li W. CD-HIT: accelerated for clustering the next-generation sequencing data. Bioinformatics. 2012;28:3150–2. doi: 10.1093/bioinformatics/bts565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xie C, Mao X, Huang J, Ding Y, Wu J, Dong S, et al. KOBAS 2.0: a web server for annotation and identification of enriched pathways and diseases. Nucl Acids Res. 2011;39:W316–W22. doi: 10.1093/nar/gkr483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Doan T, Hinterwirth A, Worden L, Arzika AM, Maliki R, Abdou A, et al. Gut microbiome alteration in MORDOR I: a community-randomized trial of mass azithromycin distribution. Nat Med. 2019;25:1370–6. doi: 10.1038/s41591-019-0533-0. [DOI] [PubMed] [Google Scholar]
- 24.Holmes I, Harris K, Quince C. Dirichlet multinomial mixtures: generative models for microbial metagenomics. PloS One. 2012;7:e30126. doi: 10.1371/journal.pone.0030126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Feng K, Peng X, Zhang Z, Gu S, He Q, Shen W, et al. iNAP: An integrated network analysis pipeline for microbiome studies. iMeta. 2022;1:e13. doi: 10.1002/imt2.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Arumugam M, Raes J, Pelletier E, Le Paslier D, Yamada T, Mende DR, et al. Enterotypes of the human gut microbiome. Nature. 2011;473:174–80. doi: 10.1038/nature09944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vandeputte D, Kathagen G, D’Hoe K, Vieira-Silva S, Valles-Colomer M, Sabino J, et al. Quantitative microbiome profiling links gut community variation to microbial load. Nature. 2017;551:507–11. doi: 10.1038/nature24460. [DOI] [PubMed] [Google Scholar]
- 28.Vieira-Silva S, Sabino J, Valles-Colomer M, Falony G, Kathagen G, Caenepeel C, et al. Quantitative microbiome profiling disentangles inflammation- and bile duct obstruction-associated microbiota alterations across PSC/IBD diagnoses. Nat Microbiol. 2019;4:1826–31. doi: 10.1038/s41564-019-0483-9. [DOI] [PubMed] [Google Scholar]
- 29.Varela JA, Puricelli M, Ortiz-Merino RA, Giacomobono R, Braun-Galleani S, Wolfe KH, et al. Origin of Lactose Fermentation in Kluyveromyces lactis by Interspecies Transfer of a Neo-functionalized Gene Cluster during Domestication. Curr Biol. 2019;29:4284–90. doi: 10.1016/j.cub.2019.10.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Navon R, Proia RL. The mutations in Ashkenazi Jews with adult GM2 gangliosidosis, the adult form of Tay-Sachs disease. Science. 1989;243:1471–4. doi: 10.1126/science.2522679. [DOI] [PubMed] [Google Scholar]
- 31.Campbell E, Nagler CR. Fe, fi, fo, fum, I smell the diet of a healthy human. Cell. 2021;184:4107–9. doi: 10.1016/j.cell.2021.07.011. [DOI] [PubMed] [Google Scholar]
- 32.Wastyk HC, Fragiadakis GK, Perelman D, Dahan D, Merrill BD, Yu FB, et al. Gut-microbiota-targeted diets modulate human immune status. Cell. 2021;184:4137–53. doi: 10.1016/j.cell.2021.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nikolova VL, Hall MRB, Hall LJ, Cleare AJ, Stone JM, Young AH. Perturbations in Gut Microbiota Composition in Psychiatric Disorders: A Review and Meta-analysis. JAMA Psychiatry. 2021;78:1343–54. doi: 10.1001/jamapsychiatry.2021.2573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mac Aogáin M, Lau KJX, Cai Z, Kumar Narayana J, Purbojati RW, Drautz-Moses DI, et al. Metagenomics Reveals a Core Macrolide Resistome Related to Microbiota in Chronic Respiratory Disease. Am J Respir Crit Care Med. 2020;202:433–47. doi: 10.1164/rccm.201911-2202OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cuthbertson L, Walker AW, Oliver AE, Rogers GB, Rivett DW, Hampton TH, et al. Lung function and microbiota diversity in cystic fibrosis. Microbiome. 2020;8:45. doi: 10.1186/s40168-020-00810-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Barberio B, Facchin S, Patuzzi I, Ford AC, Massimi D, Valle G, et al. A specific microbiota signature is associated to various degrees of ulcerative colitis as assessed by a machine learning approach. Gut Microbes. 2022;14:2028366. doi: 10.1080/19490976.2022.2028366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chen C, Liao J, Xia Y, Liu X, Jones R, Haran J, et al. Gut microbiota regulate Alzheimer’s disease pathologies and cognitive disorders via PUFA-associated neuroinflammation. Gut. 2022;71:2233–52. doi: 10.1136/gutjnl-2021-326269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wu S, Rhee K-J, Albesiano E, Rabizadeh S, Wu X, Yen H-R, et al. A human colonic commensal promotes colon tumorigenesis via activation of T helper type 17 T cell responses. Nat Med. 2009;15:1016–22. doi: 10.1038/nm.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chu H, Khosravi A, Kusumawardhani IP, Kwon AHK, Vasconcelos AC, Cunha LD, et al. Gene-microbiota interactions contribute to the pathogenesis of inflammatory bowel disease. Science. 2016;352:1116–20. doi: 10.1126/science.aad9948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mazmanian SK, Liu CH, Tzianabos AO, Kasper DL. An immunomodulatory molecule of symbiotic bacteria directs maturation of the host immune system. Cell. 2005;122:107–18. doi: 10.1016/j.cell.2005.05.007. [DOI] [PubMed] [Google Scholar]
- 41.Mohebali N, Ekat K, Kreikemeyer B, Breitrück A. Barrier Protection and Recovery Effects of Gut Commensal Bacteria on Differentiated Intestinal Epithelial Cells In Vitro. Nutrients. 2020;12:2251. doi: 10.3390/nu12082251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nakajima A, Sasaki T, Itoh K, Kitahara T, Takema Y, Hiramatsu K, et al. A Soluble Fiber Diet Increases Bacteroides fragilis Group Abundance and Immunoglobulin A Production in the Gut. Appl Environ Microbiol. 2020;86:e00405–20. [DOI] [PMC free article] [PubMed]
- 43.Yoshida N, Emoto T, Yamashita T, Watanabe H, Hayashi T, Tabata T, et al. Bacteroides vulgatus and Bacteroides dorei Reduce Gut Microbial Lipopolysaccharide Production and Inhibit Atherosclerosis. Circulation. 2018;138:2486–98. doi: 10.1161/CIRCULATIONAHA.118.033714. [DOI] [PubMed] [Google Scholar]
- 44.Yoshida N, Yamashita T, Osone T, Hosooka T, Shinohara M, Kitahama S, et al. spp. promotes branched-chain amino acid catabolism in brown fat and inhibits obesity. iScience. 2021;24:103342. doi: 10.1016/j.isci.2021.103342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dziarski R, Park SY, Kashyap DR, Dowd SE, Gupta D. Pglyrp-Regulated Gut Microflora Prevotella falsenii, Parabacteroides distasonis and Bacteroides eggerthii Enhance and Alistipes finegoldii Attenuates Colitis in Mice. PloS One. 2016;11:e0146162. doi: 10.1371/journal.pone.0146162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.López-Contreras BE, Morán-Ramos S, Villarruel-Vázquez R, Macías-Kauffer L, Villamil-Ramírez H, León-Mimila P, et al. Composition of gut microbiota in obese and normal-weight Mexican school-age children and its association with metabolic traits. Pediatr Obes. 2018;13:381–8. doi: 10.1111/ijpo.12262. [DOI] [PubMed] [Google Scholar]
- 47.López-Almela I, Romaní-Pérez M, Bullich-Vilarrubias C, Benítez-Páez A, Gómez Del Pulgar EM, Francés R, et al. Bacteroides uniformis combined with fiber amplifies metabolic and immune benefits in obese mice. Gut Microbes. 2021;13:1–20. doi: 10.1080/19490976.2020.1865706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tankou SK, Regev K, Healy BC, Tjon E, Laghi L, Cox LM, et al. A probiotic modulates the microbiome and immunity in multiple sclerosis. Ann Neurol. 2018;83:1147–61. doi: 10.1002/ana.25244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kimura I, Ozawa K, Inoue D, Imamura T, Kimura K, Maeda T, et al. The gut microbiota suppresses insulin-mediated fat accumulation via the short-chain fatty acid receptor GPR43. Nat Commun. 2013;4:1829. doi: 10.1038/ncomms2852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Laverde Gomez JA, Mukhopadhya I, Duncan SH, Louis P, Shaw S, Collie-Duguid E, et al. Formate cross-feeding and cooperative metabolic interactions revealed by transcriptomics in co-cultures of acetogenic and amylolytic human colonic bacteria. Environ Microbiol. 2019;21:259–71. doi: 10.1111/1462-2920.14454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nicholson JK, Holmes E, Kinross J, Burcelin R, Gibson G, Jia W, et al. Host-gut microbiota metabolic interactions. Science. 2012;336:1262–7. doi: 10.1126/science.1223813. [DOI] [PubMed] [Google Scholar]
- 52.Boets E, Gomand SV, Deroover L, Preston T, Vermeulen K, De Preter V, et al. Systemic availability and metabolism of colonic-derived short-chain fatty acids in healthy subjects: a stable isotope study. J Physiol. 2017;595:541–55. doi: 10.1113/JP272613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shipman JA, Berleman JE, Salyers AA. Characterization of four outer membrane proteins involved in binding starch to the cell surface of Bacteroides thetaiotaomicron. J Bacteriol. 2000;182:5365–72. doi: 10.1128/JB.182.19.5365-5372.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Brigham CJ, Malamy MH. Characterization of the RokA and HexA broad-substrate-specificity hexokinases from Bacteroides fragilis and their role in hexose and N-acetylglucosamine utilization. J Bacteriol. 2005;187:890–901. doi: 10.1128/JB.187.3.890-901.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Liu B, Hong C, Huang RK, Yu Z, Steitz TA. Structural basis of bacterial transcription activation. Science. 2017;358:947–51. doi: 10.1126/science.aao1923. [DOI] [PubMed] [Google Scholar]
- 56.Otte C, Gold SM, Penninx BW, Pariante CM, Etkin A, Fava M, et al. Major depressive disorder. Nat Rev Dis Prim. 2016;2:16065. doi: 10.1038/nrdp.2016.65. [DOI] [PubMed] [Google Scholar]
- 57.Kessler RC, Bromet EJ. The epidemiology of depression across cultures. Annu Rev Public Health. 2013;34:119–38. doi: 10.1146/annurev-publhealth-031912-114409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mueller S, Saunier K, Hanisch C, Norin E, Alm L, Midtvedt T, et al. Differences in fecal microbiota in different European study populations in relation to age, gender, and country: a cross-sectional study. Appl Environ Microbiol. 2006;72:1027–33. doi: 10.1128/AEM.72.2.1027-1033.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The metagenomic sequencing data were deposited in the China National GeneBank DataBase (CNGBdb) (https://db.cngb.org/; project ID: CNP0001162).
All the source codes are available at https://github.com/mokmoksunshine/changes_of_gut_microbiota_severity.