Summary
Development of new non-addictive analgesics requires advanced strategies to differentiate human pluripotent stem cells (hPSCs) into relevant cell types. Following principles of developmental biology and translational applicability, here we developed an efficient stepwise differentiation method for peptidergic and non-peptidergic nociceptors. By modulating specific cell signaling pathways, hPSCs were first converted into SOX10+ neural crest, followed by differentiation into sensory neurons. Detailed characterization, including ultrastructural analysis, confirmed that the hPSC-derived nociceptors displayed cellular and molecular features comparable to native dorsal root ganglion (DRG) neurons, and expressed high-threshold primary sensory neuron markers, transcription factors, neuropeptides, and over 150 ion channels and receptors relevant for pain research and axonal growth/regeneration studies (e.g., TRPV1, NAV1.7, NAV1.8, TAC1, CALCA, GAP43, DPYSL2, NMNAT2). Moreover, after confirming robust functional activities and differential response to noxious stimuli and specific drugs, a robotic cell culture system was employed to produce large quantities of human sensory neurons, which can be used to develop nociceptor-selective analgesics.
Keywords: iPS cells, cell differentiation, neural crest, sensory neuron, nociceptor, drug testing, CEPT cocktail, analgesics, pain, opioid crisis
Graphical abstract
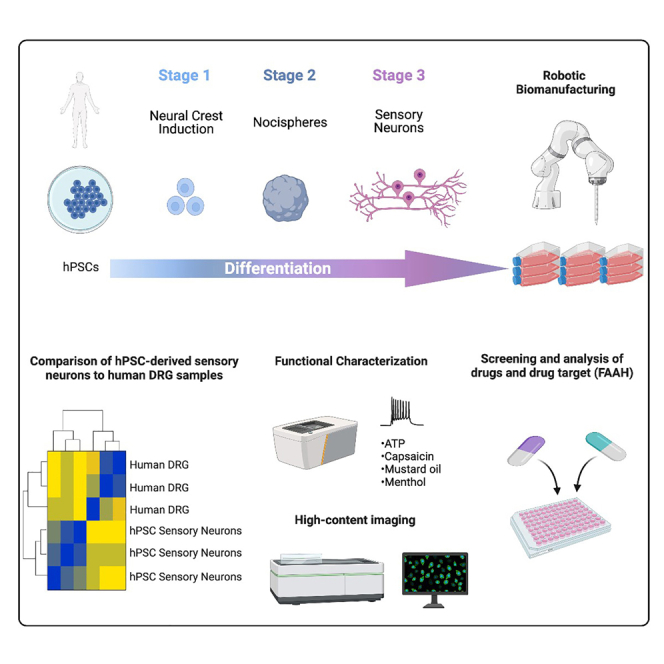
Highlights
-
•
Efficient method for neural crest and sensory neuron differentiation from hPSCs
-
•
Ultrastructural analysis of peptidergic and non-peptidergic neurons
-
•
Gene expression analysis shows high degree of similarity to native DRG neurons
-
•
Proof-of-concept for drug screening utilizing sensory neurons
Singeç and colleagues (Deng et al.) present a robust and reproducible sensory neuron differentiation method for human pluripotent stem cells that enable translational studies. Generation of pseudo-unipolar sensory neurons is scalable with a robotic platform and extensive molecular and functional characterization and comparison to DRG neurons and other published datasets establish an invaluable resource for pain research, disease modeling, and drug discovery.
Introduction
Chronic pain affects over 30% of the world population representing a major unmet medical need (Cohen et al., 2021; Yekkirala et al., 2017). Pain is a highly subjective experience and difficult to model during preclinical development. Traditional cellular models in pain research include primary rodent cells, heterologous expression of ion channels in non-neuronal cells (e.g., cancer cells, Xenopus oocytes), and in a few cases human postmortem dorsal root ganglion (DRG) neurons (Davidson et al., 2014; Kim et al., 2018; Usoskin et al., 2015). Animal models do not completely phenocopy human physiology and this may in part explain the poor efficacy or unexpected toxicity of many pain drug candidates in clinical trials (Jayakar et al., 2021; Mogil, 2009). Recent studies identified various species-specific differences at the molecular and cellular levels in human and mouse DRG neurons, suggesting that the organizational principles of the sensory system in mouse and humans are substantially different (Middleton et al., 2021; Shiers et al., 2020, 2021; Tavares-ferreira et al., 2022).
Nociceptors are specialized cell types of the peripheral nervous system (PNS) and critical for transmitting information on the presence of noxious stimuli from the body to the spinal cord and higher brain centers of the somatosensory system (Basbaum et al., 2009). The cell bodies of nociceptors are localized to DRGs and one split axon of each nociceptor projects to the periphery as well as to the spinal cord where the first synaptic contact of the nociceptive pathway is established with dorsal horn neurons as postsynaptic partners. Unlike neurons in the central nervous system (CNS) such as multipolar cortical neurons with elaborate basal and apical dendritic trees, nociceptors are anatomically unique in that they exhibit a pseudo-unipolar morphology and lack dendrites. Nociceptors express many ion channels, play a central role in inflammatory and neuropathic pain, and other chronic conditions due to congenital, metabolic, or iatrogenic causes (e.g., pain due to channelopathies, diabetic polyneuropathy, chemotherapy-induced polyneuropathy).
Self-renewing human pluripotent stem cell (hPSC) lines, including embryonic stem cells (hESCs) and induced pluripotent stem cells (iPSCs) represent an attractive source for generating human sensory neurons (Chambers et al., 2012; Desiderio et al., 2019; Eberhardt et al., 2015; Jayakar et al., 2021; McDermott et al., 2019; Namer et al., 2019; Saito-Diaz et al., 2021; Young et al., 2014). Alternative approaches are based on forced expression of transcription factors and lineage reprograming in mature cells to generate nociceptor-like cells (Blanchard et al., 2015; Wainger et al., 2015). Despite this progress, currently available methods are variable, difficult to scale up, and generate neurons that display only some aspects of human nociceptor physiology. Here, we present an efficient and reproducible differentiation method for peptidergic and non-peptidergic nociceptors derived from hPSCs. These cells can be generated in large quantities, cryopreserved using the CEPT small molecule cocktail (Chen et al., 2021), and used for translational studies and drug screens.
Results
Scalable production of human sensory neurons from hPSCs
All hPSCs used in this study were routinely cultured under feeder-free chemically defined conditions using E8 medium and vitronectin as a coating substrate. To ensure stress-free cell expansion, cells were passaged using EDTA, and treated with the CEPT small molecule cocktail for 24 h, which promotes cell viability and fitness (Chen et al., 2021). To differentiate neurons, hPSCs were plated at 1.5 × 105 cells/cm2 and treated with A83-01 (transforming growth factor-β inhibitor) and CHIR98014, a new GSK-3β inhibitor that activates the WNT signaling pathway (Figure 1A). Of note, when using the HotSpot kinase assay to test target specificity against 374 human kinases representing all major human kinase families (Anastassiadis et al., 2011), we found that CHIR98014 was more potent than CHIR99021, although both compounds showed identical target selectivity profiles (Figures S1A, and S1B and Table S1). Introducing CHIR98014 as a superior WNT agonist is important because it efficiently activates the WNT signaling pathway at lower concentrations than CHIR99021, which can be cytotoxic in the absence of knockout serum replacement (Tchieu et al., 2017). When hPSCs growing in adherent cultures were treated with A83-01 and CHIR98014 for 10 days, a mixture of SOX10+ and SOX10− cells emerged. Importantly, SOX10+ cells were exclusively present in areas where cells had spontaneously aggregated and formed three-dimensional (3D) structures (Figure S2A). These spheres also contained cells expressing the transcription factor ISL1 and the neuronal marker TUJ1 (TUBB3) (Figure S2A). When spheres were manually transferred and attached to Geltrex-coated plates, cells migrated out of the spheres and developed typical neuronal morphologies (Figures S2B). Based on these observations, we decided to dissociate the differentiating cells at day 3 and plate them into ultra-low attachment plates (AggreWell plates) to promote sphere formation (Figure 1A). These structures, which we termed “nocispheres,” were then treated until day 14 with a combination of four factors including CHIR98014, A83-01, DBZ (γ-secretase inhibitor), and PD173074 (Figures 1A and S2C). Of note, early dissociation of the nocispheres at day 8 demonstrated that virtually all cells expressed SOX10 (Figure S2D). The HotSpot kinase assay revealed that PD173074 potently and specifically inhibited the fibroblast growth factor (FGF) receptors 1/2/3. In contrast, the more commonly used SU5402 blocked FGFR1/2/3 but showed even stronger off-target activity against the TrkB and TrkC neurotrophin receptors (Figures S1C and S1D and Table S1). On day 14, nocispheres were dissociated into single cells and plated on Geltrex-coated dishes for further differentiation and maturation in a medium consisting of PD0332991 (CDK4/6 inhibitor) and a combination of neurotrophic factors (Figure 1A). Over the course of 2 weeks, the number of SOX10+ precursor cells decreased, whereas the number of neuronal cells expressing BRN3A (POU4F1), a specific transcription factor expressed by nociceptors, increased strongly (Figures 1B–1D). Accordingly, the number of Ki-67+ proliferating cells decreased during the differentiation of SOX10+ neural crest into neurons (Figure S3A). Since large quantities of human neurons are needed for high-throughput screening projects, the cell differentiation protocol was automated by using the CompacT SelecT platform (Figure S3B), a robotic cell culture system that allows reproducible scale-up and biomanufacturing of human cells (Tristan et al., 2021). By culturing cells in three bar-coded large T175 flasks and with minimal manual intervention, more than 150 million sphere-derived cells could be generated in 14 days in one experimental run (Figure S3B). Because the robotic platform can culture 90 flasks in parallel (Tristan et al., 2021), cell numbers can theoretically be increased 30-fold. At day 14, large quantities of cells can be cryopreserved using the CEPT cocktail or further differentiated until day 28 and used for the experiments described below.
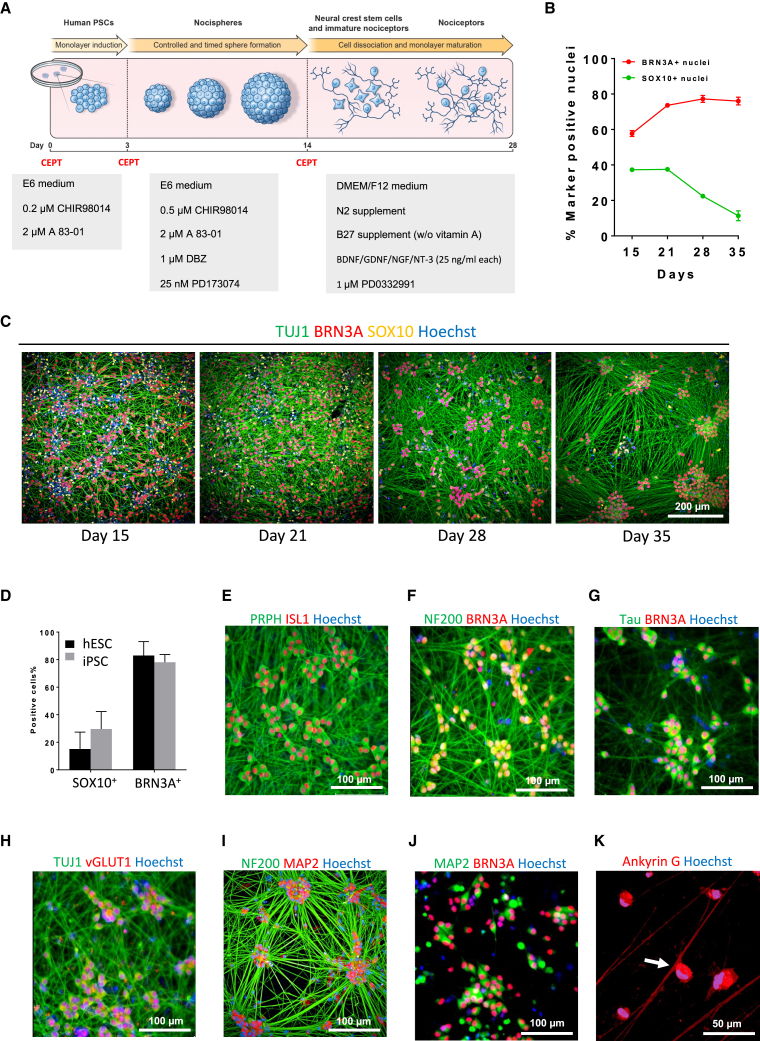
Figure 1.
Directed differentiation of hPSC into pseudo-unipolar nociceptors
(A) Schematic overview of nociceptor differentiation method.
(B) Quantitative analysis of cells expressing SOX10 and BRN3A during cell differentiation. Note that most cells express the pan-sensory marker BRN3A at days 28 and 35 (n = 3, mean ± SD). Data shown are from three different wells of one representative experiment (WA09) from multiple repeats.
(C) Representative images of differentiating cells (WA09) at different timepoints immunolabeled for neural crest marker SOX10 and neuronal markers TUJ1 and BRN3A.
(D) Quantification of cells expressing SOX10 or BRN3A derived from hESCs (WA09) and iPSCs (LiPSC-GR1.1) showing consistency of the differentiation method (n = 3, mean ± SD). Data shown are from three different wells of one representative experiment from multiple repeats.
(E–G) Representative images of nociceptor cultures (WA09) at day 28 expressing the axonal markers PRPH (peripherin), NF200, and transcription factors ISL1 and BRN3A.
(H) Sensory neurons derived from hESCs (WA09) expressing vGLUT1 consistent with their glutamatergic phenotype. See also Figure S3C.
(I and J) Immunostaining for NF200, MAP2, and BRN3A at day 28 (WA09). Note that MAP2 immunoreactivity is confined to cell bodies.
(K) Immunostaining for Ankyrin G and confocal microscopy showing axon splitting of nociceptors (WA09) (see also Figures S3F–S5H for confocal z stack images).
Morphological characterization of sensory neurons
Immunocytochemical analysis demonstrated that hPSC-derived nociceptors expressed neuronal markers TUJ1, peripherin, neurofilament 200 (NEFH), Tau (MAPT), and transcription factors BRN3A and ISL1 (Figures 1E–1G). Like DRG neurons, in vitro-generated nociceptors expressed vesicular glutamate transporter 1 (VGLUT1) and were immunoreactive for glutamate (Figures 1H and S3C). In these cultures, gamma-aminobutyric acid (GABA) neurons were only found very sporadically and cells expressing tyrosine hydroxylase, the rate-limiting enzyme for dopamine synthesis and a feature of low-threshold C-fiber mechanoreceptors (Lou et al., 2013), were not detected (Figures S3D and S3E). Neurons are polarized cells and MAP2 is a typical marker to label the somato-dendritic compartment of neurons in the CNS. Accordingly, immunostaining for MAP2 only labeled the cell bodies of nociceptors (Figures 1I and 1J). This is consistent with the pseudo-unipolar anatomy of DRG neurons, which develop a split axon but are devoid of dendrites. The peripheral part of the split axon extends to the periphery, such as the skin, as free nerve endings, whereas the central part of the same axon projects to the dorsal horn to establish synaptic contacts with spinal cord neurons. By using confocal microscopy and immunostaining for the axon initial segment marker ankyrin G (Rasband, 2010), we detected axon splitting in hPSC-derived sensory neurons (Figures 1K and S3F–S3H).
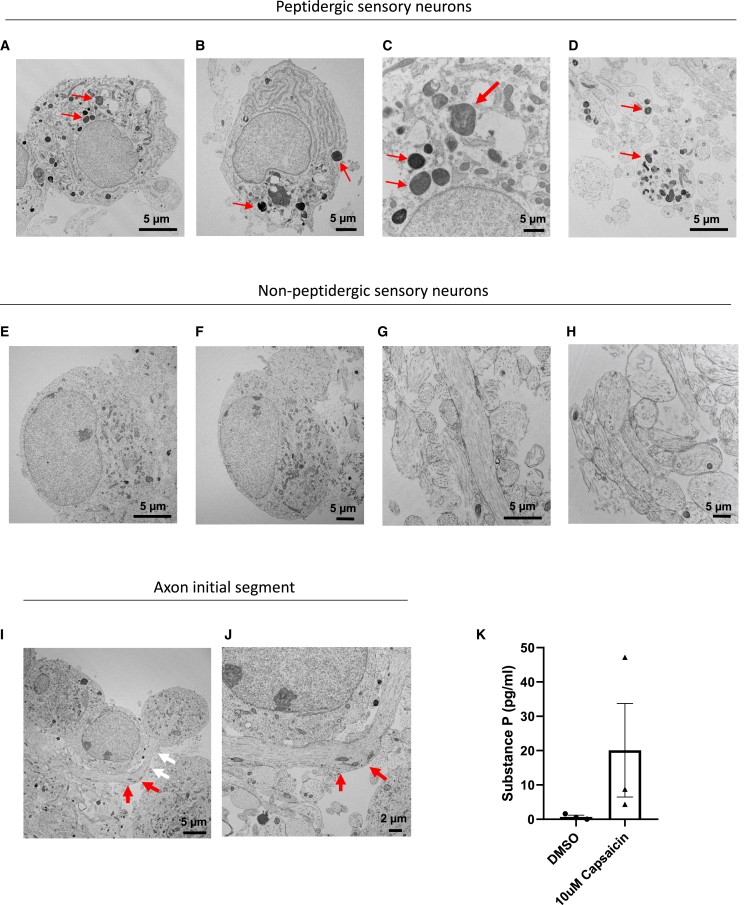
Next, to further characterize hPSC-derived sensory neurons, we performed electron microscopic analyses using day 28 cultures (Figure 2). At the ultrastructural level, two types of sensory neurons could be distinguished based on their morphology. The first presumably peptidergic cell type displayed irregularly shaped nuclei with nucleolemma invaginations, rich endoplasmic reticulum, and numerous large dense core vesicles (LDCVs) that were present in the soma as well as axons (Figures 2A–2D). The second presumably non-peptidergic cell type exhibited round nuclei of various sizes, numerous mitochondria, and was devoid of LDCVs (Figures 2E–2H). Notably, while large numbers of axonal profiles with and without LDCVs were detected in these cultures, dendritic structures were absent (Figures 2D, 2G, and 2H). Focusing on the axo-somatic region and axon initial segment, we found ultrastructural evidence for an axon directly emanating from the neuronal cell body (Figures 2I and 2J). To further validate the presence and functionality of the peptidergic cells, we treated day 28 nociceptors with capsaicin and confirmed that substance P was released into the cell culture medium as measured by an enzyme-linked immunosorbent assay (ELISA) (Figure 2K). Together, these findings provide evidence for efficient and scalable differentiation of hPSCs into sensory neurons with typical in vivo-like anatomic and immunophenotypic features.
Figure 2.
Ultrastructural analysis of peptidergic and non-peptidergic sensory neurons derived from hESCs (WA09)
(A–D) Electron microscopic images showing peptidergic nociceptors. Prominent large dense core vesicles (LDCVs; red arrows) are present in cell bodies (A–C) and in axonal profiles sectioned in different planes (D).
(E and F) Cell bodies of non-peptidergic nociceptor soma are devoid of LDCVs.
(G and H) Typical axonal profiles of non-peptidergic neurons lacking LDCVs.
(I) Overview image showing nociceptor cell bodies, axonal structures, and LDCVs. White arrows indicate the axon initial segment, which emanates from the cell soma. Red arrows point to more distal portions of the same axon.
(J) Higher magnification of the axon initial segment shown in (I).
(K) ELISA for substance P detection in the supernatant of day 28 nociceptors after DMSO and 10 μM capsaicin treatment. Data shown are from three different wells of one representative experiment using iPSC-derived nociceptors (NCRM5 cell line).
Transcriptomic analysis of directed differentiation
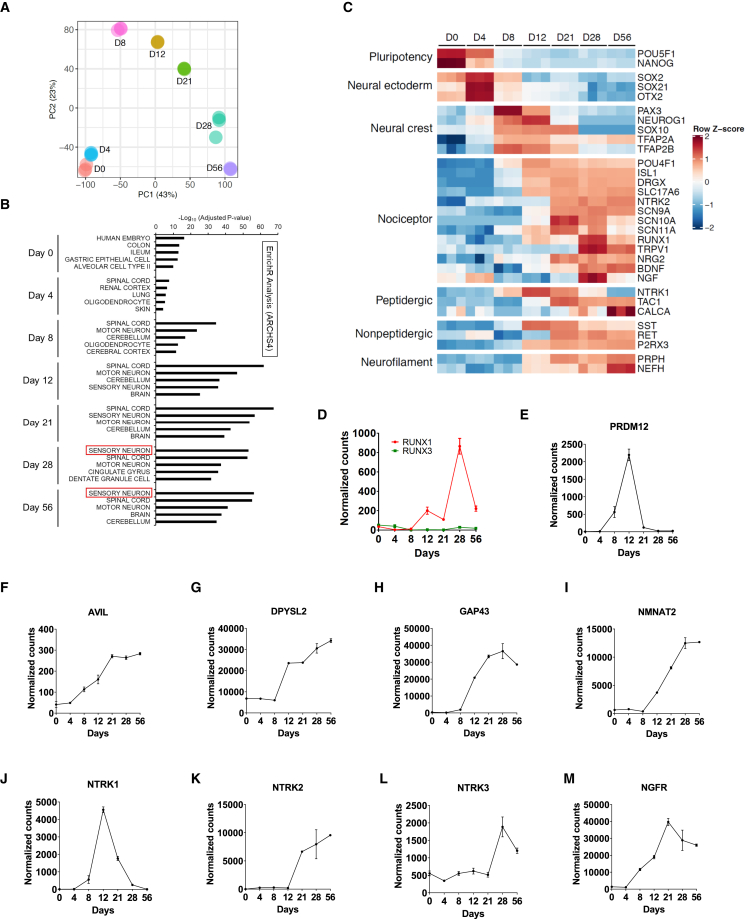
Time-course transcriptomic profiling (RNA sequencing [RNA-seq]) was performed to generate datasets that characterize the differentiation trajectory from pluripotent cells to neural crest and sensory neurons (Figure 3). Principal-component analysis (PCA) showed distinct molecular signatures of pluripotent cells (day 0) and of the differentiating cells harvested at day 4, 8, 12, 21, 28, and 56 (Figure 3A). Next, an unbiased comparison of transcriptomes was performed by using ARCHS4, which is an RNA-seq database including more than 84,863 human samples, followed by gene enrichment analysis (ENRICHR) (Lachmann et al., 2018). This approach revealed that “sensory neuron” was the top category at day 28 and 56, supporting the notion that the correct cell type was generated during in vitro differentiation (Figure 3B). Furthermore, gene expression changes were investigated at different timepoints by using a heatmap analysis (Figure 3C). Specific genes representing distinct stages and cell types were expressed or downregulated in a time-dependent fashion, consistent with neural crest lineage specification. Hence, the directed differentiation led to a downregulation of pluripotency-associated genes OCT4 (POU5F1) and NANOG and the induction of neural crest genes including SOX10, SOX21, and PAX3 (Figures 3C, S4A, and S4B). Upon further differentiation, these genes were then downregulated, followed by strong induction of non-specific neuronal genes as well as nociceptor-specific genes including RUNX1, BRN3A, ISL1, TRPV1, and other genes that indicated the presence of both peptidergic and non-peptidergic subpopulations (Figures 3C, 3D, and S4C–S4G). RUNX3 is important for the development of proprioceptive neurons (Levanon et al., 2002) and was absent throughout the entire differentiation period (Figure 3D). The transcription factor PRDM12 is important for specifying nociceptors and genetic mutations in this gene can lead to congenital insensitivity to pain (Bartesaghi et al., 2019; Desiderio et al., 2019). Transcriptomic analysis showed that expression of PRDM12 was strongly induced, reached maximum expression at day 12, and then sharply declined by day 21 to 28, consistent with a stage-specific developmental gene expression (Figure 3E). The sensory neuron-specific actin-binding protein advillin (AVIL) (Chuang et al., 2018) was upregulated during the differentiation (Figure 3F). Three genes with diverse functions in cell polarity and axonal growth and with important roles in neurological disorders and Wallerian degeneration (DPYSL2, GAP43, NMNAT2) were induced and strongly expressed across the differentiation period (Figures 3G–3I). Last, a time-dependent regulation of several neurotrophin receptors (NTRK1, NTRK2, NTRK3, NGFR) and three receptors for glial cell-derived neurotrophic factor (GFRA1, GFRA2, GFRA3) was confirmed (Figures 3J–3M and S4H–S4J), consistent with sensory neuron development in vivo (Ernsberger, 2009; Salio et al., 2014). To confirm temporal gene expression of nociceptor markers found in bulk RNA-seq experiments, we performed additional RT-qPCR experiments using three different hPSC lines and harvested samples at days 0, 14, and 28. The neural crest marker SOX10 was strongly expressed by day 14 in all cell lines (Figure S4K). Similarly, sensory neuron markers (ISL1, POU4F1, RUNX1, TRPV1) were strongly expressed at days 14 and 28 (Figure S4L). Finally, the peptidergic marker TAC1, non-peptidergic marker P2RX3, and PNS neuronal cytoskeletal protein PRPH (peripherin) were strongly expressed by day 28 (Figures S4M–S4O).
Figure 3.
Transcriptomic analysis of the differentiation of hPSCs (WA09) into neural crest and nociceptors
(A) PCA plot of RNA-seq experiment showing distinct molecular signatures at each timepoint.
(B) Gene enrichment analysis using ENRICHR and ARCHS4 database identify “sensory neuron” as the top category at days 28 and 56.
(C) Heatmap analysis showing stepwise differentiation of pluripotent cells into neural crest and nociceptors (n = 3). Data shown are from three different wells of one representative experiment.
(D–M) Time-course expression profile of different genes with importance in sensory neuron development.
Comparison of in vitro-generated neurons to human DRGs
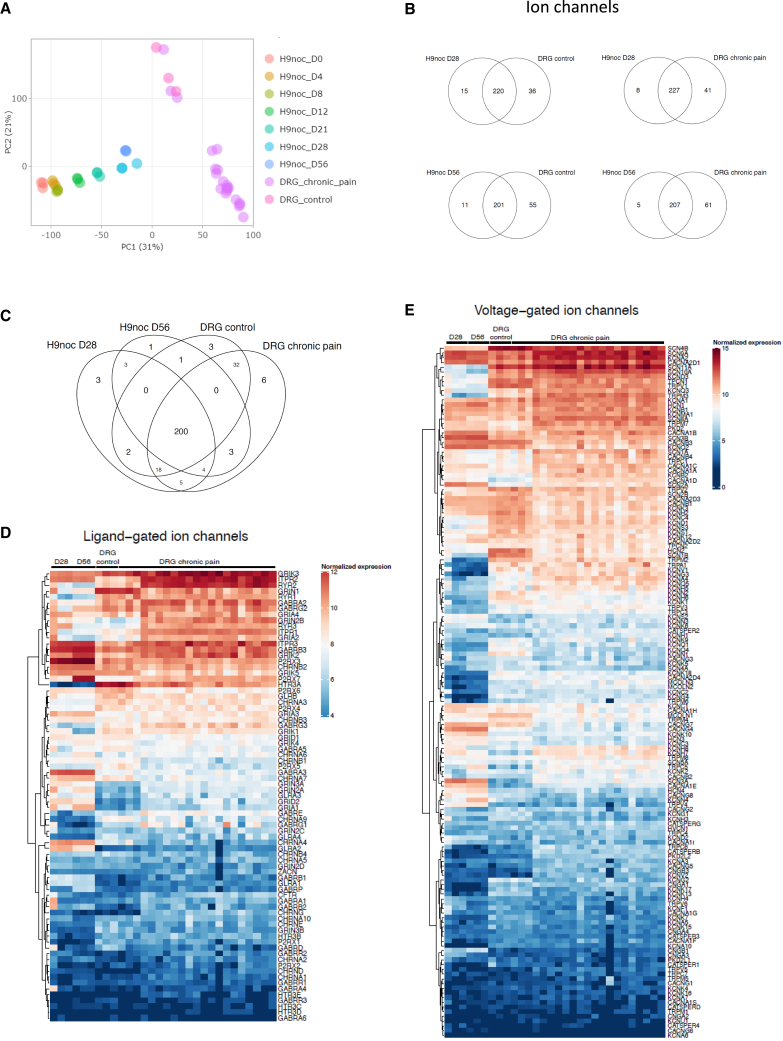
To compare the gene expression signature of hPSC-derived nociceptors to in vivo tissues, we used datasets of human DRGs established by the GTEx consortium (Aguet et al., 2017). Pluripotent and differentiating cells, harvested at different timepoints, were compared with adult human DRG samples. Because pain is associated with structural and functional plasticity (Jayakar et al., 2021), we also included tissue samples obtained from individuals with a clinical history of chronic pain. PCA plots showed that DRG samples clustered together and were distinct from cultured cells (Figure 4A). However, while the pluripotent state was most distant from DRGs, differentiating cells followed a differentiation trajectory toward the DRG signatures. Hence, the transcriptome of nociceptors at days 28 and 56 were closest to DRG samples (Figure 4A). Nociceptors are known to express large numbers of ion channels and receptors (Young et al., 2014). To establish a resource for disease modeling and drug discovery, we performed proof-of-principle analyses to validate the molecular signature of hPSC-derived nociceptors by profiling the expression of various gene families with a special focus on ligand- and voltage-gated ion channels (potassium, sodium, calcium, chloride) and other cell membrane proteins (porins, gap junction proteins). This systematic comparison revealed a striking overlap of genes expressed by derived nociceptors (days 28 and 56) and primary DRG neurons (Figures 4B–4E and S5). The high similarity of expressed genes in nociceptors and in vivo tissues provided additional evidence for the validity of this in vitro model. Next, we performed additional independent bulk RNA-seq experiments to compare the transcriptome of nociceptors generated with our method to commercially available human iPSC-derived nociceptors (Axol Bioscience), and to datasets from a recently published study (Zeidler et al., 2021). Heatmap analysis showed similarity between day 28 nociceptors and commercial nociceptors (Figure S6A) but upon closer analysis, differences were found regarding the expression levels of specific genes including RUNX1, NRG2, TAC1, PIEZO2, NGF, TRPC5, and TRPV1 (Figures S6B–S6H). Although a large fraction of genes (n = 188) was found to be expressed both by sensory neurons generated with the present method as well as the commercially obtained cells (Figure S7A), 47 genes were exclusively expressed by the nociceptors generated with the present method. Conversely, five genes were expressed in commercial cells but were absent from our nociceptors. Comprehensive gene expression heatmaps comparing all detected channel genes were prepared for future data mining (Figures S7B–S7I). To provide further molecular characterization, we compared the transcriptomic profiles of sensory neurons derived with our method to samples from the Nociceptra platform (Zeidler et al., 2021). After normalization by the RUVseq method (Risso et al., 2014), hPSC-nociceptors (days 28 and 56) were comparable to Nociceptra samples and clustered together in the PCA plot (Figure S8A). Furthermore, to find out if our dataset can be used to gain insights into the generation of specific DRG neuronal subpopulations, we performed targeted analyses based on a set of markers from a recent study that provided detailed profiling of primary human DRG neuronal subtypes (Tavares-Ferreira et al., 2022). Notably, this study found considerable differences between the classification system established based on previous rodent studies vs. DRGs from non-human primates and humans (Tavares-Ferreira et al., 2022). Considering both studies, our analysis revealed that markers of A-fiber neurons, including nociceptors (PLXNA2, TRAPPC3L), different types of Aβ low-threshold mechanoreceptors (KCNAB1, CALB2, NRG1, NTNG2, PPP1R1A), Aβ high-threshold mechanoreceptors (MCTP1, SYT6), and proprioceptors (ASIC1, ADGRG2) were expressed at higher levels in our hPSC-derived sensory neuron cultures as compared with the Nociceptra resource, whereas similar expression levels were observed in C-fiber markers including TRPM8 and PENK (proenkephalin) (Figure S8B). Together, these experiments validate the DRG-like identity of sensory neurons derived with our protocol and establish a valuable resource to further analyze the heterogeneity and polymodal nature of human sensory neurons.
Figure 4.
Molecular comparison of hPSC-derived nociceptors (WA09/H9) and human DRG samples
(A) PCA of differentiating cells harvested at different timepoints and DRG samples (normal and chronic pain patient).
(B and C) Venn diagrams comparing expressed ion channels in nociceptors (days 28 and 56) and DRG samples.
(D and E) Heatmap comparison of ligand- and voltage-gated ion channels expressed by in vitro-generated nociceptors and DRGs (n = 3). See Figure S5 for detailed analysis of other gene families. Data shown are from three different wells of one representative experiment.
Molecular targets for pain research and drug development
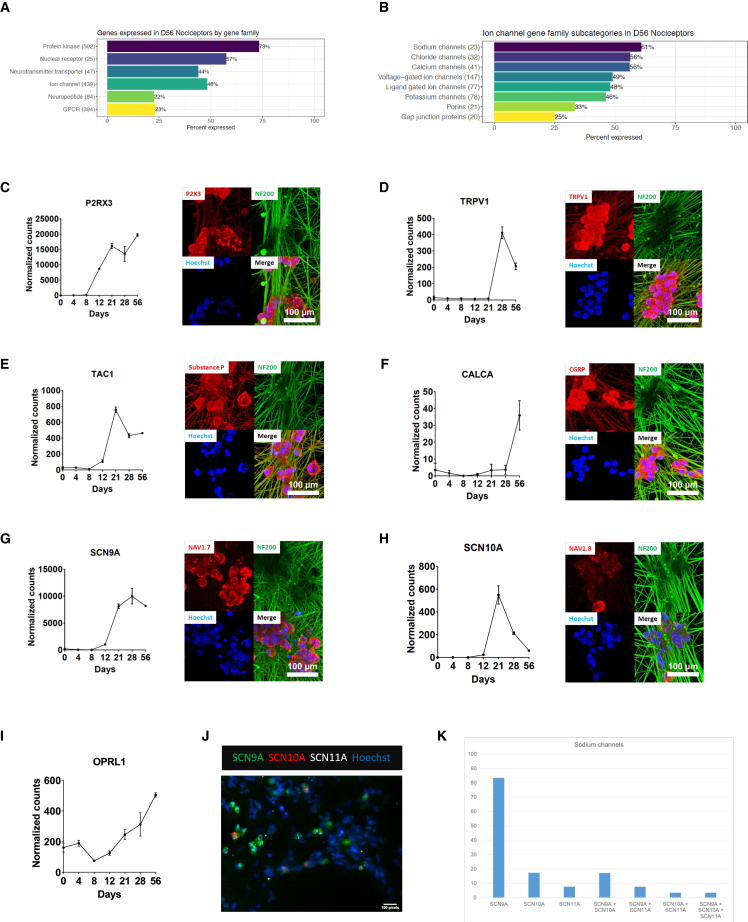
To generate hPSC-derived neurons for translational research, it is necessary to demonstrate consistent and reproducible derivation of large quantities of human nociceptors that express functional targets. We therefore performed a focused analysis of the transcriptome of hPSC-derived nociceptors and validated the expression of genes relevant for biomedical research. Using this approach, genes of interest expressed by nociceptors could be categorized into the following gene families: protein kinase, nuclear receptor, neurotransmitter transporter, ion channel, neuropeptide, and GPCR (Figure 5A). For instance, 502 protein kinases were expressed by nociceptors, which represents 73% of all protein kinases encoded by the human genome (Figure 5A). Notably, the nociceptors expressed 44%, 48%, and 22% of all known human neurotransmitter transporters, ion channels, and neuropeptides, respectively (Figure 5A). Moreover, the subcategories of expressed ion channels including sodium, chloride, and calcium channels were measured as well (Figure 5B). Next, we analyzed the expression of other biologically diverse targets upregulated during the differentiation process using RNA-seq and immunocytochemical analysis. Among these expressed targets were P2RX3, TRPV1, TAC1, CALCA, NaV1.7 (SCN9A), NaV1.8 (SCN10A), and the nociceptin receptor OPRL1 (Figures 5C–5I and Table S2). Expression of the sodium channel NaV1.9 (SCN11A) was detected on the transcript level (Table S2A) and additional experiments using quantitative in situ hybridization (RNAscope) provided insights into NaV1.7, 1.8, and 1.9 expression and co-localization at the cellular level (Figures 5J and 5K).
Figure 5.
Analysis of gene families and specific targets expressed by hPSC-derived nociceptors (WA09)
(A and B) Overview of genes and gene families expressed by hPSC-derived nociceptors (RNA-seq).
(C–I) Pain research-relevant targets expressed by nociceptors, as confirmed at the transcript and protein level (RNA-seq and immunocytochemistry) (n = 3, mean ± SD). Data shown are from three different wells of one representative experiment.
(J and K) Quantitative in situ hybridization (RNAscope) and analysis of sodium channel expression and co-expression of Nav 1.7, 1.8, and 1.9 in nociceptors.
Functional characterization of nociceptors
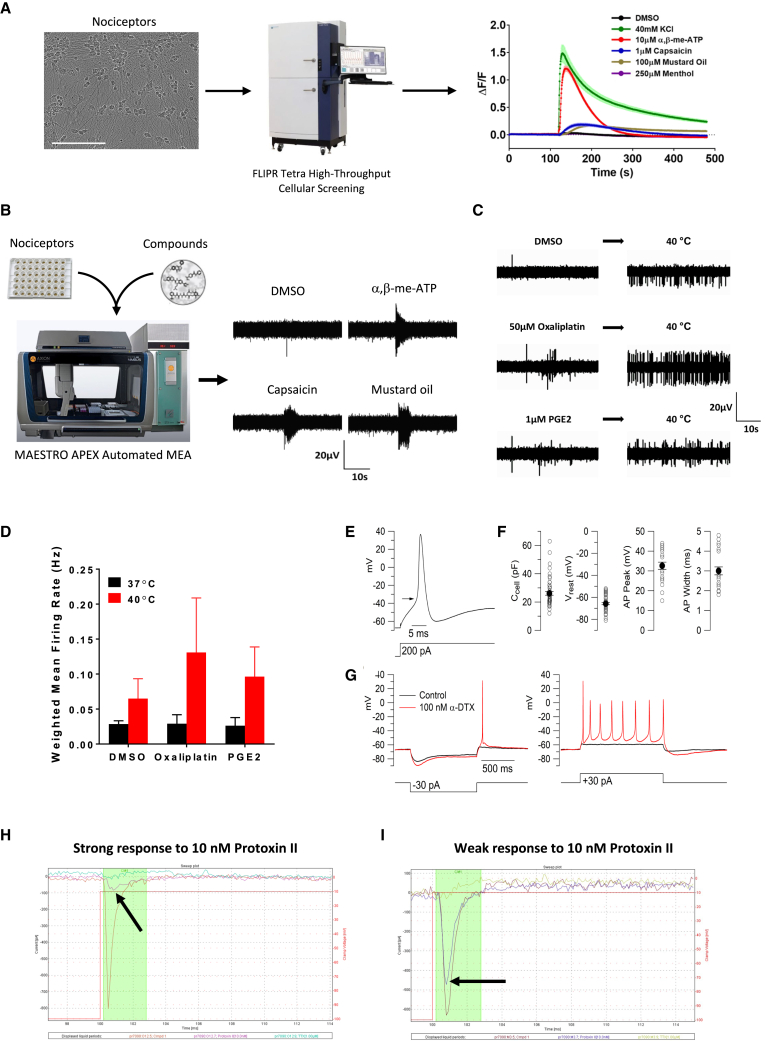
Unlike most neurons, primary nociceptors are not excited by synaptic release of transmitters from presynaptic neurons, but by specialized receptors that depolarize the neuron in response to noxious heat or cold, or molecules like ATP released during tissue damage (Caterina and Julius, 1999; Bautista et al., 2006; Woolf and Ma, 2007; Julius, 2013). As expected, morphological evidence for synapses was absent in the hPSC-derived nociceptors, but neurons could be excited by agonists for nociceptor-associated receptors. To investigate the functional properties of hPSC-derived nociceptors, we performed experiments using various methods, specific agonists, and stimulation protocols. First, calcium imaging was carried out using a cellular screening platform and nociceptors were exposed to DMSO (control), 40 mM KCl, 10 μM ATP, 1 μM capsaicin, 100 μM mustard oil, and 250 μM menthol (Figure 6A), with agonist concentrations chosen to ensure that only the respective cognate receptors were activated (Bautista et al., 2006; Wainger et al., 2015). The strongest and longest-lasting signal was elicited by 40 mM KCl, consistent with the expectation that this produces a large non-desensitizing depolarization of the neurons. Substantial responses were produced by α,β-methylene ATP (P2X receptors), capsaicin (TRPV1), and mustard oil (TRPA1) (Figure 6A). The different magnitudes and time-courses of activity evoked by the different agonists are consistent with similar experiments in native DRG neurons from mouse (Teichert et al., 2015; DuBreuil et al., 2021) and human (Enright et al., 2016) and suggest different levels of functional expression of the individual receptors. Further functional experiments using an automated multi-electrode array platform (Maestro APEX) confirmed that electrical activity of nociceptors could be evoked by specific ligands including α,β-methylene-ATP, capsaicin, and mustard oil (Figure 6B), similar to responses of mature native mouse and human nociceptors (Enright et al., 2016; Black et al., 2018). In independent experiments, when the temperature in the automated MEA system was increased from 37°C to 40°C, enhanced neuronal excitability was detected, suggesting the presence of temperature-sensitive receptors (Figures 6C and 6D). A typical feature of nociceptors is sensitization by inflammatory mediators and by chemotherapeutic drugs so that previously innocuous stimuli become painful (Gold and Gebhart, 2010). To test whether hPSC-derived nociceptors could be sensitized similar to a previous report (Wainger et al., 2015), we treated cells with the chemotherapeutic drug oxaliplatin and the inflammatory mediator PGE2 (prostaglandin E2) for 10 min (Figures 6C and 6D). To stimulate nociceptors, the temperature was then increased in the MEA system from 37°C to 40°C. Cells treated with oxaliplatin and PGE2 were more excitable when compared with DMSO controls, indicating that nociceptors could be sensitized in this in vitro assay (Figures 6C and 6D).
Figure 6.
Functional characterization of hPSC-derived nociceptors (WA09)
(A) Calcium flux analysis after stimulation of nociceptors with KCL, ATP, capsaicin, mustard oil, and menthol (n = 8, mean ± SD). Data shown are from eight different wells of one representative experiment.
(B) Automated MEA showing that nociceptors are activated by specific ligands.
(C and D) MEA experiment showing that nociceptors are stimulated by a temperature increase from 37 to 40°C and are presensitized after treatment with oxaliplatin or PGE2 (n = 3, mean ± SD). Data shown are from three different wells of one representative experiment.
(E) Typical action potential in nociceptors evoked by current injection. Arrow indicates the threshold voltage (−35 mV).
(F) Collected results for passive and active electrical properties of the neurons. Open circles show measurements from individual cells and closed circles show mean ± SEM (n = 53 for cell capacitance and resting potential; n = 25 for action potential peak and width).
(G) Enhancement of excitability by the Kv1-inhibitor α-dendrotoxin (DTX).
(H) Automated single-cell patch-clamp (Sophion Qube) recording showing effect of 10 nM Protoxin-II and 1 μM TTX (in the continuing presence of Protoxin-II) on sodium current in an hPSC-derived nociceptor.
(I) Collected results for percentage inhibition of sodium current by Protoxin-II alone and by Protoxin-II plus TTX (217 neurons analyzed).
To further investigate the electrical properties of hPSC-derived sensory neurons, we performed recordings using patch-clamp electrophysiology (Figures 6E–6G). Neurons had well-polarized resting potentials, with an average value of −65.7 ± 1.0 mV (mean ± SEM, n = 53) and had relatively high input resistances (391 ± 23 MOhms, n = 53), indicative of cell health. Almost all neurons (25 of 26) tested in current clamp mode with current injections of increasing size fired robust action potentials, with an average action potential height of 97 ± 2 mV (n = 25), reaching a peak voltage of +33 ± 2 mV (n = 25), with an average width of 3.0 ± 0.2 ms (measured at half-amplitude, n = 25). Most neurons (20 of 25) showed strong adaptation of action potential firing, firing a single action potential in response to current injections of up to 300 pA, similar to a subset of mature mouse primary nociceptors (Zheng et al., 2019). As with many native mouse DRG neurons, the strong adaptation appears to be conferred in part by the expression of Kv1 family potassium channels, which are highly effective in limiting firing in response to maintained current injections to a single action potential (Zheng et al., 2019). As with rodent nociceptors displaying this characteristic, the Kv1 inhibitor α-dendrotoxin enhanced the excitability of the neurons, decreasing the threshold current needed to evoke an action potential and causing repetitive firing during current injections (Figure 6G). Enhanced repetitive firing and decreased rheobase current were seen in four of five cells tested with 100 nM α-dendrotoxin.
Nociceptor-selective sodium channels are promising targets to develop new analgesics (Bennett et al., 2019; Jayakar et al., 2021). To test whether hPSC-derived nociceptors could be used in high-throughput fashion, we performed a functional screen using an automated patch-clamp system (Qube 384, Sophion Bioscience). To this end, day-28 neurons were dissociated and plated into single-hole 384-well QChips for patch-clamp recording. Voltage-dependent sodium currents were evoked by voltage steps to −10 mV from a holding potential of −100 mV. Application of 10 nM Protoxin-II (ProTx-II), a selective inhibitor of voltage-gated Nav1.7 channels (Schmalhofer et al., 2008), inhibited an average of 46.2% ± 15.7% (mean ± SD; n = 201) of the total sodium current and with subsequent application of 1 μM TTX together with Protoxin-II, an average of 88.5% ± 9.8% (mean ± SD, n = 191) of the total initial sodium current was inhibited (Figures 6H and 6I).
Elucidating drug effects using human nociceptors
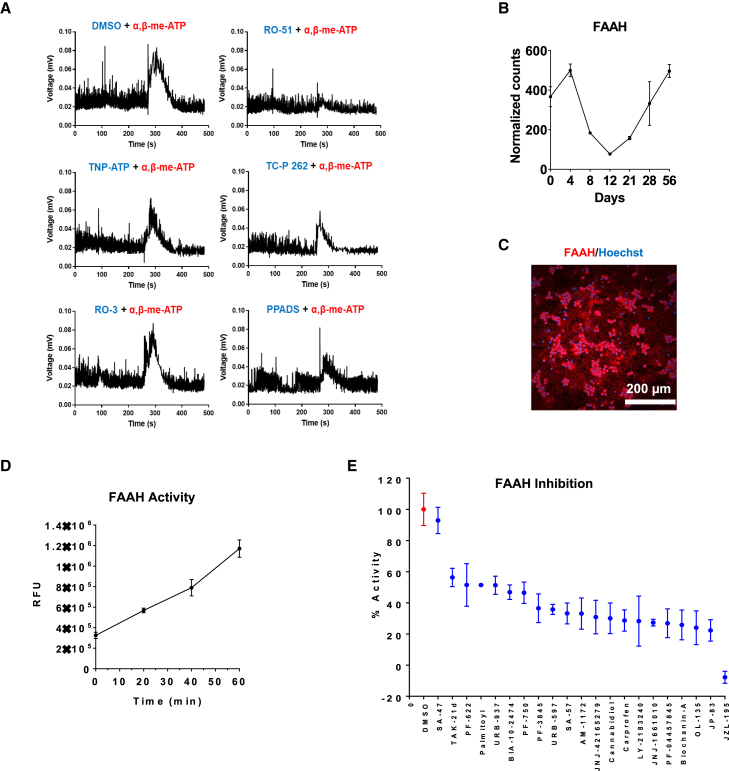
To further demonstrate translational utility of hPSC-derived nociceptors, we asked whether specific drugs could be studied in relevant assays. Such experiments are important because they provide information about the quality of human cells and molecular targets of interest, but also help characterize the efficacy and safety of established drugs or drug candidates in preclinical development. First, we tested several commercially available P2RX3 antagonists to find out whether it might be possible to identify the most potent inhibitor using a phenotypic readout. P2RX3 is a subtype of ionotropic purinergic receptors expressed specifically by nociceptors and involved in nociceptive transmission (Jahangir et al., 2009). Strong expression of P2RX3 was detected by RNA-seq, as described above (Figure 5C). The human nociceptors were pretreated with DMSO (control), four selective and potent inhibitors of P2RX3 (RO-51, TNP-ATP, TC-P 262, RO-3), and a non-selective P2 purinergic antagonist (PPADS) for 30 min, followed by stimulation with α,β-methylene-ATP, a specific purinergic receptor agonist. Automated MEA analysis revealed that RO-51 was the most potent inhibitor, completely blocking the effect of α,β-methylene-ATP in hPSC-derived nociceptors (Figure 7A).
Figure 7.
Drug testing using hPSC-derived nociceptors (WA09)
(A) MEA experiment comparing the potency of various P2RX3 inhibitors in human nociceptor cultures.
(B) Gene expression of FAAH during cell differentiation into nociceptors (RNA-seq) (n = 3, mean ± SD). Data shown are from three different wells of one representative experiment.
(C) Immunocytochemical analysis of FAAH expression in nociceptors (day 28).
(D) Real-time fluorescent assay monitoring FAAH activity in nociceptors (n = 8, mean ± SD). Data shown are from eight different wells of one representative experiment.
(E) FAAH inhibition assay testing the relative efficacy of 21 known inhibitors in day-28 nociceptor cultures (n = 8, mean ± SD). Data shown are from eight different wells of one representative experiment.
Rare genetic mutations that cause congenital insensitivity to pain can inform about new drug targets (Cohen et al., 2021). Fatty acid amide hydrolase (FAAH) is a critical enzyme responsible for the breakdown of several bioactive lipids such as anandamide, which act as endogenous ligands for cannabinoid receptors. FAAH knockout mice and patients with hypomorphic SNPs or mutations show hypoalgesia (Cravatt et al., 2001). Therefore, inhibition of FAAH emerged as a target for modulating pain but clinical trials have been unsuccessful thus far (Huggins et al., 2012; Kerbrat et al., 2016). Using the hPSC-derived nociceptors, we first confirmed that FAAH was expressed at the transcript and protein levels (Figures 7B and 7C). Next, we established an enzymatic assay to evaluate how FAAH catalyzes the hydrolysis of AAMCA (Arachidonyl 7-amino, 4-methly coumarin amide) into arachidonic acid and AMC (7-Amino 4 methyl coumarin) (Ramarao et al., 2005). Because AMC is fluorescent, the activity of FAAH can be continuously monitored over time in the human neurons (Figure 7D). Using this screening-compatible assay, we tested a panel of known FAAH inhibitors, including those that were unsuccessful in clinical trials (e.g., PF-04457845, BIA 10–2474, JNJ-42165279). When these compounds were administered to day 28 nociceptors, JZL-195 was identified as most efficient in blocking FAAH activity. Interestingly, SA-47 was previously reported to be a selective and potent FAAH inhibitor in rodent cells (Zhang et al., 2007) but showed poor efficacy in this assay using human cells. All the other inhibitors showed differential effects on FAAH activity (Figure 7E). In summary, this screen demonstrated that the in vitro-generated human nociceptors can play an important role in drug development since they express cell type-specific targets and enable the characterization of drugs, which offers new opportunities for human biology-based phenotypic drug screens.
Discussion
Development of new analgesics has largely failed over the past decades and the current opioid crisis in the United States underscores the need for effective and safe pain-relieving drugs without abuse liability, tolerance, and other adverse effects (Jayakar et al., 2021). Generation of well-characterized human nociceptors from hPSCs as an inexhaustible source is urgently needed for basic and translational pain research and drug development. Here, we developed a new method for the efficient production of sensory neurons from hPSCs. These in vitro-generated neurons were extensively characterized and displayed the unique structural, molecular, and functional properties of native sensory neurons of the DRG. Importantly, nociceptors showed robust expression of numerous molecular targets and ion channels. Screening experiments demonstrated utility in translational assays relevant for drug testing. The extensive datasets and systematic comparisons to DRG neurons represent an invaluable resource and should leverage disease modeling and toxicology studies (e.g., genetic diseases, chemotherapy-induced neuropathy). The development of our cell differentiation strategy was strongly guided by requirements for translational applicability, reproducibility, and scalability, which are all critical elements for rigorous assay development in pharmaceutical drug discovery. Because our method does not depend on additional cell sorting strategies, nociceptors can be efficiently generated at scale from different iPSC lines, cryopreserved, and used in a high-throughput fashion. Moreover, unlike previously published studies, our simplified method does not require the widely used dual-SMAD inhibition strategy, administration of “5i” inhibitors at high concentrations to sensitive cells or the ectopic expression of transcription factors (Chambers et al., 2012; Eberhardt et al., 2015; Nickolls et al., 2020; Saito-Diaz et al., 2021; Schwartzentruber et al., 2018; Tchieu et al., 2017). In our protocol, the use of small molecules at non-toxic concentrations also obviates the need for serum or knockout serum replacement (KSR). A previous report by Studer and colleagues also acknowledged that removal of KSR and using serum-free conditions required re-titration of the concentration of the widely used WNT agonist CHIR99021 (Tchieu et al., 2017). To further refine WNT pathway activation using the HotSpot kinase assay, we report that CHIR98014 has identical target specificity as CHIR99021 but is more potent and can therefore be used at lower concentrations (Figure S1). Similarly, we discovered that the FGF receptor inhibitor PD173074 used at a low concentration (25 nM) was more potent than the widely used SU5402 (0.5 μM). Of particular interest is the finding that SU5402, but not PD173074, showed off-target effects and inhibited TrkB and TrkC receptors (Figure S3). Because neutrophins and their receptors play important roles in early sensory neuron specification, and have been used for cell sorting (Ernsberger, 2009; Saito-Diaz et al., 2021), we caution that the use of SU5402 may compromise sensory neuron differentiation.
It is remarkable that neurons with a pseudo-unipolar morphology can be generated in vitro since positional molecular cues and secreted factors from the microenvironment, including chemo-attractants and chemorepellents, that typically guide axonal growth and pathfinding in the developing embryo are absent ex vivo. A previous report ectopically expressed Brn3a with either Ngn1 or Ngn2 in human and mouse fibroblasts and reported the emergence of pseudo-unipolar sensory neurons (Blanchard et al., 2015). However, reprogramming efficiency was low and definitive evidence and ultrastructural analysis were not provided. Similarly, other studies describing the differentiation of hPSCs into sensory neurons (Chambers et al., 2012; Eberhardt et al., 2015; Saito-Diaz et al., 2021) did not report the generation of neurons with distinct pseudo-unipolar anatomy. The structure-function relationship is particularly important for polarized neuronal cells and expression and localization of ion channels have distinctive patterns in cell bodies and different axon segments (e.g., proximal vs. distal, peripheral vs. central projection). Currently, little information is available on ion channel distribution in the DRG system (Jayakar et al., 2021). Further optimization of the presented protocol to streamline the production of pseudo-unipolar sensory neurons from human iPSCs should help to better understand ion channel biology and neuronal excitability with the goal of developing nociceptor-selective analgesics. The expression of marker genes that define a heterogeneous population of primary human DRG neurons, namely, A-fibers including nociceptors, Aβ low- and high-threshold mechanoreceptors and proprioceptors, show that this system more closely resembles an in vivo DRG-like model system, which can be used to comprehensively access drug effects on multiple cell types simultaneously. The robustness and scalability of the human sensory neuron model presented here should also pave the way for elucidating the precise molecular mechanisms of axonal degeneration in sensory neuropathies and regeneration after injury in future studies. Last, we anticipate that iPSC-based sensory neuron cultures will prove useful for the systematic testing of cell-autonomous effects and identification of the molecular substrates that contribute to gender-specific differences in pain biology.
Experimental procedures
Resource availability
Corresponding author
Further information and requests for resources and reagents should be directed to and will be fulfilled by the corresponding author, Ilyas Singeç (ilyassingec@gmail.com).
Materials availability
All materials and reagents generated in this study will be made available on request following NIH guidelines.
Data and code availability
Data are presented as the mean ± SD or SEM as indicated in figure legends. Statistical analyses (GraphPad Prism) were performed using different tests as appropriate and as described in figure legends. RNA-seq data have been deposited to the Sequence Read Archive (SRA) under Bioproject PRJNA783035 (https://www.ncbi.nlm.nih.gov/sra/?term=PRJNA783035). Data analysis scripts are available at https://github.com/cemalley/Deng_methods. Data representing human dorsal root ganglia were accessed through dbGaP (phs001158.v2.p1).
Cell culture
All hESCs (WA09; WiCell) and hiPSCs (LiPSC-GR1.1, NCRM5, CMT2A1.1, CMT2A2.1. CMT2A3.1) were maintained under feeder-free conditions in Essential 8 (E8) cell culture medium (Thermo Fisher Scientific) and vitronectin (Thermo Fisher Scientific) coated microplates or T175 flasks. Cells were passaged every 3 days. The hPSC colonies were treated with 0.5 mM EDTA in PBS without calcium or magnesium (Thermo Fisher Scientific) for 5 to 6 min to dissociate the hPSC colonies. The resulting cell clumps were counted using the Nexcelom Cellometer automated cell counter. The clumps were then plated at a density of 1.5 × 105 cells per cm2 in E8 cell culture medium supplemented with the CEPT cocktail (Chen et al., 2021) for the first 24 h. The CEPT cocktail, consisting of 50 nM Chroman 1 (#HY-15392; MedChem Express), 5 μM Emricasan (#S7775; Selleckchem), Polyamine supplement (#P8483, 1:1000 dilution; Sigma-Aldrich), and 0.7 μM Trans-ISRIB (#5284; Tocris), was used to improve cell survival and provide cytoprotection during cell passaging. Cells were maintained in a humidified 5% CO2 atmosphere at 37°C.
Automated cell culture
Scalable robotic cell culture and differentiation were carried out using the CompacT SelecT platform as previously described (Tristan et al., 2021) and following the steps described above. For sphere formation, T175 flasks were pretreated with anti-adherence solution (STEMCELL Technologies).
HotSpot kinase inhibitor profiling
Human kinase profiling against a panel of 374 wild-type kinases was performed by by Reaction Biology (Malvern, PA). Determination of half maximal inhibitory concentration (IC50) of CHIR99021, CHIR98014, SU5402, and PD173074 and other experimental details are presented in Table S1.
Author contributions
T.D., V.M.J., C.A.T., and I.S. conceived the study and experiments. T.D., V.M.J., C.W., P.C., J.I., S.R., J.Y., J.F.D.S., P.O., P.T., C.S., J.S., S.J., H.B.Z, S.J., W.Y., and C.A.T. performed experiments. T.D., V.M.J., C.A.T., T.C.V., A.S., W.Y., B.P.B., C.J.W., and I.S. contributed to data analysis and discussions. I.S. wrote the manuscript with contributions from T.D., V.M.J., C.A.T., B.P.B., and C.J.W.
Acknowledgments
We thank Paul Shinn, Misha Itkin, Zina Itkin, Carleen Klumpp-Thomas, John Braisted, Charles Bonney, Yeliz Gedik, and Steve Pittenger for their support throughout this work. We are grateful to Alan Hoofring and Ethan Tyler from the NIH Medical Arts Design Section for their technical expertise. The graphical abstract was created by using BioRender. Thanks to Mei Zhang and Sung Hoon Park at Sophion Bioscience for help with the automated patch-clamp experiments. The authors dedicate this work to their late colleague Paul Shinn. This work was funded by the NIH Common Fund (Regenerative Medicine Program), NIH HEAL Initiative, and the intramural research program of the National Center for Advancing Translational Sciences (NCATS), as well as the Defense Advanced Research Projects Agency (HR0011-19-2-0022, C.J.W./B.PB.), NIH NINDS (R35 NS105076, C.J.W.) and the Bertarelli Foundation (B.P.B., C.J.W.).
Conflict of interests
T.D., A.S., and I.S. are co-inventors on a US Department of Health and Human Services patent covering the nociceptor differentiation method and its utilization.
Published: April 11, 2023
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.stemcr.2023.03.006.
Supplemental information
Note that the WNT agonist CHIR98014 is more potent than CHIR99021. Similarly, PD173074 is more potent and more selective than SU5402 in inhibiting FGFR 1/2/3. See also Figure S1.
Time-course RNA-seq experiment and raw data of expressed transcripts in three different replicates per timepoint showing consistency of directed differentiation. Data shown are from three different wells of one representative experiment.
References
- Aguet F., Brown A.A., Castel S.E., Davis J.R., He Y., Jo B., Mohammadi P., Park Y.S., Parsana P., Segrè A.V., et al. Genetic effects on gene expression across human tissues. Nature. 2017;550:204–213. doi: 10.1038/nature24277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anastassiadis T., Deacon S.W., Devarajan K., Ma H., Peterson J.R. Comprehensive assay of kinase catalytic activity reveals features of kinase inhibitor selectivity. Nat. Biotechnol. 2011;29:1039–1045. doi: 10.1038/nbt.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black B.J., Atmaramani R., Kumaraju R., Plagens S., Romero-Ortega M., Dussor G., Price T.J., Campbell Z.T., Pancrazio J.J. Adult mouse sensory neurons on microelectrode arrays exhibit increased spontaneous and stimulus-evoked activity in the presence of interleukin-6. J. Neurophysiol. 2018;120:1374–1385. doi: 10.1152/jn.00158.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartesaghi L., Wang Y., Fontanet P., Wanderoy S., Berger F., Wu H., Akkuratova N., Bouçanova F., Médard J.J., Petitpré C., et al. PRDM12 is required for initiation of the nociceptive neuron lineage during neurogenesis. Cell Rep. 2019;26:3484–3492.e4. doi: 10.1016/j.celrep.2019.02.098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basbaum A.I., Bautista D.M., Scherrer G., Julius D. Cellular and molecular mechanisms of pain. Cell. 2009;139:267–284. doi: 10.1016/j.cell.2009.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bautista D.M., Jordt S.E., Nikai T., Tsuruda P.R., Read A.J., Poblete J., Yamoah E.N., Basbaum A.I., Julius D. TRPA1 mediates the inflammatory actions of environmental irritants and proalgesic agents. Cell. 2006;124:1269–1282. doi: 10.1016/j.cell.2006.02.023. [DOI] [PubMed] [Google Scholar]
- Bennett D.L., Clark A.J., Huang J., Waxman S.G., Dib-Hajj S.D. The role of voltage-gated sodium channels in pain signaling. Physiol. Rev. 2019;99:1079–1151. doi: 10.1152/physrev.00052.2017. [DOI] [PubMed] [Google Scholar]
- Blanchard J.W., Eade K.T., Szűcs A., Lo Sardo V., Tsunemoto R.K., Williams D., Sanna P.P., Baldwin K.K. Selective conversion of fibroblasts into peripheral sensory neurons. Nat. Neurosci. 2015;18:25–35. doi: 10.1038/nn.3887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caterina M.J., Julius D. Sense and specificity: a molecular identity for nociceptors. Curr. Opin. Neurobiol. 1999;9:525–530. doi: 10.1016/S0959-4388(99)00009-4. [DOI] [PubMed] [Google Scholar]
- Chambers S.M., Qi Y., Mica Y., Lee G., Zhang X.J., Niu L., Bilsland J., Cao L., Stevens E., Whiting P., et al. Combined small-molecule inhibition accelerates developmental timing and converts human pluripotent stem cells into nociceptors. Nat. Biotechnol. 2012;30:715–720. doi: 10.1038/nbt.2249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Tristan C.A., Chen L., Jovanovic V.M., Malley C., Chu P.H., Ryu S., Deng T., Ormanoglu P., Tao D., et al. A versatile polypharmacology platform promotes cytoprotection and viability of human pluripotent and differentiated cells. Nat. Methods. 2021;18:528–541. doi: 10.1038/s41592-021-01126-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuang Y.C., Lee C.H., Sun W.H., Chen C.C. Involvement of advillin in somatosensory neuron subtype-specific axon regeneration and neuropathic pain. Proc. Natl. Acad. Sci. USA. 2018;115:E8557–E8566. doi: 10.1073/pnas.1716470115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen S.P., Vase L., Hooten W.M. Chronic pain: an update on burden, best practices, and new advances. Lancet. 2021;397:2082–2097. doi: 10.1016/S0140-6736(21)00393-7. [DOI] [PubMed] [Google Scholar]
- Cravatt B.F., Demarest K., Patricelli M.P., Bracey M.H., Giang D.K., Martin B.R., Lichtman A.H. Supersensitivity to anandamide and enhanced endogenous cannabinoid signaling in mice lacking fatty acid amide hydrolase. Proc. Natl. Acad. Sci. USA. 2001;98:9371–9376. doi: 10.1073/pnas.161191698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson S., Copits B.A., Zhang J., Page G., Ghetti A., Gereau R.W. Human sensory neurons: membrane properties and sensitization by inflammatory mediators. Pain. 2014;155:1861–1870. doi: 10.1016/j.pain.2014.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desiderio S., Vermeiren S., Van Campenhout C., Kricha S., Malki E., Richts S., Fletcher E.V., Vanwelden T., Schmidt B.Z., Henningfeld K.A., et al. Prdm12 directs nociceptive sensory neuron development by regulating the expression of the NGF receptor TrkA. Cell Rep. 2019;26:3522–3536.e5. doi: 10.1016/j.celrep.2019.02.097. [DOI] [PubMed] [Google Scholar]
- DuBreuil D.M., Chiang B.M., Zhu K., Lai X., Flynn P., Sapir Y., Wainger B.J. A high- content platform for physiological profiling and unbiased classification of individual neurons. Cell Rep. Methods. 2021;1:100004. doi: 10.1016/j.crmeth.2021.100004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eberhardt E., Havlicek S., Schmidt D., Link A.S., Neacsu C., Kohl Z., Hampl M., Kist A.M., Klinger A., Nau C., et al. Pattern of functional TTX-resistant sodium channels reveals a developmental stage of human iPSC- and ESC-derived nociceptors. Stem Cell Rep. 2015;5:305–313. doi: 10.1016/j.stemcr.2015.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enright H.A., Felix S.H., Fischer N.O., Mukerjee E.V., Soscia D., Mcnerney M., Kulp K., Zhang J., Page G., Miller P., et al. Long-term non-invasive interrogation of human dorsal root ganglion neuronal cultures on an integrated microfluidic multielectrode array platform. Analyst. 2016;141:5346–5357. doi: 10.1039/c5an01728a. [DOI] [PubMed] [Google Scholar]
- Ernsberger U. Role of neurotrophin signalling in the differentiation of neurons from dorsal root ganglia and sympathetic ganglia. Cell Tissue Res. 2009;336:349–384. doi: 10.1007/s00441-009-0784-z. [DOI] [PubMed] [Google Scholar]
- Gold M.S., Gebhart G.F. Nociceptor sensitization in pain pathogenesis. Nat. Med. 2010;16:1248–1257. doi: 10.1038/nm.2235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huggins J.P., Smart T.S., Langman S., Taylor L., Young T. An efficient randomised, placebo-controlled clinical trial with the irreversible fatty acid amide hydrolase-1 inhibitor PF-04457845, which modulates endocannabinoids but fails to induce effective analgesia in patients with pain due to osteoarthritis of the knee. Pain. 2012;153:1837–1846. doi: 10.1016/j.pain.2012.04.020. [DOI] [PubMed] [Google Scholar]
- Jahangir A., Alam M., Carter D.S., Dillon M.P., Bois D.J.D., Ford A.P.D.W., Gever J.R., Lin C., Wagner P.J., Zhai Y., et al. Identification and SAR of novel diaminopyrimidines. Part 2: the discovery of RO-51, a potent and selective, dual P2X3/P2X2/3 antagonist for the treatment of pain. Bioorg. Med. Chem. Lett. 2009;19:1632–1635. doi: 10.1016/j.bmcl.2009.01.097. [DOI] [PubMed] [Google Scholar]
- Jayakar S., Shim J., Jo S., Bean B.P., Singeç I., Woolf C.J. Developing nociceptor-selective treatments for acute and chronic pain. Sci. Transl. Med. 2021;13:eabj9837. doi: 10.1126/scitranslmed.abj9837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Julius D. TRP channels and pain. Annu. Rev. Cell Dev. Biol. 2013;29:355–384. doi: 10.1146/annurev-cellbio-101011-155833. [DOI] [PubMed] [Google Scholar]
- Kerbrat A., Ferré J.C., Fillatre P., Ronzière T., Vannier S., Carsin-Nicol B., Lavoué S., Vérin M., Gauvrit J.-Y., Le Tulzo Y., et al. Acute neurologic disorder from an inhibitor of fatty acid amide hydrolase. N. Engl. J. Med. 2016;375:1717–1725. doi: 10.1056/NEJMoa1604221. [DOI] [PubMed] [Google Scholar]
- Kim J.-Y., Lam T., Torck A., Quigley L., Kim T.H., Dussor G., Price T.J., Neiman M., Rao C., Ray P., et al. Comparative transcriptome profiling of the human and mouse dorsal root ganglia. Pain. 2018;159:1325–1345. doi: 10.1097/j.pain.0000000000001217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lachmann A., Torre D., Keenan A.B., Jagodnik K.M., Lee H.J., Wang L., Silverstein M.C., Ma’ayan A. Massive mining of publicly available RNA-seq data from human and mouse. Nat. Commun. 2018;9:1366. doi: 10.1038/s41467-018-03751-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levanon D., Bettoun D., Harris-Cerruti C., Woolf E., Negreanu V., Eilam R., Bernstein Y., Goldenberg D., Xiao C., Fliegauf M., et al. The Runx3 transcription factor regulates development and survival of TrkC dorsal root ganglia neurons. EMBO J. 2002;21:3454–3463. doi: 10.1093/emboj/cdf370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lou S., Duan B., Vong L., Lowell B.B., Ma Q. Runx1 controls terminal morphology and mechanosensitivity of VGLUT3-expressing C-mechanoreceptors. J. Neurosci. 2013;33:870–882. doi: 10.1523/JNEUROSCI.3942-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDermott L.A., Weir G.A., Themistocleous A.C., Segerdahl A.R., Blesneac I., Baskozos G., Clark A.J., Millar V., Peck L.J., Ebner D., et al. Defining the functional role of Na V 1.7 in human nociception. Neuron. 2019;101:905–919.e8. doi: 10.1016/j.neuron.2019.01.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Middleton S.J., Barry A.M., Comini M., Li Y., Ray P.R., Shiers S., Themistocleous A.C., Uhelski M.L., Yang X., Dougherty P.M., et al. Studying human nociceptors: from fundamentals to clinic. Brain. 2021;144:1312–1335. doi: 10.1093/brain/awab048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mogil J.S. Animal models of pain: progress and challenges. Nat. Rev. Neurosci. 2009;10:283–294. doi: 10.1038/nrn2606. [DOI] [PubMed] [Google Scholar]
- Namer B., Schmidt D., Eberhardt E., Maroni M., Dorfmeister E., Kleggetveit I.P., Kaluza L., Meents J., Gerlach A., Lin Z., et al. Pain relief in a neuropathy patient by lacosamide: proof of principle of clinical translation from patient-specific iPS cell-derived nociceptors. EBioMedicine. 2019;39:401–408. doi: 10.1016/j.ebiom.2018.11.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nickolls A.R., Lee M.M., Espinoza D.F., Szczot M., Lam R.M., Wang Q., Beers J., Zou J., Nguyen M.Q., Solinski H.J., et al. Transcriptional programming of human mechanosensory neuron subtypes from pluripotent stem cells. Cell Rep. 2020;30:932–946.e7. doi: 10.1016/j.celrep.2019.12.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramarao M.K., Murphy E.A., Shen M.W.H., Wang Y., Bushell K.N., Huang N., Pan N., Williams C., Clark J.D. A fluorescence-based assay for fatty acid amide hydrolase compatible with high-throughput screening. Anal. Biochem. 2005;343:143–151. doi: 10.1016/j.ab.2005.04.032. [DOI] [PubMed] [Google Scholar]
- Rasband M.N. The axon initial segment and the maintenance of neuronal polarity. Nat. Rev. Neurosci. 2010;11:552–562. doi: 10.1038/nrn2852. [DOI] [PubMed] [Google Scholar]
- Risso D., Ngai J., Speed T.P., Dudoit S. Normalization of RNA-seq data using factor analysis of control genes or samples. Nat. Biotechnol. 2014;32:896–902. doi: 10.1038/nbt.2931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito-Diaz K., Street J.R., Ulrichs H., Zeltner N. Derivation of peripheral nociceptive, mechanoreceptive, and proprioceptive sensory neurons from the same culture of human pluripotent stem cells. Stem Cell Rep. 2021;16:446–457. doi: 10.1016/j.stemcr.2021.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salio C., Ferrini F., Muthuraju S., Merighi A. Presynaptic modulation of spinal nociceptive transmission by glial cell line-derived neurotrophic factor (GDNF) J. Neurosci. 2014;34:13819–13833. doi: 10.1523/JNEUROSCI.0808-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmalhofer W.A., Calhoun J., Burrows R., Bailey T., Kohler M.G., Weinglass A.B., Kaczorowski G.J., Garcia M.L., Koltzenburg M., Priest B.T. ProTx-II , a selective inhibitor of Na V 1 . 7 sodium channels , blocks action potential propagation in nociceptors. Mol. Pharmacol. 2008;74:1476–1484. doi: 10.1124/mol.108.047670. [DOI] [PubMed] [Google Scholar]
- Schwartzentruber J., Foskolou S., Kilpinen H., Rodrigues J., Alasoo K., Knights A.J., Patel M., Goncalves A., Ferreira R., Benn C.L., et al. Molecular and functional variation in iPSC-derived sensory neurons. Nat. Genet. 2018;50:54–61. doi: 10.1038/s41588-017-0005-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiers S., Klein R.M., Price T.J. Quantitative differences in neuronal subpopulations between mouse and human dorsal root ganglia demonstrated with RNAscope in situ hybridization. Pain. 2020;161:2410–2424. doi: 10.1097/j.pain.0000000000001973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiers S.I., Sankaranarayanan I., Jeevakumar V., Cervantes A., Reese J.C., Price T.J. Convergence of peptidergic and non-peptidergic protein markers in the human dorsal root ganglion and spinal dorsal horn. J. Comp. Neurol. 2021;529:2771–2788. doi: 10.1002/cne.25122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tavares-Ferreira D., Shiers S., Ray P.R., Wangzhou A., Jeevakumar V., Sankaranarayanan I., Cervantes A.M., Reese J.C., Chamessian A., Copits B.A., et al. Spatial transcriptomics of dorsal root ganglia identifies molecular signatures of human nociceptors. Sci. Transl. Med. 2022;14:eabj8186. doi: 10.1126/scitranslmed.abj8186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tchieu J., Zimmer B., Fattahi F., Amin S., Zeltner N., Chen S., Studer L. A modular platform for differentiation of human PSCs into all major ectodermal lineages. Cell Stem Cell. 2017;21:399–410.e7. doi: 10.1016/j.stem.2017.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teichert R.W., Schmidt E.W., Olivera B.M. Constellation pharmacology: a new paradigm for drug discovery. Annu. Rev. Pharmacol. Toxicol. 2015;55:573–589. doi: 10.1146/annurev-pharmtox-010814-124551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tristan C.A., Ormanoglu P., Slamecka J., Malley C., Chu P.H., Jovanovic V.M., Gedik Y., Jethmalani Y., Bonney C., Barnaeva E., et al. Robotic high-throughput biomanufacturing and functional differentiation of human pluripotent stem cells. Stem Cell Rep. 2021;16:3076–3092. doi: 10.1016/j.stemcr.2021.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Usoskin D., Furlan A., Islam S., Abdo H., Lönnerberg P., Lou D., Hjerling-Leffler J., Haeggström J., Kharchenko O., Kharchenko P.V., et al. Unbiased classification of sensory neuron types by large-scale single-cell RNA sequencing. Nat. Neurosci. 2015;18:145–153. doi: 10.1038/nn.3881. [DOI] [PubMed] [Google Scholar]
- Wainger B.J., Buttermore E.D., Oliveira J.T., Mellin C., Lee S., Saber W.A., Wang A.J., Ichida J.K., Chiu I.M., Barrett L., et al. Modeling pain in vitro using nociceptor neurons reprogrammed from fibroblasts. Nat. Neurosci. 2015;18:17–24. doi: 10.1038/nn.3886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woolf C.J., Ma Q. Nociceptors--noxious stimulus detectors. Neuron. 2007;55:353–364. doi: 10.1016/j.neuron.2007.07.016. [DOI] [PubMed] [Google Scholar]
- Yekkirala A.S., Roberson D.P., Bean B.P., Woolf C.J. Breaking barriers to novel analgesic drug development. Nat. Rev. Drug Discov. 2017;16:545–564. doi: 10.1038/nrd.2017.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young G.T., Gutteridge A., Fox H.D., Wilbrey A.L., Cao L., Cho L.T., Brown A.R., Benn C.L., Kammonen L.R., Friedman J.H., et al. Characterizing human stem cell-derived sensory neurons at the single-cell level reveals their ion channel expression and utility in pain research. Mol. Ther. 2014;22:1530–1543. doi: 10.1038/mt.2014.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeidler M., Kummer K.K., Schöpf C.L., Kalpachidou T., Kern G., Cader M.Z., Kress M. NOCICEPTRA: gene and microRNA signatures and their trajectories characterizing human iPSC-derived nociceptor maturation. Adv. Sci. 2021;8:e2102354. doi: 10.1002/advs.202102354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D., Saraf A., Kolasa T., Bhatia P., Zheng G.Z., Patel M., Lannoye G.S., Richardson P., Stewart A., Rogers J.C., et al. Fatty acid amide hydrolase inhibitors display broad selectivity and inhibit multiple carboxylesterases as off-targets. Neuropharmacology. 2007;52:1095–1105. doi: 10.1016/j.neuropharm.2006.11.009. [DOI] [PubMed] [Google Scholar]
- Zheng Y., Liu P., Bai L., Trimmer J.S., Bean B.P., Ginty D.D. Deep sequencing of somatosensory neurons reveals molecular determinants of intrinsic physiological properties. Neuron. 2019;103:598–616.e7. doi: 10.1016/j.neuron.2019.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Note that the WNT agonist CHIR98014 is more potent than CHIR99021. Similarly, PD173074 is more potent and more selective than SU5402 in inhibiting FGFR 1/2/3. See also Figure S1.
Time-course RNA-seq experiment and raw data of expressed transcripts in three different replicates per timepoint showing consistency of directed differentiation. Data shown are from three different wells of one representative experiment.
Data Availability Statement
Data are presented as the mean ± SD or SEM as indicated in figure legends. Statistical analyses (GraphPad Prism) were performed using different tests as appropriate and as described in figure legends. RNA-seq data have been deposited to the Sequence Read Archive (SRA) under Bioproject PRJNA783035 (https://www.ncbi.nlm.nih.gov/sra/?term=PRJNA783035). Data analysis scripts are available at https://github.com/cemalley/Deng_methods. Data representing human dorsal root ganglia were accessed through dbGaP (phs001158.v2.p1).