Abstract
In the early stage, N4-acetylcytidine (ac4C) was regarded as a conservative nucleoside present on tRNA and rRNA. Recently, studies have shown that ac4C also exists in human and yeast mRNA. N-Acetyltransferase-like protein 10 (NAT10) is the first enzyme to be found to catalyze ac4C production in eukaryotic RNA and has acetyltransferase activity and RNA-binding activity. Here, we first describe the structure and cellular localization of NAT10. Then, we conclude the active roles of NAT10 as the ac4C “writer” in mRNA stability and translation efficiency, oocyte maturation, bone remodeling, and fatty acid metabolism. With respect to disease, we focused on the promoting functions of NAT10 in proliferation, metastasis, and apoptosis in multiple tumors. The immune regulatory role of NAT10 in systemic lupus erythematosus and the maintenance role of NAT10 in virus RNA stability and replication in influenza A virus are also introduced. This review identifies NAT10 as a potential target for diagnosis, therapy, and prognosis in clinical application.
Keywords: MT: RNA/DNA Editing, NAT10, ac4C, acetyltransferase, writer, RNA modification
Graphical abstract
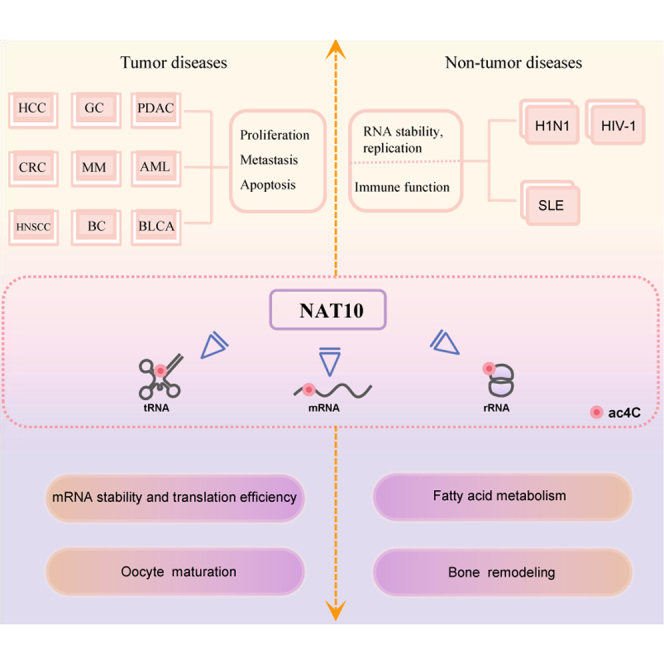
Chen and colleagues summarize characteristics and biological functions of NAT10 as an RNA acetyltransferase participating in N4-acetylcytidine modification, and focus on recent advances in NAT10 effects in diseases especially cancer, which provides a new perspective for NAT10 to be a potential diagnosis and treatment target in human diseases.
Introduction
To date, an increasing number of RNA modifications have been discovered,1,2 such as N6 methyladenosine (m6A),3 pseudouridine, 5-methylcytidine, and N4-acetylcytidine (ac4C), which are essential in mRNA stability, transcription, and translation, affecting a variety of cellular and biological processes.1,4,5 ac4C was recently identified as an mRNA modification with important roles in mRNA stability and translation,6 and was initially detected in bacterial transfer RNA (tRNA) anticodons7 and subsequently elaborated in eukaryotic serine and leucine tRNAs and 18S ribosomal RNA (rRNA).8 Arango et al. performed a localization analysis by using a transcriptome-wide approach and found that ac4C was mainly enriched at the 5′ end of the coding sequence (CDS), and a few were distributed at the 3′ end of the untranslated region (UTR). Further analysis showed that ac4C was strongly enriched at the swing site by using acRIP-seq which revealed the codon composition.9
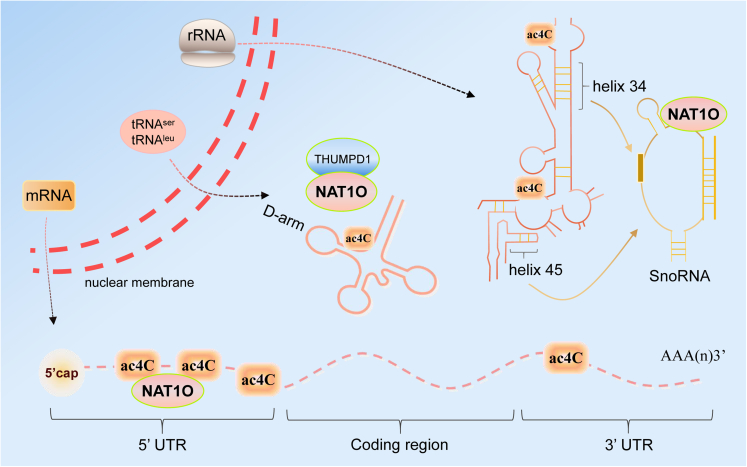
ac4C modification of mRNA, tRNA, and 18s rRNA is catalyzed by NAT10 or its homologs in both eukaryotes and prokaryotes (Figure 1). Initially, ac4C was found in the wiggle site of E. coli tRNAMet. Cytidine acetyltransferase (TmcA) catalyzes the formation of ac4C in the presence of acetyl coenzyme A (acetyl-CoA) and ATP,10 which prevents misreading of AUA’s isoleucine codon during protein biosynthesis and stabilizes the tertiary structure of tRNA molecules.11,12 Kre 33 acetyltransferase, another ac4C catalytic enzyme interacts with the conserved adaptor TAN 1 to catalyze ac4C modification at the C12 site of yeast tRNALeu and tRNASer, thereby promoting correct translation.13,14 In D. discoideum, ac4C is present at position 1844 in the 3′-terminal region of 18S rRNA.15 In S. cerevisiae, rRNA cytidyl acetyltransferase 1 (Rra1p) catalyzes the formation of ac4C 1773 using acetyl-CoA and ATP as substrates.16 NAT10 is a eukaryotic RNA acetyltransferase that was first discovered as a “writer” for the synthesis of ac4C.9 By binding to the D-arm structure of tRNASer and tRNALeu, the THUMP structural domain of THUMPD1 facilitates NAT10-catalyzed synthesis of the ac4C modification.13 NAT10 is a direct homolog of bacterial TmcA and yeast Rra1p in humans and mice, which catalyzes the formation of ac4C at the terminal helical position 1842 of mammalian 18S rRNA with the presence of acetyl-CoA and ATP, and participates in rRNA processing and ribosome formation. Knockout of the NAT10 gene leads to high-level accumulation of the 30S precursor of 18S rRNA, which results in human cell growth retardation.17 In addition, NAT10 is associated with U3 small nucleolar RNA and acetylates upstream binding factors to activate rRNA transcription.18
Figure 1.
NAT10 binds to cofactors to catalyze ac4C modification in mRNA, tRNA, and 18s rRNA in eukaryotic and prokaryotic cells
The antisense sequence of short nucleolar RNA (snoRNA) is necessary for NAT10 to bind to the target sequence of the ac4C modification of 18rRNA. When tRNA undergoes ac4C modification, THUMPD1 attaches to the tRNA and aids NAT10 to catalyze the formation of ac4C modification in the D-arm structure of tRNASer and tRNALeu.
However, whether or how ac4C production in NAT10-catalyzed mRNAs requires any other cofactors remains unknown. Oocyte maturation, bone remodeling, mRNA stability, and translation efficiency are only some of the many biological processes that are affected by NAT10-mediated ac4C alteration. This mechanism is strongly linked to tumor initiation, progression, and prognosis. The biological functions of NAT10 is discussed first in this review. Next, we provide a detailed description of the role that NAT10 plays in diseases especially cancer.
Roles of NAT10 in normal physiology and development
NAT10 structure and cellular localization
NAT10 is a vital member of the Gcn5-related N-acetyltransferase (GNAT) family and contains 1,025 amino acids with a molecular weight of 116 kDa. There are three conserved structural domains in the NAT10 protein: the N-terminal acetylase structural domain, the ATP/guanosine triphosphate (GTP) binding motif, and the ATPase structural domain.19 RNA ac4C modification requires acetyl-CoA for the provision of acetyl groups and ATP/GTP hydrolysis for energy.
NAT10 is commonly expressed in lymphoid tissue, kidney, liver, cerebellum, cerebral cortex, and the central nervous system during the embryonic period. Normal tissue cells express NAT10 in the nucleus, while tumor cells translocate NAT10 to the cytoplasm, nucleoplasm, and nuclear membrane, affecting tumor formation. For instance, hepatocellular carcinoma is characterized by an accumulation of NAT10 in both the nucleus and the cytoplasm.20 Recent investigations have demonstrated that NAT10 localization in distinct subcells depends on its NLS sequence, and deletion of approximately 30 conserved residues in the NAT10 CTE affects nuclear localization, leading to cytoplasmic accumulation.21
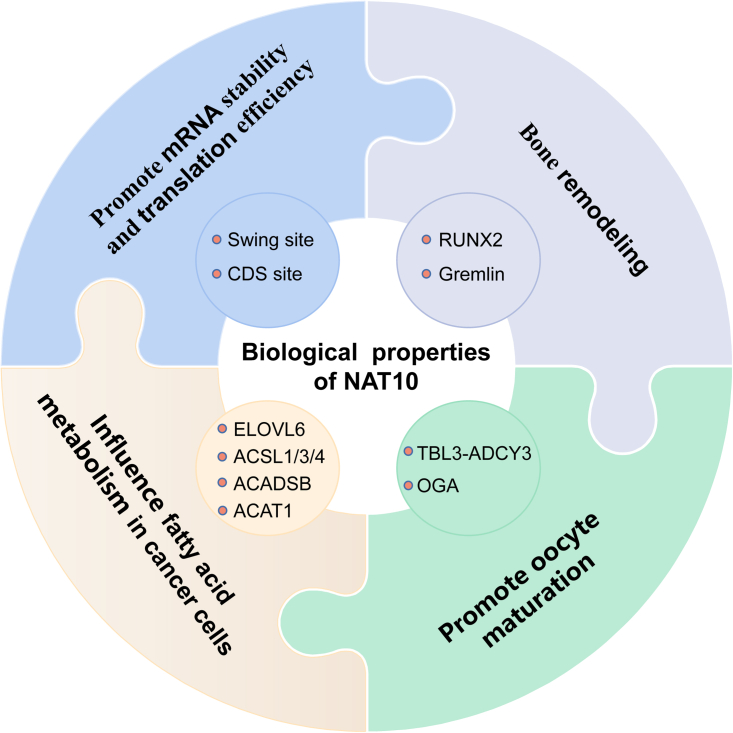
The biological functions of NAT10 in ac4C modification are mostly focused on four aspects: promotion of mRNA stability and translation efficiency, oocyte maturation, bone remodeling, and fatty acid metabolism (Figure 2).
Figure 2.
Biological function of NAT10
NAT10-modified mRNA ac4C enriched in the CDS region increases mRNA stability, and modifies the mRNA wobble site to promote translation efficiency; NAT10 can influence bone remodeling by altering ac4C modification of RUNX2 mRNA or Gremlin mRNA; NAT10-modified OGA mRNA ac4C influenced meiosis by TBL3-ADCY3 to promote oocyte maturation; fatty acid metabolism in cancer cells is modified by NAT10 through modifying ELOVL6, ACSL1, ACSL3, ACSL4, ACADSB, and ACAT1 mRNA ac4C.
Regulatory function of NAT10 in mRNA stability and translation efficiency
First, NAT10-mediated mRNA ac4C modification affects mRNA stability. When NAT10 is knocked out, ac4C in the CDS region is decreased, and the half-life of mRNA is considerably shortened, which further impacts the stability of mRNA. NAT10-modified mRNA ac4C is mostly concentrated in the CDS region. This is associated with the nucleic acid exonuclease Xrn1, as the major 5′ to 3′ exonuclease activity in cells. Xrn1 can degrade in-vitro-transcribed radiolabeled reporters that are generated in the presence or absence of ac4C. ac4C increases mRNA stability and promotes mRNA expression by separating it from nucleic acid exonuclease resistance.9 Since higher mRNA stability leads to enhanced translation, which in turn enhances mRNA stability,22 we know that mRNA degradation and translation are interrelated. Kumbhar et al. demonstrated that ac4C in the swing site stimulated translation in vitro and in vivo, and the acetylation of swing cytosine in tRNAMet prevented Watson Crick base-pairing site shielding to ensure strong association with guanosine, therefore correctly decoding methionine in bacteria.23 The efficiency of decoding in the human should be enhanced by mRNA acetylation, such as tRNA acetylation, which aids in the identification of mRNA codons in E. coli. Thus, cytidine inside the swing site is substantially enriched within the ac4C peak, as shown in the research on human HeLa mRNA, suggesting a direct involvement of ac4C in ribosomal decoding and facilitating translation efficiency.9 However, the impact of ac4C on translation occurs in a position-dependent manner. Arango et al. reported that ac4C within AUG-flanking Kozak sequences reduced the initiation and efficiency of translation in NAT10−/− HeLa cells.24
Promotion function of NAT10 in oocyte maturation
In regard to oocyte maturation, mRNA stability and posttranscriptional regulation are two of the most important factors that determine gene expression.25 Recent research has revealed that ac4C is also an important factor in determining the outcome of posttranscriptional regulation.9 It was demonstrated that siRNA-mediated NAT10 suppression in foaming-phase oocytes caused a reduction in ac4C modification, a decrease in the pace at which the first polar body is extruded, and a considerable delay in the maturation of meiotic cells in vitro. Oocyte maturation is a biological process that is linked to a protein called transducin beta-like protein 3 (TBL3), which has the capacity to connect to the ac4C molecule.26 Through its interaction with NAT10, TBL3 is able to modulate the activity of downstream cellular processes. However, it has not yet been identified whether TBL3 binds directly to the ac4C locus or whether it acts as a mediator of ac4C activity in oocytes. ac4C regulation is regulated by ADCY3 as a NAT10-mediated transcript.27 ADCY3 also shows a downregulation consistent with its trend and is therefore likely to be a NAT10-regulated downstream gene that affects meiosis.26 This is because, during oocyte maturation, the expression levels of both ac4C and NAT10 show a significant downward trend, moving from immature to mature oocytes. During this period, oocytes progress from immature to mature.
In addition, studies have revealed that NAT10 controls oocyte maturation by modifying ac4C to keep OGA mRNA stable.28 The lack of oocyte maturation due to a lack of OGA caused oscillations in O-GlcNAc maturation in vitro (IVM). Reducing OGA ac4C with NAT10 knockdown also lowered the stability of O-GlcNAc IVM, downregulated Rsph6a, Gm7788, and Gm41780, and upregulated Trpc7, Gm29036, and Gm47144. It is possible that Rsph6a and Trpc7 are significant downstream genes during NAT10-mediated OGA-ac4C modulation of oocyte maturation due to their essential roles in fertility.
Repair function of NAT10 in bone remodeling
Disorders of the skeletal metabolism are controlled by the GNAT family. N-Acetyltransferase 1 (NAT1), for instance, is overexpressed in luminal breast cancer (LBC), produces LBC bone metastases, and promotes osteoclast development. However, N-acetyltransfer protein 2 SNPs are linked to osteosarcoma progression and metastasis.29 Recent research has discovered that NAT10 acts as an anti-osteoporotic factor by encouraging the development of mesenchymal stem cells (MSCs) from bone marrow into osteoblasts. Silencing NAT10 decreased the level of ac4C modification in bone marrow mesenchymal stem cells (BMSCs) and caused them to form fewer calcium nodules in vitro, whereas overexpressing NAT10 caused them to form more calcium nodules.30 NAT10 expression and ac4C levels of total RNA were reduced in bone tissue from patients with osteoporosis. Runt-related transcription factor 2 (RUNX2) is a runt-related transcription factor that is important for osteogenic differentiation and has an effect on osteoblast development in BMSCs. It was discovered that the ac4C alteration of NAT10 could regulate the ac4C level of RUNX2, which in turn promoted osteogenic differentiation of BMSCs. BMSCs that were cultivated in osteogenic medium showed enhanced levels of RUNX2 mRNA, elevated RUNX2 mRNA half-life and protein expression, and improved the differentiation of BMSCs into osteoblasts.30
In addition, MSCs can benefit from NAT10’s ability to spur their development toward bone. Accelerating Gremlin 1 mRNA degradation by increasing NAT10 expression and increasing ac4C enrichment at the 3′ UTR of Gremlin 1 mRNA activates the BMP/Smad pathway, where BMPs (including BMP2, BMP4, BMP7, and BMP9) bind to BMPR1 or BMPR2,31 which then phosphorylates and activates downstream Smad1/5/9, regulating the osteogenic differentiation ability.32 Research has shown that RNA modifications lead to accelerated mRNA decay. The 3′ UTR m6A alteration of the MYB and MYC mRNAs speeds up their degradation and encourages leukemogenesis.33 Therefore, it may be tied to the 3′ UTR ac4C modification site, which is mediated through NAT10 in the gene gremlin 1 gene.
Improvement role of NAT10 in fatty acid metabolism
The protein known as NAT10 is significantly expressed in a number of different tumors, and has been correlated with the expansion and proliferation of cancer cells. In turn, biomolecules such as nucleic acids, proteins, and lipids are required for the development and multiplication of cancer cells.34,35 Triglyceride, diacylglycerol, cholesterol, phospholipids, and fatty acids are all examples of lipid species that are necessary for the production of energy and the maintenance of the structural integrity of membranes in both normal and cancerous cells.36,37 Furthermore, lipids play a role as signaling molecules in biological processes such as the maintenance of cell life, the progression of cell differentiation, and cell proliferation. Palmitate-loaded cells that were transfected with NAT10 siRNA were found to have a lower level of ac4C overall at the 5′ UTR.24 The CXX repeat sequence TCCDSCT was reported to be highly enriched inside the ac4C locus, as was predicted by sequencing analysis of the ac4C peak. Deletion of NAT10 in cancer cells significantly reduces the ac4C levels of ELOVL6, ACSL1, ACSL3, ACSL4, ACADSB, and ACAT1 mRNAs, which affects fatty acid synthesis, uptake, degradation, and storage.38 This dysfunctional fatty acid metabolism ultimately leads to cell death. Therefore, inhibiting NAT10 to improve fatty acid metabolism is an attractive cancer treatment option.
Role of NAT10 in tumors
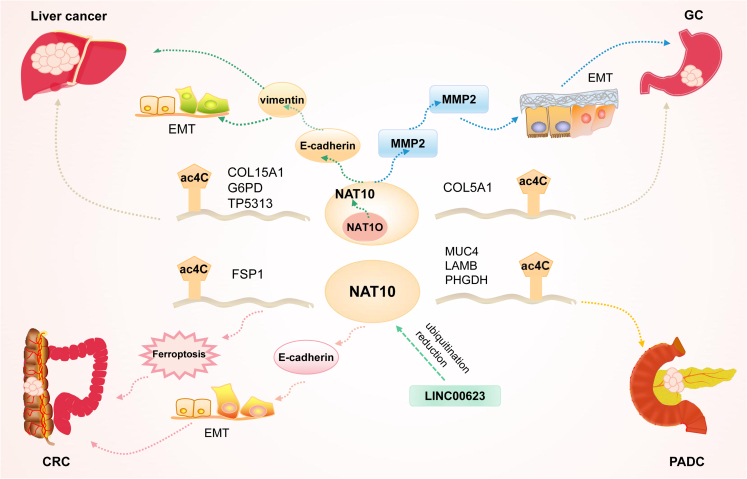
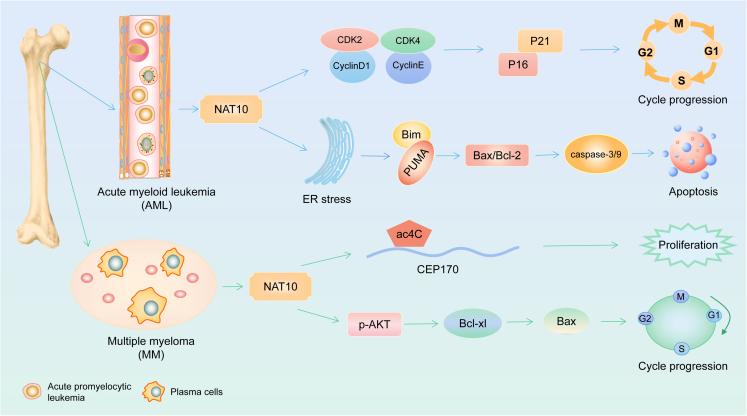
Many reports have shown that NAT10 plays a key role in gastrointestinal tumors especially by influencing epithelial-mesenchymal transformation (EMT) (Figure 3). NAT10 also plays an important role in the progression of hematological malignancies mainly through the regulation of cell-cycle progression, apoptosis, and cell proliferation (Figure 4).
Figure 3.
The mechanism of NAT10 in gastrointestinal tumors
NAT10 is extensively expressed in gastrointestinal cancers, and regulates ac4C modification of the associated RNA to influence carcinogenesis and progression.
Figure 4.
The mechanism of NAT10 in hematological malignancies
In AML, NAT10 influences the cell cycle by increasing the expression of p16 and p21 through cyclins like CDK2, CDK4, cyclin D1, and cyclin E; NAT10 causes endoplasmic reticulum stress, activates Bim and PUMA, interferes the Bax/Bcl-2-caspase-3/9 pathway, and promotes apoptosis. In MM, NAT10 influences CEP170 mRNA ac4C to enhance proliferation, and stimulates the PI3K-AKT-Bcl-xl-Bax pathway to alter the cell cycle.
Liver cancer
NAT10 is translocated from the nucleolus to the cytoplasm and cell membrane in hepatocellular carcinoma cells, which promotes invasive migration. Patient prognosis is strongly correlated with the degree to which NAT10 is redistributed from the nucleolus to the nuclear membrane, the cytoplasm, or the cell membrane.21 Upregulated expression of NAT10 in hepatocellular carcinoma interacts with and improves the stability of mutant p53, resulting in increased cell proliferation and higher tumorigenic activity in hepatocellular carcinoma cells, all of which contribute to a poor prognosis for patients.39 The EMT of hepatocellular carcinoma cells was altered and tumor accretion and metastasis were inhibited when NAT10 expression was downregulated, thereby upregulating the expression of the epithelial marker E-cadherin, and downregulating the expression of the mesenchymal marker Vimentin.40 Biological information analysis revealed that NAT10 causes ac4C changes in COL15A1, G6PD, and TP53I3, with the alterations being concentrated in the 5′ UTR and CDS regions.9 This risk model is applicable not only to predict stem cell and TME distribution and survival outcomes in patients with liver cancer but also in patients with lung and pancreatic cancer.41
Gastric cancer
Gastric cancer (GC) is one of the most common malignancies, and deep local infiltration or distant metastasis is usually associated with poor treatment and poor prognosis.42 It was discovered that GC tissues express NAT10 at considerably higher levels, which aids in invasion and migration, connecting with the role of NAT10 in EMT, since increased NAT10 expression stimulates VIM and MMP2 expression.5 COL5A1 is a tumor cell marker for EMT-II and has been shown to enhance EMT in a direct fashion.43,44 Overexpression of NAT10 was found in subsequent investigations to increase COL5A1 expression. In addition, COL5A1 overexpression in GC cells counteracted the effect of NAT10 in promoting tumor invasion and migration by downregulating EMT. Additional research using acRIP-seq and RIP-seq studies revealed that COL5A1 is a direct target of NAT10-mediated mRNA ac4C modification and that NAT10 controls the ac4C modification of the COL5A1 mRNA 3′ UTR by direct contact.45 Because of its function in controlling the COL5A1 mRNA ac4C pathway, NAT10 is very important for both GC transfer and EMT of GC cells.
Pancreatic ductal adenocarcinoma
LncRNAs can usually interact with regulatory molecules such as proteins, miRNAs, and DNA to exert biological effects.46 A recent study found that lncRNA LINC0062 expression was upregulated in pancreatic ductal adenocarcinoma (PDAC) tissues, promoting tumor migration growth, and influencing tumorigenesis development.47 Subcellular localization enrichment analysis (GO-CC) showed NAT10 as a potential protein bound to LINC00623. LINC00623 RNA interacts with NAT10, prevents its degradation and reduces its ubiquitination. The protein level of NAT10 was drastically lowered when LINC00623 was silenced and elevated when LINCO0623 was overexpressed. Foci formation and BxPC-3 cell migration were both drastically decreased when NAT10 was knocked down. Overexpressing NAT10, however, reversed the effects of LINC00623 knockdown on foci formation and migration.47 Most locations enriched in CDS for NAT10 regulation in PDAC are located in genes involved in cell proliferation, cell adhesion, and extracellular matrix organization (MUC4, LAMB3, and PHGDHR). Reduced expression and a decrease in ac4C alterations were observed for MUC4, LAMB3, and PHGDHR after NAT10 was knocked down. There was considerably greater expression of MUC4, LAMB3, and PHGDHac4C in tumor tissues than in normal tissues.48,49,50,51 As a result, NAT10, which remodels ac4C alterations in PDAC, keeps oncogenic mRNAs stable, and boosts their translation efficiency, is critical for the action of LINC00623 on PDAC malignancies.
Colorectal cancer
Colorectal cancer (CRC) cells are able to migrate and invade more easily, which might be the result of upregulated NAT10 expression in CRC.52 Importantly, miR-6716-5p downregulates E-calmodulin levels by suppressing NAT10 expression, which aids in the EMT and promotes CRC cell motility and invasion.53 Overall survival is reduced when NAT10 is underexpressed in CRC cells but is increased if NAT10 is overexpressed. In-depth examination of the role of NAT10 in colon cancer cells found that it regulates iron death suppressor protein 1 (FSP1) mRNA stability and expression. The fact that NAT10 controls acetylation of ac4C on FSP1 mRNA and blocks iron death raises the possibility that NAT10 might be utilized as a prognostic and therapeutic target for CRC.54
Multiple myeloma
The multiple myeloma (MM) clinical database analysis revealed that elevated NAT10 expression leads to poor prognosis in MM patients and promotes MM cell proliferation in vitro. NAT10 regulates the acetylation status of total RNA. Increased NAT10 raises ac4C levels, whereas NAT10 silencing decreases ac4C acetylation in MM cells. The mRNA acetylation regulator NAT10 improves translation efficiency in cells, which in turn stimulates cell growth.55 CEP170 is a centrosomal protein that promotes the development and spread of cancer, which was stabilized and its translation efficiency was increased due to high expression of NAT10 and enrichment of ac4C acetylation levels at the C terminus of the mRNA encoding CEP170.56,57,58 Notably, mitosis relies on NAT10,59,60 which is connected with the nuclear membrane and plays a significant role in cell division. Cell proliferation in MM has been studied extensively, and MTT assays have proven that CEP170 is a key factor in this growth promotion. Due to its interaction with CEP170, NAT10 increases CIN and hastens the development of MM.56 The PI3K-AKT pathway and the Bcl-2 family of proteins work together functionally to inhibit apoptosis and promote cancer.61,62 Human malignancies frequently exhibit abnormalities in the cell cycle, and apoptosis is intrinsically linked to the cell cycle. Increasing expression of cyclin-dependent kinases 4 and 6 (CDK4 and CDK6) and a reduction in the G0/G1 phase have been linked to NAT10 overexpression in MM cells. Likewise, upstream p-AKT is enhanced to activate the PI3K-AKT pathway and promote MM cell proliferation,55 yet increasing anti-apoptotic BCL-XL suppresses the downstream apoptotic protein BAX.
Acute myeloid leukemia
NAT10 plays a significant oncogenic role in numerous cancers. As a predictive and therapeutic biomarker for AML,63 NAT10 is upregulated in acute myeloid leukemia (AML) cells with poor survival along with the elevated expression of p16 and p21, while the expression of CDK2, CDK4, cyclin D1, and cyclin E was downregulated when NAT10 expression was low. Inhibition of NAT10-mediated cell proliferation and cell-cycle progression in AML cells occurs with overexpression of p16 and p21 tumor suppressors and downregulation of cell-cycle checkpoint proteins. Classical apoptosis was activated after NAT10 was inhibited by its inhibitor Remodelin or by deletion of NAT10.64 Activation of the Bax/Bcl-2 axis has been correlated with NAT10 inhibition-induced apoptosis in AML cells. Bax/bcl-2 apoptotic signaling is activated in response to endoplasmic reticulum stress,65 while the BH3 proteins Bim and PUMA not only drive endoplasmic reticulum-induced apoptotic signaling66,67 but are also required for Bax/Bak activation.68 The loss of NAT10 caused cells to experience endoplasmic reticulum stress, leading to an increase in the expression of Bim and PUMA as well as the activation of the Bax/Bcl-2 axis-mediated caspase-3/9. Therefore, increasing endoplasmic reticulum stress in AML cells is one way to inhibit cell growth and promote death,64 which can be achieved by targeting NAT10. Theoretically, NAT10’s oncogenic activities in AML warrant further investigation as a possible therapeutic target in AML.
Human head and neck squamous carcinoma
Studies have shown that NAT10 is more highly expressed in human head and neck squamous carcinoma (HNSCC) tumor cells than in normal epithelial cells. High levels of NAT10 expression are connected to a worse prognosis for patients, as measured by overall survival.69 Remodelin or siRNA, which suppresses NAT10 expression, prevents HNSCC cell lines from replicating and stops them from migrating and invading. siRNA-mediated knockdown of NAT10 led to more prominent S/G2 phase cell-cycle arrest.69 The NAT10 genome is enriched for MYC, E2F, G2M checkpoints, mTORC1, DNA repair, and oxidative phosphorylation, as shown by a genomic enrichment analysis.70 More research is needed to show whether NAT10 in HNSCC also influences cell-cycle pathways via acetylation of mTORC1. The MYCT1/NAT10 axis has been found to increase laryngeal cancer cell motility,71 and MYC targets have the greatest normalized enrichment score across all genomes. Specifically, Remodelin suppresses NAT10 expression in HNSCC cell lines, downregulates MYC expression, and upregulates LDHA expression to facilitate tumor spread.69 Thus, NAT10 is a significant player in HNSCC and a promising predictive biomarker for HNSCC patients.
Breast cancer
NAT10, which is highly expressed in breast cancer, acetylates MORC2 at the K767 site regulating DNA damage-induced dephosphorylation of H3T11 and transcriptional repression of CDK1 and cell-cycle protein B1.72 Acetylated MORC2 binds to histone H3 phosphorylation at threonine 11 (H3T11P) (phosphorylation of histone H3 on threonine 11), reduces DNA damage-induced H3T11P, represses transcription of its downstream target genes CDK1 and cyclin B1, and activates the DNA damage-induced G2 checkpoint. Revealing the role of NAT10-mediated MORC2 acetylation in regulating the DNA damage-induced G2 checkpoint provides a potential therapeutic strategy to sensitize breast cancer cells to chemotherapy and radiotherapy for DNA damage by targeting NAT10. In hepatocellular carcinoma, NAT10 can alter EMT and enhance hepatocellular carcinoma cell metastasis. In BC, NAT10 inhibition reverses EMT and reduces adriamycin resistance. The suppression of NAT10 by Remodelin increases the expression of E-cadherin, a classic marker of EMT, while decreasing the expression of waveform proteins.73 Human THUMP structural domain protein 1 (THUMPD1) interacts with NAT10, and THUMPD1 decreases E-cadherin production via the AKT-GSK3-Snail pathway, facilitating breast cancer cell invasion and migration. It is suggested that THUMPD1 NAT10 binding might synergistically enhance cancer cell invasion, or that THUMPD1 may operate as a NAT10 downstream factor to promote tumor invasion.74
Bladder urothelial carcinoma
NAT10 is abundantly expressed in bladder urothelial carcinoma (BLCA) and has been linked to a poor prognosis with lymph node or distant metastases. NAT10 shRNA knockdown or NAT10 expression decrease by Remodelin reduces mRNA ac4C levels, slows tumor cell growth, and promotes apoptosis. According to acRIP-seq investigation, NAT10 directly influences cancer pathway (BCL9L), cell cycle (AKT1), and stem cell maintenance (SOX4) transcription, and acetylation of the overswing site enhances its stability and translation efficiency. These targets are dramatically decreased by NAT10 knockdown, presumably boosting the BLCA process.75 Both NAT10-specific antagonists and NAT10 knockdown can slow BLCA growth, suggesting that targeting NAT10 may be a novel technique for BLCA therapy.
NAT10 in non-tumor diseases
Systemic lupus erythematosus
Epigenetic changes can have a direct impact on the autoimmune response by causing immune cell malfunction, particularly in CD4+ T cells.76 Epigenetic alterations are now relevant in systemic lupus erythematosus (SLE). The expression of the acetyltransferase NAT10, as well as the total ac4C level, is considerably decreased in SLE patients’ CD4+ T cells. Furthermore, ac4C peaks were shown to be abundant in mRNA CDS and 3′ UTR regions, indicating that ac4C is a conserved internal transcriptional alteration.77 In contrast, ac4C sites were found in HeLa cells around the translation start site (5′ UTR and CDS sections),9 implying that ac4C modification localization in SLE may be a disease-related characteristic. Indeed, the top motifs of ac4C peaks in lupus CD4+ T cells, “CRGRA” and “CCRCCRC,” can be meaningful targets. ac4C peaks in HeLa cells display four forms of ac4C peaks separated by two nonspecific nucleotides, which differs to some extent from the ac4C modification pattern in SLE. These discrepancies in alteration locations might be constrained by sample size and variance.
It has been theorized that pathogenic factors that govern SLE are linked to alterations in the ac4C sequence of mRNA. The results from GO analysis linked USP18, GPX1, and RGL1 to SLE, with GPX1 specifically being linked to lupus endothelial dysfunction.78,79 According to recent research,79 rGL1 is a new target of lymphocyte network inflammation-related pathways. To learn more about the causes of SLE, ac4C modification may be applied to it. Insight into the pathogenic processes of ac4C may proceed with optimism from this finding.
Influenza A virus
RNA modification is a novel element of host-virus interactions that elucidates viral life-cycle processes. The presence of ac4C in viral transcripts enhances RNA stability.80 NAT10 is downregulated in influenza A and interacts with PB1, NP, NA, and M1 viral proteins to promote ac4C acetylation in many gene areas.81 One study found that, in influenza A, the DAZAP1 gene ac4C was enriched in the UTR employing ac4C antibodies and fragmented RNA samples from virus-infected cells for RIP (UTR). The anti-ac4C antibody enriched a region around nucleotide position 150 in the PB1 fragment’s negative strand and a region near nucleotide position 350 in the NA fragment’s negative strand. A very substantial enrichment was also observed for approximately 400 bases near the 5′ end of the HA fragment’s negative strand. Furthermore, knocking down the host component inhibits viral development, implying antiviral action.81
HIV-1
HIV-1 has a high level of NAT10 expression. Once NAT10 expression is reduced, a similar reduction in viral gene RNA ac4C and HIV-1 gene expression might occur. Similarly, loss of ac4C in viral transcripts owing to NAT10 deletion suppresses HIV-1 replication by decreasing viral RNA stability.82 The NAT10 inhibitor Remodelin suppresses HIV-1 replication while having little effect on cellular function, indicating that ac4C might be a promising target for antiviral medication development.80 Using PA-ac4Cseq analysis of HIV-1 gRNA, researchers discovered that ac4C residues on HIV-1-infected cellular mRNA were mostly found in the CDS, with some also found in the 3′ UTR. As a result, uninfected and HIV-1-infected CEM cells had remarkably comparable C- and U-rich common sequences with core “UCU” motifs. However, ac4C sites were found only in the gag, pol, and env portions of intracellular RNA, as these areas are eliminated by splicing in many intracellular viral RNAs.81
Conclusions and prospects
In this review, we comprehensively summarize the normal physiological properties of NAT10 and its functions in biological development such as oocyte maturation and bone remodeling. In addition, we focus on the latest studies on the effects of NAT10 in the progression of various diseases especially cancer, which provides a new perspective from which to focus on the potential possibility of NAT10 as a diagnostic and treatment target in clinical applications.
NAT10 functions as an important writer for ac4C, identifying ac4C-modified target genes and controlling their expression through translation or RNA stability. Changes in target genes are linked to the course of many diseases, including cancers, although further research into the processes of some diseases is still needed. As a result, we summarized the mechanism and relevance of NAT10-mediated ac4C alterations in human diseases in this study. According to current research, NAT10 plays a key role in the course of many illnesses by reducing translation efficiency and controlling mRNA stability mostly through ac4C alterations at various loci, hence influencing the expression of target genes. In malignancies, NAT10 influences prognosis, proliferation, invasion and migration, tumor stem cell development, and metabolism. Furthermore, by recognizing its target genes, NAT10 regulates non-tumor diseases through immunological function and viral infections. In cancer research, the NAT10 inhibitor Remodelin has been shown to enhance apoptosis, reverse EMT, alter drug resistance, and reduce lipid buildup, and can potentially act as an anti-cancer medication. For example, in AML cell lines treated with Remodelin, anti-apoptotic genes (including CDK2, CDK4, cyclin D1 and cyclin E, and BCL2) decreased while pro-apoptotic genes (such p16 and p21) increased.64 Remodelin reversed EMT and decreased adriamycin resistance in hypoxia-induced EMT in breast and hepatocellular cancer by boosting the expression of E-calcineurin and lowering the expression of Vimentin and TWIST.40,73,83 Reducing mitochondrial lipid metabolism in cancer cells treated with Remodelin lowered the expression of ECHS1 and MECR.84
In summary, we determined the biological properties of NAT10 in normal physiology and development, the promotion functions of NAT10 in proliferation, metastasis, and apoptosis in tumors, and the regulatory roles of NAT10 in immunity, RNA stability, and replication regulation in other non-tumor diseases. These results provide novel insights into NAT10 as a promising disease biomarker for detection, progression, and prognosis, as well as a potential target for disease therapy by altering ac4C alterations. However, more clinical data are still needed to further confirm the clinical potential value of NAT10. In addition, few inhibitors targeting NAT10 in addition to Remodelin have been reported to date. Potent and selective compounds as NAT10 inhibitors with fewer side effects are to be developed, which may be an interesting and meaningful research field for future studies on NAT10.
Acknowledgments
This review is supported by research grants from the National Natural Science Foundation of China (82100537) and the Project of Hunan Health Committee (202110002229).
Author contributions
L.X. and X.Z. collected relevant literature and wrote the manuscript. L.C. and X.Z. conceptualized the main structure of this review, and revised and validated the final version. J.L. and W.C. contributed to the literature analysis and manuscript editing.
Declaration of interests
The authors declare no competing interests.
Contributor Information
Xuyu Zu, Email: zuxuyu0108@hotmail.com.
Ling Chen, Email: chenling@usc.edu.cn.
References
- 1.Nachtergaele S., He C. Chemical modifications in the life of an mRNA transcript. Annu. Rev. Genet. 2018;52:349–372. doi: 10.1146/annurev-genet-120417-031522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Roignant J.Y., Soller M. m(6)A in mRNA: an ancient mechanism for fine-tuning gene expression. Trends Genet. 2017;33:380–390. doi: 10.1016/j.tig.2017.04.003. [DOI] [PubMed] [Google Scholar]
- 3.Sun T., Wu R., Ming L. The role of m6A RNA methylation in cancer. Biomed. Pharmacother. 2019;112:108613. doi: 10.1016/j.biopha.2019.108613. [DOI] [PubMed] [Google Scholar]
- 4.Morena F., Argentati C., Bazzucchi M., Emiliani C., Martino S. Above the epitranscriptome: RNA modifications and stem cell identity. Genes. 2018;9:329. doi: 10.3390/genes9070329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Roundtree I.A., Evans M.E., Pan T., He C. Dynamic RNA modifications in gene expression regulation. Cell. 2017;169:1187–1200. doi: 10.1016/j.cell.2017.05.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dominissini D., Rechavi G. N(4)-acetylation of cytidine in mRNA by NAT10 regulates stability and translation. Cell. 2018;175:1725–1727. doi: 10.1016/j.cell.2018.11.037. [DOI] [PubMed] [Google Scholar]
- 7.Stern L., Schulman L.H. The role of the minor base N4-acetylcytidine in the function of the Escherichia coli noninitiator methionine transfer RNA. J. Biol. Chem. 1978;253:6132–6139. [PubMed] [Google Scholar]
- 8.Boccaletto P., Machnicka M.A., Purta E., Piatkowski P., Baginski B., Wirecki T.K., de Crécy-Lagard V., Ross R., Limbach P.A., Kotter A., et al. MODOMICS: a database of RNA modification pathways. Nucleic Acids Res. 2018;46:D303–D307. doi: 10.1093/nar/gkx1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Arango D., Sturgill D., Alhusaini N., Dillman A.A., Sweet T.J., Hanson G., Hosogane M., Sinclair W.R., Nanan K.K., Mandler M.D., et al. Acetylation of cytidine in mRNA promotes translation efficiency. Cell. 2018;175:1872–1886.e24. doi: 10.1016/j.cell.2018.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ikeuchi Y., Kitahara K., Suzuki T. The RNA acetyltransferase driven by ATP hydrolysis synthesizes N4-acetylcytidine of tRNA anticodon. EMBO J. 2008;27:2194–2203. doi: 10.1038/emboj.2008.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Knockenhauer K.E., Schwartz T.U. The nuclear pore complex as a flexible and dynamic gate. Cell. 2016;164:1162–1171. doi: 10.1016/j.cell.2016.01.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Solsbacher J., Maurer P., Vogel F., Schlenstedt G. Nup2p, a yeast nucleoporin, functions in bidirectional transport of importin alpha. Mol. Cell Biol. 2000;20:8468–8479. doi: 10.1128/mcb.20.22.8468-8479.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sharma S., Langhendries J.L., Watzinger P., Kötter P., Entian K.D., Lafontaine D.L.J. Yeast Kre33 and human NAT10 are conserved 18S rRNA cytosine acetyltransferases that modify tRNAs assisted by the adaptor Tan1/THUMPD1. Nucleic Acids Res. 2015;43:2242–2258. doi: 10.1093/nar/gkv075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Johansson M.J.O., Byström A.S. The Saccharomyces cerevisiae TAN1 gene is required for N4-acetylcytidine formation in tRNA. RNA. 2004;10:712–719. doi: 10.1261/rna.5198204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Johansen T., Johansen S., Haugli F.B. Nucleotide sequence of the Physarum polycephalum small subunit ribosomal RNA as inferred from the gene sequence: secondary structure and evolutionary implications. Curr. Genet. 1988;14:265–273. doi: 10.1007/BF00376747. [DOI] [PubMed] [Google Scholar]
- 16.Ito S., Akamatsu Y., Noma A., Kimura S., Miyauchi K., Ikeuchi Y., Suzuki T., Suzuki T. A single acetylation of 18 S rRNA is essential for biogenesis of the small ribosomal subunit in Saccharomyces cerevisiae. J. Biol. Chem. 2014;289:26201–26212. doi: 10.1074/jbc.M114.593996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ito S., Horikawa S., Suzuki T., Kawauchi H., Tanaka Y., Suzuki T., Suzuki T. Human NAT10 is an ATP-dependent RNA acetyltransferase responsible for N4-acetylcytidine formation in 18 S ribosomal RNA (rRNA) J. Biol. Chem. 2014;289:35724–35730. doi: 10.1074/jbc.C114.602698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kong R., Zhang L., Hu L., Peng Q., Han W., Du X., Ke Y. hALP, a novel transcriptional U three protein (t-UTP), activates RNA polymerase I transcription by binding and acetylating the upstream binding factor (UBF) J. Biol. Chem. 2011;286:7139–7148. doi: 10.1074/jbc.M110.173393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sleiman S., Dragon F. Recent advances on the structure and function of RNA acetyltransferase Kre33/NAT10. Cells. 2019;8:1035. doi: 10.3390/cells8091035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang X.M., Liu J.M., Yan S., Huang K., Bai Y.F., Zheng S.S. High expression of N-acetyltransferase 10: a novel independent prognostic marker of worse outcome in patients with hepatocellular carcinoma. Int. J. Clin. Exp. Pathol. 2015;8:14765–14771. [PMC free article] [PubMed] [Google Scholar]
- 21.Tan Y., Zheng J., Liu X., Lu M., Zhang C., Xing B., Du X. Loss of nucleolar localization of NAT10 promotes cell migration and invasion in hepatocellular carcinoma. Biochem. Biophys. Res. Commun. 2018;499:1032–1038. doi: 10.1016/j.bbrc.2018.04.047. [DOI] [PubMed] [Google Scholar]
- 22.Hanson G., Coller J. Codon optimality, bias and usage in translation and mRNA decay. Nat. Rev. Mol. Cell Biol. 2018;19:20–30. doi: 10.1038/nrm.2017.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kumbhar B.V., Kamble A.D., Sonawane K.D. Conformational preferences of modified nucleoside N(4)-acetylcytidine, ac4C occur at "wobble" 34th position in the anticodon loop of tRNA. Cell Biochem. Biophys. 2013;66:797–816. doi: 10.1007/s12013-013-9525-8. [DOI] [PubMed] [Google Scholar]
- 24.Arango D., Sturgill D., Yang R., Kanai T., Bauer P., Roy J., Wang Z., Hosogane M., Schiffers S., Oberdoerffer S. Direct epitranscriptomic regulation of mammalian translation initiation through N4-acetylcytidine. Mol. Cell. 2022;82:2797–2814.e11. doi: 10.1016/j.molcel.2022.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ivanova I., Much C., Di Giacomo M., Azzi C., Morgan M., Moreira P.N., Monahan J., Carrieri C., Enright A.J., O'Carroll D. The RNA m6A reader YTHDF2 is essential for the post-transcriptional regulation of the maternal transcriptome and oocyte competence. Mol. Cell. 2017;67:1059–1067.e4. doi: 10.1016/j.molcel.2017.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xiang Y., Zhou C., Zeng Y., Guo Q., Huang J., Wu T., Liu J., Liang Q., Zeng H., Liang X. NAT10-mediated N4-acetylcytidine of RNA contributes to post-transcriptional regulation of mouse oocyte maturation in vitro. Front. Cell Dev. Biol. 2021;9:704341. doi: 10.3389/fcell.2021.704341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Khannpnavar B., Mehta V., Qi C., Korkhov V. Structure and function of adenylyl cyclases, key enzymes in cellular signaling. Curr. Opin. Struct. Biol. 2020;63:34–41. doi: 10.1016/j.sbi.2020.03.003. [DOI] [PubMed] [Google Scholar]
- 28.Lin J., Xiang Y., Huang J., Zeng H., Zeng Y., Liu J., Wu T., Liang Q., Liang X., Li J., Zhou C. NAT10 maintains OGA mRNA stability through ac4C modification in regulating oocyte maturation. Front. Endocrinol. 2022;13:907286. doi: 10.3389/fendo.2022.907286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Huang Z., Yuan L., Jiang Z., Wang D. Associations of polymorphisms in NAT2 gene with risk and metastasis of osteosarcoma in young Chinese population. OncoTargets Ther. 2015;8:2675–2680. doi: 10.2147/OTT.S92275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yang W., Li H.Y., Wu Y.F., Mi R.J., Liu W.Z., Shen X., Lu Y.X., Jiang Y.H., Ma M.J., Shen H.Y. ac4C acetylation of RUNX2 catalyzed by NAT10 spurs osteogenesis of BMSCs and prevents ovariectomy-induced bone loss. Mol. Ther. Nucleic Acids. 2021;26:135–147. doi: 10.1016/j.omtn.2021.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhu Z., Xing X., Huang S., Tu Y. NAT10 promotes osteogenic differentiation of mesenchymal stem cells by mediating N4-acetylcytidine modification of Gremlin 1. Stem Cell. Int. 2021;2021:8833527. doi: 10.1155/2021/8833527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Carreira A.C.O., Zambuzzi W.F., Rossi M.C., Astorino Filho R., Sogayar M.C., Granjeiro J.M. Bone morphogenetic proteins: promising molecules for bone healing, bioengineering, and regenerative medicine. Vitam. Horm. 2015;99:293–322. doi: 10.1016/bs.vh.2015.06.002. [DOI] [PubMed] [Google Scholar]
- 33.Weng H., Huang H., Wu H., Qin X., Zhao B.S., Dong L., Shi H., Skibbe J., Shen C., Hu C., et al. METTL14 inhibits hematopoietic stem/progenitor differentiation and promotes leukemogenesis via mRNA m(6)A modification. Cell Stem Cell. 2018;22:191–205.e9. doi: 10.1016/j.stem.2017.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ferraro G.B., Ali A., Luengo A., Kodack D.P., Deik A., Abbott K.L., Bezwada D., Blanc L., Prideaux B., Jin X., et al. Fatty acid synthesis is required for breast cancer brain metastasis. Nat. Can. (Ott.) 2021;2:414–428. doi: 10.1038/s43018-021-00183-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Harjes U., Kalucka J., Carmeliet P. Targeting fatty acid metabolism in cancer and endothelial cells. Crit. Rev. Oncol. Hematol. 2016;97:15–21. doi: 10.1016/j.critrevonc.2015.10.011. [DOI] [PubMed] [Google Scholar]
- 36.Jin Z.N., Chai Y.D., Hu S. Fatty acid metabolism and cancer. Adv. Exp. Med. Biol. 2021;1280:231–241. doi: 10.1007/978-3-030-51652-9_16. [DOI] [PubMed] [Google Scholar]
- 37.Luo Y., Wang H., Liu B., Wei J. Fatty acid metabolism and cancer immunotherapy. Curr. Oncol. Rep. 2022;24:659–670. doi: 10.1007/s11912-022-01223-1. [DOI] [PubMed] [Google Scholar]
- 38.Dalhat M.H., Mohammed M.R.S., Alkhatabi H.A., Rehan M., Ahmad A., Choudhry H., Khan M.I. NAT10: an RNA cytidine transferase regulates fatty acid metabolism in cancer cells. Clin. Transl. Med. 2022;12:e1045. doi: 10.1002/ctm2.1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li Q., Liu X., Jin K., Lu M., Zhang C., Du X., Xing B. NAT10 is upregulated in hepatocellular carcinoma and enhances mutant p53 activity. BMC Cancer. 2017;17:605. doi: 10.1186/s12885-017-3570-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ma R., Chen J., Jiang S., Lin S., Zhang X., Liang X. Up regulation of NAT10 promotes metastasis of hepatocellular carcinoma cells through epithelial-to-mesenchymal transition. Am. J. Transl. Res. 2016;8:4215–4223. [PMC free article] [PubMed] [Google Scholar]
- 41.Liu S., Zhang Y., Qiu L., Zhang S., Meng Y., Huang C., Chen Z., Zhang B., Han J. Uncovering N4-acetylcytidine-related mRNA modification pattern and landscape of stemness and immunity in hepatocellular carcinoma. Front. Cell Dev. Biol. 2022;10:861000. doi: 10.3389/fcell.2022.861000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Xia Y., Wei Z., Huang W., Wei X., He Y. Trim47 overexpression correlates with poor prognosis in gastric cancer. Neoplasma. 2021;68:307–316. doi: 10.4149/neo_2020_200708N706. [DOI] [PubMed] [Google Scholar]
- 43.Vittal R., Fan L., Greenspan D.S., Mickler E.A., Gopalakrishnan B., Gu H., Benson H.L., Zhang C., Burlingham W., Cummings O.W., Wilkes D.S. IL-17 induces type V collagen overexpression and EMT via TGF-β-dependent pathways in obliterative bronchiolitis. Am. J. Physiol. Lung Cell Mol. Physiol. 2013;304:L401–L414. doi: 10.1152/ajplung.00080.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kinker G.S., Greenwald A.C., Tal R., Orlova Z., Cuoco M.S., McFarland J.M., Warren A., Rodman C., Roth J.A., Bender S.A., et al. Pan-cancer single-cell RNA-seq identifies recurring programs of cellular heterogeneity. Nat. Genet. 2020;52:1208–1218. doi: 10.1038/s41588-020-00726-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang Y., Jing Y., Wang Y., Tang J., Zhu X., Jin W.L., Wang Y., Yuan W., Li X., Li X. NAT10 promotes gastric cancer metastasis via N4-acetylated COL5A1. Signal Transduct. Targeted Ther. 2021;6:173. doi: 10.1038/s41392-021-00489-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Beermann J., Piccoli M.T., Viereck J., Thum T. Non-coding RNAs in development and disease: background, mechanisms, and therapeutic approaches. Physiol. Rev. 2016;96:1297–1325. doi: 10.1152/physrev.00041.2015. [DOI] [PubMed] [Google Scholar]
- 47.Feng Z., Li K., Qin K., Liang J., Shi M., Ma Y., Zhao S., Liang H., Han D., Shen B., et al. The LINC00623/NAT10 signaling axis promotes pancreatic cancer progression by remodeling ac4C modification of mRNA. J. Hematol. Oncol. 2022;15:112. doi: 10.1186/s13045-022-01338-9. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 48.Li J.T., Yin M., Wang D., Wang J., Lei M.Z., Zhang Y., Liu Y., Zhang L., Zou S.W., Hu L.P., et al. BCAT2-mediated BCAA catabolism is critical for development of pancreatic ductal adenocarcinoma. Nat. Cell Biol. 2020;22:167–174. doi: 10.1038/s41556-019-0455-6. [DOI] [PubMed] [Google Scholar]
- 49.Zhang H., Pan Y.Z., Cheung M., Cao M., Yu C., Chen L., Zhan L., He Z.W., Sun C.Y. LAMB3 mediates apoptotic, proliferative, invasive, and metastatic behaviors in pancreatic cancer by regulating the PI3K/Akt signaling pathway. Cell Death Dis. 2019;10:230. doi: 10.1038/s41419-019-1320-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kirane A., Ludwig K.F., Sorrelle N., Haaland G., Sandal T., Ranaweera R., Toombs J.E., Wang M., Dineen S.P., Micklem D., et al. Warfarin blocks Gas6-mediated Axl activation required for pancreatic cancer epithelial plasticity and metastasis. Cancer Res. 2015;75:3699–3705. doi: 10.1158/0008-5472.CAN-14-2887-T. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Deng J., Kang Y., Cheng C.C., Li X., Dai B., Katz M.H., Men T., Kim M.P., Koay E.A., Huang H., et al. DDR1-induced neutrophil extracellular traps drive pancreatic cancer metastasis. JCI Insight. 2021;6:e146133. doi: 10.1172/jci.insight.146133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhang H., Hou W., Wang H.L., Liu H.J., Jia X.Y., Zheng X.Z., Zou Y.X., Li X., Hou L., McNutt M.A., Zhang B. GSK-3β-regulated N-acetyltransferase 10 is involved in colorectal cancer invasion. Clin. Cancer Res. 2014;20:4717–4729. doi: 10.1158/1078-0432.CCR-13-3477. [DOI] [PubMed] [Google Scholar]
- 53.Liu Z., Liu X., Li Y., Ren P., Zhang C., Wang L., Du X., Xing B. miR-6716-5p promotes metastasis of colorectal cancer through downregulating NAT10 expression. Cancer Manag. Res. 2019;11:5317–5332. doi: 10.2147/CMAR.S197733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zheng X., Wang Q., Zhou Y., Zhang D., Geng Y., Hu W., Wu C., Shi Y., Jiang J. N-acetyltransferase 10 promotes colon cancer progression by inhibiting ferroptosis through N4-acetylation and stabilization of ferroptosis suppressor protein 1 (FSP1) mRNA. Cancer Commun. 2022;42:1347–1366. doi: 10.1002/cac2.12363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhang Y., Deng Z., Sun S., Xie S., Jiang M., Chen B., Gu C., Yang Y. NAT10 acetylates BCL-XL mRNA to promote the proliferation of multiple myeloma cells through PI3K-AKT pathway. Front. Oncol. 2022;12:967811. doi: 10.3389/fonc.2022.967811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wei R., Cui X., Min J., Lin Z., Zhou Y., Guo M., An X., Liu H., Janz S., Gu C., et al. NAT10 promotes cell proliferation by acetylating CEP170 mRNA to enhance translation efficiency in multiple myeloma. Acta Pharm. Sin. B. 2022;12:3313–3325. doi: 10.1016/j.apsb.2022.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Akimova E., Gassner F.J., Schubert M., Rebhandl S., Arzt C., Rauscher S., Tober V., Zaborsky N., Greil R., Geisberger R. SAMHD1 restrains aberrant nucleotide insertions at repair junctions generated by DNA end joining. Nucleic Acids Res. 2021;49:2598–2608. doi: 10.1093/nar/gkab051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pihan G.A., Wallace J., Zhou Y., Doxsey S.J. Centrosome abnormalities and chromosome instability occur together in pre-invasive carcinomas. Cancer Res. 2003;63:1398–1404. [PubMed] [Google Scholar]
- 59.Chi Y.H., Haller K., Peloponese J.M., Jr., Jeang -K.T. Histone acetyltransferase hALP and nuclear membrane protein hsSUN1 function in de-condensation of mitotic chromosomes. J. Biol. Chem. 2007;282:27447–27458. doi: 10.1074/jbc.M703098200. [DOI] [PubMed] [Google Scholar]
- 60.Gassmann R., Henzing A.J., Earnshaw W.C. Novel components of human mitotic chromosomes identified by proteomic analysis of the chromosome scaffold fraction. Chromosoma. 2005;113:385–397. doi: 10.1007/s00412-004-0326-0. [DOI] [PubMed] [Google Scholar]
- 61.Plas D.R., Talapatra S., Edinger A.L., Rathmell J.C., Thompson C.B. Akt and Bcl-xL promote growth factor-independent survival through distinct effects on mitochondrial physiology. J. Biol. Chem. 2001;276:12041–12048. doi: 10.1074/jbc.M010551200. [DOI] [PubMed] [Google Scholar]
- 62.Busca A., Saxena M., Iqbal S., Angel J., Kumar A. PI3K/Akt regulates survival during differentiation of human macrophages by maintaining NF-κB-dependent expression of antiapoptotic Bcl-xL. J. Leukoc. Biol. 2014;96:1011–1022. doi: 10.1189/jlb.1A0414-212R. [DOI] [PubMed] [Google Scholar]
- 63.Liang P., Hu R., Liu Z., Miao M., Jiang H., Li C. NAT10 upregulation indicates a poor prognosis in acute myeloid leukemia. Curr. Probl. Cancer. 2020;44:100491. doi: 10.1016/j.currproblcancer.2019.06.006. [DOI] [PubMed] [Google Scholar]
- 64.Zi J., Han Q., Gu S., McGrath M., Kane S., Song C., Ge Z. Targeting NAT10 induces apoptosis associated with enhancing endoplasmic reticulum stress in acute myeloid leukemia cells. Front. Oncol. 2020;10:598107. doi: 10.3389/fonc.2020.598107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhu X., Huang L., Gong J., Shi C., Wang Z., Ye B., Xuan A., He X., Long D., Zhu X., et al. NF-κB pathway link with ER stress-induced autophagy and apoptosis in cervical tumor cells. Cell Death Dis. 2017;3:17059. doi: 10.1038/cddiscovery.2017.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Puthalakath H., O'Reilly L.A., Gunn P., Lee L., Kelly P.N., Huntington N.D., Hughes P.D., Michalak E.M., McKimm-Breschkin J., Motoyama N., et al. ER stress triggers apoptosis by activating BH3-only protein Bim. Cell. 2007;129:1337–1349. doi: 10.1016/j.cell.2007.04.027. [DOI] [PubMed] [Google Scholar]
- 67.Wali J.A., Rondas D., McKenzie M.D., Zhao Y., Elkerbout L., Fynch S., Gurzov E.N., Akira S., Mathieu C., Kay T.W.H., et al. The proapoptotic BH3-only proteins Bim and Puma are downstream of endoplasmic reticulum and mitochondrial oxidative stress in pancreatic islets in response to glucotoxicity. Cell Death Dis. 2014;5:e1124. doi: 10.1038/cddis.2014.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kim H., Tu H.C., Ren D., Takeuchi O., Jeffers J.R., Zambetti G.P., Hsieh J.J.D., Cheng E.H.Y. Stepwise activation of BAX and BAK by tBID, BIM, and PUMA initiates mitochondrial apoptosis. Mol. Cell. 2009;36:487–499. doi: 10.1016/j.molcel.2009.09.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Tao W., Tian G., Xu S., Li J., Zhang Z., Li J. NAT10 as a potential prognostic biomarker and therapeutic target for HNSCC. Cancer Cell Int. 2021;21:413. doi: 10.1186/s12935-021-02124-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hutter C., Zenklusen J.C. The cancer genome atlas: creating lasting value beyond its data. Cell. 2018;173:283–285. doi: 10.1016/j.cell.2018.03.042. [DOI] [PubMed] [Google Scholar]
- 71.Zhang Z.X., Zhang W.N., Sun Y.Y., Li Y.H., Xu Z.M., Fu W.N. CREB promotes laryngeal cancer cell migration via MYCT1/NAT10 axis. OncoTargets Ther. 2018;11:1323–1331. doi: 10.2147/OTT.S156582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Liu H.Y., Liu Y.Y., Yang F., Zhang L., Zhang F.L., Hu X., Shao Z.M., Li D.Q. Acetylation of MORC2 by NAT10 regulates cell-cycle checkpoint control and resistance to DNA-damaging chemotherapy and radiotherapy in breast cancer. Nucleic Acids Res. 2020;48:3638–3656. doi: 10.1093/nar/gkaa130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wu J., Xu J., Liu B., Yao G., Wang P., Lin Z., Huang B., Wang X., Li T., Shi S., et al. Inhibition of N-acetyltransferase 10 using remodelin attenuates doxorubicin resistance by reversing the epithelial-mesenchymal transition in breast cancer. Am J Transl Res. 2018;557:256–260. [PMC free article] [PubMed] [Google Scholar]
- 74.Zhang X., Jiang G., Sun M., Zhou H., Miao Y., Liang M., Wang E., Zhang Y. Cytosolic THUMPD1 promotes breast cancer cells invasion and metastasis via the AKT-GSK3-Snail pathway. Oncotarget. 2017;8:13357–13366. doi: 10.18632/oncotarget.14528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wang G., Zhang M., Zhang Y., Xie Y., Zou J., Zhong J., Zheng Z., Zhou X., Zheng Y., Chen B., Liu C. NAT10-mediated mRNA N4-acetylcytidine modification promotes bladder cancer progression. Clin. Transl. Med. 2022;12:e738. doi: 10.1002/ctm2.738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wardowska A. The epigenetic face of lupus: focus on antigen-presenting cells. Int. Immunopharm. 2020;81:106262. doi: 10.1016/j.intimp.2020.106262. [DOI] [PubMed] [Google Scholar]
- 77.Guo G., Shi X., Wang H., Ye L., Tong X., Yan K., Ding N., Chen C., Zhang H., Xue X. Epitranscriptomic N4-acetylcytidine profiling in CD4+ T cells of systemic lupus erythematosus. Front. Cell Dev. Biol. 2020;8:842. doi: 10.3389/fcell.2020.00842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Huang H., Liu X., Chen D., Lu Y., Li J., Du F., Zhang C., Lu L. Melatonin prevents endothelial dysfunction in SLE by activating the nuclear receptor retinoic acid-related orphan receptor-α. Int. Immunopharm. 2020;83:106365. doi: 10.1016/j.intimp.2020.106365. [DOI] [PubMed] [Google Scholar]
- 79.Kirkby N.S., Lundberg M.H., Wright W.R., Warner T.D., Paul-Clark M.J., Mitchell J.A. COX-2 protects against atherosclerosis independently of local vascular prostacyclin: identification of COX-2 associated pathways implicate Rgl1 and lymphocyte networks. PLoS One. 2014;9:e98165. doi: 10.1371/journal.pone.0098165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Tsai K., Jaguva Vasudevan A.A., Martinez Campos C., Emery A., Swanstrom R., Cullen B.R. Acetylation of cytidine residues boosts HIV-1 gene expression by increasing viral RNA stability. Cell Host Microbe. 2020;28:306–312.e6. doi: 10.1016/j.chom.2020.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Furuse Y. RNA modifications in genomic RNA of Influenza A Virus and the relationship between RNA modifications and viral infection. Int. J. Mol. Sci. 2021;22:9127. doi: 10.3390/ijms22179127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.McIntyre W., Netzband R., Bonenfant G., Biegel J.M., Miller C., Fuchs G., Henderson E., Arra M., Canki M., Fabris D., Pager C.T. Positive-sense RNA viruses reveal the complexity and dynamics of the cellular and viral epitranscriptomes during infection. Nucleic Acids Res. 2018;46:5776–5791. doi: 10.1093/nar/gky029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Zhang X., Chen J., Jiang S., He S., Bai Y., Zhu L., Ma R., Liang X. N-acetyltransferase 10 enhances doxorubicin resistance in human hepatocellular carcinoma cell lines by promoting the epithelial-to-mesenchymal transition. Oxid. Med. Cell. Longev. 2019;2019:7561879. doi: 10.1155/2019/7561879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Dalhat M.H., Mohammed M.R.S., Ahmad A., Khan M.I., Choudhry H. Remodelin, a N-acetyltransferase 10 (NAT10) inhibitor, alters mitochondrial lipid metabolism in cancer cells. J. Cell. Biochem. 2021;122:1936–1945. doi: 10.1002/jcb.30155. [DOI] [PubMed] [Google Scholar]