Olivas et al. demonstrate that ATG9 vesicles are the membrane seed for mammalian autophagosomes using nanodisc technology to determine co-residency of ATG9 and LC3 on autophagosomes and their precursor membranes. This work establishes a key piece of the model for autophagosome expansion via direct lipid transfer.
Abstract
As the autophagosome forms, its membrane surface area expands rapidly, while its volume is kept low. Protein-mediated transfer of lipids from another organelle to the autophagosome likely drives this expansion, but as these lipids are only introduced into the cytoplasmic-facing leaflet of the organelle, full membrane growth also requires lipid scramblase activity. ATG9 harbors scramblase activity and is essential to autophagosome formation; however, whether ATG9 is integrated into mammalian autophagosomes remains unclear. Here we show that in the absence of lipid transport, ATG9 vesicles are already competent to collect proteins found on mature autophagosomes, including LC3-II. Further, we use styrene–maleic acid lipid particles to reveal the nanoscale organization of protein on LC3-II membranes; ATG9 and LC3-II are each fully integrated into expanding autophagosomes. The ratios of these two proteins at different stages of maturation demonstrate that ATG9 proteins are not continuously integrated, but rather are present on the seed vesicles only and become diluted in the expanding autophagosome membrane.
Introduction
Macroautophagy, herein referred to as autophagy, is the process of clearing large cytoplasmic debris such as protein aggregates and even whole dysfunctional organelles in order to maintain cellular homeostasis. This is mediated by the autophagosome—an organelle characterized by its de novo formation around targeted cytosolic cargoes, encapsulating and subsequently trafficking them to the lysosome for degradation. As autophagosomes are largely separate from classic vesicle trafficking pathways, how these organelles form de novo has been an area of intense exploration. Intriguingly, recent studies implicate an entirely novel method of membrane expansion: the direct delivery of lipid from a donor membrane to the autophagosome via protein-mediated lipid transport through the autophagy protein ATG2 (Maeda et al., 2019; Osawa et al., 2019; Valverde et al., 2019).
ATG2 is among a new class of bulk lipid transport proteins characterized by an N-terminal chorein domain and a long hydrophobic channel that makes it capable of transporting tens of lipids at once (Leonzino et al., 2021). This higher capacity to move large quantities of lipids suggests that bulk lipid flow may be suitable to grow an organelle; however, lipid transfer alone cannot complete the process. In order for the membrane to expand, there must also be a mechanism to populate the inner leaflet of the bilayer with lipids. The presence of a lipid floppase or scramblase, proteins capable of moving lipids across the bilayer from the outer to inner leaflet, would fulfill this requirement. Recently, ATG9, the only transmembrane protein in the core autophagy machinery, was defined to function as a scramblase (Ghanbarpour et al., 2021; Maeda et al., 2020; Matoba et al., 2020), and to bind ATG2 proteins directly to support autophagosome expansion (Ghanbarpour et al., 2021; Gómez-Sánchez et al., 2018; Kotani et al., 2018; Tang et al., 2019). This makes ATG9 an excellent candidate to be the phagophore scramblase. However, it remains unclear whether ATG9 is ever resident on the mammalian autophagosome.
Recent structural studies identified a hydrophilic channel in ATG9 (Guardia et al., 2020; Maeda et al., 2020; Matoba et al., 2020), which in vitro assays on liposomes confirmed supported lipid scrambling activity (Ghanbarpour et al., 2021; Maeda et al., 2020; Matoba et al., 2020). Isolated Atg9 vesicles from yeast can also support the lipid-conjugation of the autophagy protein Atg8 in vitro, when all the conjugation machinery is supplied exogenously, indicating the surface of these vesicles is autophagy-competent (Sawa-Makarska et al., 2020). Critically, ATG9 must be incorporated into the membrane of the growing autophagosome to support the membrane expansion model. Biochemically isolated autophagosomes from yeast appear to have a small number of Atg9 proteins (Yamamoto et al., 2012), but in mammals evidence for ATG9 on the autophagosome has been lacking. ATG9 is found on vesicles throughout the endolysosomal pathway (Imai et al., 2016; Puri et al., 2013; Zhou et al., 2022), as well as apposed to several membrane compartments including lipid droplets (Mailler et al., 2021), and even transit through the plasma membrane (Claude-Taupin et al., 2021). ATG9 vesicles also travel to and shuttle around phagophores when autophagy is initiated (Mari et al., 2010; Orsi et al., 2012; Young et al., 2006) but these vesicles appear to remain proximal to rather than integrated in growing phagophores (Orsi et al., 2012). Indeed, detection of any transmembrane proteins on the completed autophagosome is challenging (Fengsrud et al., 2000). Intriguingly, a consequence of driving membrane expansion predominantly through protein-mediated lipid transport rather than vesicle fusion is that transmembrane proteins would be largely excluded from the final mature organelle. Thus, ATG9 proteins might never accumulate at the autophagosome beyond whatever small number of proteins are present before lipid transport is activated. If ATG9-related vesicles comprised a seed membrane for autophagosome biogenesis, then the density of ATG9 proteins would be highest in the moments just before lipid transport occurs.
We previously generated cells with both forms of human ATG2 (ATG2A and ATG2B) knocked out (Valverde et al., 2019), which we speculate would accumulate the precursor to the phagophore membrane. Here we show that these cells harbor a pre-ATG2 compartment that is significantly enriched for ATG9 and LC3B-II. We then employ styrene maleic acid copolymer nanodiscs to capture small spans of membrane that biochemically show that ATG9 and LC3 are on the same membrane in cells where this compartment forms and also are present together on autophagosomes collected from wild-type cells. Together, these data establish that ATG9 is resident on the phagophore membrane, positioning it to receive lipids from ATG2 and populate the inner leaflet with lipid during autophagosome expansion (Fig. 1 A).
Figure 1.
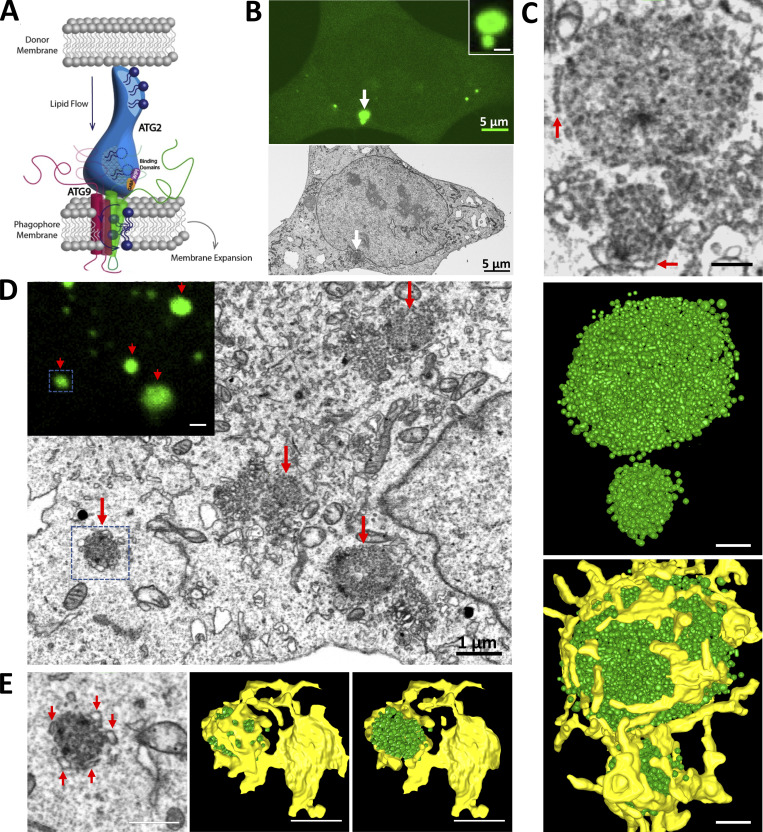
LC3B-positive vesicles accumulate in large clusters in ATG2 DKO cells. (A) Model for autophagosome growth via direct lipid transport by ATG2. ATG2 moves lipids from a donor membrane to the phagophore membrane, where ATG9 scrambles the lipids to populate both leaflets of the growing bilayer. This model requires that ATG9 be integrated in the expanding phagophore. (B) CLEM combined with FIB-SEM in ATG2 DKO HEK293 cells revealed two accumulations of GFP-LC3B signal (white arrow and inset) that correlated to the site of accumulated vesicles (white arrow on the cell of FIB-SEM image). Scale bars: 5 µm. (C) 3D segmentation of two vesicle clusters. Top: Two vesicle clusters from FIB-SEM (B) at high magnification showing ER structures (red arrows) surrounding the vesicle clusters. Middle: Segmented small vesicle clusters. Bottom: Vesicle clusters with abundant ER contacts (yellow). Scale bar: 250 nm. (D) More FIB-SEM-imaged small vesicle clusters (red arrows) are correlated to the GFP-LC3B positive signals (inset, red arrows). Scale bar: 1 µm. Inset scale bar: 1 µm. (E) 3D segmentation of the vesicle accumulation from the blue box in D. Left: Abundant ER structures are closely surrounding the vesicle cluster (red arrows) on the FIB-SEM image. Middle and right: 3D models are showing the vesicle cluster encaged by the ER structure. Scale bars: 500 nm.
Results
Large vesicle clusters enriched in early autophagy proteins accumulate in mammalian ATG2-depleted cells
ATG2-mediated lipid transport is essential to autophagosome growth, suggesting a simple model where ATG2 delivers lipids in bulk to a pre-existing progenitor or seed membrane (Fig. 1 A). To look for these progenitors, we removed ATG2 from the cell and examined what structures accumulate at the resulting sites of frustrated autophagosome biogenesis. Gene-edited HEK293 cells lacking both mammalian ATG2 genes (ATG2 double knockout or ATG2 DKO) accumulate abnormally large LC3B-positive immunofluorescent structures but no mature autophagosomes (Valverde et al., 2019). To determine the ultrastructure of these LC3B-positive structures, we employed correlative light electron microscopy (CLEM) combined with focused ion beam scanning electron microscopy (FIB-SEM) on ATG2 DKO cells expressing GFP-LC3B. 3D modeling of FIB-SEM volumes revealed that GFP-LC3B structures corresponded to large clusters of hundreds of small vesicles (Fig. 1, B and C; and Video 1). Elements of ER were generally observed to be in closest proximity, such that in some cases the ER appeared to partially encapsulate a cluster (Fig. 1, C–E).
Video 1.
FIB-SEM revealed a spherical compartment of vesicles in ATG2 DKO cells. FIB-SEM from Fig. 1 moving down the Z axis of the cluster of vesicles in ATG2 DKO HEK293 cells. Each frame is a FIB-SEM slice. Scale bar: 500 nm. FIB-SEM slice thickness: 7 nm.
Conventional transmission electron microscopy, which yields higher resolution images, revealed that clusters were primarily represented by a mixture of heterogeneously sized vesicular structures (Fig. 2 A). Vesicles within the core of the clusters were very small (<40 nm range) and interspersed within what appeared to be a proteinaceous matrix. Vesicles at the periphery of the clusters were larger (diameters ∼80 nm) and frequently adopt a cup-like morphology (red arrows). Interspersed within these cup-like vesicles were focal accumulation of tiny electron-dense particles, which likely correspond to ferritin (yellow asterisk; Kishi-Itakura et al., 2014). Consistent with FIB-SEM 3D volumes, ER elements partially surrounding the clusters were observed (black arrows).
Figure 2.
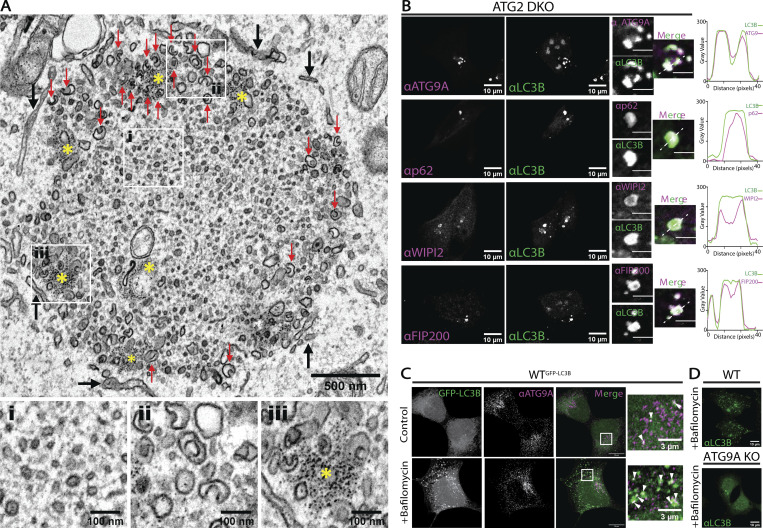
Clustering of autophagy initiation factors, ATG9A, and LC3B-II to vesicles and cup-like pre-phagophores in the absence of ATG2. (A) TEM reveals morphologies of clustered membranes in ATG2 DKO cells and nearby surrounding ER (black arrows). Zoomed fields at the bottom highlight critical features. Left: Heterogenous small vesicles embedded in a dense matrix in the center of the cluster. Center: Cup-shaped membranes at the periphery of the cluster (red arrows in the main field). Right: Focal accumulation of ferritin particles around peripheral vesicles (yellow asterisks in the main field). Scale bar: 500 nm. Inset scale bars: 100 nm. (B) Immunofluorescence of ATG2 DKO cells comparing the distribution of autophagy biogenesis factors ATG9A, p62, WIPI2, and FIP200 with LC3B. ATG9A, p62, and LC3B span the breadth of each fluorescence cluster as demonstrated with individual line scans, while WIPI2 and FIP200 are enriched at the periphery of each fluorescence cluster. Single slices from confocal Z-stacks. All line scans correspond to the white dashed line in the insets. Scale bars: 10 µm. Inset scale bars: 3 µm. (C) Immunofluorescence of GFP-LC3B expressing WT HEK293 cells showing endogenous localization of ATG9A in relation to LC3B (white arrows) in control and bafilomycin A1 treated cells (0.1 µM for 2 h). Maximum intensity projections of AiryScan confocal images. Scale bars: 10 µm. Inset scale bars: 3 µm. (D) Immunofluorescence of endogenous LC3B in WT and ATG9A KO HEK293 cells. The ATG9A KO does not show bafilomycin A1-dependent accumulation of autophagosomes, nor support LC3B-II formation (Fig. S2 A). Maximum intensity projections of confocal images. Scale bars: 10 µm.
Cup-shaped vesicles (Bieber et al., 2022; Knorr et al., 2012), proximity to ferritin clusters (Kishi-Itakura et al., 2014), and proximity to ER (Axe et al., 2008; Nascimbeni et al., 2017; Polson et al., 2010; Uemura et al., 2014) are each common elements in phagophore formation, suggesting these structures at the periphery of the clusters might be pre-phagophores. Their accumulation in an ATG2-depleted background implies that at least the initial stages of cup formation is independent of ATG2, but that growth to full size phagophores or autophagosomes requires ATG2 and presumably lipid transport. Furthermore, similar structures were recently observed in ATG2 DKO mouse embryonic fibroblasts (published while this work was in revision [Gudmundsson et al., 2022]), indicating the formation of “pre-phagophores” and the essential role for ATG2 in membrane expansion is conserved across cell types.
We next used fluorescence microscopy to explore how factors involved in initiation and formation of the phagophore distribute across this compartment. We detected ATG9A, p62, WIPI2, and FIP200 (Fig. 2 B) colocalizing with the LC3B-positive puncta, consistent with previous reports indicating most autophagosome biogenesis proteins collect in large puncta when ATG2 is depleted (Bozic et al., 2020; Stanga et al., 2019; Tamura et al., 2017; Tang et al., 2017; Tang et al., 2019; Valverde et al., 2019; Velikkakath et al., 2012). Because most of the LC3B appear to be lipidated in these cells (Valverde et al., 2019), LC3 is likely bound to membranes in this compartment. Likewise, as a transmembrane protein, ATG9A must be embedded within one or more vesicle population found here. Furthermore, virtually all of the detectable ATG9A and LC3B are found together in these structures, in stark contrast to the situation in WT cells where each protein is found in puncta throughout the cytoplasm but only rarely colocalized (Fig. 2 C and [Mari et al., 2010]).
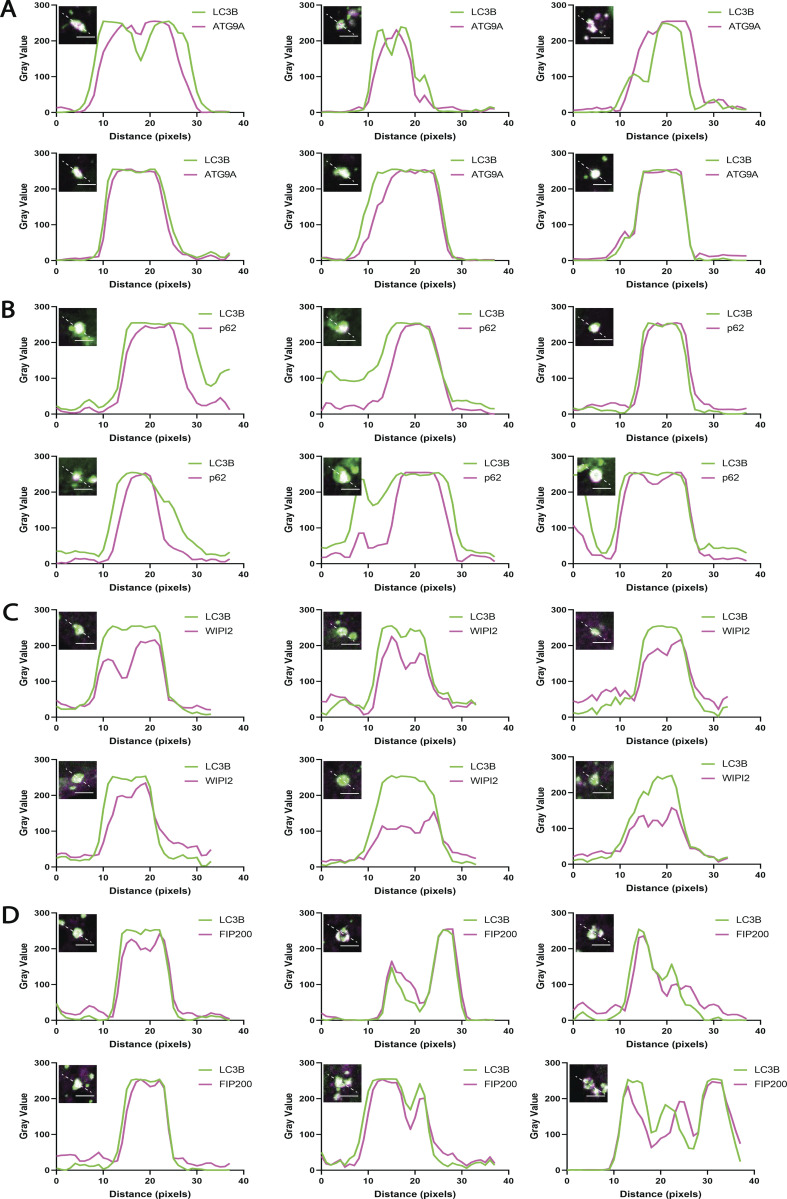
Within each structure, ATG9A and p62 each appeared to broadly colocalize with LC3B throughout the whole of the structure (Fig. 2 B). The distribution of each marker is not always homogeneous however (Fig. S1, A and B), and at this resolution we cannot conclude whether ATG9A and LC3B-II decorate the same membranes or closely adjacent different membranes. In contrast, WIPI2, a PI3P-binding protein implicated in early biogenesis events including recruitment of ATG16L1 (Dooley et al., 2014; Dooley et al., 2015; Polson et al., 2010; Zhao et al., 2017), was excluded from the core of the puncta and instead found around the periphery (Fig. 2 B and Fig. S1 C). The autophagic scaffold and initiation protein FIP200 were present in a less discernible pattern, but also tended to be near the edges of large clusters (Fig. 2 B and Fig. S1 D). Thus, initiation factors for autophagosome formation arrive at this site and tend to be enriched at the periphery where cup-like membranes are concentrated and where the ER is in closest proximity. The presence of lipidated LC3B suggests that these accumulating membranes have the characteristics of putative phagophore seed vesicles—membranes with the capacity to carry out the early events in autophagosome biogenesis. Whether ATG9A is a core component of these seeds, or functions peripherally to organize autophagy proteins is not deducible from imaging alone.
Figure S1.
The pre-ATG2 compartment is an organized collection of pre-autophagosomal proteins and membranes. Line scans corresponding to six individual LC3B-positive structures from ATG2 DKO HEK293 cells per early autophagy factor. (A) ATG9A signal largely colocalizes with LC3B but in the largest structures is also found enriched in the compartment core. (B) p62 consistently colocalizes with LC3B with strongest signal in the center of the compartment in the majority of sampled cells. (C) WIPI2 signal is consistently found on the outside of the LC3B signal, colocalizing with LC3B at the rim of the compartment. (D) FIP200 colocalizes with LC3B, with both direct overlap and adjacent localization. Inset images are single slices from confocal Z-stacks. Inset scale bars: 3 µm.
Knockout of ATG9A results in little to no LC3B lipidation
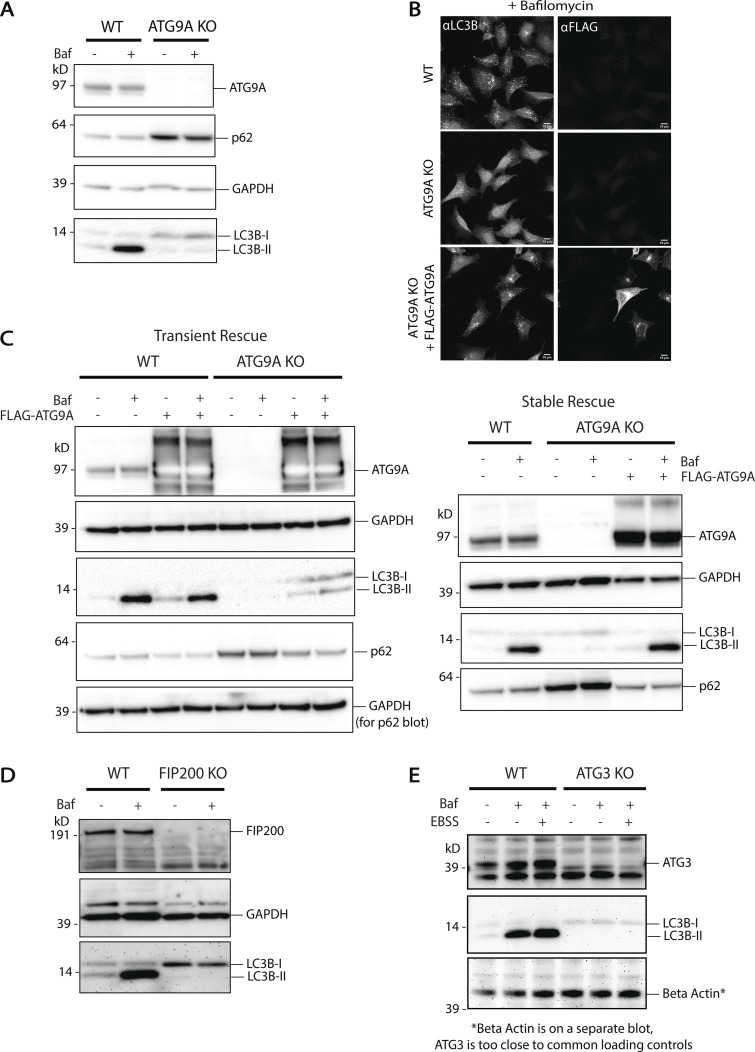
Like ATG2, ATG9 is essential for autophagosome biogenesis (He et al., 2008; Orsi et al., 2012; Yamamoto et al., 2012). To determine where in the autophagosome biogenesis pathway ATG9A is needed, we used gene-editing to remove ATG9A in HEK293 cells (ATG9A KO). ATG9A KO cells did not form LC3B-positive punctate structures even with bafilomycin A1 treatment to accumulate autophagosomes (Fig. 2 D). They also did not support autophagic flux; these cells had an accumulation of p62 and did not accumulate LC3B-II under bafilomycin A1 treatment (Fig. S2 A). These phenotypes can be partially rescued by transient expression of exogenous FLAG-tagged ATG9A (Fig. S2, B and C), and stable expression of FLAG-ATG9A fully restored bafilomycin A1-dependent LC3B-II accumulation (Fig. S2 C). Thus, in these cells, loss of ATG9A correlated with a loss of autophagy-related LC3B-II formation. An inability to drive LC3-II production in ATG9A KO cells is a similar phenotype to that observed in cells where autophagy initiation factors like FIP200 are depleted (Fig. S2 D) or where the lipidation machinery is removed (Fig. S2 E), but notably this is different from ATG2 DKO cells where LC3-II actually accumulates. Thus, although many reports have noted that ATG9 and ATG2 work together to support lipid movement during autophagosome biogenesis, ATG9 is required for LC3-II formation and ATG2 is not, suggesting that ATG9 either resides on the target membrane of LC3-II or locally coordinates the machinery that supports LC3 lipidation.
Figure S2.
ATG9A knockout displays little to no lipidation. (A) Immunoblot showing the loss of ATG9A protein in ATG9A KO HEK293 cells. LC3B lipidation is blocked and p62 accumulates, suggesting no autophagic flux. Loaded protein: 10 µg. (B) Immunofluorescence of WT HEK293 cells compared to ATG9A KO HEK293 cells and ATG9A KO cells rescued with transient expression of FLAG-ATG9A. Rescue of the ATG9A KO shows restoration of bafilomycin A1-dependent accumulation of autophagosomes as in the WT. Maximum intensity projections of confocal images. Scale bars: 10 µm. (C) Immunoblots showing LC3B lipidation, p62 turnover, and bafilomycin A1-induced accumulation of lipidated LC3B is restored by addition of exogenous ATG9A in both transient (left) and stable (right) expression in ATG9A KO cells. Loaded protein: 10 µg. (D) Immunoblot showing the loss of FIP200 protein in FIP200 KO HEK293 cells. LC3B lipidation is blocked, suggesting no autophagic turnover. Loaded protein: 10 µg. (E) Immunoblot showing the loss of ATG3 protein in ATG3 KO HEK293 cells. Loss of LC3B lipidation suggests no autophagic turnover. Loaded protein: 15 µg.
ATG9A and LC3B reside in the same membrane fraction after density gradient fractionation
If ATG9 vesicles are integrated into developing autophagosomes at any point, ATG9 and LC3B-II will be found on the same membranes. If so, these markers should co-enrich during membrane fractionation. We developed a workflow to isolate autophagosomes and other membranes at a scale appropriate for both membrane fractionation and the subsequent nanodomain isolation used throughout the rest of this report (Fig. S3). To first characterize potential co-enrichment of ATG9 and LC3-II, our workflow starts with a well-established density gradient membrane fractionation protocol (Strømhaug et al., 1998) to isolate LC3B-positive membranes. This technique involves several centrifugation steps to separate membrane compartments from one another (Fig. 3 A). We initially applied this approach to WT HEK293 cells that were also stably expressing GFP-LC3B to facilitate subsequent immunoisolation studies. Autophagosomes are relatively scarce and not protein dense, as such, <1% of total lysate protein is ultimately recovered in the autophagosome (AP) fraction (fraction 6 in Fig. 3 A) of starved and bafilomycin A1-treated WT cells, and the yield drops to only 0.1% for WT cells grown under basal conditions (Table S1).
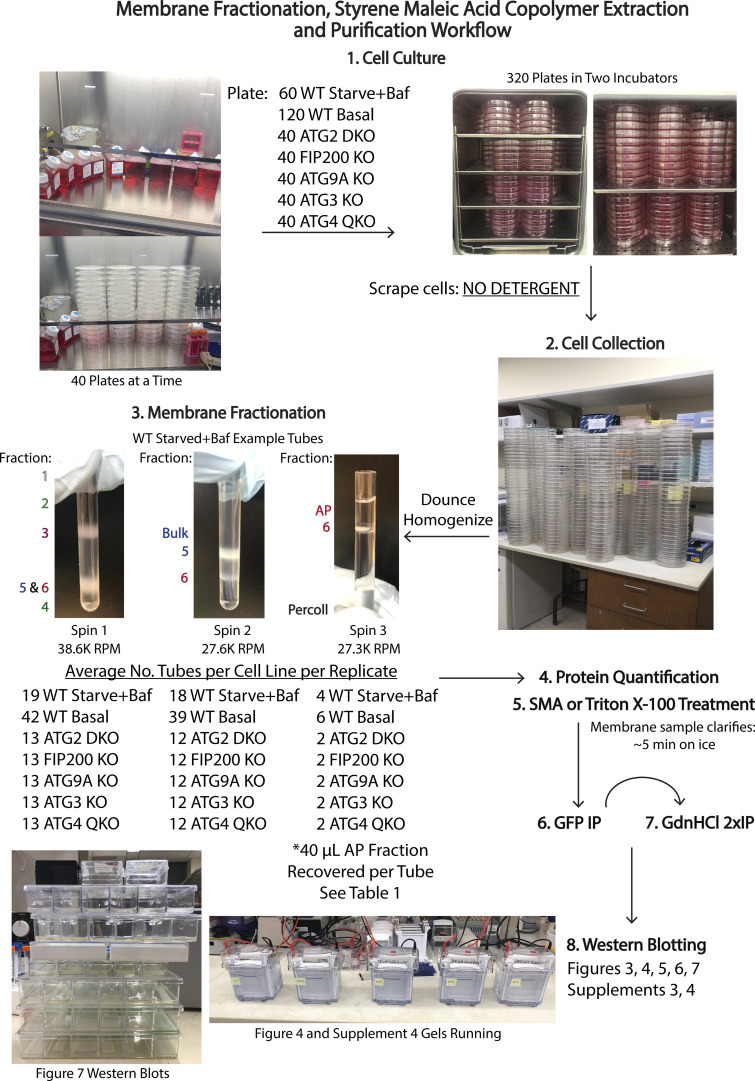
Figure S3.
Workflow for membrane collection and separation for use in this study. Autophagosomes make up only a small percentage of total membrane in most cells and thus approaches to isolate autophagosome-enriched material typically require significant starting biomass. To isolate autophagosomes, we are using a density gradient centrifugation approach originally applied to rat tissue, where biomass is plentiful (Strømhaug et al., 1998), which we later adapted for use in cell culture (Jeong et al., 2009). In this illustrated workflow, we start with various cell lines grown in culture on 15-cm plates which we then scrape and Dounce homogenize before putting through a density centrifugation fractionation protocol. Our ultimate goal in this paper is to then isolate nanoscopic regions of autophagic membrane in SMA-derived nanodiscs and subject this material to various further protein–protein interrogations including a denaturant step. Each of these steps leads to a loss of material, and thus necessitates a very significant initial investment in cell culture mass. In this illustrated workflow, the numbers of plates, tubes, etc. are detailed for one replicate per cell line.
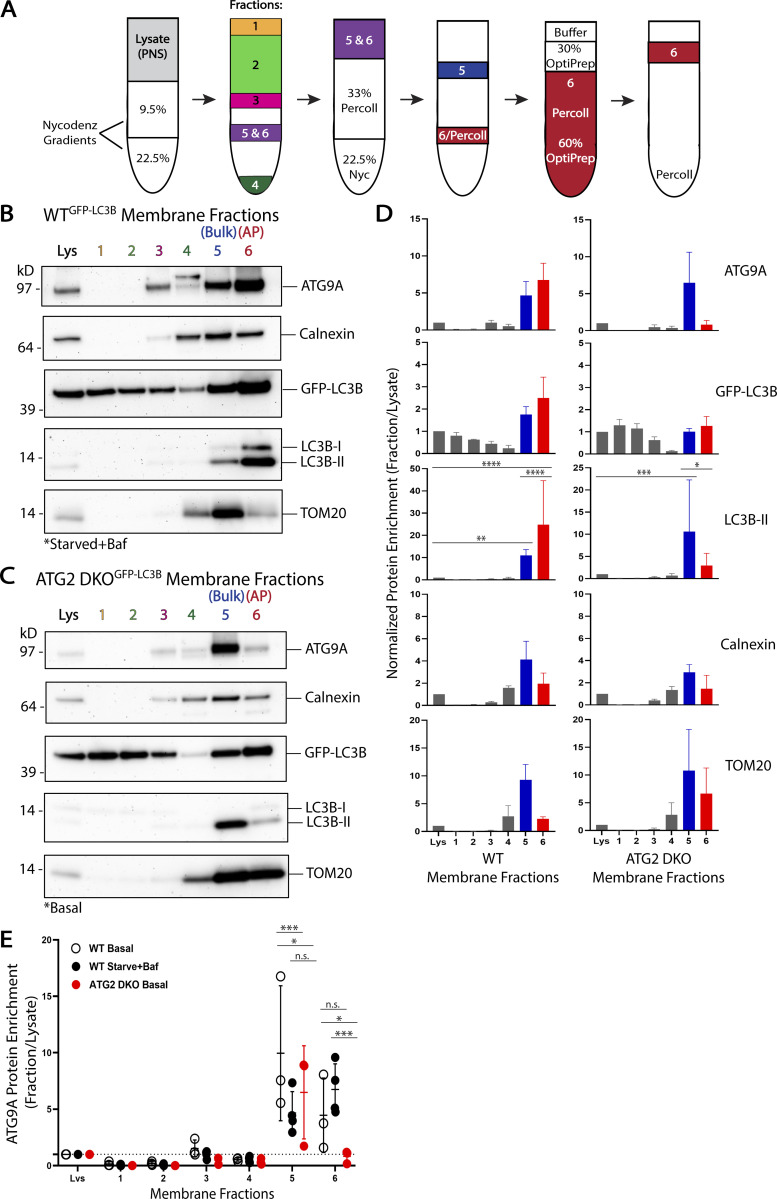
Figure 3.
ATG9A and LC3B reside in the same membrane fractions enriched for autophagosomes or their precursor membranes. (A) Experimental scheme for density gradient membrane fractionation protocol (see Materials and methods and [Strømhaug et al., 1998]). (B) Immunoblot of cell membrane fractions from GFP-LC3B expressing WT HEK293 cells showing enrichment of ATG9A and LC3B together in fractions 5 and 6, fraction 6 being reportedly enriched for autophagosome membranes. Cells were grown in DMEM before being starved in EBSS and treated with 0.1 µM bafilomycin A1 for 4–6 h. Loaded protein: 3 µg. (C) Immunoblot of cell membrane fractions from GFP-LC3B expressing ATG2 DKO HEK293 cells showing enrichment of ATG9A and LC3B together in fraction 5, while fraction 6 is largely depleted of both proteins in this cell line blocking autophagosome formation. Cells were grown in DMEM under basal conditions (ATG2 DKO cells accumulate early autophagic factors without starvation or bafilomycin A1 treatment [Valverde et al., 2019]). Loaded protein: 3 µg. (D) Densitometric quantification of ATG9A, GFP-LC3B, LC3B-II, Calnexin, and TOM20 in each cell line membrane fractionation displayed as average ± SD. The intensity of the bands in B and C were normalized to the lysate, and statistical significance was assessed by two-way ANOVA. *, adjusted P value <0.05. **, adjusted P value <0.01. ***, adjusted P value <0.001. ****, adjusted P value <0.0001. WT, n = 4 biological replicates; ATG2 DKO, n = 3 biological replicates. (E) Densitometric quantification of ATG9A in each fraction compared between the two cells lines and with WT cells grown under basal conditions displayed as average ± SD. The intensity of the bands in B and C were normalized to the lysate, and statistical significance was assessed by two-way ANOVA. *, adjusted P value <0.05. ***, adjusted P value <0.001. WT Basal, n = 3 biological replicates; WT Starved+Baf, n = 4 biological replicates; ATG2 DKO, n = 3 biological replicates. More quantifications are shown in Fig. S5. Basal WT membrane fractions are shown in Fig. S4. Baf, bafilomycin A1.
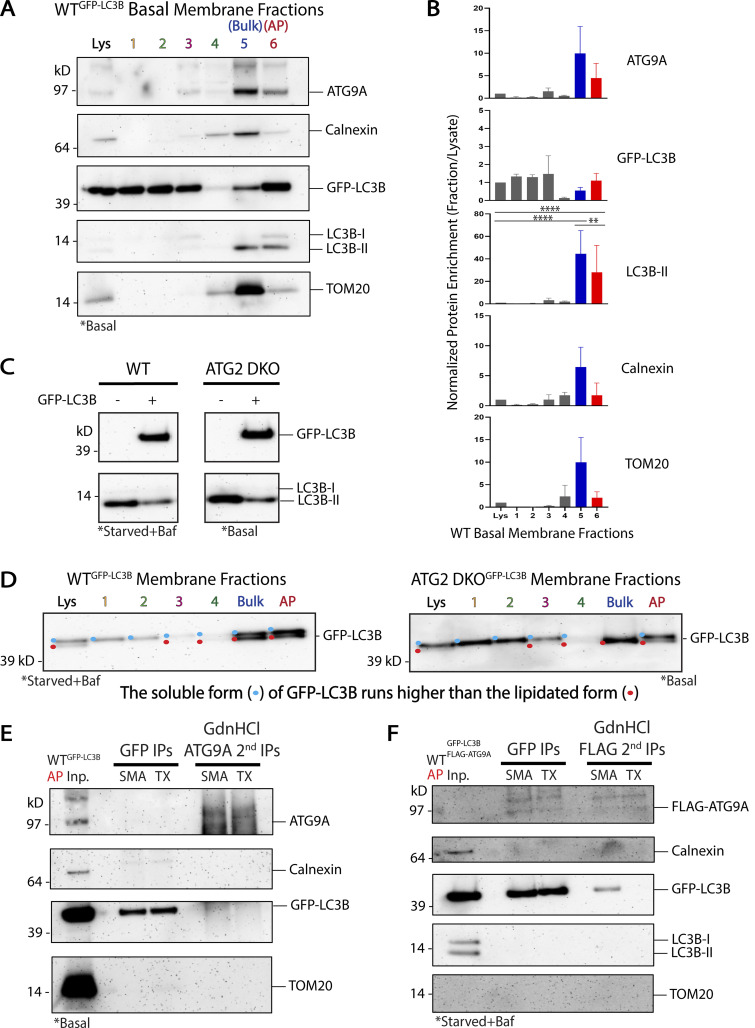
To measure enrichment of particular proteins in a given fraction, we load gels with equal amounts of total protein from each fraction (typically 3 µg) and use densitometry following immunoblot detection of individual proteins. Bands that are more intense in a fraction than the corresponding band in the lysate are said to be enriched. Consistent with Strømhaug et al. (1998) and our own previous results (Jeong et al., 2009; Nguyen et al., 2020; Strømhaug et al., 1998), GFP-LC3B-II and LC3B-II were strongly enriched in the AP fraction in WT cells grown either under basal conditions (Fig. S4, A and B) or starved and treated with bafilomycin A1 to accumulate autophagosomes (Fig. 3, B and D; and Fig. S5). Strikingly, ATG9A was also robustly enriched in this fraction in both conditions (Fig. 3, B, D, and E; and Fig. S4, A and B). Despite this enrichment, the vast majority of ATG9A was present in other fractions, including especially fraction 5 where other transmembrane proteins such as TOM20 and Calnexin were also enriched (Fig. 3, B and D; and Fig. S5) consistent with the predominantly non-autophagosomal distribution of ATG9A described by microscopy. Thus, biochemical enrichment revealed that a small fraction of the ATG9A pool strongly co-enriches with the autophagosome membrane population.
Figure S4.
Additional considerations for ATG9A and LC3B distribution and co-residency. (A) Immunoblot of cell membrane fractions from GFP-LC3B expressing WT HEK293 cells showing enrichment of ATG9A and LC3B together in fraction 5 and 6. Cells were grown in DMEM under basal conditions like the KO cells lines in Fig. 4. Loaded protein: 3 µg. (B) Densitometric quantification of ATG9A, GFP-LC3B, LC3B-II, Calnexin, and TOM20 in WT Basal membrane fractionation displayed as average ± SD. The intensity of the bands in A were normalized to the lysate, and statistical significance was assessed by two-way ANOVA. **, adjusted P value <0.01. ****, adjusted P value <0.0001. n = 3 biological replicates, except TOM20 signal was oversaturated in fraction 5 in one replicate, so n = 2 included biological replicates for TOM20 in fraction 5. (C) Immunoblot comparing endogenous levels of LC3B-II in HEK293 cells with or without overexpression of GFP-LC3B. The endogenous LC3B pool in ATG2 DKO cells is mostly but not completely in form II, perhaps because the ability to lipidate LC3B is saturated with GFP-LC3B overexpression. WT and ATG2 DKO samples are from the same blot. Loaded protein: 10 µg. (D) Immunoblots from membrane fractions from both WT and ATG2 DKO HEK293 cells were run on the Bolt SDS-PAGE system, which allows larger fusion proteins to run far enough to separate protein conjugated to lipid, to distinguish GFP-LC3B-I and GFP-LC3B-II. Colored dots indicate whether the non-lipidated (blue) and/or lipidated (red) forms of GFP-LC3B were detectable in each lane. Loaded protein: 3 µg. (E) Immunoblot of sequential IPs as in Fig. 6 F, first with GFP-LC3B and then with endogenous ATG9A, from GFP-LC3B expressing WT HEK293 cells under basal conditions. After the first IP, the sample was treated with 6 M GdnHCl before diluting over 25X and performing a second IP against ATG9A. Recovery of GFP-LC3B after the second IP is more robust in SMALPs than in soluble Triton-X micelles. Input protein for GFP IPs: 50 µg. Input for ATG9A IPs: 98% of the material collected from the GFP IPs. Loaded input protein: 5 µg (10%). Loaded GFP IPs: 1% of the total collected from the beads. Loaded ATG9A IPs: 50% of the total collected from the beads after GdnHCl treatment. (F) Immunoblot of sequential IPs as in Fig. 6 F, using WT HEK293 cells expressing both GFP-LC3B and FLAG-ATG9A. After the first IP, the sample was treated with 6 M GdnHCl before diluting over 25X and performing a second IP against FLAG. Recovery of GFP-LC3B after the second IP is more robust in SMALPs than in soluble Triton-X micelles. Input protein for GFP IPs: 35 µg. Input for FLAG IPs: 98% of the material collected from the GFP IPs. Loaded input protein: 0.35 µg (1%). Loaded GFP IPs: 1% of the total collected from the beads. Loaded ATG9A IPs: 50% of the total collected from the beads after GdnHCl treatment.
Figure S5.
Further quantifications and in vitro controls. (A–D) Densitometric quantification of GFP-LC3B, LC3B-II, Calnexin, and TOM20 from the density gradient membrane fractions in Figs. 3 and S4 displayed as average ± SD. The intensity of the bands were normalized to the lysate, and statistical significance was assessed by two-way ANOVA. *, adjusted P value <0.05. **, adjusted P value <0.01. ***, adjusted P value <0.001. ****, adjusted P value <0.0001. WT Basal, n = 3 biological replicates; WT Starved+Baf, n = 4 biological replicates; ATG2 DKO, n = 3 biological replicates. TOM20 signal was oversaturated in fraction 5 of the WT Basal in one replicate, so n = 2 included biological replicates for TOM20 in WT Basal fraction 5. Comparisons are nonsignificant unless noted otherwise in this figure. (E–I) Densitometric quantification of the GFP-LC3B IPs in Fig. 6 comparing protein levels from ATG9A/LC3B-enriched fractions between WT and ATG2 DKO HEK293 cells displayed as average ± SD. The intensity of the bands were normalized to the input, and statistical significance was assessed by two-way ANOVA. All comparisons between WT and ATG2 DKO IPs were nonsignificant. Both cell lines, n = 3 biological replicates, except WT Triton X-100 IPs where n = 2 biological replicates. (J and K) In vitro SMA isolation of GFP-LC3B and LC3B decorated liposomes reveal low frequency of co-residency in SMALPs of randomly distributed proteins. 25 nm liposomes were lipidated as described before (Kauffman et al., 2018), with both LC3B and GFP-LC3B. (J) Coomassie-stained SDS-PAGE gel showing IPs of lipidated liposomes treated with various SMA concentrations or Triton X-100. The 2.5% SMA condition mimics that used on the natural membrane SMA IPs in Fig. 6. The in vitro results show that recovery of two distinct peripheral proteins together is relatively rare and further these results closely mirror the recovery of these two proteins from natural sources as shown with the SMA titrations in Fig. 5 C on levels of GFP-LC3B and LC3B recovery. (K) Densitometric quantification of the LC3B:GFP-LC3B ratio between IPs displayed as average ± SD. Statistical significance was assessed by one-way ANOVA. *, adjusted P value <0.05. **, adjusted P value <0.01. n = 3 in vitro biological replicates.
In ATG2 DKO cells, most of the LC3B is form II independent of starvation (Valverde et al., 2019 and Fig. S4, C and D) and by microscopy, all of this LC3B appears to colocalize with ATG9A in the pre-ATG2 compartment (Fig. 2). If LC3B is directly lipidated on ATG9A vesicles in this compartment, then it would indicate ATG9 vesicles are fully competent to promote the downstream biochemistry involved in the lipidation reaction as would be expected of a seed membrane. To study whether ATG9A and LC3B are on biochemically separable membranes in these cells, we again utilized stable expression of GFP-LC3B. In ATG2 DKO cells, where autophagosome formation is largely disrupted, neither LC3B-II nor ATG9A enriched in the AP fraction (Fig. 3, C–E). Instead, virtually all of the ATG9A, LC3B-II, and GFP-LC3B-II were recovered in fraction 5, (Fig. 3, C–E and Fig. S5), which was also enriched for ER and mitochondrial markers. This fraction was originally described by Strømhaug et al. (1998) as an “ER” fraction, but in our hands we also observe significant accumulation of mitochondria suggesting less specific bulk membrane fractionates here. However, as autophagy markers in both the WT and ATG2 DKO samples also accumulate in this fraction (Fig. 3, B–D), its composition is consistent with the presence of omegasomes or phagophores expected to be proximal to the ER, and thus we consider this to also be a pre-autophagosome-enriched fraction.
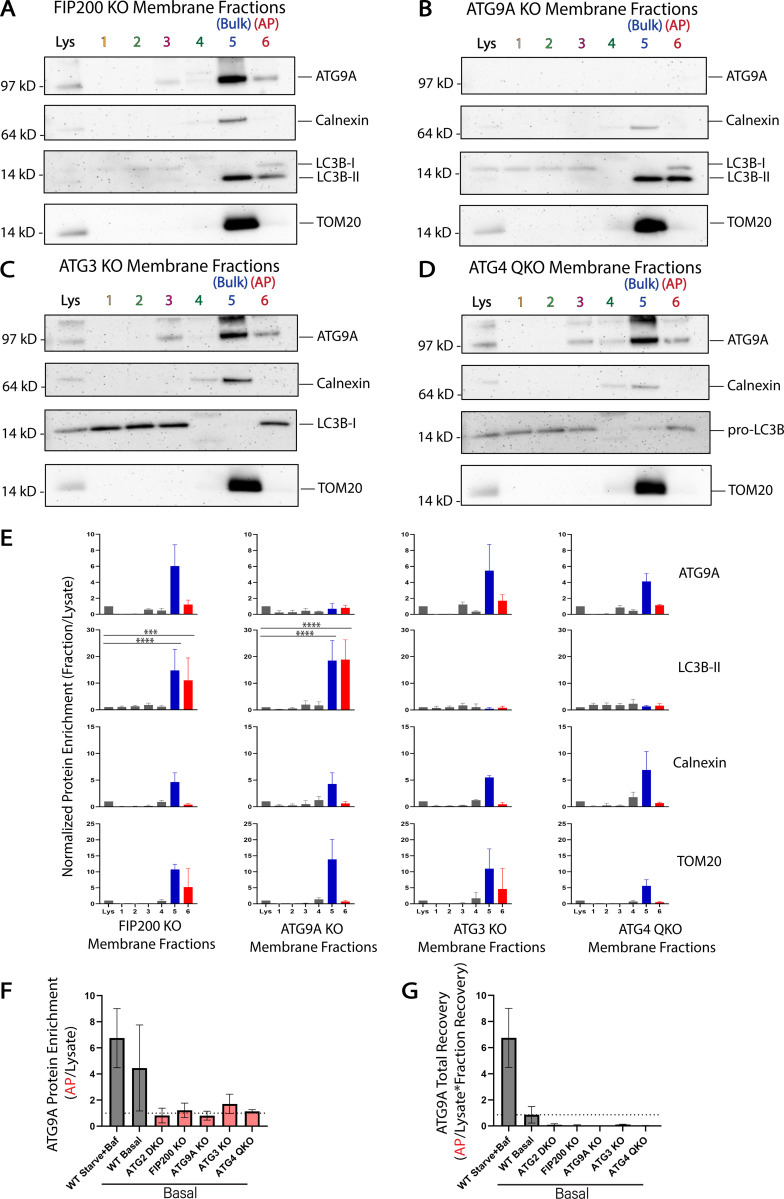
We tested whether other genes in the autophagy pathway were also essential to drive enrichment of ATG9A into the AP fraction. CRISPR knockout of genes involved in initiation (FIP200), lipidation of LC3B (ATG3), or of all four ATG4 genes which disrupts both LC3B-lipidation (Kauffman et al., 2018) and trafficking of ATG9 proteins (Nguyen et al., 2021), each eliminated enrichment of ATG9A in the AP fraction (Fig. 4, A–F). Furthermore, total recovery of protein material in this fraction was dramatically reduced in each of these cell lines (Table S1), confirming that the relationship between the density of these membranes and the enrichment of specific proteins are both directly connected to autophagosome formation. Indeed, when accounting for how little total protein is in these fractions, ATG9A is essentially excluded from the fraction (Fig. 4 G). Likewise, knockout of ATG9A itself reduces the total recovery of protein in this fraction from 0.1 to 0.01% of lysate protein (Table S1), suggesting that ATG9A membranes and the autophagy pathway in general account for ∼90% of the material recovered in this fraction under basal conditions.
Figure 4.
Enrichment of ATG9A in the autophagosome fraction is dependent on autophagosome biogenesis machinery. HEK293 cells were gene-edited to knockout autophagy genes involved in autophagosome biogenesis and then fractionated as in Fig. 3. (A–D) Immunoblot of cell membrane fractions from indicated knockout cells. Cells in A–D were grown in DMEM under basal conditions. Loaded protein: 3 µg. (E) Densitometric quantification of ATG9A, LC3B-II, Calnexin, and TOM20 in each cell line membrane fractionation displayed as average ± SD. The intensity of the bands in A–D were normalized to the lysate, and statistical significance was assessed by two-way ANOVA. ***, adjusted P value <0.001. ****, adjusted P value <0.0001. All KO cell lines, n = 3 biological replicates. TOM20 signal was oversaturated in fraction 5 of the FIP200 KO in one replicate, so n = 2 included biological replicates for TOM20 in FIP200 KO fraction 5. ATG9A is enriched in fraction 5 when initiation or lipidation is inhibited but not enriched over lysate in the AP fraction. Very little AP fraction was recovered for any sample (Table S1) and total LC3B-II in lysate is minimal (A and B) or lost (C and D). Where trace LC3B-II is present, there is a clear enrichment in the AP fraction. (F) Densitometric quantification of ATG9A enrichment in fraction 6 compared across all cell lines tested displayed as average ± SD. These values were taken directly from E, Fig. 3, B and C; and Fig. S4 B. WT Starved+Baf, n = 4 biological replicates; all other cell lines, n = 3 biological replicates. (G) Densitometric quantification of ATG9A total recovery in fraction 6 compared across all cell lines tested with the WT Starved+Baf condition as the reference value (100% recovery) displayed as average ± SD. These values were generated by multiplying the values in F by the fraction recovery highlighted in Table S1. WT Starved+Baf, n = 4 biological replicates; all other cell lines, n = 3 biological replicates.
Thus in WT cells, ATG9A and LC3B are each associated with membranes of comparable density and further, when autophagosome biogenesis is disrupted, they each redistribute to the same new density, strongly suggesting that in WT cells they were present on the same structure. However, the limitations of density gradient isolation are also obvious as both Calnexin and TOM20 can be readily detected in each LC3B-II fraction, necessitating a “higher resolution” biochemistry approach.
Nanodiscs reveal that ATG9A and LC3B-II are on the same vesicle membrane in the pre-ATG2 compartment
Styrene maleic acid (SMA) copolymer is an anionic hypercoiling polymer that has drawn attention for its ability to stabilize small, intact regions of membrane for downstream analysis (Dörr et al., 2016). In an aqueous environment, the copolymer extended chain collapses into an uncharged coil exposing hydrophobic styrenes such that the copolymer behaves as an amphipathic molecule (Tonge and Tighe, 2001). The styrene intercalates between the bilayer acyl chains similarly to cholesterol, destabilizing the membrane and capturing lipids in nanodiscs ∼10 nm in diameter on average (Jamshad et al., 2015) when no protein is present. Captured proteins are accommodated for their size by SMA copolymer, with larger nanodiscs formed around larger proteins (Walker et al., 2023 Preprint). This is possible because unlike other nanodiscs of distinct size, SMA self-assembles into heterogenous nanoparticles (Vargas et al., 2015). In addition, SMA also differs from other membrane encapsulating nanodiscs because at no time does SMA extraction require detergent, therefore maintaining the integrity of the isolated membrane.
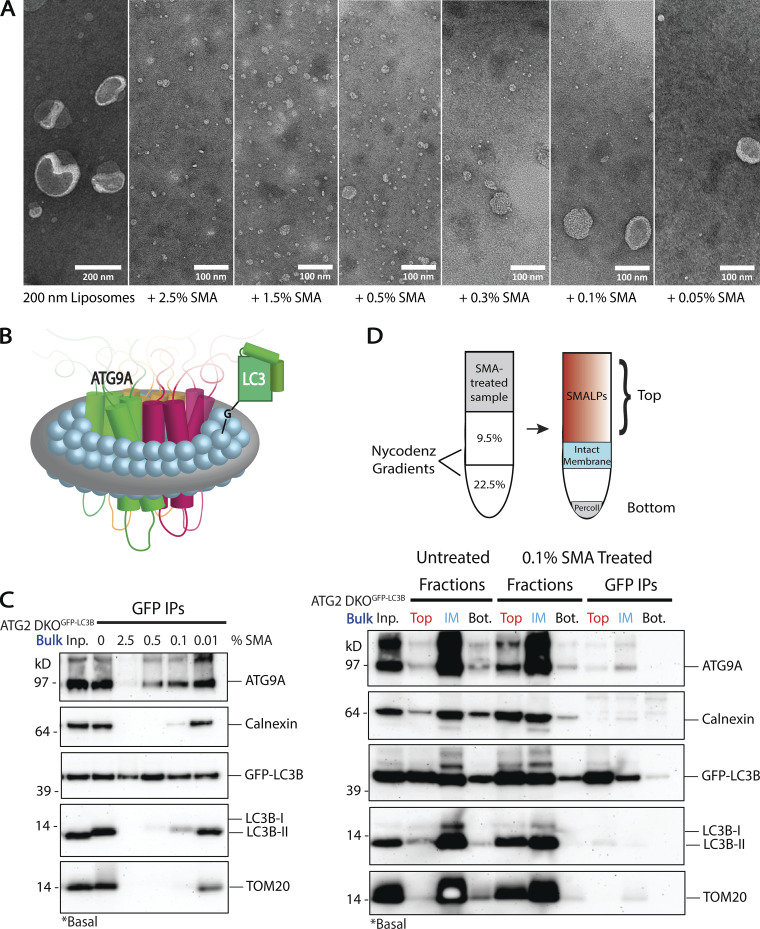
SMA is most commonly used as a preparative tool for the study of membrane-embedded proteins from native sources during structure analysis (Dörr et al., 2016; Esmaili and Overduin, 2018; Postis et al., 2015). Under those working conditions, membranes are fully solubilized into nanodiscs called SMA lipid particles (SMALPs). With artificial liposomes as a substrate, we observed this complete solubilization at 2.5% (w/v) SMA (Fig. 5 A). At lower concentrations of SMA, SMALPs of comparable dimensions still form (e.g., particles in 0.05% SMA), but largely intact membranes also persist. This demonstrates the utility of SMA to “punch out” single nanodomains from otherwise intact membrane, each estimated to comprise about 140 lipids on average (Jamshad et al., 2015).
Figure 5.
Styrene maleic acid (SMA) copolymer nanodiscs capture ATG9A and LC3B together in nanoscale membranes. (A) Transmission electron microscopy (TEM) of 200 nm liposomes treated with varying concentrations of SMA copolymer. The titration reveals the formation of SMA lipid particles (SMALPs) even at low SMA concentrations, with complete solubilization of liposomes at 1.5–2.5% SMA. Scale bar in first panel: 200 nm. Scale bars in SMA treated panels: 100 nm. (B) Cartoon of the transmembrane domains of the ATG9 trimer in a SMALP beside a lipidated LC3B. (C) Immunoblot of the bulk membrane fraction from GFP-LC3B expressing ATG2 DKO HEK293 cells. SMA was titrated in leading to increasing solubilization of the material, and then GFP-LC3B was isolated by immunoprecipitation (IP). ATG9A and GFP-LC3B co-IP with little to no membrane contaminants at higher concentrations of SMA. At lower concentrations of SMA, where membranes are incompletely solubilized into nanodiscs, the IP continues to pull-down all components. Input protein for each IP: 200 µg. Loaded input protein: 3 µg (1.5%). Loaded IPs: 5% of the total collected beads. (D) Immunoblot of the separation of intact membranes from SMALPs via density gradient fractionation (cartoon above) after treatment with a low concentration of SMA (0.1%) on the bulk membrane fraction from GFP-LC3B expressing ATG2 DKO HEK293 cells. Input protein on gradient: 600 µg. Input for each IP: 90% of the material collected from the gradient. Loaded pre-SMA input protein: 3 µg (0.5%). Loaded gradient protein: 5% of the total collected. Loaded IPs: 10% of the total collected beads.
The capability of SMA to punch out sections of membrane and preserve the bilayer provides a mechanism to biochemically establish whether ATG9A and LC3B are present on the same membrane. ATG9A is likely a trimer in the membrane (Guardia et al., 2020; Maeda et al., 2020; Matoba et al., 2020). The ATG9A trimer is ∼12 nm across (Maeda et al., 2020), thus an ATG9A trimer can be captured in a larger SMALP with enough space left for lipids including those that may be conjugated to LC3B if both proteins are on the same span of membrane (Fig. 5 B).
To develop SMA as a tool to detect protein co-residency on natural membranes, SMA was titrated onto membranes enriched in the bulk fraction containing pre-autophagosomal material from ATG2 DKO cells, and then GFP-LC3B was immunoisolated. We started with this bulk material because ATG9A is highly enriched here in ATG2 DKO cells, and because significantly more of this material is recovered during membrane fractionation than any other membrane isolate. In the absence of SMA, GFP-LC3B immunoisolation pulls down essentially the same distribution of all proteins as the starting bulk fraction (Fig. 5 C, compare Inp. to 0), suggesting elements of this compartment remains organized and associated with the ER during the pull-down. SMA-treated immunoisolates contained both ATG9A and GFP-LC3B; at low SMA concentrations, these immunoisolates harbor both SMALPs and incompletely solubilized membranes, and thus Calnexin and TOM20 are recovered. Crucially at higher concentrations of SMA, contaminating ER and mitochondrial membrane markers are lost, but GFP-LC3B and ATG9A continue to co-purify along with some detectable endogenous LC3B-II (Fig. 5 C) suggesting these molecules are co-resident in individual SMALPs.
High SMA concentrations fully solubilize membranes, potentially raising questions about the integrity of the membranes from which the individual SMALPs arise. At low SMA, each SMALP is more akin to a single bite from an apple and may better represent the natural protein distribution in the original membrane. To characterize SMALPs generated at low SMA concentrations, separate from the remaining largely intact membrane, we devised a density gradient membrane isolation that separates SMALPs from intact membranes. ATG2 DKO bulk fraction was treated with 0.1% SMA and applied to the same gradient as the first step in Fig. 3 A. We discovered that SMALPs efficiently solubilized transmembrane proteins into particles that collect at the top of the gradient like cytosolic protein while intact membranes appear to continue to concentrate at a lower interface (Fig. 5 D). Immunoisolation from this SMALP-containing top fraction showed co-purification of ATG9A, GFP-LC3B and some endogenous LC3B away from other membrane contaminants. Thus, immunoisolation with either high or low SMA percentages capture nanomembrane expanses that are populated by both ATG9A and lipidated forms of LC3B.
ATG9A and LC3B are co-resident on the autophagosome
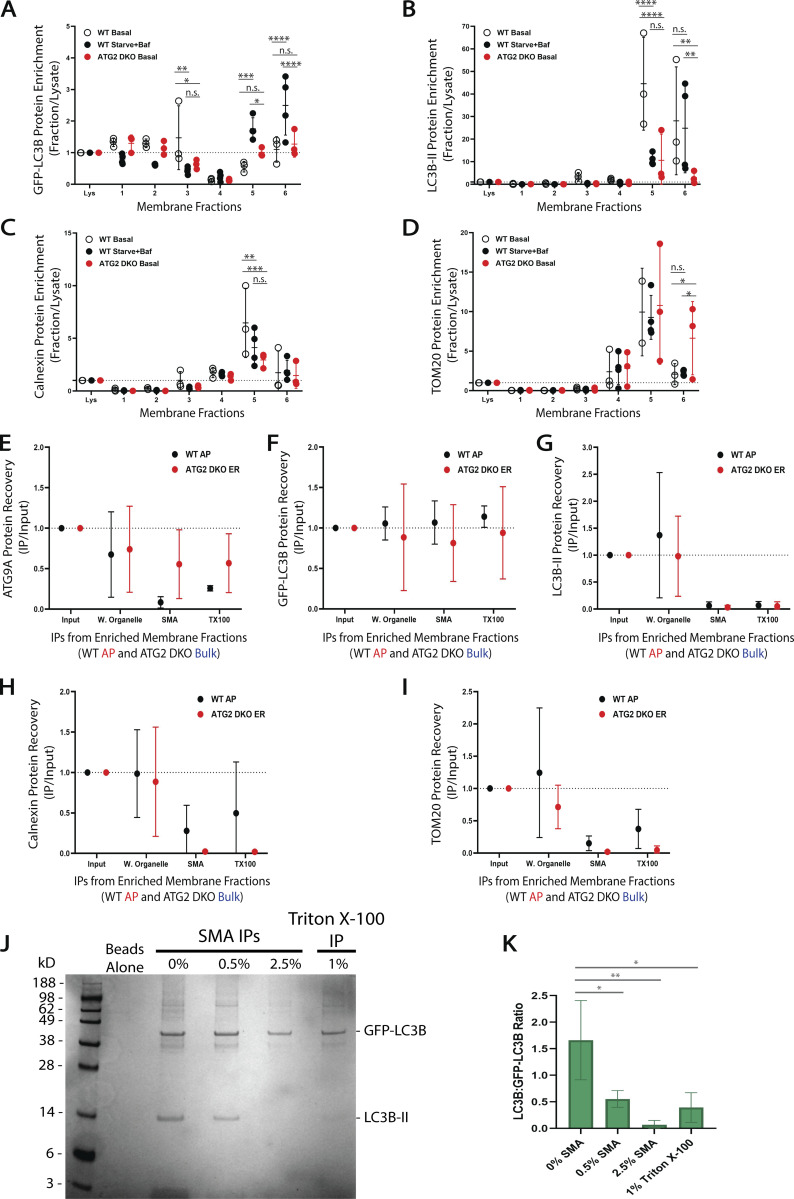
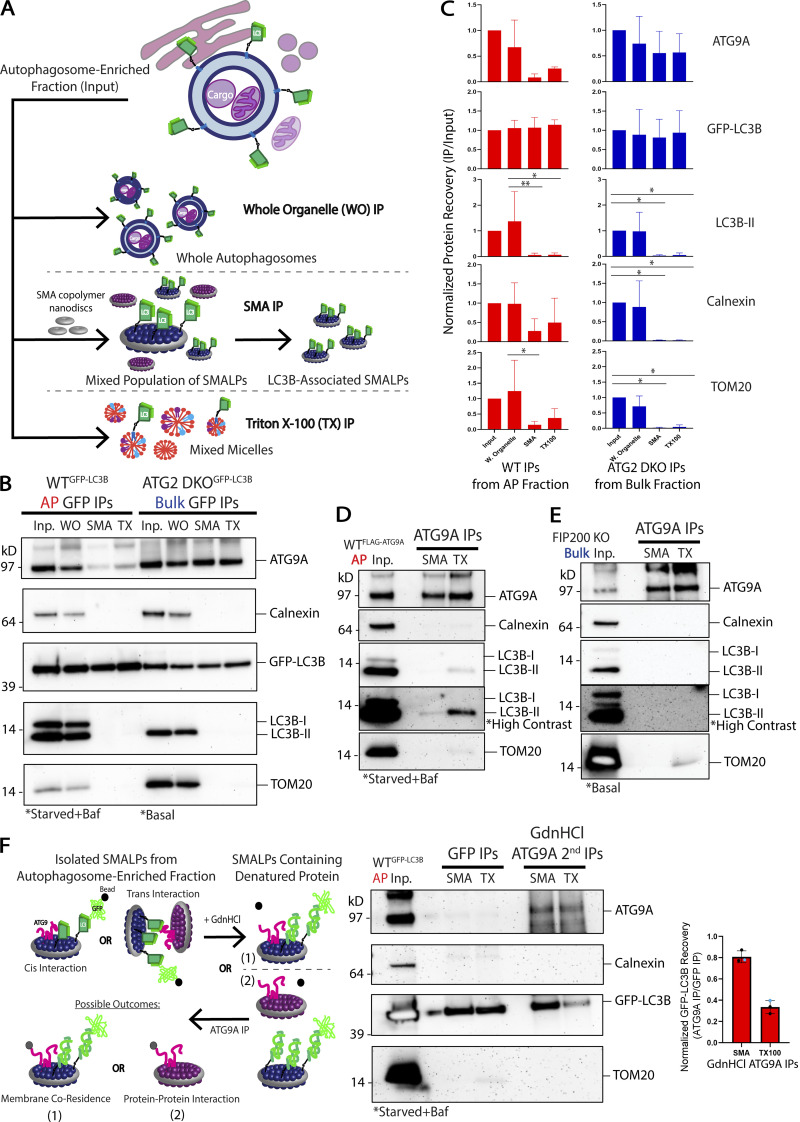
We next used SMA isolations to compare how the co-residency of ATG9A and LC3B proteins varies between the pre-autophagosomal compartment in ATG2 DKO cells and the autophagosome-enriched fraction from WT cells (Fig. 6, A–C). In both cases, we found that immunoprecipitation of whole organelles (WO) without SMA polymer did not allow for significant purification of autophagy proteins away from contaminants. Immunoisolation of SMA-treated samples lead to the specific capture of ATG9A with GFP-LC3B. Interestingly, the ratio of ATG9A to GFP-LC3B was about 7 times lower in mature autophagosomes (Fig. 6 C and Fig. S5). This suggests that as autophagosomes mature the surface density of ATG9A comes down relative to LC3B surface densities—precisely what would be expected if the membrane was expanding by lipid transfer which would facilitate more LC3B lipidation without introducing more ATG9A.
Figure 6.
Cis-interacting ATG9A and LC3B are co-resident on autophagosomes and putative autophagosome precursor membranes accumulating in ATG2 DKO cells. (A) Schematic of the three types of IPs performed to look for ATG9A associating with GFP-LC3B in membrane fractions collected as in Fig. 3. (B) Immunoblot of GFP IPs performed on membranes in the AP fraction collected from WT cells or the bulk membrane fraction collected from ATG2 DKO cells. WO (untreated samples pulling down Whole Organelles); SMA (samples incubated with 12:1 SMA:protein (∼2.5%); TX (samples treated with 1% Triton X-100). Input protein for each IP: 50 µg. Loaded input protein: 3 µg (6%). Loaded IPs: 10% of the total collected beads. (C) Densitometric quantification of ATG9A, GFP-LC3B, LC3B-II, Calnexin, and TOM20 in each type of IP displayed as average ± SD. The intensity of the bands in B were normalized to the input, and statistical significance was assessed by two-way ANOVA. *, adjusted P value <0.05. **, adjusted P value <0.01. Both cell lines, n = 3 biological replicates, except for WT Triton X-100 IPs where n = 2 biological replicates. Quantifications between cell lines are shown in Fig. S3. (D) Immunoblot of reverse IP confirming ATG9A can pull down LC3B-II. The AP fraction from WT cells expressing FLAG-ATG9A was solubilized with either 12:1 SMA:protein (∼2.5%) or 1% Triton X-100. Input protein for IPs: 100 µg. Loaded input protein: 10 µg (10%). Loaded IPs: 25% of the total collected beads. (E) Immunoblot of reverse IP demonstrating that ATG9A cannot pull down LC3B-II in autophagy deficient cells. Bulk membrane fraction from FIP200 KO cells was solubilized with either 12:1 SMA:protein (∼2.5%) or 1% Triton X-100. Input protein for IPs: 100 µg. Loaded input protein: 10 µg (10%). Loaded IPs: 25% of the total collected beads. (F) Left: schematic outlining how protein denaturation can be used to distinguish proteins interacting in trans (in separate SMALPs) vs in cis (within the same SMALP). ATG9A and GFP-LC3B are in the same SMALP only if they continue to co-purify after denaturation with guanidine hydrochloride (GdnHCl). Middle: immunoblot of denaturation experiment conducted with the AP fraction from WT cells expressing GFP-LC3B. Shown are sequential IPs, first with GFP-LC3B, then the sample was treated with 6 M GdnHCl before diluting over 25X and performing a second IP against ATG9A. Input protein for GFP IPs: 50 µg. Input for ATG9A IPs: 98% of the material collected from the GFP IPs. Loaded input protein: 5 µg (10%). Loaded GFP IPs: 1% of the total collected from the beads. Loaded ATG9A IPs: 50% of the total collected from the beads after GdnHCl treatment. Right: densitometric quantification of GFP-LC3B recovered in the final IP after GdnHCl treatment displayed as average ± SD. Two biological replicates are shown with 300 μl of anti-GFP beads (black) and one biological replicate with only 100 μl of anti-GFP beads (blue).
Because SMALPs carry only ∼140 lipids, the capture of two separate proteins in a single disc is unlikely to occur at random. For example, on liposomes reconstituted with both GFP-LC3B-II and LC3B-II, the frequency of capturing both proteins is very low (Fig. S5, J and K). ATG9A co-isolates with GFP-LC3 in SMALPs much more efficiently than would be expected if they were randomly distributed and furthermore, ATG9A and GFP-LC3B also co-isolate together in a classic Triton X-100-mediated detergent immunoisolation (Fig. 6, B and C). We also confirmed this interaction in a reverse pull-down using antibodies against ATG9A to capture endogenous LC3B-II via either SMA or Triton X-100 (Fig. 6 D). Notably, we do not observe any co-IP when FIP200 KO cells are used (Fig. 6 E), despite both proteins enriching in the bulk fraction. These results suggest these proteins either directly interact or both participate in a larger protein complex, but only in cells where autophagy can proceed at least part of the way to a phagophore. A recent preprint article has also eluded to such a direct interaction (Zhang et al., 2020 Preprint), but an ATG9-LC3 binding interaction did not surface in a previous proteomic study (Behrends et al., 2010).
Formally, such a complex could form between membranes rather than on a single membrane and thus it became important to determine whether the interaction was cis on one SMALP or trans on two SMALPs. We employed a protein denaturation protocol to disrupt any protein–protein interactions that could support a multi-SMALP trans complex (Fig. 6 F). We used WT HEK293 cell lines labeled with GFP-LC3B and enriched for autophagosomes as shown in Fig. 3. Samples were then treated with either SMA or Triton X-100 and immunoisolated for GFP-LC3B before being treated with guanidine hydrochloride (GdnHCl) to denature possible protein interactions. Treatment with 6 M GdnHCl did not change the buffered neutral pH of the sample solution, importantly maintaining favorable conditions for the pH-sensitive SMA copolymer (Dörr et al., 2016; Scheidelaar et al., 2016). After diluting the samples containing GdnHCl at least 25 times, the samples were put through a second round of immunoisolation targeting ATG9A. ATG9A continued to robustly pull-down GFP-LC3B in the SMA-treated samples (Fig. 6 F), as expected if they were each independently anchored in the same nanomembrane, while co-precipitation in the detergent sample was much less effective. We observed similar results with the AP fraction from WT cells collected under basal conditions (Fig. S4 E), though total recovery was limited due to less starting material. Likewise, we observed robust recovery of GFP-LC3 after denaturation onto FLAG beads in cells expressing FLAG-ATG9A, but again only in the SMA not Triton X-100 samples (Fig. S4 F). Thus, ATG9A and LC3B-II can be found on the same nanodomains within the confines of SMALP nanodiscs.
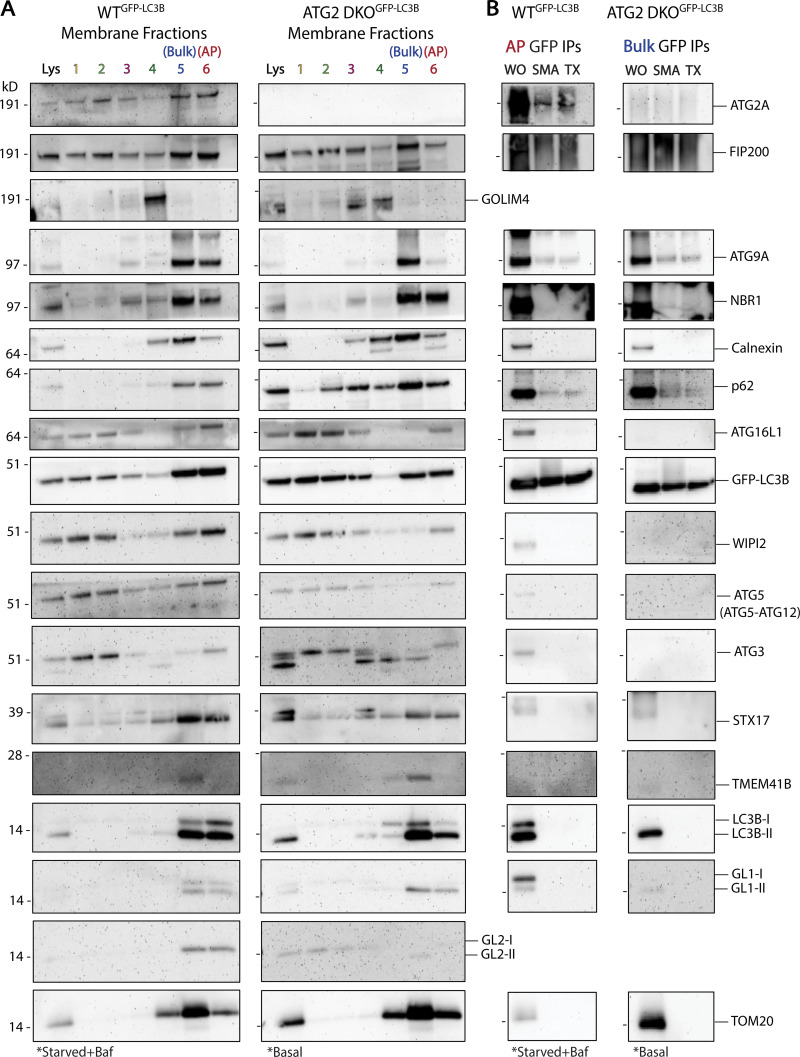
We have focused on LC3B-II and ATG9A because they are fully integrated into membranes, either as a protein–lipid conjugate or an integral membrane protein, and thus are the strongest evidence of co-residency on a common bilayer. To better evaluate whether these structures are related to autophagy, and to test the limits of nanodomains, we conducted extensive immunoblot analyses against our samples (Fig. 7). As expected, membranes, isolated in the AP fraction from WT cells (Fig. 7 A, left), show enrichment for 14 of the 15 autophagy proteins tested (compare intensity of bands in fraction 6 to intensity in lysate). One exception is TMEM41B which is an integral membrane protein of the ER involved in lipid transport and is not expected to integrate into autophagosomes themselves. The AP fraction does not enrich for proteins from the Golgi (GOLIM4), the ER (Calnexin), and only weakly for the mitochondrial marker TOM20. In fraction 5, the bulk membranes containing pre-autophagosomal material, we see similar enrichment of most autophagy factors as well as Calnexin and TMEM41B. In ATG2 DKO cells, we collect very little AP fraction, but in that material we do see autophagy proteins (Fig. 7 A, right). Only NBR1 and possibly GL1-II appear to be enriched over lysate. Instead, all of the autophagy proteins, including ATG9A and LC3B-II, are found in the bulk membrane fraction containing pre-autophagosomal material. IPs of GFP-LC3B from either of these fractions, using either SMA or Triton X-100, efficiently recover ATG9A, p62, and in WT cells, ATG2A (Fig. 7 B). There is also trace recovery of NBR1, but most peripheral proteins are either excluded from these structures or lost during the strenuous washing conditions used.
Figure 7.
Recovery of other autophagy proteins in membrane fractions and in GFP-LC3B immunoprecipitations. (A) Immunoblots of cell membrane fractions from GFP-LC3B expressing WT cells (left) and GFP-LC3B expressing ATG2 DKO cells (right) showing enrichment of autophagy factors in WT fractions 5 and 6 and in ATG2 DKO fraction 5. Cells were grown in indicated conditions as in Fig. 3. Loaded protein: 5 µg. (B) Immunoblots of GFP IPs performed as in Fig. 6 B. Input protein for each IP: 50 µg. Loaded input protein: 5 µg (10%). Loaded IPs: 10% of the total collected beads.
Collectively, this work demonstrates biochemically that mammalian ATG9A is incorporated onto the autophagosome membrane. Furthermore, in the absence of ATG2-mediated membrane expansion, LC3B still finds ATG9A vesicles and/or pre-phagophore like cupped membranes at a site proximal to the ER and undergoes lipid conjugation. Taken together, these data suggest that ATG9 vesicles are the seed membrane for the autophagosome.
Discussion
ATG9 vesicles are required for autophagosome formation, but precisely where they function has been challenging to establish (Feng and Klionsky, 2017; He et al., 2008; He et al., 2006; Karanasios et al., 2016; Kishi-Itakura et al., 2014; Mari et al., 2010; Orsi et al., 2012; Yamamoto et al., 2012; Young et al., 2006). In particular, in mammals the majority of ATG9 vesicles are not localized to the site of autophagosome biogenesis at any given time, and fluorescence imaging during autophagosome biogenesis appeared to favor a peripheral rather than integrated role for these vesicles as autophagosomes grew (Claude-Taupin et al., 2021; Mailler et al., 2021; Mattera et al., 2017; Orsi et al., 2012; Puri et al., 2013; Young et al., 2006). In contrast, in yeast, membrane fractionation proved to be a valuable approach to capture Atg9 within autophagosomes (Yamamoto et al., 2012). They have been shown to interact at the ER preceding the formation of the phagophore, suggesting they could be involved in the seeding event of the autophagosome (Karanasios et al., 2016), and isolated natural ATG9 vesicles have proven to be competent for lipidation in vitro (Sawa-Makarska et al., 2020). For ATG9 vesicles to be the seed membranes for autophagosomes, ATG9 must be shown to be incorporated in nascent autophagosome membranes. Here, we have re-engineered the structural biology tool styrene maleic acid nanodiscs to develop a nanoscale tool for capturing proteins jointly embedded in a common membrane. We used this device to formally establish that both ATG9A and LC3B-II reside in the same membranes, both in purified autophagosomes and in ER-rich membranes replete with pre-autophagosomal material.
Further, we show that in the absence of the lipid transport protein ATG2, ATG9 vesicles collect near the ER but remain competent to collect lipidated LC3B. How the vesicle cluster is maintained in its organization is unclear. The core of the cluster is composed of hundreds or even thousands of very small vesicles which adopt an irregular shape (Fig. 2 A) and appear to be in close proximity to a broader possibly proteinaceous matrix. It has been reported that p62 is capable of phase separation both in vitro and in vivo (Sun et al., 2018), so it is possible that p62 provides a matrix that holds this compartment together, but more work is needed. Interestingly, ATG9A vesicles can also be driven into similar large phase-separated clusters by proteins regulating synaptic vesicle organization (Park et al., 2023) suggesting they may generally be substrates for these types of adaptors, though in that case the vesicles appear to retain a roughly circular shape. Given the composition of the pre-ATG2 compartment, we propose that it captures the initiation stage of autophagosome biogenesis before membrane expansion can occur and by extension traps the seed membrane for the autophagosome. Indeed, we also observe pre-phagophore-like cups and ferritin clusters decorating these vesicle accumulations, suggesting that the first stages of autophagosome biogenesis including perhaps the fundamental shaping of the phagophore, are independent of ATG2-dependent lipid transfer, but growth of the organelle is almost entirely restricted without ATG2-mediated lipid transport. These results are consistent with many other studies implicating recruitment of ATG9 proteins to areas near the ER ahead of eventual ATG2-dependent activity during autophagosome biogenesis (Gómez-Sánchez et al., 2018; Hamasaki et al., 2013; Karanasios et al., 2016; Maeda et al., 2019; Osawa et al., 2019; Tang et al., 2019; Valverde et al., 2019).
Our findings offer one of the final pieces to the phagophore half of the lipid transport facilitated expansion model of autophagosome biogenesis (Fig. 1 A; Ghanbarpour et al., 2021; Maeda et al., 2020; Matoba et al., 2020; Melia and Reinisch, 2022; Noda, 2021). ATG2 binds the phagophore, where it is held in position by peripheral protein receptors including GABARAP/L1 and WIPI4 (Bozic et al., 2020; Chowdhury et al., 2018; Gómez-Sánchez et al., 2018; Kotani et al., 2018; Maeda et al., 2019). Our data presented here put ATG9 also on the phagophore membrane, where its scramblase function can populate the inner leaflet of the growing organelle as it is fed lipid by ATG2. Various studies indicate that ATG2 proteins directly engage ATG9 (Ghanbarpour et al., 2021; Gómez-Sánchez et al., 2018; van Vliet et al., 2022) forming a scramblase–transporter complex. Intriguingly, the other side of ATG2 is bound to the ER through interactions that include additional scramblases (Ghanbarpour et al., 2021) suggesting there may be a broad benefit to coupling bulk lipid transport with scrambling activity. Future work must still define the driving force behind unidirectional lipid flow through ATG2 in order to grow the autophagosome.
Materials and methods
Plasmids and reagents
1,2-dioleoyl-sn-glycero-3-phosphoethanolamine (850725C; DOPE), 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (850457C; POPC), L-α-phosphatidylinositol (from bovine liver—blPI; 840042C), 1,2-dioleoyl-sn-glycero-3-phosphocholine (850375C; DOPC), 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine-N-(lissamine rhodamine B sulfonyl; 810150C; Rhod-PE) were purchased from Avanti Polar Lipids. ATP (AB00162), DTT (AB00490), isopropyl β-D-thiogalactopyranoside (AB00841), NaCl (AB01915), sucrose (AB01900), and Tris HCl (AB02005) were purchased from AmericanBio. 100% glycerol, anhydrous (2136), CaCl2 (1332), EDTA (8993), and MgCl2 (2444) were purchased from J.T. Baker. 0.05% Trypsin-EDTA 1X (25300054), Dulbecco’s phosphate buffered saline (DPBS) 10X (14200075), and DMSO (D2650) were purchased from Thermo Fisher Scientific. Borosilicate glass tubes 10 × 75 mm (47729568) were purchased from VWR.
For transient rescue of the ATG9A KO cell line, pCMV-10-3xFLAG-ATG9A (described in Ghanbarpour et al., 2021) was used. For stable rescue, 3xFLAG-ATG9A was subcloned from pCMV-10-3xFLAG-ATG9A into pLVX-puro (632159; Clontech) for stable cell line creation. pLVX-GFP-LC3B was generated by Gibson assembly using pLVX-Puro backbone, and gel-purified PCR products of GFP-6xGS linker and LC3B to form the GFP-LC3B insert. GFP-LC3B was then cut from the pLVX-puro backbone using EcoRI and BamHI restriction sites and inserted into the digested and gel-purified backbone of pLVX-EGFP-IRES-Neo from R. Sobol (University of Pittsburgh School of Medicine, Pittsburgh, PA; 128660; Addgene) for double-labeled stable cell line creation.
Antibodies used for immunofluorescence in this study include anti-LC3B (rabbit; 1:500; PM036; MBL), anti-LC3B, clone 2G6 (mouse; 1:100; 0260100; Nanotools), anti-ATG9A (rabbit; 1:250; ab108338; Abcam), anti-p62 (mouse; 1:250; 610832; BD Biosciences), anti-WIPI2, clone 2A2 (mouse; 1:250; MABC91; Millipore), anti-RB1CC1 (rabbit; 1:100; Thermo Fisher Scientific; 172501AP), Alexa Fluor 405 goat anti-rabbit IgG (A31556; Thermo Fisher Scientific), Alexa Fluor 405 goat anti-mouse IgG (A31553; Thermo Fisher Scientific), Alexa Fluor 488 goat anti-rabbit IgG (A11008; Thermo Fisher Scientific), Alexa Fluor 594 goat anti-rabbit IgG (A11037; Thermo Fisher Scientific), and Alexa Fluor 594 donkey anti-mouse IgG (A21203; Thermo Fisher Scientific). Antibodies used for immunoblotting in this study include anti-LC3B (3868S; Cell Signaling Technology; rabbit), anti-GFP (2956S; Cell Signaling Technology; rabbit), anti-ATG9A (ab108338; Abcam; rabbit), anti-Calnexin (610524; BD Biosciences; mouse), anti-TOM20 (42406S; Cell Signaling Technology; rabbit), anti-p62 (610832; BD Biosciences; mouse), anti-GAPDH (ab9484; Abcam; mouse), anti-RB1CC1 (172501AP; Thermo Fisher Scientific; rabbit), anti-ATG5 (2630S; Cell Signaling Technology; rabbit), anti-ATG16L1 (8089S; Cell Signaling Technology; rabbit), anti-ATG3 (A3231; Sigma-Aldrich; rabbit), anti-ATG2A (15011S; Cell Signaling Technology; rabbit), anti-GL1 (26632S; Cell Signaling Technology; rabbit), anti-GL2 (14256S; Cell Signaling Technology; rabbit), anti-NBR1 (ab126175; Abcam; rabbit), anti-Syntaxin17 (178151AP; Thermo Fisher Scientific; rabbit), anti-TMEM41B (HPA014946; Sigma-Aldrich; rabbit), anti-WIPI2, clone 2A2 (MABC91; Millipore; mouse), anti-GOLIM4 (LS-C307726; LS Bio; mouse), ECL anti-rabbit IgG horseradish peroxidase-linked (NA934V; GE Healthcare), and ECL anti-mouse IgG horseradish peroxidase-linked (NA931V; GE Healthcare).
Cell culture
HEK293 cells were cultured at 37°C and 5% CO2 in DMEM (11965092; Thermo Fisher Scientific) supplemented with 10% FBS (10438062; Thermo Fisher Scientific) and 1% penicillin-streptomycin (15140122; Thermo Fisher Scientific). For experiments to collect autophagosome membranes, cells were starved by incubation in Earle’s Balanced Salt Solution (24010043; Thermo Fisher Scientific) and treated with 0.1 µM bafilomycin A1 (Enzo; BML-CM110-0100) for 4–6 h.
Lentivirus production and transduction
HEK293 cells were seeded into a 10-cm plate. At 70% confluence, cells were transfected with 4.3 µg psPAX2 (12260; Addgene), 0.43 µg pCMV-VSV-G (8454; Addgene), and 4.3 µg target plasmid using 36 μl Lipofectamine 3000 (L3000008; Thermo Fisher Scientific). DNA was added to 500 μl Opti-MEM (31985070; Thermo Fisher Scientific) and Lipofectamine 3000 to 500 μl Opti-MEM in separate 1.5 ml microcentrifuge tubes. The tubes were then mixed and left for 15 min before adding the mixture dropwise to cells in fresh DMEM. After overnight incubation at 37°C, the medium was replaced with fresh DMEM. Cell medium was then collected every 24 h for 2 d and filtered with a 0.45-µm syringe filter (4184; Pall). Generated virus was stored at 4°C overnight the first day to pool both collections and was then either used immediately or was aliquoted and stored at −80°C.
HEK293 cells to be transduced were seeded into a 6-well plate. Undiluted virus was added dropwise to cells with 10 µg/ml polybrene (TR1003G; Sigma-Aldrich) and incubated at 37°C for 24 h. The medium was replaced with fresh DMEM and left for another 24 h. The cells were then treated with 2 µg/ml puromycin (Clontech; 631306) under selection for at least 1 wk. In the case of double-labeled cells, the process was completed again and selection was performed by treating cells with 2 mg/ml Geneticin (10131035; Thermo Fisher Scientific) for at least 2 wk.
Focused ion beam scanning electron microscopy and correlative light electron microscopy
For transmission electron microscopy (TEM) CLEM, GFP-LC3B over-expressed ATG2 DKO HEK293 cells were plated on 35-mm MatTek dish (P35G-1.5-14-CGRD). Cells were pre-fixed in 4% PFA + 0.25% glutaraldehyde then washed before fluorescence light microscopy imaging. Regions of interest were selected and their coordinates on the dish were identified using phase contrast. Cells were further fixed with 2.5% glutaraldehyde in 0.1 M sodium cacodylate buffer, postfixed in 2% OsO4 and 1.5% K4Fe(CN)6 (Sigma-Aldrich) in 0.1 M sodium cacodylate buffer, en bloc stained with 2% aqueous uranyl acetate, dehydrated, embedded in Embed 812, and polymerized at 60°C for 48 hrs. Cells of interest were relocated based on the pre-recorded coordinates. Ultrathin sections (50–60 nm) were observed in a Talos L 120C TEM microscope at 80 kV; images were taken with Velox software and a 4k × 4K Ceta CMOS camera (Thermo Fisher Scientific).
For focused ion beam scanning electron microscopy (FIB-SEM) CLEM, Epon block was glued onto the SEM sample mounting aluminum stub and platinum en bloc coating on the sample surface was carried out with the sputter coater (Ted Pella, Inc.). The cell of interest was relocated under SEM imaging based on pre-recorded coordinates and FIB-SEM imaged in a Crossbeam 550 FIB-SEM workstation operating under SmartSEM (Carl Zeiss Microscopy GmbH) and Atlas 5 engine (Fibics, Inc.). The imaging resolution was set at 7 nm/pixel in the X, Y axis with milling being performed at 7 nm/step along the Z axis to achieve an isotropic resolution of 7 nm/voxel. Images were aligned and exported with Atlas 5 (Fibics, Inc.), further processed and segmented with DragonFly Pro software (Object Research Systems [ORS], Inc.).
Immunofluorescence and confocal microscopy
HEK293 cells were seeded onto coverslips in a 24-well plate. If cells were treated with bafilomycin A1, 0.1 µM was used for 2 h. Cells were rinsed in PBS, fixed in 4% PFA (15710; Electron Microscopy Sciences), washed three times with PBS, and permeabilized in ice-cold methanol by dipping the coverslip in it 20 times followed by blocking in 3% BSA (A9647; Sigma-Aldrich) in PBS for 15 min (Fig. 2 B and Fig. S1). Alternatively, cells were permeabilized and blocked in 0.1% saponin (47036; Sigma-Aldrich) and 3% BSA in PBS instead of methanol permeabilization (Fig. 2, C and D). Cells were then incubated in primary antibody at indicated concentrations (see Reagents) overnight at 4°C. Methanol-permeabilized cells were washed three times in PBS and saponin-permeabilized cells were washed three times in PBS containing 0.1% saponin and 3% BSA. Secondary antibody was applied in a 1:600 dilution for 1 h at room temperature in the dark. Cells were washed again three times and mounted on pre-cleaned microscope slides with ProLong Gold antifade reagent (P36934; Thermo Fisher Scientific) or Fluoromount-G (0100-01; Southern Biotech) when using GFP-labeled cells.
Imaging was performed at the Center for Cellular and Molecular Imaging Facility at Yale. The 63× oil-immersion objective was used on an inverted Zeiss LSM 880 laser scanning confocal microscope with AiryScan, using Zen acquisition software. Images in Fig. 2 C taken using AiryScan were pre-processed at the microscope using the AiryScan processing presets in Zen Black. Images were processed using ImageJ software. Images in Fig. 2 B and Fig. S1 depict one slice from a Z-stack. Images in Fig. 2, C and D, and Fig. S2 B are maximum intensity projections.
Generation of knockouts using CRISPR/Cas9
CRISPR guide RNAs (gRNAs) targeting the third, sixth, seventh, and eighth exons of ATG9A were accessed online from the Toronto KnockOut Library (University of Toronto; TKOv3, [Hart et al., 2017]). The target sequences were 5′-TATAGGAGGCCTCTAGGCGC-3′ (TKO1), 5′-TAGTGAAGGCAACCACAAAG-3′ (TKO2), 5′-GAAGCTGTCTTCTTCACCCG-3′ (TKO3), and 5′-AGATGAACTTGATAAGCCGG-3′ (TKO4). Complementary sequence to the overhangs created in the vector backbone after digestion was added to the 5′ ends of the forward and reverse gRNA primers. The gRNAs were cloned into pX458 (48138; Addgene) using BpiI (FD1014; Thermo Fisher Scientific) as described previously (Ran et al., 2013). HEK293 cells were transiently transfected with the constructs containing gRNAs using Lipofectamine 3000 according to the manufacturer’s suggestion. After 48 h, cells were sorted into a 96-well plate for single clones using a BD FACSAria cell sorter at the Flow Cytometry Facility at Yale. The cells were validated by genotyping, Western blotting, and immunofluorescence. For genotyping, briefly, genomic DNA from single clones was extracted using QuickExtract DNA extraction solution (QE0905T; Lucigen) according to the manufacturer’s suggestion. PCR products were generated using the following surveyor primers: TKO1-F, 5′-CAGGAAAGCAGCAGTAGACAC-3′; TKO1-R, 5′-GAGACATTAAAGGTCCCAGAG-3′; TKO2-F, 5′-GTAGTGGTGGCTGAGGTTACC-3′; TKO2-R, 5′-CTTGGCAGAGACGCTGCTATC-3′; TKO3-F, 5′-GAGGCAACAACCCCACCTTC-3′; TKO3-R, 5′-CCATAGAGTGACCAGCAGCG-3′; TKO4-F, 5′-CTTGAGTTCTGCTAGCAGGTG-3′; TKO4-R, 5′-GACCCCTTTGCCCTATATTAG-3′. Surveyor primers were designed to be unique to the target gene using NCBI Primer-BLAST and tested on WT HEK293 cell genomic DNA for easily excisable bands from a 1% agarose gel before using them on ATG9A KO candidate genomic DNA. The 400–600 bp PCR products were gel-purified and sequenced. The chosen ATG9A KO HEK293 cell line was cut with gRNA TKO1, creating an in-frame stop codon after 36 bases making the expressed ATG9A peptide fragment only 12 amino acids long. ATG9A KO cell line was used at low passage number for all experiments.
ATG3 and FIP200 KOs were generated in the same manner using the protocol above. CRISPR gRNAs for ATG3 were accessed online from Synthego, and gRNAs for FIP200 were accessed online from the Toronto KnockOut Library. ATG3 target sequences were 5′-TGTTTGCACCGCTTATAGCA-3′, 5′-TTATAGTGCCGTGCTATAAG-3′, 5′-CTATAAGCGGTGCAAACAGA-3′, and 5′-GCTATCATTGAAGAAGATGA-3′. FIP200 target sequences were 5′-GATGAACATCTTCAACACCA-3′, 5′-AGGAGAGAGCACCAGTTCAG-3′, 5′-AGGAATGGCTGAACTTCCGA-3′, and 5′-TGAAGATCGGCTCTACGCCC-3′. ATG3 KO cells were validated by genotyping, Western blotting, and immunofluorescence. FIP200 KO cells were validated by Western blotting and immunofluorescence. Both cell lines were used at low passage number for all experiments.
ATG2 DKO HEK293 cells were generated previously in a similar manner using puromycin selection with pX459 (62988; Addgene) instead of GFP (Valverde et al., 2019), as well as ATG4 QKO HEK293 cells (Kauffman et al., 2018; Nguyen et al., 2020).
Density gradient membrane fractionation
Membrane fractionation was modified from Strømhaug et al. (1998). In summary, GFP-LC3B expressing WT HEK293 cells were either seeded into 60 15-cm plates for starvation/bafilomycin treatment or seeded into 120 15-cm plates for basal conditions. GFP-LC3B expressing ATG2 DKO HEK293 cells (Valverde et al., 2019), FIP200 KO HEK293 cells, ATG9A KO HEK293 cells, ATG3 KO HEK293 cells, and ATG4 QKO HEK293 cells were seeded into 40 15-cm plates. All cell lines were grown to 80–90% confluency. WT cells were either left under basal conditions or were starved in EBSS and treated with 0.1 µM bafilomycin A1 for 4–6 h before harvesting to accumulate autophagosomes, and ATG2 DKO cells were collected untreated (autophagy factors accumulate without intervention [Valverde et al., 2019]). All other KO cell lines were also collected under basal conditions to match the ATG2 DKO. Cells were scraped from the plates, resuspended in 1 ml sucrose homogenization buffer (SHB; 50 mM Tris pH 7.4, 150 mM NaCl, 10% sucrose, and 1X EDTA-free protease inhibitor cocktail [Sigma-Aldrich; 11873580001]), and lysed with a 2-ml Dounce homogenizer (DWK8853000001; Sigma-Aldrich). The lysates were centrifuged at 4,000 rpm for 2 min at 4°C to pellet the nuclei and collect the post-nuclear supernatant (PNS). PNS fractions were loaded onto a Nycodenz (1002424; Accurate Chemical and Scientific Corp) gradient (110 μl of 22.5% Nycodenz on bottom, 270 μl of 9.5% Nycodenz layered next, and 225 μl of PNS on top) in a thin-walled ultracentrifuge tube (355090; Beckman Coulter) and centrifuged at 38,600 rpm with an SW55 rotor (342194; Beckman Coulter) for 1 h at 4°C. Fractions collected are shown in Fig. 3 A and Fig. S3, and were recovered in the following volumes: F1, 50 μl; F2, 50 μl; F3, 80 μl; F4, 30 μl; F5/6, 120 μl. Fraction 5/6 (found at the interface between 22.5 and 9.5% Nycodenz and enriched in ER and autophagosome membranes [Strømhaug et al., 1998]) was diluted 1.25X and loaded on a Percoll (P4937; Sigma-Aldrich)/Nycodenz gradient (110 μl of 22.5% Nycodenz on bottom, 300 μl of 33% Percoll layered next, and 200 μl of Fraction 5/6 on top) in an ultracentrifuge tube and centrifuged at 27,600 rpm for 30 min at 4°C. The two fractions were recovered in the following volumes: F5, 80 μl; F6/Percoll, 80 μl. For every 240 μl of F6/Percoll collected, 168 μl of 60% (w/v) OptiPrep density gradient medium (D1556; Sigma-Aldrich) was added to the fraction. This membrane/Percoll/1OptiPrep sample was loaded on the final OptiPrep gradient (408 μl of F6/Percoll/OptiPrep on bottom, 75 μl 30% OptiPrep layered next, and 120 μl of 1X SHB on top) in an ultracentrifuge tube and centrifuged at 27,300 rpm for 30 min at 4°C. Fraction 6 free of Percoll was collected in a volume of 40 μl from the 30% OptiPrep/buffer interface. All gradient mediums were diluted with 5X SHB to final concentrations in 1X SHB in 10 ml. All fractions were collected and pooled from multiple gradients.
Lysis, gel electrophoresis, immunoblotting, and Coomassie staining
Nearly all experiments required maintenance of membrane integrity, so cells were Dounce homogenized for lysis without detergent. For KO cell line validation (Fig. S2, A, D, and E) and ATG9A KO rescue (Fig. S2 C), cells were lysed in 400 μl lysis buffer (50 mM Tris pH 7.4, 150 mM NaCl, 1% Triton X-100 [AB02025; AmericanBio], 1X EDTA-free protease inhibitor cocktail), incubated on ice for 5 min, centrifuged at 14,000 rpm for 10 min at 4°C, and the supernatant was collected and used immediately or stored at −20°C short term. Protein concentration was determined using protein assay dye reagent (5000006; Bio-Rad) and a Bio-Rad SmartSpec Plus spectrophotometer. Samples were prepared in 1X LDS loading buffer (NP0007; Thermo Fisher Scientific) with 15 mM DTT and then boiled at 98°C for 5 min. Samples were electrophoresed on 12% Bis-Tris precast gels (NP0341BOX or NP0342BOX; Thermo Fisher Scientific) with a SeeBlue Plus2 pre-stained protein standard (LC5925; Thermo Fisher Scientific), typically at 150 V for 75 min in 1X MOPS running buffer (NP0001; Thermo Fisher Scientific). For separation of GFP-LC3B-I from GFP-LC3B-II, samples were electrophoresed on 8% Bolt Bis-Tris Plus precast gels (NW00080BOX; Thermo Fisher Scientific) in 1X Bolt MOPS running buffer (B0001; Thermo Fisher Scientific), typically at 180 V for ∼60 min to run the GFP band to the bottom of the gel. Gels were subsequently transferred to Immobilon-FL PVDF membranes (IPFL00010; Sigma-Aldrich) in 1X transfer buffer (NP00061; Thermo Fisher Scientific) containing 20% methanol (179337; Sigma-Aldrich) at 30 V for 75 min.
For immunoblotting, membranes were blocked with 5% Omniblok (AB10109; AmericanBio) in PBST (PBS containing 0.1% Tween 20 [AB02038; AmericanBio]) for 1 h at room temperature. Membranes were then rinsed with PBST, and incubated with primary antibody (see Reagents) diluted 1:1,000 in PBST containing 5% BSA and 0.02% sodium azide (S2002; Sigma-Aldrich) overnight at 4°C. Membranes were washed three times in PBST before incubation with secondary antibody (GENA931 or GENA934; Sigma-Aldrich) diluted 1:5,000 in PBST containing 5% Omniblok for 1 h at room temperature. Membranes were then washed three times in PBST and treated with SuperSignal West Femto substrate (34096; Thermo Fisher Scientific) for 5 min before imaging with the Bio-Rad VersaDoc imaging system.
For in vitro lipidation followed by IP, gels were electrophoresed in 1X MES running buffer (NP0002; Thermo Fisher Scientific), typically at 200 V for 55 min. The gels were stained for protein by incubation in Imperial Protein Stain (24615; Thermo Fisher Scientific) for 1 h at room temperature. Gels were then destained in water overnight at room temperature before imaging with the Bio-Rad VersaDoc imaging system.
Densitometry and quantification
Densitometry quantifications of both immunoblots and Coomassie protein-stained gels were performed using ImageJ software. For density-gradient membrane fractionation (Figs. 3, 4, S4, and S5), the band intensity of each fraction was normalized to that of the lysate, setting the lysate as 1. Mean values in each fraction for each protein and for each cell line were compared for enrichment, and statistical significance was determined by two-way ANOVA followed by Holm-Šídák’s multiple comparisons test (used for all two-way ANOVA analyses). Mean values for WT HEK293 cell membrane fractions (treated and untreated) were compared to ATG2 DKO HEK293 cell membrane fractions (untreated), and statistical significance was determined by two-way ANOVA. For immunoprecipitation (IP) experiments (Figs. 6 and S5), the band intensity of each IP was normalized to that of the input, setting the input as 1. The mean values were first compared within cell type between different IP treatments, and statistical significance was determined by two-way ANOVA. The mean values were then compared between WT and ATG2 DKO values for each IP treatment, and statistical significance was determined by two-way ANOVA. For in vitro lipidation followed by IP, the band intensity of LC3B-II was plotted as a ratio against GFP-LC3B to compare their recovery in each treatment (Fig. S5). Statistical significance was determined by one-way ANOVA. Data distributions were assumed to be normal. Most data are n = 3, except for WT membrane fractions (Figs. 3 and S5) that are n = 4, WT Triton X-100 IPs (Figs. 6 and S5) that are n = 2, WT Basal TOM20 in fraction 5 (Fig. S4) that is n = 2, and FIP200 KO TOM20 in fraction 5 (Fig. 4) that is n = 2. All n values represent biological replicates. All data were plotted with mean ± SD, and all significance values were considering adjusted P value based on grouped analyses in Prism 9 (GraphPad). Asterisks indicate significance: *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001.
All blots were adjusted in contrast for better visualization of the bands in the figures.
Dialysis of styrene maleic acid (SMA)
SMA in NaOH (XIRAN SL30010P20; Polyscope) was dialyzed in SMA buffer (50 mM Tris pH 7.4 and 150 mM NaCl) before use in experiments. Briefly, 2 ml of SMA was injected into a 20 kD dialysis cassette (66003; Thermo Fisher Scientific) and placed in 4 liters of SMA buffer with gentle mixing for 5 h at room temperature. Buffer was replaced with 4 liters of fresh SMA buffer and incubated with gentle mixing overnight at room temperature. The buffer was again changed, and the cassette incubated in 4 liters of SMA buffer with gentle mixing for 5 h at room temperature. Dialyzed SMA was carefully removed from the cassette, and the percent concentration (w/v) was determined using an Abbe 3L refractometer (Bausch & Lomb). The calculation using refractive indices is as follows: (SampleRI—BufferRI)/2 × 1,000 = % SMA.
For in vitro lipidation followed by IP, ready-to-use SMA (SMALP 30010P; Orbiscope) was used instead of dialyzed stock.
Liposome preparation
To prepare liposomes, lipids in chloroform were mixed in the noted compositions and dried to a thin film under nitrogen gas for ∼10 min. The composition of liposomes for testing SMA (Fig. 5 A) was 30 mol percent DOPE, 60 mol percent POPC, and 10 mol percent PI. The composition of liposomes for in vitro lipidation assays (Fig. S5 J) was 55 mol percent DOPE, 34.75 mol percent DOPC, 10 mol percent PI, and 0.25% rhodamine DOPE. The lipid film was further dried under vacuum for 1 h. The lipids were reconstituted in SMA buffer (Fig. 5 A) or SN buffer (Fig. S5; 20 mM Tris pH 8, 100 mM NaCl, and 5 mM MgCl2) and subjected to seven cycles of flash-freezing in liquid nitrogen and thawing in a 37°C bath. After freeze-thaw, liposomes were extruded with the LipSoFast-Basic extruder (Avestin) 21 times through two polycarbonate membranes (WHA10417004 or WHA110405; Sigma-Aldrich). For testing SMA, liposomes were extruded to 200 nm. For lipidation reactions, liposomes were first extruded to 100 nm and then were sonicated using the VirSonic 600 (VirTis) microtip sonicator to a size of 25 nm immediately prior to the lipidation reaction.
Negative stain transmission electron microscopy
For negative staining SMA-treated liposomes, 400 mesh Formvar carbon-coated copper grids (FCF400CU; Electron Microscopy Sciences) were glow discharged using a PELCO easiGlow glow discharge cleaning system (Ted Pella, Inc.). Samples were diluted 1:10 in buffer before being applied to the grid and stained with 2% uranyl formate. Imaging was performed on a JEOL JEM-1400 Plus microscope operated at 80 kV with a bottom-mount 4k × 3k CCD camera (Advanced Microscopy Technologies).
Styrene maleic acid membrane extraction, titration, and immunoprecipitation
SMA membrane extraction occurs spontaneously after addition to membranes (artificial or natural from cells). After addition of SMA in any concentration, samples were mixed and incubated for 5 min on ice to ensure complete extraction.
Titration was performed on 200 nm liposomes and on ATG2 DKO fraction 5 (Fig. 3) due to plentiful membrane material. For liposome-based SMA titration, 50 mM liposomes were treated with 2.5, 1.5, 0.5, 0.3, 0.1, and 0.01% SMA. They were then negatively stained as described previously. For SMA titration onto natural membranes, 200 µg of protein was treated with 2.5, 0.5, 0.1, and 0.01% SMA and incubated on ice for 5 min. The SMA-to-lipid ratios were about 12:1, 1:1.5, 1:3, and 1:33, estimating that the protein-to-lipid ratio in the ER is around 1:1. Autophagosomes have a lower protein to lipid ratio than the ER, so herein it was referred to as the SMA-to-protein ratio. Samples were then immunoisolated with GFP-LC3B using 50 μl of GFP microbeads from a GFP isolation kit (130091125; Miltenyi Biotec) added to each sample. Samples were incubated with beads on ice for 2 h with gentle mixing every 15 min. The beads were then run through MACS 25 MS separation columns (130042201; Miltenyi Biotec) on the MACS MultiStand magnetic stand (Miltenyi Biotec). Columns were primed with 500 μl 1X SHB, followed by the entire volume of samples containing beads, and then the beads were washed three times with 500 μl SHB. The beads were then eluted from the column by removing the column from the stand, holding it over a collection tube on ice, and adding 500 μl 1X SHB 100 µl at a time and allowing elution to occur by gravity. After the last 100 μl, any liquid left in the column was pushed through with a plastic plunger that comes with the column. Due to the large volume eluted, protein was precipitated from the beads using either methanol/chloroform precipitation (Fig. 5 C) or acetone precipitation (all other IP experiments).
For separation of whole organelles from SMA lipid particles (SMALPs) following treatment with low concentration SMA (Fig. 5 D), 600 µg of ATG2 DKO fraction 5 protein was treated with 0.1% SMA and incubated on ice for 5 min. SMA-treated sample and untreated control were loaded onto a Nycodenz gradient (110 μl of 22.5% Nycodenz on bottom, 270 μl of 9.5% Nycodenz layered next, and 100 μl of sample on top) in thin-walled ultracentrifuge tubes and centrifuged at 38,600 rpm for 1 h at 4°C. The entire top fraction was carefully collected away from the visible intact membrane band (∼400 μl), intact membranes were collected in 50 μl, as well as 50 μl from the bottom of the tube. The SMA-treated samples were then immunoisolated with GFP-LC3B using GFP microbeads as described above.
SMA was used at a concentration of 2.5% per the literature. The SMA to protein ratio in using 2.5% SMA was calculated after the first set of IPs to be 12:1 SMA/protein, and was kept consistent for all following experiments.
Three strategies for immunoprecipitation
For IPs against GFP-LC3B (Fig. 6 B), 50 µg of protein from either WT Starve+Baf fraction 6 or ATG2 DKO fraction 5 (Fig. 3) was either left untreated for whole organelle isolation, treated with a SMA to protein ratio of 12:1 SMA/protein (2.5%), or treated with 1% Triton X-100 for standard detergent-based IP (Fig. 6 A). Samples were incubated on ice for 5 min. All samples were then immunoisolated with GFP-LC3B using GFP microbeads as described in the previous section.
Methanol/chloroform protein precipitation
For precipitating protein from immunoisolated beads, 200 μl of methanol (106035; Millipore) was added to 50 μl of sample (10% of the beads) and vortexed well. 50 μl of chloroform (650498; Sigma-Aldrich) was then added, the sample was vortexed, 150 μl of ddH2O (115333; Millipore) was added, the sample was vortexed again, and was then centrifuged at 14,000 × g for 2 min at room temperature. The top aqueous layer was removed, 200 μl of methanol was added to the sample, the sample was vortexed, and was then centrifuged at 14,000 × g for 3 min at room temperature. Methanol was removed carefully, and the sample was dried under vacuum for 2 min. The pellet was then resuspended in 1X LDS, boiled at 98°C for 10 min, centrifuged at 14,000 rpm for 10 min, beads were removed on a magnetic stand, and finally 15 mM DTT was added. Samples were then used for SDS-PAGE and immunoblotting.
Acetone protein precipitation
For precipitating protein from immunoisolated beads, four times the sample volume of ice-cold acetone was added to 100 μl of sample (20% of the beads) and vortexed well. Acetone samples were incubated at −20°C for 60 min. Samples were then centrifuged at 14,000 rpm for 10 min at 4°C. Acetone was removed carefully, and samples were either dried uncapped under the fume hood for 30 min or under vacuum for 2–5 min. The pellet was then resuspended in 1X LDS, boiled at 98°C for 10 min, centrifuged at 14,000 rpm for 10 min, beads were removed on a magnetic stand, and finally, 15 mM DTT was added. Samples were then used for SDS-PAGE and immunoblotting.
Endogenous immunoprecipitation
For IPs against ATG9A (Fig. 6, D and E), 100 µg of protein from either FLAG-ATG9A expressing WT Starve+Baf fraction 6 or FIP200 KO fraction 5 (Fig. 4) was treated with an SMA to protein ratio of 12:1 SMA/protein (2.5%) or treated with 1% Triton X-100 for standard detergent-based IP (Fig. 6 A). Samples were incubated on ice for 5 min. Samples were then treated with 1 µg ATG9A antibody overnight at 4°C with mixing. Immunoisolation was carried out using Pierce protein A/G magnetic beads (88802; Thermo Fisher Scientific). 50 μl of beads per IP were washed three times with 1X SHB on a magnetic stand, added to each sample, incubated at room temperature for 60 min with mixing, and washed again three times on a magnetic stand with 1X SHB. Samples were then eluted in 100 μl 1X LDS at room temperature for 10 min, the beads were removed on a magnetic stand, and 15 mM DTT was added. Samples were heated at 98°C for 5 min and then used for SDS-PAGE and immunoblotting.
Protein denaturation using guanidine hydrochloride and sequential immunoprecipitation
To make the determination between cis or trans protein–protein interaction (Fig. 6 F), WT GFP-LC3B expressing HEK293 cells were used for sequential IP. 50 µg of WT Starve+Baf fraction 6 was treated with either SMA in a ratio of 12:1 SMA:protein or 1% Triton X-100 and immunoisolated with GFP-LC3B using 300 μl GFP microbeads, otherwise as described previously before protein immunoprecipitation. The eluted beads were then treated with 6 M guanidine hydrochloride (GdnHCl; [24115; Thermo Fisher Scientific]) and incubated for 2 h at room temperature with mixing. Beads were removed by running the sample through a second MACS 25 MS separation column and collecting the eluate. The sample was then diluted to 225 mM GdnHCl in 1X SHB and treated with 1 µg ATG9A antibody before being incubated at 4°C overnight with mixing. The sample was then applied to pre-washed Pierce protein A/G magnetic beads in 50 ml tubes for 60 min at room temperature with mixing. The 50 ml tubes were centrifuged at 4,000 rpm to pellet the beads, and the flow-through was removed. The beads were then transferred to 1.5 ml microcentrifuge tubes and washed three times with 1X SHB on a magnetic stand. The protein was eluted by adding 1X LDS at room temperature for 10 min, removing the beads on a magnetic stand, and adding 15 mM DTT before heating for 5 min at 98°C. Samples were then used for SDS-PAGE and immunoblotting. The same procedure was applied to WT material under basal conditions (Fig. S4 E), and similar protocol was applied to WT HEK293 cells expressing FLAG-ATG9A as well as GFP-LC3B (Fig. S4 F). WT FLAG-ATG9A expressing HEK293 cells were transduced with GFP-LC3B to create a double-labeled (DL) cell line for sequential IP. 35 µg of DL WT fraction 6 protein was treated with either SMA in a ratio of 12:1 SMA:protein or 1% Triton X-100 and immunoisolated with GFP-LC3B using 100 μl GFP microbeads, otherwise as described previously before protein precipitation. GdnHCl treatment was performed the same as above. The sample was then diluted to 225 mM GdnHCl in 1X SHB and applied to pre-washed Pierce anti-FLAG magnetic agarose beads (A36797; Thermo Fisher Scientific) in 50 ml tubes for 30 min at room temperature with mixing. The 50 ml tubes were centrifuged at 400 rpm to pellet the beads, and the flow-through was removed. The beads were then transferred to 1.5 ml microcentrifuge tubes and washed three times with 1X SHB on a magnetic stand. The protein was eluted by adding 1X LDS, boiling the samples at 98°C for 10 min, removing the beads on a magnetic stand, and adding 15 mM DTT. Samples were then used for SDS-PAGE and immunoblotting.
It is important to note that the dominant mechanism by which GdnHCl denatures protein is reported to be electrostatic interactions between the guanidinium ion and charged residues as well as the peptide backbone (O’Brien et al., 2007). For this reason, GdnHCl should unfold proteins without threatening the captured lipids or SMA copolymer itself. The SMA copolymer is reportedly pH-sensitive (Dörr et al., 2016), and the aqueous GdnHCl solution is weakly acidic. SMALP samples were buffered at pH 7.4 and the addition of GdnHCl to a final concentration of 6 M only shifted the pH to 7.2, remaining in the neutral range. In any case, SMA 2:1, which is the copolymer ratio we use for this study, functions at the widest pH range and remains soluble at a pH as low as 5.0 (Scheidelaar et al., 2016).
Recombinant protein expression and purification
Mouse ATG3, human LC3B, and human GFP-LC3B were expressed and purified as previously described (Kauffman et al., 2018; Nath et al., 2014). For all in vitro experiments, LC3B and GFP-LC3B refer to protein expressed and purified from constructs ending at the reactive glycine (G120) of LC3B, and thus no ATG4-mediated pre-processing was needed for lipidation. In summary, human LC3B and GFP-LC3B were cloned in the pGEX-2T GST vector, and mouse ATG3 was cloned into pGEX-6p GST vector. All three proteins were expressed in BL21-Gold (DE3) competent cells (Agilent Technologies; 230132). The cells were cultured in 4 liters of Luria Bertani broth (LB) medium with 50 mg/ml carbenicillin (C1389; Sigma-Aldrich) and induced with 0.5 mM isopropyl β-D-thiogalactopyranoside (IPTG). Bacterial pellets were treated with EDTA-free protease inhibitor tablets in either thrombin buffer (20 mM Tris pH 7.5, 100 mM NaCl, 5 mM MgCl2, 2 mM CaCl2, 1 mM DTT) for LC3B and GFP-LC3B or preScission protease buffer (50 mM Tris pH 7.5, 150 mM NaCl, 1 mM EDTA, 1 mM DTT) for ATG3. The cells were broken in a cell disrupter, and the lysate was incubated with pre-washed glutathione agarose beads (G4501; Sigma-Aldrich) for 3 h at 4°C. The beads were washed three times and then incubated with thrombin (for LC3B/GFP-LC3; 10 μl of thrombin [T6884; Sigma-Aldrich], 500 μl of thrombin buffer, 0.5 μl of DTT, 500 μl of beads) or preScission protease (for ATG3; 25 μl of preScission protease [GE27084301; Sigma-Aldrich], 500 μl of preScission protease buffer, 0.5 μl of DTT, 500 μl of beads) to cut the proteins from GST tags overnight at 4°C. Purified proteins were stored in 20% glycerol at −80°C.
Protein expression and purification for human ATG7
Human ATG7 was expressed and purified as previously described (Choy et al., 2012). In summary, the ATG7-containing plasmid was transformed into Bacmid DNA. 2 µg of DNA was used to infect 8 × 105 SF9 cells by using Cellfectin II (10362100; Thermo Fisher Scientific) two times to increase viral titer to 5 × 106 plaque-forming units/ml. 1 × 108 plaque-forming unit/ml of SF9 cells were infected with virus and grew for 72 h. The cells were treated with PIC tablets in the lysis buffer (20 mM Tris pH 8, 500 mM NaCl, 20 mM imidazole, 1 mM DTT, 10% glycerol), sonicated with the Virsonic 600 (VirTis) microtip sonicator for 3 min (30 s on, 30 s off; intensity 3.5), and centrifuged at 18,000 rpm for 1 h. The lysate was incubated with 1 ml of nickel resin (nickel–nitrilotriacetic acid–agarose) for 2 h at 4°C. The beads were washed with the wash buffer (20 mM Tris pH 8, 300 mM NaCl, 20 mM imidazole [I5513; Sigma-Aldrich], 1 mM DTT) three times and eluted with the elution buffer (20 mM Tris pH 7.5, 300 mM NaCl, 500 mM imidazole, 1 mM DTT). Purified proteins were stored in 20% glycerol at −80°C.
Lipidation assay, float up, and immunoprecipitation following SMA extraction
Recombinant LC3B and GFP-LC3B underwent the lipidation process to be coupled to liposomes at the same time. In short, LC3B protein (30 µM), GFP-LC3B protein (15 µM), ATG3 (16 µM), ATG7 (4 µM), and sonicated liposomes (8 mM) were mixed with DTT (1 mM) in SN buffer. Lipidation was initiated by adding ATP (1 mM), the reaction was incubated at 37°C for 1 h, 1 mM of ATP was added again to the reaction, and the reaction was incubated at 37°C for 1 h.
Protein-coupled liposomes were floated up by loading them onto Nycodenz gradients (150 μl of lipidation reaction mixed with 150 μl of 80% Nycodenz [40% Nycodenz layer] on bottom, 250 μl of 30% Nycodenz layered next, and 50 μl of 1X SN buffer on top) in a thin-walled ultracentrifuge tube (355090; Beckman Coulter), and centrifuging them at 48,000 rpm for 4 h at 4°C. Protein-coupled liposomes were recovered in 40 μl from the interface between 30% Nycodenz and buffer. Nycodenz was diluted with 5X SN buffer to final concentrations in 1X SN buffer in 10 ml. Protein-coupled liposomes were collected and pooled.
To validate natural membrane results with these liposomes decorated with only LC3B and GFP-LC3B, 37.5 μl of lipidated material was treated with 2.5% SMA, 0.5% SMA, or 1% Triton X-100. Samples were immunoprecipitated with GFP-LC3B using GFP microbeads as described in previous sections.
Online supplemental material
Fig. S1 shows line scans of more LC3B-positive vesicle clusters in ATG2 DKO cells. Fig. S2 shows lipidation and p62 turnover defects in ATG9A KO cells, as well as the lipidation defects in FIP200 KO and ATG3 KO cells. Fig. S3 is an annotated workflow for the isolation of autophagosomes. Fig. S4 shows the distribution of ATG9A, GFP-LC3B, LC3B, Calnexin, and TOM20 in basal WT membrane fractions. It also shows the distribution of lipidated LC3B and GFP-LC3B in WT and ATG2 DKO cells, and contains more GdnHCl sequential IPs under basal conditions and using FLAG-ATG9A pull down instead of endogenous ATG9A. Fig. S5 compares basal WT, starved and bafilomycin A1-treated WT, and basal ATG2 DKO cell membrane fractions and the IPs in Fig. 6 between the two cell lines. Additionally, it includes an in vitro experiment of SMA-treated liposomes decorated with GFP-LC3B and LC3B. Table S1 is the total recovery of ATG9A in autophagosomes between WT and all KO cell lines.
Supplementary Material
shows total protein recovery in the autophagosome fraction.
Acknowledgments
We thank Chenxiang Lin for access to the transmission electron microscope at the Yale West Campus.
This work was supported by grants from the National Institutes of Health (R01 GM100930 to T.J. Melia; R01 GM141192 to K. Gupta; R01 NS036251 and P30 DK045735 to P. De Camilli), predoctoral training grants from the China Scholarship Council to S. Yu and from the National Institutes of Health to T.J. Olivas (T32 GM007223), and a Ruth L. Kirschstein National Research Service Award Individual Predoctoral Fellowship to Promote Diversity in Health-Related Research from the National Institutes of Health to T.J. Olivas (F31 GM142294). We are grateful to the facilities that supported this work including the Yale Center for Cellular and Molecular Imaging (both the fluorescence and electron microscopy facilities) and the Yale Diabetes Research Center.
Author contributions: T.J. Olivas designed and carried out the immunofluorescence experiments, creation of the ATG9A KO and FIP200 KO, density gradient membrane fractionations, and all assays using SMA copolymer nanodiscs except for performance of the SMA isolations from liposomes carried out by P. Choi. S. Nag prepared the reconstituted proteins for the lipidation of the liposomes in that experiment. S. Yu discovered the pre-ATG2 compartment in ATG2 DKO cells and initiated the collaboration with Y. Wu who performed the FIB-SEM, TEM, and following electron microscopy analysis. E.D. Guinn created and validated the ATG3 KO. L. Luan performed the initial 3D segmentation of the FIB-SEM results, later updated by Y. Wu. T.J. Melia designed the project and supervised it with collaboration from P. De Camilli and K. Gupta. The manuscript was prepared by T.J. Melia and T.J. Olivas with input from all authors.
Data availability
Tabular data underlying Fig. 3, D and E, 4, E–G, 6, C and F, S4 B, and S5, A–I and K are openly available in Zenodo at https://doi.org/10.5281/zenodo.7799942.
References
- Axe, E.L., Walker S.A., Manifava M., Chandra P., Roderick H.L., Habermann A., Griffiths G., and Ktistakis N.T.. 2008. Autophagosome formation from membrane compartments enriched in phosphatidylinositol 3-phosphate and dynamically connected to the endoplasmic reticulum. J. Cell Biol. 182:685–701. 10.1083/jcb.200803137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behrends, C., Sowa M.E., Gygi S.P., and Harper J.W.. 2010. Network organization of the human autophagy system. Nature. 466:68–76. 10.1038/nature09204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bieber, A., Capitanio C., Erdmann P.S., Fiedler F., Beck F., Lee C.W., Li D., Hummer G., Schulman B.A., Baumeister W., and Wilfling F.. 2022. In situ structural analysis reveals membrane shape transitions during autophagosome formation. Proc. Natl. Acad. Sci. USA. 119:e2209823119. 10.1073/pnas.2209823119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bozic, M., van den Bekerom L., Milne B.A., Goodman N., Roberston L., Prescott A.R., Macartney T.J., Dawe N., and McEwan D.G.. 2020. A conserved ATG2-GABARAP family interaction is critical for phagophore formation. EMBO Rep. 21:e48412. 10.15252/embr.201948412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chowdhury, S., Otomo C., Leitner A., Ohashi K., Aebersold R., Lander G.C., and Otomo T.. 2018. Insights into autophagosome biogenesis from structural and biochemical analyses of the ATG2A-WIPI4 complex. Proc. Natl. Acad. Sci. USA. 115:E9792–E9801. 10.1073/pnas.1811874115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choy, A., Dancourt J., Mugo B., O’Connor T.J., Isberg R.R., Melia T.J., and Roy C.R.. 2012. The Legionella effector RavZ inhibits host autophagy through irreversible Atg8 deconjugation. Science. 338:1072–1076. 10.1126/science.1227026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claude-Taupin, A., Jia J., Bhujabal Z., Garfa-Traoré M., Kumar S., da Silva G.P.D., Javed R., Gu Y., Allers L., Peters R., et al. 2021. ATG9A protects the plasma membrane from programmed and incidental permeabilization. Nat. Cell Biol. 23:846–858. 10.1038/s41556-021-00706-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dooley, H.C., Razi M., Polson H.E.J., Girardin S.E., Wilson M.I., and Tooze S.A.. 2014. WIPI2 links LC3 conjugation with PI3P, autophagosome formation, and pathogen clearance by recruiting Atg12-5-16L1. Mol. Cell. 55:238–252. 10.1016/j.molcel.2014.05.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dooley, H.C., Wilson M.I., and Tooze S.A.. 2015. WIPI2B links PtdIns3P to LC3 lipidation through binding ATG16L1. Autophagy. 11:190–191. 10.1080/15548627.2014.996029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dörr, J.M., Scheidelaar S., Koorengevel M.C., Dominguez J.J., Schäfer M., van Walree C.A., and Killian J.A.. 2016. The styrene-maleic acid copolymer: A versatile tool in membrane research. Eur. Biophys. J. 45:3–21. 10.1007/s00249-015-1093-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esmaili, M., and Overduin M.. 2018. Membrane biology visualized in nanometer-sized discs formed by styrene maleic acid polymers. Biochim. Biophys. Acta Biomembr. 1860:257–263. 10.1016/j.bbamem.2017.10.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng, Y., and Klionsky D.J.. 2017. Autophagic membrane delivery through ATG9. Cell Res. 27:161–162. 10.1038/cr.2017.4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fengsrud, M., Erichsen E.S., Berg T.O., Raiborg C., and Seglen P.O.. 2000. Ultrastructural characterization of the delimiting membranes of isolated autophagosomes and amphisomes by freeze-fracture electron microscopy. Eur. J. Cell Biol. 79:871–882. 10.1078/0171-9335-00125 [DOI] [PubMed] [Google Scholar]
- Ghanbarpour, A., Valverde D.P., Melia T.J., and Reinisch K.M.. 2021. A model for a partnership of lipid transfer proteins and scramblases in membrane expansion and organelle biogenesis. Proc. Natl. Acad. Sci. USA. 118:e2101562118. 10.1073/pnas.2101562118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gómez-Sánchez, R., Rose J., Guimarães R., Mari M., Papinski D., Rieter E., Geerts W.J., Hardenberg R., Kraft C., Ungermann C., and Reggiori F.. 2018. Atg9 establishes Atg2-dependent contact sites between the endoplasmic reticulum and phagophores. J. Cell Biol. 217:2743–2763. 10.1083/jcb.201710116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guardia, C.M., Tan X.F., Lian T., Rana M.S., Zhou W., Christenson E.T., Lowry A.J., Faraldo-Gómez J.D., Bonifacino J.S., Jiang J., and Banerjee A.. 2020. Structure of human ATG9A, the only transmembrane protein of the core autophagy machinery. Cell Rep. 31:107837. 10.1016/j.celrep.2020.107837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gudmundsson, S.R., Kallio K.A., Vihinen H., Jokitalo E., Ktistakis N., and Eskelinen E.L.. 2022. Morphology of phagophore precursors by correlative light-electron microscopy. Cells. 11:3080. 10.3390/cells11193080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamasaki, M., Furuta N., Matsuda A., Nezu A., Yamamoto A., Fujita N., Oomori H., Noda T., Haraguchi T., Hiraoka Y., et al. 2013. Autophagosomes form at ER-mitochondria contact sites. Nature. 495:389–393. 10.1038/nature11910 [DOI] [PubMed] [Google Scholar]
- Hart, T., Tong A.H.Y., Chan K., Van Leeuwen J., Seetharaman A., Aregger M., Chandrashekhar M., Hustedt N., Seth S., Noonan A., et al. 2017. Evaluation and design of genome-wide CRISPR/SpCas9 knockout screens. G3. 7:2719–2727. 10.1534/g3.117.041277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He, C., Baba M., Cao Y., and Klionsky D.J.. 2008. Self-interaction is critical for Atg9 transport and function at the phagophore assembly site during autophagy. Mol. Biol. Cell. 19:5506–5516. 10.1091/mbc.e08-05-0544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He, C., Song H., Yorimitsu T., Monastyrska I., Yen W.L., Legakis J.E., and Klionsky D.J.. 2006. Recruitment of Atg9 to the preautophagosomal structure by Atg11 is essential for selective autophagy in budding yeast. J. Cell Biol. 175:925–935. 10.1083/jcb.200606084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai, K., Hao F., Fujita N., Tsuji Y., Oe Y., Araki Y., Hamasaki M., Noda T., and Yoshimori T.. 2016. Atg9A trafficking through the recycling endosomes is required for autophagosome formation. J. Cell Sci. 129:3781–3791. 10.1242/jcs.196196 [DOI] [PubMed] [Google Scholar]
- Jamshad, M., Grimard V., Idini I., Knowles T.J., Dowle M.R., Schofield N., Sridhar P., Lin Y.P., Finka R., Wheatley M., et al. 2015. Structural analysis of a nanoparticle containing a lipid bilayer used for detergent-free extraction of membrane proteins. Nano Res. 8:774–789. 10.1007/s12274-014-0560-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong, H., Then F., Melia T.J. Jr, Mazzulli J.R., Cui L., Savas J.N., Voisine C., Paganetti P., Tanese N., Hart A.C., et al. 2009. Acetylation targets mutant huntingtin to autophagosomes for degradation. Cell. 137:60–72. 10.1016/j.cell.2009.03.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karanasios, E., Walker S.A., Okkenhaug H., Manifava M., Hummel E., Zimmermann H., Ahmed Q., Domart M.C., Collinson L., and Ktistakis N.T.. 2016. Autophagy initiation by ULK complex assembly on ER tubulovesicular regions marked by ATG9 vesicles. Nat. Commun. 7:12420. 10.1038/ncomms12420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kauffman, K.J., Yu S., Jin J., Mugo B., Nguyen N., O’Brien A., Nag S., Lystad A.H., and Melia T.J.. 2018. Delipidation of mammalian Atg8-family proteins by each of the four ATG4 proteases. Autophagy. 14:992–1010. 10.1080/15548627.2018.1437341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kishi-Itakura, C., Koyama-Honda I., Itakura E., and Mizushima N.. 2014. Ultrastructural analysis of autophagosome organization using mammalian autophagy-deficient cells. J. Cell Sci. 127:4089–4102. 10.1242/jcs.164293 [DOI] [PubMed] [Google Scholar]
- Knorr, R.L., Dimova R., and Lipowsky R.. 2012. Curvature of double-membrane organelles generated by changes in membrane size and composition. PLoS One. 7:e32753. 10.1371/journal.pone.0032753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotani, T., Kirisako H., Koizumi M., Ohsumi Y., and Nakatogawa H.. 2018. The Atg2-Atg18 complex tethers pre-autophagosomal membranes to the endoplasmic reticulum for autophagosome formation. Proc. Natl. Acad. Sci. USA. 115:10363–10368. 10.1073/pnas.1806727115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonzino, M., Reinisch K.M., and De Camilli P.. 2021. Insights into VPS13 properties and function reveal a new mechanism of eukaryotic lipid transport. Biochim. Biophys. Acta Mol. Cell Biol. Lipids. 1866:159003. 10.1016/j.bbalip.2021.159003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda, S., Otomo C., and Otomo T.. 2019. The autophagic membrane tether ATG2A transfers lipids between membranes. Elife. 8:e45777. 10.7554/eLife.45777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda, S., Yamamoto H., Kinch L.N., Garza C.M., Takahashi S., Otomo C., Grishin N.V., Forli S., Mizushima N., and Otomo T.. 2020. Structure, lipid scrambling activity and role in autophagosome formation of ATG9A. Nat. Struct. Mol. Biol. 27:1194–1201. 10.1038/s41594-020-00520-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mailler, E., Guardia C.M., Bai X., Jarnik M., Williamson C.D., Li Y., Maio N., Golden A., and Bonifacino J.S.. 2021. The autophagy protein ATG9A enables lipid mobilization from lipid droplets. Nat. Commun. 12:6750. 10.1038/s41467-021-26999-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mari, M., Griffith J., Rieter E., Krishnappa L., Klionsky D.J., and Reggiori F.. 2010. An Atg9-containing compartment that functions in the early steps of autophagosome biogenesis. J. Cell Biol. 190:1005–1022. 10.1083/jcb.200912089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matoba, K., Kotani T., Tsutsumi A., Tsuji T., Mori T., Noshiro D., Sugita Y., Nomura N., Iwata S., Ohsumi Y., et al. 2020. Atg9 is a lipid scramblase that mediates autophagosomal membrane expansion. Nat. Struct. Mol. Biol. 27:1185–1193. 10.1038/s41594-020-00518-w [DOI] [PubMed] [Google Scholar]
- Mattera, R., Park S.Y., De Pace R., Guardia C.M., and Bonifacino J.S.. 2017. AP-4 mediates export of ATG9A from the trans-Golgi network to promote autophagosome formation. Proc. Natl. Acad. Sci. USA. 114:E10697–E10706. 10.1073/pnas.1717327114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melia, T.J., and Reinisch K.M.. 2022. A possible role for VPS13-family proteins in bulk lipid transfer, membrane expansion and organelle biogenesis. J. Cell Sci. 135:jcs259357. 10.1242/jcs.259357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nascimbeni, A.C., Giordano F., Dupont N., Grasso D., Vaccaro M.I., Codogno P., and Morel E.. 2017. ER-plasma membrane contact sites contribute to autophagosome biogenesis by regulation of local PI3P synthesis. EMBO J. 36:2018–2033. 10.15252/embj.201797006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nath, S., Dancourt J., Shteyn V., Puente G., Fong W.M., Nag S., Bewersdorf J., Yamamoto A., Antonny B., and Melia T.J.. 2014. Lipidation of the LC3/GABARAP family of autophagy proteins relies on a membrane-curvature-sensing domain in Atg3. Nat. Cell Biol. 16:415–424. 10.1038/ncb2940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen, N., Olivas T.J., Mires A., Jin J., Yu S., Luan L., Nag S., Kauffman K.J., and Melia T.J.. 2020. The insufficiency of ATG4A in macroautophagy. J. Biol. Chem. 295:13584–13600. 10.1074/jbc.RA120.013897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen, T.N., Padman B.S., Zellner S., Khuu G., Uoselis L., Lam W.K., Skulsuppaisarn M., Lindblom R.S.J., Watts E.M., Behrends C., and Lazarou M.. 2021. ATG4 family proteins drive phagophore growth independently of the LC3/GABARAP lipidation system. Mol. Cell. 81:2013–2030.e9. 10.1016/j.molcel.2021.03.001 [DOI] [PubMed] [Google Scholar]
- Noda, N.N. 2021. Atg2 and Atg9: Intermembrane and interleaflet lipid transporters driving autophagy. Biochim. Biophys. Acta Mol. Cell Biol. Lipids. 1866:158956. 10.1016/j.bbalip.2021.158956 [DOI] [PubMed] [Google Scholar]
- O’Brien, E.P., Dima R.I., Brooks B., and Thirumalai D.. 2007. Interactions between hydrophobic and ionic solutes in aqueous guanidinium chloride and urea solutions: Lessons for protein denaturation mechanism. J. Am. Chem. Soc. 129:7346–7353. 10.1021/ja069232+ [DOI] [PubMed] [Google Scholar]
- Orsi, A., Razi M., Dooley H.C., Robinson D., Weston A.E., Collinson L.M., and Tooze S.A.. 2012. Dynamic and transient interactions of Atg9 with autophagosomes, but not membrane integration, are required for autophagy. Mol. Biol. Cell. 23:1860–1873. 10.1091/mbc.e11-09-0746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osawa, T., Kotani T., Kawaoka T., Hirata E., Suzuki K., Nakatogawa H., Ohsumi Y., and Noda N.N.. 2019. Atg2 mediates direct lipid transfer between membranes for autophagosome formation. Nat. Struct. Mol. Biol. 26:281–288. 10.1038/s41594-019-0203-4 [DOI] [PubMed] [Google Scholar]
- Park, D., Wu Y., Wang X., Gowrishankar S., Baublis A., and De Camilli P.. 2023. Synaptic vesicle proteins and ATG9A self-organize in distinct vesicle phases within synapsin condensates. Nat. Commun. 14:455. 10.1038/s41467-023-36081-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polson, H.E., de Lartigue J., Rigden D.J., Reedijk M., Urbé S., Clague M.J., and Tooze S.A.. 2010. Mammalian Atg18 (WIPI2) localizes to omegasome-anchored phagophores and positively regulates LC3 lipidation. Autophagy. 6:506–522. 10.4161/auto.6.4.11863 [DOI] [PubMed] [Google Scholar]
- Postis, V., Rawson S., Mitchell J.K., Lee S.C., Parslow R.A., Dafforn T.R., Baldwin S.A., and Muench S.P.. 2015. The use of SMALPs as a novel membrane protein scaffold for structure study by negative stain electron microscopy. Biochim. Biophys. Acta. 1848:496–501. 10.1016/j.bbamem.2014.10.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puri, C., Renna M., Bento C.F., Moreau K., and Rubinsztein D.C.. 2013. Diverse autophagosome membrane sources coalesce in recycling endosomes. Cell. 154:1285–1299. 10.1016/j.cell.2013.08.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ran, F.A., Hsu P.D., Wright J., Agarwala V., Scott D.A., and Zhang F.. 2013. Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 8:2281–2308. 10.1038/nprot.2013.143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawa-Makarska, J., Baumann V., Coudevylle N., von Bülow S., Nogellova V., Abert C., Schuschnig M., Graef M., Hummer G., and Martens S.. 2020. Reconstitution of autophagosome nucleation defines Atg9 vesicles as seeds for membrane formation. Science. 369:eaaz7714. 10.1126/science.aaz7714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheidelaar, S., Koorengevel M.C., van Walree C.A., Dominguez J.J., Dörr J.M., and Killian J.A.. 2016. Effect of polymer composition and pH on membrane solubilization by styrene-maleic acid copolymers. Biophys. J. 111:1974–1986. 10.1016/j.bpj.2016.09.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanga, D., Zhao Q., Milev M.P., Saint-Dic D., Jimenez-Mallebrera C., and Sacher M.. 2019. TRAPPC11 functions in autophagy by recruiting ATG2B-WIPI4/WDR45 to preautophagosomal membranes. Traffic. 20:325–345. 10.1111/tra.12640 [DOI] [PubMed] [Google Scholar]
- Strømhaug, P.E., Berg T.O., Fengsrud M., and Seglen P.O.. 1998. Purification and characterization of autophagosomes from rat hepatocytes. Biochem. J. 335:217–224. 10.1042/bj3350217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, D., Wu R., Zheng J., Li P., and Yu L.. 2018. Polyubiquitin chain-induced p62 phase separation drives autophagic cargo segregation. Cell Res. 28:405–415. 10.1038/s41422-018-0017-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura, N., Nishimura T., Sakamaki Y., Koyama-Honda I., Yamamoto H., and Mizushima N.. 2017. Differential requirement for ATG2A domains for localization to autophagic membranes and lipid droplets. FEBS Lett. 591:3819–3830. 10.1002/1873-3468.12901 [DOI] [PubMed] [Google Scholar]
- Tang, Z., Takahashi Y., Chen C., Liu Y., He H., Tsotakos N., Serfass J.M., Gebru M.T., Chen H., Young M.M., and Wang H.G.. 2017. Atg2A/B deficiency switches cytoprotective autophagy to non-canonical caspase-8 activation and apoptosis. Cell Death Differ. 24:2127–2138. 10.1038/cdd.2017.133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang, Z., Takahashi Y., He H., Hattori T., Chen C., Liang X., Chen H., Young M.M., and Wang H.G.. 2019. TOM40 targets Atg2 to mitochondria-associated ER membranes for phagophore expansion. Cell Rep. 28:1744–1757.e5. 10.1016/j.celrep.2019.07.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tonge, S.R., and Tighe B.J.. 2001. Responsive hydrophobically associating polymers: A review of structure and properties. Adv. Drug Deliv. Rev. 53:109–122. 10.1016/S0169-409X(01)00223-X [DOI] [PubMed] [Google Scholar]
- Uemura, T., Yamamoto M., Kametaka A., Sou Y.S., Yabashi A., Yamada A., Annoh H., Kametaka S., Komatsu M., and Waguri S.. 2014. A cluster of thin tubular structures mediates transformation of the endoplasmic reticulum to autophagic isolation membrane. Mol. Cell. Biol. 34:1695–1706. 10.1128/MCB.01327-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valverde, D.P., Yu S., Boggavarapu V., Kumar N., Lees J.A., Walz T., Reinisch K.M., and Melia T.J.. 2019. ATG2 transports lipids to promote autophagosome biogenesis. J. Cell Biol. 218:1787–1798. 10.1083/jcb.201811139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Vliet, A.R., Chiduza G.N., Maslen S.L., Pye V.E., Joshi D., De Tito S., Jefferies H.B.J., Christodoulou E., Roustan C., Punch E., et al. 2022. ATG9A and ATG2A form a heteromeric complex essential for autophagosome formation. Mol. Cell. 82:4324–4339.e8. 10.1016/j.molcel.2022.10.017 [DOI] [PubMed] [Google Scholar]
- Vargas, C., Arenas R.C., Frotscher E., and Keller S.. 2015. Nanoparticle self-assembly in mixtures of phospholipids with styrene/maleic acid copolymers or fluorinated surfactants. Nanoscale. 7:20685–20696. 10.1039/C5NR06353A [DOI] [PubMed] [Google Scholar]
- Velikkakath, A.K., Nishimura T., Oita E., Ishihara N., and Mizushima N.. 2012. Mammalian Atg2 proteins are essential for autophagosome formation and important for regulation of size and distribution of lipid droplets. Mol. Biol. Cell. 23:896–909. 10.1091/mbc.e11-09-0785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker, G., Brown C., Ge X., Kumar S., Muzumdar M.D., Gupta K., and Bhattacharyya M.. 2023. Determination of oligomeric organization of membrane proteins from native membranes at nanoscale-spatial and single-molecule resolution. bioRxiv. (Preprint posted February 19, 2023). 10.1101/2023.02.19.529138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto, H., Kakuta S., Watanabe T.M., Kitamura A., Sekito T., Kondo-Kakuta C., Ichikawa R., Kinjo M., and Ohsumi Y.. 2012. Atg9 vesicles are an important membrane source during early steps of autophagosome formation. J. Cell Biol. 198:219–233. 10.1083/jcb.201202061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young, A.R.J., Chan E.Y., Hu X.W., Köchl R., Crawshaw S.G., High S., Hailey D.W., Lippincott-Schwartz J., and Tooze S.A.. 2006. Starvation and ULK1-dependent cycling of mammalian Atg9 between the TGN and endosomes. J. Cell Sci. 119:3888–3900. 10.1242/jcs.03172 [DOI] [PubMed] [Google Scholar]
- Zhang, T., Guo L., and Yang Y.. 2020. Mammalian ATG9s drive the autophagosome formation by binding to LC3. bioRxiv. (Preprint posted May 12, 2020). 10.1101/2020.05.12.091637 [DOI] [Google Scholar]
- Zhao, Y.G., Chen Y., Miao G., Zhao H., Qu W., Li D., Wang Z., Liu N., Li L., Chen S., et al. 2017. The ER-localized transmembrane protein EPG-3/VMP1 regulates SERCA activity to control ER-isolation membrane contacts for autophagosome formation. Mol. Cell. 67:974–989.e6. 10.1016/j.molcel.2017.08.005 [DOI] [PubMed] [Google Scholar]
- Zhou, C., Wu Z., Du W., Que H., Wang Y., Ouyang Q., Jian F., Yuan W., Zhao Y., Tian R., et al. 2022. Recycling of autophagosomal components from autolysosomes by the recycler complex. Nat. Cell Biol. 24:497–512. 10.1038/s41556-022-00861-8 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
shows total protein recovery in the autophagosome fraction.
Data Availability Statement
Tabular data underlying Fig. 3, D and E, 4, E–G, 6, C and F, S4 B, and S5, A–I and K are openly available in Zenodo at https://doi.org/10.5281/zenodo.7799942.