Abstract
Whole slide images (WSI) based survival prediction has attracted increasing interest in pathology. Despite this, extracting prognostic information from WSIs remains a challenging task due to their enormous size and the scarcity of pathologist annotations. Previous studies have utilized multiple instance learning approach to combine information from several randomly sampled patches, but this approach may not be adequate as different visual patterns may contribute unequally to prognosis prediction. In this study, we introduce a multi-head attention mechanism that allows each attention head to independently explore the utility of various visual patterns on a tumor slide, thereby enabling more comprehensive information extraction from WSIs. We evaluated our approach on four cancer types from The Cancer Genome Atlas database. Our model achieved an average c-index of 0.640, outperforming three existing state-of-the-art approaches for WSI-based survival prediction on these datasets. Visualization of attention maps reveals that the attention heads synergistically focus on different morphological patterns, providing additional evidence for the effectiveness of multi-head attention in survival prediction.
Keywords: multiple instance learning, neural networks, digital pathology, cancer prognosis
1. Introduction
Each year, nearly 2 million people living in the United States are diagnosed with cancer; one-third of these patients will die within five years1. The accurate and timely prediction of patient survival is crucial for shared clinical decision-making, treatment planning, and patient psychological adjustments2. While both host- and environment-related factors can affect the survival of the cancer patient, the tumor itself is the most fundamental prognostic factor3. Pathological examination is a routine procedure at the time of diagnosis to determine the type and malignancy of tumors. Moreover, the histopathological features of cancer, including tumor size, lymph node involvement, and metastasis, are commonly used in survival prediction models4,5, proving the prognostic value of morphological features of tumors. However, the massive amount of information in histology slides presents formidable challenges. Manual information extraction from the microscopic examination is time-consuming, and the amount of information structurally collected in pathology reports is often limited to predetermined criteria and may vary by cancer types6,7. There has been a strong need for a tool to maximize the utility of histopathological features and to make accurate prognosis predictions for cancer patients.
To solve this problem, a growing body of work has focused on applying deep learning methods for predicting prognosis using whole-slide images (WSIs) 8–15. WSIs are the digitized versions of the complete microscope slides scanned at high resolution. A typical WSI can occupy several gigabytes of storage and contain billions of pixels, while the lesions may only cover a small region on the whole slide. Considering all regions with equal importance could miss the areas that are critical for prognosis. It has been reported that incorporating region-of-interest (ROI) annotations on WSIs could improve prognosis prediction performance16; however, such annotations require extensive time and expertise and are usually not readily available. Furthermore, prognostic information is not confined to tumor tissues. Lymphocyte aggregation, angiogenesis, and stroma are all found to be related to cancer progression and prognosis14,17–19. The lack of meaningful ROI annotations on WSIs limits the application of machine learning prognostic tools in clinical practice. Besides, using WSIs for prognosis prediction poses several other challenges: 1) accurate survival prediction requires both a holistic representation of a WSI to evaluate the extent of the tumor and detailed features at the cellular level to assess tumor characteristics, which are the opposite ends of the spectrum in image analysis; and 2) the whole slide datasets with survival information are usually of limited sample size, while the problem worsens as outcome information is often only available for a portion of the participants because either the study ended before the event occurred or the participants left during follow-up (i.e., right censoring). The insufficient number of events makes a deep learning model overfit easily and the evaluation of the model performance dependent on data partitions.
The malignancy of a tumor is affected by many factors, such as the size of the tumor and the mitotic activity of cells within the tumor. Focusing only on the tumor tissue can provide a better assessment of the cancer cells’ characteristics. However, this approach may overlook some important global features, such as the size of the tumor. On the other hand, although the relative size of the tumor on the slide can be reflected by the intensity of the activation signal from the neural networks when taking the entire slide into account, such averaging operation assigns identical attention to all tissue types, which inevitably dilute the impact of individual tumor cells. Therefore, to reach a more accurate prediction of the patient’s survival, it is necessary to inspect the histopathology slide from various aspects.
Recent studies have focused on using attention mechanisms to automatically determine the importance of each patch for prognosis prediction tasks20,21. Within this framework, each instance is assigned an attention weight, and their respective feature vectors are combined by taking a weighted average over all instances. However, as the importance of each instance can only be evaluated by a single scale, the model may be limited in learning alternative patterns that are also significant for prognosis prediction. Meanwhile, in the field of sequence modeling, by using multiple layers of multi-head self-attention modules, the transformer model outperforms previous methods by a significant margin22. Despite its success, applying the transformer model directly to whole-slide images remains a challenging task, given the large size of individual whole-slide images and the significant amount of data and computational resources required to train a transformer model from scratch.
In light of the challenges described above, this paper proposes MHAttnSurv, a multi-head attention framework for cancer survival prediction using WSIs. Inspired by the powerful multi-head attention mechanism introduced in the transformer model, we modified this mechanism innovatively to allow it to work efficiently in the field of multiple instance learning (MIL). Specifically, the transformer model learns a context-sensitive representation for each element in the input sequence. This requires attention between every pair of elements and the computational cost increases quadratically with input length. However, for the prognostic information extraction task, only the global representation of the input sequence is of interest. This allows us to remove the nonessential but computationally expensive pairwise attentions among the input sequence and only keep the attentions between global query and local keys. With this modification, the computational cost scales linearly with input length. We compare our method with current state-of-the-art methods and perform extensive experiments on 4 cancer types from The Cancer Genome Atlas (TCGA) database to demonstrate its superiority. Our contributions can be summarized as follows:
We present an efficient and flexible attention-based framework with multiple attention heads for survival prediction.
We performed rigorous experiments using nested cross-validation. The experiments demonstrated that our proposed method performs better than existing state-of-the-art approaches, and is easier to implement and adapt for various tasks.
By visualizing the attention map and inspecting the prediction ability of each attention head, we have demonstrated that our approach incorporates comprehensive morphological patterns into survival prediction.
The rest of the paper is structured as follows: Section 2 provides an overview of related work. Section 3 outlines the details of our proposed method. Section 4 presents the experimental setup and results. Section 5 offers a discussion of the findings. Finally, Section 6 concludes our work.
2. Related work
2.1. Manually generated features
Early studies have relied on manually extracted features for survival analysis. Tumor size, grade, and stage are commonly used in statistical models for prognosis prediction23,24. These features are routinely collected by pathologists and are available from pathology reports. Despite their high prognostic values, these variables can only provide crude partitions for the population and cannot further stratify the risk to guide individual prognostic estimates. More specific biomarkers, such as mitotic index (MI), have been shown to be an independent predictor of long-term survival of cancer patients25. While these biomarkers usually have strong clinical significance, obtaining them requires extensive inputs from domain experts. This process can be time-consuming and subject to human discretion. Moreover, prognostic pathological factors might only be specific to certain cancer types. Another related method is to use high-throughput cell image analysis tools (e.g., CellProfiler) to extract predefined cell features automatically, such as size, shape, intensity, and texture26,27. However, these handcrafted features are usually limited and relatively redundant28, and may not contain high-level prognostic information.
2.2. Automatic feature extraction with ROI annotation
With the evolution of computer vision techniques, automatic feature extraction using the latest deep learning models has become the standard practice. Nevertheless, training a deep convolutional neural network (CNN) model using the entire WSI is not a computationally feasible task given the enormous size of WSIs. As a result, WSIs are broken into smaller images, called patches, with standard image sizes for typical computer vision tasks. This approach treats the survival prediction problem as a multiple instance learning problem and does not consider the sequence or spatial location of the patches. As patches usually contain redundant information, a sampling approach is used to select a subset of patches for model training and evaluation. The DeepConvSurv model developed by Zhu et al. sampled patches of size 1024×1024 from the ROIs of WSIs29. These sampled patches were processed with a CNN model so the local features within the patches could be extracted for survival prediction. Another method by Mobadersany et al. sampled one patch from the ROI region as the input for a CNN model10. During prediction time, multiple patches were sampled from each ROI region, and the median risk score was computed. For patients with multiple ROI regions, the second largest risk score was used as the final risk score for the patient. Nevertheless, these methods still rely on expensive and subjective ROI annotations from pathologists, which limits their future applications.
In another study, a separate model was trained to predict whether each patch belongs to ROI or not30. The predicted ROI probability was used as the weight to average patches. During inference time, the pre-trained ROI differentiation model predicts the probability that each patch belongs to the ROI. This approach only requires the ROI annotation at the training stage but not the inference stage. A similar approach was used in other studies but with hard segmentation instead of probability weighting13,16. Despite a significant reduction in ROI annotation costs, these methods remain susceptible to incomplete prognostic information, as morphological features beyond the tumor region may also contribute to risk prediction.
2.3. Automatic feature extraction without ROI annotation
Recent studies have focused on predicting the survival status of patients without ROI annotation. Wulczyn et al. used a weakly supervised approach to randomly sample several patches (e.g., 16 patches per case/iteration) and feed them to a CNN model12. The underlying assumption is that when a sufficiently large number of patches are sampled, the chance of having discriminative patches approaches 1. Average pooling was used to aggregate the feature vectors from multiple patches, and the resulting global feature vector was used for survival analysis. This entire system (i.e., feature extraction + survival prediction) was trained end-to-end. This method was also used in a prior study for predicting prognosis and genetic profiling of lower-grade gliomas and has achieved satisfying results31. Nevertheless, because this method does not distinguish discriminative patches from non-discriminative ones, there is still room for further improvement.
In order to select the discriminative patches for survival prediction, the WSISA model resizes the patches into smaller thumbnails and uses K-means to group them into several clusters8. They then fit separate DeepConvSurv models for each cluster and select the clusters with high prognosis accuracy. Despite being able to identify discriminative patch clusters, this method consists of several steps and needs to train multiple separate models. As a result, the interplay among different feature types cannot be captured, which limits its performance.
2.4. Attention-based studies
Another approach to bypass the ROI annotation requirement for survival analysis is to let the model choose the discriminative patches using the attention mechanism. Attention was first designed for the sequence-to-sequence (seq2seq) models, such as natural language machine translation, to help memorize long source sentences32. The attention mechanism was soon brought to the computer vision field to generate captions for a given image by sequentially focusing on different locations of the image33. In the digital pathology field, a study by Tomita et al. used a 3-D convolutional layer to derive an attention map for aggregating feature vectors from all patches34. 64 such filters were used to increase the model’s capacity to recognize more complex patterns. A later method, clustering-constrained attention multiple instance learning (CLAM), uses an attention network to aggregate patch features for slide-level classification. The attention output can be further used by clustering layers to facilitate the separation between positive and negative classes35. While these methods are promising for the whole slide classification task, they cannot be adopted for survival prediction, given the lack of positive/negative patch distinction.
The graph convolutional neural network (GCN) model has also been explored to represent the topological features of WSIs together with the attention mechanism. The DeepGraphSurv model36 constructs graphs from the feature representations of randomly sampled patches on WSIs. A parallel attention network helps with the selection of discriminative patches. A recent method, Patch-GCN37, constructs graphs based on WSIs by connecting each patch to the proximal patches, which allows the model to learn context-aware features. The GCN method also has been used in pancreatic cancer for histology pattern detection38.
As for the MIL-based analysis, Ilse et al. proposed an attention mechanism to adaptively aggregate multiple instances20. This attention mechanism was incorporated into the WSISA model by a later method called Deep Attention Multiple Instance Survival Learning, or DeepAttnMISL21. In this study, the patches were first grouped into multiple clusters (e.g., 8) using K-means. For each patient, the cluster-specific features were obtained from patches belonging to the same cluster. Finally, these multiple cluster-specific features were aggregated using an attention layer. This approach achieved promising results for survival prediction. However, as the patch clusters were determined beforehand, they may not directly relate to prognosis. Besides, as the clustering algorithm blocks the gradient flow from patches to the final predictions, it cannot be trained end-to-end.
A recent breakthrough in sequence analysis, the transformer model, is based solely on the attention mechanism22. The attention mechanism used in the transformer is also called Query-Key-Value (QKV) attention, following the terminology in database systems. Briefly, each record is stored as a key-value pair, where the value is the actual content. The query token works as the “search” term and will be compared with the key token to determine the relevance of each record. The self-attention algorithm allows each element to “attend” to every other component in the sequence. By integrating information from each other, a better representation could be obtained. The transformer model has achieved exciting results in both natural language processing39 and computer vision tasks40. A recent method, Multimodal Co-Attention Transformer (MCAT), combines genomic-guided co-attention with set-based MIL transformers for survival prediction35. Despite the success of this method, it remains a challenging task to expand the usage of the transformer model for general survival prediction purposes given the large number of parameters and relatively small sample size of labeled WSI datasets.
3. Methods
For each patient i in our study, denotes all the ci patches that belong to this patient, and the number of patches ci differs across patients. Under the framework of MIL, the order of the patches does not matter as P is considered as a bag of instances41,42. The observed follow-up time is denoted as ti and the status at the end of follow-up is denoted as δi. For a patient who died during the follow-up, and ti is the survival time of this patient since cancer diagnosis. Otherwise indicating the patient was censored. Our goal is to learn a function that estimates the risk score (RS) from P for every patient, so the lower this score is, the longer this patient is expected to survive after a cancer diagnosis.
Our proposed MHAttnSurv method is illustrated in Figure 1. Briefly, we first use a backbone ResNet model to extract features from randomly sampled WSI patches. Then we project the feature map into values and keys. We split the value and key matrix and the learnable query vector into several chunks. Within each chunk, the attention process works in parallel to explore and identify the discriminative regions. The result from each attention map will then be concatenated for survival prediction.
Figure 1.
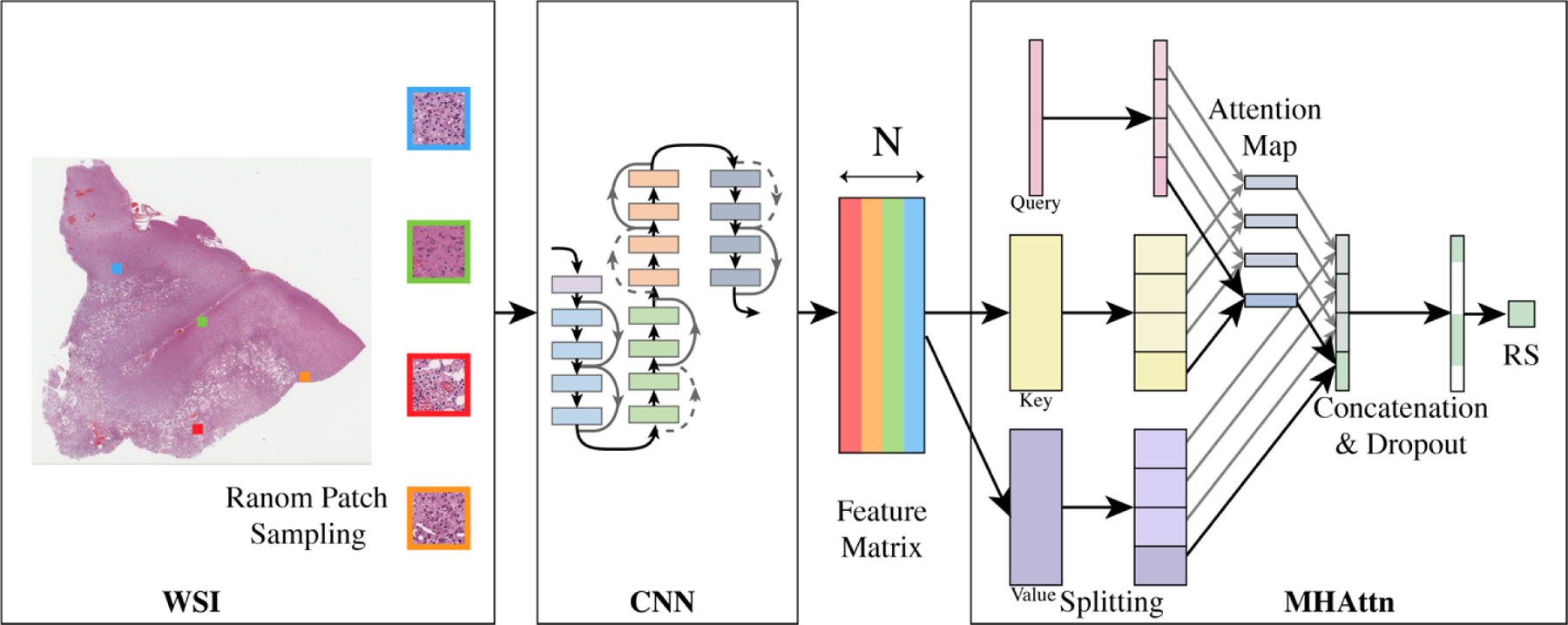
Overview of the model structure of MHAttnSurv. From each WSI, 𝑁 patches are randomly sampled. 4 heads are shown in the multi-head attention part. WSI: whole slide image; CNN: convolutional neural network; MHAttn: multi-head attention; RS: risk score estimate.
3.1. Sampling and feature extraction
Because each WSI typically contains thousands of patches and each patient can have multiple WSIs, it is not feasible to feed all of them to a model at once. Instead, we use a sampling approach to select N patches randomly during each iteration.
The ResNet model pre-trained on ImageNet provides good visual feature representations and is widely used in biomedical domains43–45. We use a ResNet model with 18 layers (i.e., ResNet-18) for feature extraction from the randomly sampled patches. By using the pre-trained backbone model, we can save time and computational resources by skipping the step of training a feature extractor from scratch and focus on developing the survival prediction model. The feature vectors from the ResNet-18 model are of size 512, and they are used as the input for the attention model.
3.2. Multi-Head Attention
Our proposed multi-head attention mechanism consists of three components analogous to self-attention in a transformer model, i.e., query (Q), key (K), and value (V). After applying the ResNet model, the feature embedding of a WSI with N sampled patches is denoted as , where d is the embedding dimension of each patch (i.e., 512 as ResNet-18 is used as the backbone). 𝑿 is then projected into K and V. Specifically, for V, we simply use an identity function, as the derived feature vectors from the pretrained ResNet model have already been adequate feature representations. The projection function for the K consists of a matrix multiplication with WK, a dropout layer46, and a ReLU47 activation function. is a trainable matrix. Its elements are initialized randomly using standard Gaussian distribution and will be updated during the training. The output K can be written as,
| (1) |
is a learnable vector with random initialization, where each element is sampled from uniform distribution . The attention aggregated feature representation can be written as
| (2) |
where the softmax normalization is applied on the raw attention scores along the patch dimension so that the attention weights A of the N patches will sum to 1. As A contains the attention weights of all the sampled patches, we will also refer to it as the attention map where applicable. The output aggregates information from N sampled patches and is used as the feature representation of the whole slide.
When expanding the attention layer from one to H heads, we split the Q, K, and V into chunks of size d/H along the embedding dimension. We then calculate the attention scores within each attention head and concatenate the final representation vectors. Formally, the multi-head attention can be expressed as
| (3) |
Where and Q(ℎ) is the ℎth chunk of Q and so on. Then we can predict the patient’s risk score as
| (4) |
where is the weight for the final layer. In the evaluation mode, the dropout layer is disabled. So, the predicted risk score R can be written as
| (5) |
| (6) |
This decomposes the multi-head attention risk score into the sum of head-wise predictions, where head-wise predictions can be obtained from the weighted average of patch-level predictions. This is the basis for attention visualization as presented later in Section 4.5.
3.3. Loss function
A fully connected layer was used to estimate the risk scores of the patients. We use the negative log of Cox proportional hazards partial likelihood as the loss function48, which is defined as
| (7) |
where is the output (risk score) of a model with parameters θ and input xi, means having the event (death), and is patient i’s risk set (i.e., those who are alive at the time Ti). For each patient who is deceased (e.g., patient i) at Ti, we compare the risk score of this patient, , to the log sum exponential of those who survive longer than this patient. Patient i is expected to have a higher risk compared to patients in . Thus, if the model is working correctly, risk scores will be smaller for patients in , while will be larger, so a lower loss can be achieved.
4. Experiments and Results
4.1. Dataset description
The datasets used in our experiments are from the TCGA Program (National Cancer Institute). The TCGA is a public database with comprehensive clinicopathological data and multi-platform molecular profiles for more than 30 cancer types, with data collected from multiple institutions. In this study, we selected 4 cancer types, Urothelial Bladder Carcinoma (BLCA), Breast Invasive Carcinoma (BRCA), Colon Adenocarcinoma (COAD), and Lower Grade Glioma (LGG). For each cancer type, we acquired the digitized Hematoxylin and Eosin (H&E) stained slides, the follow-up information, and the survival status at the end of the follow-up. Both flash-frozen and Formalin-Fixed Paraffin-embedded (FFPE) slides were included for model development and evaluation except for LGG, for which we found the flash-frozen slides contained excessive artifacts. A detailed description of the cancer types included in this study is shown in Table 1. In total, 2,375 patients (with 6,162 WSIs) were included in the experiment. The largest dataset is BRCA with over 1,000 patients included, and the smallest dataset is BLCA with only 386 patients. On average, the patients were followed up by 2 years after the cancer diagnosis until they died or the last visit.
Table 1.
Descriptive statistics of the utilized datasets from TCGA.
| BLCA | BRCA | COAD | LGG | |
|---|---|---|---|---|
| Number of patients | 386 | 1,050 | 449 | 490 |
| Number of WSIs | 898 | 3,003 | 1,418 | 843 |
| Number of patches (million) | 3.05 | 6.46 | 2.97 | 3.07 |
| Median follow-up time (years) | 1.65 | 2.34 | 1.80 | 1.87 |
| Number of events (deaths) | 175 | 144 | 100 | 115 |
The WSIs were scanned at 20× or 40× magnification. We converted the WSIs to 10× magnification and cropped the entire slide into patches of size 224×224 without overlapping to reduce the computational and storage cost. Another benefit of using a relatively smaller magnification level is that each patch will cover a larger spatial area without losing significant cellular feature details. To determine whether a patch belongs to the background, we counted the number of purple pixels in each patch by thresholding the RGB channel intensity and discarded patches with less than 100 purple pixels. A total of 15.55 million foreground patches were extracted across the four cancer datasets.
4.2. Implementation details
The proposed method was implemented in Python with the PyTorch library (version 1.8.1)50. During each iteration, we randomly sampled 32 patches (with replacement) per patient for 64 patients producing 2,048 patches per batch. For a training split with 320 patients, one training epoch consists of 5 (320 patients / 64 patients per batch) parameter update steps. We evaluated the model on the validation dataset every 100 epochs (or 500 steps) to monitor the training progress, with 100 patches sampled from every patient.
Adam optimizer was used to optimize the parameters of the model51. The starting learning rate was set to 6×10-5. A cosine learning rate scheduler was used in optimization, and we reset the learning rate every 4,000 epochs.
4.3. Evaluation metrics and methods
The concordance index (c-index) was used as the primary evaluation metric in this study. It is defined as the proportion of pairs whose rank is correctly predicted among all admissible pairs52. C-index ranges from 0 to 1. A c-index of 1 means perfect prediction, while a c-index of 0.5 means no better than a random guess. We additionally evaluated the Kaplan-Meier curves, log-rank tests, and time-varying AUC for our method and baseline methods.
As the sample size of the chosen cancer types is only several hundred, if we allocate 70% of the cases for model training and validation, and use the remaining 30% for model testing (as this is typically practiced in the deep learning field), we will end up with too few death events in our test set, and the c-index could be highly stochastic and unreliable. To overcome this problem, we used the nested cross-validation (Nested-CV) method to obtain a more stable estimate of the model’s performance. The Nested-CV method makes the most use of the data while avoiding information leakage and is widely used in evaluating machine learning methods with a moderate sample size53. Our Nested-CV framework consists of 5 outer loops, and each outer loop consists of 4 inner loops (Supplementary Figure S1). The inner loop is essentially K-fold cross-validation for hyper-parameter tuning with K = 4. As the death events are only observed for part of the study participants, to avoid possible imbalance, we split the datasets by stratifying on the outcome status of the patient, i.e., whether the patient was alive or not at the last follow-up. Both data splitting and model evaluation was performed at the patient level to avoid any potential information leak.
The hyperparameters we optimized for our model include dropout rate and early stopping epoch. We monitored the c-index on the validation set to determine the early stopping epoch for each configuration. We used a grid search approach to evaluate several dropout rates, namely, {0.0, 0.2, 0.5, 0.8, 0.95}. The optimal dropout rate is determined for each outer fold independently, by comparing the average of inner-fold validation c-indices. Then we evaluated the models on the test dataset. Dropout has been proven to be useful in preventing overfitting and improving the robustness of a model46. We applied the dropout directly on the output from the multi-head attention layer. And we showed the effect of dropout on the model performance from cross-validation in Supplementary Table S1. Additionally, we observed that if we eliminate dropout from our pipeline, the average c-index on the test sets would be reduced by 0.02. BRCA is the dataset that benefits the most from dropout, with a gain in testing c-index of 0.070.
After finetuning the hyperparameters, the final set of models trained from each inner loop were evaluated on the unseen test split of the outer loop. Their predictions were averaged to calculate the c-index. 1,000 patches were randomly sampled for each patient during the testing. We repeated this process five times (i.e., the number of outer loops) and reported the average c-index across multiple outer loops. This procedure is particularly beneficial to obtaining a more stable estimate of the performance of the proposed method when only a limited number of events/deaths are available.
4.4. Quantitative comparison with other methods
Based on our ablation study (Section 4.6.1), we set the number of attention heads to 8. We compared our model with three other state-of-the-art methods, the average pooling method (AvgPool)12, DeepAttnMISL21, and Patch-GCN37. These methods were chosen because they were developed for general survival prediction purposes, and have been tested across various cancer types, which aligns with the focus of our study. For the DeepAttnMISL method, we used the same number of clusters as the number of attention heads (i.e., 8). In the original DeepAttnMISL study, the performance of the model with 8 clusters is very close to that of the best model (either 6 or 10 clusters). We used the same strategy for hyperparameter tuning and model evaluation for the baseline methods as described in Section 4.3. And we fixed the same split when repeating the nested cross-validation for different models. Because the outer loop specific c-index can be viewed as a repeated measurement of the model performance across model types, we used paired t-test to evaluate the statistical significance.
The comparison results are shown in Table 2. Our model achieved the best results for all four cancer types selected. Specifically, the differences between our method and the AvgPool method were statistically significant for the BLCA dataset (p < 0.01) and marginally significant (p < 0.1) for the LGG dataset. Overall, the average c-index of our model is 0.640, which is significantly better than the AvgPool method (c-index = 0.603, Δ = 0.037). While our method achieved a consistently higher c-index compared to the DeepAttnMISL and Patch-GCN methods, the difference was not statistically significant for individual cancer types. On average, there is a 0.021 (p = 0.16) and 0.028 (p = 0.04) improvement in c-index of our method compared to DeepAttnMISL and Patch-GCN model, respectively.
Table 2.
Comparison of model performance.
| BLCA | BRCA | COAD | LGG | ALL | |
|---|---|---|---|---|---|
| AvgPool | 0.551 | 0.575 | 0.601 | 0.685 | 0.603 |
| DeepAttnMISL | 0.594 | 0.600 | 0.581 | 0.700 | 0.619 |
| GCN | 0.573 | 0.570 | 0.632 | 0.674 | 0.612 |
| MHAttnSurv (ours) | 0.604 | 0.607 | 0.636 | 0.714 | 0.640 |
|
| |||||
| Δ1: MHAttnSurv – AvgPool | 0.054** | 0.031 | 0.035 | 0.029† | 0.037** |
| Δ2: MHAttnSurv – DeepAttnMISL | 0.010 | 0.007 | 0.055 | 0.013 | 0.021 |
| Δ3: MHAttnSurv – Patch-GCN | 0.031 | 0.037 | 0.004 | 0.039 | 0.028* |
Boldface: Best c-index.
p-value < 0.1;
p-value < 0.05;
p-value < 0.01.
We further examined the time-varying AUC to assess the performance of each model up to 5 years after diagnosis. At each assessed time point, this evaluation method considered the patients who died before this time point as cases, and patients who were still alive after this time point as controls, to measure the discrimination power of the model predictions. Inverse probability weighting is used to correct for right censoring54. Figure 2 shows that the models are typically better at predicting early events with the exception of BRCA, for which the AUC peaked at 3 years after a cancer diagnosis. For BLCA and BRCA, the MHAttnSurv method performs the best for almost all the evaluated time points. The patch-GCN method achieved the highest AUC for COAD at year 2 and 5. While for LGG, our method performs better than the baseline methods up to 3 years after diagnosis. Overall, the MHAttnSurv method is the best of the four in terms of time-varying AUC.
Figure 2.
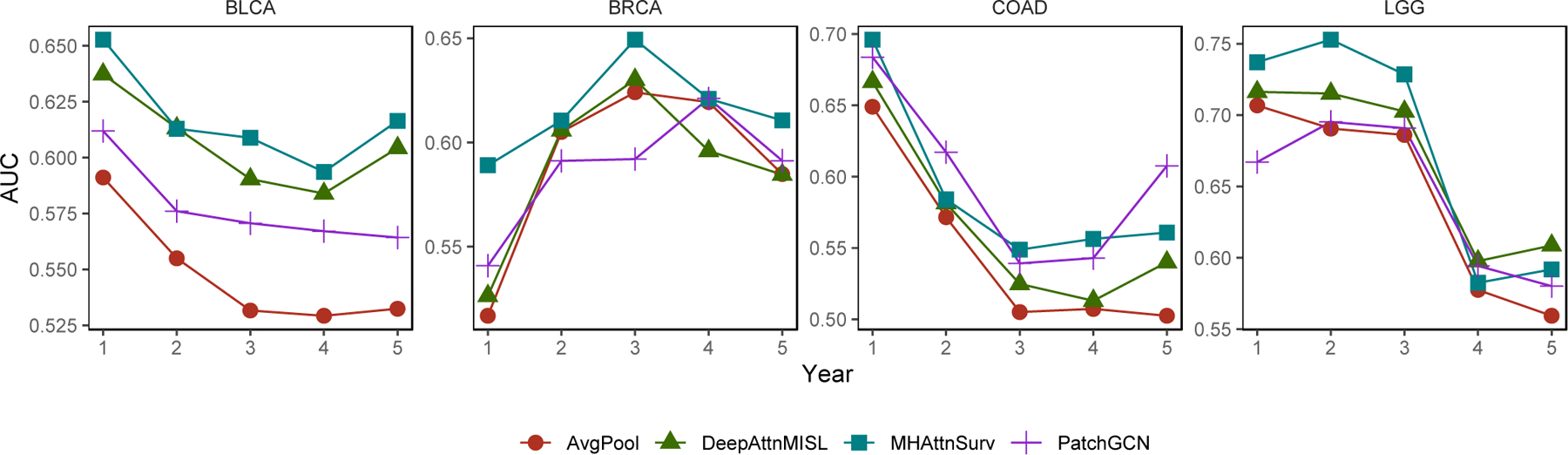
Time-varying AUC evaluated annually within 5 years after diagnosis.
Additionally, we included the Kaplan-Meier curves for MHAttnSurv in Figure 3, and additional curves for the baseline methods in Supplementary Figure S2. For the test dataset of each outer fold, we split the predicted risk score into tertiles (i.e., based on the 33rd and 67th percentiles) and denoted them as the low-risk, medium-risk, and high-risk groups. The log-rank test p-values of the MHAttnSurv method are significant for all four cancer types, and there is a discernible separation in Kaplan-Meier curves between the high-risk group and the low/medium group.
Figure 3.
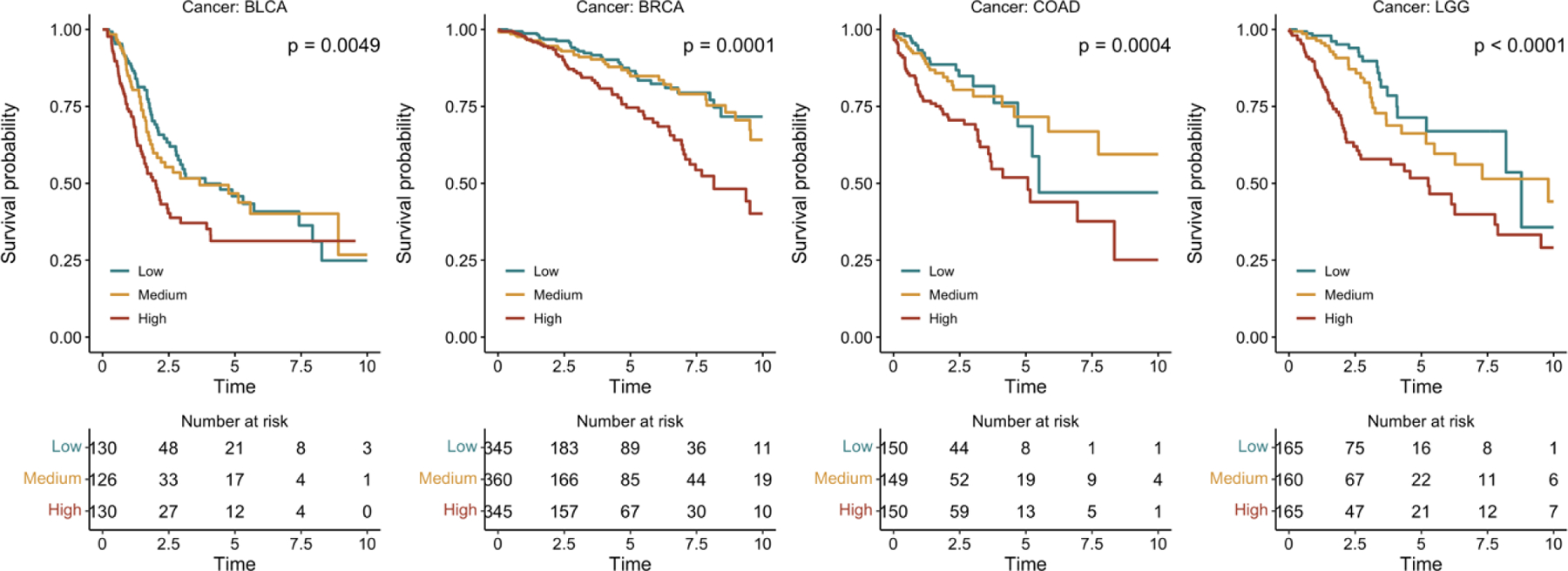
Kaplan-Meier curves of MHAttnSurv. Patients were stratified into three risk groups based on tertiles of testing c-index.
4.5. Qualitative and Quantitative Analysis of MHAttnSurv
To better understand how the attention model works, we present the connections between attention heads and patch features for one example WSI from the testing split of the LGG dataset in Figure 4. While our method does not require a clustering step, we included a clustering map in Figure 4(b) to facilitate the interpretation of the attention maps. To obtain the clustering map, we used K-means to group the patches from the selected WSI into 8 clusters and assigned each cluster to a unique color.
Figure 4.
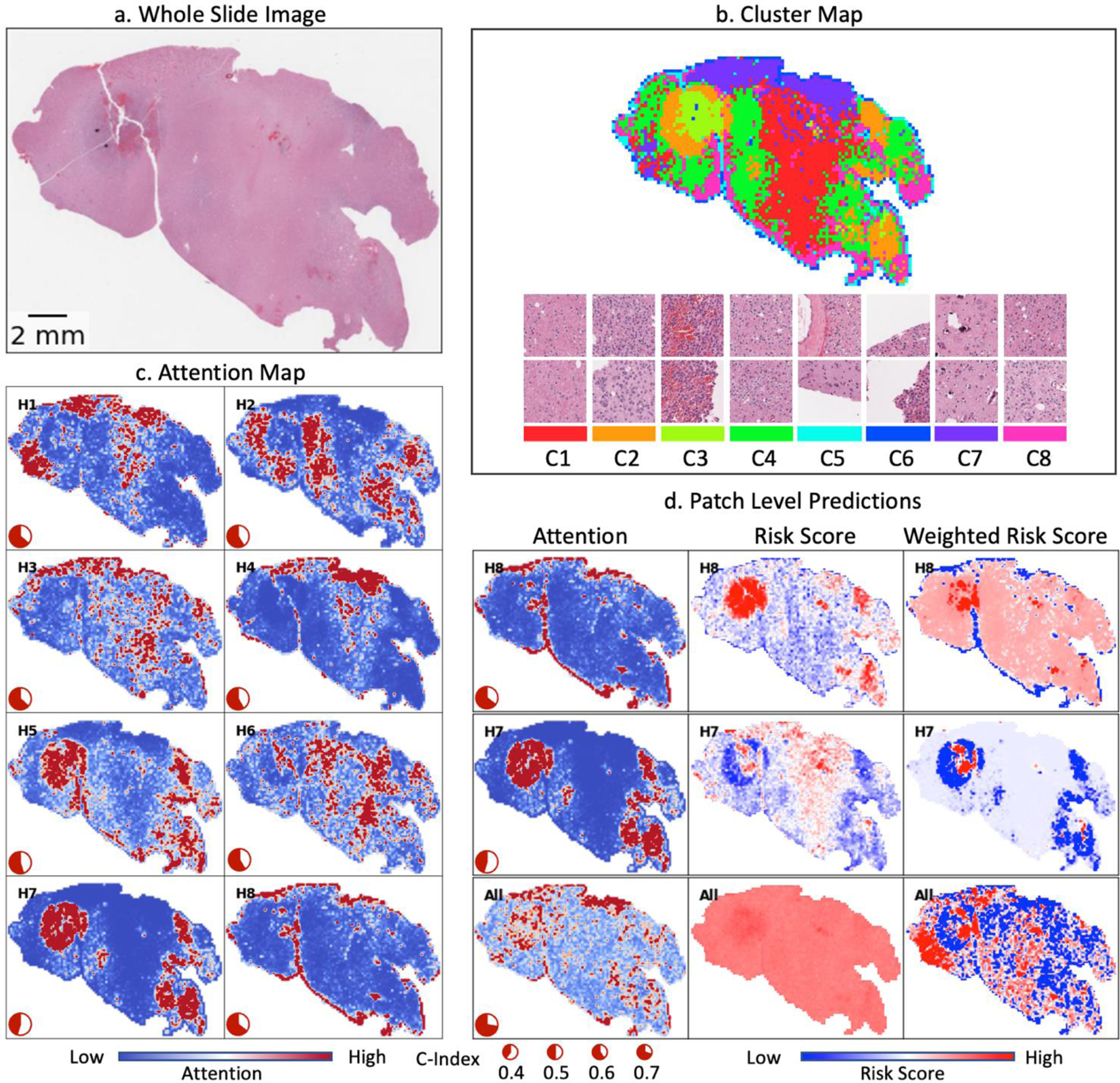
Visualization of head-wise attention map and patch clusters for one sample WSI from LGG. (a) Whole Slide Image. (b) Patch clusters on the WSI level and example patches from each cluster. (c) Head-wise attention map. Red color: rescaled-attention weights > 2; Blue color: rescaled-attention weights = 0.
Figure 4(c) illustrates the attention map for each attention head. To create these visualizations, we first processed all patches from the WSI and shuffled them 10 times. We then concatenated these 10 shuffled lists of patches to form a queue. Each time 1,000 patches are sequentially taken out of the queue to simulate the random patches sampling approaches that we took in the evaluation process. Then we predicted the attention weights for each head, and rescaled the attention weights by multiplying 1,000, so the rescaled values can be interpreted as how many times each patch is upweighted (or downweighted) compared to assigning them with equal weights. We repeated this process until the queue was depleted, ensuring that every patch from the WSI was sampled exactly 10 times. Finally, we calculated the average attention weights for each patch based on all the simulated samplings.
As derived in equation (6), we decomposed the head-wise prediction into head-specific patch level attention score and head-specific patch level risk score in Figure 4(d). We show the patch-level attention map, unweighted and attention-weighted risk score map for two selected heads, Head-8 (H8) and H7. H8 is the best performing head in this experiment with a c-index of 0.644, while H7 is the worst performing head with a c-index of 0.452. Additionally, we showed the overall attention map as calculated by taking the average of all the head-wise attention maps (Figure (4d), last row).
The attention maps exhibit significant variation across attention heads, as demonstrated in Figure 4. For example, H5 focuses mainly on regions with patches from Cluster-2 (C2), C3, and C8, characterized by high nucleus density and active angiogenesis. H7 focuses on a similar region as H5 but with stronger attention, however, the performance of this head is the worst among all the heads. A further inspection of the risk score map in Figure 4(d) reveals that H7 incorrectly generates lower risk predictions for patches from C2 and higher-risk estimates for patches from C1. While the attention suppresses much of the generated false high-risk scores from C1, it also amplifies the negative signal from C2. Notably, H8, focuses primarily on the edges where the tissues meet the background region. This attention head retains the negative predictions from the edges and achieves the best performance among the eight heads. On average, the model focuses primarily on regions covered by C2, C3, and C7. The generated rich attention patterns enable the MHAttnSurv model to comprehensively consider various morphological features which might contribute to prognosis. Additional example predictions for the other cancer types are shown in Supplementary Figures S3, S4, and S5.
We further explored the correlation among the attention heads for LGG as shown in Figure 5. We can see that the attention weights across different heads are only weakly correlated, further demonstrating that each attention head focuses on distinct patterns from a whole-slide image. On the contrary, there exist much stronger correlations among patch-level risk scores across attention heads. This is interesting as the head-wise patch-level risk scores are calculated using a distinct chunk of the extracted feature vector from the ResNet-18 model. Meanwhile, we noticed that the head-wise c-index could differ for two attention heads with a strong correlation in patch-level risk scores. Taking H2 and H7 for example, the correlation coefficients of their patch-level attention weights and patch-level risk scores are 0.24 and 0.67, respectively. Despite the high correlation coefficient in their patch-level risk score predictions, their head-wise c-indices are very different. Because H2 amplifies the negative signals from relatively normal tissues and suppresses the falsely positive signals, it still achieves a moderate c-index (0.589) compared to H7. This suggests that the attention pattern is a driving force in the performance of head-wise predictions.
Figure 5.
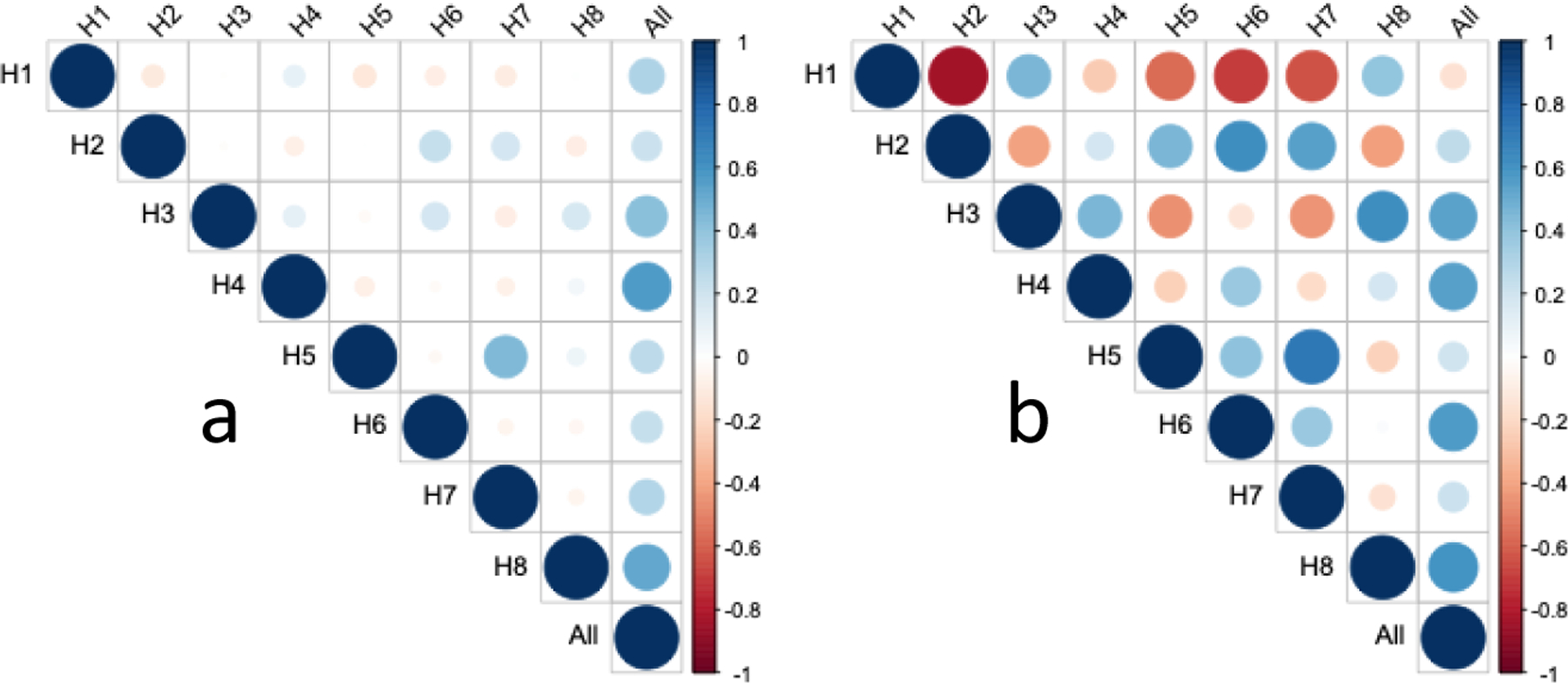
Correlation of attention weights and risk scores across attention heads for LGG. (a) correlation of attention weights. (b) correlation of patch-level risk scores.
4.6. Ablation experiment
We have conducted extensive ablation studies to investigate the effect of various design choices in our framework. This includes the number of attention heads, and the number of patches sampled during the training and evaluation phase.
4.6.1. Effects of the number of attention heads
Table 3 presents the results from the ablation study exploring whether using a different number of attention heads could affect the model performance. When using only one attention head, the average c-index is 0.609, which is comparable to (or slightly better than) the AvgPool method (c-index: 0.603). However, with 4 attention heads, the average c-index increases to 0.630. Using 8 attention heads achieves the best overall results, but using more attention heads (e.g., 16 and 32) results in performance degradation. For BLCA and COAD datasets, the best results are observed when using 8 attention heads. For BRCA and LGG, the best results occur when using 4 and 32 attention heads, respectively. Based on our results in this ablation study, starting with 8 attention heads is a reasonable choice in survival prediction studies.
Table 3.
Effect of the number of attention heads on the performance of our approach.
| Number of heads | BLCA | BRCA | COAD | LGG | ALL |
|---|---|---|---|---|---|
| 1 | 0.576 | 0.592 | 0.583 | 0.685 | 0.609 |
| 4 | 0.577 | 0.614 | 0.628 | 0.702 | 0.630 |
| 8 | 0.604 | 0.607 | 0.636 | 0.714 | 0.640 |
| 16 | 0.567 | 0.614 | 0.626 | 0.710 | 0.629 |
| 32 | 0.583 | 0.570 | 0.626 | 0.715 | 0.623 |
Boldface: Best c-index.
4.6.2. Effects of the number of patches sampled during training time
Table 4 shows the effect of the number of patches sampled during training on the model performance. Instead of running the full-scale Nested-CV which requires enormous computational resources, we restricted our experiments to 80 percent of our entire dataset (the first outer loop) and performed a 4-fold cross-validation. The presented ablation results are the average over these four validation folds.
Table 4.
Effect of the number of patches sampled during training time.
| Number of patches | BLCA | BRCA | COAD | LGG | All |
|---|---|---|---|---|---|
| 8 | 0.603 | 0.608 | 0.638 | 0.760 | 0.652 |
| 16 | 0.605 | 0.616 | 0.642 | 0.767 | 0.657 |
| 32 | 0.604 | 0.618 | 0.633 | 0.757 | 0.653 |
| 64 | 0.610 | 0.615 | 0.641 | 0.754 | 0.655 |
| 128 | 0.621 | 0.618 | 0.637 | 0.753 | 0.657 |
| 256 | 0.627 | 0.622 | 0.644 | 0.751 | 0.661 |
Boldface: Best c-index.
For BLCA, BRCA, and COAD, the model performance is the best when 256 patches are sampled. While for LGG, the best performance is achieved with 16 patches. Therefore, on average, sampling 256 patches from WSIs during the training achieves the best c-index. Of note, with more sampled patches, there are higher computation costs that limit the maximum number of sampled patches in our experiments.
The pie plot in the lower left corner shows the head-wise c-index. (d) Patch level prediction for the selected heads. Rows: best performing head, worst performing head, and all heads combined. Columns: attention map, unscaled risk score for each patch, and weighted risk scores (i.e., attention weight × risk score). “High” and “low” risk scores refer to the maximum and minimum head-wise patient-level risk
4.6.3. Effects of the number of patches sampled during testing time
We varied the number of patches sampled during the testing to illustrate its effect on the resulting c-index. The results are shown in Figure 6. When only a small number of patches are sampled, the estimates of c-index are highly volatile, with an average standard deviation of 0.017. Further increase in the number of patches stabilizes the results and improves the c-index estimate as well. When 1,000 patches are sampled, the average standard deviation declines to 0.005, while the average c-index estimate improves by 0.015 compared to sampling 32 patches. An additional increase in the number of patches still reduces the standard error in c-index estimations, but the improvement in accuracy is marginal. As a result, we kept sampling 1,000 patches in our evaluation experiments, while in practice, all the patches can be included for the most accurate results if there is no bottleneck on time or computational resources.
Figure 6.
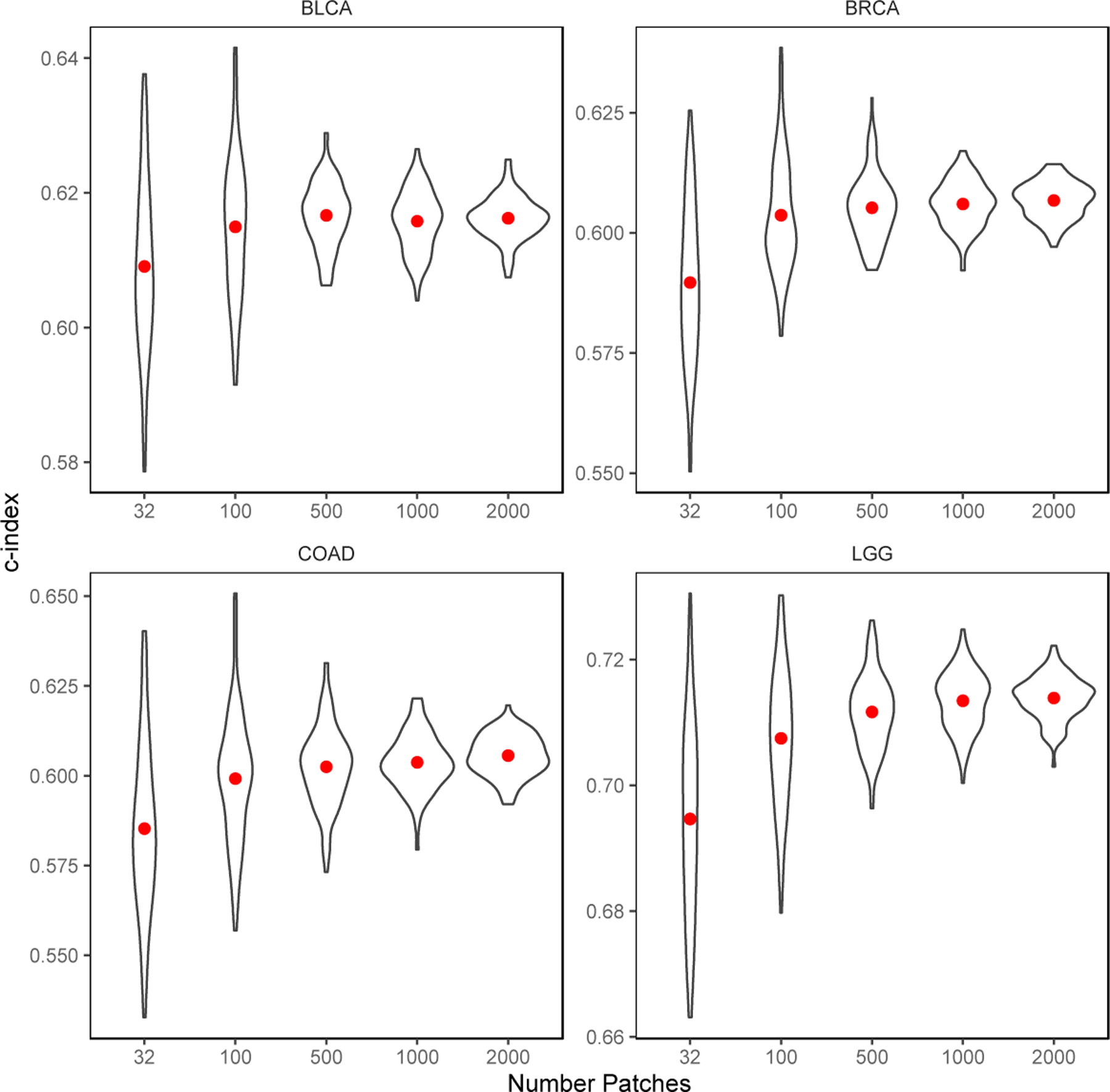
Effects of the number of patches sampled during evaluation time.
5. Discussion
In this work, we presented a novel multi-head attention approach for survival prediction on whole-slide pathology images. Accurate survival prediction and cancer risk stratification can greatly benefit cancer patients in making informed decisions regarding treatment plans and improving their physical and psychological adjustment2,55. An automated and effective prognosis prediction program will also benefit physicians and pathologists by reducing their workload and allowing them to communicate more confidently with patients when discussing treatment options56.
Prognosis prediction has been a challenging problem. Unlike the tumor subtype classification and tumor segmentation tasks, where the histopathological aspects are well-defined, making accurate prognosis predictions based on histology slides is difficult. A comprehensive prognosis evaluation requires careful consideration of both the global and local structures of the pathology slides. This problem is exacerbated by the large size of WSIs. Most of the deep learning methods developed so far either require ROI annotations10,29 or make strong assumptions that every patch contributes equally to the prognosis prediction12. ROI annotations are costly to obtain, and the annotation process is subjective to pathologists’ discretion. While for approaches that assign equal weights to each patch, the cancer malignancy signal carried by the tumor tissue can be easily diluted by the large amount of normal tissues that coexist in the same slide.
Our proposed multi-head attention mechanism was effective in the experimental results, outperforming three powerful deep learning baseline approaches: AvgPool, DeepAttnMISL, and Patch-GCN. The improvement in the performance of our model can be explained by the following reasons. Firstly, our model employs the attention mechanism to combine patches based on their relevance to the overall task. This strategy could avoid assigning the same weight (as in the AvgPool method) to patches that are not prognostically relevant. Secondly, the multiple attention heads can achieve the same effect as the Siamese model used in the DeepAttnMISL. Each attention head can provide some unique aspects regarding how to combine the different patches. But unlike the fixed clustering algorithm used in DeepAttnMISL, the attention mechanism used in our model is more flexible. The different attention heads are not determined ahead of time from the extracted patch features. Instead, each head is allowed to focus freely on patterns that collectively benefit most to the prognosis prediction. In addition, in DeepAttnMISL, patches from the same cluster share the same attention weights, while our method does not have such constraints. In contrast, the attention weight of each patch is determined independently across multiple attention heads.
Visualizing the attention maps demonstrates that each attention head acts relatively independently in exploring the regions that would benefit the overall prognosis prediction (Figure 4). While linking the attention maps to the head-wise c-index, we note that even an attention head that focuses primarily on the normal tissues (e.g., H1) can still provide some prognostic merits. One possible explanation is that the presence of normal tissue on a WSI is a positive sign for better survival. This also highlights the importance of a holistic view of the WSI to achieve more accurate prognosis prediction accuracy. In this study, we observe a 0.037 improvement in the c-index comparing our method to the AvgPool approach. On the contrary, by focusing only on the patches within the ROI region, the c-index was improved less than 0.010 in a previous study16. This implies that even with expensive ROI annotation, our results will likely be worse without using the MHAttnSurv approach. In fact, when only one attention head is used, the average c-index of our method is only slightly better than the AvgPool approach. This observation suggests that the monochromatic ROI annotation is less effective than the multi-head attention approach for a complex task such as prognosis prediction, and further justifies our design of the multi-head attention mechanism.
Additionally, some attention heads in Figure 4 appear similar, such as H1 and H3. Such overlap is not unexpected since each attention head operates in parallel and we did not impose any constraints on the attention maps to prevent it. Suppose a particular tissue type is very important for the prognosis prediction task, multiple attention heads may focus on this tissue type because doing so could benefit the objective of the model. Despite the overlap, it is still beneficial to keep all attention heads as they learn slightly different representations. Also note that our model structure splits the representation vector of each patch into multiple chunks during multi-head attention. Deleting one attention head would result in information loss from the corresponding chunk of the representation vector.
For a more comprehensive comparison of our methods to the existing methods, we evaluated multiple metrics, including the standard c-index and Kaplan-Meier curves with log-rank test, as well as time-varying AUC. While the c-index provides a single number summary of the model’s discrimination capability, it does not measure the model’s performance at a particular time point. This limitation can be addressed by time-varying AUC. A notable finding is that, while AUC tends to decrease over time, the highest AUC is observed at year 3 after cancer diagnosis for the BRCA dataset. Usually, survival prediction models are better at predicting short-term events than long term events. This is not only because those long-term events are of less certainty than short-term events, but also due to the status of long-term survivors, which is more likely to be missing (i.e., right censoring). In the case of BRCA, we noticed that all the risk groups have relatively high survival rates compared to other cancer types (Figure 3). This indicates the BRCA patients usually have a longer survival time, thus fewer events at the beginning of the follow-up period. In addition, BRCA patients usually have a longer follow-up time compared to other cancer patients. This might explain why the AUC increases within the first few years for BRCA patients.
As for limitations, we only evaluated the multi-head attention mechanism on one outcome type, which is survival after a cancer diagnosis. The utility of this method for other outcome types has not been explored. Furthermore, the number of patches randomly selected from a WSI in our pipeline is considered a hyperparameter that requires further exploration. Our ablation studies have shown that a larger number of patches sampled during training and evaluation is associated with an upward trend in model performance. Although the random sampling approach could be modified to include all patches, this would require considerably more computational resources while the improvements may only be marginal. Future studies could explore alternative sampling strategies to optimize the tradeoff between computational costs and performance.
In addition, we experimented with whether finetuning the backbone model can further improve the model performance. But we did not observe any noticeable improvement in the validation c-index when finetuning the backbone model. Based on our observations, the model quickly overfitted. Overfitting persists even when only finetuning the last few convolutional layers of ResNet. This suggests that although our model is trainable end-to-end, it requires a much larger dataset for effective finetuning of the backbone model.
In future work, we plan to evaluate our MHAttnSurv method with other patient outcome types, such as binary outcomes using cross-entropy loss (e.g., gene mutation status) or continuous outcomes using mean squared error loss (e.g., gene expression level). We also plan to evaluate the performance of our model by training it end-to-end on larger datasets.
6. Conclusion
In this paper, we proposed a multi-head attention mechanism to extract prognostic information from whole-slide images that does not require the resource-intensive region of interest annotations. Compared to three existing state-of-the-art methods, our model achieved better performance on four TCGA datasets in terms of both c-index and time-varying AUC. Moreover, our method does not require the clustering step and is easy to implement. Visualization of the attention maps learned by our method demonstrated that each attention head focuses differently on the same whole-slide while together they work synergistically to achieve a comprehensive representation for prognosis prediction. We expect future studies to validate our method with larger datasets to further demonstrate its potentials.
Supplementary Material
Multi-head attention based multiple instance learning used in cancer prognosis prediction.
Heads attend to various morphological patterns for prognostic features extraction.
Achieved outstanding performance on 4 cancer datasets in c-index and time-varying AUC.
Acknowledgments
We thank Naofumi Tomita and Joseph DiPalma for providing revision suggestions to improve the manuscript. We thank Lamar D. Moss for proofreading the article.
Funding
This research was supported in part by grants from the US National Library of Medicine (R01LM012837 and R01LM013833) and the US National Cancer Institute (R01CA249758).
Footnotes
Credit Authorship Contribution Statement
Shuai Jiang: Conceptualization, Methodology, Software, Investigation, Data Curation, Writing – Original Draft, Writing – Review & Editing, Visualization; Arief A. Suriawinata: Writing – Review & Editing, Visualization; Saeed Hassanpour: Conceptualization, Methodology, Writing – Review & Editing, Supervision, Project administration, Funding acquisition
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of Competing Interests
No conflict of interest.
References
- 1.American Cancer Society. American Cancer Society: Cancer Facts and Figures 2021, Atlanta Am Cancer Soc 2021;
- 2.Hagerty RG, Butow PN, Ellis PM, Dimitry S, Tattersall MHN. Communicating prognosis in cancer care: a systematic review of the literature., Ann Oncol Off J Eur Soc Med Oncol 2005. Jul;16(7):1005–53. [DOI] [PubMed] [Google Scholar]
- 3.Gospodarowicz M, O’Sullivan B. Prognostic factors in cancer., Semin Surg Oncol 2003;21(1):13–8. [DOI] [PubMed] [Google Scholar]
- 4.Phung MT, Tin Tin S, Elwood JM. Prognostic models for breast cancer: a systematic review, BMC Cancer 2019. Mar 14;19(1):230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Feng Q, May MT, Ingle S, Lu M, Yang Z, Tang J. Prognostic Models for Predicting Overall Survival in Patients with Primary Gastric Cancer: A Systematic Review, Nishida T, editor. Biomed Res Int 2019;2019:5634598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Morris EJA, Maughan NJ, Forman D, Quirke P. Who to treat with adjuvant therapy in Dukes B/stage II colorectal cancer? The need for high quality pathology, Gut 2007/05/09. 2007 Oct;56(10):1419–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Betge J, Pollheimer MJ, Lindtner RA, Kornprat P, Schlemmer A, Rehak P, Vieth M, Hoefler G, Langner C. Intramural and extramural vascular invasion in colorectal cancer: prognostic significance and quality of pathology reporting., Cancer 2012. Feb;118(3):628–38. [DOI] [PubMed] [Google Scholar]
- 8.Zhu X, Yao J, Zhu F, Huang J. WSISA: Making survival prediction from whole slide histopathological images, Proc - 30th IEEE Conf Comput Vis Pattern Recognition, CVPR 2017 2017;2017-Janua:6855–63.
- 9.Cheerla A, Gevaert O. Deep learning with multimodal representation for pancancer prognosis prediction, In: Bioinformatics 2019. p. i446–54. [DOI] [PMC free article] [PubMed]
- 10.Mobadersany P, Yousefi S, Amgad M, Gutman DA, Barnholtz-Sloan JS, Velázquez Vega JE, Brat DJ, Cooper LAD, Designed LADC, Performed LADC. Predicting cancer outcomes from histology and genomics using convolutional networks, 2018; [DOI] [PMC free article] [PubMed]
- 11.Yao J, Zhu X, Huang J. Deep Multi-instance Learning for Survival Prediction from Whole Slide Images, 2019;
- 12.Wulczyn E, Steiner DF, Xu Z, Sadhwani A, Wang H, Flament-Auvigne I, Mermel CH, Chen PHC, Liu Y, Stumpe MC. Deep learning-based survival prediction for multiple cancer types using histopathology images, PLoS One 2020;15(6):1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Skrede OJ, De Raedt S, Kleppe A, Hveem TS, Liestøl K, Maddison J, Askautrud HA, Pradhan M, Nesheim JA, Albregtsen F, Farstad IN, Domingo E, Church DN, Nesbakken A, Shepherd NA, Tomlinson I, Kerr R, Novelli M, Kerr DJ, et al. Deep learning for prediction of colorectal cancer outcome: a discovery and validation study, Lancet 2020;395(10221):350–60. [DOI] [PubMed] [Google Scholar]
- 14.Courtiol P, Maussion C, Moarii M, Pronier E, Pilcer S, Sefta M, Manceron P, Toldo S, Zaslavskiy M, Le Stang N, Girard N, Elemento O, Nicholson AG, Blay JY, Galateau-Sallé F, Wainrib G, Clozel T. Deep learning-based classification of mesothelioma improves prediction of patient outcome, Nat Med 2019;25(10):1519–25. [DOI] [PubMed] [Google Scholar]
- 15.Abbet C, Zlobec I, Bozorgtabar B, Thiran J-P. Divide-and-Rule: Self-Supervised Learning for Survival Analysis in Colorectal Cancer BT - Medical Image Computing and Computer Assisted Intervention – MICCAI 2020, In: Martel AL, Abolmaesumi P, Stoyanov D, Mateus D, Zuluaga MA, Zhou SK, et al. , editors. Cham: Springer International Publishing; 2020. p. 480–9. [Google Scholar]
- 16.Wulczyn E, Steiner DF, Moran M, Plass M, Reihs R, Tan F, Flament-Auvigne I, Brown T, Regitnig P, Chen P-HC, Hegde N, Sadhwani A, MacDonald R, Ayalew B, Corrado GS, Peng LH, Tse D, Müller H, Xu Z, et al. Interpretable survival prediction for colorectal cancer using deep learning, npj Digit Med 2021;4(1):71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bremnes RM, Dønnem T, Al-Saad S, Al-Shibli K, Andersen S, Sirera R, Camps C, Marinez I, Busund L-T. The Role of Tumor Stroma in Cancer Progression and Prognosis: Emphasis on Carcinoma-Associated Fibroblasts and Non-small Cell Lung Cancer, J Thorac Oncol 2011;6(1):209–17. [DOI] [PubMed] [Google Scholar]
- 18.Weidner N, Semple JP, Welch WR, Folkman J. Tumor angiogenesis and metastasis-correlation in invasive breast carcinoma., N Engl J Med 1991. Jan;324(1):1–8. [DOI] [PubMed] [Google Scholar]
- 19.Mao Y, Qu Q, Chen X, Huang O, Wu J, Shen K. The Prognostic Value of Tumor-Infiltrating Lymphocytes in Breast Cancer: A Systematic Review and Meta-Analysis., PLoS One 2016;11(4):e0152500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ilse M, Tomczak JM, Welling M. Attention-based deep multiple instance learning, In: 35th International Conference on Machine Learning, ICML 2018 2018. p. 3376–91.
- 21.Yao J, Zhu X, Jonnagaddala J, Hawkins N, Huang J. Whole slide images based cancer survival prediction using attention guided deep multiple instance learning networks, Med Image Anal 2020;65:101789. [DOI] [PubMed] [Google Scholar]
- 22.Vaswani A, Shazeer N, Parmar N, Uszkoreit J, Jones L, Gomez AN, Kaiser Ł, Polosukhin I. Attention is all you need, Adv Neural Inf Process Syst 2017;2017-Decem(Nips):5999–6009.
- 23.Rosenberg J, Chia YL, Plevritis S. The effect of age, race, tumor size, tumor grade, and disease stage on invasive ductal breast cancer survival in the U.S. SEER database, Breast Cancer Res Treat 2005;89(1):47–54. [DOI] [PubMed] [Google Scholar]
- 24.Liu H, Zhou H, Yan L, Ye T, Lu H, Sun X, Ye Z, Xu H. Prognostic significance of six clinicopathological features for biochemical recurrence after radical prostatectomy: a systematic review and meta-analysis, Oncotarget 2017. Nov 6;9(63):32238–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Van Diest PJ, Van Der Wall E. Prognostic value of proliferation in invasive breast cancer: a review, J Clin Pathol 2004;57:675–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen S, Zhang N, Jiang L, Gao F, Shao J, Wang T, Zhang E, Yu H, Wang X, Zheng J. Clinical use of a machine learning histopathological image signature in diagnosis and survival prediction of clear cell renal cell carcinoma., Int J cancer 2021. Feb;148(3):780–90. [DOI] [PubMed] [Google Scholar]
- 27.Carpenter AE, Jones TR, Lamprecht MR, Clarke C, Kang IH, Friman O, Guertin DA, Chang JH, Lindquist RA, Moffat J, Golland P, Sabatini DM. CellProfiler: image analysis software for identifying and quantifying cell phenotypes, Genome Biol 2006;7(10):R100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Caicedo JC, Cooper S, Heigwer F, Warchal S, Qiu P, Molnar C, Vasilevich AS, Barry JD, Bansal HS, Kraus O, Wawer M, Paavolainen L, Herrmann MD, Rohban M, Hung J, Hennig H, Concannon J, Smith I, Clemons PA, et al. Data-analysis strategies for image-based cell profiling, Nat Methods 2017;14(9):849–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhu X, Yao J, Huang J. Deep convolutional neural network for survival analysis with pathological images, Proc - 2016 IEEE Int Conf Bioinforma Biomed BIBM 2016 2017;(1):544–7. [Google Scholar]
- 30.Saillard C, Schmauch B, Laifa O, Moarii M, Toldo S, Zaslavskiy M, Pronier E, Laurent A, Amaddeo G, Regnault H, Sommacale D, Ziol M, Pawlotsky J, Mulé S, Luciani A, Wainrib G, Clozel T, Courtiol P, Calderaro J. Predicting survival after hepatocellular carcinoma resection using deep‐learning on histological slides, Hepatology 2020;0–3. [DOI] [PubMed]
- 31.Jiang S, Zanazzi GJ, Hassanpour S. Predicting prognosis and IDH mutation status for patients with lower-grade gliomas using whole slide images, Sci Rep 2021;11(1):16849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bahdanau D, Cho KH, Bengio Y. Neural machine translation by jointly learning to align and translate, In: 3rd International Conference on Learning Representations, ICLR 2015 - Conference Track Proceedings 2015.
- 33.Xu K, Ba JL, Kiros R, Cho K, Courville A, Salakhutdinov R, Zemel RS, Bengio Y. Show, attend and tell: Neural image caption generation with visual attention, In: 32nd International Conference on Machine Learning, ICML 2015 2015. p. 2048–57.
- 34.Tomita N, Abdollahi B, Wei J, Ren B, Suriawinata A, Hassanpour S. Attention-Based Deep Neural Networks for Detection of Cancerous and Precancerous Esophagus Tissue on Histopathological Slides, JAMA Netw Open 2019;2(11):e1914645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lu MY, Williamson DFK, Chen TY, Chen RJ, Barbieri M, Mahmood F. Data-efficient and weakly supervised computational pathology on whole-slide images, Nat Biomed Eng 2021;5(6):555–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li R, Yao J, Zhu X, Li Y, Huang J. Graph CNN for survival analysis on whole slide pathological images, MICCAI 2018 2018;11071 LNCS:174–82.
- 37.Chen RJ, Lu MY, Shaban M, Chen C, Chen TY, Williamson DFK, Mahmood F. Whole Slide Images are 2D Point Clouds: Context-Aware Survival Prediction Using Patch-Based Graph Convolutional Networks BT - Medical Image Computing and Computer Assisted Intervention – MICCAI 2021, In: de Bruijne M, Cattin PC, Cotin S, Padoy N, Speidel S, Zheng Y, et al. , editors. Cham: Springer International Publishing; 2021. p. 339–49. [Google Scholar]
- 38.Wu W, Liu X, Hamilton RB, Suriawinata AA, Hassanpour S. Graph Convolutional Neural Networks for Histological Classification of Pancreatic Cancer, 2022;(603):1–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Devlin J, Chang MW, Lee K, Toutanova K. BERT: Pre-training of deep bidirectional transformers for language understanding, NAACL HLT 2019 – 2019 Conf North Am Chapter Assoc Comput Linguist Hum Lang Technol - Proc Conf 2019;1(Mlm):4171–86. [Google Scholar]
- 40.Dosovitskiy A, Beyer L, Kolesnikov A, Weissenborn D, Zhai X, Unterthiner T, Dehghani M, Minderer M, Heigold G, Gelly S, Uszkoreit J, Houlsby N. An Image is Worth 16×16 Words: Transformers for Image Recognition at Scale, 2020;
- 41.Dietterich TG, Lathrop RH, Lozano-Pérez T. Solving the multiple instance problem with axis-parallel rectangles, Artif Intell 1997;89(1):31–71. [Google Scholar]
- 42.Maron O, Lozano-Pérez T. A Framework for Multiple-Instance Learning, In: Jordan M, Kearns M, Solla S, editors. Advances in Neural Information Processing Systems MIT Press; 1998. [Google Scholar]
- 43.Deng J, Dong W, Socher R, Li L, Li K, Fei-Fei L. ImageNet: A large-scale hierarchical image database, In: 2009 IEEE Conference on Computer Vision and Pattern Recognition 2009. p. 248–55.
- 44.Wei JW, Tafe LJ, Linnik YA, Vaickus LJ, Tomita N, Hassanpour S. Pathologist-level classification of histologic patterns on resected lung adenocarcinoma slides with deep neural networks, Sci Rep 2019;9(1):1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.He K, Zhang X, Ren S, Sun J. Deep residual learning for image recognition, In: Proceedings of the IEEE Computer Society Conference on Computer Vision and Pattern Recognition 2016. p. 770–8.
- 46.Hinton GE, Srivastava N, Krizhevsky A, Sutskever I, Salakhutdinov RR. Improving neural networks by preventing co-adaptation of feature detectors, 2012. Jul 3;
- 47.Nair V, Hinton GE. Rectified linear units improve Restricted Boltzmann machines, In: ICML 2010 - Proceedings, 27th International Conference on Machine Learning 2010. p. 807–14.
- 48.Cox DR. Regression Models and Life-Tables, J R Stat Soc Ser B 1972. Mar 20;34(2):187–220. [Google Scholar]
- 49.National Cancer Institute. The Cancer Genome Atlas Program - National Cancer Institute: Https://www.cancer.gov/about-tcga,nci/organization/ccg/research/structural-genomics/
- 50.Paszke A, Gross S, Massa F, Lerer A, Bradbury Google J, Chanan G, Killeen T, Lin Z, Gimelshein N, Antiga L, Desmaison A, Xamla AK, Yang E, Devito Z, Raison Nabla M, Tejani A, Chilamkurthy S, Ai Q, Steiner B, et al. PyTorch: An Imperative Style, High-Performance Deep Learning Library, 2019;
- 51.Kingma DP, Ba JL. Adam: A method for stochastic optimization, In: 3rd International Conference on Learning Representations, ICLR 2015 - Conference Track Proceedings. 2015. [Google Scholar]
- 52.Harrell FEJ, Califf RM, Pryor DB, Lee KL, Rosati RA. Evaluating the yield of medical tests., JAMA. 1982. May;247(18):2543–6. [PubMed] [Google Scholar]
- 53.Raschka S Model Evaluation, Model Selection, and Algorithm Selection in Machine Learning: Https://arxiv.org/abs/1811.12808, arXiv; 2018.
- 54.Blanche P, Dartigues JF, Jacqmin-Gadda H. Estimating and comparing time-dependent areas under receiver operating characteristic curves for censored event times with competing risks, Stat Med 2013. Dec 30;32(30):5381–97. [DOI] [PubMed] [Google Scholar]
- 55.Weeks JC, Cook EF, O’Day SJ, Peterson LM, Wenger N, Reding D, Harrell FE, Kussin P, Dawson NV., Connors AF, Lynn J, Phillips RS. Relationship between cancer patients’ predictions of prognosis and their treatment preferences, J Am Med Assoc 1998;279(21):1709–14. [DOI] [PubMed] [Google Scholar]
- 56.Christakis NA, Iwashyna TJ. Attitude and self-reported practice regarding prognostication in a national sample of internists., Arch Intern Med 1998. Nov;158(21):2389–95. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


