Abstract
Genetic sex determination in most vertebrates is controlled by a single master sex gene, which ensures a 1:1 sex ratio. However, more complex systems abound and several have been ascribed to polygenic sex determination, in which many genes at different loci interact to produce the sexual phenotype. Here we examine claims for polygenic sex determination in vertebrates, finding that most constitute transient states during sex chromosome turnover, or aberrant systems in species hybrids. To avoid confusion about terminology we propose a consistent nomenclature for genetic sex determination systems.
Keywords: Genetic sex determination, polygenic, Sry, temperature dependent sex determination, sex chromosome, quantitative trait loci
Monogenic and polygenic sex determination
Sex determination (SD) in multicellular animals initially directs the undifferentiated bipotential embryonal gonad to develop either as testis or ovary. The initiating signal for SD (genetic, GSD, or environmental, ESD) activates a downstream regulatory network that governs male or female gonad development. As a developmental determination process the downstream network is part of the sex determination process. However, generally the term “sex determination” is used to describe the initial, most upstream trigger (see also Glossary). The regulation of sexual development after the “decision” has been made is referred to as sex differentiation[1].
In the best understood GSD systems, SD is inherited as a monogenic trait. Sex chromosomes are defined by a master sex determining locus that triggers the male or female determining regulatory network. In some systems a male-dominant gene is borne on a male-specific Y chromosome, (e.g SRY in mammals[2]) or a female dominant gene on a female-specific W chromosome (e.g. dmw in Xenopus[3]). Alternatively, a sex determining gene on the homogametic sex chromosome may be absent or inactive on the heterogametic partner, such that the sex determining process is one of gene dosage, not dominance. For instance, dosage of the Z-linked dmrt1 genes determines sex of birds [4] and the Chinese tongue sole[5].
For most species the identity of the SD gene(s) is still unknown, but genetic crosses can usually determine whether one or more independent loci are segregating, and assess the numbers of alleles at these loci. However, genetic crosses cannot exclude the possibility that a whole suite of closely linked genes on a sex chromosome synergistically generate the SD initiating trigger, so more conservative terms “unifactorial”, “bifactorial” or “multifactorial” SD (see also Glossary) could be used.
A formal alternative to monogenic SD is polygenic SD (PSD), a concept defined by Curt Kosswig[6] (Box 1). The classic definition of polygenic SD is the combined action of alleles of multiple genes at independently inherited loci in one individual to bring about a sexual phenotype. Such a mechanism for SD has since been claimed for various species of plants, insects, fish and mammals[7–10]. However, major discrepancies in the usage of the term “polygenic SD” obscure a rich diversity of distinctly different mechanisms of multifactorial SD. We critically examine vertebrate systems described as having polygenic sex determination and offer a consistent nomenclature (see Glossary).
Box 1. A historical perspective on the concept of polygenic sex determination.
Sex chromosomes were first described in 1905[50]. This marked the origin of research on monogenic SD, which has since dominated the common knowledge how genetic sex determination works. The concept of polygenic SD (PSD) was broached only about 20 years later and was almost forgotten except by a few specialists. Recently, the field of PSD has been rejuvenated by new results from analyzing the genetics of SD in various species of plants, insects, fish and mammals[7–10].
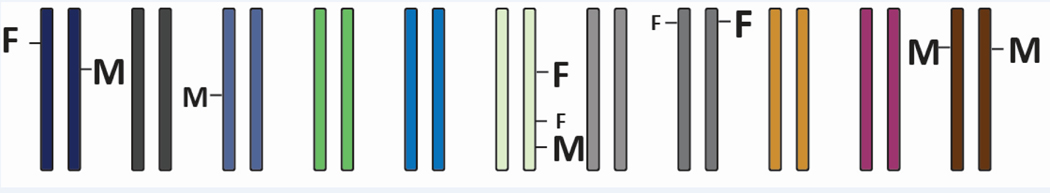
The term “polygenic sex determination” was put forward by the German geneticist Curt Kosswig in 1964 [6]. He had already developed the concept in a series of papers since the 1930s He used the inheritance of sex in hybrids of fish of the genus Xiphophorus (platyfishes and swordtails) to postulate a novel genetic mechanism for SD. In his view several male determining factors (M-factors) and female determining factors (F-factors) are distributed over the entire genome (Figure 1). The individual M and F factors can have different “strength” in initiating male or female SD, respectively. He allowed that each M and F factor could exist as different alleles in a population.
Historically, some of the confusion about what polygenic SD is, and what it is not, originated from genetic analyses of a species closely related to those that were used to establish the concept. One year after Kosswig’s 1964 paper on PSD, which was mainly built upon sex inheritance in the green swordtail, Xiphophorus hellerii, Klaus Kallman detected in another species of the same genus, the southern platyfish, Xiphophorus maculatus, the simultaneous presence of X, Y and W chromosomes in the same population [19]. Kallman clearly recognized that in this species SD is monofactorial and did not use the term “polygenic”. He even opposed the existence of polygenic sex determination in platyfish. Since then, Kosswig and Kallman debated whether sex determination in Xiphophorus species is polygenic or due to coexisting different sex chromosomes in one species, but they were clear that the two systems represented two different concepts
Kosswig’s usage of the term polyfactorial inheritance was in the sense of phenotypic genetics. He adhered to the definition of a non-Mendelian pattern of inheritance in which a particular trait is produced by the interaction of genes at many loci. The definition of polygenic inheritance that flows from this concept is that a particular trait of an individual is the resultant of the activity of several independent genes at separate loci that act additively. They must work cooperatively in the same individual, rather than merely be polymorphic between individuals in the population.
Classically, this model has been used to describe the inheritance of quantitative traits such as human height or plant leaf size. Each of these polygenes can occur as different alleles in the population so that the quantitative trait loci (QTLs) that interact to produce the phenotype are polymorphic in the population. This results in high variation of phenotypes for a single trait, typically falling into a distribution around a mean. Not all loci contribute equally to the expression of the phenotype; some are considered major QTLs and others minor. In addition, environmental influences can make decerning the polygenic system especially tricky.
One reason thought to account for the rarity of bona fide polygenic SD systems in its strict sense has been the difficulty of detecting polygenes, because loci which contribute only partially and have minor effects on the phenotype are less apparent in analyses of genetic crosses. With the new sequencing technologies, more direct approaches are possible. Methods for identifying sex-specific regions of the genome have been developed from modifications of RAD-sequencing[51] and Pool-sequencing[52–55]. Such sensitive analyses might bring surprises by detecting minor sex QTLs in systems that have so far appeared to be unifactorial with a major SD locus on a heterogametic chromosome.
Theoretical considerations
Sex determination is unique in that it channels a single embryonic tissue into one of two alternative fates. This is very different from quantitative trait loci (QTL) that vary continuously, in which many independent genes combine to deliver the phenotype, and values are distributed around a mean. Quantitative characters are typically determined in a polygenic fashion.
So firstly, how could a quantitative genetic signal in PSD produce a dichotomous qualitative output? Kosswig proposed that the sexual phenotype is determined by the additive effects of many male (M) and/or female (F) factors to reach a threshold value[6]. If the sum of “activity” of M factors is higher than that of the F factors, male development would be initiated, and vice versa (Figure 1). At the molecular level, gene activity is controlled in many ways, making gene expression usually quantitative. If a transcript threshold level is required to produce a phenotype, the output will be qualitative. Indeed, this must be the case for TSD, whereby a continuous variable (temperature of egg incubation) generates a qualitative signal to direct either male or female development of the embryo.
Figure 1:
Kosswig’s model of polygenic sex determination. M, male determining factors, F, female determining factors, size of the letter indicates male or female determining strength. If the sum of M > F, male development is initiated; if M < F, females will develop
Secondly, how stable would a PSD system be? Much theoretical work suggests that PSD is evolutionary unstable in the long run[11,12] but may be a transient state during turnover from one monogenic system to another[7]. However, there are models to explain how sexually antagonistic alleles at multiple loci could maintain a stable polymorphism[9].
One consequence of polygenic inheritance would be wide variation of sex ratio among families, depending on where the threshold was set[12]. Sex ratio parity could be achieved only by coincident evolution of allelic frequencies for the multiple sex genes and a strict threshold for the production of one sex over the other. However, if one sex is produced in excess, individuals that produce higher proportions of the rarer sex among their offspring have the advantage of leaving more grandchildren (under Fisher’s frequency dependent selection). Thus, alleles at the polygenic SD loci that favor the production of the minority sex will spread, but their selective advantage will diminish progressively until a 1:1 sex ratio is achieved. This inherently unstable system evolves to a 1:1 ratio as a single sex determining gene takes over.
Overall, evolutionary pressure on polygenic SD systems would be expected to enhance the binary nature of gonad development, because individuals with any kind of intermediate phenotype are likely to be infertile, and therefore the alleles they carry are heavily selected against. We might therefore expect monogenic systems of sex determination to be most common in vertebrates. As we show below this is, indeed, what we observe.
Fish with multiple SD genes
Much of the evidence for polygenic sex determination comes from studies of sex determination in a few fish species. A two-factor system was described for the swordtails Xiphophorus multilineatus and X. nigrensis. They have a basic XX/XY SD system, but occasionally XX males result from homozygosity of alleles of a sex modifying autosomal locus. In AA fish, XX genotypes develop normally as females, but aa determines male development of XX fish. Thus gene A/a is considered an autosomal modifier of a sex-chromosomal system[13].
In African cichlid fish, e.g. Metriaclima pyrsonotus[14], alleles at an XY locus and a WZ locus in two different linkage groups segregate independently. The presence of the W overrides the male determining effect of the Y, so ZWXY fish develop as females, constituting a bifactorial system. However, in a related species, M. mbenjii, ZWXY females have intersex-like phenotypes for some traits, and female phenotypes for others [15], suggesting that this system is not at evolutionary equilibrium.
The European sea bass (Dicentrarchus labrax) has been described as having polygenic sex determination, with more than three QTLs associated with sex-ratio [16]. The temperature to which young fish are exposed interacts with genomic determinants to promote either male or female gonad development. Intratesticular oocytes are frequent in young sea bass males [17] and there are population-specific variations of the genetic components of SD[18], again suggesting a species in transition.
Thus, in fish there are several sex determining systems controlled by more than a single factor. However, it is unclear which represent stable polygenic systems, and which transient states, a question that we examine further below.
Multiple variants at the same locus do not qualify as “polygenes”
Other cases of multiple sex chromosomes, however, do not fit the definition of PSD.
In the Southern platyfish, Xiphophorus maculatus, X, Y and W chromosomes are simultaneously present in the same population[19]. The W overrides the male determining activity of the Y so males can be either XY or YY and females XX, WX or WY. But the three sex chromosomes clearly belong to the same linkage group. Hence, we refer to this system as monofactorial (see Glossary, and also Box 1). Whether the SD loci on the three chromosomes harbor different alleles of the same gene or different genes is unknown.
Similarly, the wrinkled frog Glandirana rugosa[20] has XX/XY and ZW/ZZ populations on two different Japanese islands, with a hybrid zone on a third island where various combinations of these four sex chromosomes occur. Gene mapping shows that XY and two different WZ systems are variants of the same chromosome[21], and that the most likely SD gene SOX3 is shared [22]. Thus, we would classify this SD system as monogenic and polyallelic.
Changes in SD gene activity may be brought about by mutation at the SD locus itself (monogenic), or at another locus on the sex chromosomes or an autosome (digenic). This is the case in several rodents in which Sry is inactivated or inhibited. In six species of Patagonian akodont mice, a polymorphism for an unknown change on a variant Y* [23] produces XY* females[24]. No sequence differences in Sry were observed between XY males and XY* sex reversed females, so this may constitute a system of SD control by two separate but linked Y-borne factors (digenic). Alternatively (and perhaps more likely), Sry transcription from Y* may have been expunged by mutation in the promotor or upstream regulators that were not sequenced, which would constitute a system that is monogenic and polyallelic.
In several rodents, including the wood lemming and the African pygmy mouse, a variant X-chromosome (X*), polymorphic in the population, inhibits the male determining effect of the Y, so that X*Y animals develop as females[24,25]. The molecular identity of the X-linked locus that suppresses the action of Sry is unknown. If another locus on the X evolved to interact with SRY, we would describe the system formally as “digenic”. However, if X* has an allele of Sry or its X-borne homologue Sox3 that overrides the wildtype Sry action, this SD system would be monogenic and diallelic (polymorphic for X variants).
Thus, identifying the modifier locus, or at least documenting its independent assortment, is essential in classifying these SD systems.
Multiple genes in a biochemical pathway do not qualify as “polygenes”
Known SD master genes all enact sex determination through a complex biochemical pathway. The Y-borne mammalian SRY turns on SOX9, which in turn activates genes such as AMH and DMRT1 in the testis determining pathway. In the absence of SRY, other genes such as WNT4 and FOXL2 are activated that promote the development of an ovary. Testis- and ovary-promoting pathways interact through cross-suppression so that the outcome depends on a delicate balance that is easily upset by a mutation of any of the components, resulting in various degrees of sex reversal. In birds, dosage effects of the Z-borne SD master gene DMRT1 directs promotion of testis or ovary via alternative pathways that share many genes with those of mammals. So even regulation of sexual differentiation directed by monogenic SD depends on multiple (at least 60) genes[26,27].
However, we would not consider that these genes are acting as polygenes since they are all under the control of a single master sex determining trigger. The downstream genes do not deliver the primary signal for SD, but are the receivers of the signal.
Multiple sex chromosomes do not contain “polygenes”
In some animals – mammals, frogs and fish – as well as some plants, sex chromosomes are fused or translocated with autosomes to produce multiple sex chromosomes that segregate as meiotic chains. The male platypus has five Y and five X chromosomes, which form a meiotic chain that segregates alternately. The five X chromosomes go to one pole and the five Y chromosomes to the other, producing only two types of sperm that produce male and female offspring. Thus, they behave as a single X and Y chromosome[28] and SD is unifactorial. Identification of AMH (anti-Mullerian hormone) as candidate master SD gene on Y5 [29,30] is consistent with monogenic SD.
Similarly, the Brazilian fish piapara, Megaleporinus elongatus, has a Z1Z1Z2Z2 male/Z1W1Z2W2 female type of SD[31]. Z1-Z2 as well as W1-W2 segregate together, again signifying unifactorial (assumed monogenic) sex determination.
In several vertebrates, one of the sex chromosomes is fused with an autosome, generating composite neo-X, neo-Y or neo-W chromosomes. There has been speculation as to whether the added bits contain sex determining loci that must be inherited together as polygenes, but no evidence supports this attractive hypothesis. Indeed, comparative gene mapping shows that a large autosomal region was added to both the X and Y of eutherian mammals 105 million years ago[32] and is the origin of most human Y chromosome genes[33]. However, the 23 genes in the Y added region play no role in human SD, which is entirely controlled by SRY.
What if the chromosome complement doubles, as occurred twice in the common ancestor of all vertebrates, and again in several lineages of fish and amphibians? XY species would therefore have four sex chromosomes, and an SD gene on the Y would be present in two copies, qualifying as polygenes that segregate independently. Various combinations of gametes would generate ¼ XXXX, ½ XXXY and ¼ XXYY embryos, distorting the sex ratio or creating infertile intermediates, depending on effects of Y dosage[34]. We know of no polyploid vertebrates with duplicate sex chromosomes. Rather, the tetraploid frog Xenopus laevis solved this problem by inventing a novel ZW [35] that bears a duplicated and truncated version of DMRT1 to craft a new female dominant W that inhibits male development[3].
Thus, multiple and composite sex chromosomes generated by translocation or polyploidy do not qualify as systems of polygenic sex determination.
Species hybrids
Species hybrids may have problems with incompatible sex chromosomes. For instance, interspecies crosses often produce aberrant sex ratios[14,36]. This incompatibility is often resolved by the evolution of various parthenogenic (‘unisexual’) hybrid species amongst fishes, amphibians and reptiles[37], which reproduce as all-female biotypes.
However, species hybrids may reveal many genes with major and minor effects on sex determination. In a recent study[38] two species of catfish were crossed and genotyping of multiple SNPs used to detect 7 male-associated and 17 female-associated loci that map to different linkage groups (so are not allelic). This abnormal situation may, indeed, be polygenic.
Evolutionary replacement of a SD system
The best-known chromosomal SD systems (in mammals, birds, fruitflies) are extremely stable. However, reptiles, amphibians and fish show tremendous sex chromosome variety. Several different SD genes have been identified, often in closely related species, indicating that SD systems can frequently turn over as one monogenic system is replaced by another.
How can a new SD system take over from the old? If the emerging novel sex chromosomes evolve from a different autosome, we might expect odd mixtures of sex chromosomes, which would be disadvantageous. For instance, if a new ZW system replaced an old XY system, XXZZ individuals or XYZW might be intersex or at least infertile. Alternatively, one system might prove to be epistatic to the other. This well describes the two-factor (bifactorial) system of the African cichlid, in which alleles at an XY locus and a WZ locus on two different chromosomes segregate independently and the W factor is epistatic to the Y, and suggests that it is, indeed, a species in transition.
Modelling shows that ZW-XY transitions may evolve with changes in the threshold for the decision on male or female fate[39] without substantive genotypic innovation. The master sex gene and sex chromosome pair can be retained, the Z becoming the Y and the W becoming the X. If a nascent sex chromosome evolves from the same linkage group (for instance a W evolves from an X in a XY system and is epistatic over Y), the three sex chromosomes can coexist until the Z emerges, possibly from the now functionless Y. The unifactorial WXY systems in platyfish and the wrinkled frog may represent such a scenario.
It is interesting to consider the evolution of SD modifiers as extensions to the sex determining pathway as a first step in sex change and consider whether this situation leads to evolution of a new monogenic system. For instance, the A/a locus of Xiphophorus, and the X* repressor of Sry in lemmings may constitute such extensions of the pathway, in accordance with Wilkin’s “bottom up” hypothesis[40,41].
There are several situations (e.g. the frog Rana temporaria[42]) in which a new XY or WZ system can be detected in a population as a cline of frequencies with which different linkage groups determine sex.
Captive zebrafish colonies are particularly interesting. Many studies in independently maintained colonies gave inconsistent results for sex linkage to one of three different chromosomes; sex ratios were far from 1:1 and the environment was thought to play some role. However, studies of wild zebrafish show an unequivocal ZW system[43]. It is proposed [45] that female-to-male sex reversal of fish with the ZW genotype during repeated rounds of gynogenesis and the process of domestication led to the eventual loss of the Z chromosome. The domesticated zebrafish strains are composed of only WW genotypes, some of which become females and other become fertile neomales. Different colonies have been independently evolving novel SD systems – for only about 30 years. The sex ratios and inconsistent sex linkage suggests that downstream genes in testis and ovary pathways are battling it out for supremacy; a situation about as close to “polygenic” as we might get and a wonderful opportunity to study the first steps in evolution of new SD genes. A recent study indeed provided evidence that the domesticated zebrafish stock turn to PSD after the loss of their monogenic system[44].
In the Siamese fighting fish breeds that are the result of strong artificial selection and hybridizations, some strains are monogenic for dmrt1 as a male determiner[45] while others show polygenic sex inheritance that may denote the invasion by a novel sex determining region[46].
A loss of the W chromosome has also been documented for the Australian dragon lizard Pogona vitticeps, whose ZZ male ZW female system can be overridden at high temperatures, producing ZZ females. Mating sex reversed ZZ females to normal ZZ males produces all ZZ progeny whose sex is completely determined by incubation temperature[47]. Likewise, in the medakafish, frequent loss of the Y chromosome that bears the master male SD gene (a duplicated copy of the autosomal dmrt1a gene), was observed in laboratory populations [48]. Under environmental stress (elevated temperature), and mediated by cortisone, the autosomal dmrt1a became precociously activated in embryos and mimicked the action of the Y-chromosomal dmrt1bY[49]. As result XX fish developed as males with a novel proto-Y chromosome, which eventually supplanted XY males.
Thus, very early and transitory stages in SD turnover can qualify as polygenic SD.
Conclusions
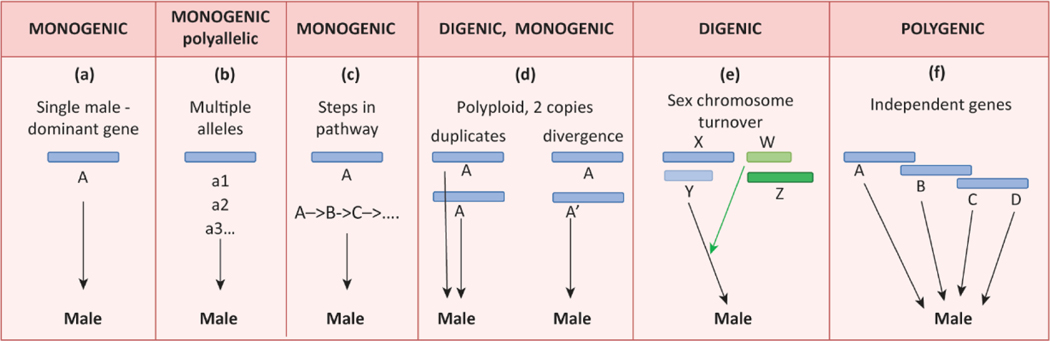
Where we find vertebrate systems with multiple sex determining factors, we must ask (a) whether there are multiple independently-inherited genes acting in the same individual, with combined effects on sexual phenotype, rather than constituting polyallelic systems or several steps in a biochemical pathway; (b) whether these genes act in the same individual or are merely polymorphic in the population; and (c) whether this is a stable relationship or a stage in turnover when one system is taking over from another. On this definition, most instances of multiple SD genes fail to qualify as polygenic sex determination (Figure 2).
Figure 2.
Monogenic, digenic and polygenic male determination. The first three panels denote monogenic sex determination, since a single gene A on a sex chromosome (blue bar) directs male determination, either by (a) its presence/absence, (b) through multiple alleles) or (c) by initiating a biochemical pathway. Panel (d) represents a tetraploid with two copies of gene A on two copies of the sex chromosome; this would produce a digenic system if both a copies were active (e.g. via A dosage), or would revert to a monogenic system if one copy A’ degenerates or changes its function. Panel (e) illustrates a digenic system in transition in which an original XY system is being taken over by a novel WZ system. Panel (f) illustrates a truly polygenic system in which four genes at distinct loci on three chromosomes contribute additively toward male determination. Blue bars, male promoting chromosome systems, green, female promoting chromosomes.
Indeed, the only examples of bona fide polygenic sex determination that we have been able to identify in vertebrates can be considered to be very young, transitioning, or very disturbed, SD systems. The digenic systems and intersex phenotypes seen in some fish may represent intermediates in turnover, and the captive zebrafish populations that accidentally lost their Z chromosome over the last few decades[43] represent an extremely early stage. The catfish species hybrids[38] represent a deranged system in which the SD genes of the two systems are fighting it out.
To broaden our knowledge about the occurrence of polygenic SD and to enhance our understanding of the evolutionary forces that lead to this system and eventually maintain it (see Outstanding questions) we need more information about the genetics and molecular developmental biology of sex in many more species. Our picture is far from complete.
Outstanding questions.
How can we use new omics techniques to identify (1) truly polygenic systems in vertebrates, (2) sex determining genes and alleles in species claimed to have a polygenic sex determining system, and (3) sex determining gene(s) in multiple sex chromosome.
Zebrafish offer a system in which an original ZW system has been disrupted recently and lost in captive colonies. What genes now contribute to sex determination and how do they interact in each colony? How do they change over time? Can different master switches be selected? Do they develop bona fide polygenic sex determination?
We need to know more about documented polygenic sex determination in microbes, plants, and invertebrates. How frequent are these systems? Are there undetected instances and how can we get a more complete picture? What genes are involved and how do they interact? Are they stable?
What are the molecular mechanisms by which quantitative signals from polygenes could integrate to elicit the binary decision towards male or female development?
Could there be adaptive value for sex determination by polygenes?
We conclude that, as predicted by evolutionary arguments, monogenic systems of sex determination predominate in vertebrates. Truly polygenic systems are rare, and represent transitions between systems, or deranged systems in species hybrids.
Highlights.
Polygenic sex determination (PSD) is defined as the determination of sexual phenotype by the combined action of two or more genes at independently inherited loci in one individual.
PSD should not be ascribed to multiple alleles at the same SD locus that may be polymorphic in the population, elements of a common biochemical pathway, and to multiple sex chromosomes that are polymorphic in a population or neo-sex chromosomes or sex chromosome chains when sexual development of the individuum is triggered by one SD locus.
A consistent nomenclature that adheres to accepted definitions of genes and alleles is recommended.
Evolutionary theory predicts that PSD is an unstable state.
Documented cases of PSD are rare. Those examined are best explained by transitionary stages during sex chromosome turnover or aberrant situations in hybrids between species with different sex chromosomes.
Acknowledgements
We thank Mateus Adolfi for discussions and critical reading of the manuscript.
Glossary and proposed nomenclature
- Sex determination (SD)
the developmental process that establishes whether the bipotential gonad primordium will become a testis or ovary. On the molecular level it tilts the balance in favor of pro-male or pro-female developmental processes in mutually antagonizing gene networks
- Genetic sex determination (GSD)
the initial trigger that activates the antagonizing gene network comes from the genome of the individual
- Environmental sex determination (ESD)
the initial trigger that activates the antagonizing gene network comes from the environment, e.g. the temperature during embryonic development of turtles or crocodiles
- Gonadal sex differentiation
the developmental process following sex determination that turns an undifferentiated gonad primordium into either a testis or an ovary
- Monogenic SD
a single gene initiates either the male or female molecular pathway of SD. The gene acts as a male- or female- dominant, or in a recessive, dosage-sensitive manner
- Digenic SD
two genes at distinct loci act together to activate the male or female sex determining pathway
- Polygenic SD
sexual phenotype determined by the combined action of alleles of multiple genes at independently-inherited loci in one individual
- Monoallelic SD
a single sex chromosome bears a single SD gene, with one allele present in a population; the inheritance of SD is monogenic
- Diallelic SD
a single sex chromosome pair bears a single SD gene, with two alleles present in a population; the inheritance of SD is monogenic
- Polyallelic SD
a single sex chromosome pair bears a single SD gene, with multiple alleles present in a population; the inheritance of SD is monogenic
- Factor
a locus that is inherited as a single unit in genetic crosses. It may consist of one or several genes
- *Unifactorial SD
genetic evidence that the SD mechanism is controlled by a single locus
- *Bifactorial SD
genetic evidence that the SD mechanism is controlled by two independently segregating loci
- *Multifactorial SD
genetic evidence that the SD mechanism is controlled by more than two separate independently segregating loci
Footnotes
*We use Greek prefixes for “genes” and “alleles” (words with Greek roots), but Latin prefixes for “factors” (word with a Latin root).
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Forconi M. et al. (2013) Characterization of sex determination and sex differentiation genes in Latimeria. PLoS One 8, e56006. 10.1371/journal.pone.0056006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Marshall Graves JA (2002) The rise and fall of SRY. Trends Genet. 18, 259–264 [DOI] [PubMed] [Google Scholar]
- 3.Yoshimoto S. et al. (2008) A W-linked DM-domain gene, DM-W, participates in primary ovary development in Xenopus laevis. Proc Natl Acad Sci U S A 105, 2469–2474. 10.1073/pnas.0712244105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Smith CA et al. (2009) The avian Z-linked gene DMRT1 is required for male sex determination in the chicken. Nature 461, 267–271 [DOI] [PubMed] [Google Scholar]
- 5.Chen S. et al. (2014) Whole-genome sequence of a flatfish provides insights into ZW sex chromosome evolution and adaptation to a benthic lifestyle. Nat Genet 46, 253–260. 10.1038/ng.2890 [DOI] [PubMed] [Google Scholar]
- 6.Kosswig C. (1964) Polygenic sex determination. Experientia 20, 190–199. 10.1007/bf02135395 [DOI] [PubMed] [Google Scholar]
- 7.Moore EC and Roberts RB (2013) Polygenic sex determination. Curr Biol 23, R510–512. 10.1016/j.cub.2013.04.004 [DOI] [PubMed] [Google Scholar]
- 8.Bachtrog D. et al. (2014) Sex determination: why so many ways of doing it? PLoS Biol 12, e1001899. 10.1371/journal.pbio.1001899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Meisel RP (2021) The maintenance of polygenic sex determination depends on the dominance of fitness effects which are predictive of the role of sexual antagonism. G3 Genes|Genomes|Genetics 11. 10.1093/g3journal/jkab149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Beukeboom LW and Perrin N. (2014) The evolution of sex determination Oxford University Press, USA [Google Scholar]
- 11.Rice WR (1986) ON THE INSTABILITY OF POLYGENIC SEX DETERMINATION: THE EFFECT OF SEX-SPECIFIC SELECTION. Evolution 40, 633–639. 10.1111/j.1558-5646.1986.tb00514.x [DOI] [PubMed] [Google Scholar]
- 12.Bull JJ (1983) Evolution of sex determining mechanisms Benjamin/Cummings Pub. Co., Advanced Book Program [Google Scholar]
- 13.Kallman KD (1984) A new look at sex determination in Poeciliid Fishes. In Evolutionary Genetics of Fishes (Turner BJ, ed), pp. 95–171, Plenum Publishing Corporation [Google Scholar]
- 14.Ser JR et al. (2010) Multiple interacting loci control sex determination in lake Malawi cichlid fish. Evolution 64, 486–501. 10.1111/j.1558-5646.2009.00871.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Moore ECC, Patrick J; Peterson Erin N.; Lamm Melissa S.; Albertson R. Craig; Roberts Reade R. (2022) Polygenic sex determination produces modular sex polymorphism in an African cichlid fish. Proccedings of the National Academy of Sciences USA in press, [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vandeputte M. and Piferrer F. (2018) Genetic and Environmental Components of Sex Determination in the European Sea Bass. In Sex Control in Aquaculture, pp. 305–325, [Google Scholar]
- 17.Piferrer F. et al. (2005) Genetic, endocrine, and environmental components of sex determination and differentiation in the European sea bass (Dicentrarchus labrax L.). General and Comparative Endocrinology 142, 102–110. 10.1016/j.ygcen.2005.02.011 [DOI] [PubMed] [Google Scholar]
- 18.Faggion S. et al. (2019) Population-specific variations of the genetic architecture of sex determination in wild European sea bass Dicentrarchus labrax L. Heredity 122, 612–621. 10.1038/s41437-018-0157-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kallman KD (1965) Genetics and geography of sex determination in the poeciliid fish, Xiphophorus maculatus. Zoologica 50, 151–190 [Google Scholar]
- 20.Ogata M. et al. (2018) Reconstruction of female heterogamety from admixture of XX-XY and ZZ-ZW sex-chromosome systems within a frog species. Mol Ecol 27, 4078–4089. 10.1111/mec.14831 [DOI] [PubMed] [Google Scholar]
- 21.Uno Y. et al. (2008) Comparative chromosome mapping of sex-linked genes and identification of sex chromosomal rearrangements in the Japanese wrinkled frog (Rana rugosa, Ranidae) with ZW and XY sex chromosome systems. Chromosome Res 16, 637–647. 10.1007/s10577-008-1217-7 [DOI] [PubMed] [Google Scholar]
- 22.Miura I. et al. (2009) The W chromosome evolution and sex-linked gene expression in the Japanese frog Rana rugosa. Sex chromosomes: genetics, abnormalities, and disorders, 123–140 [Google Scholar]
- 23.Sánchez A. et al. (2010) No Differences in the Sry Gene between Males and XY Females in Akodon (Rodentia, Cricetidae). Sexual Development 4, 155–161. 10.1159/000309780 [DOI] [PubMed] [Google Scholar]
- 24.Fredga K. (1988) Aberrant chromosomal sex-determining mechanisms in mammals, with special reference to species with XY females. Philos Trans R Soc Lond B Biol Sci 322, 83–95. 10.1098/rstb.1988.0116 [DOI] [PubMed] [Google Scholar]
- 25.Veyrunes F. et al. (2010) A novel sex determination system in a close relative of the house mouse. Proceedings of the Royal Society B: Biological Sciences 277, 1049–1056. doi: 10.1098/rspb.2009.1925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Windley SP and Wilhelm D. (2015) Signaling Pathways Involved in Mammalian Sex Determination and Gonad Development. Sexual Development 9, 297–315. 10.1159/000444065 [DOI] [PubMed] [Google Scholar]
- 27.Capel B. (2017) Vertebrate sex determination: evolutionary plasticity of a fundamental switch. Nature Reviews Genetics 18, 675–689. 10.1038/nrg.2017.60 [DOI] [PubMed] [Google Scholar]
- 28.Grützner F. et al. (2004) In the platypus a meiotic chain of ten sex chromosomes shares genes with the bird Z and mammal X chromosomes. Nature 432, 913–917. 10.1038/nature03021 [DOI] [PubMed] [Google Scholar]
- 29.Zhou Y. et al. (2021) Platypus and echidna genomes reveal mammalian biology and evolution. Nature 592, 756–762. 10.1038/s41586-020-03039-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cortez D. et al. (2014) Origins and functional evolution of Y chromosomes across mammals. Nature 508, 488–493. 10.1038/nature13151 [DOI] [PubMed] [Google Scholar]
- 31.Parise-Maltempi PP et al. (2007) Identification of a new repetitive element in the sex chromosomes of Leporinus elongatus (Teleostei: Characiformes: Anostomidae): new insights into the sex chromosomes of Leporinus. Cytogenetic and Genome Research 116, 218–223. 10.1159/000098190 [DOI] [PubMed] [Google Scholar]
- 32.Graves JA (1995) The origin and function of the mammalian Y chromosome and Y-borne genes--an evolving understanding. Bioessays 17, 311–320. 10.1002/bies.950170407 [DOI] [PubMed] [Google Scholar]
- 33.Waters PD et al. (2001) The human Y chromosome derives largely from a single autosomal region added to the sex chromosomes 80–130 million years ago. Cytogenet Cell Genet 92, 74–79. 10.1159/000056872 [DOI] [PubMed] [Google Scholar]
- 34.Wertheim B. et al. (2013) Polyploidy in Animals: Effects of Gene Expression on Sex Determination, Evolution and Ecology. Cytogenetic and Genome Research 140, 256–269. 10.1159/000351998 [DOI] [PubMed] [Google Scholar]
- 35.Session AM et al. (2016) Genome evolution in the allotetraploid frog Xenopus laevis. Nature 538, 336–343. 10.1038/nature19840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kawamura K. and Hosoya K. (2000) Masculinization mechanism of hybrids in bitterlings (Teleostei: Cyprinidae). J Hered 91, 464–473. 10.1093/jhered/91.6.464 [DOI] [PubMed] [Google Scholar]
- 37.Stöck M. et al. (2021) Sex chromosomes in meiotic, hemiclonal, clonal and polyploid hybrid vertebrates: along the extended speciation continuum’. Philosophical Transactions of the Royal Society B: Biological Sciences 376, 20200103. doi: 10.1098/rstb.2020.0103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nguyen DHM et al. (2022) Genome-Wide SNP Analysis of Hybrid Clariid Fish Reflects the Existence of Polygenic Sex-Determination in the Lineage. Front Genet 13, 789573. 10.3389/fgene.2022.789573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Quinn AE et al. (2011) Evolutionary transitions between mechanisms of sex determination in vertebrates. Biol Lett 7, 443–448. 10.1098/rsbl.2010.1126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wilkins AS (1995) Moving up the hierarchy: a hypothesis on the evolution of a genetic sex determination pathway. Bioessays 17, 71–77 [DOI] [PubMed] [Google Scholar]
- 41.Adolfi MC et al. (2021) The replaceable master of sex determination: bottom-up hypothesis revisited. Philos Trans R Soc Lond B Biol Sci 376, 20200090. 10.1098/rstb.2020.0090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rodrigues N. et al. (2016) The genetic contribution to sex determination and number of sex chromosomes vary among populations of common frogs (Rana temporaria). Heredity 117, 25–32. 10.1038/hdy.2016.22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wilson CA et al. (2014) Wild sex in zebrafish: loss of the natural sex determinant in domesticated strains. Genetics 198, 1291–1308. 10.1534/genetics.114.169284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Valdivieso A. et al. (2022) Environmentally-induced sex reversal in fish with chromosomal vs. polygenic sex determination. Environmental Research, 113549. 10.1016/j.envres.2022.113549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang L. et al. (2022) Transposon-induced epigenetic silencing in the X chromosome as a novel form of dmrt1 expression regulation during sex determination in the fighting fish. BMC Biology 20, 5. 10.1186/s12915-021-01205-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Panthum T. et al. (2022) Something Fishy about Siamese Fighting Fish (Betta splendens) Sex: Polygenic Sex Determination or a Newly Emerged Sex-Determining Region? Cells 11, 1764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Holleley CE et al. (2015) Sex reversal triggers the rapid transition from genetic to temperature-dependent sex. Nature 523, 79–82. 10.1038/nature14574 [DOI] [PubMed] [Google Scholar]
- 48.Nanda I. et al. (2003) Common spontaneous sex-reversed XX males of the medaka, Oryzias latipes. Genetics 163, 245–251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Adolfi MC et al. (2019) Increase of cortisol levels after temperature stress activates dmrt1a causing female-to-male sex reversal and reduced germ cell number in medaka. Mol Reprod Dev 86, 1405–1417. 10.1002/mrd.23177 [DOI] [PubMed] [Google Scholar]
- 50.Brush SG (1978) Nettie M. Stevens and the discovery of sex determination by chromosomes. Isis 69, 163–172. 10.1086/352001 [DOI] [PubMed] [Google Scholar]
- 51.Feron R. et al. (2020) RADSex: a computational workflow to study sex determination using Restriction Site-Associated DNA Sequencing data. bioRxiv, 2020.2004.2022.054866. 10.1101/2020.04.22.054866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Adolfi MC et al. (2021) A duplicated copy of id2b is an unusual sex-determining candidate gene on the Y chromosome of arapaima (Arapaima gigas). Sci Rep 11, 21544. 10.1038/s41598-021-01066-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kottler VA et al. (2020) Independent Origin of XY and ZW Sex Determination Mechanisms in Mosquitofish Sister Species. Genetics 214, 193–209. 10.1534/genetics.119.302698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pan Q. et al. (2019) Identification of the master sex determining gene in Northern pike (Esox lucius) reveals restricted sex chromosome differentiation. PLoS Genet 15, e1008013. 10.1371/journal.pgen.1008013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wen M. et al. (2020) Sex chromosome and sex locus characterization in goldfish, Carassius auratus (Linnaeus, 1758). BMC Genomics 21, 552. 10.1186/s12864-020-06959-3 [DOI] [PMC free article] [PubMed] [Google Scholar]