Abstract
Scientific reproducibility that effectively leverages existing study data is critical to the advancement of research in many disciplines including neuroscience, which uses imaging and electrophysiology modalities as primary endpoints or key dependency in studies. We are developing an integrated search platform called NeuroBridge to enable researchers to search for relevant study datasets that can be used to test a hypothesis or replicate a published finding without having to perform a difficult search from scratch, including contacting individual study authors and locating the site to download the data. In this paper, we describe the development of a metadata ontology based on the World Wide Web Consortium (W3C) PROV specifications to create a corpus of semantically annotated published papers. This annotated corpus was used in a deep learning model to support automated identification of candidate datasets related to neurocognitive assessment of subjects with drug abuse or schizophrenia using neuroimaging. We built on our previous work in the Provenance for Clinical and Health Research (ProvCaRe) project to model metadata information in the NeuroBridge ontology and used this ontology to annotate 51 articles using a Web-based tool called Inception. The Bidirectional Encoder Representations from Transformers (BERT) neural network model, which was trained using the annotated corpus, is used to classify and rank papers relevant to five research hypotheses and the results were evaluated independently by three users for accuracy and recall. Our combined use of the NeuroBridge ontology together with the deep learning model outperforms the existing PubMed Central (PMC) search engine and manifests considerable trainability and transparency compared with typical free-text search. An initial version of the NeuroBridge portal is available at: https://neurobridges.org/.
Introduction
Replication of published research findings and meta-analysis of experimental findings are critical for advancing scientific research (1, 2). In particular, the reuse of datasets from previous studies to test a hypothesis or replicate published results is a resource efficient approach for scientific reproducibility instead of collecting new data by recruiting new subjects and performing the previous study again. However, the reuse of existing experimental data for replicating a previous study requires the use of data generated using comparable experimental protocols, recruitment criteria, and data analysis procedures (3). There are no existing informatics tools to support systematic and accurate search of research study data from published literature; therefore, researchers with limited resources often have to manually conduct a literature survey and contact authors of previous study to get access to the study data. This data accessibility challenge is acutely felt in neuroscience research community with a large number of projects generating experiment data; for example, the National Institutes of Health (NIH) funded more than 6000 neuroimaging projects in a year and more than 19,000 full-text articles are available, including 10,000 papers describing studies using functional magnetic resonance imaging (fMRI) (4, 5). To address this growing challenge of supporting scientific reproducibility in neuroscience research, the NeuroBridge project is developing an integrated, user-friendly web portal underpinned by semantic search functionalities to enables users to efficiently find relevant datasets for their research.
To achieve its objectives, the NeuroBridge aggregates metadata or provenance information to discover datasets that match a research hypothesis with appropriate study subjects and experiment protocols. It is important to note that instead of creating a centralized warehouse of neuroscience datasets, the NeuroBridge platform aims to create a data discovery platform using neuroscience-specific provenance metadata supplied by authors of the original study. We are building on our previous work in the development of the SchizConnect platform that allowed sharing of neuroimaging, clinical and cognitive data from multiple data sources, including MCICShare, NUSDAST, NMorphCH, fBIRN-II, and COBRE (6). The MCICShare data (7) with multisite sMRI, rest-state fMRI and dMRI scans that were collected on 1.5 Tesla (1.5T) and 3T scanners. The NUSDAST data (8) include sMRI scans collected on a single Siemens 1.5T Vision scanner. The NMorphCH data (9) include sMRI, fMRI and dMRI scans collected on a single Siemens 3T Trio scanner. The FBIRN-II (10, 11) data include multisite sMRI and fMRI scans collected on a variety of 1.5T and 3T scanners. The COBR data (12) include sMRI and resting-state fMRI scans collected on a single 3T scanner.
Data on SchizConnect are modeled using a SchizConnect metadata schema to describe structural, functional, and diffusion Magnetic Resonance Imaging (MRI) data, as well as clinical and cognitive data such as demographics, psychopathological symptom ratings, and neuropsychological assessments in schizophrenia and related disorders. Metadata for individual data sources are mapped onto the SchizConnect schema, and user queries on the SchizConnect metadata are translated to queries on the individual data source in a mediator framework based on the schema maps (13). In the NeuroBridge project, we are incorporating neuroimaging and neurophysiology datasets from across a wide range of clinical neuroscience studies. However, discovery of relevant study datasets is a significant challenge; therefore, we are using the principles defined in the Findability, Accessibility, Interoperability, and Reusability (FAIR) guidelines to allow users to find relevant datasets from a large corpus of study data (14). In addition to the FAIR principles, we propose to use Selectability using similarity measures computed over the associated metadata information. Although the use of metadata information allows NeuroBridge to address many of the challenges associated with data sharing, there is significant terminological heterogeneity in the metadata terms associated with the study datasets.
For example, the names of clinical and cognitive assessment instruments vary across different studies, similarly different terms are used to describe the phenotype of the study subject, and methods used to process as well as analyze the datasets are also described using heterogeneous terms. The World Wide Web Consortium (W3C) developed the PROV specifications to standardize the representation of provenance metadata, which is used to describe the metadata associated with research studies, which can be broadly classified into three categories of: (1) Study Data, (2) Study Method, and (3) Study Tools (S3 model) (15). This classification scheme for provenance metadata associated with biomedical research studies was developed in our earlier project called the Provenance for Clinical and Health Research (ProvCaRe) (16). In the ProvCaRe project, we extended the W3C PROV specifications to create the ProvCaRe ontology for modeling the S3 terms that can be expanded to model fine-grained provenance metadata terms associated with specific biomedical research domains such as neuroscience, sleep disorder research, and substance abuse. The ProvCaRe project developed a natural language processing workflow by adapting the Clinical Text Analysis and Knowledge Extraction System (cTAKES) to extract provenance metadata from full-text articles and allowed users to search for publications related to a research hypothesis through a search engine interface (17). We leveraged the extensibility feature of the ProvCaRe ontology in the NeuroBridge project to model metadata information associated with neuroscience and substance abuse studies and the resulting NeuroBridge ontology forms the core knowledge reference model to standardize neuroscience metadata terms. The role of the NeuroBridge ontology is to support three core modules:
NeuroBridge TextMiner: This module supports the mining of full-text articles from PubMed Central (PMC) to extract metadata information associated with neuroscience studies using the NeuroBridge ontology.
NeuroBridge Data Discovery Portal: This module is an integrated search portal to allow users to search for relevant datasets based on their hypothesis-driven query (e.g., study involving schizophrenia patients with resting state fMRI and cognitive measures) using ontology for both query expansion and search.
NeuroBridge Mediator: This module extends the SchizConnect mediator to search across study datasets using metadata information modeled in the ontology.
In this paper, we describe the development and use of the NeuroBridge ontology in the NeuroBridge TextMiner module to support semantic annotation of full-text articles describing neuroscience studies related to substance abuse and schizophrenia. These annotated documents are used for training a Bidirectional Encoder Representation from Transformers (BERT) deep neural network model, which is used to mine and rank new articles in terms of their relevance to hypothesis-driven queries.
Background
Neuroscience data sharing for scientific reproducibility. There is increasing availability of multi-modal datasets in neuroscience research especially from research studies funded as part of the National Institutes of Health (NIH) Brain Research Through Advancing Innovative Neurotechnologies (BRAIN) initiative (18). However, sharing and discovery of relevant datasets remains a difficult challenge due the significant effort involved contacting individual study authors, matching terms across different datasets, and mapping variable names across studies to get datasets with comparable attributes. NIH has developed large-scale data repositories such as the National Institute of Mental Health (NIMH) Data Archive (NDA) that contains datasets from structural MRI (14,833 subjects), functional MRI (8,256 subjects), clinical phenotypes (171,116 subjects), and genomics (32,847 subjects). Similarly, our previous project SchizConnect included data from nine studies with 23,494 scans from 1392 subjects. However, the limited search functionality available in NDA portal and our own experience in the SchizConnect project highlighted a clear need to develop a metadata-based search and discovery platform, which is being implemented in the NeuroBridge platform with data from more than nine neuroscience data repositories.
W3C PROV specification and the ProvCaRe ontology. The W3C PROV specifications are formally represented in the PROV ontology that consists of three fundamental provenance terms of Entity, Activity, and Agent together with properties, which is modeled using the description logic-based Web Ontology Language (OWL) (19). In our ProvCaRe project, we extended the PROV ontology to model provenance metadata associated with biomedical research by reusing terms from existing terminologies, such the Problem/Population, Intervention, Comparison, Outcome and Time (PICOT) model used to structure clinical studies (20) and the Ontology for Clinical Research (OCRe) (21). The ProvCaRe ontology consists of 290 classes and was used to extract structured provenance metadata from 435,248 full-text articles downloaded from PMC. The ProvCaRe ontology was designed as an extensible knowledge model that can be used to represent a variety of domain-specific provenance metadata; therefore, it was extended to develop the NeuroBridge ontology in this project.
BERT model. The Bidirectional Encoder Representation from Transformers (BERT) is a deep neural network model for natural language (22). The BERT model learns from a large-scale corpus of documents to obtain the contextual representation of a word using information from all other words in a sentence. This makes BERT especially powerful in fine-grained natural language processing tasks (both at a sentence and at the word level) where nuanced syntactic and semantic understanding is critical. In this work, we fine-tune a BERT model to recognize NeuroBridge ontology concepts occurring in neuroscience literature.
Related Work. There has been unprecedented focus on improving reproducibility of biomedical research through transparency and rigor as highlighted by the NIH Rigor and Reproducibility guidelines (1, 23). Provenance metadata plays a central role in supporting reproducibility as it enables the reuse of experiment data. Provenance metadata collection, storage, and querying has been the focus of extensive research in computer science, including relational databases, scientific workflow systems, sensor networks, and Semantic Web applications (16, 24, 25). In biomedical domain, the OCRe project developed an ontology to model metadata information associated with clinical trials and an annotation workflow called Eligibility Rule Grammar and Ontology (ERGO) to semantically annotate eligibility criteria in clinical studies (21, 26). In addition, many research communities have developed guidelines to report metadata information associated research studies, including the Consolidated Standards of Reporting Trials (CONSORT) guidelines (27), and the Animals in Research: Reporting In Vivo Experiments (ARRIVE) guidelines (28). In the neuroscience research community, there are several initiatives to identify metadata information that can be used to describe the context of studies, such as the brain imaging data structure (BIDS) and its extension to represent metadata associated with neuroimaging and neurophysiology datasets (29, 30).
Method.
The NeuroBridge TextMiner component described in this paper has two primary objectives:
Given a study hypothesis, identify relevant research studies with associated experiment datasets that can be used to test a hypothesis or replicate the studies; and
Enable researchers to query and discover relevant datasets based on their hypothesis using provenance metadata associated with the research study.
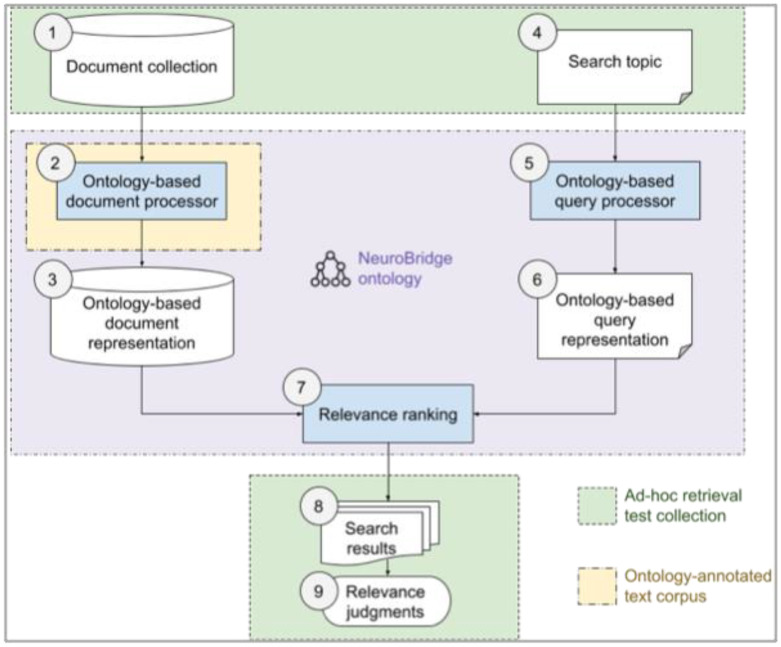
To achieve these two objectives in the NeuroBridge TextMiner, we use a multi-step approach as shown in Figure 1. Overall, our approach follows the design and evaluation methods used in semantic search engines as part of information retrieval systems. In the following section, we describe each of these components in the same order as they are numbered in Figure 1. The first component, Document collection is a subset of articles in PMC with a focus on empirical studies involving drug abuse or schizophrenia patients using neuroimaging and neurocognitive assessments. The second component, Ontology-based document processor identifies natural language expressions of concepts in the full-text article as defined in the NeuroBridge ontology and links these expressions to the ontology terms. The goal of this component is to represent a document as a collection of ontology terms. The third component, Ontology-based document representation of each article is a collection of machine-recognized ontology terms, instead of the original representation as a sequence of words. The fourth component Search topic is used by a neuroscientist in the context of a specific topic, which often consists of the clinical condition of the study subjects, imaging protocols, and neurocognitive tests. The fifth component, Ontology-based query processor interprets the search topic using standard terms in the NeuroBridge ontology. This is achieved by using a query builder in the frontend user interface of the search engine, which helps the user explore and select terms in the ontology to express the constituents of the search topic. The sixth component, Ontology-based query representation uses a Boolean expression of ontological terms. The seventh component, Relevance ranking takes ontology-based representations of a document together with query expression and uses a ranking algorithm to compute a score that estimates the relevance of the document with respect to the query. The relevance score is used by the Search results component to produce a ranked list of articles in the document collection ordered by their estimated relevance with respect to the search topic. The final component of the NeuroBridge TextMiner is called Relevance judgment, which is used for evaluation, and it consists of domain-expert assessors who are invited to judge the relevance of an article in the search results. In this work, we treat relevance as a binary status: an article is relevant if it covers all aspects of the search topic, and non-relevant otherwise. Also in Figure 1, the dashed boxes illustrate the NeuroBridge ontology and manually annotated corpora used by various parts of the TextMiner component.
Figure 1.
A diagram of methods used to implement the NeuroBridge TextMiner. Details of each numbered component are described in text.
A. NeuroBridge Ontology
The NeuroBridge ontology aims to systematically model metadata information describing neuroscience experiments, for example the number of participants in a diagnostic group, type of experiment data collected (e.g., neuroimaging, neurophysiology, and rating scales), clinical and cognitive assessment instruments. The NeuroBridge ontology extends the ProvCaRe ontology; therefore, it uses OWL constructs such as object properties to link together classes to represent metadata terms associated with neuroscience studies. In the first phase of development, we focused on studies involving mental disorder schizophrenia and substance related disorders. To model metadata associated with these studies, we extended the prov:Entity class to model a variety of neuroscience clinical findings, including neurodevelopmental disorder, mental disorders, and cognitive disorder. Following ontology engineering best practices, we reused existing ontology classes from the Systematized Nomenclature of Medicine Clinical Terms (SNOMED CT) (31) and also defined class mappings to SNOMED CT terms using OWL annotation properties (e.g., rdfs:seeAlso).
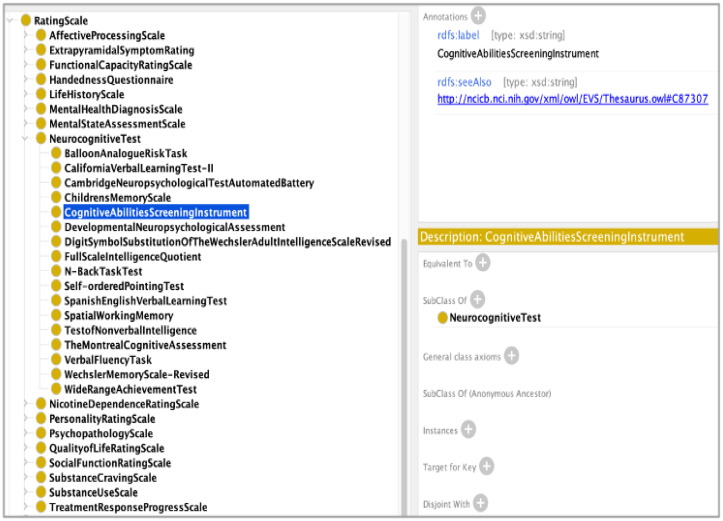
The ontology also models terms describing a variety of substance disorders, including stimulant dependence and psychoactive substance. Some research studies have focused on neurodevelopmental disorders related to substance abuse; therefore, the NeuroBridge ontology also models terms such as alcohol related neurodevelopmental disorder. To model the metadata information describing the various clinical and cognitive instruments used in neuroscience research studies, we extended the prov:Agent class to model a variety of rating scales, including substance use scales, psychopathology scales, neurocognitive scales and mental health diagnosis scale. Figure 2 shows a screenshot of the NeuroBridge ontology with the class hierarchy representing various neurocognitive scales. The latest version of the NeuroBridge ontology is available on GitHub: https://github.com/NeuroBridge/neuro-ontologies. The current version of the ontology consists of 623 classes that are used for semantic annotation of the full text articles used in this study and they are also used to support query as well as document ranking functionality of the TextMiner component.
Figure 2.
A screenshot of the NeuroBridge ontology class hierarchy representing rating scales used in neuroscience research studies.
B. Ad-hoc Retrieval Test Collection
B.1 Document collection construction. We used the following principles to guide the selection of a subset of PMC full-text articles as the document collection for this study. First, we focused on studies involving subjects with two conditions: schizophrenia and substance-related disorder, which are of increasingly interest to neuroscience researchers. Second, we selected articles that describe recent studies with empirical functional neuroimaging data collected from human subjects. We excluded meta-analysis or review papers as these types of papers do not directly collect empirical data. Third, to simplify text mining and information retrieval algorithm development during the first phase of the NeuroBridge project, the search was constrained to include only articles that are indexed by the NLM BioC collection (32). BioC offers full-text biomedical articles in formats that facilitate text mining (e.g., plain text or JSON). Following the above criteria, we issued two scoping queries using the PMC search portal in May 2021. Details of the two queries are presented in Table 1. The query identified a collection of 356 full text articles in PMC.
Table 1.
Details of two search queries used in constructing our document collection.
| Search Topic | Schizophrenia | Substance-related Disorder |
|---|---|---|
| Search string | (“functional neuroimaging”[mh]) (“schizophrenia”[mh]) NOT (meta-analysis[pt] OR review[pt]) NOT (meta-analysis[ti] or review[ti]) | (“functional neuroimaging”[mh]) (“substance-related disorders”[mh]) NOT (meta-analysis[pt] or review[pt]) NOT (meta-analysis[ti] or review[ti]) |
| PMC filters | Free full text; Time In the last 5 years; Subjects: Humans; language: English | Free full text; Time In the last 5 years; Subjects: Humans; language: English |
| # of returned articles | 335 | 200 |
| # of returned articles in BioC | 196 | 162 |
| # of articles in document collection | 196 + 162 – 2 = 356 (two articles are shared between the above two sets) | |
B.2 Test queries, search results, and evaluation of relevance. We collected a set of five search queries from neuroscience domain experts. These search queries represent typical information needs of neuroscientists who perform literature survey to identify studies that generate as well as share empirical data for secondary analysis or support replication. Table 2 shows the test queries in both natural language and using NeuroBridge ontology concepts. Each concept is an anchor text with a hyperlink pointing to the corresponding concept in the NeuroBridge ontology in GitHub. Basic statistics of relevance judgments for each query are also shown. For each query, we collected relevance judgment scores using the standard pooling procedure. More specifically, for each test query, top-k search results generated by four relevance ranking algorithms (described below) are merged into a pool of potentially relevant articles (the unordered “retrieved pool” of articles), whose relevance scores are judged by human assessors. In this preliminary work, we focus on the first page of search results, i.e., k = 10. The relevance scale is binary (either “Relevant” or “Not Relevant”). The fourth column in Table 2 shows the number of articles judged for each query, i.e., the size of the union of top-10 results retrieved by the four algorithms. Across five queries, 159 articles are judged by the human assessors for relevance.
Table 2.
Five test queries expressed in both natural language and NeuroBridge ontology concepts.
| Query ID | Expressed in natural language | Expressed in NeuroBridge ontology concepts | # of articles judged | # (%) of relevant articles |
|---|---|---|---|---|
| Q1 | Papers that had schizophrenia as some of the subject types, along with resting state fMRI, and any cognitive measure. | Schizophrenia AND (RestingStateImaging OR RestingStateImagingProtocol) AND NeurocognitiveTest | 31 | 15 (48.39%) |
| Q2 | Papers that had healthy controls (“No known disorder”), and resting state fMRI. | NoKnownDisorder AND (RestingStateImaging OR RestingStateImagingProtocol) | 40 | 27 (67.50%) |
| Q3 | Papers that had some measure of impulsivity, and structural imaging (T1-weighted) data. | ImpulsivityScale AND (T1WeightedImaging OR T1WeightedImagingProtocol) | 26 | 12 (46.13%) |
| Q4 | Papers that had subjects with alcohol use disorder, a personality questionnaire, and a task-based fMRI method. | AlcoholAbuse AND PersonalityRatingScale AND (TaskParadigmImaging OR TaskParadigmImagingProtocol) | 32 | 12 (37.50%) |
| Q5 | Papers that had subjects with cannabis use disorder, neurocognitive assessment, and resting-state fMRI. | CannabisAbuse AND NeurocognitiveTest AND (RestingStateImaging OR RestingStateImagingProtocol) | 30 | 8 (26.66%) |
For each test query, each article in the retrieved pool was redundantly judged by two expert assessors (co-authors JT and LW). If these two assessors gave different relevance labels to an article, a third domain-expert assessor was asked to judge the article and break the tie, that is, for each article in a query’s retrieved pool, at least two of three domain experts agree on its label, which becomes the gold-standard label in the subsequent evaluation of ML/NLP/IR algorithms. The fifth column in Table 2 shows the number and percentage of articles judged as “Relevant” after adjudication for each query. The two expert assessors agreed on 130 of all 159 judged articles (i.e., both agreed on an article being “Relevant” or “Not Relevant”). The interrater reliability is 0.629 (Cohen’s kappa). This level of agreement is often interpreted as “substantial” (kappa in between 0.61 and 0.80).
C. Ontology-Annotated Text Corpus
A random subset of the document collection (51 out of 356 articles) was manually annotated by project team members with ontology concepts. Two neuroscience domain experts led a team of trainees in creating these annotations. Specifically, the “Abstract” and “Methods” sections of each article were closely examined and natural language expressions of any concepts in the NeuroBridge ontology were identified and annotated with the corresponding canonical term in the ontology. The annotation team took a staged approach to this task that included training, individual annotation, and curation (quality check). During this process, the NeuroBridge ontology went through a number of revisions to resolve ambiguities and to address the need for additional concepts to modeled, which were identified by the annotation team. The Inception software was used to support the annotation workflow, team management, and data management. Table 3 shows the basic statistics of the resulting corpus. This semantically annotated corpus was subsequently used in the development of machine learning/natural language processing algorithms, i.e., concept recognition and concept linking in the ontology-based document processor.
Table 3.
Basic statistics of the ontology-annotated text corpus.
| Category | Value |
|---|---|
| Number of PMC articles annotated | 51 |
| # of sentences in all “Abstract” and “Methods” sections | 3,866 |
| # of sentences annotated with any concept | 939 (24.29%) |
| # of tokens in all “Abstract” and “Methods” sections | 118,709 |
| # of tokens annotated with any concept | 5,803 (4.89%) |
| # of tokens per concept instance | 3.44 |
| # of annotated concept instances | 1,688 |
| # of distinct concepts annotated | 94 |
| # of distinct concepts annotated only once in the corpus | 19 (20.21%) |
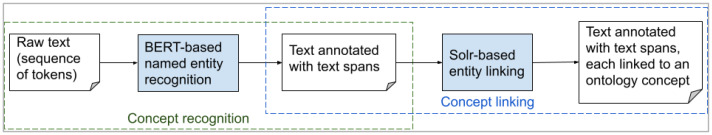
D. Ontology-based Document Processor
The goal of ontology-based document processor is to represent a document as a collection of ontology terms. This allows users to search articles using ontology concepts without spending their efforts in reformulating each concept into its synonyms and related concepts (hypernyms and hyponyms). We follow the standard entity recognition and linking pipeline approach to identify ontology terms in an article. As shown in Figure 3, the pipeline contains two stages: concept recognition and concept linking. Given raw text (sequence of tokens in the Abstract and Methods sections of an article), the concept recognition stage is responsible for tagging text spans that may mention any ontology term. Then, given a tagged text span, the concept linking stage is responsible for relating it to one of the 623 terms in the ontology. Both stages of the pipeline are developed using the ontology-annotated text corpus. We split the 51 annotated articles into 70% training (35 articles), 10% validation (5 articles), and 20% test (11 articles).
Figure 3.
Two-stage architecture of the ontology-based document processor. (a) In the first stage, a piece of plain text is input into the binary NER model, which will detect spans relevant to concepts regardless of their types. (b) In the second stage, linker will assign these spans with their corresponding types.
The rationale behind this two-stage pipeline instead of an end-to-end model (which simultaneously tags text spans and classifies the text span into one of 623 classes, or ontology terms) is due to insufficient training data. The majority of ontology terms have few annotated instances to train our model. In our pilot studies (not reported), an end-to-end model substantially underperformed a two-stage model.
D.1 Concept Recognition. We formulate the concept recognition stage as a binary sequence tagging task, in which the model only needs to determine whether a text span should be recognized as any concept or not, regardless of the concept that it is linked to in this stage (this issue is resolved in the next stage). We employed the BERT model with a conditional random fields (CRF) output layer as the binary sequence tagging model. We evaluated the BERT concept recognizer by applying it on the 11 test articles. These articles contain 367 ground-truth concept instances. The BERT concept recognizer proposed 443 text spans as concepts, 226 of which matched the ground truth. In terms of span-level named entity recognition performance, the recognizer achieves 52.2% precision (226/433), 61.6% recall (226/367), and 56.5% F1-score (harmonic mean of precision and recall). Figure 3(a) shows a sample output of the BERT model. Overall, the model was able to identify a relatively wide range of text spans potentially referring to ontology concepts.
D.2 Concept Linking. Given the tagged text spans from the concept recognition stage, the concept linking stage is to map the text span to the most relevant concept in the ontology. We estimate concept relevance as follows. For each concept, we construct a “concept document” by concatenating its textual labels in the NeuroBridge ontology, its synonyms in the Unified Medical Language system (UMLS), and its associated text spans in the training and validation data. The relevance of a text span with respect to a concept is measured by the textual similarity between the text span and the “concept document”. To accelerate the textual similarity calculation, we use Apache Solr to index all “concept documents”. A text span is treated as a free-text query and the BM25 relevance model is used to rank concepts. In case Solr returns no result for a given text span, we use fuzzy string matching between the text span and a concept as a fallback strategy to rank concepts. It measures the similarity between two strings by the Jaccard similarity of two sets of letter trigrams. We evaluated the Solr-based concept linker by applying it on text spans generated by the BERT concept recognizer on the 11 test articles. These articles contain 367 ground-truth concept instances. The concept linker proposed one concept for each of the 443 text spans tagged by the BERT concept recognizer, 134 of which matched the ground truth. In terms of span-level entity linking performance, the linker achieves 30.9% precision (134/433), 36.5% recall (134/367), and 34.4% F1-score. Figure 3(b) shows a sample output of the concept linker.
D.3. Document representation evaluation. After going through the two stages, an article is associated with a set of concepts in our NeuroBridge ontology. For each of the 11 test articles, we evaluated the set of machine-generated concepts against the set of human-annotated ones using precision, recall, and F1 score. Note that unlike the span-level evaluation for concept recognition and linking, the article-level evaluation here ignores the position of concepts. Averaged across 11 test articles, the two-stage pipeline achieved 44.5% precision, 92.4% recall, and 60.0% F1-score. Considering the low-resource nature in this task, we believe the document processor achieved an appropriate performance in this preliminary study. A high level of recall (92.4%) implies that in the subsequent retrieval stage, the system is able to retrieve almost all articles related to a query concept despite having some false positives (44.5% precision). This is desirable if the user aims to find all published articles and datasets relevant to a search criterion, that is, this represents the results of a comprehensive search in NeuroBridge.
E. Relevance Ranking
Our end goal is to improve the performance of neuroscience literature search according to FAIR data management principles. We applied the document processor on the “Abstract” and “Methods” sections of all articles in the document collection (except for the 40 training articles for the document processor), which generated a set of ontological concepts for each of the 316 articles (316 = 356 - 40). We evaluate the following approaches to result ranking. Except for the PMC approach, all other approaches are implemented using Apache Solr.
Free text: A query is represented as free-text terms (natural language words). A document is represented as a bag of free-text terms in the “Abstract” and “Methods” sections. Document relevance model is BM25.
PMC: The PMCsearch engine is used to rank documents given a query. To facilitate a fair comparison, we restrict the ranked list of documents to be within the 316 articles in our document collection.
NeuroBridge Concept (NBC): A query is represented as a disjunction of ontological terms (listed in Table 2). A document is represented as a set of machine-generated ontological terms. Document relevance model is BM25.
Expanded NBC (ExNBC): A query is represented as a disjunction of ontological terms, where each term is expanded to include their immediate parent and children in the NeuroBridge ontology. A document is represented as a set of machine-generated ontological terms. Document relevance model is BM25.
Table 4 shows the ranking performance of four retrieval methods. We use precision@10, recall@10 and mean average precision as the evaluation metrics.
Table 4.
Ranking performance of four retrieval methods.
| Precision@10 (%) | Recall@10 (%) | Mean average precision (%) | |
|---|---|---|---|
| Free text | 64.0 | 33.16 | 37.25 |
| PMC | 44.0 | 23.15 | 22.64 |
| NBC | 40.0 | 18.24 | 33.44 |
| ExNBC | 52.0 | 23.44 | 34.55 |
Discussion
Results interpretation. The ranking evaluation results show a few trends that are of significance to the overall goal of the NeuroBridge portal. First, within our proposed methods, ExNBC outperforms NBC. This highlights the importance of leveraging the ontological structure in query representation. For example, if the query contains the term “NeurocognitiveTest”, ExNBC knows that “VerbalFluencyTask” is a subtype of “NeurocognitiveTest”, and an article that contains “VerbalFluencyTask” would satisfy the term “NeurocognitiveTest”. In contrast, NBC does not have this knowledge without using ontology reasoning. Second, ExNBC outperforms PMC. This shows the proposed method has the potential to better serve the special information needs in the neuroscience domain, which is not well-covered by a general search service like PMC. Finally, ExNBC does not perform as well as the simple free-text approach, which we analyze in the error analysis below. On the other hand, our methods have two critical advantages over the free-text approach.
Trainability: Our machine learning-based method is capable of further improvements when more ontology-annotated articles are available. In comparison, the performance of the free-text approach is fixed.
Transparency: Our method is able to explain which concepts in the query have a match in the article, and where the match is. In contrast, the free-text approach is only able to show lexical matches without semantic understanding (e.g., a lexical matching system does not know that two terms are almost synonymous despite having different forms).
Error analysis. There exist three major types of errors in the system: errors in concept recognition, linking, and results ranking:
Concept recognition errors: For example, in Figure 2, “Philips Achieva” was recognized as a concept. However, the phrase itself refers to the brand of a neuroimaging device, not an imaging modality or protocol. This phrase was subsequently linked to a wrong concept “WideRangeAchievementTest”.
Concept linking errors: For example, in Figure 2, “echo planar imaging (EPI) sequence” was recognized as a concept, which is reasonable since EPI is a neuroimaging modality. However, it is linked to a wrong concept “MolecularImaging”.
Results ranking errors: For example, for a query that is a conjunction of three terms, a relevant article should contain all three terms simultaneously. However, the current ranking function of the system is a vector-space model that gives “partial credit” to each matched term between the query and an article, even if the article only contains two of the three query terms. The rationale behind this strategy is that the document processor may only recognize some (but not all) query terms in a relevant article, and these articles should still receive partial credits. However, this strategy also increases the chance of retrieving non-relevant articles that do not match all query terms. This can be attributed to the usage of BM25, in which the number of detected terms is also taken into consideration.
Conclusions and Future Work
By taking advantage of NeuroBridge ontology as a reference knowledge model, the NeuroBridge search engine demonstrates a potential for interpretable search results and a potential for iterative boosting. It can be regarded as a cornerstone of our further step focusing on the improvement of the concept processor. In future work, we will proceed to improve the system in the following directions. First, we will continue to improve the ontology-based document processor pipeline. Second, on our analysis of the ranking error implies that the BM25 vector-space model may lower the precision of retrieved results by introducing false positives. In our future iterations of this research, we will improve our document relevance model to enforce concept-based match between documents and queries. This can be implemented by a concept-based Boolean relevance model. As the new model retrieves new results per test query, additional relevance judgments will need to be collected to evaluate the new relevance model. Third, we envision a live literature search system using FAIR principles. Future evaluation of the system will involve real users interacting with a live search system that has an updated index of documents, an interactive search interface, an ontology-based query builder, a search result presentation page, and mechanisms for users to save searches and provide feedback.
Figures & Table
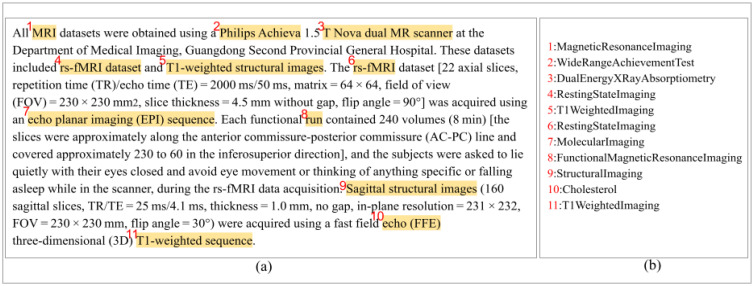
Figure 4.
This is “Section 2.3. Data acquisition” of a PMC article (Hua et al. (33)). Of the 11 concept instances detected, nine are unique concepts. The nine unique concepts are included as part of the document representation for the search system.
References
- 1.Collins FS, Tabak L.A. Policy: NIH plans to enhance reproducibility. Nature. 2014;505:612–3. doi: 10.1038/505612a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Koden N. Nihon Koden Neurology. Available from: http://www.nkusa.com/neurology_cardiology/. Retrieved on July 28th, 2022.
- 3.Nosek BA, Alter G., Banks G.C., et al. Promoting an open research culture. Science. 2015;348(6242):1422–5. doi: 10.1126/science.aab2374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.NIH RePORTER. Available from: https://projectreporter.nih.gov/reporter.cfm. Retrieved on July 28th, 2022.
- 5.Clinical Trials. Available from: http://clinicaltrials.gov/. Retrieved on July 28th, 2022.
- 6.Wang L, Alpert K.I., Calhoun V.D., et al. SchizConnect: mediating neuroimaging databases on schizophrenia and related disorders for large-scale integration. Neuroimage. 2016;124:1155–67. doi: 10.1016/j.neuroimage.2015.06.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gollub RL, Shoemaker J.M., King M.D., et al. The MCIC collection: a shared repository of multi-modal, multi-site brain image data from a clinical investigation of schizophrenia. Neuroinformatics. 2013;11(3):367–88. doi: 10.1007/s12021-013-9184-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kogan A, Alpert K., Ambite J.L., Marcus D.S., Wang L. Northwestern University schizophrenia data sharing for SchizConnect: A longitudinal dataset for large-scale integration. Neuroimage. 2016;124:1196–201. doi: 10.1016/j.neuroimage.2015.06.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Alpert K, Kogan A., Parrish T., Marcus D., Wang L. The Northwestern University Neuroimaging Data Archive (NUNDA) Neuroimage. 2016;124(0):1131–6. doi: 10.1016/j.neuroimage.2015.05.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Glover GH, Mueller B.A., Turner J.A., et al. Function biomedical informatics research network recommendations for prospective multicenter functional MRI studies. Journal of Magnetic Resonance Imaging. 2012;36(1):39–54. doi: 10.1002/jmri.23572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Potkin SG, Turner J.A., Brown G.G., et al. Working memory and DLPFC inefficiency in schizophrenia: the FBIRN study. Schizophrenia bulletin. 2009;35(1):19–31. doi: 10.1093/schbul/sbn162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cetin MS, Christensen F., Abbott C.C., et al. Thalamus and posterior temporal lobe show greater inter-network connectivity at rest and across sensory paradigms in schizophrenia. Neuroimage. 2014;97:117–26. doi: 10.1016/j.neuroimage.2014.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ambite JL, Tallis M., Alpert K., et al. International Conference on Data Integration in the Life Sciences. Springer, Cham; 2015. SchizConnect: virtual data integration in neuroimaging; pp. 37–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wilkinson MD DM, Aalbersberg IJ, Appleton G, et al. The FAIR Guiding Principles for scientific data management and stewardship. Scientific data. 2016;3(1):1–9. doi: 10.1038/sdata.2016.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Valdez J, Kim M., Rueschman M., Redline S., Sahoo S.S. International Provenance Annotation Workshop (IPAW) London, UK: Springer; 2018. Classification of Provenance Triples for Scientific Reproducibility: A Comparative Evaluation of Deep Learning Models in the ProvCaRe Project. [Google Scholar]
- 16.Sahoo SS, Valdez J., Kim M., Rueschman M., Redline S. ProvCaRe: Characterizing Scientific Reproducibility of Biomedical Research Studies using Semantic Provenance Metadata. International Journal of Medical Informatics. 2019;121:10–8. doi: 10.1016/j.ijmedinf.2018.10.009. doi: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sahoo SS, Valdez J., Rueschman M., Kim M. Semantic Provenance Graph for Reproducibility of Biomedical Research Studies: Generating and Analyzing Graph Structures from Published Literature. Studies in health technology and informatics. 2019. pp. 328–32. [DOI] [PMC free article] [PubMed]
- 18.Brain Research through Advancing Innovative Neurotechnologies (BRAIN). Washington, D.C. 2013.
- 19.Lebo T, Sahoo S.S., McGuinness D. PROV-O: The PROV Ontology. World Wide Web Consortium W3C. 2013.
- 20.Huang X, Lin J., Demner-Fushman D. Evaluation of PICO as a knowledge representation for clinical questions. AMIA Annual Symposium Proceedings. 2006. pp. 359–63. [PMC free article] [PubMed]
- 21.Sim I, Tu S.W., Carini S., Lehmann H.P., Pollock B.H., Peleg M., Wittkowski K.M. The Ontology of Clinical Research (OCRe): an informatics foundation for the science of clinical research. Journal of Biomedical Informatics. 2014;52:78–91. doi: 10.1016/j.jbi.2013.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Devlin J, Chang M.W., Lee K., Toutanova K. Bert: Pre-training of deep bidirectional transformers for language understanding. arXiv preprint arXiv:181004805. 2019.
- 23.NIH Principles and Guidelines for Reporting Preclinical Research 2016. Available from: https://www.nih.gov/research-training/rigor-reproducibility/principles-guidelines-reporting-preclinical-research. Retrieved on July 28th, 2022.
- 24.Aggarwal C. Trio: A System for Data, Uncertainty, and Lineage. In: Widom J., editor; Managing and Mining Uncertain Data. Springer; 2008. [Google Scholar]
- 25.Moreau L, Ludascher B., Altintas I., et al. The Provenance Challenge Concurr Comput. Pract Exper. 2008;20(5):409–18. [Google Scholar]
- 26.ERGO. A Template-Based Expression Language for Encoding Eligibility Criteria. Available from: http://128.218.179.58:8080/homepage/ERGO_Technical_Documentation.pdf. Retrieved on July 28th, 2022.
- 27.Schulz KF, Altman D.G., Moher D. CONSORT Group. CONSORT 2010 statement: updated guidelines for reporting parallel group randomised trials. Journal of Clinical Epidemiology. 2010;63(8):834–40. doi: 10.1016/j.jclinepi.2010.02.005. [DOI] [PubMed] [Google Scholar]
- 28.Kilkenny C, Browne W.J., Cuthill I.C., Emerson M., Altman D.G. Improving Bioscience Research Reporting: The ARRIVE Guidelines for Reporting Animal Research. PLoS Biology. 2010;8(6):e1000412. doi: 10.1371/journal.pbio.1000412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gorgolewski KJ, Auer T., Calhoun V.D., Craddock R.C., Das S., Duff E.P., Flandin G., Ghosh S.S., Glatard T., Halchenko Y.O., Handwerker D.A. The brain imaging data structure, a format for organizing and describing outputs of neuroimaging experiments. Scientific Data. 2016;3(1):1–9. doi: 10.1038/sdata.2016.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Poldrack RA, Baker C.I., Durnez J., Gorgolewski K.J., Matthews P.M., Munafò M.R., Nichols T.E., Poline J.B., Vul E., Yarkoni T. Scanning the horizon: towards transparent and reproducible neuroimaging research. Nature reviews neuroscience. 2017;18(2):115–26. doi: 10.1038/nrn.2016.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Donnelly K. SNOMED-CT: The advanced terminology and coding system for eHealth. Studies in health technology and informatics. 2006. p. 279. [PubMed]
- 32.Comeau DC, Wei C.H., Islamaj Doğan R., Lu Z. PMC text mining subset in BioC: about three million full-text articles and growing. Bioinformatics. 2019;35(18):3533–5. doi: 10.1093/bioinformatics/btz070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hua K, Wang T., Li C., Li S., Ma X., Li C., Li M., Fu S., Yin Y., Wu Y., Liu M. Abnormal degree centrality in chronic users of codeine-containing cough syrups: A resting-state functional magnetic resonance imaging study. Neuroimage: Clinical. 2018;19:775–81. doi: 10.1016/j.nicl.2018.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]