Abstract
Patients diagnosed with systemic lupus erythematosus (SLE) suffer from a decreased quality of life, an increased risk of medical complications, and an increased risk of death. In particular, approximately 50% of SLE patients progress to develop lupus nephritis, which oftentimes leads to life-threatening end stage renal disease (ESRD) and requires dialysis or kidney transplant1. The challenge is that lupus nephritis is diagnosed via a kidney biopsy, which is typically performed only after noticeable decreased kidney function, leaving little room for proactive or preventative measures. The ability to predict which patients are most likely to develop lupus nephritis has the potential to shift lupus nephritis disease management from reactive to proactive. We present a clinically useful prediction model to predict which patients with newly diagnosed SLE will go on to develop lupus nephritis in the next five years.
Introduction
Systemic lupus erythematosus (SLE) is an autoimmune disease that can disrupt multiple organ functions and ultimately be life-threatening. SLE is a minority health issue, as there is a higher prevalence among black women. It has a female to male ratio of 10-15:1 and is more than three times more prevalent in blacks than whites2,3. SLE has symptoms ranging from mild skin and joint problems to more serious cardiac, neurologic, and renal complications, leading to decreased quality of life and increased risk of death.
With no definitive diagnostic test, its similarity to other autoimmune diseases, and the range of symptoms with which patients may present, SLE diagnosis is challenging. Therefore, the American College of Rheumatology developed a standard list of criteria for clinical diagnosis, including symptoms such as malar or “butterfly” rash, photosensitivity, oral ulcers, and pleuritis4. When SLE is suspected, clinicians will typically order laboratory tests that assess a patient’s antibody profile2. However, because SLE may mimic other autoimmune diseases, patients typically hop around to several providers prior to obtaining an official diagnosis, with an average of 3.5 years from the time they first seek medical attention until they are formally diagnosed5. This delay prolongs time without treatment and allows the disease to worsen. There has been recent research suggesting there exists a set of 14 genes that are differentially expressed in patients with SLE, which could be used to confirm an SLE diagnosis early6.
To add to the mystery of this disease, the causes of SLE are also currently unknown, and thought to be a combination of genetic, hormonal and environmental factors. There is also currently no cure for SLE, but instead is typically managed through both lifestyle interventions, and medical interventions such as anti-inflammatories and steroids. However, complications do arise, and these complications tend to be extremely severe. Patients can have complications manifest in the kidneys, heart, and neural system, among others. This paper focuses on the most common serious manifestation of SLE, known as lupus nephritis, which is a complication that arises in the kidneys. About 50% of patients with SLE will go on to develop lupus nephritis, and of those, roughly 10-30% of patients will develop ESRD, a disease of the kidneys that leads to renal failure and requires dialysis or kidney transplant1,7. Not only does lupus nephritis have a long term impact on morbidity with enormous costs ranging from $43K to $107K per patient, but ESRD is its “most important and costly complication” and a profound influence on a patient’s life8.
Contrary to SLE, diagnosing lupus nephritis is more straightforward. Though clinical presentation of lupus nephritis varies, there are key indicators such as presence of hypertension, foamy urine, and lab measurements for urinalysis, urine protein excretion, etc. that signal possible presence of lupus nephritis. Even with these clinical characteristics, lupus nephritis can only be definitively diagnosed by performing a kidney biopsy, which is usually taken after there is noticeable decrease in kidney function. The international society of nephrology has outlined six classes of lupus nephritis based on the results of the biopsy, with class VI being the most aggressive form of lupus nephritis9. Given that there is no cure for lupus nephritis, the current “gold standard” for treatment is high-dose corticosteroids and high-intensity immunosuppressive agents. However, the current treatment landscape may soon shift as the FDA recently approved novel therapeutics belimumab and voclosporin for the treatment of lupus nephritis. With these therapeutics and more in the pipeline, there is potential for more targeted, proactive treatment for patients with lupus nephritis, and in a manner that does not have the adverse side effects associated with high-dose corticosteroids and immunosuppressants.
The current challenge is to identify which patients will present with lupus nephritis and when. Research has shown that there is benefit of early treatment with immunosuppressive agents and prednisone in the long-term prognosis of lupus nephritis10. That is, if patients could be diagnosed and treated with these agents earlier, they might fare better in the long-run. The challenge and opportunity detailed above motivates the development of a prediction model designed to clinically predict which patients are most likely to develop lupus nephritis of those newly diagnosed with SLE. To scope our effort, we attempt to identify patients who are likely to develop lupus nephritis within five years of index date. Five years was selected as it provided a reasonable number of positive outcomes and provided ample time for intervention.
There is existing work to predict the development of ESRD, though these models are not specific to SLE or lupus nephritis11. Other previous similar work consists of one study assessing factors associated with an increased risk of future lupus nephritis development at the time of SLE diagnosis. This study performed an associative analysis investigating factors associated with higher risk of future lupus nephritis development rather than a true prediction model. The study results concluded that a low albumin-to-globulin ratio was strongly associated with an increased risk of future lupus nephritis development12. Though the sample size was less than 300 patients, predictors identified in this research such as albumin, complement C3 levels, complement C4 levels, etc. were candidates to be included in our model.
The goal of the current work is to compare two approaches for lupus nephritis prediction, assess model performance for both, and determine the superior approach for learning such a clinical prediction model. We will compare models learned using the 1) existing feature space by leveraging the Observational Health Data Sciences and Informatics (OHDSI) tools as well as by applying domain expertise to guide feature selection, with models learned using a 2) learned feature space via representation learning13. OHDSI is a free, open-source “collaborative to bring out the value of health data through large-scale analytics”14. Additionally, we aim to provide an example of the advantage of building prediction models using a representation scheme learned via pre-training on a large unlabeled dataset.
Methods
Data.
All experiments were conducted on de-identified Stanford Electronic Health Record data from both Stanford Hospital and Lucile Packard Children’s Hospital15. Stanford’s STARR-OMOP dataset, which is Stanford Electronic Health Record data that is standardized in an Observational Medical Outcomes Partnership (OMOP) Common Data Model (CDM) was used in model development and validation16. Patients from Stanford Hospital (2009-2021) were used for model training, internal validation and final internal evaluation on a held out test set. For each particular patient, all data prior to index date (time of SLE diagnosis) was utilized to make a clinical prediction at index date.
Cohort definition.
ATLAS, an OHDSI application designed to “provide a unified interface to patient level data and analytics”, was used to translate our SLE patient phenotype into a cohort definition14. The target cohort consisted of patients with the presence of condition occurrence of SLE, which included not only the occurrence of SLE, but also all possible descendants. The index date is the date the patient was first diagnosed with SLE - given the data is limited to STARR-OMOP data, a proxy of the first instance of SLE condition occurrence present in STARR-OMOP data was used. The patient cohort was limited to patients with at least 365 days or 1 year of continuous observation prior to index date to ensure the patient had clinical events and data in STARR-OMOP data from which the model could use to make predictions at index date. Patients with prior diagnoses of diseases that mimic SLE like rheumatoid arthritis, systemic sclerosis, dermatomyositis, and vasculitis were excluded. Due to the challenging nature of SLE diagnosis, SLE condition was also confirmed using clinical notes data from STARR-OMOP’s NOTE_NLP processed note table to identify patients with an SLE concept that was present, positive and occurring to the patient at the time of the clinical note. For clinical utility purposes, patients with a lupus nephritis diagnosis prior to or on the same day as SLE diagnosis were removed from the cohort. The outcome cohort was a subset of the target cohort with the additional criteria of condition occurrence of lupus nephritis. Patients were limited to ages 14 years old and older due to the differences in clinical presentation and applicable measurement thresholds in younger children. The final target cohort consisted of 2853 patients with SLE. See Table 1 for details on cohort demographics.
Table 1.
Cohort demographic characteristics of the SLE Cohort.
| SLE Cohort | ||
|---|---|---|
| Feature | Did not develop lupus nephritis within 5 years (n = 2608) | Developed lupus nephritis within 5 years (n = 155) |
| Sex, n(%) | ||
| Female | 2367 (91) | 133 (86) |
| Male | 241 (9) | 22 (14) |
| Race, n(%) | ||
| White/Caucasian | 1245 (48) | 36 (23) |
| Asian | 440 (17) | 51 (33) |
| Black/African American | 209 (8) | 15 (10) |
| Native Hawaiian or Other Pacific Islander | 47 (2) | 3 (2) |
| American Indian or Alaska Native | 13 (<1) | 2 (1) |
| Unknown | 654 (25) | 48 (31) |
| Ethnicity, n(%) | ||
| Non-Hispanic/Non-Latino | 1945 (75) | 111 (72) |
| Hispanic/Latino | 482 (18) | 42 (27) |
| Unknown | 181 (7) | 2 (1) |
| Age (years), mean | 47 | 38 |
Model training, tuning, and feature selection.
STARR-OMOP data was randomly split into stratified training, validation and test sets by patient. 20% of the original dataset was initially removed to be the held-out test set. The remaining data was split 75% and 25% for training and validation, respectively. We learned models using two feature spaces: 1) Using existing features with OHDSI’s Patient Level Prediction and Feature Extraction package and attempting manual feature engineering with binary and categorical features. 2) Using learned representations via pre-training on unlabeled data (called CLMBR)17.
Models using existing feature space.
The first learned model using the existing feature space was developed using OHDSI’s Patient Level Prediction and Feature Extraction R packages. Feature Extraction was used to generate the features (covariates) of the cohort identified, and Patient Level Prediction (PLP) was used to build and validate the patient-level predictive models. After feature extraction, there were ~26,000 covariates with ~861,000 non-zero covariate values. During model training, covariates were normalized, and 12 redundant and ~8200 infrequent covariates were removed. Two models were trained and tuned on the cohort using the validation dataset - a lasso logistic regression and gradient boosting treemodel. Both models ran 5-fold cross validation and performed hyperparameter tuning. The best performing logistic regression used a L1 regularization, and the best performing gradient boosting model had 100 trees, a learning rate of 0.5, and a maximum depth of 1.0.
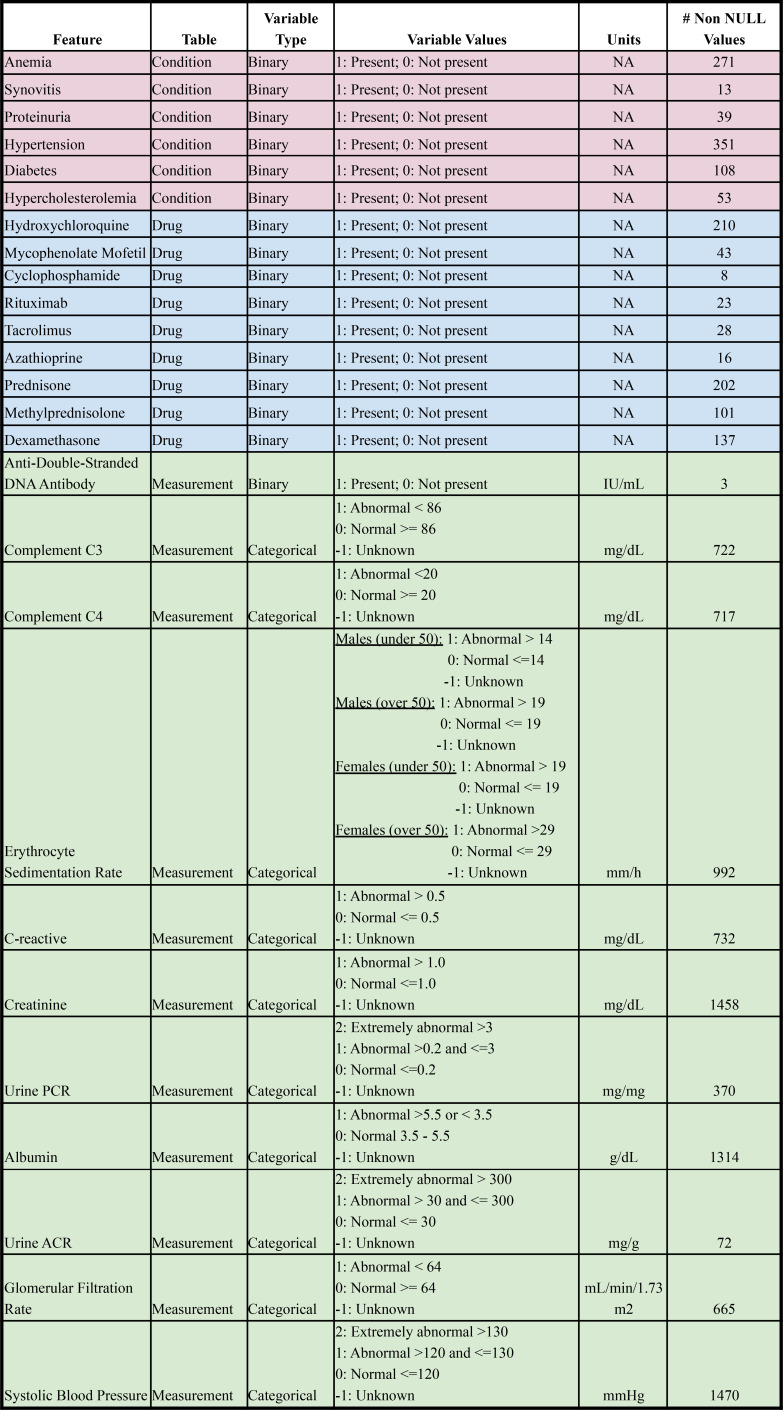
While the OHDSI model described above extracted features automatically according to a pre-developed pipeline, we also wanted to explore if a feature engineered model but with added domain expertise would result in a higher performing model. In consultation with a clinical expert, a rheumatology fellow from Stanford Hospital, we selectively constructed a machine learning model with attentive feature curation and engineering. The features chosen were selected a priori and representative of variables that a physician might consciously or subconsciously take note of that might indicate a patient’s systemic lupus is abnormally active or exhibiting flare-ups. Some features included drugs like prednisone and hydroxychloroquine, abnormal lab values of albumin, urine protein to creatinine ratio, and indirect predictors of lupus nephritis such as hypertension, diabetes, and hypercholesterolemia. ATHENA, an OHDSI tool containing standardized vocabularies, was used to appropriately select all features in STARR OMOP data15. Drug and condition feature values were derived from any time point of a patient’s history prior to the patient’s index date. Condition occurrences and drug administrations were transformed to binary representations to indicate if a patient presented with a condition or received a particular drug. Measurement features were limited to the last time point a measurement value was taken from a patient prior to index date. Labs and other continuous measurement values were binned into “abnormal” and “normal” categorical representations. The bin threshold values were established with physician consultation and hospital resources. Missing measurement values were binned into an “unknown” categorical representation as the clinical implication would suggest the patient did not require a particular lab or measurement test, which is an important value to feed into the model. Traditional patient demographics like sex, age, race, and ethnicity (see Table 1) were also included. One hot encoding was used to represent measurement categorical variables and race/ethnicity categorical variables. A total of 59 features were used. See Table 2 for a full feature set.
Table 2.
Description of feature set for manual feature engineering model.
We trained a logistic regression CV, a logistic regression with L1 regularization, a random forest, and a gradient boosting tree model on this curated feature space. All models were trained with hyperparameter tuning 5-fold cross-validation when applicable. The random forest hyperparameters included number of estimators, maximum number of features, maximum tree depth, minimum samples split, minimum samples leaf, and bootstrap. Gradient boosting hyperparameters included maximum number of features, maximum tree depth, and number of trees. Due to the extreme class imbalance with incidence of positive cases of less than 6%, optimization for metrics such as F1-score, area under the receiving-operator curve (AUROC), and area under the precision-recall curve (AUPRC) were the primary focus. We also explored two data augmentation methods to combat class imbalance: 1) Performed a grid search to manually identify the optimal weight for the minority class, and 2) Used the Synthetic Minority Oversampling Technique (SMOTE) to oversample the minority class18. After comparing all models developed, including those with data augmentation performed prior to training, the best performing lasso logistic regression, logistic regression CV, gradient boosting, and random forest models were selected for internal evaluation on the held-out test set.
Models using learned feature space.
For our second model approach, we explored using a learned feature space to represent patients. CLMBR, developed by Steinberg, et. al, is a representation learning technique that uses pre-trained medical language models to learn fixed length representations for medical timelines17. We pre-trained CLMBR on our unlabeled medical dataset after carefully excluding our held-out test set, and then trained a logistic regression CV classifier with 5-fold cross-validation, L2 regularization, and an LBFSG solver on the resulting fixed length representations.
Results
Results of models learned in existing feature space.
The OHDSI PLP models saw relatively high scores across most metrics. The best performing logistic regression used a L1 regularization, and the best performing gradient boosting model had 100 trees, a learning rate of 0.5, and a maximum depth of 1.0. For both model validation and evaluation, the gradient boosting model slightly outperformed the lasso logistic regression model in terms of AUROC, AUPRC, and F1-score. All metrics were computed with the default threshold of 0.5. The gradient boosting model achieved an AUROC of 0.81, AUPRC of 0.33, and F1-score of 0.04 on validation data and an AUROC of 0.79, AUPRC of 0.28, and F1-score of 0.11 during evaluation on the held-out test set. Meanwhile, the lasso logistic regression model achieved slightly lower AUROC, AUPRC, and F1-scores during validation of 0.79, 0.29, and 0.13, respectively. During evaluation on the held-out test set, the lasso regression model achieved an AUROC of 0.76, AUPRC of 0.27, and F1-score of 0.06.
The models using manually constructed features generated a mix of favorable and unfavorable results. Again, all metrics were computed with the default threshold of 0.5. We opted to run both a logistic regression CV model and a logistic regression model with L1 regularization as they would be most comparable to CLMBR and OHDSI lasso logistic regression, respectively. The best logistic regression model was the logistic regression CV model with an AUROC of 0.73, an AUPRC of 0.23, and an F1-score of 0.0. Both logistic regression models displayed weaker performance compared to their counterparts of OHDSI and CLMBR. The highest performing model leveraging domain expertise was the baseline gradient boosting machine model with default hyperparameters with an AUROC of 0.72, an AUPROC of 0.10, and an F1-score of 0.05. As the random forest was the lowest performing model, we also explored if this model might perform better on data with oversampling performed on the minority class, using SMOTE. The results proved relatively similar with an AUROC of 0.63, AUPRC of 0.10, and F1-score of 0.05 and indicated that model performance was not significantly altered even with data augmentation to address class imbalance.
Results of learning a model using a learned feature space.
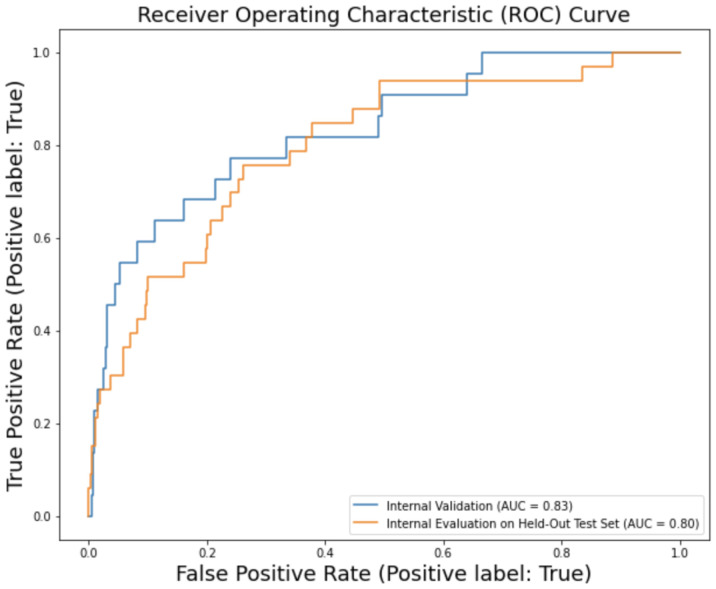
The model trained and tuned using CLMBR also had superior performance to the manually constructed model, and ultimately had the highest performance in terms of metrics we considered across all models learned. All metrics were computed with the default threshold of 0.5. For the validation dataset, CLMBR’s logistic regression had an AUROC of 0.83, AUPRC of 0.30, and F1-score of 0.08. For the evaluation on the held-out test set, CLMBR ran with an AUROC of 0.80, AUPRC of 0.30, and F1-score of 0.15. See Figure 2 for details. A comparison of the three models and their performance in model validation and evaluation is seen in Table 3.
Figure 2.
CLMBR performance on internal validation dataset and held-out test set.
Table 3.
Summary of model performance on internal validation data and held-out test set. The highest performing model results are in bold.
| AUROC | AUPRC | F1 Score | |||||
|---|---|---|---|---|---|---|---|
| Approach | Model | Validation | Test | Validation | Test | Validation | Test |
| Existing Feature Space | OHDSI Lasso Logistic Regression CV | 0.79 | 0.76 | 0.29 | 0.27 | 0.13 | 0.06 |
| OHDSI Gradient Boosting Machine | 0.81 | 0.79 | 0.33 | 0.28 | 0.04 | 0.11 | |
| DE Logistic Regression CV | 0.74 | 0.73 | 0.19 | 0.23 | 0.06 | 0.00 | |
| DE Logistic Regression L1 | 0.73 | 0.73 | 0.19 | 0.21 | 0.06 | 0.00 | |
| DE Gradient Boosting Machine | 0.75 | 0.72 | 0.27 | 0.16 | 0.12 | 0.15 | |
| DE Random Forest | 0.73 | 0.63 | 0.24 | 0.10 | 0.15 | 0.05 | |
| DE Random Forest* | 0.70 | 0.63 | 0.20 | 0.10 | 0.19 | 0.05 | |
| Learned Feature Space | CLMBR Logistic Regression CV | 0.83 | 0.80 | 0.30 | 0.30 | 0.08 | 0.16 |
| DE = Domain Expertise model; * denotes used dataset with SMOTE performed prior to model training and tuning | |||||||
Discussion
Systemic lupus erythematosus can lead to several irreversible health complications, a decreased quality of life, and an increased risk of death. Lupus nephritis is one such complication that could benefit from targeted prevention of more tailored treatments at a more optimal time of intervention. Preventing ESRD and the need for dialysis or kidney transplant has the potential to save both exorbitant costs and high quality life years. To date, there has been substantial research into the causes of ESRD but there exists no prediction model within the lupus community for lupus nephritis prediction. The recent discovery of a potential genetic biomarker that can confirm early diagnosis of SLE (thus improving the timing of the cohort selected for this model), and the recent approvals of novel therapeutics for treatment of lupus nephritis, makes anticipatory treatment more plausible without as dire adverse effects - illustrating the great potential for the implementation of one such model as a step towards precision medicine6.
The promising results from the CLMBR-based model argue for the use of learned representations for building prediction models for clinical outcomes. The method to learn the representation was created without knowledge of the lupus nephritis prediction as a use case, and has now been successfully used for predicting other low prevalence outcomes with similar positive results. Likewise, the results using models developed using the OHDSI framework further endorse OHDSI’s mission to generalize models for multi-site testing and collaboration. There is an opportunity for future work to evaluate this model across other health care systems that support the OMOP CDM. We could test this specific clinical prediction model with minimal, if any additional data manipulation or augmentation. The entire methodology of the OHDSI model, from cohort definition using ATLAS, feature extraction using the Feature Extraction package, and outcome prediction using Patient Level Prediction can be easily reproduced given the standardization and structure OHDSI has implemented. Our code is available from github at https://github.com/bechlerk/ln_prediction.
Despite these areas for promise, there is additional work that should be done to validate the results and contribute to future research. We must consider that these models were trained and tested on a relatively small sample size with a high class imbalance, which can often have misleading results. Specifically, this was exemplified in the manually constructed model and its more unfavorable results, despite the domain expertise leveraged and curation that went into feature engineering. While we cannot determine the exact nuances that led to the lower performance on this particular model, we can hypothesize it is due to the limited selection of features included that provided only sparse data to leverage in the model. It could be beneficial to identify the features of high importance in the other models as these could be features that either might not be visible to a physician or might be overlooked. Moreover, with observational data, and specifically electronic health record data, there is always discretion in a phenotype identification, which comes with the possibility of index misspecification of the target or outcome. This, in turn, could make a patient’s timeline more vague in regards to timepoints of particular feature values, and risk recapitulating the diagnostic process. A future model might consider removing the features specifically used to screen for lupus nephritis from consideration to see what other latent features we can learn to form a representation of risk. A deeper manual chart review into patients defined into a cohort would be best, though not necessarily reasonable, nor ideal with standardized applications and tools such as OHDSI’s ATLAS.
Finally, there is a need to establish clinical utility of the model and to assess the feasibility of its implementation in clinical workflow. Some of this could be accomplished through physician interviews and stakeholder consults, where we could discuss particular nuances of what thresholds might be sufficient for utility and a cost-benefit analysis of potential implementation and integration into the physician workflow. For this particular indication, we focused on optimizing the F1-score, which takes into consideration both precision and recall. From a clinical perspective, we wanted to avoid 1) false negatives, and not monitoring patients close enough or not giving pre-emptive treatment that could save both future cost and high quality life years, and 2) false positives, and performing unnecessary biopsies and burden the patient with unnecessary costs. We argue, however, that false positives would be preferred to other outcomes. Performing additional screenings and closer monitoring for patients of high risk would be relatively inexpensive and yet could have tangible health implications.
To establish clinical utility a model must be actionable, usable and safe19. Given a prediction time of five years, this algorithm is actionable as physicians can make appropriate treatment decisions and perform closer monitoring with ample time to be of consequence. This model is also usable - the actionable decisions are not time-sensitive and it would be sufficient to identify high-risk patients on a weekly or even monthly basis and then direct this information to physicians. In this scenario, it is also important to address the question of cost and methodology of model implementation20. Given the nature of prediction and time horizons involved, it can be easily deployed on a research data warehouse without disruption to clinical workflow. The workflow burden is restricted to a patient and rheumatologist. The safety, alluded to above when describing the impact of false positives and false negatives, will be proven over time as the model is utilized.
Conclusion
In conclusion, the CLMBR and OHDSI based models show promise for identifying future occurrence of lupus nephritis for patients with SLE. Specifically, these models have potential to accurately predict which patients are at high risk to develop lupus nephritis in the next five years at time of diagnosis. This ability could help shift lupus nephritis treatment from management to prevention, as additional screenings performed at more optimal times of intervention could save patients costs and quality life years. Before this model can become a part of routine care, it is necessary to address the applicable thresholds of clinical utility, identify the best path forward for implementation based on a cost-benefit analysis, and assess the long-term safety of the model.
Author Contributions
KB contributed to data collection, data analysis, review of literature, evaluation of results, conclusion and manuscript writing. LS contributed clinical expertise. ES contributed CLMBR expertise. JP contributed STARR-OMOP and OHDSI expertise. EM contributed medical expertise. NS was the primary advisor of this work. All authors have read and approved of the manuscript.
Figures & Table
References
- 1.Almaani S, Meara A, Rovin BH. Update on lupus nephritis. CJASN [Internet] 2017 May 8 [cited 2022 Feb 26];12(5):825–35. doi: 10.2215/CJN.05780616. Available from: https://cjasn.asnjournals.org/lookup/doi/10.2215/CJN.05780616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dall’Era M. Chapter 21. Systemic lupus erythematosus. In: Imboden JB, Hellmann DB, Stone JH, editors. CURRENT Diagnosis & Treatment: Rheumatology [Internet] 3rd ed. New York, NY: The McGraw-Hill Companies; 2013 [cited 2022 Feb 26]. Available from: accessmedicine.mhmedical.com/content.aspx?aid=57272268. [Google Scholar]
- 3.Hoover PJ, Costenbader KH. Insights into the epidemiology and management of lupus nephritis from the u. S. Rheumatologist’s perspective. Kidney Int [Internet] 2016 Sep [cited 2022 Mar 4];90(3):487–92. doi: 10.1016/j.kint.2016.03.042. Available from: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5679458/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aringer M, Costenbader K, Daikh D, Brinks R, Mosca M, Ramsey‐Goldman R, et al. 2019 european league against rheumatism/american college of rheumatology classification criteria for systemic lupus erythematosus. Arthritis Rheumatol [Internet] 2019 Sep [cited 2022 Feb 26];71(9):1400–12. doi: 10.1002/art.40930. Available from: https://onlinelibrary.wiley.com/doi/10.1002/art.40930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Al Sawah S, Daly RP, Foster S, Naegeli A, Benjamin K, Doll H, et al. Sat0423 understanding delay in diagnosis, access to care and satisfaction with care in lupus: findings from a cross-sectional online survey in the united states. Ann Rheum Dis [Internet] 2015 Jun [cited 2022 Feb 26];74(Suppl 2):812.3–812. Available from: https://ard.bmj.com/lookup/doi/10.1136/annrheumdis-2015-eular.1159. [Google Scholar]
- 6.Haynes WA, Haddon DJ, Diep VK, Khatri A, Bongen E, Yiu G, et al. Integrated, multicohort analysis reveals unified signature of systemic lupus erythematosus. JCI Insight [Internet] 2020 Feb 27 [cited 2022 Feb 26];5(4) doi: 10.1172/jci.insight.122312. Available from: https://insight.jci.org/articles/view/122312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wong T, Goral S. Lupus nephritis and kidney transplantation: where are we today? Adv Chronic Kidney Dis. 2019 Sep;26(5):313–22. doi: 10.1053/j.ackd.2019.08.007. [DOI] [PubMed] [Google Scholar]
- 8.Tektonidou MG, Dasgupta A, Ward MM. Risk of end-stage renal disease in patients with lupus nephritis, 1971-2015: a systematic review and bayesian meta-analysis: esrd risk in lupus nephritis. Arthritis & Rheumatology [Internet] 2016 Jun [cited 2022 Feb 26];68(6):1432–41. doi: 10.1002/art.39594. Available from: https://onlinelibrary.wiley.com/doi/10.1002/art.39594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sharma M, Das HJ, Doley PK, Mahanta PJ. Clinical and histopathological profile of lupus nephritis and response to treatment with cyclophosphamide: A single center study. Saudi Journal of Kidney Diseases and Transplantation [Internet] 2019 Mar 1 [cited 2022 Feb 26];30(2):501. doi: 10.4103/1319-2442.256857. Available from: https://www.sjkdt.org/article.asp?issn=1319-2442;year=2019;volume=30;issue=2;spage=501;epage=507;aulast=Sharma;type=0. [DOI] [PubMed] [Google Scholar]
- 10.Esdaile JM, Joseph L, MacKenzie T, Kashgarian M, Hayslett JP. The benefit of early treatment with immunosuppressive agents in lupus nephritis. J Rheumatol. 1994 Nov;21(11):2046–51. [PubMed] [Google Scholar]
- 11.Bundy JD, Mills KT, Anderson AH, Yang W, Chen J, He J, et al. Prediction of end-stage kidney disease using estimated glomerular filtration rate with and without race: a prospective cohort study. Ann Intern Med [Internet] 2022 Jan 11 [cited 2022 Feb 26]. pp. M21–2928. Available from: https://www.acpjournals.org/doi/10.7326/M21-2928. [DOI] [PMC free article] [PubMed]
- 12.Kwon OC, Lee JS, Ghang B, Kim Y-G, Lee C-K, Yoo B, et al. Predicting eventual development of lupus nephritis at the time of diagnosis of systemic lupus erythematosus. Seminars in Arthritis and Rheumatism [Internet] 2018 Dec [cited 2022 Feb 26];48(3):462–6. doi: 10.1016/j.semarthrit.2018.02.012. Available from: https://linkinghub.elsevier.com/retrieve/pii/S0049017217307229. [DOI] [PubMed] [Google Scholar]
- 13.Reps JM, Schuemie MJ, Suchard MA, Ryan PB, Rijnbeek PR. Design and implementation of a standardized framework to generate and evaluate patient-level prediction models using observational healthcare data. Journal of the American Medical Informatics Association [Internet] 2018 Aug 1 [cited 2022 Feb 26];25(8):969–75. doi: 10.1093/jamia/ocy032. Available from: https://academic.oup.com/jamia/article/25/8/969/4989437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ohdsi – observational health data sciences and informatics [Internet]. [cited 2022 Feb 27] Available from: https://www.ohdsi.org/
- 15.Datta S, Posada J, Olson G, Li W, O’Reilly C, Balraj D, et al. A new paradigm for accelerating clinical data science at Stanford Medicine. arXiv:200310534 [cs] [Internet] 2020 Mar 17 [cited 2022 Feb 26]. Available from: http://arxiv.org/abs/2003.10534.
- 16.Summary [Internet]. Observational Medical Outcomes Partnership. [cited 2022 Mar 2] Available from: https://med.stanford.edu/starr-omop/summary.html.
- 17.Steinberg E, Jung K, Fries JA, Corbin CK, Pfohl SR, Shah NH. Language models are an effective representation learning technique for electronic health record data. Journal of Biomedical Informatics [Internet] 2021 Jan [cited 2022 Feb 26];113:103637. doi: 10.1016/j.jbi.2020.103637. Available from: https://linkinghub.elsevier.com/retrieve/pii/S1532046420302653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim Y-T, Kim D-K, Kim H, Kim D-J. A comparison of oversampling methods for constructing a prognostic model in the patient with heart failure. 2020 International Conference on Information and Communication Technology Convergence (ICTC) 2020. pp. 379–83.
- 19.Seneviratne MG, Shah NH, Chu L. Bridging the implementation gap of machine learning in healthcare. BMJ Innovations [Internet] 2020 Apr 1 [cited 2022 Mar 3];6(2) Available from: https://innovations.bmj.com/content/6/2/45. [Google Scholar]
- 20.Morse KE, Bagley SC, Shah NH. Estimate the hidden deployment cost of predictive models to improve patient care. Nat Med [Internet] 2020 Jan [cited 2022 Mar 3];26(1):18–9. doi: 10.1038/s41591-019-0651-8. Available from: https://www.nature.com/articles/s41591-019-0651-8. [DOI] [PubMed] [Google Scholar]
- 21.Banda JM, Sarraju A, Abbasi F, Parizo J, Pariani M, Ison H, et al. Finding missed cases of familial hypercholesterolemia in health systems using machine learning. npj Digit Med [Internet] 2019 Apr 11 [cited 2022 Mar 3];2(1):1–8. doi: 10.1038/s41746-019-0101-5. Available from: https://www.nature.com/articles/s41746-019-0101-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shi K, Ho V, Song J, Bechler K, Chen J. Predicting unplanned 7-day intensive care unit readmissions with machine learning models for improved discharge risk assessment. Stanford University School of Medicine. 2022. [PMC free article] [PubMed]