Abstract
In this paper, a new cohort identification system that exploits the semantic hierarchy of SNOMED CT is proposed to overcome the limitations of supervised machine learning-based approaches. Eligibility criteria descriptions and free-text clinical notes from the 2018 National NLP Clinical Challenge (n2c2) were processed to map to relevant SNOMED CT concepts and to measure semantic similarity between the eligibility criteria and patients. The eligibility of a patient was determined if the patient had a similarity score higher than a threshold cut-off value. The performance of the proposed system was evaluated for three eligibility criteria. The performance of the current system exceeded the previously reported results of the 2018 n2c2, achieving the average F1 score of 0.933. This study demonstrated that SNOMED CT alone can be leveraged for cohort identification tasks without referring to external textual sources for training.
Introduction
Selecting appropriate subjects and sample population is a critical step to a successful clinical trial. Clinical trial designers need to ensure that recruited patients satisfy the inclusion and exclusion criteria of the trial to eliminate confounding factors and to avoid the study being underpowered. However, one of the major challenges to the timely conduct of research is patient recruitment for clinical trials1–3. Because of the insufficient number of participating patients, recruitment difficulties often end up with many abandoned or underpowered clinical trials4.
To fill the gap between strict inclusion/exclusion criteria and difficulties in the recruitment of patients, efforts have been made to identify eligible patients from electronic health records (EHRs)5–7. A key challenge, however, is that more detailed information of medical conditions is often embedded in the extensive occurrence of clinical narratives in EHRs in the form of unstructured text8–10. Some kinds of patient information, such as reason for prescription, are accessible exclusively through the unstructured parts of EHRs, and the manual workload burden for manual screening of patient records is one of the major obstacles to accrual success.
To reduce the workload and cost of the labor-intensive manual screening, many automated tools to identify patient cohorts from free-text clinical records have been developed11,12. Various natural language processing (NLP) techniques have been proposed to process unstructured texts and improve the accuracy of patient identification from a large collection of clinical notes in EHRs. To date, supervised machine learning NLP models which learn relationships between words from large corpora of documents have been rigorously studied13–15. We define supervised learning here as models trained by learning a mapping between input examples and the target variable (i.e., label) which was annotated by human experts. Supervised learning approaches are successfully used when labels of each data point are available. However, some important issues exist with the supervised learning NLP when it comes to cohort selection tasks.
First, a large amount of time and labor required to annotate and train a machine learning model is an ongoing concern for the NLP community16. Traditional supervised learning requires an extensive annotation and labeling of training data. A large amount of human labor and cost for annotation has limited the size of training data available, which significantly hampers the validity of supervised machine learning models16.
Second, supervised learning NLP techniques offer no hard evidence about the transferability of the models. As the provenance of data available for training is confined to only a small number of contributing institutions, the trained model is often not readily generalizable to other institutions. It has repeatedly been reported that there has often been a significant drop in performance when a system is trained in one institution and tested in another17,18.
One of the credible alternatives to supervised machine learning for cohort identification tasks is an ontology-based approach. While the machine learning approach extracts information directly from historical data and extrapolates it to make predictions, the knowledge-based approach tries to encode and construct a large number of properties of the world. Machine learning models are highly dependent on collective intelligence that is stored at the present point in the general public who are often not expertized in the field, whereas ontologies are developed by a team of experts and maintained over many years.
Although ontologies have contributed to many NLP systems, no previous systems made exclusive use of ontology for cohort identification from free-text clinical notes. Rather, ontologies had mostly been exploited for terminology or feature standardization in NLP tasks. This paper examines if semantic relations of ontologies can unilaterally be leveraged throughout the entire process of cohort identification tasks.
In this study, SNOMED CT provided by the semantic network layer of the Unified Medical Language System (UMLS) is used as a working ontology system. SNOMED CT has recently been adopted as a tool to code and classify unstructured medical narratives, working alongside various NLP and machine learning techniques in broad fields of biomedicine owing to its broad coverage of biomedical entities19. The comprehensiveness and granularity of SNOMED CT will make the proposed framework generalizable to a broad spectrum of clinical specialties and institutions.
Background and Related Works
The Track 1 Shared Task of the 2018 National NLP Clinical Challenges (n2c2) was cohort identification to identify patients who meet eligibility criteria from narrative medical records20. The task required participating systems to provide decisions on each patient’s eligibility for 13 clinical trials.
Among the participants of the 2018 n2c2, Chen et al.21 developed a task-oriented, rule-based NLP system, whose performance was compared with a hybrid general NLP system the authors previously built for general medical information extraction. Their system was plugged in with lexical-, syntactic, and meta-level evidence to indicate the presence of the target concepts, to validate the relations between the core concepts and their modification attributes, and to use section-level, note-level, document-level, and patient-level information such as patient’s gender and date of record. The rule-based and hybrid systems obtained the F-scores of 0.9028 and 0.8145, respectively.
Another team, Segura-Bedmar and Raez22, exploited several deep learning architectures such as a simple convolutional neural network (CNN), a deep CNN, a recurrent neural network (RNN), and CNN-RNN hybrid architecture for the 2018 n2c2 shared task on cohort selection for clinical trials. Unlike other existing deep learning systems, the authors used a fully connected feedforward (FFF) layer before the classification layer for all four deep learning models. With micro-F1 around 78% and macro-F1 around 49%, the RNN and hybrid designs delivered the best overall results.
Methods
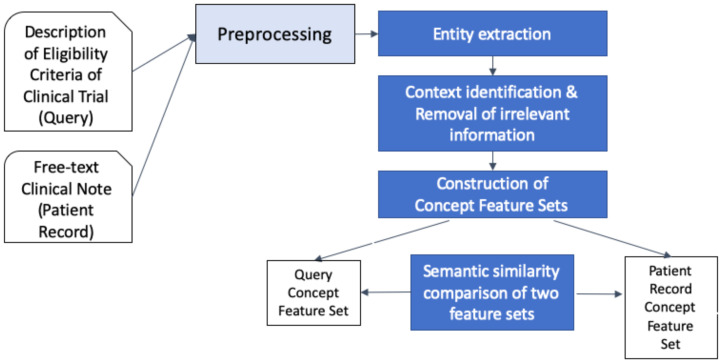
In this study, concepts and concept relations encoded in SNOMED CT within the UMLS were leveraged to prompt concept features of eligibility criteria and each patient record. Figure 1 outlines the high-level steps of the proposed framework.
Figure 1.
Architecture of the proposed cohort identification framework.
1. Dataset Free-text clinical notes from the 2018 National NLP Clinical Challenge (n2c2) Shared-Task and Workshop on Cohort Selection for Clinical Trials20 were used for the construction of the framework and evaluation of performance. The dataset consisted of records for 288 patients, of which 202 were made available as the train set; the remaining 86 were set aside as the test set. Each of the 288 patient records aggregated longitudinal records of 2-5 visits in chronological order, and patient-level eligibility for each of the 13 criteria was presented as “met” or “not met” at the end of the patient record. Since the purpose of the current study is to exploit SNOMED CT’s semantic structure for knowledge acquisition from free-text clinical notes about symptoms and signs, disease, procedures, medications, and other clinical contexts, only three highly medical and knowledge-intensive criteria— “ABDOMINAL,” “MAJOR-DIABETES,” and “ADVANCED-CAD (coronary artery disease)”—was used in this study (Table 1).
Table 1.
Clinical trials and their eligibility criteria used in this study.
| Clinical Trial Name | Criteria | Number of records | |
|---|---|---|---|
| Eligible (met) | Not eligible (not met) | ||
| ABDOMINAL | History of intra-abdominal surgery, small or large intestine resection, or small bowel obstruction | 107 | 181 |
| ADVANCED-CAD | Advanced cardiovascular disease, defined as having 2 or more of the following:
|
170 | 118 |
| MAJOR-DIABETES | Major diabetes-related complication, defined as any of the following that are a result of (or strongly correlated with) uncontrolled diabetes:
|
156 | 132 |
2. Preprocessing Many parts of the preprocessing were adopted from the work by Spasic et al.23 who preprocessed narrative medical records using regular expressions and a rule-based text mining approach to expand acronyms and abbreviations and to eliminate information irrelevant to the current status of the patient. The trade names of drugs were converted into their generic names (i.e., Lipitor becomes atorvastatin) using Drugs@FDA Data Files24 (Table 2a). To separate the unstructured record by section (e.g., Chief Complaint, Present Illness, Past Medical History, etc.), header terms that were embedded at the first position of a line were identified and forced to be separated from that line if they contained word parts such as history, exam, lab, med, allerg, or plan (Table 2b). After document parsing, any entities that referred to family members (e.g., mother, daughter, etc.) were converted into the word family member (Table 2c). Those sentences that included the word family member were deleted at a later stage.
Table 2.
Examples of preprocessed sentences and phrases.
| Original text | Preprocessed text |
|---|---|
| a. Converting drug brand names to generic names | |
| Cortril 5mg q.d. | hydrocortisone 5 mg qd |
| b. Section headers | |
| Allergy: the patient has no known allergy. | allergy: the patient has no known allergy. |
| Family history Mother was diagnosed with breast cancer at age 57. |
family history: family member was diagnosed with breast cancer at age 57. |
| c. Replacement with family members | |
| His mother was diagnosed with breast ca. but hasn’t undergone treatment. | his family member was diagnosed with breast cancer but has not undergone treatment. |
3. Section-Level Information Identification Section identification is useful for filtering out false or noisy information about a patient. For instance, the mention of “calcium” in the laboratory exam section is highly likely to indicate it was calcium content in the serum, while that in the “medication” section is likely to belong to dietary supplements. Rules and regular expressions were used to identify section headers and normalize section header terms (Table 2b). Identified header texts were assigned to one of the following standardized section header terms: “chief complaint,” “history of present illness,” “past medical history (includes past surgical history),” “family history,” “social history,” “review of systems,” “physical examination,” “allergies,” “medications,” “laboratory examinations,” “radiological examinations,” “problem list,” “assessment,” and “plans.” By sorting information by sections in which it appears, users can adapt sections needed for a specific cohort identification task. The “allergies” section, for example, may not be used in most cohort identification tasks, while the biomedical entities contained in the “problem list” section provide the most decisive clues to the patient’s eligibility in most cases.
4. Concept Extraction MetaMap25, an off-the-shelf biomedical NLP tool that automatically maps clinical free texts into SNOMED CT concepts, was used for concept extraction from free-text clinical records and was configured to map to the SNOMED CT terms only. Every occurrence of a SNOMED CT concept was assigned a Concept Unique Identifier (CUI) of the UMLS by MetaMap. The extracted CUIs were converted to the corresponding Atom Unique Identifiers (AUIs) since the UMLS defines semantic relations between concepts by AUIs only. More than two AUIs may be mapped from a single CUI. As MetaMap may suggest multiple concepts (CUIs and AUIs) for the same term, all extracted concepts, irrespective of their semantic tags, were considered for document expansion at a later stage. For example, MataMap extracts the concepts A2878436 |Amputation (procedure |, A3241080 |Amputation – action (qualifier value)|, and A2991260|Amputated structure (morphologic abnormality)| from the text string “amputation due to diabetic foot.” In this case, all three concepts were considered and passed to the next step.
5. Document Expansion with Concept Features In this step, the extracted AUIs replaced the corresponding trigger terms in the sentence. In this way, the syntactic structure of the patient record could be preserved, while the relations among biomedical entities extracted from the free text could be machine-processable. Table 3a demonstrates an example of a sentence in which trigger term strings were replaced by UMLS AUIs. As mentioned earlier, multiple AUIs extracted by MetaMap can be used in place of a trigger term, irrespective of its sort of semantic tags, for document expansion.
Table 3.
Examples of original text, text processed to replace trigger terms by UMLS AUIs, and Patient Concept Feature Set constructed from the AUI-replaced text. A3501627 |Hypertensive disorder, systemic arterial (disorder)|, A2928669 |Diabetes mellitus (disorder)|, A2992622 |Anxiety disorder (disorder)|, A2878587 |Anxiety (finding)|, A2890018 |Panic attack (finding)|, A2984272 |Type B viral hepatitis (disorder)|, A3070494 |Sexually transmitted disease (disorder)|, A3762230 |aspirin (substance)|
| Original text | Trigger terms replaced by AUIs | Patient Concept Feature Set |
|---|---|---|
| (a) She has a history of diabetes, anxiety, panic attacks, and hepatitis B. | she has a history of A2928669, A2992622 A2878587, A2890018, and A2984272. | {A2928669 A2992622 A2878587 A2890018 A2984272} |
| (b) The patient denied a past history of sexually transmitted disease. | the patient denied a past history of A3070494 | { } |
| (c) Aspirin was recommended to the patient for the prevention of future stroke. | A2878765 was recommended to the patient for the prevention of future stroke | { } |
| (d) Initially, the patient was stable | A3034375, the A2885192was A2921263 | { } |
6. Constructing Patient Concept Feature Set AUIs were extracted from the AUI-replaced text and stored in the Patient Concept Feature Set. Biomedical entities (i.e., AUIs) that were irrelevant to the current situation of a patient were detected and eliminated using regular expressions. Negation cues such as no, without, deny, rule out, negative, and free of were detected, and AUIs that were located adjacent to them were removed by empirical rules. AUIs that were not related to the rationale for current treatment were also removed by detecting modalities that were associated with the prevention or prophylaxis of diseases. Planned procedures were eliminated if the corresponding AUIs followed or were followed by the word plan or schedule. In the end, the Patient Concept Feature Set contained AUIs that were relevant to the current situation of the patient (Table 3a, 3b, 3c).
One challenge with MetaMap is that it extracts unspecific and general concepts from free texts as well. Important concepts that carry more clinically important meanings, such as symptoms, signs, anatomical sites, diseases, procedures, and medications, were grouped and assigned “quantified importance,” with 0 being the least and 1 being the most clinically meaningful. The quantified importance of a semantic group was assigned manually by the authors such that the current framework can best identify the most commonly used data elements such as diagnoses, procedures, and medications. Even though a concept was lexically meaningful, it received less importance if it was not subsumed by biomedically important supertype (i.e., hypernym or ancestor) concepts. In the current framework, those concepts whose quantified importance is less than 0.5 were discarded (Table 3d). The examples of the description of concepts, their corresponding semantic group, and quantified importance are shown below:
Concept – Quantified Importance << Supertype (SEMANTIC GROUP)
Therapeutic radiology procedure – 0.8 << Therapeutic procedure (PROC)
Penicillin – 0.8 << Chemical (MED)
Initially – 0.0 << Event orders (GEN)
Stable – 0.2 << Descriptor (DESC)
7. Constructing Query Concept Feature Set Sets of concepts extracted by MetaMap from each free-text eligibility criterion were further refined to best represent the eligibility criterion using AUIs. Those AUIs consisted of Query Concept Feature Set.
In the case of the ABDOMINAL criterion, the definition of intra-abdominal surgery is broad and ambiguous, as it does not explicitly dictate whether it includes, for example, intra-pelvic surgeries such as prostatectomy. To figure out the annotation intention of 2018 n2c2, we examined AUIs extracted from the training dataset and compared those extracted from the “met” patient records with those from the “not met” records. From this, it was found that procedures such as prostatectomy, dilation and curettage, and inguinal hernia repair were not considered intra-abdominal surgery by the annotators of the 2018 n2c2 dataset. In compliance with these findings, the final set of query concept features was constructed as the pseudocode below:
ABDOMINAL:IntraAbdominalSurgery
equivalentTo sct:OperationOnAbdominalRegion
and not (sct:OperativeProcedureOnMaleGenitourinaryTract
or sct:EndometrialScraping
or sct:AbdominalWallProcedure)
Query Concept Features Sets were built and expanded based on medical expert knowledge. For example, the Query Concept Feature Set for the sub-criteria of the ADVANCED-CAD criterion, “Taking 2 or more medications to treat CAD,” was defined with the AUIs A2884643 |Nitroglycerin|, A3651057 |Antiplatelet agent|, A3483678 |HMG-CoA reductase inhibitor|, A3802645 |Beta-blocking agent|, etc.
8. Defining is_a relations among concepts To exploit semantic relations among UMLS concepts, a full version of 2016AB release files was imported using MetamorphoSys, a UMLS installation wizard, and Metathesaurus customization tool, to a local machine. Microsoft MySQL 8.0.22 was used to load MRCONSO and MRHIER relational tables up in the MySQL database. The MRCONSO table contains information about a concept’s CUI, AUI, Lexical Unique Identifier (LUI), String Unique Identifier (SUI), and descriptions (i.e., fully specified name, synonym, and preferred term). MRHIER defines a concept’s is_a hierarchical relation back to the root to define all of its supertype concepts. A concept may have more than one is_a route to the root.
9. Similarity measurement between eligibility criteria and patient records In general, patient records hold a disproportionally more specific and greater amount of information than eligibility criteria, or queries, do. To address this, a new similarity measure is proposed as Equations (1) and (2). The proposed similarity measure makes use of a new weight metric for a concept that appeared in a patient record with respect to an individual query concept as follows:
| (1) |
wtq(p) is the weight of a SNOMED CT concept p in a patient record (i.e., Patient Concept Feature Set) with respect to a SNOMED CT concept q in a query (i.e., Query Concept Feature Set). depth(x) is the minimum number of nodes in the path from a concept x, to the root of the SNOMED CT taxonomy. The weight is larger than 0 only if the concept from the patient record is a subtype of the concept from the query. By considering the subtypes of a query concept only, it eliminates from consideration noisy concept features (i.e., concepts located outside the query concept’s hierarchy). If the patient concept p is not subsumed by the query concept q, the similarity is 0. depth(q)/depth(p) estimates how much specific p is in relation to q, and is less than or equal to 1 in most cases because query terms are usually more general (located at a lower depth in the SNOMED CT hierarchy) than the terms in patient records. If there are multiple is_a paths from q to p, the shortest path is considered. |subtypes(p)| is the number of all subtypes of patient record concept p. It is assumed that a concept with many subtype concepts is less specific than that with a smaller number of subtype concepts26.
The final similarity score between a query and a patient record is the sum of every weight of the patient record concept with respect to each query concept.
| (2) |
sim(P,Q) is the similarity score between Query Concept Feature Set Q, which contains n concepts, and Patient Concept Feature Set P, which contains m concepts, where usually m >> n. freq(pi) is the number of utterances of the i-th concept in the patient record. The larger the similarity score, the more semantically similar the query and the patient record are. Even though a single biomedical entity in a patient record is represented by multiple AUIs, only those which are the subtypes of the query concept contribute to the similarity score.
10. Determining eligibility of a patient Since the ground truth labels of the eligibility of each patient record in the test dataset of 2018 n2c2 is binary (“met” vs. “not met”), a cut-off similarity value needed to be established to determine whether the patient is eligible. Patient records were sorted by descending order of similarity score calculated in the previous section. A cut-off similarity score, above which patients were deemed “eligible,” was established so that the best F1 score could be achieved. Those patients whose similarity scores were greater than or equal to the cut-off similarity were predicted to be “eligible” by the system and the rest “not eligible.”
11. Evaluation The binary classification of patient eligibility predicted in the previous step resulted in the number of true positives, true negatives, false positives, and false negatives. The performance of the current system was evaluated with recall, precision, and F1 score on the 2018 n2c2 test dataset which is comprised of 86 patient records.
The 2018 n2c2 Shared-Task Track 1 has disclosed the performance of participating cohort identification systems in terms of precision, recall, and F1 score. Using the same data set and evaluation methods allows a legitimate evaluation of the current system’s performance by comparing it to those submitted by participants of the 2018 n2c2.
Results
The experimental results, along with the best F1 score obtained by the participants of 2018 n2c2 in the three selection criteria, are displayed in Table 4. After removing patient records with spurious labeling, the F1 scores for ABDOMINAL, ADVANCED-CAD, and MAJOR-DIABETES were 0.931, 0.894, and 0.974, respectively. The performance of the proposed system was higher than the best-performing systems of the n2c2 throughout the three selection criteria. Compared to the best-performing systems of the 2018 n2c2 Shared Task, the current system improved the average F1 score by 5.0%.
Table 4.
System’s overall performance on test data set and its comparison with n2c2 submissions (n2c2 performance data from Stubbs et al.20).
| Criteria | Current System | Best n2c2 submission | Median n2c2 submission* | |||
|---|---|---|---|---|---|---|
| Precision | Recall | F1 (before eliminating patient records with spurious labeling) | F1 (after eliminating patient records with spurious labeling) | F1 | F1 | |
| ABDOMINAL | 0.964 | 0.900 | 0.931 | 0.931 | 0.912 | 0.889 |
| ADVANCED-CAD | 0.823 | 0.933 | 0.875 | 0.894 | 0.870 | 0.780 |
| MAJOR-DIABETES | 0.974 | 0.884 | 0.927 | 0.974 | 0.884 | 0.831 |
| Average | 0.933 | 0.889 | 0.833 | |||
Median among top 10 performers
1. ABDOMINAL Before spurious data points were eliminated, there were one false-positive case and three false-negative cases among the 86 patient records in test data. In the false-negative case, transurethral resection of the prostate in the patient record was not recognized by the system as an intra-abdominal surgery because the system was configured to exclude Operative procedure on male genitourinary tract. In another false-positive case, the system regarded myomectomy as an intra-abdominal surgery.
2. ADVANCED-CAD There were 9 false-positive cases and 3 false-negative cases wrongly predicted by the current system before eliminating spurious data points. The error analysis of three false-positive cases showed that simple utterances of coronary artery disease were considered by the system to be myocardial infarction, though the two entities are clinically different. In another case, the system failed to determine if diseases were present, conditional, suspected, or hypothetical; a suspicious clinical problem was incorrectly identified by the system to be a present problem. A case in which a patient received coronary artery bypass graft surgery, which indicates severe myocardial infarction, was not labeled as “met” by the annotators of the 2018 n2c2 dataset (spurious labeling). In a false-negative case, the system failed to extract the concept of non-ST elevation myocardial infarction from the sentence “she was completed a cardiac stress test as recommended after suffering NSTEMI during her last hospitalization.” Further scrutiny revealed that the January 2016 version of SNOMED CT concept A3473213 |Acute non-ST segment elevation myocardial infarction| was only partially matched, but not exactly matched, to the string “NSTEMI.” A spurious labeling error (falsely labeled as “met”) was suspected in another false-negative case in which no evidence of advanced coronary artery disease was found.
3. MAJOR-DIABETES There were one false-positive case and 5 false-negative cases before removing spurious data. In the false-positive case, the system identified an (age-related) macular degeneration that was not associated with diabetes. Many false negatives (4 patient records) were cases where coronary artery disease was considered by the annotators of the 2018 n2c2 dataset to be a major complication of diabetes. Although the MAJOR-DIABETES criterion defined major diabetes-related complications to be microvascular complications such as nephropathy, retinopathy, and neuropathy, there may have been an implicit agreement among the annotators of the 2018 n2c2 dataset to include macrovascular complications such as coronary artery disease in their definition of major diabetes-related complications. In another false-negative case, the system could not infer that the patient had diabetic retinopathy from the sentence “was recently hold he needs laser surgery.”
Discussion
This study demonstrated that the semantic structures of a biomedical ontology such as SNOMED CT can be utilized to identify eligible patients for clinical trials from free-text clinical narratives, especially to query symptoms, diseases, procedures, and medications. This is not surprising given that SNOMED CT has ubiquitously been leveraged for storing and retrieving symptoms, disorders, tests, medications, and procedures in various types of clinical data models and data repositories.
The conventional use of SNOMED CT involves the retrieval of patient data using SNOMED CT codes input by clinicians; this use case is limited in patient data retrieval in cases of legacy systems or institutions in which SNOMED CT codes were not used. Since SNOMED CT has recently been adopted as a standardized terminology for EHRs only since 2013 in the U.S. and is still not available in many countries, SNOMED CT codes input by practicing clinicians are not readily available for patient data retrieval in many cases. When SNOMED CT concepts were extracted from free-text clinical narratives, however, the advantage of using SNOMED CT for cohort identification tasks can extend to environments where the input of SNOMED CT codes is not supported.
The extraction of SNOMED CT concepts from the free text also allowed for more sophisticated queries of patients. From free-text clinical narratives, one can query diseases that a patient had been diagnosed with or procedures that they had undergone but did not seem important enough to be codified by clinicians. Procedures such as small bowel obstruction can be seemingly unrelated to the current situation of a patient whose main problem was asthma, for example, and would be unlikely to be included on the patient’s problem list by a doctor; such comments might only be embedded in other parts of free-text clinical notes. Since those minor problems can be identified from free-text clinical notes only, cohort identification could be more sophisticated if SNOMED CT codes could be mapped from free-text clinical notes for cohort identification tasks.
Another benefit of using SNOMED CT, as shown in this study, is that it can be employed to measure similarities between queries (eligibility criteria) and patient records. This is of substantial advantage to clinical trial recruiters, because applying a quantitative measure of fitness to the selection criteria of a patient provides more information to them than a binary determination of eligibility does. By calculating similarity and quantitative measures of eligibility, clinical trial recruiters can take control of how extensively they will include potential subjects.
Error analysis showed that most errors were made by the discordance between the authors’ and the data annotators’ medical knowledge. For example, we did not accept myocardial infarction as a major complication of diabetes while some n2c2 annotators inconsistently did so. Although this may cause a prima facie drop in the performance, it does not imply a flaw in the system: a user may choose to include coronary artery disease in the definition of major complications of diabetes and, in that case, the performance of the system may seem to improve. Though knowledge engineering for the construction of Query Concept Feature Set is outside the current research’s focus, it would be worth additional independent research.
In some cases, modifying adjectives of a disease, which often radically change the original meaning of the concept, could not be recognized by the system. For example, the phrase “nonobstructive coronary artery disease” in the free-text patient record was transformed into A7873496 |Coronary arteriosclerosis|, losing the contextual adjective nonobstructive. This caused a patient record containing that phrase to be mistakenly predicted to be eligible for the ADVANCED-CAD criterion, though nonobstructive coronary artery disease would not be classified as an advanced cardiovascular disease by experts.
Another limitation of the current study is that the proposed system can be used to query symptoms, procedures, medications, and other patient situations that can be searchable within SNOMED CT’s semantic structure only. The ontology-based system has limitations in inferring patient context, and those tasks such as identifying patients who speak English from free-text clinical notes would be better performed by machine learning-based approaches. Future work will provide a hybrid system in which SNOMED CT is employed along with supervised machine learning methods to demonstrate its performance in other NLP components of cohort identification tasks.
Conclusion
SNOMED CT can be leveraged for cohort identification from free-text clinical notes, without referring to training data that requires extensive annotation and labeling. The hierarchical semantic relations of SNOMED CT can measure the semantic similarity between eligibility criteria and each patient record, and quantify how well the patient fits the eligibility criteria. Future research is suggested to develop a hybrid system that integrates ontology and machine learning-based approaches to enhance other NLP components such as inference in cohort identification tasks.
Figures & Table
References
- 1.Fletcher B, Gheorghe A, Moore D, Wilson S, Damery S. Improving the recruitment activity of clinicians in randomised controlled trials: A systematic review. BMJ Open. 2012;2(1):e000496. doi: 10.1136/bmjopen-2011-000496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mitchell AP, Hirsch BR, Abernethy AP. Lack of timely accrual information in oncology clinical trials: A cross-sectional analysis. Trials. 2014;15:92. doi: 10.1186/1745-6215-15-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Treweek S, Lockhart P, Pitkethly M, et al. Methods to improve recruitment to randomised controlled trials: Cochrane systematic review and meta-analysis. BMJ Open. 2013;3(2) doi: 10.1136/bmjopen-2012-002360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schroen AT, Petroni GR, Wang H, et al. Preliminary evaluation of factors associated with premature trial closure and feasibility of accrual benchmarks in phase III oncology trials. Clin Trials. 2010;7(4):312–21. doi: 10.1177/1740774510374973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hernandez AF, Fleurence RL, Rothman RL. The ADAPTABLE trial and PCORnet: Shining light on a new research paradigm. Ann Intern Med. 2015;163(8):635–6. doi: 10.7326/M15-1460. [DOI] [PubMed] [Google Scholar]
- 6.Jensen PB, Jensen LJ, Brunak S. Mining electronic health records: Towards better research applications and clinical care. Nat Rev Genet. 2012;13(6):395–405. doi: 10.1038/nrg3208. [DOI] [PubMed] [Google Scholar]
- 7.Mc Cord KA, Hemkens LG. Using electronic health records for clinical trials: Where do we stand and where can we go? CMAJ. 2019;191(5):E128–33. doi: 10.1503/cmaj.180841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Small AM, Kiss DH, Zlatsin Y, et al. Text mining applied to electronic cardiovascular procedure reports to identify patients with trileaflet aortic stenosis and coronary artery disease. J Biomed Inform. 2017;72:77–84. doi: 10.1016/j.jbi.2017.06.016. [DOI] [PubMed] [Google Scholar]
- 9.Afzal N, Mallipeddi VP, Sohn S, et al. Natural language processing of clinical notes for identification of critical limb ischemia. Int J Med Inform. 2018;111:83–9. doi: 10.1016/j.ijmedinf.2017.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ford E, Carroll JA, Smith HE, Scott D, Cassell JA. Extracting information from the text of electronic medical records to improve case detection: A systematic review. J Am Med Inform Assoc. 2016;23(5):1007–15. doi: 10.1093/jamia/ocv180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ni Y, Wright J, Perentesis J, et al. Increasing the efficiency of trial-patient matching: Automated clinical trial eligibility pre-screening for pediatric oncology patients. BMC Med Inform Decis Mak. 2015;15:28. doi: 10.1186/s12911-015-0149-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Beauharnais CC, Larkin ME, Zai AH, Boykin EC, Luttrell J, Wexler DJ. Efficacy and cost-effectiveness of an automated screening algorithm in an inpatient clinical trial. Clin Trials. 2012;9(2):198–203. doi: 10.1177/1740774511434844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Köpcke F, Lubgan D, Fietkau R, et al. Evaluating predictive modeling algorithms to assess patient eligibility for clinical trials from routine data. BMC Med Inform Decis Mak. 2013;13:134. doi: 10.1186/1472-6947-13-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ni Y, Bermudez M, Kennebeck S, Liddy-Hicks S, Dexheimer J. A real-time automated patient screening system for clinical trials eligibility in an emergency department: Design and evaluation. JMIR Med Inform. 2019;7(3):e14185. doi: 10.2196/14185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chakrabarti S, Sen A, Huser V, et al. An interoperable similarity-based cohort identification method using the OMOP Common Data Model version 5.0. J Healthc Inform Res. 2017;1(1):1–18. doi: 10.1007/s41666-017-0005-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Spasic I, Nenadic G. Clinical text data in machine learning: Systematic review. JMIR Med Inform. 2020;8(3):e17984. doi: 10.2196/17984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Napolitano G, Marshall A, Hamilton P, Gavin AT. Machine learning classification of surgical pathology reports and chunk recognition for information extraction noise reduction. Artif Intell Med. 2016;70:77–83. doi: 10.1016/j.artmed.2016.06.001. [DOI] [PubMed] [Google Scholar]
- 18.Ye Y, Wagner MM, Cooper GF, et al. A study of the transferability of influenza case detection systems between two large healthcare systems. PLoS ONE. 2017;12(4):e0174970. doi: 10.1371/journal.pone.0174970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chang E, Mostafa J. The use of SNOMED CT, 2013-2020: A literature review. J Am Med Inform Assoc. 2021;28(9):2017–26. doi: 10.1093/jamia/ocab084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stubbs A, Filannino M, Soysal E, Henry S, Uzuner Ö. Cohort selection for clinical trials: n2c2 2018 shared task track 1. J Am Med Inform Assoc. 2019;26(11):1163–71. doi: 10.1093/jamia/ocz163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen L, Gu Y, Ji X, et al. Clinical trial cohort selection based on multi-level rule-based natural language processing system. J Am Med Inform Assoc. 2019;26(11):1218–26. doi: 10.1093/jamia/ocz109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Segura-Bedmar I, Raez P. Cohort selection for clinical trials using deep learning models. J Am Med Inform Assoc. 2019;26(11):1181–8. doi: 10.1093/jamia/ocz139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Spasic I, Krzeminski D, Corcoran P, Balinsky A. Cohort selection for clinical trials from longitudinal patient records: text mining approach. JMIR Med Inform. 2019;7(4):e15980. doi: 10.2196/15980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.U.S. Food & Drug Administration. Drugs@FDA Data Files [Internet] 2017. [cited 2021 Apr 5]; Available from: https://www.fda.gov/drugs/drug-approvals-and-databases/drugsfda-data-files.
- 25.Aronson AR. Effective mapping of biomedical text to the UMLS Metathesaurus: The MetaMap program. Proc AMIA Symp 2001. pp. 17–21. [PMC free article] [PubMed]
- 26.Hadj Taieb MA, Ben Aouicha M, Ben Hamadou A. Ontology-based approach for measuring semantic similarity. Eng Appl Artif Intell. 2014;36:238–61. [Google Scholar]