Abstract
Most biomedical information extraction (IE) approaches focus on entity types such as diseases, drugs, and genes, and relations such as gene-disease associations. In this paper, we introduce the task of methodological IE to support fine-grained quality assessment of randomized controlled trial (RCT) publications. We draw from the Ontology of Clinical Research (OCRe) and the CONSORT reporting guidelines for RCTs to create a categorization of relevant methodological characteristics. In a pilot annotation study, we annotate a corpus of 70 full-text publications with these characteristics. We also train baseline named entity recognition (NER) models to recognize these items in RCT publications using several training sets with different negative sampling strategies. We evaluate the models at span and document levels. Our results show that it is feasible to use natural language processing (NLP) and machine learning for fine-grained extraction of methodological information. We propose that our models, after improvements, can support assessment of methodological quality in RCT publications. Our annotated corpus, models, and code are publicly available at https://github.com/kellyhoang0610/RCTMethodologyIE.
Introduction
Evidence-based medicine (EBM) brings together research evidence, clinical judgment, and patient values/preferences to support decision-making in patient care1. A core component of EBM is the evidence synthesis process, which aims to retrieve, assess, and synthesize research evidence from the scientific literature for translation to bedside2. Methodological quality assessment of clinical studies (i.e., the rigor of the study design and conduct) is a critical yet challenging step in evidence synthesis3.
Randomized controlled trials (RCTs) are a cornerstone of EBM. A properly designed and rigorously conducted RCT is the most robust method to determine the effectiveness of a therapeutic intervention4. Despite their advantages, RCTs are often poorly designed and executed, rendering their findings unreliable, potentially harming patients, and wasting research investment5. Evidence synthesis from RCT publications relies on “risk of bias” assessment, a type of methodological quality assessment that focuses on the internal validity of the study6. The primary tool used for risk of bias assessment, the Cochrane Risk of Bias tool6, considers study characteristics such as randomization, blinding, and allocation concealment in determining the risk of bias for a given study (low, high, unclear risk). While some automated tools have been developed for risk of bias assessment (e.g., RobotReviewer7), the process remains mostly manual in practice.
While biomedical natural language processing (NLP) is increasingly used for extracting information from scientific publications (most frequently topical information including diseases, drugs, and genes, as well as their relationships, such as adverse drug events)8, research focusing on extraction of methodological aspects remains scarce. Methodological weaknesses and inconsistencies of a study can render its claims questionable, even invalid. With the increasing use of literature mining in downstream applications, such as pharmacovigilance9, it becomes important to link the claims extracted via literature mining with the underlying methodology. Otherwise, the findings from low-quality studies can mislead, as we witnessed with the methodologically suspect studies recommending drugs such as ivermectin for COVID-1910 during the pandemic. Most NLP research that has considered methodological aspects has focused on PICO elements (Participants, Interventions, Comparison, Outcomes)11–13 or text classification for risk of bias7,14,15. A comprehensive and fine-grained characterization and extraction of methodological information can improve the utility of the IE models focusing on claims by providing context for their interpretation.
In this study, we propose a fine-grained view of methodological IE focusing on RCT publications. We illustrate methodological IE with an example sentence below taken from the abstract of an RCT publication.
The randomisation sequence was computer-generated by blocks, stratified by centre, disease severity (ALSFRS-R cut-off score of 33) and onset (spinal or bulbar).
-
Sequence Generation Method: computer generated
Randomization Type: {block, stratified}
Stratification Criteria: {centre, disease severity (ALSFRS-R cut-off score of 33), disease onset (spinal or bulbar)}
A methodological IE model can extract the template shown in (2) from (1), indicating, for example, that two types of randomization techniques were used in the study: block randomization and stratified randomization. This information can be used to summarize the methodology of the study in a structured format.
Toward this objective, we report a pilot annotation study in which methodological characteristics from RCT publications were captured at fine granularity. We also trained baseline named entity recognition (NER) models based on the annotated corpus. Our results demonstrate the feasibility of capturing and extracting such information at a granular level. We propose that, with additional training data and further performance improvements, our models could help contextualize the claims made in a publication and support evidence synthesis pipelines.
Related Work
Models of RCT study characteristics
Several models to represent RCT study characteristics have been proposed. The PICO framework (Participants, Intervention, Comparison, Outcomes)16 is widely used in EBM and by the systematic review community to formulate clinical questions11,17. Variations of PICO, such as PICOT (T for timeframe)18, have also been proposed.
RCT Schema19 goes beyond the PICO framework and captures details about the administration, design, execution, and results of a trial in a hierarchical model. A follow-up ontology, the Ontology of Clinical Research (OCRe)20, attempts to develop a more comprehensive model for clinical research, including classes such as interventional and observational study design and various randomization types (e.g., block randomization, minimization).
To improve reporting quality and transparency of RCT publications, CONSORT reporting guidelines have been proposed4,21. CONSORT consists of a 25-item checklist, some of which focus on methodological information (e.g., study design, blinding, randomization, statistical methods).
Corpora of RCT methodology
Several corpora based on PICO or similar characterizations have been reported11–13,22–24, differing in their granularity (sentence vs. phrases), focus (abstract vs. full-text), the categories considered, and how they are generated (manual, crowdsourcing, automatic). We recently reported the CONSORT-TM corpus, in which 37 fine-grained CONSORT checklist items were manually annotated in full-text RCT papers25.
NLP for methodological information extraction
NLP for methodological information often takes the form of sentence classification or data extraction (i.e., text snippets)26. As might be expected, most methodological IE research has focused on PICO elements using the available corpora. Some studies explored types beyond PICO, such as study design and sample size27,28. Depending on the corpora and PICO elements considered, rule-based methods11, text classification12,24,29 or NER models13,30, as well as hybrid approaches have been reported11,27,28,31. Multi-task learning has been used to develop automatic risk of bias assessment models which classify the publication as low or high risk for a bias category and simultaneously extract supporting sentences7. We reported BERT-based sentence classification models for classification of CONSORT methodology sentences25,32.
To a large extent, these approaches are coarse-grained (sentence level) and address a limited range of methodological quality characteristics. PICO elements do not focus on methodological quality. Automatic risk of bias assessment models7,14 ultimately make quality judgments but their predictions are opaque as they do not explicitly focus on IE. Extracting granular methodological information and accounting for the diversity of expressions (e.g., “open label” and “no blinding” are synonymous) not only can help create more transparent evidence quality assessment tools but also can help computers reason about the strength of evidence and risk of bias in a study and identify methodological weaknesses and inconsistencies20. In this pilot study, we aimed to fill this gap by creating a data model, an annotated corpus, and NLP models to identify fine-grained methodological quality information from RCT publications.
Methods
In this section, we first describe the data model that we developed for annotation. Next, we report the annotation process. Lastly, we describe the baseline NLP models we developed and our evaluation scheme.
Data Model
Our data model was largely drawn from OCRe20, an ontology that was designed to represent planning, execution, and analysis steps of clinical research. OCRe provides relevant definitions and categorizations for methodological aspects of clinical research studies, including but not limited to RCTs. We also used CONSORT reporting guidelines to identify additional characteristics that are relevant to assessing methodological quality.
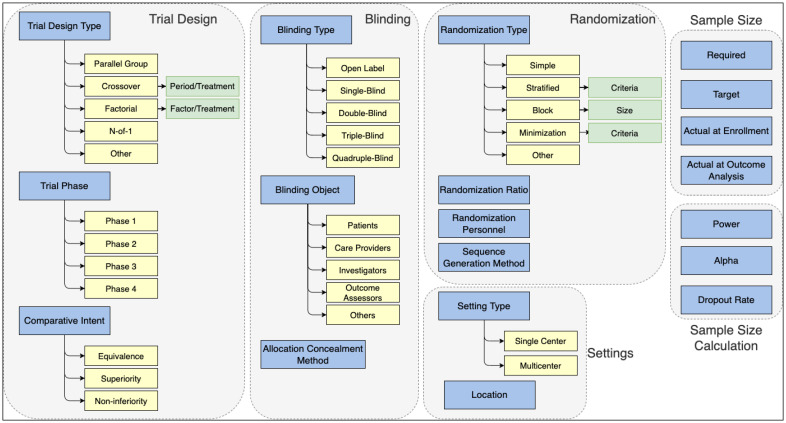
In OCRe, we primarily focused on subclasses of Interventional Study Design and Study Design Characteristic classes, including Blinding Type and Randomization Type. Some relevant characteristics were derived from data properties (e.g., Planned Sample Size and Actual Sample Size). Additional characteristics relevant for methodological quality were drawn from the CONSORT methodology checklist. For example, fine-grained information related to Sample Size Calculation (CONSORT item 7a) and Trial Settings (4b) were included, such as Power and Alpha values, and Multicenter vs. Single-center distinction. The main criterion for inclusion was whether the characteristic provides any information about methodological quality, which we ensured through literature review, and whether they can be identified in RCT publications. We also noted that some characteristics have properties whose values can be important in interpreting them (e.g., Block Size for Block Randomization). These properties were included in the data model, as well. The resulting data model is provided in Figure 1.
Figure 1:
Our proposed data model to capture methodological characteristics from RCT publications. Domains (Trial Design, Blinding, etc.) are shown in gray boxes. Top-level characteristics are shown in blue rectangles. Their subtypes are shown in yellow, and properties relevant to the subtypes are shown in green.
Corpus Annotation
After the preliminary data model was developed, we annotated several articles to assess the feasibility of annotating the items in the model and to draft annotation guidelines. As a result, the data model was also refined. Next, we conducted a pilot study to annotate 70 RCT articles in three phases. First, 10 articles were annotated by three annotators. Disagreements and inconsistencies were discussed and resolved. Data model and annotation guidelines were refined. In the second phase, three annotators annotated 20 articles. Next, inter-annotator agreement was calculated, annotation guidelines were further revised for clarity, and disagreements were resolved. In the final phase, two annotators with the highest agreement in the previous phase individually labeled 25 and 15 articles each. The annotators are the authors of this paper, two PhD students and a biomedical NLP expert with experience with biomedical literature. We used brat annotation tool for annotation33.
For annotation, we collected a set of RCT publications from PubMed Central Open Access Subset. 25 publications came from the CONSORT-TM corpus25. We collected another set of 45 articles by issuing a search query that limited by the publication type “Randomized Controlled Trials” and full-text availabilitya. We eliminated publications reporting study protocols or multiple RCT studies from the search results. From the remaining articles, we randomly selected 45 articles, for a total of 70 articles.
We adopted a minimal annotation approach, focusing on annotating the shortest meaningful text spans for a given item, often a clause. The categories that have subclasses (e.g., Patient, Investigator, etc. for Blinding Object) were annotated at the finest granularity justified by the text span. We focused primarily on abstracts and Methods sections, as they were most likely to contain methodological information. Some categories, particularly those related to Sample Size, were also annotated in the Results sections, where they were often reported. During the annotation process, the annotators were instructed to annotate a mention only once for a particular characteristic in a given section, although different mentions corresponding to the same concept (e.g., no blinding and open label for the Open Label category) were expected to be annotated. This helped reduce annotation burden while generating a diverse set of examples. After the manual annotation was completed, we generated additional annotations by automatically labeling all instances of the mentions that already appear in the same section of the document in the manually annotated set. Some of these automatic annotations were incorrect and we manually removed them (e.g., not all instances of the mention blind are about the blinding type of the study). Automatic annotation helped us increase the number of examples in the dataset.
Inter-annotator Agreement
We calculated inter-annotator agreement at the span and document levels. For span level agreement, we used exact match for all categories considered. Following earlier work, we used F1 score for span level agreement, considering annotations from one annotator to be the ground truth and those from the other as predictions34. Document level agreement was calculated for items with subcategories: Trial Design Type, Phase, Comparative Intent, Blinding Type, Randomization Type, and Setting. In this case, we examined whether two annotators agreed on whether the publication reported a particular study characteristic (e.g., Double-Blind as the Blinding Type). We used both Cohen’s κ and F1 score for inter-annotator agreement at the document level.
NER models
The task of extracting methodological information at the span level can be formulated as a typical NER task. In this study, we experimented with NER models based on current baseline neural network architectures. We applied BIO tag scheme to represent token labels in sentences. We used PubMedBERT (base-uncased-abstract-fulltext model)35 as the sentence encoder and experimented with two different classification layers: a fully-connected token classification layer and a a classification layer based on conditional random field (CRF)36 that captures the dependencies between the labels in a sequence. The following experimental settings were used: batch size of 4, Adam optimizer, learning rates of 1e-5, 2e-5, 3e-5. and 5e-5, and number of epochs of 10, 20, 30. For final training, we used the learning rate of 5e-5 for the token classification model and 3e-5 for the CRF-based model and 20 epochs for both models, which yielded the best performances.
Methodological information that we model generally occurs over a handful of sentences in a RCT publication. Including all sentences of the publication in training leads to a very imbalanced dataset. To address this problem, we adopted four strategies to sample sentences for inclusion in training:
Positive sentences only: Only sentences that include at least one annotated span are included.
Random sampling: Positive sentences + a random sentence with no annotations (i.e., negative sentence) for each positive sentence
Similarity sampling: Positive sentences + negative sentence with the highest cosine similarity with positive sentence
Random+Similarity sampling: Positive sentences + random sampling for half of the positive sentences + similarity sampling for the other half.
For cosine similarity calculation, we generated vector representations of the sentences using PubMedBERT. The dataset was split into training and test sets (55 articles/15 articles). We report the results on the test set.
Evaluation
The PubMedBERT-based NER models were evaluated at the span and document levels. In all cases, we used precision, recall, and their harmonic mean, F1 score, as the evaluation metrics. We performed two types of evaluation at the span level: strict and partial. In strict evaluation, for a prediction to count as a true positive, its span and category needs to exactly match a ground truth annotation. In partial evaluation, the predicted span and the ground truth span can overlap but a type match is still required. Document level evaluation is only applied to items with subtypes (Blinding Type, Phase, etc.).
Results
In this section, we first provide descriptive statistics about the annotated corpus. Next, we present inter-annotator agreement results. Lastly, we report the performance of NER models trained on the corpus.
Annotated Corpus
We annotated a total of 70 RCT articles in this pilot study. Table 1 shows the descriptive statistics of the annotated corpus. Among the top level categories, Sample Size had the highest number of annotations (426) followed by Randomization (384), Blinding (302), Trial Design (339), and Settings (145). Allocation Concealment Method was rarely discussed (12 instances). At the fine-grained level, Parallel Group (179), Actual Sample Size (135), and Double-Blind (117) were annotated most frequently. Although we represented some characteristics in the data model to maintain consistency with OCRe, we did not find any instances of these in the corpus: N-of-1, Factorial Factor/Treatment, Triple-Blind, Quadruple-Blind.
Table 1:
Statistical information of the annotated corpus.
| Statistics | Complete Corpus | Train Set | Test Set |
|---|---|---|---|
| Number of articles | 70 | 55 | 15 |
| Number of sentences | 10,225 | 8,715 | 1,510 |
| Number of sentences with annotations | 837 | 658 | 179 |
| Number of tokens | 312,361 | 262,647 | 49,714 |
| Number of annotated tokens | 5,758 | 4,317 | 1,441 |
| Number of annotations | 1,734 | 1,356 | 378 |
Inter-annotator agreement
Table 2 shows pair-wise inter-annotator agreement results obtained on 20 articles at span and document levels using Cohen’s κ and F1 score. The results show overall high agreement. Our Cohen’s κ scores indicate substantial to perfect agreement between the annotators (0.74-0.83)37. F1 score agreement is over 0.9 in all cases. Overall, annotators 1 and 2 achieved higher agreement at both span and document levels. These two annotators annotated the last 40 articles.
Table 2:
Pair-wise agreement at span and document levels. Document level agreement is calculated for categories with sub-classes only.
| Ann1 vs. Ann2 | Ann2 vs. Ann 3 | Ann1 vs. Ann3 | ||||
|---|---|---|---|---|---|---|
| Cohen’s κ | F 1 | Cohen’s κ | F 1 | Cohen’s κ | F 1 | |
| Span level | 0.94 | 0.90 | 0.90 | |||
| Document level | 0.83 | 0.95 | 0.74 | 0.92 | 0.79 | 0.93 |
NER models
For each NER approach (token classification vs. CRF-based), we developed four models each corresponding to a sampling strategy for training: Positive Sentences, Random Sampling, Similarity Sampling, and Random+Similarity Sampling. Table 3 shows the performances of the NER models at the span level. In both strict and partial evaluation, CRF-based classification using Similarity Sampling achieved the best F1 scores. The results with token classification are similar to CRF-based results. While using Positive Sentences only for training yields lowest F1 results, its recall is the highest. Sampling strategy has a more significant effect on precision than on recall. Token-level evaluation results (data not shown due to limited space) show the same patterns as the entity-level results shown here.
Table 3:
Model performances at the span level with four sampling strategies for training. Best precision, recall, and F1 score for strict and partial evaluation scenarios are in bold.
| Sampling Strategy | Token classification | CRF-based | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Strict | Partial | Strict | Partial | |||||||||
| P | R | F1 | P | R | F1 | P | R | F1 | P | R | F1 | |
| Positive Sentences | 0.24 | 0.65 | 0.36 | 0.29 | 0.78 | 0.42 | 0.22 | 0.64 | 0.33 | 0.27 | 0.80 | 0.42 |
| Random Sampling | 0.44 | 0.62 | 0.51 | 0.54 | 0.77 | 0.63 | 0.41 | 0.62 | 0.49 | 0.52 | 0.78 | 0.63 |
| Similarity Sampling | 0.49 | 0.62 | 0.55 | 0.56 | 0.75 | 0.65 | 0.48 | 0.64 | 0.55 | 0.57 | 0.78 | 0.66 |
| Random + Similarity Sampling | 0.48 | 0.59 | 0.53 | 0.59 | 0.72 | 0.65 | 0.46 | 0.61 | 0.53 | 0.57 | 0.76 | 0.65 |
Table 4 shows the performances of the NER models at the document level, which are largely consistent with the results at the span level. CRF-based models consistently outperform the token classification counterparts. Similarity Sampling yields highest F1 score and precision performance, while its recall is lower than of the Positive Sentence sampling. Since we are ultimately interested in summarizing methodological characteristics of a study at the document level, we consider document-level evaluation results as the main results for this study.
Table 4:
Document-level performances of four models using different classification layers. Best precision, recall, and F1 score are in bold.
| Sampling Strategy | Token classification | CRF-based | ||||
|---|---|---|---|---|---|---|
| P | R | F1 | P | R | F1 | |
| Positive Sentences | 0.65 | 0.89 | 0.76 | 0.80 | 0.84 | 0.82 |
| Random Sampling | 0.82 | 0.89 | 0.85 | 0.88 | 0.82 | 0.85 |
| Similarity Sampling | 0.89 | 0.86 | 0.88 | 0.95 | 0.85 | 0.90 |
| Random + Similarity Sampling | 0.94 | 0.83 | 0.88 | 0.89 | 0.81 | 0.88 |
We analyzed the results of the best-performing model (PubMedBERT with CRF layer trained with Similarity Sampling strategy) in more detail. These results, obtained with span level evaluation, are shown in Table 5. The results show that the results vary widely among different characteristics. Trial Phase can be recognized perfectly, which is not surprising given that it is often indicated using exact phrases, such as Phase 1 or Phase IIb. Other characteristics recognized relatively well are randomization Ratio (0.97 F1), Power and Alpha for sample size calculation (0.95 and 0.86 F1, respectively), Stratification Criteria for stratified randomization (0.81 F1), and Blinding Type (0.78 F1). Except Stratification Criteria, these characteristics are generally expressed in a small number of ways in publications, which may explain the higher performance. Another factor is that these characteristics are relatively frequent in the dataset. The model fails on several characteristics, such as Allocation Concealment Method which only had a few examples in the dataset. In contrast, the model had more success with some other characteristics, which also had few examples, such as Comparative Intent. This can also be attributed to the fact that the expressions for these characteristics are less diverse than those for, say, Randomization Personnel, which include a wide range of expressions such as individuals not associated with study conduct or separate unblinded statistical team. For some items, strict vs. partial evaluation results are the same (e.g., Comparative Intent), while there is a significant different for others that involve numbers, which merits further investigation (e.g., Block Size, randomization Sequence Generation).
Table 5:
Performance of the best model (CRF-based model trained with Similarity Sampling) at the span level. Characteristics with * next to their name are fine-grained items, while others have subtypes. For characteristics with subtypes, the results are aggregated for brevity. For example, Trial Design:Type results include predictions for Parallel Group, Factorial, etc. Similarly, Sample Size:Type aggregates the results for different sample size calculations: Required, Targeted, Actual at Enrollment, and Actual at Outcome Analysis.
| Domain | Characteristics | Strict | Partial | ||||
|---|---|---|---|---|---|---|---|
| P | R | F1 | P | R | F1 | ||
| Trial Design | Type | 0.43 | 0.46 | 0.44 | 0.41 | 0.87 | 0.55 |
| Phase | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | |
| Comparative Intent | 0.33 | 0.60 | 0.63 | 0.33 | 0.60 | 0.63 | |
| *Crossover Period/Treatment | 0.67 | 1.00 | 0.80 | 0.67 | 1.00 | 0.80 | |
| *Factorial Factor/Treatment | 0.43 | 0.60 | 0.50 | 0.43 | 0.60 | 0.50 | |
| Blinding | Type | 0.72 | 0.75 | 0.73 | 0.76 | 0.80 | 0.78 |
| Objects | 0.32 | 0.50 | 0.39 | 0.35 | 0.54 | 0.43 | |
| Randomization | Type | 0.64 | 0.38 | 0.48 | 0.79 | 0.49 | 0.62 |
| *Block Size | 0.20 | 0.33 | 0.25 | 0.40 | 0.67 | 0.50 | |
| *Minimization Criteria | 0.67 | 0.50 | 0.57 | 0.83 | 0.63 | 0.71 | |
| *Stratification Criteria | 0.71 | 0.83 | 0.77 | 0.75 | 0.88 | 0.81 | |
| *Personnel | 0.00 | 0.00 | 0.00 | 0.13 | 0.40 | 0.20 | |
| *Ratio | 0.93 | 0.88 | 0.90 | 1.00 | 0.94 | 0.97 | |
| *Sequence Generation | 0.32 | 0.43 | 0.36 | 0.68 | 0.89 | 0.77 | |
| Sample Size | Type | 0.42 | 0.65 | 0.51 | 0.52 | 0.81 | 0.63 |
| *Alpha | 0.42 | 0.50 | 0.45 | 0.79 | 0.95 | 0.86 | |
| *Dropout Rate | 0.27 | 0.43 | 0.33 | 0.45 | 0.71 | 0.56 | |
| *Power | 0.91 | 1.00 | 0.95 | 0.91 | 1.00 | 0.95 | |
| Settings | Type | 0.61 | 0.69 | 0.65 | 0.69 | 0.78 | 0.74 |
| *Location | 0.37 | 0.69 | 0.48 | 0.45 | 0.85 | 0.59 | |
| Allocation Concealment | *Allocation Concealment Methods | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 |
| OVERALL | 0.48 | 0.64 | 0.55 | 0.57 | 0.78 | 0.66 | |
Discussion
We presented a pilot annotation study and baseline NER models for recognizing methodological characteristics in RCT publications. We focused on characteristics that may affect the methodological quality of and strength of evidence from a RCT study. To our knowledge, this is the first study to focus on representing, annotating, and extracting these methodological characteristics at a fine-grained level and in a comprehensive manner. Our work complements the PICO-based characterizations which, while very important, do not address methodological quality, and automated risk of bias assessment models, which focus on classification rather than IE and thus, do not provide granular information.
Our data model was adopted from OCRe and CONSORT. OCRe, by formalizing various aspects of clinical studies, and CONSORT, by detailing the characteristics of a RCT study that needs to be reported in a publication for transparency, provide a solid foundation for methodological IE.
Our annotation study showed that annotating RCT methodological items at the span level was feasible. We obtained high inter-annotator agreement, indicating that these characteristics can be more or less reliably annotated. Several items were challenging. For example, Parallel Group is easy to annotate when it is explicit (e.g., parallel-group). However, it is often implicit and can only be determined from the description of the intervention (e.g., intravenous rhEPO 40 000 IU or placebo fortnightly). While annotators were instructed to annotate such implicit cases in the annotation guidelines, their annotations were less consistent for these cases.
CRF-based models performed comparatively better than token classification models in NER, indicating that capturing label sequences is important for methodological IE. This is not surprising, since sentences where many methodological characteristics of the RCT are mentioned together are common (e.g., This phase 2b, double-blind, placebo-controlled, parallel-group, dose-ranging randomized clinical trial… ) and capturing such patterns may benefit the models. Since only a small number of sentences in each article was annotated, we sampled unannotated sentences to increase the training set size. Similarity-based sampling yielded the best results overall, indicating that providing more difficult negative examples to the training procedure is beneficial. We observed that some characteristics appear easy to identify using simple lexical rules (e.g., Comparative Intent types such as Superiority, Non-inferiority). We created a set of 16 lexical rules based on regular expressions for characteristics that can be categorized into subtypes (e.g., Blinding or Randomization Types). Although it covers only a subset of the items, this method yielded comparable results to token classification model for the characteristics that it covered (results not shown). This indicates that an expanded set of such rules may be effective in methodological IE, although this involves some manual effort and requires expertise.
Limitations and Future Work
Our study has limitations. First, the annotated corpus is small and needs to be expanded to be more broadly useful. We anticipate that NER models would benefit from additional training data, as well. This study showed the feasibility of reliably annotating methodological characteristics at the span level and we plan to expand our corpus in future work. It would be particularly important to capture a larger number of infrequently discussed characteristics, such as allocation concealment methods, since the current models fail at recognizing them.
We only experimented with baseline NER models. While they yield promising results, more advanced NER methods can be applied (e.g., BERT with BiLSTM+CRF layers38). We experimented with the learning rate hyperparameter, but tuning other hyperparameters could also be beneficial. A simple rule-based approach seems adequate for some items and it may be worthwhile to use them, especially for the items that do not have sufficient training examples.
Conclusion
In this study, we proposed a data model that captures methodological characteristics of RCTs at fine-grained level, annotated a corpus of 70 RCT articles with these characteristics, and demonstrated the feasibility of using NER methods to automatically extract them from these publications. While there is much room for NER model improvement, we believe that the methodological IE is promising in providing methodological context for the interpretation of study findings/claims and the basis for reasoning about methodological weaknesses and inconsistencies.
Footnotes
PubMed search query: “randomized controlled trial”[Publication Type]) AND (fft[Filter])
Figures & Table
References
- 1.Sackett DL. Seminars in perinatology. vol. 21. Elsevier; 1997. Evidence-based medicine; pp. 3–5. [DOI] [PubMed] [Google Scholar]
- 2.Slavin RE. Best evidence synthesis: an intelligent alternative to meta-analysis. Journal of clinical epidemiology. 1995;48(1):9–18. doi: 10.1016/0895-4356(94)00097-a. [DOI] [PubMed] [Google Scholar]
- 3.Zeng X, Zhang Y, Kwong JS, Zhang C, Li S, Sun F, et al. The methodological quality assessment tools for preclinical and clinical studies, systematic review and meta-analysis, and clinical practice guideline: a systematic review. Journal of evidence-based medicine. 2015;8(1):2–10. doi: 10.1111/jebm.12141. [DOI] [PubMed] [Google Scholar]
- 4.Moher D, Hopewell S, Schulz KF, Montori V, Gøtzsche PC, Devereaux PJ, et al. CONSORT 2010 Explanation and Elaboration: updated guidelines for reporting parallel group randomised trials. BMJ. 2010;340 doi: 10.1136/bmj.c869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chalmers I, Glasziou P. Avoidable waste in the production and reporting of research evidence. The Lancet. 2009;374(9683):86–9. doi: 10.1016/S0140-6736(09)60329-9. [DOI] [PubMed] [Google Scholar]
- 6.Higgins JP, Altman DG, Gøtzsche PC, Ju¨ni P, Moher D, Oxman AD, et al. The Cochrane Collaboration’s tool for assessing risk of bias in randomised trials. BMJ. 2011;343 doi: 10.1136/bmj.d5928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Marshall IJ, Kuiper J, Wallace BC. RobotReviewer: evaluation of a system for automatically assessing bias in clinical trials. Journal of the American Medical Informatics Association. 2016;23(1):193–201. doi: 10.1093/jamia/ocv044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huang CC, Lu Z. Community challenges in biomedical text mining over 10 years: success, failure and the future. Briefings in bioinformatics. 2016;17(1):132–44. doi: 10.1093/bib/bbv024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Harpaz R, Callahan A, Tamang S, Low Y, Odgers D, Finlayson S, et al. Text mining for adverse drug events: the promise, challenges, and state of the art. Drug safety. 2014;37(10):777–90. doi: 10.1007/s40264-014-0218-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lawrence JM, Meyerowitz-Katz G, Heathers JAJ, Brown NJL, Sheldrick KA. The lesson of ivermectin: meta-analyses based on summary data alone are inherently unreliable. Nature Medicine. 2021. [DOI] [PubMed]
- 11.Demner-Fushman D, Lin J. Answering Clinical Questions with Knowledge-Based and Statistical Techniques. Computational Linguistics. 2007;33(1):63–103. [Google Scholar]
- 12.Wallace BC, Kuiper J, Sharma A, Zhu M, Marshall IJ. Extracting PICO sentences from clinical trial reports using supervised distant supervision. The Journal of Machine Learning Research. 2016;17(1):4572–96. [PMC free article] [PubMed] [Google Scholar]
- 13.Nye B, Li JJ, Patel R, Yang Y, Marshall I, Nenkova A, et al. Proceedings of the 56th Annual Meeting of the Association for Computational Linguistics (Volume 1: Long Papers) Melbourne, Australia: Association for Computational Linguistics; 2018. A Corpus with Multi-Level Annotations of Patients, Interventions and Outcomes to Support Language Processing for Medical Literature; pp. 197–207. [PMC free article] [PubMed] [Google Scholar]
- 14.Millard LA, Flach PA, Higgins JP. Machine learning to assist risk-of-bias assessments in systematic reviews. International journal of epidemiology. 2016;45(1):266–77. doi: 10.1093/ije/dyv306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Marshall IJ, Nye B, Kuiper J, Noel-Storr A, Marshall R, Maclean R, et al. Trialstreamer: A living, auto-matically updated database of clinical trial reports. Journal of the American Medical Informatics Association. 2020;27(12):1903–12. doi: 10.1093/jamia/ocaa163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Richardson WS, Wilson MC, Nishikawa J, Hayward RS, et al. The well-built clinical question: a key to evidence-based decisions. ACP Journal Club. 1995;123(3):A12–3. [PubMed] [Google Scholar]
- 17.Mavergames C, Oliver S, Becker L. Systematic Reviews as an Interface to the Web of (Trial) Data: using PICO as an Ontology for Knowledge Synthesis in Evidence-based Healthcare Research. SePublica. 2013;994:22–6. [Google Scholar]
- 18.Riva JJ, Malik KM, Burnie SJ, Endicott AR, Busse JW. What is your research question? An introduction to the PICOT format for clinicians. The Journal of the Canadian Chiropractic Association. 2012;56(3):167. [PMC free article] [PubMed] [Google Scholar]
- 19.Sim I, Olasov B, Carini S. An ontology of randomized controlled trials for evidence-based practice: content specification and evaluation using the competency decomposition method. Journal of Biomedical Informatics. 2004;37(2):108–19. doi: 10.1016/j.jbi.2004.03.001. [DOI] [PubMed] [Google Scholar]
- 20.Sim I, Tu SW, Carini S, Lehmann HP, Pollock BH, Peleg M, et al. The Ontology of Clinical Research (OCRe): an informatics foundation for the science of clinical research. Journal of biomedical informatics. 2014;52:78–91. doi: 10.1016/j.jbi.2013.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schulz KF, Altman DG, Moher D. CONSORT 2010 Statement: updated guidelines for reporting parallel group randomised trials. BMJ. 2010;340:c332. doi: 10.1136/bmj.c332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dawes M, Pluye P, Shea L, Grad R, Greenberg A, Nie JY. The identification of clinically important elements within medical journal abstracts: Patient population problem, exposure intervention, comparison, outcome, duration and results (PECODR) Journal of Innovation in Health Informatics. 2007;15(1):9–16. [PubMed] [Google Scholar]
- 23.Kim SN, Mart´ınez D, Cavedon L, Yencken L. Automatic classification of sentences to support Evidence Based Medicine. BMC Bioinformatics. 2011;12(S-2):S5. doi: 10.1186/1471-2105-12-S2-S5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jin D, Szolovits P. PICO Element Detection in Medical Text via Long Short-Term Memory Neural Networks. Proceedings of the BioNLP 2018 workshop. 2018. pp. 67–75.
- 25.Kilicoglu H, Rosemblat G, Hoang L, Wadhwa S, Peng Z, Malicˇki M, et al. Toward assessing clinical trial publications for reporting transparency. Journal of Biomedical Informatics. 2021;116:103717. doi: 10.1016/j.jbi.2021.103717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Marshall IJ, Wallace BC. Toward systematic review automation: a practical guide to using machine learning tools in research synthesis. Systematic reviews. 2019;8(1):1–10. doi: 10.1186/s13643-019-1074-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.De Bruijn B, Carini S, Kiritchenko S, Martin J, Sim I. AMIA Annual Symposium Proceedings. vol. 2008. American Medical Informatics Association; 2008. Automated information extraction of key trial design elements from clinical trial publications; p. 141. [PMC free article] [PubMed] [Google Scholar]
- 28.Kiritchenko S, De Bruijn B, Carini S, Martin J, Sim I. ExaCT: automatic extraction of clinical trial characteristics from journal publications. BMC medical informatics and decision making. 2010;10(1):1–17. doi: 10.1186/1472-6947-10-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hassanzadeh H, Groza T, Hunter J. Identifying scientific artefacts in biomedical literature: The evidence based medicine use case. Journal of biomedical informatics. 2014;49:159–70. doi: 10.1016/j.jbi.2014.02.006. [DOI] [PubMed] [Google Scholar]
- 30.Brockmeier AJ, Ju M, Przybyła P, Ananiadou S. Improving reference prioritisation with PICO recognition. BMC Medical Informatics and Decision Making. 2019;19(1):256. doi: 10.1186/s12911-019-0992-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhao J, Bysani P, Kan MY. AMIA Annual Symposium Proceedings. vol. 2012. American Medical Informatics Association; 2012. Exploiting classification correlations for the extraction of evidence-based practice information; p. 1070. [PMC free article] [PubMed] [Google Scholar]
- 32.Hoang L, Jiang L, Kilicoglu H. AMIA Informatics Summit 2022. vol. 2022. American Medical Informatics Association; 2022. Investigating the impact of weakly supervised data on text mining models of publication transparency: a case study on randomized controlled trials; pp. 254–63. [PMC free article] [PubMed] [Google Scholar]
- 33.Stenetorp P, Pyysalo S, Topic´ G, Ohta T, Ananiadou S, Tsujii J. brat: a Web-based Tool for NLP-Assisted Text Annotation. Proceedings of the Demonstrations at the 13th Conference of the European Chapter of the Association for Computational Linguistics. 2012. pp. 102–7.
- 34.Hripcsak G, Rothschild AS. Agreement, the f-measure, and reliability in information retrieval. Journal of the American medical informatics association. 2005;12(3):296–8. doi: 10.1197/jamia.M1733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gu Y, Tinn R, Cheng H, Lucas M, Usuyama N, Liu X, et al. Domain-specific language model pretraining for biomedical natural language processing. ACM Transactions on Computing for Healthcare (HEALTH) 2021;3(1):1–23. [Google Scholar]
- 36.Lafferty J, McCallum A, Pereira FC. Conditional random fields: Probabilistic models for segmenting and labeling sequence data. Proceedings of the Eighteenth International Conference on Machine Learning. 2001. pp. 282–289.
- 37.Landis JR, Koch GG. An application of hierarchical kappa-type statistics in the assessment of majority agreement among multiple observers. Biometrics. 1977. pp. 363–74. [PubMed]
- 38.Lample G, Ballesteros M, Subramanian S, Kawakami K, Dyer C. Neural Architectures for Named Entity Recognition. Proceedings of the 2016 Conference of the North American Chapter of the Association for Computational Linguistics: Human Language Technologies. 2016. pp. 260–70.