Abstract
Organoids are a reliable method for modeling organ tissue due to their self-organizing properties and retention of function and architecture after propagation from primary tissue or stem cells. This method of organoid generation forgoes single-cell differentiation through multiple passages and instead uses differential centrifugation to isolate mammary epithelial organoids from mechanically and enzymatically dissociated tissues. This protocol provides a streamlined technique for rapidly producing small and large epithelial organoids from both mouse and human mammary tissue in addition to techniques for organoid embedding in collagen and basement extracellular matrix. Furthermore, instructions for in-gel fixation and immunofluorescent staining are provided for the purpose of visualizing organoid morphology and density. These methodologies are suitable for myriad downstream analyses, such as co-culturing with immune cells and ex vivo metastasis modeling via collagen invasion assay. These analyses serve to better elucidate cell-cell behavior and create a more complete understanding of interactions within the tumor microenvironment.
Introduction
The ability to model epithelial cells in vitro has been the foundation of modern biomedical research because it captures cellular features that are not accessible in vivo. For instance, growing epithelial cell lines in a two-dimensional plane can provide an assessment of the molecular changes that occur in an epithelial cell during proliferation1. Furthermore, measuring the dynamic regulation between signaling and gene expression is limited in in vivo systems2. In cancer research, cancer epithelial cell line modeling has enabled the identification of molecular drivers of disease progression and potential drug targets3. However, growing cancer epithelial cell lines on a two-dimensional plane has limitations, as most are genetically immortalized and modified, often clonal in nature, selected for their ability to grow in non-physiologic conditions, limited in their assessment of three-dimensional (3D) tumor tissue architecture, and do not adequately model microenvironment interactions within a realistic tissue environment4. These constraints are particularly evident in modeling metastasis, which in vivo includes several distinct biological stages, including invasion, dissemination, circulation, and colonization at the distant organ site5.
Cancer epithelial organoids have been developed to better recapitulate the 3D environment and behavior of tumors6,7,8. Organoids were first developed from single LRG5+ intestinal crypt cells and differentiated to represent the 3D structure of crypt-villus units that maintained the hierarchical structure of the small intestine in vitro9. This approach permitted real-time visualization and characterization of self-organizing tissue architecture under homeostatic and stress conditions. As a natural extension, cancer epithelial organoids were developed to model many different cancer types, including colorectal10, pancreatic11, breast12, liver13, lung14, brain15, and gastric cancers16. Cancer epithelial organoids have been exploited to characterize cancer evolution17,18 and metastatic spatiotemporal behaviors19,20 and interrogate tumor heterogeneity21, and test chemotherapies22. Cancer epithelial organoids have also been isolated and collected during ongoing clinical trials to predict patient response to anticancer agents and radiation therapy ex vivo8,23,24,25.
Furthermore, systems incorporating cancer epithelial organoids can be combined with other non-cancer cells, such as immune cells, to form a more comprehensive model of the tumor microenvironment to visualize interactions in real-time, uncover how cancer epithelial cells change the fundamental nature of cytotoxic effector immune cells such as natural killer cells, and test potential immunotherapies and antibody-drug dependent cytotoxic activity26,27,28. This article demonstrates a method of generating epithelial organoids without passaging and embedding in collagen and basement extracellular matrix (ECM). Additionally, techniques for downstream imaging of isolated organoids are also shared.
Protocol
All mouse tissue utilized in this manuscript has been ethically collected in accordance with the Institutional Animal Care and Use Committee (IACUC) regulations and guidelines of the University of Texas Southwestern Medical Center. Likewise, all the patients consented prior to tissue donation under the oversight of an Institutional Review Board (IRB), and the samples were deidentified.
NOTE: This protocol describes the generation of organoids from primary tissue.
1. Overnight preparation of materials
For embedding in basement extracellular matrix (BECM), thaw the BECM aliquot by leaving it at 4 °C.
2. Preparing collagenase and bovine serum albumin (BSA) coating solution
Prepare 2.64% BSA/PBS coating solution (BSA Solution): Sterile filter 50 mL of phosphate-buffered saline (PBS) and 4.10 mL of 30% BSA solution using a 50 mL/0.2 μm filter flask. This BSA solution can be filtered and used again.
Prepare collagenase solution for mouse mammary tissue: Sterile filter 27 mL of basal cell medium, 1.5 mL of fetal bovine serum (FBS), 30 μL of 50 mg/mL gentamicin, 15 μL of 10 mg/mL insulin, 600 μL of 0.1 g/mL collagenase A, and 600 μL of 0.1 g/mL trypsin using a 50 mL/0.2 μm filter flask. Allocate between 10 mL and 30 mL of collagenase solution per mouse, depending on tissue mass.
Prepare collagenase solution for human breast tissue: Sterile filter 18 mL of RPMI-1640, 200 μL of 100x penicillin-streptomycin solution (pen/strep), 200 μL of 10 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES) Buffer, 1 mL of FBS, and 400 μL of 0.1 g/mL Collagenase A using a 50 mL/0.2 μm filter flask. Allocate between 10 mL and 20 mL per tissue sample, depending on the tissue mass.
3. Preparing media
Prepare organoid media: Sterile filter basal cell medium with 1% pen/strep, and 1% Insulin-Transferrin-Selenium (ITS) using a 50 mL/0.2 μm filter flask. To make organoid growth factor media, add basic fibroblast growth factor (bFGF) to a final concentration of 2.5 nM.
Prepare mammary epithelial media: To make 100 mL of mammary epithelial media, sterile filter high-glucose common basal medium with 1 mL of 100x glutamine supplement, 1 mL of pen/strep, 1 mL of 10 mM HEPES buffer (7.3 pH), 250 μL of 30% BSA stock, 1 μL of 1 mg/ mL cholera toxin stock, 1 mL of 50 μg/mL hydrocortisone stock in PBS, 50 μL of 10 mg/mL human insulin solution, 10 μL of 50 μg/mL epidermal growth factor (EGF) stock, and 1% FBS using a 150 mL/0.2 μm filter flask. Store the media at 4 °C and use for up to 2 weeks.
Prepare amphotericin wash: Sterile filter PBS with 2% pen/strep and 2% amphotericin B. Store at 4 °C.
4. Collecting and digesting tissue
- Mouse mammary tissue
- Euthanize the mouse by placing in a CO2 chamber for 2–5 min, and then follow with cervical dislocation. With the ventral side facing upward, splay mouse limbs and use four 19 G needles to pin the mouse by the paws to a board covered with an absorbent pad. Spray with 70% ethanol to smooth the fur down and sanitize the skin. Wipe away feces with a gauze pad or tissue.
- Starting just above the anogenital region, cut upward from the midline using surgical scissors, taking care not to pierce through the peritoneum. Upon reaching the chin, make lateral cuts down both clavicles and down hind legs, and pin the skin of the mouse taut to the board to expose mammary fat pads.
- Locate the inguinal mammary fat pads on wild-type mice by identifying the inguinal lymph node located near the hindlimbs. Locate the thoracic mammary fat pads on wild-type mice as the thicker, vascularized tissue beneath the front limbs.
- Use forceps to elevate the mammary fat pad. Avoid collecting underlying muscle. Using the blunt end of sharp-blunt scissors, create a pocket underneath the mammary fat pad, away from the skin. Cut away the mammary fat pad in one complete piece.
-
Once mammary fat pads have been removed, rinse in PBS before placing them in a sterile tissue culture dish. Quickly transfer to a tissue culture hood.NOTE: For mammary tumors, use a cotton swab wetted with PBS to gently roll the tumor away from the skin. Be certain to avoid darkened or soft areas, as dark discoloration indicates necrosis, which will not yield healthy organoids.
-
Mince the mammary tumors with a #10 or #11 scalpel to loosen tissue until it reaches a paste-like consistency. Transfer the minced tissue into a conical tube containing 10–30 mL of collagenase solution using a scalpel. To ensure all tissue is collected, pipette 1 mL of collagenase solution onto the tissue culture plate and back into the conical tube.NOTE: To produce larger organoids, mechanical digestion should be minimized, leaving some small visible pieces of tissue intact. Alternatively, tissue can be stored frozen for later organoid preparation with minimal loss in viability. To do so, wash tissue in a solution of PBS or basal cell media + 1% FBS, and then coarsely mince to increase surface area (keeping the tissue in one or two solid pieces). Move the tissue to a cryo-vial, top with freezing media (90% FBS + 10% dimethyl sulfoxide (DMSO)). Freeze the vials in a controlled-rate freezing container overnight before moving to storage in liquid nitrogen.
- Place the conical tube into a 37 °C benchtop shaking incubator at 180 RPM until the tissue becomes stringy and the collagenase solution becomes cloudy. For example, a large tissue mass (around 500–800 mg) from a tumor takes 30–60 min. A smaller tissue mass (around 100–300 mg) from wild-type mammary fat pads takes about 20–30 min. If uncertain, check at 5 min intervals.
- After incubation, centrifuge the conical tube for 10 min at 550 x g at room temperature (RT).
-
Carefully aspirate the supernatant from the conical tube.NOTE: Moving forward, pre-coat all pipette tips, serological pipets, and conical tubes with BSA solution to avoid loss of organoids due to adhesion to plastic.
- Add 8 mL of basal cell media and 80 μL of DNase solution [2 U/μL] to the tube. Invert gently for 1–3 min.
- Add 12 mL of basal cell media to the tube and carefully mix by pipetting or gently rotate the tube 15 times to mix.
- Centrifuge for 10 min at 550 x g at RT.
- Aspirate the supernatant, and then resuspend the pellet in 12 mL of basal cell media.
- Let the heaviest tissue fragments settle to the bottom. Collect the supernatant with a serological pipet and transfer to a 15 mL conical tube. This suspension contains epithelial organoids and stromal/immune cells.
- Human breast tissue
- Process the human breast tissue samples for organoids or store them within 24 h of collection. Store the samples at 4 °C in CO2-Independent media with 1% 100x antibiotic-antimycotic.
- Transfer the tissue into a tissue culture plate. Tilt the plate and aspirate the excess storage media, and then rinse the tissue with 5 mL of the amphotericin B wash. Remove the amphotericin B wash immediately.
-
Mince the tissue sample with a #10 or #11 scalpel to loosen the tissue until it reaches a paste-like consistency. Transfer the minced tissue into a conical tube containing 10–30 mL of collagenase solution using a scalpel. To ensure all tissue is collected, pipette 1 mL of collagenase solution onto the tissue culture plate and return to the conical tube.NOTE: To produce larger organoids, mechanical digestion should be minimized, leaving some small visible pieces of tissue intact. Starting tissue mass for this protocol ranged between 100–250 mg. Alternatively, tissue can be stored frozen for later organoid preparation with minimal loss in viability after step 4.2.2 in 90% FBS + 10% DMSO as above.
- Place the conical tube into a 37 °C benchtop shaking incubator at 180 RPM until the tissue becomes smaller and collagenase becomes cloudy. Check the tissue at 5 min intervals. Collagenase dissociation of human samples takes roughly 5–20 min.
- After incubation, spin down the conical tube for 10 min at 550 x g at RT.
-
Carefully aspirate the supernatant from the conical tube.NOTE: Moving forward, pre-coat all the pipette tips, serological pipets, and conical tubes with BSA solution to avoid loss of organoids due to adhesion to plastic.
- Add 4 mL of basal cell media and 40 μL of DNase solution [2 U/μL] to the conical tube. Invert gently for 3 min.
- Add 6 mL of basal cell media to the tube and carefully mix by pipetting or gently rotate the conical tube 15 times to mix.
- Centrifuge for 10 min at 550 x g at RT.
- Aspirate the supernatant, and then resuspend the pellet in 10 mL of basal cell media.
- Let the heaviest tissue fragments settle to the bottom. Collect the supernatant and transfer it to a 15 mL conical tube. This suspension contains epithelial organoids and stromal/immune cells.
5. Differential centrifugation
Pulse to 550 x g for 3–4 s at RT, aspirate the supernatant, and resuspend the pellet in 10 mL of basal cell media. Repeat this step three more times (a total of four spins).
After each centrifugation, ensure that the pellet becomes increasingly opaque. This remaining pellet will be epithelial organoids.
6. Collecting small organoids
Pulse to 80–100 x g for 3 s at RT. Collect the supernatant into a fresh BSA-coated tube, and then pulse to 550 x g for 3 s at RT to pellet the small organoids.
7. Embedding organoids in BECM
-
Aliquot appropriate suspension of organoids into microcentrifuge tubes according to the organoid density relative to the amount of BECM per well (Table 1). For example, use 50–100 organoids for 20 μL of BECM in one well of a 96-well plate.
NOTE: Organoid density can be counted using the following formula: ((Average number of organoids)/50 μL) x dilution factor.
Perform all the steps on ice. Place the thawed BECM on ice in a tissue culture hood. While preparing the hood, place all the pipette tips on ice to cool.
-
Centrifuge the microcentrifuge tubes for 10 min at 300 x g at RT. Discard the supernatant from the tubes. Move the tubes with organoid pellets to ice and add the appropriate volume of BECM to each microcentrifuge tube (Table 1).
NOTE: Since BECM is viscous, add in additional volumes (approximately 10%−20% extra of dome volume) of BECM to account for the loss in the pipet tip.
Gently pipette up and down to resuspend the organoids in BECM, taking care not to produce bubbles.
Slowly and carefully pipette the BECM-suspended organoids onto the plating surface. While pipetting, slowly bring up the pipette to create a dome. Fill all the empty wells with PBS to maintain humidity.
Place the plate into a 37 °C incubator for 1 h to allow BECM to solidify, and then cover with the appropriate volume of media (Table 1).
Table 1:
Recommended volume of ECM components for domes, density of organoids, and volume of media needed per well on varying culture plates.
| Culture plate | ECM dome Volume | Recommended number of organoids |
Media volume |
|---|---|---|---|
| 6-well plate | 200 μL | 300 | 3 mL |
| 12-well plate | 150 μL | 225 | 2 mL |
| 24-well plate | 100 μL | 150 | 1 mL |
| 48-well plate | 40 μL | 60 | 300 μL |
| 96-well plate | 20 μL | 30 | 150 μL |
8. Embedding organoids in collagen
Perform all the steps on ice. Prepare the collagen solution by combining, in sequential order, 375 μL of 10x DMEM, 100 μL of 1 N sodium hydroxide (NaOH), and 3 mL of rat tail collagen I solution in a 15 mL conical tube. Pipette mix, taking care not to make bubbles. Titrate the solution to a pH of 7.2–7.4 with small amounts (increments of 1–3 μL) of NaOH or 10x DMEM as needed.
Coat the bottom of the wells with the minimum amount of collagen required to fully cover the bottom of the well. One approach is to place a small amount of collagen and rock the plate from side to side to coat. Allow the collagen underlays to set at 37 °C for 30 min-2 h.
Aliquot the appropriate suspension of organoids into microcentrifuge tubes according to the organoid density relative to the amount of collagen per well (Table 1). Place in a 37 °C incubator.
-
Store the collagen solution at 4 °C and monitor polymerization by checking for fiber formation under a microscope every 10 min. Collagen will reach appropriate polymerization between 30 min-2 h.
NOTE: Proper polymerization can be determined by the presence of fibers branching throughout the matrix. Proceed to step 8.5 immediately once a few fibers are observed in the field of view.
-
Centrifuge the microcentrifuge tubes at 300 x g for 10 min at RT. Discard the supernatant from the tubes. Move the organoid pellets to ice and add the appropriate volume of collagen to each microcentrifuge tube (Table 1).
NOTE: Since collagen is viscous, add in additional volumes (approximately 10%−20% extra of dome volume) of collagen to account for the loss in the pipet tip.
Gently pipette up and down to resuspend organoids in collagen, taking care not to produce bubbles.
Slowly and carefully pipette the collagen-suspended organoids onto the plating surface. While pipetting, slowly bring up the pipette to create a dome. Fill all the empty wells with PBS to maintain humidity.
Place the plate into a 37 °C incubator for 1 h to allow collagen to solidify, and then cover with the appropriate volume of media (Table 1).
NOTE : Organoids maintain viability in 3D matrix for up to 7 days. If imaging is being used for analysis, proceed to steps 9 and 10.
9. Fixing embedded organoids
Use a pipette to remove all media from the wells without making contact with ECM domes.
Fix with 4% paraformaldehyde (PFA)/PBS solution for 5 min.
Wash two to three times by applying PBS to wells after placing them on a slowly moving shaker. After washes, apply fresh PBS to the domes and store the plate at 4 °C.
10. Immunofluorescent staining of embedded organoids
- Preparing solutions for immunofluorescent staining
- Permeabilization buffer: Prepare a PBS solution with 10% Triton X-100.
- Blocking buffer: Prepare a PBS solution with 10% FBS and 0.2% Triton X-100.
- Antibody dilution buffer: Prepare a PBS solution with 2% FBS and 0.2% Triton X-100.
- Permeabilization and blocking
- Pipette enough permeabilization buffer to cover the ECM domes. Incubate for 1 h at RT on a slowly moving shaker.
- Remove the permeabilization buffer with a pipette and apply the same volume of blocking buffer for 3 h.
- Primary antibody incubation
- Remove the blocking buffer with a pipette and apply the antibody dilution buffer containing the primary antibody at manufacturer-specified concentrations. Incubate the sample at 4 °C for 12–16 h on a slowly-moving shaker.
- Remove the primary antibody solution and wash the samples three times for 10 min with PBS at RT on a slowly moving shaker.
- Secondary antibody incubation and nuclear stain
- Aspirate the remaining PBS wash and apply the antibody dilution buffer containing secondary antibody at manufacturer-specified concentrations. Additionally, add DAPI or Hoechst (to label nuclei) or phalloidin (to label actin) to the buffer at a 1:250 ratio. Incubate for 2–4 h at RT on a slowly-moving shaker.
- Remove the secondary antibody solution and wash samples three times for 10 min in PBS at RT on a shaker. Store the samples in PBS at 4 °C until imaging.
Representative Results
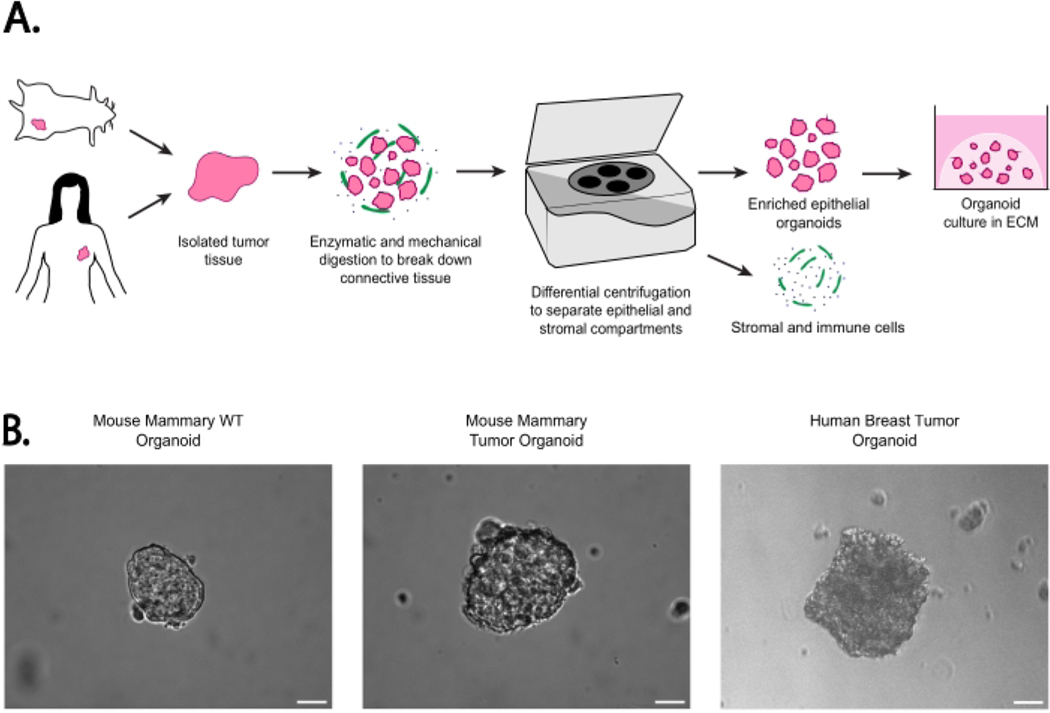
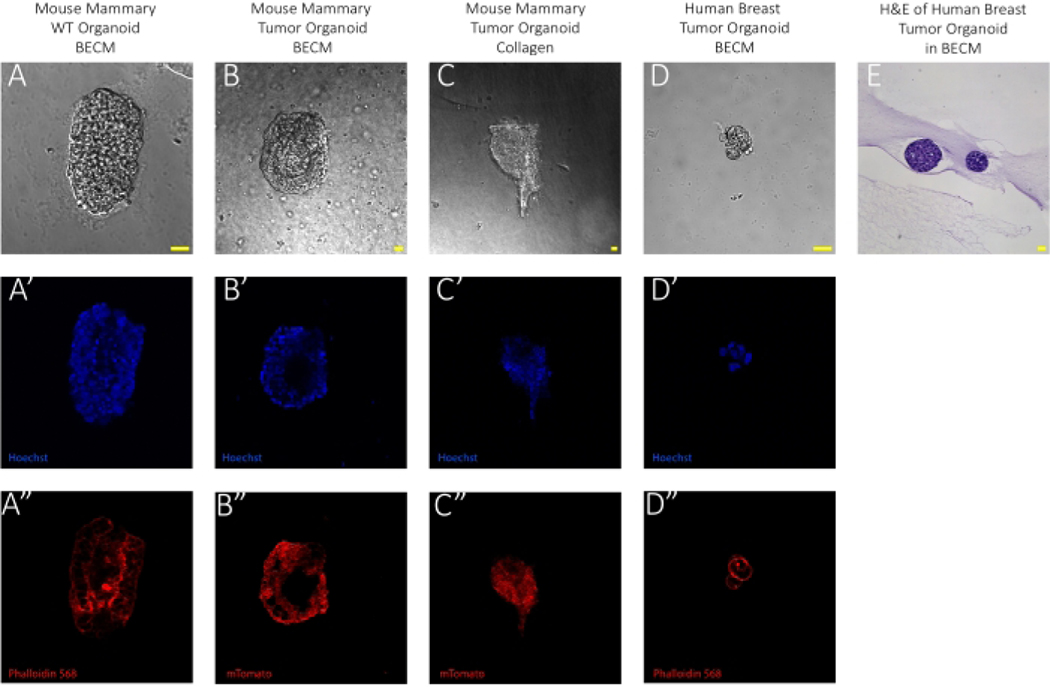
The images featured in Figure 1 provide an example of wild-type and tumorous mammary epithelial organoids from human and mouse tissues. An at-a-glance illustration of the method for isolating epithelial organoids through differential centrifugation is provided in the cartoon workflow in Figure 1A, showing that primary tissues from different species can be processed in near-identical ways while yielding epithelial tissue as shown in the brightfield images (Figure 1B). Furthermore, these inter-species tissue composition similarities can be seen in the immunofluorescent images of embedded organoids in either basement extracellular matrix or collagen displayed in Figure 2A–D. The organoid structure can be visualized through either membrane Tomato (mTomato)-labeling or phalloidin staining of actin. These figures also demonstrate the expected organoid composition and size using this method. Organoids embedded in collagen matrix can be used for an invasion assay and analyzed by tracking the expansion of tendrils branching out from the organoid itself, as seen in Figure 2C. Lastly, hematoxylin and eosin (H&E) staining of paraffin-embedded organoids shows that organoids maintain the same histology of breast cancer (Figure 2E).
Figure 1: Workflow for the epithelial organoid generation with example organoids.
(A) Schema of workflow for organoid generation from mouse or human tissue without passaging. (B) Representative images of isolated epithelial organoids from mouse WT mammary glands, mouse mammary tumors, and human breast tumors in media following isolation. Each image was adjusted individually for brightness and contrast for enhanced visualization. Images were taken in brightfield on an inverted epi-fluorescent microscope at 10x magnification. Scale bar represents 20 μm. Mammary tumors were isolated from MMTV-PyMT mice; normal mammary tissue was isolated from FVB mice. Mice ranged from 8–14 weeks of age29.
Figure 2: Imaging organoids in the extracellular matrix.
Each image was adjusted individually for brightness and contrast for enhanced visualization for this collection. Prior to imaging, samples were fixed with 4% paraformaldehyde. Wild-type samples were stained with phalloidin 568 to visualize the cell membrane and all samples were stained with Hoechst to visualize nuclei, showing organoid morphology and density. Images were taken at 10x magnification on a confocal microscope and further enlarged with a scanning zoom of 3.003. The laser wavelength used to detect DAPI was 405 nm with a power of 5 and the laser wavelength used to detect phalloidin was 561.0 nm for channel 3 with a power of 0.5. Confocal pinhole size was maintained at 19.16 for all images. (A) Representative brightfield image of mouse mammary WT organoid embedded in BECM at Day 0. (A’) 1:250 Hoechst labeling nuclei and (A’’) 1:250 phalloidin labeling actin immunofluorescent images of A. Scale bar represents 20 μm. Normal mammary tissue was isolated from FVB mice. Mice ranged from 8–12 weeks of age. (B) Representative brightfield image of mouse mammary tumor organoid embedded in BECM at Day 0. (B’) 1:250 Hoechst labeling nuclei and (B’’) mTomato-labeled immunofluorescent image of B. Scale bar represents 20 μm. Mammary tumors were isolated from MMTV-PyMT mice. Mice ranged from 12–14 weeks of age. (C) Representative brightfield image of mouse mammary tumor organoid embedded in Collagen I at Day 3. The image represents an organoid demonstrating “invasive” property. (C’) 1:250 Hoechst labeling nuclei and (C’) mTomato-labeled immunofluorescent image of C. Scale bar represents 20 μm. Mammary tumors were isolated from MMTV-PyMT mice. Mice ranged from 12–14 weeks of age. (D) Representative brightfield image of human breast tumor organoid embedded in BECM at Day 0. (D’) Hoechst labeling nuclei and (D’’) 1:250 Actin-targeting phalloidin-labeled immunofluorescent image of D. Scale bar represents 20 μm. (E) Sectioned image of an organoid embedded in BECM and stained with H&E taken at 20x magnification. Scale bar represents 20 μm. H&E staining was performed by the University of Texas Southwestern Tissue Management Shared Resource30.
Discussion
Different methods have been described in the literature to generate tumor organoids. This protocol highlights a method for generating tumor organoids directly from the tumor without passaging. Using this method, tumor organoids are producible within hours of initiating the procedure and generate close to 100% viable organoids compared to 70% reported in the literature31. In comparison, other methods require serial passaging of cells into organoids over several weeks. Thus, the downstream applications, such as determining and visualizing immune cell interactions with matched organoid and immune samples from the same host without the impact of long-term culture, become more feasible. Further, as highlighted in Figure 2C, embedding tumor organoids in different extracellular matrices can permit the identification of key phenotypes throughout the metastatic cascade, such as invasion out of the primary tumor. Other downstream applications include phenotypic assays of branching morphogenesis32, invasion, dissemination, and colony formation33 to assess for various epithelial cell behaviors. Immune interactions can also be functionally and visually captured with these organoid-based assays. Further, gels can be dissolved to isolate embedded cells for downstream analysis of genetic and protein content using standard biochemical and flow-based assays. Finally, because large quantities of organoids can be quickly generated from tumor tissue, these assays can be scaled for drug screening applications and integration into clinical trial workflows.
There are several key steps that are critical to this protocol. First, the amount of time required for collagenase digestion is dependent on the tissue composition and amount of tissue being digested. For example, when working with smaller pieces of tissue, such as human breast tumor surgical samples (on average 100–250 mg), a shorter time of digestion is required. However, mammary tumors harvested from a mouse are much larger in size (500–800 mg) and may require 30–60 min of enzymatic digestion. Secondly, to ensure a maximum yield of epithelial organoids, it is also important to coat all pipette tips and serological pipets with BSA to avoid loss from cell adhesion to plastic. Third, a brief differential centrifugation time is crucial for eliminating non-epithelial tissue components. This approach permits heavier epithelial organoids to pellet while lighter stromal and immune compartments remain in the supernatant. For creating invasion assays by embedding organoids in collagen, it is critical to allow for proper polymerization of the collagen before embedding organoids. This step should be checked visually by confirming collagen fiber formation under a light microscope. Finally, for best imaging results, take care to avoid producing bubbles when resuspending organoids in ECM and plating. Bubbles will obscure organoids within the gel and distort images. Table 2 lists potential problems that have been encountered and solutions to overcome these challenges.
Table 2:
Table of potential problems, causes, and solutions.
| Problem | Potential cause | Solution |
|---|---|---|
| ECM domes have collapsed. | The plate was shaken, the ECM volume was too large for the dome shape to be sustained, or the domes were touching the edge of the well, causing them to stick and fall. | Avoid moving the plate too vigorously before domes have set, reduce the volume of ECM used for the resuspension of organoids, or be very careful to pipette the domes in the center of the plate. |
| There is tissue that does not look like organoids. | Muscle tissue or nerve may have been collected in the process of harvesting the mammary fat pad. | Only cut away what is clearly a mammary fat pad. Avoid removing the tissue that is adhered tightly to the skin. |
| There are many dead organoids. | Necrotic tumor tissue was harvested. | Avoid collecting the tissue from tumors that are necrotic, cystic, excessively soft, or darker in color. Tumor tissue should be firm. |
| There are more single cells than organoids. | Tissue was minced too finely, or collagenase digestion was run for too long. | Avoid over-mincing the tissue or reduce the collagenase incubation time. |
| There are bubbles in ECM. | Resuspension by pipetting was too vigorous, and ejection while pipetting domes was too fast. | Slowly resuspend the organoids in the ECM and slowly pipette them into domes. If the issue persists, avoid going to the second stop while pipetting. |
| There is no invasion of organoids within collagen. | Collagen was not properly polymerized or over-polymerized. | Do not resuspend the organoid in collagen until properly polymerized. Check every 30 min. If no polymerization is visible, resuspend at the 2 h mark and plate immediately. |
| There are no collagen fibers visible while watching for polymerization. | Microscope settings were not favorable. | Adjust the phase contrast for maximum darkness, then bring the brightness of the microscope all the way up. Additionally, increasing the magnification may enhance viewing. |
| ECM domes have dissolved. | Fixation time was too long, or PFA was not thoroughly removed from the ECM-embedded samples. | Keep fixation of ECM domes time below 5 min and follow up with five washes on a rotating shaker for 5 min each. |
Certain steps in the protocol permit modifications to customize the size of organoids generated or reduce the time required to execute the protocol. For example, increasing the duration of mechanical digestion time can result in a shorter collagenase digestion time, smaller organoids, and more individual cells. Centrifugation following collagenase digestion can be shortened to 5 min if working with a large amount of tumor tissue. Media used for culturing and growing organoids can be prepared a day prior to save time during the organoid generating steps. Similarly, tumor tissue can be stored for up to 24 h in appropriate media before organoid preparation. If time is extremely limited, this protocol includes pausing steps by freezing tumor tissue on the day of collection. Then, these frozen tissues can be used to generate viable organoids at a later date. Approximately 90% of organoids derived from the frozen tissue were viable, confirmed visually under a light microscope and with a trypan blue solution.
There are some limitations to this protocol. While this approach generates viable organoids quickly, the quantity of organoids generated is limited by the amount of tumor tissue. This limitation becomes especially apparent when working with clinical samples in which the amount of tumor tissue is less or, at times, even restricted to a few cells. In those extreme cases where only a few cells can be recovered as starting material, passaging may be a better option. Another limitation is the reductionist approach of this method. Removal of stromal compartments such as fibroblasts or endothelial cells enrich epithelial organoid generation. However, these cell populations are critical to the function of the tumor. Therefore, their removal limits the interpretation of tumor biology that is derived solely from epithelial organoid models. In conclusion, this protocol provides an approach for the quick generation of epithelial organoids for immediate use in downstream imaging, functional (including immune interactions), and drug-screening applications.
Supplementary Material
Acknowledgments
This study was supported by funding provided by METAvivor, the Peter Carlson Trust, Theresa’s Research Foundation, and the NCI/UTSW Simmons Cancer Center P30 CA142543. We acknowledge the assistance of the University of Texas Southwestern Tissue Management Shared Resource, a shared resource at the Simmons Comprehensive Cancer Center, which is supported in part by the National Cancer Institute under award number P30 CA142543. Special thanks to all members of the Chan Lab.
Footnotes
A complete version of this article that includes the video component is available at http://dx.doi.org/10.3791/64626.
Disclosures
The authors declare no conflicts of interest.
References
- 1.Ghandi M.et al. Next-generation characterization of the cancer cell line encyclopedia. Nature. 569 (7757), 503–508 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Roarty K, Echeverria GV Laboratory models for investigating breast cancer therapy resistance and metastasis. Frontiers in Oncology. 11, 645698 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hanahan D.Hallmarks of cancer: New dimensions. Cancer Discovery. 12 (1), 31–46 (2022). [DOI] [PubMed] [Google Scholar]
- 4.Gillet JP, Varma S, Gottesman MM The clinical relevance of cancer cell lines. Journal of the National Cancer Institute. 105 (7), 452–458 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lambert AW, Pattabiraman DR, Weinberg RA Emerging biological principles of metastasis. Cell. 168 (4), 670–691 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lo YH, Karlsson K, Kuo CJ Applications of organoids for cancer biology and precision medicine. Nature Cancer. 1 (8), 761–773 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Drost J, Clevers H.Organoids in cancer research. Nature Reviews Cancer. 18 (7), 407–418 (2018). [DOI] [PubMed] [Google Scholar]
- 8.Tuveson D, Clevers H.Cancer modeling meets human organoid technology. Science. 364 (6444), 952–955 (2019). [DOI] [PubMed] [Google Scholar]
- 9.Fujii M.et al. A colorectal tumor organoid library demonstrates progressive loss of niche factor requirements during tumorigenesis. Cell Stem Cell. 18 (6), 827–838 (2016). [DOI] [PubMed] [Google Scholar]
- 10.van de Wetering M.et al. Prospective derivation of a living organoid biobank of colorectal cancer patients. Cell. 161 (4), 933–945 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Boj SF et al. Organoid models of human and mouse ductal pancreatic cancer. Cell. 160 (1–2), 324–338 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sachs N.et al. A living biobank of breast cancer organoids captures disease heterogeneity. Cell. 172 (1–2), 373–386 (2018). [DOI] [PubMed] [Google Scholar]
- 13.Broutier L.et al. Human primary liver cancer-derived organoid cultures for disease modeling and drug screening. Nature Medicine. 23 (12), 1424–1435 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim M.et al. Patient-derived lung cancer organoids as in vitro cancer models for therapeutic screening. Nature Communications. 10 (1), 3991 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jacob F.et al. A patient-derived glioblastoma organoid model and biobank recapitulates inter- and intra-tumoral heterogeneity. Cell. 180 (1), 188–204 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yan HHN et al. A comprehensive human gastric cancer organoid biobank captures tumor subtype heterogeneity and enables therapeutic screening. Cell Stem Cell. 23 (6), 882–897 (2018). [DOI] [PubMed] [Google Scholar]
- 17.Njoroge RN et al. Organoids model distinct vitamin E effects at different stages of prostate cancer evolution. Scientific Reports. 7 (1), 16285 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee SH et al. Tumor evolution and drug response in patient-derived organoid models of bladder cancer. Cell. 173 (2), 515–528 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cheung KJ, Gabrielson E, Werb Z, Ewald AJ Collective invasion in breast cancer requires a conserved basal epithelial program. Cell. 155 (7), 1639–1651 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wrenn ED et al. Regulation of collective metastasis by nanolumenal signaling. Cell. 183 (2), 395–410 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kopper O.et al. An organoid platform for ovarian cancer captures intra- and interpatient heterogeneity. Nature Medicine. 25 (5), 838–849 (2019). [DOI] [PubMed] [Google Scholar]
- 22.Vlachogiannis G.et al. Patient-derived organoids model treatment response of metastatic gastrointestinal cancers. Science. 359 (6378), 920–926 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yao Y.et al. Patient-derived organoids predict chemoradiation responses of locally advanced rectal cancer. Cell Stem Cell. 26 (1), 17–26 (2020). [DOI] [PubMed] [Google Scholar]
- 24.Yao J.et al. A pancreas tumor derived organoid study: from drug screen to precision medicine. Cancer Cell International. 21 (1), 398 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vlachogiannis G.et al. Patient-derived organoids model treatment response of metastatic gastrointestinal cancers. Science. 359 (6378), 920–926 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chan IS et al. Cancer cells educate natural killer cells to a metastasis-promoting cell state. Journal of Cell Biology. 219 (9), e202001134 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chan IS, Ewald AJ Organoid co-culture methods to capture cancer cell-natural killer cell interactions. Methods in Molecular Biology. 2463, 235–250 (2022). [DOI] [PubMed] [Google Scholar]
- 28.Chan IS, Ewald AJ The changing role of natural killer cells in cancer metastasis. The Journal of Clinical Investigation. 132 (6), e143762 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Guy CT, Cardiff RD, Muller WJ Induction of mammary tumors by expression of polyomavirus middle T oncogene: a transgenic mouse model for metastatic disease. Molecular and Cellular Biology. 12 (3), 954–961 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Feldman AT, Wolfe D.Tissue processing and hematoxylin and eosin staining. Methods in Molecular Biology. 1180, 31–43 (2014). [DOI] [PubMed] [Google Scholar]
- 31.LeSavage BL, Suhar RA, Broguiere N, Lutolf MP, Heilshorn SC Next-generation cancer organoids. Nature Materials. 21 (2), 143–159 (2022). [DOI] [PubMed] [Google Scholar]
- 32.Nguyen-Ngoc KV et al. 3D culture assays of murine mammary branching morphogenesis and epithelial invasion. Methods in Molecular Biology. 1189, 135–162 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Padmanaban V.et al. Organotypic culture assays for murine and human primary and metastatic-site tumors. Nature Protocols. 15 (8), 2413–2442 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.