Abstract
Current transcriptome-wide analyses have identified a growing number of regulatory RNA with expression that is characterized in a cell-type specific manner. Herein, we describe RNA metabolic labeling with improved cell-specificity utilizing the in vivo expression of an optimized uracil phosphoribosyltransferase (UPRT) enzyme. We demonstrate improved selectivity for metabolic incorporation of a modified nucleobase (5-vinyuracil) into nascent RNA, using a battery of tests. The selective incorporation of vinyl-U residues were demonstrated in 3xUPRT LM2 cells through validation with dot blot, qPCR, LC-MS/MS and microscopy analysis. We also report using this approach in a metastatic human breast cancer mouse model for profiling cell-specific nascent RNA.
During the last decade, RNA has been recognized as a macromolecule with diverse biological functions important in distinctive cell-type specific processes1. Identifying regulatory RNA in a specific cell type remains particularly challenging within in vivo experimental models due to the presence of many cell types. To capture RNA from cell-types of interest, extensive dissociation and isolation steps are required prior to isolating RNA for analysis2. These approaches rely on mechanical separation, enzymatic digestion and flow cytometry which are all known to introduce transcriptional artifacts, as the cellular transcriptome is transformed when the cells are removed from the original whole-organism environmental niche3. Recapitulation of RNA expression pathways from intact, living cells within model organisms are difficult to analyze with these current techniques due to these challenges.
RNA metabolic labeling methods tag nascent RNA within living cells which provide opportunities for capturing transcriptional information present during key biological events. These methods expose cells to non-canonical nucleobase or nucleosides derivatized with chemical handles for subsequent metabolic incorporation into newly polymerizing RNA4. Isolation of total RNA includes a portion of metabolically-tagged nascent transcripts that are reactive to biotin-conjugated, orthogonal chemical reagents for subsequent streptavidin affinity-based enrichment and analysis. Attenuating RNA metabolic labeling to specific cell-types is achieved by expressing exogenous metabolic enzymes not active in mammalian cells that will selectively catalyze nucleobase analogs.
Our lab has demonstrated cell-specific RNA metabolic labeling with uracil phosphoribosyltransferase (UPRT) expression (from Toxoplasma gondii), TgUPRT, paired with 5-ethynyluracil (5-eu) consisting of a more stable, bioorthogonal chemical handle for imaging experiments with fluorophores and biochemical affinity-based separation of nascent transcripts5–6. RNA labeled with ethynyl groups reacts with biotin-azide through Cu(I)-catalyzed azide-alkyne addition (CuAAC) that is known to deteriorate RNA integrity due to the production of metal-ion radicals7. Recent findings identified endogenous metabolic enzymes in the de novo pyrimidine biosynthetic pathway, notably uridine monophosphate synthase (UMPS), is capable of catalyzing 5-eu in mammalian cell types independent of UPRT expression limiting the applications for in vivo cell-specific RNA metabolic labeling8. These results necessitated the development of a more stringent enzyme-analog pair appropriate for cell-specific RNA metabolic labeling by the elimination of background, non-specific RNA labeling in other cell types.
To overcome challenges with 5-eu background RNA metabolic labeling in other mammalian cell types, the active site of TgUPRT was engineered to generate triple-mutant TgUPRT (3xUPRT) capable of catalyzing 5-vinyuracil (5-vu) into nascent, vinyl-labeled RNA without background labeling in wild-type cells9. Advantages of using 5-vu include increased cell-specific stringency of labeling, as 5-vu is not recognized as a metabolic substrate by UMPS9. Additionally, the vinyl handle reacts selectively with tetrazines through an inverse electron-demand Diels-Alder reaction which does not affect RNA integrity9. Finally, 5-vinyluridine (5-vuD) treatment in cells is shown to be less detrimental to cell proliferation at longer incubation times compared with 5-ethynyluridine for RNA metabolic labeling applications in cells treated with each analog for >12 hours10. These findings support use of the 3xUPRT enzyme paired with 5-vu for improved cell-type specific RNA labeling within a broad range of applications in animal models and for maintaining the level of RNA integrity necessary for downstream sequencing analysis of enriched, labeled transcripts.
We predicted the 3xUPRT/5-vu cell-specific RNA labeling approach would be advantageous for profiling RNA in animal models where it is difficult to capture nascent RNA expression within specific cell types without inducing transcriptional artifacts. Additionally, these technical improvements improve the flexibility of these protocols to include longer incubations beyond a few minutes and at higher concentrations which could be necessary in the larger context of a whole organism for labeling a specific cell population. To test this, 3xUPRT was stably and constitutively expressed in highly metastatic MDA-MB-231 LM2 (LM2) cells using CRISPR/Cas9 genome-editing11. LM2 human breast cancer cells have shown preferential metastases into the lungs of mouse xenograft models without overburdening mice with the primary tumor size12. Before applying this approach to in vivo models, in vitro RNA metabolic labeling was characterized with dot blot analysis. LM2 wild-type (WT) and (+)-3xUPRT cells were incubated with 5-eu or 5-vu at 1 mM for 3 hours. As a positive control for RNA metabolic labeling, 5-ethynyluridine (5-euD) and 5-vinyluridine (5-vuD) were included in these experiments which label RNA in all cells regardless of 3xUPRT expression. Finally, DMSO was used as a negative control in cell treatments as the solvent of choice for these water-insoluble molecules. The chemiluminescent signal from these dot blots was quantified with ImageJ to compare the differences in signal-to-noise between 5-eu and 5-vu treatments in (+)-3xUPRT LM2 cells compared with WT cells. These results show a significantly favorable signal-to-noise ratio with 5-vu/3xUPRT when compared to 5-eu/3xUPRT due to the prevalent background labeling in WT cells treated with 5-eu. To verify 3xUPRT expression in LM2 cells, mCherry was co-expressed with 3xUPRT, whereas GFP is expressed in all the LM2 cell types in this study12 (Figure 1). Prior to establishing xenograft mouse models, optimization for in vivo RNA metabolic labeling protocols was tested in (WT) C57BL/6 mice. Rigorous testing of the solubility of these compounds in DMSO was shown to be maximized at 500 mM resulting in using this concentration for intraperitoneal (IP) injections. Initially, 5-vinyluridine (5-vuD) was tested in two sets of WT mice which demonstrated strong reproducibility of labeling across different tissue types depicted with RNA dot blot analysis (Figure S1). Next, RNA labeling was performed in mice treated with uracil analogs, 5-eu and 5-vu, with uridine analogs, 5-euD and 5-vuD, as positive controls. These results clearly indicate stronger background labeling with 5-eu treatment compared with 5-vu over a 24-hour treatment period, indicating 5-vu is advantageous for improved cell-specificity of RNA labeling in mice (Figure S2).
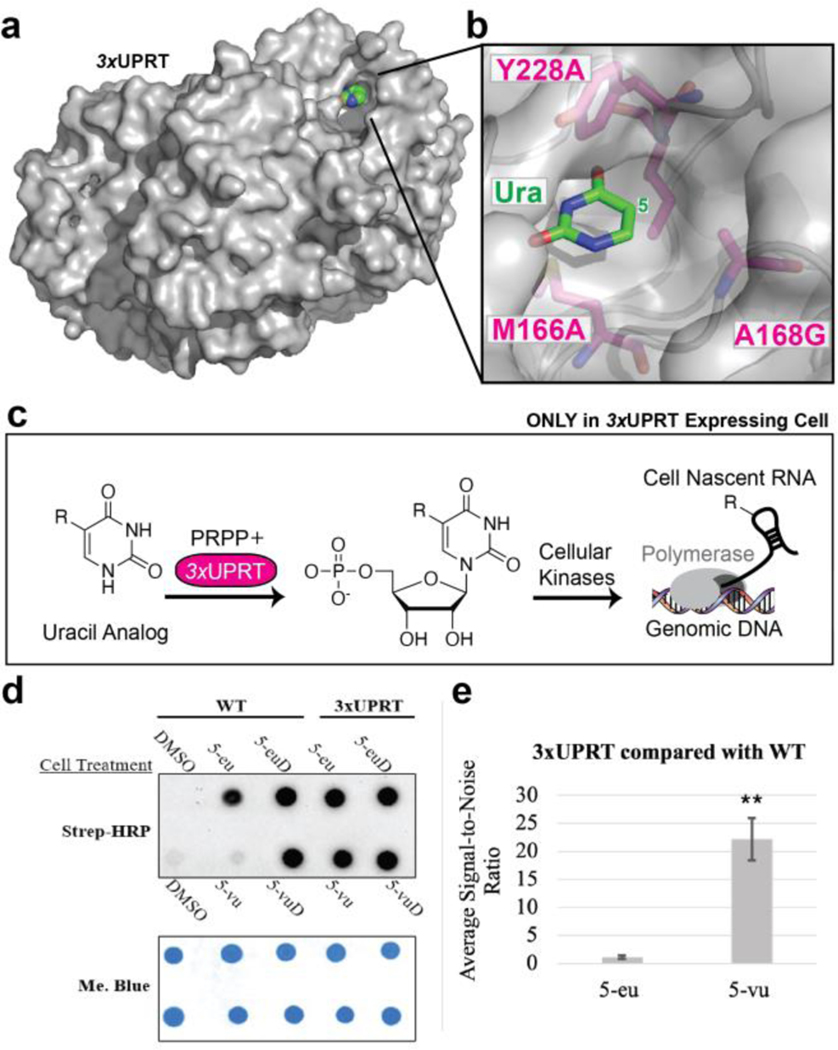
Figure 1. Characterizing cell-specific RNA metabolic labeling in MDA-MB-231 LM2 cells.
3-hour cell treatment with 1 mM of DMSO (negative control), uracil analog, or uridine analog (positive control) in WT or (+)-3xUPRT cell lines. a. Crystal structure of TgUPRT enzyme (PDB 1bd4). b. Close-up view of TgUPRT active site. Positions chosen for mutagenesis are labeled. c. Schematic of (−) TgUPRT versus TgUPRT expressing cell that enable cell-specific metabolic labeling of RNA. d. RNA dot blot is performed after reacting with biotin-azide (for ethynyl RNA) or biotin-tetrazine (for vinyl RNA). RNA was crosslinked to membrane and incubated with streptavidin-horseradish peroxidase (Strep-HRP) followed by staining with methylene blue (Me. Blue) as a loading control. 5-eu= 5-ethynyluracil, 5-euD= 5-ethynyluridine, 5-vu= 5-vinyluracil, 5-vuD=5-vinyluridine. e. ImageJ quantification of chemiluminescence was used to calculate signal-to-noise ratios. Statistical significance relative to WT signal was determined using a one-tailed Student’s t test indicated as follows: P<0.01; **.
In order to determine the range of background labeling with 5-vu, two biological replicates of WT mice were tested with increasing time points and number of injections after which organs were surveyed with RNA dot blot analysis. Although the pancreas-derived RNA was extensively degraded due to high concentrations of endonucleases13, the majority of the RNA was compared effectively with this study to conclude that single-injection treatments at 1-hr have significantly reduced background labeling compared to >5-hour time points with >2 injections (Figure S3). Although the source of 5-vu background labeling was not investigated, we chose the 3-hour time point and 500 mM injection [5-vu dose is 150 mg/kg and 5-vuD dose is 300 mg/kg] for testing cell-specificity of RNA labeling in mouse tumor xenografts.
Using the stable LM2 WT (negative control) and (+)-3xUPRT cells, mouse xenografts were generated through mammary-fat pad implantation into NSG female mice. After 3–4 weeks, tumors were visible and mice were treated for 3 hours with 500 mM 5-eu or 5-vu in triplicate. 5-euD and 5-vuD were each used as positive control treatments into one replicate mouse with WT LM2 tumors. After sacrificing mice, tumor and organ RNA was isolated and reacted with biotin-tetrazine and analyzed with RNA dot blot to determine the extent of labeling (Figure 2A and S4). Signal-to-noise ratios of the chemiluminescent signal in mouse xenografts were lower with more variability across 5-eu treatments in WT tumors compared with 5-vu treatments with overall significantly greater signal-to-noise (Figure 2B). To determine if 5-vu background could be reduced to any extent, 2-fold reduced titrations were included in (+)-3xUPRT tumor containing mice (Figure S5A). Ex vivo mCherry fluorescence showed similar size and consistent expression of 3xUPRT across these tumors (Figure S5B). Flow cytometry was used to quantify GFP fluorescence in all LM2 cells used within mouse xenografts and mCherry fluorescence to verify numbers of the (+)-3xUPRT positive cell populations prior to transplantation (Figure S6).
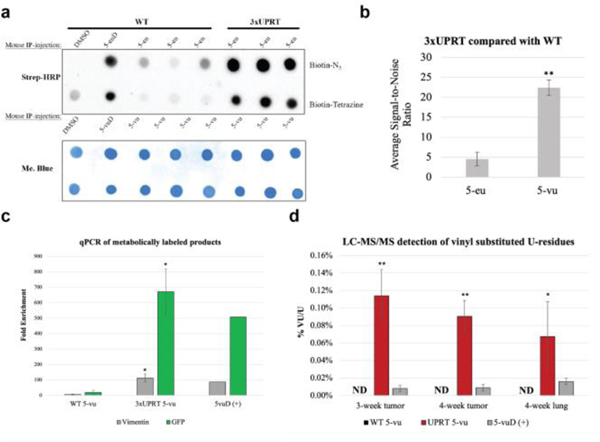
Figure 2. Cell-specific RNA metabolic labeling in mouse xenograft WT and (+)-3xUPRT LM2 primary tumors.
3-week xenografted mice were IP-injected with 500 mM of each indicated substance for 3 hours in biological triplicates for uracil analog treatments. a. RNA was extracted from tumors, reacted with biotin-N3 with ethynyl-RNA and biotin-tetrazine with vinyl-RNA prior to dot blot analysis. b. ImageJ quantification of chemiluminescence was used to calculate signal-to-noise ratios. Statistical significance relative to WT signal was determined using a one-tailed Student’s t test indicated as follows: P<0.01; **. 5-eu= 5-ethynyluracil, 5-euD= 5-ethynyluridine, 5-vu= 5-vinyluracil, 5-vuD=5-vinyluridine. c. Streptavidin bead enrichment of biotin-RNA:cDNA for qPCR analysis. Biotinylated RNA was reverse transcribed to make intact RNA:cDNA which were subsequently enriched with streptavidin beads, eluted by RNase hydrolysis and quantified with qPCR. Fold enrichment was determined through 2-dCT standardized to the untreated mouse for vimentin and GFP labeled RNA. Statistical significance relative to enrichment from untreated mice was determined using a one-tailed Student’s t test indicated as follows: P<0.1; *. WT=wild-type, UPRT=3xUPRT, 5-vu=5-vinyluracil, 5vuD=5-vinyluridine. d. LC-MS/MS for analysis of vinyl-U modification from tumors and metastatic lungs. Purified RNA was analyzed for % of vinyl-U substation of total U normalized to untreated mouse tissue. 3-week xenograft mouse lungs did not produce metastases and were not included in the analysis. Biological triplicates were used in this data set. Statistical significance relative to WT tumors and metastases was determined using a one-tailed Student’s t test indicated as follows: P<0.05; **, P<0.5 *. WT=wild-type, UPRT=3xUPRT, 5-vu=5-vinyluracil, 5vuD=5-vinyluridine, ND=not detected.
To quantify the biochemical enrichment of metabolically labeled transcripts from WT and (+)-3xUPRT tumors, RNA was subjected to reverse transcription after reaction with tetrazine-biotin. Bypassing the RNAse-H step, RNA:cDNA hybrids were incubated with MyOne C1 streptavidin magnetic beads for separation and subsequent elution of cDNA from beads by RNAse hydrolysis and heat; eluted cDNA was quantified with qPCR. Endogenous vimentin, known to be highly expressed in MDA-MB-231 cells14, and GFP, which is exogenously expressed within all MDA-MB-231 LM2 cells12 in these experiments were both quantified with qPCR. Untreated mouse RNA was included as a negative control and 5-vinyluridine treated WT tumors were used as a positive control for enrichment. The fold enrichment was calculated by 2-dCT with normalization to the untreated mouse enrichment levels for each detected gene8. A minimum of 10-fold enrichment for both genes of interest was reproducibly detected in (+)-3xUPRT/5-vu treated tumor samples (Figure 2C).
To quantify vinyl-substituted RNA, LC-MS/MS was used to determine the level of vinyl-U relative to total-U in tumor and lung samples. Both WT and (+)-3xUPRT tumor RNA from 3-week and 4-week xenografts were quantified to determine % vinyl substitution in uridine residues with metabolic labeling. Due to appreciable levels of lung metastases detected in 4-week xenografts, only 4-week lung RNA was included in this analysis. The ratio of 5-vu/total uracil residues was used to determine the % vinyl substitution rates across biological triplicates. WT tumor and lung-derived RNA from 5-vuD treated mice were used as a positive control. These samples were normalized to RNA from mouse tissues without any analog treatment (Figure 2D). The percent incorporation of 5-vu/total uracil residues is an average of 0.1% which is remarkable considering in vitro vinyl-incorporation was 0.8% for 5-vuD10 RNA labeling in cells that were subjected to higher local analog concentrations and incubation times compared with conditions in these mouse xenograft studies.
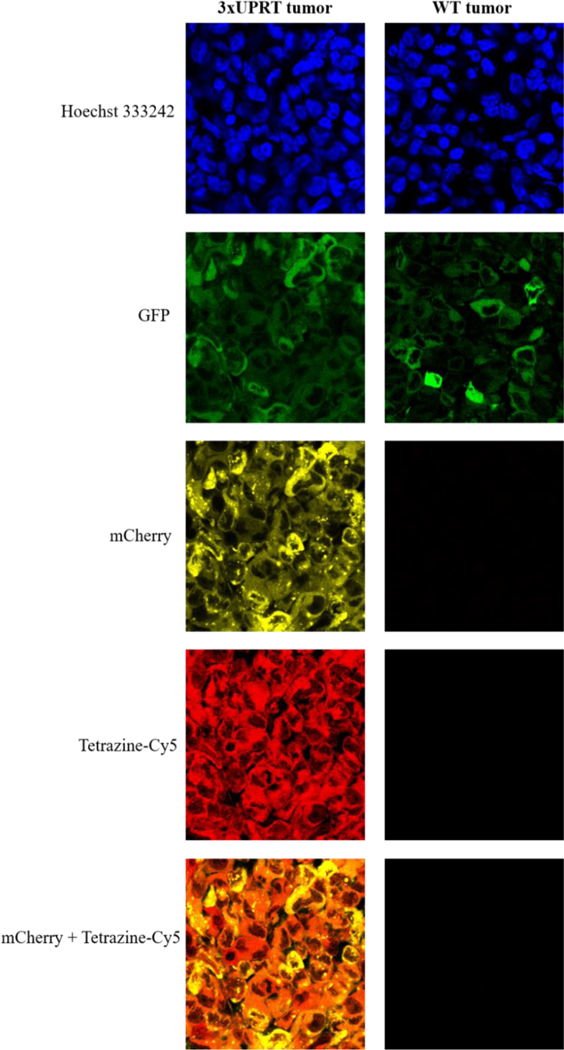
For imaging nascent RNA, tetrazine-Cy5 was used to conjugate nascent, vinyl-labeled RNA in mouse tumor and lung tissue sections. Tumor sections show uniformity of mCherry expression indicating cells with 3xUPRT expression. 4-week lung sections demonstrate dispersed metastatic (+)-3xUPRT LM2 cells within lung tissue with the co-localization of Cy5 signal demonstrating the level of cell-specificity achieved when captured through confocal microscopy. No background signal was detected in WT tumor sections reacted with tetrazine-Cy5 or the surrounding cell types in the lung tissue. Although overall fluorescent dye signal was reduced in lung tissue compared to tumor tissue, this can be attributed to the lungs being further from the IP-injection site which may have reduced the bioavailability of 5-vu. These results clearly show that only (+)-3xUPRT cells in mouse tissues label nascent RNA with 5-vu, which can be detected with tetrazine-Cy5. These findings support the utility for 3xUPRT/5-vu for in vivo imaging of cell-specific nascent RNA (Figure 3 and Figure S7).
Figure 3. Metabolic labeling in (+)-3xUPRT-mCherry MDA-MB-231 LM2 cells in tumor tissues.
Vinyl-containing RNA are fluorophore-conjugated with tetrazine-Cy5. GFP signal is present in WT MDA-MB-231 LM2 cells (parental line). mCherry signal indicates (+)-3xUPRT MDA-MB-231 cells.
The cell-specific RNA metabolic labeling method described herein has the potential to transform the ability to analyze nascent RNA in vivo using widely available laboratory techniques. The most technically challenging aspect involved is to generate the stable expression of 3xUPRT into cell-types of interest which can be performed with CRISPR/Cas9 genome-editing strategies. Secondly, 5-vu, biotin-tetrazine, and Cy5-tetrazine are commercially available for researchers to independently access if they lack resources to synthesize these reagents. Lastly, through the expression of downstream gene-specific promoters, 3xUPRT expression labels RNA when a gene-of-interest is actively expressed thereby extending the utility of these tools to capture and visualize nascent RNA in a gene-specific and cell-specific context.
As a proof-of-principle, we generated MDA-MB-231 LM2 cells that stably express 3xUPRT driven by the endogenous vimentin promoter. We chose to generate vimentin-3xUPRT for the purpose of labeling RNA important for the maintenance of mesenchymal cell states which occurs during the epithelial-to-mesenchymal transition in early metastatic progression 14 (Figure S8). The extent of labeling in vimentin-3xUPRT LM2 cells is overall comparable in chemiluminescent signal to the constitutively-expressed (+)-3xUPRT LM2 cells used throughout this study and to the 5-vuD positive control signal. These exciting results support using this experimental approach for analyzing actively transcribed RNA pertinent to the epithelial-to-mesenchymal transition in greater detail by co-expressing 3xUPRT with other relevant genes and transcription factors of interest with the expectation of reduced non-specific, background RNA labeling shown throughout this work.
In conclusion, these results have demonstrated the potential for researchers to design unique in vivo cell-specific RNA metabolic labeling experiments for model organisms of interest in order to elucidate complex transcriptional pathways in important cell-types.
Supplementary Material
ACKNOWLEDGMENT
We thank members of the Spitale lab for their careful reading and critique of the manuscript. Spitale lab work in this manuscript is supported by the University of California, Irvine and the Pew Biomedical Scholar program (R.C.S.).
Funding Sources
No competing financial interests have been declared.
ABBREVIATIONS
- Strep-HRP
Streptavidin-Horseradish Peroxidase
- Me.-Blue
Methylene Blue
- 5-eu
5-ethynyluracil
- 5-euD
5-ethynyluridine
- 5-vu
5-vinyluracil
- 5-vuD
5-vinyluridine
- WT
wild-type
- 3xUPRT
triple-mutant uracil phosphoribosyltransferase (Toxoplasma gondii
- UMPS
uridine monophosphate synthase
Footnotes
Supporting Information
Supporting information figures and methods are available online.
REFERENCES
- 1.Wei J-W; Huang K; Yang C; Kang C-S Non-Coding RNAs as Regulators in Epigenetics. Oncology Reports 2017, 37 (1), 3–9. [DOI] [PubMed] [Google Scholar]
- 2. Richardson GM; Lannigan J; Macara IG Does FACS Perturb Gene Expression? Cytometry 2015, 87 (2), 166–175. [DOI] [PubMed] [Google Scholar]
- 3.Pfister G; Toor SM; Sasidharan Nair V; Elkord E. An Evaluation of Sorter Induced Cell Stress (SICS) on Peripheral Blood Mononuclear Cells (PBMCs) after Different Sort Conditions - Are Your Sorted Cells Getting SICS? Journal of Immunological Methods 2020, 487, 112902. [DOI] [PubMed] [Google Scholar]
- 4. Singha M; Spitalny L; Nguyen K; Vandewalle A; Spitale RC Chemical Methods for Measuring RNA Expression with Metabolic Labeling. WIREs RNA 2021, 12 (5). [DOI] [PubMed] [Google Scholar]
- 5. Nguyen K; Fazio M; Kubota M; Nainar S; Feng C; Li X; Atwood SX; Bredy TW; Spitale RC Cell-Selective Bioorthogonal Metabolic Labeling of RNA. J. Am. Chem. Soc 2017, 139 (6), 2148–2151. [DOI] [PubMed] [Google Scholar]
- 6.Zajaczkowski EL; Zhao Q-Y; Zhang ZH; Li X; Wei W; Marshall PR; Leighton LJ; Nainar S; Feng C; Spitale RC; Bredy TW Bioorthogonal Metabolic Labeling of Nascent RNA in Neurons Improves the Sensitivity of Transcriptome-Wide Profiling. ACS Chem. Neurosci 2018, 9 (7), 1858–1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Paredes E; Das SR Click Chemistry for Rapid Labeling and Ligation of RNA. ChemBioChem 2011, 12 (1), 125–131. [DOI] [PubMed] [Google Scholar]
- 8. Nainar S; Cuthbert BJ; Lim NM; England WE; Ke K; Sophal K; Quechol R; Mobley DL; Goulding CW; Spitale RC An Optimized Chemical-Genetic Method for Cell-Specific Metabolic Labeling of RNA. Nat Methods 2020, 17 (3), 311–318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Nguyen K; Kubota M; Arco J. del; Feng C; Singha M; Beasley S; Sakr J; Gandhi SP; Blurton-Jones M; Fernández Lucas J; Spitale RC A Bump-Hole Strategy for Increased Stringency of Cell-Specific Metabolic Labeling of RNA. ACS Chem. Biol 2020, 15 (12), 3099–3105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kubota M; Nainar S; Parker SM; England W; Furche F; Spitale RC Expanding the Scope of RNA Metabolic Labeling with Vinyl Nucleosides and Inverse Electron-Demand Diels–Alder Chemistry. ACS Chem. Biol 2019, 14 (8), 1698–1707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Suzuki K; Tsunekawa Y; Hernandez-Benitez R; Wu J; Zhu J; Kim EJ; Hatanaka F; Yamamoto M; Araoka T; Li Z; Kurita M; Hishida T; Li M; Aizawa E; Guo S; Chen S; Goebl A; Soligalla RD; Qu J; Jiang T; Fu X; Jafari M; Esteban CR; Berggren WT; Lajara J; Nuñez-Delicado E; Guillen P; Campistol JM; Matsuzaki F; Liu G-H; Magistretti P; Zhang K; Callaway EM; Zhang K; Belmonte JCI In Vivo Genome Editing via CRISPR/Cas9 Mediated Homology-Independent Targeted Integration. Nature 2016, 540 (7631), 144–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Minn AJ; <Gupta GP; Siegel PM; Bos PD; Shu W; Giri DD; Viale A; Olshen AB; Gerald WL; Massagué J. Genes That Mediate Breast Cancer Metastasis to Lung. Nature 2005, 436 (7050), 518–524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Azevedo-Pouly ACP; Elgamal OA; Schmittgen TD RNA Isolation from Mouse Pancreas: A Ribonuclease-Rich Tissue. JoVE 2014, No. 90, 51779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Satelli A; Li S. Vimentin in Cancer and Its Potential as a Molecular Target for Cancer Therapy. Cell. Mol. Life Sci 2011, 68 (18), 3033–3046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Neef AB, Pernot L, Schreier VN, Scapozza L, and Luedtke NW (2015) A Bioorthogonal Chemical Reporter of Viral Infection. Angew. Chem 127 (27), 8022–8025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Beasley S; Vandewalle A; Singha M; Nguyen K; England W; Tarapore E; Dai N; Corrêa IR; Atwood SX; Spitale RC Exploiting Endogenous Enzymes for Cancer-Cell Selective Metabolic Labeling of RNA in Vivo. J. Am. Chem. Soc 2022, 144 (16), 7085–7088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang G-L; Zhang Y; Cao K-X; Wang X-M Orthotopic Injection of Breast Cancer Cells into the Mice Mammary Fat Pad. JoVE 2019, No. 143, 58604. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.