Abstract
Background:
Suppression of p53 is an important mechanism in Epstein-Barr virus associate-tumors and described as EBNA1-USP7 which is a key axis in p53 suppression. Thus, in this study, we aimed to evaluate the function of EBNA1 on the expression of p53-inhibiting genes including HDAC-1, MDM2, MDM4, Sirt-3, and PSMD10 and the influence of USP7 inhibition using GNE-6776 on p53 at protein/mRNA level.
Methods:
The electroporation method was used to transfect the BL28 cell line with EBNA1. Cells with stable EBNA1 expression were selected by Hygromycin B treatment. The expression of seven genes, including PSMD10, HDAC-1, USP7, MDM2, P53, Sirt-3, and MDM4, was evaluated using a real-time PCR assay. For evaluating the effects of USP7 inhibition, the cells were treated with GNE-6776; after 24 hours and 4 days, the cells were collected and again expression of interest genes was evaluated.
Results:
MDM2 (P=0.028), MDM4 (P=0.028), USP7 (P=0.028), and HDAC1 (P=0.015) all showed significantly higher expression in EBNA1-harboring cells compared to control plasmid transfected cells, while p53 mRNA expression was only marginally downregulated in EBNA1 harboring cells (P=0.685). Four-day after treatment, none of the studied genes was significantly changed. Also, in the first 24-hour after treatment, mRNA expression of p53 was downregulated (P=0.685), but after 4 days it was upregulated (P=0.7) insignificantly.
Conclusion:
It seems that EBNA1 could strongly upregulate p53-inhibiting genes including HDAC1, MDM2, MDM4, and USP7. Moreover, it appears that the effects of USP7 suppression on p53 at protein/mRNA level depend on the cell nature; however, further research is needed.
Key Words: Epstein-Barr virus, EBNA1, P53, USP7, P53-inhibiting Genes
Introduction
The Epstein-Barr virus (EBV) was the first recognized human virus associated with human cancer (1). The EBV is a member of the Herpesviridae family, subfamily Gammaherpesvirinae, and genus Lymphocryptovirus. The link between Burkitt Lymphoma (BL) and EBV infection has been frequently recognized (2). BL is an aggressive non-Hodgkin’s lymphoma (NHL) that affects B cells, with EBV infection accounting for 95% of endemic BL cases (3). In BL, EBV may initially induce proliferation of B cells resulting in oncogene activation (e.g., c-myc in BL) and chromosomal translocation (4).
Different EBV-encoded products during viral latency may relate to oncogenesis process in EBV-associated malignancies (5). Epstein-Barr virus nuclear antigen 1 (EBNA-1) is one of them and is present in all EBV-associated malignancies (6); it is also the only viral protein expressed in latency type I, such as BL (7). EBNA1 binds to the host and viral DNA and works as a transcriptional regulator of both host and viral promoter (6) as well as episome replication and maintenance in host cells (7).
A de-ubiquitin enzyme called ubiquitin-specific protease 7 (USP7) is crucial for releasing ubiquitin from its substrates, including the tumor suppressor p53 (8,9). Numerous reports have revealed that USP7 paradoxically controls the actions of p53 (10). According to certain research, specific USP7 inhibition causes cancer cell death through a p53-dependent mechanism (11,12). On the other hand, it's been reported that EBNA1 interacts with USP7 and effectively breaks down the interaction between p53 and USP7, leading to the degradation of p53 (13).
In cancers harboring the wild-type p53 gene, disruption of p53 at expression or function levels contributes to the progression of cancer, and it was found that in some BL cells harboring a wild-type p53 gene, the function of p53 protein was compromised by the overexpression of Mouse double minute 2 homolog (MDM2) (14-16). Additionally, p53 gene mutations are rare in EBV-associated gastric cancer tissues (17). Thus, overexpression of some p53-inhibiting genes such as Histone deacetylases1(HDAC1), Mouse double minute 2(MDM2), MDM4, Sirtuin 3(SIRT3), and proteasome 26S subunit, non-ATPase 10 (PSMD10) might be an important way in suppression of wild-type p53.
HDAC1 deacetylates p53 and affects its function on cell development and apoptosis as a class I histone deacetylase (18). MDM2 protein (also known as HDM2 in humans) has been discovered to bind and inhibit p53 (19). MDM4 is a homolog of MDM2 (20). Like MDM2, MDM4 is a crucial p53 negative regulator (21). In cancer, SIRT3 has the ability to function as both an oncogene and a tumor suppressor, controlling cell death by focusing on a variety of important modulators and their associated pathways (22). SIRT3 has the potential to deacetylate p53 as well (23). PSMD10 (or Gankyrin) inhibits apoptosis by destroying p53 and, consequently, p53-dependent gene transcription (24-26).
We therefore sought to assess the expression of p53-inhibiting genes such as HDAC-1, MDM2, MDM4, SIRT3, and PSMD10 in BL28-harboring EBNA1 as well as the impact of USP7 inhibition using GNE-6776 on p53 suppression.
Materials and Methods
Cell culture, electroporation transfection, and clonal selection by hygromycin B
The BL28 cell line that harbors p53 wild type and is negative for EBV was obtained from the Pasteur Institute of Iran's Cell Bank. PCEP4 plasmid (Invitrogen, USA) harboring EBNA1 gene (strain B95.8) and EBV replication origin (OriP), as well as a control plasmid, were transfected into BL28 cells. The transfection was carried out using the electroporation technique (Gene Pulser Electro protocols for mammalian and suspension cell). After 24 hours, Hygromycin B (Sigma Aldrich, Germany) (350 µg/mL) was added to the cell culture for 16 days to select the transfected cells with stable EBNA1 expression.
Quantitative reverse transcription PCR (qRT-PCR) assay and validation of EBNA1 expression
Total RNA was extracted from BL28 cell culture, using an RNA Isolation Kit (Dena Zist, Mashhad, Iran). The EasycDNA Synthesis Kit (Parstos, Mashhad, Iran) was used for RT-PCR, and 1000 ng/µL of total RNA from each specimen was employed. To remove PCEP4 plasmid contamination, total RNA from BL28 harboring EBNA1 was treated with RNase-free DNase Kit (Sinaclon, Tehran, Iran). We synthesized cDNA and used Real-time PCR to confirm that EBNA1 was expressed. Additionally, DNase-treated total RNA was used as a negative control.
qRT-PCR based on SYBR green I fluorescence was used to assess the expression of 7 genes including HDAC-1, MDM2, MDM4, USP7, Sirt-3, P53, and PSMD10. For the gene expression assay, the beta-actin gene was used as a reference gene (13). Each mixture with 15 μl final volume included 7.5 μL of 2x SYBR Green Master Mix (Ampliqon Inc., Denmark), 4.7 of water, 0.4 μL of each unique primer pair (Table 1), and 2 μL of cDNA. Next, the qRT-PCR was carried out on an ABI 7500 instrument (Applied Biosystems, Grand Island, NY, USA), using the following thermal cycling procedure: initial denaturation for 15 min at 95 °C, followed by 40 cycles of 15-sec denaturation at 95 °C, annealing/extension at 62 °C (for SIRT3 gene at 58 °C) for 1 min.
Table 1.
The sequences of primers used in Real-time PCR test.
| Gene name | Sequences | Product Size (bp) |
|---|---|---|
| TP53 | 5΄-GATAGCGATGGTCTGGC-3΄ | 117 |
| 5΄-CGGCTCATAGGGCACC-3΄ | ||
| SIRT3 | 5΄-ACTCCCATTCTTCTTTCACAAC-3΄ | 176 |
| 5΄-GGATGCCCGACACTCT-3΄ | ||
| USP7 | 5΄-TGGTGGAGCGATTACAAGA-3΄ | 100 |
| 5΄-TCCTCTGCGACTATCTGC-3΄ | ||
| EBNA-1 | 5΄-GGGTGGTTTGGAAAGCATCG-3΄ | 156 |
| 5΄-CTTACTACCTCCATATACGAACACA-3΄ | ||
| PSMD10 | 5΄-CTACTAGAACTGACCAGGACA-3΄ | 145 |
| 5΄-GCCGCAATATGAAGAGGAG-3΄ | ||
| MDM2 | 5΄-AACCACCTCACAGATTCCA-3΄ | 87 |
| 5΄-GCACCAACAGACTTTAATAACTTC-3΄ | ||
| MDM4 | 5΄-GCCTGCCTTGGTGGTT-3΄ | 160 |
| 5΄-CCTAACTGCTCTGATACTGACTC-3΄ | ||
| HDAC1 | 5΄-GACGGTAGGGACGGGAG-3΄ | 203 |
| 5΄-GGCTTTGTGAGGGCGATAG-3΄ | ||
| Beta actin (28) | 5΄-GCCTTTGCCGATCCGC-3΄ | 90 |
| 5΄-GCCGTAGCCGTTGTCG-3΄ |
Morphological evaluation and immunocytochemistry analysis
BL28 cells were harvested after 20 days of transfection with EBNA1 and control plasmids, and pathological staining was used to analyze the morphological changes. In addition, immunocytochemistry staining was performed, using kit (Samatashkhis, Tehran, Iran), according to the manufacturer’s instruction.
GNE-6776 toxicity evaluation using trypan blue exclusion test and propidium iodide spectrofluorometric assay
According to the manufacturer’s instructions, GNE-6776 suppresses both endogenous and recombinant cellular USP7 at a 15 µM concentration. We performed two independent tests including PI spectrofluorometry and trypan blue exclusion test to confirm that GNE-6776 at a 15 µM concentration is not toxic and does not induce necrosis instead of apoptosis in BL28 cells as cell death, it is known necrosis causes bias in p53-dependent cell death evaluation (14).
USP7 inhibitor treatment
For evaluating the effects of USP7 inhibition on p53, 5×105 cells of EBNA1-transfected cells, control plasmid and non-transfected cells were seeded into 6-well plates, separately. After 24 hours, the GNE-6776 (MedChemExpress, USA), as USP7 inhibitor with 15 µM final concentration, was utilized. Finally, the cells were collected at two different time points (1 and 4 days later).
Acridine orange and propidium iodide staining for cell viability analysis following GNE-6776 treatment
In a 12-well plate, 2 × 105 cells of the EBNA1 transfected, control plasmid, and non-transfected cells were seeded into 6-well plate separately per well. After 24 hours, each well was treated with GNE6776 in 15 µM final concentration and incubated for another 24 hours. Finally, the cells in each well received 5 µg of acridine orange/propidium iodide (AO/PI) reagent, and the cells were incubated for 10 minutes at room temperature (RT). For evaluating the results, an invert fluorescence microscope (FLUOstar Omega, BMG LABTECH), Germany, was used.
Data analysis
The CtNorm technique was used to normalize all Ct values acquired throughout the qRT-PCR test (the CtNormsoftware is available online at http://ctnorm.sums.ac.ir/). In Microsoft Excel, the normalized values were then computed (2-ΔΔCT). The Mann–Whitney U test was used to compare the means of groups using the GraphPad Prism version 5.0 software. Finally, a statistically significant difference was defined as a P value of less than 0.05.
Results
The p53-inhibiting genes expression in BL28 cell harboring EBNA1 plasmid
The qRT-PCR assay showed that the expression of the three genes of interest including MDM2 (P=0.028), MDM4 (P=0.028), and HDAC1 (P=0.015) was significantly higher in EBNA1-harboring cells than the control plasmid transfected cells (Fig. 1). Although the mRNA expression levels of SIRT3 (P=0.342) and PSMD10 (P=0.9) were upregulated in the EBNA1-harboring cell compared with control plasmid transfected cell, these upregulations were not statistically significant. mRNA expression of USP7 (P=0.028) in EBNA1-transfected cells was significantly upregulated compared with control cells. mRNA expression level of p53 was downregulated insignificantly in EBNA1-harboring cell compared with the control cells (P=0.685) (Fig. 1).
Fig. 1.
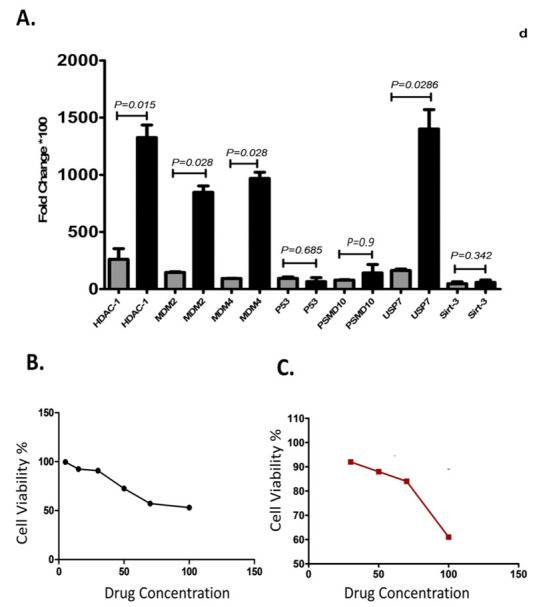
Thep53 inhibitor genes expression and toxicity assays of GNE6776: A) The p53 inhibitor genes expression in BL28 cell harboring EBNA1 plasmid; B) Toxicity evaluation using trypan blue exclusion test; C) Propidium iodide spectrofluorometric assay.
Altered expression of p53-inhibiting genes at mRNA level following GNE6776 treatment
Analysis of mRNA expression levels after 24 hours from GNE6776 treatment revealed that the expression levels of all genes in the EBNA1-harboring cell were higher than the untreated EBNA1-harboring cell that only MDM2 mRNA expression was significant (Fig. 2A). Also, after 24 hours, the mRNA expression of p53 (P=0.685) in the treated EBNA1-harboring cells was lower than the untreated EBNA1-harboring cells (Fig. 2A). After 4 days, in the EBNA1-harboring cell that were treated with GNE6776, the expression of MDM2 (P=0.1), MDM4 (P=0.7), PSMD10 (P=0.375), and SIRT3 (P=0.76) was higher than untreated the EBNA1-tranfected cells although they were not statistically significant. p53 mRNA expression was upregulated after four days insignificantly (P=0.7) (Fig. 2A). Alteration expression of p53-inhibiting genes at mRNA level in the control plasmid transfected cell following GNE6776 treatment and the EBNA1 harboring cell vs. control plasmid transfected cell following GNE6776 treatment were shown in Figures 2B and 3, respectively.
Fig. 2.
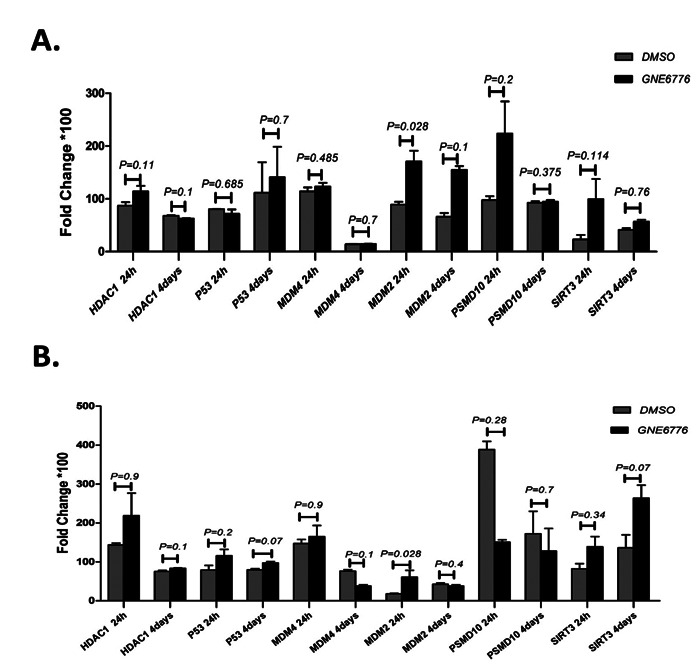
Thep53 inhibitor genes expression: A) Expression of p53 inhibitor genes at mRNA level in EBNA1 harboring cell following GNE6776 treatment; B) Expression of p53 inhibitor genes at mRNA level in control plasmid harboring cell following GNE6776 treatment.
Fig. 3.
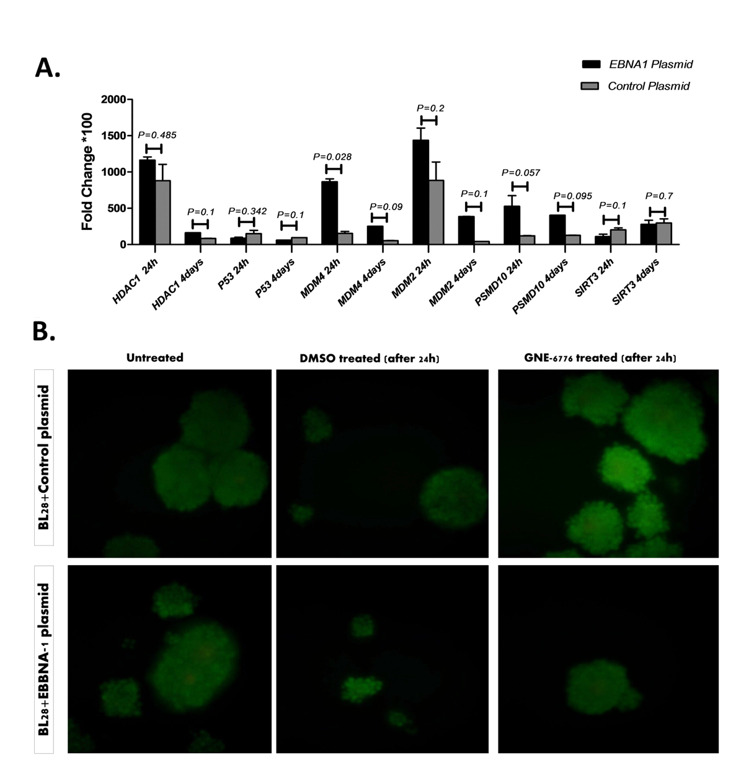
Effect of GNE6776 treatment on EBNA1 harboring cell vs. the control plasmid transfected cell and apoptosis analysis: A) Effect of GNE6776 treatment on the EBNA1-harboring cell versus the control plasmid transfected cell; B) Acridine orange and Propidium iodide staining for cell apoptosis analysis after GNE-6776 treatment.
Morphologic examination and immunocytochemistry assay for p53 expression
After selecting hygromycin B-resistant clones, no pathological changes were seen in the morphology of the EBNA1-transfected cells. Moreover, immunocytochemistry test showed that p53 expression in the EBNA1-transfected cell, EBNA1 transfected cell+ GNE6776, and control plasmid transfected cell were 10-15%, 20-25% and 10-15%, respectively.
Acridine orange and propidium iodide staining for cell apoptosis analysis after GNE-6776 treatment
In acridine orange and propidium iodide staining, each fluorescence, including red, yellow, and green, indicates late apoptosis (necrosis), early apoptosis, and live cells, respectively. After 24 hours, we did not observe any sign of apoptosis in the cells treated with GNE-6776. After GNE-6776 treatment, apoptosis was not seen in both control plasmid and EBNA1plasmid transfected cells.
Discussion
Based on our findings, the expression of HDAC1 gene increased significantly in the EBNA1-expressing BL28 cells compared to the control plasmid transfected cell. HDACs are enzymes that remove the acetyl groups from non-histone and histone proteins and are important regulators of chromatin architecture and gene expression (27). The acetylation of p53 modulates its transcriptional activity, which is important for the elimination of cancer cells (28). It is also involved in the regulation of autophagy and apoptosis, among other things (28). HDAC1 overexpression has been found in the EBV-positive gastric adenocarcinoma cell line, and it may have a role in epigenetic control during tumor development (29). In AGS-EBV cell line, HDAC1 was discovered to be an EBV-specific upstream regulator, indicating the change to epigenetic regulation during tumor development (29). Accordingly, EBV-EBNA1 might be involved in BL pathogenesis by increasing the expression of HDAC1, resulting in deacetylation of p53 gene that might decrease its expression and inhibit apoptosis of the infected cells.
In the present study, MDM2 mRNA expression was significantly upregulated in the EBNA1- harboring cells in comparison with control plasmid transfected cells. MDM2 has two key mechanisms for inhibiting the p53 activation. It may bind to p53's transactivation domain, preventing it from causing transcription of its targets, and it can also operate as an E3 ubiquitin ligase, reducing the capacity of p53 to induce transcription of its targets (30, 31). AlQarni et al. reported that some MDM2 isoforms overexpressed in the EμEBNA1 transgenic mice tumor cells, and expression of MDM2 was related to EBNA1 expression (14). Additionally, using 4 independent MDM2 inhibitors, they showed the tumor cells were dependent upon MDM2 protein for survival (14). Renouf et al., by using nutlin-3 as a p53–MDM2 interaction inhibitor in combination with different chemotherapeutic drugs, showed that p53 was stabilized and activated (15). In addition, they reported EBV negative or latency I EBV BL cells were sensitive to these drugs, but latency III EBV cells were not affected by them (15). Similarly, in a previous study, we observed that expression of MDM2 was upregulated significantly in MKN-45 cell line harboring EBNA1 plasmid (unpublished data). Based on our and other studies, EBNA1 might induce the overexpression of MDM2 gene in the BL cells, inhibit the p53 function, prevent the infected cell apoptosis, and support the pathogenesis of EBV in BL. Additionally, the SNP rs2279744 in MDM2, which is connected to the p53 activation pathway, has also been linked to an increased risk of leukemia (32).
In our work, MDM4 was found to be significantly overexpressed in BL28 cells that have EBNA1 compared to the control plasmid transfected cells. MDM4 is overexpressed in a variety of malignancies that, nonetheless, have wild type p53 (33). A study based on CRISPR/Cas9 screens revealed that MDM4 and MDM2 were critical for the survival of EBV-transformed B cells (34). Also, in our previous study on MKN-45 cell line harboring EBNA1 plasmid, MDM4 was significantly overexpressed (unpublished data). Taken together, overexpression of MDM4 by EBNA1 might be related to p53 suppression in BL cancer cells harboring wild type p53.
In present study, the expression of USP7 gene in the EBNA1-expressing BL28 cells was increased significantly compared to the control plasmid transfected cell. USP7 is a de-ubiquitin enzyme that plays an essential role in cleaving ubiquitin from its substrates and is most typically related to cellular protein stability (35). USP7 regulates p53 functions in a paradoxical way via several ways (10). In a context-dependent way, HAUSP works as both a tumor suppressor and an oncogene, and its substrates play a key role in either tumor suppression or oncogenesis (36). According to the findings, EBNA1 might interact with USP7, which facilitates p53 degradation and prevents apoptosis (13). However, USP7-mediated stabilization of MDM2 (as a p53-inhibiting protein) results in p53 degradation, preventing the cell cycle arrest and death and boosting tumor cell proliferation (10, 12). Additionally, it was reported that USP7 might influence gene expression via modulating the transcription factors (9). He et al. reported inhibiting USP7 reduced the MDM2 expression (as a p53-inhibiting gene) and increased the p53 expression (37).
In non-small cell lung cancer (NSCLC), USP7 plays a significant role in carcinogenesis through p53-dependent pathways (10). It was reported that USP7 overexpression, through changes in some processes such as apoptosis, cell cycle control, and DNA damage response, contributes to the progression of tumor. For instance, it was shown that USP7 was upregulated in CLL (38). It is possible that the EBNA1 protein functions in tumor progression by inducing USP7 expression, but it needs to be studied further.
Our findings demonstrated that although SIRT3 expression was higher in the BL28 cell line containing EBNA1 than in the controls, the difference was insignificant. Expression of SIRT3 was not significantly higher in the EBNA1-harboring cells than the control plasmid transfected cells. The function of SIRT3 varies depending on the tumor and cell type; as a result, SIRT3 might operate as a tumor suppressor or an oncogene (22). It was reported that there was a strong relationship between SIRT3 and p53(39). SIRT3 is a target in cancer therapy with both tumor-suppressive and oncogenic properties (23). Previously, it was reported that SIRT inhibition might prevent p53 deacetylation by viruses, and hyperacetylated p53 induces cell death and virus eradication (40). Our finding showed that SIRT3 might have not key role in p53 inhibition in the cell harboring EBNA1, like EBV-associated BL.
In our study, PSMD10 expression was shown to be greater in BL28 cell lines containing EBNA1 than in the control cells, albeit this difference was not statistically significant. Increased Gankyrin production reduces apoptosis by degrading p53 and, as a result, the transcription of p53-dependent genes (24). In vivo and in vitro experiments show that Gankyrin binds to MDM2, boosting p53–MDM2 interaction, resulting in p53 ubiquitylation and subsequent proteasomal destruction (41). Several studies have shown that Gankyrin is overexpressed in a variety of malignancies, contributing to tumor development as well as poor prognosis (24, 42). Kashyapetal. reported EBV and Helicobacter pylori coinfection by regulating Gankyrin that might induce gastric cancer progression (43). They reported that the expression of Gankyrin might be directly linked with EBNA3C (43). In our previous research on MKN-45 cells that harbored EBNA1 plasmid, PSMD10 expression was increased significantly (unpublished data). It seems PSMD10 might not be involved in the pathogenesis of EBV in BL through EBNA1, but it might contribute to the pathogenesis of EBV in stomach cancer; therefore, based on cell nature, the effect of EBNA1 on PSMD10 gene expression maybe different.
Results also showed that p53 transcription was downregulated following EBNA1 transfection, but not significantly. Moreover, immunocytochemistry assay showed no changes at the protein level. Likewise, Ribeiro et al. reported that EBV-associated gastric carcinomas had p53 wild type reduction in p53 mRNA level with high p53 protein in IHC assay (44). We previously demonstrated in MKN-45 cell line that harboring EBNA1, p53 transcription was downregulated but slightly upregulated at the protein levels (unpublished data). One interpretation of this event maybe that EBNA1 affects the expression of p53 at the mRNA level and inhibits p53 protein or its pathways by upregulating some inhibitor genes. Thus, wild type p53 has no function in these cell lines.
In the second phase of this study, GNE6776 was used to test the impact of USP7 inhibition on p53 and its inhibitor genes expression in the cells carrying the EBNA1 and control plasmids. After passing 4 days from GNE6776 treatment, none of p53-inhibiting genes was significantly elevated in the control plasmid transfected cells and transfected EBNA1 in comparison with their control that was treated with DMSO. However, when we compared the EBNA1 transfected cells with the control plasmid transfected cells treated with GNE6776, certain p53-inhibiting genes such as MDM4 and PSMD10 had significantly higher expression in the BL28 cells carrying the EBNA1 plasmid vs. the control plasmid. The difference in the effect of GNE-6776 on the EBNA1 plasmid transfected cells vs. the control plasmid transfected cells might be attributed to the presence of EBNA1 protein. Using nutlin-3, as a p53–MDM2 interaction inhibitor, Renouf et al. showed EBV negative or latency I EBV BL cells were sensitive to these drugs, but latency III EBV cells were not affected by them (15). Thus, it seems that the effect of drugs on EBV negative cells or each EBV latency is different. We performed two independent tests (PI spectrofluorometry and trypan blue exclusion test) and confirmed that GNE-6776 at a 15 µM concentration was not toxic for the BL28 cells (not inducing necrosis instead of apoptosis at this concentration). When we used GNE-6776 at a 15 µM concentration in the AO/PI assay, USP7 inhibition caused no apoptosis in both control plasmid and EBNA1 plasmid transfected cells.
In the current study, we did not find significant changes in the expression of p53 (mRNA or protein level) following USP7 inhibition. Additionally, no sign of apoptosis was seen. However, in our prior study, by USP7 inhibition in MKN-45 cell line, we found that p53 expression was upregulated at protein level, and apoptosis was induced in the MKN-45 cell line (unpublished data). It seems that the function of USP7on p53 and cancer process may be different in each cell with different biology. In other studies, it was reported in a context-dependent way that HAUSP worked as both a tumor suppressor and an oncogene, and its substrates played a key role in either tumor suppression or oncogenesis. This suggests that when USP7 inhibitors are evaluated in clinical trials, rigorous patient selection and biomarker analysis would be required (36).
In conclusion, the results indicated that EBNA1 might suppress p53 in latency type I cancers like Burkitt's lymphoma by upregulating several p53-inhibiting genes including MDM2, MDM4, and HDAC1.Upregulation of p53-inhibiting genes might occur along with other mechanisms of EBV in oncogenesis such chromosomal translocation and oncogene activation involved in the fully malignant phenotype. In addition, following USP7 inhibition by GNE6776, we did not see statistically significant changes in expression of p53 at mRNA or protein level, and it appears that the effects of USP7 suppression on p53 at protein/mRNA level depend on the cell nature. Further research is needed for clarifying these results.
Funding
The present study was financially supported by Golestan University of Medical Sciences (IR.GOUMS.REC.1400.130).
Conflicts of Interest
The authors declare no conflict of interest.
Acknowledgements
The present study was extracted from a PhD thesis written by Seyed Mohammad Ali Hashemi.
References
- 1.Grywalska E, Rolinski J. Epstein-Barr virus-associated lymphomas. Semin Oncol. . 2015;42(2):291–303. doi: 10.1053/j.seminoncol.2014.12.030. [DOI] [PubMed] [Google Scholar]
- 2.Zanella L, Riquelme I, Buchegger K, Abanto M, Ili C, Brebi P, et al. A reliable Epstein-Barr Virus classification based on phylogenomic and population analyses. Sci Rep. 2019;9(1):9829. doi: 10.1038/s41598-019-45986-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zanelli M, Sanguedolce F, Palicelli A, Zizzo M, Martino G, Caprera C, et al. EBV-Driven Lymphoproliferative Disorders and Lymphomas of the Gastrointestinal Tract: A Spectrum of Entities with a Common Denominator (Part 2). Cancers (Basel). . 2021;13(18):4527. doi: 10.3390/cancers13184578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Howley PM, Knipe DM, Cohen JL, Damania BA, editors. Fields Virology: DNA Viruses. Wolters Kluwer Health/Lippincott Williams & Wilkins;; 2021. [Google Scholar]
- 5.Stuhlmann-Laeisz C, Oschlies I, Klapper W. Detection of EBV in reactive and neoplastic lymphoproliferations in adults-when and how? . J Hematop. 2014;7(4):165–170. doi: 10.1007/s12308-014-0209-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wilson JB, Manet E, Gruffat H, Busson P, Blondel M, Fahraeus R. EBNA1: Oncogenic Activity, Immune Evasion and Biochemical Functions Provide Targets for Novel Therapeutic Strategies against Epstein-Barr Virus- Associated Cancers. . Cancers (Basel). . 2018;10(4):109. doi: 10.3390/cancers10040109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Reisman D, Yates J, Sugden B. A putative origin of replication of plasmids derived from Epstein-Barr virus is composed of two cis-acting components. Mol Cell Biol. 1985;5(8):1822–32. doi: 10.1128/mcb.5.8.1822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kessler BM, Fortunati E, Melis M, Pals CE, Clevers H, Maurice MM. Proteome changes induced by knock-down of the deubiquitylating enzyme HAUSP/USP7. . J Proteome Res. . 2007;6(11):4163–72. doi: 10.1021/pr0702161. [DOI] [PubMed] [Google Scholar]
- 9.Pozhidaeva A, Bezsonova I. USP7: Structure, substrate specificity, and inhibition. DNA repair. 2019;76:30, 9. doi: 10.1016/j.dnarep.2019.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang Z, Kang W, You Y, Pang J, Ren H, Suo Z, et al. USP7: novel drug target in cancer therapy. . Front Pharmacol. . 2019;10:427. doi: 10.3389/fphar.2019.00427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Qi SM, Cheng G, Cheng XD, Xu Z, Xu B, Zhang WD, et al. Targeting USP7-mediated deubiquitination of MDM2/MDMX-p53 pathway for cancer therapy: are we there yet? . Front Cell Dev Biol. . 2020;8:233. doi: 10.3389/fcell.2020.00233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schauer NJ, Liu X, Magin RS, Doherty LM, Chan WC, Ficarro SB, et al. Selective USP7 inhibition elicits cancer cell killing through a p53-dependent mechanism. . Sci Rep. 2020;10(1):5324. doi: 10.1038/s41598-020-62076-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Saridakis V, Sheng Y, Sarkari F, Holowaty MN, Shire K, Nguyen T, et al. Structure of the p53 binding domain of HAUSP/USP7 bound to Epstein-Barr nuclear antigen 1: implications for EBV-mediated immortalization. Mol Cell. 2005;18(1):25–36. doi: 10.1016/j.molcel.2005.02.029. [DOI] [PubMed] [Google Scholar]
- 14.AlQarni S, Al-Sheikh Y, Campbell D, Drotar M, Hannigan A, Boyle S, et al. Lymphomas driven by Epstein–Barr virus nuclear antigen-1 (EBNA1) are dependant upon Mdm2. . Oncogene. . 2018;37(29):3998–4012. doi: 10.1038/s41388-018-0147-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Renouf B, Hollville E, Pujals A, Tetaud C, Garibal J, Wiels J. Activation of p53 by MDM2 antagonists has differential apoptotic effects on Epstein–Barr virus (EBV)-positive and EBV-negative Burkitt's lymphoma cells. Leukemia. . 2009;23(9):1557–63. doi: 10.1038/leu.2009.92. [DOI] [PubMed] [Google Scholar]
- 16.Ghotaslou A, Samii A, Boustani H, Ghalesardi OK, Shahidi M. AMG-232, a New Inhibitor of MDM-2, Enhance Doxorubicin Efficiency in Pre-B Acute Lymphoblastic Leukemia Cells. . Rep Biochem Mol Biol . 2022;11(1):111–124. doi: 10.52547/rbmb.11.1.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zheng X, Wang J, Wei L, Peng Q, Gao Y, Fu Y, et al. Epstein-Barr virus microRNA miR-BART5-3p inhibits p53 expression. . Virol J. . 2018;92(23):e01022–18. doi: 10.1128/JVI.01022-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Luo J, Su F, Chen D, Shiloh A, Gu W. Deacetylation of p53 modulates its effect on cell growth and apoptosis. Nature. . 2000;408(6810):377–81. doi: 10.1038/35042612. [DOI] [PubMed] [Google Scholar]
- 19.Shadfan M, Lopez-Pajares V, Yuan ZM. MDM2 and MDMX: Alone and together in regulation of p53. Transl Cancer Res. . 2012;1(2):88–89. [PMC free article] [PubMed] [Google Scholar]
- 20.Shvarts A, Steegenga WT, Riteco N, Van Laar T, Dekker P, Bazuine M, et al. MDMX: a novel p53-binding protein with some functional properties of MDM2. The EMBO J. 1996;15(19):5349–57. [PMC free article] [PubMed] [Google Scholar]
- 21.Danovi D, Meulmeester E, Pasini D, Migliorini D, Capra M, Frenk R, et al. Amplification of Mdmx (or Mdm4) directly contributes to tumor formation by inhibiting p53 tumor suppressor activity. Mol Cell Biol. . 2004;24(13):5835, 43. doi: 10.1128/MCB.24.13.5835-5843.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen Y, Fu L, Wen X, Wang XY, Liu J, Cheng Y, et al. Sirtuin-3 (SIRT3), a therapeutic target with oncogenic and tumor-suppressive function in cancer. Cell Death Dis. . 2014;5(2):e1047. doi: 10.1038/cddis.2014.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen J, Wang A, Chen Q. SirT3 and p53 deacetylation in aging and cancer. J Cell Physiol. . 2017;232(9):2308, 11. doi: 10.1002/jcp.25669. [DOI] [PubMed] [Google Scholar]
- 24.Kashyap D, Varshney N, Parmar HS, Jha HC. Gankyrin: At the crossroads of cancer diagnosis, disease prognosis, and development of efficient cancer therapeutics. . Adv Cancer Biol Metastasis. 2021;4:100023. [Google Scholar]
- 25.Dowran R, Sarvari J, Moattari A, Fattahi MR, Ramezani A, Hosseini SY. Analysis of TLR7, SOCS1 and ISG15 immune genes expression in the peripheral blood of responder and non-responder patients with chronic Hepatitis C. . Gastroenterol Hepatol Bed Bench. . 2017;10(4):272, 277. [PMC free article] [PubMed] [Google Scholar]
- 26.Hussain H, Raj S, Ahmad S, Razak MFA, Wan Mohamud WN, Bakar J, Ghazali HM. Determination of cell viability using acridine orange/propidium iodide dual-spectrofluorometry assay. . Cogent Food & Agriculture. 2019;5(1):158239. [Google Scholar]
- 27.Hassig CA, Schreiber SL. Nuclear histone acetylases and deacetylases and transcriptional regulation: HATs off to HDACs. . Curr Opin Chem Biol. . 1997;1(3):300, 8. doi: 10.1016/s1367-5931(97)80066-x. [DOI] [PubMed] [Google Scholar]
- 28.Yamaguchi H, Woods NT, Piluso LG, Lee HH, Chen J, Bhalla KN, et al. p53 acetylation is crucial for its transcription-independent proapoptotic functions. J. . Biol. Chem. 2009;284(17):11171, 83. doi: 10.1074/jbc.M809268200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Edwards RH, Dekroon R, Raab-Traub N. Alterations in cellular expression in EBV infected epithelial cell lines and tumors. PLoS Pathog. 2019;15(10):e1008071. doi: 10.1371/journal.ppat.1008071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Momand J, Zambetti GP, Olson DC, George D, Levine AJ. The mdm-2 oncogene product forms a complex with the p53 protein and inhibits p53-mediated transactivation. . cell . 1992;69(7):1237, 45. doi: 10.1016/0092-8674(92)90644-r. [DOI] [PubMed] [Google Scholar]
- 31.Oliner JD, Pietenpol JA, Thiagalingam S, Gyuris J, Kinzler KW, Vogelstein B. Oncoprotein MDM2 conceals the activation domain of tumour suppressor p53. . Nature. . 1993;362(6423):857, 60. doi: 10.1038/362857a0. [DOI] [PubMed] [Google Scholar]
- 32.Lotfi Garavand A, Mohammadi M, Mohammadzadeh S. Evaluation of TP53 Codon 72, P21 Codon 31, and MDM2 SNP309 Polymorphisms in Iranian Patients with Acute Lymphocytic Leukemia. . Rep Biochem Mol Biol. 2020;9(1):26, 32. doi: 10.29252/rbmb.9.1.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Marine JC, Jochemsen AG. MDMX (MDM4), a Promising Target for p53 Reactivation Therapy and Beyond. . Cold Spring Harb Perspect Med. . 2016;6(7):a026237. doi: 10.1101/cshperspect.a026237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ma Y, Walsh MJ, Bernhardt K, Ashbaugh CW, Trudeau SJ, Ashbaugh IY, et al. CRISPR/Cas9 screens reveal Epstein-Barr virus-transformed B cell host dependency factors. . Cell Host Microbe. . 2017;21(5):580–91.e7. doi: 10.1016/j.chom.2017.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhou L, Ouyang T, Li M, Hong T, Mhs A, Meng W, et al. Ubiquitin-Specific Peptidase 7: A Novel Deubiquitinase That Regulates Protein Homeostasis and Cancers. Front Oncol. 2021;11:784672. doi: 10.3389/fonc.2021.784672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bhattacharya S, Chakraborty D, Basu M, Ghosh MK. Emerging insights into HAUSP (USP7) in physiology, cancer and other diseases. . Signal Transduct Target Ther. . 2018;3(1):1, 12. doi: 10.1038/s41392-018-0012-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.He Y, Li W, Lv D, Zhang X, Zhang X, Ortiz YT, et al. Inhibition of USP7 activity selectively eliminates senescent cells in part via restoration of p53 activity. . Aging Cell. . 2020;19(3):e13117. doi: 10.1111/acel.13117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Agathanggelou A, Smith E, Davies NJ, Kwok M, Zlatanou A, Oldreive CE, et al. USP7 inhibition alters homologous recombination repair and targets CLL cells independently of ATM/p53 functional status. Blood. . 2017;130(2):156, 66. doi: 10.1182/blood-2016-12-758219. [DOI] [PubMed] [Google Scholar]
- 39.Li S, Banck M, Mujtaba S, Zhou M-M, Sugrue MM, Walsh MJ, et al. p53-induced growth arrest is regulated by the mitochondrial SirT3 deacetylase. . PloS one. 2010;5(5):e10486. doi: 10.1371/journal.pone.0010486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jing H, Hu J, He B, YL NA, Stupinski J, Weiser K, et al. A SIRT2-selective inhibitor promotes c-Myc oncoprotein degradation and exhibits broad anticancer activity. Cancer cell. . 2016;29(3):297, 310. doi: 10.1016/j.ccell.2016.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Higashitsuji H, Higashitsuji H, Itoh K, Sakurai T, Nagao T, Sumitomo Y, et al. The oncoprotein gankyrin binds to MDM2/HDM2, enhancing ubiquitylation and degradation of p53. . Cancer Cell. 2005;8(1):75, 87. doi: 10.1016/j.ccr.2005.06.006. [DOI] [PubMed] [Google Scholar]
- 42.Wang C, Li Y, Chu CM, Zhang XM, Ma J, Huang H, et al. Gankyrin is a novel biomarker for disease progression and prognosis of patients with renal cell carcinoma. EBioMedicine. . 2019;39:255, 64. doi: 10.1016/j.ebiom.2018.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kashyap D, Baral B, Jakhmola S, Singh AK, Jha HC. Helicobacter pylori and Epstein-Barr Virus Coinfection Stimulates Aggressiveness in Gastric Cancer through the Regulation of Gankyrin. mSphere. . 2021;6(5):e0075121. doi: 10.1128/mSphere.00751-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ribeiro J, Malta M, Galaghar A, Silva F, Afonso LP, Medeiros R, Sousa H. P53 deregulation in Epstein-Barr virus-associated gastric cancer. Cancer Lett. 2017;404:37, 43. doi: 10.1016/j.canlet.2017.07.010. [DOI] [PubMed] [Google Scholar]


