Summary
Microglia exhibit diverse phenotypes in various central nervous system disorders and metabolic pathways exert crucial effects on microglial activation and effector functions. Here, we discovered two novel distinct microglial clusters, functionally associated with enhanced phagocytosis (PEMs) and myelination (MAMs) respectively, in human patients with multiple sclerosis by integrating public snRNA-seq data. Microglia adopt a PEMs phenotype during the early phase of demyelinated lesions, predominated in pro-inflammatory responses and aggravated glycolysis, while MAMs mainly emerged during the later phase, with regenerative signatures and enhanced oxidative phosphorylation. In addition, microglial triggering receptor expressed on myeloid cells 2 (Trem2) was greatly involved in the phenotype transition in demyelination, but not indispensable for microglia transition toward PEMs. Rosiglitazone could promote microglial phenotype conversion from PEMs to MAMs, thus favoring myelin repair. Taken together, these findings provide insights into therapeutic interventions targeting immunometabolism to switch microglial phenotypes and facilitate regenerative capacity in demyelination.
Subject areas: Neuroscience, Cellular neuroscience, Immunology, Cell biology
Graphical abstract
Highlights
-
•
Microglia loaded with lipid adopt a PEMs phenotype
-
•
Rosiglitazone helps microglia adopt MAMs phenotype
-
•
Rosiglitazone facilitates glucose metabolism conversion by enhancing PPAR-γ signaling
Neuroscience; Cellular neuroscience; Immunology; Cell biology
Introduction
Multiple sclerosis (MS) is characterized by inflammation and demyelination in the CNS.1 Microglia, the brain-resident immune cells, are crucial for MS neuropathology as active demyelination during acute attacks is associated with a microglial pro-inflammatory phenotype.2 Interestingly, microglia act as a double-edged sword in both CNS damage and repair, which shows their ability to acquire myelin microenvironment-specific activation states. Early lesions refer to the early stages of demyelination diseases. In early lesions, microglia predominantly express several differential markers associated with phagocytosis, oxidative injury, and antigen presentation, while in late stages, the major process that takes place is remyelination and tissue repair.2 In addition, microglia are increasingly recognized for their pro-remyelination properties in MS pathogenesis.3 This activation is dynamically regulated during myelin damage and repair, with a switch between pro-inflammatory/pro-demyelination states and inflammation-resolution/pro-remyelination phenotypes. However, the mechanisms underlining this plasticity in microglial activation are still being uncovered.
Emerging studies of immunometabolism have focused on regulating microglial functional states in controlling immunity and inflammation in CNS.4,5 Usually, different cell phenotypes utilize distinctive metabolic pathways in correspondence to functional alterations. An emerging theory is that these phenotypic transitions are mediated by metabolic reprogramming.6 For demyelinating lesions, it is obvious that microglia engulfing myelin debris would face an elevated lipid load to digest, pushing the catabolic machinery to, or over, their limit.7
In this study, we investigated microglia-specific immunometabolism in demyelinated lesions and its role in inflammation and remyelination. In order to comprehensively reveal the transcriptional profiles of microglia in multiple sclerosis, via integrating and re-analyzing publicly available single-nuclei RNA-seq data, we first identified an enrichment of two distinct MS-specific subgroups of human microglia from MS brain tissues, microglial subsets highly associated with enhanced phagocytosis (phagocytosis-enhanced microglia, PEMs) and myelination (myelination-associated microglia, MAMs), respectively, with differential activation of cytokine secretion, phagocytosis, glycolipid metabolism, and regenerative properties. Next, in a reverse translational approach, we provided evidence for myelin-related morphological and transcriptional changes in microglia in animal model of lysophosphatidyl choline (LPC)-induced demyelination, and observed profound intrinsic differences in the transcriptome and metabolism between the two microglial subsets. We further demonstrated that modulation of microglial metabolism could promote the switch from PEMs to MAMs. Consequently, we turned our attention to several particular metabolic processes. Since it has been found that PPAR pathways are associated with phenotypic transformation and closely related to glycolipid metabolism in microglia by RNA sequencing, we subsequently focused on PPAR-γ pathway to target microglial immunometabolism as potential therapeutic strategies for myelin repair.
Results
Specific microglial subpopulations related to enhanced phagocytosis and myelination identified in MS
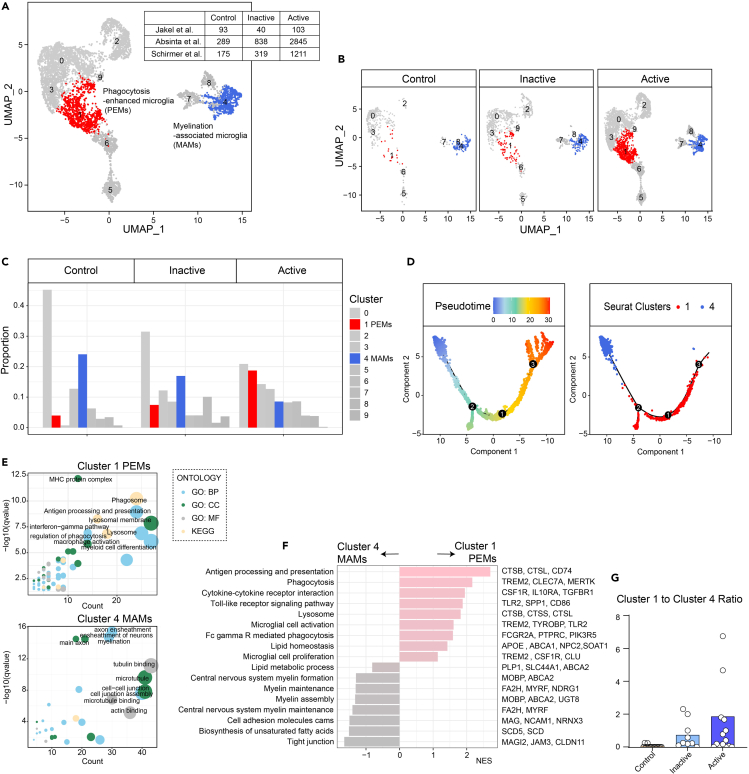
Microglial heterogeneity has been identified both in human MS and in animal models of demyelination.1,8,9,10,11 To this end, we re-analyzed single-cell gene expression profiles from three recent studies (GSE118257, GSE180759 and data from UCSC website).8,12,13 The datasets included white matter from healthy controls, inactive lesions and active lesions from individuals with MS (Cohort characteristics shown in Table S2). We combined all three datasets and analyzed them using unsupervised uniform manifold approximation and projection (UMAP) analysis. 19 clusters were generated. Microglia/macrophages belonged to cluster 3. Cluster 3 was more abundant in MS lesions, particularly the active ones (Figure S1).
Subsequently, we subclustered the population of microglia (pooled from all samples, a total of 5,913 nuclei). Unsupervised clustering yielded ten subclusters among all samples (Figure 1A). We identified two microglial subpopulations (Clusters 1 and 4) that were enriched in either active or inactive lesions (Figures 1B–1D). Cluster 1 was greatly enriched in active lesions (Figures 1B and 1C), and featured hallmark “phagocytosis”, “microglia/macrophages activation”, and “antigen processing and presentation” genes such as TREM2, APOE, CTSB, MERTK, and TLR2, indicating a specific population of microglia associated with higher levels of phagocytosis and microglial activation (Figures 1E and 1F and Table S3). In contrast, Cluster 4 showed high levels of genes involved in regulation of myelin formation and maintenance, such as MOBP, FA2H, MYRF, NDRG1, and others (Figures 1E and 1F). We designated this cluster as MAMs, considering its functions related to axon ensheathment and myelination, defined by gene set enrichment analysis (Figures 1E and 1F and Table S3). Understanding the differences among these “activated” subclasses of microglia (Clusters 1 and 4) could help better understand the phenotypic transition of differentially activated microglia. Compared with Cluster 4 “remyelination” microglia, a prominent feature of Cluster 1 microglia was upregulation of phagocytosis genes as well as cytokine pathways, and downregulation of myelin assembly genes (Figure 1F).
Figure 1.
Re-analysis of microglia in human MS single-nuclei RNA-sequencing datasets
(A) UMAP and unsupervised clustering of microglia subclusters from human MS brain single-nuclei RNA-sequencing (snRNA-seq) datasets were performed for a total 10 clusters.8,12,13 Of these, cluster 1 was designated as phagocytosis-enhanced microglia (PEMs) and cluster 4 was myelination-associated microglia (MAMs).
(B) Microglia subclusters in healthy controls, MS inactive lesions, and MS active lesions.
(C) Relative frequency of microglia subclusters in different groups.
(D) Branched pseudo-time trajectory, which contains 3 roots, each cell being colored by its pseudo-time value (left) and its Seurat clusters (right).
(E) Gene enrichment and pathway analysis of the differentially expressed genes selected by “FindAllMarkers” function are shown for Cluster 1 and Cluster 4, separately. Only significant terms are plotted in the graphs (p < 0.05, q < 0.05, correction methods “Benjamini and Hochberg FDR (BH)”).
(F) Top upregulated and downregulated pathways with featured genes in phagocytosis-enhanced microglia (PEMs) (Cluster 1) compared with myelination-associated microglia (MAMs) (Cluster 4).
(G) Ratio of microglial cluster 1 to cluster 4 in different MS groups of human postmortem tissue samples.
Notably, the ratio of Cluster 1 to 4 was elevated in active lesions compared with inactive lesions or healthy control, suggesting an underlying key role of the phenotypic transition in MS acute attacks.
In summary, these data strongly support our conclusion that specific clusters of microglia/macrophage-like cells (i.e. PEMs and MAMs) could be identified in brain tissues from patients with MS, suggesting that these microglia/macrophage subsets were highly involved in disease progression.
Microglia are irreplaceable for spontaneous remyelination in demyelinated lesions
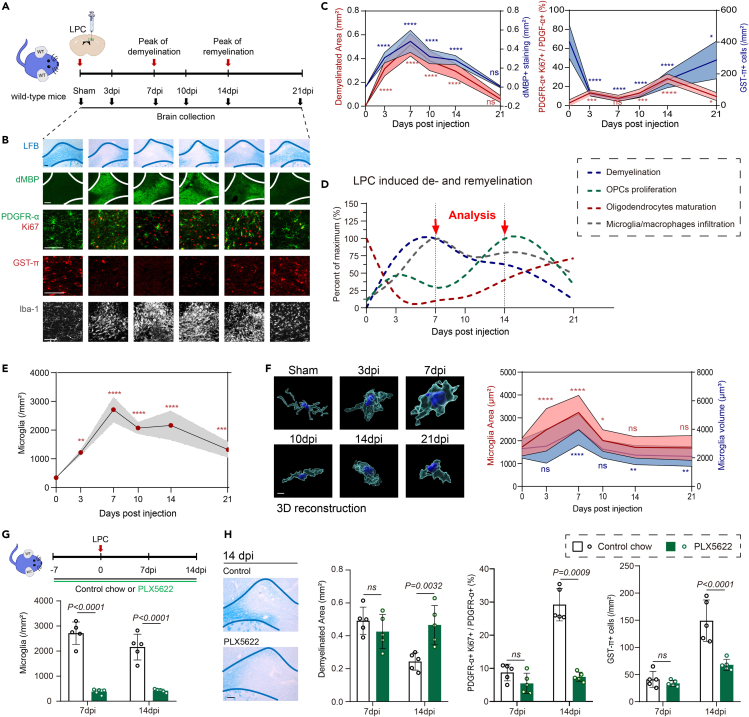
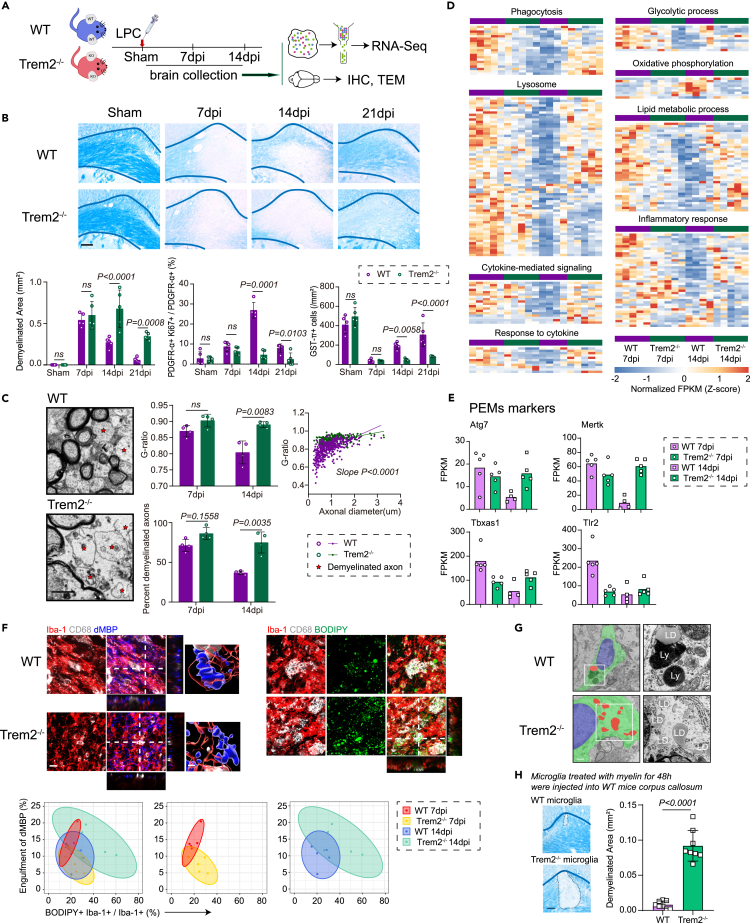
To characterize these microglial subgroups in disease progression, we utilized the LPC mouse model which induces focal demyelination lesions in the corpus callosum where regeneration occurs after demyelination mimicking acute attacks. This model’s timing also allows for clear definition of microglia activation status (Figures 2A–2F). Quantification of Luxol fast blue staining throughout the 3 weeks after LPC injection revealed significant demyelination pathology 7 days post injection (dpi), which recovered at 21 dpi (Figures 2A–2D). At peak demyelination (7 dpi), immunostaining of the microglia and macrophage marker ionized calcium-binding adaptor 1 (Iba-1) and three-dimensional (3D) surface rendering displayed the highest increase in Iba-1+ cells at the demyelinated lesions as well as morphological characteristics of significantly enlarged surface area and total volume (Figures 2B–2F). During the process of myelin repair, oligodendrocyte precursor cell (OPC) proliferation precedes oligodendrocyte differentiation and remyelination.10 PDGFR-α+ Ki67+ proliferating OPCs reached the peak at 14 dpi, accompanied with decreased microglia densities and reduced total volume compared with those at 7 dpi (Figures 2B–2D). Next, the potential role of microglia in LPC-induced myelin damage and repair was explored using microglia ablation approaches. To this end, mice were given PLX5622-formulated chow for microglia elimination14,15 or control diet continuously until 7–14 dpi. Oral PLX5622 administration led to gross microglial depletion (80%–90% reduction; Figure 2G), even with LPC treatment. However, depletion of microglia did not induce measurable effects on myelin damage at 7 dpi, but did significantly enlarge demyelinated lesions at 14 dpi, indicating that microglia are irreplaceable for spontaneous remyelination in demyelinated lesions (Figure 2H).
Figure 2.
Microglia play an essential role in CNS demyelination and remyelination
(A) Schematic depicting the experimental design for lysophosphatidyl choline (LPC) injection model and sample collection.
(B) Representative images of LFB staining, dMBP (green), PDGFR-α (green), and Ki67 (red), GST-π (red), and Iba-1 (white) immunostaining following LPC injection. Scale bar, 100 μm.
(C) Quantification of demyelinated area in LFB staining, dMBP staining intensity, the percentage of proliferating OPCs (PDGFR-α+Ki67+/PDGFR-α+ cells), and mature oligodendrocytes (GST-π+) densities. n = 5 mice per group, mean ± SD, ns not significant, ∗p < 0.05, ∗∗∗p < 0.001, and ∗∗∗∗p < 0.0001 vs. sham group, one-way ANOVA followed by Bonferroni’s post hoc test.
(D) The graphical representation of demyelination, OPCs proliferation, oligodendrocytes maturation, and microglia/macrophages infiltration of the LPC model is based on histochemical staining as shown in panel B and C.
(E) Quantification of microglia/macrophages (Iba-1+) densities. n = 5 mice per group, mean ± SD, ∗∗p < 0.01, ∗∗∗p < 0.001, and ∗∗∗∗p < 0.0001 vs. sham group, one-way ANOVA followed by Bonferroni’s post hoc test.
(F) Representative images of microglial morphology at different time points. Quantification of microglia surface area and volume. n = 5 mice per group, mean ± SD, ns not significant, ∗p < 0.05, ∗∗p < 0.01, and ∗∗∗∗p < 0.0001 vs. sham group, one-way ANOVA followed by Bonferroni’s post hoc test. Scale bar, 10 μm.
(G) Experimental design for mice given PLX5622-formulated chow or control chow. Quantification of microglia densities at 7 and 14 dpi. n = 5 mice per group, mean ± SD, two-way ANOVA followed by Bonferroni’s post hoc test.
(H) Representative images of LFB staining of brain samples treated with or without PLX5622 chow at 14 dpi. Scale bar, 100 μm n = 5 mice per group. Quantification of demyelinated area, the percentage of proliferating OPCs (PDGFR-α+Ki67+/PDGFR-α+ cells), and mature oligodendrocytes (GST-π+) densities. n = 5 mice per group, mean ± SD, two-way ANOVA followed by Bonferroni’s post hoc test.
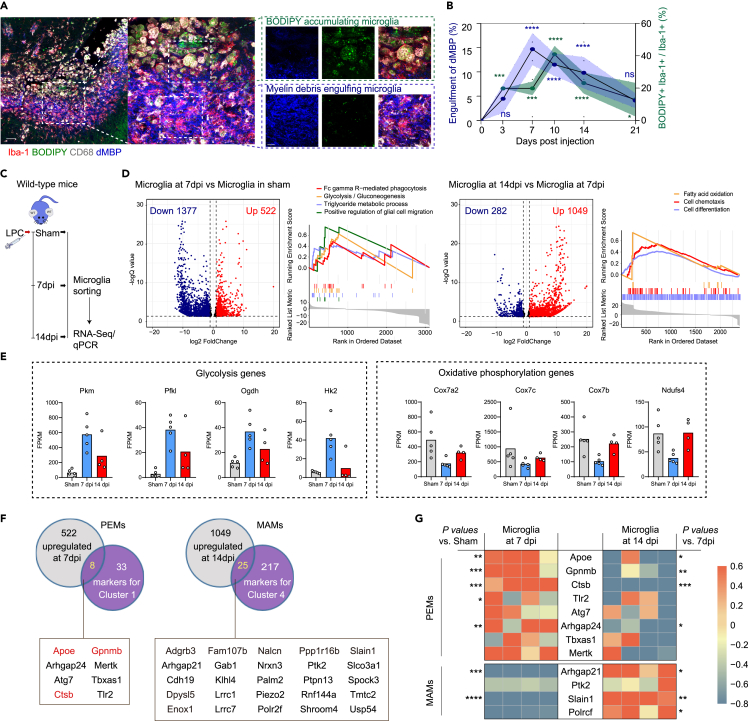
Using co-immunostaining 3D reconstruction, we further revealed that maximal amounts of dMBP+ myelin debris were engulfed by Iba-1+ cells at 7 dpi, and that lipid droplets accumulation reached the peak at 10 dpi, and then gradually degraded (Figures 3A and 3B). To determine the existence of our identified microglial clusters, PEMs and MAMs, in LPC model, we performed RNA sequencing of microglia isolated from focal demyelinated lesions at the peak of demyelination (7 dpi) and at the peak of remyelination (14 dpi) (Figures 3C–3E). During demyelination, microglia showed distinct patterns of gene expression with functions in phagocytosis, lipid metabolism, inflammation, and glycolysis, in concordance with PEMs identified in human MS lesions. For PEMs, those genes including Apoe, Ctsb, Tlr2, and Mertk were upregulated, while the anti-inflammatory gene Tgfb1 and the regenerative genes Vegfb were downregulated (Figures 3C–3G and Tables S4 and S5). Gene set enrichment analysis of the isolated microglia at 14 dpi highlighted the functional enrichment for resolution in inflammation and pro-myelination, in line with MAMs’ expression profiles from humans. The MAMs markers Arhgap21, Ptk2, Stain1, and Polrcf were upregulated, while the pro-inflammatory genes Cd68 and Il6ra, and the PEMs markers were downregulated (Figures 3C–3G and Tables S4 and S5). Notably, the predominant immunometabolism pattern of microglia at 7 dpi was glycolysis, and then switched to oxidative phosphorylation and fatty acid oxidation at 14 dpi (Figures 3D and 3E). These data raise the possibility that the two distinct microglial subsets, PEMs and MAMs, are highly related to the orchestrated formation and resolution of cellular myelin debris engulfment and lipid droplets, and the underlying metabolic alterations.
Figure 3.
Phagocytosis-enhanced microglia and myelination-associated microglia orchestrate acute demyelination and subsequent spontaneous remyelination
(A and B) Representative images of Iba-1 (red), BODIPY (green), CD68 (white), and dMBP (blue) immunostaining at 7 days post LPC injection. Scale bar, 50, 20, and 10 μm from left to right, respectively. Quantification of dMBP within microglia (blue) and the percentage of BODIPY+ Iba-1+ in Iba-1+ cells (green) after LPC injection. n = 5 mice per group, mean ± SD, ns not significant, ∗p < 0.05, ∗∗∗p < 0.001, and ∗∗∗∗p < 0.0001 vs. sham group, one-way ANOVA followed by Bonferroni’s post hoc test.
(C) Schematic depicting the experimental design. Microglia were isolated from sham control corpus callosum or focal LPC-demyelinated lesions at 7 and 14 dpi, n = 4–5 biologically independent replicates. Key time points for demyelination and remyelination, respectively, for RNA sequencing and PCR analysis.
(D) Volcano plots report differentially expressed genes for microglia at 7 dpi vs. sham, and microglia at 14 dpi vs. microglia at 7 dpi. Positively enriched gene sets identified by GSEA. Numerical data with statistics are shown in Table S4.
(E) Expression levels of glycolysis and oxidative phosphorylation-related genes in Sham, 7 days post-injury and 14 days post-injury mouse microglia.
(F) Venn diagrams indicate significantly differentially expressed genes in each comparison, and report overlapping genes as markers for PEMs and MAMs.
(G) Quantitative RT-PCR analysis of select marker expression in isolated microglia. Gene expression (fold change) was normalized to β-Actin. Each square represents data obtained from one mouse. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001 and ∗∗∗∗p < 0.0001, one-way ANOVA followed by Bonferroni’s post hoc test. Numerical data with statistics are shown in Table S5.
Metabolic reprogramming underlies microglia reactive phenotypes
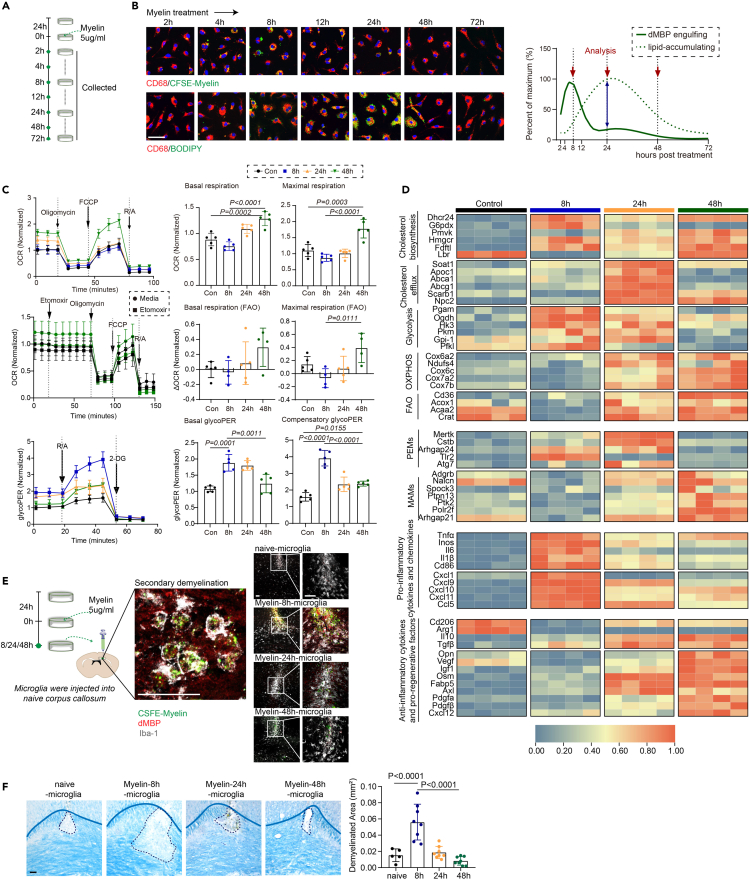
Next, we tried to explore the metabolism requirements for myelin debris phagocytosis, clearance, and remyelination. After primary microglia were exposed to purified myelin debris, debris engulfment and formation of lipid droplets were observed (Figures 4A and 4B). Myelin debris stimulation induced higher expression of “glycolysis” genes in microglia, while most of OXPHOS/FAO-related genes slightly downregulated. This led to a significant increase in the basal proton efflux rate related with glycolysis, suggesting energy dependence on glycolysis at the peak of myelin engulfment in microglia (“engulfment” microglia, 8 h after treatment) (Figures 4C and 4D and Table S6). In contrast, 48 h after myelin debris stimulation, microglia at lipid resolution stage (“resolution” microglia) showed increased expression of “OXPHOS” genes, together with increased O2 consumption rate. Concordantly, the expression levels of inflammation and immune response genes together with PEMs markers were reduced, and the expression of pro-regenerative genes and MAMs markers were elevated in “resolution” microglia compared to “engulfment” microglia (Figure 4D and Table S6). Together, our results indicate a close relationship between microglial immunometabolism balance and functional status.
Figure 4.
Metabolic reprogramming underlies the identified microglia phenotypes during demyelination
(A) Schematic depicting the strategy of myelin debris treatment and sample collection.
(B) Representative images of CFSE-Myelin (green), and BODIPY (green), co-stained with CD68 (red) in microglia with myelin debris treatment. The graphical representation of microglial myelin debris engulfment and BODIPY accumulation is based on histochemical staining (left). Scale bar, 50 μm.
(C) Oxygen consumption rate (OCR) and extracellular acidification rate (ECAR) were measured over time using a Seahorse XFe24 analyzer. Quantification of basal OCR, maximal OCR, basal glycoPER, and compensatory glycoPER. n = 4–5 biologically independent replicates, mean ± SD, one-way ANOVA followed by Bonferroni’s post hoc test.
(D) Quantitative RT-PCR analysis of gene expression of microglia treated with myelin debris at different time points (8, 24, and 48 h after myelin treatment). Each square represents data obtained from one biologically independent replicate. Numerical data with statistics are shown in Table S6.
(E and F) Schematic depicting the strategy of microglia injection. Immunostaining of demyelinated area (dMBP, red) in naive corpus callosum injected with CFSE-labeled myelin (green)-engulfed microglia (Iba-1, white). Scale bar, 50 μm. Representative images of LFB staining, and CFSE-Myelin (green), dMBP (red), and Iba-1 (white) immunostaining in corpus callosum injected with microglia at different time points after myelin debris treatment. Scale bar, 50 μm. Quantification of demyelinated area. n = 5–8 mice per group, mean ± SD, one-way ANOVA followed by Bonferroni’s post hoc test.
To understand the function of myelin-engulfed microglia, we collected myelin-treated microglia at different time points (8, 24, and 48 h) and performed cell injection into naive corpus collosum. Interestingly, we found that injection of “engulfment” microglia (8 h after myelin stimulation) led to severe demyelinated lesions enlargement (Figures 4E and 4F). In contrast, myelin-24-h microglia and myelin-48-h microglia (“resolution” microglia) only induced minor demyelination (Figures 4E and 4F). Thus, these results support a possible role of PEMs in propagation of inflammatory damage and demyelination.
Trem2-mediated microglial phagocytosis and metabolism are involved in active demyelination, but are not indispensable
Previous works have shown that triggering receptor expressed on myeloid cells 2 (Trem2) is key to microglia activation upon various neuropathological disorders.16,17 Our human single-nucleus RNA sequencing (snRNA-seq) analysis indeed revealed that Trem2 was upregulated in PEMs (Figure 1F). We compared Trem2−/− mice with the wild-type controls during LPC-induced demyelination. As expected, histological analysis showed remyelination deficits in the Trem2−/− group (Figures 5A and 5B). Proliferating OPC density was also approximately six times higher in wild-type mice when compared to Trem2−/− mice at 14 dpi (Figure 5B). Trem2−/− mice exhibited stronger inhibition of oligodendrocyte maturation (GST-π+), leading to significantly lower remyelination than wild-type mice, as shown by the G-ratio and percentage of demyelinated axons (Figures 5B and 5C).
Figure 5.
Trem2-mediated microglial phagocytosis and metabolism were deeply involved in active demyelination, but not required
(A) Schematic depicting the experimental design for LPC injection model and sample collection.
(B) Representative images of LFB staining. Scale bar, 100 μm. Quantification of demyelinated area, the percentage of proliferating OPCs (PDGFR-α+Ki67+/PDGFR-α+ cells), and mature oligodendrocytes (GST-π+) densities. n = 5 mice per group, mean ± SD, two-way ANOVA followed by Bonferroni’s post hoc test.
(C) Representative transmission electron microscopy (TEM) image of the corpus callosum in Trem2 −/− and Wild-Type (WT) mice, showing demyelinated axons (stars). Scale bar, 1 μm. Quantification of g-ratios and percentage of demyelinated axons in Trem2 −/− compared with WT mice at 14 dpi. n = 4 mice per group, mean ± SD, two-way ANOVA followed by Bonferroni’s post hoc test.
(D) Transcriptome analysis was performed using RNA sequencing of microglia sorted from demyelinated lesions in Trem2−/− and control mice at 7 and 14 dpi. Heatmap showing concurrent regulation of genes within specific functional signaling pathway. Each row represents data obtained from one mouse. n = 4–5 mice per group. Numerical data with statistics are shown in Table S4.
(E) Quantification of relative expression of PEMs markers in WT and Trem2−/− mice at 7 and 14 dpi. n = 4–5 mice per group.
(F) Representative images of Iba-1 (red), CD68 (white), dMBP (blue), and BODIPY (green) immunostaining in Trem2−/− mice and wild-type control at 7 and 14 dpi. Quantified histochemical staining for engulfment of dMBP and the percentage of BODIPY+ Iba-1+ in Iba-1+ cells is shown as 95% confidence ellipses with data points of individual animals. n = 5 mice per group. Scale bar, 20μm.
(G) Representative TEM image showing the morphology of microglia in Trem2−/− and WT mice. n = 4 mice per group. Lysosomes (Ly), and lipid droplets (LD). Scale bar, 1μm.
(H) Representative images of LFB staining in normal corpus callosum injected with Trem2−/−microglia or wild-type control microglia treated with myelin debris for 48 h. Scale bar, 100 μm. Quantification of demyelinated area. n = 8 mice per group, mean ± SD, unpaired t-test.
To comprehensively understand the consequences of Trem2 deletion during demyelination, we performed RNA sequencing of microglia sorted from Trem2-deficient and wild-type mice in sham, 7 and 14 dpi groups. We detected 541 and 2365 differentially expressed genes (adjusted p value <0.05; absolute value of log2 (Fold change) > 1) between Trem2-deficient mice and wild-type group at days 7 and 14, respectively (Figures 5D and 5E and Table S4).
In accordance, microglia containing myelin debris were much less frequently seen in Trem2−/− mice at 7 dpi, indicating that Trem2 deficiency attenuated phagocytosis during the early stage (Figure 5F). Interestingly, Trem2 deficiency induced transcriptional downregulation of genes for lipid metabolism (Figures 5D and 5E). These results suggested potential compensatory pathways of phagocytosis and impaired lipid metabolism in Trem2−/− microglia, which gradually contributed to lipid droplets accumulation at a later stage (Figures 5F and 5G). The lipid droplets-accumulating Trem2−/− cells (Figure 5G) exhibited PEMs markers and could cause secondary demyelinated injury when injecting into naive corpus callosum (Figures 5E and 5H), suggesting that Trem2 was not indispensable for microglia transition toward PEMs.
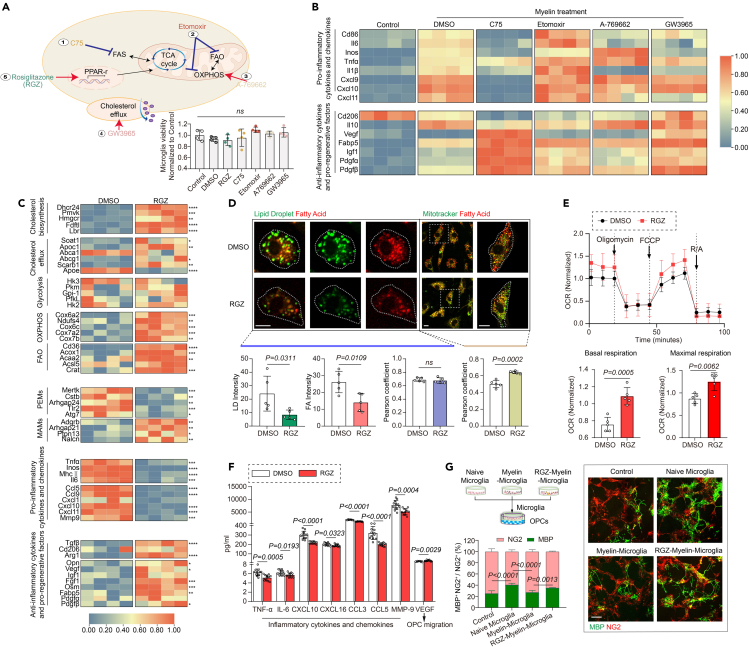
Modulating immunometabolism efficiently regulates the functional phenotype of microglia
Immune activation is usually accompanied by an increase in glycolytic flux.5 To determine whether the microglial diversity after engulfing myelin debris depends on glycolipid metabolism, we exposed debris-treated microglia to a variety of glycolipid metabolism regulators that function on different pathway components, including CPT-1 inhibitor etomoxir (which blocks mitochondrial fatty acid oxidation, FAO, and OXPHOS),18 fatty acid synthase inhibitor C75 (which blocks fatty acid synthesis, FAS),19 AMPK agonist A-769662 (which facilitates OXPHOS),20 and LXR agonist GW3965 (which facilitates lipid efflux) (Figure 6A).21,22 Transcriptional levels of genes related to microglial pro-inflammatory cytokines and chemokines, and anti-inflammatory cytokines and pro-regenerative factors were detected by qPCR. We found that blocking FAS by C75, stimulation of OXPHOS by A-769662, or enhancement of cholesterol efflux by GW3965, could suppress the expression of pro-inflammatory cytokines induced by myelin debris, and facilitate the pro-regenerative factors expression in microglia. However, inhibition of FAO and OXPHOS by etomoxir aggravated microglial inflammation (Figure 6B and Table S7). These data raise the possibility that modulation of microglia metabolism could help resolving inflammation and promoting remyelination.
Figure 6.
Modulating metabolism can efficiently regulate the functional phenotype of microglia
(A) Schematic depicting the strategy of cellular metabolism modulation in microglia treated with myelin debris for 8 h. Normalized values of CCK8 were utilized to evaluate cell viability under different treatment.
(B) Quantitative RT-PCR analysis of gene expression in microglia treated with myelin debris and different metabolism modulators: fatty acid synthase inhibitor C75, CPT1 inhibitor etomoxir, AMPK activator A-769662, and LXR agonist GW3965. Each square represents data obtained from one biologically independent replicate. Numerical data with statistics are shown in Table S7.
(C) Quantitative RT-PCR analysis of gene expression in myelin debris-treated microglia with or without rosiglitazone. Each square represents data obtained from one biologically independent replicate. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001 and ∗∗∗∗p < 0.0001, unpaired t-test. Numerical data with statistics are shown in Table S8.
(D) Representative images of lipid droplets (LD, green) or mitochondria (green) co-staining with fatty acid (FA, red) in myelin debris-treated microglia with or without rosiglitazone. Scale Bar, 10 μm. Quantification of intensity of LD and FA, and the co-localization of LD and FA, and mitochondria with FA shown as Pearson coefficient. n = 5 biologically independent replicates, mean ± SD, unpaired t-test.
(E) Oxygen consumption rate (OCR) measured over time using a Seahorse XFe24 analyzer. Quantification of basal respiration and maximal respiration. n = 5 biologically independent replicates, mean ± SD, unpaired t-test.
(F) Quantification of cytokines and chemokines measured by Luminex assay. n = 12 biologically independent replicates, mean ± SD, one-way ANOVA followed by Bonferroni’s post hoc test.
(G) Schematic indicates primary microglia were treated with myelin debris with or without rosiglitazone for 8 h and then co-cultured with OPCs for an additional 3 days. Representative images and quantification of MBP+ (green) and NG2+ cells (red) as percentages of total NG2+ cells. Scale bar, 100 μm. n = 5–6 biologically independent replicates, mean ± SD, two-way ANOVA followed by Bonferroni’s post hoc test.
Rosiglitazone promotes remyelination via enhancing microglial OXPHOS
Peroxisome proliferators-activated receptors (PPARs) could be classified into different subtypes, of which PPAR-γ is shown to influence both lipid and glucose metabolism.23 For the potential translation clinically, we asked whether the PPAR-γ agonist rosiglitazone,24 a drug widely prescribed for type 2 diabetes, might exert the effects of immunometabolism modulation in vitro and in vivo. To test this idea, we treated myelin-engulfed microglia with rosiglitazone. As expected, rosiglitazone-treated microglia exhibited a transcriptional profile of elevated MAMs markers, with enhanced “OXPHOS/FAO” genes (Figure 6C). Consistently, rosiglitazone treatment decreased lipid droplets accumulation in cultured microglia, and increased fatty acid in mitochondria (Figure 6D), with increased levels of cellular O2 consumption rate (Figure 6E). More interestingly, rosiglitazone attenuated pro-inflammatory cytokine release, and promoted the secretion of pro-remyelination factors (Figures 6C and 6F and Table S8). The impact of rosiglitazone-treated microglia on OPC differentiation was explored using a primary co-culture system (Figure 6G). Naive microglia induced the differentiation of NG2+ OPCs into MBP+ mature oligodendrocytes while the myelin-engulfed microglia failed. To note, rosiglitazone-treated microglia recovered a comparable pro-remyelination capacity similar to naive microglia (Figure 6G). These data suggest that the enhancement of mitochondrial function by rosiglitazone treatment in debris-engulfed microglia promotes the restoration of myelin regeneration capacity, further implying its therapeutic potentials in human demyelinated-related diseases.
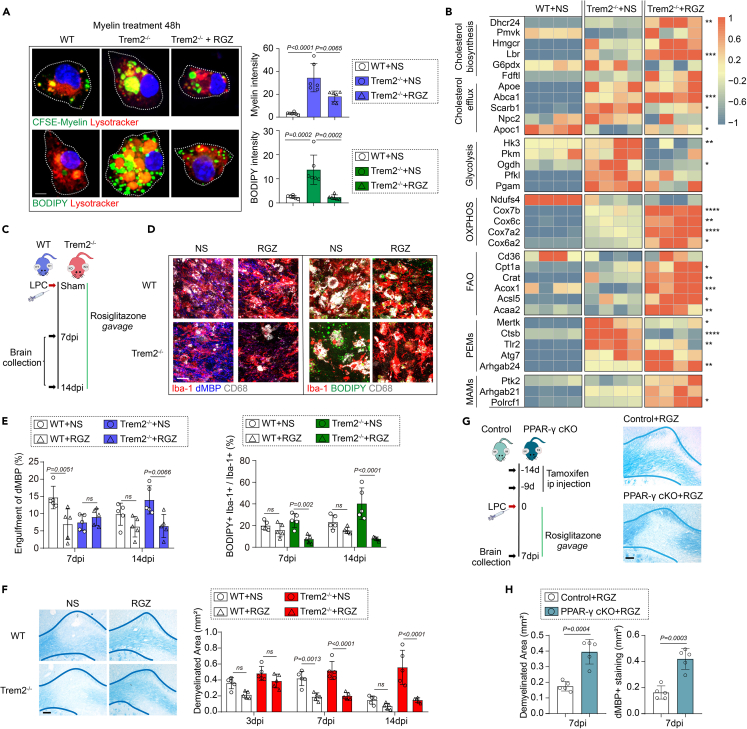
We then tested whether rosiglitazone would also be sufficient to restore the regenerative properties of Trem2-deficient microglia. Significantly higher accumulation of myelin debris and lipid droplets was observed in Trem2−/− microglia than in the control group, 48 h after myelin debris stimulation, accompanied by higher levels of “glycolysis” markers and lower expression of “OXPHOS” genes (Figures 7A and 7B and Table S9). Notably, rosiglitazone administration in Trem2-deficient microglia switched this trend from glycolysis toward OXPHOS/FAO state, as well as promoting cholesterol metabolism. In line with the process of metabolic reprogramming, rosiglitazone treatment facilitated the transition of microglia from PEMs to MAMs, and restored their regenerative capacity both in Trem2−/− microglia and wild-type controls (Figures 6C and 7B and Tables S8 and S9). Finally, rosiglitazone-treated microglia, whether Trem2 deficient or not, exhibited less myelin debris and lipid droplets accumulation both in vivo and in vitro, although without significance in wild-type groups (Figures 7C–7E). Rosiglitazone treatment facilitated remyelination in vivo, independent of Trem2 (Figure 7F), whereas microglia-specific PPAR-γ deficiency abrogated the effect of rosiglitazone on remyelination (Figures 7G and 7H). These data suggest that the mechanisms by which rosiglitazone restored microglial regenerative properties after demyelination were at least in part, through the PPAR-γ pathway.
Figure 7.
Rosiglitazone treatment restores regenerative microglial properties by enhancing OXPHOS function
(A) Representative images of Myelin (green) or BODIPY (green) co-stained with Lysotracker (red) in myelin-treated wild-type controls, or Trem2−/− microglia with or without rosiglitazone. Scale Bar, 10um. Quantification of microglial myelin debris engulfment and BODIPY accumulation. n = 6 biologically independent replicates, mean ± SD, two-way ANOVA followed by Bonferroni’s post hoc test.
(B) Quantitative RT-PCR analysis of gene expression in myelin-treated wild-type controls, or Trem2−/− microglia with or without rosiglitazone. Each square represents data obtained from one biologically independent replicate. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001 and ∗∗∗∗p < 0.0001 for Trem2−/− microglia with rosiglitazone vs. Trem2−/− microglia with NS, two-way ANOVA followed by Bonferroni’s post hoc test. Numerical data with statistics are shown in Table S9.
(C) Schematic depicting the experimental design for rosiglitazone treatment in Trem2 −/− mice or wild-type controls and sample collection, n = 5 mice per group.
(D) Representative images of Myelin (blue) or BODIPY (green) co-stained with Iba-1 (red) and CD68 (white) in Trem2−/− or wild-type mice with rosiglitazone treatment or with normal saline (NS). Scale Bar 20 μm.
(E) Quantification of dMBP within microglia and the percentage of BODIPY+Iba-1+ in Iba-1+ cells. n = 5 mice per group, mean ± SD, two-way ANOVA followed by Bonferroni’s post hoc test.
(F) Representative images of LFB staining in Trem2−/− mice or wild-type controls with or without rosiglitazone treatment at 14 dpi. Scale Bar, 100 μm. Quantification of demyelinated area. n = 5 mice per group, mean ± SD, two-way ANOVA followed by Bonferroni’s post hoc test.
(G and H) Schematic depicting the experimental design for rosiglitazone treatment in microglia-specific PPAR-γ-deficient mice (Cx3Cr1CreER/+:PPAR-γfl/fl; PPAR-γ cKO) or littermate controls and sample collection. Representative images of LFB staining. Scale bar, 100 μm. Quantification of demyelinated area. n = 5 mice per group, mean ± SD, unpaired t test.
Discussion
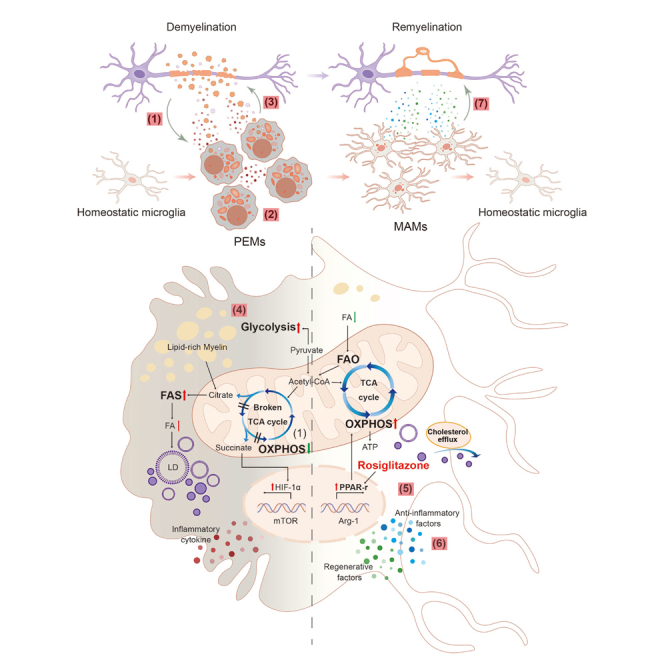
Microglia are among the first responders to demyelinated lesions and remain even when the lesions resolve or become inactive.2,25 Our study identifies an enrichment of two myelin-specific microglia subsets, PEMs and MAMs, in human MS brain lesions. PEMs were more predominantly occurred in active lesions with compromised glycolipid metabolism which might underlie excessive inflammation during demyelination (Figure 1). Consistently, these phagocytes were also required in experimentally demyelinated lesions to internalize lipid-rich myelin debris, and exhibited pro-inflammatory/destructive or anti-inflammatory/pro-regenerative phenotypes (Figures 2, 3, and 4). We further demonstrated that Trem2-mediated microglial phagocytosis and immunometabolism participated both in excessive inflammation during demyelination in the acute phase and the following pro-remyelination properties in the later stage (Figure 5). However, it was not indispensable for the presence of PEMs. Effective modulation of glycolipid remodeling, including facilitating FAO/OXPHOS, and cholesterol recycling, or activating PPAR-γ signaling via rosiglitazone administration could skew the phenotype of microglia toward an anti-inflammatory, pro-regenerative signature (Figures 6 and 7).
It has been stated that microglia are involved in many crucial CNS functions, such as myelination, possibly via secreting neurotrophic factors, morphological flexibility, and regulation of phagocytosis.26 Microglia play a pivotal role in demyelination disease progression. Microglial homeostatic marker significantly diminished in the normal-appearing white matter (NAWM) of patients with MS compared to controls, which was completely lost in active lesions.2 In comparison, phenotypic traits associated with classical activation of microglia were pronounced in the NAWM, and enhanced in active demyelinated lesions, such as CD68 and p22phox.2,27 It has also been reported that TMEM119, a cell surface protein, functions as a specific biomarker for microglia in both human and mouse.28 Studies have already shown the loss of homeostatic microglia in active MS lesions, with high expression of TMEM119 in microglia at the active lesion edge.2,29 Taken together, microglia are reportedly activated and accumulating to the demyelination lesions, which corresponds to our results that the density of microglia is increased in focal demyelination, and alterations in microglial status and functions may possibly contribute to the processes of both demyelination and remyelination in multiple sclerosis.
In the present study, we re-analyzed single-cell sequencing data from human MS samples to uncover distinct patterns of microglial diversity in different MS lesions, and two distinct myelin-specific microglial subsets, PEMs and MAMs, were defined. These two microglial subclusters exhibit different metabolic states and phagocytosing abilities, which contribute to their functional properties of either demyelination or pro-myelination in pathological processes of multiple sclerosis. Genes correlated with the PEMs subset were not only closely related to enhanced immune function and inflammatory responses but also the change in glucolipid metabolism. The MAMs subset adopted a pro-regenerative signature related to myelination followed by lipid remodeling, suggesting that this group of microglia might contribute to maintaining the homeostasis of the CNS, especially the white matter integrity.
Lloyd et al. have stated that the pro-remyelination microglia exhibited upregulation of neurotrophic factors such as IGF1, as well as some other biomarkers for classical M2 microglial phenotype, including ARG-1 and CD206. Besides these properties, all the alterations in microglia contribute to their pro-remyelination properties in MS lesions.3 Similarly, MAMs in our study also showed regenerative and anti-inflammatory properties based on RNA-seq and q-RT-PCR analysis. However, besides those aforementioned pro-remyelination and neurotrophic properties, MAMs we studied also showed enriched pathways related with glucose metabolism and lipid metabolism. The ratio of PEMs to MAMs was elevated in active demyelinated lesions compared to healthy controls and inactive lesions, suggesting differential roles of PEMs and MAMs in the pathological process. Differentially activated microglia were also found accumulated within the demyelinated lesions induced by LPC through the whole process. During demyelination, microglia/macrophages digest myelin debris, most of which is degraded into fatty acids that are deposited in lipid droplets or exported outside the cells.3,10,27 Here, we demonstrated that microglial depletion gradually resulted in larger lesions as lack of debris clearance, and less proliferation/maturation of oligodendrocytes, suggesting an irreplaceable role of microglia/macrophages in remyelination. In addition, the debris-engulfed phagocytes could be detrimental by initiating secondary demyelination themselves by the release of inflammatory factors or metabolic changes. The PEMs subset was identified at a time point that correlates with the peak of demyelination, while MAMs with the peak of remyelination, consistent with the data from human MS brain snRNA sequencing.
PEMs in human MS lesions and LPC-induced demyelinated region shared with a compartmentalized profile of highly expressed Trem2. The immune receptor Trem2 is key to the process of phagocytosis and lipid metabolism.27 Consistent with previous studies, we confirmed that Trem2 signaling plays a key role in inflammatory status during demyelination.17,30 PEMs shared several characteristics with disease-associated microglia (DAM) identified in neurodegenerative diseases.31 However, in contrast to DAM, PEMs identified in this study does not require Trem2 for activation. The selective loss of Trem2 in microglia/macrophages at sites of inflammatory demyelination transiently decreased the level of engulfed myelin debris in microglia at acute phase. Furthermore, Trem2 deficiency resulted in lipid overload gradually, and promoted the pro-inflammatory phenotype and demyelination at a later stage. Interestingly, a recent study demonstrated that Trem2 is required for cholesterol esters and lipid droplets formation, and contributes to protections required for remyelination.17 One possibility for the discrepancy is the compensatory pathways in phagocytosis and formation of lipid droplets independent of Trem2.30 Several receptors coordinate debris clearance by microglia,3 and Trem2 deficiency could induce expression of additional receptors and enzymes that facilitate this process. The excessive amounts of engulfed myelin debris accelerated lipid droplets formation, and the underlying glycolipid dysfunction facilitated phagocytes into a pathogenic role.
Recently, lipid droplets-accumulated microglia have been identified in the aging brain.32 These immune cells were phagocytosis defective and suffered from oxidative stress, representing a dysfunctional microglial state.30,32 Another type of lipid-laden phagocyte, known as foamy cells in atherosclerotic plaques, showed enrichment in lipid metabolism, regulation of oxidative stress, and anti-inflammatory profiles.33 The PEMs subset in the present study, though still engulfing debris and generating lipid droplets, mainly presented enhanced phagocytosis and pro-inflammatory signature during the initial stage. On the other hand, the MAMs subset promotes myelin formation and maintenance, and exhibited regenerative properties, with lipid droplets gradually degraded. One distinguishing feature of PEMs and MAMs appears to be their differential glycolipid metabolism, which might trigger the conversion between PEMs and MAMs in demyelinated lesions.
Recent studies have shown that cellular metabolic processes in innate immune cells may be linked with their effector function.4 Our results showed that microglia can adopt a PEMs phenotype in which immune responses are strengthened by elevated glycolysis, while MAMs showed regenerative signature with an enhancement of FAO and OXPHOS. These results are partially corroborated by two studies examining metabolic reprogramming under other conditions.4,34 Transcriptional analyses and in vitro assays revealed that microglia after engulfing debris exhibit many hallmarks of glycolysis and relatively compromised OXPHOS and FAO. Facilitating OXPHOS/FAO in microglia could result in inflammation resolution and pro-regenerative properties. Thus, understanding how demyelination affects microglial glycolipid metabolism and how to reverse these detrimental effects is key to harness regenerative potential for the treatment of progressive MS.
Modulation of immunometabolism was efficient for CNS inflammatory demyelination diseases.5,35 Rosiglitazone, which is a regulator of the PPAR-γ pathway and glyco-metabolism,36 could be one competitive player among various modulating drugs for potentially translational research. In our study, rosiglitazone was shown to ameliorate microglial hallmarks of inflammation, and facilitate secretion of pro-regenerative factors, which was associated with an enhancement in OXPHOS. More importantly, we showed that the Trem2 deficiency-induced blockage of remyelination potential was reversible, by modulation of metabolism using rosiglitazone treatment. As addition of rosiglitazone to debris-engulfed microglia led to functional remyelination, rosiglitazone could also have possibly exerted a direct effect on PEMs in vivo, which was a pre-requisite for therapies targeting microglia for remyelination. We showed a direct effect of rosiglitazone on the conversion of PEMs to MAMs, at least partially through PPAR-γ-dependent signaling pathway. The remyelination efficacy of the candidate therapeutics largely attenuated in microglial PPAR-γ-deficient mice, indicating that the pro-regenerative effect of rosiglitazone is mainly relied on microglia-specific PPAR-γ signaling pathway.
We identified phagocytosis-enhanced and myelination-associated microglial subpopulation in MS lesions, with differentially altered glycolipid metabolism, phagocytosis, inflammation responses, and regenerative properties. This study, in addition, strongly suggests that immunometabolism orchestrated by microglial phenotype in demyelinated lesions could be pharmacologically modulated. We further demonstrate that PPAR-γ agonist, rosiglitazone, is a potential candidate drug in pro-remyelination by enhancing microglial OXPHOS and facilitating its conversion from detrimental toward regenerative status.
Limitations of the study
This study has some limitations. First, single-cell transcriptomics analysis could better characterize the status and functions of different microglial clusters in murine demyelination models. Also, the in vivo microglia injection model may be different from the in vitro co-culture model, which makes the results of in vivo experiments, to some extent, not completely in accordance with the in vitro studies. We will further elucidate these shortcomings in future studies.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| CD11b-FITC (1:100) | Biolegend | Cat#101206; RRID: AB_312789 |
| CD45-APC (1:100) | Biolegend | Cat#147708; RRID: AB_2563540 |
| Iba-1 (1:500) | WAKO | Cat #019-19741; RRID: AB_839504 |
| Iba-1 (1:500) | Novus Biologicals | Cat #NB100-1028; RRID: AB_521594 |
| CD68 (1:500) | Bio-Rad | Cat #MCA1957; RRID: AB_322219 |
| NG2 (1:200) | Millipore | Cat #AB5320; RRID: AB_11213678 |
| PDGFRα (1:500) | CST | Cat #D1E1E; RRID: AB_2162345 |
| Ki67 (1:100) | Invitrogen | Cat #14-5698-37; RRID: AB_2865119 |
| GST-Π (1:200) | MBL | Cat #311; RRID: AB_591791 |
| dMBP (1:500) | Millipore | Cat #AB5864; RRID: AB_2140351 |
| Trem2 (1:100) | R&D | Cat #MAB17291-SP; RRID: AB_2208679 |
| CD11b microbeads (1:10) | Miltenyi biotec | Cat#130-093-636 |
| Chemicals, peptides, and recombinant proteins | ||
| TRIzol | ThermoFisher | Cat#15596026 |
| BODIPY | ThermoFisher | Cat #D2184 |
| BODIPY 558/568 C12 | ThermoFisher | Cat #D3835 |
| CFSE | Yeasen | Cat #40715ES25 |
| PKH26 | Sigma | Cat #MINI26 |
| Neutral red | Sigma | Cat #N4638 |
| LFB staining kit | ServiceBio | Cat #G1030 |
| PrimeScript™ RT Master Mix | TAKARA | Cat #RR036A |
| Hieff® qPCR SYBR Green Master Mix | Yeasen | Cat # 11201ES03 |
| Seahorse glycolytic rate assay | Agilent | Cat #103344-100 |
| Seahorse mitostress assay | Agilent | Cat #103015-100 |
| Seahorse fatty acid oxidation assay | Agilent | Cat #103672-100 |
| XFe testing plate | Agilent | Cat #102342-100 |
| Seahorse XF DMEM testing medium | Agilent | Cat #103575-100 |
| Seahorse XF Glucose solution | Agilent | Cat #103577-100 |
| Seahorse XF Pyruvate solution | Agilent | Cat #103578-100 |
| Seahorse XF glutamine solution | Agilent | Cat #103579-100 |
| Rosiglitazone | MCE | Cat #HY-17386 |
| C75 | MCE | Cat #HY-12364 |
| Etomoxir | MCE | Cat #HY-50202 |
| GW3965 | MCE | Cat #HY-10627 |
| A769662 | MCE | Cat #HY-50662 |
| Tamoxifen | MCE | Cat #HY-13757A |
| Lysotracker | YEASEN | Cat #40739ES50 |
| Mitotracker | YEASEN | Cat #40740ES50 |
| PLX5622 | Research Diets | Cat #28927 |
| Adult brain dissociation kit | Miltenyi Biotec | Cat #130-107-677 |
| Rneasy micro kit | Qiagen | Cat #74004 |
| L-α-Lysolecthin | Sigma | Cat #L4129 |
| 7-AAD | BD Bioscience | Cat #559925 |
| MS column | Miltenyi Biotec | Cat # 130-042-201 |
| Oligonucleotides | ||
| Primer, Table S10 | This paper | Table S10 |
| Experimental models: Organisms/strains | ||
| WT C57BL/6 mice | Hunan SJA Laboratory Animal Co. Ltd, Hunan, China. | Hunan SJA Laboratory Animal Co. Ltd, Hunan, China. |
| Trem2-/- mice | This paper | Prof. Marco Colonna, Washington University |
| Cx3cr1CreER mice(B6.129P2(Cg)-Cx3cr1tm2.1(cre/ERT2)Litt/WganJ) | The Jackson Laboratory | Stock No: 021160 |
| PPARγf/f mice(B6.129-Pparγtm2Rev/J) | The Jackson Laboratory | Stock No: 004584 |
| Software and algorithms | ||
| GraphPad Prism 9 | GraphPad Software. Inc. | https://www.graphpad.com/ |
| Imaris 9 | Bitplane | https://imaris.oxinst.com/ |
| R 4.04 | R | https://www.r-project.org/ |
| ImageJ (Fiji) | NIH | https://imagej.nih.gov/ij/ |
| R package Seurat 4 | Stuart et al., 201937 | https://satijalab.org |
| R package clusterProfiler v4.05 | Wu et al., 202138 | https://yulab-smu.top/biomedical-knowledge-mining-book/ |
| R package monocle v3 | Trapnell et al., 201439 | Monocle (cole-trapnell-lab.github.io) |
| Deposited data | ||
| GSE118257 | https://www.ncbi.nlm.nih.gov | Jaekel et al. 201912 |
| GSE180759 | https://www.ncbi.nlm.nih.gov | Absinta et al. 20218 |
| Schirmer et al. 201913 | https://cells.ucsc.edu | Schirmer et al. 201913 |
| Deposited RNA-Seq data | This paper | PRJNA941196 |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Wei Wang (wwang@tjh.tjmu.edu.cn).
Materials availability
This study did not generate new unique reagents.
Experimental model and subject details
Animals and ethics
C57BL/6 mice (wild-type, WT; 20–25g; 8-10 weeks) were obtained from Hunan SJA Laboratory Animal Co. Ltd, Hunan, China. Trem2-/- mice were kindly gifted by prof. Marco Colonna at Washington University, St. Louis. Cx3cr1CreER mice (B6.129P2(Cg)-Cx3cr1tm2.1(cre/ERT2)Litt/WganJ) and PPARγfl/fl mice(B6.129-Pparγtm2Rev/J) were obtained from The Jackson Laboratory (Bar Harbor, ME, USA). Cx3Cr1-Cre+PPARγfl/fl mice were obtained by mating the previously mentioned strains. Male and female mice were randomly grouped in the study. Altogether 191 adult animals, including 110 wild-type mice, 76 Trem2-/- mice and 5 Cx3cr1crePPARγfl/fl mice were used in this experiment. The animals were allocated into different groups randomly. Briefly, randomization was implemented with the random numbers generated by GraphPad Prism before animals receive different treatments. Basically, the sample size for each experimental group was utilized based on previous studies.40 For most of the animal experiments, n = 5 animals per group was used. For several experiments such as injecting-primary microglia-induced demyelination, n = 8 animals per group was otherwise used to maximally avoid statistical bias.
All animal surgeries were approved by the Animal Care Committee of Tongji Medical College, Huazhong University of Science and Technology. Mice were housed in a non-specific pathogen (SPF) animal facility and freely provided with water and food. It was subjected to a 12-hour cycle of alternating light and dark under 22°C and 55-60% relative humidity. All animal experiments comply with the guidelines of ARRIVE. All analyses were performed by blinded and experienced investigators.
Method details
Sn-RNA-Seq bioinformatics analysis
Public data from Jaekel et al.,12 Schirmer et al.13 and Absinta8 et al. were downloaded from the Gene Expression Omnibus (GEO, https://www.ncbi.nlm.nih.gov/geo/) and re-analyzed using RStudio (v 4.02) (for statistical analysis, RStudio Inc). Cell nuclei samples from healthy control, inactive MS lesions, and active MS lesions were enrolled for further analysis. Seurat package (v 4.04) (for single cell data analysis, New York Genome Center, Satija Lab)37 was used for subsequent analysis. Only nuclei with mitochondrial contamination less than 5% and between 200 and 2500 genes expressed were utilized. The expression matrix was normalized using the normalizeData function and the top 2000 variable genes were calculated with FindVariableFeatures. The Seurat object was then scaled with the scaleData function and principal component analysis (PCA) was further implemented. The top 20 principal components were implemented for the unsupervised clustering. Harmony package (v 0.1.0) was utilized to combine the three different datasets and remove the batch effects among them. FindNeighbors function was used to construct the k-nearest neighbors graph, and FindClusters function was used to iteratively group nuclei (resolution = 0.4). The cell type of each cluster was identified based on the cellular markers identified based on FindAllMarkers function. Totally, after removal of singletons, 19 clusters were retained and visualized by Uniform Manifold Approximation and Projection (UMAP). For the re-clustering of microglia/macrophages, top 10 principal components were used for the clustering. R package monocle (2.20.0),39 clusterProfiler (v 4.0.5),38 msigdbr (7.4.1), ggplot2 (v 3.3.5) and pheatmap (1.0.12) were utilized for subsequent analysis.
LPC-induced demyelination and rosiglitazone treatment
Experiments were conducted as previous described with some modifications.40,41 Briefly, mice were anesthetized with isoflurane and then mounted in a stereotaxic frame. Demyelination of the corpus callosum was induced by stereotactic injection of 2 μL 1% LPC (Sigma) in a 0.9% sterile saline solution at the rate of 0.4 μL /min with a 32-gauge 2-inch needle and 10μL Hamilton syringe. The injection site was 1.0 mm lateral to the bregma, 1.0mm anterior, and 2.2mm deep. Afterwards, the needle remained in situ for 10 min. Rosiglitazone (HY-17386, MCE) was administered daily at 10mg/kg by gavage after LPC injection until day 7 or day 14.
Microglia depletion
Depletion of microglia and macrophages is implemented as previously described.42 Briefly, mice were fed with PLX5622 containing chow (Research Diets) at 1200 mg/kg of chow, beginning 7 days prior to surgery and continuing throughout the experiment.
Preparation of brain samples
At 3, 7, 10, 14, and 21 days after LPC injection, mice were terminally anesthetized with isoflurane, then transcardially perfused with 30 mL cold 0.1% phosphate buffer (PBS), followed by 30 mL cold 4% PFA. The average number of mice in each group is 5-6. Brains were collected after perfusion, fixed overnight at 4°C with 4% PFA, and then dehydrated with 30% sucrose. Serial 20 μm coronal slices were cut on a frozen slicer (CryoStar NX50, Thermo) and directly thaw mounted on the slides for staining.
Luxol fast blue staining
Staining was performed as previously described.41,43 Briefly, 20μm brain slices in serial sections were stained with 0.1% LFB dye (G1030, Servicebio) at 60°C for 6-8 hours. Sections were then differentiated alternately in 0.05% lithium carbonate solution and 70% ethanol, then dehydrated stepwise with 75%, 90%, and 100% ethanol. Finally, sections were soaked in xylene for 5-10 min and sealed with neutral resin. Images were captured by a microscope (BX51, Olympus, Japan). The area of demyelination in corpus callosum were calculated by using ImageJ (NIH). On average, 2-3 20× fields were taken per mouse.
Immunofluorescent staining
For immunofluorescent staining, brain sections were permeabilized with 0.25% Triton-X100 (Beyotime) for 10 min. After blocking with QuickBlock (Beyotime) at room temperature for 15 min, sections were incubated with primary antibodies overnight at 4°C and subsequently incubated with secondary antibodies for 1 h at a dilution of 1:200 at 37°C in the dark. Details of primary and secondary antibodies are shown in the key resources table. Finally, sections were stained with 4,6-diamidino-2-phenylindole (DAPI) (10 μg/mL, Sigma-Aldrich). Images were captured with a confocal microscope (OLYMPUS, FV1200) , in which keeping the threshold values for fluorescence intensity and pixels consistent for z-stacking.
Image analysis was performed by randomly selecting one or two microscope fields in the lesion region of the corpus callosum. Captured images were loaded into ImageJ and quantified by 2 independent observers who were not informed of the group. dMBP (a marker of degraded myelin debris) staining, the aggregation and morphology of Iba1+ microglia or Ki67+ PDGFR-α+ and GST-π+ oligodendrocytes determine the degree of demyelination, microglial activation and OPC proliferation/differentiation.
BODIPY staining in vivo
After secondary antibody labeling, sections were washed three times in PBS and then incubated with BODIPY 493/503 (1:1000 from a 1 mg/ml stock solution in DMSO; Thermo Fisher) for 15 min at RT. Then tissues were washed again in 1× PBS and immediately covered. To avoid fluorescence quenching, images were captured as soon as possible.
Cell morphology and engulfment analysis
Cell morphological and engulfment analysis was performed as previously described.44,45 2-3 randomly chosen 60x fields within the demyelination lesions in 1-2 brain sections per mouse were captured with an OLYMPUS FV1200 confocal microscope. The volumes and areas of the cells were calculated by Surface function of Imaris software (Bitplane). Engulfment was calculated as the volume of engulfed dMBP divided by the volume of CD68+ Iba-1+ cells (%). 12-15 cells were randomly chosen for one sample and average % engulfment of those cells was calculated per animal. Data from 5 animals were enrolled for one group and analyzed. For the quantification of BODIPY+Iba1+ cells, 4-5 fields were chosen per animal, and the ratio of BODIPY+Iba1+/Iba+ cells was calculated per field. The average BODIPY+Iba1+/Iba+ ratio per animal were calculated and analyzed.
Single cell suspensions prepared from LPC-induced demyelination lesions
Microglia were sorted from LPC-induced demyelination lesions as previously described.46 Mice were anesthetized with isoflurane and transcardially perfused with cold and sterile PBS solutions. Briefly, 500μL of 1% neutral red solution was injected intraperitoneally 2-3 hours prior to collection of the brain to better differentiate between demyelinating lesions and relatively healthy tissue. And demyelination lesions were dissected and collected under a microscope. Single cell suspensions were prepared using an adult brain dissociation kit (Miltenyi Biotec, Germany). The lesion tissues were digested with enzyme mix containing enzyme P and enzyme A and incubated in 37°C for 30 min. Then 10 mL PBS was added to the enzyme mixture and mixed well. The mixed suspension was transferred to a 70 μm cell strainer and centrifuged at 300xg, 4°C for 5 min. Cell debris were removed by debris removal solution. After that, cell pellets were re-suspended with PBS containing 2% FBS and used for further analysis.
Fluorescence activated cell sorting (FACS)
For FACS cells sorting, Fc-Block (BD Biosciences) was added to the cell suspension at 1:50 dilution and incubated for 15 min on ice. After that, CD45-APC (Biolegend) and CD11b-FITC (Biolegend) were added to the suspension at a 1:100 dilution and incubated for 30 min on ice. The cell pellets were further washed twice with PBS. Right before FACS, 7-AAD (BD Biosciences) was added at the dilution of 1:20 and incubated at room temperature for about 5 min. 7-AAD-CD45lowCD11bhigh live microglial cells were sorted by Moflo XDP (Beckman Coulter) directly into lysis buffer (QIAGEN, Germany) for RNA extraction.
RNA-Sequencing
Total RNA from FACS-sorted microglia was extracted using a Micro RNeasy Kit (QIAGEN, Germany). Samples were then stored at -80°C for subsequent use. Total RNA samples were amplified with oligo-dT and dNTPs, and then RNA was reverse-transcribed to cDNA. cDNA libraries were further quantified using Qubit dsDNA HS Assay, and were then fragmented using fragment buffer. In the end, the amplified flowcell was sequenced single end on the BGI-500 platform (BGI-Shenzhen, China).
Magnetic activated cell sorting (MACS)
Microglia were sorted with CD11b microbeads (Miltenyi Biotec) for RT-PCR analysis following manufacturer’s instructions. Briefly, CD11b microbeads-conjugated antibodies were added to the cell suspension at a dilution of 1:10 and incubated on ice for 10-15 min. The cell suspensions were then transferred to an MS magnetic column and subsequently injected into a centrifuge tube. After centrifugation at 300xg at 4°C for 5 min, the supernatant was completely removed carefully and 75 μL RLT lysis buffer was added directly to the cell pellet. Subsequent steps of RNA extraction were the same with the process in RNA-sequencing.
Real-time PCR
Total RNA from microglia was extracted with TRIzol or Micro RNeasy Kit. cDNA was reverse transcribed from 1 μg of RNA using PrimeScript™ RT Master Mix (TAKARA, RR036A). A total of 20 μL reaction system was prepared for quantitative RT-PCR using Hieff qPCR SYBR Green Master Mix (YEASEN, 11201ES03). Real-time PCR system (CFX96, BioRad) was used for all reactions. The expression levels of target genes were normalized to β-actin and calculated using 2ΔΔCt method. Primers used is provided in Table S10.
Transmission electron microscopy
Transmission electron microscopy (TEM) was used to evaluate myelin thickness in the corpus callosum area at 7 and 14 days after LPC injection.41,47,48 Mice were anesthetized with isoflurane, then intraventricularly perfused with 2.5% glutaraldehyde and 4% paraformaldehyde (PFA). The brain was taken and cut coronally at 1 mm thickness. The corpus callosum was cut into 1 mm3 pieces and fixed with 2.5% glutaraldehyde. After that, the tissues were put into acetone:812 (1:1) overnight for osmosis, then the tissues are embedded at 60 degrees celsius for 48 hours. 60-80nm slices were obtained for subsequent analysis. The samples were observed under a 100 kV in Hitachi TEM system with a magnification of 1500x. ImageJ was used to measure the ratio of myelinated axon thickness to axon diameter (G ratio) of fibers (at least 40 axons per mouse, 4 mice per group) and percent of demyelinated axons. G values and the number of demyelinated axons were calculated.
Microglia under transmission electron microscopy were defined by the following characteristics.49 The cytoplasm of microglia is electron dense and the nucleus is lenticular. Microglia show distinct heterochromatin pattern. There is a deep band of heterochromatin with high electron density near the nuclear envelope and a dense heterochromatin network in the whole nucleus. Additionally, lipid droplets are gray and homogeneous under electron microscopy.32
Primary microglia, oligodendrocyte precursor cells (OPCs), and oligodendrocyte culture and microglia-OPCs co-culture
Primary microglia and OPCs cultures were prepared from mice using the differential attachment method.50,51 Briefly, initial mixed glia cultures were obtained from the brains of either WT and Trem2-/- P1-2 neonates. Neonates were executed under carbon dioxide anaesthesia. Their whole brains were removed, cut up and digested with trypsin into single cell suspensions. Isolated cells were cultured in poly-d-lysine (PDL) coated flask with DMEM/F12 containing 10% heat-inactivated fetal bovine serum. The cells were cultured in a humidified incubator at 37°C and 5% CO2 until fusion (12-14 days). Microglia were collected by shaking at 180 RPM for 1 h. After microglia were removed, 0.25% trypsin was used to isolate OPCs from astrocyte layers. The cell suspension was transferred to an untreated petri dish and incubated for 30 min for astrocytes and microglia to attach to the surface, while the OPCs remained suspended. OPCs were then plated on PDL coated plates and maintained in serum-free basal medium for 3-5 days (DMEM/F12, 0.1% bovine serum albumin, 1% N2, 2% B27), containing 10 ng/mL platelet derived growth factor (PDGF) and 10 ng/mL basic fibroblast growth factor (bFGF). To induce oligodendrocytes, OPCs were stimulated with triiodothyronine (T3) (50 ng/mL) and ciliary neurotrophic factor (CNTF) (10 ng/mL) or co-cultured with treated microglia for 3 d. The OPCs media: microglia culture media = 1:1. Co-cultured microglia were pre-treated with myelin with or without 50μM rosiglitazone for 8 h.
Metabolic intervention in vitro
After microglia were harvested and adhered to the bottom of plate, the cells were pretreated with different reagents (rosiglitazone (50 μM, HY-17386), C75 (50 μM, HY-12364) and GW3965 (10 μM, HY-10627) for 1 h; etomoxir (200 μM, HY-50202) and A-769662 (100 μM, HY-50662) for 3 h, and then treated with myelin (5 μg/mL) for 8 or 24 h.
Myelin purification and phagocytosis
Myelin was purified from the brains of wild-type C57BL/6 mice as previously described.16,22 Each brain was dissected and placed in 800 μL cold lysis buffer (10 mM HEPES, 5 mM EDTA, 0.3 M sucrose, protease inhibitor PMSF (BOSTER, AR1178), sliced into fine pieces with a grinding machine (Servicebio). The homogenates were transferred to 13.2 mL polypropylene tube (Beckman 331372). An equal amount of 0.32 M sucrose (1.2 mL/brain) was carefully added using sterilized Pasteur straws, followed by an equal amount of 0.85 M sucrose (1.2 mL/brain) below the two layers. The sample was centrifuged in an Optima XPN-100 supercentrifuge (Beckman) at 75000xg at 4°C for 30 min in a SW 41Ti rotor. The myelin sheath was obtained at the interface of 0.32M/0.85M sucrose, resuspended in 10 mL distilled water, and placed in another tube. The mixture was centrifuged at 75000xg at 4°C for 15 min. The pellet was resuspended with 10 mL distilled water, then spun at 75000xg, 4°C, 15 min twice. All previous steps were repeated to obtain pure myelin. After purification, myelin adjusted to 1 mg/mL protein concentration using BCA protein analysis Kit (BOSTER, AR1189). For the fluorescence labeling of myelin, purified myelin was labeled according to manufacturer's instructions using CFSE (MCE, HY-D0938) or PKH26 (Sigma, MINI26). Primary microglia were seeded at a density of 150,000 cells per well and 5 μg/mL myelin/myelin-CFSE/myelin-PKH26 was added to the cell culture medium. The cells were cultured with myelin for up to 72 h, and then fixed with 4% PFA for immunofluorescence. Images were acquired by confocal microscopy (FV1200, Olympus, Japan) and mean fluorescence of myelin was calculated. Briefly, the average fluorescence intensity was equal to the total fluorescence intensity of the myelin divided by the total area of the microglia in a filed.
Seahorse analysis of primary murine microglia
Glycolysis rates, mitostress, and fatty acid oxidation rates were measured with a XF-24 Seahorse extracellular flux analyzer (Seahorse Bioscience, CA). Briefly, primary mouse microglia were plated into XFe24 cell culture plates at a density of 80,000 cells per well. Purified myelin debris were then added to the cell culture medium and incubated for up to 48 h. For the rosiglitazone group, 50μM rosiglitazone was added at the same concentration described previously for 1 h prior to myelin treatment. 1 d before the experiment, 1 mL of XF calibration media was added the XF sensor cartridges and kept in a non-CO2 incubator. On the day of the experiment, cell culture medium was replaced with fresh running buffer (XF DMEM medium plus 10 mM glucose, 2 mM glutamine, and 1 Mm pyruvate) and incubated in a non-CO2 incubator for 60 min.
For glycolysis rate analysis, 0.5 μM rotenone plus antimycin A and 50 mM 2-DG were added to the sensor cartridges. For mitostress analysis, 1.5 μM oligomycin, 2 μM FCCP and 0.5 μM rotenone plus antimycin A were added to the sensor cartridges. For fatty acid oxidation rate analysis, 4 μM etomoxir, 1.5 μM oligomycin, 2 μM FCCP and 0.5 μM rotenone plus antimycin were added to the sensor cartridges. Once the measurements were completed, the number of cells in the plates were determined. The values of OCR and glycoPER were normalized to the cell number in each well. Finally, the levels of the first measurement points of OCR and glycoPER for control microglia were normalized to 1 according to previous study.52
Microglia injection in naïve corpus callosum
As described previously,53 after primary microglia were treated with myelin debris or CFSE-myelin debris for 8, 24, or 48 h, they were washed with PBS twice. 1x105 of microglia in 2 μL PBS were injected into the corpus callosum by a 32 needle with Hamilton syringe. Blank controls injected 2 μL PBS only. Three days after injection, then brains were collected for routine histological examination and immunostaining.
BODIPY and fatty acid in vitro staining
Both primary mouse microglia and human microglia cell lines were seeded at 15×104 cells onto PDL-coated glass coverslips. Following specific treatments, cells were incubated with BODIPY 558/568 C12 (1:1000 from a 1 mM stock solution in DMSO; Thermo Fisher) for 30 min in 37°C, then fixed with 4% PFA for 15 min, washed three times in PBS, and finally incubated in PBS with BODIPY 493/503. Eight randomly selected visual fields per coverslip were imaged (×40 magnification) using a confocal microscope (OLYMPUS, FV1200, Japan). Mean fluorescence intensity of BODIPY and BODIPY C12 was calculated.
Co-localization analysis in vitro
Sections were imaged in multiple layers with the same parameters with a confocal microscope. ImageJ was used to analyze co-localization of BODIPY or mitotracker and BODIPY C12. The Pearson coefficient value represents degree of co-localization.
Cytokine measurements by Luminex assay
Cells were cultured as described above. Cell-free and myelin-free supernatants were collected 8h after culture, then frozen at −80°C until assessment. The concentrations of CXCL1, CXCL10, CXCL16, CCL3, CCL5, TNF-α, and IL-6 were detected by Mouse Luminex Discovery Assay (R&D System). The experiment was conducted according to the mouse cytokine kit protocol provided. All cytokines were determined simultaneously from one specimen.
Quantification and statistical analysis
The number of animals and in vitro replicates for each experiment are shown in the figure legends. Our sample sizes are similar to those reported in previous publications.10,16,53,54,55 Unpaired two-sided Student’s t-test, one-way ANOVA or two-way ANOVA followed by Bonferroni test were performed by GraphPad Prism software version 9. P value < 0.05 was considered statistically significant. Statistical data with exact P values for each comparison are provided in Table S1.
Acknowledgments
We thank Prof. Marco Colonna for providing Trem2−/− mice (Washington University, St. Louis, USA). This work was supported by Ministry of Science and Technology China Brain Initiative Grant (2022ZD0204704), National Natural Science Foundation of China (Grants: 82071380, 81873743, 81801223), and Tongji Hospital (HUST) Foundation for Excellent Young Scientist (Grant No. 2020YQ06).
Author contributions
Conceptualization: C.Q., S.Y., M.C., D.S.T., W.W. Methodology: S.Y., M.C., M.H.D., L.Q.Z., Y.H.C., Z.X.S., D.B.B. Investigation: C.Q., S.Y., M.C., M.H.D., L.Q.Z., Y.H.C., Z.X.S. Visualization: L.J.W., D.S.T., W.W. Funding acquisition: C.Q., D.S.T. Project administration: C.Q., S.Y., M.C., L.J.W., D.S.T., W.W. Supervision: L.J.W., D.S.T., W.W. Writing – original draft: C.Q., S.Y., M.C., M.H.D. Writing – review & editing: D.B.B., L.J.W., D.S.T., W.W.
Declaration of interests
The authors declare no competing interests.
Published: April 11, 2023
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2023.106588.
Contributor Information
Dai-Shi Tian, Email: tiands@tjh.tjmu.edu.cn.
Wei Wang, Email: wwang@tjh.tjmu.edu.cn.
Supplemental information
Data and code availability
Tables S1, S2, S3, S4, S5, S6, S7, S8, S9 and S10 source data for figure panels are listed supplemental information.
Human single-cell/nucleus RNA sequencing data were obtained from previously published datasets (data resource provided in key resources table). All the data supporting the conclusions of the current study are presented in the figures and tables. If necessary, the raw data are available from the corresponding author upon reasonable request.
RNA-Seq data has been deposited in SRA with the accession number PRJNA941196, and will be publicly available upon publication.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- 1.Berglund R., Guerreiro-Cacais A.O., Adzemovic M.Z., Zeitelhofer M., Lund H., Ewing E., Ruhrmann S., Nutma E., Parsa R., Thessen-Hedreul M., et al. Microglial autophagy-associated phagocytosis is essential for recovery from neuroinflammation. Sci. Immunol. 2020;5:eabb5077. doi: 10.1126/sciimmunol.abb5077. [DOI] [PubMed] [Google Scholar]
- 2.Zrzavy T., Hametner S., Wimmer I., Butovsky O., Weiner H.L., Lassmann H. Loss of 'homeostatic' microglia and patterns of their activation in active multiple sclerosis. Brain. 2017;140:1900–1913. doi: 10.1093/brain/awx113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lloyd A.F., Miron V.E. The pro-remyelination properties of microglia in the central nervous system. Nat. Rev. Neurol. 2019;15:447–458. doi: 10.1038/s41582-019-0184-2. [DOI] [PubMed] [Google Scholar]
- 4.Baik S.H., Kang S., Lee W., Choi H., Chung S., Kim J.I., Mook-Jung I. A breakdown in metabolic reprogramming causes microglia dysfunction in alzheimer's disease. Cell Metab. 2019;30:493–507.e6. doi: 10.1016/j.cmet.2019.06.005. [DOI] [PubMed] [Google Scholar]
- 5.Artyomov M.N., Van den Bossche J. Immunometabolism in the single-cell era. Cell Metab. 2020;32:710–725. doi: 10.1016/j.cmet.2020.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Locatelli G., Theodorou D., Kendirli A., Jordão M.J.C., Staszewski O., Phulphagar K., Cantuti-Castelvetri L., Dagkalis A., Bessis A., Simons M., et al. Mononuclear phagocytes locally specify and adapt their phenotype in a multiple sclerosis model. Nat. Neurosci. 2018;21:1196–1208. doi: 10.1038/s41593-018-0212-3. [DOI] [PubMed] [Google Scholar]
- 7.Van Eijk M., Aerts J.M.F.G. The unique phenotype of lipid-laden macrophages. Int. J. Mol. Sci. 2021;22:4039. doi: 10.3390/ijms22084039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Absinta M., Maric D., Gharagozloo M., Garton T., Smith M.D., Jin J., Fitzgerald K.C., Song A., Liu P., Lin J.P., et al. A lymphocyte-microglia-astrocyte axis in chronic active multiple sclerosis. Nature. 2021;597:709–714. doi: 10.1038/s41586-021-03892-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Masuda T., Sankowski R., Staszewski O., Böttcher C., Amann L., Sagar, Scheiwe C., Scheiwe C., Nessler S., Kunz P., van Loo G., et al. Spatial and temporal heterogeneity of mouse and human microglia at single-cell resolution. Nature. 2019;566:388–392. doi: 10.1038/s41586-019-0924-x. [DOI] [PubMed] [Google Scholar]
- 10.Berghoff S.A., Spieth L., Sun T., Hosang L., Schlaphoff L., Depp C., Düking T., Winchenbach J., Neuber J., Ewers D., et al. Microglia facilitate repair of demyelinated lesions via post-squalene sterol synthesis. Nat. Neurosci. 2021;24:47–60. doi: 10.1038/s41593-020-00757-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Esaulova E., Cantoni C., Shchukina I., Zaitsev K., Bucelli R.C., Wu G.F., Artyomov M.N., Cross A.H., Edelson B.T. Single-cell RNA-seq analysis of human CSF microglia and myeloid cells in neuroinflammation. Neurol. Neuroimmunol. Neuroinflamm. 2020;7:e732. doi: 10.1212/NXI.0000000000000732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jäkel S., Agirre E., Mendanha Falcão A., van Bruggen D., Lee K.W., Knuesel I., Malhotra D., Ffrench-Constant C., Williams A., Castelo-Branco G. Altered human oligodendrocyte heterogeneity in multiple sclerosis. Nature. 2019;566:543–547. doi: 10.1038/s41586-019-0903-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schirmer L., Velmeshev D., Holmqvist S., Kaufmann M., Werneburg S., Jung D., Vistnes S., Stockley J.H., Young A., Steindel M., et al. Neuronal vulnerability and multilineage diversity in multiple sclerosis. Nature. 2019;573:75–82. doi: 10.1038/s41586-019-1404-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Feng X., Valdearcos M., Uchida Y., Lutrin D., Maze M., Koliwad S.K. Microglia mediate postoperative hippocampal inflammation and cognitive decline in mice. JCI Insight. 2017;2:e91229. doi: 10.1172/jci.insight.91229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Henry R.J., Ritzel R.M., Barrett J.P., Doran S.J., Jiao Y., Leach J.B., Szeto G.L., Wu J., Stoica B.A., Faden A.I., Loane D.J. Microglial depletion with CSF1R inhibitor during chronic phase of experimental traumatic brain injury reduces neurodegeneration and neurological deficits. J. Neurosci. 2020;40:2960–2974. doi: 10.1523/JNEUROSCI.2402-19.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nugent A.A., Lin K., van Lengerich B., Lianoglou S., Przybyla L., Davis S.S., Llapashtica C., Wang J., Kim D.J., Xia D., et al. TREM2 regulates microglial cholesterol metabolism upon chronic phagocytic challenge. Neuron. 2020;105:837–854.e9. doi: 10.1016/j.neuron.2019.12.007. [DOI] [PubMed] [Google Scholar]
- 17.Gouna G., Klose C., Bosch-Queralt M., Liu L., Gokce O., Schifferer M., Cantuti-Castelvetri L., Simons M. TREM2-dependent lipid droplet biogenesis in phagocytes is required for remyelination. J. Exp. Med. 2021;218:e20210227. doi: 10.1084/jem.20210227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Divakaruni A.S., Hsieh W.Y., Minarrieta L., Duong T.N., Kim K.K.O., Desousa B.R., Andreyev A.Y., Bowman C.E., Caradonna K., Dranka B.P., et al. Etomoxir inhibits macrophage polarization by disrupting CoA homeostasis. Cell Metab. 2018;28:490–503.e7. doi: 10.1016/j.cmet.2018.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Carroll R.G., Zasłona Z., Galván-Peña S., Koppe E.L., Sévin D.C., Angiari S., Triantafilou M., Triantafilou K., Modis L.K., O'Neill L.A. An unexpected link between fatty acid synthase and cholesterol synthesis in proinflammatory macrophage activation. J. Biol. Chem. 2018;293:5509–5521. doi: 10.1074/jbc.RA118.001921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Herzig S., Shaw R.J. AMPK: guardian of metabolism and mitochondrial homeostasis. Nat. Rev. Mol. Cell Biol. 2018;19:121–135. doi: 10.1038/nrm.2017.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xu X., Xiao X., Yan Y., Zhang T. Activation of liver X receptors prevents emotional and cognitive dysfunction by suppressing microglial M1-polarization and restoring synaptic plasticity in the hippocampus of mice. Brain Behav. Immun. 2021;94:111–124. doi: 10.1016/j.bbi.2021.02.026. [DOI] [PubMed] [Google Scholar]
- 22.Cantuti-Castelvetri L., Fitzner D., Bosch-Queralt M., Weil M.T., Su M., Sen P., Ruhwedel T., Mitkovski M., Trendelenburg G., Lütjohann D., et al. Defective cholesterol clearance limits remyelination in the aged central nervous system. Science. 2018;359:684–688. doi: 10.1126/science.aan4183. [DOI] [PubMed] [Google Scholar]
- 23.Montaigne D., Butruille L., Staels B. PPAR control of metabolism and cardiovascular functions. Nat. Rev. Cardiol. 2021;18:809–823. doi: 10.1038/s41569-021-00569-6. [DOI] [PubMed] [Google Scholar]
- 24.Bosch-Queralt M., Cantuti-Castelvetri L., Damkou A., Schifferer M., Schlepckow K., Alexopoulos I., Lütjohann D., Klose C., Vaculčiaková L., Masuda T., et al. Diet-dependent regulation of TGFbeta impairs reparative innate immune responses after demyelination. Nat. Metab. 2021;3:211–227. doi: 10.1038/s42255-021-00341-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zia S., Rawji K.S., Michaels N.J., Burr M., Kerr B.J., Healy L.M., Plemel J.R. Microglia diversity in health and multiple sclerosis. Front. Immunol. 2020;11:588021. doi: 10.3389/fimmu.2020.588021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Paolicelli R.C., Sierra A., Stevens B., Tremblay M.E., Aguzzi A., Ajami B., Amit I., Audinat E., Bechmann I., Bennett M., et al. Microglia states and nomenclature: a field at its crossroads. Neuron. 2022;110:3458–3483. doi: 10.1016/j.neuron.2022.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Voet S., Prinz M., van Loo G. Microglia in central nervous system inflammation and multiple sclerosis pathology. Trends Mol. Med. 2019;25:112–123. doi: 10.1016/j.molmed.2018.11.005. [DOI] [PubMed] [Google Scholar]
- 28.Bennett M.L., Bennett F.C., Liddelow S.A., Ajami B., Zamanian J.L., Fernhoff N.B., Mulinyawe S.B., Bohlen C.J., Adil A., Tucker A., et al. New tools for studying microglia in the mouse and human CNS. Proc. Natl. Acad. Sci. USA. 2016;113:E1738–E1746. doi: 10.1073/pnas.1525528113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hayashida S., Masaki K., Suzuki S.O., Yamasaki R., Watanabe M., Koyama S., Isobe N., Matsushita T., Takahashi K., Tabira T., et al. Distinct microglial and macrophage distribution patterns in the concentric and lamellar lesions in Balo's disease and neuromyelitis optica spectrum disorders. Brain Pathol. 2020;30:1144–1157. doi: 10.1111/bpa.12898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Safaiyan S., Besson-Girard S., Kaya T., Cantuti-Castelvetri L., Liu L., Ji H., Schifferer M., Gouna G., Usifo F., Kannaiyan N., et al. White matter aging drives microglial diversity. Neuron. 2021;109:1100–1117.e10. doi: 10.1016/j.neuron.2021.01.027. [DOI] [PubMed] [Google Scholar]
- 31.Deczkowska A., Keren-Shaul H., Weiner A., Colonna M., Schwartz M., Amit I. Disease-associated microglia: a universal immune sensor of neurodegeneration. Cell. 2018;173:1073–1081. doi: 10.1016/j.cell.2018.05.003. [DOI] [PubMed] [Google Scholar]
- 32.Marschallinger J., Iram T., Zardeneta M., Lee S.E., Lehallier B., Haney M.S., Pluvinage J.V., Mathur V., Hahn O., Morgens D.W., et al. Lipid-droplet-accumulating microglia represent a dysfunctional and proinflammatory state in the aging brain. Nat. Neurosci. 2020;23:194–208. doi: 10.1038/s41593-019-0566-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Willemsen L., de Winther M.P. Macrophage subsets in atherosclerosis as defined by single-cell technologies. J. Pathol. 2020;250:705–714. doi: 10.1002/path.5392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cheng S.C., Quintin J., Cramer R.A., Shepardson K.M., Saeed S., Kumar V., Giamarellos-Bourboulis E.J., Martens J.H.A., Rao N.A., Aghajanirefah A., et al. mTOR- and HIF-1alpha-mediated aerobic glycolysis as metabolic basis for trained immunity. Science. 2014;345:1250684. doi: 10.1126/science.1250684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Negrotto L., Farez M.F., Correale J. Immunologic effects of metformin and pioglitazone treatment on metabolic syndrome and multiple sclerosis. JAMA Neurol. 2016;73:520–528. doi: 10.1001/jamaneurol.2015.4807. [DOI] [PubMed] [Google Scholar]
- 36.Jucker B.M., Schaeffer T.R., Haimbach R.E., McIntosh T.S., Chun D., Mayer M., Ohlstein D.H., Davis H.M., Smith S.A., Cobitz A.R., Sarkar S.K. Normalization of skeletal muscle glycogen synthesis and glycolysis in rosiglitazone-treated Zucker fatty rats: an in vivo nuclear magnetic resonance study. Diabetes. 2002;51:2066–2073. doi: 10.2337/diabetes.51.7.2066. [DOI] [PubMed] [Google Scholar]
- 37.Stuart T., Butler A., Hoffman P., Hafemeister C., Papalexi E., Mauck W.M., 3rd, Hao Y., Stoeckius M., Smibert P., Satija R. Comprehensive integration of single-cell data. Cell. 2019;177:1888–1902.e21. doi: 10.1016/j.cell.2019.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wu T., Hu E., Xu S., Chen M., Guo P., Dai Z., Feng T., Zhou L., Tang W., Zhan L., et al. clusterProfiler 4.0: a universal enrichment tool for interpreting omics data. Innovation. 2021;2:100141. doi: 10.1016/j.xinn.2021.100141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Trapnell C., Cacchiarelli D., Grimsby J., Pokharel P., Li S., Morse M., Lennon N.J., Livak K.J., Mikkelsen T.S., Rinn J.L. The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nat. Biotechnol. 2014;32:381–386. doi: 10.1038/nbt.2859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lloyd A.F., Davies C.L., Holloway R.K., Labrak Y., Ireland G., Carradori D., Dillenburg A., Borger E., Soong D., Richardson J.C., et al. Central nervous system regeneration is driven by microglia necroptosis and repopulation. Nat. Neurosci. 2019;22:1046–1052. doi: 10.1038/s41593-019-0418-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen M., Yang L.L., Hu Z.W., Qin C., Zhou L.Q., Duan Y.L., Bosco D.B., Wu L.J., Zhan K.B., Xu S.B., Tian D.S. Deficiency of microglial Hv1 channel is associated with activation of autophagic pathway and ROS production in LPC-induced demyelination mouse model. J. Neuroinflammation. 2020;17:333. doi: 10.1186/s12974-020-02020-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sariol A., Mackin S., Allred M.G., Ma C., Zhou Y., Zhang Q., Zou X., Abrahante J.E., Meyerholz D.K., Perlman S. Microglia depletion exacerbates demyelination and impairs remyelination in a neurotropic coronavirus infection. Proc. Natl. Acad. Sci. USA. 2020;117:24464–24474. doi: 10.1073/pnas.2007814117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Qin C., Liu Q., Hu Z.W., Zhou L.Q., Shang K., Bosco D.B., Wu L.J., Tian D.S., Wang W. Microglial TLR4-dependent autophagy induces ischemic white matter damage via STAT1/6 pathway. Theranostics. 2018;8:5434–5451. doi: 10.7150/thno.27882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schafer D.P., Lehrman E.K., Kautzman A.G., Koyama R., Mardinly A.R., Yamasaki R., Ransohoff R.M., Greenberg M.E., Barres B.A., Stevens B. Microglia sculpt postnatal neural circuits in an activity and complement-dependent manner. Neuron. 2012;74:691–705. doi: 10.1016/j.neuron.2012.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Werneburg S., Jung J., Kunjamma R.B., Ha S.K., Luciano N.J., Willis C.M., Gao G., Biscola N.P., Havton L.A., Crocker S.J., et al. Targeted complement inhibition at synapses prevents microglial synaptic engulfment and synapse loss in demyelinating disease. Immunity. 2020;52:167–182.e7. doi: 10.1016/j.immuni.2019.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liu L., Besson-Girard S., Ji H., Gehring K., Bulut B., Kaya T., Usifo F., Simons M., Gokce O. Dissociation of microdissected mouse brain tissue for artifact free single-cell RNA sequencing. STAR Protoc. 2021;2:100590. doi: 10.1016/j.xpro.2021.100590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Carlström K.E., Zhu K., Ewing E., Krabbendam I.E., Harris R.A., Falcão A.M., Jagodic M., Castelo-Branco G., Piehl F. Gsta4 controls apoptosis of differentiating adult oligodendrocytes during homeostasis and remyelination via the mitochondria-associated Fas-Casp8-Bid-axis. Nat. Commun. 2020;11:4071. doi: 10.1038/s41467-020-17871-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liu K.M., Shen C.L. Ultrastructural sequence of myelin breakdown during Wallerian degeneration in the rat optic nerve. Cell Tissue Res. 1985;242:245–256. doi: 10.1007/BF00214537. [DOI] [PubMed] [Google Scholar]
- 49.Savage J.C., Picard K., González-Ibáñez F., Tremblay M.È. A brief history of microglial ultrastructure: distinctive features, phenotypes, and functions discovered over the past 60 Years by electron microscopy. Front. Immunol. 2018;9:803. doi: 10.3389/fimmu.2018.00803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shi L., Sun Z., Su W., Xu F., Xie D., Zhang Q., Dai X., Iyer K., Hitchens T.K., Foley L.M., et al. Treg cell-derived osteopontin promotes microglia-mediated white matter repair after ischemic stroke. Immunity. 2021;54:1527–1542.e8. doi: 10.1016/j.immuni.2021.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yang J., Cheng X., Shen J., Xie B., Zhao X., Zhang Z., Cao Q., Shen Y., Qiu M. A novel approach for amplification and purification of mouse oligodendrocyte progenitor cells. Front. Cell. Neurosci. 2016;10:203. doi: 10.3389/fncel.2016.00203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hu Y., Mai W., Chen L., Cao K., Zhang B., Zhang Z., Liu Y., Lou H., Duan S., Gao Z. mTOR-mediated metabolic reprogramming shapes distinct microglia functions in response to lipopolysaccharide and ATP. Glia. 2020;68:1031–1045. doi: 10.1002/glia.23760. [DOI] [PubMed] [Google Scholar]
- 53.Zhou T., Zheng Y., Sun L., Badea S.R., Jin Y., Liu Y., Rolfe A.J., Sun H., Wang X., Cheng Z., et al. Microvascular endothelial cells engulf myelin debris and promote macrophage recruitment and fibrosis after neural injury. Nat. Neurosci. 2019;22:421–435. doi: 10.1038/s41593-018-0324-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kim J.S., Kolesnikov M., Peled-Hajaj S., Scheyltjens I., Xia Y., Trzebanski S., Haimon Z., Shemer A., Lubart A., Van Hove H., et al. A binary cre transgenic approach dissects microglia and CNS border-associated macrophages. Immunity. 2021;54:176–190.e7. doi: 10.1016/j.immuni.2020.11.007. [DOI] [PubMed] [Google Scholar]
- 55.Otxoa-de-Amezaga A., Miró-Mur F., Pedragosa J., Gallizioli M., Justicia C., Gaja-Capdevila N., Ruíz-Jaen F., Salas-Perdomo A., Bosch A., Calvo M., et al. Microglial cell loss after ischemic stroke favors brain neutrophil accumulation. Acta Neuropathol. 2019;137:321–341. doi: 10.1007/s00401-018-1954-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Tables S1, S2, S3, S4, S5, S6, S7, S8, S9 and S10 source data for figure panels are listed supplemental information.
Human single-cell/nucleus RNA sequencing data were obtained from previously published datasets (data resource provided in key resources table). All the data supporting the conclusions of the current study are presented in the figures and tables. If necessary, the raw data are available from the corresponding author upon reasonable request.
RNA-Seq data has been deposited in SRA with the accession number PRJNA941196, and will be publicly available upon publication.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.