Highlights
-
•
We identified N7-methylguanosine methylation prognostic molecular subtypes and signature in osteosarcoma for the first time.
-
•
The N7-methylguanosine-related signature was effective in predicting survival and immune landscape in osteosarcoma patients.
-
•
Dual validation of N7-methylguanosine regulators with bulk and single cell transcriptome data.
-
•
EIF4E3 may promote the growth, invasion, and migration of osteosarcoma cells as verified in-vitro experiments.
Keywords: Osteosarcoma, M7G modification, EIF4E3, Immunotherapy, Single-cell analysis
Abstract
Background
Osteosarcoma is one of the most prevalent bone malignancies with a poor prognosis. The N7-methylguanosine (m7G) modification facilitates the modification of RNA structure and function tightly associated with cancer. Nonetheless, there is a lack of joint exploration of the relationship between m7G methylation and immune status in osteosarcoma.
Methods
With the support of TARGET and GEO databases, we performed consensus clustering to characterize molecular subtypes based on m7G regulators in all osteosarcoma patients. The least absolute shrinkage and selection operator (LASSO) method, Cox regression, and receiver operating characteristic (ROC) curves were employed to construct and validate m7G-related prognostic features and derived risk scores. In addition, GSVA, ssGSEA, CIBERSORT, ESTIMATE, and gene set enrichment analysis were conducted to characterize biological pathways and immune landscapes. We explored the relationship between risk scores and drug sensitivity, immune checkpoints, and human leukocyte antigens by correlation analysis. Finally, the roles of EIF4E3 in cell function were verified through external experiments.
Results
Two molecular isoforms based on regulator genes were identified, which presented significant discrepancies in terms of survival and activated pathways. Moreover, the six m7G regulators most associated with prognosis in osteosarcoma patients were identified as independent predictors for the construction of prognostic signature. The model was well stabilized and outperformed traditional clinicopathological features to reliably predict 3-year (AUC = 0.787) and 5-year (AUC = 0.790) survival in osteosarcoma cohorts. Patients with increased risk scores had a poorer prognosis, higher tumor purity, lower checkpoint gene expression, and were in an immunosuppressive microenvironment. Furthermore, enhanced expression of EIF4E3 indicated a favorable prognosis and affected the biological behavior of osteosarcoma cells.
Conclusions
We identified six prognostic relevant m7G modulators that may provide valuable indicators for the estimation of overall survival and the corresponding immune landscape in patients with osteosarcoma.
1. Introduction
Osteosarcoma is the most common primary malignant bone tumor in adolescents and children, accounting for 20% of bone malignancies worldwide [1]. It could metastasize to almost any site or organ, mainly to the lungs and occasionally to other bones or lymph nodes [2]. The first peak of osteosarcoma incidence occurs mainly in rapidly growing adolescents, and the second peak appears mainly in people over 60 years of age [3], [4]. In young patients, primary osteosarcoma predominates, often occurring in the long bone metaphysis [5]. In older patients, secondary osteosarcomas are more common, principally located in the axial skeleton with a history of radiation or underlying bone abnormality-related disease (e.g., Paget's disease) [6], [7]. In the early stages of osteosarcoma, clinical symptoms are not obvious or specific, making it difficult for patients to capture the best treatment opportunities [8]. In addition, the prognosis of patients with metastatic or recurrent osteosarcoma remains poor due to the ineffectiveness of current treatment options [9], [10]. During treatment, osteosarcoma could also acquire drug resistance through multiple mechanisms, such as activation or mutation of proto-oncogenes or altered expression levels of drug targets, as well as a highly dynamic tumor microenvironment (TME), which makes the anti-cancer efficacy of drugs progressively less effective [11]. Therefore, it is imperative to increase the understanding of the genetic and epigenetic mechanisms underlying osteosarcoma and to explore reliable prognostic biomarkers and therapeutic targets.
Dysregulation of gene expression plays a vital role in tumor biology, and RNA modification is an approach to manipulate gene expression at the post-transcriptional level [12], [13]. Numerous RNA modifications have been identified to date, such as N5-methylcytosine, N1-methyladenosine, N6-methyladenosine (m6A), and N7-methylguanosine (m7G), among others [14]. Nucleotide modifications of mRNA and non-coding RNAs are essential in regulating various aspects of RNA metabolism, including transcription, translation, splicing, and stability [15], [16], [17]. Among them, m6A is the most abundant internal modification in mRNA, which dysregulation is responsible for the development, progression, drug resistance, metastasis, and refractory treatment of various cancers [18], [19]. Distinct from m6a, m7G is one of the most prevalent methylation modifications on tRNAs contributing to the maintenance of tRNA stabilization [20]. Importantly, tRNA modification is integrally linked to codon recognition and efficient protein synthesis, affecting multiple aspects of mitochondrial disease, neurological disorders, and tumorigenesis [21]. Moreover, m7G has been found in both the 5′ caps and interior regions of eukaryotic transcripts, which could be installed by various methyltransferases into different mRNA sequences or secondary structure motifs to perform a diversity of functions, including translation effects [22], [23]. Reportedly, the metabolism of transcripts also relies on binding the m7G caps present on the 5′ terminal of mRNAs to the eukaryotic translation initiation factor eIF4E [24]. M7G regulators, including METTL1/WDR4 complex, WBSCR22/TRMT112 complex, RNMT/RAM complex, and others, enable the addition of m7G modifications to target RNAs, thereby affecting RNA production, structure, and maturation, which ultimately mediate a myriad of critical biological processes [22]. Specifically, m7G modification on tRNA is mediated by the METTL1-WDR4 complex [25]. RNMT/RAM is actively involved in the m7G modification apparatus in the 5′ cap of mRNA, while the modification on rRNA is accounted for by WBSCR22/TRMT112 [26].
There is accumulating evidence that changes in m7G modification levels are significantly associated with carcinoma progression and prognosis [23]. M7G methyltransferases are commonly aberrantly expressed in cancer and catalyze m7G modifications in tRNA, rRNA, or miRNA, eventually affecting target gene expression and regulating tumor-associated biological functions [22]. For instance, METTL1 is severely upregulated in bladder cancer (BC) tissues while also exerting oncogenic roles in BC initiation and progression by altering the m7G modification of tRNAs to specifically affect the translation of EGFR/EFEMP1 [27]. On the other hand, m7G-modified tRNAs dominated by METTL1/WDR4 could also enhance cell cycle gene expression by reshaping mRNA translation activity, thus driving the pathogenesis in acute myeloid leukemia (AML) [28]. Also, Xia et al. [29] indicated that m7G regulator WDR4 overexpression increased m7G methylation levels in Hepatocellular carcinoma (HCC) and was strongly associated with poorer clinical survival. Mechanistically, WDR4 enhances CCNB1 mRNA stability and translation by driving the binding of EIF2A to CCNB1 mRNA, thereby promoting epithelial-mesenchymal transition (EMT) and sorafenib resistance in HCC cells. The specific implications of RNA methylation modifications in osteosarcoma have also been increasingly studied in recent years. Numerous investigations have revealed that methylation modifications extensively regulate osteosarcoma proliferation, apoptosis, migration, invasion, and tumor microenvironment [30], [31], [32]. Furthermore, aberrant expression of methylation modification regulators is intimately correlated with poor prognosis and chemotherapy resistance in osteosarcoma [30], [33]. Although the functions and mechanisms of RNA modifications have been identified in the complex epigenomic and transcriptomic environment of osteosarcoma, the biological significance and essential target genes of m7G regulators in osteosarcoma remain elusive to date.
In the current study, we comprehensively analyzed single-cell RNA-seq (scRNA-seq) and bulk RNA-seq data from multiple osteosarcoma cohorts based on the GTEx, TARGET, and GEO databases. The relationship between m7G-related molecular clusters, clinical outcomes, and immune cell infiltration characteristics was investigated. Considering the heterogeneity of osteosarcoma development, we then screened prognosis-related m7G modulators and established a validated six-gene panel for predicting the prognosis, drug sensitivity, tumor immune microenvironment (TIME), and immunotherapeutic targets of individual patients. Finally, we examined the expression of crucial genes in different cell types using scRNA-seq datasets and preliminarily validated the effect of EIF4E3 on osteosarcoma cell lines in vitro.
2. Materials and methods
2.1. Cohort collection and pre-processing
Transcriptomic and clinical data for 88 osteosarcoma samples and 396 normal control samples were downloaded from the TARGET (https://ocg.cancer.gov/programs/target) and GTEx (https://gtexportal.org) databases, respectively, for screening differential genes [34], [35]. Subsequently, scRNA-seq and bulk RNA-seq data for osteosarcoma were obtained from the GEO database (https://www.ncbi.nlm.nih.gov/geo/) for the construction of the validation cohort. Among them, fifty-three osteosarcoma samples from GSE21257 and six osteosarcoma samples from GSE162454 were included in the study. All gene expression levels were normalized with log2 (FPKM+1), and combined data were batch corrected for bias using the “sva” package.
2.2. Processing of Single-Cell RNA-Seq data
The six samples in the GSE162454 dataset were all from patients with primary osteosarcoma and not receiving neoadjuvant chemotherapy. Patient details are shown in Supplementary Table S1. The final single-cell suspension of each sample was loaded onto a 10× Genomics Chromium Single-Cell Chip and sequenced on an Illumina HiSeq X Ten instrument [36]. TISCH2 (https://tisch.comp- genomics.org/home/) is a research platform that enables interactive single-cell transcriptome visualization of the TME [37]. We analyzed the GSE162454 dataset through the above online tool to investigate the heterogeneous expression pattern of m7G regulators in osteosarcoma cells and different immune cells at the single cell level. Clusters were visualized by performing uniform manifold approximation and projection (UMAP) analysis through the TISCH2 platform and annotated against the reference genome and differential gene expression.
2.3. Differential expression analysis and enrichment analysis of the m7G regulators
The m7G modulators were identified by the search method of the previous study, and the results are shown in Supplementary Table S2 [38]. Afterwards, the “limma” package was implemented to detect the differential expression of m7G regulators between normal and osteosarcoma tissues. The average value is taken as the gene expression when multiple probes correspond to the same gene. The “reshape2,” “Corrplot,” and “circlize” packages were used to determine the correlation between m7G regulators. Information regarding the location of m7G modulators on chromosomes was mapped using the “RCircos” package. Finally, we applied the “clusterProfiler,” “ggplot2,” and “enrichplot” R packages to perform Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis and visualize the results given an adjusted p-value < 0.05.
2.4. Identification of molecular subtypes defined by m7G regulators
Unsupervised clustering analysis was performed to identify m7G-associated molecular subtypes based on differentially expressed m7G modulators (DEm7Gs) by applying the “ConsensusClusterPlus” package in osteosarcoma patients from the TARGET cohort. The Single-sample Gene Set Enrichment Analysis (ssGSEA) algorithm enables accurate estimation of immune cell infiltration level based on the RNA-seq data from the osteosarcoma cohort to compare differences in the immune characteristics of distinct phenotypes. Kaplan-Meier (KM) survival curves were then used for subgroup survival analysis by the “survival” and “survminer” packages. The subset (c2.cp.kegg.v7.4.symbols.gmt) was obtained from the MSigDB database (https://www.gsea-msigdb.org/gsea/index.jsp). Then, GSEA was applied to identify differences in biological function between diverse clusters using the “GSEABase” and “GSVA” packages with p < 0.05 considered statistically significant.
2.5. Generation of m7G-regulated risk score
To further understand the molecular characteristics of each osteosarcoma sample, risk scores quantifying the m7G modification pattern of individual patients were constructed by univariate Cox regression analysis and Lasso regression analysis. Univariate Cox analysis was conducted to examine the prognostic significance of DEm7Gs in osteosarcoma. Following this, we used Lasso regression analysis to avert overfitting with 10-fold cross-validation to determine the optimal penalty parameter (λ). The m7G-related risk score (termed MRRS) was calculated as follows: MRRS = Σ (βi × Expi) (β: coefficient, Exp: expression level of m7G modulators). After calculating MRRS for each patient separately in the TARGET and GSE21257 cohort, KM analysis was applied to assess the survival outcomes of patients with distinct MRRS. The reliability of the risk score was verified by performing ROC analysis using the “timeROC” package to calculate the AUC values at different time points. Finally, PCA analysis was performed to verify the usefulness of the MRRS in differentiating individual patients.
2.6. Analysis of immune landscape and immune checkpoint genome
The CIBERSORT, ssGSEA, and ESTIMATE algorithms were used to obtain further evidence on the utility of MRRS in anticipating the immune landscape of osteosarcoma, respectively. The CIBERSORT algorithm was implemented to quantify the extent of immune infiltration of 22 immune cell subtypes in osteosarcoma samples based on bulk RNA-seq data. Pearson correlation analysis was then performed to calculate the correlation coefficient between immune cells and the MRRS. We executed the ssGSEA algorithm to analyze the differential profiles of various immune cells and functions in distinct MRRS subgroups and visualize the results by “limma,” “reshape2,” “GSEABase,” and “ggpubr” packages. The ESTIMATE algorithm is a widely used method for inferring the content of mesenchymal and immune cells in malignant tissues using gene expression features [39]. We predicted immune scores, stromal scores, and tumor purity for each osteosarcoma patient using the “estimate” package. Based on available studies, immune checkpoint expression and human leukocyte antigen (HLA) genotype may be associated with clinical outcomes of immune checkpoint inhibitor (ICI) treatment [40], [41]. Then, we applied the “limma,” “reshape2”, “plyr,” “ggplot2”, and “ggpubr” packages to investigate the divergent expression of immune checkpoint genes as well as HLA genes between high and low MRRS subgroups to predict the response to ICI in patients with osteosarcoma.
2.7. Clinical correlation analysis
Clinical information of patients with osteosarcoma in the TARGET dataset was extracted and subdivided for each clinical parameter. Specifically, two groups were classified according to age (≤18 and >18 years), gender (female and male), and metastatic status (metastatic and non-metastatic) [42]. Categorical variables were used to assess the relationship between MRRS and clinical characteristics by chi-square test. Kaplan-Meier survival analysis stratified by clinicopathological characteristics was subsequently performed. We screened for independent prognostic factors from clinical variables and MRRS by conducting univariate and multivariate Cox regression analyses.
2.8. Evaluation of drug sensitivity
The “pRRophetic” package can be implemented to predict phenotypic and drug response based on gene expression data to explore potential therapeutic agents for osteosarcoma. We calculate the half maximal inhibitory concentration (IC50) of common chemotherapeutic agents for drug sensitivity analysis in different MRRS groups of osteosarcoma patients by applying the “pRRophetic” package. The CellMiner database (https://discover.nci.nih.gov/cellminer) contains NCI-60 compound activity data and RNA-seq expression profiles from which FDA-approved drugs were screened to predict the correlation between core gene expression levels and drug sensitivity (P < 0.05 was used as a screening criterion).
2.9. Methylation modification analysis
MEXPRESS is a database used to visualize the relationship between genomic location, methylation modifications and gene expression in the TCGA database [43], [44]. We used this database (https://mexpress.be/) to analyze the expression of six signature-contained m7G modulators in relation to the methylation level of CpG in sarcoma.
2.10. Cell lines culture and quantitative real-time polymerase chain reaction (qRT-PCR)
All cell lines were obtained through the Cell Bank of the Chinese Academy of Sciences (Shanghai, China). HFOB1.19 cells were cultured in DMEM/F12 medium (BI, USA) at 33.5 °C. U2OS cells were maintained in McCoy's 5A medium (Gibco, USA). MNNG/HOS and MG63 cells were cultivated in high-glucose DMEM (Gibco, USA), while 143B cells were maintained in RPMI 1640 medium (BI, USA). All three osteosarcoma cells were incubated at 37 °C, 5% CO2. The media were supplemented with 10% fetal bovine serum (Gibco, USA) and 1% penicillin–streptomycin (Solarbio, Beijing, China).
Total RNA was prepared using RNA-easyTM isolation reagent (Vazyme Biotech Co., Ltd., Nanjing, China) and then reverse transcribed into cDNA with HiScript III RT SuperMix (Vazyme, China). Finally, qRT-PCR was performed using the UltraSYBR Mixture Kit (CWBIO, China). The relative mRNA expression of genes was calculated by 2−ΔΔCt methods and normalized to GAPDH. Primer sequences for relevant genes are described in Supplementary Table S3.
2.11. Cell proliferation assay and Transwell assay
Osteosarcoma cells were transfected with the human EIF4E3 overexpression plasmid and incubated in serum-free, antibiotic-free medium for 4 to 6 h before replacing the complete medium. Following this, cell viability was assessed by a colony formation assay in which 1 × 10 3 osteosarcoma cells were inoculated in 6-well plates and cultured for 12 days before staining with crystal violet. For the CCK-8 assay, 5 × 103 transfected osteosarcoma cells were incorporated into a 96-well culture plate for 24 h. Afterward, 10 μL of CCK-8 reagent (Yeasen, China) was added to measure the absorbance at 450 nm. Transwell assays were conducted to analyze the effect of EIF4E3 on osteosarcoma cell migration and invasion. In the upper chamber, osteosarcoma cells were inoculated at 5 × 104/well into serum-free medium, and in the lower chamber, 500 μL of medium containing 10% FBS was added. Cells were seeded into the upper chamber with precoated Matrigel For invasion. One day later, cells migrating to the subsurface of the PET membrane were immobilized with 4% paraformaldehyde and stained with crystal violet.
2.12. Statistical analysis
The statistical analysis of bioinformatics results in this study was performed by R software (v4.2.1). Univariate and Lasso Cox regression analyses were applied to determine prognostic indicators in osteosarcoma. The KM method was carried out to measure the survival discrepancy between subgroups. The Spearman correlation method was used to analyze the association between risk scores and immune cell infiltration abundance. Statistical differences between groups were calculated by t-test or one-way ANOVA using GraphPad Prism 8.0. P < 0.05 was considered statistically significant.
3. Results
3.1. The landscape of m7G modulators in osteosarcoma
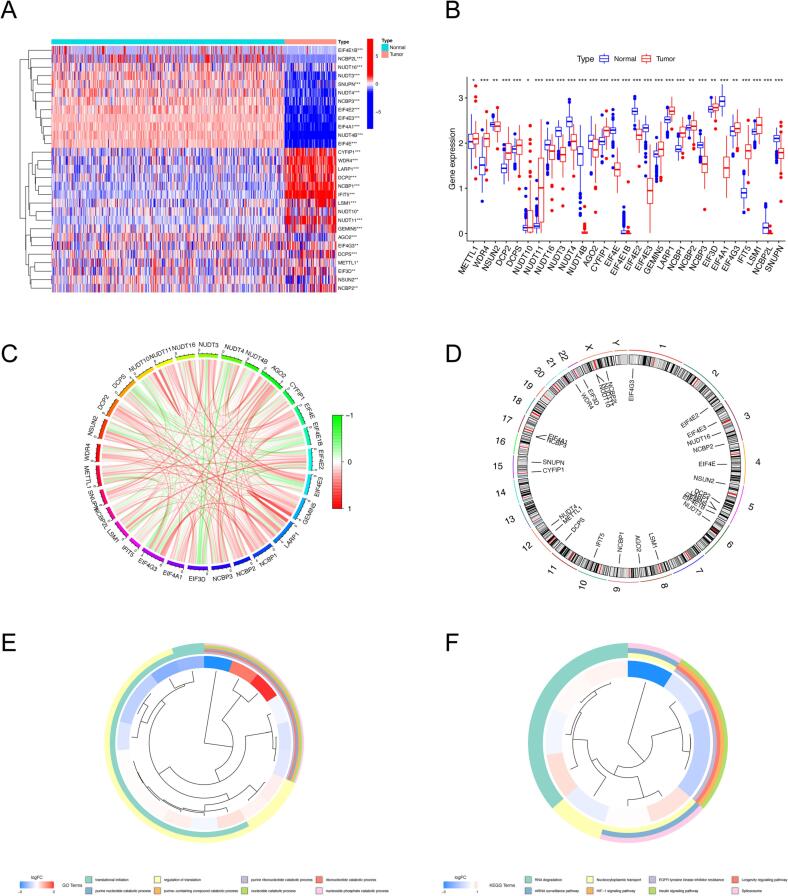
In this study, we collected m7G-related genes and proceeded to compare the expression discrepancies in osteosarcoma and normal samples before assessing their prognostic performance. The results indicated that mRNA expression of 15 m7G regulators was significantly higher in osteosarcoma tissues than in normal tissues, whereas 14 m7G regulators were downregulated (Fig. 1A, B). These DEm7Gs served as candidate genes for the construction of prognostic features. Following this, the network of m7G modulators was plotted according to the expression abundance of each regulator to illustrate their internal associations (Fig. 1C). Fig. 1D shows the detailed location of RNA methylation regulators on the chromosomes. Additionally, we performed GO and KEGG functional enrichment analysis to delve into potential biological functions. According to GO enrichment analysis, DEm7Gs are involved in several biological processes, such as translational initiation, regulation of translation, nucleotide catabolic process, and nucleobase-containing small molecule biosynthetic process; concerning cellular component, DEm7Gs were primarily enriched in RNA cap binding complex and mRNA cap binding complex. Also, these DEm7Gs have a complex set of molecular functions, including nucleotide diphosphatase activity, RNA 7-methylguanosine cap binding, and translation regulator activity (Fig. 1E, Supplementary Table S4). Moreover, KEGG enrichment results revealed that DEm7Gs are prominently involved in RNA degradation and nucleocytoplasmic transport among others (Fig. 1F, Supplementary Table S4). We accordingly hypothesized that the potential biological functions of DEm7Gs are relevant to mRNA translation, m7G modification, and epigenetic regulation. In conclusion, the above results indicate the heterogeneous expression and molecular regulatory mechanisms of m7G modulators in osteosarcoma.
Fig. 1.
Expression levels and chromosome localization of m7G modulators. (A-B) Heatmap (A) and boxplot (B) of DEm7Gs in osteosarcoma and normal tissues. (C) The association network shows the interactions between m7G modulators. (D) Location of m7G modulators on the chromosome. (E-F) GO enrichment analysis (E) and KEGG pathway analysis (F) of DEm7Gs. GO, Gene Ontology; KEGG, Kyoto Encyclopedia of Genes and Genomes; DEm7Gs, differentially expressed m7G modulators. *p < 0.05; **p < 0.01; ***p < 0.001.
3.2. Consensus clustering identified m7G modification patterns in osteosarcoma
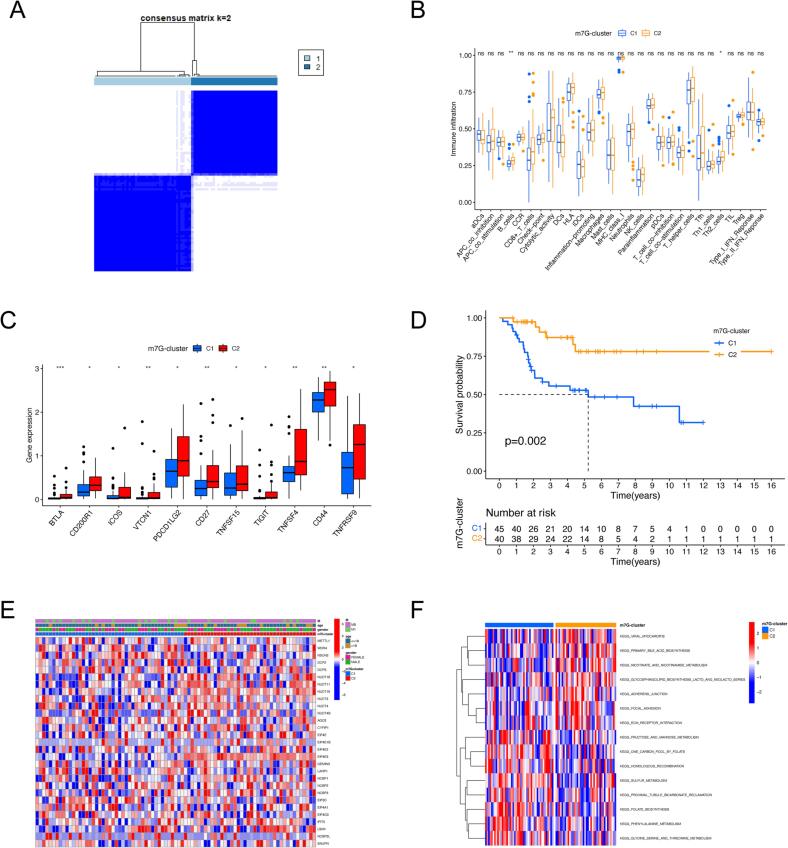
We executed an unsupervised consensus clustering to segment the osteosarcoma samples with different m7G patterns according to the expression of 29 m7G regulators, and k = 2 showed the best cluster stability (Fig. 2A). Two distinguishable m7G patterns were finally identified, named m7G-cluster 1 to 2. Then, we performed the ssGSEA algorithm to evaluate the immune microenvironment features in the two m7G modification patterns. As shown in Fig. 2B, the enrichment score of B cells and Th2 cells were significantly downregulated in m7G-cluster1compared to m7G-cluster2. In addition, there were significant discrepancies in the expression of several immune checkpoint genes between two m7G modification subtypes. The m7G-cluster1 showed impaired expression of immune checkpoint genes, including BTLA, CD200R1, ICOS, VTCN1, PDCD1LG2, CD27, TNFSF15, TIGIT, TNFSF4, CD44, and TNFRSF9 (Fig. 2C). This result suggests m7G-cluster2 is more likely to benefit from immune checkpoint inhibitor (ICI) therapy. Interestingly osteosarcoma patients in the m7G-cluster2 also showed a matched survival advantage using KM survival analysis (Fig. 2D). The correlation between clinicopathological features and m7G modification patterns is shown in Fig. 2E, but no significant discrepancies were detected. Next, the biological functions activated in distinct phenotypes were assessed by GSVA, and our results demonstrated that metabolism-related pathways, including glycine, serine, threonine, fructose, mannose, and sulfur metabolism pathways, were all significantly upregulated in the m7G-cluster1. In contrast, functions, specifically primary bile acid biosynthesis, extracellular matrix (ECM) receptor interaction, and focal adhesion, are prominently activated in m7GCluster2 (Fig. 2F, Supplementary Table S5).
Fig. 2.
Identification of m7G modification patterns in osteosarcoma by consensus clustering. (A) Two clusters identified by consistent clustering analysis (k = 2). (B) The discrepancies in the abundance of immune cell infiltration between the two m7G clusters. (C) The distinction in immune checkpoint expression levels between the two m7G clusters. (D) Kaplan-Meier survival curves of osteosarcoma patients with different m7G modification patterns. (E) Variation in clinical features and expression levels of m7G modulators between the two m7G clusters. (F) GSVA enrichment analysis of osteosarcoma patients with different m7G modification patterns for ascertaining the activation status of biological pathways. GSVA, gene set variation analysis; *p < 0.05; **p < 0.01; ***p < 0.001; NS, no significance.
3.3. Establishment and validation of m7G-related risk score
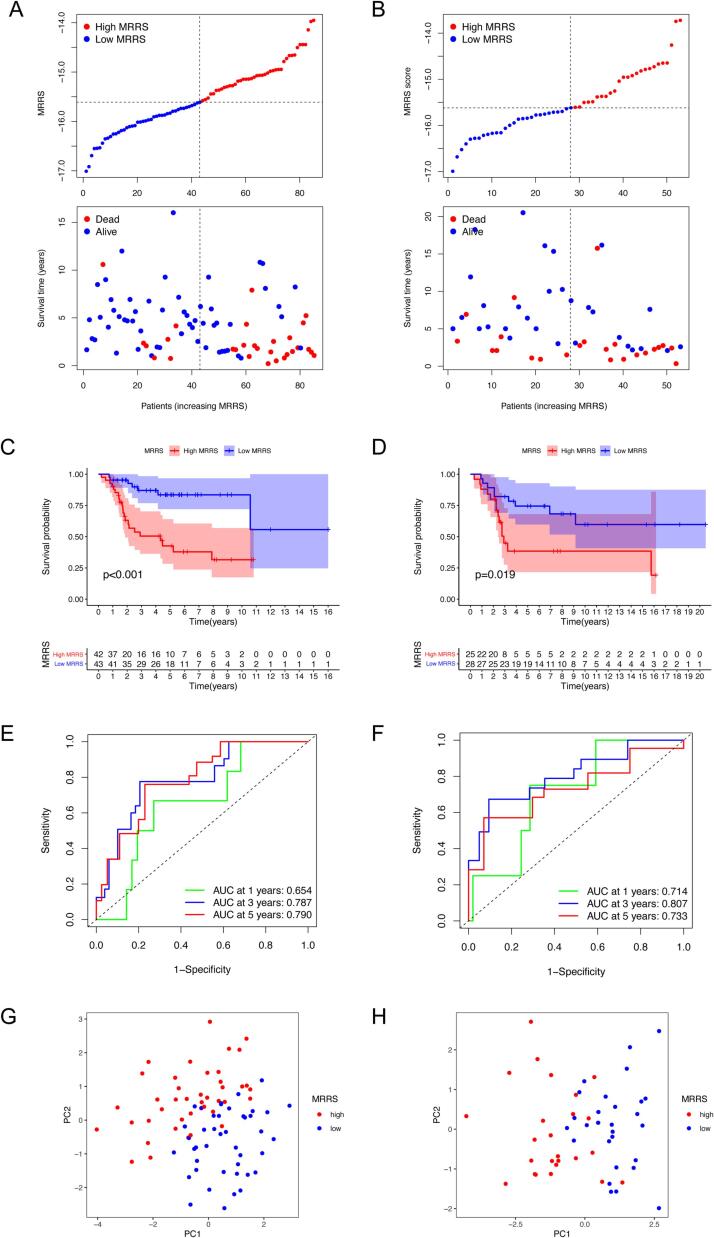
To estimate the m7G methylation modification patterns in individual patients, we obtained 22 survival-associated m7G regulators by univariate Cox regression analysis (Supplementary Fig. S1A). Subsequent Lasso regression analysis identified six m7G regulators for constructing the m7G-related risk score termed MRRS to characterize the immune microenvironment and predict overall survival (OS) (Supplementary Fig. S1B, C). The MRRS was calculated as follows: MRRS = (−0.27242 × NUDT11 expression) + (−1.21282 × NUDT16 expression) + (−3.09094 × CYFIP1 expression) + (−0.15031 × EIF4E3 expression) + (0.50841 × LARP1 expression) + (0.40932 × EIF4A1 expression). Osteosarcoma patients in the TARGET (training set) and GSE21257 (test set) cohorts were divided into low-MRRS and high-MRRS groups based on the median MRRS, and the survival status of patients with different MRRS was examined (Fig. 3A, B). Kaplan-Meier analysis revealed a significantly higher survival probability for patients with low MRRS relative to those with high MRRS in both cohorts, suggesting that MRRS contributes to differentiating the prognosis of osteosarcoma patients with various m7G modification patterns (Fig. 3C, D). ROC analysis demonstrated the robustness of the MRRS in forecasting the outcome of osteosarcoma patients in the TARGET cohort with the area under the curves (AUCs) of 0.654, 0.787, and 0.790 for 1-, 3-, and 5-year survival, respectively (Fig. 3E). Again this was also confirmed in the external validation set with AUCs greater than 0.7 for 1-, 3-, and 5-year OS (Fig. 3F). PCA mapping revealed differences in the distribution between patients with high and low MRRS in both osteosarcoma cohorts (Fig. 3G, H).
Fig. 3.
Construction and validation of the m7G-related risk score. (A-B) The MRRS distribution of osteosarcoma patients in the TARGET (A) and GSE21257 (B) cohort. (C-D) Kaplan-Meier survival curves for osteosarcoma patients in the TARGET (C) and GSE21257 (D) cohort. (E-F) ROC curves for 1-, 3-, and 5-year survival in the TARGET (E) and GSE21257 (F) cohort. (G-H) PCA analysis for patients with diverse MRRS in the TARGET (G) and GSE21257 (H) cohort. ROC, receiver operating characteristic; PCA, principal components analysis; MRRS, m7G-related risk score.
3.4. Extended application of MRRS in immune infiltration and immunotherapy
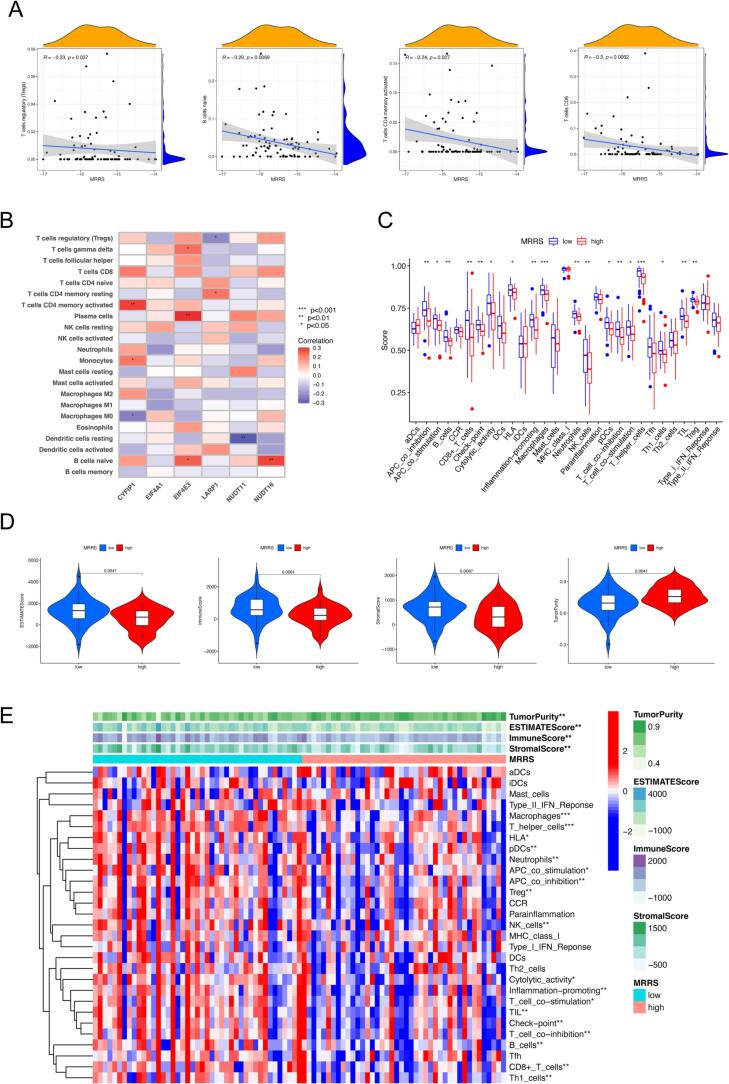
Studies have shown that osteosarcoma creates a locoregional immune-tolerant TME by regulating the recruitment and polarization of immune cells [45]. Notably, the composition and content of immune infiltrating cells in the TME are critical determinants of tumor progression and immunotherapeutic outcome [46], [47]. Therefore, we utilized a combination of the CIBERSORT, ssGSEA, and ESTIMATE algorithms to investigate the effectiveness of MRRS in predicting the immune landscape. The correlation between immune cells and MRRS was obtained by calculating the content of 22 immune cell subsets in osteosarcoma patients by the CIBERSORT algorithm. As shown in Fig. 4A, four infiltrating immune cells, including regulatory T cells (Tregs), naive B cells, activated memory CD4+ T cells, and CD8+ T cells were significantly negatively related to MRRS. In addition, we illustrate the association between the six m7G regulators involved in the MRRS and the abundance of 22 immune cells (Fig. 4B). The ssGSEA was performed to assess the relative abundance of immune cell infiltration and immune function in osteosarcoma TME. Compared with the high MRRS group, the low MRRS group exhibited generally increased levels of infiltrating immune cells, including B cells, CD8+ T cells, macrophages, neutrophils, natural killer (NK) cells, plasmacytoid dendritic cells (pDCs), T helper (Th) cells, Th1 cells, tumor-infiltrating lymphocytes (TILs), and Tregs (P < 0.05, Fig. 4C). We also examined a variety of immune-related functions, among which antigen-presenting cell (APC) co-stimulation, cytolytic activity, human leukocyte antigen, inflammation-promoting, checkpoint, T cell co-inhibition, APC co-inhibition, and T cell co-stimulation were also more active in the low MRRS (P < 0.05, Fig. 4C). Concerning the ESTIMATE algorithm, the results demonstrated that low MRRS had a higher immune score, stromal score, ESTIMATE score, and lower tumor purity compared to high MRRS, which also corresponded to the results of the CIBERSORT, ssGSEA algorithm (Fig. 4D). Considering the lower enrichment of most immune cells and functions, we hypothesized that the low MRRS group with high tumor purity might exhibit an immune-desert phenotype. Finally, we combined the above results to produce the heatmap visually expressing the differences in immune scores, interstitial scores, estimated scores, tumor purity, immune cells, and functions of different MRRS subgroups (Fig. 4E).
Fig. 4.
Extended application of MRRS in the immune landscape. (A) Correlation analysis between MRRS and immune infiltrating cells. (B) Correlation analysis between m7G modulators involved in the MRRS and immune infiltrating cells. (C) ssGSEA analysis of diverse MRRS subgroups. (D) ESTIMATE analysis of diverse MRRS subgroups. (E) Heatmap for visualization of differences in the immune score, stromal score, ESTIMATE score, tumor purity, immune cells, and function in distinct MRRS subgroups. ssGSEA, single-sample gene-set enrichment analysis; MRRS, m7G-related risk score. *p < 0.05; **p < 0.01; ***p < 0.001.
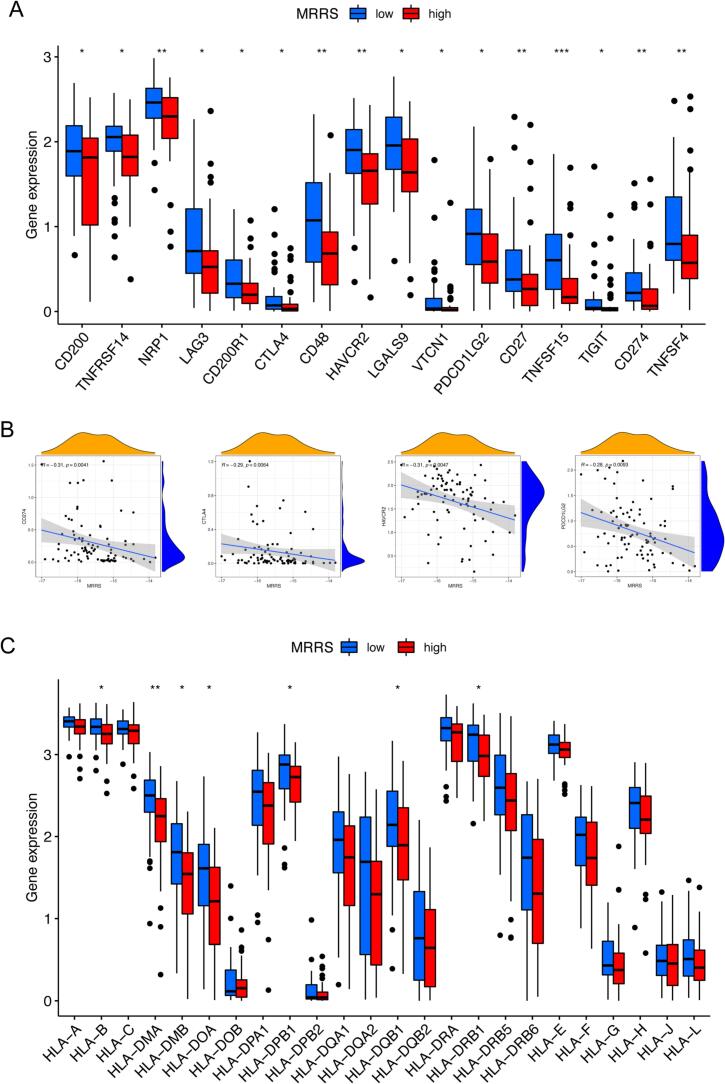
Immune checkpoints play a pivotal position in predicting the efficacy of ICI therapy as well as immune escape [48], [49]. We further analyzed the differential expression of common immune checkpoints in MRRS subgroups to assess the impact of MRRS in immunotherapy. Our investigation revealed that osteosarcoma patients with low MRRS manifested enhanced expression of CD200, TNFRSF14, NRP1, LAG3, CD200R1, CTLA4, CD48, HAVCR2, LGALS9, VTCN1, PDCD1LG2, CD27, TNFSF15, TIGIT, CD274, and TNFSF4 (P < 0.05, Fig. 5A). In addition, four immune checkpoints (CTLA4, HAVCR2, PDCD1LG2, and CD274) were negatively correlated with MRRS (P < 0.05, Fig. 5B). The structural and functional variability of HLA is the basis for an effective adaptive immune response to tumor antigens, and individual variation in HLA genotype patterns could affect the clinical outcome of ICI therapy [41], [50]. Therefore, we also evaluated the relationship between MRRS and HLA-related gene expression. Osteosarcoma patients with high MRRS exhibited elevated expression of HLA class I genes (including HLA–E, HLA–H, and HLA–L) and HLA class II genes (including HLA–DMB, HLA–DOA, HLA–DPA1, HLA–DPB1, HLA–DQA1, HLA–DQB1, HLA–DRA, HLA–DRB1, and HLA–DRB5) (P < 0.05, Fig. 5C). In conclusion, the above results suggest that osteosarcoma patients with diverse MRRS present variations in immune checkpoints, immune cells, and function. The MRRS may be instrumental in reflecting the TME infiltration characteristics of individual osteosarcoma patients and providing a valuable reference in selecting appropriate immunotherapeutic agents in the clinical treatment of osteosarcoma.
Fig. 5.
Extended application of MRRS in immunotherapy response. (A) Discrepancies in immune checkpoint gene expression between diverse MRRS subgroups. (B) Correlation analysis between MRRS and immune checkpoint genes. (C) Discrepancies in human leukocyte antigen gene expression between diverse MRRS subgroups. MRRS, m7G-related risk score. *p < 0.05; **p < 0.01; ***p < 0.001.
3.5. Characterization of the relationship between MRRS and clinical features
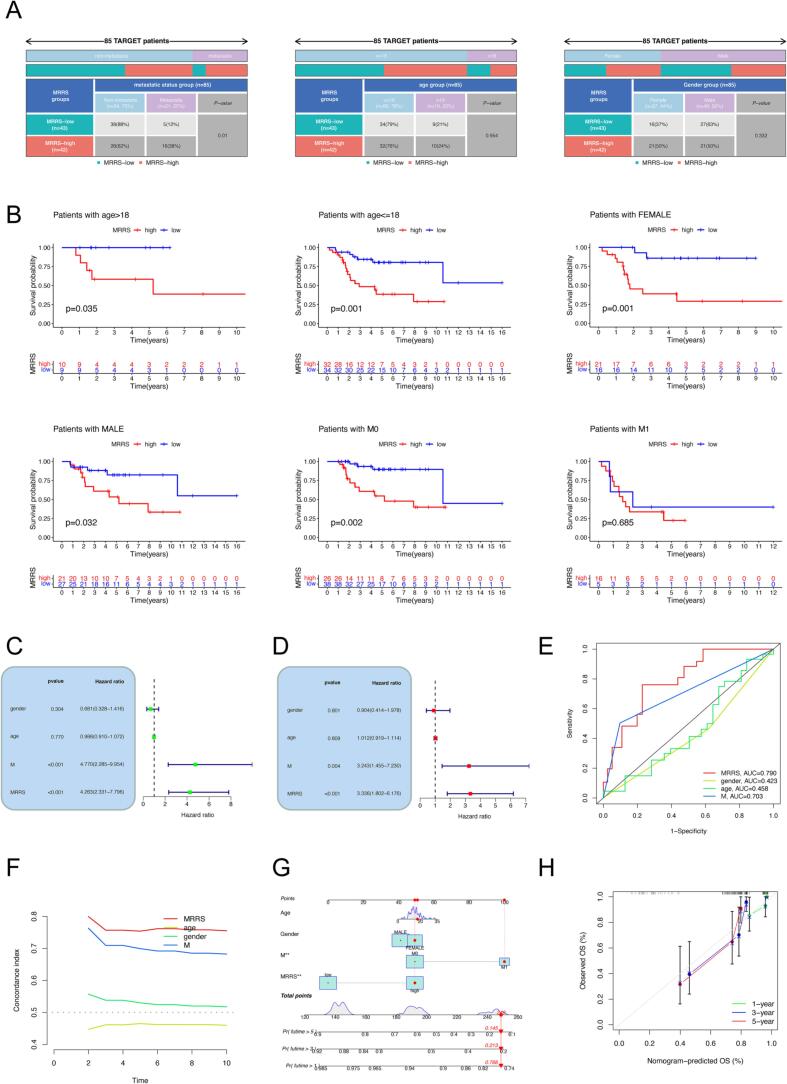
We compared the clinicopathologic features of high and low MRRS groups and determined whether MRRS could be an independent predictor for osteosarcoma. Distant metastasis was associated with high MRRS in patients with osteosarcoma but without significant differences in gender and age between MRRS subgroups (Fig. 6A). The high propensity of osteosarcoma to spread and metastasize is the most influential intrinsic factor for poor patient prognosis [51]. Again, this validates the consequences of MRRS-based survival analysis that clinical outcomes are worse in the high MRRS subgroup (Fig. 3C). Additionally, low MRRS portended a better prognosis in all subgroups, including non-metastatic subgroups, different gender and age subgroups, with the exception of metastatic subgroups (Fig. 6B). We subsequently identified MRRS and metastatic status as independent indicators for osteosarcoma patients by univariate and multivariate Cox regression analysis (Fig. 6C, D). Moreover, the 5-year AUC of MRRS was greater than the AUC of metastasis in the TARGET cohort (Fig. 6E). The C-index also indicated higher discriminatory strength of MRRS as a predictor of OS than other clinical features (Fig. 6F). Finally, we created a nomogram integrating MRRS and clinical parameters to predict the likelihood of survival in patients with osteosarcoma (Fig. 6G). Calibration curves were plotted for 1-, 3-, and 5-year OS to assess the accuracy of the nomogram (Fig. 6H).
Fig. 6.
Clinical application potential of MRRS. (A) Discrepancies in clinical features between different MRRS subgroups. (B) Kaplan-Meier survival analysis stratified by clinical characteristics. (C-D) Univariate (C) and multivariate (D) independent prognostic analysis of MRRS and clinical variables. (E) Comparison of the predictive performance of MRRS and different clinical features by ROC analysis. (F) Comparison of the C-index between MRRS and clinical variables. (G) Nomogram for predicting the overall survival of osteosarcoma patients at 1, 3, and 5 years. (H) Calibration curve of the nomogram. MRRS, m7G-related risk score.
3.6. Characterization of the relationship between MRRS and drug sensitivity
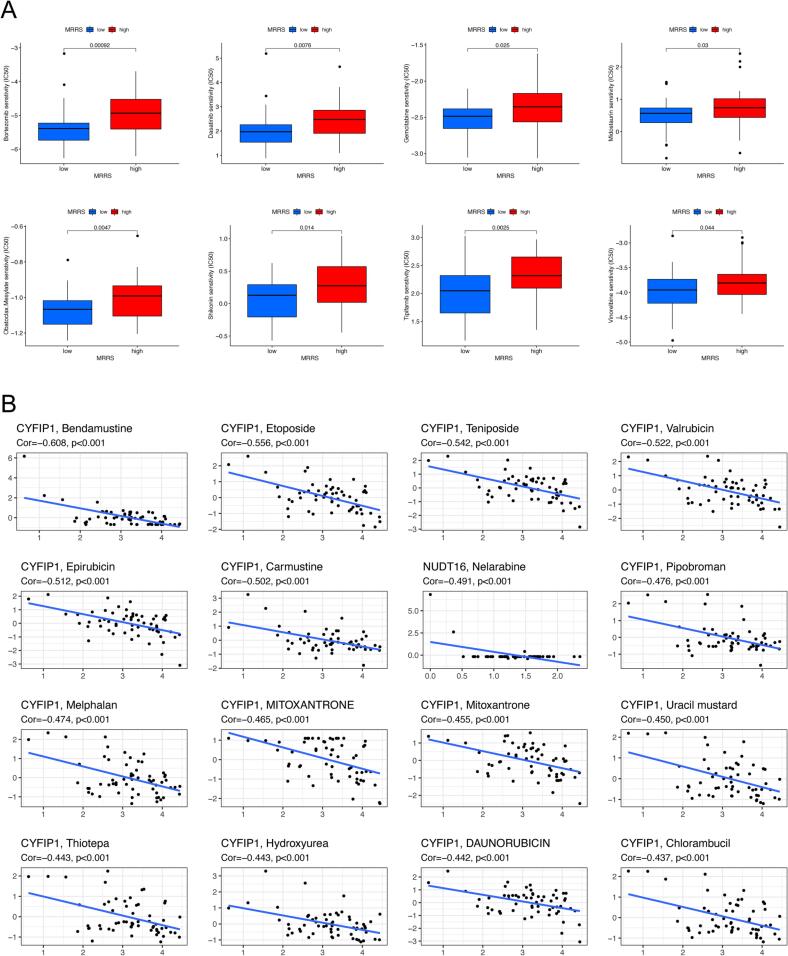
We appraised the value of MRRS in predicting drug sensitivity to common chemotherapeutic agents and molecularly targeted drugs by analyzing the IC50. Our results indicated significantly stronger response sensitivity in patients with low MRRS than in patients with high MRRS, as the IC50 of bortezomib, dasatinib, gemcitabine, midostaurin, obatoclax mesylate, shikonin, tipifarnib, and vinorelbine were more elevated in the high MRRS subgroup, suggesting a possibly more significant benefit from the above drug treatments (Fig. 7A). Furthermore, we explored the association between the expression of m7G regulators involved in MRRS and chemotherapies sensitivity. The expression of CYFIP1 and NUDT16 was negatively correlated with sensitivity to most drugs, i.e., the higher the expression of CYFIP1 and NUDT16 in patients, the more resistant they were to drugs such as bendamustine, etoposide, teniposide, valrubicin, epirubicin, and nelarabine among others (Fig. 7B). All the results are shown in Supplementary Table S6.
Fig. 7.
MRRS-based chemotherapy sensitivity analysis in osteosarcoma. (A) Discrepancies in drug sensitivity between diverse MRRS subgroups. (B) Correlation analysis of m7G modulators involved in MRRS with drug sensitivity. MRRS, m7G-related risk score.
3.7. Single-cell transcriptomic analysis of m7G regulators
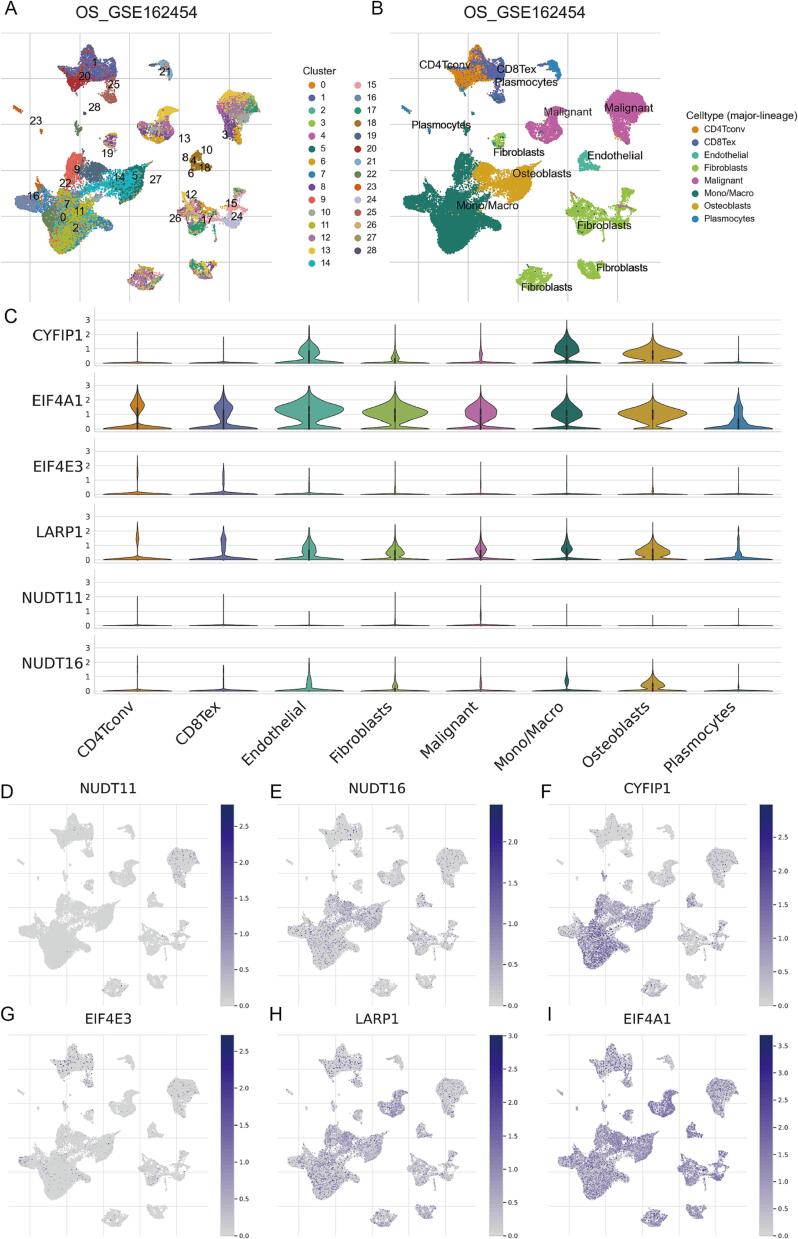
The bulk RNA-seq data has more samples and facilitates reflection of inter-population characteristics, whereas scRNA-seq data provides transcript abundance of individual cells with higher sensitivity. In this study, we also examined the expression levels of m7G regulators involved in MRRS by using the TISCH platform to analyze osteosarcoma scRNA-seq data (GSE162454). After dimensionality reduction, UMAP-based cell clustering delineates 29 major clusters (Fig. 8A). According to the lineage markers, these clusters were labeled as conventional CD4+ T cells (CD4Tconv), exhausted CD8+ T cells (CD8Tex), endothelial cells, fibroblasts, malignant cells, monocytes or macrophages (Mono/Macro), osteoblasts, and plasmocytes (Fig. 8B). It is worth mentioning that osteoblasts form four cell clusters, whereas malignant cells form two cell clusters. Genetic and phenotypic heterogeneity of osteosarcoma is widespread in terms of cell morphology, differentiation stage, microenvironment, treatment response, and function [52], [53], [54]. Hence we hypothesize that this phenomenon may be caused by intra- and inter-tumor heterogeneity in the tested osteosarcoma. Following this, we investigated the expression profiles of six m7G regulators in the above cell types. The result revealed a significant upregulation of EIF4A1 in osteosarcoma and immunocytes, while the transcript abundance of EIF4E3 was reduced in all major cell types (Fig. 8C). As shown in Fig. 8D-I, we visualized the abundance distribution of the six core m7G modulators in various cell types by UMAP plots, with CYFIP1 markedly enriched in endothelial cells, osteoblasts, monocytes, or macrophages.
Fig. 8.
Expression patterns of m7G modulators at the single-cell profiles. (A) Cells were clustered into 29 types by the UMAP dimensionality reduction algorithm, with each color representing an annotated phenotype. (B) UMAP plot of 8 predominant cell types from osteosarcoma scRNA-seq data. (C) Violin plot for displaying the expression levels of m7G modulators in all cell types. (D-H) UMAP plots for visualizing the abundance distribution of core m7G modulators, including NUDT11 (D), NUDT16 (E), CYFIP1 (F), EIF4E3 (G), LARP1 (H), and EIF4A1 (I).
3.8. Overexpression of EIF4E3 inhibits osteosarcoma cell proliferation and migration
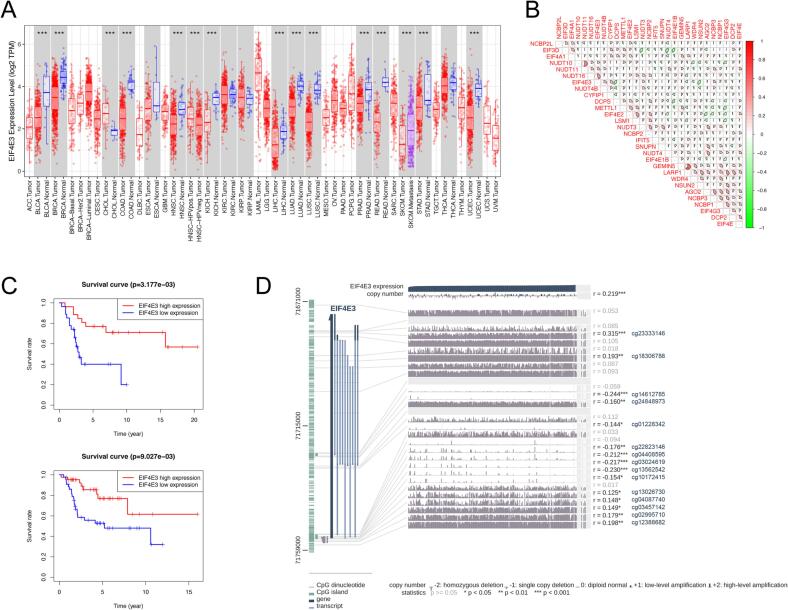
EIF4E3 belongs to the eukaryotic translation initiation factor 4E (eIF4E) family. It was previously reported that EIF4E3 acts as a specific tumor suppressor, binding to m7G caps atypically to inhibit the oncogenic transformation of cells [55]. In prostate cancer, EIF4E3 was implicated in hyaluronan-mediated invasion and metastasis of androgen-independent tumor cells [56]. However, the prognostic value of EIF4E3 and its relationship to the malignant phenotype of osteosarcoma remains virtually unknown. Our pan-cancer analysis revealed thwarted expression of EIF4E3 in most cancers, including bladder cancers, breast carcinoma, colon adenocarcinoma, and lung adenocarcinoma, among others (Fig. 9A). Moreover, the content of EIF4E3 was positively associated with the expression of NUDT10, NUDT11, NUDT16, NUDT4B, and CYFIP1, but negatively related to the expression of LARP1, WDR4, and NSUN2 in osteosarcoma (Fig. 9B). Following this, survival curves showed that osteosarcoma patients in the EIF4E3 high-expression group exhibited superior OS in both cohorts (Fig. 9C). In addition, fifteen methylation CpG sites were associated with EIF4E3 expression in MEXPRESS (Fig. 9D). Regarding the level of immune cell infiltration in osteosarcoma, EIF4E3 was positively correlated with the abundance of gamma delta T cells, plasma cells, and naive B cells (Fig. 4B).
Fig. 9.
Expression level and survival analysis of EIF4E3 in osteosarcoma. (A) Pan-cancer analysis of differential expression EIF4E3 in different cancers. (B) Correlation analysis of the expression of EIF4E3 and other m7G regulators in osteosarcoma. (C) Overall survival analysis of osteosarcoma patients with diverse EIF4E3 expression levels in the GSE21257 (top) and TARGET cohort (bottom). (D) Associations between EIF4E3 expression and methylation CpG sites in MEXPRESS. *p < 0.05; **p < 0.01; ***p < 0.001.
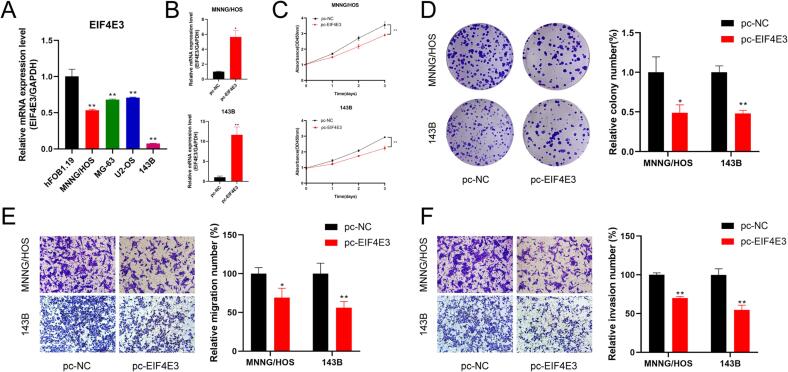
We further validated the expression levels of EIF4E3 in osteosarcoma by qRT-PCR. EIF4E3 exhibited poor expression in all osteosarcoma cell lines compared to osteoblasts, consistent with our comprehensive analysis derived from scRNA-seq and bulk RNA-seq data (Fig. 10A). Subsequently, we overexpressed EIF4E3 in 143B and MNNG/HOS cells to further determine the impact of EIF4E3 on the biological function of osteosarcoma (Fig. 10B). Cell viability was verified by subjecting the transfected 143B and MNNG/HOS cells to CCK-8 assay and clone formation assay, which suggested that the upregulation of EIF4E3 inhibited cell proliferation (Fig. 10C, D). In addition, reduced migration and invasion of 143B and MNNG/HOS cells were observed after transfection with EIF4E3 compared to the control group (Fig. 10E, F). These results imply that EIF4E3 may be a potential functional biomarker for osteosarcoma.
Fig. 10.
Validation of EIF4E3 expression levels and cell functions assay. (A) Expression of EIF4E3 was detected in osteosarcoma cell lines by qRT-PCR. (B-F) Overexpression of EIF4E3 (B) inhibited the proliferation (C, D), migration (E), and invasion (F) abilities of osteosarcoma cells. *p < 0.05; **p < 0.01.
4. Discussion
Osteosarcoma typically occurs in childhood and adolescence and is now the second leading cause of tumor-related death in adolescents due to its high metastatic rate, aggressiveness, and increasing annual prevalence [57], [58]. The definition of early-stage osteosarcoma is tremendously challenging owing to the non-specific presentation, which leads to the vast majority of patients with micrometastatic lesions at the time of diagnosis [59], [60]. Remarkably, distant metastasis is a principal cause of treatment failure and death in patients with osteosarcoma [61]. Therefore, it is crucial to determine novel diagnostic markers and therapeutic targets in patients with osteosarcoma. The intimate correlation between epigenetic changes and tumor progression has been extensively characterized [62]. Although the complexity of cancer pathogenesis is implicated in cumulative genetic and epigenetic alterations, it is not negligible that epigenetic regulation occurs more frequently and significantly than somatic mutations [63], [64]. The m7G modification facilitates the modification of RNA structure and function to manipulate specific gene expression as a hotspot in recent epigenetic studies [22]. It has extensive impacts on mRNA, tRNA, and rRNA along with serving in diverse biological processes such as transcriptional elongation and pre-mRNA splicing [65]. Accumulating evidence emphasized that addressing aberrant m7G modifications in cancer is expected to be a practical therapeutic approach [66]. Therefore, we investigated m7G regulators as potentially valuable targets to enhance treatment and ameliorate prognosis in osteosarcoma.
In the present study, we first screened 29 m7G regulators differentially expressed in osteosarcoma and found them abundant in biological functions such as initiation and regulation of translation and RNA catabolic processes by enrichment analysis. In fact, m7G methylation, as a critical component of post-transcriptional modification, is involved in various aspects of cancer RNA metabolism that have been extensively characterized, including miRNA maturation, mRNA translation, and mRNA decay [22], [67], [68]. In hepatocellular carcinoma, METTL1-mediated m7G tRNA modification enhanced mRNA translation in a codon frequency-dependent manner and accelerated tumorigenesis and progression in vitro and in vivo [20]. In another study, Pandolfini et al. identified m7G-cap methylation as a novel modification pathway of miRNA structure and suggested that METTL1 promotes let-7 miRNA processing and specifically affects cell migration via m7G methylation [69]. Orellana et al. also demonstrated that increased m7G modification of Arg-TCT tRNA helps prevent mRNA decay and reduce ribosomal arrest to drive oncogenic transformation, which is associated with poor survival in human cancers [28]. Considering the extraordinary potential of these distinctively expressed m7G regulators in regulating tumor progression, we subsequently identified the expression patterns of m7G regulators in individual osteosarcoma patients in an attempt to explore their association with prognosis, immune microenvironment characteristics, drug sensitivity, along with clinical factors. Based on Cox regression analysis, we screened for prognosis-related m7G regulators and constructed MRRS. In different cohorts, KM survival analysis and ROC analysis indicated that MRRS predicts outcomes of patients with osteosarcoma effectively. In addition, we revealed that MRRS was significantly higher in osteosarcoma patients with metastasis, suggesting that MRRS may contribute to distinguishing patients with different disease states when early symptoms of osteosarcoma are insidious and not easily detectable. We also constructed a nomogram based on MRRS and different clinicopathological parameters to further improve clinical applications.
Multidrug resistance is a fundamental cause of chemotherapy failure in osteosarcoma [70]. Due to acquired or intrinsic resistance, conventional chemotherapeutic agents are inevitably limited in patients with primary and metastatic osteosarcoma [71]. Therefore, we investigated the association between MRRS and chemotherapeutic drug sensitivity in osteosarcoma to provide new insights into individualized patient management. The results showed significant discrepancies in the IC50 of numerous drugs in different MRRS subgroups. Patients with lower MRRS appeared more sensitive to these chemotherapeutic agents, including bortezomib, dasatinib, gemcitabine, midostaurin, and obatoclax mesylate, among others. The therapeutic efficacy and promise of these agents in osteosarcoma have now been elucidated in studies. For example, bortezomib is a proteasome inhibitor endorsed for treating multiple myeloma and mantle cell lymphoma [72]. Preclinical studies have documented that human osteosarcoma cell lines are susceptible to bortezomib in vitro [57]. Bortezomib induces apoptosis and autophagy in osteosarcoma cells by inhibiting extracellular regulated protein kinase phosphorylation [73]. Dasatinib is a Src family kinase and Bcr-Abl inhibitor that has demonstrated activity in preclinical models of multiple sarcomas [74]. Beck et al. also observed that the ceritinib and dasatinib combination was exceedingly safe and well tolerated in osteosarcoma patients with a long history of pulmonary metastases and that the extent of tumor necrosis was proportional to the drug concentration [75]. Gemcitabine, the most prominent cytidine analogue developed, is an effective radiosensitizer for multiple solid tumors and has synergistic activity with other chemotherapeutic agents [76], [77]. In osteosarcoma, the aerosol gemcitabine significantly inhibited the growth of the primary tumor and prevented metastatic spread without toxicity to normal tissue [78]. Midostaurin, an orally available small molecule inhibitor of FMS-like tyrosine kinase 3 (FLT3), has been shown to reduce relapse and improve overall and event-free survival in FLT3-mutant acute myeloid leukemia [79], [80]. Obatoclax mesylate, a pan-Bcl-2 inhibitor with BH3 domain mimicry, enhances the sensitivity of tumor cells to different cytotoxic drugs [81]. Our results suggest that MRRS may help to inform the appropriate chemotherapy strategy for patients. However, further studies are necessary to evaluate the effectiveness of these drugs in treating osteosarcoma.
In the last decades, there has been increasing evidence that cancer initiation and procession are related not only to the abnormal genetic and epigenetic alterations of tumor cells but also to TME [82], [83]. It is now widely recognized that an appropriate local microenvironment is non-negligible for the development of osteosarcoma [84]. The immune environment of osteosarcoma consists mainly of T lymphocytes and macrophages but also contains other subpopulations, including B lymphocytes and mast cells [85]. Osteosarcoma controls the recruitment and differentiation of immune infiltrating cells to establish a local immune-tolerant TME conducive to tumor growth, drug resistance, and metastases [86]. TME is inextricably linked to immunotherapy, and targeting TME components, such as immunoregulatory cells and their secreted factors, helps guide and ameliorate various immunotherapies [87]. We characterized the immune microenvironment of osteosarcoma, intending to provide breakthroughs in immunotherapy. Our study specifically focused on the relationship between m7G modulators and immune cell infiltration in osteosarcoma. We stratified patients according to their respective TME and found that patients with high and low MRRS exhibited a significantly distinct abundance of immune cell infiltration, such as CD8+ T cells, macrophages, NK cells, pDCs, TILs, and Tregs. Accumulation of tissue-resident memory T cells at neoplastic sites explains a more excellent prognosis and is related to the efficacy of ICI therapy in some cancers [88]. In addition, NK cells are crucial to tumor immunosurveillance, rapidly recognizing and eliminating tumor cells or influencing the activity of other immune cells by secreting various cytokines and chemokines, and their infiltration level is likewise a prognostic indicator [89]. In another study, Buddingh et al. reported that M1 macrophages are associated with low metastasis and high survival in patients with osteosarcoma, whereas M2 macrophages have the opposite effect [90]. M1-polarized macrophages have the potential to kill tumor cells and enhance the immune response; however, most tumors typically tend for M2-polarized macrophages to facilitate angiogenesis, extravasation, and immune escape, ultimately leading to tumor progression and metastasis [91]. Our results are consistent with these findings. Patients with high MRRS had lower levels of tumor immune cell infiltration and immune function enrichment, and accordingly, their survival was worse. Additionally, as the heterogeneous and highly dynamic expression of immune checkpoint genes in primary or metastatic tumors, it is necessary to detect these molecules before and during treatment to improve the therapeutic effect of ICI [92], [93]. From our analysis, MRRS could distinguish the expression levels of immune checkpoints in different osteosarcoma patients. In conclusion, MRRS helps to understand the immune profile of osteosarcoma TME, which is crucial for better predicting antitumor immune responses and selecting current immunotherapeutic approaches.
Six m7G regulators used to construct MRRS have been reported to be associated with tumors; however, the exact mechanism and functional effect of EIF4E3 in osteosarcoma remain unclear. EIF4E3 belongs to the eIF4E family capable of binding the mRNA 5′ cap and regulating proteome and cellular phenotype [94]. EIF4E family members generally recognize the m7G cap by inserting the m7G caps between two aromatic residues; however, EIF4E3 interacts more extensively with the m7G caps using an atypical cap-binding strategy of a pocket defined by Trp98, S1–S2 loop, among others [55]. EIF4E3 uses its cap-binding activity to compete with eIF4E1 for downstream target mRNAs, such as VEGF and c-MYC, to impair oncogenic transformation [55]. In another study, Landon et al. reported that EIF4E3 could maintain cell viability without rapid proliferation by regulating the translation of key transcription factors and inducing the expression of pro-proliferative genes [95]. Hu et al. also observed that MiR-584-5p targets EIF4E3 to promote DNA impairment, cell cycle arrest, and radiosensitivity in medulloblastoma [96]. This seemingly contradictory result makes us interested in the function of EIF4E3. In our study, EIF4E3 expression was downregulated in multiple cancers, including osteosarcoma. In terms of immune cell infiltration, EIF4E3 positively correlated with the abundance of gamma delta T cells, plasma cells, and naive B cells in the immune microenvironment of osteosarcoma. Gamma delta T cells are recognized as a bridge between innate and adaptive immunity, along with a widespread favorable prognostic feature in 39 malignancies [97], [98]. Numerous studies have shown gamma delta T cells can directly target tumor cells through the granule exocytosis pathway, antibody-dependent cellular cytotoxicity effect, and secretion of cytokines (interferon-γ, tumor necrosis factor-α), as well as indirectly impact antitumor immunity by activating other immune cells and coordinating downstream immune responses [99], [100]. The absence of infiltrating gamma delta T cells may be one of the reasons for the poor overall survival demonstrated by osteosarcoma patients with low expression of EIF4E3. We subsequently confirmed that overexpression of EIF4E3 inhibited the proliferation and migration of osteosarcoma in vitro, indicating a tumor suppressor role for EIF4E3. These results suggest that the function of EIF4E3 is complex, and it appears to have opposing capabilities in different tumors.
Our findings may contribute to a further understanding of the molecular mechanisms of osteosarcoma, but of course, there are some unavoidable limitations. First, key experiments to verify the correlation of m7G genes with immune phenotype are required; for example, detect T cells, B cells, HLA, CD274, and TNFSF4 in patients with different MRRS. Secondly, Predicting drugs that are more sensitive in patients with low MRRS needs to be validated by experiments using commercial cell lines or patient tissues in vitro. Moreover, we have only preliminarily explored the role of EIF4E3 in osteosarcoma, and a further understanding of the function and specific mechanisms of these prognostic m7G modulators is promising.
5. Conclusion
In summary, this study systematically designed and validated a robust signature consisting of m7G modulators, which may contribute to survival prediction and the design of personalized treatment strategies in osteosarcoma. Among them, EIF4E3 may be a potential biomarker for osteosarcoma, as its elevated expression indicated a favorable prognosis and affected the proliferation and migration of osteosarcoma cells.
Author Contributions
YZ and WG conceived the research. YZ, WG, and NR contributed to the data collection and interpretation. YZ, ZX, ZC, and WC conducted the experiments and administration. YZ and WG drafted the manuscript. HW and XZ critically revised the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This study was supported by the Clinical Frontier Technology Program of the First Affiliated Hospital of Jinan University, China (No. JNU1AF-CFTP-2022-a01204), and the Medical Scientific Research Foundation of Guangdong Province (A2018544).
Data Availability Statement
The raw RNA-seq data generated in this study are available in the TARGET, GTEX, and GEO public databases. All remaining data are available in the article, in the supplementary information file, or from the corresponding author upon reasonable request.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
The authors are very grateful to Wuwu Jiang, and Jinnan Chen for their selfless help and to the researchers who provided the datasets.
Footnotes
Supplementary data (Figure S1: Screening of prognosis-related m7G modulators; Table S1: osteosarcoma patient information for single-cell RNA-sequencing specimens in the GSE162454 dataset; Table S2:29 m7G-related genes; Table S3: Primers for qRT-PCR; Table S4: GO and KEGG enrichment analysis of differentially expressed m7G modulators; Table S5: GSVA enrichment analysis of m7G regulators-related molecular subtypes; Table S6: Characterization of the relationship between m7G regulators and drug sensitivity.) to this article can be found online at https://doi.org/10.1016/j.jbo.2023.100481.
Contributor Information
Huajun Wang, Email: hjwang@jnu.edu.cn.
Xiaofei Zheng, Email: xfzheng@jnu.edu.cn.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
osteosarcoma patient information for single-cell RNA-sequencing specimens in the GSE162454 dataset.
29 m7G-related genes.
Primers for qRT-PCR.
GO and KEGG enrichment analysis of differentially expressed m7G modulators.
GSVA enrichment analysis of m7G regulators-related molecular subtypes.
Characterization of the relationship between m7G regulators and drug sensitivity.
Screening of prognosis-related m7G modulators.
References
- 1.Khalili-Tanha G., Moghbeli M. Long non-coding RNAs as the critical regulators of doxorubicin resistance in tumor cells. Cell. Mol. Biol. Lett. 2021;26(1):39. doi: 10.1186/s11658-021-00282-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cai W., Xu Y., Zuo W., Su Z. MicroR-542-3p can mediate ILK and further inhibit cell proliferation, migration and invasion in osteosarcoma cells. Aging. 2019;11(1):18–32. doi: 10.18632/aging.101698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lu K.H., Lin R.C., Yang J.S., Yang W.E., Reiter R.J., Yang S.F. Molecular and cellular mechanisms of melatonin in osteosarcoma. Cells. 2019;8(12) doi: 10.3390/cells8121618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liu Y., Leng A., Li L., Yang B., Shen S., Chen H., Zhu E., Xu Q., Ma X., Shi P., Liu Y., Liu T., Li L., Li K., Zhang D., Xiao J. AMTB, a TRPM8 antagonist, suppresses growth and metastasis of osteosarcoma through repressing the TGFβ signaling pathway. Cell Death Dis. 2022;13(3):288. doi: 10.1038/s41419-022-04744-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fellenberg J., Lehner B., Saehr H., Schenker A., Kunz P. Tumor suppressor function of miR-127-3p and miR-376a-3p in osteosarcoma cells. Cancers. 2019;11(12) doi: 10.3390/cancers11122019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang Z., Wu B., Zhou Y., Huang X., Pan W., Liu M., Yan X., Lin N., Ye Z. Predictors of the survival of primary and secondary older osteosarcoma patients. J. Cancer. 2019;10(19):4614–4622. doi: 10.7150/jca.32627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dray M.S., Miller M.V. Paget's osteosarcoma and post-radiation osteosarcoma: secondary osteosarcoma at Middlemore Hospital, New Zealand. Pathology. 2008;40(6):604–610. doi: 10.1080/00313020802320663. [DOI] [PubMed] [Google Scholar]
- 8.Cui L., Zhang J.Y., Ren Z.P., Zhao H.J., Li G.S. APLNR promotes the progression of osteosarcoma by stimulating cell proliferation and invasion. Anticancer Drugs. 2019;30(9):940–947. doi: 10.1097/CAD.0000000000000785. [DOI] [PubMed] [Google Scholar]
- 9.Siegel H.J., Pressey J.G. Current concepts on the surgical and medical management of osteosarcoma. Expert Rev. Anticancer Ther. 2008;8(8):1257–1269. doi: 10.1586/14737140.8.8.1257. [DOI] [PubMed] [Google Scholar]
- 10.Bian Z.J., Shan H.J., Zhu Y.R., Shi C., Chen M.B., Huang Y.M., Wang X.D., Zhou X.Z., Cao C. Identification of Gαi3 as a promising target for osteosarcoma treatment. Int. J. Biol. Sci. 2022;18(4):1508–1520. doi: 10.7150/ijbs.68861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Marchandet L., Lallier M., Charrier C., Baud'huin M., Ory B., Lamoureux F. Mechanisms of resistance to conventional therapies for osteosarcoma. Cancers. 2021;13(4) doi: 10.3390/cancers13040683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Serra R.W., Fang M., Park S.M., Hutchinson L., Green M.R. A KRAS-directed transcriptional silencing pathway that mediates the CpG island methylator phenotype. Elife. 2014;3:e02313. doi: 10.7554/eLife.02313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang W., Shao F., Yang X., Wang J., Zhu R., Yang Y., Zhao G., Guo D., Sun Y., Wang J., Xue Q., Gao S., Gao Y., He J., Lu Z. METTL3 promotes tumour development by decreasing APC expression mediated by APC mRNA N(6)-methyladenosine-dependent YTHDF binding. Nat. Commun. 2021;12(1):3803. doi: 10.1038/s41467-021-23501-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Han X., Guo J., Fan Z. Interactions between m6A modification and miRNAs in malignant tumors. Cell Death Dis. 2021;12(6):598. doi: 10.1038/s41419-021-03868-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Doamekpor S.K., Grudzien-Nogalska E., Mlynarska-Cieslak A., Kowalska J., Kiledjian M., Tong L. DXO/Rai1 enzymes remove 5'-end FAD and dephospho-CoA caps on RNAs. Nucleic Acids Res. 2020;48(11):6136–6148. doi: 10.1093/nar/gkaa297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Trixl L., Amort T., Wille A., Zinni M., Ebner S., Hechenberger C., Eichin F., Gabriel H., Schoberleitner I., Huang A., Piatti P., Nat R., Troppmair J., Lusser A. RNA cytosine methyltransferase Nsun3 regulates embryonic stem cell differentiation by promoting mitochondrial activity. Cell. Mol. Life Sci. 2018;75(8):1483–1497. doi: 10.1007/s00018-017-2700-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Noma A., Sakaguchi Y., Suzuki T. Mechanistic characterization of the sulfur-relay system for eukaryotic 2-thiouridine biogenesis at tRNA wobble positions. Nucleic Acids Res. 2009;37(4):1335–1352. doi: 10.1093/nar/gkn1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sun T., Wu R., Ming L. The role of m6A RNA methylation in cancer. Biomed. Pharmacother. 2019;112 doi: 10.1016/j.biopha.2019.108613. [DOI] [PubMed] [Google Scholar]
- 19.He L., Li H., Wu A., Peng Y., Shu G., Yin G. Functions of N6-methyladenosine and its role in cancer. Mol. Cancer. 2019;18(1):176. doi: 10.1186/s12943-019-1109-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen Z., Zhu W., Zhu S., Sun K., Liao J., Liu H., Dai Z., Han H., Ren X., Yang Q., Zheng S., Peng B., Peng S., Kuang M., Lin S. METTL1 promotes hepatocarcinogenesis via m(7) G tRNA modification-dependent translation control. Clin. Transl. Med. 2021;11(12):e661. doi: 10.1002/ctm2.661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Suzuki T. The expanding world of tRNA modifications and their disease relevance. Nat. Rev. Mol. Cell Biol. 2021;22(6):375–392. doi: 10.1038/s41580-021-00342-0. [DOI] [PubMed] [Google Scholar]
- 22.Luo Y., Yao Y., Wu P., Zi X., Sun N., He J. The potential role of N(7)-methylguanosine (m7G) in cancer. J. Hematol. Oncol. 2022;15(1):63. doi: 10.1186/s13045-022-01285-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen Y., Lin H., Miao L., He J. Role of N7-methylguanosine (m(7)G) in cancer. Trends Cell Biol. 2022;32(10):819–824. doi: 10.1016/j.tcb.2022.07.001. [DOI] [PubMed] [Google Scholar]
- 24.Osborne M.J., Volpon L., Memarpoor-Yazdi M., Pillay S., Thambipillai A., Czarnota S., Culjkovic-Kraljacic B., Trahan C., Oeffinger M., Cowling V.H., Borden K.L.B. Identification and characterization of the interaction between the methyl-7-guanosine cap maturation enzyme RNMT and the cap-binding protein eIF4E. J. Mol. Biol. 2022;434(5) doi: 10.1016/j.jmb.2022.167451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cheng W., Gao A., Lin H., Zhang W. Novel roles of METTL1/WDR4 in tumor via m(7)G methylation. Mol. Ther. Oncolyt. 2022;26:27–34. doi: 10.1016/j.omto.2022.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tomikawa C. 7-Methylguanosine modifications in transfer RNA (tRNA) Int. J. Mol. Sci. 2018;19(12) doi: 10.3390/ijms19124080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ying X., Liu B., Yuan Z., Huang Y., Chen C., Jiang X., Zhang H., Qi D., Yang S., Lin S., Luo J., Ji W. METTL1-m(7) G-EGFR/EFEMP1 axis promotes the bladder cancer development. Clin. Transl. Med. 2021;11(12):e675. doi: 10.1002/ctm2.675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Orellana E.A., Liu Q., Yankova E., Pirouz M., De Braekeleer E., Zhang W., Lim J., Aspris D., Sendinc E., Garyfallos D.A., Gu M., Ali R., Gutierrez A., Mikutis S., Bernardes G.J.L., Fischer E.S., Bradley A., Vassiliou G.S., Slack F.J., Tzelepis K., Gregory R.I. METTL1-mediated m(7)G modification of Arg-TCT tRNA drives oncogenic transformation. Mol. Cell. 2021;81(16):3323–3338.e14. doi: 10.1016/j.molcel.2021.06.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xia P., Zhang H., Xu K., Jiang X., Gao M., Wang G., Liu Y., Yao Y., Chen X., Ma W., Zhang Z., Yuan Y. MYC-targeted WDR4 promotes proliferation, metastasis, and sorafenib resistance by inducing CCNB1 translation in hepatocellular carcinoma. Cell Death Dis. 2021;12(7):691. doi: 10.1038/s41419-021-03973-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wu Y., Wang Z., Shen J., Yan W., Xiang S., Liu H., Huang W. The role of m6A methylation in osteosarcoma biological processes and its potential clinical value. Hum. Genomics. 2022;16(1):12. doi: 10.1186/s40246-022-00384-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yadav P., Subbarayalu P., Medina D., Nirzhor S., Timilsina S., Rajamanickam S., Eedunuri V.K., Gupta Y., Zheng S., Abdelfattah N., Huang Y., Vadlamudi R., Hromas R., Meltzer P., Houghton P., Chen Y., Rao M.K. M6A RNA methylation regulates histone ubiquitination to support cancer growth and progression. Cancer Res. 2022;82(10):1872–1889. doi: 10.1158/0008-5472.CAN-21-2106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Huang H., Cui X., Qin X., Li K., Yan G., Lu D., Zheng M., Hu Z., Lei D., Lan N., Zheng L., Yuan Z., Zhu B., Zhao J. Analysis and identification of m(6)A RNA methylation regulators in metastatic osteosarcoma. Mol. Ther. Nucleic Acids. 2022;27:577–592. doi: 10.1016/j.omtn.2021.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang Y., Wang Y., Ying L., Tao S., Shi M., Lin P., Wang Y., Han B. Regulatory role of N6-methyladenosine (m(6)A) modification in osteosarcoma. Front. Oncol. 2021;11 doi: 10.3389/fonc.2021.683768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu D., Hu Z., Jiang J., Zhang J., Hu C., Huang J., Wei Q. Five hypoxia and immunity related genes as potential biomarkers for the prognosis of osteosarcoma. Sci. Rep. 2022;12(1):1617. doi: 10.1038/s41598-022-05103-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Thanindratarn P., Wei R., Dean D.C., Singh A., Federman N., Nelson S.D., Hornicek F.J., Duan Z. T-LAK cell-originated protein kinase (TOPK): an emerging prognostic biomarker and therapeutic target in osteosarcoma. Mol. Oncol. 2021;15(12):3721–3737. doi: 10.1002/1878-0261.13039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu Y., Feng W., Dai Y., Bao M., Yuan Z., He M., Qin Z., Liao S., He J., Huang Q., Yu Z., Zeng Y., Guo B., Huang R., Yang R., Jiang Y., Liao J., Xiao Z., Zhan X., Lin C., Xu J., Ye Y., Ma J., Wei Q., Mo Z. Single-cell transcriptomics reveals the complexity of the tumor microenvironment of treatment-naive osteosarcoma. Front. Oncol. 2021;11 doi: 10.3389/fonc.2021.709210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Y. Han, Y. Wang, X. Dong, D. Sun, Z. Liu, J. Yue, H. Wang, T. Li, C. Wang, TISCH2: expanded datasets and new tools for single-cell transcriptome analyses of the tumor microenvironment, Nucleic Acids Res. 51(D1) (2023) D1425-d1431. [DOI] [PMC free article] [PubMed]
- 38.Chen M., Nie Z., Gao Y., Cao H., Zheng L., Guo N., Peng Y., Zhang S. m7G regulator-mediated molecular subtypes and tumor microenvironment in kidney renal clear cell carcinoma. Front. Pharmacol. 2022;13 doi: 10.3389/fphar.2022.900006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yoshihara K., Shahmoradgoli M., Martínez E., Vegesna R., Kim H., Torres-Garcia W., Treviño V., Shen H., Laird P.W., Levine D.A., Carter S.L., Getz G., Stemke-Hale K., Mills G.B., Verhaak R.G. Inferring tumour purity and stromal and immune cell admixture from expression data. Nat. Commun. 2013;4:2612. doi: 10.1038/ncomms3612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu Y., Cai J., Liu W., Lin Y., Guo L., Liu X., Qin Z., Xu C., Zhang Y., Su X., Deng K., Yan G., Liang J. Intravenous injection of the oncolytic virus M1 awakens antitumor T cells and overcomes resistance to checkpoint blockade. Cell Death Dis. 2020;11(12):1062. doi: 10.1038/s41419-020-03285-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chowell D., Morris L.G.T., Grigg C.M., Weber J.K., Samstein R.M., Makarov V., Kuo F., Kendall S.M., Requena D., Riaz N., Greenbaum B., Carroll J., Garon E., Hyman D.M., Zehir A., Solit D., Berger M., Zhou R., Rizvi N.A., Chan T.A. Patient HLA class I genotype influences cancer response to checkpoint blockade immunotherapy. Science (New York, N.Y.) 2018;359:582–587. doi: 10.1126/science.aao4572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wu Z.L., Deng Y.J., Zhang G.Z., Ren E.H., Yuan W.H., Xie Q.Q. Development of a novel immune-related genes prognostic signature for osteosarcoma. Sci. Rep. 2020;10(1):18402. doi: 10.1038/s41598-020-75573-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.A. Koch, J. Jeschke, W. Van Criekinge, M. van Engeland, T. De Meyer, MEXPRESS update 2019, Nucleic Acids Res. 47(W1) (2019) W561-w565. [DOI] [PMC free article] [PubMed]
- 44.Koch A., De Meyer T., Jeschke J., Van Criekinge W. MEXPRESS: visualizing expression, DNA methylation and clinical TCGA data. BMC Genomics. 2015;16(1):636. doi: 10.1186/s12864-015-1847-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Heymann M.F., Lézot F., Heymann D. The contribution of immune infiltrates and the local microenvironment in the pathogenesis of osteosarcoma. Cell. Immunol. 2019;343 doi: 10.1016/j.cellimm.2017.10.011. [DOI] [PubMed] [Google Scholar]
- 46.Wedekind M.F., Wagner L.M., Cripe T.P. Immunotherapy for osteosarcoma: Where do we go from here? Pediatr. Blood Cancer. 2018;65(9):e27227. doi: 10.1002/pbc.27227. [DOI] [PubMed] [Google Scholar]
- 47.T. Li, J. Fu, Z. Zeng, D. Cohen, J. Li, Q. Chen, B. Li, X.S. Liu, TIMER2.0 for analysis of tumor-infiltrating immune cells, Nucleic Acids Res. 48(W1) (2020) W509-W514. [DOI] [PMC free article] [PubMed]
- 48.G.T. Gibney, L.M. Weiner, M.B. Atkins, Predictive biomarkers for checkpoint inhibitor-based immunotherapy, The Lancet. Oncology 17(12) (2016) e542-e551. [DOI] [PMC free article] [PubMed]
- 49.Choo Y.W., Kang M., Kim H.Y., Han J., Kang S., Lee J.R., Jeong G.J., Kwon S.P., Song S.Y., Go S., Jung M., Hong J., Kim B.S. M1 macrophage-derived nanovesicles potentiate the anticancer efficacy of immune checkpoint inhibitors. ACS Nano. 2018;12(9):8977–8993. doi: 10.1021/acsnano.8b02446. [DOI] [PubMed] [Google Scholar]
- 50.Pagliuca S., Gurnari C., Rubio M.T., Visconte V., Lenz T.L. Individual HLA heterogeneity and its implications for cellular immune evasion in cancer and beyond. Front. Immunol. 2022;13 doi: 10.3389/fimmu.2022.944872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bielack S.S., Kempf-Bielack B., Delling G., Exner G.U., Flege S., Helmke K., Kotz R., Salzer-Kuntschik M., Werner M., Winkelmann W., Zoubek A., Jürgens H., Winkler K. Prognostic factors in high-grade osteosarcoma of the extremities or trunk: an analysis of 1,702 patients treated on neoadjuvant cooperative osteosarcoma study group protocols. J. Clin. Oncol. 2002;20(3):776–790. doi: 10.1200/JCO.2002.20.3.776. [DOI] [PubMed] [Google Scholar]
- 52.Odri G.A., Tchicaya-Bouanga J., Yoon D.J.Y., Modrowski D. Metastatic progression of osteosarcomas: a review of current knowledge of environmental versus oncogenic drivers. Cancers. 2022;14(2) doi: 10.3390/cancers14020360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chen Y., Cao J., Zhang N., Yang B., He Q., Shao X., Ying M. Advances in differentiation therapy for osteosarcoma. Drug Discov. Today. 2020;25(3):497–504. doi: 10.1016/j.drudis.2019.08.010. [DOI] [PubMed] [Google Scholar]
- 54.Zhou Y., Yang D., Yang Q., Lv X., Huang W., Zhou Z., Wang Y., Zhang Z., Yuan T., Ding X., Tang L., Zhang J., Yin J., Huang Y., Yu W., Wang Y., Zhou C., Su Y., He A., Sun Y., Shen Z., Qian B., Meng W., Fei J., Yao Y., Pan X., Chen P., Hu H. Single-cell RNA landscape of intratumoral heterogeneity and immunosuppressive microenvironment in advanced osteosarcoma. Nat. Commun. 2020;11(1):6322. doi: 10.1038/s41467-020-20059-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.M.J. Osborne, L. Volpon, J.A. Kornblatt, B. Culjkovic-Kraljacic, A. Baguet, K.L. Borden, eIF4E3 acts as a tumor suppressor by utilizing an atypical mode of methyl-7-guanosine cap recognition, Proc. Natl. Acad. Sci. U. S. A. 110(10) (2013) 3877-82. [DOI] [PMC free article] [PubMed]
- 56.Lin S.L., Chang D., Ying S.Y. Hyaluronan stimulates transformation of androgen-independent prostate cancer. Carcinogenesis. 2007;28(2):310–320. doi: 10.1093/carcin/bgl134. [DOI] [PubMed] [Google Scholar]
- 57.Van Stiphout C.M., Luu A.K., Viloria-Petit A.M. Proteasome inhibitors and their potential applicability in osteosarcoma treatment. Cancers. 2022;14(19) doi: 10.3390/cancers14194544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Meltzer P.S., Helman L.J. New horizons in the treatment of osteosarcoma. N. Engl. J. Med. 2021;385(22):2066–2076. doi: 10.1056/NEJMra2103423. [DOI] [PubMed] [Google Scholar]
- 59.Chen K., Chen Y., Zhu X.D., Bai Y.S., Wei X.Z., Wang C.F., Chen Z.Q., Li M. Expression and significance of Kruppel-like factor 6 gene in osteosarcoma. Int. Orthop. 2012;36(10):2107–2111. doi: 10.1007/s00264-012-1626-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Harting M.T., Blakely M.L. Management of osteosarcoma pulmonary metastases. Semin. Pediatr. Surg. 2006;15(1):25–29. doi: 10.1053/j.sempedsurg.2005.11.005. [DOI] [PubMed] [Google Scholar]
- 61.Marais L.C., Bertie J., Rodseth R., Sartorius B., Ferreira N. Pre-treatment serum lactate dehydrogenase and alkaline phosphatase as predictors of metastases in extremity osteosarcoma. J Bone Oncol. 2015;4(3):80–84. doi: 10.1016/j.jbo.2015.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hogg S.J., Beavis P.A., Dawson M.A., Johnstone R.W. Targeting the epigenetic regulation of antitumour immunity. Nat. Rev. Drug Discov. 2020;19(11):776–800. doi: 10.1038/s41573-020-0077-5. [DOI] [PubMed] [Google Scholar]
- 63.Kapoor A., Goldberg M.S., Cumberland L.K., Ratnakumar K., Segura M.F., Emanuel P.O., Menendez S., Vardabasso C., Leroy G., Vidal C.I., Polsky D., Osman I., Garcia B.A., Hernando E., Bernstein E. The histone variant macroH2A suppresses melanoma progression through regulation of CDK8. Nature. 2010;468(7327):1105–1109. doi: 10.1038/nature09590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gallou-Kabani C., Vigé A., Junien C. Lifelong circadian and epigenetic drifts in metabolic syndrome. Epigenetics. 2007;2(3):137–146. doi: 10.4161/epi.2.3.4897. [DOI] [PubMed] [Google Scholar]
- 65.Cui L., Ma R., Cai J., Guo C., Chen Z., Yao L., Wang Y., Fan R., Wang X., Shi Y. RNA modifications: importance in immune cell biology and related diseases. Signal Transduct. Target. Ther. 2022;7(1):334. doi: 10.1038/s41392-022-01175-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Rong D., Sun G., Wu F., Cheng Y., Sun G., Jiang W., Li X., Zhong Y., Wu L., Zhang C., Tang W., Wang X. Epigenetics: Roles and therapeutic implications of non-coding RNA modifications in human cancers. Mol. Ther. Nucleic acids. 2021;25:67–82. doi: 10.1016/j.omtn.2021.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhang M., Song J., Yuan W., Zhang W., Sun Z. Roles of RNA methylation on tumor immunity and clinical implications. Front. Immunol. 2021;12 doi: 10.3389/fimmu.2021.641507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Chen H.M., Li H., Lin M.X., Fan W.J., Zhang Y., Lin Y.T., Wu S.X. Research progress for RNA modifications in physiological and pathological angiogenesis. Front. Genet. 2022;13 doi: 10.3389/fgene.2022.952667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Pandolfini L., Barbieri I., Bannister A.J., Hendrick A., Andrews B., Webster N., Murat P., Mach P., Brandi R., Robson S.C., Migliori V., Alendar A., d'Onofrio M., Balasubramanian S., Kouzarides T. METTL1 promotes let-7 MicroRNA processing via m7G methylation. Mol. Cell. 2019;74(6):1278–1290.e9. doi: 10.1016/j.molcel.2019.03.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Daqian W., Chuandong W., Xinhua Q., Songtao A., Kerong D. Chimaphilin inhibits proliferation and induces apoptosis in multidrug resistant osteosarcoma cell lines through insulin-like growth factor-I receptor (IGF-IR) signaling. Chem. Biol. Interact. 2015;237:25–30. doi: 10.1016/j.cbi.2015.05.008. [DOI] [PubMed] [Google Scholar]
- 71.Lilienthal I., Herold N. Targeting molecular mechanisms underlying treatment efficacy and resistance in osteosarcoma: A review of current and future strategies. Int. J. Mol. Sci. 2020;21(18) doi: 10.3390/ijms21186885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Das T.K., Esernio J., Cagan R.L. Restraining network response to targeted cancer therapies improves efficacy and reduces cellular resistance. Cancer Res. 2018;78(15):4344–4359. doi: 10.1158/0008-5472.CAN-17-2001. [DOI] [PubMed] [Google Scholar]
- 73.Lou Z., Ren T., Peng X., Sun Y., Jiao G., Lu Q., Zhang S., Lu X., Guo W. Bortezomib induces apoptosis and autophagy in osteosarcoma cells through mitogen-activated protein kinase pathway in vitro. J. Int. Med. Res. 2013;41(5):1505–1519. doi: 10.1177/0300060513490618. [DOI] [PubMed] [Google Scholar]
- 74.Shor A.C., Keschman E.A., Lee F.Y., Muro-Cacho C., Letson G.D., Trent J.C., Pledger W.J., Jove R. Dasatinib inhibits migration and invasion in diverse human sarcoma cell lines and induces apoptosis in bone sarcoma cells dependent on SRC kinase for survival. Cancer Res. 2007;67(6):2800–2808. doi: 10.1158/0008-5472.CAN-06-3469. [DOI] [PubMed] [Google Scholar]
- 75.Beck O., Paret C., Russo A., Burhenne J., Fresnais M., Steimel K., Seidmann L., Wagner D.C., Vewinger N., Lehmann N., Sprang M., Backes N., Roth L., Neu M.A., Wingerter A., Henninger N., Malki K.E., Otto H., Alt F., Desuki A., Kindler T., Faber J. Safety and activity of the combination of ceritinib and dasatinib in osteosarcoma. Cancers. 2020;12(4) doi: 10.3390/cancers12040793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Bastiancich C., Bastiat G., Lagarce F. Gemcitabine and glioblastoma: challenges and current perspectives. Drug Discov. Today. 2018;23(2):416–423. doi: 10.1016/j.drudis.2017.10.010. [DOI] [PubMed] [Google Scholar]
- 77.Mini E., Nobili S., Caciagli B., Landini I., Mazzei T. Cellular pharmacology of gemcitabine. Ann. Oncol. 2006;17(Suppl 5):v7–v. doi: 10.1093/annonc/mdj941. [DOI] [PubMed] [Google Scholar]
- 78.Gordon N., Felix K., Daw N.C. Aerosolized chemotherapy for osteosarcoma. Adv. Exp. Med. Biol. 2020;1257:67–73. doi: 10.1007/978-3-030-43032-0_6. [DOI] [PubMed] [Google Scholar]
- 79.Larson R.A., Mandrekar S.J., Huebner L.J., Sanford B.L., Laumann K., Geyer S., Bloomfield C.D., Thiede C., Prior T.W., Döhner K., Marcucci G., Voso M.T., Klisovic R.B., Galinsky I., Wei A.H., Sierra J., Sanz M.A., Brandwein J.M., de Witte T., Niederwieser D., Appelbaum F.R., Medeiros B.C., Tallman M.S., Krauter J., Schlenk R.F., Ganser A., Serve H., Ehninger G., Amadori S., Gathmann I., Döhner H., Stone R.M. Midostaurin reduces relapse in FLT3-mutant acute myeloid leukemia: the Alliance CALGB 10603/RATIFY trial. Leukemia. 2021;35(9):2539–2551. doi: 10.1038/s41375-021-01179-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Schmalbrock L.K., Dolnik A., Cocciardi S., Sträng E., Theis F., Jahn N., Panina E., Blätte T.J., Herzig J., Skambraks S., Rücker F.G., Gaidzik V.I., Paschka P., Fiedler W., Salih H.R., Wulf G., Schroeder T., Lübbert M., Schlenk R.F., Thol F., Heuser M., Larson R.A., Ganser A., Stunnenberg H.G., Minucci S., Stone R.M., Bloomfield C.D., Döhner H., Döhner K., Bullinger L. Clonal evolution of acute myeloid leukemia with FLT3-ITD mutation under treatment with midostaurin. Blood. 2021;137(22):3093–3104. doi: 10.1182/blood.2020007626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Joudeh J., Claxton D. Obatoclax mesylate: pharmacology and potential for therapy of hematological neoplasms. Expert Opin. Invest. Drugs. 2012;21(3):363–373. doi: 10.1517/13543784.2012.652302. [DOI] [PubMed] [Google Scholar]
- 82.Wu T., Dai Y. Tumor microenvironment and therapeutic response. Cancer Lett. 2017;387:61–68. doi: 10.1016/j.canlet.2016.01.043. [DOI] [PubMed] [Google Scholar]
- 83.Denton A.E., Roberts E.W., Fearon D.T. Stromal cells in the tumor microenvironment. Adv. Exp. Med. Biol. 2018;1060:99–114. doi: 10.1007/978-3-319-78127-3_6. [DOI] [PubMed] [Google Scholar]
- 84.Corre I., Verrecchia F., Crenn V., Redini F., Trichet V. The osteosarcoma microenvironment: a complex but targetable ecosystem. Cells. 2020;9(4) doi: 10.3390/cells9040976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Mori K., Rédini F., Gouin F., Cherrier B., Heymann D. Osteosarcoma: current status of immunotherapy and future trends (Review) Oncol. Rep. 2006;15(3):693–700. [PubMed] [Google Scholar]
- 86.Kansara M., Teng M.W., Smyth M.J., Thomas D.M. Translational biology of osteosarcoma. Nat. Rev. Cancer. 2014;14(11):722–735. doi: 10.1038/nrc3838. [DOI] [PubMed] [Google Scholar]
- 87.Pitt J.M., Marabelle A., Eggermont A., Soria J.C., Kroemer G., Zitvogel L. Targeting the tumor microenvironment: removing obstruction to anticancer immune responses and immunotherapy. Ann. Oncol. 2016;27(8):1482–1492. doi: 10.1093/annonc/mdw168. [DOI] [PubMed] [Google Scholar]
- 88.Corgnac S., Boutet M., Kfoury M., Naltet C., Mami-Chouaib F. The emerging role of CD8(+) tissue resident memory T (T(RM)) cells in antitumor immunity: A unique functional contribution of the CD103 integrin. Front. Immunol. 2018;9:1904. doi: 10.3389/fimmu.2018.01904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Shimasaki N., Jain A., Campana D. NK cells for cancer immunotherapy. Nat. Rev. Drug Discov. 2020;19(3):200–218. doi: 10.1038/s41573-019-0052-1. [DOI] [PubMed] [Google Scholar]
- 90.Buddingh E.P., Kuijjer M.L., Duim R.A., Bürger H., Agelopoulos K., Myklebost O., Serra M., Mertens F., Hogendoorn P.C., Lankester A.C., Cleton-Jansen A.M. Tumor-infiltrating macrophages are associated with metastasis suppression in high-grade osteosarcoma: a rationale for treatment with macrophage activating agents. Clin. Cancer Res. 2011;17(8):2110–2119. doi: 10.1158/1078-0432.CCR-10-2047. [DOI] [PubMed] [Google Scholar]
- 91.Franklin R.A., Li M.O. Ontogeny of tumor-associated macrophages and its implication in cancer regulation. Trends Cancer. 2016;2(1):20–34. doi: 10.1016/j.trecan.2015.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Taube J.M., Anders R.A., Young G.D., Xu H., Sharma R., McMiller T.L., Chen S., Klein A.P., Pardoll D.M., Topalian S.L., Chen L. Colocalization of inflammatory response with B7–h1 expression in human melanocytic lesions supports an adaptive resistance mechanism of immune escape. Sci. Transl. Med. 2012;4(127):127ra37. doi: 10.1126/scitranslmed.3003689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Sharma P., Allison J.P. The future of immune checkpoint therapy. Science (New York, N.Y.) 2015;348:56–61. doi: 10.1126/science.aaa8172. [DOI] [PubMed] [Google Scholar]
- 94.Borden K.L.B., Volpon L. The diversity, plasticity, and adaptability of cap-dependent translation initiation and the associated machinery. RNA Biol. 2020;17(9):1239–1251. doi: 10.1080/15476286.2020.1766179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Landon A.L., Muniandy P.A., Shetty A.C., Lehrmann E., Volpon L., Houng S., Zhang Y., Dai B., Peroutka R., Mazan-Mamczarz K., Steinhardt J., Mahurkar A., Becker K.G., Borden K.L., Gartenhaus R.B. MNKs act as a regulatory switch for eIF4E1 and eIF4E3 driven mRNA translation in DLBCL. Nat. Commun. 2014;5:5413. doi: 10.1038/ncomms6413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Abdelfattah N., Rajamanickam S., Panneerdoss S., Timilsina S., Yadav P., Onyeagucha B.C., Garcia M., Vadlamudi R., Chen Y., Brenner A., Houghton P., Rao M.K. MiR-584-5p potentiates vincristine and radiation response by inducing spindle defects and DNA damage in medulloblastoma. Nat. Commun. 2018;9(1):4541. doi: 10.1038/s41467-018-06808-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Saura-Esteller J., de Jong M., King L.A., Ensing E., Winograd B., de Gruijl T.D., Parren P., van der Vliet H.J. Gamma delta T-cell based cancer immunotherapy: past-present-future. Front. Immunol. 2022;13 doi: 10.3389/fimmu.2022.915837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Gentles A.J., Newman A.M., Liu C.L., Bratman S.V., Feng W., Kim D., Nair V.S., Xu Y., Khuong A., Hoang C.D., Diehn M., West R.B., Plevritis S.K., Alizadeh A.A. The prognostic landscape of genes and infiltrating immune cells across human cancers. Nat. Med. 2015;21(8):938–945. doi: 10.1038/nm.3909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Chan K.F., Duarte J.D.G., Ostrouska S., Behren A. γδ T cells in the tumor microenvironment-interactions with other immune cells. Front. Immunol. 2022;13 doi: 10.3389/fimmu.2022.894315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Silva-Santos B., Mensurado S., Coffelt S.B. γδ T cells: pleiotropic immune effectors with therapeutic potential in cancer. Nat. Rev. Cancer. 2019;19(7):392–404. doi: 10.1038/s41568-019-0153-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
osteosarcoma patient information for single-cell RNA-sequencing specimens in the GSE162454 dataset.
29 m7G-related genes.
Primers for qRT-PCR.
GO and KEGG enrichment analysis of differentially expressed m7G modulators.
GSVA enrichment analysis of m7G regulators-related molecular subtypes.
Characterization of the relationship between m7G regulators and drug sensitivity.
Screening of prognosis-related m7G modulators.
Data Availability Statement
The raw RNA-seq data generated in this study are available in the TARGET, GTEX, and GEO public databases. All remaining data are available in the article, in the supplementary information file, or from the corresponding author upon reasonable request.