Abstract
A toxin variant strain of Clostridium difficile was isolated from two patients with C. difficile-associated disease (CDAD), one of whom died from extensive pseudomembranous colitis. This strain, identified by restriction endonuclease analysis (REA) as type CF2, was not detected by an immunoassay for C. difficile toxin A. Culture supernatants of CF2 failed to elicit significant enterotoxic activity in the rabbit ileal loop assay but did produce atypical cytopathic effects in cell culture assay. Southern hybridization, PCR amplification, and DNA sequence analyses were performed on the toxin A (tcdA) and toxin B (tcdB) genes of type CF2 isolate 5340. Type CF2 5340 tcdA exhibited a 1,821-bp truncation, due to three deletions in the 3′ end of the gene, and a point mutation in the 5′ end of the gene, resulting in a premature stop codon at tcdA position 139. Type CF2 5340 tcdB exhibited multiple nucleotide base substitutions in the 5′ end of the gene compared to tcdB of the standard toxigenic strain VPI 10463. Type CF2 5340 toxin gene nucleotide sequences and deduced amino acid sequences showed a strong resemblance to those of the previously described variant C. difficile strain 1470, a strain reported to have reduced pathogenicity and no association with clinical illness in humans. REA of strain 1470 identified this strain as a distinct type (CF1) within the same REA group as the closely related type CF2. A review of our clinical-isolate collection identified five additional patients infected with type CF2, three of whom had documented CDAD. PCR amplification of the 3′ end of tcdA demonstrated identical 1.8-kb deletions in all seven type CF2 isolates. REA type CF2 is a toxin variant strain of C. difficile that retains the ability to cause disease in humans but is not detected in clinical immunoassays for toxin A.
Clostridium difficile is the most important infectious cause of nosocomial diarrhea and is responsible for clinical manifestations ranging from asymptomatic carriage to pseudomembranous colitis and death (3, 12, 13, 26, 35). Toxigenic C. difficile strains produce two major toxins, toxin A and toxin B. Toxin A has been regarded as the primary virulence factor in C. difficile-associated disease (CDAD) (17, 30). In in vivo studies of toxins purified from C. difficile strain VPI 10463, purified toxin B was unable to cause disease unless the intestinal mucosa was damaged or toxin A was present (18). In contrast, purified toxin A was capable of causing disease without the presence of toxin B (18). From these and other data, toxin A had been postulated to initiate the pathogenic process of CDAD. It has been generally accepted that toxigenic strains produce both toxin A and toxin B whereas nontoxigenic strains lack both toxins (19).
Toxin variant strains have been discovered which fail to produce detectable toxin A yet produce toxin B. The two best-characterized toxin A-negative, toxin B-positive variants are strain 1470, the reference strain for serogroup F in a well-characterized serogroup typing system (9, 10), and strain 8864 (14, 31). Both strains are truncated at the 3′ ends of their toxin A genes (tcdA), 8864 with a 6.0-kb deletion (27, 28) and 1470 with an approximately 1.7-kb deletion (21). Both of these strains also possess variations in their toxin B genes (tcdB). The variant tcdB genes produce cytotoxins which demonstrate unusual cytopathic effects (CPE) in cell culture assays which resemble the CPE caused by the lethal toxin (LT) of Clostridium sordellii (31, 33). The variant toxin B of strain 1470 appears to be a functional hybrid between the LT of C. sordellii and the toxin B of standard C. difficile strain VPI 10463 (6). In the hamster model of C. difficile disease, strain 8864 was pathogenic (5) but strain 1470 was reported as nonpathogenic (9). The clinical significance of these toxin A-negative strains is unclear. Strain 8864 was isolated from a patient specimen, but the associated clinical manifestations were not reported (14). No other isolation of a C. difficile strain with this extensive tcdA deletion has ever been reported. Strain 1470 was isolated from a healthy neonate (9), and other serogroup F strain isolates, all of which contain genetic alterations similar to those of 1470 (10), have primarily been associated with asymptomatic colonization in infants and adults (9, 16).
During a comparison study of a new immunoassay for toxin A with a cell culture assay for C. difficile cytotoxin, we have discovered a toxin A-negative, toxin B-positive C. difficile strain in two patients with unambiguous CDAD, one of whom had fatal pseudomembranous colitis. The isolates (5340 and 5362) were identical by restriction endonuclease analysis (REA) and are designated type CF2 (8). We tested type CF2 5340 in the hamster model and found that type CF2 was pathogenic by this assay, although it was less virulent in hamsters than C. difficile strains responsible for human epidemics (S. P. Sambol, M. Merrigan, S. Johnson, and D. N. Gerding, Abstr. 99th Gen. Meet. Am. Soc. Microbiol. 1999, abstr. L-3, 1999). In this study, we investigate the relationship of REA type CF2 to toxin variant strains 8864 and 1470 using the REA typing system. We examine our collection of >5,000 patient isolates for type CF2 isolates and report the clinical manifestations of the patients infected with CF2. We also analyze the cytotoxic and enterotoxic activities of type CF2 5340 and 5362 culture filtrates. Finally, we define the variant tcdA and tcdB genes of type CF2 5340 and compare the sequences to those of the standard toxigenic strain VPI 10463, variant strains 1470 and 8864, and the LT gene of C. sordellii.
MATERIALS AND METHODS
Bacterial strains.
REA type CF2 isolates 5340 and 5362 were recovered from two patients with CDAD at the Veterans Administration Medical Center in Minneapolis (MVAMC), Minn., in 1993. The additional CF2 isolates 4092, 4139, 4151, 5264, and 5265 were recovered from stool specimens of patients at the MVAMC (Table 1). Nontoxigenic REA type M3 isolate 1413 was recovered from an asymptomatic patient at the MVAMC in 1989. Toxigenic REA type G1 isolate 1077 was recovered from a patient with CDAD at the MVAMC in 1986. Strain 8864 was a clinical isolate from The General Hospital, Birmingham, United Kingdom (14). Strain VPI 10463 was a clinical isolate from the VPI Anaerobe Collection, Virginia Polytechnic Institute and State University, Blacksburg, Va. (T. D. Wilkins, personal communication). Strain 1470, obtained from the American Type Culture Collection (CCUG 43598), was recovered from an asymptomatic infant (9).
TABLE 1.
Clinical summary of patients with C. difficile toxin variant type CF2 infections
| Parameter | Value for patient:
|
||||||
|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | |
| Date of isolatea | 7-5-90 | 8-23-90 | 9-5-90 | 12-18-92 | 12-20-92 | 1-4-93 | 1-26-93 |
| Isolate no. | 4092 | 4139 | 4151 | 5265 | 5264 | 5340 | 5362 |
| Age (yr)/sexb | 71/M | 59/M | 60/M | 81/M | 62/M | 76/M | 71/M |
| Stool cytotoxin assayc | Pos. | NAe | Pos. | Pos. | Neg.f | Pos. | Pos. |
| Clinical syndrome | CDAD | NA | CDAD | CDAD | Colonization | CDAD | CDADg |
| Specific treatmentd | Metro | NA | Metro | None | None | Metro and Vanco | None |
Month-day-year.
M, male.
Pos., positive; neg., negative.
Metro, metronidazole; Vanco, vancomycin.
NA, clinical information not available.
Culture filtrate positive in cytotoxin assay.
With fatal pseudomembranous colitis.
REA.
C. difficile strains were inoculated into brain heart infusion (BHI) from a blood agar plate 48-h culture and incubated anaerobically overnight. The DNA was isolated as previously described (8) and then digested with HindIII. The fragments were separated on a 0.7% agarose gel, producing a characteristic banding pattern for each isolate. The pattern of each isolate was visually compared with the patterns of the previously identified REA types in our well-characterized library. New isolates which showed six or fewer band differences (a similarity index of >90%) from an established REA type were placed within the same group (letter designation) as the REA type (8). Only isolates with identical patterns were assigned the same type (number designation).
Phenotypic assays of toxin variant C. difficile culture filtrates. (i) Production of culture filtrates.
For the toxin A immunoassay, rabbit ileal loop assay, and OTF9-63 cytotoxicity assays, bacterial strains were inoculated into 20 ml of reduced BHI broth and incubated anaerobically at 37°C for 3 days. The cultures were centrifuged, and the supernatants were filtered through 0.45-μm-pore-size syringe filters. For the human fibroblast cytotoxicity assay, bacterial strains were inoculated into 5 ml of chopped-meat medium and incubated anaerobically at 37°C for 5 days. The supernatants were also filtered prior to being assayed as described above.
(ii) Immunoassay for toxin A.
The Tox-A Test (TechLab, Blacksburg, Va.) was used to assay culture filtrates for the presence of toxin A according to the manufacturer's instructions. This assay employs the monoclonal antibody PCG-4 as the detecting antibody and is specific for epitopes of the toxin encoded by the 3′ end of the tcdA gene.
(iii) Enterotoxicity assay.
The enterotoxic activities of culture filtrates were determined in the rabbit ileal loop assay as previously described (15). One milliliter (each) of culture filtrates from C. difficile type CF2 isolates 5340 and 5362 was injected into ligated ileal loops. Each test loop was separated by an intervening noninjected loop. Positive control loops contained culture filtrate from a standard toxigenic C. difficile strain (REA type G1) or purified toxin A at 10 μg/ml. Negative control loops contained culture filtrates from a nontoxigenic C. difficile strain (REA type M3) or BHI. The rabbit was allowed to rest for 18 h and was then sacrificed. Results were expressed as the ratio of the accumulated fluid (in milliliters) over the length (in centimeters) of the loop. Ratios of >1.0 were indicative of enterotoxic activity.
(iv) Cytotoxicity assays.
C. difficile culture filtrates were screened for cytotoxicity in a microtiter assay kit (C. difficile cytotoxicity assay kit; Bartels, Inc., Issaquah, Wash.) which utilized human fibroblasts as the target cells and specific anti-C. difficile antiserum as the neutralizing antibody. Cytotoxicity was also determined for C. difficile culture filtrates using OTF9-63 cells, a mouse teratocarcinoma cell line with increased sensitivity to the CPE of toxin A but also sensitive to toxin B (32). Test and control samples were assayed in 1-ml aliquots of culture filtrates in serial 10-fold dilutions in 24-well plates containing monolayers of OTF9-63 cells in 1 ml of tissue culture medium. Culture filtrates of a standard toxigenic C. difficile strain (REA type G1) were used as positive controls, and culture filtrates of a nontoxigenic C. difficile strain (REA type M3) and BHI broth were used as negative controls. The cytotoxic titer was reported as the reciprocal of the highest dilution to cause rounding in >90% of cells at 24 h postinoculation.
(v) Separation of toxins.
Toxins were isolated from culture supernatants of C. difficile strains VPI 10463 and CF2 5340 and 5362 over the anion-exchange resin DEAE-Sepharose CL-6B using the methods of Lyerly et al. (20). The fractions were tested for cytotoxic activity using Chinese hamster ovary (CHO-K1) cells (20).
Molecular analysis of C. difficile toxin variant genes. (i) Hybridization studies.
Total cellular DNA from C. difficile variant type CF2 isolate 5340 and control strains was isolated and digested with HindIII or PstI, and the fragments were separated on a 0.7% agarose gel as described above. The DNA was then transferred to a nylon membrane (Nytran; Schleicher & Schuell, Inc., Keene, N.H.) by the method of E. M. Southern (29). Portions of the tcdA and tcdB genes were detected with six large genomic probes spanning the major regions of tcdA and tcdB (20) and two oligonucleotide probes, AIP-1 and AIP-2, corresponding to sequences in the tcdA gene from strain VPI 10463 (11). The positions of the probes relative to tcdA and tcdB are shown in Fig. 1. The large probes were labeled with [α-32P]deoxycytidine triphosphate (NEN, Boston, Mass.) by random priming (Random Primers DNA labeling system; Gibco-BRL, Grand Island, N.Y.), and the oligoprobes were labeled with [γ-32P]adenosine triphosphate (NEN) using the RTS T4 kinase labeling system (Gibco-BRL). Prehybridization, hybridization, and wash conditions were according to the manufacturer's instructions for the Nytran membrane. Hybridized sequences were detected by autoradiography with Kodak X-Omat AR film at −70°C for 18 h.
FIG. 1.
DNA probes specific for the toxin genes of C. difficile strain VPI 10463 used in analysis of C. difficile type CF2 5340 toxin B (tcdB) and toxin A (tcdA) genes. DNA probes generated from the toxin genes of standard toxigenic C. difficile strain VPI 10463 and used in Southern blot analysis of C. difficile variant REA type CF2 are shown in relation to the pathogenicity locus (PaLoc) of strain VPI 10463. The expected sizes of HindIII-restricted DNA fragments of C. difficile strain VPI 10463 are shown above the PaLoc. The locations of primers 5AU-2 and 5AD are indicated by solid triangles. These primers were used to amplify a 4.3-kb region of the type CF2 5340 tcdA gene which hybridized to oligoprobes AIP-1 and AIP-2.
(ii) Preparation of oligonucleotide primers and probes.
Oligonucleotide primers were constructed for the purpose of toxin gene segment amplification and DNA sequence analysis. Primers for the 5′ end of tcdB (3-kb amplicon), the middle of tcdB (2.54-kb amplicon), the 3′ end of tcdB (2.5-kb amplicon), the 5′ end of tcdA (4.5- and 1.5-kb amplicons), and the 3′ end of tcdA (5- and 3-kb amplicons) were designed based on the published toxin gene sequences of strain VPI 10463 (2, 11). Nested primers for sequencing the interior of the cloned CF2 toxin gene fragments were designed based on CF2 5340 sequences determined in initial sequencing runs by a chromosome-walking technique. Oligonucleotide probes AIP-1 and AIP-2 were also designed based on the toxin gene sequences of strain VPI 10463 for the purpose of assaying the previously unhybridized portion of CF2 tcdA which lies between sequences hybridized by genomic probes 2382 and 5660 (Fig. 1). The oligonucleotide probe sequences and the primers which generated the toxin gene amplicons described above are listed in Table 2. The oligonucleotides were commercially prepared by Operon Technologies, Alameda, Calif.
TABLE 2.
Primers and oligoprobes used in PCR and sequencing studies
| Primer or oligoprobe | Primer orientation | Amplicon location | Sequence | Position of 3′ end of primer in toxin gene |
|---|---|---|---|---|
| B-u1-b | Upstream | 5′ tcdB | AGACAAGCTGTTAATAAGGCTAA | 23a |
| B-d1-b | Downstream | 5′ tcdB | TCAGTCTCTGATATGTCCTCAAA | 3067a |
| 3pB-A | Upstream | Middle tcdB | GCACTATGTGACTTAAAACAAC | 3034a |
| 3pB-C | Downstream | Middle tcdB | GATAACAGTTGAAGATATATCTC | 5411a |
| 3pB-B | Upstream | 3′ tcdB | GATGATAGTAAGCCTTCATTTG | 4930a |
| 3pB-D | Downstream | 3′ tcdB | CTATTCACTAATCACTAATTGAG | 7490a |
| ACU-2 | Upstream | 5′ tcdA | GGTTATAGAAGTGGATTTATTATC | 24b |
| 5AD-1 | Downstream | 5′ tcdA | CAATTGAGATAGATTATTTTCTGG | 1557b |
| 5AU-2 | Upstream | 5′ tcdA | GCAGCATCTGACATAGTAAG | 975c |
| 5AD | Downstream | 5′ tcdA | GGAATGTATTTGGGTTAAGAAC | 5306c |
| A-u2 | Upstream | 3′ tcdA | AATGAGTACTACCCTGAGAT | 5301c |
| A-d1-b | Downstream | 3′ tcdA | AATTTCTTAGTAGCACAGGAAT | 8316c |
| AIP-1 | Oligoprobe | Middle tcdA | CAGGCGATATAGATAATAAAG | 4358c |
| AIP-2 | Oligoprobe | Middle tcdA | GACGATGTTTTCCATTCACC | 5060c |
(iii) PCR amplification of toxin gene sequences.
Template DNA was isolated from test C. difficile strains as described above. Amplification of toxin gene sequences was carried out using a modification of the method of Cheng et al. (7). In brief, we amplified 250 ng of template DNA in a reaction mixture of 50 mM KCl, 10 mM Tris-Cl (pH 8.3), 0.01% gelatin, deoxynucleoside triphosphates (200 μM each), primers (10 pmol each), 1.8 U of AmpliTaq DNA polymerase (Perkin-Elmer-Roche, Foster City, Calif.), and 0.15 U of Pfu DNA polymerase (Stratagene, La Jolla, Calif.) in the presence of MgCl2 concentrations empirically determined for each primer pair. The cycling parameters consisted of denaturation at 94°C, annealing at the melting temperature for each primer pair minus 5°C, and extension at 68°C for 2 min per kb of amplicon.
DNA sequencing and analysis.
For DNA sequencing, amplicons of CF2 5340 and 5264 toxin gene sequences (Fig. 2) were cloned into PCR-Script Amp vectors (Stratagene) and transformed into Escherichia coli strain XL-10 Blue (Stratagene) with the PCR-Script Amp cloning system (Stratagene) according to the manufacturer's instructions. The sizes of the cloned toxin gene amplicons and their locations in relation to the pathogenicity locus of type CF2 are shown in Fig. 2. Recombinant plasmids were isolated from transformed colonies by alkaline lysis (23), purified by phenol-chloroform-isoamyl alcohol extraction, and precipitated with ethanol. The nucleotide pellets were washed with 75% ethanol and then resuspended in nuclease-free H2O at 2 mg/ml. Sequencing was performed by the Sanger method of dideoxy-mediated chain termination (24) on an ABI Prism automated DNA sequencer (Perkin-Elmer, Applied Biosystems Division, Foster City, Calif.). All sequence analyses were performed either manually or with sequence analysis software (Sequence Navigator; Perkin-Elmer). Both CF2 5340 tcdA and tcdB sequences were analyzed for homology to the corresponding genes of the standard toxigenic C. difficile strain VPI 10463 and variant strain 1470. CF2 5340 tcdB sequences were also analyzed for homology to the gene for the LT of C. sordellii and to the tcdB gene of variant C. difficile strain 8864. Toxin gene sequences for C. difficile strains VPI 10463 (accession no. X53138 and M30307), 1470 (accession no. Z23277, Y12616, and AJ132669), 8864 (accession no. AF053400), and the LT of C. sordellii (accession no. X82638) were obtained from the GenBank database at the National Center for Biotechnology Information.
FIG. 2.
Cloned fragments of toxin B (tcdB) and toxin A (tcdA) genes used for DNA sequencing of C. difficile variant type CF2 isolate 5340. The locations of the primers used to amplify gene fragments from C. difficile variant type CF2 are indicated by arrows. The sizes and locations of the cloned CF2 amplicons are shown in relation to the pathogenicity locus of type CF2 isolate 5340.
Nucleotide sequence accession numbers.
The nucleotide sequence and deduced amino acid sequence for CF2 5340 tcdB is available through the GenBank sequence database, accession no. AF217292. The nucleotide sequence and deduced amino acid sequence for the 3′ end of CF2 5340 tcdA is available through the GenBank sequence database, accession no. AF217291. The nucleotide sequence and deduced amino acid sequence for the 5′ end of CF2 5340 tcdA is available through the GenBank sequence database, accession no. AF279457.
RESULTS
Index patients.
Two clinical C. difficile isolates were identified during a comparative study of diagnostic tests for C. difficile toxins. Patient stool specimens from which these isolates were recovered were negative by immunoassay for toxin A but positive by HEp-2 cell culture cytotoxin assay (25). The first isolate (no. 5340) was recovered from a 76-year-old man with aortic valvular disease who developed diarrhea with positive stool C. difficile culture and stool cytotoxin assays following surgery and administration of several antibiotics. The diarrhea resolved on specific therapy with oral metronidazole and vancomycin. The second isolate (no. 5362) was recovered from a 71-year-old man with coronary artery disease and severe cardiomyopathy who developed diarrhea during cephalosporin treatment for pneumonia. He was readmitted to the hospital with abdominal pain and distention, decreased bowel sounds, and rebound tenderness on examination. He was thought to have ischemic bowel; therefore, C. difficile disease was not suspected at that time. His abdominal pain and diarrhea worsened, and he suffered a fatal cardiac arrest 4 days after readmission. Autopsy findings demonstrated confluent pseudomembranous colitis, and stools obtained after readmission and prior to death were positive in C. difficile culture and stool cytotoxin assays. REA typing of both isolates revealed identical HindIII restriction patterns, which were designated type CF2.
Restriction endonuclease analysis of toxin variant strains.
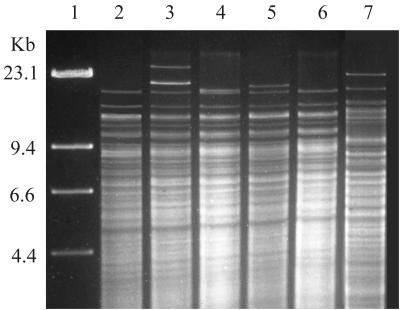
Comparison of the HindIII restriction patterns of cellular DNA from variant type CF2 isolates 5340 and 5362 to the HindIII restriction patterns from previously reported C. difficile toxin variant strains showed similarities to strain 1470 but not to strain 8864. Strain 1470 was identical to type CF1 by REA and therefore has more than 90% pattern homology to type CF2 (Fig. 3). Strain 8864 did not fit within any group previously identified in our REA typing system and has been given the new group and type designation CY1 (Fig. 3).
FIG. 3.
HindIII REA of C. difficile variant strains 1470 and 8864 compared to REA group CF types. Lane 1, lambda phage DNA ladder; lanes 2 to 5, REA group CF isolates 5922 (type CF1), 4092 (CF2), 4241 (CF3), and 5572 (CF4), respectively, from our clinical C. difficile isolate collection (8); lane 6, toxin variant strain 1470 (9, 10); lane 7, toxin variant strain 8864 (14, 31). The REA pattern of strain 1470 is identical to that of REA type CF1, but the REA pattern of type 8864 is not similar to those of REA group CF or any previously described REA group in our clinical-isolate collection.
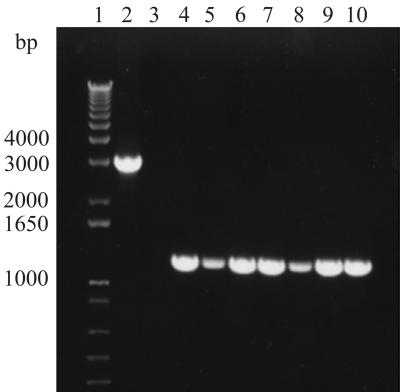
A review of our clinical C. difficile collection of >5,000 isolates from the past 17 years identified five additional type CF2 isolates (Table 1). PCR amplification of cellular DNA from all seven isolates using primers A-u2 and A-d1-b (Table 2) confirmed the presence of the same 1.8-kb deletion at the 3′ end of tcdA (Fig. 4). Five of the seven patients found to have CF2 infections had documented CDAD (Table 1). They were all elderly men, reflecting the population at the MVAMC, where these patients were hospitalized between 1990 and 1993. Patient 1 was transferred from another Veterans Administration hospital in South Dakota and had CDAD on admission to the MVAMC, indicating that this CF2 infection probably originated in South Dakota. Patients 4 and 5 were identified through a surveillance study of enteral feeding and C. difficile infection (4) and acquired type CF2 within 2 days of each other while in the same surgical intensive-care unit, suggesting nosocomial acquisition of these isolates.
FIG. 4.
PCR amplification of DNA from C. difficile variant type CF2 isolates using tcdA primers A-u2 and A-d1-b. Lane 1, 1-kb DNA ladder; lane 2, standard toxigenic strain VPI 10463; lane 3, nontoxigenic type M3; lanes 4 to 10, type CF2 isolates from patients 1 to 7 (Table 1), respectively.
Toxin activity of C. difficile variant type CF2 culture filtrates.
Culture filtrates from variant type CF2 isolates 5340 and 5362 were assayed for the presence of toxin A and enterotoxicity by immunoassay and the ileal loop assay, respectively. Both isolates were repeatedly negative by immunoassay. Culture filtrates from both type CF2 isolates failed to produce fluid accumulation responses typical for toxin A in the rabbit ileal loop assay; volume-to-length ratios of 0.09 and 0.08 were recorded for variant type CF2 isolates 5340 and 5362, respectively, whereas ratios of 1.5 and 2.36 were recorded for toxigenic C. difficile type G1 and purified toxin A (10 μg/ml in BHI). Ratios of 0.18 and 0.05 were recorded for the negative controls, nontoxigenic C. difficile type M3 and BHI alone. Culture filtrates of variant type CF2 isolates 5340 and 5362 did, however, produce a grossly hemorrhagic viscous mucus in the ileal loops compared to the clear or slightly blood-tinged fluid seen in the negative controls.
Culture filtrates of all type CF2 isolates from our clinical collection (Table 1) were also analyzed for cytotoxicity by cell culture assays. All seven isolates demonstrated CPE in the human fibroblast cytotoxicity assay that were neutralized by the specific anti-C. difficile antisera. Culture filtrates of type CF2 isolates 5340 and 5362 were also tested in the OTF9-63 cytotoxicity assay. They demonstrated cytotoxic titers of 10 in the OTF9-63 assay compared to a titer of 100 for the control toxigenic C. difficile type G1. The cytopathic effects of the CF2 isolates also differed from those of the type G1 control isolate in the OTF9-63 cytotoxicity assay: in response to type CF2 filtrates, target cells were not merely rounded but appeared collapsed and detached from the well and were floating in clumps in the cell culture media.
In addition, separation of toxins from the culture filtrates of CF2 isolates 5340 and 5362 was performed by column chromatography. DEAE-Sepharose column separation failed to elute any toxin from the CF2 culture filtrates in the linear gradient of 0.05 to 0.25 M NaCl, whereas the characteristic toxin A was eluted from strain VPI 10463 in this gradient. In contrast, fractions of CF2 culture filtrates eluted over a linear gradient of 0.25 to 0.6 M NaCl demonstrated cytotoxicity in CHO K1 cells. This elution gradient is identical to the gradient over which toxin B was isolated from strain VPI 10463.
Comparison of C. difficile variant type CF2 isolates and strain VPI 10463 toxin genes by DNA hybridization and PCR amplification.
Southern blot analysis of C. difficile variant type CF2 isolate 5340 demonstrated that the majority of tcdA and tcdB sequences are preserved in comparison to strain VPI 10463. All of the probes hybridized with HindIII-restricted cellular DNA from strain VPI 10463 and failed to hybridize to HindIII-restricted cellular DNA from nontoxigenic type M3. All of the large DNA probes specific for the tcdB and tcdA genes of strain VPI 10463 hybridized to HindIII-restricted cellular DNA from variant type CF2 except for probe J66 (specific for the 5′ end of tcdB). PCR amplifications of the 5′ region of tcdB from both VPI 10463 and type CF2 5340 were performed to investigate the failure of probe J66 to hybridize to type CF2. Amplicons generated by primers B-u1-b and B-d1-b, which bracket the first 3,000 bp of tcdB, showed no difference in size between CF2 tcdB and 10463 tcdB, indicating that the hybridization failure of probe J66 was not due to a large nucleotide deletion or insertion in this region.
In contrast to the results for CF2 5340 tcdB, probes specific for the 3′ end of tcdA (5660 and 2382) hybridized with type CF2 5340 HindIII restriction fragments which were approximately 2 kb shorter than the corresponding restriction fragments from strain VPI 10463, suggesting a possible major deletion in CF2 tcdA. Amplification of cellular DNA using primers A-u2 and A-d1-b, which bracket the last 3,000 bp of the 3′ end of strain VPI 10463 tcdA, confirmed an approximately 1.8-kb deletion in the 3′ end of CF2 tcdA. Subsequent amplification of a 4.3-kb segment of tcdA immediately upstream from the region recognized by probe 5660 was performed using primers 5AU-2 and 5AD (Fig. 1) and cellular DNA from type CF2 5340 and strain VPI 10463. This amplification yielded amplicons of 4.3 kb from both type CF2 5340 and strain VPI 10463, indicating that the deletion was downstream from this segment. Oligoprobes AIP-1 and AIP-2, specific for the interprobe region between the large genomic probes 2382 and 5660, hybridized to these amplicons from type CF2 5340 and from strain VPI 10463, implying base homology between VPI 10463 and CF2 in this region.
Southern blot analysis using PstI restriction digests of cellular DNA revealed a genetic alteration toward the 5′ end of CF2 5340 tcdA. Specifically, a PstI restriction fragment from type CF2 5340 which hybridized to probe 2382, specific for the middle sequences of tcdA, was 500 bp shorter than the corresponding fragment from strain VPI 10463. The previously described PCR amplification of the 5′ end of type CF2 5340 and strain VPI 10463 tcdA using primers 5AU-2 and 5AD indicated that the discrepancy was not due to a 500-bp deletion in this region of CF2 tcdA. Subsequent restriction of these amplicons with PstI confirmed the presence of an additional PstI site approximately 1,300 bp from the 5′ end of CF2 5340 tcdA.
DNA sequence comparison of tcdA and tcdB from C. difficile variant type CF2 isolates to other clostridial toxin genes.
Comparative analysis of the nucleotide sequences of type CF2 5340 tcdB to strain VPI 10463 tcdB sequences showed the presence of multiple nucleotide substitutions throughout CF2 tcdB, with the greatest concentration (320 substitutions) occurring in the first 2,000 bases. We also detected a 13-bp insert approximately 30 bp upstream from the tcdB open reading frame and a 3-bp (1-amino-acid) insert at position 933 of the tcdB open reading frame. Comparative analysis of the deduced amino acid sequences of type CF2 and strain VPI 10463 found 147 differences in the first 1,000 amino acid sequences (Fig. 5). Analysis of the 147 amino acid variations found 45 of the CF2 variant amino acids to be identical to those in the LT of C. sordellii, whereas 102 of the CF2 variant amino acids were unique to CF2, showing no identity with either strain VPI 10463 or C. sordellii (Fig. 5). In comparing the tcdB gene of type CF2 5340 to that of strain 1470, there were 16 nucleotide differences and 12 amino acid differences throughout the entire gene, showing 99.8% homology of nucleotide bases and 99.5% homology of amino acid sequences between the two strains. The catalytic domain of tcdB has been defined as the first 1,680 nucleotides, which code for amino acids 1 to 560 (27). When we compared the deduced amino acid sequence of the catalytic domain of type CF2 5340 tcdB to that of strain 8864, we detected 10 amino acid substitutions in this region, which shows 98.3% homology between the catalytic domains of the tcdB genes of the two strains.
FIG. 5.
Comparative amino acid analysis of toxin B from C. difficile strain VPI 10463 and toxin B from C. difficile variant type CF2 isolate 5340. The number of deduced amino acid differences between toxin B from type CF2 and the toxin B from standard toxigenic strain VPI 10463 are displayed in 200-amino-acid increments. The CF2 amino acid sequences which differed from those of strain VPI 10463 were also compared to the amino acid sequences of the LT of C. sordellii.
Comparative analysis of type CF2 5340 3′ tcdA sequences to strain VPI 10463 3′ tcdA sequences detected three large deletions (a total of 1,821 bp deleted) in this “repeating” region of tcdA (2,562 bp long in VPI 10463 tcdA). This represents a loss of 71% of the nucleotides in this region and an overall loss of 22.3% of the nucleotides in tcdA. Although severely truncated, the 3′ repeating region of CF2 tcdA still possesses at least one of each of the class I- and class II-type and -subtype regions of repeating DNA sequences found in strain VPI 10463 (2) (Fig. 6). Comparative analysis of CF2 tcdA sequences on the 3′ end to the same tcdA region of strain 1470 showed 99.6% homology of nucleotide bases.
FIG. 6.
DNA sequence deletions in the 3′ repeating region of tcdA from C. difficile variant type CF2 isolate 5340. The open boxes indicate repeating units present in the tcdA of strain VPI 10463 but absent in type CF2. Repeating units defined by Dove et al. (11) include class I (subclasses I1 through I7) and class II (subclasses IIA through IID) repeating units distinguished by length and level of nucleotide sequence similarities. Hatched boxes, repeating units present in the tcdA of both type CF2 and strain VPI 10463; vertical bars, deletion sites in type CF2 tcdA. The lower diagram shows the repeating units of type CF2 as sequenced.
Comparative analysis of the nucleotide sequences of the first 1.5 kb of type CF2 tcdA to strain VPI 10463 tcdA sequences showed occasional point mutations throughout CF2 tcdA, for a total of 1.1% substitutions. One of these substitutions, a C-to-T substitution at tcdA position 139, introduces a premature stop codon identical to that described for strain 1470 (34). Another base substitution, a C-to-T substitution at tcdA position 1317, created the new PstI site initially detected by Southern hybridization studies. Comparative analysis of the CF2 5340 5′ tcdA sequences to the same tcdA region of strain 1470 (GenBank accession no. AJ132669) showed 100% homology of deduced amino acids up to the premature stop codon and 99.6% homology of nucleotide bases throughout the 1.5-kb sequence.
DISCUSSION
In this study, we have characterized a toxin A-negative, toxin B-positive strain of C. difficile, REA type CF2, that was responsible for C. difficile-associated diarrhea in five patients, one of whom died of pseudomembrane colitis. Through REA, we have compared type CF2 to the well-characterized toxin variant strains 8864 and 1470 and found that although strain 1470 was closely related to type CF2, strain 8864 was not similar to either variant strain. Previously, there were few data to suggest that strain 8864 or strain 1470, or the related serogroup F isolates, was responsible for human disease (9, 16). In contrast, variant type CF2 is associated with typical clinical syndromes caused by standard toxigenic strains of C. difficile. Our data and the recent report of an outbreak of CDAD in a Canadian hospital caused by a toxin A-negative B-positive strain of C. difficile (1) emphasize the potential clinical importance of these toxin variant C. difficile strains.
We have previously demonstrated the ability of type CF2 5340 to cause disease in hamsters, although with significantly lower colonization efficiency and mortality than other toxigenic C. difficile isolates (Sambol et al., Abstr. 99th Gen. Meet. Am. Soc. Microbiol. 1999). Pathogenicity in hamsters has also been reported for strain 8864 (5) but not for strain 1470 (9). Other serogroup F isolates have been reported as nonpathogenic in axenic mice (10).
Comparative analysis of the nucleotide sequences of the type CF2 5340 tcdB and strain VPI 10463 tcdB sequences showed the presence of multiple nucleotide substitutions throughout CF2 5340 tcdB, with the greatest concentration (320 substitutions) occurring in the first 2,000 bases, which code for the entire catalytic portion as well as the first part of the hydrophobic domain of toxin B (27). Consequently, the deduced amino acid sequence of the catalytic portion of CF2 5340 toxin B shows only 84% homology with that of strain VPI 10463, whereas comparison of CF2 toxin B to strain 8864 toxin B shows 98.3% homology in this region. Comparison of CF2 5340 tcdB to 1470 tcdB shows greater than 99.5% homology in both nucleotide and deduced amino acid sequences.
Assays for cytotoxicity (primarily an effect of toxin B) demonstrated that CF2 5340 and 5362 culture filtrates produced atypical CPE similar to those described for strains 8864 and 1470 (5, 6, 20). Although these CPE are atypical for standard C. difficile toxin B, they are typical of the effects of the LT of C. sordellii (6, 33). Our comparative analysis of the extrapolated amino acid composition of CF2 5340 toxin B to that of VPI 10463 and the LT of C. sordellii has shown that in the first 1,000 amino acids of CF2 5340, there are 147 amino acids which differ from those in VPI 10463. Of those 147 variant amino acids, approximately one-third are identical to those in the LT of C. sordellii and two-thirds differ from both the toxin B of strain VPI 10463 and the LT of C. sordellii. This observation, together with the sequence homology of the tcdB genes of CF2 5340 and 1470, is consistent with the findings of Chaves-Olarte, et al. (6), who have found the variant form of toxin B in strain 1470 to be a functional hybrid of C. difficile toxin B and the LT of C. sordellii. They demonstrate that 1470 toxin B and the LT of C. sordellii glucosylate similar cellular substrates, including R-Ras, and postulate that R-Ras glucosylation is a key determinant in the cell rounding and detachment seen in the CPE of strain 1470 and C. sordellii.
Bioassays and immunoassays have failed to detect toxin A in CF2 culture supernatants. The ileal loop bioassay for enterotoxicity demonstrated that culture supernatants of type CF2 isolates 5340 and 5362 did not exhibit typical enterotoxic edema but did demonstrate a qualitative difference (hemorrhagic mucus) from the negative controls. This atypical reaction was also demonstrated by Borriello et al. in ileal loop assays of the toxin A-negative, toxin B-positive strain 8864 (5). DEAE-Sepharose chromatography also failed to isolate toxin A from CF2 culture supernatants. We also failed to detect toxin A in type CF2 5340 and 5362 culture filtrates using an immunoassay which employs the monoclonal antibody PCG-4, which is widely used in commercial immunoassays to detect C. difficile toxin A in stool specimens. PCG-4 immunoassays that fail to detect toxin A in variant pathogenic strains such as CF2 5340 will produce false-negative stool test results for patients infected with these toxin variant strains.
The results of these assays suggest that no toxin A is being produced by type CF2 strains, although genetic analyses of the type CF2 tcdAs have shown that more than 77% of tcdA is still present in these strains. The most obvious genetic difference between type CF2 and VPI 10463, a 1.8-kb truncation, occurs in the 3′ end of the CF2 tcdA gene. We have shown that the loss of 1,821 nucleotides is due to three separate deletions in the 3′ region of tcdA, which codes for the receptor binding portion of toxin A (2) and for the epitopes recognized by PCG-4. Despite the loss of greater than two-thirds of the sequences in the repeating region of tcdA, we have demonstrated that type CF2 5340 possesses at least one of each of the class I and class II isotypes of repeating DNA sequences found in strain VPI 10463. These data led us to speculate that CF2 tcdA was coding for a variant toxin A (with alterations in the receptor binding portion) that plays an active role in the pathogenic process yet is undetectable by standard bioassays and immunoassays. Recent data, however, indicate that a mutation in the 5′ end of tcdA of toxin variant strains leads to premature termination of toxin A translation.
Von Eichel-Streiber et al. have shown that serogroup F strain 1470 and two similar strains from serogroup X failed to produce toxin A due to a point mutation at the 5′ end of tcdA that introduces a premature stop codon at amino acid position 47 (34). They also demonstrated that the truncated polypeptide encoded by 1470 tcdA lacks enzymatic activity (34). We sequenced the first 1.5 kb of tcdA in CF2 patient isolates 5264 and 5340 to determine if this point mutation is present in REA type CF2. Both isolates demonstrated the same point mutation, a C-to-T substitution at the same position (tcdA position 139) as was reported for strain 1470. Premature termination of toxin A translation would then account for the failure of bioassays and immunoassays to detect an intact toxin A in CF2 culture supernatants.
We conclude that REA type CF2, like strain 1470, lacks a functional toxin A. This is an intriguing result in light of the association of type CF2 with both human and hamster disease and the reported role of standard toxin A in the pathogenesis of C. difficile disease. The pathogenicity of type CF2 appears to be toxin A independent, leading us to presume that either the variant cytotoxin B acts as the sole pathogenic factor, as postulated by other investigators in previous studies of strain 8864 (20, 27), or that there is an undefined virulence factor which contributes to pathogenicity. We are conducting further studies to define the virulence factors of type CF2.
In summary, the toxin B gene of type CF2 5340 has a high degree of homology (>98%) to the toxin B genes of strains 8864 and 1470, and the truncated toxin A gene of CF2 5340 is nearly identical to that of strain 1470. REA typing has also shown a close relationship between type CF2 and strain 1470. In spite of these genetic similarities to strain 1470, type CF2 is pathogenic in hamsters and in humans. Due to the current inability to synthesize artificial genetic constructs of C. difficile, a naturally occurring toxin variant such as type CF2 is a valuable tool with which to elucidate the functions of toxins and their relationship to mechanisms of pathogenicity. Furthermore, the emerging role of toxin variant strains in C. difficile disease in humans, and their ability to escape detection by commercial immunoassays for toxin A, indicates the need to investigate the prevalence of these strains in the clinical setting.
ACKNOWLEDGMENTS
We thank Kevin Kunstman, Steven Wolinsky, Sabine Simon, Lance R. Peterson, Carol J. Shanholtzer, Keith E. Willard, Connie R. Clabots, and Donna Z. Bliss.
Grant support was received from Northwestern University Medical School (Intramural Research Grant [S.J.]) and the U.S. Department of Veterans Affairs Research Service (D.N.G. and S.J.).
REFERENCES
- 1.Al-Barrak A, Embil J, Dyck B, Olekson K, Nicoll D, Alfa M, Kabani A. An outbreak of toxin A negative, toxin B positive Clostridium difficile-associated diarrhea in a Canadian tertiary-care hospital. Can Commun Dis Rep. 1999;25:7. [PubMed] [Google Scholar]
- 2.Barroso L A, Wang S Z, Phelps C J, Johnson J L, Wilkins T D. Nucleotide sequence of Clostridium difficile toxin B gene. Nucleic Acids Res. 1990;18:4004. doi: 10.1093/nar/18.13.4004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bartlett J G, Moon N, Chang T W, Taylor N, Onderdonk A B. Role of Clostridium difficile in antibiotic-associated pseudomembranous colitis. Gastroenterology. 1978;75:778–782. [PubMed] [Google Scholar]
- 4.Bliss D Z, Johnson S, Savik K, Clabots C R, Willard K, Gerding D N. Acquisition of Clostridium difficile and Clostridium difficile-associated diarrhea in hospitalized patients receiving tube feeding. Ann Intern Med. 1998;129:1012–1019. doi: 10.7326/0003-4819-129-12-199812150-00004. [DOI] [PubMed] [Google Scholar]
- 5.Borriello S P, Wren B W, Hyde S, Seddon S V, Sibbons P, Krishna M M, Tabaqchali S, Manek S, Price A B. Molecular, immunological, and biological characterization of a toxin A-negative, toxin B-positive strain of Clostridium difficile. Infect Immun. 1992;60:4192–4199. doi: 10.1128/iai.60.10.4192-4199.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chaves-Olarte E, Low P, Freer E, Norlin T, Weidmann M, von Eichel-Streiber C, Thelestam M. A novel cytotoxin from Clostridium difficile serogroup F is a functional hybrid between two other large Clostridial toxins. J Biol Chem. 1999;274:11046–11052. doi: 10.1074/jbc.274.16.11046. [DOI] [PubMed] [Google Scholar]
- 7.Cheng S, Fockler C, Barnes W M, Higuchi R. Effective amplification of long targets from cloned inserts and human genomic DNA. Proc Natl Acad Sci USA. 1994;91:5695–5699. doi: 10.1073/pnas.91.12.5695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Clabots C R, Johnson S, Bettin K M, Mathie P A, Mulligan M E, Schaberg D R, Peterson L R, Gerding D N. Development of a rapid and efficient restriction endonuclease analysis typing system for Clostridium difficile and correlation with other typing systems. J Clin Microbiol. 1993;31:1870–1875. doi: 10.1128/jcm.31.7.1870-1875.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Delmee M, Avesani V. Virulence of ten serogroups of Clostridium difficile in hamsters. J Med Microbiol. 1990;33:85–90. doi: 10.1099/00222615-33-2-85. [DOI] [PubMed] [Google Scholar]
- 10.Depitre C, Delmee M, Avesani V, L'Haridon R, Roels A, Popoff M, Corthier G. Serogroup F strains of Clostridium difficile produce toxin B but not toxin A. J Med Microbiol. 1993;38:434–441. doi: 10.1099/00222615-38-6-434. [DOI] [PubMed] [Google Scholar]
- 11.Dove C H, Wang S Z, Price S B, Phelps C J, Lyerly D M, Wilkins T D, Johnson J L. Molecular characterization of the Clostridium difficile toxin A gene. Infect Immun. 1990;58:480–488. doi: 10.1128/iai.58.2.480-488.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.George R H, Symonds J M, Dimock F, Brown J D, Arabi Y, Shinagawa N, Keighley M R, Alexander-Williams J, Burdon D W. Identification of Clostridium difficile as a cause of pseudomembranous colitis. Br Med J. 1978;1:695. doi: 10.1136/bmj.1.6114.695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gerding D N, Olson M M, Peterson L R, Teasley D G, Gebhard R L, Schwartz M L, Lee J T., Jr Clostridium difficile-associated diarrhea and colitis in adults: a prospective case-controlled epidemiologic study. Arch Intern Med. 1990;146:95–100. [PubMed] [Google Scholar]
- 14.Haslam S C, Ketley J M, Mitchell T J, Stephen J, Burdon D W, Candy D C A. Growth of Clostridium difficile and production of toxins A and B in complex and defined media. J Med Microbiol. 1986;21:293–297. doi: 10.1099/00222615-21-4-293. [DOI] [PubMed] [Google Scholar]
- 15.Johnson S, Sypura W D, Gerding D N, Ewing S L, Janoff E N. Selective neutralization of a bacterial enterotoxin by serum IgA in response to mucosal disease. Infect Immun. 1995;63:3166–3173. doi: 10.1128/iai.63.8.3166-3173.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kato H, Kato N, Watanabe K, Iwai N, Nakamura H, Yamamoto T, Suzuki K, Kim S M, Chong Y, Wasito E B. Identification of toxin A-negative, toxin B-positive Clostridium difficile by PCR. J Clin Microbiol. 1998;36:2178–2182. doi: 10.1128/jcm.36.8.2178-2182.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lyerly D M, Lockwood D E, Richardson S H, Wilkins T D. Biological activities of toxins A and B of Clostridium difficile. Infect Immun. 1982;35:1147–1150. doi: 10.1128/iai.35.3.1147-1150.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lyerly D M, Saum K E, MacDonald D K, Wilkins T D. Effects of Clostridium difficile toxins given intragastrically to animals. Infect Immun. 1985;47:349–352. doi: 10.1128/iai.47.2.349-352.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lyerly D M, Krivan H C, Wilkins T D. Clostridium difficile: its disease and toxins. Clin Microbiol Rev. 1988;1:1–18. doi: 10.1128/cmr.1.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lyerly D M, Barroso L A, Wilkins T D, Depitre C, Corthier G. Characterization of a toxin A-negative, toxin B-positive strain of Clostridium difficile. Infect Immun. 1992;60:4633–4639. doi: 10.1128/iai.60.11.4633-4639.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rupnik M, Braun V, Soehn F, Janc M, Hofstetter M, Laufenberg-Feldmann R, von Eichel-Streiber C. Characterization of polymorphisms in the toxin A and B genes of Clostridium difficile. FEMS Microbiol Lett. 1997;148:197–202. doi: 10.1111/j.1574-6968.1997.tb10288.x. [DOI] [PubMed] [Google Scholar]
- 22.Rupnik M, Avesani V, Jane M, von Eichel-Streiber C, Delmee M. A novel toxinotyping scheme and correlation of toxinotypes with serogroups of Clostridium difficile isolates. J Clin Microbiol. 1998;36:2240–2247. doi: 10.1128/jcm.36.8.2240-2247.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. pp. 1.25–1.28. [Google Scholar]
- 24.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shanholzer C J, Peterson L R, Olson M M, Gerding D N. Prospective study of gram stain stool smears in the diagnosis of Clostridium difficile. J Clin Microbiol. 1983;17:906–908. doi: 10.1128/jcm.17.5.906-908.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Siegel D L, Edelstein P H, Nachamkin I. Inappropriate testing for diarrheal diseases in the hospital. JAMA. 1990;263:979–982. [PubMed] [Google Scholar]
- 27.Soehn F, Wagenknecht-Wiesner A, Leukel P, Kohl M, Weidmann M, von Eichel-Streiber C, Braun V. Genetic rearrangements in the pathogenicity locus of Clostridium difficile strain 8864—implications for transcription, expression and enzymatic activity of toxins A and B. Mol Gen Genet. 1998;258:222–232. doi: 10.1007/s004380050726. [DOI] [PubMed] [Google Scholar]
- 28.Song K P, Bai X L, Chang S Y. Nucleotide and peptide sequences of the open reading frame encoding a truncated toxin A gene of Clostridium difficile strain CCUG 20309. DNA Seq. 1999;10:93–96. doi: 10.3109/10425179909008423. [DOI] [PubMed] [Google Scholar]
- 29.Southern E M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975;98:503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- 30.Sullivan N M, Pellett S, Wilkins T D. Purification and characterization of toxins A and B of Clostridium difficile. Infect Immun. 1982;35:1032–1040. doi: 10.1128/iai.35.3.1032-1040.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Torres J F. Purification and characterization of toxin B from a strain of Clostridium difficile that does not produce toxin A. J Med Microbiol. 1991;35:40–44. doi: 10.1099/00222615-35-1-40. [DOI] [PubMed] [Google Scholar]
- 32.Tucker K D, Carrig P E, Wilkins T D. Toxin A of Clostridium difficile is a potent cytotoxin. J Clin Microbiol. 1990;28:869–871. doi: 10.1128/jcm.28.5.869-871.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Von Eichel-Streiber C, Meyer zu Heringdorf D, Habermann E, Sartingen S. Closing in on the toxic domain through analysis of a variant Clostridium difficile cytotoxin B. Mol Microbiol. 1995;17:313–321. doi: 10.1111/j.1365-2958.1995.mmi_17020313.x. [DOI] [PubMed] [Google Scholar]
- 34.Von Eichel-Streiber C, Zec-Pirnat I, Grabnar M, Rupnik M. A nonsense mutation abrogates production of a functional enterotoxin A in Clostridium difficile toxinotype VIII strains of serogroups F and X. FEMS Microbiol Lett. 1999;178:163–168. doi: 10.1111/j.1574-6968.1999.tb13773.x. [DOI] [PubMed] [Google Scholar]
- 35.Yannelli B, Gurevich I, Schoch P E, Cunha B A. Yield of stool cultures, ova and parasite tests, and Clostridium difficile determinations in nosocomial diarrhea. Am J Infect Control. 1988;16:246–249. doi: 10.1016/s0196-6553(88)80003-8. [DOI] [PubMed] [Google Scholar]