Abstract
Diffusion magnetic resonance imaging (dMRI) tractography has yielded intriguing insights into brain circuits and their relationship to behavior in response to gene mutations or neurological diseases across a number of species. Still, existing tractography approaches suffer from limited sensitivity and specificity, leading to uncertain interpretation of the reconstructed connections. Hence, in this study, we aimed to optimize the imaging and computational pipeline to achieve the best possible spatial overlaps between the tractography and tracer-based axonal projection maps within the mouse brain corticothalamic network. We developed a dMRI-based atlas of the mouse forebrain with structural labels imported from the Allen Mouse Brain Atlas (AMBA). Using the atlas and dMRI tractography, we first reconstructed detailed node-to-node mouse brain corticothalamic structural connectivity matrices using different imaging and tractography parameters. We then investigated the effects of each condition for accurate reconstruction of the corticothalamic projections by quantifying the similarities between the tractography and the tracer data from the Allen Mouse Brain Connectivity Atlas (AMBCA). Our results suggest that these parameters significantly affect tractography outcomes and our atlas can be used to investigate macroscopic structural connectivity in the mouse brain. Furthermore, tractography in mouse brain gray matter still face challenges and need improved imaging and tractography methods.
Keywords: Diffusion MRI, C57BL/6J mouse brain, Tractography, Anatomically constrained tractography, Structural connectivity, Corticothalamic network
1. Introduction:
Understanding the structural and functional networks in the brain is one of the major areas of neuroscience research. Over the past decade, tremendous efforts have been made in mapping neural architecture at various scales, from tract tracing at the mesoscopic scale (Hunnicutt et al., 2014; Oh et al., 2014; Zingg et al., 2014) to the identification of cellular-level connections and the synaptic molecular properties (Siegle et al., 2021; Winnubst et al., 2019). Mapping of neural networks has also been reported in multiple species, ranging from C. elegans to human (Cook et al., 2019; Majka et al., 2020; Oh et al., 2014; Scheffer et al., 2020; Szczupak et al., 2021; Xu et al., 2021).
Structural connectivity in the mouse brain has been a main focus due to the large number of mouse models of neurological diseases together with accessibility of mouse genomic sequence information and a large number of publicly available resources on the mouse brain (Sandberg et al., 2000; Siddiqui et al., 2005; Zapala et al., 2005). A leading example is the Allen Mouse Brain Connectivity Atlas (AMBCA) (http://connectivity.brain-map.org), a systematic analysis of long-distance axonal connectivity using recombinant adeno-associated virus (AAV) expressing enhanced green fluorescent protein (EGFP) as the tracer. AMBCA presents a mesoscale connectome of the mouse brain (Kuan et al., 2015; Oh et al., 2014), which is also integrated with genome-scale collection of cellular resolution gene expression profiles (Goldowitz, 2010; Lein et al., 2007; Ng et al., 2009). The same group has recently expanded the AMBCA resource demonstrating cell-class-specific hierarchical organization of mouse brain corticothalamic projections (Harris et al., 2019). These and similar resources from other groups (Hunnicutt et al., 2014; Jeong et al., 2016; Zingg et al., 2014) have led to greater understanding of the mouse brain mesoscopic connectivity and its organization principles (Coletta et al., 2020; Rubinov et al., 2015).
Although the uses of chemical and viral tracers have allowed direct visualization of neural connections, one limitation of the technique is the inability to examine a large number of circuitries within a single brain. In contrast, diffusion magnetic resonance imaging (dMRI) tractography (Jbabdi et al., 2015; Mori and van Zijl, 2002) permits non-invasive mapping of multiple pathways in the entire brain. Combining advanced dMRI tractography and computational tools further allows non-invasive exploration of structural connectivity in rodent brains and alterations due to genetic modifications or pathological conditions (Arefin et al., 2021; Arefin et al., 2017; Calabrese et al., 2015; Degiorgis et al., 2022; Mechling et al., 2016; Moldrich et al., 2010; Ren et al., 2007; Wang et al., 2020a; Wang et al., 2006; White et al., 2020; Wu et al., 2022; Yee et al., 2018). However, more wide-spread applications of dMRI tractography in connectome analysis has been hindered by its relatively low sensitivity and specificity, especially in gray matter (GM) structures, (Aydogan et al., 2018; Calamante, 2019; Schilling et al., 2019; Thomas et al., 2014) due to the lack of direct links between dMRI signals and the underlying cellular structures (e.g. axons) and the low spatial resolution limitation of MRI. A recent study by Trinkle et al. (Trinkle et al., 2021) compared connectomes based on AMBCA and dMRI data and revealed considerable differences between them, in particular, the lack of connections in the corticothalamic network. However, in depth quantification of the corticothalamic circuitry that can be reconstructed using dMRI tractography as compared to the corresponding AMBCA is lacking.
For the general use of dMRI to examine connectome in normal or diseased mouse brains, it is important to obtain detailed knowledge on its sensitivity and specificity and locate optimal parameters for dMRI tractography. Therefore, we used different imaging and tractography parameters and reconstructed a detailed corticothalamic structural connectivity matrix specific for each condition. Considering the tracer-based corticothalamic projection maps from the AMBCA as ground truth, we counted the true positive (TP), true negative (TN), false positive (FP), and false negative (FN) connections from the tractography derived corticothalamic structural connectivity matrices. Additionally, we assessed voxel-wise similarities of the streamline termination points in the thalamic region between the tractography and AMBCA. Finally, we identified the imaging and tractography condition that reconstruct the corticothalamic network with improved spatial overlaps with the AMBCA using tractography. Similar studies have been carried out in the macaque monkey brain (Ambrosen et al., 2020; Girard et al., 2020; Maffei et al., 2022), which has high translational value, but the AMBCA contains a more comprehensive tracer tracing dataset.
For systematic analysis of tractography results together with resources such as AMBCA, an MRI-based atlas with structural labels that are compatible with the Allen Mouse Brain Atlas (AMBA) is necessary. To date, a number of MRI-based mouse brain atlases and datasets have been reported, covering both adult and developing mouse brains (Ali et al., 2005; Badea et al., 2007a; Badea et al., 2007b; Bock et al., 2006; Chen et al., 2006; Chuang et al., 2011; Dorr et al., 2007; Dorr et al., 2008; Johnson et al., 2007; Kovacevic et al., 2005; Lee et al., 2005; Ma et al., 2005; MacKenzie-Graham et al., 2004; Richards et al., 2011; Sharief et al., 2008; Steadman et al., 2014; Szulc et al., 2015; Ullmann et al., 2013). A summary of these atlases and datasets is provided in Supplementary Table S1. Many mouse brain atlases were created based on ex vivo T1/ T2 MRI with high spatial resolution (~ 30 um) and strong tissue contrasts in the adult mouse brain, mainly reflecting tissue water and myelin contents. Atlases based on dMRI and other MRI contrasts have emerged over the past decade (Aggarwal et al., 2009; Calabrese et al., 2015; Chuang et al., 2011; Jiang and Johnson, 2011; Szulc et al., 2015). Compared to T1-weighted/T2-weighted MRI, dMRI provides additional contrasts reflecting tissue microstructural organization (Mori and Zhang, 2006) and can be used to map macroscopic connectivity in the mouse brain (Aydogan et al., 2018; Calabrese et al., 2015; Keifer et al., 2015). The existing atlases, however, often do not have detailed cortical and subcortical structural labels, mainly because MRI data lack the resolution and tissue contrasts comparable to histology for delineation of subregions in GM such as deep thalamic nuclei or cortical layers.
To meet this demand, we have constructed a dMRI based mouse brain atlas by importing the AMBA structural labels (common coordinate framework version 3, CCFv3)(Wang et al., 2020c) in our previously published group average diffusion tensor images of the adult mouse brain (Chuang et al., 2011) and constructed a mapping between the DTI atlas and AMBA. The mapping of AMBA data to our atlas provided structural labels with high granularity and allowed us to examine mouse brain tractography results with viral tracer results in AMBCA in the same space consistently. Hence, we were able to use the node specific tracer maps as the gold standard to quantify the accuracy of tractography-based reconstruction of the corticothalamic connections in ex vivo dMRI data acquired from a separate group of mouse brains.
2. Materials and Methods:
All experimental procedures were approved by the Animal Use and Care Committee at the New York University Grossman School of Medicine.
2.1. Animals:
Adult C57BL/6J mice (males, P60-P90, n = 16) were used in this study. Six of them were used for in vivo manganese-enhanced MRI (section 2.3). The remaining ten mice were perfused transcardially with 4% paraformaldehyde in 0.1M phosphate buffer saline (PBS) and stored in 4% paraformaldehyde solution for 24 hours before transferred into PBS. Mouse brains kept in the skull were placed in 10 mL syringes filled with Fomblin (Perfluoropolyether, Solvay Solexis Inc.,Thorofare, N), a non-hydrogen containing liquid for susceptibility matching and to prevent dehydration. Detailed procedures on how to prepare the ex vivo brain sample can be found in (Arefin et al., 2021).
2.2. Diffusion-weighted MRI (DW-MRI) data acquisition:
Cohort 1:
data in this cohort were from a previous study (Chuang et al., 2011). Ex vivo imaging was performed on an 11.7 Tesla (T) NMR spectrometer (Bruker Biospin, Billerica, MA, USA) as described earlier (Chuang et al., 2011). Briefly, dMRI data was acquired using 3D diffusion-weighted multiple spin echo sequence at an isotropic spatial resolution of 0.125 mm3 with further isotropic interpolation to 0.0625 mm by zero-padding in the k-space.
Cohort 2:
High resolution ex vivo dMRI datasets were acquired on a horizontal 7T MR scanner (Bruker Biospin, Billerica, MA, USA). We used a 72-mm conventional circularly polarized birdcage radiofrequency resonator (Bruker Biospin, Ettlingen, Germany) for homogeneous transmission in conjunction with a four-channel receive-only phased array CryoProbe (CRP) anatomically shaped to tightly cover the mouse head (Bruker Biospin, Ettlingen, Germany) for high sensitivity and a modified 3D diffusion-weighted gradient-and-spin-echo (DW-GRASE) sequence (Aggarwal et al., 2010). Three sets of data were obtained with different imaging parameters (Table 1):
Table 1:
The imaging parameters of the dMRI datasets used in this study. TE and TR are abbreviations of echo time and repetition time, respectively.
| Parameters | Dataset 1 | Dataset 2 | Dataset 3 |
|---|---|---|---|
| No. of subjects | 10 | 10 | 10 |
| b-value (s/mm2) | 2000 | 5000 | 5000 |
| Number of diffusion encoding directions | 30 | 30 | 60 |
| TE/ TR (ms) | 35/400 | 35/400 | 35/400 |
| Isotropic resolution (mm) | 0.1 | 0.1 | 0.1 |
| Acquisition time (h) | ~ 5.3 | ~ 5.3 | ~ 10.6 |
2.3. Manganese-enhanced MRI (MEMRI) data acquisition:
Solution of manganese chloride (MnCl2) tetrahydrate (Sigma-Aldrich-221279) in isotonic saline (30 mM) was injected intraperitoneally to 6 animals 24 hours before MRI at a dose per weight of 0.5 mmol/kg (62.5 mg MnCl2 per kg body weight) as described in (Rallapalli et al., 2020). Animals were anesthetized using isofluorane (3% isofluorane for induction and 1–2% isofluorane during MRI mixed with compressed air at a flow rate of 1.5 L/min through a nose cone) and imaged using the horizontal 7T MRI scanner with the same 4-element phased array CRP for high sensitivity. T1-weighted images were acquired using a 3D spoiled gradient echo sequence (TE/TR = 3/30 ms, flip angle = 30°; field of view (FOV) = 1.8 × 0.9 × 1.6 cm; matrix size = 180 × 90 × 160 voxels, 0.1 mm isotropic resolution) with a total imaging time of 30 minutes. Individual subject MEMRI data were first rigidly aligned to the dMRI atlas (Chuang et al., 2011), and group average MEMRI data were computed using an iterative procedure as described in (Chuang et al., 2011). The group average MEMRI data were then aligned to the dMRI atlas using affine transformation.
A summary of the experiments carried out in this study has been provided in the Supplementary Table S2.
2.4. dMRI data processing:
Signals from non-brain tissue in the dMRI datasets were manually removed using AMIRA (ThermoFisher Scientific, https://www.thermofisher.com) and corrected for ghosting artifacts likely caused by frequency drifts using rigid alignment implemented in DTIStudio (Jiang et al., 2006). For both cohorts of mice, the six elements of diffusion tensor were determined by log-linear fitting via DTIStudio. The tensor was diagonalized to obtain three eigenvalues (λ1’ λ2’ λ3) and corresponding eigenvectors (v1, v2, v3). Fractional anisotropy (FA) was then calculated from the eigenvalues (Basser and Pierpaoli, 1996; Mori and Zhang, 2006). Finally, average diffusion-weighted images (aDWI) and FA images were normalized and averaged across all subjects of cohort 1 to create a group averaged mouse brain template (aDWI and FA, dimension: 200 × 280 × 128, resolution: 0.0625 mm) for the dMRI-based adult mouse brain atlas (Chuang et al., 2011).
2.5. Image registration pipeline for atlas construction:
Fig. 1 demonstrates the image registration pipeline used for mapping the ABMA structural labels (CCFv3) into the dMRI template space to construct the dMRI based mouse brain atlas. First, group averaged mouse brain template (DWI and FA images – Fig. 1A) and AMBA (Fig. 1B) were co-registered using landmark-based rigid transformation followed by intensity-based affine and large deformation diffeomorphic metric mapping (LDDMM) transformation implemented in DiffeoMap (www.mristudio.org) as shown in Fig. 1C–D. Second, segmentation of major brain structures (e.g., cortex, hippocampus, and cerebellum) were imported from AMBA and manually adjusted to fit corresponding structures in the dMRI data. We assigned unique intensity values to regions occupied by these major brain structures in the AMBA space and dMRI template space to compute a fine-tuned mapping to carry AMBA to the dMRI template space based on these major brain structures using LDDMM. Third, detailed AMBA structural labels were transferred to the group averaged template image (Fig. 1D) and further manually refined slice by slice along the axial orientation forfeiting attention to the other two orientations as well as to the slices preceding and following throughout the segmentation process Fig. 1E. The level of registration accuracy was measured using landmarks manually placed throughout the brain in the AMBA and dMR images resulting in an average error of 0.15±0.08 mm (Fig. 1F).
Fig. 1:
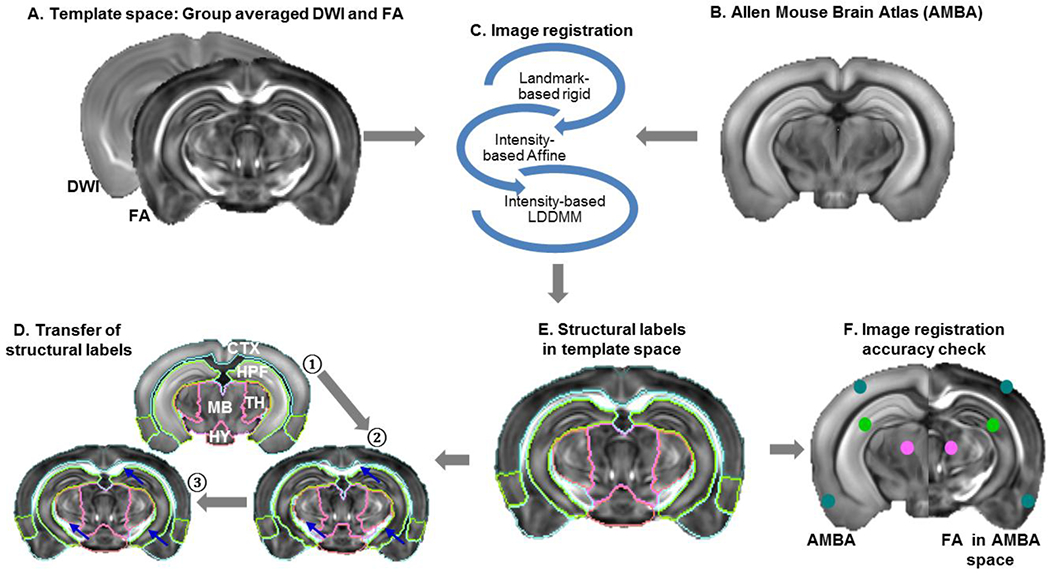
Image registration pipeline for atlas construction: A) Template space: averaged DWI and FA images of the adult mouse brain. B) A matching brain slice from the Allen Mouse Brain atlas (AMBA). C) Coregistration of MR and AMBA images using a series of image registration steps: 1. landmark based rigid transformation, 2. intensity based affine transformation, and 3. intensity based large deformation diffeomorphic metric mapping (LDDMM). D) After co-registration, structural labels of major brain compartments were transferred from AMBA (step ①) to the template space, manually edited (step ②), and a fine-tuned mapping between the template and AMBA was generated based on the labels of major brain compartments (step ③). E) The AMBA structural labels were mapped to the dMRI template using the fine-tuned mapping. F) Assessment of registration accuracy by placing landmarks manually in the template and AMBA images.
2.6. Non-constrained and anatomically constrained tractography (NCT and ACT):
dMRI datasets from cohort 2 were analyzed using MRtrix (https://www.mrtrix.org/) (Tournier et al., 2012). Non-constrained tractography (NCT) was performed using the pipeline described in (Arefin et al., 2021). In brief, using the pre-processed data, fiber orientation distribution (FOD) map was estimated for each subject (Tournier et al., 2007; Tournier et al., 2004), and a whole-brain tractogram was then generated containing a total of 5 million streamlines by seeding randomly within the whole-brain mask with minimum streamline length of 3 mm, FOD cut-off 0.05, angle 45°, step size 0.025.
For anatomically-constrained tractography (ACT), in addition to the criteria used for NCT, an image containing five labels of tissue segmentation (5tt) was used to guide the termination and acceptance/rejection criteria during tractography (Smith et al., 2012). The so-called 5tt image file contained cortical GM, sub-cortical GM, white matter (WM), cerebrospinal fluid (CSF), and a null channel (Supplementary Fig. S1 A). In ACT, any streamlines that entered the cortical GM or sub-cortical region from the WM were counted as valid and streamlines attempting to leave the pre-defined mask were terminated and accepted as valid. Any streamlines attempting to enter CSF or terminating in the WM were truncated and rerouted to find a valid termination point, as known as back-tracking (Fig. 2B–C). A whole-brain tractogram with 5 million streamline was generated using ACT for each subject with minimum streamline length of 3 mm.
Fig. 2:
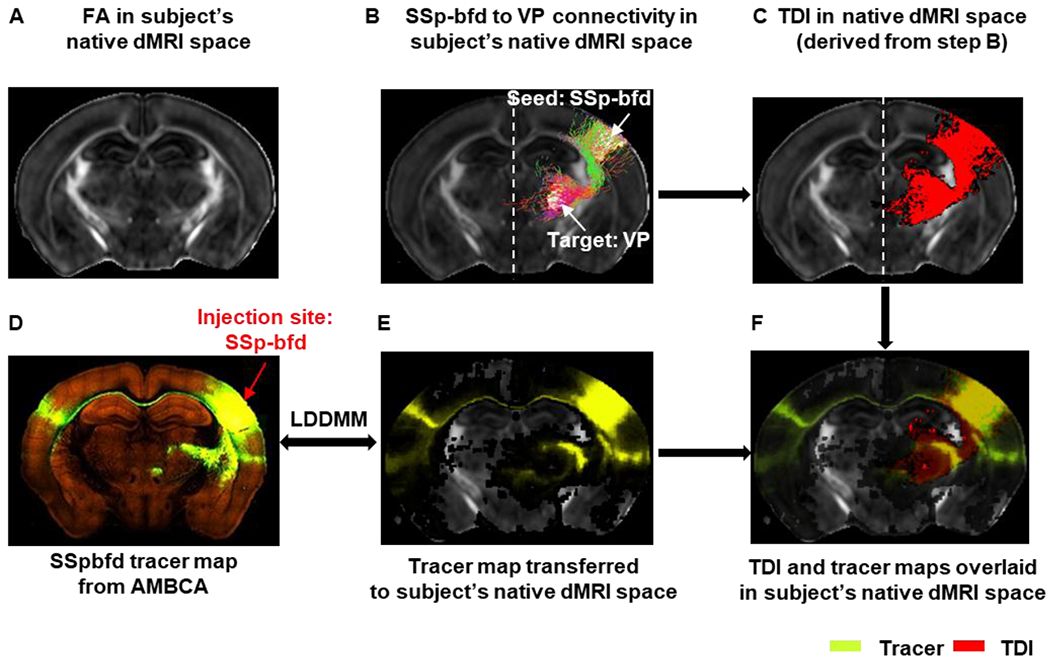
Transferring tracer map to each subject’s native dMRI space for tractography: A) Representative FA image in the native dMRI space. B) Ipsi-lateral tractogram from primary somatosensory barrel field area (SSp-bfd) to ventral posterior complex of the thalamus (VP) extracted from whole-brain tractogram shown in the native dMRI space. C) Track density image (TDI) derived from the SSp-bfd to VP streamline tractogram shown in B. D) Tract tracing image from AMBCA – selected injection site: SSp-bfd (ID:112951804). E) Tracer map transferred to the subject’s native dMRI space. F) TDI and tracer maps overlaid on subject’s native dMRI space.
Datasets acquired using three different imaging parameters (Table 1) and tractography algorithms (NCT or ACT) yielded six conditions (Table 2) that were used in the subsequent analysis.
Table 2:
The six conditions that were compared in this study.
| Conditions | b-values (s/mm2) | Number of diffusion directions | Tractography method |
|---|---|---|---|
| 1 (baseline) | 2000 | 30 | NCT |
| 2 | 5000 | 30 | NCT |
| 3 | 5000 | 60 | NCT |
| 4 | 2000 | 30 | ACT |
| 5 | 5000 | 30 | ACT |
| 6 | 5000 | 60 | ACT |
2.7. -Corticothalamic tractography:
As the injection sites in the (Harris et al., 2019; Oh et al., 2014) were chosen in the right hemisphere (RH) of the brain, to be consistent with the AMBCA, we selected 14 cortical regions from the RH (Supplementary Table S3) whose parcellations in CCFv3 showed no major difference from CCFv2 (Oh et al., 2014). For each subject, we constructed the corticothalamic connectome using two approaches as following:
Open-end tractography:
Using 14 RH cortical regions (Supplementary Table S3) we extracted individual streamline projections from the NCT and ACT whole-brain tractograms of each mouse brain.
Node-to-node tractography:
We used the previous 14 RH cortical regions and 12 RH thalamic regions (Supplementary Table S4) in AMBA and selected node-to-node streamlines from the RH of the whole-brain tractogram. Each cortical node served as the seed and a thalamic region as the target region. This approach allowed us to dissect entire circuitry into specific tracts and assess the patterns of streamline projections without superfluous connectivity to other brain regions.
2.8. Generation of the mouse brain corticothalamic structural connectome:
a. Ground truth connectivity matrix from AMBCA:
First, we constructed the tracer-based corticothalamic connectivity matrix from the AMBCA. As the atlas and the tracer maps were co-registered, we were able to define the boundary of the target regions in the thalamus. We considered two regions as ‘connected’ if a projection from the cortical seed region to the target thalamic region was detected, otherwise, ‘not connected’. Subsequent binary matrix served as the ground truth for further quantitative analysis.
b. Tractography-based connectivity matrices using ROIs from the atlas:
Next, we constructed dMRI tractography based corticothalamic connectivity matrix under the six conditions listed in Table 2. Two regions were considered to be connected if one or more streamlines had their endpoints in both regions. All detected region-to-region connections were marked as ‘1’, otherwise 0 (not connected). This resulted in six 14×12 binarized structural connectivity matrix. In these matrices, cortical regions were assigned in rows and thalamic regions in columns. We will sometimes refer these regions as nodes. Each cell of the matrix denotes whether there is a connection present in between the corresponding thalamic and cortical regions or not. Gray, green, red, and orange colors represent true negative (TN), true positive (TP), false positive (FP), and false negative (FN) connections.
2.9. Voxel-wise quantification of the level of similarities between tractography and the tracer maps: We quantified the level of spatial overlaps between tractography derived corticothalamic maps and the trace maps from AMBCA.
First, we generated high resolution tract density images (TDI) for each open-end tractogram as described in (Calamante et al., 2010).
Then we selected 14 tracer data with injection sites in cortex, identical to the cortical ROIs used for tractography. Detailed information about the injection sites is provided in Supplementary Table S5. To eliminate the spurious axonal projections, voxels with projection density >0.001 were counted as structurally connected with the injection site (seed region).
As the tractography was performed in subject’s native dMRI space, we transferred each tracer map into each subject’s native dMRI space individually using the image registration steps described earlier and an example is shown in Fig.2. The level of spatial overlaps between the tracer and tractography results was evaluated using the DICE score (Dice, 1945).
Where TP, FP, FN stand for true positive, false positive, and false negative, respectively. DICE score of 1 represents 100% overlaps/similarities between the estimated corticothalamic connection and the corresponding AMBCA results, while 0 indicates no overlap at all. We defined the level of overlaps in 3 categories: good (DICE > 0.8), moderate (0.6 < DICE < 0.8), and poor (DICE < 0.6).
For all six conditions in Table 2, we quantified the DICE scores comparing the TDI and corresponding tracer maps in 2 steps: 1) open-end tractography: whole-brain axonal projection maps for 14 cortical regions as described in previous section; 2) node-to-thalamus tractography: extracting tractography streamlines from each cortical node to the entire thalamus (for example, SSp-bfd to TH, AUDd to TH etc.) from the open-end tractography results. We examined the gross effects of imaging and tractography parameters by computing the DICE scores for each open-end tractograms generated in step 1. Then we computed the DICE for termination points in the thalamus for each cortical node-to-thalamus result (computed in step 2) and categorized the level of similarities between the tractography and tracer maps as good, moderate, or poor based on their spatial overlap in the thalamus using the DICE score. In this way, we were able to identify the conditions that significantly improve the streamline termination in valid target regions and achieve the best match with the corresponding tracer maps for the sub-networks as well as for the specific tracts within the entire corticothalamic network.
2.10. Statistical analysis:
We used Matlab (www.mathworks.com) and GraphPad Prism (Version 8.4.3 for Windows, GraphPad Software, La Jolla California USA) (www.graphpad.com) for statistical analysis. We performed Two-way ANOVA with Tukey’s multiple comparisons test at α = 0.05 to quantify the level of significance for each of the six conditions.
3. Results:
3.1. dMRI-based adult mouse forebrain atlas:
Mapping structural labels from AMBA to the dMRI-based atlas space (Fig. 3) yielded an atlas with a comprehensive set of neuroanatomical regions covering the entire mouse forebrain as mentioned in Table 3 and Fig. 4. Detailed description of the structures can be found in the supplementary material.
Fig. 3:
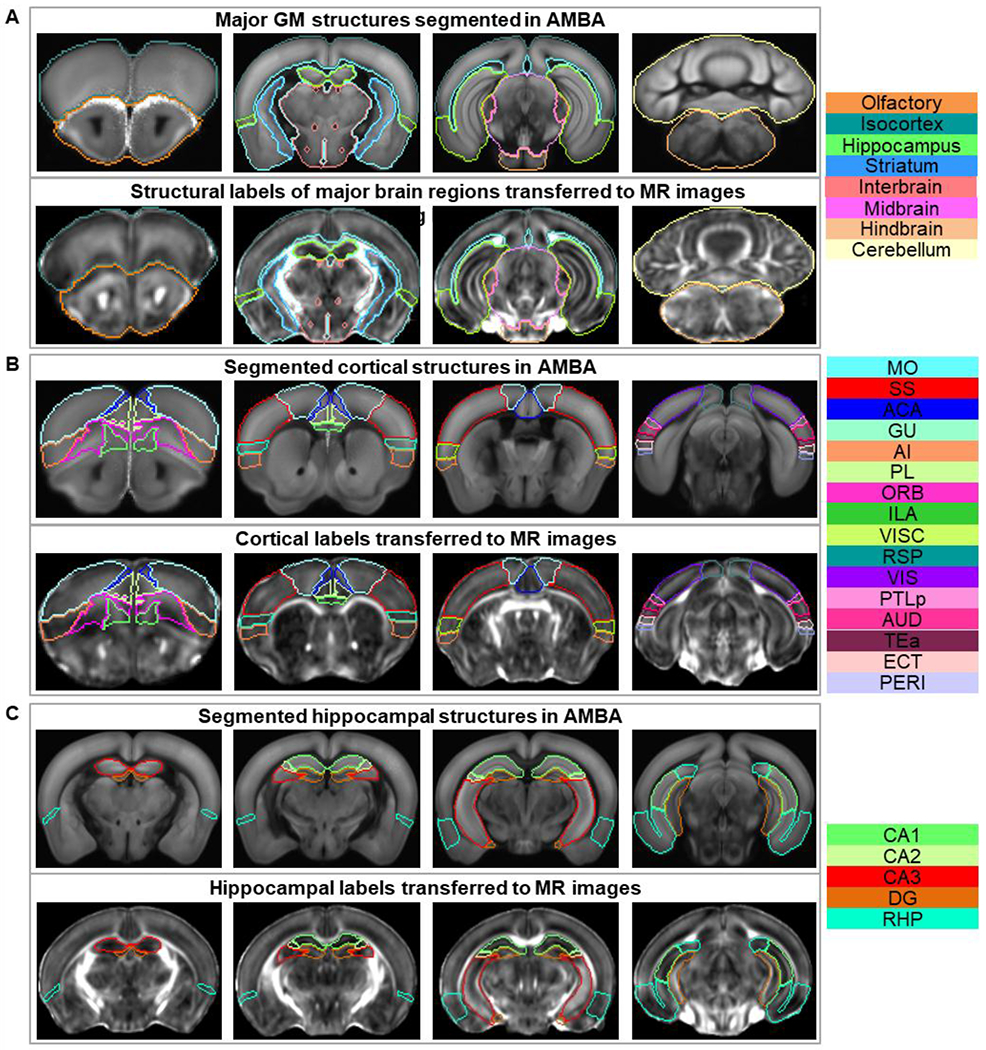
AMBA structural labels transferred to the dMRI space: A) Major GM structures. B) Cortical structures. C) Hippocampal structures. Upper and lower panels of A, B, and C show the structural labels in AMBA space and in dMRI space after coregistration respectively. The rightmost column lists the color-coded regions included in the corresponding atlas. Abbreviations of the anatomical structures are same as defined in the AMBA (https://mouse.brain-map.org/static/atlas).
Table 3:
Names and number of structures of the dMRI-based atlas.
| Parent structures | Name of the atlases | No. of structures | |
|---|---|---|---|
| Left | Right | ||
| (a) Cerebrum | Whole brain (WB) | 11 | 11 |
| (b) Cortical plate | Olfactory area (OLF) | 10 | 10 |
| Isocortex (CTX) – only major regions | 17 | 17 | |
| Isocortex (CTX) – with subregions | 40 | 40 | |
| Hippocampus (HPF) – only major regions | 5 | 5 | |
| Hippocampus (HPF) – with subregions | 9 | 9 | |
| (c) Cortical subplate | Cortical subplate (CTXsp) | 6 | 6 |
| (d) Cerebral nuclei | Striatum (STR) | 4 | 4 |
| Pallidum (PAL) | 4 | 4 | |
| (e) Brain stem | Interbrain – Thalamus (TH) | 16 | 16 |
| Interbrain – Hypothalamus (HY) | 4 | 4 | |
| Midbrain (MB) | 3 | 3 | |
| Hindbrain (HB) | 6 | 6 | |
Fig. 4:
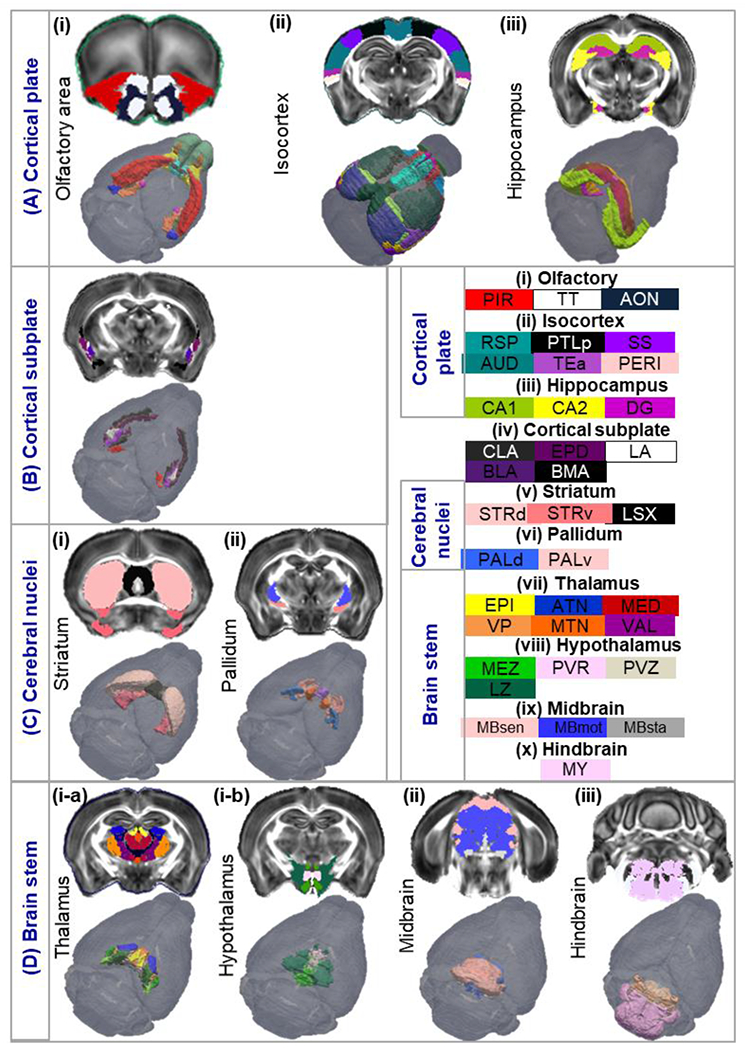
Segmentation tree illustrating the hierarchy of the atlas: A) Cortical plate is partitioned into olfactory, isocortex, and hippocampus regions. B) Cortical subplate/amygdala) and its sub-regions. C) Cerebral nuclei atlas containing the striatum and pallidum. D) Brain stem including the thalamus, hypothalamus, midbrain and hindbrain. Abbreviations of the anatomical structures are same as defined in the AMBA (https://mouse.brain-map.org/static/atlas).
Major cortical regions described in the isocortex atlas (Fig. 4A–ii) were further partitioned into 40 sub-regions in the left and right hemisphere respectively as shown in Supplementary Fig. S2A. For example, somatosensory area (SS), including both primary and secondary SS regions in Fig. 4A–ii, was separated into 7 parts in the left and right hemisphere (Supplementary Fig. S2A: primary somatosensory area – nose (SSp-n), mouth (SSp-m), upper limb (SSp-ul), lower limb (SSp-ll), barrel field (SSp-bfd), trunk (SSp-tr), and secondary somatosensory area (SSs)). Similarly, the hippocampal region was separated into 9 subregions: CA1, CA2, CA3, DG, entorhinal area (ENT), parasubiculum (PAR), postsubiculum (POST), presubiculum (PRE), and subiculum (SUB) (Supplementary Fig. S2B). Additionally, we imported 219 cortical layer labels from AMBA into our dMRI-based atlas and formed a comprehensive bi-lateral atlas of the mouse cortical layers (Supplementary Fig. S2C–D).
3.2. Comparison with MEMRI data:
As the structural labels were imported into the template space (DWI and FA), based on the tissue contrasts in the AMBA reference images (autofluorescence) and dMRI (FA and DWI, mostly linked to white matter), it is not clear whether boundaries of GM structural labels were mapped consistently as gray matter structures (e.g., cortical area and thalamic nuclei) often lack definitive contrasts in mouse MRI at 7T. Since in vivo MEMRI images show T1-enhancement in the hippocampal subregions and in many thalamic nuclei (Szulc et al., 2015), acquired datasets were mapped to the atlas following acquisition from a separate cohort to qualitatively examine the spatial agreement of several gray matter structural labels. Fig. 5A shows three axial slices (top to bottom) of an average of MEMRI images (n = 6) overlaid with structural labels mapped using affine transformations. Regions of several thalamic nuclei from the atlas (e.g., epithalamus (EPI), reticular nucleus (RT), ventral posterior complex of the thalamus (VP), dorsal part of the lateral geniculate complex (LGd), geniculate group, ventral thalamus (GENv), lateral group of the dorsal thalamus (LAT) can be distinguished in the MEMRI data based on their intensity levels (Fig. 5B). Similarly, regions of several hypothalamic nuclei from the atlas (e.g., periventricular region (PVR), medial zone (MEZ), and lateral zone (LZ)) also matched the MEMRI results (Fig. 5C – upper panel). Magnified view of the hippocampal regions (Fig. 5C) showed enhanced neuronal layers that fitted the defined boundaries of hippocampal regions (e.g., CA1, CA2, and DG in Fig. 5C). The ventral part of the hippocampus region in the atlas did not align well with the in vivo MEMRI data (Fig. 5C – lower panel), due to changes in brain morphology after death or fixation and may be corrected by nonlinear brain mappings.
Fig. 5:
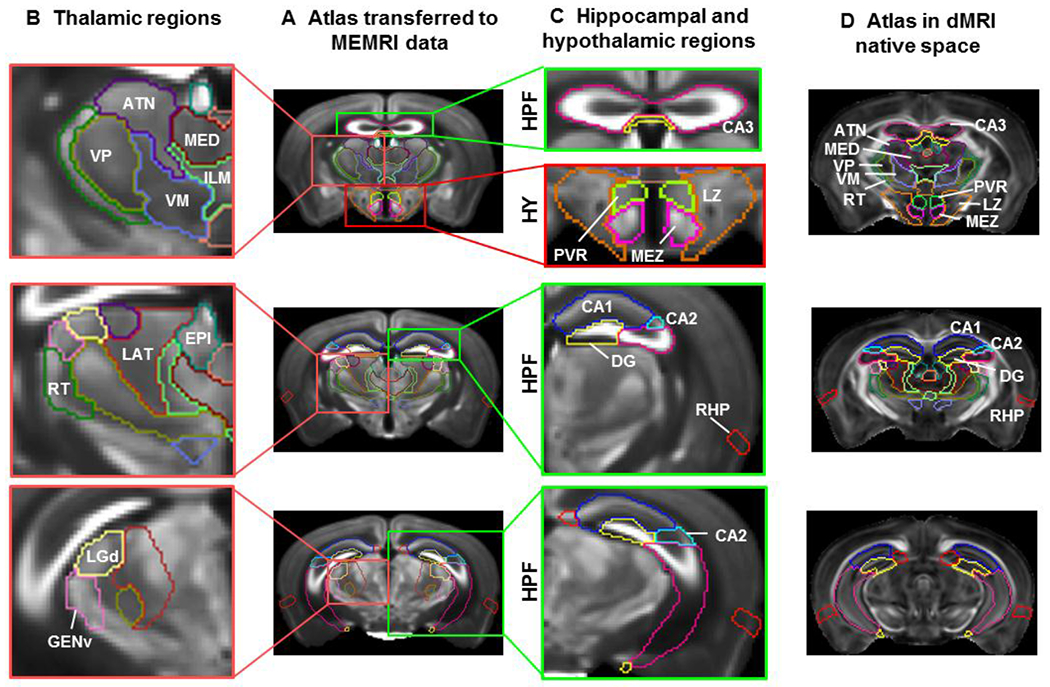
Comparisons of the dMRI-based atlas with MEMRI data: A) Representative axial slices of an in vivo MEMRI data overlaid with affine-aligned structural labels in the hippocampus (HPF, green rectangle), thalamus (pink rectangle) and hypothalamus (HY, red rectangle). B) Magnified view of the thalamic region showing several nuclei of the thalamus –reticular nucleus (RT), Ventral posterior complex of the thalamus (VP), dorsal part of the lateral geniculate complex (LGd), geniculate group, ventral thalamus (GENv), and lateral group of the dorsal thalamus (LAT). C) Magnified view of the hippocampal and hypothalamic regions, showing highlighted neuronal layers in the CA1, CA2, CA3, and dentate gyrus (DG) of the hippocampus and hypothalamic region highlighting paraventricular, lateral, and medial zone (PVR, LZ, and MEZ respectively). D) The structural labels used in B and C overlaid on FA images in the dMRI space for comparison.
3.3. Mapping mouse brain corticothalamic connectivity:
Ipsi-lateral corticothalamic connections between cortical seed regions and target regions in the thalamus were reconstructed using dMRI (Fig. 6A and C) and compared with corresponding tracer maps (Fig. 6B, injections sites identical to the seed regions) for their spatial overlaps (Fig. 6D–E).
Fig. 6:
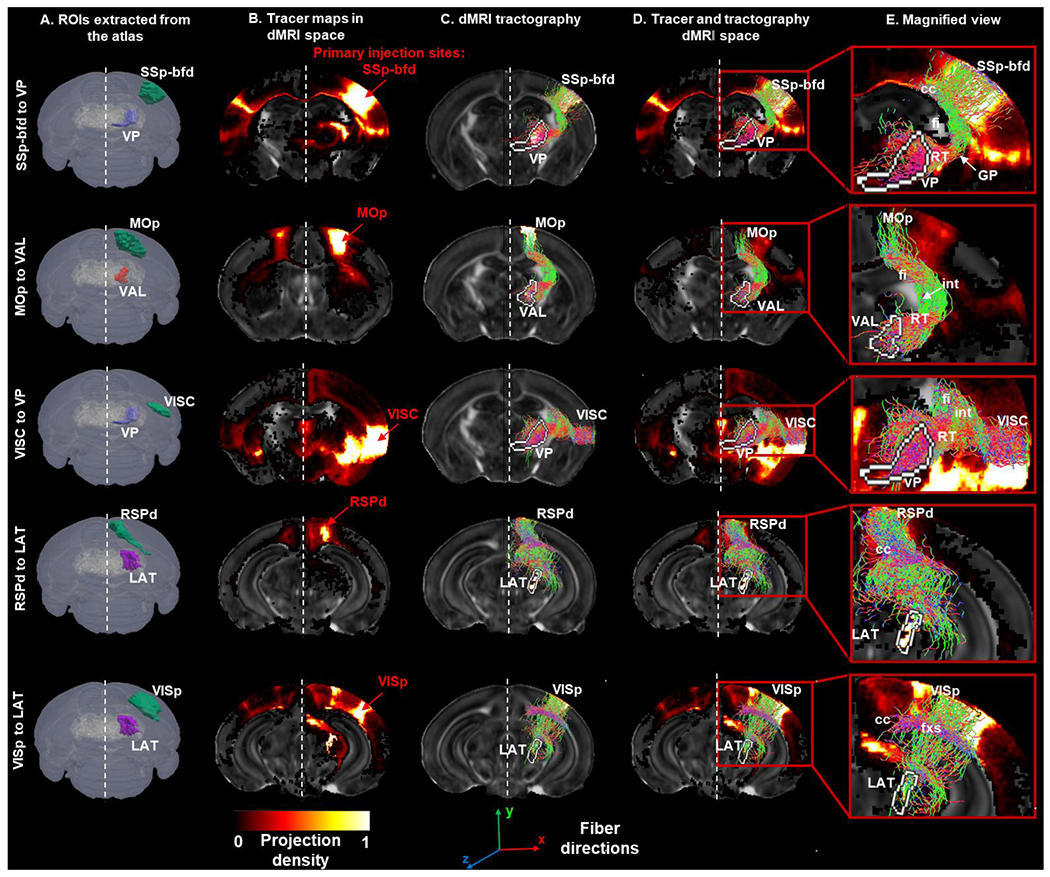
Validating dMRI tractography using tracer maps from AMBCA: A) Ipsi-lateral cortical and thalamic ROIs extracted from the dMRI-based atlas used for tractography: (from top to bottom) somatosensory barrel-field (SSp-bfd), ventral posterior complex of the thalamus (VP), primary motor area (MOp), ventral anterior-lateral complex of the thalamus (VAL), primary visual area (VISp), lateral group of the dorsal thalamus (LAT), retrosplenial area – dorsal part, and visceral area (VISC). B) Representative tracer maps for the selected injection sites (from top to bottom: SSp-bfd, MOp, VISp, RSPd, and VISC) from the AMBCA transferred to the dMRI space overlaid on FA image. C) Ipsi-lateral streamline projections from the cortical seed regions to the thalamic target regions derived via dMRI tractography: (from top to bottom) SSp-bfd to VP, MOp to VAL, VISp to LAT, RSPd to LAT, and VISC to VP. D) Tracer maps and tractography overlaid onto FA image in the dMRI space. E) Magnified view showing corticothalamic projection maps with the thalamic termination points labelled in white. Quantification of the similarities (DICE scores) between the tractography and tracer was computed within the termination regions.
The tractography results produced a binary 14×12 corticothalamic connectivity matrix. Fig. 7 shows the 14×12 corticothalamic connectivity matrices derived from the tracer data in AMBCA – ground truth (Fig. 7A) and from dMRI tractography (averaging from 10 mouse brains) under six conditions listed in table 2 (Fig. 7B–C). Supplementary Fig. S3 shows the individual tractography-based corticothalamic connectivity matrices under condition 6. The matrix generated under condition 1 (b = 2,000 s/mm2 and 30 diffusion directions using NCT) served as the baseline. Increasing the diffusion weighting to b=5,000 s/mm2 (condition 2) recovered several FN connections (blue dots in Fig. 7B) but also introduced several FP connections (black dots in Fig. 7B). Increasing the number of diffusion direction to 60 (condition 3) improved the recovery of FNs without inducing any further FPs. Based on all 10 subjects, condition 3 improved the TP rate from 67.79% to 92.37% (p < 0.0001) and reduced the FN rate from 32.2% to 7.6% (p < 0.0001) over condition 1 but at the cost of an increased FP rate (31.57% to 42.1%, p = 0.0169) (Fig. 7D top panel).
Fig. 7:
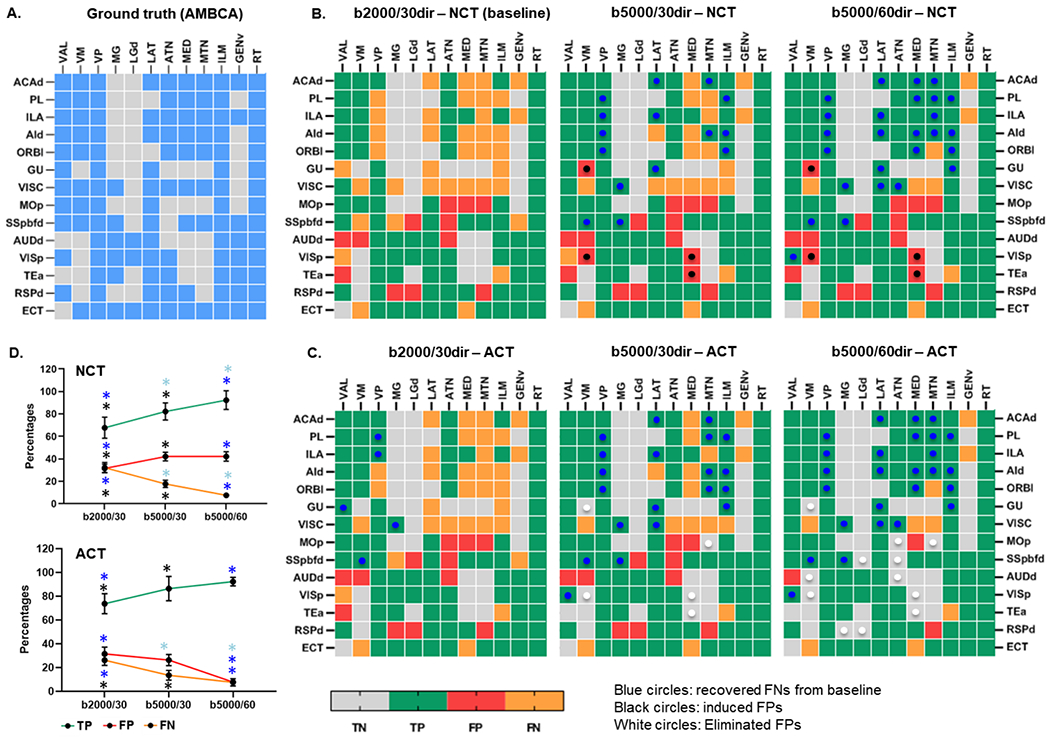
A) Ground truth corticothalamic connectivity matrix generated from Allen Mouse Brain Connectivity Atlas (AMBCA). Tractography based average (n = 10) corticothalamic matrices generated under six conditions (Table 2): B) Using conventional tractography (NCT) and C) Using anatomically constrained tractography (ACT). D) Graph showing the percentages (%) of detection of TP, FP, and FN connections using different imaging and tractography parameters.
Compared to the baseline, ACT under condition 4 reduced FN detections by 6% (blue dots) but did not eliminate FPs. Condition 5 on the other hand, increased the detection of TPs (by 18.6%) as compared with the baseline and further removed the FPs by 15.8% that were induced with higher b-values or diffusion directions using NCT (conditions 3 and 4). Notably, condition 6, i.e., b=5,000 s/mm2 and 60 diffusion directions using ACT, produced the lowest FP rates (7.89%), with a 92.37% TP rate and thus outperforms all other conditions (Fig. 7D, Two-way ANOVA with Tukey’s multiple comparisons test, α = 0.05). The summary of the findings from all six condition is summarized in Table 4.
Table 4:
Detection rates (Mean ± Standard Deviation) of TP, FP, and FN corticothalamic node-to-node connections for each condition in Table 2 computed from the binarized matrices shown in Fig. 7B–C.
| Conditions | 1: b=2000, 30 dirs, NCT | 2: b=5000, 30 dirs, NCT | 3: b=5000, 60 dirs, NCT | 4: b=2000, 30 dirs, ACT | 5: b=5000, 30 dirs, ACT | 6: b=5000, 60 dirs, ACT |
|---|---|---|---|---|---|---|
| TP rate (%) | 67.8 ± 9.3 | 82.2 ± 7.6 | 92.4 ± 8.3 | 73.7 ± 8.4 | 86.4 ± 10.2 | 92.4 ± 3.6 |
| FN rate (%) | 32.2 ± 4.5 | 17.8 ± 3.2 | 7.6 ± 2.1 | 26.3 ± 4.4 | 13.6 ± 4.1 | 7.6 ± 3.0 |
| FP rate (%) | 31.6 ± 3.7 | 42.1 ± 3.8 | 42.1 ± 4.2 | 31.6 ± 5.7 | 26.3 ± 4.7 | 7.9 ± .56 |
Visual comparisons of the NCT and ACT results showed ACT streamlines were more likely to stay in WM than NCT streamlines (Supplementary Fig. S4A). In addition, compared to NCT, the ACT results showed marked reductions in ill-defined streamline termination points, such as, streamlines that ended in the WM or CSF regions (Supplementary Fig. S4B). This difference was expected as ACT checked termination conditions, which may help ACT to identify more TP connections while minimizing the FP detections for the same imaging setups.
3.4. Spatial overlap between tractography and tracer:
While dMRI tractography can find streamlines passing through a pair of cortical and thalamic regions, the trajectories of tractography streamlines, in some cases, differed from those of the tracer results (Fig. 8). Fig. 8 illustrates some examples of the open-end streamline projection maps derived from the six conditions and quantification of the spatial overlaps with corresponding tracer data. We observed that both imaging parameters and tractography methods had effects on the level of spatial overlap between the tracer and tractography results, and the effects differed among cortico-thalamic connections. For example, both imaging parameters and tractography methods had significant effects (Two-way ANOVA with Tukey’s multiple comparisons test, α = 0.05) on the SSp-bfd projection map (Fig. 8A). However, only diffusion imaging parameters had a significant effect on the DICE score for dorsal auditory (AUDd) projection map (Fig. 8B), whereas, for the primary visual area tractography method ACT performed better than the NCT method (Fig. 8C).
Fig. 8:
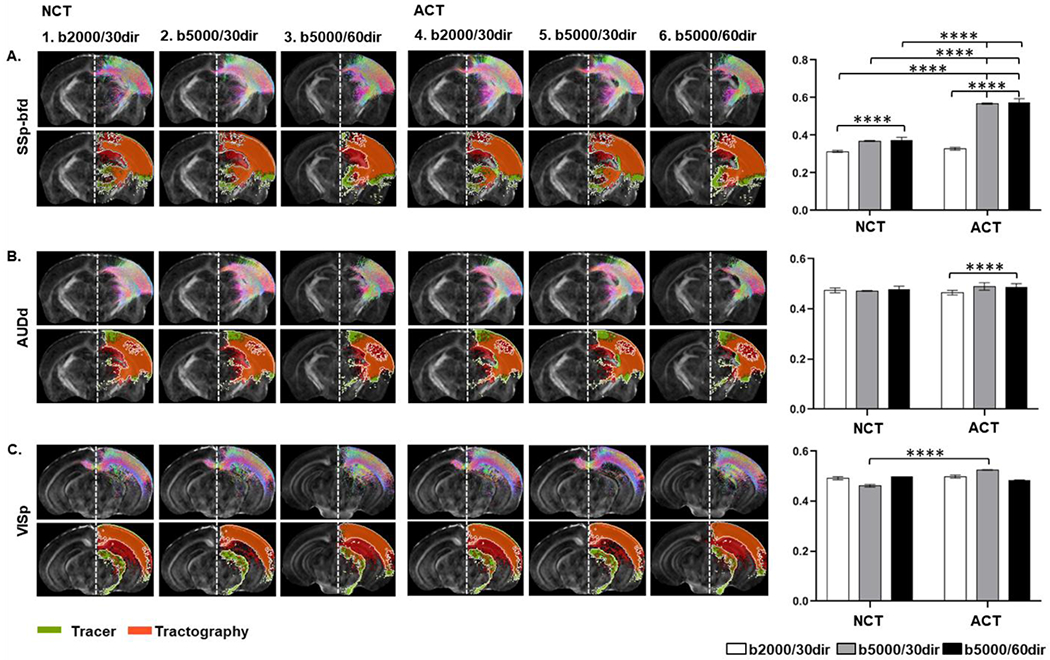
Axial slices showing open-end tractography (upper panel) and corresponding tracer maps with boundaries in white and tract density images using six conditions (table 2) overlaid on subject’s FA image: A) Somatosensory barrel field (SSp-bfd, B) Auditory cortex - dorsal area (AUDd) and C) Visual cortex – primary area (VISp). Graphs on the right panel show the level of similarities between tractography and the AMBCA (Two-way ANOVA with Tukey’s multiple comparisons test, α = 0.05)
As each tracer data may contain multiple connections within the corticothalamic network, it is difficult to segregate the tracts and form ground truth for node-to-node tractography comparisons. Instead, we compared the portion of each thalamic nuclei occupied by tracer (valid termination sites) and tractography streamlines from comparable cortical regions. Fig. 9 shows the DICE scores in detected TP connections in the corticothalamic circuitry under different conditions (1, 2, 3, 5, and 6). Green, yellow, and blue cells represent good, moderate, and poor spatial agreements in termination between the tractography and tracer data between the corresponding cortical and thalamic nodes. From cortical node-to-thalamus quantification, we identified three groups of connections (SSp-bfd to TH, MOp to TH, and VISp to TH) that showed 80% or more similarities with the corresponding tracer maps. Another three groups of connections (AUDd to TH, RSPd to TH, and TEa to TH) showed 60% to 80% similarities, and the remaining 8 group of connections showed poor agreement (below 60%) with the tracer maps. Three representative images showing quantification of the DICE scores for different tracts from three different levels have been shown in the Supplementary Fig. S5.
Fig. 9.
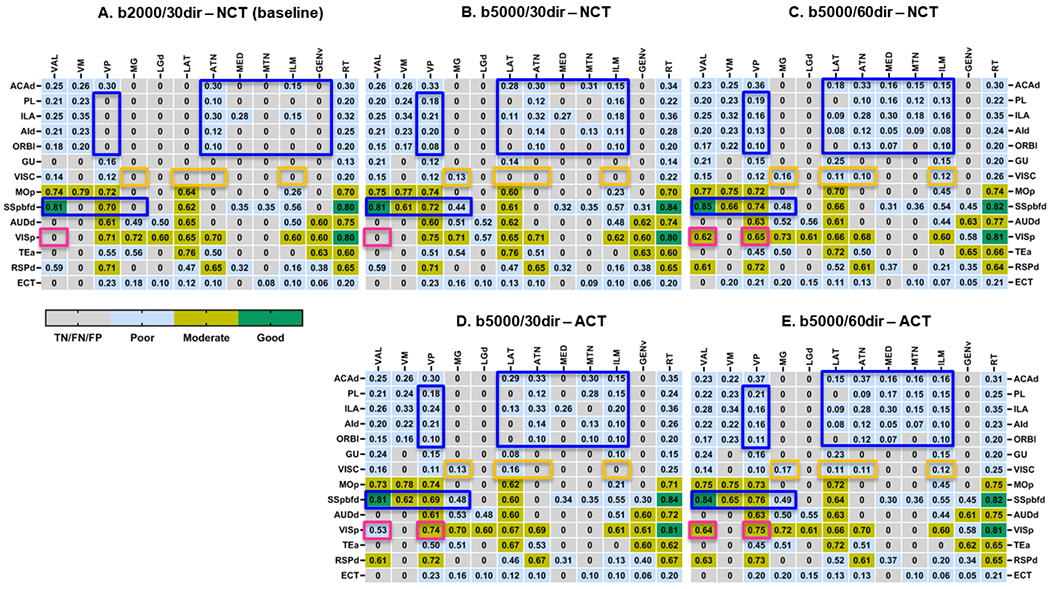
Tract specific DICE scores of thalamic termination points in the corticothalamic network computed from six conditions in Table 2. For each condition, DICE scores measuring the spatial agreement in thalamic termination points for each TP connections are displayed and classified. Dice scores of connections within the blue rectangles improved with both higher b-values and more diffusion directions, and dice scores of connections within the orange rectangles improved with either higher b-values or more diffusion directions. Dice scores of connections within the magenta rectangles improved with switching from NCT to ACT.
While the spatial agreements for thalamic termination point remained poor for most connections, we observed that the imaging parameters and tractography methods have tracts specific effects within the corticothalamic network. For example, in addition to the tractography techniques, higher b-values and diffusion directions can significantly improve the detection of TP connections along the prefrontal-thalamic and the sensory-thalamic pathways (Fig. 9 A–E, blue boxes). Changing only the b-values and diffusion directions on the other hand significantly impact the interactions between visceral to the polymodal association cortex related thalamic nuclei (LAT, ATN, ILM) and medial geniculate complex (MG) of the dorsal thalamus (MG/LGd) (Fig. 9 A–E, indicated by orange boxes), whereas tractography method shows significant effect on the visual to the ventral group of dorsal thalamic pathway (Fig. 9 A–E, magenta boxes).
Discussion:
Here, we reported an improved dMRI-based atlas of the adult C57BL/6J mouse forebrain integrated with AMBA structural labels. We demonstrated utility in reconstructing the mouse corticothalamic connectivity from dMRI data. The long-term goal of this atlas is accurate assessment of structural connectome of the mouse brain that would be useful to extend in various mouse models.
In accordance with the structural hierarchy defined in the AMBA, our dMRI-based atlas includes structural labels of the major GM regions at different cortical and subcortical levels. However, the boundaries of GM subregions can be ambiguous due to limited MRI resolution and nonspecific contrast patterns. For example, our isocortex atlas includes 17 cortical regions such as FRP, MO, SS, etc., which was additionally segmented into dorsal, ventral, anterior, and posterior regions following the AMBA, resulting a detailed atlas of the mouse cortex containing 80 structures segregated equally in both hemispheres. These regions, originally defined in the AMBA, were imported using a mapping that was computed based on segmentation of major brain compartments, not based on specific contrasts in dMRI signals. For the same reason, our atlas lacks in-depth classification of the subcortical layers and nuclei as labeled in the AMBA. Even though the comparison using the MEMRI data demonstrated good agreement in several hippocampal and thalamic regions in normal mouse brains, the accuracy of other regions has not been evaluated due to lack of good tissue contrasts that match histological methods. Applying the atlas to mutant mouse brains with altered cortical regions and cytoarchitecture or severe deformation, may result in inaccurate delineation of cortical and other GM regions.
Although major WM pathways can be mostly reconstructed using dMRI tractography, taking the advantage of the atlas, we demonstrated the possibility of seeking neural connections within largely GM structures. We focused on the corticothalamic network because it is a major pathways conserved across multiple species (Garel and Rubenstein, 2004; Leyva-Diaz and Lopez-Bendito, 2013; Rakic, 1975), and contribute in fundamental brain functions (Bagshaw et al., 2014; Sarnthein et al., 2005; Seeley et al., 2007). Understanding the extent to which the reconstructed pathways reflect realistic trajectories is important for future use of dMRI tractography to study the corticothalamic pathways or similar pathways in mouse models. Although we used post mortem mouse brains in this study as it avoids most of the sources known to degrade in vivo DW-MRI, such as low SNR, and motion artefacts (Le Bihan et al., 2006; Lori et al., 2002), we have previously demonstrated the feasibility to acquire high-resolution dMRI data from live mouse brains (Wu et al., 2014).
With dMRI datasets and AMBCA tract tracing data coregistered into the template space, we were able to perform a thorough investigation of individual node-to-node connections in the corticothalamic network. Looking at the connectivity matrix, results obtained from the baseline showed good consistency with the ground truth (Fig. 7A–B), albeit FPs and FNs were observed in several connections. FP connections (red cells in Fig. 7B) were detected in five out of fourteen cortical nodes (MOP, SSp-bfd, AUDd, Tea, and RSPd) to the thalamic network, and FN connections (orange cells in Fig. 7B) were predominantly associated with several thalamic regions (e.g., MED, MTN, ILM, LAT, and VP). Identification of FPs and FNs indicates the gap between tractography results and tracer ground truth for connections passing through gray matter regions. These findings not only suggest the importance of the validation of tractography maps by AMBCA, but also highlight the necessity of optimizing the tractography method to control the false positives or negatives.
We addressed this issue by applying additional constraints based on the anatomical knowledge of the brain tissues while generating the streamlines that infer more realistic microstructural details in relation to the inherent biology. Despite the NCT-based corticothalamic tracts seeming reasonable, we observed significant improvement in the overall connectome using ACT. In addition, varying the b-values and diffusion directions, we identified suitable imaging parameters (b-value/ number of diffusion directions) that can be used for more reliable estimation of the corticothalamic tracts. For example, increasing the imaging parameters from baseline conditions improved the rate of TP detections with trivial improvement in FP detections. However, altering both the imaging and tractography methods yielded significant improvement in TP detections with minimal FP and FN connections. These findings suggest that without anatomical priors, the NCT approach accepts biologically implausible streamline trajectories as valid termination points. In contrast, the ACT approach allows tracing the continuation of streamlines through GM regions using the acceptance/rejection criteria. Nevertheless, we could not detect all TP connections within the corticothalamic network, which still suffer from FP biases and may be likely due to the differences in FOD profiles between tractography and tracer maps that warrant further investigations.
When we further examined the spatial agreements between tractography and tracer results for each node-to-node connection, only a small number of connections showed good to moderate agreements between the two (Fig. 9). For example, a connection was found between VISC and VAL, but the trajectories of tractography and tracer results are quite different (Fig. 8), resulting in low DICE score. This demonstrates the limitation of dMRI tractography in reconstructing structural connections through mouse brain GM, which is not surprising given the complex microstructural organization often found in GM regions while it is difficult to separate axons from cell bodies and dendrites solely based on dMRI signals. With higher GM to WM compartment ratio in the mouse brain than in the human brain, this limitation may pose a big challenge for studies that use dMRI tractography to examine structural connectivity in the mouse brain. However, it is important to note that apart from the imaging and tractography parameters, achieving higher similarity index between the tractography and tracer maps also depends on the injection site (if spanning multiple regions or specific to the injection site only), injection volume, and complex fiber orientations. Therefore, we examined how these factors impact the DICE scores. AMBCA database contains axonal projection maps from tracer specific to the injection site only as well as spanning multiple regions (Gămănuţ et al., 2018). To check the effect of the injection sites chosen for DICE analysis, out of 14 cortical injection sites used in our study we identified only 4 regions (VISp, SSp-bfd, RSPd, and ACAd) where tracer did not span over multiple regions (Supplementary Table S6). We did not observe any significant improvement in the DICE scores while comparing these two conditions (Supplementary Fig. S6).
As the majority of the corticothalamic projections start from layers 5 and 6 (L5 and L6) of the cortex (Harris and Mrsic-Flogel, 2013), we next examined whether the tracer cortical projection maps that we used in this study were dominated by the neurons of the injection sites and whether neurons in L5/L6 were included. We observed that the percentage of the injection volume were higher in the injection site than in the neighboring regions (where tracer spanned upon injection) (Supplementary Table S6). Furthermore, the percentage of projection volume (PV) in L5 and L6 (calculated from the total projection volume of the injection site) were higher compared to other layers (L1-4) (Supplementary Table S7) and the axons from the injection sites passed though L5 and L6 (Supplementary Fig. S7). Although we do not have definite information on whether L5/L6 neurons were infected, these observations suggest that the tracer maps used for DICE quantification likely contained the target corticothalamic projections from the corresponding injection sites.
Acquiring high resolution MR data might be one of the solutions as it will reduce partial volume effect, so that smaller axon bundles can be more easily separated from surrounding tissues, but at an expense of longer data acquisition time (Anderson et al., 2020; Crater et al., 2022). Over the past decade, the compressed sensing (CS) technique has been widely used to speed up the MR data acquisition by under-sampling the q-space or the k-space (Aranda et al., 2015; Daducci et al., 2015; Menzel et al., 2011; Tobisch et al., 2018). Recently, Wang et al. demonstrated the acquisition of 50 μm dMRI datasets of the mouse brain using CS in the k-space (Wang et al., 2020b), and we have reported a joint q-space and k-space CS method that can reconstruct ex vivo mouse brain dMRI data with acceleration factors up to 8 (Zhang et al., 2020). Another potential solution is the development of more advanced dMRI acquisition and tractography techniques. For example, the recent reported b-tensor technique (Cottaar et al., 2020) can potentially separate axons from cell bodies by introducing spherical diffusion encoding.
Finally, our study has several limitations. For example, we collected dMRI data with low angular resolution on a single shell. Acquiring multi-shell dMRI datasets would have provided additional microstructural details for tractography. We also did not examine the effects of resolution on tractography. Recent reports suggest that tractography in general benefits from high spatial resolution (Anderson et al., 2020) but potential loss of signal-to-noise ratio at high spatial resolution may negatively impact tractography results (Ambrosen et al., 2020). In addition, differences between the mouse brain and the human brain, for instance, the absence of brain sulci and relatively short association bundles in the mouse brain, hinders translating the findings in the mouse brain to improve tractography in the human brain.
Conclusion:
In conclusion, we have developed an atlas based on dMRI with structural labels imported from the AMBA that can serve as a requisite template for cross-examination of potential disrupted connections/volumetric changes in the mouse brain. Furthermore, comparing dMRI tractography results with AMBCA tracer results revealed significant differences in the trajectories of mouse corticothalamic tracts, indicating the need for further development of dMRI tractography for accurate characterization of mouse brain structural connectivity.
Supplementary Material
Highlights.
A diffusion MRI mouse brain atlas compatible with the Allen Mouse Brain Atlas
Node-to-node mouse thalamocortical structural connectivity using tractography
Imaging and tractography parameters significantly affect tractography outcomes
Diffusion MRI tractography in mouse brain gray matter still face challenges
Acknowledgment
We would like to thank the members of the labs of Drs. Jiangyang Zhang, Daniel Turnbull, Youssef Z. Wadghiri and the team of the preclinical imaging core at the NYU Grossman School of Medicine, for their support and feedback during the evolution of our work.
Funding Sources
This work was supported by NIH grants R01NS102904 and R01HD074593. The majority of this work was performed at the NYU Langone Health Preclinical Imaging Laboratory, a shared resource partially supported by the NIH/SIG 1S10OD018337-01, the Laura and Isaac Perlmutter Cancer Center Support Grant, NIH/NCI 5P30CA016087, and the NIBIB Biomedical Technology Re-source Center Grant NIH P41 EB017183 as well as by the NYU CTSA grant UL1 TR000038 from the National Center for Advancing Translational Sciences, National Institutes of Health.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Credit Author Statement
The project was conceived by TMA. and JZ. ZL, YZW, and DHT were involved in discussion. HR and CHL acquired most MR images, and TMA performed tractography and data analysis. TMA and JZ wrote and prepared the initial draft, CHL, ZL, HR, YZW, and DHT edited the paper.
Declarations of interest: none
Data and materials availability:
The atlas and the corresponding structural labels that support the findings of this study are openly available in the respiratory: ‘dMRI_mouse_brain_atlas’ at https://github.com/Tanzil-M-Arefin/dMRI_mouse_brain_atlas
Tracer datasets used in this study were obtained from the Allen Mouse Brain Connectivity project (http://connectivity.brain-map.org).
References:
- Aggarwal M, Mori S, Shimogori T, Blackshaw S, Zhang J, 2010. Three-dimensional diffusion tensor microimaging for anatomical characterization of the mouse brain. Magn Reson Med 64, 249–261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aggarwal M, Zhang J, Miller MI, Sidman RL, Mori S, 2009. Magnetic resonance imaging and micro-computed tomography combined atlas of developing and adult mouse brains for stereotaxic surgery. Neuroscience 162, 1339–1350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ali AA, Dale AM, Badea A, Johnson GA, 2005. Automated segmentation of neuroanatomical structures in multispectral MR microscopy of the mouse brain. Neuroimage 27, 425–435. [DOI] [PubMed] [Google Scholar]
- Ambrosen KS, Eskildsen SF, Hinne M, Krug K, Lundell H, Schmidt MN, van Gerven MAJ, Morup M, Dyrby TB, 2020. Validation of structural brain connectivity networks: The impact of scanning parameters. Neuroimage 204, 116207. [DOI] [PubMed] [Google Scholar]
- Anderson RJ, Long CM, Calabrese ED, Robertson SH, Johnson GA, Cofer GP, O’Brien RJ, Badea A, 2020. Optimizing Diffusion Imaging Protocols for Structural Connectomics in Mouse Models of Neurological Conditions. Front Phys 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aranda R, Ramirez-Manzanares A, Rivera M, 2015. Sparse and Adaptive Diffusion Dictionary (SADD) for recovering intra-voxel white matter structure. Med Image Anal 26, 243–255. [DOI] [PubMed] [Google Scholar]
- Arefin TM, Lee CH, White JD, Zhang J, Kaffman A, 2021. Macroscopic Structural and Connectome Mapping of the Mouse Brain Using Diffusion Magnetic Resonance Imaging. Bio Protoc 11, e4221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arefin TM, Mechling AE, Meirsman AC, Bienert T, Hubner NS, Lee HL, Ben Hamida S, Ehrlich A, Roquet D, Hennig J, von Elverfeldt D, Kieffer BL, Harsan LA, 2017. Remodeling of Sensorimotor Brain Connectivity in Gpr88-Deficient Mice. Brain Connect 7, 526–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aydogan DB, Jacobs R, Dulawa S, Thompson SL, Francois MC, Toga AW, Dong H, Knowles JA, Shi Y, 2018. When tractography meets tracer injections: a systematic study of trends and variation sources of diffusion-based connectivity. Brain Struct Funct 223, 2841–2858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badea A, Ali-Sharief AA, Johnson GA, 2007a. Morphometric analysis of the C57BL/6J mouse brain. Neuroimage 37, 683–693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badea A, Nicholls PJ, Johnson GA, Wetsel WC, 2007b. Neuroanatomical phenotypes in the reeler mouse. Neuroimage 34, 1363–1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagshaw AP, Rollings DT, Khalsa S, Cavanna AE, 2014. Multimodal neuroimaging investigations of alterations to consciousness: the relationship between absence epilepsy and sleep. Epilepsy Behav 30, 33–37. [DOI] [PubMed] [Google Scholar]
- Basser PJ, Pierpaoli C, 1996. Microstructural and physiological features of tissues elucidated by quantitative-diffusion-tensor MRI. J Magn Reson B 111, 209–219. [DOI] [PubMed] [Google Scholar]
- Bock NA, Kovacevic N, Lipina TV, Roder JC, Ackerman SL, Henkelman RM, 2006. In vivo magnetic resonance imaging and semiautomated image analysis extend the brain phenotype for cdf/cdf mice. J Neurosci 26, 4455–4459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calabrese E, Badea A, Cofer G, Qi Y, Johnson GA, 2015. A Diffusion MRI Tractography Connectome of the Mouse Brain and Comparison with Neuronal Tracer Data. Cereb Cortex 25, 4628–4637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calamante F, 2019. The Seven Deadly Sins of Measuring Brain Structural Connectivity Using Diffusion MRI Streamlines Fibre-Tracking. Diagnostics (Basel, Switzerland) 9, 115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calamante F, Tournier JD, Jackson GD, Connelly A, 2010. Track-density imaging (TDI): super-resolution white matter imaging using whole-brain track-density mapping. Neuroimage 53, 1233–1243. [DOI] [PubMed] [Google Scholar]
- Chen XJ, Kovacevic N, Lobaugh NJ, Sled JG, Henkelman RM, Henderson JT, 2006. Neuroanatomical differences between mouse strains as shown by high-resolution 3D MRI. Neuroimage 29, 99–105. [DOI] [PubMed] [Google Scholar]
- Chuang N, Mori S, Yamamoto A, Jiang H, Ye X, Xu X, Richards LJ, Nathans J, Miller MI, Toga AW, Sidman RL, Zhang J, 2011. An MRI-based atlas and database of the developing mouse brain. Neuroimage 54, 80–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coletta L, Pagani M, Whitesell JD, Harris JA, Bernhardt B, Gozzi A, 2020. Network structure of the mouse brain connectome with voxel resolution. Sci Adv 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook SJ, Jarrell TA, Brittin CA, Wang Y, Bloniarz AE, Yakovlev MA, Nguyen KCQ, Tang LT, Bayer EA, Duerr JS, Bulow HE, Hobert O, Hall DH, Emmons SW, 2019. Whole-animal connectomes of both Caenorhabditis elegans sexes. Nature 571, 63–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cottaar M, Szczepankiewicz F, Bastiani M, Hernandez-Fernandez M, Sotiropoulos SN, Nilsson M, Jbabdi S, 2020. Improved fibre dispersion estimation using b-tensor encoding. Neuroimage 215, 116832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crater S, Maharjan S, Qi Y, Zhao Q, Cofer G, Cook JC, Johnson GA, Wang N, 2022. Resolution and b value dependent structural connectome in ex vivo mouse brain. Neuroimage 255, 119199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daducci A, Canales-Rodriguez EJ, Zhang H, Dyrby TB, Alexander DC, Thiran JP, 2015. Accelerated Microstructure Imaging via Convex Optimization (AMICO) from diffusion MRI data. Neuroimage 105, 32–44. [DOI] [PubMed] [Google Scholar]
- Degiorgis L, Arefin TM, Ben-Hamida S, Noblet V, Antal C, Bienert T, Reisert M, von Elverfeldt D, Kieffer BL, Harsan LA, 2022. Translational Structural and Functional Signatures of Chronic Alcohol Effects in Mice. Biol Psychiatry 91, 1039–1050. [DOI] [PubMed] [Google Scholar]
- Dice LR, 1945. Measures of the Amount of Ecologic Association Between Species. Ecology 26, 297–302. [Google Scholar]
- Dorr A, Sled JG, Kabani N, 2007. Three-dimensional cerebral vasculature of the CBA mouse brain: a magnetic resonance imaging and micro computed tomography study. Neuroimage 35, 1409–1423. [DOI] [PubMed] [Google Scholar]
- Dorr AE, Lerch JP, Spring S, Kabani N, Henkelman RM, 2008. High resolution three-dimensional brain atlas using an average magnetic resonance image of 40 adult C57Bl/6J mice. Neuroimage 42, 60–69. [DOI] [PubMed] [Google Scholar]
- Garel S, Rubenstein JL, 2004. Intermediate targets in formation of topographic projections: inputs from the thalamocortical system. Trends Neurosci 27, 533–539. [DOI] [PubMed] [Google Scholar]
- Girard G, Caminiti R, Battaglia-Mayer A, St-Onge E, Ambrosen KS, Eskildsen SF, Krug K, Dyrby TB, Descoteaux M, Thiran JP, Innocenti GM, 2020. On the cortical connectivity in the macaque brain: A comparison of diffusion tractography and histological tracing data. Neuroimage 221, 117201. [DOI] [PubMed] [Google Scholar]
- Goldowitz D, 2010. Allen Reference Atlas. A Digital Color Brain Atlas of the C57BL/6J Male Mouse - by H. W. Dong. Genes, Brain and Behavior 9, 128–128. [Google Scholar]
- Harris JA, Mihalas S, Hirokawa KE, Whitesell JD, Choi H, Bernard A, Bohn P, Caldejon S, Casal L, Cho A, Feiner A, Feng D, Gaudreault N, Gerfen CR, Graddis N, Groblewski PA, Henry AM, Ho A, Howard R, Knox JE, Kuan L, Kuang X, Lecoq J, Lesnar P, Li Y, Luviano J, McConoughey S, Mortrud MT, Naeemi M, Ng L, Oh SW, Ouellette B, Shen E, Sorensen SA, Wakeman W, Wang Q, Wang Y, Williford A, Phillips JW, Jones AR, Koch C, Zeng H, 2019. Hierarchical organization of cortical and thalamic connectivity. Nature 575, 195–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris KD, Mrsic-Flogel TD, 2013. Cortical connectivity and sensory coding. Nature 503, 51–58. [DOI] [PubMed] [Google Scholar]
- Hunnicutt BJ, Long BR, Kusefoglu D, Gertz KJ, Zhong H, Mao T, 2014. A comprehensive thalamocortical projection map at the mesoscopic level. Nat Neurosci 17, 1276–1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jbabdi S, Sotiropoulos SN, Haber SN, Van Essen DC, Behrens TE, 2015. Measuring macroscopic brain connections in vivo. Nat Neurosci 18, 1546–1555. [DOI] [PubMed] [Google Scholar]
- Jeong M, Kim Y, Kim J, Ferrante DD, Mitra PP, Osten P, Kim D, 2016. Comparative three-dimensional connectome map of motor cortical projections in the mouse brain. Sci Rep 6, 20072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang H, van Zijl PC, Kim J, Pearlson GD, Mori S, 2006. DtiStudio: resource program for diffusion tensor computation and fiber bundle tracking. Comput Methods Programs Biomed 81, 106–116. [DOI] [PubMed] [Google Scholar]
- Jiang Y, Johnson GA, 2011. Microscopic diffusion tensor atlas of the mouse brain. Neuroimage 56, 1235–1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson GA, Ali-Sharief A, Badea A, Brandenburg J, Cofer G, Fubara B, Gewalt S, Hedlund LW, Upchurch L, 2007. High-throughput morphologic phenotyping of the mouse brain with magnetic resonance histology. Neuroimage 37, 82–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keifer OP Jr., Gutman DA, Hecht EE, Keilholz SD, Ressler KJ, 2015. A comparative analysis of mouse and human medial geniculate nucleus connectivity: a DTI and anterograde tracing study. Neuroimage 105, 53–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovacevic N, Henderson JT, Chan E, Lifshitz N, Bishop J, Evans AC, Henkelman RM, Chen XJ, 2005. A three-dimensional MRI atlas of the mouse brain with estimates of the average and variability. Cereb Cortex 15, 639–645. [DOI] [PubMed] [Google Scholar]
- Kuan L, Li Y, Lau C, Feng D, Bernard A, Sunkin SM, Zeng H, Dang C, Hawrylycz M, Ng L, 2015. Neuroinformatics of the Allen Mouse Brain Connectivity Atlas. Methods 73, 4–17. [DOI] [PubMed] [Google Scholar]
- Le Bihan D, Poupon C, Amadon A, Lethimonnier F, 2006. Artifacts and pitfalls in diffusion MRI. J Magn Reson Imaging 24, 478–488. [DOI] [PubMed] [Google Scholar]
- Lee EF, Jacobs RE, Dinov I, Leow A, Toga AW, 2005. Standard atlas space for C57BL/6J neonatal mouse brain. Anat Embryol (Berl) 210, 245–263. [DOI] [PubMed] [Google Scholar]
- Lein ES, Hawrylycz MJ, Ao N, Ayres M, Bensinger A, Bernard A, Boe AF, Boguski MS, Brockway KS, Byrnes EJ, Chen L, Chen L, Chen TM, Chin MC, Chong J, Crook BE, Czaplinska A, Dang CN, Datta S, Dee NR, Desaki AL, Desta T, Diep E, Dolbeare TA, Donelan MJ, Dong HW, Dougherty JG, Duncan BJ, Ebbert AJ, Eichele G, Estin LK, Faber C, Facer BA, Fields R, Fischer SR, Fliss TP, Frensley C, Gates SN, Glattfelder KJ, Halverson KR, Hart MR, Hohmann JG, Howell MP, Jeung DP, Johnson RA, Karr PT, Kawal R, Kidney JM, Knapik RH, Kuan CL, Lake JH, Laramee AR, Larsen KD, Lau C, Lemon TA, Liang AJ, Liu Y, Luong LT, Michaels J, Morgan JJ, Morgan RJ, Mortrud MT, Mosqueda NF, Ng LL, Ng R, Orta GJ, Overly CC, Pak TH, Parry SE, Pathak SD, Pearson OC, Puchalski RB, Riley ZL, Rockett HR, Rowland SA, Royall JJ, Ruiz MJ, Sarno NR, Schaffnit K, Shapovalova NV, Sivisay T, Slaughterbeck CR, Smith SC, Smith KA, Smith BI, Sodt AJ, Stewart NN, Stumpf KR, Sunkin SM, Sutram M, Tam A, Teemer CD, Thaller C, Thompson CL, Varnam LR, Visel A, Whitlock RM, Wohnoutka PE, Wolkey CK, Wong VY, Wood M, Yaylaoglu MB, Young RC, Youngstrom BL, Yuan XF, Zhang B, Zwingman TA, Jones AR, 2007. Genome-wide atlas of gene expression in the adult mouse brain. Nature 445, 168–176. [DOI] [PubMed] [Google Scholar]
- Leyva-Diaz E, Lopez-Bendito G, 2013. In and out from the cortex: development of major forebrain connections. Neuroscience 254, 26–44. [DOI] [PubMed] [Google Scholar]
- Lori NF, Akbudak E, Shimony JS, Cull TS, Snyder AZ, Guillory RK, Conturo TE, 2002. Diffusion tensor fiber tracking of human brain connectivity: aquisition methods, reliability analysis and biological results. NMR Biomed 15, 494–515. [DOI] [PubMed] [Google Scholar]
- Ma Y, Hof PR, Grant SC, Blackband SJ, Bennett R, Slatest L, McGuigan MD, Benveniste H, 2005. A three-dimensional digital atlas database of the adult C57BL/6J mouse brain by magnetic resonance microscopy. Neuroscience 135, 1203–1215. [DOI] [PubMed] [Google Scholar]
- MacKenzie-Graham A, Lee EF, Dinov ID, Bota M, Shattuck DW, Ruffins S, Yuan H, Konstantinidis F, Pitiot A, Ding Y, Hu G, Jacobs RE, Toga AW, 2004. A multimodal, multidimensional atlas of the C57BL/6J mouse brain. J Anat 204, 93–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maffei C, Girard G, Schilling KG, Aydogan DB, Adluru N, Zhylka A, Wu Y, Mancini M, Hamamci A, Sarica A, Teillac A, Baete SH, Karimi D, Yeh FC, Yildiz ME, Gholipour A, Bihan-Poudec Y, Hiba B, Quattrone A, Quattrone A, Boshkovski T, Stikov N, Yap PT, de Luca A, Pluim J, Leemans A, Prabhakaran V, Bendlin BB, Alexander AL, Landman BA, Canales-Rodriguez EJ, Barakovic M, Rafael-Patino J, Yu T, Rensonnet G, Schiavi S, Daducci A, Pizzolato M, Fischi-Gomez E, Thiran JP, Dai G, Grisot G, Lazovski N, Puch S, Ramos M, Rodrigues P, Prckovska V, Jones R, Lehman J, Haber SN, Yendiki A, 2022. Insights from the IronTract challenge: Optimal methods for mapping brain pathways from multi-shell diffusion MRI. Neuroimage 257, 119327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majka P, Bai S, Bakola S, Bednarek S, Chan JM, Jermakow N, Passarelli L, Reser DH, Theodoni P, Worthy KH, Wang XJ, Wojcik DK, Mitra PP, Rosa MGP, 2020. Open access resource for cellular-resolution analyses of corticocortical connectivity in the marmoset monkey. Nat Commun 11, 1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mechling AE, Arefin T, Lee HL, Bienert T, Reisert M, Ben Hamida S, Darcq E, Ehrlich A, Gaveriaux-Ruff C, Parent MJ, Rosa-Neto P, Hennig J, von Elverfeldt D, Kieffer BL, Harsan LA, 2016. Deletion of the mu opioid receptor gene in mice reshapes the reward-aversion connectome. Proc Natl Acad Sci U S A 113, 11603–11608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menzel MI, Tan ET, Khare K, Sperl JI, King KF, Tao X, Hardy CJ, Marinelli L, 2011. Accelerated diffusion spectrum imaging in the human brain using compressed sensing. Magn Reson Med 66, 1226–1233. [DOI] [PubMed] [Google Scholar]
- Moldrich RX, Gobius I, Pollak T, Zhang J, Ren T, Brown L, Mori S, De Juan Romero C, Britanova O, Tarabykin V, Richards LJ, 2010. Molecular regulation of the developing commissural plate. J Comp Neurol 518, 3645–3661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori S, van Zijl PC, 2002. Fiber tracking: principles and strategies - a technical review. NMR Biomed 15, 468–480. [DOI] [PubMed] [Google Scholar]
- Mori S, Zhang J, 2006. Principles of diffusion tensor imaging and its applications to basic neuroscience research. Neuron 51, 527–539. [DOI] [PubMed] [Google Scholar]
- Ng L, Bernard A, Lau C, Overly CC, Dong HW, Kuan C, Pathak S, Sunkin SM, Dang C, Bohland JW, Bokil H, Mitra PP, Puelles L, Hohmann J, Anderson DJ, Lein ES, Jones AR, Hawrylycz M, 2009. An anatomic gene expression atlas of the adult mouse brain. Nat Neurosci 12, 356–362. [DOI] [PubMed] [Google Scholar]
- Oh SW, Harris JA, Ng L, Winslow B, Cain N, Mihalas S, Wang Q, Lau C, Kuan L, Henry AM, Mortrud MT, Ouellette B, Nguyen TN, Sorensen SA, Slaughterbeck CR, Wakeman W, Li Y, Feng D, Ho A, Nicholas E, Hirokawa KE, Bohn P, Joines KM, Peng H, Hawrylycz MJ, Phillips JW, Hohmann JG, Wohnoutka P, Gerfen CR, Koch C, Bernard A, Dang C, Jones AR, Zeng H, 2014. A mesoscale connectome of the mouse brain. Nature 508, 207–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rakic P, 1975. Timing of major ontogenetic events in the visual cortex of the rhesus monkey. UCLA Forum Med Sci, 3–40. [DOI] [PubMed] [Google Scholar]
- Rallapalli H, Darwin BC, Toro-Montoya E, Lerch JP, Turnbull DH, 2020. Longitudinal MEMRI analysis of brain phenotypes in a mouse model of Niemann-Pick Type C disease. Neuroimage 217, 116894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren T, Zhang J, Plachez C, Mori S, Richards LJ, 2007. Diffusion tensor magnetic resonance imaging and tract-tracing analysis of Probst bundle structure in Netrin1- and DCC-deficient mice. J Neurosci 27, 10345–10349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards K, Watson C, Buckley RF, Kurniawan ND, Yang Z, Keller MD, Beare R, Bartlett PF, Egan GF, Galloway GJ, Paxinos G, Petrou S, Reutens DC, 2011. Segmentation of the mouse hippocampal formation in magnetic resonance images. Neuroimage 58, 732–740. [DOI] [PubMed] [Google Scholar]
- Rubinov M, Ypma RJ, Watson C, Bullmore ET, 2015. Wiring cost and topological participation of the mouse brain connectome. Proc Natl Acad Sci U S A 112, 10032–10037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandberg R, Yasuda R, Pankratz DG, Carter TA, Del Rio JA, Wodicka L, Mayford M, Lockhart DJ, Barlow C, 2000. Regional and strain-specific gene expression mapping in the adult mouse brain. Proc Natl Acad Sci U S A 97, 11038–11043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarnthein J, Morel A, von Stein A, Jeanmonod D, 2005. Thalamocortical theta coherence in neurological patients at rest and during a working memory task. Int J Psychophysiol 57, 87–96. [DOI] [PubMed] [Google Scholar]
- Scheffer LK, Xu CS, Januszewski M, Lu Z, Takemura SY, Hayworth KJ, Huang GB, Shinomiya K, Maitlin-Shepard J, Berg S, Clements J, Hubbard PM, Katz WT, Umayam L, Zhao T, Ackerman D, Blakely T, Bogovic J, Dolafi T, Kainmueller D, Kawase T, Khairy KA, Leavitt L, Li PH, Lindsey L, Neubarth N, Olbris DJ, Otsuna H, Trautman ET, Ito M, Bates AS, Goldammer J, Wolff T, Svirskas R, Schlegel P, Neace E, Knecht CJ, Alvarado CX, Bailey DA, Ballinger S, Borycz JA, Canino BS, Cheatham N, Cook M, Dreher M, Duclos O, Eubanks B, Fairbanks K, Finley S, Forknall N, Francis A, Hopkins GP, Joyce EM, Kim S, Kirk NA, Kovalyak J, Lauchie SA, Lohff A, Maldonado C, Manley EA, McLin S, Mooney C, Ndama M, Ogundeyi O, Okeoma N, Ordish C, Padilla N, Patrick CM, Paterson T, Phillips EE, Phillips EM, Rampally N, Ribeiro C, Robertson MK, Rymer JT, Ryan SM, Sammons M, Scott AK, Scott AL, Shinomiya A, Smith C, Smith K, Smith NL, Sobeski MA, Suleiman A, Swift J, Takemura S, Talebi I, Tarnogorska D, Tenshaw E, Tokhi T, Walsh JJ, Yang T, Horne JA, Li F, Parekh R, Rivlin PK, Jayaraman V, Costa M, Jefferis GS, Ito K, Saalfeld S, George R, Meinertzhagen IA, Rubin GM, Hess HF, Jain V, Plaza SM, 2020. A connectome and analysis of the adult Drosophila central brain. Elife 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schilling KG, Nath V, Hansen C, Parvathaneni P, Blaber J, Gao Y, Neher P, Aydogan DB, Shi Y, Ocampo-Pineda M, Schiavi S, Daducci A, Girard G, Barakovic M, Rafael-Patino J, Romascano D, Rensonnet G, Pizzolato M, Bates A, Fischi E, Thiran JP, Canales-Rodriguez EJ, Huang C, Zhu H, Zhong L, Cabeen R, Toga AW, Rheault F, Theaud G, Houde JC, Sidhu J, Chamberland M, Westin CF, Dyrby TB, Verma R, Rathi Y, Irfanoglu MO, Thomas C, Pierpaoli C, Descoteaux M, Anderson AW, Landman BA, 2019. Limits to anatomical accuracy of diffusion tractography using modern approaches. Neuroimage 185, 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seeley WW, Menon V, Schatzberg AF, Keller J, Glover GH, Kenna H, Reiss AL, Greicius MD, 2007. Dissociable intrinsic connectivity networks for salience processing and executive control. J Neurosci 27, 2349–2356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharief AA, Badea A, Dale AM, Johnson GA, 2008. Automated segmentation of the actively stained mouse brain using multi-spectral MR microscopy. Neuroimage 39, 136–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siddiqui AS, Khattra J, Delaney AD, Zhao Y, Astell C, Asano J, Babakaiff R, Barber S, Beland J, Bohacec S, Brown-John M, Chand S, Charest D, Charters AM, Cullum R, Dhalla N, Featherstone R, Gerhard DS, Hoffman B, Holt RA, Hou J, Kuo BY, Lee LL, Lee S, Leung D, Ma K, Matsuo C, Mayo M, McDonald H, Prabhu AL, Pandoh P, Riggins GJ, de Algara TR, Rupert JL, Smailus D, Stott J, Tsai M, Varhol R, Vrljicak P, Wong D, Wu MK, Xie YY, Yang G, Zhang I, Hirst M, Jones SJ, Helgason CD, Simpson EM, Hoodless PA, Marra MA, 2005. A mouse atlas of gene expression: large-scale digital gene-expression profiles from precisely defined developing C57BL/6J mouse tissues and cells. Proc Natl Acad Sci U S A 102, 18485–18490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegle JH, Jia X, Durand S, Gale S, Bennett C, Graddis N, Heller G, Ramirez TK, Choi H, Luviano JA, Groblewski PA, Ahmed R, Arkhipov A, Bernard A, Billeh YN, Brown D, Buice MA, Cain N, Caldejon S, Casal L, Cho A, Chvilicek M, Cox TC, Dai K, Denman DJ, de Vries SEJ, Dietzman R, Esposito L, Farrell C, Feng D, Galbraith J, Garrett M, Gelfand EC, Hancock N, Harris JA, Howard R, Hu B, Hytnen R, Iyer R, Jessett E, Johnson K, Kato I, Kiggins J, Lambert S, Lecoq J, Ledochowitsch P, Lee JH, Leon A, Li Y, Liang E, Long F, Mace K, Melchior J, Millman D, Mollenkopf T, Nayan C, Ng L, Ngo K, Nguyen T, Nicovich PR, North K, Ocker GK, Ollerenshaw D, Oliver M, Pachitariu M, Perkins J, Reding M, Reid D, Robertson M, Ronellenfitch K, Seid S, Slaughterbeck C, Stoecklin M, Sullivan D, Sutton B, Swapp J, Thompson C, Turner K, Wakeman W, Whitesell JD, Williams D, Williford A, Young R, Zeng H, Naylor S, Phillips JW, Reid RC, Mihalas S, Olsen SR, Koch C, 2021. Survey of spiking in the mouse visual system reveals functional hierarchy. Nature 592, 86–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith RE, Tournier J-D, Calamante F, Connelly A, 2012. Anatomically-constrained tractography: Improved diffusion MRI streamlines tractography through effective use of anatomical information. Neuroimage 62, 1924–1938. [DOI] [PubMed] [Google Scholar]
- Steadman PE, Ellegood J, Szulc KU, Turnbull DH, Joyner AL, Henkelman RM, Lerch JP, 2014. Genetic effects on cerebellar structure across mouse models of autism using a magnetic resonance imaging atlas. Autism Res 7, 124–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szczupak D, lack PM, Liu C, Consortium IRC, Tovar-Moll F, Lent R, Silva AC, 2021. Direct Interhemispheric Cortical Communication via Thalamic Commissures: A New White-Matter Pathway in the Rodent Brain. Cereb Cortex 31, 4642–4651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szulc KU, Lerch JP, Nieman BJ, Bartelle BB, Friedel M, Suero-Abreu GA, Watson C, Joyner AL, Turnbull DH, 2015. 4D MEMRI atlas of neonatal FVB/N mouse brain development. Neuroimage 118, 49–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas C, Ye FQ, Irfanoglu MO, Modi P, Saleem KS, Leopold DA, Pierpaoli C, 2014. Anatomical accuracy of brain connections derived from diffusion MRI tractography is inherently limited. Proc Natl Acad Sci U S A 111, 16574–16579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tobisch A, Stirnberg R, Harms RL, Schultz T, Roebroeck A, Breteler MMB, Stocker T, 2018. Compressed Sensing Diffusion Spectrum Imaging for Accelerated Diffusion Microstructure MRI in Long-Term Population Imaging. Front Neurosci 12, 650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tournier JD, Calamante F, Connelly A, 2007. Robust determination of the fibre orientation distribution in diffusion MRI: non-negativity constrained super-resolved spherical deconvolution. Neuroimage 35, 1459–1472. [DOI] [PubMed] [Google Scholar]
- Tournier JD, Calamante F, Connelly A, 2012. MRtrix: Diffusion tractography in crossing fiber regions. International Journal of Imaging Systems and Technology 22, 53–66. [Google Scholar]
- Tournier JD, Calamante F, Gadian DG, Connelly A, 2004. Direct estimation of the fiber orientation density function from diffusion-weighted MRI data using spherical deconvolution. Neuroimage 23, 1176–1185. [DOI] [PubMed] [Google Scholar]
- Trinkle S, Foxley S, Wildenberg G, Kasthuri N, La Riviere P, 2021. The role of spatial embedding in mouse brain networks constructed from diffusion tractography and tracer injections. Neuroimage 244, 118576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ullmann JF, Watson C, Janke AL, Kurniawan ND, Reutens DC, 2013. A segmentation protocol and MRI atlas of the C57BL/6J mouse neocortex. Neuroimage 78, 196–203. [DOI] [PubMed] [Google Scholar]
- Wang N, Anderson RJ, Ashbrook DG, Gopalakrishnan V, Park Y, Priebe CE, Qi Y, Laoprasert R, Vogelstein JT, Williams RW, Johnson GA, 2020a. Variability and heritability of mouse brain structure: Microscopic MRI atlases and connectomes for diverse strains. Neuroimage 222, 117274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang N, White LE, Qi Y, Cofer G, Johnson GA, 2020b. Cytoarchitecture of the mouse brain by high resolution diffusion magnetic resonance imaging. Neuroimage 216, 116876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Ding SL, Li Y, Royall J, Feng D, Lesnar P, Graddis N, Naeemi M, Facer B, Ho A, Dolbeare T, Blanchard B, Dee N, Wakeman W, Hirokawa KE, Szafer A, Sunkin SM, Oh SW, Bernard A, Phillips JW, Hawrylycz M, Koch C, Zeng H, Harris JA, Ng L, 2020c. The Allen Mouse Brain Common Coordinate Framework: A 3D Reference Atlas. Cell 181, 936–953 e920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Zhang J, Mori S, Nathans J, 2006. Axonal growth and guidance defects in Frizzled3 knock-out mice: a comparison of diffusion tensor magnetic resonance imaging, neurofilament staining, and genetically directed cell labeling. J Neurosci 26, 355–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White JD, Arefin TM, Pugliese A, Lee CH, Gassen J, Zhang J, Kaffman A, 2020. Early life stress causes sex-specific changes in adult fronto-limbic connectivity that differentially drive learning. Elife 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winnubst J, Bas E, Ferreira TA, Wu Z, Economo MN, Edson P, Arthur BJ, Bruns C, Rokicki K, Schauder D, Olbris DJ, Murphy SD, Ackerman DG, Arshadi C, Baldwin P, Blake R, Elsayed A, Hasan M, Ramirez D, Dos Santos B, Weldon M, Zafar A, Dudman JT, Gerfen CR, Hantman AW, Korff W, Sternson SM, Spruston N, Svoboda K, Chandrashekar J, 2019. Reconstruction of 1,000 Projection Neurons Reveals New Cell Types and Organization of Long-Range Connectivity in the Mouse Brain. Cell 179, 268–281 e213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu D, Reisinger D, Xu J, Fatemi SA, van Zijl PC, Mori S, Zhang J, 2014. Localized diffusion magnetic resonance micro-imaging of the live mouse brain. Neuroimage 91, 12–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu D, Richards LJ, Zhao Z, Cao Z, Luo W, Shao W, Shi SH, Miller MI, Mori S, Blackshaw S, Zhang J, 2022. A diffusion MRI-based spatiotemporal continuum of the embryonic mouse brain for probing gene-neuroanatomy connections. Proc Natl Acad Sci U S A 119, e2111869119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu F, Shen Y, Ding L, Yang CY, Tan H, Wang H, Zhu Q, Xu R, Wu F, Xiao Y, Xu C, Li Q, Su P, Zhang LI, Dong HW, Desimone R, Xu F, Hu X, Lau PM, Bi GQ, 2021. High-throughput mapping of a whole rhesus monkey brain at micrometer resolution. Nat Biotechnol 39, 1521–1528. [DOI] [PubMed] [Google Scholar]
- Yee Y, Fernandes DJ, French L, Ellegood J, Cahill LS, Vousden DA, Spencer Noakes L, Scholz J, van Eede MC, Nieman BJ, Sled JG, Lerch JP, 2018. Structural covariance of brain region volumes is associated with both structural connectivity and transcriptomic similarity. Neuroimage 179, 357–372. [DOI] [PubMed] [Google Scholar]
- Zapala MA, Hovatta I, Ellison JA, Wodicka L, Del Rio JA, Tennant R, Tynan W, Broide RS, Helton R, Stoveken BS, Winrow C, Lockhart DJ, Reilly JF, Young WG, Bloom FE, Lockhart DJ, Barlow C, 2005. Adult mouse brain gene expression patterns bear an embryologic imprint. Proc Natl Acad Sci U S A 102, 10357–10362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Arefin TM, Nakarmi U, Lee CH, Li H, Liang D, Zhang J, Ying L, 2020. Acceleration of three-dimensional diffusion magnetic resonance imaging using a kernel low-rank compressed sensing method. Neuroimage 210, 116584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zingg B, Hintiryan H, Gou L, Song MY, Bay M, Bienkowski MS, Foster NN, Yamashita S, Bowman I, Toga AW, Dong HW, 2014. Neural networks of the mouse neocortex. Cell 156, 1096–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The atlas and the corresponding structural labels that support the findings of this study are openly available in the respiratory: ‘dMRI_mouse_brain_atlas’ at https://github.com/Tanzil-M-Arefin/dMRI_mouse_brain_atlas
Tracer datasets used in this study were obtained from the Allen Mouse Brain Connectivity project (http://connectivity.brain-map.org).


