Abstract
Bibenzyls, a kind of important plant polyphenols, have attracted growing attention for their broad and remarkable pharmacological activities. However, due to the low abundance in nature, uncontrollable and environmentally unfriendly chemical synthesis processes, these compounds are not readily accessible. Herein, one high-yield bibenzyl backbone-producing Escherichia coli strain was constructed by using a highly active and substrate-promiscuous bibenzyl synthase identified from Dendrobium officinale in combination with starter and extender biosynthetic enzymes. Three types of efficiently post-modifying modular strains were engineered by employing methyltransferases, prenyltransferase, and glycosyltransferase with high activity and substrate tolerance together with their corresponding donor biosynthetic modules. Structurally different bibenzyl derivatives were tandemly and/or divergently synthesized by co-culture engineering in various combination modes. Especially, a prenylated bibenzyl derivative (12) was found to be an antioxidant that exhibited potent neuroprotective activity in the cellular and rat models of ischemia stroke. RNA-seq, quantitative RT-PCR, and Western-blot analysis demonstrated that 12 could up-regulate the expression level of an apoptosis-inducing factor, mitochondria associated 3 (Aifm3), suggesting that Aifm3 might be a new target in ischemic stroke therapy. This study provides a flexible plug-and-play strategy for the easy-to-implement synthesis of structurally diverse bibenzyls through a modular co-culture engineering pipeline for drug discovery.
Key words: Bibenzyls, Modular co-culture engineering, Promiscuous enzymes, Neuroprotective agent, Aifm3, Dendrobium officinale
Graphical abstract
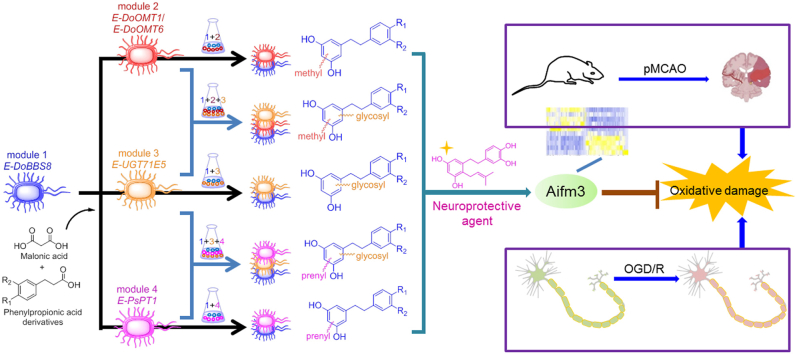
Structurally diverse bibenzyl derivatives were tandemly and divergently synthesized by modular co-culture engineering, and a potent neuroprotective agent was discovered after in vitro and in vivo activity evaluation.
1. Introduction
Bibenzyls, a class of plant polyphenols characterized by a 1,2-diphenylethyl scaffold1, have attracted considerable interest for their significant and diverse pharmacological activity, including anti-tumor2,3, anti-diabetic complications4, neuroprotective5,6, anti-oxidant7, anti-inflammatory8, anti-fungal9, anti-platelet aggregation10, and immune modulatory activities11. During the past decades, a growing number of naturally-occurred bibenzyls have been reported. And strikingly, the bibenzyls with methyl, prenyl, and/or glycosyl moieties substitutions, usually exhibit prominent biological activities. For example, many O-methylated and C-prenylated bibenzyls had good anti-tumor properties and can be used as potential anti-tumor drugs1,12, 13, 14, and several O-glycosylated bibenzyls possessed strong neuroprotective activity15. However, natural bibenzyls usually occur in trace amounts of limited plant species, and chemical synthesis encounters numerous difficulties such as the rigorous conditions, and uncontrollable and environmentally unfriendly processes. Whereas, biosynthesis via engineering artificial pathways in microbes might alleviate these disadvantages and be an alternative approach to synthesizing structurally diverse bioactive bibenzyls.
Generally, plant bibenzyls are derived from the hybrid pathways of shikimate and polyketide16. Phenylalanine is metabolized by a series of enzymes including phenylalanine ammonia-lyase (PAL), cinnamate-4-hydroxylase (C4H), double bond reductase (DBR), and 4-coumarate-CoA ligase (4CL) to generate phenylpropionyl CoA. While, malonyl-CoA is synthesized by acetyl CoA carboxylase (ACC), or malonyl CoA synthetase. Subsequently, the bibenzyl scaffold is formed through the condensation of phenylpropionyl CoA and three molecules of malonyl-CoA mediated by bibenzyl synthase (BBS)17. After that, a variety of tailoring reactions occurred on bibenzyl backbone resulting in a plethora of structurally diverse bibenzyls. Usually, the engineering of such multi-step biosynthetic pathways directly in a single microbe to produce bibenzyls will inevitably increase the metabolic burden of the host, and thus decrease the titer of the desired products. While, it could be conducted by modular co-culture engineering, in which the pathways are distributed into the backbone-producing strain and different post-modifying modular strains. In such microbial consortia, every module can be individually and functionally constructed and optimized, and easily assembled different pathways to efficiently yield a variety of compounds18, 19, 20, 21. Nowadays, the reconstruction of phenylpropionyl CoA and malonyl CoA biosynthesis have been common in microbes, however, the mining of bibenzyl backbone and post-modifying enzymes including BBSs, methyltransferases (MTs), prenyltransferases (PTs), and glycosyltransferases (GTs) responsible for the generation of different building blocks with high catalytic activities and substrate promiscuity, is essential for developing the engineered microbial consortia in further structural diversification of bibenzyls.
In the present study, we identified and functionally characterized BBSs, and OMTs from Dendrobium officinale. And, we mined and screened a novel PT with substrate promiscuity from a fungus strain Periconia sp. F-3122 and a reported promiscuous GT from Carthamus tinctorius23. Subsequently, we established a high-yield bibenzyl backbone-producing Escherichia coli strain and three types of post-modifying modular strains equipped with promiscuous OMTs, PT, and GT. A series of natural and unnatural structurally diverse bibenzyls were biosynthesized via the robust plug-and-play of modules in a variety of combinations. Especially, further in vitro and in vivo pharmacological activity evaluation for these bibenzyl derivatives exhibited that a prenylated bibenzyl (12) would be a novel and promising neuroprotective agent possibly acting via an apoptosis-inducing factor, mitochondria associated 3 (Aifm3)-medicated anti-oxidative stress for ischemia stroke therapeutics.
2. Results and discussion
2.1. Cloning and identification of DoBBSs genes from D. officinale
BBSs, as the vital enzyme for the biosynthesis of bibenzyls, only a handful have been reported and characterized such as PsBBS24 from Pinus sylvestris, pBibsy811, and pBibsy21225 from Phalaenopsis sp., with the same selectivity towards m-hydroxyphenylpropionyl-CoA. To mine BBSs with the capability of generating bibenzyls, a total of 9 candidate genes (DoBBS1−9) were screened from the transcriptome of D. officinale, a traditional Chinese rare and precious medicine with bibenzyls as main chemical constituents1,26, 27, 28. All of the predicted polypeptides encoded by these candidates were grouped into the class of BBSs through further phylogenetic analysis25,29 (Supporting Information Fig. S1), containing Cys-His-Asn catalytic-triad conserved in all known type III PKSs30, 31, 32 (Supporting Information Fig. S2). The full-length cDNAs of DoBBS1−9 were cloned and expressed in E. coli Transetta (DE3), and the recombinant DoBBSs were subjected to purification with Ni-NTA agarose chromatography for enzymatic assay after overnight induction (Supporting Information Fig. S3).
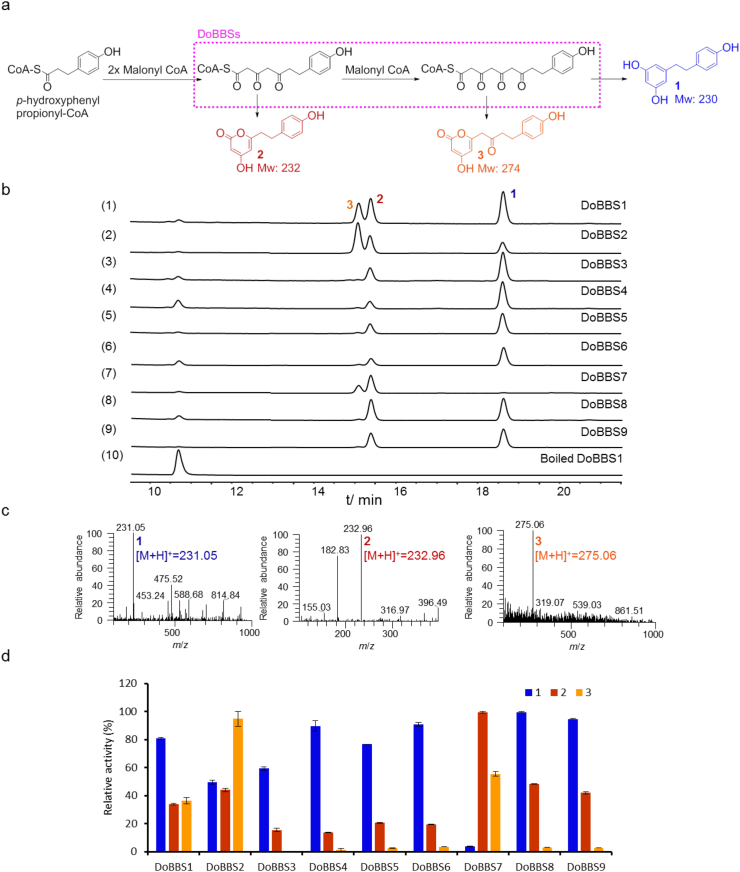
The catalytic activities of DoBBSs were biochemically assayed with p-hydroxyphenylpropionyl-CoA and malonyl-CoA as the starter and extender units, respectively. Three product peaks were observed in HPLC chromatograms of all of the reactions incubated with these recombinant DoBBSs (1–3, Fig. 1). Product 1 with [M+H]+ at m/z 231.05 in mass spectrum was in consistent with the supposed bibenzyl product, dihydro-resveratrol, and further confirmed by 1H and 13C NMR spectroscopic data analysis (Supporting Information). Whereas, the other two products with [M+H]+ at m/z 232.96 and 275.06 were in line with the lactonization products (2 and 3, Fig. 1; also see NMR spectra in Supporting Information), which were commonly observed and regarded as the derailed products in the polyketide biosynthesis33. Therefore, DoBBS1−9 possessed the ability to catalyze the synthesis of bibenzyl backbone, dihydro-resveratrol (1), accompanied by two shunt products (2 and 3). Of the nine DoBBSs, DoBBS8 exhibited the highest bibenzyl synthase activity with the Km of 61.83 μmol/L, kcat of 0.54 min−1 (Supporting Information Fig. S4) and was selected for the subsequent engineering strain construction. The investigation of the biochemical property revealed that the optimum reaction condition for recombinant DoBBS8 was at pH 8.0 and 30 °C, and independent on divalent metal ions (Fig. S4). Besides, the further substrate promiscuity analysis (Supporting Information Fig. S5) of the purified DoBBS8 demonstrated that DoBBS8 could also accept 3,4-dihydroxyphenylpropionyl-CoA, m-hydroxyphenylpropionyl-CoA, and even p-coumaroyl CoA as the starter units.
Figure 1.
Functional characterization of DoBBSs. (a) Enzymatic reaction catalyzed by DoBBSs; (b) HPLC analysis (λ = 280 nm) of the reactions with (1) DoBBS1, (2) DoBBS2, (3) DoBBS3, (4) DoBBS4, (5) DoBBS5, (6) DoBBS6, (7) DoBBS7, (8) DoBBS8, (9) DoBBS9, (10) boiled DoBBS1; (c) MS spectra of products 1–3 at positive mode; (d) Relative activity of DoBBSs for the synthesis of 1−3. For each product, the relative activity of the reaction with the highest yield was set as 100%. The relative activity represents mean ± SD (n = 2).
2.2. Assembly of bibenzyl backbone-producing strain
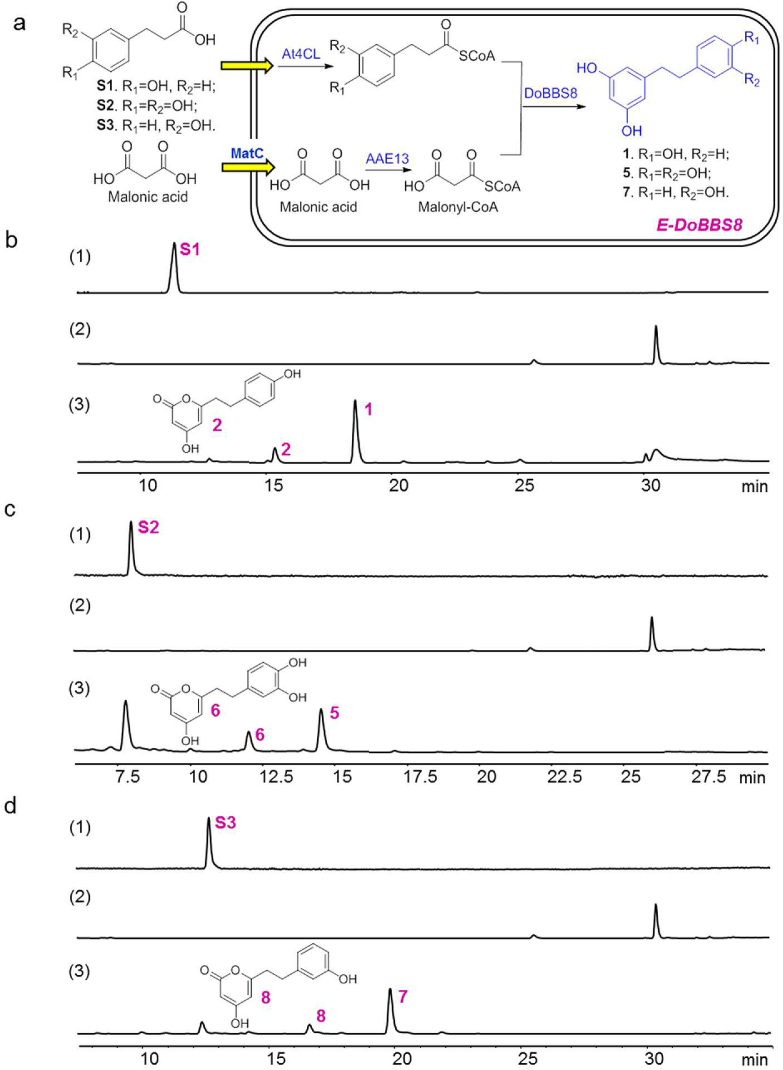
To construct a bibenzyl backbone-producing strain, the bibenzyl backbone synthetic pathway was assembled with three units, including starter unit phenylpropionyl CoA and extender unit malonyl CoA synthetic parts, together with a condensation unit with BBS to generate bibenzyl backbone. To improve the intracellular supply of malonyl-CoA, AAE13 (AAP03025.1)34 and MatC (KF765784.1)35 encoding a malonyl CoA synthetase and a malonate carrier protein, respectively, were introduced to construct the extender unit-producing plasmid. Accompanied with these, At4CL1 (AY376729.1)36 and DoBBS8 were introduced for the starter unit- and bibenzyl backbone-producing plasmids, respectively. All the above recombinant plasmids were co-transformed into E. coli BL21(DE3) to initiate a bibenzyl backbone-producing strain (Fig. 2a).
Figure 2.
Establishment of bibenzyl backbone-producing strain E-DoBBS8. (a) The reconstructed biosynthetic pathway for bibenzyl backbones in E. coli; (b–d) HPLC analysis (λ = 280 nm) of the reaction catalyzed by E-DoBBS8 strain with malonic acid and S1 (b)/S2 (c)/S3 (d) as starting materials; HPLC analysis of (1) the standards S1–S3, (2) the negative reaction without substrate, (3) the reaction with malonic acid and S1–S3 as starting materials.
To verify the capacity of the engineering strain, the whole-cell reactions were performed by feeding malonic acid and phenylpropionic acid derivatives. As shown in Fig. 2, a bibenzyl (1) accompanied by a triacetic acid lactone (2) was produced when malonic acid and p-hydroxyphenylpropionic acid (S1) were fed as starting materials. Besides, the corresponding bibenzyls (3,3′,4′,5-tetrahydroxybibenzyl, 5; 3,3′,5-tetrahydroxybibenzyl, 7) were generated when feeding 3,4-dihydroxyphenylpropionic acid (S2) and m-hydroxyphenylpropionic acid (S3) instead of S1. These results suggested that both DoBBS8 and At4CL1 exhibited substrate promiscuity in the whole cell of the engineering strain, which could produce bibenzyl backbones with different substituents, and this strain was designated as E-DoBBS8.
2.3. Screening of enzymes for bibenzyl post-modification
As mentioned previously, substituted groups such as methyl, prenyl, and/or glycosyl not only lead to the structural diversity of bibenzyls, and also play pivotal roles in their pharmacological activities12, 13, 14, 15. Thus, multi-organism-derived tailoring enzymes for the post-modification of bibenzyl backbone were probed.
The existence of numerous O-methylated bibenzyls in D. officinale inspired us to mine desired OMT from this plant species, and 11 candidate genes (DoOMT1−11) were screened from the transcriptome. Further sequence analysis revealed that DoOMT1−5 belonged to caffeic acid OMT (COMT) superfamily, and DoOMT6−11 were grouped into the caffeoyl-CoA OMT (CCOMT) superfamily (Supporting Information Fig. S6). Given the reported different properties for dependence on divalent ions and substrate preference between COMTs and CCOMTs37,38, DoOMT1−11 might exhibit divergent activities in the methylation of bibenzyl-related compounds. The full-length of DoOMT1−11 were amplified via RT-PCR from cDNA of D. officinale, and successfully expressed in E. coli Transetta (DE3). Soluble proteins except DoOMT7 were obtained from transformants of E. coli cells after overnight induction and further purified with Ni-NTA agarose affinity chromatography (Supporting Information Fig. S7). The enzymatic activities of DoOMTs were tested by using dihydro-resveratrol (1) as methyl acceptor and S-adenosylmethionine (SAM) as methyl donor. The results demonstrated that each reaction with COMTs (DoOMT1−5) generated a mono-O-methylated product, 3,4′-dihydroxy-5-methoxy-bibenzyl (4, Fig. 3 and Supporting Information Fig. S8). While, there was no product observed in the reactions with those CCOMTs, possibly because their native substrate is caffeoyl-CoA derivatives rather than bibenzyl derivatives. Among the five active COMTs, DoOMT1 exhibited the highest conversion rate of 100% when using 1 as acceptor substrate (Fig. S8). To further probe the substrate promiscuity of DoOMT1, other fourteen bibenzyl derivatives (5, 7, a–l) were used as substrates and subjected to enzyme reactions. Enzyme assay results showed that eight bibenzyls (5, 7, a–d, i, l) could be mono-O-methylated to the corresponding products by HPLC–MS analysis, suggesting a broad substrate spectra of DoOMT1 (Supporting Information Fig. S9). Accordingly, DoOMT1 was selected for the subsequent engineering methylation strain construction.
Figure 3.
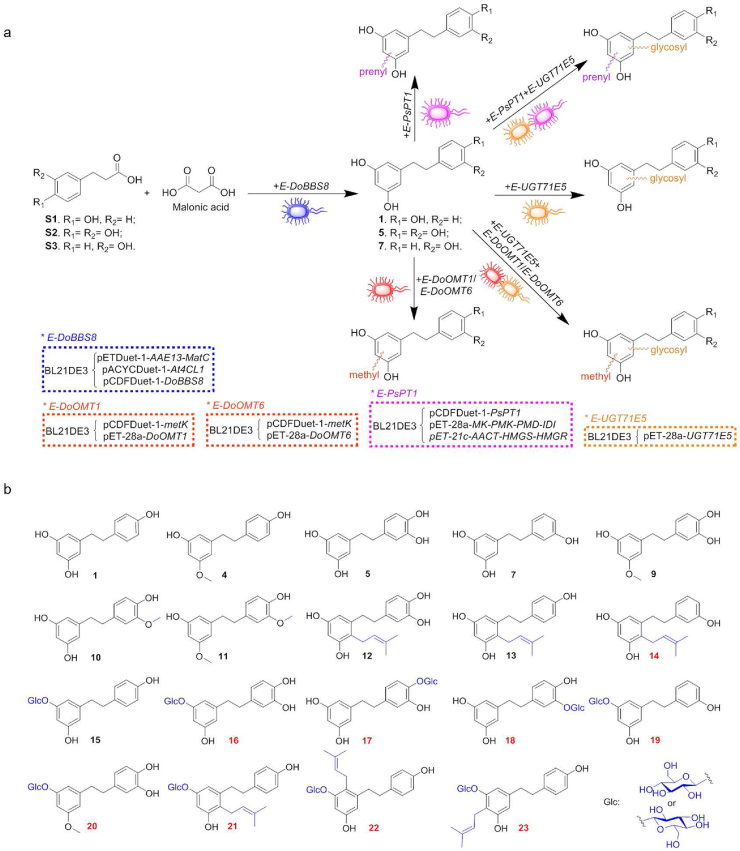
Structural diversification of bibenzyls through modular co-culture engineering. (a) The reconstructed synthetic pathway for the structural diversification of bibenzyls. (b) The structurally diverse bibenzyl derivatives prepared from the engineered E. coli modular co-culture. The compounds in red numbers are new compounds.
Meanwhile, four PTs with substrate promiscuity including AtaPT39 and newly identified PTs, were tested for the bibenzyl prenylation with 1 as the prenyl acceptor and dimethylallyl diphosphate (DMAPP) as the prenyl donor. HPLC–MS analysis (Supporting Information Fig. S10) showed that PsPT1, a novel prenyltransferase from Periconia sp. F-3122 by genome-mining exhibited the highest conversion rate of 95%. Further substrate promiscuity analysis revealed the broad substrate tolerance of PsPT1 (Supporting Information Fig. S11), in which PsPT1 could catalyze the prenylaton of eight tested acceptor substrates (5, 7, a–d, i, l). Accordingly, PsPT1 was used for the construction of the prenylation module.
Similarly, UGT71E5, a GT especially with excellent substrate promiscuity from C. tinctorius23 was screened out of 5 GTs including O-, N-, and C-GTs40, 41, 42, 43, 44, with its high conversion rate of 94% and substrate promiscuity in the reaction with 1 and various acceptors (5, 7, a–d, I, l) and uridine 5′-diphosphoglucose (UDP-Glc) by HPLC‒MS analysis (Supporting Information Figs. S12 and S13). Accordingly, UGT71E5 was used for the construction of glycosylation modular strain.
2.4. Establishment of bibenzyl post-modifying modular strains
Based on the above results, we set to conduct the biosynthesis of structurally diverse bibenzyls by the coordination of every functional component. Firstly, we attempted to construct bibenzyl derivative-producing strains through mono-culture engineering. For example, the gene DoOMT1 was introduced into the bibenzyl backbone-producing strain E-DoBBS8 to build up methylated bibenzyl-producing strain. Unfortunately, no methylated product was observed after fermentation, even though introducing a SAM synthase metK45 for the methyl donor supplement in this strain. The poor performance of the mono-culture system might be attributed to the metabolic burden and difficulties in the coordination among different functional modules in a single cell, in which each module required specialized environments or compartments for optimal function, etc. Since modular co-culture engineering18, 19, 20, 21 completely modularize and segregate a biosynthetic pathway into multiple separate cells, each of which bears a portion of the pathway and can be engineered independently to achieve optimal function of the combined pathway, and might alleviate the disadvantages of mono-culture engineering.
Therefore, we conducted to construct the post-modifying modular strains. For the methylation modular strain, the gene metK encoding SAM synthase was introduced to ensure the supply of methyl donor SAM. For example, the strain (E-DoOMT1) harboring recombinant plasmids of DoOMT1 and metK was taken as the independent methylation module. As shown in Supporting Information Fig. S14a, the methylated product 4 was produced when bibenzyl backbone 1 was fed as the starting material, and this engineered strain was designated as E-DoOMT1. Similarly, the strains E-DoOMT6−11 harboring recombinant plasmids of DoOMT6−11 and metK was constructed as additional methylation modular strains.
For the prenylation modular strain, the recombinant plasmids of pXL13 and pXL17 carrying 7 genes involved in the MVA (mevalonic acid) pathway46 to ensure the sufficient supply of prenyl donor, along with the plasmid harboring PsPT1 gene, were co-transformed into BL21(DE3), generating the engineered prenylation strain (E-PsPT1). Similarly, the prenylated product 1-P1 was produced when 1 was used as substrate (Supporting Information Fig. S14b).
Considering the relatively sufficient supply of the endogenous glycosyl donor from the E. coli host, the strain (E-UGT71E5) harboring the recombinant plasmid of UGT71E5 was directly employed as the glycosylation strain. As shown in Supporting Information Fig. S14c, substrate 1 was nearly totally conversed to the glycosylated product 1-G1 by the glycosylation modular strain E-UGT71E5.
2.5. Co-culturing mono-post-modifying modular strains with E-DoBBS8 strain
As described above, a bibenzyl backbone-producing and three types of post-modifying modular stains have been constructed, in which the crucial enzymes including BBS, OMT, PT, and GT were featured acceptor substrate promiscuity. Moreover, the donor biosynthetic pathways were enhanced (for methylation) or engineered (for prenylation), or intrinsic (for glycosylation). Thus, these functional modular strains would enable the structural diversification of bibenzyls by employing co-culture engineering with different combination modes.
To produce methylated bibenzyl derivatives, the methylation strain E-DoOMT1 was co-cultured with bibenzyl backbone-producing strain E-DoBBS8. When the malonic acid and three different p-hydroxyphenylpropionic acid derivatives (S1–S3) were individually used as the starting materials, the corresponding O-methylated bibenzyls (4, 9, 35) were generated (Fig. 3 and Supporting Information Fig. S15). Considering the predicted native substrate of CCOMT members of DoOMT6−11 and the generation of intermediates of caffeoyl-CoA derivatives by E-DoBBS8 and as well as the substrate promiscuity of DoBBS8, the methylation strains E-DoOMT6−11 were co-cultured with E-DoBBS8, respectively. Intriguingly, a mono-O-methylated derivative (10) was generated for the strain E-DoOMT6 when adding S2. Besides, when both of E-DoOMT1 and E-DoOMT6 strains were co-cultured together with E-DoBBS8 in the presence of malonic acid and S2, a di-O-methylated product (11) was observed. Further NMR spectroscopic analyses (Supporting Information) demonstrated that mono-methyls in 9/10 were attached at 3-OH/3′-OH of the bibenzyl backbone, respectively, and two methyls in 11 at 3-OH and 3′-OH. These results suggested that E-DoOMT6 might regio-specifically catalyze 3-O-methylation of caffeoyl-CoA derivative followed by BBS-mediated condensation, providing an additional alternative for structural diversification of such bibenzyls. This assumption was further confirmed by an in vitro cascade reaction catalyzed by the purified At4CL1 and DoOMT6 (Supporting Information Fig. S16), in which two peaks correspond to the methylated CoA derivatives (designated as S2-CoA-M1 and S2-CoA-M2) presented with [M+H]+ at m/z 946.05, and 946.18, respectively. Besides, the enzymatic assay in vitro also demonstrated that DoOMT6 could catalyze the methylation of 5 to generate 5-M1, corresponding to 10. That is, in such consortia, the methylation catalyzed by E-DoOMT6 not only occurred on the CoA derivatives, but also the bibenzyl, convergently resulting in the synthesis of 10. Notably, 10 (Tristin)4,47 and 11 (gigantol)2 were reported as promising antioxidative, anti-tumor, and anti-spasmodic agents.
Similarly, the prenylation modular strain E-PsPT1 was co-cultivated with E-DoBBS8 to produce the prenylated products, and the corresponding prenylated bibenzyl derivatives (12–14) were yielded when feeding with the malonic acid and different p-hydroxyphenylpropionic acid derivatives (S1–S3), respectively (Fig. 3 and Supporting Information Fig. S17). 12–14 were structurally characterized as regio-specifically 2-C-prenylated bibenzyls by MS and NMR spectroscopic data analyses after scale-up synthesis (Supporting Information).
Likewise, the co-culture of the glycosylation modular strain E-UGT71E5 and E-DoBBS8, generated the corresponding glycosides (15–19) (Fig. 3 and Supporting Information Fig. S18). When feeding with the malonic acid and S1 or S3, only one regio-specifically 3-O-glycosylated bibenzyl (15 or 19) with a higher conversion rate was yielded, respectively. While, in the case of S2, three glycosides, 3-O, 4′-O, and 3′-O-glycosylated bibenzyls (16–18) were generated. Nevertheless, the major product of 3-O-glycosides implied that UGT71E5 in the whole-cell system preferred to 3-O-glycosylation of these bibenzyls.
2.6. Co-culturing multi-post-modifying modular strains with E-DoBBS8 strain
Since the substrate promiscuity of these post-modifying enzymes/engineered modular strains, we investigated the fitness of two or more post-modifying modular strains with E-DoBBS8 for the synthesis of more diverse bibenzyls with multiple modifications. For example, apart from the basic bibenzyl backbone-related products 1–8, the corresponding bibenzyl derivatives (20, 24, and 25, Fig. 3, Supporting Information Fig. S19) with both glycosyl and methyl groups were obtained when E-UGT71E5, E-DoOMT1, and E-DoBBS8 strains were co-cultured by adding malonic acid and S1–S3. Besides, glycosylated and methylated product (26) monitored by LC–MS was generated when E-DoOMT6 instead of E-DoOMT1 was co-cultured and S2 as starter unit (Fig. S19). Moreover, co-culturing E-UGT71E5, E-PsPT1, and E-DoBBS8 strains led to the synthesis of various additional bibenzyl derivatives with both glycosyl and prenyl groups (21–23, 27–34, Fig. 3, Supporting Information Fig. S20). Intriguingly, other than mono-glycosylated and mono-prenylated products (21–23, 27–32), two products (33 and 34) undergone di-O-glucosylation and mono-prenylation were generated in this bioprocess by LC‒MS analysis (Fig. S20). A further scale-up co-cultivation with malonic acid and S1 resulted in the isolation of products 21–23. Different from the above mono-modification co-culture, prenylation occurred at C-6, C-2, and C-4 of the bibenzyl backbone in such multi-modification co-culture, further highlighting the robust prenylation promiscuity of E-PsPT1 modular strains.
Accordingly, such plug-and-play modular co-culture engineering exhibited the powerful capacity for tandemly and divergently synthesizing structurally diverse bibenzyl derivatives (Fig. 3a), and a series of natural/unnatural bibenzyl derivatives (Fig. 3b) including nine new compounds had been acquired from the selected scale-up reactions. These results implied the excellent fitness of these modular engineered strains with the capability of yielding more structurally various derivatives.
2.7. Evaluation of the neuroprotective activities of bibenzyls in vitro
Most of the obtained products were subjected to pharmacological assay, the results showed that compound 12 exhibited strong neuroprotective effect against glutamate-induced neuronal excitatory injury (Supporting Information Table S5 and Fig. S21a). By comparing the structures and activities of these compounds (Fig. 3 and Table S5), it seemed that prenyl group might play a vital role for neuroprotective effect, and 3′-, 4′-OHs in molecule could enhance the activity. Further scavenging assay showed that 12 had a stronger 1,1-diphenyl-2-picrylhydrazyl radical (DPPH·), oxidative radical and •OH scavenging ability than vitamin C (Supporting Information Fig. S22a−d), demonstrating that such neuroprotective activity of bibenzyls was related to their antioxidative stress effects48, although the intensity of 12 in scavenging O2‒· was weaker than vitamin C (Fig. S22e). Besides, 12 significantly increased the total antioxidant capacity (Fig. S21b) and decreased reactive oxygen species (ROS) levels of injured SK-N-SH cells induced by glutamate (Fig. S21c). Additionally, 12 could recover the activity changes of antioxidative enzymes including catalase (CAT), glutathione peroxidase (GSH-Px), and superoxide dismutase (SOD) (Fig. S21d−f), possibly due to compensatory response. Thus, compound 12 might be a promising neuroprotective agent.
2.8. Optimization and recycling utilization of engineered strains for the synthesis of 12
The potent neuroprotective activity of compound 12 in vitro inspired us to further explore its bioactivity in vivo and action mechanism. Thus, it's necessary to establish an approach for easy access to 12. Firstly, the whole-cell culture conditions, including the cell density of cultures, the input concentration of substrates, and the incubation time were investigated and optimized to improve the productivity of bibenzyl backbone-producing strain E-DoBBS8. The results demonstrated that cell density at OD600 of 5.0, 1.50 mmol/L p-hydroxyphenylpropionic acid with 4.50 mmol/L malonic acid, and 24 h of incubation led to the optimum yield of approximately 190 mg/L of 1 (Supporting Information Fig. S23). Moreover, considering that the engineered strains E-DoBBS8 and E-PsPT1 were generally discarded after a round of incubation, we attempted to recycle the co-cultured strains to maximize the capacity of the synthesis of 12. Interestingly, the producibility of the two co-cultured strains remained 64%, 33%, 21%, and 17% in the second to fifth batch, respectively. Accordingly, 66 mg/L of 12 was yielded after 3 cycles (Supporting Information Fig. S24), which was about two times yield of one batch. These results suggested that the efficient and sustainable production of 12 could be achieved through optimizing reaction conditions and recycling utilization of engineered strains.
2.9. Neuroprotective effect in pMCAO rats
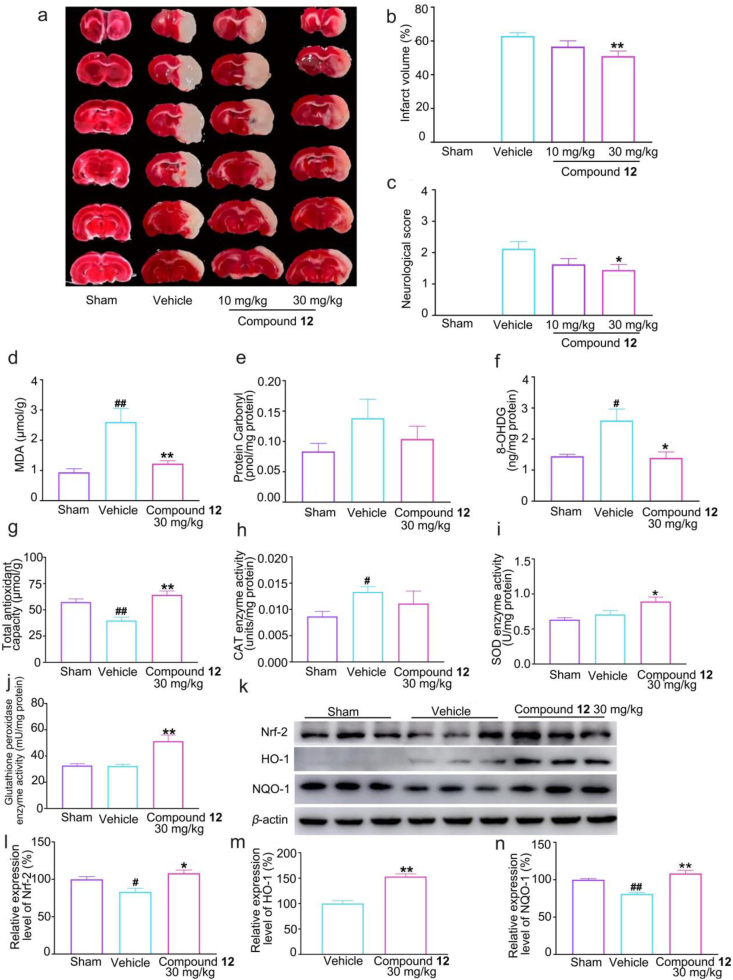
Oxidative stress plays an important role in cerebral ischemia, thus we used permanent middle cerebral artery occlusion (pMCAO) rats to explore the efficacy of compound 12. As shown in Fig. 4a–c, 12 at the dose of 30 mg/kg obviously reduced infarct volume and improved neurobehavioral scores. Meanwhile, oxidative damage of lipids, proteins, and DNA measures showed that 12 reversed ischemia-induced elevations of malondialdehyde (MDA) and 8-hydroxy-2′-deoxyguanosine (8-OHDG), nor in protein carbonylation levels (Fig. 4d–f). In addition, 12 remarkably reversed the downregulation of total antioxidant capacity (Fig. 4g), and slightly decreased CAT level (Fig. 4h). Notably, 12 resulted in significant increases in SOD and GSH-Px levels, although no significant decrease was detected in the vehicle group compared with the sham group (Fig. 4i–j). Since nuclear factor erythroid 2-related factor 2 (Nrf-2) is one of the most important regulators of the cellular defense system against oxidative stress in stroke49,50. Western-blot analysis showed that all of the pathway-related enzymes (Nrf-2, HO-1, and NQO-1) levels were significantly higher than the vehicle group (Fig. 4k–n), further suggesting the antioxidant activity of 12. Taken together, compound 12 might exert neuroprotective function by increasing antioxidative capacity in cerebral ischemic rats.
Figure 4.
Effects of compound 12 on cerebral infarction volume, neurobehavioral scores, antioxidant enzymes and oxidative damage indexes of pMCAO rats. (a–b) Cerebral infarction volume of pMCAO rats (n = 8–9); (c) Neurobehavioral scores of pMCAO rats (n = 8–9); (d–e) The levels of MDA and protein carbonylation in pMCAO rat brains (n = 10); (f) The level of 8-OHDG in pMCAO rat brains (n = 5); (g) The level of total antioxidant capacity in pMCAO rat brains (n = 5); (h) The activities of CAT in pMCAO rat brains (n = 5); (i) The activities of SOD in pMCAO rat brains (n = 10); (j) The activities of GSH-Px in pMCAO rat brains (n = 5); (k–n) The protein levels of Nrf-2, HO-1 and NQO-1 (n = 3). Data were shown as mean ± SEM, #P < 0.05, ##P < 0.01 vs. the sham group, ∗P < 0.05, ∗∗P < 0.01 vs. the vehicle group.
2.10. Molecular mechanism of neuroprotective effect in pMCAO rats
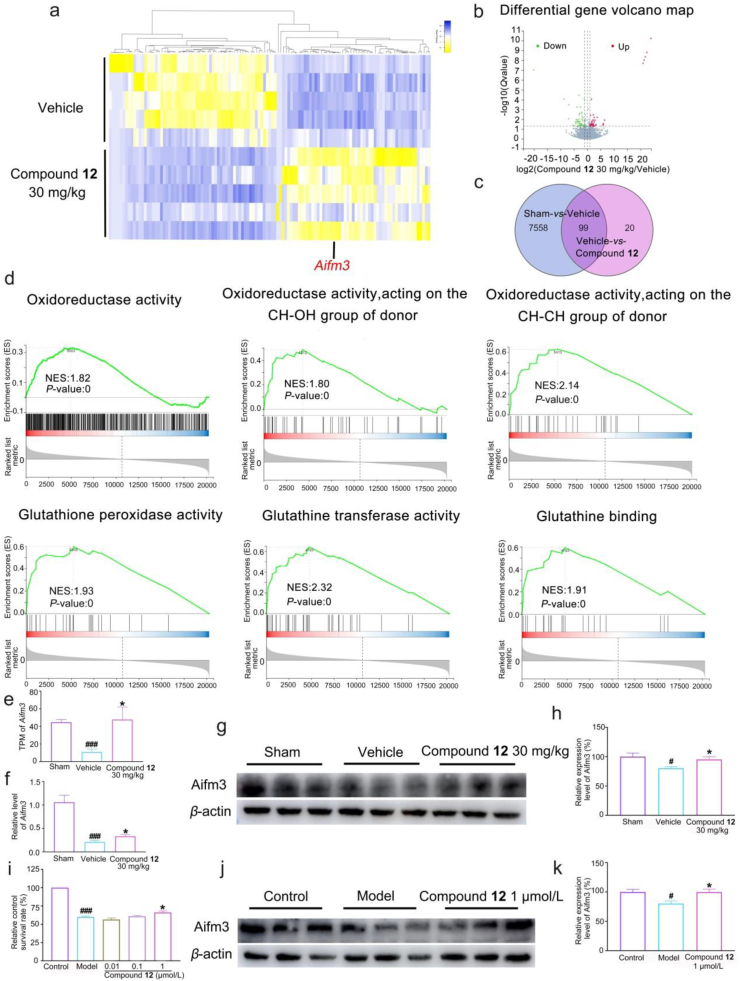
RNA-seq was exploited to pursue the potential molecular mechanism of compound 12 in pMCAO rats. As shown in Fig. 5a–c, a total of 119 differentially expressed genes were identified between the vehicle group and compound 12 group, of which 57 genes were up-regulated and 62 genes were down-regulated after treatment. Gene enrichment analysis (GSEA) indicated that 12 mainly increased gene expression of the antioxidant-related pathway, including oxidoreductase activity, GSH-Px activity, glutathione transferase activity, and glutathione binding (Fig. 5d). In each of these antioxidant related pathways, the genes in compound 12 group were upregulated. Notably, the increased GSH-Px activity had been demonstrated in the previous results, which was consistent with GSEA enrichment (Fig. 4j). Among the above genes, apoptosis-inducing factor, mitochondria associated 3 (Aifm3), reported to show REDOX activity and induce apoptosis51, was dramatically reduced after cerebral ischemia. Interestingly, 12 vigorously enhanced Aifm3 expression (Fig. 5a and e). Furthermore, quantitative RT-PCR and Western-blot analysis demonstrated that 12 could reverse ischemia-induced downregulation of Aifm3 in mRNA and protein levels (Fig. 5f–h). Furtherly, 12 exhibited a concentration-dependently protective effect on oxygen glucose deprivation/reoxygenation (OGD/R) injured neuronal cells (Fig. 5i) by improving the total antioxidant capacity and GSH-Px activity. While, there was no significant difference in SOD and CAT levels compared with the model group (Supporting Information Fig. S25a−d). Similarly, we also found that 12 raised the expression of Aifm3 after OGD/R injury (Fig. 5j−k) in consistence with in vivo data, implying that Aifm3 might be a key target in ischemic stroke therapy. In addition, we verified the effect of 12 on apoptosis by detecting the changes of mRNA levels. However, mRNA levels of apoptosis-related genes including Bax, Bcl-2, Cytochrome c, and Caspase-3 did not significantly change (Supporting Information Fig. S26a−d), further suggesting that 12 could exert antioxidant function by up-regulating the level of Aifm3.
Figure 5.
The molecular mechanisms of compound 12 in anti-ischemia. (a) RNA-seq analysis of differential genes in pMCAO rat brains tissues (n = 5). 119 differential genes were identified between the vehicle group and compound 12 group. (b) 57 genes were up-regulated and 62 genes were down-regulated after compound 12 treatment. (c) 99 genes with opposite trends in the intersection of vehicle group versus sham group and vehicle group versus compound 12 group. (d) Gene enrichment analysis (GSEA) indicated antioxidant related pathway, including oxidoreductase activity; oxidoreductase activity (acting on the CH–OH group of donors); oxidoreductase activity (acting on the CH–CH group of donors); glutathione peroxidase activity; glutathione transferase activity and glutathione binding. (e) The level of Aifm3 mRNA in RNA-seq results. (f) compound 12 increased Aifm3 mRNA level by Q-PCR verification (n = 8). (g–h) compound 12 reversed the down-regulation of Aifm3 protein expressions in pMCAO rat brains (n = 3). (i) compound 12 protected OGD/R-induced neuronal injury (n = 4). (j–k) compound 12 elevated Aifm3 protein levels after OGD/R injury (n = 3). Data were shown as mean ± SEM, #P < 0.05, ###P < 0.001 vs. the sham group or control group, ∗P < 0.05 vs. the vehicle group or model group.
3. Conclusions
In the present study, several BBSs and OMTs from D. officinale, a PT from fungus Periconia sp., and a GT from C. tinctorius have been identified and functionally characterized. Based on the high activities and substrate promiscuity of these enzymes, a high-yield bibenzyl backbone-producing modular E. coli strain and three types of post-modification including methylation, prenylation, and glycosylation modular strains were constructed. Subsequently, a versatile pipeline for structural diversification of bibenzyls was established through a plug-and-play modular co-culture engineering strategy, and a number of structurally diverse bibenzyl derivatives were tandemly and divergently synthesized. Besides, initial optimization and recycling utilization of engineered strains were achieved for the efficient synthesis of the target product. Most importantly, one prenylated product (12) was found to be a novel potential neuroprotective agent after systematic pharmacological activity evaluation in vitro and in vivo. Molecular mechanism investigation revealed that 12 might function as an antioxidant by up-regulating the level of Aifm3, which is firstly reported as a putatively key target in ischemic stroke therapy. This work not only provides a powerful and flexible approach for the synthesis of structurally diverse bibenzyl derivatives but also provides a proof-of-concept for the efficient biosynthesis of bioactive natural compounds for drug design and discovery by modular co-culture engineering.
4. Experimental
4.1. General
Chemicals and reagents were purchased from Sigma–Aldrich (St. Louis, USA), Biodee Biotech Co., Ltd. (Beijing, China), and InnoChem Science & Technology Co., Ltd. (Beijing, China), unless noted elsewhere. 4-Hydroxyphenylpropionyl-CoA and DMAPP were prepared according to the literatures52,39. Bibenzyls a–l were isolated in our previous work53. KOD-Plus neo DNA polymerase was purchased from Toyobo Biotech Co., Ltd. (Shanghai, China). ClonExpress® II One Step Cloning Kit was purchased from Vazyme Biotech Co., Ltd. (Nanjing, China). Restriction enzymes were purchased from Takara Biotech Co. Ltd. (Dalian, China). Primer synthesis and DNA sequencing were conducted at Sangon Biotech Co., Ltd. (Shanghai, China). Genes encoding 4-coumaroyl CoA ligase (At4CL1, Genbank No.: AY376729.1)36 and malonyl CoA synthase (AAE13, Genbank No.: AAP03025.1)34 from Arabidopsis thaliana were gifted from Prof. Shepo Shi (Beijing University of Chinese Traditional Medicine). Plasmids pXL13 and pXL1746 were kindly provided by Prof. Yong Wang (Institute of Plant Physiology and Ecology, Chinese Academy of Sciences). The gene encoding a dicarboxylate carrier protein (MatC, Genbank No.: KF765784.1)35 was chemically synthesized (Sangon Biotech, Shanghai, China) with a codon-optimization for E. coli expression.
The analyses of enzyme activity and the determination of conversion rates were performed on an Agilent 1200 series HPLC system (Agilent Technologies, Germany) coupled with an LCQ Fleet ion trap mass spectrometer (Thermo Electron Corp., USA) equipped with an electrospray ionization (ESI) source. HR-ESI-MS data were measured with an Agilent Technologies 6520 Accurate Mass Q-TOF LC‒MS spectrometer. 1H and 13C NMR spectra were measured on Mercury-400 and Bruker AVIIIHD spectrometers, chemical shifts (δ) were referenced to internal solvent resonances and are given in parts per million (ppm), and coupling constants (J) are given in hertz (Hz). For quantification, three parallel assays were routinely performed.
The procedures of DPPH, pyrogallol autoxidation, Fenton reaction, and oxygen radical absorbance capacity (ORAC) assay, determinations of 8-hydroxy-2′ deoxyguanosine (8-OHDG) and protein carbonyl, quantitative RT-PCR, and Western-blot were described in Supporting Information. The levels of CAT, GSH-Px, MDA, ROS, SOD, and total antioxidant capacity were detected following the manufacturer's instructions (Beyotime Biotechnology, Shanghai, China). For pharmacological experiments, all data were presented as the mean ± SEM, statistical significance was determined using unpaired Student's t-tests, all analyses were conducted using GraphPad Prism 8.0.1 software, and differences at P < 0.05 were considered statistically significant.
4.2. Plant materials
The plants of D. officinale were collected in Jiangsu Province, China, and authenticated by Prof. Xiaomei Chen, Institute of Medicinal Plant Development, Chinese Academy of Medical Sciences.
4.3. Molecular cloning and the heterologous expression of DoBBSs genes
Total RNA of the fresh stems of D. officinale was prepared using the E.Z.N.A.™ Plant RNA kit (Omega Bio-Tek, Inc. GA, USA) and reverse-transcribed to cDNA with SmartScribe reverse transcriptase (Clontech, USA) following the manufacturer's instructions. Full-length cDNAs of the DoBBSs (DoBBS1−9) were amplified from D. officinale by PCR using gene-specific primer pairs (Supporting Information Table S4). The amplified DNA fragments were inserted into pET-28a vector between the BamH I and XhoI sites. After sequence confirmation, the recombinant plasmids were transformed into E. coli Transetta (DE3) (TransGen, Beijing, China) for heterologous expression. The subsequent detailed procedures for protein expression and purification were conducted as previously reported39.
4.4. Bibenzyl synthase activity assays
The reaction mixture consisted of 1.5 mmol/L malonyl-CoA, 0.5 mmol/L p-hydroxyphenylpropionyl-CoA and 1 mg/mL DoBBSs in 100 μL of reaction buffer (50 mmol/L Tris-HCl, 1 mmol/L DTT, 1.5% glycerol (v/v), pH 7.4). After incubation at 30 °C for 12 h, the reaction was terminated by the addition of 200 μL of MeOH and vortexing for 2 min. The protein was removed by centrifugation at 12,000×g for 30 min. The supernatant was analyzed by HPLC‒MS on a Shiseido Capcell Pak C18 MGIII column (250 mm × 4.6 mm I.D., 5 μm, Shiseido Co., Ltd., Japan) at a flow rate of 1 mL/min. The mobile phase was a gradient elution of solvent A (0.1% formic acid aqueous solution) and solvent B (acetonitrile). The gradient programs used for analysis were shown in Supporting Information Table S6.
For investigation of pH effects on DoBBS8 activity, the reactions were performed in reaction buffers with pH 4.0–6.0 (50 mmol/L citric acid-sodium citrate buffer), 6.0–7.0 (50 mmol/L Bis-Tris-HCl buffer), 7.0–9.0 (50 mmol/L Tris-HCl buffer), 9.0–10.5 (50 mmol/L glycine/NaOH buffer). For temperature, the reactions were conducted at different temperatures ranging from 15–55 °C. For the dependence of divalent metal ions, the solutions of MgCl2, SrCl2, BaCl2, CaCl2, SnCl2, NiCl2, CoCl2, ZnCl2, MnCl2, CuCl2, and EDTA were added to reactions with the final concentration of 5 mmol/L, respectively. Reactions without divalent metal ions were used as a control. All assays were carried out with p-hydroxyphenylpropionyl-CoA and malonyl-CoA as substrates.
For the determination of kinetic parameters, 10 μg of recombinant DoBBS8 was incubated with various concentrations of p-hydroxyphenylpropionyl-CoA (5–800 μmol/L) and a fixed concentration of malonyl-CoA (2.0 mmol/L) in a total volume of 50 μL at 30 °C for 30 min. All of these reactions were performed in duplicate, and enzyme activity was evaluated via quantification of the corresponding bibenzyl product dihydroresveratrol (1) as nmol per mg of recombinant protein per minute. Kinetic parameters, including Michaelis–Menten constant (Km) and turnover number (kcat), were calculated by nonlinear regression analysis using GraphPad Prism 8.0.1 software.
The cascade reaction catalyzed by At4CL136, AAE1334 and DoOMT6 was firstly initiated by the At4CL1 and AAE13 reaction, the former consisted of 0.5 mmol/L S2/S3/S4, 2 mmol/L CoA, 4 mmol/L ATP, 5 mmol/L MgCl2, and 0.25 mg/mL purified At4CL1 in 100 μL of reaction buffer (50 mmol/L Tris-HCl, 1 mmol/L DTT, 1.5% glycerol (v/v), pH 7.4), while the latter consisted of 1.5 mmol/L malonic acid, and 0.25 mg/mL purified AAE13, the other same as the At4CL1. After incubation at 30 °C for 30 min, the cascade reaction was pushed by supplement of 1 mg/mL purified DoBBS8, with the reaction volume adjustment to 200 μL. The supernatant was sequentially analyzed by HPLC on a Tosoh TSK gel ODS column (250 mm × 4.6 mm I.D., 5 μm) and a Shiseido Capcell Pak C18 MGIII column at a flow rate of 1 mL/min. The mobile phase for the former was a gradient elution of solvent A (0.1% trifluoroacetic acid aqueous solution) and solvent B (acetonitrile). While for the latter, it was same as the bibenzyl derivatives. The gradient programs used for analysis were shown in Table S6.
4.5. Establishment of bibenzyl backbone-producing strain
AAE13 (AAP03025.1)34 and MatC (KF765784.1)35 were sequentially inserted into the Not I and EcoR V sites of pETDuet-1, resulting in the plasmid (designated as pETDuet-AAE13-MatC). At4CL1 (AY376729.1)36 and DoBBS8 were individually inserted into EcoR V site of pACYCDuet-1 and pCDFDuet-1, generating the plasmids pACYCDuet-At4CL1 and pCDFDuet-DoBBS8. With all of the recombinant plasmids co-transformed into E. coli BL21(DE3), the bibenzyl backbone-producing strain was generated and designated as E-DoBBS8. After induction for 16 h, the engineered E. coli cells were collected and washed twice with M9 minimal medium. Pellets were resuspended in M9 minimal medium with OD600 of 3.0. With the malonic acid and p-hydroxyphenylpropionic acid derivatives (S1–S3) feeding into the culture at a final concentration of 3.0 and 1.0 mmol/L, respectively, the whole-cell reactions were incubated at 30 °C and 200 rpm for 24 h, and analyzed by HPLC‒MS.
4.6. Screening of enzymes for bibenzyl backbone post-modification
The methyltransferase genes DoOMTs (DoOMT1−11) were amplified from D. officinale by PCR using gene-specific primer pairs (Supporting Information Table S4). The amplified DNA fragments were inserted into pET-28a vector and transformed into E. coli Transetta (DE3) for heterologous expression. For methyltransferase activity assays, the reaction mixture routinely consisted of 0.4 mmol/L methyl acceptor, 0.8 mmol/L methyl donor (SAM), 5 mmol/L MgCl2, and 1 mg/mL purified DoOMTs in 100 μL of reaction buffer (50 mmol/L Tris-HCl, 1 mmol/L DTT, 1.5% glycerol (v/v), pH 7.4). The cascade reaction catalyzed by At4CL1 and DoOMT6 was firstly initiated by the At4CL1 reaction, which consisted of 0.5 mmol/L S2, 2 mmol/L CoA, 4 mmol/L ATP, 5 mmol/L MgCl2, and 0.25 mg/mL purified At4CL1 in 100 μL of reaction buffer (50 mmol/L Tris-HCl, 1 mmol/L DTT, 1.5% glycerol (v/v), pH 7.4). After incubation at 30 °C for 30 min, the cascade reaction was pushed by supplement of 0.5 mmol/L SAM and 1 mg/mL purified DoOMT6, with the reaction volume adjustment to 200 μL. The supernatant was analyzed by HPLC‒MS on a Tosoh TSK gel ODS column (250 mm × 4.6 mm I.D., 5 μm) at a flow rate of 1 mL/min. The mobile phase was a gradient elution of solvent A (0.1% trifluoroacetic acid aqueous solution) and solvent B (acetonitrile). The gradient programs used for analysis were shown in Table S6.
The coding region of PsPT1 (GenBank No. OM809169) was amplified by RT-PCR from the cDNA of Periconia sp. F-3122 and subcloned into pET-28a vector and transformed into E. coli Transetta (DE3) for heterologous expression. The cloning and expression of GT gene UGT71E5 were conducted as previously reported23. GTs and PTs activity assays were performed similarly to DoOMTs. The reaction mixture routinely consisted of 0.4 mmol/L dihydro-resveratrol (1), 0.8 mmol/L glycosyl/prenyl donor (UDP-Glc/DMAPP) and 1 mg/mL purified GTs/PTs in 100 μL of reaction buffer [50 mmol/L Tris-HCl, 1 mmol/L DTT, 1.5% glycerol (v/v), pH 7.4].
4.7. Establishment of bibenzyl backbone post-modification modular strains
The coding region of metK45 was cloned from E. coli Transetta (DE3) and inserted into EcoR V site of pCDFDuet-1, after sequencing confirmation, the plasmid pCDFDuet-metK was then co-transformed into the E. coli BL21(DE3) with plasmids of pET-28a-DoOMTs, generating the methylation modular strains (designated as E-DoOMTs). The coding region of PsPT1 was cloned and inserted into EcoR V site of pCDFDuet-1, and the resultant plasmid pCDFDuet-PsPT1 was then co-transformed with plasmids of pXL13 and pXL1746 into BL21(DE3), generating the prenylation modular strain E-PsPT1. To establish glycosylation strain, an E. coli BL21(DE3) harboring the recombinant plasmid of pET-28a-UGT71E5 was constructed and designated as E-UGT71E5.
4.8. Structural diversification of bibenzyls through modular co-culture
Methylation modular strains E-DoOMTs, prenylation modular strain E-PsPT1, and glycosylation modular strain E-UGT71E5 were individually co-cultured with the bibenzyl backbone-producing strain E-DoBBS8 for the synthesis of corresponding mono-post-modified bibenzyls. Both E-DoOMT1 and/or E-DoOMT6 and E-UGT71E5 strains together with E-DoBBS8 were co-cultured for the synthesis of multi-post-modified bibenzyl derivatives. Similarly, E-UGT71E5 and E-PsPT1 were co-cultivated with E-DoBBS8 to yield both glycosylated and prenylated bibenzyls. Each modular strain was kept at OD600 of 5.0 and mixed up in equal volume, malonic acid and S1–S3 were fed at 4.5 and 1.5 mmol/L for E-DoBBS8. After being incubated at 30 °C and 200 rpm for 24 h, the reactions were quenched and analyzed by HPLC‒MS.
4.9. Preparative-scale reactions
The preparative-scale reactions were performed in 50 mL cultures. After incubation at 30 °C and 200 rpm for 24 h, the reaction mixture was centrifuged at 6000×g for 5 min, and the resultant supernatant was subjected to an Amberlite XAD-16 macroporous resin column chromatography and eluted with gradient ddH2O-ethanol. The 80% (v/v) ethanol fraction containing product was pooled, concentrated, and further purified by reversed-phase semi-preparative HPLC. The purified products were subjected to MS and NMR analysis. The detailed HPLC methods and spectroscopic data were shown in Supporting Information.
4.10. Optimization and recycling utilization of engineered strains for the synthesis of 12
For the optimal cell density, the reactions were performed in various cell densities with OD600 values in the range of 1.0–18.0. The concentration of S1 was fixed at 1.0 and 2.0 mmol/L, respectively, with three times malonic acid of S1, and incubated for 24 h. For the optimal concentration of substrate, the reactions were performed with 0.25–3.0 mmol/L of S1 with three times malonic acid at the cell density of OD600 5.0 for 24 h of incubation. For the optimal incubation time, the time course of the reaction in the range of 0–50 h was monitored with the cell density of 5.0, 1.0, and 3.0 mmol/L S1 and malonic acid. In the experiments of recycling utilization of co-cultured E-PsPT1 and E-DoBBS8, the culture mixtures were collected at 6000×g for 5 min after one batch of incubation, and resuspended in an equal volume M9 medium for the next batch. The supernatants were collected for HPLC analysis.
4.11. Neuroprotective assay
4.11.1. Glutamate-induced neuronal excitatory injury
Neuroblastama SK-N-SH cell line was a gift from Dr. Dennis Selkoe of Brigham and Woman's hospital, Harvard Medical School. SK-N-SH cells were cultured in 96-well microplates with DMEM media supplemented with 10% fetal bovine serum (FBS) and 1% penicillin streptomycin cocktail at a density of 7.5 × 103 cells/well. A panel of compound samples were prepared in DMSO as 100 mmol/L stock solutions. SK-N-SH cells were pre-incubated with compound samples for 4 h before 29 mmol/L glutamate to the cells. After 4 h of co-incubation, each well was added with MTT solution (final concentration 0.5 mg/mL) for another 4 h at 37 °C. After removing the culture medium, the MTT formazan crystals were solubilized with 150 μL DMSO and determined by a microplate reader at 570 nm. The cell viability was expressed as a percentage of the control group54.
4.11.2. Oxygen glucose deprivation/reoxygenation (OGD/R) injury
SK-N-SH cells were cultured in 96-well microplates at a density of 6.5 × 103 cells/well for 20 h. SK-N-SH cells were incubated with compound 12 at different concentration, and the medium were changed to low-glucose and serum free DMEM medium in hypoxia chamber containing 95% N2 and 5% CO2 at 37 °C for 4 h. Then low-glucose DMEM medium was changed to control medium for 4 h. Besides, control cells were placed in a normal incubator throughout the OGD/R process. The cell survival rate was determined by MTT method as above mentioned.
4.11.3. Permanent middle cerebral artery occlusion
Sprague–Dawley rats weighing 260–280 g were obtained from Beijing Vital River Laboratory Animal Technology Co., Ltd. (Beijing, China). Permanent middle cerebral artery occlusion (pMCAO) was induced as previously described55. Rats were anesthetized with isoflurane, and a midline incision were made in the neck. The right common carotid artery (CCA), external carotid artery (ECA), and internal carotid artery (ICA) were isolated. The proximal CCA and distal ECA were ligated. Then a cut was made on the ECA, and a 4–0 silicon coated monofilament suture (Φ 0.26 mm) was inserted from ECA to ICA until occlude the origin of the right middle cerebral artery. Sham-operated rats received the same surgical procedures without pMCAO. The pMCAO rats were randomly divided into three groups: vehicle group, compound 12 10 mg/kg group, and compound 12 30 mg/kg group. Compound 12 and control solvent were administered by intraperitoneal injections 5 min after surgery. Rats were housed in plastic cages at 23 ± 1 °C with a 12 h light/dark cycle and free access to water and food. All experiments were approved and performed in accordance with the National Institute of Health Guidelines for the Care and Use of Laboratory Animals and approved by the institutional Ethics Committee of the Experimental Animal Center of the Chinese Academy of Medical Sciences (Beijing, China).
4.11.4. Measurement of cerebral infarction
The brain samples were collected at 24 h after cerebral ischemia, and sliced into 2-mm-thick coronal slices and immediately stained with 1% triphenylte-trazoliumchloride (TTC) solution at 37 °C for 40 min. Cerebral infarction volume measurements were performed as described previously56. The infarct volume was calculated as a percentage according to Eq. (1):
| Infarct volume (%) = (area of contralateral hemisphere − area of non-infarcted region of ipsilateral hemisphere)/area of contralateral hemisphere × 100 | (1) |
4.11.5. Neurological deficit assessment
Neurological deficit assessment was performed using Bederson score system by an assessor who was blinded to the experimental groups. Bederson scoring system evaluated forelimb flexion, resistance to lateral push and circling behavior which was graded 0–4 according to following criteria: 0, no neurological deficit; 1, flexion of contralateral forelimb; 2, severe forelimb flexion and decreased resistance to lateral push without circling; 3, unidirectional circling; and 4, loss of spontaneous motor activity and a depressed level of consciousness57.
4.11.6. RNA-seq
Total RNA was extracted and the constructed cDNA library was sequenced using BGIseq 500 (BGI-Shenzhen, China). The reads were mapped to Ensembl rat (mRatBN7.2) reference genome using HISAT2 (v2.0.4). Differential expression analysis was performed using the DESeq2 (v1.4.5) with Q value less than 0.05.
4.12. Data availability
The authors declare that all data supporting the findings of this study are available within the paper and Supporting Information, including experimental details, characterization data, and NMR spectra (Supporting Information Spectra S32‒106) of all prepared compounds. All the data are available from the authors upon reasonable request. Source data are provided with this paper. Sequence data from this article can be found in GenBank under the following accession numbers: OM809166 for DoOMT1, OM809167 for DoOMT6, OM809168 for DoBBS8, OM809169 for PsPT1.
Acknowledgments
This work was supported by the National Key Research and Development Program of China (2020YFA0908000), CAMS Innovation fund for Medical Sciences (CIFMS-2021-I2M-1-028 and CIFMS-2021-I2M-1-029, China), Beijing Key Laboratory of non-Clinical Drug Metabolism and PK/PD Study (Z141102004414062, China). We are thankful for Prof. Shepo Shi (Beijing University of Chinese Traditional Medicine) to gift genes At4CL1 and AAE13, and Prof. Yong Wang (Institute of Plant Physiology and Ecology, Chineses Academy of Sciences) to kindly provide the plasmids pXL13 and pXL17.
Author contributions
Jungui Dai, Ying Peng, and Ridao Chen. conceived this project. Yuyu Liu, Ridao Chen, Dawei Chen, and Kebo Xie. conducted the cloning and expression of BBS and PT genes, protein purification, biochemical kinetics. Yuyu Liu contributed to the construction of modular strains, evaluation of the capacity of enzymes and strains, and co-culture. Yuyu Liu, Songyang Sui, and Jimei Liu contributed to biosynthesis, isolation, and structural determination of the enzymatic products. Xinnan Li, Jingshu Tang, Yuying Kang, Jiaqi Lan and Lei Wu contributed to all the pharmacological work. All authors discussed the results and wrote the manuscript.
Conflicts of interest
The authors declare no conflicts of interest.
Footnotes
Peer review under responsibility of Chinese Pharmaceutical Association and Institute of Materia Medica, Chinese Academy of Medical Sciences.
Supporting data to this article can be found online at https://doi.org/10.1016/j.apsb.2022.10.007.
Contributor Information
Ridao Chen, Email: chenridao@imm.ac.cn.
Ying Peng, Email: ypeng@imm.ac.cn.
Jungui Dai, Email: jgdai@imm.ac.cn.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.He L., Su Q., Bai L., Li M., Liu J., Liu X., et al. Recent research progress on natural small molecule bibenzyls and its derivatives in Dendrobium species. Eur J Med Chem. 2020;204:112530–112546. doi: 10.1016/j.ejmech.2020.112530. [DOI] [PubMed] [Google Scholar]
- 2.Chen H., Huang Y., Huang J., Lin L., Wei G. Gigantol attenuates the proliferation of human liver cancer HepG2 cells through the PI3K/Akt/NF-κB signaling pathway. Oncol Rep. 2017;37:865–870. doi: 10.3892/or.2016.5299. [DOI] [PubMed] [Google Scholar]
- 3.Lee E., Han A.R., Nam B., Kim Y.R., Jin C.H., Kim J.B., et al. Moscatilin induces apoptosis in human head and neck squamous cell carcinoma cells via JNK signaling pathway. Molecules. 2020;25:901–912. doi: 10.3390/molecules25040901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li X., Chen H., He Y., Chen W., Chen J., Gao L., et al. Effects of rich-polyphenols extract of Dendrobium loddigesii on anti-diabetic, anti-inflammatory, anti-oxidant, and gut microbiota modulation in db/db mice. Molecules. 2018;23:3245–3264. doi: 10.3390/molecules23123245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lai M., Liu W., Liou S., Liu I. A bibenzyl component moscatilin mitigates glycation-mediated damages in an SH-SY5Y cell model of neurodegenerative diseases through AMPK activation and RAGE/NF-κB pathway suppression. Molecules. 2020;25:4574–4589. doi: 10.3390/molecules25194574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Huang J.M., Huang F.I., Yang C.R. Moscatilin ameliorates tau phosphorylation and cognitive deficits in Alzheimer's disease models. J Nat Prod. 2019;82:1979–1988. doi: 10.1021/acs.jnatprod.9b00375. [DOI] [PubMed] [Google Scholar]
- 7.Cioffi G., Montoro P., Ugaz O.L.D., Vassallo A., Severino L., Pizza C., et al. Antioxidant bibenzyl derivatives from Notholaena nivea Desv. Molecules. 2011;16:2527–2541. doi: 10.3390/molecules16032527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Allergrone G., Pollastro F., Magagnini G., Taglialatela-Scafati J., Seegers J., Koeberle A., et al. The bibenzyl canniprene inhibits the production of pro-inflammatory eicosanoids and selectively accumulates in some Cannabis sativa strains. J Nat Prod. 2017;80:731–734. doi: 10.1021/acs.jnatprod.6b01126. [DOI] [PubMed] [Google Scholar]
- 9.Cretton S., Oyarzún A., Righi D., Sahib L., Kaiser M., Christen P., et al. A new antifungal and antiprotozoal bibenzyl derivative from Gavilea lutea. Nat Prod Res. 2018;32:695–701. doi: 10.1080/14786419.2017.1338287. [DOI] [PubMed] [Google Scholar]
- 10.Chen C., Wu L.G., Ko F.N., Teng C.M. Antiplatelet aggregation principles of Dendrobium loddigesii. J Nat Prod. 1994;57:1271–1274. doi: 10.1021/np50111a014. [DOI] [PubMed] [Google Scholar]
- 11.Khoonrit P., Mirdogan A., Dehlinger A., Mekboonsonglarp W., Likhitwitayawuid K., Priller J., et al. Immune modulatory effect of a novel 4,5-dihydroxy-3,3′,4′-trimethoxybibenzyl from Dendrobium lindleyi. PLoS One. 2020;15 doi: 10.1371/journal.pone.0238509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang H., Xie X., Li G., Chen J., Li M., Xu X., et al. Erianin inhibits human lung cancer cell growth via PI3K/Akt/mTOR pathway in vitro and in vivo. Phytother Res. 2021;35:4511–4525. doi: 10.1002/ptr.7154. [DOI] [PubMed] [Google Scholar]
- 13.Zhang C., Gao Y., Zhu R., Qiao Y., Zhou J., Zhang J., et al. Prenylated bibenzyls from the Chinese liverwort Radula constricta and their mitochondria-derived paraptotic cytotoxic activities. J Nat Prod. 2019;82:1741–1751. doi: 10.1021/acs.jnatprod.8b00897. [DOI] [PubMed] [Google Scholar]
- 14.Asakawa Y., Nagashima F., ludwiczuk A. Distribution of bibenzyls, prenyl bibenzyls, bis-bibenzyls, and terpenoids in the liverwort genus Radula. J Nat Prod. 2020;83:756–769. doi: 10.1021/acs.jnatprod.9b01132. [DOI] [PubMed] [Google Scholar]
- 15.Lee K.Y., Sung S.H., Kim Y.C. Neuroprotective bibenzyl glycosides of Stemona tuberosa roots. J Nat Prod. 2006;69:679–681. doi: 10.1021/np0504154. [DOI] [PubMed] [Google Scholar]
- 16.Chong J., Poutaraud A., Hugueney P. Metabolism and roles of stilbenes in plants. Plant Sci. 2009;177:143–155. [Google Scholar]
- 17.Friederich S., Maier U.H., Deus-Neumann B., Asakawa Y., Zenk M.H. Biosynthesis of cyclic bis(bibenzyls) in Marchantia polymorpha. Phytochemistry. 1999;50:589–598. [Google Scholar]
- 18.Zhang H., Pereira B., Li Z., Sterphanopoulos G. Engineering Escherichia coli coculture systems for the production of biochemical products. Proc Natl Acad Sci U S A. 2015;112:8266–8271. doi: 10.1073/pnas.1506781112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhou K., Qiao K., Edgar S., Stephanopoulos G. Distributing a metabolic pathway among a microbial consortium enhances production of natural products. Nat Biotechnol. 2015;33:377–383. doi: 10.1038/nbt.3095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Agapakis C., Boyle P., Silver P. Natural strategies for the spatial optimization of metabolism in synthetic biology. Nat Chem Biol. 2012;8:527–535. doi: 10.1038/nchembio.975. [DOI] [PubMed] [Google Scholar]
- 21.Jones J.A., Wang X. Use of bacterial co-cultures for the efficient production of chemicals. Curr Opin Biotechnol. 2018;53:33–38. doi: 10.1016/j.copbio.2017.11.012. [DOI] [PubMed] [Google Scholar]
- 22.Zhang D., Tao X., Chen R., Liu J., Li L., Fang X., et al. Pericoannosin A, a polyketide synthase–nonribosomal peptide synthetase hybrid metabolite with new carbon skeleton from the endophytic fungus Periconia sp. Org Lett. 2015;17:4304–4307. doi: 10.1021/acs.orglett.5b02123. [DOI] [PubMed] [Google Scholar]
- 23.Xie K., Chen R., Chen D., Li J., Wang R., Yang L., et al. Enzymatic N-glycosylation of diverse arylamine aglycones by a promiscuous glycosyltransferase from Carthamus tinctorius. Adv Synth Catal. 2017;359:603–608. [Google Scholar]
- 24.Fliegmann J., Schröder G., Schanz S., Britsch L., Schröder J. Molecular analysis of chalcone and dihydropinosylvin synthase from Scots pine (Pinus sylvestris), and differential regulation of these and related enzyme activities in stressed plants. Plant Mol Biol. 1992;18:489–503. doi: 10.1007/BF00040665. [DOI] [PubMed] [Google Scholar]
- 25.Preisigmuller R., Gnau P., Kindl H. The inducible 9,10-dihydrophenanthrene pathway: characterization and expression of bibenzyl synthase and S-adenosylhomocysteine hydrolase. Arch Biochem Biophys. 1995;317:201–207. doi: 10.1006/abbi.1995.1154. [DOI] [PubMed] [Google Scholar]
- 26.Ng T.B., Liu J., Wong J.H., Ye X., Sze S.C.W., Tong Y., et al. Review of research on Dendrobium, a prized folk medicine. Appl Microbiol Biotechnol. 2012;93:1795–1803. doi: 10.1007/s00253-011-3829-7. [DOI] [PubMed] [Google Scholar]
- 27.Li L., Jiang Y., Liu Y., Niu Z., Xue Q., Liu W., et al. The large single-copy (LSC) region functions as a highly effective and efficient molecular marker for accurate authentication of medicinal Dendrobium species. Acta Pharm Sin B. 2020;10:1989–2001. doi: 10.1016/j.apsb.2020.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Niu Z., Zhu F., Fan Y., Li C., Zhang B., Zhu S., et al. The chromosome-level reference genome assembly for Dendrobium officinale and its utility of functional genomics research and molecular breeding study. Acta Pharm Sin B. 2021;11:2080–2092. doi: 10.1016/j.apsb.2021.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Reinecke T., Kindl H. Characterization of bibenzyl synthase catalysing the biosynthesis of phytoalexins of orchids. Phytochemistry. 1993;35:63–66. [Google Scholar]
- 30.Flores-Sanchez I.J., Verpoorte R. Plant polyketide synthases: a fascinating group of enzymes. Plant Physiol Biochem. 2009;47:167–174. doi: 10.1016/j.plaphy.2008.11.005. [DOI] [PubMed] [Google Scholar]
- 31.Austin M.B., Bowman M.E., Ferrer J.L., Schröder J., Noel J.P. An aldol switch discovered in stilbene synthases mediates cyclization specificity of type III polyketide synthases. Chem Biol. 2004;11:1179–1194. doi: 10.1016/j.chembiol.2004.05.024. [DOI] [PubMed] [Google Scholar]
- 32.Abe I., Morita H. Structure and function of the chalcone synthase superfamily of plant type III polyketide synthases. Nat Prod Rep. 2010;27:809–838. doi: 10.1039/b909988n. [DOI] [PubMed] [Google Scholar]
- 33.Waki T., Mameda R., Nakano T., Yamada S., Terashita M., Ito K., et al. A conserved strategy of chalcone isomerase-like protein to rectify promiscuous chalcone synthase specificity. Nat Commun. 2020;11:1–14. doi: 10.1038/s41467-020-14558-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen H., Kim H.U., Weng H., Browse J. Malonyl-CoA synthetase, encoded by acyl activating enzyme 13, is essential for growth and development of Arabidopsis. Plant Cell. 2011;23:2247–2262. doi: 10.1105/tpc.111.086140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.An J.H., Kim Y.S. A gene cluster encoding malonyl-CoA decarboxylase (MatA), malonyl-CoA synthetase (MatB) and a putative dicarboxylate carrier protein (MatC) in Rhizobium trifolii: cloning, sequencing, and expression of the enzymes in Escherichia coli. Eur J Biochem. 1998;257:395–402. doi: 10.1046/j.1432-1327.1998.2570395.x. [DOI] [PubMed] [Google Scholar]
- 36.Ehlting J., Büttner D., Wang Q., Douglas C.J., Somssich I.E., Kombrink E. Three 4-coumarate: coenzyme A ligases in Arabidopsis thaliana represent two evolutionarily divergent classes in angiosperms. Plant J. 1999;19:9–20. doi: 10.1046/j.1365-313x.1999.00491.x. [DOI] [PubMed] [Google Scholar]
- 37.Lam K.C., Ibrahim R.K., Behdad B., Dayanandan S. Structure, function, and evolution of plant O-methyltransferases. Genome. 2007;50:1001–1013. doi: 10.1139/g07-077. [DOI] [PubMed] [Google Scholar]
- 38.Liscombe D.K., Louie G.V., Noel J.P. Architectures, mechanisms and molecular evolution of natural product methyltransferases. Nat Prod Rep. 2012;29:1238–1250. doi: 10.1039/c2np20029e. [DOI] [PubMed] [Google Scholar]
- 39.Chen R., Gao B., Liu X., Ruan F., Zhang Y., Lou J., et al. Molecular insights into the enzyme promiscuity of an aromatic prenyltransferase. Nat Chem Biol. 2017;13:226–234. doi: 10.1038/nchembio.2263. [DOI] [PubMed] [Google Scholar]
- 40.Xie K., Chen R., Chen D., Li J., Wang R., Yang L., et al. Exploring the catalytic promiscuity of a new glycosyltransferase from Carthamus tinctorius. Org Lett. 2014;16:4874–4877. doi: 10.1021/ol502380p. [DOI] [PubMed] [Google Scholar]
- 41.Chen D., Chen R., Wang R., Li J., Xie K., Bian C., et al. Probing the catalytic promiscuity of a regio- and stereospecific C-glycosyltransferase from Mangifera indica. Angew Chem Int Ed. 2015;127:12869–12873. doi: 10.1002/anie.201506505. [DOI] [PubMed] [Google Scholar]
- 42.Chen D., Fan S., Chen R., Xie K., Yin S., Sun L., et al. Probing and engineering key residues for bis-C-glycosylation and promiscuity of a C-glycosyltransferase. ACS Catal. 2018;8:4917–4927. [Google Scholar]
- 43.Sui S., Guo R., Xie K., Yang L., Dai J. UGT88B2: a promiscuous O-glycosyltransferase from Carthamus tinctorius. Chin Herb Med. 2020;12:440–445. doi: 10.1016/j.chmed.2020.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chen D., Sun L., Chen R., Xie K., Yang L., Dai J. Enzymatic synthesis of acylphloroglucinol 3-C-glucosides from 2-O-glucosides using a C-glycosyltransferase from Mangifera indica. Chem Eur J. 2016;22:5873–5877. doi: 10.1002/chem.201600411. [DOI] [PubMed] [Google Scholar]
- 45.Markham G.D., DeParasis J., Gatmaitan J. The sequence of metK, the structural gene for S-adenosylmethionine synthetase in Escherichia coli. J Biol Chem. 1984;259:14505–14507. [PubMed] [Google Scholar]
- 46.Wang J., Li S., Xiong Z., Wang Y. Pathway mining-based integration of critical enzyme parts for de novo biosynthesis of steviolglycosides sweetener in Escherichia coli. Cell Res. 2016;26:258–261. doi: 10.1038/cr.2015.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yang M.H., Chin Y.W., Yoon K.D., Kim J. Phenolic compounds with pancreatic lipase inhibitory activity from Korean yam (Dioscorea opposita) J Enzym Inhib Med Chem. 2014;29:1–6. doi: 10.3109/14756366.2012.742517. [DOI] [PubMed] [Google Scholar]
- 48.Wang S., Han Q., Zhou T., Zhang C., Zhu C., Zhou X., et al. A bibenzyl compound 20C protects rats against 6-OHDA-induced damage by regulating adaptive immunity associated molecules. Int Immunopharmacol. 2020;91 doi: 10.1016/j.intimp.2020.107269. [DOI] [PubMed] [Google Scholar]
- 49.Yang T., Sun Y., Mao L., Zhang M., Li Q., Zhang L., et al. Brain ischemic preconditioning protects against ischemic injury and preserves the blood‒brain barrier via oxidative signaling and Nrf2 activation. Redox Biol. 2018;17:323–337. doi: 10.1016/j.redox.2018.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang Y., Shi Z., Zhou Y., Xiao Q., Wang H., Peng Y. Emerging substrate proteins of Kelch-like ECH associated protein 1 (Keap1) and potential challenges for the development of small-molecule inhibitors of the keap1-nuclear factor erythroid 2-related factor 2 (Nrf2) protein-protein interaction. J Med Chem. 2020;63:7986–8002. doi: 10.1021/acs.jmedchem.9b01865. [DOI] [PubMed] [Google Scholar]
- 51.Novo N., Ferreira P., Medina M. The apoptosis-inducing factor family: moonlighting proteins in the crosstalk between mitochondria and nuclei. IUBMB Life. 2021;73:568–581. doi: 10.1002/iub.2390. [DOI] [PubMed] [Google Scholar]
- 52.Wang J., Wang X., Liu X., Li J., Shi X., Song Y., et al. Synthesis of unnatural 2-substituted quinolones and 1,3-diketones by a member of type III polyketide synthases from Huperzia serrata. Org Lett. 2016;18:3550–3553. doi: 10.1021/acs.orglett.6b01501. [DOI] [PubMed] [Google Scholar]
- 53.Sun J., Liu J., Chen R., Liu Y., Li Y., Cen S., et al. Study on chemical bibenzyls in Dendrobium gratiosissimum. China J Chin Mater Med. 2020;45:4929–4937. doi: 10.19540/j.cnki.cjcmm.20200523.201. [DOI] [PubMed] [Google Scholar]
- 54.Wang R., Chen R., Li J., Liu X., Xie K., Chen D., et al. Regiospecific prenylation of hydroxyxanthones by a plant flavonoid prenyltransferase. J Nat Prod. 2016;79:2143–2147. doi: 10.1021/acs.jnatprod.6b00417. [DOI] [PubMed] [Google Scholar]
- 55.Feng H., Hu L., Zhu H., Tao L., Wu L., Zhao Q., et al. Repurposing antimycotic ciclopirox olamine as a promising anti-ischemic stroke agent. Acta Pharm Sin B. 2020;10:434–446. doi: 10.1016/j.apsb.2019.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Peng Y., Xu S., Wang L., Feng Y., Wang X. Effects of chiral NBP on cerebral infarct volume due to transient focal cerebral ischemia. Chin J New Drugs. 2005;14:420–423. [Google Scholar]
- 57.Bederson J.B., Pitts L.H., Tsuji M., Nishimura M.C., Davis R.L., Bartkowski H. Rat middle cerebral artery occlusion: evaluation of the model and development of a neurologic examination. Stroke. 1986;17:472–476. doi: 10.1161/01.str.17.3.472. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The authors declare that all data supporting the findings of this study are available within the paper and Supporting Information, including experimental details, characterization data, and NMR spectra (Supporting Information Spectra S32‒106) of all prepared compounds. All the data are available from the authors upon reasonable request. Source data are provided with this paper. Sequence data from this article can be found in GenBank under the following accession numbers: OM809166 for DoOMT1, OM809167 for DoOMT6, OM809168 for DoBBS8, OM809169 for PsPT1.