Abstract
Exosome is an excellent vesicle for in vivo delivery of therapeutics, including RNAi and chemical drugs. The extremely high efficiency in cancer regression can partly be attributed to its fusion mechanism in delivering therapeutics to cytosol without endosome trapping. However, being composed of a lipid-bilayer membrane without specific recognition capacity for aimed-cells, the entry into nonspecific cells can lead to potential side-effects and toxicity. Applying engineering approaches for targeting-capacity to deliver therapeutics to specific cells is desirable. Techniques with chemical modification in vitro and genetic engineering in cells have been reported to decorate exosomes with targeting ligands. RNA nanoparticles have been used to harbor tumor-specific ligands displayed on exosome surface. The negative charge reduces nonspecific binding to vital cells with negatively charged lipid-membrane due to the electrostatic repulsion, thus lowering the side-effect and toxicity. In this review, we focus on the uniqueness of RNA nanoparticles for exosome surface display of chemical ligands, small peptides or RNA aptamers, for specific cancer targeting to deliver anticancer therapeutics, highlighting recent advances in targeted delivery of siRNA and miRNA that overcomes the previous RNAi delivery roadblocks. Proper understanding of exosome engineering with RNA nanotechnology promises efficient therapies for a wide range of cancer subtypes.
KEY WORDS: RNA interference, RNA nanotechnology, Endolysosome trapping, Exosome engineering, Targeted delivery, Chemical drug delivery
Graphical abstract
This review illustrated the RNA-decorated exosome to display specific ligand/marker for targeted delivery of therapeutics to cells, thus ensuring safety & reducing cytotoxicity.

1. Introduction
RNA interference (RNAi) has held great potential for therapeutic applications by specific gene suppression since the inception of the concept1,2. Small interfering RNA (siRNA) is advantageous over small molecules and monoclonal antibodies because siRNA executes its function by complete Watson–Crick base pairing with mRNA. In contrast, small molecule and monoclonal antibody drugs need to recognize the complicated spatial conformations of the target proteins1. Last few decades have resulted in the first approved siRNA therapy Onpattro (patisiran), for treating hereditary amyloidogenic transthyretin (hATTR) amyloidosis with polyneuropathy in adults3,4. Nanotechnology applied to therapeutics is leading to a new era of cancer treatment by reducing systemic toxicity, overcoming drug resistance, and providing improved platforms for combination therapy5. However, delivery strategies of RNAi to cancer cells include cationic lipids, cationic liposomes, and cationic polymers, but specific targeting is still challenging6. Moreover, the therapeutic efficiency of nanoparticle-mediated delivery is often compromised due to endocytosis. Endocytosis often leads to nanoparticle entrapment followed by endolysosomal degradation.
Exosomes have recently been highlighted as an excellent therapeutic vesicle for in vivo delivery of therapeutics, including RNAi and chemical drugs, for the treatment of diseases. It is extremely efficient in cancer regression. However, without specific recognition capacity for cancer cells, the entry to nonspecific cells can lead to potential side effects and toxicity. It is desirable to provide a targeted ability to deliver therapeutics to specific cells. Approaches with chemical modification in vitro and genetic engineering in cells have been reported to decorate exosomes with targeting ligands. Due to the advantage of negative charge and multivalency, RNA nanoparticles have been used to harbor tumor-specific ligands displayed on the surface of the exosome7.
In most nano-delivery systems, endocytosis mediates cellular uptake of RNAi therapeutics (miRNA/anti-miRNA/siRNA). However, endosome entrapment of RNAi prevents interactions with RNA-induced silencing complex (RISC) and its targeting of mRNA located in the cytosol, which is one of the most well-recognized setbacks for the success of RNAi therapeutics8,9. An appealing strategy to address endosome entrapment is to learn from nature. Nature has evolved intelligent nanomachines such as exosomes as a class of extracellular vesicles (EVs). Exosomes are derived from the late endosomal route/multivesicular body (MVB). Typically, these nanovesicles range from 30 to 150 nm in diameter10,11. Recent interest has been focused on harnessing exosomes as nanocarriers for RNAi delivery. These nanovesicles show unique membrane properties (chemical composition such as lipid, peptides, protein, etc.), offering the natural ability to fuse with the recipient membrane of the cellular organelles12, 13, 14. However, their lack of cell targetability/specificity has led to poor therapeutic efficacy and potential toxicity13,14. Exosome specificity can be enhanced for cancer treatment by embracing RNA nanotechnology.
In this review, we will discuss the unique properties of RNA nanotechnology and summarize its potential application in exosome display for specific targeting. We will focus on combining the advantages of two worlds: i) RNA nanotechnology for specific targeting and ii) exosome for highly efficient cell entry and endosome escape. Here, we demonstrate the advancement of targeted drug delivery to cancer cells using RNA-ligand displaying exosomes. This review broadly addresses two therapeutic challenges in cancer: the lack of specific therapeutic targeting to diseased cells and endolysosome entrapment of treatment payload after cell entry.
2. Methods for displaying ligands on the exosome surface
The idea of exosomes as a biological marker for the long-distance organ communication system has generated substantial interest in the scientific community. Exosomes have been shown to naturally traffic miRNA cargos to deliver to other cells during intracellular communications15, 16, 17. Generally, cancer cells navigate their metastatic materials to increase the burden of metastatic progression18. Readers are referred to other review papers addressing exosome biogenesis, biochemical characteristics, or stimulus condition19, 20, 21, 22. In this section, we will briefly highlight the most recent research on engineered exosomes in cancer therapy. Exosomes have great potential as delivery vectors because they are easy to extract, well-tolerated in vivo, and nano-sized to cross biological barriers13,14,23,24. These vesicles can become immunogenically inert and fuse directly with cell membranes through tetraspanin domains. Exosomes can also merge with endosome membranes following receptor-mediated endocytosis. The immense possibility exhibited by the exosome is that on clinicaltrials.gov, almost 100 clinical trials are currently being conducted worldwide targeting different diseases. The most recent use of exosomes in cancer research involves cancer diagnosis, identification of cancer type, and targeted drug delivery as therapy. For example, exosome protein profiling fairly separates patients of all stages, cancer types, and histological subtypes in cancer diagnosis25,26. Exosomes could also be utilized as a potential strategy for future therapeutic tumor vaccination. These nanoparticles can induce tumor-specific immunity and prevent tumor development, and might well provide an optimized, individual-specific antigen source27, 28, 29, 30, 31. Exosomes can be engineered to express a cell-surface-specific ligand that can be used to achieve targeted delivery to cancer cells. In order to reprogram exosomes for specific applications, two approaches are mainly employed to design and manipulate the membrane surface of exosomes. The modifications include (i) in vitro approach with chemical attachment on the surface of the EVs or (ii) genetic engineering directly into cells to express EVs with desired molecules via gene transfection (see graphical abstract)32.
2.1. In vitro chemical approach for displaying ligands on the exosome surface
The in vitro approaches include the use of RNA/DNA aptamers (see Section 3.2), chemical ligands, or short peptides (see below) as ligands to display on the exosome surface. The general methods of exosome loading, and surface decoration are depicted in Fig. 1. It has been shown that many chemicals, such as folate33 have specificity in binding to cell receptors. Thus, chemicals have been used as ligands that are conjugated to RNA nanoparticles for display on the surface of the exosome6,7,34, 35, 36, 37. In vitro approach starts with the harvested EVs from immortalized cells or plant sources. Exosome sources are critical for better production, purification, and reproducible yield. Followed by exosome collection from the culture medium, ultracentrifugation or filtration yields pure exosomes. Most frequently, density gradient ultracentrifugation is used to collect desired-sized exosomes. The strategies commonly practiced for producing exosomes are highlighted in Fig. 2i and ii. Readers are advised to read other literature on detailed exosome production and purification procedures38,39. After collecting and purifying the EVs from cell culture media, chemical modification is done on the exosome surface. Chemical modifications of exosomes utilize conjugation reactions such as click chemistry, hydrophobic insertion, receptor-ligand binding, etc. Chemical modification of the exosome surface presented a synergistic therapeutic effect and allowed mechanistic studies both in vitro and in vivo. Examples include CD63 specific exosome marker peptide CP0540, neuropilin-1-targeted peptide RGE41, immunomodulatory protein FasL42, bone marrow stromal cell (BMSC)-specific aptamer43. Chemically modified exosomes are reported to successfully deliver therapeutic cargos such as curcumin41,44, chemotherapeutic drugs45, 46, 47, 48, photosensitizer49, and quantum dots photothermal agents50. For example, Jia et al.41 used a safe blood‒brain barrier (BBB)-traversing drug delivery system for glioma treatment. In this study, superparamagnetic iron oxide nanoparticles and curcumin were loaded into exosomes. Then the exosome membrane was conjugated with neuropilin-1-targeted peptide (RGERPPR, RGE) by click chemistry. Thus glioma-targeting exosomes with imaging and therapeutic functions were obtained. This approach significantly improved diagnostic and therapeutic effects on glioma while reducing the side effects.
Figure 1.
Exosome loading and decoration process. For simplifying the discussion, RNA nanoparticles were used to depict the general process. Applying different techniques, exosomes can be loaded with anticancer drugs/RNAi while surface displayed with ligands/markers (figure prepared with Biorender.com).
Figure 2.
Exosome production and administration. Schematic presentation of the production, harvest, and administration of exosomes for targeted anticancer drug/siRNA delivery. Shown here is i) in vitro exosome production, ii) in vitro exosome membrane decoration, and iii) genetic engineering approach on cells commonly used in research laboratories (figure prepared with Biorender.com).
2.2. Genetic engineering approach in cells for displaying ligands/markers on the exosome surface
Genetic engineering as an indirect approach is another way of efficient exosome modification. Although challenging, transformation with targeting molecules can be achieved by engineering exosome-producing cells via transfection with plasmid vectors (Fig. 2iii). Typically, cells or tissues are collected directly from patients, murine models, or immortalized cells. These cells are then harvested in desired media specific to the experiment, such as cerebrospinal fluid and fetal bovine/calf serum. After certain confluency on the culture plate, the cells are transfected with plasmids encoding molecules particular to the cell membrane or intracellular organelle binding moieties. There are mainly two different categories of transmembrane proteins commonly found on the surface of the exosomes. Lysosome-associated membrane protein 2b (Lamp2b) is the most frequently found nonspecific protein compound on the surface of the exosome32. Alvarez et al.51 demonstrated a source of ‘self’ exosomes to load them with therapeutics, such as chemically modified siRNAs, and the ability to introduce targeting moieties. In the study, plasmids encoding the Lamp2b constructs were transfected into the dendritic cells four days before exosome purification. Thus, FLAG-Lamp2b was strongly expressed in dendritic cells and was incorporated into the dendritic cell-derived exosomes. Tetraspanin superfamily CD63/CD9/CD81 is another class of primary compounds observed on the exosome surface32. Studies reported that the T7 peptide-decorated exosomes and RAGE-binding peptide to an exosome membrane integral protein Lamp2b exhibited a higher intracellular delivery efficacy compared to unmodified exosomes52,53. Sutaria et al.54 have engineered HEK293T cells to secrete EVs by overexpressing Lamp-2a protein containing liver cancer targeting peptide PC94 and therapeutic pre-miR-199. The novel feature of the system was that both the cargo and the microvesicles were synthesized by the same cells, thus did not require the loading of synthetic oligonucleotides into EVs. The data demonstrates that stably transfecting HEK293T cells with vectors expressing therapeutic miRNAs dramatically increases their loading into EVs and thus improve anticancer efficacy54. Li et al.55 engineered exosomes for RNA loading by fusing CD9 with an RNA binding protein HuR that interacts with miR-155 with a relatively high affinity. The study revealed that the engineered exosomes could efficiently accumulate in the recipient cells with a higher recognition of the endogenous targets. Literature also reports decoy exosomes inserting TNFR1-EC at the 2nd extracellular loop of CD63. The incorporation of TNFR1-EC antagonizes the activity of TNFα in cellular inflammation models explicitly56. Other approaches including receptor membrane proteins present on specific exosomes such as epidermal growth factor receptor (EGFR)57, Glycosylphosphatidylinositol (GPI)57, HER258, platelet-derived growth factor receptor (PDGFR)59, the C1C2 domain of lactadherin44,60, also reported to exhibit enhanced binding to cancer cells, and improved targeting capabilities in RNAi mediated cancer diagnosis and treatment.
Taken together, the future of this burgeoning field demonstrates tremendous potential in cancer diagnosis and treatment. For example, pancreatic cancer is one of the most lethal malignancies, with an overall 5-year survival rate of approximately 9%61. Fortunately, with the aid of exosome engineering, we can approach almost all incurable diseases under treatment. Kamerkar et al.62 demonstrated an approach for direct and specific targeting of oncogenic KRAS in pancreatic tumors. For this work, exosomes were derived from normal fibroblast-like mesenchymal cells. Thus, the exosomes were engineered to carry siRNA or shRNA specific to oncogenic KrasG12D. The study reported that the engineered exosomes show CD47-dependent efficacy specific to these mutated pancreatic cancer cells62. CRISPR/Cas9 mediated genome editing is a promising platform in cancer therapy parallel to RNAi-mediated gene silencing. Engineered exosomes also show promise in CRISPR/Cas9 genome editing for targeted therapy. Exosomes loaded with CRISPR/Cas9 can target the mutant oncogenic allele in cancer cells. This parallel approach can suppress cancer proliferation and inhibit tumor growth63, 64, 65.
3. Advantages and approaches for using RNA nanotechnology to display ligands on the exosome surface
This section will highlight the advantages and unique properties of RNA nanotechnology and summarizes its potential application in nanomedicine, serving as a bridge between RNA nanotechnology and ligands displayed on the surface of the exosome.
3.1. RNA nanoparticles are stable in vivo & allow the construction of nanoparticles with different sizes and shapes
RNA nanotechnology, conceived in 199866, is the construction of nanoparticles composed of short oligo strands primarily of ribonucleic acid, including the core framework and functional modules67, 68, 69. Since its discovery, the field has expanded toward building RNA nanoparticles with the potential for clinical applications. RNA nanoparticles for in vivo use necessitate high thermodynamic stability to not dissociate in harsh conditions or within the body during circulation. For example, the ideal Cas9: sgRNA ratio for systemic Cas9-mediated editing of endothelial cells depends on the two molecules' relative stability70. RNA in the biological cell is typically a single-stranded molecule. The 2ʹ-OH on the ribose sugar is susceptible to hydrolysis and degradation, making the RNA less stable. Fortunately, the advantage of RNA over other nanoparticle platforms is that the bioavailability and stability could be modulated to achieve the desired function. The Zamore laboratory demonstrated that the stability of PIWI-interacting RNAs (piRNAs), that silence transposons71, depend on 3ʹ terminal trimming and 2ʹ-O-methylation72. According to the study, complementarity-dependent destabilization of piRNAs is blocked by 3ʹ terminal 2ʹ-O-methylation and does not require base pairing to both the piRNA seed and the 3ʹ sequence. Barnaby et al.73 have found a rapid endonuclease hydrolysis at specific sites near gold nanoparticle-facing terminus of siRNA. For their experiment, a spherical nucleic acid conjugate was prepared using gold nanoparticles with siRNA chemisorbed on the surface. The study indicates that the chemical environment of siRNA on a nanoparticle surface can alter the recognition of siRNA by serum nucleases and change the inherent stability of the nucleic acid. However, incorporation of 2′-O-methyl RNA nucleotides at sites of nuclease hydrolysis on siRNA results in a 10-fold increase in siRNA lifetime73. The Guo laboratory has discovered a thermostable high melting temperature (Tm) three-way junction (3WJ) that has allowed for the construction of tens of highly stable RNA nanoparticles (Fig. 3)74,75. Furthermore, inclusion of 2ʹ-fluoro (2ʹ-F) or 2ʹ-O-methyl (2ʹ-OMe)37 modified nucleotides on RNA nanoparticles results in the generation of stable and homogeneous shape without aggregation; and are not susceptible to RNase and serum degradation75. As a result, the RNA oligonucleotides used in RNA nanoparticle construction act as natural, versatile, anionic polymers that act like Lego blocks to build self-assembling nano-sized particles.
Figure 3.
Different sizes and shaped thermodynamically stable nanoparticles constructed from core packaging RNA-3-way junction (pRNA-3WJ) motif. RNA strands act like a Lego building block to make this structural variation possible. Images were collected with AFM and 3D structures from the Cryo-EM instrument. Reproduced with permission from Ref. 74. Copyright © 2011, Nature Publishing Group. Reproduced with permission from Refs. 75, 76. Copyright © 2014 +++Khisamutdinov et al. under CC By-NC 4.0 https://creativecommons.org/licenses/by/4.0/. Copyright © 2020, Ghimire et al., American Chemical Society.
3.2. RNA aptamers are efficient ligands for receptor binding
RNA aptamers are short oligonucleotide sequences capable of targeting high affinity and specificity, like antigen-antibody binding76,77. However, monoclonal antibodies have limited abilities as detection reagents in biotechnology due to selection and preparation difficulties, high costs of production, stability, and cross-reactivity issues78. On the contrary, aptamers offer substantial advantages over antibodies such as stability79,80, longer shelf life81, smaller in size77,82,83, inexpensive, and faster processing and generation time84. Aptamer–siRNA chimeras proven to be dual-functioning antiviral therapy. They may provide sufficient rationale for applying against the SARS-CoV-285. For discovering actionable biomarkers and therapeutic applications in cancer therapeutics; researchers are attempting to use cell-SELEX (Systematic Evolution of Ligands by Exponential Enrichment) to the generation of oligonucleotide aptamers86. In short, this process utilizes random RNA libraries. Through several rounds of binding selections, it isolates a small number of RNA sequences to become aptamers with high binding affinity for their targeted receptors. For example, aptamer development has been reported to differentiate high-metastatic and low-metastatic cancer cells of different tumor subtypes. This includes colorectal cancer87, breast cancer88,89, osteosarcoma90, prostate cancer89, hepatocellular carcinoma91,92, and colon cancer93,94. The prostate specific membrane antigen (PSMA) RNA aptamer has become one of the most successful and widely used aptamer families to date95,96. Apart from safe and efficient delivery of RNAi payloads to specific target cells, leukocytes are notoriously hard to transfect97. Rajagopalan et al.98 demonstrated an approach of using aptamer-based targeting moiety to deliver siRNAs and silence a receptor to modulate immune function of circulating CD8+ T cells. The data show that systemic administration of the 4-1BB aptamer-CD25 siRNA conjugate downregulated CD25 mRNA only in 4-1BB-expressing CD8+ T cells promoting their differentiation into memory cells. Thus, inducing T cell differentiation to memory precursor effector cells yields an intense and prolonged immune response to antigen-presenting cells.
3.3. Conjugation of chemical ligands & membrane anchoring agents to RNA nanoparticles is simple
RNA nanotechnology has garnered popularity due to the ease of manipulation at the molecular level. With site-specific conjugation via click chemistry99,100 or enzymatic approach101,102, various functionalities can be introduced to the strands. Functional moieties range from anticancer drugs, imaging agents, and multiple probes that alter RNA chemistry which is crucial for nanoparticle application. Chemical conjugation or post-synthetic modification now provides the means to modify almost any RNA of interest. Guo et al.69 utilized RNA modification techniques to conjugate chemical drugs, RNAi nucleic acid sequences, ribozymes, targeting aptamers and chemical ligands, and fluorescent probes. The group studied the effects of small molecule conjugation on RNA nanoparticles for biodistribution, pharmacokinetics, and therapeutic efficacies34,103,104. These works showed improved drug loading and targeted the cell of interest, optimizing theranostic applications. Afonin group also reported a strategy to design 3D nanoscale scaffolds that can be self-assembled from RNA with precise control over their shape, size, and composition105,106. These short oligonucleotide cubic nanostructures are amenable to chemical synthesis, point modifications, and further functionalization.
3.4. RNA nanoparticles are safe for in vivo application
The use of nanoparticles must be safe for in vivo applications. So far, nucleic acid (NA) nanoparticles have demonstrated the ability to selectively target and bind cells by including targeting ligands, as discussed above103,107, 108, 109, 110, 111, 112, 113. Additionally, through their conjugation to RNA nanoparticles, the limitation of poor aqueous solubility of chemotherapeutics can be resolved, without compromising tumor cell-specific delivery103. The RNA nanoparticles with cell-specific targeting ligands could deliver the hydrophobic chemotherapeutics to cancer cells and demonstrated improved efficacies with fewer side effects. For example, poor solubility of Camptothecin (CPT) in an aqueous medium was improved dramatically by conjugating 7-CPT molecules to 3WJ-RNA nanoparticles as prodrugs using click chemistry34. Here, folic acid was used as a cancer cell targeting ligand. The tumor growth was effectively inhibited by CPT-RNA nanoparticles in the KB tumor xenograft (Fig. 4A)34. Fluorescence colocalization assays confirmed fluorescent RNA nanoparticles' efficient binding and internalization into KB cells. FA-7CPT-3WJ-Alexa647 treated cells show high intracellular fluorescence signals. On the contrary, a low signal was detected for the control group of RNA nanoparticles without Folate (FA) ligand. The poor water solubility of another common anticancer drug paclitaxel (PTX) has led to an unsatisfactory biodistribution profiles, undesired side effects, and cellular toxicity. Conjugating paclitaxel to RNA nanoparticles yielded enhanced water solubility (Fig. 4B). While the water solubility of RNA conjugated PTX increased 32,000-fold, the free PTX at the same concentration showed turbidity103. Self-assembly of four single-stranded RNA carrying six PTX molecules yielded 4WJ-X (X-shape) with an increased drug loading capacity. Due to high water solubility, the homogenous assembly of 4WJ-X-24 PTXs could carry 24 PTXs drugs. Again, the particle sizes of the nanoparticles remain less than 10 nm. This small particle diameter allows the particle to pass through leaky vasculature to carry drug molecules to target sites. The rubber-like property114 of RNA nanoparticles resulted in a substantial accumulation of RNA nanoparticles in tumor vasculature by EPR effect as the nanoparticles could deform their shape to squeeze through leaky vasculature (Fig. 4C)115. There were initial concerns about the RNA nanoparticles becoming toxic once injected intravenously because the loaded PTX and CPT were active drugs. However, initial toxicity studies demonstrated that RNA nanoparticles harboring 24 copies of paclitaxel resulted in no signs of toxicity, or immunogenic responses (Fig. 4D), and mice showed no gross weight changes indicating no significant toxicity34,103,113. The figure shows no considerable elevation of most chemokines production (only IL-6 data shown for simplicity) observed for the 4WJ-X-24 PTXs nanoparticles. This high tumor accumulation also indicates low drug toxicity, as the RNA nanoparticles have limited healthy organ buildup (Fig. 4D). Nanoparticles are excreted from the body within a short time. Moreover, due to the presence of phosphate moieties on the RNA macromolecules, RNA nanoparticles are negatively charged polymers that hold self-assembly properties into perfectly controllable nanoscale-sized and shaped nanostructures. An overall negative charge on the RNA nanoparticle adds further advantage by preventing nonspecific binding to negatively charged cell membranes. This resulting repulsion from nontargeted and immune cells provides favorable biodistribution in vivo.
Figure 4.
Unique properties of pRNA nanoparticles for improving drug solubility, cancer cell-targeted delivery, minimal immunogenicity, and low in vivo cytotoxicity. (A) In-vivo distribution of Camptothecin (CPT) conjugated with 3WJ-RNA. Reproduced with permission from Ref. 34. Copyright © 2019 Piao et al. under CC By-NC 4.0 https://creativecommons.org/licenses/by/4.0/ (B) Improved solubility of Paclitaxel (PTX) after conjugation to RNA nanoparticle. Reproduced with permission from Ref. 104. Copyright © 2020 Guo et al. under CC By-NC 4.0 https://creativecommons.org/licenses/by/4.0/ (C) Rubbery property of RNA nanoparticle help improved extravasation via EPR effect. Reproduced with permission from Ref. 114. Copyright © 2020 American Chemical Society (D) In vitro immunogenicity of RNA-MTX NPs. LPS was used as a positive control. Reproduced with permission from Ref. 103. Copyright © Guo et al. under CC By-NC 4.0 https://creativecommons.org/licenses/by/4.0/.
3.5. RNA nanoparticles can carry multiple components
RNA nanoparticles are constructed via bottom-up self-assembly from multiple component strands suitable for incorporating various functional groups into RNA nanoparticles with defined structure and stoichiometry. RNA nanoparticles' branched nature allows for functional moieties at each helical branch region through simple sequence extension of 5′/3′ end chemical modifications. Additionally, nucleic acid chemistry has generated a wide array of chemically modified nucleic acid phosphoramidites that can be utilized during the chemical synthesis of RNA component strands. These chemical modifications, such as –NH2, –COOH, maleimide, –SH, alkyne, and azide throughout the RNA strand, allow for downstream conjugation of other chemical groups onto the RNA nanoparticles116,117. As a result, RNA nanoparticles can harbor a wide variety of conjugated groups while generally remaining thermodynamically stable and retaining the functionalities of each conjugated component without effecting the core motif structuring. RNA nanoparticles can carry chemotherapeutic drugs and ligands for targeted drug delivery to cancer sites or fluorescence dyes for live-cell imaging purposes.
In summary, RNA decorated exosomes can improve their target specificity and reduce nonspecific cell membrane interactions. Exosomes decorated with RNA nanoparticles will protect the loaded exosome cargos from circulatory exonuclease degradation. These nanovesicles will also safeguard the payloads from nonspecific protein binding during circulation. This feature could be beneficial for therapeutic RNA or RNAi delivery. Ligand displaying exosomes could target cancer cells via ligand-receptor interaction and deliver the therapeutic molecules to the cytosol. RNA nanoparticles also help exosome loading as an added advantage. After exosome-mediated targeted delivery via probable cell membrane fusion, RNA nanoparticles will be eliminated rapidly via the excretory system. The unique rubber-like property of the RNA nanoparticle reduces toxicity by rapid elimination from the body. Moreover, the RNA-ligand display technology on exosome surface could be utilized to track the fate of the RNA/exosome complex and to follow up on the consequence of therapy. Taken together, the low toxicity, fast renal clearance, and functionalized targeting molecules make RNA nanoparticles an ideal candidate for exosome engineering via chemical modification in RNAi-mediated cancer treatment.
4. Construction of pRNA-ligand complex for exosome displaying
RNA nanotechnology has now developed into an emerging field of nanomaterials with precise control over design118. The bacteriophage phi29 motor packaging RNA (pRNA-3WJ) can intrinsically rearrange with three angles of 60°, 120°, and 180° between helical regions (Fig. 5A)7. The modules remain functional in vitro and in vivo. The functionality of the nanoparticle assembly suggests that the 3WJ core can be used as a platform for building a variety of multifunctional nano-sized particles. Each RNA oligomer can contain functional domains such as a receptor-binding ligand, aptamer, short interfering RNA, or ribozymes (Fig. 5B). For example, the pRNA-3WJ was extended into an arrow-shaped structure by incorporating an RNA aptamer as a targeting ligand for binding to specific receptors overexpressed on cancer cells7. Cholesterol on the arrowhead promotes the entry of RNA nanoparticles into EVs. On the other hand, the two arms of the arrowtail configuration that form a 60° angle act like a hook to lock the RNA nanoparticle in place. This hook structure with the cholesterol tag naturally comprises and prevents the RNA nanoparticle from entering the exosomes. As a result, the RNA nanoparticle is displayed on the surface allowing the exosomes to project out targeting ligands (see Fig. 1). The idea of using cholesterol for systemic siRNA delivery was reported decades ago119. One study stated cholesterol conjugation dramatically improved the pharmacokinetic parameters of siRNA in serum120. However, cholesterol and other lipid-conjugated oligonucleotides could bind to circulating lipoproteins such as low-density lipoprotein (LDL)121. Interestingly, the same off-target binding of cholesterol could be translated as a beneficial method for decorating small molecules, ligands, and fluorescent dyes on the surface of the exosome. For example, Zhuang et al.122 designed valency-controlled tetrahedral DNA nanostructures. For the study, the DNA nanostructures were conjugated with DNA aptamer. Then the aptamer was loaded on the EV surface via cholesterol anchoring for specific cell targeting. This module has efficiently facilitated the tumor-specific accumulation of the EVs in HepG2 cells in vitro and human primary liver cancer-derived organoids and xenograft tumor models. Lin et al.123 have successfully delivered an anticancer agent (imperialine) against non-small cell lung cancer (NSCLC). The integrin α3β1-binding octapeptide cNGQGEQc was modified onto the exosome like vesicle platform for tumor targeting using cholesterol as an anchor. This strategy was reported to significantly improve imperialine tumor accumulation and retention onto the NSCLC cells. He et al.124,125 have shown a direct method for accurate quantification of exosomes using cholesterol as an anchor. For one of these studies, exosomes were specifically captured by immunomagnetic beads, and then a bivalent-cholesterol-labeled DNA anchor with high affinity was spontaneously inserted into the exosome membrane. Detection of the exosomes were based on the color change of horseradish peroxidase -catalyzed H2O2-mediated oxidation of 3,3′,5,5′-tetramethyl benzidine. The oxidation could be conveniently observed by the naked eye and monitored by UV–Vis spectrometry.
Figure 5.
Displaying RNA nanoparticle (RNPs) on EVs for Targeted siRNA Delivery. (A) AFM image showing planar structure of pRNA-3WJ. There are two possible pRNA-3WJ motifs, arrowhead and arrowtail, in exosome displaying and cargo loading. Reproduced with permission from Ref. 7. Copyright © 2017 Pi et al. (B) Sequence of RNA nanoparticles scaffold in RNA nanoparticle carrying different chemical moieties. Shown here is RNA-3WJ-18PTX-Ligand-Fluorescent dye (figure prepared with Biorender.com).
The multivalency of RNA allows to harbor anticancer agents, ligands, or fluorescent dyes on the self-assembled RNA nanoparticles7. By conjugating cholesterol to arrowtail of the 3WJ-pRNA allows the entry of the nanoparticles on the lipid-rich exosome surface (Fig. 5B). The fishhook structure prevents the hooked RNA from passing through the membrane. By this hooked RNA, cancer cell-specific ligands can be displayed on the surface of the exosomes. The chemically modified (2′-F U/C) 3WJ-pRNA complex is also resistant to degradation compared to the unmodified RNA, which degrades within a few minutes7. This added advantage leads the exosome-mediated cancer cell-specific drug delivery attainable.
5. Construction of therapeutics payload for exosome loading
The construction of therapeutic payloads and exosome loading is one of the most crucial factors in exosome-mediated cancer therapy. Among different exosome loading strategies, mechanical extrusion126, electroporation51,127, saponin permeabilization128, sonications129, direct incubation with exosome128,130, and repeated freeze-thaw cycles128 are some widely verified strategies126,131. Depending on the nature of the exosome loading, these strategies can be categorized into active or passive loading. While passive loading involves the loading of therapeutic cargo into EVs through diffusion, active loading consists of the membrane disruption of EVs, allowing the cargo to enter the EVs132. Each approach has its advantage and drawbacks. For example, passive loading with direct incubation suffers from long incubation time and low loading capacity due to the nonspecific hydrophobic binding between cargos and exosomes. However, Haraszti et al.133 presented a framework for successful hydrophobic modification as a strategy for productive loading of RNA cargo onto extracellular vesicles. The study shows that the linker chemistry influences the efficiency of cholesterol-mediated loading of siRNAs onto extracellular vesicles. The study also reported that the hydrophobic modifications of siRNA enable the association of a large number of RNA molecules per extracellular vesicle. However, destabilization of the linker or overloading could impair the silencing activity of extracellular vesicles. Active loading techniques for hydrophilic nucleic acids also suffer from scalability and product quality. So, optimizing suitable strategies could improve exosome loading. At the same time, it could maintain the physical properties of the nanoparticles without compromising their functionality or stability. A promising approach of passive loading into the exosomes is the exosome engineering by plasmid insertion into cells. This approach produces exosomes with labels and cargos in a natural biogenesis pathway. By transfecting adipose tissue-derived mesenchymal stem cells with a microRNA expression plasmid promoted chemosensitivity of hepatocellular carcinoma cells for chemotherapy134, 135, 136, 137, 138, 139. These studies demonstrated that the transfected mesenchymal cells effectively packaged the microRNA into secreted exosomes and enhanced anticancer efficacy of chemotherapeutic agents in vivo mediated by altering target gene expression.
Huang and Leonard140 investigated the general biophysical properties that impact the active loading and delivery of RNA by EVs. For the study, MS2 bacteriophage protein and vesicular stomatitis virus glycoprotein (VSVG) were fused to EV-associated proteins. With this Targeted and Modular EV Loading (TAMEL) approach, a 40-fold enhanced cargo RNA loading efficacy was observed. However, inefficient endosomal fusion or escape still represents a limiting barrier to EV-mediated RNA transfer140. de Abreu et al.141 found that the loading efficiency with ExoFect is 50% higher than electroporation, heat-shock, or saponin-mediated permeabilization. An essential finding of the study is that the membrane of ExoFect-loaded small EVs was altered compared with native EVs. Due to this altered membrane configuration, the internalization of ExoFect-loaded EVs to the target cells was enhanced. These modulated EVs also decreased the interaction between the exosomes and lysosomes.
To evaluate in vivo prostate, breast, lung, and colon cancer inhibition efficacy, Guo lab prepared siRNA loaded EVs with targeting moieties combined with survivin siRNA loaded into the EVs (like Fig. 1)7,35. In brief, RNA nanoparticles were incubated with ExoFect transfection system to evaluate loading efficiency, followed by a heat-shock protocol. Subsequently, the cholesterol-modified RNA nanoparticles were incubated with siRNA-loaded EVs to display respective cancer ligands. Then the mixture was allowed to cool to prepare the RNA-decorated EVs. An iodixanol cushion was used to achieve reproducible and homogenous particle size. Loading efficiencies were determined to be between 70% and 80%. The stability of the therapeutic payload is also essential in retaining its activity in cancer gene silencing. To improve siRNA's stability in vivo, Guo lab used 2′F-modified pyrimidines on the passenger strand, providing RNase resistance7,35. In contrast, the guide strand was kept unmodified7,35.
6. RNA-ligand displaying exosome for cytosol delivery without endosome entrapment
So far, we have discussed the possible exosome surface decoration and the mechanisms of cargo loading in previous sections. We have highlighted how RNA nanoparticles could be used to decorate the exosome surface with different chemical moieties. In this section, the endosome trapping is explained in detail. Endosome trapping is one of the significant bottlenecks for RNAi-mediated cancer treatment. Here we will discuss how RNA-nanoparticles harboring ligands could be used to deliver the cargos directly into the cytosol by displaying them on the surface of the exosomes.
6.1. Cellular internalization pathway, endosome trapping, and its impact on gene/drug delivery
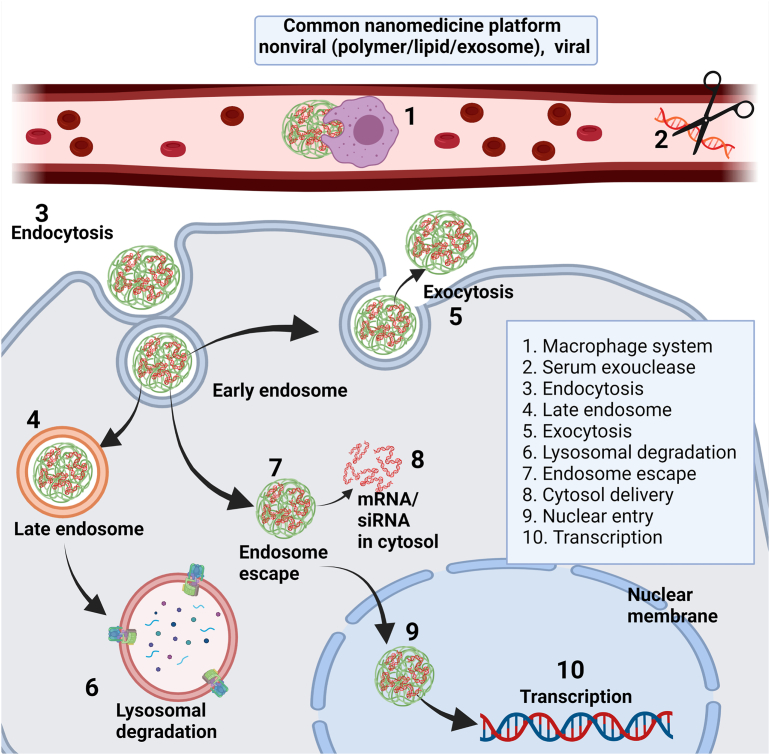
A successful drug/gene delivery carrier should protect the cargo until reaching its target, i.e., the tumor microenvironment, and withstand all barriers before reaching the target and showing its therapeutic action. Fig. 6 demonstrates the most common obstacles to a nanoparticle drug delivery system. Circulatory enzymes and macrophage systems pose the first obstacle for nanoparticles after they enter the circulatory system142. Cellular entry or endocytosis pathways are other barriers preventing the therapeutic molecules' successful cytosolic release. Nanocarriers can be internalized via clathrin, caveolae, flotillin, phagocytosis, or micropinocytosis pathways and typically entrapped into the early endosome after cellular internalization143,144. Early endosomes mature into late endosomes with a pH change to ∼5.5–6. Most late endosomes fuse with lysosomes with luminal pH ∼4.5–5.5. Lysosomal enzymes degrade most of the nanoparticles that are entrapped in the lysosome. Moreover, many nanoparticles that enter the cell result in reduced therapeutic efficacies due to being eliminated via exocytosis. The destination for RNAi/mRNA ends after reaching the cytosol, while DNA should still migrate to the nucleus.
Figure 6.
Common barriers to intracellular trafficking of the nanomaterials with nonviral or viral delivery systems. For the simplicity of the discussion, polymeric nonviral delivery system was used throughout the endocytosis pathway (figure prepared with Biorender.com).
Although the mechanism is unclear, it is thought that the tiny fraction of the RNAi that escapes the endolysosomal entrapment is sufficient to show its therapeutic action in the cytosol8. This complex cellular internalization is the bottleneck for nanoparticle-mediated cargo delivery and successful anticancer therapy using RNAi. The endolysosomal trap could also hamper the reproducibility of in vivo experiments. To address these issues, most nonviral therapy using polymers or lipids as carriers incorporate membrane fusogenic145 and endosomolytic peptides146 or chemical modifications of polymers8,147. These modifications aim to protect the cargo from endolysosomal degradation and enhance delivery8,148. Some molecular modifications/post-polymerization modifications could induce receptor-mediated endocytosis147. Even though the literature reports orchestrating chemicals to withstand a lysosomal environment, the overall success rate, with some exception149,150, in vivo, is still insufficient to translate the work into the clinic. However, there is always room for modulating newer formulations development and understanding the principles of RNAi delivery to cancer cells. For example, it could be helpful to combine ionizable lipid/polymeric materials with exosomes and cell membranes derived from the human body to deliver RNAi, or siRNA nanogels which may reduce side effects and improve therapeutic effects151,152. To increase the accumulation in vivo and evade the mononuclear phagocyte system from circulation, biomimetic materials such as erythrocyte membrane, platelet-derived extracellular vesicles, macrophage membranes, and macrophage-derived exosomes could also be incorporated153.
6.2. RNA-ligand displaying exosome helps escape endosome trapping
Above, we discussed endosome entrapment, a significant setback for cytosolic delivery of RNAi in cancer treatment. Endosome entrapment channels the internalized cargo into a lysosome and degrades. This lysosomal degradation is one of the leading causes showing minimal or no response of the therapeutic molecules. The routes and mechanisms of extracellular vesicle uptake may depend on proteins and glycoproteins found on the surface of both the vesicle and the target cell154. Most research describes the uptake mechanism involving endocytosis pathways because the heterogeneous population of EVs may gain entry into a cell via more than one route154, 155, 156, 157. For example, Joshi et al.158 show that endocytosis is the primary exosome uptake pathway. For the study, a membrane-bound EV cargo (GFP-CD63) was used to increase the chance of detecting EV cargo exposure to the cytosol of acceptor cells. By forming mCherry puncta in mCherry-tagged GFP antibody-expressing HEK293T cells upon their exposure to GFP-CD63 EVs, cargo delivery to the cytosol was achieved. However, the study does not rule out the possibility of a direct fusion of EVs with the plasma membrane. Meng et al.159 also demonstrated that genetically programmable fusion cellular vesicles significantly increase the phagocytosis of cancer cells by macrophages thus promotes antigen presentation and activates antitumor T-cell immunity. However, as the endocytosis is the bottleneck for RNAi-mediated anticancer therapy, identifying the EV components that enable functional cargo transfer to the cytosol will allow the development of more efficient therapeutics in the future157. Ligand displaying RNA nanoparticles prevent the exosome from off-target binding and targets specific cancer cells via ligand-receptor interaction. The net negative charge on the exosome displayed with pRNA-3WJ repels the cell membrane protein until the ligand binds to the receptor on the targeted cell surface. Zheng et al.6 demonstrated the efficient cancer suppression with the folic acid (FA)-displaying exosome. Due to the receptor-mediated cytosol delivery of the siRNA payload without endosome trapping, cancer regression was enhanced. Following binding to the specific Folate receptors, the FA displaying-exosomes released its payload siRNA to the cytosol by fusing the cell membrane. The folate decorated exosomes demonstrated a significant enhancement in gene knockdown efficacy both in vitro and in vivo6. Shown in Fig. 7A is the tumor size regression with FA-displaying exosome compared to the non-displaying exosome and the control group. Folate displaying exosome keeps the tumor size in check (knockdown of survivin expression by 60%) while the control groups show no noticeable gene expression change over twelve days6. Fig. 7B shows the confocal microscopy image of the cargo distribution within the cell. For the study, KB cells were incubated with folate-exosome complexes labeled on the exosome membrane where the RNA was red (Alexa 647) and/or cytosol was green (Cy3) while the nucleus was dyed blue. The cargo showed an even distribution inside the cell. The red color represents the exosome membrane located on the membrane of the cells instead of the cytoplasm. The dye on the exosome surface was retained on the cell membrane during cellular uptake, distinct from cargo located evenly in the cytosol. These observations suggest that the folate displaying exosomes is more likely to enter the cancer cell through direct membrane fusion.
Figure 7.
Active delivery of siRNA therapeutics by RNA nanoparticles to disease sites. (A) Folate displaying exosome show regression in survivin gene expression, but the control group does not. (B) RNA nanoparticles facilitate endosomal escape and enhanced cytosol delivery. Here, RNA is red (Alexa 647) and/or cytosol is green (Cy3) while the nucleus is blue. (C) Illustration of membrane fusion mechanism in two possible pathways. 1) Direct membrane fusion and 2) Reverse fusion via endocytosis. (D) Exosome displayed with folate enables delivery of siRNA into cell cytosol without endosome trapping. Shown here is Folate-Survivin-Alexa 647 and Folate/Exosome/ds-Survivin-Alexa 647 (E) Colocalization indicates the siRNA delivered by exosome can avoid endosome trapping. Shown here are Folate-Survivin-Alexa 647 and Folate/Exosome/Survivin-Alexa 647. [All siRNA is labeled with red dye Alexa 647. Immunofluorescence staining of organelle markers are shown in green. EEA1 was used as a marker for early endosomes and LAMP1 for lysosomes. DAPI staining nucleus (blue) while colocalization are displayed in yellow.] Reproduced with permission from Ref. 6. Copyright © 2019 Elsevier B.V.
6.3. Discovery of the exosome fusion mechanism that led to cytosol delivery without endosome trapping
The cellular uptake and internalization mechanism of exosomes is still under extensive scrutiny. Exosome cargos can enter the cytosol via direct membrane fusion with the outer layer of the cell membrane. Or exosomes can enter by reverse fusion with the endosomal membrane after receptor-mediated endocytosis (Fig. 7C). The EV membrane surface can trigger signaling through interaction with receptors-ligands on the cell surface without EV entry and thus could determine their functional effects21. If the EVs enter via endocytosis, their cargo must exit the inherently degradative lysosomal pathway. Or EVs could be ejected out again through the MVB-plasma membrane fusion pathway21, also discussed in section 6.1 (Fig. 6). Despite endocytosis-mediated uptake of the EVs into target cells, extracellular vesicle-derived mRNA expression in recipient cells was found negligible, suggesting that endosomal escape is also critical to RNA function140. Most research on extracellular vesicle uptake mechanism focuses on endocytosis pathways because of the difficulty in assessing fusion events. However, previously published reports suggest that one of the significant mechanisms of exosome delivery is via direct membrane fusion160,161, although the mechanism could vary depending on the recipient cell162. Direct cell membrane fusion could be achieved by incorporating fusion proteins into the EV surface161,163. Some examples of fusion protein include syncytin 1, syncytin 2, epithelial fusion failure 1 protein, etc. Glycoproteins could also be harbored on the extracellular vesicles to promote membrane fusion by incorporating gap junction proteins (e.g., connexin 43, glycans, or vesicular stomatitis virus)164, 165, 166.
Proteins of the extracellular vesicle membranes can structurally rearrange themselves. Their hydrophobic sequences are inserted into the target cell surface, which undergoes lipid reorganization, protein restructuring, and membrane dimpling, allowing membrane fusion161. The study shown in Fig. 7B demonstrates that the folate displaying exosome membrane using RNA-3WJ nanoparticles located on the cell surface. The study used exosomes that were fully decorated with FA where the folate receptor-mediated endocytosis might take over and play a role in cellular uptake. As discussed in section 6.2, no red dye was seen inside the cytoplasm and remains on the cell surface. On the other hand, the cargo with green dye is distributed across the cell cytosol, supporting the literature on membrane fusion. Fig. 7D,E further reinforces the exosome uptake via the membrane fusion process. When folate decorated exosomes delivered the siRNA, an even distribution was observed in the cytoplasm, with a little overlap of endosome/lysosome staining in cells. The images suggest an effective cytosol delivery property using exosome combined with folate as a targeting molecule. Taken together, it is hypothesized that the smoothness of the exosome surface is more prone to endocytosis. Unless decorated with fusogenic membrane agents, exosomes will be endocytosed, channeling to lysosomal degradation. However, a cell can incorporate phagocytosis or micropinocytosis to internalize the exosome into the cell. And then, it might deliver the cargo to the cytosol via back fusion. Or even the exosome could end up merging with the lysosome and degrade. Interestingly, when the EV surface is decorated with ligand-RNA nanoparticles conjugate, the surface becomes rough, preventing typical natural cellular endocytosis. The ligand-receptor-mediated cell surface attachment will be longer than the normal process due to the roughness of the EV surface. This event, followed by the proximity of the exosome surface to the cell membrane, will initiate membrane-specific fusion as a major EV uptake pathway. So, among the two possible mechanisms, the data support that direct membrane fusion is the most probable route for internalizing RNA-ligand displaying exosome to the cytosol. However, further research is needed to reach a conclusive decision.
7. Processing and releasing of the RNAi from RNA nanoparticles after delivery by exosome
Recently Binzel et al.167 have aimed to demonstrate the efficient processing of siRNA from RNA nanoparticles for high gene silencing while providing a comprehensive, mechanistic understanding of siRNA Dicer processing from RNA nanoparticles. Loading Dicer substrate RNA into exosomes and decorating ligand displaying pRNA nanoparticles allowed direct visualization of the in vitro siRNA processing from RNA nanoparticles. The exosomes were decorated with pRNA nanoparticles harboring folate ligand to provide specific targeting and binding to its receptor-expressing HT29 cells. This nanovesicle allowed the 3WJ-Luciferase-siRNA beacon nanoparticles to be efficiently distributed to the cytosol of the cancer cells for siRNA processing by Dicer (Fig. 8A). The processing of siRNA from the 3WJ scaffold was reflected by Cy5 fluorescence gradual increase in signal over time in the confocal microscopy image (Fig. 8B). The data demonstrate that Dicer can bind and cleave siRNA incorporated into RNA nanoparticles. This work proves that the RNA nanoparticle motif does not interfere with Dicer siRNA processing due to steric hindrance or processing issues due to the nanoparticles' branched nature. The incorporation of BBQ650 resulted in high quenching of the Cy5 signal (Fig. 8C). Shown in Fig. 8D is the prostate cancer trial in nude mice. At one dose every three days for six doses, prostate cancer treatment with PSMAapt/EV/survivin siRNA (siSurvivin) completely suppressed in vivo tumor growth compared to the control and scramble group7. Across three tumor models (prostate, NSCLC, TNBC), exosomes modified with RNA nanoparticles can deliver Survivin siRNA that is then processed in the tumors, resulting in significant tumor inhibition (Fig. 8E)7,167,168. These findings suggest that integration of length, nucleotide components, structure, and RNA nanoparticles into the designs of RNAi efficiently silence the targeted genes in an in vivo cancer therapy setting. The work proved that pRNA nanoparticles do not interfere with Dicer's ability to read and process siRNA sequences while also providing design principles to incorporate Dicer-substrate siRNA onto the nanoparticles.
Figure 8.
Controlled release of Dicer-substrate siRNA harbored in phi29 pRNA-based nanoparticles. (A) Folate exosome designs carrying RNA nanoparticle beacons (3WJ/LUC2-siRNA-truncated beacon, 3WJ/LUC2-siRNA beacon, 3WJ/LUC2-siRNA/Cy5); red star: Cy5 fluorophore, black circle: BBQ650 quencher (B) Intracellular imaging of Dicer processing of 3WJ/LUC2-siRNA delivered by exosomes after 2-h incubation with HT29 cells. (C) Assembly of 3WJ/LUC2-siRNA molecular beacon assayed by 15% native polyacrylamide gel electrophoresis (PAGE) (green: ethidium bromide [EB], red: Cy5, yellow: overlap). (D) The figure show cancer regression in prostate cancer mouse trials in vivo. Clearly, ligand displaying exosome show a noticeable difference in cancer regression compared to the control group. Reproduced with permission from Ref. 7. Copyright © 2017 Pi et al. under CC By-NC 4.0 https://creativecommons.org/licenses/by/4.0/. (E) Summary of previous animal trials to elucidate the processing and releasing of Survivin siRNA incorporated in the RNA nanoparticles. Reprinted with permission from Ref. 168. Copyright © 2021, Binzel et al. under CC By-NC 4.0 https://creativecommons.org/licenses/by/4.0/.
8. Conclusions
Exosomes can potentially address roadblocks in cancer therapy by enhancing anticancer drugs or siRNA delivery to the target cells. However, the issues with target specificity are the key hindrance to exosomes' poor efficacy. This review explained that the RNA nanoparticles displayed with cell-specific ligand would potentially rejuvenate the research that it could serve as a powerful platform for efficient siRNA delivery. The multivalency, anionic nature, and rubbery properties for rapid cancer accumulation with fast renal clearance make the RNA nanoparticles a unique therapeutic tool. Applying RNA nanotechnology on the exosome surface could serve as an excellent remedy for cancer. One of the major bottlenecks in cancer therapy with RNAi is endolysosomal entrapment. We demonstrate that ligand displaying RNA nanotechnology delivers the siRNA with target specificity and enhances efficacy by delivering the cargo into the cytosol. We also emphasize incorporating robust imaging techniques to evaluate a detailed mechanism of EV-associated membrane fusion. Overall, this review ushers the tremendous possibility of RNA nanotechnology as the base for translating exosome biology into RNAi-based cancer therapies.
Acknowledgments
The research in authors office was supported in part by NIH grants U01CA207946 and R01EB019036 to Peixuan Guo and NIH grant R01CA257961 to Dan Shu and Daniel W. Binzel. Peixuan Guo's Sylvan G. Frank Endowed Chair position in Pharmaceutics and Drug Delivery is funded by the CM Chen Foundation. Confocal images presented in this report were granted with the instruments and services at the Campus Microscopy and Imaging Facility, The Ohio State University. This facility is supported in part by Grant P30CA016058, National Cancer Institute, Bethesda, MD.
Footnotes
Peer review under responsibility of Chinese Pharmaceutical Association and Institute of Materia Medica, Chinese Academy of Medical Sciences.
Author contributions
Nasir Uddin: wrote and revised the article, Daniel Binzel: edited the article, Tian-Min Fu: edited the article, Dan Shu: reviewed and edited the article, Peixuan Guo: Supervised, reviewed, and edited the article.
Conflicts of interest
Peixuan Guo is the licensor, grantee and consultant of Oxford Nanopore Technologies, the cofounder of Shenzhen P&Z Bio-medical Co. Ltd, as well as the cofounder and Board member of the ExonanoRNA, LLC.
References
- 1.Hu B., Zhong L., Weng Y., Peng L., Huang Y., Zhao Y., et al. Therapeutic siRNA: state of the art. Signal Transduct Target Ther. 2020;5:101. doi: 10.1038/s41392-020-0207-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Whitehead K.A., Langer R., Anderson D.G. Knocking down barriers: advances in siRNA delivery. Nat Rev Drug Discov. 2009;8:129–138. doi: 10.1038/nrd2742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Garber K. Alnylam launches era of RNAi drugs. Nat Biotechnol. 2018;36:777–778. doi: 10.1038/nbt0918-777. [DOI] [PubMed] [Google Scholar]
- 4.Weng Y., Xiao H., Zhang J., Liang X.-J., Huang Y. RNAi therapeutic and its innovative biotechnological evolution. Biotechnol Adv. 2019;37:801–825. doi: 10.1016/j.biotechadv.2019.04.012. [DOI] [PubMed] [Google Scholar]
- 5.Yao Y., Zhou Y., Liu L., Xu Y., Chen Q., Wang Y., et al. Nanoparticle-based drug delivery in cancer therapy and its role in overcoming drug resistance. Front Mol Biosci. 2020;7:193. doi: 10.3389/fmolb.2020.00193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zheng Z., Li Z., Xu C., Guo B., Guo P. Folate-displaying exosome mediated cytosolic delivery of siRNA avoiding endosome trapping. J Control Release. 2019;311–2:43–49. doi: 10.1016/j.jconrel.2019.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pi F., Binzel D.W., Lee T.J., Li Z., Sun M., Rychahou P., et al. Nanoparticle orientation to control RNA loading and ligand display on extracellular vesicles for cancer regression. Nat Nanotechnol. 2018;13:82–89. doi: 10.1038/s41565-017-0012-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pei D., Buyanova M. Overcoming endosomal entrapment in drug delivery. Bioconjug Chem. 2019;30:273–283. doi: 10.1021/acs.bioconjchem.8b00778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Smith S.A., Selby L.I., Johnston A.P.R., Such G.K. The endosomal escape of nanoparticles: toward more efficient cellular delivery. Bioconjug Chem. 2019;30:263–272. doi: 10.1021/acs.bioconjchem.8b00732. [DOI] [PubMed] [Google Scholar]
- 10.Doyle L.M., Wang M.Z. Overview of extracellular vesicles, their origin, composition, purpose, and methods for exosome isolation and analysis. Cells. 2019;8:727. doi: 10.3390/cells8070727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang Y., Liu Y., Liu H., Tang W.H. Exosomes: biogenesis, biologic function and clinical potential. Cell Biosci. 2019;9:1–18. doi: 10.1186/s13578-019-0282-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.El-Andaloussi S., Lee Y., Lakhal-Littleton S., Li J., Seow Y., Gardiner C., et al. Exosome-mediated delivery of siRNA in vitro and in vivo. Nat Protoc. 2012;7:2112–2126. doi: 10.1038/nprot.2012.131. [DOI] [PubMed] [Google Scholar]
- 13.El-Andaloussi S., Lakhal S., Mager I., Wood M.J. Exosomes for targeted siRNA delivery across biological barriers. Adv Drug Deliv Rev. 2013;65:391–397. doi: 10.1016/j.addr.2012.08.008. [DOI] [PubMed] [Google Scholar]
- 14.van Dommelen S.M., Vader P., Lakhal S., Kooijmans S.A., van Solinge W.W., Wood M.J., et al. Microvesicles and exosomes: opportunities for cell-derived membrane vesicles in drug delivery. J Control Release. 2012;161:635–644. doi: 10.1016/j.jconrel.2011.11.021. [DOI] [PubMed] [Google Scholar]
- 15.Valadi H., Ekstrom K., Bossios A., Sjostrand M., Lee J.J., Lotvall J.O. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol. 2007;9:654–659. doi: 10.1038/ncb1596. [DOI] [PubMed] [Google Scholar]
- 16.Skog J., Würdinger T., van Rijn S., Meijer D.H., Gainche L., Sena-Esteves M., et al. Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nat Cell Biol. 2008;10:1470–1476. doi: 10.1038/ncb1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Al-Nedawi K., Meehan B., Micallef J., Lhotak V., May L., Guha A., et al. Intercellular transfer of the oncogenic receptor EGFRvIII by microvesicles derived from tumour cells. Nat Cell Biol. 2008;10:619–624. doi: 10.1038/ncb1725. [DOI] [PubMed] [Google Scholar]
- 18.Costa-Silva B., Aiello N.M., Ocean A.J., Singh S., Zhang H., Thakur B.K., et al. Pancreatic cancer exosomes initiate pre-metastatic niche formation in the liver. Nat Cell Biol. 2015;17:816–826. doi: 10.1038/ncb3169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu J., Ren L., Li S., Li W., Zheng X., Yang Y., et al. The biology, function, and applications of exosomes in cancer. Acta Pharm Sin B. 2021;11:2783–2797. doi: 10.1016/j.apsb.2021.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Adriano B., Cotto N.M., Chauhan N., Jaggi M., Chauhan S.C., Yallapu M.M. Milk exosomes: Nature's abundant nanoplatform for theranostic applications. Bioact Mater. 2021;6:2479–2490. doi: 10.1016/j.bioactmat.2021.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Abels E.R., Breakefield X.O. Introduction to extracellular vesicles: biogenesis, RNA cargo selection, content, release, and uptake. Cell Mol Neurobiol. 2016;36:301–312. doi: 10.1007/s10571-016-0366-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mathieu M., Martin-Jaular L., Lavieu G., Théry C. Specificities of secretion and uptake of exosomes and other extracellular vesicles for cell-to-cell communication. Nat Cell Biol. 2019;21:9–17. doi: 10.1038/s41556-018-0250-9. [DOI] [PubMed] [Google Scholar]
- 23.Melo S.A., Sugimoto H., O'Connell J.T., Kato N., Villanueva A., Vidal A., et al. Cancer exosomes perform cell-independent microRNA biogenesis and promote tumorigenesis. Cancer Cell. 2014;26:707–721. doi: 10.1016/j.ccell.2014.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zomer A., Vendrig T., Hopmans E.S., van Eijndhoven M., Middeldorp J.M., Pegtel D.M. Exosomes: fit to deliver small RNA. Commun Integr Biol. 2010;3:447–450. doi: 10.4161/cib.3.5.12339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sandfeld-Paulsen B., Jakobsen K.R., Bæk R., Folkersen B.H., Rasmussen T.R., Meldgaard P., et al. Exosomal proteins as diagnostic biomarkers in lung cancer. J Thorac Oncol. 2016;11:1701–1710. doi: 10.1016/j.jtho.2016.05.034. [DOI] [PubMed] [Google Scholar]
- 26.Hoshino A., Kim H.S., Bojmar L., Gyan K.E., Cioffi M., Hernandez J., et al. Extracellular vesicle and particle biomarkers define multiple human cancers. Cell. 2020;182:1044–1061. doi: 10.1016/j.cell.2020.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gu X., Erb U., Büchler M.W., Zöller M. Improved vaccine efficacy of tumor exosome compared to tumor lysate loaded dendritic cells in mice. Int J Cancer. 2015;136:74–84. doi: 10.1002/ijc.29100. [DOI] [PubMed] [Google Scholar]
- 28.Altieri S.L., Khan A.N., Tomasi T.B. Exosomes from plasmacytoma cells as a tumor vaccine. J Immunother. 2004;27:282–288. doi: 10.1097/00002371-200407000-00004. [DOI] [PubMed] [Google Scholar]
- 29.Lee Y.S., Kim S.H., Cho J.A., Kim C.W. Introduction of the CIITA gene into tumor cells produces exosomes with enhanced anti-tumor effects. Exp Mol Med. 2011;43:281–290. doi: 10.3858/emm.2011.43.5.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tian X., Zhu M., Nie G. How can nanotechnology help membrane vesicle-based cancer immunotherapy development? Hum Vaccin Immunother. 2013;9:222–225. doi: 10.4161/hv.22130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gehrmann U., Hiltbrunner S., Näslund T.I., Gabrielsson S. Potentiating antitumor immunity with αGC-loaded exosomes. Oncoimmunology. 2013;2 doi: 10.4161/onci.26261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen H., Wang L., Zeng X., Schwarz H., Nanda H.S., Peng X., et al. Exosomes, a new star for targeted delivery. Front Cell Dev Biol. 2021;9 doi: 10.3389/fcell.2021.751079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cresswell G.M., Wang B., Kischuk E.M., Broman M.M., Alfar R.A., Vickman R.E., et al. Folate receptor beta designates immunosuppressive tumor-associated myeloid cells that can be reprogrammed with folate-targeted drugs. Cancer Res. 2021;81:671–684. doi: 10.1158/0008-5472.CAN-20-1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Piao X., Yin H., Guo S., Wang H., Guo P. RNA nanotechnology to solubilize hydrophobic antitumor drug for targeted delivery. Adv Sci. 2019;6 doi: 10.1002/advs.201900951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li Z., Wang H., Yin H., Bennett C., Zhang H-g, Guo P. Arrowtail RNA for ligand display on ginger exosome-like nanovesicles to systemic deliver siRNA for cancer suppression. Nat Sci Rep. 2018;8 doi: 10.1038/s41598-018-32953-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lee T.J., Yoo J.Y., Shu D., Li H., Zhang J., Yu J.G., et al. RNA nanoparticle-based targeted therapy for glioblastoma through inhibition of oncogenic miR-21. Mol Ther. 2017;25:1544–1555. doi: 10.1016/j.ymthe.2016.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Abdelmawla S., Guo S., Zhang L., Pulukuri S.M., Patankar P., Conley P., et al. Pharmacological characterization of chemically synthesized monomeric phi29 pRNA nanoparticles for systemic delivery. Mol Ther. 2011;19:1312–1322. doi: 10.1038/mt.2011.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lane R.E., Korbie D., Anderson W., Vaidyanathan R., Trau M. Analysis of exosome purification methods using a model liposome system and tunable-resistive pulse sensing. Nat Sci Rep. 2015;5:7639. doi: 10.1038/srep07639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang J., Ma P., Kim D.H., Liu B.F., Demirci U. Towards microfluidic-based exosome isolation and detection for tumor therapy. Nano Today. 2021;37 doi: 10.1016/j.nantod.2020.101066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gao X., Ran N., Dong X., Zuo B., Yang R., Zhou Q., et al. Anchor peptide captures, targets, and loads exosomes of diverse origins for diagnostics and therapy. Sci Transl Med. 2018;10 doi: 10.1126/scitranslmed.aat0195. [DOI] [PubMed] [Google Scholar]
- 41.Jia G., Han Y., An Y., Ding Y., He C., Wang X., et al. NRP-1 targeted and cargo-loaded exosomes facilitate simultaneous imaging and therapy of glioma in vitro and in vivo. Biomaterials. 2018;178:302–316. doi: 10.1016/j.biomaterials.2018.06.029. [DOI] [PubMed] [Google Scholar]
- 42.Yerneni S.S., Lathwal S., Shrestha P., Shirwan H., Matyjaszewski K., Weiss L., et al. Rapid on-demand extracellular vesicle augmentation with versatile oligonucleotide tethers. ACS Nano. 2019;13:10555–10565. doi: 10.1021/acsnano.9b04651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Luo Z.W., Li F.X., Liu Y.W., Rao S.S., Yin H., Huang J., et al. Aptamer-functionalized exosomes from bone marrow stromal cells target bone to promote bone regeneration. Nanoscale. 2019;11:20884–20892. doi: 10.1039/c9nr02791b. [DOI] [PubMed] [Google Scholar]
- 44.Tian T., Cao L., He C., Ye Q., Liang R., You W., et al. Targeted delivery of neural progenitor cell-derived extracellular vesicles for anti-inflammation after cerebral ischemia. Theranostics. 2021;11:6507–6521. doi: 10.7150/thno.56367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kim M.S., Haney M.J., Zhao Y., Yuan D., Deygen I., Klyachko N.L., et al. Engineering macrophage-derived exosomes for targeted paclitaxel delivery to pulmonary metastases: in vitro and in vivo evaluations. Nanomed NBM. 2018;14:195–204. doi: 10.1016/j.nano.2017.09.011. [DOI] [PubMed] [Google Scholar]
- 46.Zou J., Shi M., Liu X., Jin C., Xing X., Qiu L., et al. Aptamer-functionalized exosomes: elucidating the cellular uptake mechanism and the potential for cancer-targeted chemotherapy. Anal Chem. 2019;91:2425–2430. doi: 10.1021/acs.analchem.8b05204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wan Y., Wang L., Zhu C., Zheng Q., Wang G., Tong J., et al. Aptamer-conjugated extracellular nanovesicles for targeted drug delivery. Cancer Res. 2018;78:798–808. doi: 10.1158/0008-5472.CAN-17-2880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lee J., Lee H., Goh U., Kim J., Jeong M., Lee J., et al. Cellular engineering with membrane fusogenic liposomes to produce functionalized extracellular vesicles. ACS Appl Mater Interfaces. 2016;8:6790–6795. doi: 10.1021/acsami.6b01315. [DOI] [PubMed] [Google Scholar]
- 49.Cheng H., Fan J.H., Zhao L.P., Fan G.L., Zheng R.R., Qiu X.Z., et al. Chimeric peptide engineered exosomes for dual-stage light guided plasma membrane and nucleus targeted photodynamic therapy. Biomaterials. 2019;211:14–24. doi: 10.1016/j.biomaterials.2019.05.004. [DOI] [PubMed] [Google Scholar]
- 50.Cao Y., Wu T., Zhang K., Meng X., Dai W., Wang D., et al. Engineered exosome-mediated near-infrared-II region V(2)C quantum dot delivery for nucleus-target low-temperature photothermal therapy. ACS Nano. 2019;13:1499–1510. doi: 10.1021/acsnano.8b07224. [DOI] [PubMed] [Google Scholar]
- 51.Alvarez-Erviti L., Seow Y., Yin H., Betts C., Lakhal S., Wood M.J.A. Delivery of siRNA to the mouse brain by systemic injection of targeted exosomes. Nat Biotechnol. 2011;29:341–345. doi: 10.1038/nbt.1807. [DOI] [PubMed] [Google Scholar]
- 52.Kim G., Lee Y., Ha J., Han S., Lee M. Engineering exosomes for pulmonary delivery of peptides and drugs to inflammatory lung cells by inhalation. J Control Release. 2021;330:684–695. doi: 10.1016/j.jconrel.2020.12.053. [DOI] [PubMed] [Google Scholar]
- 53.Kim G., Kim M., Lee Y., Byun J.W., Hwang D.W., Lee M. Systemic delivery of microRNA-21 antisense oligonucleotides to the brain using T7-peptide decorated exosomes. J Control Release. 2020;317:273–281. doi: 10.1016/j.jconrel.2019.11.009. [DOI] [PubMed] [Google Scholar]
- 54.Sutaria D.S., Jiang J., Elgamal O.A., Azevedo-Pouly A.-C.P., Pavlovicz R.E., Li C., et al. Abstract 2068: engineering of hairpin loop enhances the loading of endogenously expressed pre-miRNA into extracellular vesicles. Cancer Res. 2016;76:2068. [Google Scholar]
- 55.Li Z., Zhou X., Wei M., Gao X., Zhao L., Shi R., et al. In vitro and in vivo RNA inhibition by CD9-HuR functionalized exosomes encapsulated with miRNA or CRISPR/dCas9. Nano Lett. 2019;19:19–28. doi: 10.1021/acs.nanolett.8b02689. [DOI] [PubMed] [Google Scholar]
- 56.Duong N., Curley K., Brown A., Campanelli A., Do M.A., Levy D., et al. Decoy exosomes as a novel biologic reagent to antagonize inflammation. Int J Nanomed. 2019;14:3413–3425. doi: 10.2147/IJN.S196975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kooijmans S.A., Aleza C.G., Roffler S.R., van Solinge W.W., Vader P., Schiffelers R.M. Display of GPI-anchored anti-EGFR nanobodies on extracellular vesicles promotes tumour cell targeting. J Extracell Vesicles. 2016;5 doi: 10.3402/jev.v5.31053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Molavipordanjani S., Khodashenas S., Abedi S.M., Moghadam M.F., Mardanshahi A., Hosseinimehr S.J. Tc-radiolabeled HER2 targeted exosome for tumor imaging. Eur J Pharm Sci. 2020;148 doi: 10.1016/j.ejps.2020.105312. 99m. [DOI] [PubMed] [Google Scholar]
- 59.Shi X., Cheng Q., Hou T., Han M., Smbatyan G., Lang J.E., et al. Genetically engineered cell-derived nanoparticles for targeted breast cancer immunotherapy. Mol Ther. 2020;28:536–547. doi: 10.1016/j.ymthe.2019.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kamińska A., Enguita F.J., Stępień E. Lactadherin: an unappreciated haemostasis regulator and potential therapeutic agent. Vascul Pharmacol. 2018;101:21–28. doi: 10.1016/j.vph.2017.11.006. [DOI] [PubMed] [Google Scholar]
- 61.Siegel R.L., Miller K.D., Jemal A. Cancer statistics, 2020. CA Cancer J Clin. 2020;70:7–30. doi: 10.3322/caac.21590. [DOI] [PubMed] [Google Scholar]
- 62.Kamerkar S., LeBleu V.S., Sugimoto H., Yang S., Ruivo C.F., Melo S.A., et al. Exosomes facilitate therapeutic targeting of oncogenic KRAS in pancreatic cancer. Nature. 2017;546:498–503. doi: 10.1038/nature22341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.McAndrews K.M., Xiao F., Chronopoulos A., LeBleu V.S., Kugeratski F.G., Kalluri R. Exosome-mediated delivery of CRISPR/Cas9 for targeting of oncogenic Kras(G12D) in pancreatic cancer. Life Sci Alliance. 2021;4 doi: 10.26508/lsa.202000875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lentsch E., Li L., Pfeffer S., Ekici A.B., Taher L., Pilarsky C., et al. CRISPR/Cas9-mediated knock-out of Kras(G12D) mutated pancreatic cancer cell lines. Int J Mol Sci. 2019;20 doi: 10.3390/ijms20225706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kim S.M., Yang Y., Oh S.J., Hong Y., Seo M., Jang M. Cancer-derived exosomes as a delivery platform of CRISPR/Cas9 confer cancer cell tropism-dependent targeting. J Control Release. 2017;266:8–16. doi: 10.1016/j.jconrel.2017.09.013. [DOI] [PubMed] [Google Scholar]
- 66.Guo P., Zhang C., Chen C., Garver K., Trottier M. Inter-RNA interaction of phage Phi29 pRNA to form a hexameric complex for viral DNA transportation. Mol Cell. 1998;2:149–155. doi: 10.1016/s1097-2765(00)80124-0. [DOI] [PubMed] [Google Scholar]
- 67.Guo P. The emerging field of RNA nanotechnology. Nat Nanotechnol. 2010;5:833–842. doi: 10.1038/nnano.2010.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Jasinski D., Haque F., Binzel D.W., Guo P. Advancement of the emerging field of RNA nanotechnology. ACS Nano. 2017;11:1142–1164. doi: 10.1021/acsnano.6b05737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Binzel D.W., Li X., Burns N., Khan E., Lee W.J., Chen L.C., et al. Thermostability, tunability, and tenacity of RNA as rubbery anionic polymeric materials in nanotechnology and nanomedicine-specific cancer targeting with undetectable toxicity. Chem Rev. 2021;121:7398–7467. doi: 10.1021/acs.chemrev.1c00009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sago C.D., Lokugamage M.P., Paunovska K., Vanover D.A., Monaco C.M., Shah N.N., et al. High-throughput in vivo screen of functional mRNA delivery identifies nanoparticles for endothelial cell gene editing. Proc Natl Acad Sci U S A. 2018;115:E9944–E9952. doi: 10.1073/pnas.1811276115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zheng K., Xiol J., Reuter M., Eckardt S., Leu N.A., McLaughlin K.J., et al. Mouse MOV10L1 associates with Piwi proteins and is an essential component of the Piwi-interacting RNA (piRNA) pathway. Proc Natl Acad Sci U S A. 2010;107:11841–11846. doi: 10.1073/pnas.1003953107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Gainetdinov I., Colpan C., Cecchini K., Arif A., Jouravleva K., Albosta P., et al. Terminal modification, sequence, length, and PIWI-protein identity determine piRNA stability. Mol Cell. 2021;81:4826–4842. doi: 10.1016/j.molcel.2021.09.012. e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Barnaby S.N., Lee A., Mirkin C.A. Probing the inherent stability of siRNA immobilized on nanoparticle constructs. Proc Natl Acad Sci U S A. 2014;111:9739. doi: 10.1073/pnas.1409431111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Shu D., Shu Y., Haque F., Abdelmawla S., Guo P. Thermodynamically stable RNA three-way junction for constructing multifunctional nanoparticles for delivery of therapeutics. Nat Nanotechnol. 2011;6:658–667. doi: 10.1038/nnano.2011.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Khisamutdinov E.F., Jasinski D.L., Guo P. RNA as a boiling-resistant anionic polymer material to build robust structures with defined shape and stoichiometry. ACS Nano. 2014;8:4771–4781. doi: 10.1021/nn5006254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Germer K., Leonard M., Zhang X. RNA aptamers and their therapeutic and diagnostic applications. Int J Biochem Mol. 2013;4:27–40. [PMC free article] [PubMed] [Google Scholar]
- 77.Tuerk C., Gold L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science. 1990;249:505–510. doi: 10.1126/science.2200121. [DOI] [PubMed] [Google Scholar]
- 78.Thiviyanathan V., Gorenstein D.G. Aptamers and the next generation of diagnostic reagents. Proteomics Clin Appl. 2012;6:563–573. doi: 10.1002/prca.201200042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kumar Kulabhusan P., Hussain B., Yüce M. Current perspectives on aptamers as diagnostic tools and therapeutic agents. Pharmaceutics. 2020;12:646. doi: 10.3390/pharmaceutics12070646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Chávez J.L., Lyon W., Kelley-Loughnane N., Stone M.O. Theophylline detection using an aptamer and DNA-gold nanoparticle conjugates. Biosens Bioelectron. 2010;26:23–28. doi: 10.1016/j.bios.2010.04.049. [DOI] [PubMed] [Google Scholar]
- 81.Bruno J.G. Long shelf life of a lyophilized DNA aptamer beacon assay. J Fluoresc. 2017;27:439–441. doi: 10.1007/s10895-016-2014-x. [DOI] [PubMed] [Google Scholar]
- 82.Zhou J., Rossi J. Aptamers as targeted therapeutics: current potential and challenges. Nat Rev Drug Discov. 2017;16:181–202. doi: 10.1038/nrd.2016.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Liu M., Wang L., Lo Y., Shiu S.C.-C., Kinghorn A.B., Tanner J.A. Aptamer-enabled nanomaterials for therapeutics, drug targeting and imaging. Cells. 2022;11:159. doi: 10.3390/cells11010159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Lakhin A.V., Tarantul V.Z., Gening L.V. Aptamers: problems, solutions and prospects. Acta Nat. 2013;5:34–43. [PMC free article] [PubMed] [Google Scholar]
- 85.Khanali J., Azangou-Khyavy M., Asaadi Y., Jamalkhah M., Kiani J. Nucleic acid-based treatments against COVID-19: potential efficacy of aptamers and siRNAs. Front Microbiol. 2021;12 doi: 10.3389/fmicb.2021.758948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Shigdar S., Agnello L., Fedele M., Camorani S., Cerchia L. Profiling cancer cells by cell-SELEX: use of aptamers for discovery of actionable biomarkers and therapeutic applications thereof. Pharmaceutics. 2021;14:28. doi: 10.3390/pharmaceutics14010028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Yuan B., Jiang X., Chen Y., Guo Q., Wang K., Meng X., et al. Metastatic cancer cell and tissue-specific fluorescence imaging using a new DNA aptamer developed by Cell-SELEX. Talanta. 2017;170:56–62. doi: 10.1016/j.talanta.2017.03.094. [DOI] [PubMed] [Google Scholar]
- 88.Li W.M., Zhou L.L., Zheng M., Fang J. Selection of metastatic breast cancer cell-specific aptamers for the capture of CTCs with a metastatic phenotype by Cell-SELEX. Mol Ther Nucleic Acids. 2018;12:707–717. doi: 10.1016/j.omtn.2018.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Speransky S., Serafini P., Caroli J., Bicciato S., Lippman M.E., Bishopric N.H. A novel RNA aptamer identifies plasma membrane ATP synthase beta subunit as an early marker and therapeutic target in aggressive cancer. Breast Cancer Res Treat. 2019;176:271–289. doi: 10.1007/s10549-019-05174-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Wang L., Li P., Xiao X., Li J., Li J., Yang H.H., et al. Generating lung-metastatic osteosarcoma targeting aptamers for in vivo and clinical tissue imaging. Talanta. 2018;188:66–73. doi: 10.1016/j.talanta.2018.05.011. [DOI] [PubMed] [Google Scholar]
- 91.Wang F.B., Rong Y., Fang M., Yuan J.P., Peng C.W., Liu S.P., et al. Recognition and capture of metastatic hepatocellular carcinoma cells using aptamer-conjugated quantum dots and magnetic particles. Biomaterials. 2013;34:3816–3827. doi: 10.1016/j.biomaterials.2013.02.018. [DOI] [PubMed] [Google Scholar]
- 92.Rong Y., Chen H., Zhou X.F., Yin C.Q., Wang B.C., Peng C.W., et al. Identification of an aptamer through whole cell-SELEX for targeting high metastatic liver cancers. Oncotarget. 2016;7:8282–8294. doi: 10.18632/oncotarget.6988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Li W.M., Bing T., Wei J.Y., Chen Z.Z., Shangguan D.H., Fang J. Cell-SELEX-based selection of aptamers that recognize distinct targets on metastatic colorectal cancer cells. Biomaterials. 2014;35:6998–7007. doi: 10.1016/j.biomaterials.2014.04.112. [DOI] [PubMed] [Google Scholar]
- 94.Yoon S., Armstrong B., Habib N., Rossi J.J. Blind SELEX approach identifies RNA aptamers that regulate EMT and inhibit metastasis. Mol Cancer Res. 2017;15:811–820. doi: 10.1158/1541-7786.MCR-16-0462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Ptacek J., Zhang D., Qiu L., Kruspe S., Motlova L., Kolenko P., et al. Structural basis of prostate-specific membrane antigen recognition by the A9g RNA aptamer. Nucleic Acids Res. 2020;48:11130–11145. doi: 10.1093/nar/gkaa494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Rockey W.M., Hernandez F.J., Huang S.Y., Cao S., Howell C.A., Thomas G.S., et al. Rational truncation of an RNA aptamer to prostate-specific membrane antigen using computational structural modeling. Nucleic Acid Ther. 2011;21:299–314. doi: 10.1089/nat.2011.0313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Ng B.D., Peer D. Orchestrating a symphony on a single conjugate: aptamer targeting, gene silencing, and immunomodulation to enhance antitumor response. Mol Ther. 2017;25:5–7. doi: 10.1016/j.ymthe.2016.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Rajagopalan A., Berezhnoy A., Schrand B., Puplampu-Dove Y., Gilboa E. Aptamer-targeted attenuation of IL-2 signaling in CD8(+) T cells enhances antitumor immunity. Mol Ther. 2017;25:54–61. doi: 10.1016/j.ymthe.2016.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Sun S., Wu P. Mechanistic insights into Cu(I)-catalyzed azide−alkyne “click” cycloaddition monitored by real time infrared spectroscopy. J Phys Chem A. 2010;114:8331–8336. doi: 10.1021/jp105034m. [DOI] [PubMed] [Google Scholar]
- 100.Mushtaq S., Yun S.J., Jeon J. Recent advances in bioorthogonal click chemistry for efficient synthesis of radiotracers and radiopharmaceuticals. Molecules. 2019;24:3567. doi: 10.3390/molecules24193567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Thomas C., Rusanov T., Hoang T., Augustin T., Kent T., Gaspar I., et al. One-step enzymatic modification of RNA 3ʹ termini using polymerase θ. Nucleic Acids Res. 2019;47:3272–3283. doi: 10.1093/nar/gkz029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Dai W., Li A., Yu N.J., Nguyen T., Leach R.W., Wühr M., et al. Activity-based RNA-modifying enzyme probing reveals DUS3L-mediated dihydrouridylation. Nat Chem Biol. 2021;17:1178–1187. doi: 10.1038/s41589-021-00874-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Guo S., Vieweger M., Zhang K., Yin H., Wang H., Li X., et al. Ultra-thermostable RNA nanoparticles for solubilizing and high-yield loading of paclitaxel for breast cancer therapy. Nat Commun. 2020;11:972. doi: 10.1038/s41467-020-14780-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Jasinski D.L., Yin H., Li Z., Guo P. Hydrophobic effect from conjugated chemicals or drugs on in vivo biodistribution of RNA nanoparticles. Hum Gene Ther. 2018;29:77–86. doi: 10.1089/hum.2017.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Afonin K.A., Bindewald E., Yaghoubian A.J., Voss N., Jacovetty E., Shapiro B.A., et al. In vitro assembly of cubic RNA-based scaffolds designed in silico. Nat Nanotechnol. 2010;5:676–682. doi: 10.1038/nnano.2010.160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Afonin K.A., Grabow W.W., Walker F.M., Bindewald E., Dobrovolskaia M.A., Shapiro B.A., et al. Design and self-assembly of siRNA-functionalized RNA nanoparticles for use in automated nanomedicine. Nat Protoc. 2011;6:2022–2034. doi: 10.1038/nprot.2011.418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Shu D., Li H., Shu Y., Xiong G., Carson W.E., 3rd, Haque F., et al. Systemic delivery of anti-miRNA for suppression of triple negative breast cancer utilizing RNA nanotechnology. ACS Nano. 2015;9:9731–9740. doi: 10.1021/acsnano.5b02471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Binzel D.W., Shu Y., Li H., Sun M., Zhang Q., Shu D., et al. Specific delivery of miRNA for high efficient inhibition of prostate cancer by RNA nanotechnology. Mol Ther. 2016;24:1267–1277. doi: 10.1038/mt.2016.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Yin H., Xiong G., Guo S., Xu C., Xu R., Guo P., et al. Delivery of anti-miRNA for triple-negative breast cancer therapy using RNA nanoparticles targeting stem cell marker CD133. Mol Ther. 2019;27:1252–1261. doi: 10.1016/j.ymthe.2019.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Zhang Y., Leonard M., Shu Y., Yang Y., Shu D., Guo P., et al. Overcoming tamoxifen resistance of human breast cancer by targeted gene silencing using multifunctional pRNA nanoparticles. ACS Nano. 2017;11:335–346. doi: 10.1021/acsnano.6b05910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Rychahou P., Haque F., Shu Y., Zaytseva Y., Weiss H.L., Lee E.Y., et al. Delivery of RNA nanoparticles into colorectal cancer metastases following systemic administration. ACS Nano. 2015;9:1108–1116. doi: 10.1021/acsnano.5b00067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Lee T.J., Haque F., Shu D., Yoo J.Y., Li H., Yokel R.A., et al. RNA nanoparticle as a vector for targeted siRNA delivery into glioblastoma mouse model. Oncotarget. 2015;6:14766–14776. doi: 10.18632/oncotarget.3632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Wang H., Ellipilli S., Lee W.J., Li X., Vieweger M., Ho Y.S., et al. Multivalent rubber-like RNA nanoparticles for targeted co-delivery of paclitaxel and MiRNA to silence the drug efflux transporter and liver cancer drug resistance. J Control Release. 2020;330:173–184. doi: 10.1016/j.jconrel.2020.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Ghimire C., Wang H., Li H., Vieweger M., Xu C., Guo P. RNA nanoparticles as rubber for compelling vessel extravasation to enhance tumor targeting and for fast renal excretion to reduce toxicity. ACS Nano. 2020;14:13180–13191. doi: 10.1021/acsnano.0c04863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Matsumura Y., Maeda H. A new concept for macromolecular therapeutics in cancer chemotherapy: mechanism of tumoritropic accumulation of proteins and the antitumor agent smancs. Cancer Res. 1986;46:6387–6392. [PubMed] [Google Scholar]
- 116.Bartosik K., Debiec K., Czarnecka A., Sochacka E., Leszczynska G. Synthesis of nucleobase-modified rna oligonucleotides by post-synthetic approach. Molecules. 2020;25:3344. doi: 10.3390/molecules25153344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Paredes E., Evans M., Das S.R. RNA labeling, conjugation and ligation. Methods. 2011;54:251–259. doi: 10.1016/j.ymeth.2011.02.008. [DOI] [PubMed] [Google Scholar]
- 118.Shu Y., Haque F., Shu D., Li W., Zhu Z., Kotb M., et al. Fabrication of 14 different RNA nanoparticles for specific tumor targeting without accumulation in normal organs. RNA. 2013;19:767–777. doi: 10.1261/rna.037002.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Osborn M.F., Khvorova A. Improving siRNA delivery in vivo through lipid conjugation. Nucleic Acid Ther. 2018;28:128–136. doi: 10.1089/nat.2018.0725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Soutschek J., Akinc A., Bramlage B., Charisse K., Constien R., Donoghue M., et al. Therapeutic silencing of an endogenous gene by systemic administration of modified siRNAs. Nature. 2004;432:173–178. doi: 10.1038/nature03121. [DOI] [PubMed] [Google Scholar]
- 121.Wolfrum C., Shi S., Jayaprakash K.N., Jayaraman M., Wang G., Pandey R.K., et al. Mechanisms and optimization of in vivo delivery of lipophilic siRNAs. Nat Biotechnol. 2007;25:1149–1157. doi: 10.1038/nbt1339. [DOI] [PubMed] [Google Scholar]
- 122.Zhuang J., Tan J., Wu C., Zhang J., Liu T., Fan C., et al. Extracellular vesicles engineered with valency-controlled DNA nanostructures deliver CRISPR/Cas9 system for gene therapy. Nucleic Acids Res. 2020;48:8870–8882. doi: 10.1093/nar/gkaa683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Lin Q., Qu M., Zhou B., Patra H.K., Sun Z., Luo Q., et al. Exosome-like nanoplatform modified with targeting ligand improves anti-cancer and anti-inflammation effects of imperialine. J Control Release. 2019;311–312:104–116. doi: 10.1016/j.jconrel.2019.08.037. [DOI] [PubMed] [Google Scholar]
- 124.He F., Liu H., Guo X., Yin B.-C., Ye B.-C. Direct exosome quantification via bivalent-cholesterol-labeled DNA anchor for signal amplification. Anal Chem. 2017;89:12968–12975. doi: 10.1021/acs.analchem.7b03919. [DOI] [PubMed] [Google Scholar]
- 125.Tian Y.-F., Ning C.-F., He F., Yin B.-C., Ye B.-C. Highly sensitive detection of exosomes by SERS using gold nanostar@Raman reporter@nanoshell structures modified with a bivalent cholesterol-labeled DNA anchor. Analyst. 2018;143:4915–4922. doi: 10.1039/c8an01041b. [DOI] [PubMed] [Google Scholar]
- 126.Xu M., Yang Q., Sun X., Wang Y. Recent advancements in the loading and modification of therapeutic exosomes. Front Bioeng Biotechnol. 2020;8 doi: 10.3389/fbioe.2020.586130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Wahlgren J., Statello L., Skogberg G., Telemo E., Valadi H. Delivery of small interfering RNAs to cells via exosomes. Methods Mol Biol. 2016;1364:105–125. doi: 10.1007/978-1-4939-3112-5_10. [DOI] [PubMed] [Google Scholar]
- 128.Haney M.J., Klyachko N.L., Zhao Y., Gupta R., Plotnikova E.G., He Z., et al. Exosomes as drug delivery vehicles for Parkinson's disease therapy. J Control Release. 2015;207:18–30. doi: 10.1016/j.jconrel.2015.03.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Sutaria D.S., Badawi M., Phelps M.A., Schmittgen T.D. Achieving the promise of therapeutic extracellular vesicles: the devil is in details of therapeutic loading. Pharm Res. 2017;34:1053–1066. doi: 10.1007/s11095-017-2123-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Sun D., Zhuang X., Xiang X., Liu Y., Zhang S., Liu C., et al. A novel nanoparticle drug delivery system: the anti-inflammatory activity of curcumin is enhanced when encapsulated in exosomes. Mol Ther. 2010;18:1606–1614. doi: 10.1038/mt.2010.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Zhu Q., Heon M., Zhao Z., He M. Microfluidic engineering of exosomes: editing cellular messages for precision therapeutics. Lab Chip. 2018;18:1690–1693. doi: 10.1039/c8lc00246k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Han Y., Jones T.W., Dutta S., Zhu Y., Wang X., Narayanan S.P., et al. Overview and update on methods for cargo loading into extracellular vesicles. Processes. 2021;9:356. doi: 10.3390/pr9020356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Haraszti R.A., Miller R., Didiot M.-C., Biscans A., Alterman J.F., Hassler M.R., et al. Optimized cholesterol-siRNA chemistry improves productive loading onto extracellular vesicles. Mol Ther. 2018;26:1973–1982. doi: 10.1016/j.ymthe.2018.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Lou G., Song X., Yang F., Wu S., Wang J., Chen Z., et al. Exosomes derived from miR-122-modified adipose tissue-derived MSCs increase chemosensitivity of hepatocellular carcinoma. J Hematol Oncol. 2015;8:122. doi: 10.1186/s13045-015-0220-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Lou G., Chen L., Xia C., Wang W., Qi J., Li A., et al. MiR-199a-modified exosomes from adipose tissue-derived mesenchymal stem cells improve hepatocellular carcinoma chemosensitivity through mTOR pathway. J Exp Clin Cancer Res. 2020;39:4. doi: 10.1186/s13046-019-1512-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Zhang K., Shao C.-X., Zhu J-d, Lv X.-L., Tu C.-Y., Jiang C., et al. Exosomes function as nanoparticles to transfer miR-199a-3p to reverse chemoresistance to cisplatin in hepatocellular carcinoma. Biosci Rep. 2020;40 doi: 10.1042/BSR20194026. BSR20194026. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 137.Katakowski M., Buller B., Zheng X., Lu Y., Rogers T., Osobamiro O., et al. Exosomes from marrow stromal cells expressing miR-146b inhibit glioma growth. Cancer Lett. 2013;335:201–204. doi: 10.1016/j.canlet.2013.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Xu H., Zhao G., Zhang Y., Jiang H., Wang W., Zhao D., et al. Mesenchymal stem cell-derived exosomal microRNA-133b suppresses glioma progression via Wnt/β-catenin signaling pathway by targeting EZH2. Stem Cell Res Ther. 2019;10:381. doi: 10.1186/s13287-019-1446-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Li R., Zhao K., Ruan Q., Meng C., Yin F. Bone marrow mesenchymal stem cell-derived exosomal microRNA-124-3p attenuates neurological damage in spinal cord ischemia-reperfusion injury by downregulating Ern1 and promoting M2 macrophage polarization. Arthritis Res Ther. 2020;22:75. doi: 10.1186/s13075-020-2146-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Hung M.E., Leonard J.N. A platform for actively loading cargo RNA to elucidate limiting steps in EV-mediated delivery. J Extracell Vesicles. 2016;5 doi: 10.3402/jev.v5.31027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.de Abreu R.C., Ramos C.V., Becher C., Lino M., Jesus C., da Costa Martins P.A., et al. Exogenous loading of miRNAs into small extracellular vesicles. J Extracell Vesicles. 2021;10 doi: 10.1002/jev2.12111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Tang Y., Wang X., Li J., Nie Y., Liao G., Yu Y., et al. Overcoming the reticuloendothelial system barrier to drug delivery with a “Don't-Eat-Us” strategy. ACS Nano. 2019;13:13015–13026. doi: 10.1021/acsnano.9b05679. [DOI] [PubMed] [Google Scholar]
- 143.Harisa G.I., Badran M.M., Alanazi F.K., Attia S.M. An overview of nanosomes delivery mechanisms: trafficking, orders, barriers and cellular effects. Artif Cells Nanomed Biotechnol. 2018;46:669–679. doi: 10.1080/21691401.2017.1354301. [DOI] [PubMed] [Google Scholar]
- 144.Mayor S., Pagano R.E. Pathways of clathrin-independent endocytosis. Nat Rev Mol. 2007;8:603–612. doi: 10.1038/nrm2216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Suzuki M., Iwaki K., Kikuchi M., Fujiwara K., Doi N. Characterization of the membrane penetration-enhancing peptide S19 derived from human syncytin-1 for the intracellular delivery of TAT-fused proteins. Biochem Biophys Res Commun. 2022;586:63–67. doi: 10.1016/j.bbrc.2021.11.065. [DOI] [PubMed] [Google Scholar]
- 146.Erazo-Oliveras A., Najjar K., Dayani L., Wang T.Y., Johnson G.A., Pellois J.P. Protein delivery into live cells by incubation with an endosomolytic agent. Nat Methods. 2014;11:861–867. doi: 10.1038/nmeth.2998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Uddin N., Warriner L.W., Pack D.W., DeRouchey J.E. Enhanced gene delivery and CRISPR/Cas9 homology-directed repair in serum by minimally succinylated polyethylenimine. Mol Pharm. 2021;18:3452–3463. doi: 10.1021/acs.molpharmaceut.1c00368. [DOI] [PubMed] [Google Scholar]
- 148.Chen X., Liu H., Li A., Ji S., Fei H. Hydrophobicity-tuned anion responsiveness underlies endosomolytic cargo delivery mediated by amphipathic vehicle peptides. J Biol Chem. 2021;297 doi: 10.1016/j.jbc.2021.101364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Liu C., Zhou Q., Li Y., Garner L.V., Watkins S.P., Carter L.J., et al. Research and development on therapeutic agents and vaccines for COVID-19 and related human coronavirus diseases. ACS Cent Sci. 2020;6:315–331. doi: 10.1021/acscentsci.0c00272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Heinz F.X., Stiasny K. Distinguishing features of current COVID-19 vaccines: knowns and unknowns of antigen presentation and modes of action. npj Vaccines. 2021;6:104. doi: 10.1038/s41541-021-00369-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Xiao Y., Tang Z., Huang X., Chen W., Zhou J., Liu H., et al. Emerging mRNA technologies: delivery strategies and biomedical applications. Chem Soc Rev. 2022;51:3828–3845. doi: 10.1039/d1cs00617g. [DOI] [PubMed] [Google Scholar]
- 152.Huang X., Wu G., Liu C., Hua X., Tang Z., Xiao Y., et al. Intercalation-driven formation of siRNA nanogels for cancer therapy. Nano Lett. 2021;21:9706–9714. doi: 10.1021/acs.nanolett.1c03539. [DOI] [PubMed] [Google Scholar]
- 153.Chen W., Schilperoort M., Cao Y., Shi J., Tabas I., Tao W. Macrophage-targeted nanomedicine for the diagnosis and treatment of atherosclerosis. Nat Rev Cardiol. 2022;19:228–249. doi: 10.1038/s41569-021-00629-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Mulcahy L.A., Pink R.C., Carter D.R.F. Routes and mechanisms of extracellular vesicle uptake. J Extracell Vesicles. 2014;3 doi: 10.3402/jev.v3.24641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Gurung S., Perocheau D., Touramanidou L., Baruteau J. The exosome journey: from biogenesis to uptake and intracellular signalling. Cell Commun Signal. 2021;19:47. doi: 10.1186/s12964-021-00730-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Heath N., Osteikoetxea X., de Oliveria T.M., Lázaro-Ibáñez E., Shatnyeva O., Schindler C., et al. Endosomal escape enhancing compounds facilitate functional delivery of extracellular vesicle cargo. Nanomed J. 2019;14:2799–2814. doi: 10.2217/nnm-2019-0061. [DOI] [PubMed] [Google Scholar]
- 157.Costa Verdera H., Gitz-Francois J.J., Schiffelers R.M., Vader P. Cellular uptake of extracellular vesicles is mediated by clathrin-independent endocytosis and macropinocytosis. J Control Release. 2017;266:100–108. doi: 10.1016/j.jconrel.2017.09.019. [DOI] [PubMed] [Google Scholar]
- 158.Joshi B.S., de Beer M.A., Giepmans B.N.G., Zuhorn I.S. Endocytosis of extracellular vesicles and release of their cargo from endosomes. ACS Nano. 2020;14:4444–4455. doi: 10.1021/acsnano.9b10033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Meng Q.F., Zhao Y., Dong C., Liu L., Pan Y., Lai J., et al. Genetically programmable fusion cellular vesicles for cancer immunotherapy. Angew Chem Int Ed Engl. 2021;60:26320–26326. doi: 10.1002/anie.202108342. [DOI] [PubMed] [Google Scholar]
- 160.Meldolesi J. Exosomes and ectosomes in intercellular communication. Curr Biol. 2018;28:R435–R444. doi: 10.1016/j.cub.2018.01.059. [DOI] [PubMed] [Google Scholar]
- 161.Prada I., Meldolesi J. Binding and fusion of extracellular vesicles to the plasma membrane of their cell targets. Int J Mol Sci. 2016;17:1296. doi: 10.3390/ijms17081296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Horibe S., Tanahashi T., Kawauchi S., Murakami Y., Rikitake Y. Mechanism of recipient cell-dependent differences in exosome uptake. BMC Cancer. 2018;18:47. doi: 10.1186/s12885-017-3958-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.O'Brien K., Breyne K., Ughetto S., Laurent L.C., Breakefield X.O. RNA delivery by extracellular vesicles in mammalian cells and its applications. Nat Rev Mol. 2020;21:585–606. doi: 10.1038/s41580-020-0251-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Soares A.R., Martins-Marques T., Ribeiro-Rodrigues T., Ferreira J.V., Catarino S., Pinho M.J., et al. Gap junctional protein Cx43 is involved in the communication between extracellular vesicles and mammalian cells. Sci Rep. 2015;5 doi: 10.1038/srep13243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Meyer C., Losacco J., Stickney Z., Li L., Marriott G., Lu B. Pseudotyping exosomes for enhanced protein delivery in mammalian cells. Int J Nanomed. 2017;12:3153–3170. doi: 10.2147/IJN.S133430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Williams C., Pazos R., Royo F., González E., Roura-Ferrer M., Martinez A., et al. Assessing the role of surface glycans of extracellular vesicles on cellular uptake. Sci Rep. 2019;9 doi: 10.1038/s41598-019-48499-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Binzel D.W., Guo S., Yin H., Lee T.J., Liu S., Shu D., et al. Rational design for controlled release of Dicer-substrate siRNA harbored in phi29 pRNA-based nanoparticles. Mol Ther Nucleic Acids. 2021;25:524–535. doi: 10.1016/j.omtn.2021.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Li Z., Yang L., Wang H., Binzel D.W., Williams T.M., Guo P. Non-small-cell lung cancer regression by siRNA delivered through exosomes that display EGFR RNA aptamer. Nucleic Acid Ther. 2021;31:364–374. doi: 10.1089/nat.2021.0002. [DOI] [PMC free article] [PubMed] [Google Scholar]