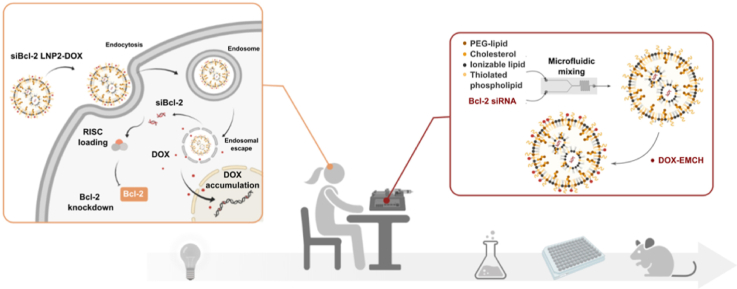
Abstract
Evasion of apoptosis is a hallmark of cancer, attributed in part to overexpression of the anti-apoptotic protein B-cell lymphoma 2 (Bcl-2). In a variety of cancer types, including lymphoma, Bcl-2 is overexpressed. Therapeutic targeting of Bcl-2 has demonstrated efficacy in the clinic and is the subject of extensive clinical testing in combination with chemotherapy. Therefore, the development of co-delivery systems for Bcl-2 targeting agents, such as small interfering RNA (siRNA), and chemotherapeutics, such as doxorubicin (DOX), holds promise for enabling combination cancer therapies. Lipid nanoparticles (LNPs) are a clinically advanced nucleic acid delivery system with a compact structure suitable for siRNA encapsulation and delivery. Inspired by ongoing clinical trials of albumin-hitchhiking doxorubicin prodrugs, here we developed a DOX-siRNA co-delivery strategy via conjugation of doxorubicin to the surface of siRNA-loaded LNPs. Our optimized LNPs enabled potent knockdown of Bcl-2 and efficient delivery of DOX into the nucleus of Burkitts’ lymphoma (Raji) cells, leading to effective inhibition of tumor growth in a mouse model of lymphoma. Based on these results, our LNPs may provide a platform for the co-delivery of various nucleic acids and DOX for the development of new combination cancer therapies.
Key words: Lipid nanoparticles, Doxorubicin, Bcl-2, siRNA delivery, Chemotherapy, Lymphoma
Graphical abstract
LNPs have been shown to efficiently encapsulate siRNA and enable potent intracellular delivery for gene knockdown. Doxorubicin-conjugated siRNA LNPs were engineered to achieve potent chemo- and RNA interference therapy.
1. Introduction
The World Health Organization estimated that 10 million cancer deaths occurred worldwide in 2020, an increase from 9.4 million cancer-related cases in 20181,2. Cancer treatment typically involves the use of systemically administered therapies, such as chemotherapy or immunotherapy, or localized therapies (e.g., surgery) used separately or in combination3. For patients with non-Hodgkin’s lymphoma, chemotherapy is a major treatment option4. However, many cancer patients do not respond to chemotherapy or lose responsiveness over time, in part due to cancer cell resistance to apoptosis5.
Recent studies indicate that evasion of apoptosis occurs through several mechanisms, including the enhancement of DNA repair by nonhomologous end-joining proteins or the overexpression of anti-apoptotic proteins, which can act individually or synergistically6. One major anti-apoptotic factor is the B cell lymphoma 2 (Bcl-2) family of proteins, discovered almost 30 years ago in non-Hodgkin’s lymphoma7,8. Bcl-2 is incorporated into the membranes of mitochondria and endoplasmic reticulum, which can prevent the release of apoptosis-inducing factor and cytochrome c, inhibiting caspase-mediated cell apoptosis9. Due to the important role of Bcl-2 in the apoptotic pathway, several therapeutic strategies have been developed to inhibit or downregulate the Bcl-2 protein10. For example, Bcl-2-targeting antisense oligonucleotides (ASOs)—such as Oblimersen, SPC2996 LNA gapmer, or PNT2258—were developed and tested in clinical trials against various types of cancers including multiple myeloma and leukemia11,12. In 2016, a small molecule inhibitor of Bcl-2, Venetoclax, was approved as a monotherapy by the US Food and Drug Administration (FDA) for treating patients with chronic lymphocytic leukemia13. However, due to the limited benefit of this monotherapy, many clinical trials are currently ongoing to evaluate combination therapies with various chemotherapeutics, such as doxorubicin (DOX)14. DOX acts as a pro-apoptotic drug which can cause cell death15,16. However, the overexpression of Bcl-2 by cancer cells can largely counteract the pharmacological effect of DOX17. Therefore, development of a clinically advanced platform to co-deliver Bcl-2-targeting therapeutics and DOX would significantly advance combination cancer therapies18,19.
Recent studies have shown that co-delivery of DOX and other nucleic acids can enhance the therapeutic effect against many cancer types. Studies have reported promising results using fibroin to deliver survivin siRNA and DOX for breast cancer therapy, as well as short hairpin RNA and DOX against gastric cancer20,21. Additionally, CRISPR/Cas9 and shRNA-expressing plasmids have been synthesized for potential co-delivery with DOX as a promising strategy for cancer therapy22. In recent years, efforts have been made to co-deliver Bcl-2 siRNA (siBcl-2) and DOX for combination cancer therapies against ovarian cancer, glioma, and other cancer types23,24,25. So far, nanoparticles are the most widely used drug carriers for these applications due to their multi-functionality and ability to either encapsulate or conjugate drugs26. However, most of these delivery systems are non-degradable, complex, and suffer from insufficient biological activity27. Lipid nanoparticles (LNPs) are a promising platform for the co-delivery of Bcl-2 siRNA and DOX, as they are a clinically advanced delivery system shown to effectively encapsulate siRNA and facilitate its intracellular delivery28. The success of LNPs lies in their ability to remain stable at physiologically relevant neutral pH but become ionized under acidic conditions—such as the endosomal compartment of the cell—which enables LNPs to escape from the endosome and release siRNA into the cytosol29,30. Over the last two decades, there has been significant progress in the development of LNP-based nucleic acid delivery systems31. The first LNP-based siRNA therapeutic, Onpattro, was approved by the FDA in 2018 for the treatment of hereditary transthyretin-mediated polyneuropathy32. More recently, two LNP-based COVID-19 mRNA vaccines received emergency use authorization33,34.
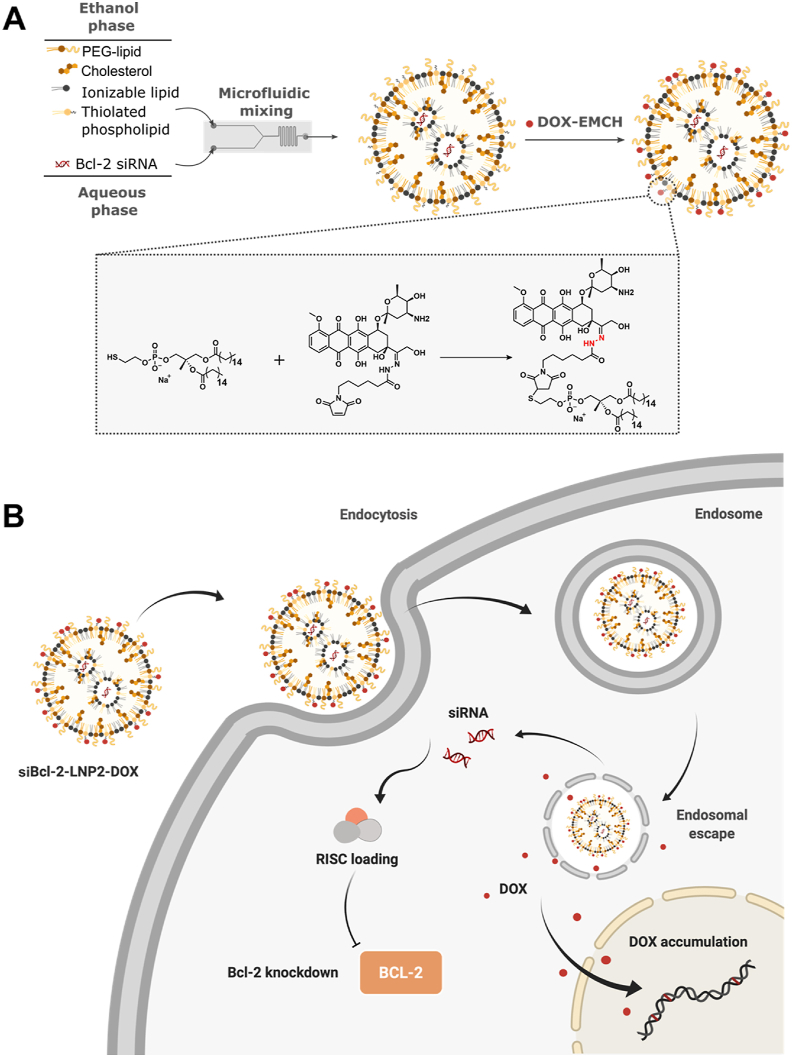
Unlike liposomes, such as those used in the FDA-approved liposomal doxorubicin drug Doxil, LNPs have a compact spherical structure and are highly optimized for siRNA delivery35,36. Therefore, incorporation of chemotherapeutic drugs into siRNA-LNPs by physical encapsulation may disrupt its highly organized structure and cause potential issues, such as low encapsulation or destabilization of its spherical shape due to accumulation in the lipid bilayer37. To circumvent these issues, we developed a strategy to conjugate doxorubicin directly to LNPs. We modified formulated siRNA-LNPs using a thiol-maleimide Michael addition click reaction to conjugate a (6-maleimidocaproyl) hydrazone derivative of DOX (DOX-EMCH) to the LNP surface. DOX-EMCH is a clinical prodrug of DOX possessing a maleimide group and an acid-cleavable hydrazone bond, which can bind to albumin following intravenous injection to prolong its circulation half-life and release parental drug in an acidic environment38. Inspired by this process, we formulated a thiolated LNP with a sulfur-containing phospholipid to conjugate DOX-EMCH to the surface of LNPs. Our optimized LNP formulation, siBcl-2 LNP2-DOX, achieved potent knockdown of Bcl-2 in Burkitts’ lymphoma (Raji) cells while also successfully delivering DOX to the cell nucleus. We also provide in vivo evidence that siBcl-2 LNP2-DOX can effectively inhibit tumor growth in a NSG mouse model of lymphoma. These results suggest that our LNPs may provide a platform for the co-delivery of various nucleic acids and DOX, which could be used to develop combination therapies for a wide range of cancers.
2. Materials and methods
2.1. Materials
Aldoxorubicin (DOX-EMCH) was purchased from MedChem Express (Monmouth Junction, NJ, USA). Cholesterol and dimethyl sulfoxide (DMSO) were purchased from Sigma-Aldrich (St. Louis, MO, USA). 1,2-Dimyristoyl-sn-glycero-3-phosphoethanolamine-N-[methoxy (polyethylene glycol)-2000] (ammonium salt, C14-PEG2000), 1,2-dipalmitoyl-sn-glycero-3-phosphathioethanol (sodium salt, PTE) and 1,2-distearoyl-sn-glycero-3-phosphocholine (DSPC) were purchased from Avanti (Alabaster, AL, USA). The ionizable lipid 1,1′-((2-(4-(2-((2-(bis(2-hydroxydodecyl)amino)ethyl) (2 hydroxydodecyl) amino)ethyl)piperazin-1-yl)ethyl)azanediyl)bis (dodecan-2-ol) (C12-200) was synthesized as previously described39. Human ON-TARGETplus SMARTpool Bcl-2 siRNA and Luciferase siRNA-1 were purchased from Horizon Discovery (Waterbeach, UK).
2.2. Formulation of LNPs
The ionizable lipid C12-200 was diluted in ethanol with cholesterol, DSPC or PTE, and C14-PEG-2000 at a 50:38.5:10:1.5 M ratio in a total volume of 100 μL. Luciferase (siLuc) or Bcl-2 (siBcl-2) siRNA was dissolved in 10 mmol/L citrate buffer (pH 3.0) at weight ratios of 5:1 or 15:1 (ionizable lipid:siRNA), in a total volume of 300 μL. The aqueous and ethanol phases were combined via mixing in a microfluidic device at a volume ratio of 3:1 (citrate buffer:ethanol, v/v)40. Two LNPs with different ionizable lipid:siRNA weight ratios (LNP1 5:1 and LNP2 15:1) were prepared. LNPs were dialyzed against 1 × PBS for 2 h before sterile filtration via 0.22 μm syringe filters (Genesee Scientific, El Cajon, CA, USA). Next, DOX-EMCH (1.2 mmol) was added to LNPs (1 mmol PTE) and incubated with shaking (250 rpm) for 2 h. Afterwards, the mixture was dialyzed against 1 × PBS for 2 h, sterilized through 0.22 μm syringe filters, and kept at 4 °C. The coupling product, DOX-EMCH-PTE, was characterized using 1H NMR spectrum, performing on a NEO 400 MHz spectrometer. A modified Quant-iT RiboGreen assay (ThermoFisher, Waltham, MA, USA) was used to obtain the concentration and encapsulation efficiency of siRNA in LNPs by comparing fluorescence intensity (λex = 490 nm, λem = 530 nm) in the presence and absence of Triton X-10041. A spectrofluorimetric measurement was used to determine the concentration of DOX conjugated onto LNPs.
2.3. Physicochemical characterization of LNPs
20 μL of LNP solution was combined with 780 μL of H2O in 4 mL disposable cuvettes or DTS1070 zeta cuvettes for dynamic light scattering (DLS) and zeta potential (ζ) measurements. Measurements were performed using a Zetasizer Nano (Malvern Instruments, Malvern, UK) in triplicate, and reported as average diameter (z-average) ± SD and polydispersity index (PDI) from three experimental runs. The DLS measurements for final LNP2 and LNP2-DOX were taken on Days 1 and 7. Light absorption spectra were obtained in a wide wavelength range of 350–650 nm with 2 nm intervals at 24 °C using an Infinite M Plex (Tecan, Morrisville, NC, USA) absorbance plate reader. Measurements were done in 100 μL of 1 PBS (pH 7.4) containing the same concentration of LNP2, DOX, and LNP2 conjugated with DOX (LNP2-DOX) using a transparent 96-well plate.
2.4. Transmission electron microscopy (TEM)
A JEOL 1010 electron microscope system (Jeol, Tokyo, Japan) was used to acquire transmission electron microscopy (TEM) images. 10 μL of LNP2-DOX suspension was deposited on thin carbon films supported by copper grids (Ted Pella Inc., Redding, CA, USA) and was dried at 37 °C. The sample was then stained with 2% uranyl acetate (Electron Microscopy Sciences, Hatfield, PA, USA) for 10 min. TEM was operated under a 80 kV voltage mode.
2.5. Doxorubicin release profile
To assess DOX release in a physiological and slightly acidic environment, LNP2-DOX was suspended in 150 μL of 1 × PBS buffer solution adjusted to pH 7.4 or 5.1. The suspensions were placed into dialysis cassettes (MWCO: 20 kDa), and subsequently placed into a beaker with 100 mL of PBS buffer at the same pH values at 37 °C. Beakers were constantly stirred in the dark at 250 rpm for 24 h. 10 μL of the solution inside the dialysis cassettes was collected at the indicated time points and fluorescence (λex = 470 nm, λem = 595 nm) of the samples was detected using an Infinite M Plex (Tecan, Morrisville, NC, USA) fluorescence plate reader to determine the amount of DOX released.
2.6. Cell culture
The human Burkitt lymphoma (Raji) cell line and luciferase-expressing Raji cell line (Raji Luc +) were purchased from American Type Culture Collection (ATCC, Manassas, VA, USA). Both cell lines were cultured in RPMI medium (ThermoFisher, Waltham, MA, USA) supplemented with 10% (v/v) fetal bovine serum (FBS) and 100 U/mL/100 μg/mL penicillin/streptomycin. Cells were maintained at 37 °C in a humidified atmosphere with 5% CO2. Cells were passaged every 2–3 days after reaching 70% confluence.
2.7. In vitro cytotoxicity and luciferase knockdown assays
Luc + Raji cells were plated at 1 × 104 cells per well in 96-well plates in 100 μL of RPMI media. Cells were then treated with LNPs at a siRNA dose of 50 nmol/L diluted in RPMI. For dose-dependent cytotoxicity analysis, cells were treated with LNPs at concentrations ranging from 10 to 100 nmol/L of siRNA. DharmaFECT 1 Transfection Reagent (Horizon, Cambridge, UK) was used as a positive control of knockdown according to the manufacturer’s protocol. Cell viability was measured using a CellTiter-Glo Luminescent Cell Viability assay according to the manufacturer’s protocol. Luciferase expression was measured using Luciferase1000 according to the manufacturer’s protocol. Cell luminescence was quantified using an Infinite M Plex plate reader (Tecan, Morrisville, NC, USA) and normalized to untreated cells. IC50 for siRNA knockdown was determined using GraphPad Prism 8 software through non-linear regression.
2.8. Confocal microscopy
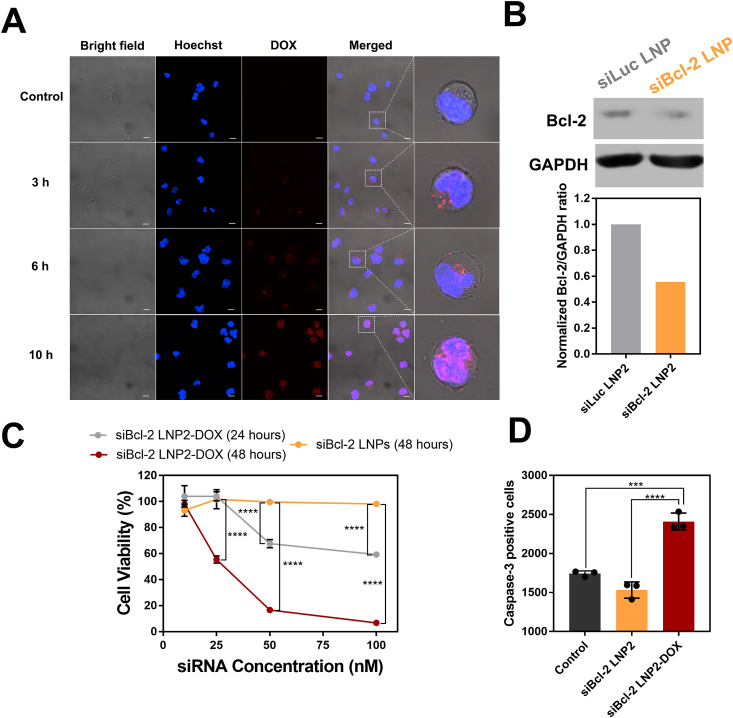
Raji cells were plated at 5 × 105 cells per well in a 6-well plate in 1 mL of RPMI media and incubated with 50 nmol/L of LNP2-DOX for 3, 6, and 10 h at 37 °C. Nuclei were stained using Hoechst 33342 (10 μg/mL). Cells were transferred into glass-bottom chambers and imaged using a confocal laser scanning microscope Zeiss LSM 710 (Zeiss, White Plains, NY, USA) with a 63 × oil immersion lens. For siBcl-2-DOX-LNPs, the excitation wavelength was set to 479 nm and emission was detected at 595 nm.
2.9. Western blot
Raji cells were plated at 5 × 105 cells per well in a 6-well plate in 1 mL of RPMI media and incubated with 50 nmol/L of Bcl-2 siRNA-encapsulating LNP2 (siBcl-2-LNP2) and Luc siRNA-encapsulating LNP2 (siLuc-LNP2) for 48 h at 37 °C. Cells were then collected, and total cell protein was extracted. Cells were lysed using lysis buffer, and the concentrations of total protein were measured using a BCA assay (Thermo Scientific). 40 μg of protein was then separated using 10% SDS-polyacrylamide gel electrophoresis (SDS-PAGE), and protein was then transferred to PVDF membranes. After blocking with 5% non-fat milk, the blots were incubated overnight with anti-Bcl-2 primary antibodies (R&D Systems) at 4 °C. After repeated washing, the blots were incubated with secondary antibodies at room temperature for 1 h. Protein expression was detected using an enhanced chemiluminescence reagent (Thermo Scientific). GAPDH was used as an internal control.
2.10. Caspase-3 assay
Caspase-3 activity in Raji cells was measured using a Caspase-3 Assay Kit (ab39401, Abcam, Waltham, MA, USA). Raji cells were seeded in a 6-well plate (5 × 105 cells/well) and then treated with 50 nmol/L of siBcl-2-LNP2 or siBcl-2-LNP2-DOX for 6 h. Raji cells were collected and incubated at 37 °C in the dark for 1 h with a fluorescein isothiocyanate conjugate of caspase inhibitor FITC-DEVD-FMK. Caspase-3 signal was quantified using flow cytometry according to the manufacturer’s protocol.
2.11. Animal experiments
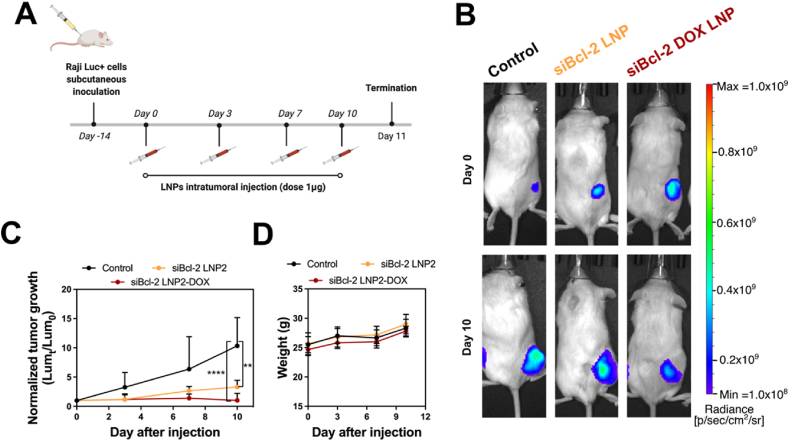
All animal use was in accordance with the guidelines and approval from the University of Pennsylvania Institution of Animal Care and Use Committee. Male NSG mice aged 6–8 weeks were inoculated with 2 × 106 Raji Luc + cells per mouse by subcutaneous injection. After two weeks, mice were separated randomly into the following treatment groups: PBS, siBcl-2-LNP2, and siBcl-2-LNP2-DOX treatment (n = 5). PBS and LNP formulations were administered via intratumoral injection every three or four days for 11 days. Mice were treated with LNPs at an siRNA dose of 1 μg/mouse (DOX concentration: 80 μg/mL). Tumor growth curves were plotted based on normalized luminescence intensity to day 0 (Lumt/Lum0). Bioluminescence imaging was performed using an IVIS Spectrum Imaging system (Caliper Life Sciences, Waltham, MA, USA) after mice were injected with 150 mg/kg of D-luciferin potassium salt.
2.12. Statistical analysis
Data represent the mean ± SD for triplicate measurements in each experiment. The results were evaluated statistically with GraphPad Prism 8 software. One-way variance analysis (ANOVA) followed by the post-hoc RIR Tukey’s test was applied. Significance level was established at P < 0.05.
3. Results and discussion
3.1. Formulation of LNPs encapsulating siRNA
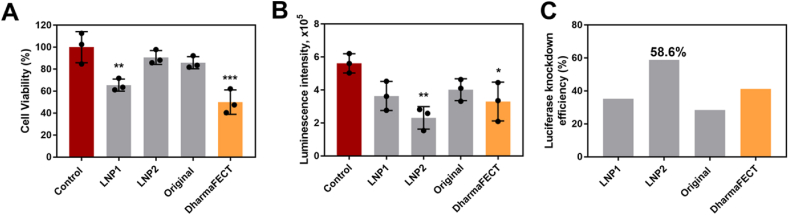
In this study, LNPs were chosen for the co-delivery of siRNA and DOX to lymphoma cells due to their physicochemical properties and in vivo potency. We first formulated two siRNA-encapsulating LNPs (LNP1 and LNP2) comprising four components: a gold standard ionizable lipid (C12-200), cholesterol, a thiolated phospholipid (PTE), and a lipid-anchored polyethylene glycol (PEG) conjugate (C14-PEG-2000)42,43. The ionizable lipid to siRNA weight ratios were 5:1 and 15:1 for LNP1 and LNP2, respectively (Table 1). The LNPs were formulated by microfluidic mixing at a component molar ratio of 50 ionizable lipid:38.5 cholesterol:10 PTE:1.5 PEG (Fig. 1A)44. These molar ratios were selected based on previously optimized data for efficient siRNA delivery45. The original C12-200 LNP incorporating DSPC was also formulated to serve as a positive control. Resulting LNPs were 65–95 nm in terms of z-average diameter with siLuc encapsulation efficiencies between 48% and 72% (Table 1). Additionally, PDIs of the original C12-200 LNP and LNP2 were low (0.032 and 0.078, respectively), while the PDI of LNP1 was equal to 1.0. These PDI values suggested that the original C12-200 LNP and LNP2 formulations were monodisperse. The high PDI value for LNP1 indicated the formulation was polydisperse, and thus suggested that these LNPs had a broad size distribution. All LNPs were neutral or slightly negatively charged, which is consistent with previous studies46. All three LNPs loaded with siRNA against luciferase (siLuc) were then evaluated for their in vitro toxicity and ability to knockdown luciferase in a Raji Luc + cell line. After treatment with LNP1 and DharmaFECT (a commercial siRNA transfection reagent), cell viability was significantly reduced compared to control samples (untreated group, Fig. 2A). By contrast, LNP2 and the original LNP induced minimal toxic effects at the same concentration. Furthermore, treatment with LNPs led to a decrease in luminescence signal intensity in all groups. However, LNP1, DharmaFECT, and the original LNP were less effective than LNP2 at silencing luciferase expression (Fig. 2B). LNP2 was the most effective formulation, with approximately 60% luciferase silencing efficacy (Fig. 2C). Collectively, these results suggested that LNP2 was the optimal formulation for siRNA knockdown, and thus was selected for subsequent DOX conjugation.
Table 1.
LNP characterization data consisting of hydrodynamic diameter, polydispersity index (PDI), encapsulation efficiency (EE), and zeta potential of each LNP formulation (±standard deviation).
| LNP | Phospholipid | EE (%) | Ionizable lipid: siRNA (wt:wt) | Size (nm) | PDI | Zeta potential (mV) |
|---|---|---|---|---|---|---|
| Original | DSPC | 71.80 | 5:1 | 65.59 ± 1.85 | 0.032 | −4.96 ± 1.13 |
| LNP1 | PTE | 48.25 | 5:1 | 95.06 ± 5.29 | 1.000 | −13.83 ± 3.73 |
| LNP2 | PTE | 65.06 | 15:1 | 69.88 ± 4.16 (Day 1) 71.91 ± 3.18 (Day 7) |
0.078 (Day 1) 0.030 (Day 7) |
−5.02 ± 1.68 |
| LNP2-DOX | PTE | 65.00 | 15:1 | 72.60 ± 1.49 (Day 1) 79.18 ± 1.55 (Day 7) |
0.120 (Day 1) 0.174 (Day 7) |
−6.42 ± 0.27 |
Figure 1.
(A) A schematic illustrating the preparation of doxorubicin-conjugated Bcl-2 siRNA-loaded lipid nanoparticles (siBcl-2 LNP2-DOX). LNPs were formulated by microfluidic mixing, followed by conjugation with DOX through a thiol-maleimide Michael addition click reaction between DOXO-EMCH and thiolated phospholipid PTE (B) Schematic diagram of intracellular co-delivery of siBcl2 and DOX enabled by siBcl-2 LNP2-DOX.
Figure 2.
LNP-mediated luciferase knockdown in vitro (A) Viability of Raji cells treated with 50 nmol/L siLuc for 48 h using different LNP formulations. Data are plotted as mean ± SD, n = 3; ∗∗P < 0.05, ∗∗∗P < 0.001 vs. Control (B) Luciferase expression in Raji cells after treatment with different LNP formulations for 48 h at a dose of 50 nmol/L siLuc. Data are plotted as mean ± SD, n = 3; ∗P < 0.05, ∗∗P < 0.005 vs. Control (C) Average luciferase knockdown efficiency in Raji cells. Data are plotted as average mean vs. Control.
3.2. Synthesis and characterization of DOX-conjugated siRNA LNPs
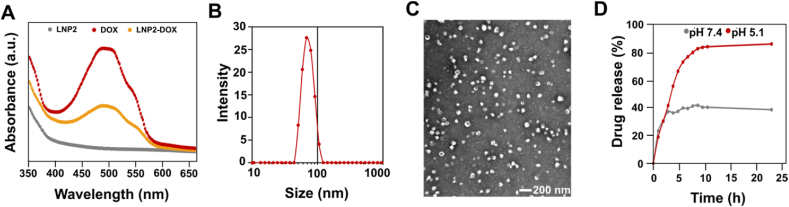
After the initial screening, DOXO-EMCH, a clinically tested maleimide-containing prodrug, was chosen for conjugation to LNPs. DOXO-EMCH contains a pH-sensitive hydrazone bond, which is relatively stable in blood plasma, but cleavable under slightly acidic conditions such as those in the endosome47. To synthesize LNP2-DOX, we utilized a well-known click chemistry reaction between thiol and maleimide groups that results in a thiosuccinimide product (Fig. 1A)48. DOXO-EMCH was incubated with LNP2 containing free thiol groups. The reaction product was then dialyzed against 1 × PBS, sterilized using syringe filters, and characterized. Examination of the 1H NMR spectra (Supporting Information Fig. S1) reveals the disappearance of alkene resonance from the maleimide group (δ = 6.94 ppm) and the appearance of assignable resonances (δ = 3.73 and 3.85 ppm) from the succinimide functionality, demonstrating the successful conjugation of DOX-EMCH onto the phospholipid. DOX conjugation was further examined by absorbance spectroscopy. The UV–Vis absorption spectrum of free DOX along with the spectra of LNP2 and LNP2-DOX are depicted in Fig. 3A. The LNP2-DOX spectrum show the characteristic shape of the DOX spectrum, indicating successful conjugation. Importantly, the reaction with DOXO-EMCH does not affect the encapsulation efficiency of siRNA in LNPs (Table 1).
Figure 3.
Physicochemical characterization of DOX-conjugated siRNA LNPs (A) UV–Vis spectra of LNP2, DOX, and LNP2-DOX in PBS (B) The size (z-average) distribution of a representative sample of siBcl-2 LNP2-DOX, revealing a diameter of 70 nm using DLS (C) TEM image of siBcl-2 LNP2-DOX. Scale bar = 200 nm (D) The time-dependent release profiles of DOX from Bcl-2 LNP-DOX at pH 5.1 and pH 7.4.
We then measured the z-average diameter and PDI of LNP2-DOX, and no significant changes in z-average diameter or PDI were observed compared to LNP2 (Fig. 3B). Nonsignificant changes were observed in z-average diameter and PDI 7 days post LNP2-DOX formulation. Additionally, the surface charge of LNP2-DOX was similar to that of unconjugated LNP2. Next, the size of LNP2-DOX and its morphology were confirmed by TEM. The TEM image of LNP2-DOX shows a monodisperse spherical structure with an average size of ∼70 nm, which was comparable with DLS results (Fig. 3C). Since the DOXO-EMCH used for LNP2-DOX synthesis is an acid-sensitive prodrug, we investigated the DOX release from LNP2-DOX via hydrolysis of the hydrazone bond (Supporting Inforamtion Fig. S2)49. We performed a drug release experiment in 1 × PBS buffer solution adjusted to pH 5.1, which corresponds to the pH of the late endosome/lysosome (Fig. 3D)50. At pH 5.1, 80% of DOX was released within 8 h, and reached almost 90% at 24 h. In comparison, only 40% of DOX was released from LNP2-DOX at pH 7.4 within 8 h. These results suggest that DOX can successfully be conjugated to the LNP2 formulation and achieve drug release under acidic conditions.
3.3. Bcl-2 knockdown and DOX cytotoxicity in vitro
A key aspect of siRNA and DOX action is cellular uptake and release into cancer cells (Fig. 1B). To investigate the intracellular distribution of LNP2-DOX, LNP-treated Raji cells were assessed using confocal microscopy. After 3 h of incubation, strong punctate signals indicative of DOX fluorescence appeared in the cytoplasm (Fig. 4A). These puncta indicate that LNP2-DOX was taken up through the endocytic pathway and trapped initially in endosomal compartments, similar to other LNPs reported previously51. Based on pH-dependent drug release results, it was expected that accelerated DOX release can be achieved in the acidic endosome. After 6 h of incubation, a weak red fluorescent signal appeared in the nucleus, suggesting that DOX can be successfully released from LNP2-DOX and accumulate in the nucleus. At 10 h post LNP2-DOX treatment, the DOX fluorescence signal was predominantly localized in the nucleus, where DOX can intercalate within DNA and induce cell apoptosis52.
Figure 4.
Intracellular delivery, Bcl-2 knockdown, and caspase-3 activity in vitro using DOX-conjugated siRNA LNPs (A) Confocal microscopy images of Raji cancer cells. Cells were incubated with siBcl-2 LNP2-DOX for 3, 6, and 10 h. Scale bar = 10 μm (B) Detection of Bcl-2 knockdown by western blot (C) Viability of Raji cells treated at a range of concentrations (0–100 nmol/L) for 24 or 48 h using different LNP formulations. Data are plotted as mean ± SD, n = 3; ∗∗P < 0.005, ∗∗∗∗P < 0.0001 vs. Control (D) Caspase-3 activity assessed by a FITC-DEVD-FMK probe after 6 h of incubation with 50 nM of Bcl-2 LNP and Bcl-2 LNP2-DOX. The mean fluorescence intensity of FITC-DEVD-FMK-stained cells was presented. Data are plotted as mean ± SD, n = 3; ∗∗∗P < 0.001, ∗∗∗∗P < 0.0001, vs. indicated.
An important consideration for the combination of DOX and Bcl-2 knockdown is to enhance cytotoxicity when cellular anti-apoptotic defense is dampened. First, knockdown of anti-apoptotic Bcl-2 protein by siBcl-2 LNP was confirmed by western blot. The results showed that after treatment with siBcl-2 LNP2, Bcl-2 was down-regulated to 55% of that of the control group (Fig. 4B). However, based on viability measurements of siBcl-2 LNP2-treated cells, knockdown of Bcl-2 alone was unable to trigger cell death after 48 h at siRNA doses ranging from 10 to 100 nmol/L (Fig. 4C). These results motivated us to explore the cytotoxicity of siBcl-2 LNP2-DOX. Raji cells incubated with a high dose of siBcl-2 LNP2-DOX for 24 h showed a significant reduction in cell viability, but no obvious toxicity was observed in low dose-treated cells. This is most likely due to insufficient Bcl-2 knockdown and DOX accumulation at this short incubation time point. To test this, we measured cell viability at 48 h post-treatment. As expected, siBcl-2 LNP2-DOX exhibited toxic effects at the lowest tested concentration (25 nmol/L), and reduced cell viability to 60%. The percentage of live cells was further reduced to 20% when cells were treated with 50 nmol/L of siBcl-2 LNP2-DOX and reduced further to 6% at the highest dose (100 nmol/L). Based on these results, the half-maximal inhibitory concentration (IC50) of siBcl-2 LNP2-DOX at 48 h was calculated to be 25.64 nmol/L. Prior investigations have shown that DOX is a pro-apoptotic anticancer drug53. Therefore, we measured caspase-3 activity—a crucial mediator of apoptosis—to examine apoptotic pathways54. Cells treated with siBcl-2 LNP2 had lower levels of caspase-3 activity compared to that of the control group (Fig. 4D and Supporting Information Fig. S3), further suggesting that knockdown of Bcl-2 alone is not enough to trigger caspase 3-mediated apoptosis. However, caspase-3 activity was significantly higher in cells treated with Bcl-2 LNP2-DOX than other groups. Collectively, these results indicate that treatment with siBcl-2 LNP2-DOX can activate caspase-3 in Raji cells to induce apoptosis.
3.4. Tumor growth inhibition by siBcl-2 and DOX co-delivered LNPs
To evaluate the therapeutic potential of siBcl-2 LNP2-DOX, we performed an in vivo experiment in Raji Luc + tumor-bearing NSG mice. As a proof-of-concept, different LNP formulations (siBcl-2 LNP2 and siBcl-2 LNP2-DOX) were intratumorally administered into mice at a dose of 1 μg of siRNA (Fig. 5A). Tumor growth was measured every 3 days by bioluminescence imaging (Fig. 5B). Tumors in the non-treated control group grew rapidly, causing the tumor luminescence signal on Day 11 to be 10 times higher than the tumor luminescence signal on Day 0 (Fig. 5C). Interestingly, tumor growth for the siBcl-2 LNP2 treatment group was significantly reduced. While no cytotoxic effect for siBcl-2 LNP2 was observed in vitro—potentially due to the lack of pro-apoptotic triggers—such significant in vivo anti-tumor activity may be ascribed to existing intrinsic pro-apoptotic pressures such as hypoxia and nutrient shortage55. Importantly, tumor growth was inhibited completely after treatment with siBcl-2 LNP2-DOX. After 11 days, tumors had no significant increase in size, suggesting that a combination of siBcl-2 siRNA and DOX was the most effective treatment. In addition to tumor growth examination, body weight was monitored to study the potential toxic effects of various treatment regimens. As shown in Fig. 5D, all groups exhibited no significant weight loss within the 11 days of treatment, and no abnormal changes in mouse behavior was observed. Together, these results suggest that DOX-conjugated siBcl-2 LNP2 can serve as a promising regimen for enhanced LNP-based combination chemotherapy and RNAi therapy.
Figure 5.
In vivo delivery of DOX-conjugated siRNA LNPs to treat a mouse model of lymphoma (A) Drug treatment schedules for in vivo experiments. The treatment doses for Bcl-2 siRNA and DOX were 1 μg and 0.1 μg per mouse, respectively (B) Representative bioluminescence images of tumor-bearing mice (C) Tumor growth curves for different treatment groups. All data is normalized to bioluminescence radiant efficiency (photons/sec/cm2/sr) on day 0. Data are plotted as mean ± SD, n = 5; ∗∗P < 0.005, ∗∗∗∗P < 0.0001 vs. Control (D) Average body weight of different treatment groups during the treatment schedule.
4. Conclusions
In summary, we have developed a DOX-conjugated siRNA LNP platform for combination chemotherapy and RNAi therapy. The optimized LNP achieved efficient Bcl-2 silencing while also delivering DOX in a controlled manner. This dual delivery system exhibits strong toxicity in lymphoma cells, due to increased apoptosis. Finally, we demonstrate that introducing DOX into LNPs enhanced Bcl-2-targeted RNAi therapy, and inhibited tumor growth in a mouse model of lymphoma. Therefore, our DOX conjugation strategy holds great promise in augmenting the anti-tumor efficacy of LNP-based siRNA therapy. Together, these results suggest that DOX-conjugated LNPs are a promising candidate for combination cancer therapies, and in the future could potentially be used in the treatment of many types of cancer.
Acknowledgments
Michael J. Mitchell acknowledges support from a US National Institutes of Health (NIH) Director’s New Innovator Award (DP2 TR002776, USA), a Burroughs Wellcome Fund Career Award at the Scientific Interface (CASI), a grant from the American Cancer Society (129784-IRG-16-188-38-IRG, USA), and the National Institutes of Health (NCI R01 CA241661, NCI R37 CA244911, and NIDDK R01 DK123049, USA). Kamila Butowska acknowledges support from Polish National Agency for Academic Exchange (No. PNN/IWA/2019/00057, Poland). Rebecca M. Haley was supported by the National Science Foundation Graduate Research Fellowship Program (NSF-GRFP) under Grant No. DGE-1845298. Any opinions, finding, and conclusions or recommendations expressed in this material are those of the authors and do not necessarily reflect the views of the NSF.
Author contributions
Kamila Butowska and Xuexiang Han contributed equally to this work. Kamila Butowska, Xuexiang Han, and Michael J. Mitchell conceived the project and designed the experiments. The experiments were performed by Kamila Butowska, Xuexiang Han, Ningqiang Gong, Lulu Xue, and Wenqun Zhong and interpreted by all authors. Kamila Butowska, Xuexiang Han, and Michael J. Mitchell wrote the manuscript. Rebecca M. Haley, Rakan El-Mayta, and Karin Wang contributed to manuscript revision. Kamila Butowska designed and prepared the figures. All authors edited the manuscript and figures and approved the final version for submission.
Conflicts of interest
The authors declare the following competing financial interest(s): Michael J. Mitchell, Kamila Butowska, and Xuexiang Han are inventors on a patent related to this work filed by the Trustees of the University of Pennsylvania.
Footnotes
Peer review under responsibility of Chinese Pharmaceutical Association and Institute of Materia Medica, Chinese Academy of Medical Sciences.
Supporting data to this article can be found online at https://doi.org/10.1016/j.apsb.2022.07.011.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Bray F., Ferlay J., Soerjomataram I., Siegel R.L., Torre L.A., Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA A Cancer J Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Sung H., Ferlay J., Siegel R.L., Laversanne M., Soerjomataram I., Jemal A., et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA A Cancer J Clin. 2021;71:209–249. doi: 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- 3.Tavan H., Azadi A., Veisani Y. Return to work in cancer patients: a systematic review and meta-analysis. Indian J Palliat Care. 2019;25:147–152. doi: 10.4103/IJPC.IJPC_114_18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.American Cancer Society . American Cancer Society; Atlanta: 2019. Cancer treatment & survivorship facts & figures 2019‒2021. [Google Scholar]
- 5.Lage H. An overview of cancer multidrug resistance: a still unsolved problem. Cell Mol Life Sci. 2008;65:3145–3167. doi: 10.1007/s00018-008-8111-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gillet J.P., Gottesman M.M. In: Multi-drug resistance in cancer. Methods in molecular biology (methods and protocols) Zhou J., editor. Humana Press; New York: 2010. Mechanism of multidrug resistance in cancer; pp. 47–76. [DOI] [PubMed] [Google Scholar]
- 7.Tsujimoto Y., Cossman J., Jaffe E., Croce C. Involvement of the Bcl-2 gene in human follicular lymphoma. Science. 1985;228:1440–1443. doi: 10.1126/science.3874430. [DOI] [PubMed] [Google Scholar]
- 8.Yip K.W., Reed J.C. Bcl-2 family proteins and cancer. Oncogene. 2008;27:6398–6406. doi: 10.1038/onc.2008.307. [DOI] [PubMed] [Google Scholar]
- 9.Brunelle J.K., Letai A. Control of mitochondrial apoptosis by the Bcl-2 family. J Cell Sci. 2009;122:437–441. doi: 10.1242/jcs.031682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Adams C.M., Clark-Garvey S., Porcu P., Eischen C.M. Targeting the Bcl-2 family in B cell lymphoma. Front Oncol. 2019;8:636. doi: 10.3389/fonc.2018.00636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wei Y., Cao Y., Sun R., Cheng L., Xiong X., Jin X., et al. Targeting Bcl-2 proteins in acute myeloid leukemia. Front Oncol. 2020;10:2137. doi: 10.3389/fonc.2020.584974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xiong H., Veedu R.N., Diermeier S.D. Recent advances in oligonucleotide therapeutics in oncology. Int J Mol Sci. 2021;22:3295. doi: 10.3390/ijms22073295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.DiNardo C.D., Konopleva M.Y. A venetoclax bench-to-bedside story. Nat Can (Que) 2021;2:3–5. doi: 10.1038/s43018-020-00165-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Knox J.J., Chen X.E., Feld R., Nematollahi M., Cheiken R., Pond G., et al. A phase I‒II study of oblimersen sodium (G3139, Genasense) in combination with doxorubicin in advanced hepatocellular carcinoma (NCI #5798) Invest N Drugs. 2008;26:193–194. doi: 10.1007/s10637-007-9104-1. [DOI] [PubMed] [Google Scholar]
- 15.Mitchell M.J., Chen C.S., Ponmudi V., Hughes A.D., King M.R. E-selectin liposomal and nanotube-targeted delivery of doxorubicin to circulating tumor cells. J Control Release. 2012;160:609–617. doi: 10.1016/j.jconrel.2012.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mitchell M.J., Castellanos C.A., King M.R. Nanostructures surfaces to target and kill circulating tumor cells while repelling leukocytes. J Nanomater. 2012;2012:1–10. doi: 10.1155/2012/831263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guimaraes P.P.G., Gaglione S., Sewastianik T., Carrasco R.D., Langer R., Mitchell M.J. Nanoparticles for immune cytokine TRIAL-based cancer therapy. ACS Nano. 2018;12:912–931. doi: 10.1021/acsnano.7b05876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Marcucci G., Byrd J.C., Dai G., Klisovic M.I., Kourlas P.J., Young D.C., Cataland S.R., et al. Phase 1 and pharmacodynamic studies of G3139, a Bcl-2 antisense oligonucleotide, in combination with chemotherapy in refractory or relapsed acute leukemia. Blood. 2003;101:425–432. doi: 10.1182/blood-2002-06-1899. [DOI] [PubMed] [Google Scholar]
- 19.Walker A.R., Marcucci G., Yin J., Blum W., Stock W., Kohlschmidt J., et al. Phase 3 randomized trial of chemotherapy with or without oblimersen in older ALM patients: CALGB 10201 (Alliance) Blood Adv. 2021;13:2775–2787. doi: 10.1182/bloodadvances.2021004233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li Z., Zhang L., Tang C. Co-delivery of doxorubicin and survivin shRNA-expressing plasmid via microenvironment-responsive dendritic mesoporous silica nanoparticles for synergistic cancer therapy. Pharm Res (N Y) 2017;34:2829–2841. doi: 10.1007/s11095-017-2264-6. [DOI] [PubMed] [Google Scholar]
- 21.Peng Z., Wang C., Fang E., Lu X., Wang E., Lu X., et al. Co-delivery of doxorubicin and SATB1 shRNA by thermosensitive magnetic cationic liposomes for gastric cancer therapy. PLoS One. 2014;9 doi: 10.1371/journal.pone.0092924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li Q., Lv X., Tang C., Yin C. Co-delivery of doxorubicin and CRISPR/Cas9 or RNAi-expressing plasmid by chitosan-based nanoparticle for cancer therapy. Carbohydr Polym. 2022;287 doi: 10.1016/j.carbpol.2022.119315. [DOI] [PubMed] [Google Scholar]
- 23.Chen A.M., Zhang M., Wei D., Stueber D., Taratula O., Minko T., et al. Co-delivery of doxorubicin and Bcl-2 siRNA by mesoporous silica nanoparticles enhances the efficacy of chemotherapy in multidrug-resistant cancer cells. Small. 2009;5:2673–2677. doi: 10.1002/smll.200900621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cheng D., Cao N., Chen J., Yu X., Shuai X. Multifunctional nanocarrier mediated co-delivery of doxorubicin and siRNA for synergistic enhancement of glioma apoptosis in rat. Biomaterials. 2012;33:1170–1179. doi: 10.1016/j.biomaterials.2011.10.057. [DOI] [PubMed] [Google Scholar]
- 25.Sun W., Chen X., Xie C., Wang Y., Lin L., Zhu K., et al. Co-delivery of doxorubicin and anti-BCL-2 siRNA by pH-responsive polymeric vector to overcome drug resistance in in vitro and in vivo HepG2 hepatoma model. Biomacromolecules. 2018;19:2248–2256. doi: 10.1021/acs.biomac.8b00272. [DOI] [PubMed] [Google Scholar]
- 26.Mitchell M.J., Billingsley M.M., Haley R.M., Wechsler M.E., Peppas N.A., Langer R. Engineering precision nanoparticles for drug delivery. Nat Rev Drug Discov. 2021;20:101–124. doi: 10.1038/s41573-020-0090-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fenton O.S., Olafson K.N., Pillai P.S., Mitchell M.J., Langer R. Advances in biomaterials for drug delivery. Adv Mater. 2018;30 doi: 10.1002/adma.201705328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hou X., Zaks T., Langer R., Dong Y. Lipid nanoparticle for mRNA delivery. Nat Rev Mater. 2021;170:83–112. doi: 10.1038/s41578-021-00358-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Swingle K.L., Hamilton A.G., Mitchell M.J. Lipid nanoparticle-mediated delivery of mRNA therapeutics and vaccines. Trends Mol Med. 2021;27:616–617. doi: 10.1016/j.molmed.2021.03.003. [DOI] [PubMed] [Google Scholar]
- 30.Han X., Zhang H., Butowska K., Swingle K.L., Alameh M.G., Weissman D., et al. An ionizable lipid toolbox for RNA delivery. Nat Commun. 2021;12:7222. doi: 10.1038/s41467-021-27493-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Thi T.T.H., Suys E.J., Lee J.S., Nguyen D.H., Park K.D., Truong N.P. Lipid-based nanoparticle in the clinic and clinical trials: from cancer nanomedice to COVID-19 vaccines. Vaccines. 2021;9:359. doi: 10.3390/vaccines9040359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Akin A., Maier M.A., Manoharan M., Fitzgerald K., Jayaraman M., Barros S., et al. The Onpattro story and the clinical translation of nanomedicines containing nucleic acid-based drugs. Nat Nanotechnol. 2019;14:1084–1087. doi: 10.1038/s41565-019-0591-y. [DOI] [PubMed] [Google Scholar]
- 33.Andreadakis Z., Kumar A., Roman R.G., Tollefsen S., Saville M., Mayhew S. The COVID-19 vaccine development landscape. Nat Rev Drug Discov. 2020;19:305–306. doi: 10.1038/d41573-020-00073-5. [DOI] [PubMed] [Google Scholar]
- 34.Baden L.R., El Sahly H.M., Essink B., Kotloff K., Frey S., Novak R., et al. Efficacy and safety of the mRNA-1273 SARS-CoV-2 vaccine. N Engl J Med. 2021;384:403–416. doi: 10.1056/NEJMoa2035389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Evers M.J.W., Kulkarni J.A., van der Meel R., Cullis P.R., Vader P., Schiffelers R.M. State-of-the-art design and rapid-mixing production techniques of lipid nanoparticles for nucleic acid delivery. Small Methods. 2018;2 [Google Scholar]
- 36.Krohn-Grimberghe M., Mitchell M.J., Schloss M.J., Khan O.F., Courties G., Guimaraes P.P.G., et al. Nanoparticle-encapsulated siRNA for gene silencing in the haematopoietic stem-cell niche. Nat Biomed Eng. 2020;4:1076–1089. doi: 10.1038/s41551-020-00623-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ickenstein L.M., Garidel P. Lipid-based nanoparticles formulations for small molecules and RNA drugs. Expet Opin Drug Deliv. 2019;16:1205–1226. doi: 10.1080/17425247.2019.1669558. [DOI] [PubMed] [Google Scholar]
- 38.Kratz F. DOXO-EMCH (INNO-206): the first albumin-binding prodrug of doxorubicin to enter clinical trials. Expet Opin Invest Drugs. 2007;16:855–866. doi: 10.1517/13543784.16.6.855. [DOI] [PubMed] [Google Scholar]
- 39.Billingsley M.M., Singh N., Ravikumar P., Zhang R., June C.H., Mitchell M.J. Ionizable lipid nanoparticle-mediated mRNA delivery for human CAR T engineering. Nano Lett. 2020;20:1578–1589. doi: 10.1021/acs.nanolett.9b04246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shepherd S.J., Warzecha C.C., Yadavali S., El-Mayta R., Alameh M.G., Wang L., et al. Scalable mRNA and siRNA lipid nanoparticle production using a parallelized microfluidic device. Nano Lett. 2021;21:5671–5680. doi: 10.1021/acs.nanolett.1c01353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Riley R.S., Kashyap M.V., Billingsley M.M., White B., Alameh M.G., Bose S.K., et al. Ionizable lipid nanoparticle for in utero mRNA delivery. Sci Adv. 2021;7 doi: 10.1126/sciadv.aba1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.El-Mayta R., Zhang R., Shepherd S.J., Wang F., Billingsley M.M., Dudkin V., et al. A nanoparticle platform for accelerated in vivo oral delivery screening of nucleic acids. Adv Ther. 2021;4 [Google Scholar]
- 43.Zhang R., El-Mayta R., Murdoch T.J., Warzecha C.C., Billingsley M.M., Shepherd S.J., et al. Helper lipid structure influences protein adsorption and delivery of lipid nanoparticles to spleen and liver. Biomater Sci. 2021;9:1449–1463. doi: 10.1039/d0bm01609h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Guimaraes P.P.G., Zhang R., Spektor R., Tan M., Chung A., Billingsley M.M., et al. Ionizable lipid nanoparticles encapsulating barcoded mRNA for accelerated in vivo delivery screening. J Control Release. 2019;316:404–417. doi: 10.1016/j.jconrel.2019.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Whitehead K.A., Dorkin J.R., Vegas A.J., Chang P.H., Veiseh O., Matthews J., et al. Degradable lipid nanoparticles with predictable in vivo siRNA delivery activity. Nat Commun. 2014;5:4277. doi: 10.1038/ncomms5277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Whitehead K.A., Matthews J., Chang P.H., Niroui F., Dorkin J.R., Severgnini M., Anderson D.G. In vitro‒in vivo translation of lipid nanoparticles for hepatocellular siRNA delivery. ACS Nano. 2012;6:6922–6929. doi: 10.1021/nn301922x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Walker J.A., Sorkin M.R., Alabi C.A. In: Quantitative analysis of cellular drug transport, disposition, and delivery. Rosania G.R., Thurber G.M., editors. Humana Press; New York: 2021. Quantitative determination of intracellular bond cleavage; pp. 305–330. [Google Scholar]
- 48.Northrop B.H., Frayne S.H., Choudhary U. Thiol-maleimide “click” chemistry: evaluating the influence of solvent, initiator, and thiol on the reaction mechanism, kinetics, and selectivity. Polym Chem. 2015;6:3415–3430. [Google Scholar]
- 49.Butowska K., Woziwodzka A., Borowik A., Piosik J. Polymeric nanocarriers: a transformation in doxorubicin therapies. Materials. 2021;14:2135. doi: 10.3390/ma14092135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.White K.A., Grillo-Hill B.K., Barber D.L. Cancer cell behaviors mediated by dysregulated pH dynamics at a glance. J Cell Sci. 2017;130:663–669. doi: 10.1242/jcs.195297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yu B., Wang X., Zhou C., Teng L., Ren W., Yang Z., et al. Insight into mechanism of cellular uptake of lipid nanoparticles and intracellular release of small RNAs. Pharm Res (N Y) 2014;31:2685–2695. doi: 10.1007/s11095-014-1366-7. [DOI] [PubMed] [Google Scholar]
- 52.Patel A.G., Kaufmann S.H. Cancer: how does doxorubicin work? Elife. 2012;1 doi: 10.7554/eLife.00387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Decaudin D., Geley S., Hirsch T., Castedo M., Marchetti P., Macho A., et al. Bcl-2 and Bcl-XL antagonize the mitochondrial dysfunction preceding nuclear apoptosis induces by chemotherapeutic agents. Cancer Res. 1997;57:62–67. [PubMed] [Google Scholar]
- 54.Porter A.G., Jänicke R.U. Emerging roles of caspase-3 in apoptosis. Cell Death Differ. 1999;6:99–104. doi: 10.1038/sj.cdd.4400476. [DOI] [PubMed] [Google Scholar]
- 55.Shimizu A., Eguchi Y., Kamiike W., Itoh Y., Hasegawa J., Yamabe K., et al. Induction of apoptosis as well as necrosis by hypoxia and predominant prevention of apoptosis by Bcl-2 and Bcl-XL. Cancer Res. 1996;56:2161–2166. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.