Abstract
There are a variety of regulatory systems in bacteria, among which the two-component system (TCS) can sense external environmental changes and make a series of physiological and biochemical reactions, which is crucial for the life activities of bacteria. As a member of TCS, SaeRS is considered to be an important virulence factor in Staphylococcus aureus, but its function in tilapia (Oreochromis niloticus)-derived Streptococcus agalactiae remains unknown. To explore the role of SaeRS in regulating virulence in the two-component system (TCS) of S. agalactiae from tilapia, ΔSaeRS mutant strain and CΔSaeRS complementary strain were constructed by homologous recombination. The results showed that the abilities of growth and biofilm formation of ΔSaeRS strain were significantly decreased when cultured in a brain heart infusion (BHI) medium (P < 0.01). Also, the survival rate of the ΔSaeRS strain in blood was decreased when compared with the wild strain S. agalactiae THN0901. Under the higher infection dose, the accumulative mortality of tilapia caused by the ΔSaeRS strain was significantly decreased (23.3%), of which THN0901 and CΔSaeRS strains were 73.3%. The results of competition experiments in tilapia showed that the invasion and colonization abilities of the ΔSaeRS strain were also dramatically lower than those of the wild strain (P < 0.01). Compared with the THN0901, the mRNA expression levels of virulence factors (fbsB, sip, cylE, bca, etc.) in the ΔSaeRS strain were significantly down-regulated (P < 0.01). SaeRS is one of the virulence factors of S. agalactiae. It plays a role in promoting host colonization and achieving immune evasion during the infection of tilapia, which provides a basis for exploring the pathogenic mechanism of S. agalactiae infected with tilapia.
Keywords: Streptococcus agalactiae, two-component system, SaeRS, virulence, colonization
1. Introduction
Streptococcus agalactiae, or group B Streptococcus (GBS), is a major bacterial species of the genus Streptococcus (Tavares et al., 2019), which can cause invasive infection in a range of hosts, such as humans (Furfaro et al., 2018), cattle (Lyhs et al., 2016), and fish (Boonyayatra et al., 2020). GBS infection of tilapia (Oreochromis niloticus) is associated with septicemia and meningoencephalitis (Evans et al., 2008). In addition, GBS can cause enormous economic losses to the tilapia industry (Liu et al., 2016). For example, outbreaks in which up to 30% mortality in tilapia culture have been reported in Thailand (Naraid et al., 2008; Kayansamruaj et al., 2014) and many other countries (Anshary et al., 2014; Aisyhah et al., 2015). The exact and complete pathogenic mechanism of GBS on tilapia has not been clarified, so it is necessary to explore its virulence factors. The two-component system (TCS) in bacteria is used to make a series of responses for adaptive regulation in environment changes (Groisman, 2016), and is consisted of two main components, a histidine kinase and a response regulator (Capra and Laub, 2012; Christopher et al., 2016). Histidine kinase can identify specific stimulus signals and activate response regulators through auto-phosphorylation and participate in gene transcription, resulting in adaptation to the particular environment by binding to specific DNA sequences (Mitrophanov and Groisman, 2008). TCS is required for bacterial growth and fitness (Mitrophanov and Groisman, 2008) and drug resistance (Tierney and Rather, 2019) and is also associated with the induction of virulence (Dagmar and Roy, 2006), and biofilm-forming ability (Badal et al., 2020). A previous study demonstrated that inactivation of the SaeRS system in Staphylococcus aureus significantly decreased apoptosis or death of lung epithelial cells (A549), and attenuated virulence in a murine infection model (Liang et al., 2006). A previous study in Staphylococcus epidermidis showed that the deletion of SaeRS affected the expression levels of genes with a variety of functions, including bacterial autolysis (lrgA, arlR, and lytS), biofilm formation (ebhA), leucine biosynthesis (leuD), protein hydrolysis (clpP), stress resistance (asp23), and cell viability (yycH) (Lou et al., 2011).
In S. agalactiae strains isolated from humans, plasminogen binding protein (pbsP) and other factors can promote host colonization of GBS through the up-regulation of SaeRS in TCS by the mouse vaginal colonization model (Cook et al., 2018). In S. aureus, SaeRS plays an important role in the regulation of virulence factors and pathogenicity. Such as regulation of relevant virulence factors α-Hemolysin, lipase, coagulase and adhesin, etc., (Ericson et al., 2017; Liu et al., 2020, 2021). Studies on S. epidermidis showed that the deletion of SaeRS would affect genes with multiple functions, including bacterial autolysis (lrgA, arlR, and lytS), biofilm formation (ebhA), leucine biosynthesis (leuD), protein hydrolysis (clpP), stress resistance (asp23) and cell viability (yycH) (Lou et al., 2011).
SaeRS plays a key role in the pathogenicity of S. agalactiae. However, the characteristics and virulence of GBS affecting tilapia are still unknown. To study the role of SaeRS in GBS, a ΔSaeRS mutant was constructed with deletion of the genes that encode both the histidine kinase (SaeS) and the response regulator (SaeR) and its corresponding complementary strain CΔSaeRS, and the role of the gene deletion mutant was assessed under in vivo and in vitro conditions.
2. Materials and methods
2.1. Bacteria and animals
Streptococcus agalactiae THN0901, a wild-type virulent strain, was isolated from tilapia infected with GBS obtained from an intensive tilapia farm, Hainan, China. GBS strains were cultured aerobically overnight at 28°C, 180 rpm in a brain heart infusion (BHI) bath and inoculated into a BHI medium with a diluted rate of 1:100 (vol/vol) for 12 h under the same conditions. Our laboratory has previously sequenced the whole genome of THN0901, so the primer design in this experiment is derived from the genome of THN0901. Tilapia (25 ± 5 g of body weight) were purchased from a farm in Guangzhou, Guangdong, China, which were acclimated with flowing water at 28 ± 0.5°C for 2 weeks and fed with commercial feed twice a day (1% body weight every time). All protocols involved in the care of fish were approved by the Institutional Animal Care and Use Committees at Sun Yat-sen University (SYSU-IACUC-2020-B0740).
2.2. Strain construction
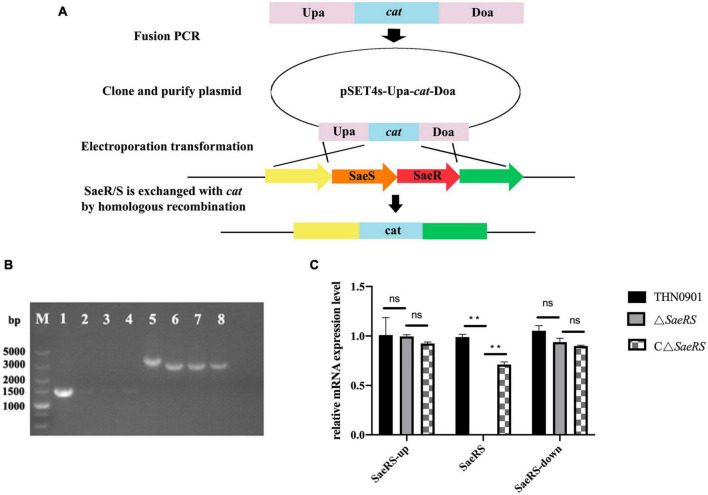
To generate a functional deletion mutant of SaeRS, a 1,257-bp fragment upstream of SaeRS and a downstream homologous sequence (1,042 bp) from S. agalactiae THN0901 strain were amplified with the primers Sae-up-F/R, Sae-down-F/R, and cat-F/R (Table 1). The chloramphenicol resistant gene (cat) (1,056 bp) was acquired from the plasmid pSET5s using the primers cat-F and cat-R (Table 2). The amplified products were cloned into pMD-19T plasmid to construct recombinant fragment Upa-cat-Doa replacing the sequence of SaeRS with cat gene, which was ligated with cleaved thermo-sensitive suicide vector pSET4s (Miaoling Biological Co., Ltd., China) showing spectinomycin resistance (Figure 1A). The correct orientation insert and sequence of pSET4s-Upa-cat-Doa plasmid were confirmed by DNA sequencing and then transformed into THN0901 wild strain via electroporation (Takamatsu et al., 2001). The proposed strains were selected at 28°C and cultured at 37°C in a BHI bath with chloramphenicol (10 μg/mL) several times to contribute to homologous recombination. The mutant strain ΔSaeRS was achieved by selection for sensitive to chloramphenicol resistance (Figure 1B), and it was confirmed by DNA sequencing and quantitative polymerase chain reaction (qPCR) analysis using the SaeRS gene-specific primers (Table 1). Using the sae-HB-F and sae-HB-R primers, the SaeRS and its upstream promoter sequence were linked together into the pSET2s expression vector to construct a complementing expression vector, which was electroporated into the ΔSaeRS mutant strain and verified by PCR, sequencing and quantitative reverse transcription PCR (RT-qPCR) to get the complementary strain (Figure 1C).
TABLE 1.
List of primers mutant strain validation.
| Name | Primers (5′–3′) |
| Sae-up-F | CCGGAATTCGGTGGAGATGGCATGTTAA |
| Sae-down-R | CCCAAGCTTATTGGATGATCAGTCATCGT |
| Sae-F | AGAAGCACTCAAAGAGGTAG |
| Sae-R | TTATCCCATAGCCTTGGTC |
| Sae-up-qp-F | CCTTAAAGCTTCAGAGGGAGCA |
| Sae-up-qp-R | CCTGTCGCAAAGGCTAGACTAA |
| Sae-qp-F | CAGACACGGCTGAACCTCAT |
| Sae-qp-R | CGAAACATCTGGCGCAACAT |
| Sae-down-qp-F | CCTTAAAGCTTCAGAGGGAGCA |
| Sae-down-qp-R | CCTGTCGCAAAGGCTAGACTAA |
| Cat-F | AGACGTTATCTAGAGCAAGCCGGGATCCGCACCGAACT AGAGCTTGATGA |
| Cat-R | CATTCAGTTGATGTGCAAGTTGCACTGCAGTGCATAAT TCGATGGGTTCC |
| Sae-HB-F | CGCGGATCCGCAAATGTCATCACAAGCC |
| Sae-HB-R | AACTGCAGGTTGGCAGTTCCTTACAATC |
The underline is the restriction site.
TABLE 2.
Cumulative survival of tilapia challenged.
| Bacterial strain | Infection dose (CFU/fish) |
Total fish | Survival fish | Motility rate |
| THN0901 | 3.5 × 108 | 30 | 8 | 73.3% |
| 3.5 × 107 | 30 | 14 | 53.3% | |
| 3.5 × 106 | 30 | 20 | 33.3% | |
| ΔSaeRS | 3.5 × 108 | 30 | 23 | 23.3% |
| 3.5 × 107 | 30 | 27 | 10% | |
| 3.5 × 106 | 30 | 28 | 6.7% | |
| CΔSaeRS | 3.5 × 108 | 30 | 8 | 73.3% |
| 3.5 × 107 | 30 | 14 | 53.3% | |
| 3.5 × 106 | 30 | 16 | 46.7% |
FIGURE 1.
(A) Construction of the SaeRS gene deletion strain of S. agalactiae THN0901 by homologous recombination. (B) Detection of the ΔSaeRS mutant by the polymerase chain reaction (PCR). Lanes 1–4 using the primers Sae-F and Sae-R; Lanes 5–8 using the primers Sae-up-F and Sae-down-R; The primer sources used in lanes 1–4 are upstream homologous arm and downstream homologous arm. The primer sources used in lanes 5–8 are WP-000703870.1 and AK1948 13.1. Lane M is DL5000 DNA marker; Lane 1 and 5 is S. agalactiae THN0901, Lane 2 and 6 is 5th-generation ΔSaeRS; Lane 3, and 7 is 10th-generation ΔSaeRS, Lane 4 and 8 is 15th-generation ΔSaeRS, respectively. (C) Relative messenger RNA (mRNA) expression level of gene expression up and down stream of SaeRS among three strains. Each experiment was performed in triplicate, and data were presented as mean ± standard deviation (SD), *: 0.01 < P < 0.05, **: P < 0.01.
2.3. Growth analysis
The wild strain THN0901, the mutant strain ΔSaeRS and complementary strain CΔSaeRS were cultured to analyze the bacterial growth characteristics. Proliferation was assessed in BHI liquid medium and strains were first cultured at 28°C overnight and then inoculate into 100 mL fresh BHI medium at a ratio of 1/50 (vol/vol) by shaking at 180 rpm. It is worth mentioning that the mutant and complementary strains were always cultured in the presence of antibiotics. The optical density at 600 nm (OD600) value of the culture was measured in 1-h intervals to describe the growth curve of the above strains.
2.4. Determination of biofilm formation
The biofilm formation of GBS was quantified as described by a previous study (O’Toole and Kolter, 1998) with slight modification. First, fresh overnight cultures of S. agalactiae wild strain THN0901, mutant strain ΔSaeRS and complementary strain CΔSaeRS were taken and adjusted to OD600 = 1.0 by adding BHI medium, and then 200 μL was added to each well of a 96-well plate. A fresh BHI medium was used as a negative control. The supernatant of the wells was carefully discarded and washed twice with 1 × phosphate-buffered saline (PBS) to remove non-adherent cells. Then, the biofilm-forming ability was assessed by staining with 50 μL crystal violet at 0.1% for 20 min at room temperature. The supernatant was discarded and washed twice. The plate was dried at room temperature, and 100 μL of ethanol: acetone (8:2) was added to each well. The absorbance at 575 nm (A575) was measured. Each experiment was performed in four independent experiments. Two-tailed Student’s t-test was performed to analyze the mean ± standard deviation (SD).
2.5. Mixed competition experiments in vitro and in vivo
For in vitro experiments, the overnight cultures of THN0901 strain and ΔSaeRS mutant strain were adjusted to the same OD value, and then combined in equal volume in a microcentrifuge tube, which was diluted in 50 mL of BHI medium at 1:100 (vol/vol). Diluted and incubated aerobically at 28°C in a shaker. Serially diluted bacterial solutions were then plated onto BHI plates after 2, 4, 6, 9, 12, and 24 h (Patterson et al., 2012). The competition index (CI), the ratio of the ΔSaeRS mutant to the total bacterial count (TBC), was determined by counting the TBC of the samples on the BHI plate and identifying the bacteria on the BHI plate with 20 μg/mL chloramphenicol. For in vivo experiment, the concentration is 1.0 × 107 (CFU)/mL mutant and 1.0 × 107 CFU/mL of wild strains were mixed in equal volume, and a total of 20 Nile tilapia were intraperitoneally injected with the same amount of mixed bacterial solution (100 μL per fish). After 3, 6, and 9 h, the brain, liver, and spleen were aseptically removed from the tilapia. Ten-fold serial dilutions of whole tissue and blood samples were prepared in PBS and then plated onto BHI plates to determine CI values. The CI value was calculated in the same way as done for in vitro experiments. After counting, we dissected the spleen of tilapia and used the plate with antibiotics to verify the accuracy of the mutant strain. Each experiment was performed in three independent experiments. Two-tailed Student’s t-test was performed to analyze the mean ± SE. The null hypothesis was defined as follows: mean index is not significantly different from 1.0 (*: 0.01 < P < 0.05, **: P < 0.01) (Macho et al., 2010).
2.6. Challenge experiment
A total of 300 fish were equally divided into 10 groups and then cultured under laboratory conditions in flowing water at 30°C. Nine experimental groups were injected intraperitoneally with THN0901, ΔSaeRS and CΔSaeRS strains at doses of 3.5 × 108, 3.5 × 107, and 3.5 × 106 CFU per fish, respectively. The control group was injected with sterile PBS (pH = 7.4). Mortality of challenged fish was recorded daily for 2 weeks. The mortality rate is the number of fish killed divided by the total number of fish.
2.7. Whole blood-killing assays
The whole blood killing method in this study refers to a previous study (Yao et al., 2015). Collect blood from tail vein of tilapia with anticoagulant tube. Fresh bacterial liquid of THN0901, ΔSaeRS, and CΔSaeRS strains with OD600 = 0.4 was collected and diluted 10 times with sterile PBS. Then, 10 μL of the above dilutent was added, mixed with 300 μL of tilapia blood, incubated in a water bath at 30°C for 1 h, serially diluted, and then spread on BHI plates. Each experiment was repeated three times. After overnight culture at 37°C, the survival index was calculated as follows: the ratio of the number of colonies at the end to the number of colonies at the beginning.
2.8. Real-time fluorescence quantitative PCR
Total RNA was isolated from S. agalactiae THN0901, ΔSaeRS, and CΔSaeRS strains using the Trizol reagent, and complementary DNA (cDNA) was synthesized using Evo M-MLV RT Kit (Vazyme, Nanjing, China) following the manufacturer’s protocol. The primers used in the study (Table 3) were designed using Primer 5.0. PCR conditions for SYBR Green RT-PCR were as follows: 95°C for 3 min, followed by 40 cycles of 95°C for 5 s and 60°C for 30 s. Melting curve analysis was performed at the end of the RT-qPCR cycle to confirm the PCR specificity. Each group had three replications, and each sample was amplified in triplicate. The expression level of the target gene relative to the 16s rRNA gene was quantified by the 2–ΔΔCt method. The RT-qPCR data were analyzed using a one-way analysis of variance (ANOVA).
TABLE 3.
Primers for S. agalactiae virulence factors.
| Name | Primers (5′–3′) | GeneBank ID |
| 16srRNA-F | TTATGACCTGGGCTACAC | NP_687037.1 |
| 16srRNA-R | CCTACAATCCGAACTGAGA | |
| fbsB-F | AGTTGCGCAAACTTCTGTCC | AM050622.1 |
| fbsB-R | TTTCCGCAGTTGTTACACCG | |
| bibA-F | CAAGCTCATCAACTTGACTCTTA | CP003810.1 |
| bibA-R | TAGGCACATGGCTCAAAATGACG | |
| pavA-F | TACGGAAAATACAATCACCTACC | CHZE01000013.1 |
| pavA-R | GCTTATGTTGTTTATCATGTGCGCG | |
| Srr-1-F | TCATTCCCAGTTTTATCGCTTGC | AY669067.1 |
| Srr-1-R | TCGGAGTTACAGACTTCCAAAAT | |
| bac-F | TTGGACAAGCAGTATTTACATCA | AY672152.1 |
| bac-R | ACTCTTTCGTCGTTACTTCCTTG | |
| sodA-F | TCAACTGCCAATCAAGATACTCC | DQ232581.1 |
| sodA-R | GCTTTGATGTAGTTAGGACGAACA | |
| sip-F | AATTCAGTACATACCGTGCGGGAGA | HQ878436.1 |
| sip-R | GTTATTTGCTGCCATATTTTGTG | |
| cpsD-F | GTAGGTCGTAATGGTAGGAT | NC_021195.1 |
| cpsD-R | TTCTAGGATCATCGTCTAACTT | |
| cpsG-F | TAGAGATTTGATTGGGTCAGA | KC290918.1 |
| cpsG-R | AGTTACCACTGTCATAGGAAT | |
| cylE-F | ATTCTCCTCCTGGCAAAGCC | AF157015.2 |
| cylE-R | TGACGCTTGGTAGTTGCTGT | |
| bca-F | AATACTATGGGGATGTTTCTCAG | M97256.1 |
| bca-R | TAACTTCTTCAATCTTATCCCTC | |
| hylB-F | GAATAACTACTTCACTGACGCTG | JN247784.1 |
| hylB-R | AACGCGCCCCATATCTACTA | |
| cfb-F | TAGCTTAGTTATCCCAAATCCC | HQ148672.1 |
| cfb-R | TAAAGACTTCATTGCGTGCC | |
| ponA-F | GCTCCTGATGAAAACTTTGTCGG | KX078639.1 |
| ponA-R | AGAGCCCTTCTGGCATTGTC | |
| cspA-F | TGCACGTAACCAGTATCGCA | A964_2021 |
| cspA-R | GCACCGAGTTTAACGGCATC |
2.9. Statistical analysis
The results were presented as mean ± standard deviation (SD). P < 0.05 (*) was considered as being significant, and 0.05 < P < 0.01 (**) was considered highly significant. These date were analyzed by one-way analysis of variance (ANOVA). Statistical analysis was performed in SPSS (SPSS Inc., Chicago, IL, USA), and graphs were generated using GraphPad Prism 8.0 (GraphPad Software, Inc., San Diego, CA, USA).
3. Results
3.1. Construction of SaeRS deletion mutant strain and the complementary strain
To explore the role of SaeRS gene cluster in S. agalactiae, the mutant was constructed with deletion of a 710 bp fragment (Figure 1B). The SaeRS deletion strain was observed by PCR using the primers, including Sae-up-F, Sae-down-R, and Sae-F/R (Table 2), generating amplicons of 3,834 and 3,127 bp for the ΔSaeRS mutant strain and THN0901 wild strain, respectively (Figure 1B). DNA sequencing confirmed that the desired SaeRS gene cluster deletion had occurred by homologous recombination. The complementary strain CΔSaeRS was also verified by PCR and DNA sequencing. The RT-qPCR analysis showed that the relative mRNA expression levels of SaeRS genes in the ΔSaeRS was not expressed at all, and the CΔSaeRS strain was recovered compared with the wild strain THN0901 (p < 0.05) (Figure 1C). However, SaeRS gene deletion had little effect on the expression levels of upstream and downstream genes (Figure 1C). The results indicated that the ΔSaeRS mutant and complementary CΔSaeRS strain was successfully constructed.
3.2. Growth curve
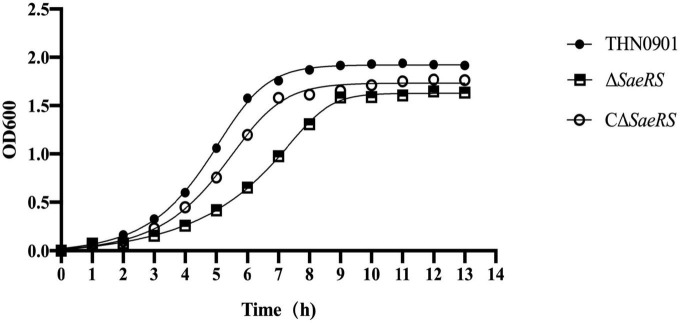
According to the growth curves in BHI, there were some differences in the growth of the S. agalactiae THN0901, ΔSaeRS, and CΔSaeRS strains under the same culture conditions. The growth rates of the ΔSaeRS and the THN0901 strains were significantly different. The logarithmic growth phase of the ΔSaeRS strain was delayed, and the time to reach the plateau phase was 2 h longer than the THN0901 strain. Also, the OD600 [1OD = log10 (1/trans), where trans is the transmittance T value of the test substance] of the THN0901 strain in the plateau phase was 0.333 higher than that of the ΔSaeRS strain, which means that the bacterial population of the mutant strain decreases in the plateau phase. The growth rate of the complementary strain CΔSaeRS was observed between the wild strain THN0901 and the mutant strain ΔSaeRS, which implies that the growth rate of the strain is restored after complementing the SaeRS gene (Figure 2).
FIGURE 2.
Growth curve of S. agalactiae THN0901, ΔSaeRS, and CΔSaeRS strains in brain heart infusion (BHI) medium. The growth curve was checked spectrophotometrically at 600 nm.
3.3. Biofilm formation
In the biofilm formation experiment, the average optical density at 575 nm (OD575) of S. agalactiae wild strains THN0901, mutant strains ΔSaeRS, and complementary strains CΔSaeRS was 0.576 ± 0.06, 0.328 ± 0.02, and 0.475 ± 0.05, respectively, showing that the biofilm-forming ability of the ΔSaeRS strain was significantly lower than that of the THN0901 and the CΔSaeRS strains (P < 0.01) (Figure 3). It means the deletion of SaeRS significantly reduced the ability of biofilm formation.
FIGURE 3.
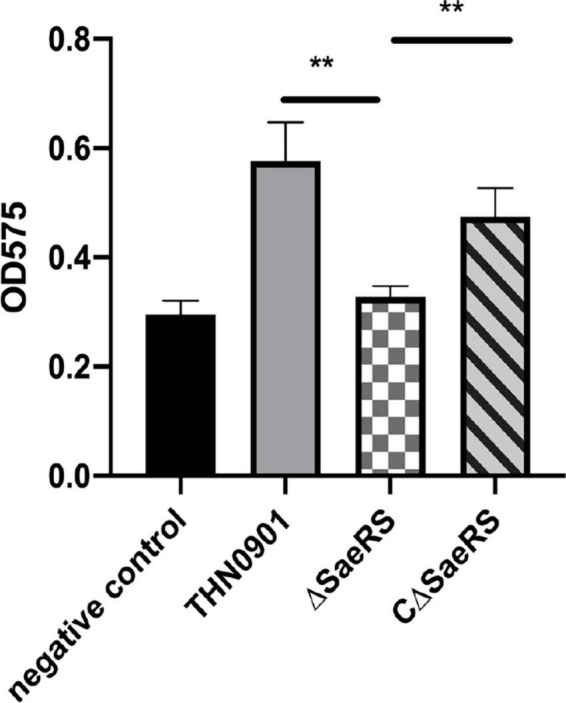
Biofilm formation of three S. agalactiae strains by microtiter plate assays. Each experiment was performed in triplicate, and data were presented as mean ± standard deviation (SD), *: 0.01 < P < 0.05, **: P < 0.01.
3.4. Mixed competition in vivo and in vitro
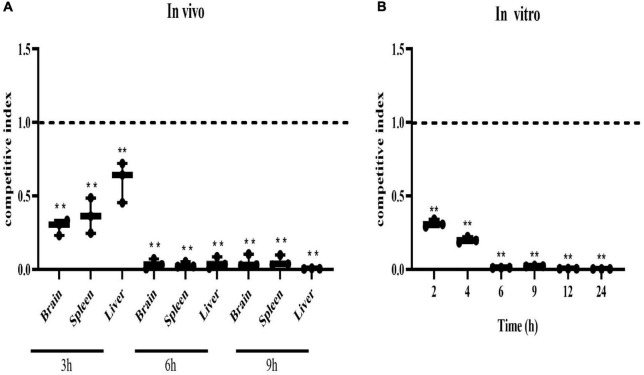
In vivo competition experiment, the CI values of the mutant strain ΔSaeRS in the brain, liver, and spleen of tilapia were significantly lower than 1 and close to 0 at 6 and 9 h, indicating that the in vivo colonization ability of the mutant strain ΔSaeRS on tilapia was significantly lower than the wild strain THN0901 (Figure 4A). The competitiveness of the mutants gradually decreased with the increase of time, and the CI value of the mutant was significantly less than 1 and close to 0 at 6 h (Figure 4B). The competitiveness of the mutant strain ΔSaeRS was significantly lower than that of the wild strain THN0901 both in vivo and in vitro.
FIGURE 4.
Competitive index of the ΔSaeRS mutant vs. total bacterial count. In vivo, the brain, spleen, and liver of tilapia (Oreochromis niloticus) infected with the mixed inoculum of the S. agalactiae THN0901 and ΔSaeRS strains were sampled and counted after 3, 6, and 12 h, respectively (A). In vitro, the mixture of S. agalactiae THN0901 and ΔSaeRS stains was inoculated in a fresh brain heart infusion (BHI) medium, and the bacterial liquid was sampled and counted after 2, 4, 6, 9, 12, and 24 h, respectively. Samples were tested in triplicate. A competitive index of 0.5 indicated that the ΔSaeRS mutant strain with THN0901 strain was proliferating equally (B). Each experiment was performed in triplicate, and data were presented as mean ± standard deviation (SD), *: 0.01 < P < 0.05, **: P < 0.01.
3.5. Challenge test
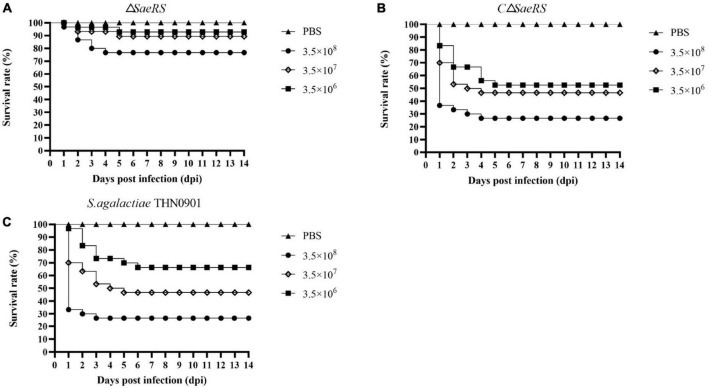
Tilapia were injected intraperitoneally with different concentration gradients of S. agalactiae THN0901, ΔSaeRS, and CΔSaeRS strains to detect the virulence. The cumulative survival results of tilapia after the challenge are shown in Table 2, and the related survival curve results are shown in Figure 5. The time of death in tilapia was concentrated within 6 days after the challenge, and the death was stable. Under high-dose infection, the fatality rate of wild strain THN0901 and complementary strain CΔSaeRS were both 73.3%, which was significantly higher than the mutant strain ΔSaeRS (33.3%). At low dose infection, the mortality rate of S. agalactiae THN0901, ΔSaeRS, and CΔSaeRS strains to tilapia were 33.3, 46.7, and 6.7%, respectively. The virulence of the ΔSaeRS mutant strain was significantly lower than that of the wild strain THN0901 and complementary strain CΔSaeRS.
FIGURE 5.
Survival curves of tilapia (Oreochromis niloticus) after intraperitoneal infection of S. agalactiae with three strains at the same dose. The mutant strain ΔSaeRS (A). The complementary strain CΔSaeRS (B). The wild-type strain THN0901 (C).
3.6. Whole blood-killing assays
In this experiment, the survival index was the ratio of the colony count at the end to the initial colony count. Among them, the survival index of S. agalactiae wild strain THN0901 was 3.81 ± 0.207, and the ΔSaeRS mutant strain was 1.39 ± 0.025. The survival index of the CΔSaeRS complementary strain was significantly higher than the mutant strain, which was 4.03 ± 0.551 (Figure 6). Therefore, the SaeRS genes are involved in survival of S. agalactiae in tilapia blood.
FIGURE 6.
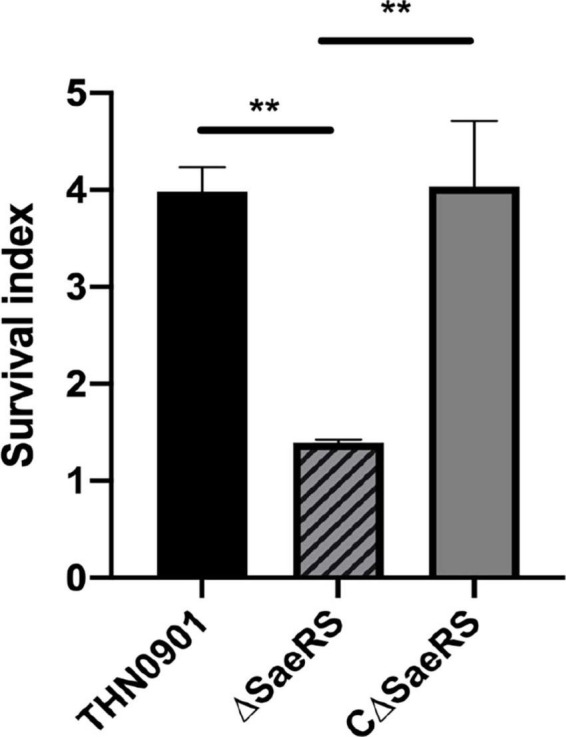
Whole blood-killing assay. The survival index was calculated. Each experiment was performed in triplicate, and data were presented as mean ± standard deviation (SD), *: 0.01 < P < 0.05, **: P < 0.01.
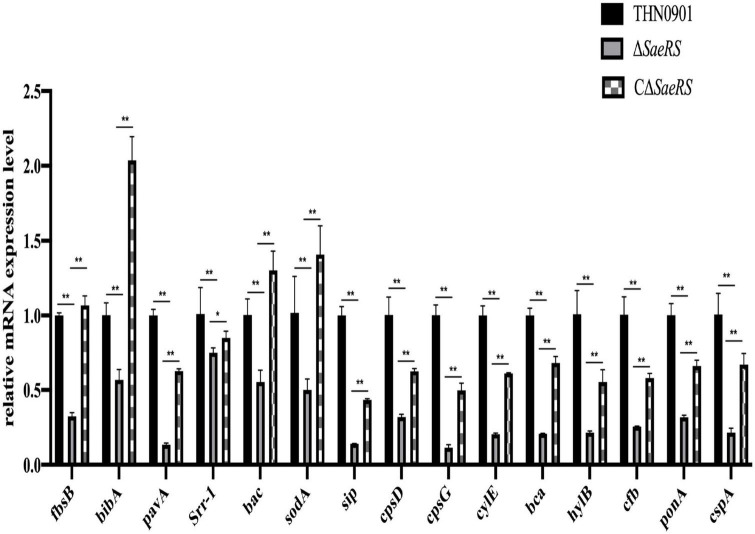
3.7. Virulence factor gene expression
The virulence factors related to adhesion, immune evasion, and bacterial invasion of the mutants were down-regulated compared with the wild strain THN0901 (Figure 7). Of these, the expression levels of fbsB, pavA, sip, cpsG, cylE, and bca genes in the ΔSaeRS mutant strain were significantly down-regulated compared with those in the wild strain THN0901 (P < 0.01), which decreased by 70, 90, 90, 90, 80, and 80%, respectively. After complement, the expression levels of related genes were all recovered, and most of the related virulence genes were lower than those of the wild strains. The expression levels of fbsB, bibA, bac, and sodA in the complementary strain CΔSaeRS were all higher than those of the wild strain THN0901.
FIGURE 7.
Relative messenger RNA (mRNA) expression level of virulence factors of S. agalactiae among three strains. Each experiment was performed in triplicate, and data were presented as mean ± standard deviation (SD), *: 0.01 < P < 0.05, **: P < 0.01.
4. Discussion
Group B Streptococcus has at least 22 different TCSs (Faralla et al., 2014), which are almost twice as many as other streptococci. For example, there are 13 species identified in S. pneumoniae and S. pyogenes (Vega et al., 2016; Alejandro et al., 2017). GBS has a higher capability of monitoring environmental conditions and reacting to changing stimuli (Thomas and Cook, 2020). SaeRS is a pair of TCS, and the signal is introduced by the histidine protein kinase and transferred through a series of phosphorylation events, phosphorylating the response protein and affecting a series of physiological and biochemical reactions of bacteria (Liao et al., 2021). In genome annotation, SaeS and SaeR are considered histidine kinase and response regulator, respectively. Most of the current research on SaeRS of TCS focus on S. aureus, but the TCS of S. agalactiae is homologous to the well-characterized SaeR (48% identical) and SaeS (34% identical) TCS in S. aureus (Cook et al., 2018). To explore the function of SaeRS in S. agalactiae, the biological properties of wild strain THN0901, mutant strain ΔSaeRS, and complementary strain CΔSaeRS were evaluated.
A previous study of Moraxella catarrhalis found that the inactivation of a gene (mesR) encoding a predicted response regulator of a TCS in M. catarrhalis yielded a mutant unable to grow in liquid media, which means that the response regulator is essential for growth (Joslin et al., 2015). Based on our experimental evidence, it appears that the SaeRS genes participate in growth regulation in S. agalactiae. Future research will focus on whether additional genes are upregulated in the SaeRS mutant to account for the observed growth defect.
SaeRS affects biofilm synthesis. Biofilms are heterogeneous aggregates of surface-associated microorganisms encapsulated in a self-produced polymeric matrix composed of polysaccharides, protein, and DNA, providing an ideal environment to protect bacteria from phagocytosis and harmful molecules (Kumar et al., 2017; Pang et al., 2019). In S. aureus Newman, a point mutation in SaeS resulted in the substitution of proline for leucine at amino acid 18, and the S. aureus Newman was unable to form a robust biofilm. RNA-Seq results showed that Sae regulated many genes that might affect the biofilm-forming ability of S. aureus Newman, such as lytS, lrgA, atlR, atlA, arlR, and aaa, they all directly or indirectly affect the formation of biofilm (Cue et al., 2015). But, deletion of SaeRS in S. epidermidis increased biofilm-forming ability, which was associated with increased environmental DNA (eDNA) release and upregulated assembly-activating protein (Aap) expression (Lou et al., 2011). Our experimental results are similar to those of S. aureus Newman. The biofilm-forming ability of the mutant strain was significantly reduced than that of the wild strain. The deletion of the SaeRS gene affects the expression of downstream biofilm-related genes, which leads to weakened biofilm-forming ability.
In S. aureus, SaeRS regulates bacterial survival in blood in a coagulase-dependent manner, and deletion of the SaeRS gene enhances the bacterial survival in human blood, which is contrary to the results of this study. Blood contains a variety of antibacterial substances, such as antibodies, phagocytes, and complements, so it has certain antibacterial functions (Ramsey, 2016). Our study showed that the survival index of the ΔSaeRS mutant strain in fresh tilapia blood was 1.39 ± 0.025, which was significantly lower than that of the wild strain and the complementary strain. The mutant strain had a reduced survival index through immune evasion in the blood. Some virulence factors related to immune evasion were also verified by RT-qPCR. The Cβ protein (bac) is one of the immunodominant components on the surface of S. agalactiae and can bind to the fragment crystallizable region (Fc) portion of immunoglobulin A (IgA). It may play an important role in resistance to immune defense mechanisms (Mawn et al., 1993; Schalen, 1993; Kreikemeyer and Jerlstrom, 1999; Berner et al., 2002; Das and Bishayi, 2009; Munzenmayer et al., 2016). Sip protein is a highly conserved surface protein in S. agalactiae with strong immunogenicity (Brodeur et al., 2000; Manning et al., 2006; Xue et al., 2010). CPS is a specific polysaccharide antigen in the outer capsule of the bacterial cell wall. The extra capsule of GBS is rich in sialic acid and can easily form oligosaccharides, which can inhibit the activation of the host’s alternative pathway of complement and significantly reduce the deposition of complement compound 3 (C3) and opsonin in cells, hindering complement-mediated opsonization and evading phagocytosis by the immune system (Marques et al., 1992). The results of q-PCR showed that the mRNA expression levels of sip, bac, cpsD, and cpsG in the mutant strain were significantly lower than those in the wild strain and recovered after complementary, which indicates that deletion of the SaeRS helps the immune escape of the S. agalactiae in blood.
In human strains of S. agalactiae, Cook showed through a mouse vaginal colonization model that pbsP and other potential genes promoted host colonization by upregulating SaeRS in TCS (Cook et al., 2018). Also, in S. aureus, inactivation of the SaeRS system stopped staphylococcal adhesion and lung epithelial internalization (Liang et al., 2006). The above results are consistent with the results of adhesion and colonization in this study. To explore the differences in colonization and invasion abilities between mutant strain ΔSaeRS and wild strain THN0901, in vivo competition experiment was conducted on tilapia. The colonization ability of the mutant strain in tilapia was significantly weaker than that of the wild strain, and the same results were also obtained in the in vitro competition. In S. aureus, SaeRS-regulated genes were associated with adhesion and invasion (Liang et al., 2006). We measured the mRNA expression of some colonization and invasion-related genes by RT-qPCR. Of these, srr-1, bibA, and bac genes are involved in the adhesion between S. agalactiae and host cells (Sundaresan et al., 2011; Souza et al., 2013; Manne et al., 2020), while hylB and fbsB genes are involved in the infection and spread of GBS in host cells (Pritchard et al., 1994; Gutekunst et al., 2004; Bobadilla et al., 2021). The decreased mRNA expression of these adhesion and invasion factors may be the reason for the in vivo competitive ability of mutant strain.
In S. aureus, mutation of SaeRS attenuates virulence in a mouse model of postoperative osteomyelitis, mainly due to reduced production of S. aureus virulence factors (Mashruwala et al., 2017). In the tilapia challenge experiment, the mortality of the mutant strain was significantly lower than that of the wild strain, which is consistent with the results of the mutants’ biofilm-forming ability and in vitro competition ability. The results of S. agalactiae colonization assays and biofilm-forming ability are closely related to bacterial virulence (Casadevall and Pirofski, 2001; Tajbakhsh et al., 2016). Attenuated virulence of mutant strain may be due to multiple effects of the above phenotypic changes, which requires further study.
In this study, the growth ability, biofilm-forming ability, in vivo competition ability of the SaeRS deletion of S. agalactiae THN0901 strain were lower than those of the wild strain. Therefore, SaeRS as a regulator of the virulence of S. agalactiae, plays a key role in promoting host colonization and realizing immune escape in tilapia infection. In future research, SaeRS can be used as the target of attenuated vaccine in production practice to enhance the immune protection effect of tilapia.
Data availability statement
The original contributions presented in the study are included in the article/supplementary material, further inquiries can be directed to the corresponding author. The genbank ID of the primers used in the article has been supplemented in Table 3.
Ethics statement
The animal study was reviewed and approved by the Sun Yat-sen University Animal Laboratory Center.
Author contributions
WL conceived the study. SL, QL, and JC performed the experiments. HL analyzed the data. SL, WL, and ZL provided the reagents and technical assistance and contributed to the completion of the study. SL drafted the manuscript. AL reviewed and finalized the manuscript. All authors reviewed the results and approved the final version of the manuscript.
Acknowledgments
We thank support of the “Development and Application of Immune Control Technology for Streptococcal Diseases of Tilapia.”
Funding Statement
This study was funded by the Agricultural Technology Promotion Center of Maoming City for this work.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
- Aisyhah M. A. S., Amal M. N. A., Zamri-Saad M., Siti-Zahrah A., Shaqinah N. N. (2015). Streptococcus agalactiae isolates from cultured fishes in Malaysia manifesting low resistance pattern towards selected antibiotics. J. Fish Dis. 38 1093–1098. 10.1111/jfd.12351 [DOI] [PubMed] [Google Scholar]
- Alejandro G., Gustavo G., Sven H. (2017). Streptococcus pneumoniae two-component regulatory systems: The interplay of the pneumococcus with its environment. Int. J. Med. Microbiol. 308 722–737. 10.1016/j.ijmm.2017.11.012 [DOI] [PubMed] [Google Scholar]
- Anshary H., Kurniawan R. A., Sriwulan S., Ramli R., Baxa D. V. (2014). Isolation and molecular identification of the etiological agents of streptococcosis in Nile tilapia (Oreochromis niloticus) cultured in net cages in Lake Sentani, Papua, Indonesia. Springerplus 3:627. 10.1186/2193-1801-3-627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badal D., Jayarani A. V., Kollaran M. A., Kumar A., Singh V. (2020). Pseudomonas aeruginosa biofilm formation on endotracheal tubes requires multiple two-component systems. J. Med. Microbiol. 69 906–919. 10.1099/jmm.0.001199 [DOI] [PubMed] [Google Scholar]
- Berner R., Ruess M., Bereswill S., Brandis M. (2002). Polymorphisms in the cell wall-spanning domain of the C protein beta-antigen in clinical Streptococcus agalactiae isolates are caused by genetic instability of repeating DNA sequences. Pediatr. Res. 51 106–111. 10.1203/00006450-200201000-00019 [DOI] [PubMed] [Google Scholar]
- Bobadilla F. J., Novosak M. G., Cortese I. J., Delgado O. D., Laczeski M. E. (2021). Prevalence, serotypes and virulence genes of Streptococcus agalactiae isolated from pregnant women with 35-37 weeks of gestation. BMC Infect. Dis. 21:73. 10.1186/s12879-020-05603-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boonyayatra S., Wongsathein D., Tharavichitkul P. (2020). Genetic relatedness among Streptococcus agalactiae isolated from cattle, fish, and humans. Foodborne Pathog. Dis. 17 137–143. 10.1089/fpd.2019.2687 [DOI] [PubMed] [Google Scholar]
- Brodeur B. R., Boyer M., Charlebois I., Hamel J., Couture F., Rioux C. R. (2000). Identification of group B streptococcal Sip protein, which elicits cross-protective immunity. Infect. Immun. 68 5610–5618. 10.1128/IAI.68.10.5610-5618.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capra E. J., Laub M. T. (2012). Evolution of two-component signal transduction systems. Annu. Rev. Microbiol. 66 325–347. 10.1146/annurev-micro-092611-150039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casadevall A., Pirofski L. (2001). Host-pathogen interactions: The attributes of virulence. J. Infect. Dis. 184 337–344. 10.1086/322044 [DOI] [PubMed] [Google Scholar]
- Christopher P. Z., Victoria K., Hendrik S. (2016). Molecular mechanisms of two-component signal transduction. J. Mol. Biol. 428 3752–3775. 10.1016/j.jmb.2016.08.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook L., Hu H., Maienschein-Cline M., Federle M. J. (2018). A vaginal tract signal detected by the group B Streptococcus SaeRS system elicits transcriptomic changes and enhances murine colonization. Infect. Immun. 86:e00762-17. 10.1128/IAI.00762-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cue D., Junecko J. M., Lei M. G., Blevins J. S., Smeltzer M. S., Lee C. Y. (2015). SaeRS-dependent inhibition of biofilm formation in Staphylococcus aureus Newman. PLoS One 10:e0123027. 10.1371/journal.pone.0123027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dagmar B., Roy G. (2006). Regulation of bacterial virulence by two-component systems. Curr. Opin. Microbiol. 9 143–152. 10.1016/j.mib.2006.01.005 [DOI] [PubMed] [Google Scholar]
- Das D., Bishayi B. (2009). Staphylococcal catalase protects intracellularly survived bacteria by destroying H2O2 produced by the murine peritoneal macrophages. Microb. Pathog. 47 57–67. 10.1016/j.micpath.2009.04.012 [DOI] [PubMed] [Google Scholar]
- Ericson M., Subramanian C., Frank M., Rock C. (2017). Role of fatty acid kinase in cellular lipid homeostasis and SaeRS-dependent virulence factor expression in Staphylococcus aureus. mBio 8:e00988-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans J. J., Bohnsack J. F., Klesius P. H., Whiting A. A., Garcia J. C., Shoemaker C. A., et al. (2008). Phylogenetic relationships among Streptococcus agalactiae isolated from piscine, dolphin, bovine and human sources: A dolphin and piscine lineage associated with a fish epidemic in Kuwait is also associated with human neonatal infections in Japan. J. Med. Microbiol. 57(Pt 11), 1369–1376. [DOI] [PubMed] [Google Scholar]
- Faralla C., Metruccio M. M., De Chiara M., Mu R., Patras K. A., Muzzi A., et al. (2014). Analysis of two-component systems in group B Streptococcus shows that RgfAC and the novel FspSR modulate virulence and bacterial fitness. mBio 5:e00870-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furfaro L. L., Chang B. J., Payne M. S. (2018). Perinatal Streptococcus agalactiae epidemiology and surveillance targets. Clin. Microbiol. Rev. 31:e00049-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groisman E. A. (2016). Feedback control of two-component regulatory systems. Annu. Rev. Microbiol. 70 103–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutekunst H., Eikmanns B. J., Reinscheid D. J. (2004). The novel fibrinogen-binding protein FbsB promotes Streptococcus agalactiae invasion into epithelial cells. Infect. Immun. 72 3495–3504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joslin S. N., Pybus C., Labandeira-Rey M., Evans A. S., Attia A. S., Brautigam C. A., et al. (2015). A Moraxella catarrhalis two-component signal transduction system necessary for growth in liquid media affects production of two lysozyme inhibitors. Infect. Immun. 83 146–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kayansamruaj P., Pirarat N., Kondo H., Hirono I., Rodkhum C. (2014). Draft genome sequences of Streptococcus agalactiae strains isolated from Nile Tilapia (Oreochromis niloticus) farms in Thailand. Genome Announc. 2:e01300-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreikemeyer B., Jerlstrom P. G. (1999). An Escherichia coli-Enterococcus faecalis shuttle vector as a tool for the construction of a group B Streptococcus heterologous mutant expressing the beta antigen (Bac) of the C protein complex. FEMS Microbiol. Lett. 180 255–262. [DOI] [PubMed] [Google Scholar]
- Kumar A., Alam A., Rani M., Ehtesham N. Z., Hasnain S. E. (2017). Biofilms: Survival and defense strategy for pathogens. Int. J. Med. Microbiol. 307 481–489. [DOI] [PubMed] [Google Scholar]
- Liang X., Yu C., Sun J., Liu H., Landwehr C., Holmes D., et al. (2006). Inactivation of a two-component signal transduction system, SaeRS, eliminates adherence and attenuates virulence of Staphylococcus aureus. Infect. Immun. 74 4655–4665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao B., Ye X., Chen X., Zhou Y., Cheng L., Zhou X., et al. (2021). The two-component signal transduction system and its regulation in Candida albicans. Virulence 12 1884–1899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu G., Zhu J., Chen K., Gao T., Yao H., Liu Y., et al. (2016). Development of Streptococcus agalactiae vaccines for tilapia. Dis. Aquat. Organ. 122 163–170. [DOI] [PubMed] [Google Scholar]
- Liu L., Shen X., Yu J., Cao X., Zhan Q., Guo Y., et al. (2020). Subinhibitory concentrations of fusidic acid may reduce the virulence of S. Aureus by down-regulating sarA and saeRS to reduce biofilm formation and α-toxin expression. Front. Microbiol. 11:25. 10.3389/fmicb.2020.00025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Gao W., Yang J., Guo H., Zhang J., Ji Y. (2021). Contribution of coagulase and its regulator SaeRS to lethality of CA-MRSA 923 bacteremia. Pathogens 10:1396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lou Q., Zhu T., Hu J., Ben H., Yang J., Yu F., et al. (2011). Role of the SaeRS two-component regulatory system in Staphylococcus epidermidis autolysis and biofilm formation. BMC Microbiol. 11:146. 10.1186/1471-2180-11-146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyhs U., Kulkas L., Katholm J., Waller K. P., Saha K., Tomusk R. J., et al. (2016). Streptococcus agalactiae serotype IV in humans and cattle, Northern Europe1. Emerg. Infect. Dis. 22 2097–2103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macho A. P., Zumaquero A., Ortizmartín I., Beuzón C. R. (2010). Competitive index in mixed infections: a sensitive and accurate assay for the genetic analysis of Pseudomonas syringae-plant interactions. Mol. Plant Pathol. 8, 437–450. 10.1111/j.1364-3703.2007.00404.x [DOI] [PubMed] [Google Scholar]
- Manne K., Chattopadhyay D., Agarwal V., Blom A. M., Khare B., Chakravarthy S., et al. (2020). Novel structure of the N-terminal helical domain of BibA, a group B Streptococcus immunogenic bacterial adhesin. Acta Crystallogr. D Struct. Biol. 76(Pt 8), 759–770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manning S. D., Wood S., Kasha K., Martin D., Rioux S., Brodeur B., et al. (2006). Naturally occurring antibodies for the group B streptococcal surface immunogenic protein (Sip) in pregnant women and newborn babies. Vaccine 24 6905–6912. [DOI] [PubMed] [Google Scholar]
- Marques M. B., Kasper D. L., Pangburn M. K., Wessels M. R. (1992). Prevention of C3 deposition by capsular polysaccharide is a virulence mechanism of type III group B streptococci. Infect. Immun. 60 3986–3993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mashruwala A., Gries C., Scherr T., Kielian T., Boyd J. (2017). SaeRS is responsive to cellular respiratory status and regulates fermentative biofilm formation in Staphylococcus aureus. Infect. Immun. 85:e00157-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mawn J. A., Simpson A. J., Heard S. R. (1993). Detection of the C protein gene among group B streptococci using PCR. J. Clin. Pathol. 46 633–636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitrophanov A. Y., Groisman E. A. (2008). Signal integration in bacterial two-component regulatory systems. Genes Dev. 22 2601–2611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munzenmayer L., Geiger T., Daiber E., Schulte B., Autenrieth S. E., Fraunholz M., et al. (2016). Influence of Sae-regulated and Agr-regulated factors on the escape of Staphylococcus aureus from human macrophages. Cell. Microbiol. 18 1172–1183. [DOI] [PubMed] [Google Scholar]
- Naraid S., Fanrong K., Danny K., Gwendolyn L. G., Kidchakan S. (2008). Occurrence of rare genotypes of Streptococcus agalactiae in cultured red tilapia Oreochromis sp. and Nile tilapia O. niloticus in Thailand—relationship to human isolates? Aquaculture 284 35–40. [Google Scholar]
- O’Toole G. A., Kolter R. (1998). Flagellar and twitching motility are necessary for Pseudomonas aeruginosa biofilm development. Mol. Microbiol. 30 295–304. [DOI] [PubMed] [Google Scholar]
- Pang Z., Raudonis R., Glick B., Lin T., Cheng Z. (2019). Antibiotic resistance in Pseudomonas aeruginosa: Mechanisms and alternative therapeutic strategies. Biotechnol. Adv. 37 177–192. [DOI] [PubMed] [Google Scholar]
- Patterson H., Saralahti A., Parikka M., Dramsi S., Trieu-Cuot P., Poyart C., et al. (2012). Adult zebrafish model of bacterial meningitis in Streptococcus agalactiae infection. Dev. Comp. Immunol. 38, 447–455. 10.1016/j.dci.2012.07.007 [DOI] [PubMed] [Google Scholar]
- Pritchard D. G., Lin B., Willingham T. R., Baker J. R. (1994). Characterization of the group B streptococcal hyaluronate lyase. Arch. Biochem. Biophys. 315 431–437. [DOI] [PubMed] [Google Scholar]
- Ramsey G. (2016). Hemostatic efficacy of pathogen-inactivated blood components. Semin. Thromb. Hemost. 42 172–182. [DOI] [PubMed] [Google Scholar]
- Schalen C. (1993). Prevalence of IgA receptors in clinical isolates of Streptococcus pyogenes and Streptococcus agalactiae: Serologic distinction between the receptors by blocking antibodies. FEMS Immunol. Med. Microbiol. 7 39–45. [DOI] [PubMed] [Google Scholar]
- Souza V. C., Kegele F. C., Souza S. R., Neves F. P., de Paula G. R., Barros R. R. (2013). Antimicrobial susceptibility and genetic diversity of Streptococcus agalactiae recovered from newborns and pregnant women in Brazil. Scand. J. Infect. Dis. 45 780–785. [DOI] [PubMed] [Google Scholar]
- Sundaresan R., Samen U., Ponnuraj K. (2011). Expression, purification, crystallization and preliminary X-ray diffraction studies of the human keratin 4-binding domain of serine-rich repeat protein 1 from Streptococcus agalactiae. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 67(Pt 12), 1582–1585. 10.1107/S1744309111040413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tajbakhsh E., Ahmadi P., Abedpour-Dehkordi E., Arbab-Soleimani N., Khamesipour F. (2016). Biofilm formation, antimicrobial susceptibility, serogroups and virulence genes of uropathogenic E. coli isolated from clinical samples in Iran. Antimicrob. Resist. Infect. Control 5:11. 10.1186/s13756-016-0109-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takamatsu D., Osaki M., Sekizaki T. (2001). Thermosensitive suicide vectors for gene replacement in Streptococcus suis. Plasmid 46 140–148. [DOI] [PubMed] [Google Scholar]
- Tavares G. C., Pereira F. L., Barony G. M., Rezende C. P., Da S. W., de Souza G., et al. (2019). Delineation of the pan-proteome of fish-pathogenic Streptococcus agalactiae strains using a label-free shotgun approach. BMC Genomics 20:11. 10.1186/s12864-018-5423-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas L., Cook L. (2020). Two-component signal transduction systems in the human pathogen Streptococcus agalactiae. Infect. Immun. 88: e00931-19. 10.1128/IAI.00931-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tierney A. R., Rather P. N. (2019). Roles of two-component regulatory systems in antibiotic resistance. Future Microbiol. 14 533–552. 10.2217/fmb-2019-0002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vega L., Malke H., McIver K. (2016). Virulence-related transcriptional regulators of Streptococcus pyogenes. Streptococcus pyogenes: Basic biology to clinical manifestations. Oklahoma City, OK: University of Oklahoma Health Sciences Center. [PubMed] [Google Scholar]
- Xue G., Yu L., Li S., Shen X. (2010). Intranasal immunization with GBS surface protein Sip and ScpB induces specific mucosal and systemic immune responses in mice. FEMS Immunol. Med. Microbiol. 58 202–210. 10.1111/j.1574-695X.2009.00623.x [DOI] [PubMed] [Google Scholar]
- Yao X., Li M., Wang J., Wang C., Hu D., Zheng F., et al. (2015). Isolation and characterization of a native avirulent strain of Streptococcus suis serotype 2: A perspective for vaccine development. Sci. Rep. 5:9835. 10.1038/srep09835 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The original contributions presented in the study are included in the article/supplementary material, further inquiries can be directed to the corresponding author. The genbank ID of the primers used in the article has been supplemented in Table 3.