Abstract
Influx of sodium ions through voltage-gated sodium channels in cardiomyocytes is essential for proper electrical conduction within the heart. Both acquired conditions associated with sodium channel dysfunction (myocardial ischaemia, heart failure) as well as inherited disorders secondary to mutations in the gene SCN5A encoding for the cardiac sodium channel Nav1.5 are associated with life-threatening arrhythmias. Research in the last decade has uncovered the complex nature of Nav1.5 distribution, function, in particular within distinct subcellular subdomains of cardiomyocytes. Nav1.5-based channels furthermore display previously unrecognized non-electrogenic actions and may impact on cardiac structural integrity, leading to cardiomyopathy. Moreover, SCN5A and Nav1.5 are expressed in cell types other than cardiomyocytes as well as various extracardiac tissues, where their functional role in, e.g. epilepsy, gastrointestinal motility, cancer and the innate immune response is increasingly investigated and recognized. This review provides an overview of these novel insights and how they deepen our mechanistic knowledge on SCN5A channelopathies and Nav1.5 (dys)function.
This article is part of the theme issue ‘The heartbeat: its molecular basis and physiological mechanisms’.
Keywords: electrophysiology, arrhythmia, ion channels, genetics, SCN5A
1. Introduction
Influx of sodium ions through voltage-gated sodium channels in cardiomyocytes initiates the cardiac action potential and is essential for excitability of myocardial cells and proper electrical conduction within the heart. The importance of cardiac sodium channels is underscored by the occurrence of potentially lethal arrhythmias in the setting of acquired conditions associated with sodium channel dysfunction (myocardial ischaemia, heart failure) as well as inherited disorders secondary to mutations in the gene SCN5A encoding for the cardiac sodium channel Nav1.5 [1]. While genetic, electrophysiological and molecular studies have provided insight into the (dys)function and (dys)regulation of SCN5A and Nav1.5, it has become increasingly clear that sodium channel distribution, function and regulation is more complicated than traditionally assumed, in particular within distinct subcellular subdomains of cardiomyocytes [2]. Nav1.5-based channels furthermore display previously unrecognized non-electrogenic actions and may impact on cardiac structural integrity, thereby also potentially affecting arrhythmogenesis [3]. Moreover, SCN5A and Nav1.5 are expressed in cell types other than cardiomyocytes as well as various extracardiac tissues, where their functional role is increasingly investigated and recognized. SCN5A mutations have now been associated with (sudden unexpected death in) epilepsy and gastrointestinal disorders, and a role for Nav1.5 in smooth muscle cell (SMC) function, cancer, innate immune response and inflammation has been reported. This review provides an overview of these novel insights and how they deepen our mechanistic knowledge on sodium channel (dys)function and their role in cardiac disorders, ultimately facilitating development of novel strategies for diagnosis, risk stratification and treatment in patients with SCN5A channelopathies.
2. Sodium channel structure, distribution and function
(a) . Cardiac sodium channel structure and mode of action
The sodium channel family comprises a total of nine genes (SCN1A-SCN5A, SCN7A-SCN11A), of which the SCN5A gene located on human chromosome 3p22 encodes Nav1.5, the pore-forming alpha subunit of the cardiac sodium channel. Nav1.5 is made up of a cytoplasmic N terminus, four internally homologous domains (DI-DIV; consisting of six transmembrane α-helical segments, S1–S6) interconnected by cytoplasmic linkers, and a cytoplasmic C terminal domain [4]. The DI-DIV domains fold around an ion-conducting pore; during membrane depolarization, outward movement of the positively charged S4 segments (which acts as voltage sensor) results in the opening of the channel pore, allowing for sodium influx [5]. As a result, the membrane is further depolarized (phase 0 of the cardiac action potential), leading to L-type calcium channel activation, calcium influx and contraction. Subsequent fast and slow inactivation closes the channel pore, until membrane repolarization allows the channel to recover from inactivation and once again become available for activation [6,7]. During physiological conditions, activation and inactivation of sodium channels is closely regulated, but alterations in these processes in the setting of acquired and inherited sodium channel dysfunction may set the stage for electrical disturbances and arrhythmias (as discussed in more detail in the next sections).
(b) . SCN5A/Nav1.5 distribution in heart and subcellular domains of cardiomyocytes
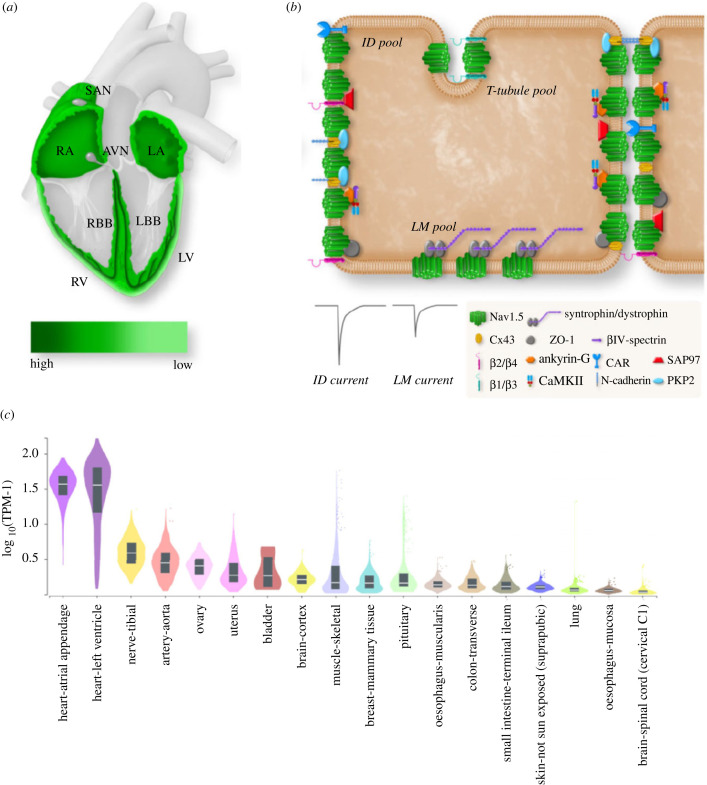
SCN5A/Nav1.5 expression is high in atrial and ventricular myocardium, His bundle, bundle branches and Purkinje fibres, but low to absent in the central sino-atrial and atrio-ventricular nodes [8]. Furthermore, SCN5A/Nav1.5 displays a transmural gradient in ventricular myocardium, showing higher abundance in subepicardium when compared with subendocardium (figure 1a) [8]. Within cardiomyocytes, Nav1.5 is localized in a distinct subcellular pattern, with a relatively low density at the crests, grooves and T-tubules at the lateral membrane (LM) versus an enrichment in the intercalated disc (ID) region (figure 1b) [9]. Accordingly, sodium currents measured at the ID are larger than at the LM [10], and ID-based Nav1.5 is considered especially relevant for fast propagation of electrical signals [11]. Nevertheless, loss of Nav1.5 at the LM leads to conduction slowing [12], and hence is also functionally relevant in this subcellular domain.
Figure 1.
(a) Expression levels of Nav1.5 throughout various regions of the heart. (b) Cardiomyocyte subcellular distribution of Nav1.5 and interacting proteins in lateral membrane (LM), T-tubule and intercalated disc (ID) (from [3], with permission). (c) Expression levels of SCN5A in various human tissues (bulk tissue gene expression for ENSG00000183873.15, logarithmic scale; data source: GTEx Analysis release V8 (dbGaP accession: phs000424.v8.p2)).
(c) . Nav1.5 interacting proteins and macromolecular complex
Sodium channels are not isolated units within the myocyte membrane; instead, Nav1.5 forms a macromolecular complex with interacting proteins, an assortment of proteins which regulate Nav1.5 trafficking and localization as well as sodium current biophysical properties [9,13–15]. Through this complex, Nav1.5 associates not only with various isoforms of the accessory β-subunits, but also with proteins that participate in cell adhesion, signal transduction, and cytoskeleton anchoring [3,15]. Several of these Nav1.5 interacting proteins are specifically localized at or enriched in distinct subcellular domains within cardiomyocytes (figure 1b) [3,15]. At the LM, Nav1.5 interacts with the dystrophin–syntrophin complex, the calcium/calmodulin-dependent serine protein kinase (CASK) and caveolin-3 [16], whereas at the ID Nav1.5 associates with N-cadherin, connexin-43, βIV-spectrin and desmosomal proteins such as plakophilin-2 and desmoglein-2 [15]. Nav1.5 interacting proteins likely play an essential role in the specific subcellular localization of Nav1.5 within cardiomyocytes, and their regional variation is considered to underlie at least in part the observed differences in Nav1.5 expression, sodium current density and kinetics between distinct subcellular microdomains [15]. As further discussed below, mutations in genes encoding a number of these interacting proteins have been associated with cardiac electrical disorders and/or Nav1.5 dysfunction.
(d) . Transcriptional and post-translational regulation of SCN5A and Nav1.5
Following transcription, alternative splicing of the SCN5A gene produces different transcript variants. The neonatal SCN5A-001 transcript, which is most abundant during embryonic development, is replaced by the mature transcript SCN5A-003 after birth, but may be upregulated during pathophysiological conditions [17]. These two isoforms differ in exon 6 (exon 6b in SCN5A-003 and exon 6a in SCN5A-001) leading to a difference of seven amino acids and distinct gating properties of Nav1.5 [17,18]. This has been shown to be of potential functional relevance; for instance, an SCN5A mutation may display more severe biophysical effects in the presence of the neonatal isoform resulting in a highly malignant fetal LQTS phenotype [19]. As reviewed by Schroeter et al., additional splice variants exist, displaying unique biophysical properties, tissue- and species-specific expression, and developmental regulation [17]. Following translation, Nav1.5 channels are assembled in the endoplasmic reticulum, transported to the Golgi apparatus and targeted to the membrane via the microtubule network (reviewed by Balse et al. [20]). Phosphorylation, glycosylation, S-nitrosylation, ubiquitination and methylation are important post-translational regulatory mechanisms impacting on Nav1.5 trafficking, function and degradation [21]. Notably, phosphorylation of Nav1.5 by PKA, PKC and calcium/calmodulin-dependent protein kinase II (CamKII) has been shown to modulate Nav1.5 trafficking as well as (late) sodium current magnitude [22,23]. Ubiquitylation of Nav1.5 mediates its internalization and subsequent degradation, and hence is an important determinant of channel density at the membrane [24]. Nav1.5 function is furthermore regulated by numerous factors, including intracellular calcium levels, reactive oxygen species and temperature [25].
(e) . SCN5A/Nav1.5 expression in extracardiac tissue and other cell types in the heart
In addition to heart, SCN5A and Nav1.5 are expressed in multiple extracardiac tissues including brain, (smooth) muscle, nerves, intestine and artery (figure 1c). Moreover, their expression has been found in both excitable and non-excitable cell types, including SMCs, neurons, (myo)fibroblasts, endothelial cells, macrophages, thymocytes and cancer cells. The potential functional role of SCN5A/Nav1.5 in these tissues and cell types will be discussed in separate sections of this review.
3. Electrophysiological consequences of SCN5A dysfunction
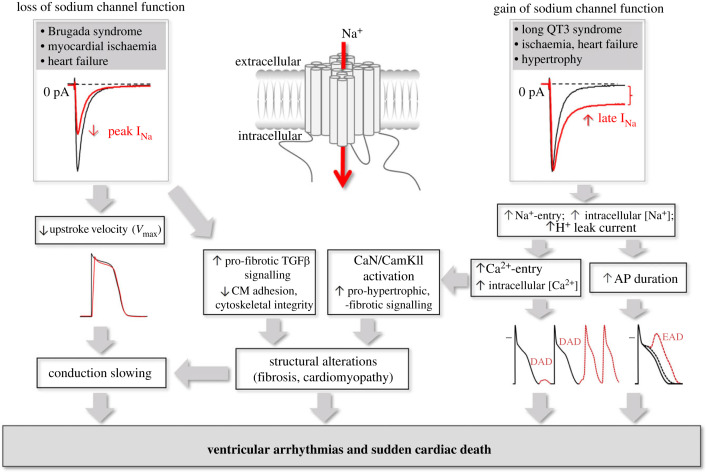
Distinct alterations in gating and other biophysical properties of the sodium channel may have various electrophysiological consequences [26]. These can be divided into alterations causing a loss of sodium channel function and consequent depolarization disturbances, and those leading to an enhanced late sodium current and consequent prolonged repolarization (gain of function). These electrophysiological changes may, through distinct mechanisms, set the stage for potentially life-threatening ventricular arrhythmias (figure 2). The associated clinical syndromes are discussed in more detail in the next section.
Figure 2.
Pro-arrhythmic consequences of gain and loss of sodium channel function, including electrogenic effects (conduction slowing, action potential prolongation) as well as effects on cardiac structure with proposed mechanisms (CM, cardiomyocyte; CaN, calcineurin; CamKII, calmodulin-dependent protein kinase II; EAD, early afterdepolarization; DAD, delayed afterdepolarization).
(a) . Causes and consequences of reduced sodium channel availability
Reduced sodium channel availability may occur in the setting of both inherited and acquired pathological conditions. Decreased peak inward sodium current and consequent lower influx of sodium ions into the cardiomyocyte reduces upstroke velocity of the action potential and slows propagation. Depending on the underlying cause and location involved, slowing of conduction through parts of the specialized conduction system may occur (sino-atrial node, atrio-ventricular node, His–Purkinje) and (heterogeneous) conduction slowing throughout the ventricular myocardium may set the stage for re-entrant ventricular arrhythmias. Acquired conditions include myocardial ischaemia, during which metabolic changes within the myocardium lead to inactivation of sodium channels, and heart failure, where alterations in SCN5A expression and post-translational regulation affect sodium current [27]. In inherited arrhythmia syndromes (see below), loss of function mutations in SCN5A can lead to a decreased number of functional channels on the membrane due to misfolding of the channel and/or altered trafficking. Alternatively, sodium channels may be present on the membrane with decreased functionality secondary to reduced conductivity, disruption of activation, accelerated inactivation or impaired recovery from inactivation [28,29].
(b) . Causes and consequences of increased late sodium current
The initial peak sodium current is for the most part rapidly inactivated, but a small fraction of the current may persist throughout the duration of the action potential plateau phase. While this late sodium current is small during physiological conditions, it may be enhanced in the setting of acquired (ischaemia, hypertrophy and heart failure) and inherited disease (e.g. SCN5A mutations) [30,31]. Irrespective of the underlying cause, delayed repolarization and action potential prolongation increase the occurrence of early after-depolarizations which can trigger torsades de pointes arrhythmias and sudden death (figure 2). Enhanced late sodium current may furthermore dysregulate intracellular sodium and calcium homeostasis: increased intracellular sodium induces a secondary rise in intracellular calcium via the sodium–calcium exchanger thereby inducing calcium-dependent pro-arrhythmic events including delayed after-depolarizations [32]. During acquired conditions such as heart failure, post-translational modulation of sodium channels by calcium-dependent pathways (such as CamKII) may increase late sodium current magnitude [33]. In the setting of inherited disorders (see below), gain of function SCN5A mutations often disrupt fast inactivation, allowing for sodium channels to re-open during the action potential plateau phase thereby increasing late sodium current, or alternatively may lead to slowed inactivation (resulting in channel openings of longer duration), faster recovery from inactivation, or a shift in voltage dependence of inactivation [25,34]. Certain mutations may lead to a so-called overlap syndrome, causing biophysical defects leading to both gain and loss of function of Nav1.5 (see below).
4. Disorders associated with Nav1.5 dysfunction
(a) . Inherited disorders associated with SCN5A mutations
Mutations in SCN5A have been implicated in multiple inherited disorders, which each display distinct phenotypical characteristics. These mutations may lead to a gain or a loss of sodium channel function, or a combination of both. Long QT syndrome (LQTS) is characterized by prolonged QT intervals on the ECG, and increased risk for sudden death due to ventricular tachyarrhythmias, in particular torsades de pointes. Various subtypes of LQTS exist, caused by mutations in different genes and displaying distinct clinical features. Patients with LQTS type 3 (LQT3) are often bradycardic and have a high risk of ventricular arrhythmias which occur predominantly during rest or sleep (at slow heart rates) [35]. LQT3 is caused by gain of function SCN5A mutations which prolong repolarization and set the stage for torsades de points arrhythmias (see previous section). Brugada syndrome (BrS) is associated with the occurrence of ventricular arrhythmias and sudden cardiac death occurring mostly in males, predominantly during rest and sleep [36]. The hallmark ECG pattern of BrS comprises a coved ST-segment in the right-precordial leads V1 to V3, which may be variably present and can be unmasked or increased after administration of sodium channel blocking drugs (ajmaline, flecainide), or during fever or exercise [36]. BrS patients frequently display low voltages, fractionated late potentials and (subtle) structural abnormalities such as fibrosis in the epicardial layer of the right ventricular outflow tract (RVOT), and arrhythmias are often inducible in the RVOT of affected individuals [37]. Loss of function SCN5A mutations are identified in approximately 20% of BrS patients, while mutations in other ion channel and non-ion channel genes are sporadically found [38,39]. In addition, a more complex oligogenic or polygenic inheritance is now also recognized, with multiple genetic modifiers (common or rare) defining the genetic basis [38,39]. Loss of function SCN5A mutations leading to reduced sodium channel availability have also been identified in patients with inherited sick sinus syndrome as well as progressive cardiac conduction defect (PCCD), also called Lenègre or Lev disease, an inherited disorder characterized by progressive conduction slowing through the His–Purkinje system, (complete) AV block, syncope and sudden death [40,41]. Mutations in SCN5A have furthermore been associated with atrial fibrillation in young patients with structurally normal hearts. Both loss of function and gain of function mutations have been described in this familial form, which may induce AF by decreasing atrial conduction velocity and/or increasing atrial excitability [42,43]. In some instances, one single SCN5A mutation can result in a ‘sodium channel overlap syndrome', with multiple disease phenotypes consistent with both gain and loss of sodium channel function occurring even within one affected family. For instance, carriers of the SCN5A-1795insD mutation within in a large Dutch family present with extensive variability in type and severity of symptoms, including sinus node dysfunction, bradycardia, conduction disease, BrS, LQT3 (either in isolation or in combinations thereof), and an increased risk for nocturnal sudden death [44]. Electrophysiological analysis of transgenic mice carrying the heterozygous Scn5a-1798insD/+ mutation (the mouse homologue of the human mutation) demonstrated that the mutation caused a reduced peak sodium current density, a delayed time course of fast inactivation and a small late sodium current, explaining the observed multiple phenotypes [45]. Similar clinical and electrophysiological overlap has been reported for other SCN5A mutations, although a clear parallel between the mixed clinical phenotype of a certain SCN5A mutation and its biophysical properties is not always observed; here, factors such as age and genetic modifiers may play a modulatory role [46,47]. Finally, SCN5A mutations are increasingly associated with dilated and arrhythmogenic cardiomyopathy (ACM), which will be discussed in more detail in the section below.
(b) . Arrhythmia disorders associated with mutations in Nav1.5-interacting proteins
As mentioned above, Nav1.5 forms part of a macromolecular complex through which it interacts with a large a number of associated proteins. Crucially, mutations in these interacting proteins have been associated with sodium channel dysfunction and arrhythmia syndromes (table 1). For instance, mutations in the sodium channel accessory β-subunits have been identified in patients with BrS, LQTS, atrial fibrillation and idiopathic ventricular fibrillation [46]. Furthermore, mutations in the Nav1.5 interacting proteins alpha-1-syntrophin and caveolin-3, which are located at the LM, have been reported in LQTS patients and have been demonstrated to increase late sodium current [48,49], while mutations in ankyrin-B have been associated with sinus node dysfunction, idiopathic ventricular fibrillation, LQTS and catecholaminergic polymorphic ventricular tachycardia [50,51]. Moreover, the absence of dystrophin in Duchenne muscular dystrophy is associated with arrhythmias in affected patients and a reduced sodium current is observed in mdx mouse cardiomyocytes [52]. Finally, mutations in genes encoding desmosomal proteins located at the ID region have been associated with ACM [53]. Crucially, a number of these proteins interact with Nav1.5 and decreased sodium current has been observed in ACM models, even prior to development of cardiomyopathic changes [54,55]. Interestingly, as discussed in more detail below, SCN5A mutations have now also been implicated in ACM.
Table 1.
Nav1.5 interacting proteins, their (preferential) localization in adult cardiomyocytes (ID, intercalated discs; LM, lateral membrane), and associated inherited cardiac disorders (LQTS, long QT syndrome; BrS, Brugada syndrome; AF, atrial fibrillation; CCD, cardiac conduction disease; SND, sinus node dysfunction; CPVT, catecholaminergic polymorphic ventricular tachycardia; IVF, idiopathic ventricular fibrillation; ACM, arrhythmogenic cardiomyopathy; DCM, dilated cardiomyopathy).
| gene | protein | subdomain | associated cardiac disease |
|---|---|---|---|
| SCN1B | β1 | LM, ID | BrS, AF, CCD |
| SCN2B | β2 | ID | AF |
| SCN3B | β3 | LM | BrS, AF |
| SCN4B | β4 | LM, ID | LQTS, AF |
| SNTA1 | α1-syntrophin | LM | LQTS |
| CASK | calcium/calmodulin-dependent serine protein kinase (CASK) | LM | – |
| ANK2 | ankyrin-B | LM | LQTS, SND, CPVT, IVF |
| CAV3 | caveolin-3 | LM | LQTS |
| FGF13 | fibroblast growth factor 13 | LM, ID | – |
| ANK3 | ankyrin-G | LM, ID | – |
| PKP2 | plakophilin-2 | ID | ACM |
| DSG2 | desmoglein-2 | ID | ACM, DCM |
| CDH2 | N-cadherin | ID | ACM |
| SPTBN4 | βIV-spectrin | ID | – |
| DCTN2 | dynactin subunit 2 (p50/dynamitin) | ID | – |
| SAP97 | SAP97; DLG1 | ID | – |
| GPD1L | glycerol-3-phosphate dehydrogenase 1-like | unknown | BrS |
(c) . Acquired disorders associated with Nav1.5 dysfunction
As briefly mentioned above, in addition to inherited disorders, sodium channel dysfunction also occurs in the setting of a number of acquired conditions. Apart from alterations in (late) sodium current and consequent deleterious effects on conduction, repolarization and calcium homeostasis in the setting of myocardial ischaemia, Nav1.5 dysfunction is also observed in metabolic disorders such as diabetes and obesity [56]. Here, both loss and gain of sodium channel function have been observed; a reduced SCN5A expression and smaller peak sodium current leading to conduction slowing [57], as well as enhanced late sodium current. The latter is the consequence of metabolic dysregulation, including alterations in cytosolic calcium and CaMKII [58,59], and may ultimately lead to the occurrence of calcium-dependent pro-arrhythmic events. Furthermore, a crucial role for Nav1.5 has been described in tumour growth, invasiveness and metastasis of certain cancers; this topic will be discussed in more detail in a later section of this review.
5. Cardiomyopathy and SCN5A channelopathy: mechanisms and consequences
(a) . SCN5A channelopathy, cardiac structural alterations and cardiomyopathy
In addition to primary electrical disorders, SCN5A mutations have also been associated with cardiac structural alterations such as myocardial fibrosis and (dilated) cardiomyopathy [60–64]. In BrS patients, ventricular hypertrophy has been reported in addition to increased collagen content and fibrosis, particularly in the subepicardium of the RVOT [64]. Furthermore, familial forms of dilated cardiomyopathy (DCM) have been reported in patients with SCN5A mutations, often presenting in combination with conduction disease, atrial arrhythmias and/or fibrillation [62,65]. Interestingly, biophysical properties consistent with both loss and gain of sodium channel function have been observed in SCN5A mutations associated with DCM [65,66]. A specific subtype, characterized by DCM and multifocal ectopic Purkinje-related premature contractions (MEPPC), is associated with gain of function SCN5A mutations which lead to hyperexcitability of the fascicular–Purkinje system [67,68]. SCN5A mutations are now increasingly identified in DCM patients [69], and can present with a broad range of electrical disorders ranging from conduction abnormalities to atrial and ventricular arrhythmias [70]. More recently, an overlap between BrS and arrhythmogenic (right ventricular) cardiomyopathy (ARVC/ACM) has been described; clinical features of both syndromes were observed in patients that did not carry mutations in ARVC/ACM-related desmosomal proteins, but were found to have a (loss of function) SCN5A mutation [38,53]. ARVC and BrS both affect predominantly the right ventricle with involvement of cardiac cell–cell junctions, and demonstrate pathophysiological, genetic, structural and electrophysiological overlap [71]. While cardiac structural alterations were originally considered to occur secondary to repetitive, long-standing arrhythmias in affected patients, they are now increasingly recognized as a more direct consequence of Nav1.5 dysfunction in the setting of SCN5A mutations. This is further strengthened by the age-dependent development of structural remodelling in mice with heterozygous Scn5a deficiency [72]. Clearly, development of cardiac structural derangements will contribute to arrhythmogenesis in affected patients. For instance, in the SCN5A-1795insD family, we found that most mutation carriers who suffered ventricular arrhythmias above the age of 40 years (despite pacemaker treatment), had ventricular hypertrophy [44,73]. Although the latter could be explained by hypertension in these patients, studies in mice carrying the mouse homologue mutation demonstrated age-dependent development of hypertrophy and fibrosis, indicating a direct effect of Nav1.5 dysfunction, as discussed in more detail below [44].
(b) . SCN5A/Nav1.5 and cardiac development
The observation that both gain and loss of function mutations are associated with cardiomyopathy suggests that Nav1.5 may also impact on cardiac structure in a non-electrogenic manner, independent from its ion-conducting properties. Nav1.5 is present at early developmental stages but is not yet capable of generating sodium currents because of the relatively depolarized resting membrane potential [74]. In zebrafish, the presence of Scn5a homologues was found to be essential for normal cardiac development at an early stage, before sodium channels impact on the electrophysiological properties of the heart; pharmacological inhibition of sodium current did not have the same effect, indicating that the early embryonic lethality occurred independently of electrogenic effects of Nav1.5 [75]. Similarly, Scn5a-deficient and mutant mice die at an early embryonic stage [76,77], and we recently demonstrated abnormal development and cardiac structural abnormalities in homozygous Scn5a-1798insD embryos at an early stage before sodium channels become functionally relevant for cardiac electrical activity, providing further evidence for a non-electrogenic role for Nav1.5 [78].
(c) . Mechanisms underlying cardiac structural alterations secondary to Nav1.5 dysfunction
Cardiac structural remodelling secondary to SCN5A mutations may be explained by various mechanisms (figure 2). In the setting of gain of function mutations, enhanced late sodium current and consequent increased sodium influx will result in elevated intracellular calcium levels by way of the sodium–calcium exchanger. Increased calcium concentrations may activate calcium/calmodulin-dependent protein kinase II (CAMKII) and protein phosphatase calcineurin (CaN), signalling pathways controlling pro-hypertrophic and pro-fibrotic gene transcription [3,79]. This has indeed been confirmed in studies in murine LQT3 models, which have also demonstrated the beneficial effects of the late sodium current inhibitor ranolazine on calcium homeostasis, pro-arrhythmia and cardiac structural alterations [32,44,80,81]. Certain DCM-related SCN5A mutations have been shown to induce a proton leak current which leads to intracellular sodium overload via the sodium–hydrogen exchanger, ultimately resulting in intracellular calcium overload [82,83]. Ionic imbalances and intracellular acidosis may not only impact on arrhythmogenesis and pro-fibrotic pathways, but also potentially impair excitation–contraction and myofilament function [83]. With respect to structural consequences of loss of sodium channel function, electrical activity-dependent stimulation of pro-fibrotic factors of the transforming growth factor β (TGFβ) pathway has been proposed [84], with inhibition of sodium current leading to increased levels of TGFβ1 receptors [85]. Interestingly, both fibrosis and TGFβ1 transcript levels were found to be increased in ventricles of aged heterozygous Scn5a-deficient mice, suggesting development of TGFβ1-mediated structural abnormalities secondary to sodium channel dysfunction [86]. A potential non-electrogenic mechanism of loss of function SCN5A mutations on cardiac structure involves protein–protein interactions. As described in previous sections, Nav1.5 interacts with a variety of proteins, including cytoskeletal proteins, components of the extracellular matrix and adhesion- and desmosomal proteins at the ID. The latter function as mechanical linkers between cardiomyocytes, and are vital for myocardial structure. It is therefore tempting to speculate that Nav1.5 dysfunction impairs the function or localization of these mechanical linkers or contractile proteins, destabilizing intercellular adhesion and/or cytoskeletal integrity. Indeed, loss of Nav1.5 in HL1 cells has been shown to reduce intercellular adhesion strength [87]. However, while it is clear that Nav1.5 interacting proteins modulate sodium channel function [9,13–15], there is currently limited experimental evidence to support the notion that modulation also occurs in the opposite direction, and the impact of different SCN5A mutations on interacting proteins remains to investigated. Overall, it is clear that Nav1.5 not only determines electrophysiological characteristics of the myocardium, but also exerts effects on myocardial structure and function through various mechanisms (figure 2).
6. Role of SCN5A/Nav1.5 in cancer
(a) . Impact of SCN5A/Nav1.5 on tumour growth and metastasis
Increased expression of sodium channels in tumours is well established, with different sodium channel isoforms found in distinct tumour types [88]. Nav1.5 is present in many types of tumour tissue, including breast, colon, lymphoma, neuroblastoma, non-small cell lung cancer, ovarian, small cell lung cancer, with the highest overexpression levels observed in metastatic breast cancer, colon cancer and ovarian cancer [89,90]. Moreover, Nav1.5 represents the most highly expressed subunit in lymphoma and breast cancer cells [90]. While the adult isoform of SCN5A is found in colon cancer cells, in breast and prostate cancer cells the neonatal SCN5A splice variant is mostly expressed [89,90]. Overall, increased expression of Nav1.5 is associated with more aggressive tumour characteristics, including lymph node invasion, recurrence of metastasis and reduced survival [91]. This has been observed for breast cancer, colon cancer and ovarian cancer, but not for gliomas or lung cancer cells [90]. Silencing either adult or neonatal SCN5A has been shown to reduce invasion and migration of MDA-MB-231 breast cancer cells [92,93], and MDA-MB-231 cells with downregulated Nav1.5 orthotopically implanted into mice showed reduced tumour growth and local invasion in vivo [94].
(b) . Mechanisms underlying Nav1.5-mediated alterations in tumour growth and metastasis
Nav1.5 regulates a number of processes in tumour cells which can affect its metastatic properties, including galvanotaxis (directed movement in response to electrical current), migration, invasion, adhesion, and gene expression [90]. In tumour cells, sodium channels are particularly enriched in invadopodia, plasma membrane protrusions involved in extracellular matrix degeneration and therefore relevant for migration and invasion [95,96]. In breast cancer cells, Nav1.5 channels co-localize and coimmunoprecipitate with the nitrogen–hydrogen exchanger 1 (NHE-1) and increase its activity [95,97,98], enhancing influx of sodium and efflux of hydrogen. This results in acidification (and thus a lower pH) of the extracellular environment and an alkalization of the intracellular environment. This acidification of the extracellular space increases cathepsin activity, enhances invadopodial formation and activity and promotes invasion through extracellular matrix degeneration, enabling the tumour to metastasize [95,97]. Increased expression of (neonatal) Nav1.5 in cancer cells may also lead to increased late sodium current and subsequent increased intracellular calcium levels, which in turn may activate SNARE-mediated vesicle fusion and stimulate the activity of podosomes, actin-rich plasma membrane components that facilitate cell motility, adhesion and migration of tumour cells [90,97,99]. Indeed, abnormally high sodium concentrations are found in several types of cancer cells [100], and non-invasive imaging of sodium accumulation in breast tissue (23Na MRI) may provide a useful diagnostic and prognostic biomarker [101].
(c) . Therapeutic considerations
Given the deleterious effects of overexpression of Nav1.5 in tumour cells, pharmacological inhibition of Nav1.5 would be expected to be beneficial. Indeed, tetrodotoxin (TTX) has been reported to inhibit Nav1.5, impeding sodium influx, phagocytosis and endosomal acidification in breast cancer cells and reducing their invasion and migration [95]. Similarly, Nav1.5 blockade with phenytoin reduced invasion of breast cancer cells in vitro [91], and reduced tumour growth, invasion, proliferation and metastasis of breast cancer cells in vivo in mice [102]. In addition, ranolazine inhibited Nav1.5-mediated breast cancer cell invasiveness in vitro and attenuated metastatic lung colonization by breast cancer cells in mice [103]. It is important to note that, since the expression of sodium channel isoforms other than Nav1.5 is also increased in certain types of cancer cells [88], (part of) the observed effects of sodium channel blockers may also be due to their action on these other isoforms. Theoretically, patients using such sodium channel blocking drugs (for instance, for epilepsy) who subsequently develop cancer, may have a less aggressive tumour with less extensive metastasis. However, such a beneficial effect has as yet not been observed [104], and prospective studies will be required to investigate this further. Moreover, it should be noted that the use of sodium blocking drugs carries a potential risk of side effects, including cardiac conduction slowing and arrhythmias. More recently, small molecule blockers of neonatal Nav1.5 were designed, which reduced invasion of breast cancer cells in vitro, providing a promising novel therapeutic approach [105].
7. SCN5A/Nav1.5 (dys)function in brain and neuronal tissue
(a) . SCN5A/Nav1.5 expression in neuronal tissue
Neuronal tissues express a wide range of sodium channel isoforms displaying distinct biophysical properties and TTX sensitivity, and their differential expression have been shown to mediate neuron subtype-specific firing patterns [106]. In addition to neuronal sodium channel isoforms, several studies have also detected the presence of SCN5A and Nav1.5 in rodent and human brain, ganglia and neurons. In the mouse brain, Nav1.5 protein has been observed in the cortex, thalamus, hypothalamus, basal ganglia, cerebellum and brain stem, clustering in axons [107]. In the frontal lobe cortex of the human brain, Nav1.5 was found to be predominantly located in neuronal cell bodies, axons and dendrites, but to a far lesser extent in glial components [108]. On the mRNA level, SCN5A showed a restricted expression pattern in rat forebrain with selective localization in limbic regions, including the pirifom cortex and subcortical limbic nuclei [109]. These regions include projections to and from the amygdala, hippocampus and hypothalamus, indicating that SCN5A may play a role in or modulate various functions such as olfactory perception and autonomic responses [109]. SCN5A/Nav1.5 expression has furthermore been observed in dorsal root ganglia (DRG), vestibular ganglion neurons, olfactory sensory neurons and intracardiac neurons [110–113], with electrophysiological studies indicating a functional role for Nav1.5 in neuronal firing in a number of these tissues [110,112]. Crucially, distinct SCN5A splice isoforms appear relevant for neuronal function. For instance, in mouse DRG the exon 18-deleted Nav1.5a is the predominant isoform, which has a 53-amino acid truncation in the intracellular loop between DII and III and consequently altered inactivation properties [110]. By contrast, wild-type Nav1.5 in addition to the splice variants Nav1.5c (containing the variant Q1077) and Nav1.5e (the embryonic or neonatal variant containing exon 6A instead of 6) were found to be expressed in human brain cortex [108]. While the overall expression of SCN5A/Nav1.5 in neuronal tissue is clearly lower than in the heart, their increasingly reported involvement in several neurological conditions underscores their functional relevance in the brain and neurons.
(b) . SCN5A and epilepsy
Mutations in various ion channels are well known to be associated with various forms of epilepsy. It is, however, becoming increasingly clear that patients with inherited cardiac arrhythmia syndromes such as LQTS may present with seizures, and they are often (initially) misdiagnosed with epilepsy [114]. In a cohort of 610 LQTS patients, 11% displayed seizures or seizure-like episodes, the majority of which had a diagnosis of LQTS2 associated with a mutation in KCNH2 [115]. Nevertheless, in addition to mutations in neuronal sodium channels, a potential role for SCN5A/Nav1.5 in epilepsy has been described. In a rat model of temporal lobe epilepsy, increased expression of Scn5a and Nav1.5 was observed in the brain, including the hippocampus [116]. In addition, a number of case reports and studies have described the identification of SCN5A mutations in patients with epilepsy, mostly co-occurring with other SCN5A-related cardiac disorders. Epileptic seizures have been reported as the initial clinical presentation in patients with BrS [117,118]. In one family, three males carrying the SCN5A-p.W1095X mutation were initially treated for epileptic seizures in childhood before being diagnosed with BrS during adulthood [119]. In a child with LQTS and neonatal seizures, the SCN5A mutation p.R1623Q was found, which is known to be associated with a severe LQTS phenotype [120]. In a further study of 21 infants with LQTS, a high incidence of epilepsy and neurodevelopmental disorders was observed in patients with perinatal LQTS, whereas this did not occur in those with non-perinatal LQTS [121]. Of the perinatal LQTS patients suffering epileptic seizures, two patients carried SCN5A mutations (G1631D and P1332L) [121]. In an infant presenting with QT-prolongation and self-terminating torsades de pointes shortly after birth, a generalized tonic–clonic seizure (in the setting of a normal heart rhythm) occurred 2 days after birth, and the infant was found to carry the SCN5A-M1766L mutation [122]. While hypoperfusion of the brain consequent to arrhythmias may increase susceptibility to epilepsy in LQTS patients, some reports demonstrated the occurrence of seizures in the absence of cerebral abnormalities, indicating a more direct mechanistic link [121]. From a biophysical point of view, gain of function SCN5A mutations may lead to persistent depolarization of neurons, increased firing frequency and potentially epileptiform bursting behaviour; in contrast, it is difficult to predict how loss of function mutations observed in BrS could lead to seizures. Interestingly, as indicated above, SCN5A is expressed in limbic regions such as the piriform cortex; the latter area is known to have a low threshold for epileptogenesis [109]. Nevertheless, functional proof of a direct link between SCN5A mutations and epilepsy is as yet still lacking.
(c) . SCN5A and sudden unexpected death in epilepsy
In recent years, it has become increasingly clear that epilepsy patients are at increased risk of sudden death, a condition known as sudden unexpected death in epilepsy (SUDEP), with the incidence of SUDEP in the general epilepsy population being around 1.2–1.3 per 1000 person-years [123–125]. SUDEP is defined as ‘sudden, unexpected, witnessed or unwitnessed, non-traumatic and non-drowning death, occurring in benign circumstances, in an individual with epilepsy, with or without evidence for a seizure and excluding documented status epilepticus (seizure duration ≥30 min or seizures without recovery in between), in which post-mortem examination does not reveal a cause of death' [126]. Clinical risk factors for SUDEP include recent seizures, increased seizure frequency and nocturnal seizures [124,126]. The pathophysiology of SUDEP is likely heterogeneous and includes ictal and peri-ictal respiratory dysfunction and apnoea, autonomic nervous system dysregulation, arousal system dysfunction, neurotransmitter dysbalance and ion channel dysfunction [125–127]. SUDEP may occur during or immediately following a generalized tonic–clonic seizure, but is also observed without any immediately prior visible seizures, indicating a potential role for other pathomechanisms, including cardiac [126]. Indeed, the genetic spectrum of SUDEP not only includes epilepsy genes and genes involved in respiration and arousal, but variants in arrhythmia genes are also increasingly recognized, including KCNH2, KCNQ1, RYR2 and SCN5A [128,129]. Genetic testing using post-mortem blood collected from 48 definite or possible SUDEP cases revealed, in addition to variants in KCNQ1 and KCNH2, six synonymous and three non-synonymous variants in SCN5A; the latter were all previously associated with LQTS [129]. In another cohort of 61 SUDEP cases, two SCN5A variants (Ile397Val and Val223Gly) were identified [130]. Interestingly, Ile397Val and Val223Leu (but not Val223Gly as in the SUDEP case) were previously reported in BrS patients [29,131]. A case report furthermore described a female patient diagnosed with epilepsy but who died 1 year later in her sleep; post-mortem genetic analysis revealed the presence of the SCN5A-R523C mutation [132], which was previously reported in an LQTS patient [133]. To gain further insight into the potential pathogenicity of these last three variants, Soh and colleagues investigated their biophysical properties in CHO cells and observed a reduced peak current for Val223Gly as well as changes in gating properties for all three variants, underlining their potential functional impact [134]. Hence, the (co-)occurrence of SCN5A mutations in these patients may have contributed to the development of cardiac arrhythmias and potentially SUDEP. However, some of these patients were also treated with anti-epileptic drugs, many of which are in fact sodium channel blockers, which may by themselves, or in co-occurrence with SCN5A variants, decrease peak sodium current and increase the risk for arrhythmias [135,136], thereby providing an additional factor underlying and/or contributing to SUDEP.
(d) . SCN5A and other neurological disorders
In addition to epilepsy, SCN5A and Nav1.5 have been implicated in other neurological disorders. The expression of SCN5A/Nav1.5 was found to be downregulated in DRG neurons and axons of peripheral sensory neurons in rats with spared nerve injury, a model of neuropathic pain [137], but the functional implications of this observation remains unknown. A potential role for SCN5A in schizophrenia has also been proposed, possibly by altering the activity of the limbic system [138]. Interestingly, Blom and colleagues observed an increased prevalence of a Brugada ECG pattern in patients with schizophrenia when compared with similarly aged controls [139]. While these findings suggest a higher prevalence of BrS in schizophrenia patients (which in turn also have an increased risk for SCD), other factors may also play a confounding role. For instance, drugs used in the management of schizophrenia may also block cardiac sodium channels, and as such increase the risk for arrhythmias [140]. However, the observed increased prevalence of a Brugada ECG was independent of sodium channel blocker use [139], suggesting a more direct interaction. Indeed, ion channels have been implicated in mediating schizophrenia risk [141], and hence further investigation of a potential role for SCN5A is warranted. Finally, alterations in Nav1.5 have been demonstrated in multiple sclerosis (MS), a condition associated with destruction of myelin sheaths and axonal damage. Upregulated Nav1.5 expression was observed in and around MS lesions, in particular within reactive astrocytes, which play a prominent role in the response of the nervous system to injury [142]. This upregulation was proposed to provide a compensatory mechanism in order to maintain ionic homeostasis and prevent injury-induced calcium dysregulation [142,143]. Indeed, deletion of Nav1.5 from astrocytes in mice significantly worsened outcomes in an experimental model of MS, with increased inflammatory infiltrate in both early and late stages of the disease [144]. Nav1.5 expression was furthermore observed in macrophages within active MS lesions (predominantly in late endosomes and phagolysosomes), suggesting a potential role in phagocytic myelin degradation [145].
8. SCN5A channelopathies and gastrointestinal disorders
(a) . Expression and function of Nav1.5 in the gastrointestinal tract
Interstitial cells of Cajal (ICC) and intestinal SMCs are essential for gastrointestinal motility. Smooth muscle contractility is governed by excitability and changes in membrane potential, and as such is regulated by a wide variety of ion channels, including sodium channels [146]. Of the latter, Nav1.5 is the predominant isoform in the gastrointestinal tract, with functional expression of SCN5A/Nav1.5 having been demonstrated in rat and human jejunal and colon circular SMCs [147–149]. While calcium is classically considered key for SMC function, a role for sodium channels is increasingly recognized [150]. Nav1.5 activation may impact on gastrointestinal motility by modulating electrical slow waves in ICC, and by regulating calcium entry and subsequent contraction in SMC [150,151]. Crucially, Nav1.5 in ICC is mechanosensitive: it is activated by shear stress, with stretch increasing slow wave frequency in human ICC, while sodium current inhibition by lidocaine decreased slow wave frequency [151]. Moreover, ranolazine was found to decrease peak sodium current and prevent shear stress-induced increase in sodium current in human colon SMCs, in addition to reducing contractility of human colon muscle strips [148]. The latter observation has been suggested to underlie the known side-effect of obstipation reported with use of ranolazine [148].
(b) . Gastrointestinal disorders associated with SCN5A mutations
In 2006, the first potential association between SCN5A mutations and gastrointestinal disorders was presented. Of 31 SCN5A mutation carriers, more than half self-reported abdominal pain and/or other gastrointestinal symptoms; most of these patients had a gain of function mutation and a clinical LQT3 phenotype [152]. Given the localization of Nav1.5 in the gastrointestinal tract, and the fact that a genetic basis has been proposed for irritable bowel syndrome (IBS), Saito et al. investigated the presence of SCN5A mutations in 49 IBS patients [153]. In one patient, they identified the SCN5A-G298S mutation which was shown to lead to a loss of sodium channel function [153]. Subsequent studies in larger IBS cohorts confirmed this initial observation, reporting the presence of an SCN5A mutation in approximately 2% of IBS patients [154,155], with the majority of SCN5A mutation carriers suffering from constipation-predominant IBS [154]. Most SCN5A mutations identified in IBS patients have been shown to lead to loss of sodium channel function [154,155]. The SCN5A-A997T mutation, identified in a patient suffering from constipation-predominant IBS, was found to almost completely abolish peak sodium current, which could be rescued by chronic incubation with mexiletine; interestingly, mexiletine treatment of the patient successfully reduced IBS symptoms [154]. In addition, identified mutations also impaired mechanosensitive function of Nav1.5 [155]. In fact, the IBS-associated mutation SCN5A-G615E was observed to have limited impact on baseline sodium current function, but mostly caused disruptions in mechanosensitivity and mechano-electrical feedback [156]. While this observation may suggest that reduced mechanosensitivity underlies smooth muscle pathophysiology caused by SCN5A mutations, it will require further mechanistic investigation.
9. SCN5A/Nav1.5 (dys)function, immune response and inflammation
(a) . SCN5A/Nav1.5 in macrophages
In 2007, work by Carrithers et al. revealed the presence of Nav1.5 on the late endosome of human monocyte-derived macrophages [157]. They furthermore demonstrated a functional role for Nav1.5 in regulating phagocytosis and endosomal acidification, providing evidence that lipopolysaccharide-induced activation of Nav1.5 leads to sodium efflux and consequently decreased intraendosomal pH [157]. A subsequent study by the same group showed that Nav1.5 also regulates phagocytosis and intracellular processing of mycobacteria by human macrophages [158]. Human macrophage SCN5A was furthermore found to act as a pathogen sensor, initiating signalling and transcription of immune response genes for antiviral host defence, enhancing the anti-inflammatory profile of macrophages [159]. More recently, it was concluded that human macrophage SCN5A mediates an innate immune signalling pathway that limits DNA damage through increased expression of PPP1R10, a regulator of phosphatase activity and DNA repair [160]. As discussed in a previous section, Nav1.5 expression has been found in late endosomes and phagolysosomes of macrophages within active MS lesions, where they likely participate in the phagocytic pathway of myelin degradation [145]; moreover, transfer of macrophages expressing Nav1.5 into mice with experimentally induced autoimmune encephalomyelitis promoted recovery [161].
(b) . SCN5A/Nav1.5 in thymocytes and T cells
Thymocytes, immune cells in the thymus, transform into mature T cells through a selection process by which peripheral T cells are formed that are able to respond to foreign pathogens but do not target the body's own antigens. Lo and colleagues demonstrated that SCN5A mediates calcium entry into CD4+CD8+ double-positive thymocytes, thereby regulating positive selection of CD4+ T cells in the thymus [162]. Moreover, they showed that overexpression of SCN5A in peripheral T cells resulted in altered CD4+ T-cell sensitivity and T-cell receptor signalling, in addition to an impaired response during infection [163]. Since SCN5A is normally no longer detectable in T cells following the CD4+ T-cell selection stage, the authors hypothesized that this repression of SCN5A is essential to prevent unwanted T-cell response and consequent auto-immunity [163].
(c) . Potential implications for arrhythmogenesis
Macrophages may play an important role in arrhythmogenesis via a number of mechanisms, including secretion of pro-inflammatory cytokines, alterations in cell-to-cell coupling, and induction of electrical, structural and autonomic remodelling [164]. Hence, alterations in macrophage function consequent to alterations in SCN5A may well lead to an abnormal immune response, autoimmune disorder and ultimately pro-arrhythmia. Although there is as yet little direct evidence for such a disease mechanism, signs of myocardial inflammation have been reported in a large proportion of BrS patients, in particular in the RVOT [165]. Moreover, autoantibodies directed at a number of cardiac proteins have been detected in plasma samples of BrS patients [166]. Recently, extensive myocardial inflammation in the absence of myocarditis or sarcoidosis was found in a patient who was successfully resuscitated for ventricular fibrillation, and who was found to carry a novel likely pathogenic SCN5A variant [167]. Moreover, myocardial inflammation is commonly observed in patients with ACM [53], which has been associated with SCN5A variants (see earlier sections).
10. Other non-cardiomyocyte effects of SCN5A/Nav1.5
(a) . SCN5A/Nav1.5 in (vascular) smooth muscle cells
As discussed in the previous section, Nav1.5 is expressed in SMCs of the gastrointestinal tract where they are thought to regulate motility. Some studies have also reported its (functional) presence in SMCs of other tissues, where they may contribute to the generation and/or propagation of spontaneous electrical activity underlying myogenic contractions [168]. SCN5A expression and functional Nav1.5 activity was detected in airway SMCs from rabbit bronchi [168]. Although the authors were not able to demonstrate a potential impact on myogenic contraction, they hypothesized that this is likely due to the fact that Nav1.5 channels are inactivated at the relatively depolarized membrane potential of these cells under normal conditions, but that they may still become functionally relevant during pathophysiological conditions and contribute to e.g. bronchospasm [168]. In rat femoral artery, Nav1.5 was found to be expressed in endothelial cells and SMCs of the media [169]. Exposure to the Nav1.5 activator veratridine or hypoxia caused vasocontraction, which was prevented by the potent Nav1.5 channel blocker F-15845; SMCs rather than endothelial cells were shown to be involved in these effects [169]. In bovine pulmonary artery SMCs, Nav1.5 expression was observed in the cell membrane and caveolae, where they were shown to mediate the effects of the vasoconstrictor endothelin-1 on pulmonary vascular tone [170]. Taken together, these studies suggest a potential modulatory role of Nav1.5 in SMCs with putative functional impact on various (patho)physiological processes, but the evidence is as yet limited and further exploration is warranted.
(b) . SCN5A/Nav1.5 in endothelial cells
In cultured endothelial cells derived from rat interlobar artery and human umbilical vein, a TTX-resistant sodium current was identified with properties similar to Nav1.5 [171]. Similarly, a TTX-resistant sodium current was observed in endothelial cells cultured from human saphenous vein, in addition to expression of SCN5A [172]. As mentioned above, Nav1.5 was also found to be expressed in endothelial cells of rat femoral artery, but no functional impact was observed [169]. Human umbilical vein endothelial cells (HUVECs) express both Nav1.5 and Nav1.7 and display a mostly TTX-resistant current; inhibition of the latter (i.e. inhibition of Nav1.5) inhibited HUVEC angiogenesis activity, increased HUVEC adhesion, and reduced HUVEC proliferation induced by vascular endothelial growth factor (VEGF) [173]. Moreover, Nav1.5 was shown to potentiate VEGF-induced ERK1/2 activation by attenuating membrane depolarization, altering intracellular calcium kinetics and PKCα activity [173]. These findings identified Nav1.5 as a regulator of angiogenic function in endothelial cells and a potential novel strategy for controlling angiogenesis.
(c) . SCN5A/Nav1.5 in (myo)fibroblasts
Fibroblasts play an important role in extracellular matrix formation and production of various paracrine and autocrine factors. Although they are non-excitable, fibroblasts can couple with other cell types including cardiomyocytes and affect their electrophysiological properties [174]. In human atrial fibroblast cultures, fast inward sodium current appeared de novo upon differentiation into myofibroblasts, which was predominantly TTX-resistant with properties similar to Nav1.5; in addition, SCN5A was the most abundant transcript identified [175]. Similarly, cultured fibroblasts isolated from right atrial tissue from patients with chronic atrial fibrillation demonstrated an increase in functional Nav1.5 channels upon differentiation into myofibroblasts [176]. A large window current was observed in myofibroblasts, which would be expected to lead to substantial sodium entry and consequently calcium influx [175]. While the latter could theoretically enhance myofibroblast proliferation, direct evidence is lacking; moreover, it remains to be investigated whether sodium channels promote fibroblasts differentiation into myofibroblasts and/or modulate myofibroblast secretion properties [177]. If so, it would open up potential novel avenues for therapeutic strategies to prevent fibrosis and ultimately arrhythmias.
11. Conclusion: insight gained and remaining questions
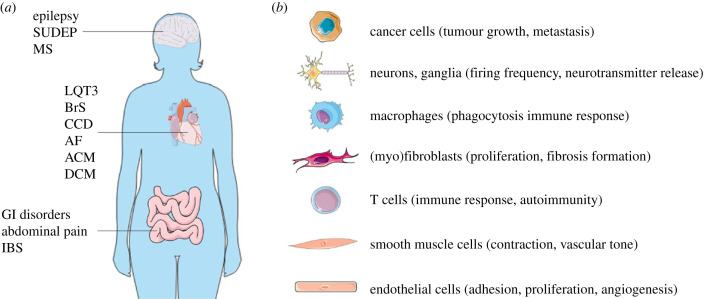
SCN5A/Nav1.5 (dys)function is clearly highly complex on multiple levels, not only in distinct cardiomyocyte subcellular microdomains, but also in other cell types within the heart (figure 3). While the consequences of SCN5A mutations are increasingly investigated in non-ventricular cardiomyocytes (e.g. atrial, conduction system), their potential impact on cells in the heart other than cardiomyocytes (e.g. SMCs, fibroblasts, neurons) has hardly been explored. Moreover, given the functional role of Nav1.5 in many non-cardiac tissues (figure 3), SCN5A mutations may result in extracardiac consequences which may or may not in turn exacerbate cardiac dysfunction and/or arrhythmias. Conversely, these non-cardiac clinical phenotypes may also provide potential new ways to diagnose and risk stratify patients with SCN5A mutations; for instance, one could also consider using biopsy material from extracardiac tissues (e.g. gastrointestinal) to study patient-specific impact of sodium channel dysfunction and potential therapeutic approaches. Regarding the latter, it should be taken into account that (pharmacological) interventions targeting Nav1.5 will impact not only the intended organ, but also other tissues including the heart. Hence, novel Nav1.5 modulators developed, for example, neurological and gastrointestinal disorders as well as cancer may inadvertently lead to an increased risk for arrhythmias. Mechanistically, knowledge obtained on the effects of Nav1.5 (dys)function in non-excitable cells and tumour cells provides insight into the non-electrogenic role of the channel (including mechanosensitivity), which is increasingly recognized to contribute to the development of cardiomyopathy and other structural alterations secondary to SCN5A mutations. Regardless of the underlying mechanism, the latter will further predispose to arrhythmogenesis, contributing significantly to disease severity, progression and prognosis. Thus, therapeutic strategies aimed at preventing structural abnormalities secondary to SCN5A mutations may prove a promising approach for certain patients.
Figure 3.
(a) Disorders in brain, heart and intestine associated with SCN5A mutations/variants (SUDEP, sudden unexplained death in epilepsy; MS, multiple sclerosis; LQT3, Long QT syndrome type 3; BrS, Brugada syndrome; AF, atrial fibrillation; CCD, cardiac conduction disease; ACM, arrhythmogenic cardiomyopathy; DCM, dilated cardiomyopathy; GI, gastrointestinal; IBS, irritable bowel syndrome). (b) Non-cardiomyocyte cell types expressing SCN5A/Nav1.5 with proposed function.
Taken together, research discussed in this review demonstrates that SCN5A channelopathies are more complex than previously appreciated, and raises many new questions, such as:
-
•
Do SCN5A mutations modulate invasiveness of tumours, and as such are these patients at higher risk for metastasis and worse outcome when they get cancer?
-
•
Do patients using sodium channel blocking drugs (e.g. for epilepsy) who subsequently develop cancer, have a less aggressive tumour with less extensive metastasis?
-
•
Do SCN5A mutations lead to dysregulation of the autonomic nervous system, thereby potentially impacting on arrhythmogenesis?
-
•
Are variants in SCN5A associated with worse outcome in MS?
-
•
Are SCN5A mutations associated with an increased risk for schizophrenia?
-
•
Can clinical parameters of, e.g. gastrointestinal or autonomic nervous system (dys)function be used for risk prediction in SCN5A mutation carriers?
-
•
Do SCN5A mutations impact on vascular SMC function, thereby potentially increasing the risk for hypertension and/or (subclinical) ischaemia?
-
•
Do SCN5A mutations affect (myo)fibroblast (dys)function, potentially inducing cardiac fibrosis?
-
•
Do SCN5A mutations affect macrophage function and/or innate immune response, leading to inflammation and/or inappropriate auto-immunity?
Clearly, SCN5A channelopathies need to be approached in a more holistic, multidisciplinary manner, with collaboration between different specialists to improve patient management. Similarly, basic and translational research into SCN5A mutations and Nav1.5 dysfunction should address the various roles of this channel in different cell types and tissues using appropriate human and animal models. Ultimately, a multidisciplinary approach combining basic, translational, genetic and clinical studies should lead to improved diagnosis, risk stratification, treatment and outcome in patients with SCN5A channelopathy.
Data accessibility
This article has no additional data.
Conflict of interest declaration
I declare I have no competing interests.
Funding
This work was supported by the Netherlands CardioVascular Research Initiative (CVON): the Dutch Heart Foundation, Dutch Federation of University Medical Centers, the Netherlands Organisation for Health Research and Development, and the Royal Netherlands Academy of Sciences (CVON-PREDICT2 2018-30), by the Innovational Research Incentives Scheme Vidi grant from The Netherlands Organisation for Health Research and Development (ZonMw; 91714371), and by Fondation Leducq (17CVD02).
References
- 1.Remme CA, Bezzina CR. 2010. Sodium channel (dys)function and cardiac arrhythmias. Cardiovasc. Ther. 28, 287-294. ( 10.1111/j.1755-5922.2010.00210.x) [DOI] [PubMed] [Google Scholar]
- 2.Marchal GA, Remme CA. 2023. Subcellular diversity of Nav1.5 in cardiomyocytes: distinct functions, mechanisms and targets. J. Physiol. 601, 941–960. ( 10.1113/JP283086) [DOI] [PubMed] [Google Scholar]
- 3.Rivaud MR, Delmar M, Remme CA. 2020. Heritable arrhythmia syndromes associated with abnormal cardiac sodium channel function: ionic and non-ionic mechanisms. Cardiovasc. Res. 116, 1557-1570. ( 10.1093/cvr/cvaa082) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gellens ME, George AL, Chen LQ, Chahine M, Horn R, Barchi RL, Kallen RG. 1992. Primary structure and functional expression of the human cardiac tetrodotoxin-insensitive voltage-dependent sodium channel. Proc. Natl Acad. Sci. USA 89, 554-558. ( 10.1073/pnas.89.2.554) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Catterall WA. 2000. From ionic currents to molecular mechanisms: the structure and function of voltage-gated sodium channels. Neuron 26, 13-25. ( 10.1016/S0896-6273(00)81133-2) [DOI] [PubMed] [Google Scholar]
- 6.Balser JR. 2001. The cardiac sodium channel: gating function and molecular pharmacology. J. Mol. Cell. Cardiol. 33, 599-613. ( 10.1006/jmcc.2000.1346) [DOI] [PubMed] [Google Scholar]
- 7.Goldin AL. 2003. Mechanisms of sodium channel inactivation. Curr. Opin. Neurobiol. 13, 284-290. ( 10.1016/S0959-4388(03)00065-5) [DOI] [PubMed] [Google Scholar]
- 8.Remme CA, et al. 2009. The cardiac sodium channel displays differential distribution in the conduction system and transmural heterogeneity in the murine ventricular myocardium. Basic Res. Cardiol. 104, 511-522. ( 10.1007/s00395-009-0012-8) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Agullo-Pascual E, et al. 2014. Super-resolution imaging reveals that loss of the C-terminus of connexin43 limits microtubule plus-end capture and NaV1.5 localization at the intercalated disc. Cardiovasc. Res. 104, 371-381. ( 10.1093/cvr/cvu195) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rivaud MR, Agullo-Pascual E, Lin X, Leo-Macias A, Zhang M, Rothenberg E, Bezzina CR, Delmar M, Remme CA. 2017. Sodium channel remodeling in subcellular microdomains of murine failing cardiomyocytes. J. Am. Heart Assoc. 6, e007622. ( 10.1161/JAHA.117.007622) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kucera JP, Rohr S, Rudy Y. 2002. Localization of sodium channels in intercalated disks modulates cardiac conduction. Circ. Res. 91, 1176-1182. ( 10.1161/01.RES.0000046237.54156.0A) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shy D, et al. 2014. PDZ domain-binding motif regulates cardiomyocyte compartment-specific Nav1.5 channel expression and function. Circulation 130, 147-160. ( 10.1161/CIRCULATIONAHA.113.007852) [DOI] [PubMed] [Google Scholar]
- 13.Petitprez S, et al. 2011. SAP97 and dystrophin macromolecular complexes determine two pools of cardiac sodium channels Nav1.5 in cardiomyocytes. Circ. Res. 108, 294-304. ( 10.1161/CIRCRESAHA.110.228312) [DOI] [PubMed] [Google Scholar]
- 14.Abriel H. 2010. Cardiac sodium channel Nav1.5 and interacting proteins: physiology and pathophysiology. J. Mol. Cell. Cardiol. 48, 2-11. ( 10.1016/j.yjmcc.2009.08.025) [DOI] [PubMed] [Google Scholar]
- 15.Shy D, Gillet L, Abriel H. 2013. Cardiac sodium channel NaV1.5 distribution in myocytes via interacting proteins: the multiple pool model. Biochim. Biophys. Acta Mol. Cell Res. 1833, 886-894. ( 10.1016/j.bbamcr.2012.10.026) [DOI] [PubMed] [Google Scholar]
- 16.Eichel CA, et al. 2016. Lateral membrane-specific MAGUK CASK down-regulates NaV1.5 channel in cardiac myocytes. Circ. Res. 119, 544-556. ( 10.1161/CIRCRESAHA.116.309254) [DOI] [PubMed] [Google Scholar]
- 17.Schroeter A, Walzik S, Blechschmidt S, Haufe V, Benndorf K, Zimmer T. 2010. Structure and function of splice variants of the cardiac voltage-gated sodium channel Nav1.5. J. Mol. Cell. Cardiol. 49, 16-24. ( 10.1016/j.yjmcc.2010.04.004) [DOI] [PubMed] [Google Scholar]
- 18.Onkal R, Mattis JH, Fraser SP, Diss JKJ, Shao D, Okuse K, Djamgoz MBA. 2008. Alternative splicing of Nav1.5: an electrophysiological comparison of ‘neonatal’ and ‘adult’ isoforms and critical involvement of a lysine residue. J. Cell. Physiol. 216, 716-726. ( 10.1002/jcp.21451) [DOI] [PubMed] [Google Scholar]
- 19.Murphy LL, Moon-Grady AJ, Cuneo BF, Wakai RT, Yu S, Kunic JD, Benson DW, George AL. 2012. Developmentally regulated SCN5A splice variant potentiates dysfunction of a novel mutation associated with severe fetal arrhythmia. Hear. Rhythm 9, 590-597. ( 10.1016/j.hrthm.2011.11.006) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Balse E, Steele DF, Abriel H, Coulombe A, Fedida D, Hatem S. 2012. Dynamic of ion channel expression at the plasma membrane of cardiomyocytes. Physiol. Rev. 92, 1317-1358. ( 10.1152/physrev.00041.2011) [DOI] [PubMed] [Google Scholar]
- 21.Rook MB, Evers MM, Vos MA, Bierhuizen MFA. 2012. Biology of cardiac sodium channel Nav1.5 expression. Cardiovasc. Res. 93, 12-23. ( 10.1093/cvr/cvr252) [DOI] [PubMed] [Google Scholar]
- 22.Herren AW, Bers DM, Grandi E. 2013. Post-translational modifications of the cardiac Na channel: contribution of CaMKII-dependent phosphorylation to acquired arrhythmias. Am. J. Physiol. Heart Circ. Physiol. 305, H431-H445. ( 10.1152/ajpheart.00306.2013) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Marionneau C, Abriel H. 2015. Regulation of the cardiac Na+ channel NaV1.5 by post-translational modifications. J. Mol. Cell. Cardiol. 82, 36-47. ( 10.1016/j.yjmcc.2015.02.013) [DOI] [PubMed] [Google Scholar]
- 24.Abriel H, Staub O. 2005. Ubiquitylation of ion channels. Physiology (Bethesda) 20, 398-407. [DOI] [PubMed] [Google Scholar]
- 25.Remme CA. 2013. Cardiac sodium channelopathy associated with SCN5A mutations: electrophysiological, molecular and genetic aspects. J. Physiol. 591, 4099-4116. ( 10.1113/jphysiol.2013.256461) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Remme CA, Wilde AAM, Bezzina CR. 2008. Cardiac sodium channel overlap syndromes: different faces of SCN5A mutations. Trends Cardiovasc. Med. 18, 78-87. ( 10.1016/j.tcm.2008.01.002) [DOI] [PubMed] [Google Scholar]
- 27.Shang LL, et al. 2007. Human heart failure is associated with abnormal C-terminal splicing variants in the cardiac sodium channel. Circ. Res. 101, 1146-1154. ( 10.1161/CIRCRESAHA.107.152918) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Viswanathan PC, Balser JR. 2004. Inherited sodium channelopathies a continuum of channel dysfunction. Trends Cardiovasc. Med. 14, 28-35. ( 10.1016/j.tcm.2003.10.001) [DOI] [PubMed] [Google Scholar]
- 29.Kapplinger JD, et al. 2010. An international compendium of mutations in the SCN5A-encoded cardiac sodium channel in patients referred for Brugada syndrome genetic testing. Hear. Rhythm 7, 33-46. ( 10.1016/j.hrthm.2009.09.069) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Remme CA, Wilde AAM. 2013. Late sodium current inhibition in acquired and inherited ventricular (dys)function and arrhythmias. Cardiovasc. Drugs Ther. 27, 91-101. ( 10.1007/s10557-012-6433-x) [DOI] [PubMed] [Google Scholar]
- 31.Portero V, et al. 2017. Anti-arrhythmic potential of the late sodium current inhibitor GS-458967 in murine Scn5a-1798insD+/- and human SCN5A-1795insD+/- iPSC-derived cardiomyocytes. Cardiovasc. Res. 113, 829-838. ( 10.1093/cvr/cvx077) [DOI] [PubMed] [Google Scholar]
- 32.Rivaud MR, Baartscheer A, Verkerk AO, Beekman L, Rajamani S, Belardinelli L, Bezzina CR, Remme CA. 2018. Enhanced late sodium current underlies pro-arrhythmic intracellular sodium and calcium dysregulation in murine sodium channelopathy. Int. J. Cardiol. 263, 54-62. ( 10.1016/j.ijcard.2018.03.044) [DOI] [PubMed] [Google Scholar]
- 33.Macleod KT. 2022. Changes in cellular Ca2+ and Na+ regulation during the progression towards heart failure. J. Physiol. 598, 1339-1359. ( 10.1113/JP283082) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Veerman CC, Wilde AAM, Lodder EM. 2015. The cardiac sodium channel gene SCN5A and its gene product NaV1.5: role in physiology and pathophysiology. Gene 573, 177-187. ( 10.1016/j.gene.2015.08.062) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schwartz PJ. 2006. The congenital long QT syndromes from genotype to phenotype: clinical implications. J. Intern. Med. 259, 39-47. ( 10.1111/j.1365-2796.2005.01583.x) [DOI] [PubMed] [Google Scholar]
- 36.Marsman EMJ, Postema PG, Remme CA. 2022. Brugada syndrome: update and future perspectives. Heart 108, 668-675. ( 10.1136/heartjnl-2020-318258) [DOI] [PubMed] [Google Scholar]
- 37.Lambiase PD, Providência R. 2020. Epicardial ablation in Brugada syndrome. Card. Electrophysiol. Clin. 12, 345-356. ( 10.1016/j.ccep.2020.04.006) [DOI] [PubMed] [Google Scholar]
- 38.Cerrone M, Remme CA, Tadros R, Bezzina CR, Delmar M. 2019. Beyond the one gene–one disease paradigm: complex genetics and pleiotropy in inheritable cardiac disorders. Circulation 140, 595-610. ( 10.1161/CIRCULATIONAHA.118.035954) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Behr ER, Ben-Haim Y, Ackerman MJ, Krahn AD, Wilde AAM. 2021. Brugada syndrome and reduced right ventricular outflow tract conduction reserve: a final common pathway? Eur. Heart J. 44, 1-12. [DOI] [PubMed] [Google Scholar]
- 40.Benson DW, Wang DW, Dyment M, Knilans TK, Fish FA, Strieper MJ, Rhodes TH, George AL. 2003. Congenital sick sinus syndrome caused by recessive mutations in the cardiac sodium channel gene (SCN5A). J. Clin. Invest. 112, 1019-1028. ( 10.1172/JCI200318062) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wolf CM, Berul CI. 2006. Inherited conduction system abnormalities—one group of diseases, many genes. J. Cardiovasc. Electrophysiol. 17, 446-455. ( 10.1111/j.1540-8167.2006.00427.x) [DOI] [PubMed] [Google Scholar]
- 42.Darbar D, Kannankeril PJ, Donahue BS, Kucera G, Stubblefield T, Haines JL, George AL, Roden DM. 2008. Cardiac sodium channel (SCN5A) variants associated with atrial fibrillation. Circulation 117, 1927-1935. ( 10.1161/CIRCULATIONAHA.107.757955) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Makiyama T, et al. 2008. A novel SCN5A gain-of-function mutation M1875T associated with familial atrial fibrillation. J. Am. Coll. Cardiol. 52, 1326-1334. ( 10.1016/j.jacc.2008.07.013) [DOI] [PubMed] [Google Scholar]
- 44.Rivaud MR, et al. 2018. A common co-morbidity modulates disease expression and treatment efficacy in inherited cardiac sodium channelopathy. Eur. Heart J. 39, 2898-2907. ( 10.1093/eurheartj/ehy247) [DOI] [PubMed] [Google Scholar]
- 45.Remme CA, et al. 2006. Overlap syndrome of cardiac sodium channel disease in mice carrying the equivalent mutation of human SCN5A-1795insD. Circulation 114, 2584-2594. ( 10.1161/CIRCULATIONAHA.106.653949) [DOI] [PubMed] [Google Scholar]
- 46.Remme CA. 2018. Cardiac sodium channel (dys)function and inherited arrhythmia syndromes. Cardiac Vasc. Biol. 6, 9-45. ( 10.1007/978-3-319-77812-9_2) [DOI] [Google Scholar]
- 47.Remme CA, Wilde AAM. 2008. SCN5A overlap syndromes: no end to disease complexity? Europace 10, 1253-1255. ( 10.1093/europace/eun267) [DOI] [PubMed] [Google Scholar]
- 48.Wu G, et al. 2008. Alpha-1-syntrophin mutation and the long-QT syndrome: a disease of sodium channel disruption. Circ. Arrhythm. Electrophysiol. 1, 193-201. ( 10.1161/CIRCEP.108.769224) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vatta M, et al. 2006. Mutant caveolin-3 induces persistent late sodium current and is associated with long-QT syndrome. Circulation 114, 2104-2112. ( 10.1161/CIRCULATIONAHA.106.635268) [DOI] [PubMed] [Google Scholar]
- 50.Mohler PJ, et al. 2003. Ankyrin-B mutation causes type 4 long-QT cardiac arrhythmia and sudden cardiac death. Nature 421, 634-639. ( 10.1038/nature01335) [DOI] [PubMed] [Google Scholar]
- 51.Schott JJ, et al. 1995. Mapping of a gene for long QT syndrome to chromosome 4q25-27. Am. J. Hum. Genet. 57, 1114-1122. [PMC free article] [PubMed] [Google Scholar]
- 52.Marchal GA, Van Putten M, Verkerk AO, Casini S, Putker K, Van Amersfoorth SCM, Aartsma-Rus A, Lodder EM, Remme CA. 2021. Low human dystrophin levels prevent cardiac electrophysiological and structural remodelling in a Duchenne mouse model. Sci. Rep. 11, 1-11. ( 10.1038/s41598-021-89208-1) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Van Der Voorn SM, Te Riele ASJM, Basso C, Calkins H, Remme CA, Van Veen TAB. 2020. Arrhythmogenic cardiomyopathy: pathogenesis, pro-arrhythmic remodeling, and novel approaches for risk stratification and therapy. Cardiovasc. Res. 116, 1571-1584. ( 10.1093/cvr/cvaa084) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rizzo S, et al. 2012. Intercalated disc abnormalities, reduced Na+ current density, and conduction slowing in desmoglein-2 mutant mice prior to cardiomyopathic changes. Cardiovasc. Res. 95, 409-418. ( 10.1093/cvr/cvs219) [DOI] [PubMed] [Google Scholar]
- 55.Cerrone M, et al. 2012. Sodium current deficit and arrhythmogenesis in a murine model of plakophilin-2 haploinsufficiency. Cardiovasc. Res. 95, 460-468. ( 10.1093/cvr/cvs218) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Remme CA. 2022. Sudden cardiac death in diabetes and obesity: mechanisms and therapeutic strategies. Can. J. Cardiol. 38, 418-426. ( 10.1016/j.cjca.2022.01.001) [DOI] [PubMed] [Google Scholar]
- 57.Yang KC, Kyle JW, Makielski JC, Dudley SC. 2015. Mechanisms of sudden cardiac death: oxidants and metabolism. Circ. Res. 116, 1937-1955. ( 10.1161/CIRCRESAHA.116.304691) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hegyi B, Ko CY, Bossuyt J, Bers DM. 2021. Two-hit mechanism of cardiac arrhythmias in diabetic hyperglycaemia: reduced repolarization reserve, neurohormonal stimulation, and heart failure exacerbate susceptibility. Cardiovasc. Res. 117, 2781-2793. ( 10.1093/cvr/cvab006) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Banerjee D, Grammatopoulos TN, Palmisciano A, Klinger JR, Krishnan I, Whittenhall M, Zhou A, Dudley S, Ventetuolo CE. 2020. Alternative splicing of the cardiac sodium channel in pulmonary arterial hypertension. Chest 158, 735-738. ( 10.1016/j.chest.2019.12.052) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Frustaci A, Priori SG, Pieroni M, Chimenti C, Napolitano C, Rivolta I, Sanna T, Bellocci F, Russo MA. 2005. Cardiac histological substrate in patients with clinical phenotype of Brugada syndrome. Circulation 112, 3680-3687. ( 10.1161/CIRCULATIONAHA.105.520999) [DOI] [PubMed] [Google Scholar]
- 61.Coronel R, et al. 2005. Right ventricular fibrosis and conduction delay in a patient with clinical signs of Brugada syndrome: a combined electrophysiological, genetic, histopathologic, and computational study. Circulation 112, 2769-2777. ( 10.1161/CIRCULATIONAHA.105.532614) [DOI] [PubMed] [Google Scholar]
- 62.Mcnair WP, et al. 2011. SCN5A mutations associate with arrhythmic dilated cardiomyopathy and commonly localize to the voltage-sensing mechanism. J. Am. Coll. Cardiol. 57, 2160-2168. ( 10.1016/j.jacc.2010.09.084) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wilde AAM, Amin AS. 2018. Clinical spectrum of SCN5A mutations: long QT syndrome, Brugada syndrome, and cardiomyopathy. JACC Clin. Electrophysiol. 4, 569-579. ( 10.1016/j.jacep.2018.03.006) [DOI] [PubMed] [Google Scholar]
- 64.Oliva A, et al. 2022. Structural heart alterations in Brugada syndrome: is it really a channelopathy? A systematic review. J. Clin. Med. 11, 4406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bezzina CR, Remme CA. 2008. Dilated cardiomyopathy due to sodium channel dysfunction: what is the connection? Circ. Arrhythm. Electrophysiol. 1, 80-82. ( 10.1161/CIRCEP.108.791434) [DOI] [PubMed] [Google Scholar]
- 66.Asatryan B. 2019. Cardiac sodium channel dysfunction and dilated cardiomyopathy: a contemporary reappraisal of pathophysiological concepts. J. Clin. Med. 8, 1029. ( 10.3390/jcm8071029) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Doisne N, et al. 2020. A novel gain-of-function mutation in SCN5A responsible for multifocal ectopic Purkinje-related premature contractions. Hum. Mutat. 41, 850-859. ( 10.1002/humu.23981) [DOI] [PubMed] [Google Scholar]
- 68.Laurent G, et al. 2012. Multifocal ectopic Purkinje-related premature contractions: a new SCN5A-related cardiac channelopathy. J. Am. Coll. Cardiol. 60, 144-156. ( 10.1016/j.jacc.2012.02.052) [DOI] [PubMed] [Google Scholar]
- 69.Jordan E, et al. 2021. Evidence-based assessment of genes in dilated cardiomyopathy. Circulation 144, 7-19. ( 10.1161/CIRCULATIONAHA.120.053033) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Peters S, Thompson BA, Perrin M, James P, Zentner D, Kalman JM, Vandenberg JI, Fatkin D. 2022. Arrhythmic phenotypes are a defining feature of dilated cardiomyopathy-associated SCN5A variants: a systematic review. Circ. Genomic Precis. Med. 15, E003432. [DOI] [PubMed] [Google Scholar]
- 71.Ben-Haim Y, Asimaki A, Behr ER. 2021. Brugada syndrome and arrhythmogenic cardiomyopathy: overlapping disorders of the connexome? Europace 23, 653-664. ( 10.1093/europace/euaa277) [DOI] [PubMed] [Google Scholar]
- 72.Royer A, et al. 2005. Mouse model of SCN5A-linked hereditary Lenègre's disease: age-related conduction slowing and myocardial fibrosis. Circulation 111, 1738-1746. ( 10.1161/01.CIR.0000160853.19867.61) [DOI] [PubMed] [Google Scholar]
- 73.Verkerk AO, Amin AS, Remme CA. 2018. Disease modifiers of inherited SCN5A channelopathy. Front. Cardiovasc. Med. 5, 137. ( 10.3389/fcvm.2018.00137) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Sasse P, Zhang J, Cleemann L, Morad M, Hescheler J, Fleischmann BK. 2007. Intracellular Ca2+ oscillations, a potential pacemaking mechanism in early embryonic heart cells. J. Gen. Physiol. 130, 133-144. ( 10.1085/jgp.200609575) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Chopra SS, Stroud DM, Watanabe H, Bennett JS, Burns CG, Wells KS, Yang T, Zhong TP, Roden DM. 2010. Voltage-gated sodium channels are required for heart development in zebrafish. Circ. Res. 106, 1342-1350. ( 10.1161/CIRCRESAHA.109.213132) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Nuyens D, et al. 2001. Abrupt rate accelerations or premature beats cause life-threatening arrhythmias in mice with long-QT3 syndrome. Nat. Med. 7, 1021-1027. ( 10.1038/nm0901-1021) [DOI] [PubMed] [Google Scholar]
- 77.Papadatos GA, et al. 2002. Slowed conduction and ventricular tachycardia after targeted disruption of the cardiac sodium channel gene Scn5a. Proc. Natl Acad. Sci. USA 99, 6210-6215. ( 10.1073/pnas.082121299) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Marchal GA, Verkerk AO, Mohan RA, Wolswinkel R, Boukens BJD, Remme CA. 2020. The sodium channel NaV1.5 impacts on early murine embryonic cardiac development, structure and function in a non-electrogenic manner. Acta Physiol. 230, e13493. ( 10.1111/apha.13493) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Molkentin JD, Lu J-R, Antos CL, Markham B, Richardson J, Robbins J, Grant SR, Olson EN. 1998. A calcineurin-dependent transcriptional pathway for cardiac hypertrophy. Cell 93, 215-228. ( 10.1016/S0092-8674(00)81573-1) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lindegger N, Hagen BM, Marks AR, Lederer WJ, Kass RS. 2009. Diastolic transient inward current in long QT syndrome type 3 is caused by Ca2+ overload and inhibited by ranolazine. J. Mol. Cell. Cardiol. 47, 326-334. ( 10.1016/j.yjmcc.2009.04.003) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Yao L, et al. 2011. Nav1.5-dependent persistent Na+ influx activates CaMKII in rat ventricular myocytes and N1325S mice. Am. J. Physiol. Cell Physiol. 301, C577-C586. ( 10.1152/ajpcell.00125.2011) [DOI] [PubMed] [Google Scholar]
- 82.Gosselin-Badaroudine P, Keller DI, Huang H, Pouliot V, Chatelier A, Osswald S, Brink M, Chahine M. 2012. A proton leak current through the cardiac sodium channel is linked to mixed arrhythmia and the dilated cardiomyopathy phenotype. PLoS ONE 7, e38331. ( 10.1371/journal.pone.0038331) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Moreau A, Chahine M. 2018. A new cardiac channelopathy: from cinical phenotypes to molecular mechanisms associated with Nav1.5 gating pores. Front. Cardiovasc. Med. 5, 139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Leask A. 2007. TGFβ, cardiac fibroblasts, and the fibrotic response. Cardiovasc. Res. 74, 207-212. ( 10.1016/j.cardiores.2006.07.012) [DOI] [PubMed] [Google Scholar]
- 85.Ugarte G, Brandan E. 2006. Transforming growth factor beta (TGF-beta) signaling is regulated by electrical activity in skeletal muscle cells. TGF-beta type I receptor is transcriptionally regulated by myotube excitability. J. Biol. Chem. 281, 18 473-18 481. ( 10.1074/jbc.M600918200) [DOI] [PubMed] [Google Scholar]
- 86.Hao X, et al. 2011. TGF-β1-mediated fibrosis and ion channel remodeling are key mechanisms in producing the sinus node dysfunction associated with SCN5A deficiency and aging. Circ. Arrhythmia Electrophysiol. 4, 397-406. ( 10.1161/CIRCEP.110.960807) [DOI] [PubMed] [Google Scholar]
- 87.Leo-Macias A, et al. 2016. Nanoscale visualization of functional adhesion/excitability nodes at the intercalated disc. Nat. Commun. 7, 10342. ( 10.1038/ncomms10342) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Patel F, Brackenbury WJ. 2015. Dual roles of voltage-gated sodium channels in development and cancer. Int. J. Dev. Biol. 366, 357-366. ( 10.1387/ijdb.150171wb) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Hernandez-Plata E, Ortiz CS, Marquina-Castillo B, Medina-Martinez I, Alfaro A, Berumen J, Rivera M, Gomora JC. 2012. Overexpression of NaV1.6 channels is associated with the invasion capacity of human cervical cancer. Int. J. Cancer 130, 2013-2023. ( 10.1002/ijc.26210) [DOI] [PubMed] [Google Scholar]
- 90.Brackenbury WJ. 2012. Voltage-gated sodium channels and metastatic disease. Channels (Austin) 6, 352-361. ( 10.4161/chan.21910) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Luiz AP, Wood JN. 2016. Sodium channels in pain and cancer: new therapeutic opportunities. Adv. Pharmacol. (San Diego, Calif.) 75, 153-178. [DOI] [PubMed] [Google Scholar]
- 92.Gillet L, Roger S, Besson P, Lecaille F, Gore J, Bougnoux P, Lalmanach G, Le Guennec J-Y. 2009. Voltage-gated sodium channel activity promotes cysteine cathepsin-dependent invasiveness and colony growth of human cancer cells. J. Biol. Chem. 284, 8680-8691. ( 10.1074/jbc.M806891200) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Brackenbury WJ, Chioni AM, Diss JKJ, Djamgoz MBA. 2007. The neonatal splice variant of Nav1.5 potentiates in vitro invasive behaviour of MDA-MB-231 human breast cancer cells. Breast Cancer Res. Treat. 101, 149-160. ( 10.1007/s10549-006-9281-1) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Nelson M, Yang M, Millican-Slater R, Brackenbury WJ. 2015. Nav1.5 regulates breast tumor growth and metastatic dissemination in vivo. Oncotarget 6, 32 914-32 929. ( 10.18632/oncotarget.5441) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Besson P, Driffort V, Bon É, Gradek F, Chevalier S, Roger S. 2015. How do voltage-gated sodium channels enhance migration and invasiveness in cancer cells? Biochim. Biophys. Acta Biomembr. 1848, 2493-2501. ( 10.1016/j.bbamem.2015.04.013) [DOI] [PubMed] [Google Scholar]
- 96.Black JA, Waxman SG. 2013. Noncanonical roles of voltage-gated sodium channels. Neuron 80, 280-291. ( 10.1016/j.neuron.2013.09.012) [DOI] [PubMed] [Google Scholar]
- 97.Roger S, Gillet L, Le Guennec JY, Besson P. 2015. Voltage-gated sodium channels and cancer: is excitability their primary role? Front. Pharmacol. 6, 1-22. ( 10.3389/fphar.2015.00152) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Brisson L, et al. 2013. NaV1.5 Na+ channels allosterically regulate the NHE-1 exchanger and promote the activity of breast cancer cell invadopodia. J. Cell Sci. 126, 4835-4842. [DOI] [PubMed] [Google Scholar]
- 99.Carrithers MD, Chatterjee G, Carrithers LM, Offoha R, Iheagwara U, Rahner C, Graham M, Waxman SG. 2009. Regulation of podosome formation in macrophages by a splice variant of the sodium channel SCN8A. J. Biol. Chem. 284, 8114-8126. ( 10.1074/jbc.M801892200) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Leslie TK, Brackenbury WJ. 2022. Sodium channels and the ionic microenvironment of breast tumours. J. Physiol. ( 10.1113/JP282306) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.James AD, et al. 2022. Sodium accumulation in breast cancer predicts malignancy and treatment response. Br. J. Cancer 127, 337-349. ( 10.1038/s41416-022-01802-w) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Nelson M, Yang M, Dowle AA, Thomas JR, Brackenbury WJ. 2015. The sodium channel-blocking antiepileptic drug phenytoin inhibits breast tumour growth and metastasis. Mol. Cancer 14, 1-7. ( 10.1186/s12943-014-0277-x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Driffort V, et al. 2014. Ranolazine inhibits NaV1.5-mediated breast cancer cell invasiveness and lung colonization. Mol. Cancer 13, 1-16. ( 10.1186/1476-4598-13-264) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Fairhurst C, Watt I, Martin F, Bland M, Brackenbury WJ. 2015. Sodium channel-inhibiting drugs and survival of breast, colon and prostate cancer: a population-based study. Sci. Rep. 5, 1-9. ( 10.1038/srep16758) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Dutta S, Charcas OL, Tanner S, Gradek F, Driffort V, Roger S, Selander K, Velu SE, Brouillette W. 2018. Discovery and evaluation of nNaV1.5 sodium channel blockers with potent cell invasion inhibitory activity in breast cancer cells. Bioorg. Med. Chem. 26, 2428-2436. ( 10.1016/j.bmc.2018.04.003) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Bean BP. 2007. The action potential in mammalian central neurons. Nat. Rev. Neurosci. 8, 451-465. ( 10.1038/nrn2148) [DOI] [PubMed] [Google Scholar]
- 107.Wu L, Nishiyama K, Hollyfield JG, Wang Q. 2002. Localization of Nav1.5 sodium channel protein in the mouse brain. Neuroreport 13, 2547-2551. ( 10.1097/00001756-200212200-00033) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Wang J, Ou SW, Zhang ZY, Qiu B, Wang YJ. 2018. Molecular expression of multiple Nav1.5 splice variants in the frontal lobe of the human brain. Int. J. Mol. Med. 41, 915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Hartmann HA, Colom LV, Sutherland ML, Noebels JL. 1999. Selective localization of cardiac SCN5A sodium channels in limbic regions of rat brain. Nat. Neurosci. 2, 593-595. ( 10.1038/10147) [DOI] [PubMed] [Google Scholar]
- 110.Kerr NCH, Gao Z, Holmes FE, Hobson S-A, Hancox JC, Wynick D, James AF. 2007. The sodium channel Nav1.5a is the predominant isoform expressed in adult mouse dorsal root ganglia and exhibits distinct inactivation properties from the full-length Nav1.5 channel. Mol. Cell. Neurosci. 35, 283-291. ( 10.1016/j.mcn.2007.03.002) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Liu X-P, Wooltorton JRA, Gaboyard-Niay S, Yang F-C, Lysakowski A, Eatock RA. 2016. Sodium channel diversity in the vestibular ganglion: NaV1.5, NaV1.8, and tetrodotoxin-sensitive currents. J. Neurophysiol. 115, 2536-2555. ( 10.1152/jn.00902.2015) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Frenz CT, Hansen A, Dupuis ND, Shultz N, Levinson SR, Finger TE, Dionne VE. 2014. NaV1.5 sodium channel window currents contribute to spontaneous firing in olfactory sensory neurons. J. Neurophysiol. 112, 1091-1104. ( 10.1152/jn.00154.2014) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Scornik FS, Desai M, Brugada R, Guerchicoff A, Pollevick GD, Antzelevitch C, Pérez GJ. 2006. Functional expression of ‘cardiac-type’ Nav1.5 sodium channel in canine intracardiac ganglia. Hear. Rhythm 3, 842-850. ( 10.1016/j.hrthm.2006.03.021) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Johnson JN, Hofman N, Haglund CM, Cascino GD, Wilde AAM, Ackerman MJ. 2009. Identification of a possible pathogenic link between congenital long QT syndrome and epilepsy. Neurology 72, 224-231. ( 10.1212/01.wnl.0000335760.02995.ca) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Anderson JH, Bos JM, Cascino GD, Ackerman MJ. 2014. Prevalence and spectrum of electroencephalogram-identified epileptiform activity among patients with long QT syndrome. Hear. Rhythm 11, 53-57. ( 10.1016/j.hrthm.2013.10.010) [DOI] [PubMed] [Google Scholar]
- 116.Zhou Y, Luo Z, Bi F, Li J, Xiao B. 2019. Expression of Nav1.5 in the pathogenesis of temporal lobe epilepsy. Cell. Mol. Biol. 65, 58-62. ( 10.14715/cmb/2019.65.2.9) [DOI] [PubMed] [Google Scholar]
- 117.Foley J, Burnham V, Tedoldi M, Danial NN, Yellen G. 2018. BAD knockout provides metabolic seizure resistance in a genetic model of epilepsy with sudden unexplained death in epilepsy. Epilepsia 59, e1-e4. ( 10.1111/epi.13960) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Abdelghani MS, Chapra A, Asaad N, Hayat SA. 2020. Epilepsy and Brugada syndrome: association or uncommon presentation? Heart Views 21, 114. ( 10.4103/HEARTVIEWS.HEARTVIEWS_34_20) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Parisi P, et al. 2013. Coexistence of epilepsy and Brugada syndrome in a family with SCN5A mutation. Epilepsy Res. 105, 415-418. ( 10.1016/j.eplepsyres.2013.02.024) [DOI] [PubMed] [Google Scholar]
- 120.Heron SE, Hernandez M, Edwards C, Edkins E, Jansen FE, Scheffer IE, Berkovic SF, Mulley JC. 2010. Neonatal seizures and long QT syndrome: a cardiocerebral channelopathy? Epilepsia 51, 293-296. ( 10.1111/j.1528-1167.2009.02317.x) [DOI] [PubMed] [Google Scholar]
- 121.Miyazaki A, et al. 2016. Comorbid epilepsy and developmental disorders in congenital long QT syndrome with life-threatening perinatal arrhythmias. JACC. Clin. Electrophysiol. 2, 266-276. ( 10.1016/j.jacep.2015.10.010) [DOI] [PubMed] [Google Scholar]
- 122.Valdivia CR, Ackerman MJ, Tester DJ, Wada T, McCormack J, Ye B, Makielski JC. 2002. A novel SCN5A arrhythmia mutation, M1766L, with expression defect rescued by mexiletine. Cardiovasc. Res. 55, 279-289. ( 10.1016/S0008-6363(02)00445-5) [DOI] [PubMed] [Google Scholar]
- 123.Kløvgaard M, Sabers A, Ryvlin P. 2022. Update on sudden unexpected death in epilepsy. Neurol. Clin. 40, 741-754. ( 10.1016/j.ncl.2022.06.001) [DOI] [PubMed] [Google Scholar]
- 124.O'Neal TB, Shrestha S, Singh H, Osagie I, Ben-Okafor K, Cornett EM, Kaye AD. 2022. Sudden unexpected death in epilepsy. Neurol. Int. 14, 600-613. ( 10.3390/neurolint14030048) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Friedman D. 2022. Sudden unexpected death in epilepsy. Curr. Opin. Neurol. 35, 181-188. ( 10.1097/WCO.0000000000001034) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Goldman AM. 2015. Mechanisms of sudden unexplained death in epilepsy. Curr. Opin. Neurol. 28, 166-174. ( 10.1097/WCO.0000000000000184) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Bagnall RD, Crompton DE, Semsarian C. 2017. Genetic basis of sudden unexpected death in epilepsy. Front. Neurol. 8, 348. ( 10.3389/fneur.2017.00348) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Goldman AM, Behr ER, Semsarian C, Bagnall RD, Sisodiya S, Cooper PN. 2016. Sudden unexpected death in epilepsy genetics: molecular diagnostics and prevention. Epilepsia 57(Suppl. 1), 17-25. ( 10.1111/epi.13232) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Tu E, Bagnall RD, Duflou J, Semsarian C. 2011. Post-mortem review and genetic analysis of sudden unexpected death in epilepsy (SUDEP) cases. Brain Pathol. 21, 201-208. ( 10.1111/j.1750-3639.2010.00438.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Bagnall RD, et al. 2016. Exome-based analysis of cardiac arrhythmia, respiratory control, and epilepsy genes in sudden unexpected death in epilepsy. Ann. Neurol. 79, 522-534. ( 10.1002/ana.24596) [DOI] [PubMed] [Google Scholar]
- 131.Le Scouarnec S, et al. 2015. Testing the burden of rare variation in arrhythmia-susceptibility genes provides new insights into molecular diagnosis for Brugada syndrome. Hum. Mol. Genet. 24, 2757-2763. ( 10.1093/hmg/ddv036) [DOI] [PubMed] [Google Scholar]
- 132.Aurlien D, Leren TP, Taubøll E, Gjerstad L. 2009. New SCN5A mutation in a SUDEP victim with idiopathic epilepsy. Seizure 18, 158-160. ( 10.1016/j.seizure.2008.07.008) [DOI] [PubMed] [Google Scholar]
- 133.Berge KE, et al. 2008. Molecular genetic analysis of long QT syndrome in Norway indicating a high prevalence of heterozygous mutation carriers. Scand. J. Clin. Lab. Invest. 68, 362-368. ( 10.1080/00365510701765643) [DOI] [PubMed] [Google Scholar]
- 134.Soh MS, Bagnall RD, Semsarian C, Scheffer IE, Berkovic SF, Reid CA. 2022. Rare sudden unexpected death in epilepsy SCN5A variants cause changes in channel function implicating cardiac arrhythmia as a cause of death. Epilepsia 63, e57-e62. [DOI] [PubMed] [Google Scholar]
- 135.Bardai A, Blom MT, Van Noord C, Verhamme KM, Sturkenboom MCJM, Tan HL. 2015. Sudden cardiac death is associated both with epilepsy and with use of antiepileptic medications. Heart 101, 14-22. ( 10.1136/heartjnl-2014-305664) [DOI] [PubMed] [Google Scholar]
- 136.Jia L, Eroglu TE, Wilders R, Verkerk AO, Tan HL. 2022. Carbamazepine increases the risk of sudden cardiac arrest by a reduction of the cardiac sodium current. Front. cell Dev. Biol. 10, 891996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Wang J, Ou SW, Bai YF, Wang YJ, Xu ZQD, Luan GM. 2018. Downregulation of adult and neonatal Nav1.5 in the dorsal root ganglia and axon of peripheral sensory neurons of rats with spared nerve injury. Int. J. Mol. Med. 41, 2225-2232. [DOI] [PubMed] [Google Scholar]
- 138.Roberts E. 2006. GABAergic malfunction in the limbic system resulting from an aboriginal genetic defect in voltage-gated Na+-channel SCN5A is proposed to give rise to susceptibility to schizophrenia. Adv. Pharmacol. 54, 119-145. ( 10.1016/S1054-3589(06)54006-2) [DOI] [PubMed] [Google Scholar]
- 139.Blom MT, Cohen D, Seldenrijk A, Penninx BWJH, Nijpels G, Stehouwer CDA, Dekker JM, Tan HL. 2014. Brugada syndrome ECG is highly prevalent in schizophrenia. Circ. Arrhythm. Electrophysiol. 7, 384-391. ( 10.1161/CIRCEP.113.000927) [DOI] [PubMed] [Google Scholar]
- 140.Kim DH, Lee SJ, Hahn SJ, Choi JS. 2016. Trifluoperazine blocks the human cardiac sodium channel, Nav1.5, independent of calmodulin. Biochem. Biophys. Res. Commun. 479, 584-589. ( 10.1016/j.bbrc.2016.09.115) [DOI] [PubMed] [Google Scholar]
- 141.Askland K, Read C, O'connell C, Moore JH. 2012. Ion channels and schizophrenia: a gene set-based analytic approach to GWAS data for biological hypothesis testing. Hum. Genet. 131, 373-391. ( 10.1007/s00439-011-1082-x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Black JA, Newcombe J, Waxman SG. 2010. Astrocytes within multiple sclerosis lesions upregulate sodium channel Nav1.5. Brain 133, 835-846. ( 10.1093/brain/awq003) [DOI] [PubMed] [Google Scholar]
- 143.Pappalardo LW, Samad OA, Black JA, Waxman SG. 2014. Voltage-gated sodium channel NaV1.5 contributes to astrogliosis in an in vitro model of glial injury via reverse Na+/Ca2+ exchange. Glia 62, 1162-1175. ( 10.1002/glia.22671) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Pappalardo LW, Samad OA, Liu S, Zwinger PJ, Black JA, Waxman SG. 2018. Nav1.5 in astrocytes plays a sex-specific role in clinical outcomes in a mouse model of multiple sclerosis. Glia 66, 2174-2187. ( 10.1002/glia.23470) [DOI] [PubMed] [Google Scholar]
- 145.Black JA, Newcombe J, Waxman SG. 2013. Nav1.5 sodium channels in macrophages in multiple sclerosis lesions. Mult. Scler. 19, 532-542. ( 10.1177/1352458512460417) [DOI] [PubMed] [Google Scholar]
- 146.Farrugia G. 1999. Ionic conductances in gastrointestinal smooth muscles and interstitial cells of Cajal. Annu. Rev. Physiol. 61, 45-84. ( 10.1146/annurev.physiol.61.1.45) [DOI] [PubMed] [Google Scholar]
- 147.Ou Y, et al. 2002. SCN5A is expressed in human jejunal circular smooth muscle cells. Neurogastroenterol. Motil. 14, 477-486. ( 10.1046/j.1365-2982.2002.00348.x) [DOI] [PubMed] [Google Scholar]
- 148.Neshatian L, et al. 2015. Ranolazine inhibits voltage-gated mechanosensitive sodium channels in human colon circular smooth muscle cells. Am. J. Physiol. Gastrointest. Liver Physiol. 309, G506-G512. ( 10.1152/ajpgi.00051.2015) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Beyder A, et al. 2016. Expression and function of the Scn5a-encoded voltage-gated sodium channel NaV1.5 in the rat jejunum. Neurogastroenterol. Motil. 28, 64-73. ( 10.1111/nmo.12697) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Joshi V, Strege PR, Farrugia G, Beyder A. 2021. Mechanotransduction in gastrointestinal smooth muscle cells: role of mechanosensitive ion channels. Am. J. Physiol. Gastrointest. Liver Physiol. 320, G897-G906. ( 10.1152/ajpgi.00481.2020) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Strege PR, Ou Y, Sha L, Rich A, Gibbons SJ, Szurszewski JH, Sarr MG, Farrugia G. 2003. Sodium current in human intestinal interstitial cells of Cajal. Am. J. Physiol. Gastrointest. Liver Physiol. 285, G1111-G1121. ( 10.1152/ajpgi.00152.2003) [DOI] [PubMed] [Google Scholar]
- 152.Locke GR, Ackerman MJ, Zinsmeister AR, Thapa P, Farrugia G. 2006. Gastrointestinal symptoms in families of patients with an SCN5A-encoded cardiac channelopathy: evidence of an intestinal channelopathy. Am. J. Gastroenterol. 101, 1299-1304. ( 10.1111/j.1572-0241.2006.00507.x) [DOI] [PubMed] [Google Scholar]
- 153.Saito YA, et al. 2009. Sodium channel mutation in irritable bowel syndrome: evidence for an ion channelopathy. Am. J. Physiol. Gastrointest. Liver Physiol. 296, G211-G218. ( 10.1152/ajpgi.90571.2008) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Beyder A, et al. 2014. Loss-of-function of the voltage-gated sodium channel NaV1.5 (channelopathies) in patients with irritable bowel syndrome. Gastroenterology 146, 1659-1668. ( 10.1053/j.gastro.2014.02.054) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Strege PR, et al. 2018. Irritable bowel syndrome patients have SCN5A channelopathies that lead to decreased NaV1.5 current and mechanosensitivity. Am. J. Physiol. Gastrointest. Liver Physiol. 314, G494-G503. ( 10.1152/ajpgi.00016.2017) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Strege PR, Mercado-Perez A, Mazzone A, Saito YA, Bernard CE, Farrugia G, Beyder A. 2019. SCN5A mutation G615E results in NaV1.5 voltage-gated sodium channels with normal voltage-dependent function yet loss of mechanosensitivity. Channels (Austin) 13, 287-298. ( 10.1080/19336950.2019.1632670) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Carrithers MD, Dib-Hajj S, Carrithers LM, Tokmoulina G, Pypaert M, Jonas EA, Waxman SG. 2007. Expression of the voltage-gated sodium channel NaV1.5 in the macrophage late endosome regulates endosomal acidification. J. Immunol. 178, 7822-7832. ( 10.4049/jimmunol.178.12.7822) [DOI] [PubMed] [Google Scholar]
- 158.Carrithers LM, Hulseberg P, Sandor M, Carrithers MD. 2011. The human macrophage sodium channel NaV1.5 regulates mycobacteria processing through organelle polarization and localized calcium oscillations. FEMS Immunol. Med. Microbiol. 63, 319-327. ( 10.1111/j.1574-695X.2011.00853.x) [DOI] [PubMed] [Google Scholar]
- 159.Jones A, Kainz D, Khan F, Lee C, Carrithers MD. 2014. Human macrophage SCN5A activates an innate immune signaling pathway for antiviral host defense. J. Biol. Chem. 289, 35 326-35 340. ( 10.1074/jbc.M114.611962) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.White CR, Dungan M, Carrithers MD. 2019. Activation of human macrophage sodium channels regulates RNA processing to increase expression of the DNA repair protein PPP1R10. Immunobiology 224, 80-93. ( 10.1016/j.imbio.2018.10.005) [DOI] [PubMed] [Google Scholar]
- 161.Rahgozar K, Wright E, Carrithers LM, Carrithers MD. 2013. Mediation of protection and recovery from experimental autoimmune encephalomyelitis by macrophages expressing the human voltage-gated sodium channel NaV1.5. J. Neuropathol. Exp. Neurol. 72, 489-504. ( 10.1097/NEN.0b013e318293eb08) [DOI] [PubMed] [Google Scholar]
- 162.Lo W-L, Donermeyer DL, Allen PM. 2012. A voltage-gated sodium channel is essential for the positive selection of CD4(+) T cells. Nat. Immunol. 13, 880-887. ( 10.1038/ni.2379) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Milam AAV, et al. 2018. Tuning T cell signaling sensitivity alters the behavior of CD4+ T cells during an immune response. J. Immunol. 200, 3429-3437. ( 10.4049/jimmunol.1701422) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Xia R, Tomsits P, Loy S, Zhang Z, Pauly V, Schüttler D, Clauss S. 2022. Cardiac macrophages and their effects on arrhythmogenesis. Front. Physiol. 13, 1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Pieroni M, et al. 2018. Electroanatomic and pathologic right ventricular outflow tract abnormalities in patients with Brugada syndrome. J. Am. Coll. Cardiol. 72, 2747-2757. ( 10.1016/j.jacc.2018.09.037) [DOI] [PubMed] [Google Scholar]
- 166.Chatterjee D, et al. 2020. An autoantibody profile detects Brugada syndrome and identifies abnormally expressed myocardial proteins. Eur. Heart J. 41, 2878-2890A. ( 10.1093/eurheartj/ehaa383) [DOI] [PubMed] [Google Scholar]
- 167.Poller W, et al. 2022. Missense variant E1295K of sodium channel SCN5A associated with recurrent ventricular fibrillation and myocardial inflammation. JACC. Case Rep. 4, 280-286. ( 10.1016/j.jaccas.2022.01.016) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Bradley E, Webb TI, Hollywood MA, Sergeant GP, Mchale NG, Thornbury KD. 2013. The cardiac sodium current Nav1.5 is functionally expressed in rabbit bronchial smooth muscle cells. Am. J. Physiol. Cell Physiol. 305, C427-C435. ( 10.1152/ajpcell.00034.2013) [DOI] [PubMed] [Google Scholar]
- 169.Bocquet A, Sablayrolles S, Vacher B, Le Grand B. 2010. F 15845, a new blocker of the persistent sodium current prevents consequences of hypoxia in rat femoral artery. Br. J. Pharmacol. 161, 405-415. ( 10.1111/j.1476-5381.2010.00912.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Sarkar J, Chakraborti T, Pramanik PK, Ghosh P, Mandal A, Chakraborti S. 2021. PKCζ-NADPH oxidase-PKCα dependent Kv1.5 phosphorylation by endothelin-1 modulates Nav1.5-NCX1-Cav1.2 axis in stimulating Ca2+ level in caveolae of pulmonary artery smooth muscle cells. Cell Biochem. Biophys. 79, 57-71. ( 10.1007/s12013-020-00954-x) [DOI] [PubMed] [Google Scholar]
- 171.Gordienko DV, Tsukahara H. 1994. Tetrodotoxin-blockable depolarization-activated Na+ currents in a cultured endothelial cell line derived from rat interlobar artery and human umbilical vein. Pflugers Arch. 428, 91-93. ( 10.1007/BF00374756) [DOI] [PubMed] [Google Scholar]
- 172.Gosling M, Harley SL, Turner RJ, Carey N, Powell JT. 1998. Human saphenous vein endothelial cells express a tetrodotoxin-resistant, voltage-gated sodium current. J. Biol. Chem. 273, 21 084-21 090. ( 10.1074/jbc.273.33.21084) [DOI] [PubMed] [Google Scholar]
- 173.Andrikopoulos P, et al. 2011. Angiogenic functions of voltage-gated Na+ Channels in human endothelial cells: modulation of vascular endothelial growth factor (VEGF) signaling. J. Biol. Chem. 286, 16 846-16 860. ( 10.1074/jbc.M110.187559) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Jousset F, Maguy A, Rohr S, Kucera JP. 2016. Myofibroblasts electrotonically coupled to cardiomyocytes alter conduction: insights at the cellular level from a detailed in silico tissue structure model. Front. Physiol. 7, 496. ( 10.3389/fphys.2016.00496) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Chatelier A, et al. 2012. A distinct de novo expression of Nav1.5 sodium channels in human atrial fibroblasts differentiated into myofibroblasts. J. Physiol. 590, 4307-4319. ( 10.1113/jphysiol.2012.233593) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Poulet C, Künzel S, Büttner E, Lindner D, Westermann D, Ravens U. 2016. Altered physiological functions and ion currents in atrial fibroblasts from patients with chronic atrial fibrillation. Physiol. Rep. 4, e12681. ( 10.14814/phy2.12681) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Zhou X, Dobrev D. 2012. Voltage-gated Na+ channels: novel players in fibroblast-to-myofibroblast transition with a potential role in atrial arrhythmogenesis? J. Physiol. 590, 4975. ( 10.1113/jphysiol.2012.241455) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
This article has no additional data.