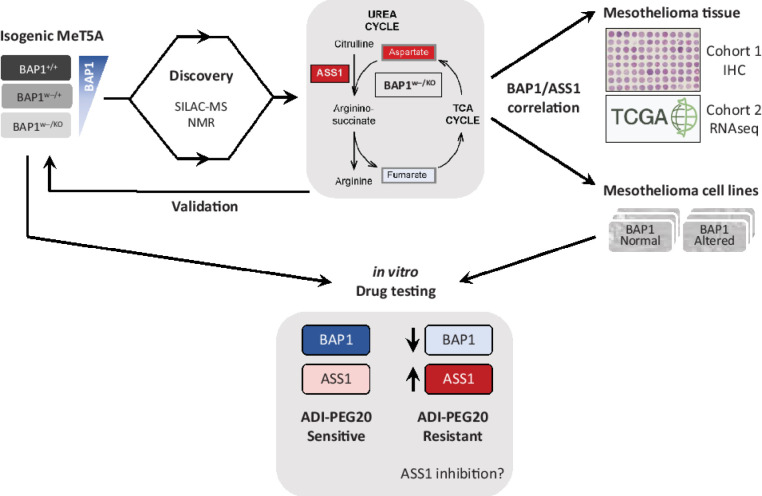
Abstract
The nuclear deubiquitylase BRCA1-associated protein 1 (BAP1) is frequently inactivated in malignant pleural mesothelioma (MPM) and germline BAP1 mutation predisposes to cancers including MPM. To explore the influence on cell physiology and drug sensitivity, we sequentially edited a predisposition mutation (w-) and a promoter trap (KO) into human mesothelial cells. BAP1w-/KO MeT5A cells express less BAP1 protein and phenocopy key aspects of BAP1 loss in MPM. Stable isotope labeling with amino acids in cell culture–mass spectrometry revealed evidence of metabolic adaptation, with concomitant alteration of cellular metabolites. In MeT5A, BAP1 deficiency reduces glycolytic enzyme levels but increases enzymes involved in the tricarboxylic acid cycle and anaplerotic pathways. Notably both argininosuccinate synthase 1 (ASS1), essential for cellular synthesis of arginine, and its substrate aspartate, are elevated in BAP1w-/KO MeT5A cells. Likewise, ASS1 expression is higher in BAP1-altered MPM cell lines, and inversely correlates with BAP1 in The Cancer Genome Atlas MESO dataset. Elevated ASS1 is also evident by IHC staining in epithelioid MPM lacking nuclear BAP1 expression, with improved survival among patients with BAP1-negative/ASS1-expressing tumors. Alterations in arginine metabolism may sensitize cells to metabolic drugs and we find that BAP1-negative/ASS1-expressing MPM cell lines are more sensitive to ASS1 inhibition, although not to inhibition of purine synthesis by mizoribine. Importantly, BAP1w-/KO MeT5A become desensitized to arginine deprivation by pegylated arginine deiminase (ADI-PEG20), phenocopying BAP1-negative/ASS1-expressing MPM cell lines.
Implications:
Our data reveal an interrelationship between BAP1 and arginine metabolism, providing a potential means of identifying patients with epithelioid MPM likely to benefit from ADI-PEG20.
Visual Overview
Introduction
Malignant pleural mesothelioma (MPM) originates in the mesothelial lining of the pleural cavity and is strongly associated with asbestos exposure several decades prior to diagnosis. It remains a disease of significant unmet need with high global mortality rates and rising incidence, despite asbestos bans in many countries (1). Exposure to similarly sized carbon nanotubes also initiates MPM in mice, raising future concerns (2). Histologically, MPM are classified as epithelioid, sarcomatoid, and biphasic, which encompass a continuous spectrum of disease (3). Treatment with cisplatin and pemetrexed, with or without the VEGF antibody bevacizumab (4), results in only modest improvement in survival. Recent randomized trials have shown survival benefit for both immune checkpoint inhibitors (5) and arginine deprivation (6, 7), offering new promise for patients with MPM. However, benefits vary widely with histologic and genetic features in this heterogeneous disease. More complete understanding of MPM biology may help broaden the options available and allow selection of the most appropriate therapies for individual patients.
MPM has few actionable oncogenic driver mutations but is instead characterized by tumor suppressor inactivation. One common example is BRCA1-associated protein 1 (BAP1), an early truncal change that may occur with or without inactivation of neurofibromin-2 (NF2) and/or cyclin-dependent kinase inhibitor 2A/2B (CDKN2A/B; refs. 8–10). BAP1 mutation typically results in catalytic inactivation or loss of BAP1 nuclear localization. It is predominantly associated with epithelioid MPM, but also occurs in biphasic and occasionally sarcomatoid subtypes. Although the reported frequency for somatic BAP1 loss-of-function mutation varies, IHC studies show loss of nuclear BAP1 (nBAP1) in up to 60% of all patients with MPM and 80% of epithelioid cases (11–14). Germline BAP1 mutation underpins a cancer predisposition syndrome where family members develop multiple cancers, most commonly MPM, peritoneal mesothelioma, and uveal melanoma (13, 15).
Despite initial identification as a tumor suppressor in lung and breast cancer (16), somatic BAP1 mutation is infrequent in epithelial cancers. Indeed, CRISPR/Cas9 screens show BAP1 is an essential/fitness gene (17), suggesting cancers may need to reprogram essential pathways to tolerate BAP1 loss of function. One explanation for the limited palette of tumors promoted by BAP1 loss is that it activates a specific apoptotic program in other cell backgrounds (18). Indeed, roles for BAP1 appear to be highly complex and context dependent. For example, BAP1 expression can promote pro-oncogenic pathways, including breast cancer cell proliferation and radioresistance in head and neck cancer (19, 20). BAP1 depletion has also been shown to reduce cell proliferation through S-phase accumulation in MPM (14), while it is suggested to function primarily as a metastasis suppressor in uveal melanoma (21). Clinically, systematic analyses show that although BAP1 loss of function is associated with worse prognosis in uveal melanoma and renal clear cell carcinoma (RCCC), it predicts better prognosis in patients with MPM (22).
Despite these complexities, genetically engineered mouse models (GEMM) provide fundamental evidence that BAP1 is tumor suppressor. Mice with heterozygous germline Bap1 mutation spontaneously develop tumors of the ovary, lung, or breast and, in response to asbestos exposure, develop MPM more frequently and rapidly than wild-type mice (23–25). More recently, deletion of Bap1 in the mesothelial lining of the thoracic cavity was shown to require Nf2 and Cdkn2ab inactivation to develop tumors that recapitulate key histologic, inflammatory, and gene expression features of human MPM (26). However, the critical roles of BAP1 as a tumor suppressor in MPM remain incompletely understood, and utilization of BAP1 status for therapeutic stratification has not yet been realized. In addition to GEMMs, modification of human cell lines by reexpression or genetic deletion of BAP1 can provide useful insights, although to date cancer cell lines with heterogenous genetic alterations have been employed (27–29).
At a molecular level, BAP1 encodes a nuclear deubiquitylase that regulates processes including the cell cycle, DNA damage repair, and metabolism, through fundamental roles in protein stabilization, histone modification, and transcriptional regulation (13, 30), suggesting potential targets for therapeutic intervention in MPM with BAP1 loss. Recent clinical studies investigating enhancer of zeste 2 polycomb repressive complex 2 subunit (EZH2) and PARP inhibitors suggest some clinical efficacy for both (7, 31). However, the tazemetostat study only recruited BAP1-negative patients and rucaparib efficacy did not relate to BAP1 status. Taken together with the relatively low efficacy of tazemetostat, this highlights the need for further study into understanding and targeting BAP1 dependencies.
Here we functionally interrogated cellular adaptation to BAP1 deficiency, to define the consequences for cell physiology and potential therapy. To provide a clean model for acquisition of BAP1 alterations, we generated isogenic mesothelial cells with sequential introduction of clinically relevant BAP1 mutations. These BAP1w-/KO cells express sufficient low-level BAP1 to support viability, but effectively mimic BAP1-mutated MPM. We employed proteomic and metabolomic analyses to profile BAP1 dependencies, which included metabolic reprogramming. Notably, upregulation of argininosuccinate 1 synthase (ASS1), an enzyme essential for cellular synthesis of arginine and its downstream metabolites, was identified in BAP1w-/KO cells and in BAP1-deficient MPM cells or tumor samples. Importantly, we found that BAP1 status is associated with sensitivity to metabolic drugs including pegylated arginine deiminase (ADI-PEG20), and thus may be exploited in patient stratification for relevant therapeutic interventions such as arginine deprivation.
Materials and Methods
Cell culture
MeT5A cells (ATCC, obtained 2012, RRID:SCR_003193) were cultured in Media 199 (1 g/L glucose) supplemented as described previously (32) with selection antibiotics as appropriate. MPM cell lines (for RRID, see Supplementary Table S1) were sourced from ATCC (NCI or MSTO, obtained 2012/2104) or Mesobank (33) [MESO (34) obtained 2017, and MPM# (35) obtained 2015; www.mesobank.com]. MPM cells were cultured in RPMI supplemented with 10% FBS (2 g/L glucose), and for MESO cell lines 20 ng/mL EGF (Peprotech), 1 μg/mL hydrocortisone (Sigma-Aldrich), and 2 μg/mL heparin (Sigma-Aldrich). All cells were grown in a humidified incubator at 37°C and 5% CO2. Cells were routinely verified as Mycoplasma free during culture, using EZ-PCR Mycoplasma (Geneflow) or e-Myco Plus PCR (Bulldog bio) detection kits. All cells were authenticated by short tandem repeat (STR) profiling using the GenePrint10 system (Promega). This was last performed in September 2018; all lines achieved a DSMZ score >0.8 (Supplementary Table S1) and were batch frozen, with all experiments performed between passage 3 and 20 after thawing the STR-profiled cells.
Genome editing of BAP1
The BAP1 locus was edited using recombinant adeno-associated virus (rAAV) homologous recombination strategies designed by Horizon Discovery to sequentially target alleles with a predisposition point mutation (w-) and a promoter trap (KO). HEK293T cells (RRID:SCR_003193) were cotransfected with the genome-editing cassette and AAV-2 helper plasmid (Plasmid Factory) using Lipofectamine LTX (Invitrogen) and rAAVs extracted 48 hours later using Virakit (Virapur); the viral titre was estimated by real-time PCR. To establish the MeT5A-BAP1w-/+ line, clonal MeT5A-BAP1+/+(C2) cells were infected with purified rAAV-0365 for 72 hours, then seeded into 96-well plates at 10–1,000 cells/well in selection media (0.7 μg/mL puromycin, Sigma-Aldrich). To establish MeT5A-BAP1w-/KO cells, the MeT5A-BAP1w-/+ line was infected with rAAV-0612 and selected in 0.7 μg/mL puromycin and 0.1 mg/mL G418 (ForMedia). After 2 weeks, colonies were lysed in Direct PCR Lysis Reagent (Viagen Biotech) and screened with primers that distinguished the edited locus using GoTaq Flexi (Promega). Positive colonies were single cell diluted and rescreened to establish clones, which were sequence verified (Dundee Sequencing Services) and authenticated by STR profiling.
Protein extraction and immunoblotting
Whole-cell extracts were prepared by hot lysis in Laemmli buffer, with protein concentrations determined and immunoblotting performed as described previously (36). Primary antibodies were mouse anti-ASS1 (2C10 MABN704, Sigma-Aldrich; RRID:AB_2927674), BAP1 (C-4 sc-28383, Santa Cruz Biotechnology; RRID:AB_626723), PGK1 (22C5D8 ab113687, Abcam; RRID:AB_10861977), and β-actin (ab6276, Abcam; RRID:AB_2223210); rabbit anti-AK4 (HPA049461, Sigma-Aldrich; RRID:AB_2680776), DHFR (ab124814, Abcam; RRID:AB_10975115), ENO2 (#9536, Cell Signaling Technology; RRID:AB_2099308), GLS (ab156876, Abcam; RRID:AB_2721038), histone H2A (07-146, Upstate; RRID:AB_310394), H2A-Ub (#8240, Cell Signaling Technology; RRID:AB_10891618), MDH2 (HPA019716, Sigma-Aldrich; RRID:AB_1853680), OGT (#5368S, Cell Signaling Technology; RRID:AB_11217621), PGAM1 (ab96622, Abcam; RRID:AB_10687155), PGM2 (ab151746, Abcam; RRID:AB_2927675), PYGL (HPA004119, Sigma-Aldrich; RRID:AB_1079723), RRM1 (3388, Cell Signaling Technology; RRID:AB_2180382) and β-actin (A2066, Sigma-Aldrich; RRID:AB_476693), or goat anti-ALDOC (D-14 sc-12066, Santa Cruz Biotechnology; RRID:AB_2226594), IDH3B (ab118287, Abcam; RRID:AB_10899317), and SUCLG2 (A-15 sc-99646, Santa Cruz Biotechnology; RRID:AB_10843272). Proteins were visualized using secondary antibodies conjugated to IRDyes and the LI-COR Odyssey 2.1 system; 16-bit images were quantified using ImageStudioLite (RRID:SCR_013715).
Cell proliferation and therapeutic efficacy assays
MeT5A cells were seeded at suitable density and monitored for up to 5 days. The relative cell number was determined by direct counting, cell growth by CellTiter-Glo assay (Promega), or cell confluence by live imaging using an IncuCyte S3 (Essen Bioscience; RRID:SCR_019874). For drug assays, cells were seeded in 96-well plates at appropriate densities for each cell line between 1 × 103 and 4.5 × 103 cells/well. After 24 hours, cells were treated with vehicle, α-methyl-DL-aspartic acid (αMDLA, Biosynth AG), mizoribine (Sigma-Aldrich), or ADI-PEG20 (Polaris Pharmaceuticals Inc) at the indicated doses, with viability (CellTiter-Glo) and/or confluence (IncuCyte) monitored for up to 4 days.
Cell-cycle analysis
Exponentially growing cells were fixed prior to 7-aminoactinomycin D (Invitrogen) staining to determine DNA content. The fluorescence of single cells was quantified using a Attune NxT system and analyzed using FlowJo v10.8.1 software (RRID:SCR_008520).
Mass spectrometry
For stable isotope labeling with amino acids in cell culture (SILAC), cells were maintained for 6 passages in arginine/lysine-free media with normal supplements and amino acids (Sigma-Aldrich) in light, medium, or heavy configurations (37) at final concentrations of 84 mg/L arginine, 146 mg/L lysine, and 200 mg/L proline. Labeling efficiency was >98% with <2% proline conversion. For in-gel digestion and peptide extraction, lysates from MeT5A-BAP1+/+(C2), BAP1w-/+, and BAP1w-/KO (C3.1 or C5.1) were mixed (1:1:1, 90 mg total) and separated by SDS-PAGE. Proteins were extracted as described previously (36). Briefly, gel pieces were de-stained with 50% acetonitrile/50% 100 mmol/L NH4HCO3, reduced with 10 mmol/L dithiothreitol and alkylated with 50 mmol/L iodoacetamide. Following acetonitrile dehydration and incubation with Trypsin Gold (Promega), peptides were extracted in acetonitrile, vacuum dried and resuspended in 1% formic acid. For filter-aided sample preparation (FASP), MESO-8T, MESO-12T, and MeT5A-BAP1+/+(C2) lysates were mixed (1:1:1, 300 mg total) and diluted in 8 mol/L urea in 50 mmol/L Tris and 75 mmol/L NaCl. Samples were centrifuged for 20 minutes at 8,000 × g in Amicon Ultra 0.5 mL 10 kDa molecular weight cut-off filters (Merck). Following two washes with 8 mol/L urea buffer and two with 50 mmol/L NH4HCO3, samples were reduced and alkylated with 10 mmol/L tris(2-carboxyethyl)phosphine and 40 mmol/L 2-chloroacetamide in 50 mmol/L NH4HCO3 for 30 minutes then washed twice with 50 mmol/L NH4HCO3. Samples were digested with Lys-C (0.5 mg/100 mg protein) for 2 hours at room temperature and Trypsin Gold (2 mg/100 mg protein) overnight at 37°C before peptides were eluted by centrifugation. Mass spectrometry (MS) analysis of the isogenic cell panel was performed in-house as described previously (36) with two replicate runs, or by the Proteomics Research Technology Platform (University of Warwick, Coventry, United Kingdom) for samples prepared via FASP. All MS RAW files were analyzed using MaxQuant (RRID:SCR_014485) equipped with the Andromeda search engine, and further processed using Microsoft Excel (RRID:SCR_016137) and Perseus (RRID:SCR_015753).
Metabolomics
Pellets of MeT5A-BAP1+/+(C2) and BAP1w-/KO (C5.1) cells were collected in triplicate for three biological replicates. For metabolite extraction, cells were mixed with ice-cold 50% acetonitrile/water and subject to 3×30 seconds sonication (23 KHz, 10 μm amplitude) with 30-second intervals, then centrifuged for 10 minutes at 4°C and 12,000 × g. Supernatants were lyophilized for 16 hours and mixed with 190 μL buffer [100 mmol/L sodium phosphate (pH7.4), 0.1 mmol/L 3-(Trimethylsilyl)propionic-2,2,3,3-d4 acid sodium salt (Sigma-Aldrich), 1.2 mmol/L sodium azide (Sigma-Aldrich)]. Spectra were acquired in 3 mm nuclear magnetic resonance (NMR) tubes using a Bruker Avance III 700 MHz NMR spectrometer fitted with a 5 mm [1H, 15N, 13C]-TCI Cryoprobe and SampleJet. All 1H NMR spectra were acquired at 25°C (± 0.1°C) and processed (phase and baseline correction) using automation for consistency. CPMG and 1D NOE pulse sequences were used for acquisition, with a spectral width of 30 ppm, 48K complex points and 32 scans (1D NOE) or 256 scans (CPMG). Acquired spectra that passed quality checks were used in statistical analysis and metabolite identification using in-house standards library and the Chenomx NMR Suite (RRID:SCR_014682). For statistical analysis, each peak was individually bucketed. Prior to analysis each spectrum was centred to the median and samples were pareto scaled to each other. Principal component analysis (PCA) and t tests were performed.
RNA extraction and qRT-PCR
Total RNA was extracted and qRT-PCR performed using SYBR Green supermix as described previously (36). Primer sequences were: ACTB (For: 5′-CACCTTCTACAATGAGCTGCGTGTG-3′, Rev: 5′-ATAGCACAGCCTGGATAGCAACGTAC-3′), and ASS1 (For: 5′-GAAGTGCGCAAAATCAAACAAG-3′, Rev: 5′-GATGTACACCTGGCCCTTG). ASS1 was normalized to ACTB and represented as 2−[ΔΔCt] relative to MeT5A-BAP1+/+.
IHC staining and scoring
NHS Health Research Authority National Research Ethics Service approval, conforming to the principles of the Declaration of Helsinki and with written informed patient consent, was conferred by the supplying tissue banks. Antibodies were optimized on whole MPM sections from Royal Papworth Hospital Research Tissue bank (NHS REC 18/EE/0269). Tissue microarrays (TMA) containing 3- to 4-μm-thick and 800 μm diameter cores from MPM samples (4 cores/patient) were obtained from Mesobank (ref. 33; http://www.mesobank.com/; NHS REC 18/EE/0161). Antigen retrieval was performed using the PT Link Pretreatment module (Agilent Technologies). Slides were incubated at high-pH (pH9.0; BAP1, pan-cytokeratin) or low-pH (pH6.0; ASS1) at 96°C for 20 minutes. IHC was performed using the DAKO Envision FLEX Kit (Agilent). Slides were incubated for 30 minutes with mouse anti-pan-cytokeratin (1:80; MNF116/MA1-26237; Thermo Fisher Scientific; RRID:AB_794730), anti-BAP1 (1:400; C-4/sc-28383; Santa Cruz Biotechnology; RRID:AB_626723), or rabbit anti-ASS1 (1:200; HPA020934; Sigma-Aldrich; RRID:AB_1845118), and counterstained with hematoxylin. Tumor cell nuclear immunoreactivity for BAP1 (13) was scored by three independent observers blinded to clinical and/or pathologic data; non-MPM cells acted as internal positive controls. ASS1 stained slides were scanned (x40, .svs format) using a Leica Aperio CS2 for and QuPath Bioimage analysis v.0.2.0 (https://QuPath.github.io/; RRID:SCR_018257; ref. 38) used to generate tumor H-scores. There was some sample attrition as cores that were missing, had insufficient tumor tissue, were nonepithelioid or not scorable for BAP1, were excluded from analysis. Cell detection identified all cells within a core, then a two-way random tree classifier trained the software to distinguish tumor and non-tumor cells. Staining intensity for each tumor cell within a core was determined and assigned to an intensity group following manual calibration of thresholds (0 = none, 1 = weak, 2 = moderate, 3 = strong) with H-Score calculated as: [1×(% weakly stained) + 2×(% moderately stained) + 3×(% strongly stained)]. The mean tumor H-Score was calculated for each epithelioid patient with >2 scorable cores for BAP1/ASS1 (n = 164). These samples included 125 male and 33 female patients (6 no gender recorded) with a median age of 71 (range, 53–88); both gender and age distribution are in keeping with patient demographics.
Bioinformatics and statistical analysis
Proteins modulated >1.5-fold in C5.1 BAP1w-/KO cells were functionally annotated using the WEB-based GEne SeT AnaLysis (GESA) Toolkit (http://www.webgestalt.org; RRID:SCR_006786; ref. 39) for GESA of Hallmark-50 pathways (40) and Over-representation analysis of Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways. The Cancer Genome Atlas (TCGA) Pan-Cancer dataset for mesothelioma (10) was analyzed using cBioportal (https://www.cbioportal.org; RRID:SCR_014555; ref. 41) or exported for analysis. Morpheus (https://software.broadinstitute.org/morpheus; RRID:SCR_017386) was used to generate heatmaps and perform unsupervised hierarchical clustering.
Biochemical measurements represent several thousand cells and are represented as the mean value from at least three independent experiments, with error bars showing SD. No statistical method was used to predetermine sample size. All statistical tests were performed using GraphPad Prism version 9 (RRID:SCR_002798). Distribution of data was assessed by the D'Agostino & Pearson omnibus normality test and variance equivalency between samples by a variance homogeneity test. Data were analyzed by parametric or nonparametric tests as appropriate (see figure legends). In power calculations for TMA analyses, 200 patient samples were estimated to provide power of 75% to 80% to detect an OR of approximately 3.5, with 1:1–2:1 phenotypic ratio. Survival estimations were made using Kaplan–Meier method and plotted in IBM SPSS v26 (RRID:SCR_016479) or with Cox proportional hazards models in R v4 (RRID:SCR_017301; see Supplementary Materials and Methods). P values less than 0.05 were considered significant.
Data availability
The data generated in this study are available within the article and its Supplementary Data. In this study, there are also analyses based upon datasets generated by TCGA Research Network (https://www.cancer.gov/tcga; RRID:SCR_003193).
Results
To explore whether BAP1 mutation leads to adaptations in MPM that may influence therapeutic strategies, we used genome editing to create isogenic cell lines with clinically relevant monoallelic and biallelic BAP1 mutations on a mesothelial background. Transformed normal human pleural mesothelial MeT5A cells were selected as they have a near diploid genome (42). A clonal MeT5A-BAP1+/+ parental line was established, which retains two copies of the BAP1 locus (Supplementary Fig. S1) and expresses catalytically active BAP1 (Supplementary Fig. S2). Although SV40-transformed, MeT5A are not tumorigenic (42) and inactivation of P53 by the SV40 T-antigen is consistent with the genetic alteration of CDKN2AB or TP53 commonly found in clinical MPM (9, 10). MeT5A-BAP1+/+ therefore enabled sequential BAP1 mutation, to generate a novel in vitro model of BAP1 loss-of-function mutations in mesothelial cells. Genome editing with rAAV was used to first introduce a cancer-predisposition mutation on one BAP1 allele, generating MeT5A-BAP1w-/+ cells. This splice site A-to-G mutation, reported in the w-family germline, leads to exon 7 skipping, premature translational termination (Pro147fsX48), and nonsense mediated decay (15). A promoter trap was then introduced on the second BAP1 allele to mimic biallelic inactivation in MPM, generating two independent MeT5A-BAP1w-/KO clones referred to as C3.1 and C5.1.
Targeted sequencing confirmed gene editing of each allele (Fig. 1A), while analysis of BAP1 transcripts confirmed altered splicing driven by the w-mutation and neomycin expression from within the promoter trap (Supplementary Fig. S3A and S3B). Surprisingly, the w-mutation increased overall BAP1 transcription and only partially abrogated normal BAP1 splicing (Supplementary Fig. S3C) so that mRNA encoding wild-type BAP1 reduced by approximately 70% to 80% in BAP1w-/KO clones, with constitutive expression of catalytically active BAP1 protein reduced to approximately 60% in C3.1 and approximately 40% in C5.1, relative to parental MeT5A cells (Fig. 1B; Supplementary Fig. S3D). Of several thousand clones screened, only these two BAP1w-/KO clones correctly integrated the promoter trap. Consistent with identification of BAP1 in CRISPR/Cas9 screens for fitness genes (17) and evidence that BAP1 loss promotes apoptosis on many cell backgrounds (18), our data suggest that complete deletion of BAP1 may be incompatible with MeT5A cell survival. To investigate further, we performed whole-genome sequencing (WGS). This reconfirmed introduction of the correct BAP1 mutations by genome editing and demonstrated little genomic divergence between cell lines (Supplementary Fig. S4; Supplementary Table S2). The number of differential SNPs relative to parental MeT5A cells was only 0.28% of that for MeT5A relative to the human reference genome. Between 16 and 19 differential SNPs were predicted to have potentially deleterious effects on an encoded protein, but only two were common between all cell lines (Supplementary Table S2). While we cannot rule out a contribution to survival of BAP1-mutated MeT5A cells, neither proteasome activator subunit 4 (PSME4) nor ATPase phospholipid transporting 11A (ATP11A) are obvious candidates.
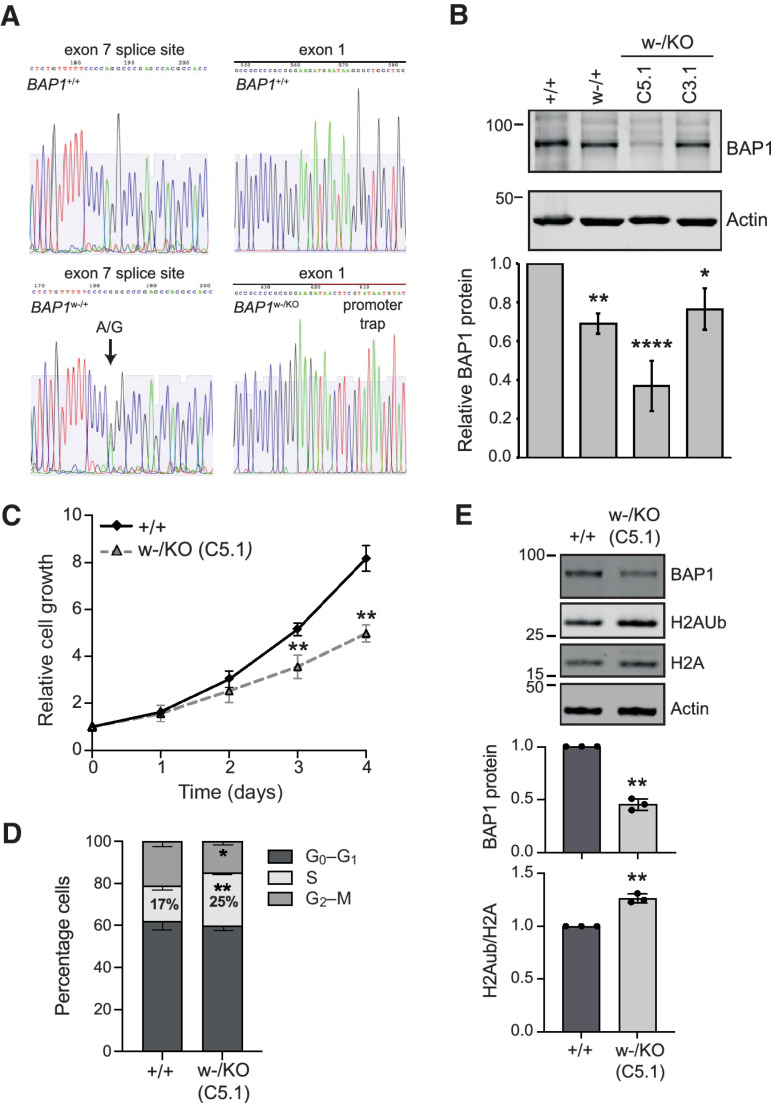
Figure 1.
MeT5A BAP1w-/KO cells recapitulate phenotypic aspects of BAP1-deficient MPM cells. A, Sequence confirmation of edited alleles: introduction of the w-family splice site mutation to induce exon 7 skipping (left) and a promoter trap to knockout (KO) BAP1 expression (right) in the isogenic MeT5A cells. B, BAP1 protein levels are constitutively reduced in BAP1w-/+ and BAP1w-/KO cells. Representative immunoblot and quantification relative to actin, mean of three independent experiments, error bars SD, one-way ANOVA with Dunnett post hoc test. *, P < 0.05; **, P < 0.01; ****, P < 0.0001. C, BAP1 deficiency slows proliferation of MeT5A cells. ATP-luciferase assay for BAP1w-/KO C5.1; mean of three independent experiments, error bars SD, t test. **, P < 0.01 compared with MeT5A-BAP1+/+. D, BAP1 deficiency causes MeT5A cells to accumulate in S-phase. Cell-cycle distribution determined by flow cytometry for BAP1+/+ and BAP1w-/KO C5.1 (supporting data, Supplementary Fig. S5E). Mean of three independent experiments, error bars SD; unpaired t test; *, P = 0.0303; **, P = 0.0014. E, H2A ubiquitylation is increased in BAP1-mutated MeT5A cells. Representative immunoblot and quantification, mean of three independent experiments, error bars SD, one-sample t test. **, P < 0.01.
Importantly, BAP1w-/KO clones phenocopied the effect of BAP1 depletion on growth characteristics of MPM cells (14), with reduced proliferation and accumulation in S-phase (Fig. 1C and D; Supplementary Fig. S5). We also observed moderately increased ubiquitylation of histone H2A K119, a major and abundant target of BAP1 (18), in the BAP1w-/KO C5.1 cells commensurate with their residual BAP1 (Fig. 1E). To further explore the effects of BAP1 deficiency, we used SILAC-MS in two triplex-configured experiments to compare the proteome of the MeT5A isogenic cell panel (Fig. 2A). Overall, we identified more than 2,300 individual proteins (Supplementary Table S3), 75% of which were detected in both experiments (Fig. 2B). Compared with either parental or haploinsufficient cells, 22% of detected proteins increased or decreased by more than 1.5-fold in the BAP1w-/KO clones (Fig. 2C), with sequential gene editing leading to stepwise divergence in the proteome (Fig. 2D). Consistent with residual BAP1 levels in the BAP1w-/KO clones (Fig. 2A), more proteins were modulated in C5.1 than C3.1 (Fig. 2D) and most of the proteins modulated in both BAP1w-/KO clones showed greater differential expression in C5.1 (Fig. 2E). We therefore focused on investigating adaptation of BAP1w-/KO C5.1 cells compared with BAP1+/+ cells (Fig. 2F).
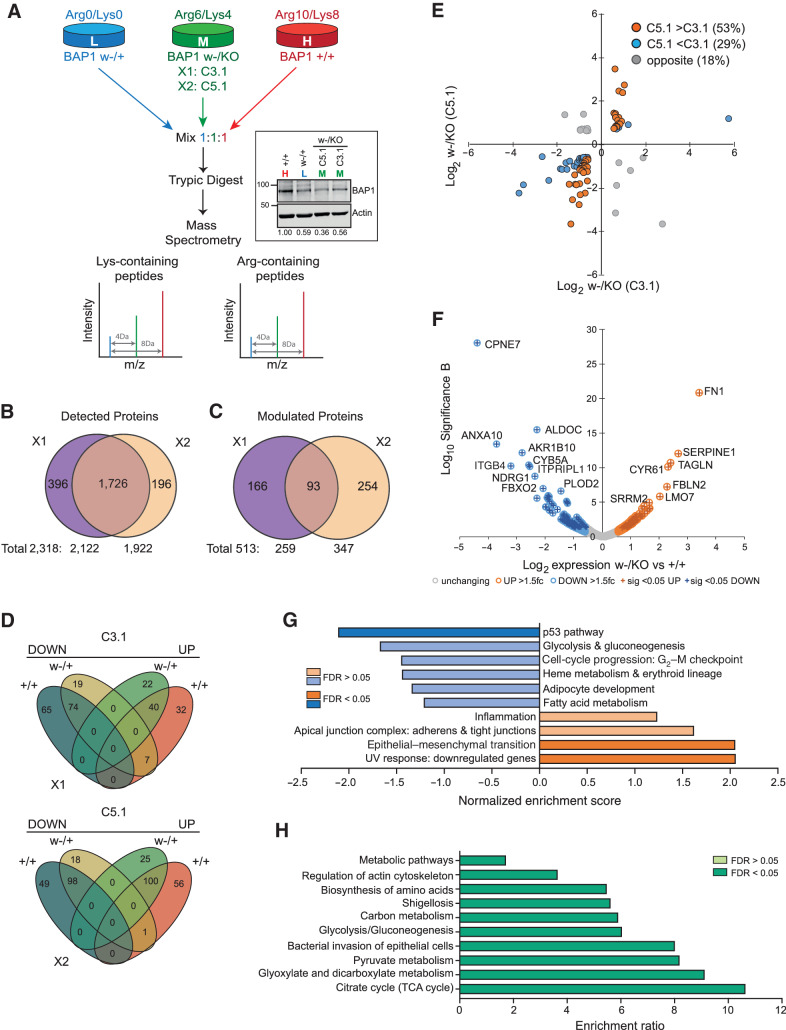
Figure 2.
BAP1 deficiency reprograms the proteome of MeT5A cells. A, Experimental strategy for triplex SILAC-MS analysis of isogenic MeT5A cells, two alternative configurations (X1 and X2) used either the MeT5A-BAP1w-/KO C5.1 or C3.1 clones. Inset shows stable BAP1 expression after 6 passages in SILAC-labeling media; numbers indicate mean expression relative to BAP1+/+ cells from three independent analyses. B, Summary of proteins identified by MS across two experiments, based on gene name identifiers; supporting data, Supplementary Table S3. C, Summary of proteins modulated >1.5-fold in BAP1w-/KO relative to either BAP1+/+ or BAP1w-/+ cells. D, Overview of directional expression changes in each experimental configuration for the 513 proteins modulated by >1.5-fold. E, Relative expression of the 93 proteins modulated >1.5-fold in both BAP1w-/KO clones. F, Volcano plot showing proteins modulated >1.5-fold (colored circles) with P < 0.05 (crossed circles) in C5.1 BAP1w-/KO cells. G and H, Heatmaps summarizing functional annotation of enriched pathways for proteins modulated by >1.5-fold in C5.1 BAP1w-/KO cells. Gene set enrichment analysis showing weighted set cover for Hallmark-50 pathways: blue, C5.1 downregulated proteins; orange, C5.1 upregulated proteins (G); supporting data in Supplementary Fig. S7A and S7B. Overrepresentation analysis for KEGG pathways among all C5.1 modulated proteins (H).
Functional annotation of proteins modulated in BAP1w-/KO C5.1 cells revealed marked enrichment of gene ontology (GO) terms and pathways involved in cancer cell biology, metabolism, and movement of cells (Fig. 2G and H; Supplementary Fig. S6). None of the 18 differential deleterious SNPs identified by WGS in the BAP1w-/KO C5.1 clone mapped to relevant pathways or biological processes (Supplementary Table S2). GESA identified hallmark pathways associated with cancer including cell-cycle progression, the P53 pathway, metabolism, and epithelial–mesenchyme transition (EMT; Fig. 2G; Supplementary Fig. S7A and S7B), highlighting the disease relevance of our BAP1w-/KO mesothelial cell model. Highly modulated EMT markers included fibulin 2 (FBLN2), a paralog of the MPM biomarker FBLN3 (43), and transglutaminase 2 (TGM2), a cancer stem cell survival protein required for MPM tumor formation (44). Indeed, fibronectin (FN1) and plasminogen activator inhibitor 1 (SERPINE1, PAI), the most significantly upregulated proteins in C5.1 BAP1w-/KO cells (Fig. 2F), are associated with EMT. However, while FN1 and PAI1 validated by immunoblotting (Supplementary Fig. S7C–S7E), they did not correlate with BAP1 status in an MPM cell panel (Supplementary Fig. S7F–S7H). We therefore focused our attention on the metabolic reprogramming highlighted by both pathway and GO term analyses. Glycolysis/gluconeogenesis and tricarboxylic acid (TCA) cycle KEGG pathways were overrepresented (Fig. 2H), with proteins downregulated in C5.1 enriched for glycolysis/gluconeogenesis (Fig. 2G; Supplementary Fig. S7A) and those upregulated in C5.1 enriched for TCA metabolic processes (Supplementary Fig. S6).
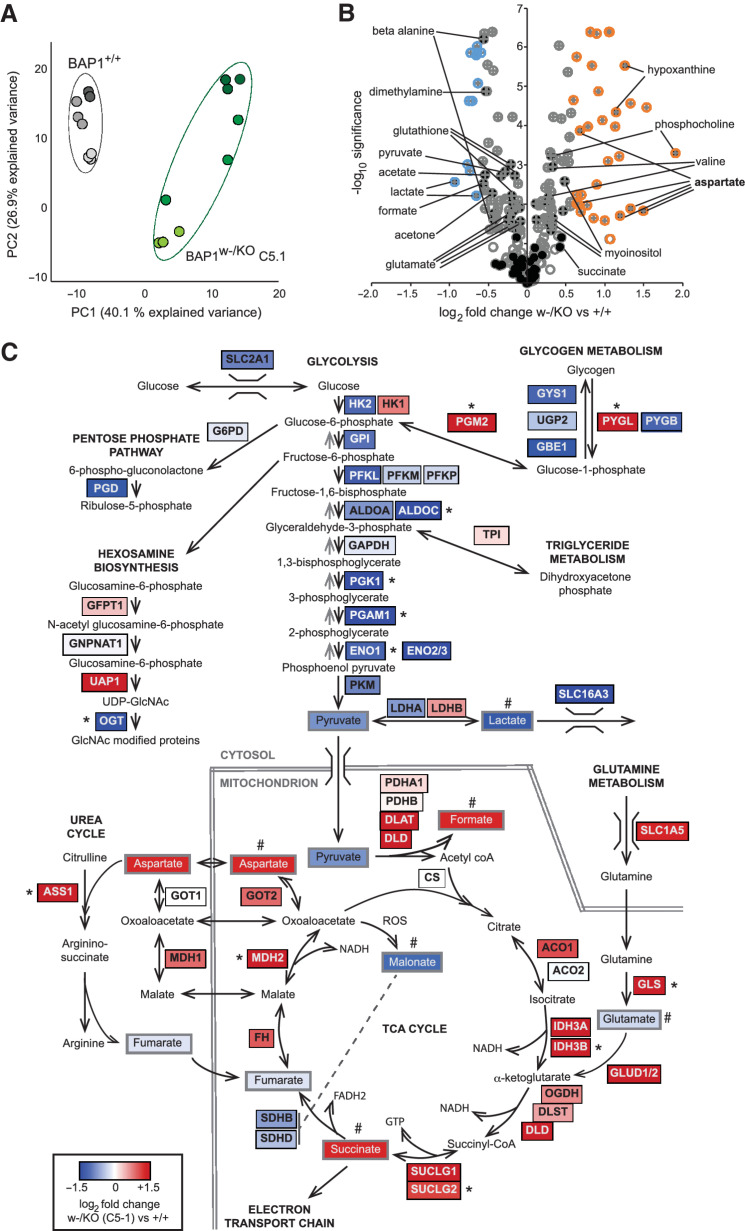
To investigate further, we interrogated BAP1w-/KO and BAP1+/+ MeT5A cells using NMR metabolomics (Supplementary Table S4). PCA showed clear separation of the cell lines according to PC1, which accounted for 40% of the variance among samples (Fig. 3A), and 82 metabolite peaks in the NMR spectra were formally identified (Fig. 3B). To integrate protein and metabolite data, we mapped both datasets onto key metabolic pathways (Fig. 3C). In BAP1w-/KO cells, differential expression of around 60 proteins was associated with the most enriched metabolic processes. Particularly striking was reduced expression of multiple glycolysis/gluconeogenesis enzymes, accompanied by downregulation of glucose and lactate transporters. Notably, levels of lactate and pyruvate were also low, potentially consistent with reduced glycolysis (Fig. 3B and C; Supplementary Fig. S8). In contrast, there was increased expression of enzymes promoting glycogen or glutamine metabolism, the TCA cycle and anaplerotic pathways, including the urea cycle. Correspondingly, levels of identified TCA intermediates were also higher in BAP1w-/KO MeT5A and aspartate, which feeds into the urea cycle, was prominently upregulated (Fig. 3B and C; Supplementary Fig. S8). For selected metabolic enzymes identified by MS, their expression was cross-validated by immunoblotting in independent unlabeled MeT5A lysates (Fig. 4A, compare columns 1 and 2). In general, where protein expression decreased in BAP1w-/KO cells by MS, this was confirmed by immunoblotting, including for the glycolysis enzymes ALDOC, PGAM1, and ENO2, as well as AK4 and O-linked N-acetylglucosamine transferase (OGT; Fig. 4A; Supplementary Fig. S9). PGM2 trended toward higher expression in BAP1w-/KO cells (Fig. 4A; Supplementary Fig. S9) while ASS1 levels were most significantly increased (Fig. 4A). Thus, metabolic reprogramming in BAP1w-/KO MeT5A may signal reliance on alternative energy sources and anaplerotic pathways.
Figure 3.
BAP1-deficient MeT5A cells have altered levels of metabolites and metabolic enzymes. A and B, NMR analysis reveals altered metabolites in C5.1 BAP1w-/KO relative to BAP1+/+ MeT5A. PCA plot for samples from three independent experiments analyzed in triplicate (A). Volcano plot of metabolites modulated >1.5-fold (colored circles) with P < 0.05 (crossed circles), named metabolites (filled black circles; B). C, Overview of metabolic adaptation for key pathways in BAP1-deficient MeT5A cells combining proteomic and metabolomic data, color scale represents log2 fold change in C5.1 MeT5A-BAP1w-/KO versus BAP1+/+ cells. *Enzymes investigated further. #Significantly modulated metabolites (supporting data, Supplementary Table S4; Supplementary Fig. S8).
Figure 4.
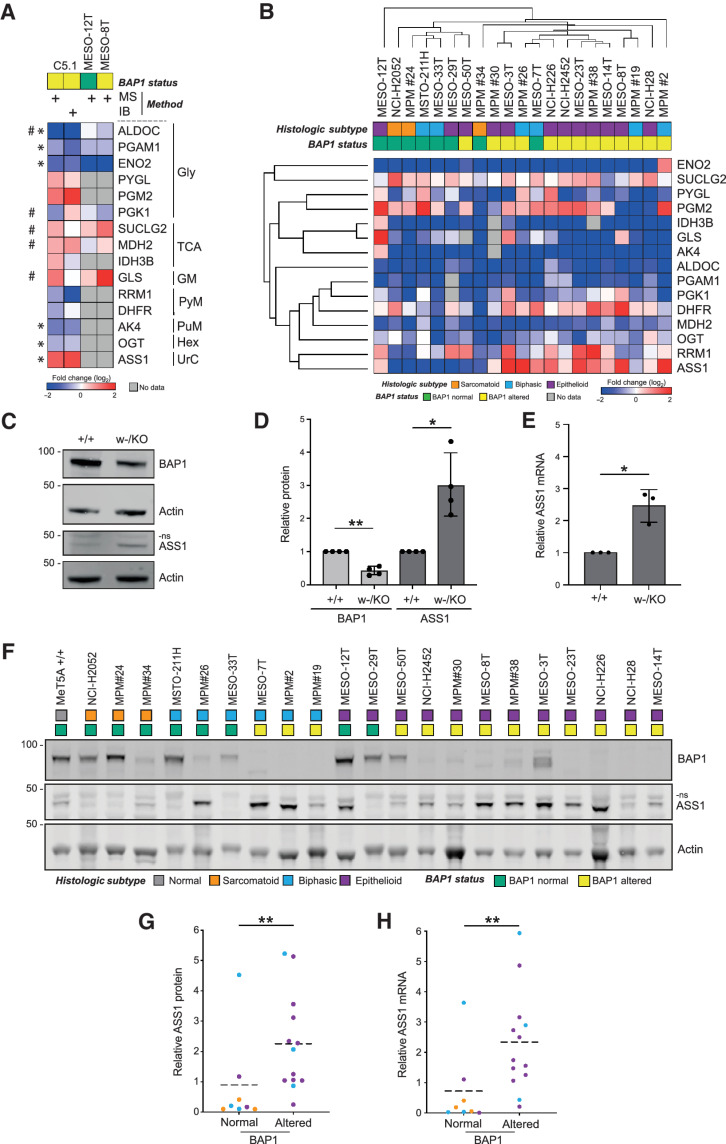
ASS1 expression is a metabolic BAP1 dependency in isogenic MeT5A and MPM cell lines. A, Heatmap comparing expression in BAP1w-/KO C5.1 cells determined by SILAC-MS or immunoblotting (IB, three independent experiments, supporting data in Supplementary Fig. S9), with SILAC-MS data (n = 1; supporting data, Supplementary Table S3) for two epithelioid MPM cell lines: MESO-8T (BAP1-altered), MESO-12T (BAP1-normal). *Significant modulation confirmed in MeT5A by immunoblotting, #data for MESO-8T versus MESO-12T consistent with BAP1 dependency in MeT5A. Gly, glycolysis; TCA, tricarboxylic acid cycle; GM, glycogen metabolism; PyM, pyrimidine metabolism; PuM, purine metabolism; Hex, hexosamine biosynthesis; UrC, urea cycle. B, Unsupervised hierarchical clustering for expression in 21 MPM cell lines determined by immunoblotting. Heatmap shows protein levels for selected metabolic enzymes in cell lines relative to MeT5A BAP1+/+. Key indicates histologic subtype and BAP1 status, supporting data in Supplementary Figs. S10 and S11. ASS1 protein levels increase in MeT5A BAP1w-/KO cells. Representative immunoblot with anti-ASS1 antibody (Millipore; C) and quantification (D). Mean of four independent experiments, error bars SD; Welch t test; *, P = 0.026; **, P = 0.003; ns, non-specific band. E,ASS1 mRNA expression increases in MeT5A BAP1w-/KO cells. ASS1 qRT-PCR normalized to ACTB and GAPDH; mean of three independent experiments, error bars SD; Welch t test; *, P = 0.021. F–H, ASS1 protein and mRNA expression are significantly higher in BAP1-altered than BAP1-normal MPM cell lines. Representative immunoblot for the cell panel (F); histologic subtype and BAP1 status indicated; ns, non-specific band. Mean ASS1 protein level from three independent experiments (G); normalized to actin and relative to MeT5A BAP1+/+, population mean (dashed line), Mann–Whitney **, P = 0.006. Mean ASS1 mRNA determined by qRT-PCR (H); normalized to ACTB and GAPDH relative to MeT5A BAP1+/+ from three independent experiments, population mean (dashed line), Mann–Whitney **, P = 0.006. For G and H, histologic subtype is indicated in F; supporting data, Supplementary Fig. S12.
To determine whether metabolic adaptions to BAP1 mutation in the isogenic MeT5A model were also evident in BAP1-altered MPM, we utilized a panel of 21 patient-derived cell lines, representing the histologic and genetic heterogeneity of the disease. These were characterized for BAP1 expression, catalytic activity, and cellular localization, with confirmatory targeted sequencing where indicated, to stratify the panel according to BAP1 status (Supplementary Fig. S10). Two epithelioid cell lines were chosen that differed in BAP1 status but were otherwise closely matched in terms of the originating laboratory, growth media, presence of MPM markers and lack of NF2/CDKN2A expression. The MESO-8T (BAP1-altered) and MESO-12T (BAP1-normal) cells were initially compared with parental BAP1+/+ MeT5A cells by triplex SILAC-MS (Supplementary Table S3; Fig. 4A columns 3 and 4). Seven metabolic enzymes of interest were detected in both MPM cell lines, with the majority showing the highest differential expression in MESO-8T, consistent with BAP1 dependency predicted from the MeT5A model (Fig. 4A).
As it is difficult to generalize from two cell lines, fuller coverage of metabolic enzymes of interest across the MPM cell panel was obtained by immunoblotting (Fig. 4B; Supplementary Fig. S11A). Levels of several enzymes (e.g., ALDOC, PGAM1, ENO2) were consistently reduced in all MPM cell lines, irrespective of BAP1 status, compared with the normal mesothelial BAP1+/+ MeT5A (Fig. 4B). Intriguingly BAP1w-/KO MeT5A cells largely recapitulate these cancer-associated expression changes (Fig. 4A) suggesting BAP1 mutation may be sufficient, although not required, for adaptation. Metabolic enzymes including OGT, DFHR, and RRM1 exhibited variable expression amongst the MPM cell lines (Fig. 4B). DHFR and RRM1 are linked with sensitivity to therapeutics used in MPM, pemetrexed, and gemcitabine, respectively; however, they did not stratify by BAP1 status (Supplementary Fig. S11B), implying more complex regulation of their expression in cancer. We did however observe a striking correlation between BAP1 status and levels of ASS1 (Fig. 4B), an essential enzyme in arginine synthesis that may predict sensitivity to certain therapeutics targeting metabolism. We also examined the relationship between expression of these metabolic transcripts and BAP1 genetic alteration, transcript expression, or protein levels in TCGA MESO dataset (ref. 10; Supplementary Fig. S11C). Although limited association was evident for transcripts including OGT, only ASS1 significantly correlated with all three molecular measures of BAP1 status, reinforcing data from the MPM cell lines.
We investigated the BAP1 dependence of ASS1 expression in more depth using two validated antibodies (Supplementary Fig. S12A and S12B). Significantly higher levels of ASS1 protein were confirmed in BAP1w-/KO compared with the parental BAP1+/+ MeT5A cell line (Fig. 4C and D; Supplementary Fig. S12C and S12D). Analysis of ASS1 mRNA in parallel samples demonstrated that this was driven by a transcriptional response to BAP1 mutation (Fig. 4E). Similarly, both protein and mRNA expression of ASS1 were significantly higher in MPM cell lines with altered BAP1 (Fig. 4F–H; Supplementary Fig. S12E–S12G). ASS1 mRNA and protein were highly correlated across the cell lines (Supplementary Fig. S12H) suggesting that transcript expression is a useful surrogate for protein levels. Consistent with a transcriptional response to BAP1 inactivation, ASS1 mRNA expression was also inversely correlated with that of BAP1 in the MPM cell lines (Supplementary Fig. S12I).
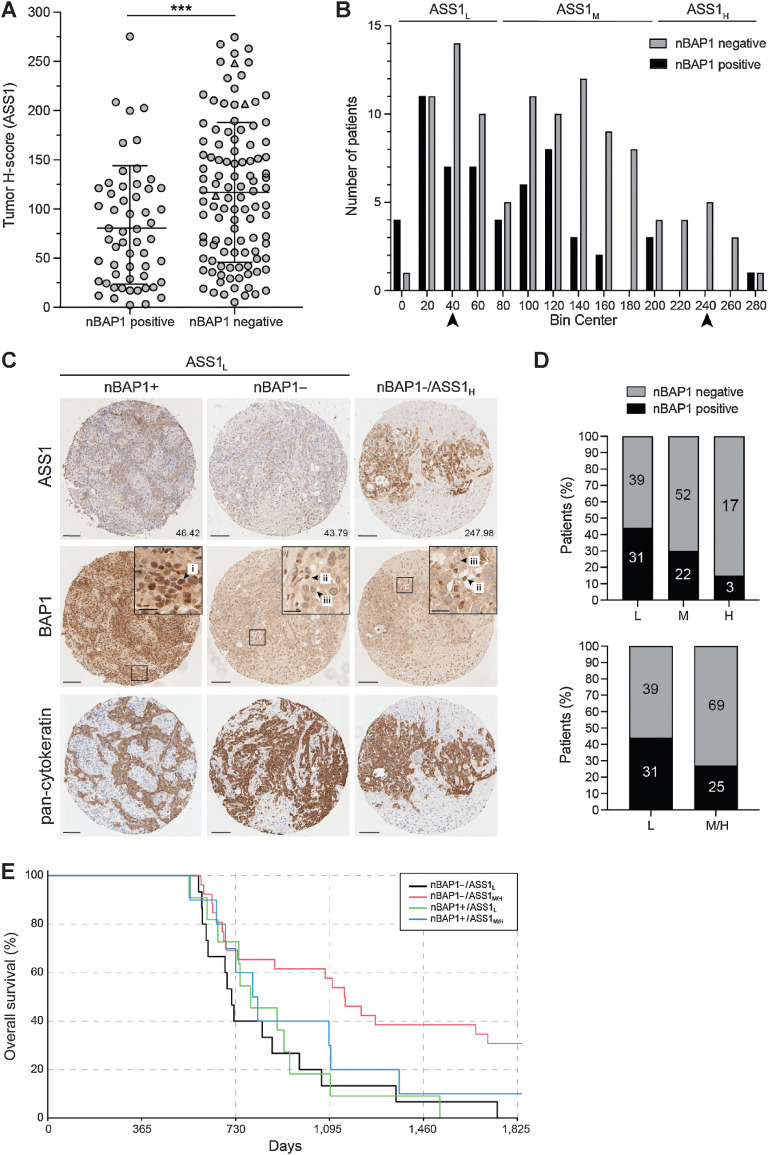
To further explore this relationship, ASS1 protein levels were assessed in MPM tissue samples, which were also stained for nBAP1, as a surrogate for BAP1 mutation and loss of function, as well as pan-cytokeratin as a tumor cell marker (Fig. 5). In line with previous studies (11–13), loss of nBAP1 was most common among patients with epithelioid MPM in our cohort. Given the confounding effects of histologic subtypes for digital pathology and the relative frequency of BAP1 mutations, we focused on the 164 epithelioid cases, 66% of which lacked nBAP1 (Supplementary Table S5). Digital quantification of total cellular ASS1 protein revealed significantly higher ASS1 in tumors without nBAP1 (n = 108) compared with those with nBAP1 (n = 56, Mann–Whitney P = 0.001; Fig. 5A). The tumor H-score distribution defined three groups as ASS1 low (ASS1L), moderate (ASS1M), or high (ASS1H). The proportion of nBAP1-negative cases significantly increased across these groups, so that ASS1H cases were almost exclusively nBAP1-negative (Fig. 5B–D).
Figure 5.
ASS1 protein levels are elevated in epithelioid MPM samples with loss of nBAP1. nBAP1 staining was scored as positive (n = 56) or negative (n = 108) by three independent observers, and mean ASS1 tumor H-scores digitally determined for 164 epithelioid samples (2 to 4 cores per patient). A, ASS1 expression is higher in epithelioid MPM without nBAP1; bars indicate mean and 1 SD; Mann–Whitney test, ***, P = 0.001. B, Frequency distribution of ASS1 tumor H-scores defines three groups: ASS1L (low ASS1, tumor H-score <80); ASS1M (moderate ASS1, tumor H-score ≥80–200); ASS1H (high ASS1, tumor H-score ≥200). C, Images of MPM cores representing the mid-point of the ASS1L and ASS1H groups (indicated by arrow heads in B). The tumor H-score for ASS1 is shown; magnified insets indicate (i) nBAP1-positive tumor cells, (ii) nBAP1-positive stromal cells, (iii) nBAP1-negative tumor cells. Scale bars, 100 or 20 μm in insets. D, Loss of nBAP1 is more frequent in groups with higher ASS1 expression. Comparison of the three ASS1 groups χ2 test, ***, P < 0.0001 (top); and ASS1 L group compared to the combined ASS1M/H groups, Fisher exact test, *, P = 0.018 (below). The numbers of nBAP1-positive or nBAP1-negative patients in each group are indicated. E, Improved survival of patients with nBAP1-negative ASS1M/H tumors. Kaplan–Meier curve is shown at 18-month landmark. Cox model: nBAP1-negative ASS1M/H (n = 26) versus ASS1L (n = 15) P = 0.003; nBAP1-positive ASS1M/H (n = 10) versus ASS1L (n = 11) P = 0.383; supporting data in Supplementary Table S5 and Supplementary Fig. S13.
While there were no statistically significant differences in overall survival (OS) between nBAP1-positive and -negative groups, or based on ASS1 H-score, we observed a late separation in the curves (Supplementary Fig. S13A–S13D). Therefore, further landmark analyses were performed at 12, 18, and 24 months, where differences in survival with ASS1 H-score were evident (Supplementary Table S5; Supplementary Fig. S13E–S13G) consistent with previous reports (6). Notably however, the best outcomes were seen for BAP1-negative ASS1M/H cases (Fig. 5E; Supplementary Table S5). This was a small subgroup of patients with a very late separation in the curves and does not suggest a clinically significant difference in outcome. However, the cosegregation of the biomarkers suggests a potentially distinct biology for this subgroup of MPM.
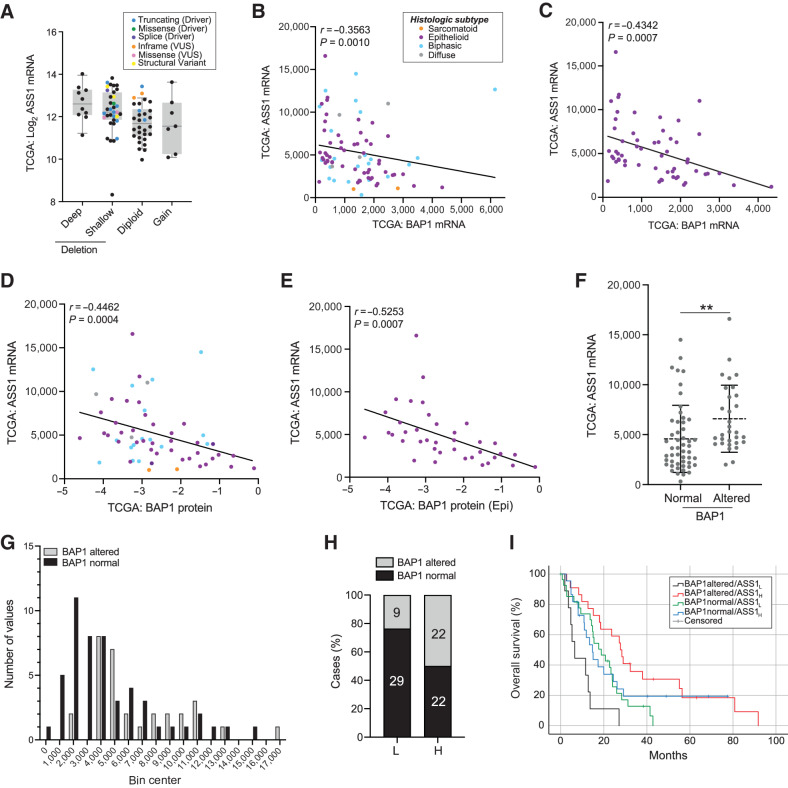
To explore whether the relationship between ASS1 and BAP1 was a general feature of cancer, we examined TCGA pan-cancer datasets. Although most significant in MPM, inverse correlation was evident in the other cancers with common BAP1 inactivation (cholangiocarcinoma, uveal melanoma, and RCCC) but relatively uncommon among other cancers (Supplementary Fig. S14), suggesting cell context specificity. As ASS1 transcript inversely correlated with BAP1 genetic alteration, transcript expression, and protein levels in TCGA MESO dataset, particularly among the epithelioid subtype (Fig. 6A–E), we used this independent cohort to confirm findings from our IHC study. On stratification by BAP1 status, ASS1 transcript was significantly higher in BAP1-altered tumors (Mann–Whitney, P = 0.0016; Fig. 6F), while the distribution of ASS1 expression was similarly shifted for BAP1-altered cases in both cohorts (Figs. 5B and 6G). Further stratification of TCGA MESO cohort by ASS1 Z-scores showed marked enrichment of BAP1-normal cases within the ASS1-low group (Fisher exact test P = 0.0002; Fig. 6H). Notably, the BAP1-altered ASS1-low cases had the worst outcomes in TCGA cohort, with longer survival for BAP1-altered ASS1-high cases (Fig. 6I) similar to our IHC study (Fig. 5E).
Figure 6.
Association between BAP1 status, ASS1 transcript expression and survival in TCGA MESO cohort. A,ASS1 transcript levels are higher in cases with deletion and/or mutation of BAP1. Putative BAP1 copy-number variations from GISTIC; ASS1 mRNA expression log2 batch normalized RSEM, colored by BAP1 mutation. B–E, Inverse correlation of ASS1 mRNA (batch normalized RSEM) with BAP1 colored by histologic type as indicated in B. BAP1 mRNA (for the cohort, n = 82 (B) or only the epithelioid cases, n = 58 (C), and BAP1 protein (RPPA) for the cohort, n = 59 (D) or only the epithelioid cases, n = 38 (E); Spearman correlations shown. F,ASS1 mRNA is higher in BAP1-altered cases (deletion and/or mutation); n = 82, bars indicate mean and 1 SD; Mann–Whitney test, **, P = 0.0016. G, Frequency distribution of ASS1 mRNA stratified by BAP1 status. H,BAP1 alteration is more frequent in tumors with higher ASS1 expression. ASS1 transcript stratified by z-scores: below zero (ASS1-low) or above zero (ASS1-high); Fisher exact test, ****, P = 0.0002; the numbers of BAP1-normal or BAP1-altered patients in each group are indicated. I, Improved survival of patients with BAP1-altered ASS1-high tumors. Kaplan–Meier analysis for 69 uncensored patients according to groups in H. Median survival (lower and upper bound): BAP1-altered ASS1-high 21.6–33.9 months (n = 19), BAP1-altered ASS1-low 2.7–10.2 months (n = 9), BAP1-normal ASS1-high 10.1–19.3 months (n = 17), BAP1-normal ASS1-low 11.8–25.8 months (n = 24); Log-rank (Mantel–Cox) P = 0.001. The results shown in this figure are based upon data generated by TCGA Research Network: https://www.cancer.gov/tcga.
BAP1 IHC is robust, binary, and increasingly adopted into routine clinical pathology, facilitating its use to inform patient care (13). This presents an opportunity to use BAP1 IHC to help stratify patients for therapeutic approaches that target ASS1 or downstream metabolic pathways, or that cause arginine deprivation. In other cancers, high ASS1 is associated with poor prognosis and increased purine synthesis, potentially sensitizing cells to mizoribine (45). However, in MPM, we find high ASS1 predominantly associated with nBAP1 loss and better prognosis (Fig. 5). Indeed, BAP1w-/KO MeT5A cells with elevated ASS1 were more resistant to mizoribine (Supplementary Fig. S15A). In MPM cell lines, the LC50 ranged from 9.7 μmol/L to >1.2 mmol/L (Supplementary Fig. S15B); however, mizoribine sensitivity appeared unrelated to ASS1 expression or BAP1 status, correlating most closely with proliferative capacity (Supplementary Fig. S15C) and suggesting MPM are biologically distinct from other ASS1-high cancers (45).
ASS1 function is blocked by αMDLA which can reduce growth of lung or colorectal cancer cells with high ASS1 (45, 46). Although elevated in BAP1w-/KO MeT5A, the BAP1+/+ counterpart also expresses ASS1, and treatment with αMDLA similarly reduced growth of both cell lines (Supplementary Fig. S16A and S16B). This suggests MeT5A mesothelial cells may be able to utilize extracellular arginine when ASS1 is inhibited. To assess utility of αMDLA in MPM cancer cells, we selected the paired MESO-8T (BAP1-altered, ASS1-moderate) and MESO-12T (BAP1-normal, ASS1-low) cells, and another pair of low passage epithelioid cells that have similar tumor suppressor status (lacking CDKN2A but not NF2, Supplementary Fig. S10) and mizoribine sensitivity (Supplementary Fig. S15): MESO-29T (BAP1-normal, ASS1-undetectable) and MESO-23T (BAP1-altered, ASS1-moderate). The BAP1-altered MPM cell lines appeared moderately more sensitive to αMDLA when grown in full media. A significant reduction in cell confluency was visible after treatment with 10 mmol/L αMDLA, a dose commonly used in the literature, although an ATP-dependent viability assay appeared less reliable in this context (Supplementary Fig. S16C–S16F).
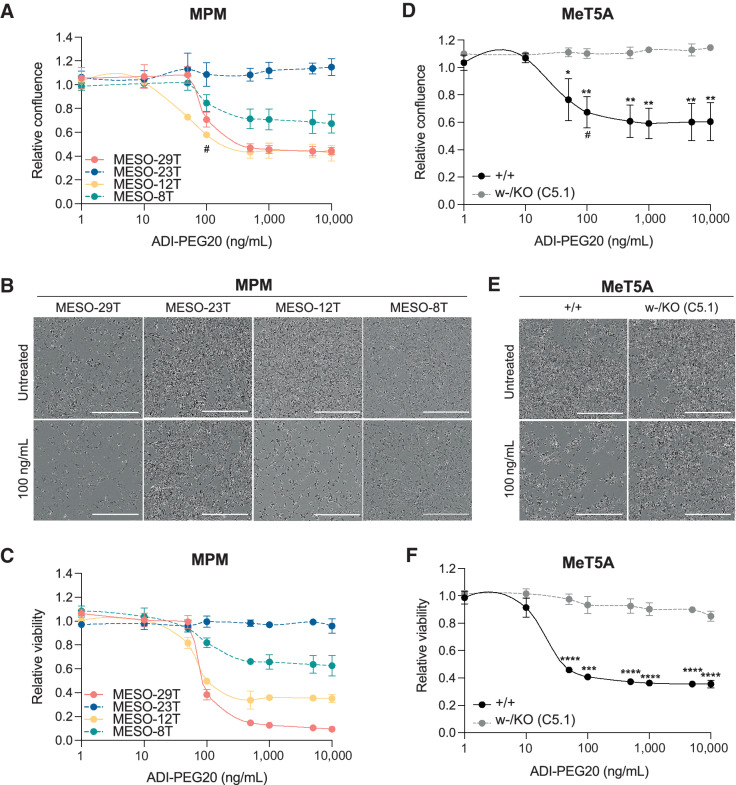
In an inverse approach, arginine deprivation by ADI-PEG20 is undergoing clinical trials in ASS1-deficient cancers that depend upon uptake of exogenous arginine, including nonepithelioid MPM (6). As expected, the BAP1 normal cell lines MESO-29T and MESO-12T were sensitive, while BAP1-altered cell lines MESO-23T and MESO-8T were more resistant (Fig. 7A–C). Overall, the MPM cell lines showed inverse sensitivity to αMDLA and ADI-PEG20, that related to their ASS1 expression and BAP1 status (Supplementary Fig. S16F). To investigate whether BAP1 mutation influenced ADI-PEG20 sensitivity, we compared dose responses in the isogenic MeT5A cells (Fig. 7D–F). BAP1+/+ MeT5A were sensitive to ADI-PEG20 with a similar LC50 to MESO-29T, while strikingly BAP1w-/KO MeT5A were resistant to ADI-PEG20, phenocopying MESO-23T. Thus, BAP1 mutation is sufficient to induce resistance, suggesting that BAP1 status may be driving the survival benefit previously reported in patients with epithelioid MPM treated with ADI-PEG20 (6).
Figure 7.
The influence of BAP1 status on response to arginine deprivation with ADI-PEG20. Paired MPM cell lines MESO-29T (BAP1-normal/ASS1-low) and MESO-23T (BAP1-altered/ASS1-high), MESO-12T (BAP1-normal/ASS1-low) and MESO-8T (BAP1-altered/ASS1-high; A–C) or MeT5A BAP1+/+ and BAP1w-/KO cells (D–F) were treated for 96 hours with ADI-PEG20; mean of three independent experiments, error bars SD; t test; *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001. Dose response for relative confluence assessed by live imaging at 96 hours (A and D), # indicates 100 ng/mL ADI-PEG20, the approximate LC50 for sensitive cells, for which representative images at 96 hours are shown (B and E). ATP-based luciferase assay conducted following imaging at 96 hours (C and F). Summary data, Supplementary Fig. S16F.
Discussion
BAP1 loss of function is an early event in MPM tumorigenesis and defines a molecular subtype of MPM with likely distinct drug sensitivities. A number of previous studies, mainly utilizing MPM cell lines with differing BAP1 status, have suggested specific sensitivities, for example targeting TNF-related apoptosis inducing ligand (TRAIL; ref. 28), FGFR (47), histone deacetylases (HDAC; ref. 32), EZH2 (48) or more controversially PARP (29, 49). However, to date relevant agents have typically shown either poor efficacy or limited association with BAP1 status in clinical trials. For example, a recent trial of the PARP inhibitor rucaparib reported benefits for some patients with MPM, but this was not predicted by BAP1 status (31). While the reasons remain opaque, this is likely in part due to heterogeneity of cell lines with the presence of confounding mutations and adaptations. We therefore sought to develop an isogenic model on a mesothelial background to remove as many confounding factors as possible. We knocked in a mutation that predisposes to familial mesothelioma (15) and asbestos-induced tumors in mice (24), combined with knockout of the other BAP1 allele. MeT5A-BAP1w-/KO have stably reduced BAP1 expression and, given that BAP1 mutation is an early truncal change (8), represent a more relevant model to study tumor biology and drug sensitivity than modulation of BAP1 in established cancer cell lines that already harbor a spectrum of tumorigenic mutations.
We profiled MeT5A-BAP1w-/KO cells by SILAC-MS to investigate alternative BAP1 dependencies that may confer therapeutic sensitivity. No protein expression changes were evident amongst epigenetic regulators or DNA repair processes, instead cytoskeletal organization and metabolic processes were significantly enriched. Similar pathways were identified by proteomics following reintroduction of BAP1 into BAP1-null NCI-H226 lung cancer cells (27); however, the specific proteins and cellular responses differ, reinforcing the context-specific nature of BAP1 function.
Among the enriched metabolic pathways, downregulation of glycolysis enzymes and upregulation of TCA cycle enzymes were evident in MeT5A-BAP1w-/KO. However, for most of these proteins, their levels were not closely linked to BAP1 status in MPM cell lines, suggesting potential convergent evolution during tumorigenesis. Interestingly, glycolytic enzymes were mostly reduced compared with wild-type MeT5A in both MeT5A-BAP1w-/KO and the MPM cell lines. While functional analysis would be required to draw firm conclusions, this appears to be at odds with the Warburg effect, where cancer cells rely more heavily on glycolysis for energy production. Germline BAP1 mutation was previously shown to induce a Warburg effect in fibroblasts from affected BAP1+/− individuals (50), but in contrast an in vivo proteomic study of Neucode-labeled Bap1 knockout mice revealed metabolic changes that included decreased glycolysis/gluconeogenesis (51). Interestingly, OGT, which posttranslationally modifies proteins by addition of N-Acetylglucosamine (GlcNAc), was downregulated in MeT5A-BAP1w-/KO cells. OGT was previously identified as an interactor and substrate of BAP1 (52) that by modifying PGC-1α with GlcNAc, facilitates its stabilization by BAP1 to promote gluconeogenesis (53). Thus, BAP1 can interplay with glycolysis/gluconeogenesis with context-specific consequences.
The most notable novel BAP1 dependency identified in our study was ASS1, an enzyme essential for cellular synthesis of arginine. MeT5A-BAP1w-/KO cells had elevated ASS1 mRNA and protein, which was recapitulated in BAP1-altered patient-derived MPM cell lines and tissues samples. In all cases, this appears to be a transcriptional response to BAP1 loss, implying that BAP1 normally represses ASS1 transcription, either directly or indirectly. While we have not formally demonstrated higher ASS1 enzymatic activity, it is generally accepted that increased transcription of ASS1 is indirect evidence for the cellular requirement to generate argininosuccinate endogenously. BAP1 operates as a transcriptional activator through its role in deubiquitylating H2A (18), so likely regulates ASS1 through an alternative mechanism. Although defining this is beyond the scope of the current study, we found no evidence for BAP1 recruitment to the ASS1 gene promoter in publicly available chromatin immunoprecipitation sequencing data, and one possibility is that BAP1 interplays with the methylation of CpG islands that restrict ASS1 expression (6).
ASS1 is often silenced in cancers, including MPM, conferring reliance upon extracellular arginine, and hence susceptibility to arginine deprivation using ADI-PEG20. However, elevated ASS1 mRNA and protein have also been reported in MPM three-dimensional spheroid models and patient samples (54). We confirm that ASS1 can be highly expressed in MPM and show for the first time that this is often associated with BAP1 loss. This elevated ASS1 expression was associated with resistance to ADI-PEG20, an indirect measure of ASS1 enzymatic activity. In some cancers, high ASS1 is associated with poor prognosis (45); however, like BAP1 loss (22), elevated ASS1 is associated with better OS in patients with MPM (6). This was recapitulated in our study among the patients surviving beyond the 18-month landmark and was significantly associated with nBAP1-negative status. Inhibition of ASS1 can reduce proliferation of breast or colorectal cancer (45, 46) and moderately affected BAP1-altered MPM cells expressing ASS1, consistent with the idea that ASS1 can support MPM cell survival (54). In breast cancer, high ASS1 is associated with increased purine synthesis, potentially sensitizing cells to mizoribine (45), an immunosuppressive drug used in renal transplantation; however, in MPM we found no link between mizoribine sensitivity and either ASS1 or BAP1 status. ASS1 inhibition with αMDLA in the presence of extracellular arginine, was not influenced by BAP1 mutation in MeT5A cells. However, BAP1-altered MPM that are more resistant to arginine deprivation with ADI-PEG20, were more sensitive to αMDLA. Thus, these cancer cells may be less able to switch to using extracellular arginine, or have higher arginine demand, than MeT5A cells. Further analysis will be required to dissect out other potential contributory factors, such as extracellular arginine concentration or genetic background, to assess whether ASS1 inhibition could be a useful therapeutic approach in BAP1-altered MPM.
While the ADAM and TRAP trials showed promise for ADI-PEG20 in ASS1-deficient epithelioid and nonepithelioid MPM, respectively (6, 55, 56), subsequent clinical development of ADI-PEG20 has focused on patients with nonepithelioid MPM, as high-frequency ASS1 loss in this group avoids the need for screening in patients, reflecting the lack of a validated companion IHC diagnostic for ASS1. The recently announced results of the randomized phase II/III ATOMIC-Meso trial validated this strategy with a 1.64-month median OS benefit [HR = 0.71; 95% confidence interval (CI), 0.55–0.93] in nonepithelioid MPM with combined ADI-PEG20 and cisplatin/pemetrexed compared with standard chemotherapy; supporting imminent regulatory submission (https://polarispharma.com/2022/09/21/20220921001/?lang=en). Meanwhile, the CheckMate-743 study, which initiated concurrently with ATOMIC-Meso and assessed ipilimumab/nivolumab versus standard chemotherapy, reported earlier and led to adoption of ipilimumab/nivolumab as a first-line standard of care (5). Notably, the study showed a striking 9.3-month improvement in OS in the nonepithelioid group (HR = 0.46; 95% CI, 0.31–0.68), while in contrast, the absolute benefit of combination immunotherapy for epithelioid disease was substantially lower at 2.7 months (HR = 0.86; 95% CI, 0.69–1.08), highlighting the need for further treatment options in this group, and supporting reconsideration of ADI-PEG20.
We found that the association between BAP1 status and ASS1 expression was most notable in epithelioid MPM and, as loss of BAP1 expression is associated with resistance to ADI-PEG20, propose that as a validated tool in MPM histopathology, BAP1 IHC will be useful in stratifying epithelioid MPM for ADI-PEG20 treatment. While the benefit of therapeutic stratification by BAP1 status has yet to be realized, our study provides a new avenue for exploration. Further bridging preclinical investigation, beyond the scope of this study, is planned to explore this hypothesis and may reveal additional molecular modulators to further refine predictive models.
Supplementary Material
Supplementary Methods, References and List of Figures and Tables
Fig. S1. SNP6.0 analysis of parental MeT5A-BAP1+/+ cells.Fig. S2. Expression of catalytically active BAP1 in parental MeT5A.Fig. S3. Characterisation of BAP1 expression in gene-edited MeT5A.Fig. S4. Whole genome sequencing of gene-edited MeT5A.Fig. S5. Proliferative profiles for gene-edited MeT5A.Fig. S6. Enriched GO terms in gene-edited MeT5A SILAC-MS.Fig. S7. Enriched KEGG pathways in gene-edited MeT5A SILAC-MS include EMT.Fig. S8. Metabolite responses to BAP1 mutation in isogenic MeT5A.Fig. S9. Immunoblotting for differentially expressed metabolic enzymes identified by SILAC-MS in isogenic MeT5A cell lines.Fig. S10. Characterisation of BAP1-status for MPM cell panel.Fig. S11. Evaluating correlation between BAP1 and selected metabolic enzymes in a panel of MPM cell lines and the TCGA MESO pan-cancer dataset.Fig. S12. Validation of ASS1 response to BAP1 alteration.Fig. S13. Improved prognosis for epithelioid MPM patients with loss of nBAP1 and increased expression of ASS1.Fig. S14. Relationship between BAP1 and ASS1 transcripts in the TCGA Pan-Cancer datasets for other cancer types.Fig. S15. The influence of BAP1-status on response to inhibition of purine metabolism.Fig. S16. The influence of BAP1-status on response to ASS1 inhibition.
STR profiling MPM cell lines
WGS MeT5A isogenics
Proteomics MeT5A isogenics and MPM cell lines
Metabolomics MeT5A isogenics
Patient scoring and survival summary
Acknowledgments
This work was funded by the British Lung Foundation (now Asthma + Lung UK) grant references APG12-9 (J. Kenyani) and MPG16-5 (S.E. Barnett, F. Querques), Wellcome grant reference 102382/Z/13/Z (Z. Butt), NC3R grant reference NC/T001631/1 (L. Shaw), Fight for Sight grant reference 5109/5119 (L.C. Silva), and North West Cancer Research PhD studentship (M. Tripari). This research was funded in whole, or in part, by the Wellcome Trust (102382/Z/13/Z). For the purpose of Open Access, the author has applied a CC BY public copyright license to any Author Accepted Manuscript version arising from this submission.
The authors wish to thank the following for their valuable advice and experimental training: Dr Simon Oliver, Dr Rob Howes and Horizon Discovery (genome-editing), Dr Emma Rusilowicz (mass spectrometry), Dr Kathy Till (flow cytometry), Dr Helen Kalirai (immunohistochemistry) and Dr Anthony Evans (QuPath). Sean Donahue and Joshua Charkin assisted with Ub-VME assays and ASS1 knockdown, respectively. Polaris Pharmaceuticals provided ADI-PEG20 for this research. We acknowledge use of the Centre for Cell Imaging (CCI) provided by Liverpool Shared Research Facilities, Faculty of Health and Life Sciences, University of Liverpool.
The publication costs of this article were defrayed in part by the payment of publication fees. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
This article is featured in Highlights of This Issue, p. 387
Footnotes
Note: Supplementary data for this article are available at Molecular Cancer Research Online (http://mcr.aacrjournals.org/).
Authors' Disclosures
S.E. Barnett reports grants from British Lung Foundation and NC3Rs during the conduct of the study. J. Kenyani reports grants from British Lung Foundation during the conduct of the study. M. Tripari reports grants from North West Cancer Research during the conduct of the study. Z. Butt reports grants from Wellcome Trust during the conduct of the study. F. Querques reports grants from British Lung Foundation during the conduct of the study. L. Shaw reports grants from NC3Rs during the conduct of the study. L.C. Silva reports grants from Fight For Sight during the conduct of the study. P.W. Szlosarek reports grants from Polaris Pharmaceuticals during the conduct of the study. J.J. Sacco reports grants from British Lung Foundation (now Asthma + Lung UK), National Centre for the Replacement, Refinement and Reduction of Animals in Research (NC3Rs), and North West Cancer Research during the conduct of the study; grants and personal fees from Immunocore, other support from Replimune, non-financial support from MSD, grants and non-financial support from BMS, grants from AstraZeneca, and personal fees from Delcath outside the submitted work. J.M. Coulson reports grants from British Lung Foundation, Wellcome, NC3Rs, Fight for Sight, North West Cancer Research, and non-financial support from Polaris during the conduct of the study. No disclosures were reported by the other authors.
Authors' Contributions
S.E. Barnett: Data curation, formal analysis, investigation, visualization, methodology, writing–review and editing. J. Kenyani: Data curation, formal analysis, investigation, visualization, methodology, writing–review and editing. M. Tripari: Data curation, formal analysis, investigation, visualization, methodology, writing–review and editing. Z. Butt: Formal analysis, investigation, visualization, methodology, writing–review and editing. R. Grosman: Data curation, formal analysis, investigation, visualization, methodology, writing–review and editing. F. Querques: Investigation, methodology. L. Shaw: Investigation. L.C. Silva: Formal analysis, investigation. Z. Goate: Investigation. S.J. Marciniak: Resources, writing–review and editing. D.M. Rassl: Resources, formal analysis, methodology. R. Jackson: Formal analysis, visualization, methodology, writing–review and editing. L.-Y. Lian: Supervision, methodology, writing–review and editing. P.W. Szlosarek: Resources, writing–review and editing. J.J. Sacco: Conceptualization, supervision, funding acquisition, writing–review and editing. J.M. Coulson: Conceptualization, data curation, formal analysis, supervision, funding acquisition, visualization, methodology, writing–original draft, project administration, writing–review and editing.
References
- 1. Takahashi K, Landrigan PJ, Collegium R. The global health dimensions of asbestos and asbestos-related diseases. Ann Glob Health 2016;82:209–13. [DOI] [PubMed] [Google Scholar]
- 2. Chernova T, Murphy FA, Galavotti S, Sun XM, Powley IR, Grosso S, et al. Long-fiber carbon nanotubes replicate asbestos-induced mesothelioma with disruption of the tumor suppressor gene Cdkn2a (Ink4a/Arf). Curr Biol 2017;27:3302–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Blum Y, Meiller C, Quetel L, Elarouci N, Ayadi M, Tashtanbaeva D, et al. Dissecting heterogeneity in malignant pleural mesothelioma through histo-molecular gradients for clinical applications. Nat Commun 2019;10:1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Zalcman G, Mazieres J, Margery J, Greillier L, Audigier-Valette C, Moro-Sibilot D, et al. Bevacizumab for newly diagnosed pleural mesothelioma in the mesothelioma avastin cisplatin pemetrexed study (MAPS): a randomised, controlled, open-label, phase 3 trial. Lancet 2016;387:1405–14. [DOI] [PubMed] [Google Scholar]
- 5. Baas P, Scherpereel A, Nowak AK, Fujimoto N, Peters S, Tsao AS, et al. First-line nivolumab plus ipilimumab in unresectable malignant pleural mesothelioma (CheckMate 743): a multicentre, randomised, open-label, phase 3 trial. Lancet 2021;397:375–86. [DOI] [PubMed] [Google Scholar]
- 6. Szlosarek PW, Steele JP, Nolan L, Gilligan D, Taylor P, Spicer J, et al. Arginine deprivation with pegylated arginine deiminase in patients with argininosuccinate synthetase 1-deficient malignant pleural mesothelioma: a randomized clinical trial. JAMA Oncol 2017;3:58–66. [DOI] [PubMed] [Google Scholar]
- 7. Zauderer MG, Szlosarek PW, Le Moulec S, Popat S, Taylor P, Planchard D, et al. EZH2 inhibitor tazemetostat in patients with relapsed or refractory, BAP1-inactivated malignant pleural mesothelioma: a multicentre, open-label, phase 2 study. Lancet Oncol 2022;23:758–67. [DOI] [PubMed] [Google Scholar]
- 8. Zhang M, Luo JL, Sun Q, Harber J, Dawson AG, Nakas A, et al. Clonal architecture in mesothelioma is prognostic and shapes the tumour microenvironment. Nat Commun 2021;12:1751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Bueno R, Stawiski EW, Goldstein LD, Durinck S, De Rienzo A, Modrusan Z, et al. Comprehensive genomic analysis of malignant pleural mesothelioma identifies recurrent mutations, gene fusions and splicing alterations. Nat Genet 2016;48:407–16. [DOI] [PubMed] [Google Scholar]
- 10. Hmeljak J, Sanchez-Vega F, Hoadley KA, Shih J, Stewart C, Heiman D, et al. Integrative molecular characterization of malignant pleural mesothelioma. Cancer Discov 2018;8:1548–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Forest F, Patoir A, Dal Col P, Sulaiman A, Camy F, Laville D, et al. Nuclear grading, BAP1, mesothelin and PD-L1 expression in malignant pleural mesothelioma: prognostic implications. Pathology 2018;50:635–41. [DOI] [PubMed] [Google Scholar]
- 12. McGregor SM, Dunning R, Hyjek E, Vigneswaran W, Husain AN, Krausz T. BAP1 facilitates diagnostic objectivity, classification, and prognostication in malignant pleural mesothelioma. Hum Pathol 2015;46:1670–8. [DOI] [PubMed] [Google Scholar]
- 13. Carbone M, Harbour JW, Brugarolas J, Bononi A, Pagano I, Dey A, et al. Biological mechanisms and clinical significance of BAP1 mutations in human cancer. Cancer Discov 2020;10:1103–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Bott M, Brevet M, Taylor BS, Shimizu S, Ito T, Wang L, et al. The nuclear deubiquitinase BAP1 is commonly inactivated by somatic mutations and 3p21.1 losses in malignant pleural mesothelioma. Nat Genet 2011;43:668–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Testa JR, Cheung M, Pei J, Below JE, Tan Y, Sementino E, et al. Germline BAP1 mutations predispose to malignant mesothelioma. Nat Genet 2011;43:1022–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Jensen DE, Proctor M, Marquis ST, Gardner HP, Ha SI, Chodosh LA, et al. BAP1: a novel ubiquitin hydrolase which binds to the BRCA1 RING finger and enhances BRCA1-mediated cell growth suppression. Oncogene 1998;16:1097–112. [DOI] [PubMed] [Google Scholar]
- 17. Hart T, Chandrashekhar M, Aregger M, Steinhart Z, Brown KR, MacLeod G, et al. High-resolution CRISPR screens reveal fitness genes and genotype-specific cancer liabilities. Cell 2015;163:1515–26. [DOI] [PubMed] [Google Scholar]
- 18. He M, Chaurushiya MS, Webster JD, Kummerfeld S, Reja R, Chaudhuri S, et al. Intrinsic apoptosis shapes the tumor spectrum linked to inactivation of the deubiquitinase BAP1. Science 2019;364:283–5. [DOI] [PubMed] [Google Scholar]
- 19. Liu X, Kumar M, Yang L, Molkentine DP, Valdecanas D, Yu S, et al. BAP1 Is a novel target in HPV-negative head and neck cancer. Clin Cancer Res 2018;24:600–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Qin J, Zhou Z, Chen W, Wang C, Zhang H, Ge G, et al. BAP1 promotes breast cancer cell proliferation and metastasis by deubiquitinating KLF5. Nat Commun 2015;6:8471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Szalai E, Wells JR, Ward L, Grossniklaus HE. Uveal melanoma nuclear BRCA1-associated protein-1 immunoreactivity is an indicator of metastasis. Ophthalmology 2018;125:203–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Luchini C, Veronese N, Yachida S, Cheng L, Nottegar A, Stubbs B, et al. Different prognostic roles of tumor suppressor gene BAP1 in cancer: a systematic review with meta-analysis. Genes Chromosomes Cancer 2016;55:741–9. [DOI] [PubMed] [Google Scholar]
- 23. Napolitano A, Pellegrini L, Dey A, Larson D, Tanji M, Flores EG, et al. Minimal asbestos exposure in germline BAP1 heterozygous mice is associated with deregulated inflammatory response and increased risk of mesothelioma. Oncogene 2016;35:1996–2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kadariya Y, Cheung M, Xu J, Pei J, Sementino E, Menges CW, et al. Bap1 is a bona fide tumor suppressor: genetic evidence from mouse models carrying heterozygous germline Bap1 mutations. Cancer Res 2016;76:2836–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Xu J, Kadariya Y, Cheung M, Pei J, Talarchek J, Sementino E, et al. Germline mutation of Bap1 accelerates development of asbestos-induced malignant mesothelioma. Cancer Res 2014;74:4388–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Badhai J, Pandey GK, Song JY, Krijgsman O, Bhaskaran R, Chandrasekaran G, et al. Combined deletion of Bap1, Nf2, and Cdkn2ab causes rapid onset of malignant mesothelioma in mice. J Exp Med 2020;217:e20191257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hebert L, Bellanger D, Guillas C, Campagne A, Dingli F, Loew D, et al. Modulating BAP1 expression affects ROS homeostasis, cell motility and mitochondrial function. Oncotarget 2017;8:72513–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kolluri KK, Alifrangis C, Kumar N, Ishii Y, Price S, Michaut M, et al. Loss of functional BAP1 augments sensitivity to TRAIL in cancer cells. Elife 2018;7:e30224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Rathkey D, Khanal M, Murai J, Zhang J, Sengupta M, Jiang Q, et al. Sensitivity of mesothelioma cells to PARP inhibitors is not dependent on BAP1 but is enhanced by temozolomide in cells with high-schlafen 11 and low-O6-methylguanine-DNA methyltransferase expression. J Thorac Oncol 2020;15:843–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Masclef L, Ahmed O, Estavoyer B, Larrivee B, Labrecque N, Nijnik A, et al. Roles and mechanisms of BAP1 deubiquitinase in tumor suppression. Cell Death Differ 2021;28:606–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Fennell DA, King A, Mohammed S, Branson A, Brookes C, Darlison L, et al. Rucaparib in patients with BAP1-deficient or BRCA1-deficient mesothelioma (MiST1): an open-label, single-arm, phase 2a clinical trial. Lancet Respir Med 2021;9:593–600. [DOI] [PubMed] [Google Scholar]
- 32. Sacco JJ, Kenyani J, Butt Z, Carter R, Chew HY, Cheeseman LP, et al. Loss of the deubiquitylase BAP1 alters class I histone deacetylase expression and sensitivity of mesothelioma cells to HDAC inhibitors. Oncotarget 2015;6:13757–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Rintoul RC, Rassl DM, Gittins J, Marciniak SJ, MesobanK collaborators. MesobanK UK: an international mesothelioma bioresource. Thorax 2016;71:380–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Chernova T, Sun XM, Powley IR, Galavotti S, Grosso S, Murphy FA, et al. Molecular profiling reveals primary mesothelioma cell lines recapitulate human disease. Cell Death Differ 2016;23:1152–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Al-Taei S, Salimu J, Lester JF, Linnane S, Goonewardena M, Harrop R, et al. Overexpression and potential targeting of the oncofoetal antigen 5T4 in malignant pleural mesothelioma. Lung Cancer 2012;77:312–8. [DOI] [PubMed] [Google Scholar]
- 36. Fielding AB, Concannon M, Darling S, Rusilowicz-Jones EV, Sacco JJ, Prior IA, et al. The deubiquitylase USP15 regulates topoisomerase II alpha to maintain genome integrity. Oncogene 2018;37:2326–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Hammond DE, Mageean CJ, Rusilowicz EV, Wickenden JA, Clague MJ, Prior IA. Differential reprogramming of isogenic colorectal cancer cells by distinct activating KRAS mutations. J Proteome Res 2015;14:1535–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Bankhead P, Loughrey MB, Fernandez JA, Dombrowski Y, McArt DG, Dunne PD, et al. QuPath: open source software for digital pathology image analysis. Sci Rep 2017;7:16878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Wang J, Vasaikar S, Shi Z, Greer M, Zhang B, WebGestalt 2017: a more comprehensive, powerful, flexible and interactive gene set enrichment analysis toolkit. Nucleic Acids Res 2017;45:W130–W7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Liberzon A, Birger C, Thorvaldsdottir H, Ghandi M, Mesirov JP, Tamayo P. The molecular signatures database (MSigDB) hallmark gene set collection. Cell Syst 2015;1:417–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov 2012;2:401–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Ke Y, Reddel RR, Gerwin BI, Reddel HK, Somers AN, McMenamin MG, et al. Establishment of a human in vitro mesothelial cell model system for investigating mechanisms of asbestos-induced mesothelioma. Am J Pathol 1989;134:979–91. [PMC free article] [PubMed] [Google Scholar]
- 43. Tsim S, Alexander L, Kelly C, Shaw A, Hinsley S, Clark S, et al. Serum proteomics and plasma fibulin-3 in differentiation of mesothelioma from asbestos-exposed controls and patients with other pleural diseases. J Thorac Oncol 2021;16:1705–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Adhikary G, Grun D, Alexander HR, Friedberg JS, Xu W, Keillor JW, et al. Transglutaminase is a mesothelioma cancer stem cell survival protein that is required for tumor formation. Oncotarget 2018;9:34495–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Keshet R, Lee JS, Adler L, Iraqi M, Ariav Y, Lim LQJ, et al. Targeting purine synthesis in ASS1-expressing tumors enhances the response to immune checkpoint inhibitors. Nat Cancer 2020;1:894–908. [DOI] [PubMed] [Google Scholar]
- 46. Bateman LA, Ku WM, Heslin MJ, Contreras CM, Skibola CF, Nomura DK. Argininosuccinate synthase 1 is a metabolic regulator of colorectal cancer pathogenicity. ACS Chem Biol 2017;12:905–11. [DOI] [PubMed] [Google Scholar]
- 47. Quispel-Janssen JM, Badhai J, Schunselaar L, Price S, Brammeld J, Iorio F, et al. Comprehensive pharmacogenomic profiling of malignant pleural mesothelioma identifies a subgroup sensitive to FGFR inhibition. Clin Cancer Res 2018;24:84–94. [DOI] [PubMed] [Google Scholar]
- 48. LaFave LM, Beguelin W, Koche R, Teater M, Spitzer B, Chramiec A, et al. Loss of BAP1 function leads to EZH2-dependent transformation. Nat Med 2015;21:1344–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Yu H, Pak H, Hammond-Martel I, Ghram M, Rodrigue A, Daou S, et al. Tumor suppressor and deubiquitinase BAP1 promotes DNA double-strand break repair. Proc Natl Acad Sci U S A 2014;111:285–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Bononi A, Yang H, Giorgi C, Patergnani S, Pellegrini L, Su M, et al. Germline BAP1 mutations induce a Warburg effect. Cell Death Differ 2017;24:1694–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Baughman JM, Rose CM, Kolumam G, Webster JD, Wilkerson EM, Merrill AE, et al. NeuCode proteomics reveals bap1 regulation of metabolism. Cell Rep 2016;16:583–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Dey A, Seshasayee D, Noubade R, French DM, Liu J, Chaurushiya MS, et al. Loss of the tumor suppressor BAP1 causes myeloid transformation. Science 2012;337:1541–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Ruan HB, Han X, Li MD, Singh JP, Qian K, Azarhoush S, et al. O-GlcNAc transferase/host cell factor C1 complex regulates gluconeogenesis by modulating PGC-1alpha stability. Cell Metab 2012;16:226–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Barbone D, Van Dam L, Follo C, Jithesh PV, Zhang SD, Richards WG, et al. Analysis of gene expression in 3D spheroids highlights a survival role for ASS1 in mesothelioma. PLoS One 2016;11:e0150044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Beddowes E, Spicer J, Chan PY, Khadeir R, Corbacho JG, Repana D, et al. Phase 1 dose-escalation study of pegylated arginine deiminase, cisplatin, and pemetrexed in patients with argininosuccinate synthetase 1-deficient thoracic cancers. J Clin Oncol 2017;35:1778–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Szlosarek PW, Baas P, Ceresoli GL, Fennell DA, Gilligan D, Johnston A, et al. ATOMIC-Meso: A randomized phase 2/3 trial of ADI-PEG20 or placebo with pemetrexed and cisplatin in patients with argininosuccinate synthetase 1-deficient non-epithelioid mesothelioma. J Clin Oncol 35:15s, 2017. (suppl; abstr TPS8582). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Methods, References and List of Figures and Tables
Fig. S1. SNP6.0 analysis of parental MeT5A-BAP1+/+ cells.Fig. S2. Expression of catalytically active BAP1 in parental MeT5A.Fig. S3. Characterisation of BAP1 expression in gene-edited MeT5A.Fig. S4. Whole genome sequencing of gene-edited MeT5A.Fig. S5. Proliferative profiles for gene-edited MeT5A.Fig. S6. Enriched GO terms in gene-edited MeT5A SILAC-MS.Fig. S7. Enriched KEGG pathways in gene-edited MeT5A SILAC-MS include EMT.Fig. S8. Metabolite responses to BAP1 mutation in isogenic MeT5A.Fig. S9. Immunoblotting for differentially expressed metabolic enzymes identified by SILAC-MS in isogenic MeT5A cell lines.Fig. S10. Characterisation of BAP1-status for MPM cell panel.Fig. S11. Evaluating correlation between BAP1 and selected metabolic enzymes in a panel of MPM cell lines and the TCGA MESO pan-cancer dataset.Fig. S12. Validation of ASS1 response to BAP1 alteration.Fig. S13. Improved prognosis for epithelioid MPM patients with loss of nBAP1 and increased expression of ASS1.Fig. S14. Relationship between BAP1 and ASS1 transcripts in the TCGA Pan-Cancer datasets for other cancer types.Fig. S15. The influence of BAP1-status on response to inhibition of purine metabolism.Fig. S16. The influence of BAP1-status on response to ASS1 inhibition.
STR profiling MPM cell lines
WGS MeT5A isogenics
Proteomics MeT5A isogenics and MPM cell lines
Metabolomics MeT5A isogenics
Patient scoring and survival summary
Data Availability Statement
The data generated in this study are available within the article and its Supplementary Data. In this study, there are also analyses based upon datasets generated by TCGA Research Network (https://www.cancer.gov/tcga; RRID:SCR_003193).