Whole-genome sequencing of Hodgkin lymphoma identifies driver events and reconstructs their acquisition timing during B-cell ontogenesis and oncogenesis.
Abstract
The rarity of malignant Hodgkin and Reed Sternberg (HRS) cells in classic Hodgkin lymphoma (cHL) limits the ability to study the genomics of cHL. To circumvent this, our group has previously optimized fluorescence-activated cell sorting to purify HRS cells. Using this approach, we now report the whole-genome sequencing landscape of HRS cells and reconstruct the chronology and likely etiology of pathogenic events leading to cHL. We identified alterations in driver genes not previously described in cHL, APOBEC mutational activity, and the presence of complex structural variants including chromothripsis. We found that high ploidy in cHL is often acquired through multiple, independent chromosomal gains events including whole-genome duplication. Evolutionary timing analyses revealed that structural variants enriched for RAG motifs, driver mutations in B2M, BCL7A, GNA13, and PTPN1, and the onset of AID-driven mutagenesis usually preceded large chromosomal gains. This study provides a temporal reconstruction of cHL pathogenesis.
Significance:
Previous studies in cHL were limited to coding sequences and therefore not able to comprehensively decipher the tumor complexity. Here, leveraging cHL whole-genome characterization, we identify driver events and reconstruct the tumor evolution, finding that structural variants, driver mutations, and AID mutagenesis precede chromosomal gains.
This article is highlighted in the In This Issue feature, p. 171
INTRODUCTION
Classic Hodgkin lymphoma (cHL) is characterized by a unique pathologic composition where a small fraction of Hodgkin and Reed Sternberg (HRS) tumor cells (∼1%) are surrounded by an extensive and complex immune and stromal infiltrate (1). The paucity of HRS cells in tumor tissue has precluded the genomic investigation of cHL using standard platforms. Our group has optimized fluorescence-activated cell sorting (FACS) to isolate HRS cells and intratumor B and T cells and perform whole-exome sequencing (WES; ref. 2). Data on HRS exomes from our group and others have revealed mutations in critical driver genes involved in the NF-κB and JAK/STAT signaling pathways as well as genes involved in immune escape (2–9). These investigations, however, were limited to captured coding sequences, and therefore not able to comprehensively decipher the genomic complexity of cHL. Furthermore, the chronologic order in which somatic alterations are acquired is largely unknown in cHL, limiting our understanding of the earliest drivers/initiating events.
Whole-genome sequencing (WGS) has the potential to fully characterize the somatic genomic landscape including: (i) the catalog of coding and noncoding mutations, (ii) large and focal copy-number alterations (CNA), (iii) structural variants (SV) including complex events, and (iv) mutational processes involved in cancer pathogenesis (10). Here we performed WGS on FACS-isolated HRS cells and matched normal tissue from 25 patients with cHL and WES from an additional 36 cases. Combining CNA, single-nucleotide variant (SNV), and SV analyses, we reveal insights into the pathogenesis of cHL, including drivers, genomic mechanisms, and mutational processes, and we provide a comprehensive overview of how these events are acquired over time.
RESULTS
Mutational Burden and Single-Nucleotide Variants in Classic Hodgkin Lymphoma Driver Genes
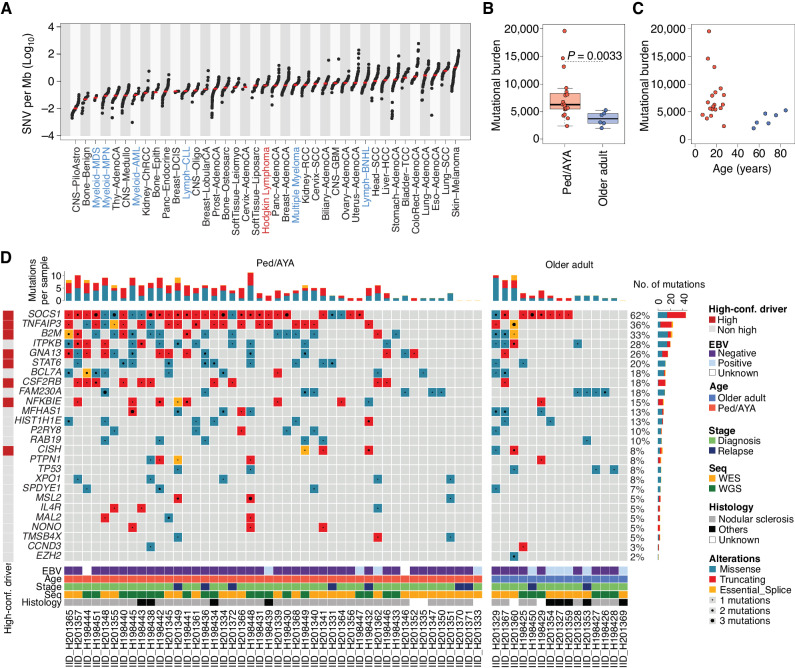
To evaluate the genome of HRS cells, HRS and intratumoral T cells were isolated from cHL biopsies using FACS as previously described (11). Intratumoral T cells from each case were used as the germline control. We interrogated WGS from 25 cases of cHL, including tumors from pediatric and adult patients. Two age groups emerged in our cohort, consistent with the epidemiologic age peaks for cHL: one group between ages 7 and 27 years, which we defined as pediatric, adolescent, and young adults (ped/AYA); another between ages 55 and 85, which we defined as “older adults” (Supplementary Table S1). Twenty-two (88%) of the cases were obtained at the time of diagnosis and 3 (12%) were obtained at the time of relapse. Given the low DNA input from HRS cells (median 13.6 ng; range, 4.2–226 ng), sequencing data were generated by amplifying the DNA (median 10 amplification cycles, range, 7–15). To ensure a robust quality control, we explored possible amplification-based mutational artifacts across the genome, observing an enrichment of single base substitution (SBS) within distinct trinucleotide context reflecting palindromic artifacts (Supplementary Fig. S1A–S1C; Supplementary Table S2; ref. 12). After having removed these amplification-induced palindromic sequencing artifacts, we observed a median SBS and indel burden of 5,279 per genome (range, 1,880–18,883) and 342 (range, 108–953), respectively. This mutational burden is within the range of what has been observed in other malignancies (Fig. 1A; ref. 13). Coverage, cancer cell fraction, and mutational burden for each case are detailed in Supplementary Table S1. Pediatric, adolescent, and young adult patients had a higher genome-wide SBS and indel burden than older adults (median 6,270 vs. 3,723, P = 0.0033 using Wilcoxon test; Fig. 1B and C). This difference was observed considering SBS and indels separately and was independent from sequencing coverage, histology, and cancer cell fraction (Supplementary Fig. S2A–S2I). As Epstein-Barr Virus (EBV)+ HL is suggested to have a lower mutational burden than EBV− HL (9), we repeated the analysis after removing the small number of EBV+ cases (n = 3) in our cohort. For this analysis, EBV+ cases were defined by EBER in situ hybridization. When restricted to EBV+ cases, we again observed an increased mutational burden among pediatric/AYA patients, suggesting that the effect is independent of EBV status (P = 0.001, Wilcoxon test; Supplementary Fig. S2H). The small number of EBV+ cases precluded an analysis of genome-wide mutational burden in EBV+ versus EBV− HL; however, this will be important to study in larger series.
Figure 1.
cHL mutational landscape. A, WGS mutational burden comparison between cHL (this study) and other cancers included in the PCAWG (n = 2,780) and in MM (n = 71) WGS studies. cHL is highlighted in red, and other hematologic cancers in blue. For each cancer type other than cHL, the total SNV mutational burden was taken from publicly available data (13, 93). The median of each tumor type is annotated with a red line. Tumors are presented in ascending order based on the median mutational burden. B, Whole-genome mutational burden comparison between Ped/AYA (n = 19) and older adults with cHL (n = 6). P value was calculated using the Wilcoxon rank-sum test. Box plots show the median and interquartile range. C, Correlation between mutational burden and age among Ped/AYA (n = 19, red dot) and older adult (n = 6, blue dot) cases evaluated by WGS. D, Oncoplot summarizing the 26 mutated driver genes/hot spots across 61 cHL patients with available WGS or WES data. A high-confidence (conf.) driver is defined as a gene that was extracted by at least two of the three driver discovery tools used (OncodriveFML, MutSigCV, and dndscv). EBV, Epstein-Barr Virus; Seq, sequencing.
To increase the sample size, we performed WES on HRS cells from additional independent 36 cHL cases (Supplementary Table S3), 10 of which have been previously reported (2). Using the same WGS pipeline, we identified and removed the amplification-based palindromic artifact from the WES data. When comparing the WES mutational burden between age groups, we did not observe a significant difference once corrected for coverage (Supplementary Fig. S2J and S2K), suggesting that noncoding alterations may be driving the differences in mutational burden observed in our genome cohort. We combined the two cohorts [WGS (n = 25); WES (n = 36)] to perform a driver mutation discovery analysis. For this analysis, 23 of our 25 WGS cases also had WES available. In cases with WES and WGS, WGS was used as the primary source. Unique WES mutations in cases sequenced with both WES and WGS were included only in the driver mutation discovery analysis after manual inspection.
The full list of nonsynonymous mutations in the combined WES and WGS cohorts is provided in Supplementary Table S4. To identify genes that are hit by nonsynonymous mutations more frequently than what would be expected by chance (i.e., genes under positive selection), we ran MutSigCV, OncodriveFML, and dndsv (Supplementary Data S1; refs. 14–16). Overall, 26 genes were identified as significantly enriched for nonsynonymous mutations, 8 of which were called by two or more driver discovery tools, which we have defined as “high confidence drivers” (Fig. 1D; Supplementary Tables S5 and S6). Ninety-five percent of cHL cases had at least one of these driver genes mutated, with a median of 4 (range, 0–14) mutations in driver genes per sample. Among the 23 patients with WGS and WES, only 15% (n = 20) of drivers were identified by WES alone. No distinct gene emerged as differentially mutated among different clinical groups including when grouped by age, histology, or stage (Supplementary Table S7). None of the driver genes was associated with the high mutational burden (Supplementary Table S7). The only significant correlation observed was an increased rate of mutations in GNA13 among EBV-negative cases (10/35 vs. 0/10; P = 0.008, in Fisher exact test). To determine if coverage was affecting the number of driver genes identified in each sample, we compared coverage and number of driver mutations, and the distribution of mutations in driver genes between WES and WGS and identified no significant differences (Supplementary Fig. S2L; Supplementary Table S7).
The most common driver alterations were in SOCS1 (62% of cases), TNFAIP3 (36%), and B2M (32%). We observed alterations in 8 driver genes not previously described in cHL, some of which are known to be altered in other B-cell lymphomas, such as primary mediastinal B-cell lymphoma or diffuse large B-cell lymphoma (CISH, FAM230A, MSL2, and TMSB4X), and some of which have not been previously reported in lymphoma (MAL2, NONO, RAB19, SPDYE1; Supplementary Tables S5 and S6; refs. 2, 7–9, 17–33). Cytokine-inducible SH2-containing protein (CISH) is a member of the SOCS family that regulates cytokine receptor signaling through the JAK/STAT pathway (34). Interleukin 4 receptor (IL4R) is also linked to JAK/STAT signaling, which is known to be dysregulated in cHL. Other novel drivers are involved in chromatin modifications, including MSL2, which is responsible for histone 4 lysine 16 acetylation (H4K16) and has been reported as altered in marginal zone lymphoma. Hypoacetylation of H4K16 is associated with defective DNA damage repair. A significant dnds hotspot mutation in the histone methyltransferase EZH2 was identified in one patient in our cohort (Supplementary Data S1; Fig. 1D). Although the EZH2 hotspot mutations were identified only in one patient, this was a significant enrichment compared with the dnds background model and has been reported in other HL patient cohorts (7, 8, 35, 36). This alteration is of particular interest as a therapeutic target given that EZH2 inhibitors are in clinical development (37) and the EZH2 inhibitor tazemetostat is now FDA approved in follicular lymphoma (38). Further annotation of the novel drivers, their function, and association with other cancers is provided in Supplementary Table S6 (39).
EBV Genome Analysis in cHL
To evaluate EBV genomes in the cases for which we had WGS, we aligned to the Gr37 reference including HHV-4 (EBV) as a decoy sequence. Three of 25 cases demonstrated >95% HHV-4 genomic coverage consistent with the presence of the full-length EBV genome (Supplementary Fig. S3). All three cases had been identified previously as EBV+ based on EBER in situ hybridization. We also evaluated EBV strain and found all three cases to be type 1/A as defined by alignment of EBNA2 with the NC-07605 type A genome and the absence of the Zp-V3 polymorphism, which is a characteristic of type 2/B. Lastly, to determine if there were any integration events of EBV into the host genome we performed integration analysis using VirusBreakend, SvABA, and manual review of read mate-pairs. No integration of EBV into the host genome was identified.
Mutational Signatures in cHL
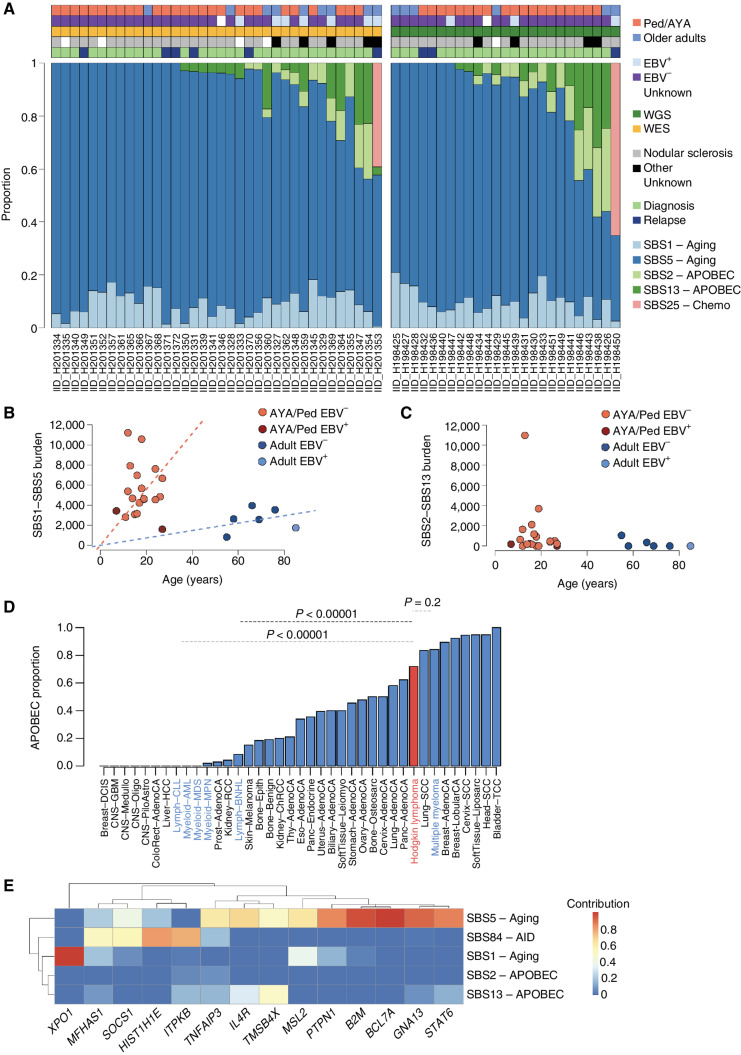
To investigate which mutational processes are involved in shaping the cHL mutational profile, we used both SigProfiler and the hierarchical Dirichlet process (hdp) identifying 5 mutational signatures (or SBS signatures), all previously reported in cHL (9, 13, 40). Subsequently, mmsig was used to accurately estimate the activity and contribution of each SBS signature (Fig. 2A; Supplementary Fig. S4A–S4D; Supplementary Table S8; ref. 41). Although SigProfiler did not detect the sequencing artifact signatures SBS45 and SBS49 (Supplementary Fig. S1A), those signatures were previously extracted when the palindromic mutations were included. Therefore, to increase the quality and accuracy of our SNV calls, we forced and included SBS45 and SBS49 in the mmsig fitting analysis, which identified a minimal proportion of mutations assigned to these two artifactual SBS signatures [5.3% (range, 0–26.5%]. These two artifactual SBS signatures were independent of patient age and did not affect the difference in SBS burden observed between pediatric/AYA and older adults (Supplementary Fig. S4B and S4C). In cases with both WGS and WES, the mutational signature distribution was similar when examined by genomes or exomes (Supplementary Fig. S5).
Figure 2.
Mutational signatures in cHL. A, Contribution of mutational signatures across 25 and 36 cHL cases with available WGS and WES data, respectively. B and C, Linear regression showing the association between age and SBS1/SBS5 mutational burden (top) and SBS2/SBS13 (bottom) among the 25 cases evaluated by WGS. D, Proportion of patients with any APOBEC activity across cHL and other cancers included in the PCAWG and MM WGS studies. cHL is highlighted in red, and other hematologic cancers in blue. E, Mutational signatures contribution among all coding mutations occurring within recurrently mutated driver genes with at least 10 SBS (n = 61 cases evaluated by either WGS or WES).
SBS1 and SBS5, the so-called clocklike mutational processes, were detected in all patients. The proportional contribution of SBS1 and SBS5 was similar in pediatric/AYA patients and older adults (Fig. 2A; Supplementary Table S9). The SBS1 and SBS5 mutational processes are known to act at a constant rate over time across different cancers and normal cells, including naïve and memory B cells (13, 42). Although the SBS1 and SBS5 mutational rate of older adults in our cohort is in line with other lymphoproliferative disorders (41, 43), the rate among pediatric/AYA patients is increased up to 6-fold (P = 0.003 using the Wilcoxon test; Fig. 2B; Supplementary Data S2).
Across the WES and WGS cohorts, 64% of patients had evidence of APOBEC mutational activity (SBS2 and SBS13; Fig. 2A). This high prevalence of APOBEC is similar to what has been reported based on HL WES data (9). APOBEC absolute and relative contribution was not different between pediatric/AYA and older adult patients and was slightly enriched at relapse (Fig. 2C; Supplementary Table S9). No APOBEC mutational burden difference was observed between age groups or between EBV+ and EBV− cases (P > 0.05, Wilcoxon test). When compared with APOBEC contribution evaluated using the same approach in other hematologic malignancies, APOBEC in cHL was similar to multiple myeloma (P = 0.2403, Fisher exact test) and significantly higher than non-Hodgkin lymphoma (P < 0.00001) and chronic lymphocytic leukemia (P < 0.00001; Fig. 2D). Overall, this suggests a pathogenic role for APOBEC in cHL. Among the WGS cohort, four patients (16%) showed a particularly high APOBEC contribution mostly driven by a major APOBEC3A activity compared with APOBEC3B (hyper APOBEC), in line with what has been reported in other cancers (41, 44). The other 14 (84%) cHL WGS cases were characterized an APOBEC3A:3B ratio ∼1 (“canonical” APOBEC). Overall, these data showed that the high mutational burden observed among pediatric/AYA patients with cHL is driven by SBS1 and SBS5 clock-like mutational processes.
Two out of eight patients whose sample was collected at relapse (IID_H198450 and IID_H201353) showed a clear presence of SBS25 (Fig. 2A; Supplementary Fig. S4D), an SBS signature previously linked to a still unknown chemotherapy agent in cHL cell lines (9, 13). One of the cases with SBS25 was from the WES cohort (IID_H201353). The mutational signatures fitting tool mmsig has been shown to have high accuracy in detecting distinct SBS signatures in WES/WGS with more than 100 SBSs (45). The HL whole-exome case with SBS25 had 175 SNVs, enough to confidentially call SBS25 (Supplementary Fig. S4D). This report of SBS25 in relapsed cHL demonstrates that, similar to other cancers, cHL can acquire hundreds of SBSs after exposure to distinct chemotherapy agents (13, 41, 46). To investigate the potential chemotherapy agent responsible for SBS25, we retrieved the treatment history, which was available for seven of eight relapsed cases in our cohort and for two of the four HL cell lines known to have SBS25 (Supplementary Table S10). Three of the four cases/cell lines with SBS25 had a known exposure to procarbazine/dacarbazine-containing regimens. Treatment for the fourth case with SBS25 is unknown (IID_H198450). Procarbazine and dacarbazine have been linked with intrinsic mutagenic activities (47–49); however, the existence of a distinct SBS signature has never been demonstrated. In contrast, other chemotherapy agents to which these cases were exposed including vincristine, bleomycin, and doxorubicin do not have any known direct mutagenic activity in vivo and in vitro (41, 50–52). The presence of SBS25 in cell lines and cases exposed to procarbazine/dacarbazine and the lack of a mutational signature associated with the other agents in these cases suggest a link between procarbazine/dacarbazine and SBS25.
Given the high mutational burden and the activity of multiple mutational processes, we investigated the mutational density and contribution of each SBS signature to each driver gene (Fig. 2E). B2M, BCL7A, GNA13, ITPKB, and SOCS1 tended to have more than one nonsynonymous mutation in the same patient (Fig. 1C). Using the WGS and WES mutational distribution on the entire footprint of all driver genes, we observed that BCL7A, SOCS1, TMSB4X, IL4R, and ITPKB showed a higher noncoding:coding mutation ratio compared with other driver genes (FDR <0.1, Fisher exact test; Supplementary Table S11). Most of these SBSs were compatible with the AID mutational signature (AID/SBS84 COSMIC signature; Fig. 2E; Supplementary Figs. S6 and S7; refs. 40, 53–55). The presence of localized AID hypermutation activity, likely related to somatic hypermutation (SHM), provides a mechanistic explanation of why these genes tend to be mutated multiple times in the same patient. In contrast, B2M and GNA13 were recurrently mutated more than once without enrichment for either intronic or SHM/AID SBS (Supplementary Figs. S6 and S7). When adding SBS84-AID to mmsig, we identified SBS84-AID activity within the footprint of seven driver genes (Fig. 2E; Supplementary Fig. S6; Supplementary Table S12).
Copy-Number Alterations
Combining WGS and WES data, we investigated the somatic copy-number alteration (CNA) landscape in cHL. We observed high ploidy (median 2.95; range, 1.66–5.33), which is well known in cHL. Of note, not all patients had whole-genome duplication (WGD, 59%; Supplementary Table S13; ref. 56). The presence/absence of WGD was validated by FISH in six patients with available material (Supplementary Fig. S8 and Supplementary Table S13). This suggests that multinucleation is not universally associated with WGD in cHL (57, 58).
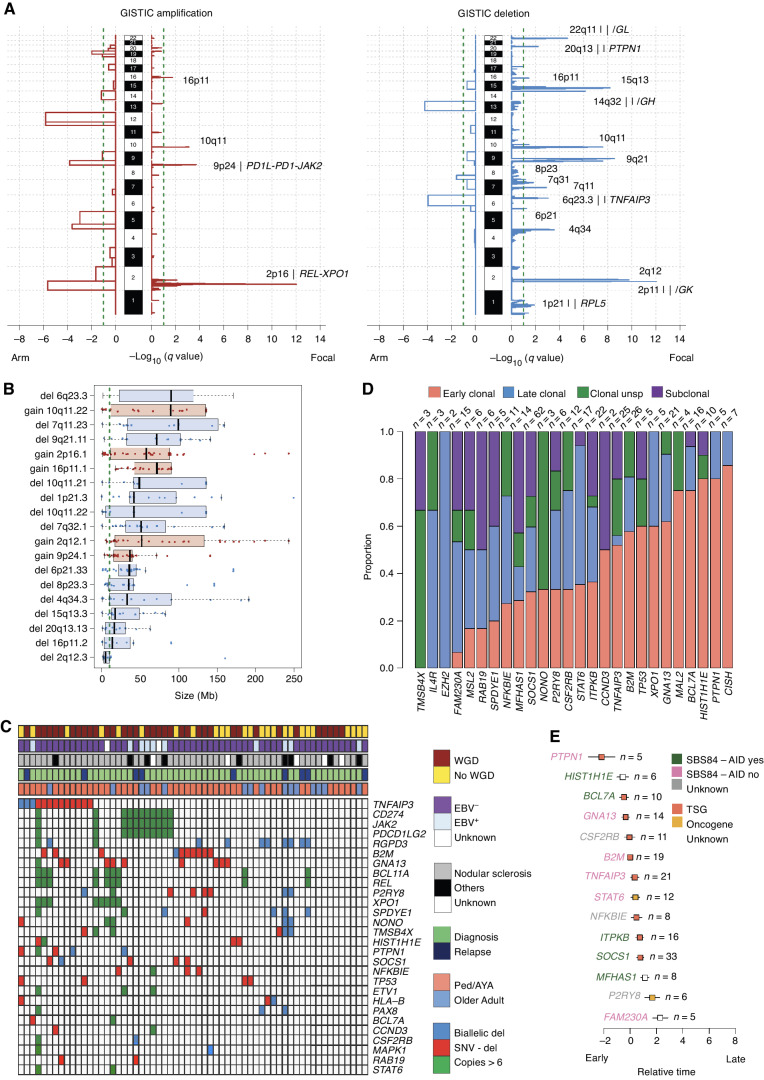
Running GISTIC2.0 (59), which identifies genes and chromosomal segments recurrently targeted by somatic CNA, we identified 19 recurrent CNA peaks: 5 gains and 14 losses (Fig. 3A; Supplementary Fig. S9A and S9B; Supplementary Table S14). The majority of these CNAs were caused by either large (>10 Mb) or whole chromosome/arm events (95.6%; Fig. 3B; Supplementary Fig. S9). Matching GISTIC CNA peaks with genes that are cHL driver genes and/or reported in the COSMIC census, we identified 7 genes recurrently involved in amplifications and 24 by deletions. Del 6p21.33 and del 6q23.3 (TNFAIP3) were the most frequently deleted peaks (52% and 44% of all patients, respectively), and 81% and 77% of deletions were either large (>10 Mb) or whole arm/chromosome loss, respectively. The HLA locus is approximately 100 kb from the loss-of-function GISTIC peak 6p21.33. Most of the patients with this GISTIC lesion had a deletion that also involved the HLA locus (Supplementary Fig. S9). The most frequently amplified loci were 9p24.1 (PDCD1LG2/JAK2/CD274) and 2p16.1 (REL/XPO1), (67% and 68% of patients, respectively; Supplementary Fig. S9). These amplifications were caused mostly by large and whole chromosome/arm duplications (78.5% and 85%, respectively). Overall, our data suggest that known cHL driver genes are recurrently involved in large CNA, and, in a smaller proportion, by focal events. We also observed a high prevalence of high-level gains [>6 copies) in 2p16.1—REL/XPO1 (median 11 copies; range, 7–31; n = 10, 16%) and 9p24.1—PDCD1LG2/JAK2/CD274 (median 8 copies; range, 6.3–13; n = 11, 18%) Fig. 3C]. When comparing CNAs across age groups, gain at 2q12.1 was the only GISTIC event to be significantly enriched in pediatric/AYA cases compared with older adults (P = 0.018 in Fisher exact test; Supplementary Table S15).
Figure 3.
CNA in cHL. A, Significant CNA GISTIC2.0 peaks and involved genes. B, Size of all CNAs involving GISTIC peaks. The gray dashed line represents 10 Mb, and the threshold used to differentiate focal and large events. Black bars represent the standard error of the mean. C, Heat map summarizing biallelic events and high-level gains (>6 copies) involving driver genes extracted by GISTIC (n = 21) and by dndscv (n = 26) in at least two cases. D, Proportion of early clonal (duplicated clonal mutations within duplicated copy number), clonal unsp. (unspecified, clonal mutations within diploid or deleted copy-number segments), late clonal (nonduplicated clonal mutations within duplicated copy-number segment), subclonal mutations in cHL driver genes. E, Chronological order of mutations in 14 cHL driver genes observed in 5 or more patients. Relative order of mutation acquisition is based on pairwise precedence among driver genes and their relative timing (from D) shown in the Bradley–Terry plot. X axis reflects the relative order of acquisition combining all cases. Red dots indicate the point estimate and black lines indicate 95% confidence intervals of gene ordering in time. Genes are positioned along the y axis on the basis of their relative order. Because this analysis is based on pairwise precedence among driver genes, not all mutations involving driver genes were included. For example, if gene A is earlier than B in one patient, both mutations will be included in the model. In contrast, if in a patient it is impossible to define the chronological order of A and B, these mutations will not be included. Asterisks represent genes involved by SHM (i.e., significant ratio between coding and noncoding mutations; Supplementary Table S11). Genes with less than 10 SBS were not included in the individual driver gene SBS signature analysis and are reported as “unknown.” TSG, tumor suppressor gene.
We combined CNA data and nonsynonymous mutations to investigate biallelic events involving driver genes extracted by GISTIC2.0 (n = 21) and by dndscv (n = 26). TNFAIP3 (n = 13; 21%), B2M (n = 10; 16%), and GNA13 (n = 10; 16%) were the most common driver genes with biallelic inactivation. This was mostly driven by deletion on one allele and a nonsynonymous mutation on the other (Fig. 3C).
Timing of cHL Driver Alterations
We next investigated the relative timing of driver mutation acquisition with respect to chromosomal gains. For this analysis, we included chromosomal gains and copy-neutral loss of heterozygosity (LOH) of any size and level. By leveraging the high cHL ploidy and high prevalence of chromosomal gains, we performed a comprehensive investigation of the relative timing of driver mutations using the approach previously reported by the pan-cancer analysis of genomes (PCAWG) project (41, 43, 60). In this workflow, mutations in driver genes are divided into subclonal and clonal according to their cancer cell fraction and phylogeny using the Dirichlet process (60). Clonal mutations are then further divided into three groups according to their copy-number status and purity-corrected variant allelic frequencies (VAF). Specifically, clonal mutations are defined as (i) “late clonal” if the mutation was detected within chromosomal gains with VAF ≤33% reflecting mutations acquired either on one of the two duplicated alleles after the gain or on the minor allele nonduplicated allele; (ii) “early clonal” if the mutation was acquired within chromosomal gains with VAF >66% reflecting mutations acquired on the duplicated allele before the duplication; or (iii) “clonal unspecified” if the mutation was detected outside of chromosomal gains. Across 313 nonsynonymous mutations in driver genes identified in our HL genomes, 53 (17%) were subclonal, 133 (42.5%) early clonal, 85 (27%) late clonal, and 42 (13.5%) clonal unspecified (Fig. 3D). Similar to the PCAWG, these data suggest that a large fraction of mutations in driver genes is acquired prior to chromosomal gains (42). The proportion of duplicated clonal mutations in driver genes within chromosomal gains in HL (133/218; 61%) was significantly higher than what is observed in multiple myeloma where large chromosomal gains have an established early pathogenetic role (25/65, 38%; P = 0.001 in Fisher exact test; ref. 55). Among loss-of-function mutations involved by LOH, 88% (49/56) were duplicated and therefore acquired before the duplication. This proportion was significantly higher than that observed for mutations in driver genes within non-LOH chromosomal gains (84/162; 52%; P < 0.00001, in Fisher exact test).
Next, we estimated the relative chronological order of mutations in cHL driver genes (Fig. 3E). Temporal estimates were generated by the Bradley–Terry model based on the PCAWG temporal model summarize above (55, 61). Mutations in PTPN1, GNA13, and HIST1H1E emerged as early drivers occurring prior to other mutations and prior to chromosomal gain events, while SOCS1, NFKBIE, and STAT6 tended to be acquired later. In contrast to other tumors such as multiple myeloma where AID has an established initiating role and its activity is restricted to the earliest phases, SBS84-AID mutational activity was neither enriched nor limited to any distinct phase of cancer development in our HL cohort (Fig. 3E; refs. 41, 55).
Investigating the relative activity of different SBS signatures, we observed an enrichment among the APOBEC nonduplicated SBS compared with the APOBEC duplicated reflecting a later role in cancer pathogenesis (i.e., after the gain; P = 0.002 using paired Wilcoxon test). However, in contrast to other cancers like multiple myeloma (41, 43), APOBEC activity was also detectable before the chromosomal gains, suggesting that large chromosomal duplications often occur in a clone in which APOBEC is already active. Based on this, we conclude that APOBEC mutational activity is an intermediate/late event in cHL similar to what is observed in other tumor types (41, 62).
Molecular Timing of Multichromosomal Gain Events
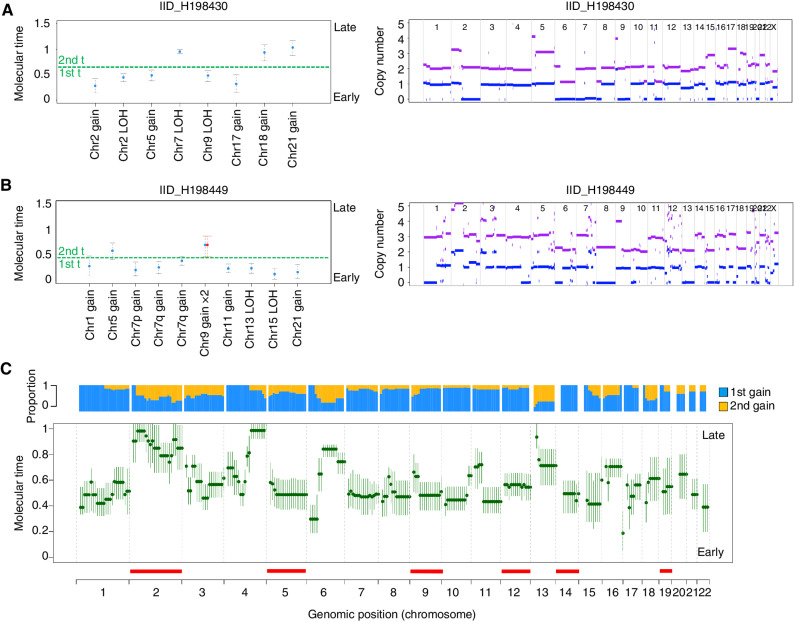
cHL is characterized by high ploidy and multiple chromosomal gains (e.g., WGD). Although most of these events are clonal, they may not have been acquired at the same time. To explore the temporal pattern of acquisition of chromosomal gains, including WGD, in cHL, we evaluated the corrected proportion of duplicated and nonduplicated clonal mutations within large chromosomal gains (i.e., molecular time approach; ref. 43). In line with our previous work (60), we restricted our analysis to clonal chromosomal trisomy, tetrasomy, gains, and copy-neutral LOH larger than 1 Mb, with more than 50 clonal SNVs after having removed immunoglobulin loci and all localized hypermutated events (i.e., kataegis). Overall, in 62% of gains, the molecular time was higher than 0.5 (median 0.58; range, 0.12–1.00), reflecting an intermediate–late acquisition pattern (Fig. 4A and B; ref. 43). Copy-neutral LOH showed a lower molecular time compared with non-LOH gains (P = 0.02 using Wilcoxon test). There were distinct events that were acquired particularly late, such as large gains on chromosome 2—REL/XPO1, 6q, and 4q (Fig. 4C). Other recurrent CNAs such as 9p24.1—PDCD1LG2/JAK2/CD274—were intermediate. In 12 of 20 patients (60%), the final clonal copy-number profile was acquired through at least two independent events (i.e., two groups of large gains with different molecular time; Fig. 4A–C; Supplementary Fig. S10). This suggests that HRS cells have a predisposition over time to acquire additional chromosomal gains.
Figure 4.
Molecular time and in cHL. A and B, Two examples showing molecular time estimates for two cases with multiple gains. Left, the molecular time (blue dots) was estimated for each clonal gain and copy-neutral LOH with more than 50 clonal SNVs. Red dots represent the molecular time of a second gain occurring on a previous one. The dashed green line divides the two independent time windows in which different chromosomal gains were acquired (first t: first-time window; second t: second-time window. Right, standard copy-number profile of 2 cHL cases. Horizontal purple and blue lines represent the total copy number and minor allele, respectively. C, Median molecular time estimates for chromosomal gains across each chromosome in cHL. Each chromosome was divided into 10-mb bins. Each green dot reflects the median molecular time across different patients with the gained bin. CI were generated using the median of CI molecular time estimate for each bin. Horizontal red line reflects GISTIC large amplifications. The blue/yellow plot represents the distribution of chromosomal gains acquired within either the first- or second-time window for each locus.
As validation of our temporal model based on molecular time, we leveraged the concept of chemotherapy signatures as single-cell barcodes (63) estimating the SBS25 contribution before and after the three large chromosomal gains in IID_H198450. Chemotherapy-related SBS were detected both before and after the large chromosomal gains indicating that these CNA events were acquired after exposure to chemotherapy, in line with the late molecular time (Supplementary Fig. S11A–S11E).
SV and Complex Events
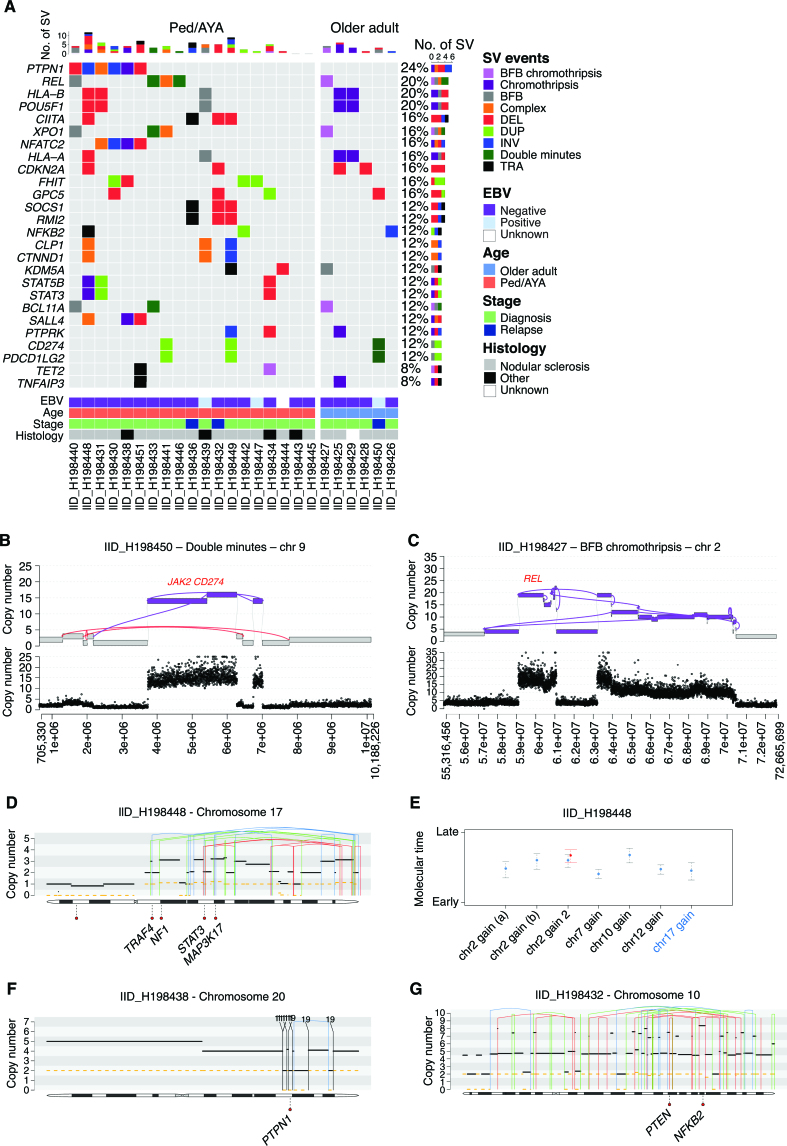
The high resolution of WGS allows us to perform the first characterization of the landscape of SVs in cHL (Supplementary Table S16; refs. 64, 65). Integrating JaBbA and our SV annotation, we were able to infer SV and CNA junction-balanced genome graphs with high fidelity, allowing a detailed characterization of both single and complex events (Supplementary Figs. S12 and S13; refs. 64, 65). Overall, we observed at least one complex events in all but two cases (92%). Chromothripsis and breakage–fusion–bridge (BFB) were the most recurrent complex events detected in 32% and 28% cases, respectively (Supplementary Fig. S12). Additional four cases (16%) had complex events including evidence of both BFB and chromothripsis. The role of single and complex events in the acquisition of CNAs involving distinct drivers emerged as heterogeneous (Fig. 5A). For example, although most of the PDCD1LG2/JAK2/CD274 high-level gains were caused by large whole-arm chromosomal gains or single events, in one patient these genes had multiple copies as a consequence of double minutes (DM) (IID_H198450; Fig. 5B). Similarly, in IID_H198427, a BFB event was responsible for the acquisition of 18 copies of XPO1/REL (Fig. 5C). The most frequently cHL driver genes involved by SV and complex events were PTPN1 (24%), REL (20%), HLA-B (20%), and POU5F1 (20%; Fig. 5A). When comparing SVs in our cohort to what has been previously described (66), we did not observe PDL1 and CIITA fusions, and we observed only one patient with SV involving SOCS1 (32). SV and the prevalence of complex events were not affected by EBV status, stage, or histology. Pediatric/AYA cases were slightly enriched for single deletions and duplications (P = 0.04 and P = 0.01, respectively, using Wilcoxon test; Supplementary Table S17).
Figure 5.
SV and complex event in cHL. A, Oncoplot showing cHL driver genes involved by SV and complex events. B and C, Example of complex events involving key cHL drivers. D and E, Example of chromothripsis event in which it was possible to estimate the molecular time. Case IID_H198448 had an intrachromosomal chromothripsis event responsible for multiple large/intermediate chromosomal gain on chromosome 17 (E). The molecular time of this event was similar to other gains acquired in the first/earliest-time window (E). Based on this link, we could estimate that the chromothripsis event was acquired together with the other chromosomal gains (WGD). F and G, Examples of two chromothripsis events that occurred before WGD. The two chromothripsis events were each responsible for a multiple copy-number jump from 2:2 to 2:0. In D, F, and G, the black and dashed yellow horizontal line represent the total number copy number and the minor allele, respectively. The blue, red, green, and black vertical lines represent inversion, deletion, tandem duplication, and translocation, respectively. The partner of each translocation is reported on the top of the vertical black line.
To estimate the timing of loss-of-function events and the acquisition of distinct SVs, we utilized two approaches (60): we linked SV breakpoints to the molecular time of chromosomal gains caused by the same SV (Fig. 5D and E); and we estimated the relative time of SVs that occurred within large chromosomal gains based on the copy number of the SV breakpoint (Fig. 5F and G; Supplementary Fig. S14A–S14J). Applying these approaches, we were able to time the acquisition of single and complex events observing two relevant SV/CNA temporal patterns. In the first, chromothripsis emerged as an early event in cHL pathogenesis preceding WGD in three of three patients in which this analysis was possible. (Fig. 5D–G). In the second temporal pattern, we observed early acquisition of PTPN1 deletion (i.e., before chromosomal gains/WGD) in four of five patients, suggesting the early driver role of this gene in cHL pathogenesis and in line with the PTPN1 nonsynonymous mutation early temporal estimates (Fig. 3D and E). Combining SV and SNVs in driver genes, PTPN1 was involved in 36% (9/25) of cHL, either as early deletion (n = 4) or SBS (n = 5), each duplicated by a subsequent chromosomal gain.
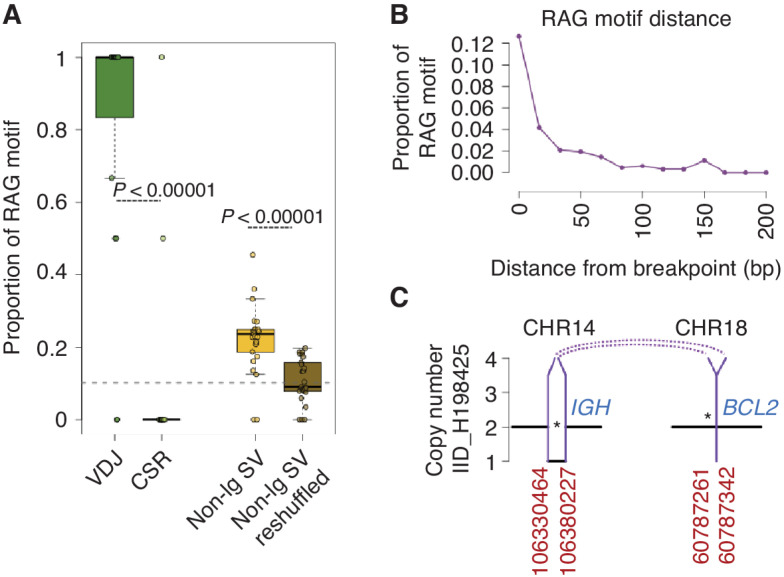
According to our relative temporal estimate, AID mutational activity emerged as a key mutational process involved in introducing mutations in driver genes whose activity is not restricted to the earliest phase of tumor development. This suggests that, in some patients, other drivers can be acquired before the first encounter with the GC and AID SHM exposure. To further explore the acquisition of alterations prior to the GC, we evaluated for the presence of SVs with breakpoints enriched for recombinase activating gene (RAG)-motifs (67). Some GC-derived lymphomas are known to be initiated by RAG-mediated SV events which occur exclusively in the bone marrow during the early phases of B-cell development before the GC encounter (67, 68). These events include BCL2::IGH translocations in follicular lymphoma (67) and CCND1::IGH in mutated IGHV mantle cell lymphoma (69). To evaluate for the presence of potential RAG-mediated pre-GC SVs in cHL, we estimated the recombination signal sequence (RSS) score of each SV breakpoint. Overall, 25% of non-VDJ and 86% of VDJ SV breakpoints were linked to RAG (Fig. 6A and B). This proportion was significantly higher than what was expected by chance (P < 0.000001 in Fisher exact test) and higher than the genomic background rate of RAG motifs (P < 0.000001 in Fisher exact test). Among these breakpoints enriched for RAG motifs, a total of 63 involved distinct cHL genomic drivers. As an emblematic example of a known and established RAG-mediated SV, we detected a BCL2::IGH translocation with RAG motifs and evidence of TdT activity (Fig. 6C). The proportion of RAG-associated SVs was not correlated with age, stage, EBV status, or histology (Supplementary Table S18). Similar to other lymphoproliferative malignancies (67–69), the presence of potential RAG-mediated SVs suggests that cHL initiation can begin in the bone marrow before the first GC encounter in subset of patients.
Figure 6.
cHL SVs are enriched for RAG motifs. A, Proportion of RAG motif-enriched SVs across SVs within the VDJ (VDJ), SVs within the class switch recombination (CSR), SVs outside of the immunoglobulin genes (Non-Ig SV), and reshuffling all the nonimmunoglobulin SVs across the genome (Non-Ig SV reshuffled). The gray dashed line represents the genomic background rate of RAG motifs. Each dot represents the proportion of RAG-positive breakpoints for each patient. B, The proportion of SV with an RSS (RAG) motif as a function of distance from their breakpoint. C, Example of a likely RAG-mediated BCL2::IGH translocation in one HL case.
Using Mutational Signatures to Identify the HRS Cell-of-Origin
The HRS cell-of-origin is suspected to be a B-cell that has entered the GC and is unable to fully mature due to an unproductive B-cell receptor (BCR; refs. 9, 17, 70–73). This model has been supported by the detection of AID-mediated somatic hypermutation (SHM) on an unproductive VDJ, and nonsynonymous mutations involving AID off-target genes. To molecularly evaluate this model and the relationship between the GC and HRS cells, we explored the SBS signature within the immunoglobulin loci (Ig) and within the footprint of genes known to be involved by AID off-target activity (Supplementary Table S19). To generate this list, we used a catalog of AID off-target genes in normal B-cell, and hematologic malignancies (40, 55, 74, 75). These regions showed clear activity of AID/SBS84 (Supplementary Fig. S15A–S15C). The presence of AID/SBS84 on Ig and on AID off-target genes has been reported in GC-derived malignancies including in genes that are functionally validated as drivers such as HISTH1E in DLBCL (13, 41, 76, 77).
In tumors with AID/SBS84, this signature is thought to always cooccur with a genome-wide mutational process historically called “noncanonical” AID (SBS9) and thought to result from SHM. Contrary to this model, we did not observe any evidence of genome-wide SBS9 activity in AID/SBS84+ HRS cells (Supplementary Fig. S15D; refs. 13, 41, 54, 60). This demonstrates that SBS84 and SBS9 are likely driven by two different mutational processes, consistent with recent data in normal B cells (42).
Next, to reconstruct the V(D)J of HRS cells, we ran IgCaller (78) and confirmed that the VDJ rearrangements were unproductive in all but 3 patients (Supplementary Fig. S15E; Supplementary Table S20). To investigate when these unproductive BCRs were affected by SHM/AID, we focused on Ig and AID off-target genes involved by large chromosomal gains. We observed both duplicated and nonduplicated AID mutations (Supplementary Fig. S15F), suggesting that chromosomal duplications are often acquired on a B cell that has already been exposed to the GC.
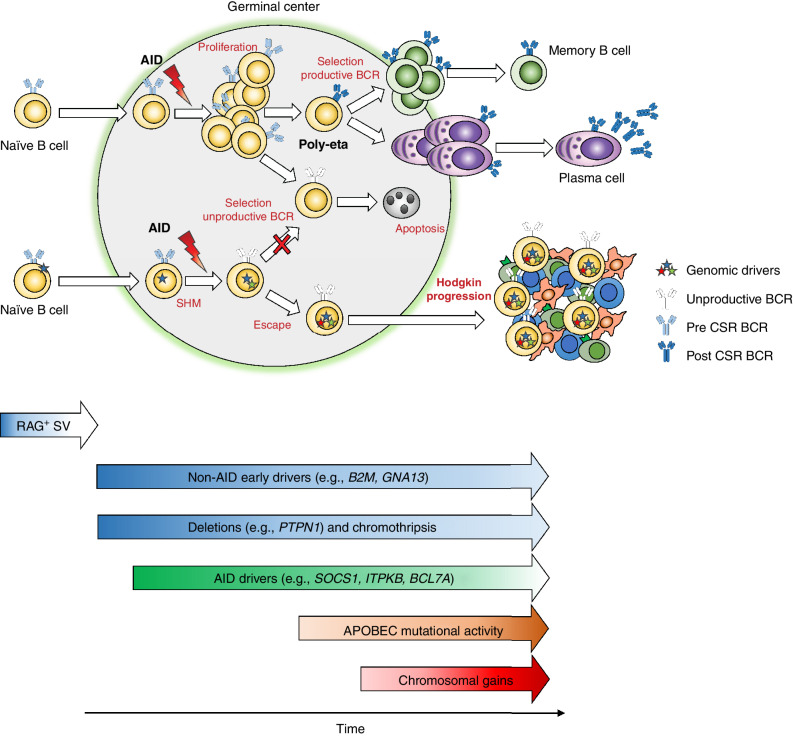
Altogether, these data suggest a pathogenetic model for cHL in which, for a fraction of patients, the initiating event can occur in a B cell prior to entry into the GC. Entering into the GC the premalignant clone is exposed to AID and SHM but fails to finalize its maturation, potentially due to an unproductive BCR. The acquisition of distinct genomic drivers acquired before (e.g., RAG-mediated events in the bone marrow) and during the GC reaction (e.g., AID) likely allows the pre-HL clone to escape negative selection in the GC. After being established and immortalized, in order to progress into cHL, the pre-cHL clone acquires additional driver events over time, in particular complex genomic events such as aneuploidies, WGD, and SVs (Fig. 7).
Figure 7.
Proposed model of the molecular timing of events leading to classic Hodgkin lymphoma. Our data suggest that the initiating event in cHL can occur in a naïve B cell prior to entry in the germinal center as evidenced by potential RAG-mediated SVs. Other early events include alterations in non–AID-mediated drivers such as B2M and GNA13 as well as deletions in PTPN1 and chromothripsis. In the GC, the premalignant cell undergoes somatic hypermutation without finalizing its maturation for unproductive BCR in most cases. Acquisition of AID-mediated events in off-target genes is not limited to early phases of cancer development, and it might be proceeded by other drivers. APOBEC mutational activity and chromosomal gains are both present in cHL and occur as intermediate/late events in cHL pathogenesis.
DISCUSSION
Human cancers are suspected to arise from a premalignant clone that evolves over time and often can be detected years before diagnosis, in some cases even in the perinatal period (43). Understanding the chronology of mutational processes leading to malignancy can help guide novel diagnostic strategies and treatment approaches directed at early driver events. In this study, we leveraged WGS resolution to elucidate a pathogenetic model for cHL whereby tumor development is shaped by the acquisition and selection of multiple drivers across different time windows (Fig. 7). Although our coverage and sequencing approach does not allow for a comprehensive analysis of subclonal and late events, the high ploidy in cHL allows us to define the relative timing of clonal events. In this model, distinct driver mutations (e.g., PTPN1 and GNA13) emerged as early events as did SVs enriched for RAG motifs, deletions in PTPN1, and complex SV events such as chromothripsis. Although some of these early events are likely acquired within the GC before large chromosomal gains, evidence of potential RAG-mediated events suggests that a fraction of cHL can be initiated in the bone marrow. RAG-mediated SV events involving oncodrivers have been already reported in HL (79–82) and have recently been reported in normal B cells, suggesting that RAG-mediated events other than IGH translocations can be detected in B-naïve cells (42, 67, 68). Overall, most of the observed early events occur before large chromosomal duplications which are often acquired at an intermediate/late timepoint. This temporal pattern is different from what is observed in other hematologic malignancies, such as acute lymphoblastic leukemia, multiple myeloma, and chronic lymphocytic leukemia, where large chromosomal duplications tend to be early events, potentially playing a cancer-initiating role (41, 43, 83).
Hodgkin lymphoma has long been described as a malignancy with karyotype instability and ongoing chromosomal alterations (70, 84). Our data support this model as evidenced by high genomic complexity that is acquired over time including WGD, chromothripsis, breakage–bridge fusions, and multiple aneuploidies. In addition, we specifically observed events that were acquired between diagnosis and relapse, for example, IID_H198450, where large trisomies were acquired after exposure to chemotherapy.
We also observed several key differences in the mutational landscape of HRS cells across age groups. When compared with older adults ages 55 to 85, pediatric and AYA patients ages 7 to 27 were found to have a higher mutational burden genome-wide. The high mutational burden in pediatric/AYA cases was driven mostly by the clocklike signatures SBS1 and SBS5. These two mutational processes are known to act in a constant way since the fertilized egg in all mammals and in most of the nonhypermutated tumors (13, 41, 43, 53, 85). Although genetic conditions have been associated with enrichment for distinct mutational signatures in normal cells such as in patients with MBD4 (SBS1), BRCA (SBS3), and POLE (SBS10) deficiencies, respectively (13, 86, 87), these signatures and known genomic events were not observed in our cohort. Furthermore, in contrast to SBS1, the mechanisms behind SBS5 mutagenesis are largely unknown, and no genomic lesion has been known to increase the rate in normal or tumor cells. Considering these details, an exponential increase of both mutational processes due to the presence of at least two different genetic mechanisms is unlikely, suggesting a potential somatic etiology for the increased rate observed in pediatric/AYA cases. Although the SBS1–SBS5 distribution in pediatric/AYA can be explained by linearity (Supplementary Data S2), this pattern might also be compatible with an early acceleration in the SBS1–SBS5 mutation rate over time (i.e., piecewise linear model). However, considering the relatively small sample set, the lack of multiple samples from the same individual collected at different time points, the dramatically increased pediatric/AYA mutational burden, and the impossibility to estimate when the mutational rate accelerated diverging from normal B-cell mutation rate, it is impossible to establish how early this acceleration occurred with this cohort. Except for age, no other clinical or genomic feature was associated with an increased SBS1/SBS5 mutational burden. These data suggest that the biology and the conditions in which cHL develop across these two age peaks might be distinct. We did not observe differences in distinct driver genes between age groups or between EBV+ and EBV− tumors; however, our cohort was enriched for pediatric/AYA and EBV− patients, which may limit the ability to detect more subtle differences in comparison with older adults and EBV+ cases. Additional and larger studies leveraging new low-input WGS approaches are needed to define biological differences between these groups and to investigate if the characteristics observed in younger patients may be due to accelerated B-cell aging.
Combining WES, WGS, and clinical data, we were able to indirectly correlate the SBS25 mutational signature to procarbazine/dacarbazine exposure. Of note, SBS25 was not detectable in 6 of 8 cases of relapsed cHL, despite exposure to procarbazine/dacarbazine in 5 of 6 of these cases. The absence of SBS25 in samples exposed to procarbazine/dacarbazine, it is likely due to lack of ”single-cell expansion” (46, 88). In this model, chemotherapy signatures can be seen as a single-cell barcode. Each tumor (and normal) cell will acquire hundreds of private SBS as a consequence of the chemotherapy exposure. Therefore, to detect this mutational signature in bulk sequencing, one single cell would need to take clonal dominance. In the presence of polyclonal relapse (e.g., refractory disease), tumor progression will be driven by multiple clones with their private chemotherapy-related SBS, and the resolution of these SBS will be under the level of WGS/WES detectability. This scenario is common, especially in early relapse and primary refractory disease (41, 46, 89). In our cohort, all 5 of 6 cHL relapsed cases without SBS25 progressed <12 months after the completion of chemotherapy (range, 4–8 months; Supplementary Table S10), suggesting polyclonal relapsed disease. The concept of polyclonal disease is further supported in case IID_H201372, where the patient was treated with both dacarbazine- and platinum-based regimens before the sample collection. Platinum is one of the strongest mutagenic chemotherapy agents, and its SBS signature (SBS31 and SBS35) can be found in all platinum-exposed cells (13, 46). The absence of SBS31 and SBS35, in this case, can only be explained by the absence of a single-cell expansion. In line with this concept, this case did not show any evidence of SBS25.
The HRS cell mutational signature landscape revealed robust confirmation that the SBS9 signature represents a distinct GC mutational process independent from AID and SHM. The original term “noncanonical-AID” was first reported because, across the lymphoproliferative disorders tested (follicular lymphoma, CLL, multiple myeloma, Burkitt, and diffuse large B-cell lymphomas), SBS9 always co-occurred with AID/SBS84 and SHM (13, 40, 41, 54, 76). This model has been recently challenged by the observations that (i) AID/SBS84 can be active in CSR loci in CLL with unmutated immunoglobulin heavy-chain variable region gene in the absence of SBS9, (ii) analysis of SBS9 genomic distribution in both tumor and normal B-cell genomes, and (iii) by the fact that SBS9 profile seems to reflect polymerase-eta repair activity (41, 42, 54). In this study, the presence of all the GC hallmarks of SHM and AID activity with the absence of SBS9 in cHL fills an important gap in our understanding of which mutational processes are involved in both normal and pathologic activity within the GC.
Lastly, with WGS comprehensive resolution, we confirmed and expanded the cHL pathogenic model proposed 30 years ago by Küppers and colleagues (70–73). We identified that the initiating event in cHL can occur before the GC. The subsequent interaction between the premalignant cells and the GC emerged as a critical phase for cHL development. During this encounter, the premalignant cell survives and escapes the negative selection despite its unproductive BCR, potentially due to early and concomitant acquisition of genomic drivers. Overall, this study provides a critical new perspective on cHL pathogenesis and sheds light into which mutational processes are involved in the interaction between the GC and B cells.
METHODS
Tumor Collection
Hodgkin lymphoma biopsies were collected fresh or were viably cryopreserved at centers throughout the United States. Specimens were then shipped to Memorial Sloan Kettering Cancer Center for sorting. All specimens were collected under IRB approval with waiver of consent at Memorial Sloan Kettering Cancer (protocol X17-027) and the local institution.
Sorting
The HRS, B-, and T-cell sorting was performed as described previously (2, 11). Briefly, single-cell suspensions from cHL tumors containing up to 1 × 108 cells were either taken fresh or rapidly defrosted at 37°C and washed in 50 mL of RPMI-1640/20% fetal bovine serum solution. The cells were stained with an antibody cocktail composed of CD64 FITC (22; Beckman Coulter, BC), CD30-PE (HRS4; BC), CD5-BV510 (L17F12; Beckton-Dickinson, BD); CD40-PerCP-eFluor 710 (5C3; eBioscience); CD20-PC7 (B9E9; BC); CD15-APC (HI98; BD); CD71 APC-A700 (YDJ1.2.2, BC); CD45 APC-H7 (2D1; BD), and CD95-Pacific Blue (DX2; Life Technologies). All sorting experiments were performed on an FACSAria-Fusion special-order research sorter using a 130-μm nozzle at 12 psi, acquiring up to 5 × 107 cells and collecting HRS, B, and T cells from the tumor using a 3-way sort. A representative gating strategy is shown in Supplementary Fig. S16A–S16F.
WGS
Sample library construction was performed using the Kapa HyperPlus Kits with enzymatic fragmentation (Roche). Fragmented gDNA was used to perform end repair, A-tailing, and adapter ligation following the manufacturer's instruction. The indexed library construct was split into two fractions, one fraction was used for WGS on NovaSeq6000 at PE2 × 150 cycles (Illumina), and another fraction was normalized and pooled at four samples from tumor and four samples from germline. Pooled sample libraries were hybridized with SeqCap EZ Human Exome v3.0 probes (Roche) for WES. The pooled, indexed, and captured final libraries were used to sequence on Illumina HiSeq4000 sequencer at 2 × 100 cycles pair-end reads. The raw sequencing reads in BCL format were processed through bcl2fastq 2.19 (Illumina) for FASTQ conversion and demultiplexing for downstream data analysis.
To determine if our tumor coverage (∼27×) affected the detection of clonal events and the estimation of their VAF, we down-sampled to 30× the five HL genomes in our cohort with coverage >45× (IID_H198431, IID_H198434, IID_H198438, IID_H198440, and IID_H198442). We then evaluated mutations in driver genes within chromosomal gains and their purity-corrected VAFs at the two coverage depths to determine if reduced coverage affected the corrected VAF. All of the 23 duplicated and 7 nonduplicated driver mutations identified at >45× were also detected at 30X. The corrected VAF at 30X highly correlated with the corrected VAF at >45× (P < 0.00000001; Supplementary Fig. S17A). Importantly, the duplication status of all mutations in driver genes (duplicated/early clonal vs. nonduplicated) was confirmed for all mutations (Supplementary Fig. S17B). This suggests that lower coverage did not significantly limit our ability to detect monoallelic drivers within copy-number gains.
Processing of WGS Data
Overall, the median sequence coverage was 27.5× (range, 15–55×; Supplementary Data S1). Short insert paired-end reads/FASTQ files were aligned to the reference human genome (GRCh37) using Burrows–Wheeler Aligner, BWA (v0.5.9). All samples were uniformly analyzed by the whole-genome analysis bioinformatic pipeline developed at Memorial Sloan Kettering Cancer Center (41). Specifically, CaVEMan was used for SNVs and indels were analyzed with Pindel. To further increase the quality of our calls, Mutect and Strelka annotations and flags were used to filter out low-quality variants called by Caveman and Pindel. CNAs were explored by Battenberg. To determine the tumor clonal architecture, and to model clusters of clonal and subclonal point mutations, the Dirichlet process was applied. BRASS and JaBba (64) were used to detect SVs through discordantly mapping paired-end reads (large inversions and deletions, translocations, and internal tandem duplication). Complex events such as chromothripsis, chromoplexy, DM, BFB and templated insertions were defined and validated after manual inspection as previously described (64, 65). All SVs not part of a complex event were defined as single. Immunoglobulin VDJ, HCDR3, CRS, and productivity were defined using IgCaller (78). WGD was defined if the fraction of the autosomal tumor genome with a major copy number of two or greater was higher than 50% as previously described (Supplementary Table S13; ref. 56).
RAG Analysis
We assessed the enrichment of V(D)J recombination mediated by RAG binding and endonuclease activity by looking for RAG motifs within a window of 50 bp (before and after) each SV breakpoint, as previously described (42), using the RSSsite web server (http://www.itb.cnr.it/rss). RSSsite consists of a web-accessible database for the identification of precomputed potential RSSs, which are composed of seven conserved nucleotides, a space of 12 or 23 poorly conserved nucleotides, and one conserved nonamer (68, 90). The tool can identify RRSs by exploiting both poorly conserved spaces. The results are filtered using the Recombination Information Content score, calculated by the tool, applying the default values. To assess the reliance on the calls, we implemented a reshuffle analysis exploiting each SV breakpoint and performed the same analysis as described above. The RAG genomic background rate was estimated as described by Machado and colleagues (42). The distance in bp between each breakpoint and the closest RAG motif was estimated by centering each SV breakpoint at zero, then calculating its distance in bp from the nearest RAG motifs (using a window of 200 bp at 5′ and 3′; ref. 42).
WES
WES data reads were aligned to the reference human genome (GRCh37) using the Burrows–Wheeler Alignment tool (bwa mem v0.7.12). PCR duplicate read removal, InDel realignment, fixing mates, and base quality score recalibration were applied to the aligned bams using PICARD tools or the Genome Analysis Toolkit (GATK) according to GATK best practices. Samples in the selected cohort had an average coverage of 76× in tumors and 49× in germline (range, 15×–135×). The tumor purities were inferred using TITAN and ranged between 31% and 91%. WES somatic calls were performed using CaVEMan for SNVs, Pindel for indels, and Facets for CNA.
Considering the hyperamplification and the lower coverage, we applied a stringent quality control on our data to minimize miscalling of SNVs. Specifically, we first ran Caveman and then used mutect and strelka germline and artifact flags to further clean the output (i.e., we kept only SNV called by caveman and not flagged as germline/artifacts by strelka and mutect).
Mutational Signatures
Analysis of SBS signatures was performed following our published workflow based on three main steps: (i) de novo extraction, (ii) assignment, and (iii) fitting (40). For the de novo extraction of mutational signatures, we ran two independent algorithms; SigProfiler and the hierarchical Dirichlet process (hdp; refs. 13, 41). Next, each extracted process was assigned to one or more mutational signatures included in the latest COSMIC v3.2 catalog (https://cancer.sanger.ac.uk/signatures/sbs/). Lastly, mmsig, a fitting algorithm designed for hematologic cancers, was applied to accurately estimate the contribution of each mutational signature in each sample (45). For individual driver gene mutational signature contribution, we collapsed all mutations occurring within each gene footprint and ran mmsig, excluding driver genes with fewer than 10 mutations (75). AID-SBS84 activity was defined as present if the gene's low 95% confidence of interval (CI) SBS84 contribution was higher than 0.01%.
PCAWG mutational signatures data generated using SigProfiler were downloaded from the ICGC public repository (https://www.synapse.org/#!Synapse:syn11801889).
Timing Copy-Number and Structural Variant Events
The relative timing of each multichromosomal duplication event was estimated using the R package mol_time (DOI: 10.5281/zenodo.4542145; refs. 41, 60). This approach allows the estimation of the relative timing of acquisition of large (>1 Mb) and clonal chromosomal gains (3;1 or 4:1), trisomies (3:1), copy-neutral LOH (2:0), and tetrasomies (4:1) using the purity-corrected ratio between duplicated clonal mutations (defined by DP). Clonal SNVs were defined using the Dirichlet process (60) and divided into duplicated or not according to the VAF corrected for the cancer purity that was estimated by combining purity estimates from both Battenberg and from the SNV VAF density and distribution within clonal diploid regions. Overall, after removing immunoglobulin loci and kataegis events, only CNA segments with more than 50 clonal mutations were considered (41, 60). Tetrasomies, with both alleles duplicated (2:2), were removed given the impossibility of defining whether the two chromosomal gains occurred in close temporal succession or not (41, 60). Using this approach, we were able to estimate the relative molecular time of each gained CNA segment allowing to define if different chromosomal gains were acquired in the same or different time windows. To define if different gains occurred in one single time window or in different independent events, we used a multiple hierarchical clustering approach for each single bootstrap solution (hclust R function; www.r-project.org) and we integrated the most likely results with the Battenberg CNA changes over the time.
To estimate the timing of loss-of-function events and the acquisition of distinct SVs, we utilized two previously reported approaches (60): (1) We linked SV breakpoints to the molecular time of chromosomal gains caused by the same SV; (2) we estimated the relative time of SVs that occurred within large chromosomal gains based on the copy number of the SV breakpoint. Approach (2) is based on the concept that any SV involving a large gain can produce three different copy-number scenarios: (i) SV-associated loss occurred on the allele involved by the duplication before the duplication. In this scenario, the deleted segment will not be duplicated creating a copy-number jump of at least 2 copies with the adjacent duplicated segments (e.g., 3:1 and 1:0); (ii) the SV is responsible for a loss (or gain) on one of the two duplicated alleles (e.g., from 2:1 to 1:1), suggesting that the SV event occurred after the chromosomal gain; (iii) SV causes the loss (or gain) of part of the minor allele creating a copy-neutral LOH (e.g., from 2:1 to 2:0). In this last scenario, it is not possible to establish if the SV occurred before or after the chromosomal duplication.
The chronological acquisition order of recurrent mutations in driver genes was estimated by combining the Battenberg cancer cell fraction and pre/post gain data into a Bradley–Terry model as previously described, including just the earliest sample of each patient (60).
EBV Analysis
Integration analysis was performed via VirusBreakend (91). Internal to the VirusBreakend algorithm, bams aligned to human hg19 sequences along with HHV-4 decoy sequences were realigned to candidate viral references, taxonomic classification was performed, and SVs representing candidate integration events were identified using the GRIDSS2 SV caller. Secondary candidate integration events were identified using SvABA, a local-assembly–based tool. Unfiltered SvABA outputs were filtered to identify candidate integration events between the hg19 chromosomal and HHV4 contigs. Both VirusBreakend and SvABA candidate integrations were manually reviewed for read-level evidence underlying the tool-assisted structural variant call. Finally, the frequency and distribution of hg19-HHV4 split-reads were examined at 200-bp resolution across the entirety of the 172-kb HHV4 genome to manually identify candidate sites of integration potentially missed by junction callers.
Identification of EBV type/strain was performed by manual examination of reads aligned to an EBV type 1 reference. Reads were examined for overhanging bases and incomplete alignment consistent with a known 16-bp deletion in EBV type 2 (92). Manual review of polymorphic sites at the BZLF1 promoter was also conducted for the presence of the Zp-V3 polymorphism, which is characteristic of type 2 EBV.
Combined Immunophenotyping and FISH Analysis
After immunophenotyping with mouse anti-CD30 antibody labeled with AMCA (blue), the relevant tissue slides with coverslip were briefly reviewed and recorded under a fluorescence microscope for the intensity and quality of CD30 staining. After removing the coverslip, the slides were washed in 2× SSC at room temperature for 5 minutes and then fixed with 10% neutral buffered formalin for 20 minutes, followed by dehydrating for 5 minutes each in a series of 70%, 85%, and 100% of alcohol. Slides were then pretreated for 10 minutes with 20 mmol/L citrate buffer/1% NP-40 mixture, pH 6.0–6.5, followed by protease treatment at 40°C for 10 to 15 minutes (Abbott Molecular), then by dehydrating 2 minutes each in a series of 70%, 85%, and 100% of alcohol. Relevant FISH probes were selected based on WGS ploidy findings and primarily were those used in clinical diagnosis tests, including CDK2 and CEP12, PDL1 and PDL2 and CEP9, and 19p and 19q, and EWSR1 (Abbott Molecular; Genomic Empire). These probes were labeled in spectrum orange, green, or aqua, respectively. After applying the FISH probes to the tissue areas, both tissue and probes were codenatured at 94°C for 7 minutes, and then incubated at 37°C overnight, followed by posthybridization washing in 2× SSC/0.3% NP-40 at 77°C for 1 minute. Tissue sections were counterstained with antifade medium without DAPI (Vector Laboratories).
The slides were evaluated under a fluorescence microscope coupled with appropriate filters for CD30 immunophenotype and the relevant probes. Signal analysis was performed in combination with tissue structure and cell morphology correlation and was focused on the interested tissue areas with strong CD30-positive cells only. The copy numbers of individual probes were counted in each case.
Data Analysis and Statistics
Data analysis was carried out in R version 3.6.1. Standard statistical tests are mentioned consecutively in the manuscript while more complex analyses are described above. Wilcoxon rank-sum test between three groups was run using pairwise.wilcox.test R function with all P value adjusted for FDR. All reported P values are two-sided, with a significance threshold of <0.05.
Data Availability
Data from this original work are deposited in the European Genome Phenome Archive (EGA) under ID EGAS00001006884. The other already published data are deposited in the EGA and dbGap database under the following accession numbers: EGAS00001001692: PCAWG cohort (https://ega-archive.org/studies/EGAS00001001692); EGAD00001003309, 67 WGS raw data from 30 patients with MM and myeloma precursor conditions (https://www.ebi.ac.uk/ega/datasets/EGAD00001003309); phs000348.v2.p1, 22 WGS raw data from patients with MM (https://www.ncbi.nlm.nih.gov/projects/gap/cgi-bin/study.cgi?study_id=phs000348.v2.p1). All these data are available under restricted access; access can be obtained by contacting the public depository.
Supplementary Material
All supplementary figures S1-S17
R code of our analysis for driver mutation discovery in Hodgkin lymphoma WGS and WES.
R code of our analysis clock like mutational signatures in Hodgkin lymphoma WGS and WES.
Supplementary Tables S1-S20
Acknowledgments
This work was supported by the Children's Oncology Group, the Hartwell Foundation, the Gant Family Foundation, the Sylvester Comprehensive Cancer Center NCI Core Grant (P30 CA 240139), and the Memorial Sloan Kettering Cancer Center NCI Core Grant (P30 CA 008748). F. Maura is supported by the American Society of Hematology. F. Maura and O. Landgren are supported by the Riney Family Foundation. L. Giulino-Roth is supported by the NIH (K08CA219473), the Gant Family Foundation, and the Hartwell Foundation.
The publication costs of this article were defrayed in part by the payment of publication fees. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Footnotes
Note: Supplementary data for this article are available at Blood Cancer Discovery Online (https://bloodcancerdiscov.aacrjournals.org/).
Authors’ Disclosures
M.J. Oberley reports other support from Caris Life Sciences outside the submitted work. O. Landgren reports grants and personal fees from Amgen, Janssen, and Bristol Myers Squibb, other support from Takeda and Janssen, personal fees from Karyopharm, personal fees from Celgene, Adaptive Biotech, Binding Site outside the submitted work. M. Imielinski reports personal fees from ImmPACT Bio outside the submitted work. O. Elemento reports other support from OneThree Bio, grants, personal fees, and other support from Volastra Therapeutics, other support from Owkin, Freenome, personal fees from Champions Oncology, and personal fees and other support from Pionyr during the conduct of the study. M. Roshal reports other support from Auron, grants from Roche/Genetech, Cellularity, and NGM outside the submitted work. L. Giulino-Roth reports grants from Hartwell Foundation, Gant Family Foundation, and NIH during the conduct of the study. No disclosures were reported by the other authors.
Authors’ Contributions
F. Maura: Conceptualization, data curation, formal analysis, writing–original draft, project administration, writing–review and editing. B. Ziccheddu: Formal analysis. J.Z. Xiang: Data curation, formal analysis. B. Bhinder: Formal analysis. J. Rosiene: Data curation, formal analysis. F. Abascal: Formal analysis. K.H. Maclachlan: Formal analysis. K.W. Eng: Formal analysis. M. Uppal: Formal analysis. F. He: Formal analysis. W. Zhang: Formal analysis. Q. Gao: Data curation. V.D. Yellapantula: Formal analysis. V. Trujillo-Alonso: Data curation. S.I. Park: Resources. M.J. Oberley: Resources. E. Ruckdeschel: Resources. M.S. Lim: Resources. G.B. Wertheim: Resources. M.J. Barth: Resources. T.M. Horton: Supervision. A. Derkach: Formal analysis. A.E. Kovach: Resources, data curation. C.J. Forlenza: Resources, data curation. Y. Zhang: Data curation, formal analysis. O. Landgren: Formal analysis. C.H. Moskowitz: Supervision, funding acquisition. E. Cesarman: Formal analysis, supervision. M. Imielinski: Conceptualization, data curation, formal analysis, supervision. O. Elemento: Conceptualization, data curation, formal analysis, supervision. M. Roshal: Conceptualization, resources, data curation, formal analysis, supervision, writing–original draft, project administration, writing–review and editing. L. Giulino-Roth: Conceptualization, resources, data curation, formal analysis, supervision, funding acquisition, validation, writing–original draft, project administration, writing–review and editing.
References
- 1. Mathas S, Hartmann S, Kuppers R. Hodgkin lymphoma: pathology and biology. Semin Hematol 2016;53:139–47. [DOI] [PubMed] [Google Scholar]
- 2. Reichel J, Chadburn A, Rubinstein PG, Giulino-Roth L, Tam W, Liu Y, et al. Flow sorting and exome sequencing reveal the oncogenome of primary Hodgkin and Reed-Sternberg cells. Blood 2015;125:1061–72. [DOI] [PubMed] [Google Scholar]
- 3. Green MR, Monti S, Rodig SJ, Juszczynski P, Currie T, O'Donnell E, et al. Integrative analysis reveals selective 9p24.1 amplification, increased PD-1 ligand expression, and further induction via JAK2 in nodular sclerosing Hodgkin lymphoma and primary mediastinal large B-cell lymphoma. Blood 2010;116:3268–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Joos S, Kupper M, Ohl S, von Bonin F, Mechtersheimer G, Bentz M, et al. Genomic imbalances including amplification of the tyrosine kinase gene JAK2 in CD30+ Hodgkin cells. Cancer Res 2000;60:549–52. [PubMed] [Google Scholar]
- 5. Joos S, Menz CK, Wrobel G, Siebert R, Gesk S, Ohl S, et al. Classical Hodgkin lymphoma is characterized by recurrent copy number gains of the short arm of chromosome 2. Blood 2002;99:1381–7. [DOI] [PubMed] [Google Scholar]
- 6. Kato M, Sanada M, Kato I, Sato Y, Takita J, Takeuchi K, et al. Frequent inactivation of A20 in B-cell lymphomas. Nature 2009;459:712–6. [DOI] [PubMed] [Google Scholar]
- 7. Spina V, Bruscaggin A, Cuccaro A, Martini M, Di Trani M, Forestieri G, et al. Circulating tumor DNA reveals genetics, clonal evolution, and residual disease in classical Hodgkin lymphoma. Blood 2018;131:2413–25. [DOI] [PubMed] [Google Scholar]
- 8. Tiacci E, Ladewig E, Schiavoni G, Penson A, Fortini E, Pettirossi V, et al. Pervasive mutations of JAK-STAT pathway genes in classical Hodgkin lymphoma. Blood 2018;131:2454–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Wienand K, Chapuy B, Stewart C, Dunford AJ, Wu D, Kim J, et al. Genomic analyses of flow-sorted Hodgkin Reed-Sternberg cells reveal complementary mechanisms of immune evasion. Blood Adv 2019;3:4065–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Consortium ITP-CAWG. Pan-cancer analysis of whole genomes. Nature 2020;578:82–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Reichel JB, McCormick J, Fromm JR, Elemento O, Cesarman E, Roshal M. Flow-sorting and exome sequencing of the Reed-Sternberg cells of classical Hodgkin lymphoma. J Vis Exp 2017(124):54399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Tanaka N, Takahara A, Hagio T, Nishiko R, Kanayama J, Gotoh O, et al. Sequencing artifacts derived from a library preparation method using enzymatic fragmentation. PLoS One 2020;15:e0227427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Alexandrov LB, Kim J, Haradhvala NJ, Huang MN, Tian Ng AW, Wu Y, et al. The repertoire of mutational signatures in human cancer. Nature 2020;578:94–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lawrence MS, Stojanov P, Polak P, Kryukov GV, Cibulskis K, Sivachenko A, et al. Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature 2013;499:214–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Martincorena I, Raine KM, Gerstung M, Dawson KJ, Haase K, Van Loo P, et al. Universal patterns of selection in cancer and somatic tissues. Cell 2017;171:1029–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Mularoni L, Sabarinathan R, Deu-Pons J, Gonzalez-Perez A, Lopez-Bigas N. OncodriveFML: a general framework to identify coding and non-coding regions with cancer driver mutations. Genome Biol 2016;17:128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Mottok A, Renne C, Seifert M, Oppermann E, Bechstein W, Hansmann ML, et al. Inactivating SOCS1 mutations are caused by aberrant somatic hypermutation and restricted to a subset of B-cell lymphoma entities. Blood 2009;114:4503–6. [DOI] [PubMed] [Google Scholar]
- 18. Hang Q, Fei M, Hou S, Ni Q, Lu C, Zhang G, et al. Expression of Spy1 protein in human non-Hodgkin's lymphomas is correlated with phosphorylation of p27 Kip1 on Thr187 and cell proliferation. Med Oncol 2012;29:3504–14. [DOI] [PubMed] [Google Scholar]
- 19. de Miranda NF, Georgiou K, Chen L, Wu C, Gao Z, Zaravinos A, et al. Exome sequencing reveals novel mutation targets in diffuse large B-cell lymphomas derived from Chinese patients. Blood 2014;124:2544–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Balinas-Gavira C, Rodriguez MI, Andrades A, Cuadros M, Alvarez-Perez JC, Alvarez-Prado AF, et al. Frequent mutations in the amino-terminal domain of BCL7A impair its tumor suppressor role in DLBCL. Leukemia 2020;34:2722–35. [DOI] [PubMed] [Google Scholar]
- 21. Morton LM, Purdue MP, Zheng T, Wang SS, Armstrong B, Zhang Y, et al. Risk of non-Hodgkin lymphoma associated with germline variation in genes that regulate the cell cycle, apoptosis, and lymphocyte development. Cancer Epidemiol Biomarkers Prev 2009;18:1259–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Mottok A, Hung SS, Chavez EA, Woolcock B, Telenius A, Chong LC, et al. Integrative genomic analysis identifies key pathogenic mechanisms in primary mediastinal large B-cell lymphoma. Blood 2019;134:802–13. [DOI] [PubMed] [Google Scholar]
- 23. Arthur SE, Jiang A, Grande BM, Alcaide M, Cojocaru R, Rushton CK, et al. Genome-wide discovery of somatic regulatory variants in diffuse large B-cell lymphoma. Nat Commun 2018;9:4001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Vigano E, Gunawardana J, Mottok A, Van Tol T, Mak K, Chan FC, et al. Somatic IL4R mutations in primary mediastinal large B-cell lymphoma lead to constitutive JAK-STAT signaling activation. Blood 2018;131:2036–46. [DOI] [PubMed] [Google Scholar]
- 25. Zani VJ, Asou N, Jadayel D, Heward JM, Shipley J, Nacheva E, et al. Molecular cloning of complex chromosomal translocation t(8;14;12)(q24.1;q32.3;q24.1) in a Burkitt lymphoma cell line defines a new gene (BCL7A) with homology to caldesmon. Blood 1996;87:3124–34. [PubMed] [Google Scholar]
- 26. Mosquera Orgueira A, Ferreiro Ferro R, Diaz Arias JA, Aliste Santos C, Antelo Rodriguez B, Bao Perez L, et al. Detection of new drivers of frequent B-cell lymphoid neoplasms using an integrated analysis of whole genomes. PLoS One 2021;16:e0248886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ennishi D, Jiang A, Boyle M, Collinge B, Grande BM, Ben-Neriah S, et al. Double-hit gene expression signature defines a distinct subgroup of germinal center B-cell-like diffuse large B-cell lymphoma. J Clin Oncol 2019;37:190–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Mareschal S, Dubois S, Viailly PJ, Bertrand P, Bohers E, Maingonnat C, et al. Whole exome sequencing of relapsed/refractory patients expands the repertoire of somatic mutations in diffuse large B-cell lymphoma. Genes Chromosomes Cancer 2016;55:251–67. [DOI] [PubMed] [Google Scholar]
- 29. Rossi D, Trifonov V, Fangazio M, Bruscaggin A, Rasi S, Spina V, et al. The coding genome of splenic marginal zone lymphoma: activation of NOTCH2 and other pathways regulating marginal zone development. J Exp Med 2012;209:1537–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ng SB, Chung TH, Kato S, Nakamura S, Takahashi E, Ko YH, et al. Epstein-Barr virus-associated primary nodal T/NK-cell lymphoma shows a distinct molecular signature and copy number changes. Haematologica 2018;103:278–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Camus V, Viennot M, Lequesne J, Viailly PJ, Bohers E, Bessi L, et al. Targeted genotyping of circulating tumor DNA for classical Hodgkin lymphoma monitoring: a prospective study. Haematologica 2021;106:154–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Desch AK, Hartung K, Botzen A, Brobeil A, Rummel M, Kurch L, et al. Genotyping circulating tumor DNA of pediatric Hodgkin lymphoma. Leukemia 2020;34:151–66. [DOI] [PubMed] [Google Scholar]
- 33. Gomez F, Mosior M, McMichael J, Skidmore ZL, Duncavage EJ, Miller CA, et al. Ultra-deep sequencing reveals the mutational landscape of classical Hodgkin lymphoma. medRxiv [Preprint]. 2021. Available from: http://medrxiv.org/content/early/2021/07/04/2021.06.25.21258374.abstract. [DOI] [PMC free article] [PubMed]
- 34. Trengove MC, Ward AC. SOCS proteins in development and disease. Am J Clin Exp Immunol 2013;2:1–29. [PMC free article] [PubMed] [Google Scholar]
- 35. Dukers DF, van Galen JC, Giroth C, Jansen P, Sewalt RG, Otte AP, et al. Unique polycomb gene expression pattern in Hodgkin's lymphoma and Hodgkin's lymphoma-derived cell lines. Am J Pathol 2004;164:873–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Sobesky S, Mammadova L, Cirillo M, Drees EEE, Mattlener J, Dorr H, et al. In-depth cell-free DNA sequencing reveals genomic landscape of Hodgkin's lymphoma and facilitates ultrasensitive residual disease detection. Med (N Y) 2021;2:1171–93. [DOI] [PubMed] [Google Scholar]
- 37. Gulati N, Beguelin W, Giulino-Roth L. Enhancer of zeste homolog 2 (EZH2) inhibitors. Leuk Lymphoma 2018;59:1574–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Morschhauser F, Tilly H, Chaidos A, McKay P, Phillips T, Assouline S, et al. Tazemetostat for patients with relapsed or refractory follicular lymphoma: an open-label, single-arm, multicentre, phase 2 trial. Lancet Oncol 2020;21:1433–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Krishnan V, Chow MZ, Wang Z, Zhang L, Liu B, Liu X, et al. Histone H4 lysine 16 hypoacetylation is associated with defective DNA repair and premature senescence in Zmpste24-deficient mice. Proc Natl Acad Sci U S A 2011;108:12325–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Maura F, Degasperi A, Nadeu F, Leongamornlert D, Davies H, Moore L, et al. A practical guide for mutational signature analysis in hematological malignancies. Nat Commun 2019;10:2969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Rustad EH, Yellapantula V, Leongamornlert D, Bolli N, Ledergor G, Nadeu F, et al. Timing the initiation of multiple myeloma. Nat Commun 2020;11:1917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Machado HE, Mitchell E, Obro NF, Kubler K, Davies M, Leongamornlert D, et al. Diverse mutational landscapes in human lymphocytes. Nature 2022;608:724–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Gerstung M, Jolly C, Leshchiner I, Dentro SC, Gonzalez S, Rosebrock D, et al. The evolutionary history of 2,658 cancers. Nature 2020;578:122–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Chan K, Roberts SA, Klimczak LJ, Sterling JF, Saini N, Malc EP, et al. An APOBEC3A hypermutation signature is distinguishable from the signature of background mutagenesis by APOBEC3B in human cancers. Nat Genet 2015;47:1067–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Rustad EH, Nadeu F, Angelopoulos N, Ziccheddu B, Bolli N, Puente XS, et al. mmsig: a fitting approach to accurately identify somatic mutational signatures in hematological malignancies. Commun Biol 2021;4:424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Pich O, Muinos F, Lolkema MP, Steeghs N, Gonzalez-Perez A, Lopez-Bigas N. The mutational footprints of cancer therapies. Nat Genet 2019;51:1732–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Clive D, Turner N, Krehl R. Procarbazine is a potent mutagen at the heterozygous thymidine kinase (tk +/-) locus of mouse lymphoma assay. Mutagenesis 1988;3:83–7. [DOI] [PubMed] [Google Scholar]
- 48. Mudipalli A, Nadadur SS, Maccubbin AE, Gurtoo HL. Mutations induced by dacarbazine activated with cytochrome P-450. Mutat Res 1995;327:113–20. [DOI] [PubMed] [Google Scholar]
- 49. Pletsa V, Valavanis C, van Delft JH, Steenwinkel MJ, Kyrtopoulos SA. DNA damage and mutagenesis induced by procarbazine in lambda lacZ transgenic mice: evidence that bone marrow mutations do not arise primarily through miscoding by O6-methylguanine. Carcinogenesis 1997;18:2191–6. [DOI] [PubMed] [Google Scholar]
- 50. Jain MD, Ziccheddu B, Coughlin CA, Faramand R, Griswold AJ, Reid K, et al. Genomic drivers of large B-cell lymphoma resistance to CD19 CAR-T therapy. Blood 2021;138. doi 10.1182/blood-2021-148605. [DOI] [Google Scholar]
- 51. Kucab JE, Zou X, Morganella S, Joel M, Nanda AS, Nagy E, et al. A compendium of mutational signatures of environmental agents. Cell 2019;177:821–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Pich O, Cortes-Bullich A, Muinos F, Pratcorona M, Gonzalez-Perez A, Lopez-Bigas N. The evolution of hematopoietic cells under cancer therapy. Nat Commun 2021;12:4803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Alexandrov LB, Jones PH, Wedge DC, Sale JE, Campbell PJ, Nik-Zainal S, et al. Clock-like mutational processes in human somatic cells. Nat Genet 2015;47:1402–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Kasar S, Kim J, Improgo R, Tiao G, Polak P, Haradhvala N, et al. Whole-genome sequencing reveals activation-induced cytidine deaminase signatures during indolent chronic lymphocytic leukaemia evolution. Nat Commun 2015;6:8866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Maura F, Rustad EH, Yellapantula V, Luksza M, Hoyos D, Maclachlan KH, et al. Role of AID in the temporal pattern of acquisition of driver mutations in multiple myeloma. Leukemia 2020;34:1476–80. [DOI] [PubMed] [Google Scholar]
- 56. Bielski CM, Zehir A, Penson AV, Donoghue MTA, Chatila W, Armenia J, et al. Genome doubling shapes the evolution and prognosis of advanced cancers. Nat Genet 2018;50:1189–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Martin-Subero JI, Klapper W, Sotnikova A, Callet-Bauchu E, Harder L, Bastard C, et al. Chromosomal breakpoints affecting immunoglobulin loci are recurrent in Hodgkin and Reed-Sternberg cells of classical Hodgkin lymphoma. Cancer Res 2006;66:10332–8. [DOI] [PubMed] [Google Scholar]
- 58. Roemer MG, Advani RH, Ligon AH, Natkunam Y, Redd RA, Homer H, et al. PD-L1 and PD-L2 genetic alterations define classical hodgkin lymphoma and predict outcome. J Clin Oncol 2016;34:2690–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Mermel CH, Schumacher SE, Hill B, Meyerson ML, Beroukhim R, Getz G. GISTIC2.0 facilitates sensitive and confident localization of the targets of focal somatic copy-number alteration in human cancers. Genome Biol 2011;12:R41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Maura F, Bolli N, Angelopoulos N, Dawson KJ, Leongamornlert D, Martincorena I, et al. Genomic landscape and chronological reconstruction of driver events in multiple myeloma. Nat Commun 2019;10:3835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Papaemmanuil E, Gerstung M, Bullinger L, Gaidzik VI, Paschka P, Roberts ND, et al. Genomic classification and prognosis in acute myeloid leukemia. N Engl J Med 2016;374:2209–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Dentro SC, Leshchiner I, Haase K, Tarabichi M, Wintersinger J, Deshwar AG, et al. Characterizing genetic intra-tumor heterogeneity across 2,658 human cancer genomes. Cell 2021;184:2239–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Landau HJ, Yellapantula V, Diamond BT, Rustad EH, Maclachlan KH, Gundem G, et al. Accelerated single cell seeding in relapsed multiple myeloma. Nat Commun 2020;11:3617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Hadi K, Yao X, Behr JM, Deshpande A, Xanthopoulakis C, Tian H, et al. Distinct classes of complex structural variation uncovered across thousands of cancer genome graphs. Cell 2020;183:197–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Rustad EH, Yellapantula VD, Glodzik D, Maclachlan KH, Diamond B, Boyle EM, et al. Revealing the impact of structural variants in multiple myeloma. Blood Cancer Discov 2020;1:258–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Steidl C, Shah SP, Woolcock BW, Rui L, Kawahara M, Farinha P, et al. MHC class II transactivator CIITA is a recurrent gene fusion partner in lymphoid cancers. Nature 2011;471:377–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Lieber MR. Mechanisms of human lymphoid chromosomal translocations. Nat Rev Cancer 2016;16:387–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Papaemmanuil E, Rapado I, Li Y, Potter NE, Wedge DC, Tubio J, et al. RAG-mediated recombination is the predominant driver of oncogenic rearrangement in ETV6-RUNX1 acute lymphoblastic leukemia. Nat Genet 2014;46:116–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Nadeu F, Martin-Garcia D, Clot G, Diaz-Navarro A, Duran-Ferrer M, Navarro A, et al. Genomic and epigenomic insights into the origin, pathogenesis, and clinical behavior of mantle cell lymphoma subtypes. Blood 2020;136:1419–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Kanzler H, Kuppers R, Hansmann ML, Rajewsky K. Hodgkin and Reed-Sternberg cells in Hodgkin's disease represent the outgrowth of a dominant tumor clone derived from (crippled) germinal center B cells. J Exp Med 1996;184:1495–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Kuppers R, Rajewsky K, Zhao M, Simons G, Laumann R, Fischer R, et al. Hodgkin disease: Hodgkin and Reed-Sternberg cells picked from histological sections show clonal immunoglobulin gene rearrangements and appear to be derived from B cells at various stages of development. Proc Natl Acad Sci U S A 1994;91:10962–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Kuppers R, Rajewsky K, Zhao M, Simons G, Laumann R, Fischer R, et al. Hodgkin's disease: clonal Ig gene rearrangements in hodgkin and reed-sternberg cells picked from histological sections. Ann N Y Acad Sci 1995;764:523–4. [DOI] [PubMed] [Google Scholar]
- 73. Kuppers R, Zhao M, Hansmann ML, Rajewsky K. Tracing B cell development in human germinal centres by molecular analysis of single cells picked from histological sections. EMBO J 1993;12:4955–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Alvarez-Prado AF, Perez-Duran P, Perez-Garcia A, Benguria A, Torroja C, de Yebenes VG, et al. A broad atlas of somatic hypermutation allows prediction of activation-induced deaminase targets. J Exp Med 2018;215:761–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Chapuy B, Stewart C, Dunford AJ, Kim J, Kamburov A, Redd RA, et al. Molecular subtypes of diffuse large B cell lymphoma are associated with distinct pathogenic mechanisms and outcomes. Nat Med 2018;24:679–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Hubschmann D, Kleinheinz K, Wagener R, Bernhart SH, Lopez C, Toprak UH, et al. Mutational mechanisms shaping the coding and noncoding genome of germinal center derived B-cell lymphomas. Leukemia 2021;35:2002–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Yusufova N, Kloetgen A, Teater M, Osunsade A, Camarillo JM, Chin CR, et al. Histone H1 loss drives lymphoma by disrupting 3D chromatin architecture. Nature 2021;589:299–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Nadeu F, Mas-de-Les-Valls R, Navarro A, Royo R, Martin S, Villamor N, et al. IgCaller for reconstructing immunoglobulin gene rearrangements and oncogenic translocations from whole-genome sequencing in lymphoid neoplasms. Nat Commun 2020;11:3390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Poppema S, Kaleta J, Hepperle B. Chromosomal abnormalities in patients with Hodgkin's disease: evidence for frequent involvement of the 14q chromosomal region but infrequent bcl-2 gene rearrangement in Reed-Sternberg cells. J Natl Cancer Inst 1992;84:1789–93. [DOI] [PubMed] [Google Scholar]
- 80. Stetler-Stevenson M, Crush-Stanton S, Cossman J. Involvement of the bcl-2 gene in Hodgkin's disease. J Natl Cancer Inst 1990;82:855–8. [DOI] [PubMed] [Google Scholar]
- 81. Szymanowska N, Klapper W, Gesk S, Kuppers R, Martin-Subero JI, Siebert R. BCL2 and BCL3 are recurrent translocation partners of the IGH locus. Cancer Genet Cytogenet 2008;186:110–4. [DOI] [PubMed] [Google Scholar]
- 82. Wlodarska I, Dierickx D, U. P, Doms K, Graux C, Van Hoof A, et al. IGH-mediated translocations, recurrent in classic Hodgkin lymphoma, frequently correlate with an aggressive behavior. Blood 2016;128:2922. [Google Scholar]
- 83. Paulsson K, Lilljebjorn H, Biloglav A, Olsson L, Rissler M, Castor A, et al. The genomic landscape of high hyperdiploid childhood acute lymphoblastic leukemia. Nat Genet 2015;47:672–6. [DOI] [PubMed] [Google Scholar]
- 84. Weber-Matthiesen K, Deerberg J, Poetsch M, Grote W, Schlegelberger B. Numerical chromosome aberrations are present within the CD30+ hodgkin and reed-sternberg cells in 100% of analyzed cases of Hodgkin's disease. Blood 1995;86:1464–8. [PubMed] [Google Scholar]
- 85. Cagan A, Baez-Ortega A, Brzozowska N, Abascal F, Coorens THH, Sanders MA, et al. Somatic mutation rates scale with lifespan across mammals. Nature 2022;604:517–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Robinson PS, Coorens THH, Palles C, Mitchell E, Abascal F, Olafsson S, et al. Increased somatic mutation burdens in normal human cells due to defective DNA polymerases. Nat Genet 2021;53:1434–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Sanders MA, Chew E, Flensburg C, Zeilemaker A, Miller SE, Al Hinai AS, et al. MBD4 guards against methylation damage and germ line deficiency predisposes to clonal hematopoiesis and early-onset AML. Blood 2018;132:1526–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Maura F, Weinhold N, Diamond B, Kazandjian D, Rasche L, Morgan G, et al. The mutagenic impact of melphalan in multiple myeloma. Leukemia 2021;35:2145–50. [DOI] [PubMed] [Google Scholar]
- 89. Poos AM, Giesen N, Catalano C, Paramasivam N, Huebschmann D, John L, et al. Comprehensive comparison of early relapse and end-stage relapsed refractory multiple myeloma. Blood 2020;136. doi 10.1182/blood-2020-141611. [DOI] [Google Scholar]
- 90. Merelli I, Guffanti A, Fabbri M, Cocito A, Furia L, Grazini U, et al. RSSsite: a reference database and prediction tool for the identification of cryptic Recombination Signal Sequences in human and murine genomes. Nucleic Acids Res 2010;38( Web Server issue):W262–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Cameron DL, Jacobs N, Roepman P, Priestley P, Cuppen E, Papenfuss AT. VIRUSBreakend: viral integration recognition using single breakends. Bioinformatics 2021;37:3115–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Frank D, Cesarman E, Liu YF, Michler RE, Knowles DM. Posttransplantation lymphoproliferative disorders frequently contain type A and not type B Epstein-Barr virus. Blood 1995;85:1396–403. [PubMed] [Google Scholar]
- 93. Oben B, Froyen G, Maclachlan KH, Leongamornlert D, Abascal F, Zheng-Lin B, et al. Whole-genome sequencing reveals progressive versus stable myeloma precursor conditions as two distinct entities. Nat Commun 2021;12:1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
All supplementary figures S1-S17
R code of our analysis for driver mutation discovery in Hodgkin lymphoma WGS and WES.
R code of our analysis clock like mutational signatures in Hodgkin lymphoma WGS and WES.
Supplementary Tables S1-S20
Data Availability Statement
Data from this original work are deposited in the European Genome Phenome Archive (EGA) under ID EGAS00001006884. The other already published data are deposited in the EGA and dbGap database under the following accession numbers: EGAS00001001692: PCAWG cohort (https://ega-archive.org/studies/EGAS00001001692); EGAD00001003309, 67 WGS raw data from 30 patients with MM and myeloma precursor conditions (https://www.ebi.ac.uk/ega/datasets/EGAD00001003309); phs000348.v2.p1, 22 WGS raw data from patients with MM (https://www.ncbi.nlm.nih.gov/projects/gap/cgi-bin/study.cgi?study_id=phs000348.v2.p1). All these data are available under restricted access; access can be obtained by contacting the public depository.