Abstract
Rapid, sensitive, simultaneous quantification of multiple biomarkers in point-of-care (POC) settings could improve the diagnosis and management of sepsis, a common, potentially life-threatening condition. Compared to high-end commercial analytical systems, POC systems are often limited by low sensitivity, limited multiplexing capability, or low throughput. Here, we report an ultra-sensitive, multiplexed plasmonic sensing technology integrating chemifluorescence signal enhancement with plasmon-enhanced fluorescence detection. Using a portable imaging system, the dual chemical and plasmonic amplification enabled rapid analysis of multiple cytokine biomarkers in 1 hr with sub-pg/mL sensitivities. Furthermore, we also developed a plasmonic sensing chip based on nanoparticle-spiked gold nanodimple structures fabricated by wafer-scale batch processes. We used the system to detect six cytokines directly from clinical plasma samples (n = 20) and showed 100% accuracy for sepsis detection. The described technology could be employed in rapid, ultrasensitive, multiplexed plasmonic sensing in POC settings for myriad clinical conditions.
Keywords: sepsis, cytokines, plasmonics, chemifluorescence, point-of-care
Graphical Abstract
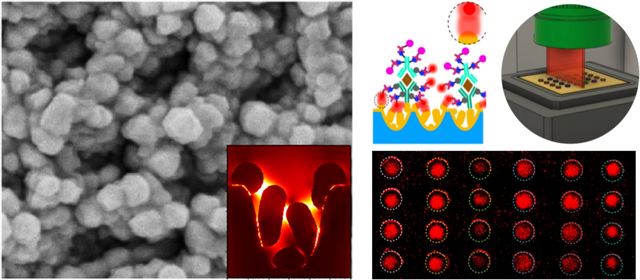
Sepsis remains a life-threatening medical emergency and the leading cause of mortality in patients with bacterial infections. In 2017, there were 48.9 million sepsis cases estimated worldwide, which accounted for 20% of all global deaths with a 50% mortality rate; the highest burden occurred in low- and medium-income countries.1,2 In North America, sepsis accounts for approximately 215,000 deaths annually and incurs $20 billion in hospital costs.3,4 Unfortunately, the diagnosis of sepsis often occurs too late and is primarily based on clinical findings and vital signs rather than molecular signatures. Late diagnosis invariably leads to multiorgan failure5 with soaring healthcare costs and abysmal prognoses. Therefore, it is critical to develop tools to identify patients at risk of sepsis or worse outcomes, particularly in resource-limited, point-of-care (POC) settings.
Current sepsis research has focused mainly on soluble biomarkers and specific pro-inflammatory and anti-inflammatory cytokines as readouts.6–9 By serially measuring the expression levels of multiple cytokines, the progression of sepsis can be better tracked.10–13 Various commercial systems allow for such multiplexed cytokine measurements, including Luminex (fluorescent magnetic bead-based assay),14–16 Quanterix (chemiluminescence assay),17 Mesoscale (electrochemiluminescence assay),14 among others. These assays and systems achieve sub pg/mL sensitivity but at the expense of assay time, typically more than 3 hours, and high equipment costs, making them less suitable in resource-limited POC settings.
POC technologies, which are low-cost and easy to use with minimal infrastructure requirements, could provide rapid actionable information in real-time and on-site for sepsis evaluation and treatment.18,19 POC systems to detect single analytes have been developed, such as blood lactate (fingerpick sampling),20 procalcitonin (microfluidics),21,22 presepsin (chemiluminescence assay),23 interleukin 6 (IL-6, Coulter counting),24 interleukin 3 (IL-3, magneto-electrochemical sensor),25 among others. In general, single-analyte methods are less sensitive and specific for predicting the onset of cytokine storm, a multifactorial process. Multiplexed cytokine detection has been demonstrated using a single-step sandwich assay on epoxy slides,26,27 achieving a 2.5-hour assay time and limit of detection (LOD) in the range of tenths of pg/mL to μg/mL (depending on the targets). The assay time could not be further shortened because it would sacrifice the LOD28,29, as demonstrated in the total internal reflection fluorescence microarray biochip, having an assay time of 25 min but a LOD in the range of ng/mL.30 Multiplexed cytokine detection was presented using a plasmonic gold substrate, achieving high sensitivity in sub-pg/mL, but the reported assay time was ~7.5 hrs.31 As a result, a POC device capable of multiplexed cytokine detection with high sensitivity in sub-pg/mL with a short assay time is still highly desired.32 This requires a robust signal amplification method that can achieve sufficient detectable signals in a short assay time.
Here, we report on the development and validation of a dual-enhanced plasmonic sensing technology exploiting both plasmonic enhancement from nanoplasmonic gold substrates and chemifluorescence enhancement via tyramine signal amplification to achieve sub-pg/mL sensitivities of sepsis-associated cytokines in 1 hr. We also developed low-cost plasmonic chips fabricated on a wafer scale and an automated, portable laser line-scanning imaging system to facilitate microarray-type analysis in a POC setting. The assay and imaging system could be well poised for longitudinal monitoring and managing septic patients.
RESULTS AND DISCUSSION
Dual-enhanced plasmonic sensing technology
The detection technology is based on plasmon-enhanced fluorescence detection and chemifluorescence amplification (Figure 1a) strategies, called DuPLUS (dual-enhanced plasmonic ultrasensitive sensing). We first created a plasmonic sensing chip with nanoparticles spiked on gold nanodimple arrays (NPOD) to excite localized surface plasmon resonances (LSPRs) and amplify fluorescence signals for immunoassay. We also employed tyramide chemifluorescence signal amplification33 to further improve the assay’s sensitivity. Tyramide chemifluorescence signal amplification is a catalyzed reporter deposition technique used to detect a low abundance of antigens. This uses horseradish peroxide (HRP) to catalyze the deposition of tyramide molecules on or near the antigens. Subsequently, fluorophores are bound on the deposited tyramide molecules for fluorescence detection. In the DuPLUS assay, capture antibodies of respective cytokines or chemokines were first functionalized on the NPOD substrate. After incubation with target molecules, respective biotinylated detection antibodies were incubated, followed by the binding of streptavidin-HRP. With the presence of HRP and hydrogen peroxide, the activation of biotinylated tyramide molecules was catalyzed to covalently bind to tyrosine residues on antibodies and proteins near the HRP. Finally, streptavidin-bound AF647 fluorophore molecules were covalently bound to the available biotin molecules. The fluorescence emission from the fluorophore AF647 was enhanced by the localized plasmonic effect of the underlying NPOD structures, in which the emission spectrum achieves high electric field enhancements by plasmonic coupling.34 The dual-enhancement could greatly improve the detection sensitivity while keeping total assay times around 1 hr.
Figure 1. Dual-enhanced plasmonic ultrasensitive sensing (DuPLUS) for multiplexed cytokine analysis.
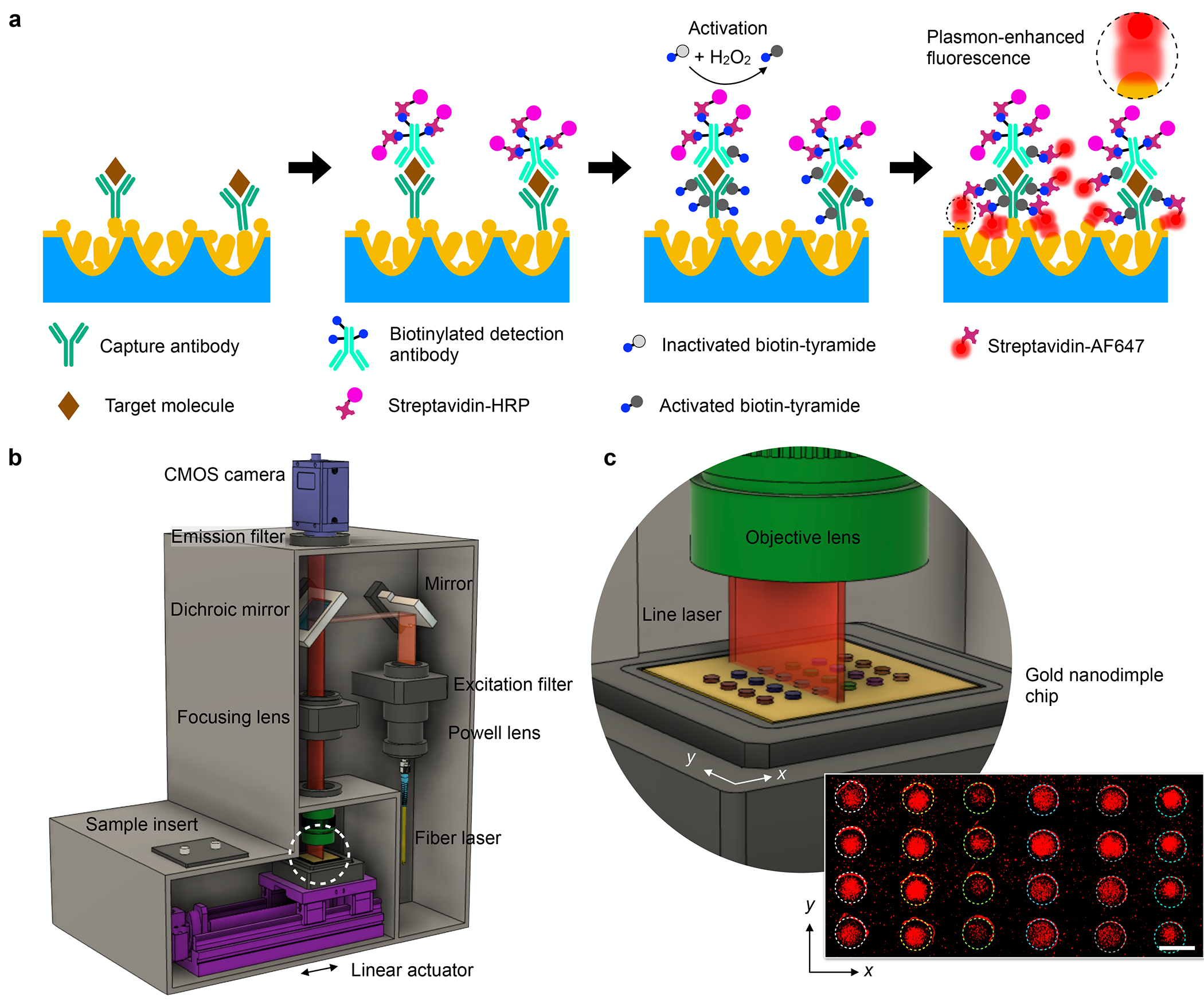
(a) Schematic of DuPLUS assay integrating a plasmon-enhanced chemifluorescence assay with tyramide signal amplification. (b) Schematic of a line-scanning laser imaging system to measure multiple sensing arrays in a single chip. (c) Zoomed-in schematic showing scanning of sensing arrays using a line laser indicated by a white dashed circle in (b). A linear actuator moves a chip along the x-direction to scan multiple sensing arrays. The inset shows the scanned fluorescence image of multiplexed cytokine measurement. Scale bar: 2 mm.
For point-of-care analysis, we developed an automatic line-scanning imaging system (Figures 1b and S1). The system has a dimension of (W × H × D) 24 cm × 36 cm × 32 cm and weighs about 12 kg. A 638-nm laser was coupled onto fiber optics and converted to a line laser using a Powell lens. The line laser passed through an excitation filter and then was reflected by a mirror into the dichroic mirror and focused by an objective lens to excite fluorophores bound on the NPOD chip. The fluorescence emission was detected by a CMOS camera placed on top of the imaging system. The NPOD chip (30 mm × 30 mm) was placed on a chip holder and sat on a stage fixed on a linear actuator (Figure S1a). The NPOD chip was scanned by moving the stage in the x-direction (Figures 1c and S1c). The line-scanning imaging system has a scanning length of 12 mm in the y-direction, defined by the length of the line laser and camera’s field of view, but a much larger scanning length of more than 30 mm in the x-direction, facilitated by the linear actuator. A representative image (12 mm × 24 mm) of the detection of 6 different cytokines with quadruplicate measurements on a single NPOD chip was shown as an inset in Figure 1c.
Wafer-scale batch fabrication of NPOD chip
Low-cost, high-throughput chip fabrication often becomes a bottleneck in translating plasmonic sensors for clinical applications. To address this limitation, we designed and fabricated large-size NPOD chips through batch processes (Figures 2a–b). In brief, we first formed high-density nanodimple structures by oxygen ion beam sputtering of a polyethylene naphthalate (PEN) film (9 cm × 9 cm, Figure S2a)35 and subsequently depositing a 100 nm-thick Au layer. The thickness of the Au layer was determined to satisfy the signal enhancement of the Au nanostructures while avoiding optical losses and Au nanoparticle aggregation. These processes produced uniform sizes of Au nanodimples (104 ±18 nm, Figures 2c–d and S2b–c) across the entire PEN film. To create additional plasmonic hotspots, we coated the Au surface with a self-assembled monolayer of 1H,1H,2H,2H-perfluorodecanethiol (PFDT). Herein, the 1 nm-thick self-assembled PFDT layer provides extremely low surface energy compared to other spacing materials (Table S1), having 100-fold lower surface energy than Au (0.015 J/m2 for PFDT and 1.54 J/m2 for Au). We previously showed that Au deposition on the low-energy PFDT surface self-forms Au nanoparticles from Au deposition.34 Similarly, additional Au deposition on the PFDT-coated surface enabled the growth of Au nanoparticles at defect sites, which resulted in the formation of spherical Au islands by the Volmer–Weber mode36 (Figures 2e–f and S3). We can control the size of Au nanoparticles formed on the nanodimples from the deposited Au film thicknesses (Figures 2g–h, Figure S4). The fabricated 3D structures possess plasmonic hotspots made at the nanogaps between adjacent AuNPs as well as between the AuNP and Au nanodimples. Figures 2i–j show the E-field distributions inside the nanodimple structure calculated using the finite-difference time-domain (FDTD) method with normal incident light at 638 nm. It shows that strong E-fields were confined between the Au nanostructures in proximity (i.e., plasmonic hotspots), for instance, between the adjacent AuNPs (sub-10 nm gaps) or between the AuNPs and Au nanodimples via PFDT layer (1 nm gaps). The curved nanodimple structures provide a high-density hotspot in the 3D space.37
Figure 2. Fabrication and characterization of Au nanoparticle on nanodimples (NPOD) chip.
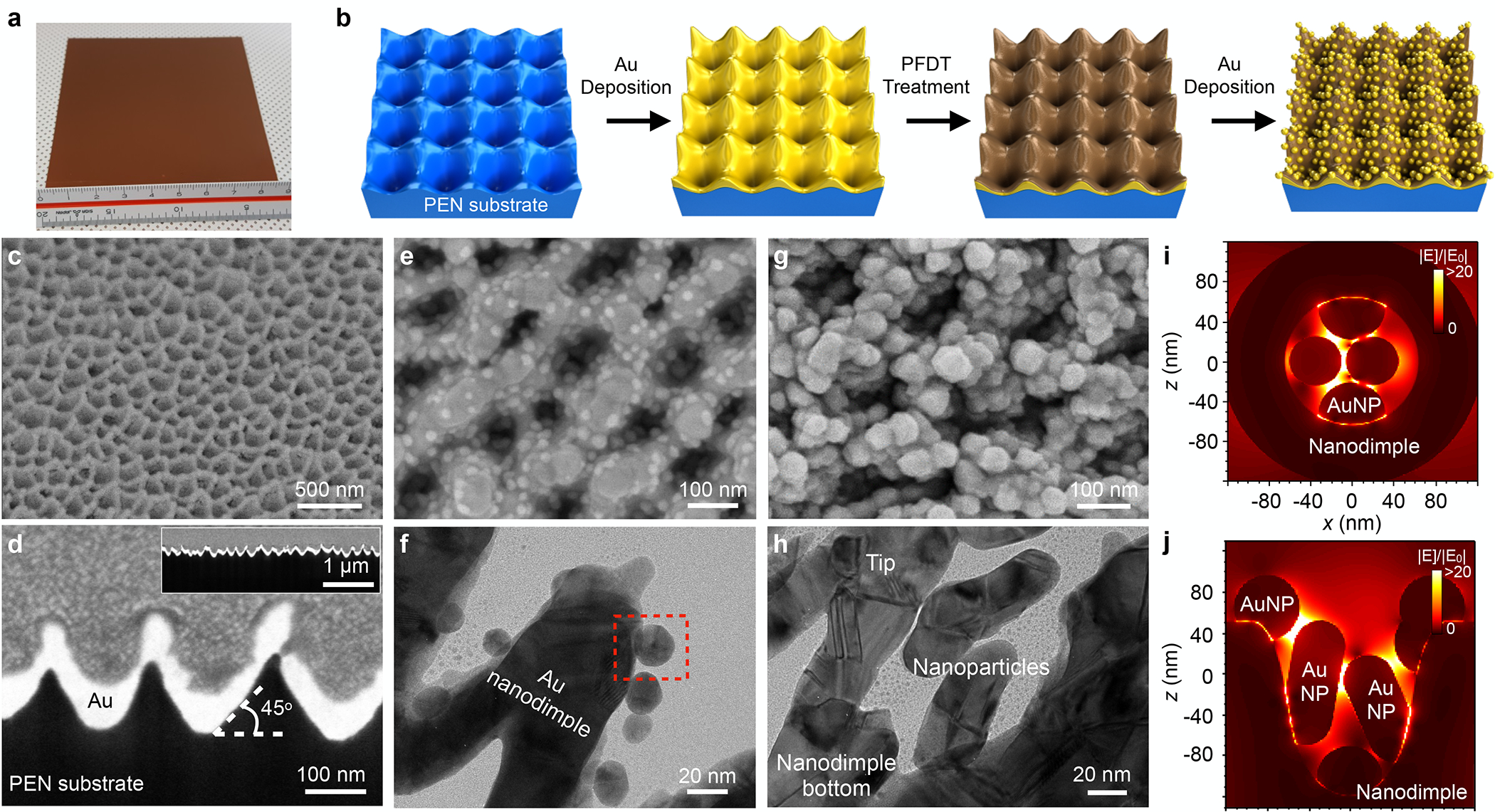
(a) Photograph of fabricated NPOD chip in 9 cm × 9 cm. The chip can be cut into smaller sizes. (b) The schematic diagram for the fabrication procedure of the NPOD chip. Reactive ion-etching of a polyethylene naphthalate (PEN) substrate and subsequent Au deposition make high-density gold nanodimple structures. The self-assembled monolayer coating of 1H,1H,2H,2H-perfluorodecanethiol (PFDT) and additional Au deposition produce NPOD chips. (c-d) Scanning electron micrographs (SEM) of Au nanodimple structures on top (c) and cross-section (e) views. The inset of (d) shows the cross-section in low magnification to show the uniformity of the high-density nanodimple structures. (e-f) SEM (e) and transmission electron micrograph (TEM, f) of NPOD structures after 10 nm Au deposition on PFDT coating. A zoomed-in image of a red dashed box region is shown in Figure S3. (g-h) Top view of SEM (g) and TEM (h) of NPOD structures after 80 nm Au deposition on PFDT coating. (i-j) 3D finite-difference time-domain (FDTD) simulations of NPOD structures with 80 nm Au deposition in the top (i) and side (j) views, showing plasmonic hotspots between Au nanoparticles as well as Au nanoparticle and nanodimple structures.
We next characterized the optical properties of the NPOD chip. First, we measured Rayleigh scattering spectra of the NPOD chips with varying thicknesses of deposited Au (i.e., varying sizes of AuNPs) ranging from 40 to 100 nm using dark-field spectroscopy (Figure S5). By increasing the deposition thickness from 40 to 80 nm, the scattering of the NPOD chip became more intense, indicating the stronger enhancement of electromagnetic fields in the narrower gaps. However, further increasing the Au deposition thickness to 100 nm led to a lower scattering signal than 80 nm. The reduced scattering could be due to the aggregation of AuNPs. It can be observed from the scattering spectra that the resonant peak shifted to a longer wavelength when the Au deposition thickness increased. The resonant peak was approximately 650 nm for the 80-nm Au deposition, exhibiting a good agreement with the true color of the NPOD chip, as shown in the inset in Figure S5a. We further investigated the optimal Au deposition thickness for Au nanoparticle formation on the nanodimples using surface-enhanced Raman scattering (SERS). We measured Raman spectra of methylene blue molecules coated on the NPOD chip surfaces made with different Au thicknesses (40, 60, 80, and 100 nm). We obtained Raman spectra at the incident wavelengths of 638 and 785 nm, as shown in Figures S5b and 5c, respectively. Based on the SERS spectra, Raman scattering was greatly enhanced with incident light at 638 nm compared to those with incident light at 785 nm. For both cases, the most intense Raman scattering signal was observed from the 80 nm-thick NPOD chip, achieving 106 and 5 × 105 counts per second (CPS) with an incident wavelength of 638 and 785 nm, respectively. Those results were coincident with the scattering properties (Figure S5a). Compared to Au nanodimple structures without Au nanoparticles, the NPOD chip showed an additional 250-fold Raman enhancement at 1625 cm−1 with the incident wavelength at 638 nm by forming Au nanoparticles on the nanodimple structures (Figure S5d). The Raman measurements also show good uniformity of fabricated NPOD substrates (Figure S6), showing a coefficient of variation of less than 10% from 36 random spot measurements across a 9 cm × 9 cm area. It also shows good inter-chip uniformity with a coefficient of variation of 7.1% for ten random spot measurements of chips from four different batches (Figure S7).
Characterization of DuPLUS assay on NPOD chip
We characterized the fluorescence signal enhancement of the NPOD chip as compared to the glass slide and plain Au substrate. Figure 3a shows fluorescence images of assay signals performed on different substrates. The chemifluorescence assay on the glass slide enhanced the signal by 3-fold, whereas the plasmonic effect further enhanced the signal by 15.3-fold on plain Au and ultimately by 895.5 on the NPOD chip (Figure 3b). We also compared the fluorescence signal enhancement before and after spiking nanoparticles. A 30% higher fluorescence signal was measured by adding the spiked nanoparticles on nanodimple arrays (P < 0.0001) without a significant difference in the background signal (Figure S8). The assay showed good uniformity and reproducibility between measurements within the same batch, and different batches made 2 months apart in a total period of 6 months. The coefficients of variance (COV) were less than 10.1% within the same batch and 8.2% between batches with no significant difference (P = 0.64, Kruskal-Wallis test, Figure 3c).
Figure 3. DuPLUS assay characterization.
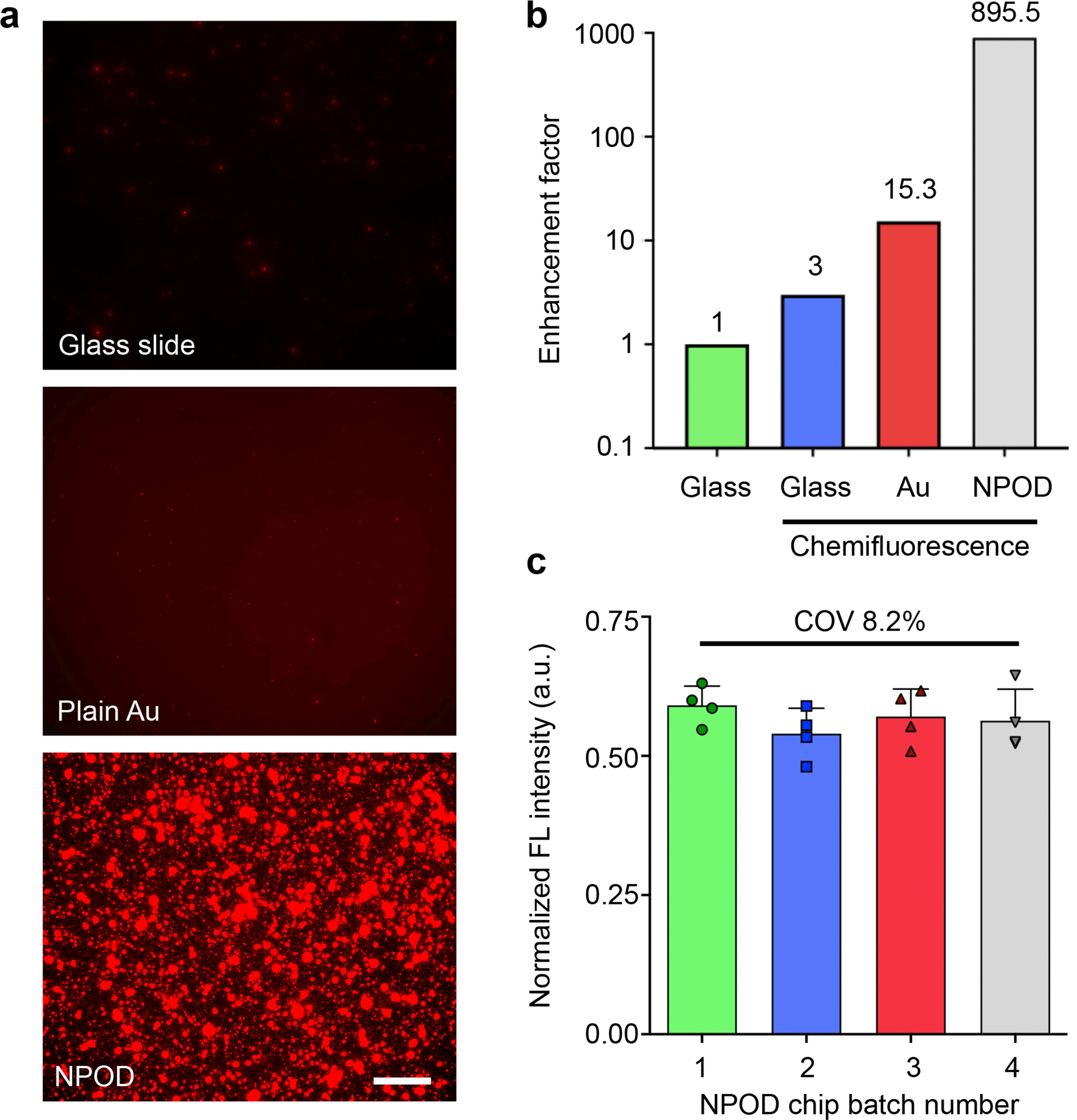
(a) Representative fluorescence images of DuPLUS assay using different substrates: glass, plain Au, and NPOD substrates. Scale bar: 20 μm. (b) Comparison of signal-to-noise ratio in different substrates and assays: fluorescence-based assay on a glass slide, chemifluorescence assay on a glass slide, a plain Au chip, and an NPOD chip. A dual plasmon-enhanced NPOD chip achieved an enhancement of 896-fold as compared to a fluorescence-based assay on a glass slide. (c) Uniformity and reproducibility using different batches of NPOD chips. Each batch was made 2 months apart in a total period of 6 months. The coefficient of variation is less than 8.2%.
The high signal enhancement and uniformity achieved in the NPOD chip highlighted the platform’s promise for molecular profiling. We further characterized the DuPLUS assay with the NPOD chip by comparing it with commercial enzyme-linked immunosorbent assay (ELISA) kits in detecting the chemokine MCP-1. The results show that the DuPLUS assay signal is enhanced significantly (120-fold with 100 pg/mL of MCP-1) compared to the conventional ELISA kit. Thus the limit of detection (LOD) is improved 35-fold, down to 0.1 pg/mL (Figure 4a). The LOD of 0.1 pg/mL was maintained for targets spiked in human plasma samples (Figure S9). It shows that the DuPLUS assay is capable of detecting target molecules directly from human plasma samples. Subsequently, we applied the DuPLUS assay to detect six different cytokines, including IL-3, IL-6, IL-10, IL-1β, TNF-α, and MCP-1. Figure S1a shows spotting 4 arrays of capture antibody spots per target onto the NPOD chip to measure the six cytokines in quadruplicates; the array is scanned and imaged by the line-scanning imaging system. The DuPLUS assay demonstrated sub-pg/mL sensitivities for all six cytokines (Figures 4b and 4c). Compared to previously reported detection platforms (Table 1), the DuPLUS assay demonstrated higher sensitivity than most platforms but slightly lower than the plasmonic gold film31 (e.g., 0.5 pg/mL vs. 0.06 pg/mL for IL-6). However, among those with sub-pg/mL sensitivities, the DuPLUS assay showed the shortest assay time (1 h), which is essential for a clinical workflow of rapid treatment. The assay also showed excellent specificity among the six targets with negligible cross-reactivity in detecting the six cytokines (Figure 4d).
Figure 4. Measurements of cytokines using the DuPLUS assay.
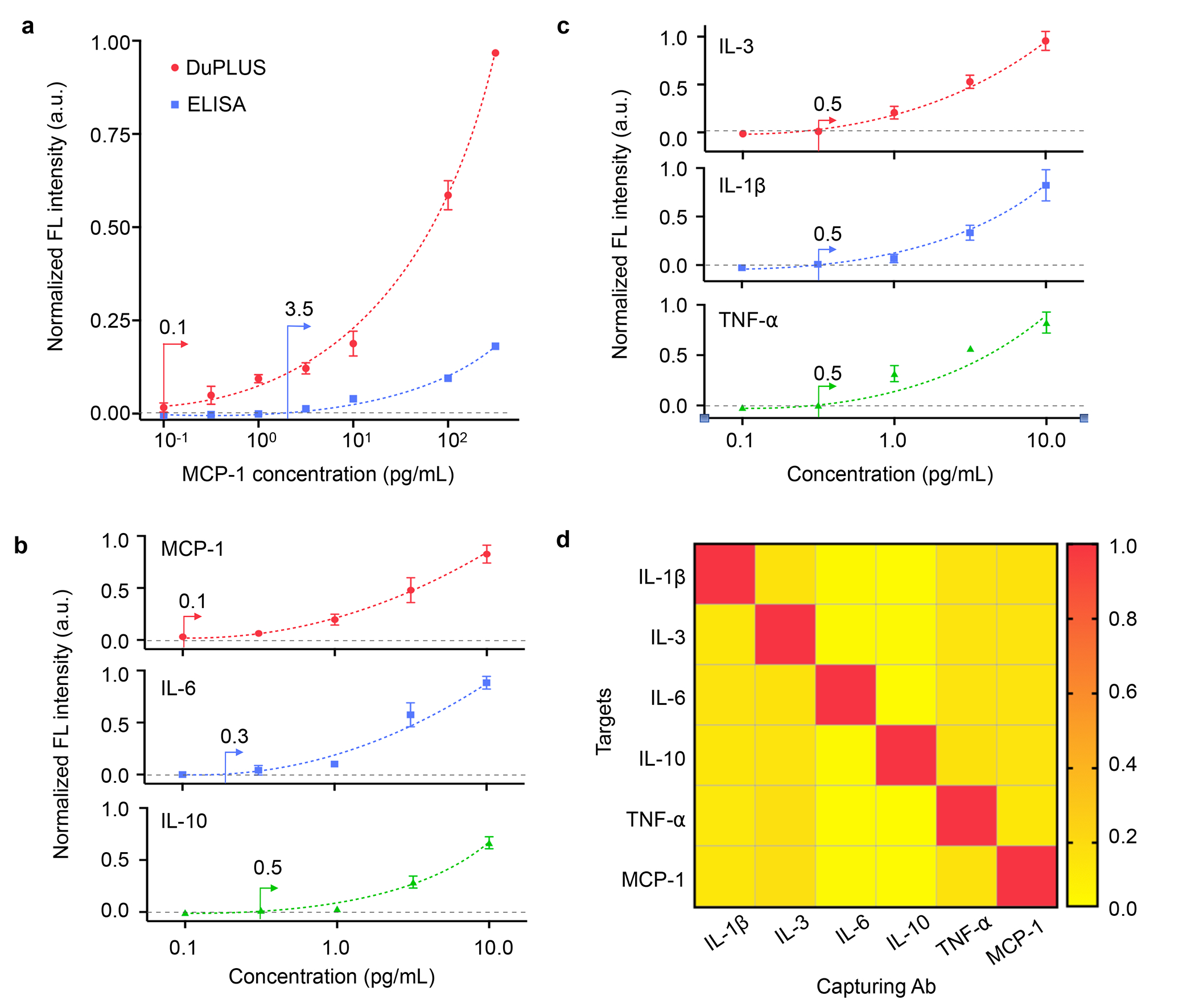
(a) Comparison of MCP-1 detection using DuPLUS and conventional ELISA. (b) Titration curves of MCP-1, IL-6, and IL-10 in diluted human plasma. (c) Titration curves of IL-3, IL-1β, and TNF-α in diluted human plasma. (d) Specificity test on all 6 cytokine/chemokine detection.
Table 1.
Comparison of detection specifications of various platforms for the same targets
| Platforms | Targets | LOD (pg/mL) |
LOQ (pg/mL) |
Assay Time (hr) |
|---|---|---|---|---|
| Luminex [14] | IL-10 | - | 11.6 | 4 |
| TNFα | - | 5.9 | ||
| Mesoscale [14] | IL-10 | - | 3.6 | 3.5 |
| TNFα | - | 3.2 | ||
| Quanterix (from Vendor datasheet) | IL-1β | 0.66 | 6.3 | 2.5 |
| IL-6 | 1.52 | 4.8 | ||
| TNF | 1.67 | 24.2 | ||
| Biochip Coulter counter [24] | IL-6 | 127 | - | 3.5 |
| Magneto-electrochemical sensor [25] | IL-3 | 5 | - | 1 |
| Magneto-fluorescence sensor [26] | IL-6 | 1.2 | 7 | 2.5 |
| Fluorescence-based sandwich assay [27] | IL-6 | 15000 | - | 2.5 |
| IL-10 | 65000 | - | ||
| TNFα | 40000 | - | ||
| Total internal reflection fluorescence [30] | IL-6 | 90 | - | 0.42 |
| Plasmonic gold substrates [31] | IL-1β | 0.07 | - | 7.5 |
| IL-6 | 0.06 | - | ||
| TNF | 0.47 | - | ||
| DuPLUS (our work) | IL-3 | 0.5 | 1.31 | 1 |
| IL-1β | 0.5 | 2.76 | ||
| IL-6 | 0.3 | 0.87 | ||
| IL-10 | 0.5 | 1.30 | ||
| TNFα | 0.5 | 1.32 | ||
| MCP-1 | 0.1 | 0.24 |
Clinical sample testing
Following the assay validation, we analyzed clinical plasma samples from 20 human subjects, 15 patients with sepsis, and 5 age-matched controls. The demographic summary of septic and healthy control patients is shown in Tables S2 and S3. The test was done in a double-blinded manner. Table S4 shows the threshold levels of the six cytokines, in which the values were set based on the LODs of commercial ELISA kits. In each sample, we included three standards - one negative and two positives at 1× and 2× thresholds. We first validated the DuPLUS assay on an NPOD chip against fluorescence-based commercial ELISA kits by measuring the IL-6 expression levels of all clinical plasma samples. The measured fluorescence signal of each clinical plasma sample using the DuPLUS assay and the commercial ELISA kits showed an excellent correlation (Spearman r = 0.9674, P < 0.0001; Figure S10).
Figure 5a shows the measured expression levels of the six cytokines in the clinical plasma samples as waterfall plots. The data show that IL-6 was elevated across all patients with sepsis but only marginally in some patients. Additional cytokines were also elevated in some but not all patients. A combined value integrating the measured cytokines (i.e., a linear sum of six cytokine signals) yielded the most discriminatory power between septic and control patients (Figure 5b). We determined cut-off values of individual markers from receiver operating characteristic curves that maximize the classification accuracies. Then, the cut-off of combined markers (4.1) was determined from a sum of the cut-off values of individual markers. With the cut-off value, all sepsis cases were accurately detected from the combined scores (Figure 5c).
Figure 5. Sepsis detection from clinical samples.
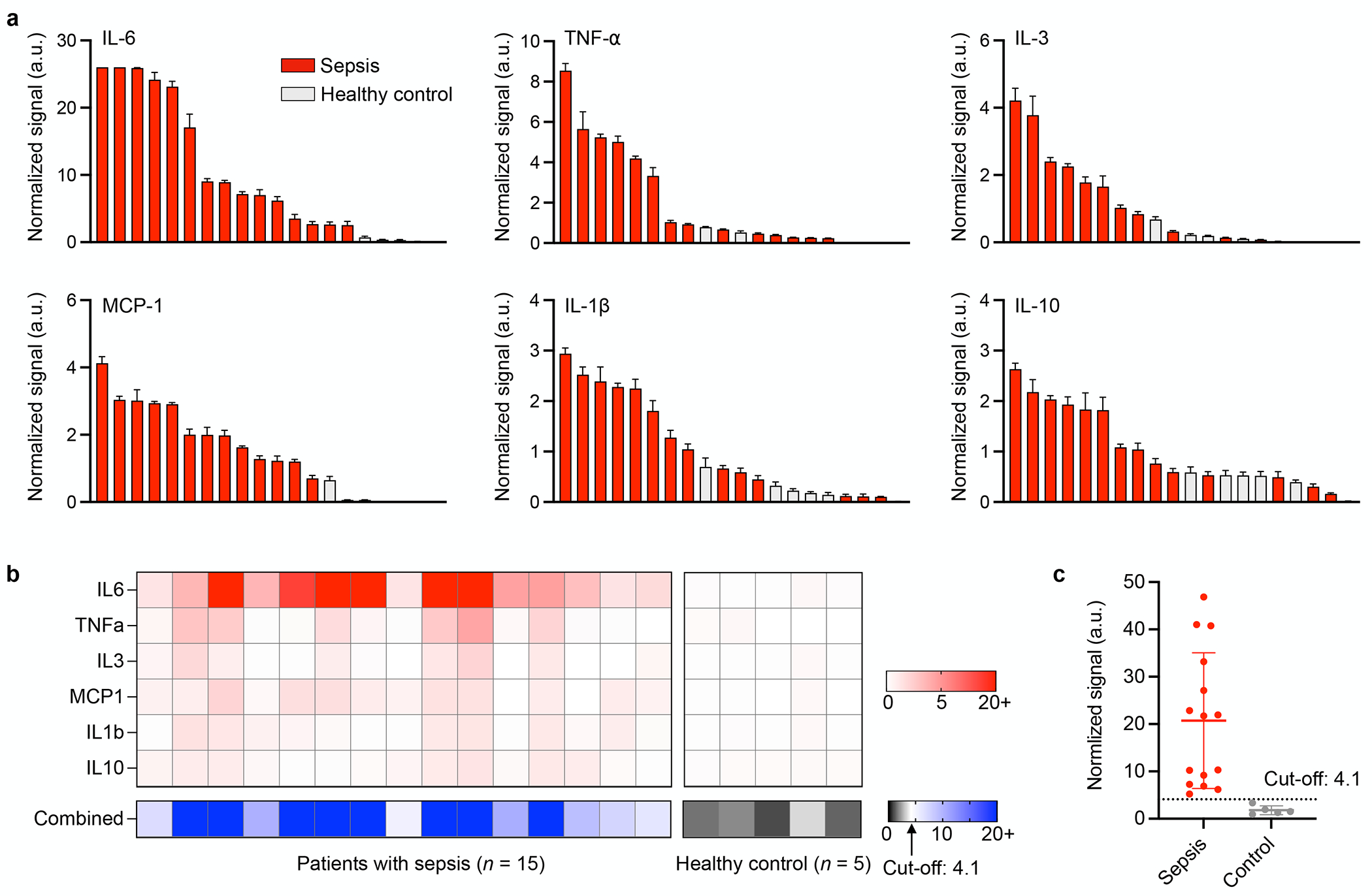
(a) Waterfall plots of 6 cytokine and chemokine levels of clinical samples from 15 septic patients and 5 healthy controls. Fluorescence signals are normalized based on the threshold value of each cytokine or chemokine. (b) Heatmap analysis of Individual markers and a marker combination for sepsis detection. The cut-off value was determined by the receiver operating characteristics of individual markers. (c) A combined score shows good discrimination between septic patients and healthy controls.
CONCLUSIONS
We developed an ultra-sensitive plasmonic platform based on a chemifluorescence assay for multiplexed molecular profiling of cytokines in plasma samples. The platform exploits the synergy of two signal enhancement techniques, i.e., tyramide chemifluorescence signal amplification and localized plasmonic coupling. Biotinylated tyramide molecules were activated by HRPs to covalently bind to tyrosine residues on antibodies and target molecules, followed by the binding of streptavidin-bound AF647 fluorophore molecules. The fluorescence emission from the fluorophore AF647 was then further enhanced by LSPRs of the NPOD substrate. The coupled effect between the nanoplasmonic enhancement and chemifluorescence signal amplification created the DuPLUS assay for multiplexed molecular profiling with sub-pg/mL sensitivity in 1 hr. We have demonstrated the detection of 6 different cytokines in a small cohort of septic and non-septic patients as a proof of concept. This assay has the potential to further increase the multiplexed capabilities of the DuPLUS assay to include other key cytokines for sepsis monitoring. The DuPLUS assay’s high sensitivity and short assay time could lead to subsequent clinical studies. These include early diagnosis and longitudinal monitoring of septic patients following intensive care unit management and treatment, investigation of cytokine levels in COVID-19 and cancer patients who suffer from cytokine release syndrome.
Plasmonic technologies, including commercial systems, have shown great potential for sensing applications in research settings, but their translation for clinical applications, especially in POC settings, has not yet been widely realized.38 While the “translational valley of death” commonly happens in many sensing technologies, in plasmonics, the hindrance often comes from the lack of a high-throughput fabrication method, assays with good accuracies and reproducibility, a portable readout system, or a combination of these. Here, we address these issues with an integrated approach by developing a plasmonic substrate, a sensitive detection assay, and an automatic, portable imaging system. The fabrication process of the NPOD substrate is simple, reproducible, inexpensive, and, more importantly, can be easily scaled. Large sizes of NPOD chips could enable a microarray-type assay. For instance, an integrated open reservoir fluidic chamber was designed to perform multiplexed cytokine analysis, including positive and negative controls on a single chip (Figure S11). Alternatively, larger NPOD substrates that could accommodate a 96-well plate platform are also feasible; in this case, a commercial microplate reader commonly found in clinical analytical laboratories could be used. The dual enhancements in the DuPLUS assay allow for rapid, highly sensitive, multiplexed profiling of cytokines routinely tested in clinics. These capabilities could vastly facilitate the longitudinal cytokine profiling of patients, assisting healthcare providers in determining treatment plans and analyzing their patients’ responses. Finally, the automated imaging system enables quantitative measurements over a large area (12 mm × 24 mm). Combined with the large size of NPOD chips, the imaging system allows us to incorporate multiple sensing arrays with positive and negative controls on a single chip. This configuration is essential for clinical testing to ensure that a given assay is adequately performed.
In summary, the developed DuPLUS assay based on the NPOD chip is a highly sensitive and specific molecular profiling method. With the potential to be adapted to a microplate platform, the method enables rapid, multiplexed cytokine profiling in human plasma for broad applications in addition to sepsis monitoring, including virus infection such as SARS-CoV-2,39–41 cancer therapy,42–44 metabolism disorders such as diabetes.45,46
MATERIALS AND METHODS
NPOD chip fabrication
A 125-μm-thick polyethylene naphthalate (PEN; Dupont Teijin Films, USA) with 90 × 90 mm2 was used as a substrate. After removing its protective film, it was loaded on a linear moving stage in an oxygen (O2) ion beam chamber. The linear O2 ion beams with a 300 mm width were generated at a working pressure of 9.0 × 10−4 Torr and an O2 flow rate of 70 sccm. The surface of PEN was modified by an O2 ion beam bombardment incident in a normal direction. Subsequently, a 100 nm-thick Au layer was deposited on the PEN nanodimples using a thermal evaporator (LAT, South Korea). The working pressure and deposition rate were maintained at 1.8 × 10−5 Torr and 1.8 Å/s, respectively. To render a space layer, 1H,1H,2H,2H-perfluorodecanethiol (PFDT; Sigma Aldrich Korea) was vapor-deposited on the Au/PEN nanodimples for 2 hr at room temperature. Lastly, an Au layer was thermally deposited on the PFDT-covered sample at a deposition rate of 0.3 Å/s.
Antibodies and recombinant human proteins
Antibodies used in the study were purchased from R&D systems, including monoclonal capture antibodies: IL-1β (MAB601), IL-3 (MAB603R), IL-6 (MAB206), IL-10 (MAB2172), TNF-α (MAB610) and MCP-1 (MAB679); biotinylated polyclonal detection antibodies: IL-1β (BAF201), IL-3 (BAF203), IL-6 (BAF206), IL-10 (BAF217), TNF-α (BAF210) and MCP-1 (BAF279). Respective recombinant human proteins for spiking experiments were purchased from Peprotech: IL-1β (200-01B), IL-3 (200-03), IL-6 (200-06), IL-10 (200-10), TNF-α (300-01A) and MCP-1 (300-04).
DuPLUS assay
The NPOD chips were first cleaned in ethanol. A mixture of 5 mM 11-Mercaptoundecanoic acid (Sigma-Aldrich, 450561) and 15 mM 1-octanethiol (Sigma-Aldrich, 471836) in ethanol was added to the chips for overnight incubation. Then, the chips were washed with 0.1 M 2-(N-morpholino) ethanesulfonic acid (MES) buffered saline, pH 4.7 (ThermoFisher, 28390) three times. Subsequently, the chips were incubated for 15 min in a solution of 20 mM 1-ethyl-3-(3-dimethylaminopropyl) carbodiimide (EDC, Thermo Fisher, PG82073) and 50 mM sulfo-N-hydroxysulfosuccinimide (sulfo-NHS, Thermo Fisher, PG82071) in MES buffer. The chips were washed three times in MES buffer, and then respective capture antibody (8 μg mL−1) in phosphate-buffered saline (PBS, ThermoFisher, 70011044) solution was added for 30 min incubation at room temperature. The same process was performed to coat capture antibodies onto plain gold chips. The following surface modification processes were used for immobilizing capture antibodies onto glass chips: First, 4% (3-Mercaptopropyl) trimethoxysilane (Sigma-Aldrich, 175617) in ethanol was added onto the chips for 30 min incubation. After washing three times in ethanol, 2.5 mM GMBS (ThermoFisher, 22309) was added for 30 min incubation. Subsequently, the chips were washed three times in PBS buffer, and then respective capture antibodies (8 μg mL−1 in PBS) were added for 30 min incubation at room temperature. The chips were immersed in PBS solution for blocking with 2% bovine serum albumin (BSA, ThermoFisher, 37525) and 0.05% Tween20 (Sigma-Aldrich, P9416) for 30 min. Once the functionalized chip is ready, it could be stored in PBS at 4 °C until used or lyophilized for more extended storage.47
For spiking experiments, a serial dilution standard of respective proteins in 50% human plasma (Rockland, D519-04-0050) and 50% sample diluent (Abcam, ab193972) solution were added to each chip for 20 min incubation at room temperature. Then, respective detection antibody (1.5 μg mL−1 in 0.2% BSA-0.05%Tween20-PBS (PBST) solution) was added for 15 min incubation at room temperature. Subsequently, 1:8000 streptavidin-horseradish peroxidase (HRP, ThermoFisher, 21130) diluted in PBST was then added to the chips for 10 min at room temperature, followed by incubating 5 μg mL−1 biotinylated tyramide (Sigma-Aldrich, SML2135) in amplification diluent (0.1 M borate and 0.003% H2O2, pH 8.5 with NaOH) for another 10 min at room temperature. Finally, 1 μg mL−1 streptavidin-AF647 (BioLegend, 405237) was added for 10 min incubation at room temperature. Washing was performed after each step using PBST for three times.
ELISA
ELISA was performed based on the vendor-recommended protocol (ThermoFisher). Respective capture antibody (2 μg mL−1) in PBS was added to an ELISA plate (NUNC, Maxisorp) for 2 hr incubation at room temperature. For blocking, the chips were immersed in PBS solution with 2% bovine serum albumin and 0.05% Tween20 for 1 hr. For spiking experiments, a serial dilution standard of respective proteins in PBST was added to each well for 1 hr incubation at room temperature. Then, the respective detection antibody (0.4 μg mL−1) was added for 1 hr incubation at room temperature. Subsequently, streptavidin-HRP in PBST was then added to the chips for 30 min at room temperature, followed by incubating biotinylated tyramide in an amplification diluent for another 10 min at room temperature. Finally, streptavidin-AF647 was added for 30 min incubation at room temperature. Washing was performed after each step using PBST for three times.
Multiplexed chip
To facilitate multiplexed measurement of 6 cytokines on a single NPOD chip, the capture antibodies were spotted (4 spots per antibody for quadruplicate measurements) onto the NPOD chip using a microspotting machine (DigiLab). After EDC and sulfo-NHS incubation, the chip was washed with MES buffer for three times and then dried with air nitrogen before antibody spotting. In the case with open reservoir fluidic integration, a 3D printed part (Formlabs, Form 2) was adhered onto the NPOD chip after antibody spotting using a 3M VHB acrylic tape.
Microscopy
Fluorescence images were acquired on a Nikon Eclipse Ti inverted microscope with a 20× objective lens (CFI Plan Apochromat Lambda, NA 0.75). Data were analyzed using ImageJ software (v2.0).
Simulation
Numerical simulations of the E-field distribution around the 3D NPOD substrates were conducted using Lumerical FDTD (Lumerical Inc.). The geometric models of the 3D NPOD substrates were based on the TEM image of the structures (Figure 2h). All boundary conditions were modeled as perfectly matched layers, while the width of the cell was set to 0.5 nm for the x-, y-, and z-directions. The light source was modeled as a plane wave with a wavelength of 638 nm or 785 nm, polarized in the x-direction and incident on the structure along the z-direction.
Clinical study
The clinical study was approved by the Institutional Review Board of Massachusetts General Hospital (Protocol #: 2019P003446, PI: Hyungsoon Im). Plasma samples of septic patients were collected from Hôpitaux Universitaire Saint-Louis, France (Benjamin Chousterman). Sample collection and processing were approved by the Institutional Review Board (Hôpitaux Universitaire Saint-Louis, France), and all patients signed informed consent. Plasma samples of healthy patients, who were not suffering from inflammatory conditions, were obtained from the Biobanks of Massachusetts General Hospital, which was approved by the Institutional Review Board (Massachusetts General Hospital, United States). Plasma samples were diluted by mixing with 50% sample diluent solution (Abcam, ab193972) and then processed using the DuPLUS assay on NPOD chips. To validate and correlate the measured expression using NPOD chips with the gold standard, fluorescence-based ELISA kits (Abcam, ab229434) for IL6 measurement were used.
Statistics
Statistical analyses and data plotting were performed in GraphPad Prism 8.
Supplementary Material
ACKNOWLEDGMENTS
We would like to acknowledge extensive discussions with K. Yang on chemifluorescence assays. This work was supported in part by National Institute of Health grants (R00CA201248, R21CA217662, R01GM138778) and the Fundamental Research Program (PNK 9350) of the Korea Institute of Materials Science (KIMS) and National R&D Program through the National Research Foundation of Korea (NRF) funded by Ministry of Science and ICT (NRF-2020R1A5A1018052 and NRF-2021R1A2C2011048).
Footnotes
Conflict of interest
HI is a consultant to Aikili Biosystems, Cellkey, and Noul and receives research support from Canon USA, CytoGen, Healcerion, and Noul. RW is a consultant to ModeRNA, Tarveda Pharmaceuticals, Lumicell, Seer, Earli, and Accure Health. CMC is a consultant to Aikili Biosystems, Qiagen, Teladoc, and InfiniteMD. The remaining authors report no competing interests.
Supporting Information Available: In-house chip and system development (Figure S1); Characterization of PEN and Au nanodimples (Figure S2); Transmission electron micrographs (TEMs) of Au nanoparticles formed on the nanodimple structure (Figure S3); Au Nanoparticle on nanodimple substrates with varying thicknesses of Au deposited on 1H,1H,2H,2H-perfluorodecanethiol (PFDT)-coated gold nanodimple structures (Figure S4); Optical characteristics of NPOD chip (Figure S5); SERS measurements of benzenethiol (BT)-treated NPOD substrates (Figure S6); Batch-to-batch reproducibility of NPOD substrates (Figure S7); Comparison between Au nanodimple arrays without spiked nanoparticles and with spiked nanoparticles (Figure S8); Comparison of the DuPLUS assay for detecting MCP-1 spiked in 1% BSA in PBS and diluted human plasma (Figure S9); Correlation of IL-6 measurements on septic samples between DuPLUS assay and commercial fluorescence-based ELISA kits (Figure S10); Integrated open reservoir fluidic chip. (Figure S11); Surface energy of spacer materials (Table S1); Demographic summary of septic patients (Table S2); Demographic summary of healthy controls (Table S3); Threshold levels for cytokines and chemokine (Table S4).
Data availability
Materials are available upon request by contacting the corresponding authors.
REFERENCES
- (1).Rudd KE; Johnson SC; Agesa KM; Shackelford KA; Tsoi D; Kievlan DR; Colombara DV; Ikuta KS; Kissoon N; Finfer S; Fleischmann-Struzek C; Machado FR; Reinhart KK; Rowan K; Seymour CW; Watson RS; West TE; Marinho F; Hay SI; Lozano R et al. Global, Regional, and National Sepsis Incidence and Mortality, 1990–2017: Analysis for the Global Burden of Disease Study. Lancet 2020, 395, 200–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Angus DC; van der Poll T Severe Sepsis and Septic Shock. N. Engl. J. Med. 2013, 369, 840–851. [DOI] [PubMed] [Google Scholar]
- (3).Perman SM; Goyal M; Gaieski DF Initial Emergency Department Diagnosis and Management of Adult Patients with Severe Sepsis and Septic Shock. Scand. J. Trauma Resusc. Emerg. Med. 2012, 20, 41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Singer M; Deutschman CS; Seymour CW; Shankar-Hari M; Annane D; Bauer M; Bellomo R; Bernard GR; Chiche JD; Coopersmith CM; Hotchkiss RS; Levy MM; Marshall JC; Martin GS; Opal SM; Rubenfeld GD; van der Poll T; Vincent JL; Angus DC The Third International Consensus Definitions for Sepsis and Septic Shock (Sepsis-3). JAMA 2016, 315, 801–810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Probst L; Schalk E; Liebregts T; Zeremski V; Tzalavras A; von Bergwelt-Baildon M; Hesse N; Prinz J; Vehreschild JJ; Shimabukuro-Vornhagen A; Eichenauer DA; Garcia Borrega J; Kochanek M; Böll B Working Party on Intensive Care Medicine in Hematologic and Oncologic Patients (iCHOP) of the German Society of Hematology and Medical Oncology, D. G. H. O. Prognostic Accuracy of Sofa, Qsofa and Sirs Criteria in Hematological Cancer Patients: A Retrospective Multicenter Study. J. Intensive Care 2019, 7, 41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Chaudhry H; Zhou J; Zhong YIN; Ali MM; McGuire F; Nagarkatti PS; Nagarkatti M Role of Cytokines as a Double-Edged Sword in Sepsis. In vivo 2013, 27, 669–684. [PMC free article] [PubMed] [Google Scholar]
- (7).Kumar V Immunometabolism: Another Road to Sepsis and Its Therapeutic Targeting. Inflammation 2019, 42, 765–788. [DOI] [PubMed] [Google Scholar]
- (8).van der Poll T; van de Veerdonk FL; Scicluna BP; Netea MG The Immunopathology of Sepsis and Potential Therapeutic Targets. Nat. Rev. Immunol. 2017, 17, 407–420. [DOI] [PubMed] [Google Scholar]
- (9).Rello J; Valenzuela-Sánchez F; Ruiz-Rodriguez M; Moyano S Sepsis: A Review of Advances in Management. Adv. Ther. 2017, 34, 2393–2411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Andaluz-Ojeda D; Bobillo F; Iglesias V; Almansa R; Rico L; Gandía F; Resino S; Tamayo E; de Lejarazu RO; Bermejo-Martin JF A Combined Score of Pro- and Anti-Inflammatory Interleukins Improves Mortality Prediction in Severe Sepsis. Cytokine 2012, 57, 332–336. [DOI] [PubMed] [Google Scholar]
- (11).Faix JD Biomarkers of Sepsis. Crit. Rev. Clin. Lab. Sci. 2013, 50, 23–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Matsumoto H; Ogura H; Shimizu K; Ikeda M; Hirose T; Matsuura H; Kang S; Takahashi K; Tanaka T; Shimazu T The Clinical Importance of a Cytokine Network in the Acute Phase of Sepsis. Sci. Rep. 2018, 8, 13995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Tipoe TL; Wu WKK; Chung L; Gong M; Dong M; Liu T; Roever L; Ho J; Wong MCS; Chan MTV; Tse G; Wu JCY; Wong SH Plasminogen Activator Inhibitor 1 for Predicting Sepsis Severity and Mortality Outcomes: A Systematic Review and Meta-Analysis. Front. Immunol. 2018, 9, 1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Chowdhury F; Williams A; Johnson P Validation and Comparison of Two Multiplex Technologies, Luminex and Mesoscale Discovery, for Human Cytokine Profiling. J. Immunol. Methods 2009, 340, 55–64. [DOI] [PubMed] [Google Scholar]
- (15).Leligdowicz A; Conroy AL; Hawkes M; Zhong K; Lebovic G; Matthay MA; Kain KC Validation of Two Multiplex Platforms to Quantify Circulating Markers of Inflammation and Endothelial Injury in Severe Infection. PLoS One 2017, 12, e0175130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Mosevoll KA; Skrede S; Markussen DL; Fanebust HR; Flaatten HK; Aßmus J; Reikvam H; Bruserud Ø Inflammatory Mediator Profiles Differ in Sepsis Patients with and Without Bacteremia. Front. Immunol. 2018, 9, 691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Smith N; Goncalves P; Charbit B; Grzelak L; Beretta M; Planchais C; Bruel T; Rouilly V; Bondet V; Hadjadj J; Yatim N; Pere H; Merkling SH; Ghozlane A; Kernéis S; Rieux-Laucat F; Terrier B; Schwartz O;Mouquet H; Duffy D et al. Distinct Systemic and Mucosal Immune Responses During Acute SARS-CoV-2 Infection. Nat. Immunol. 2021, 22, 1428–1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Oeschger T; McCloskey D; Kopparthy V; Singh A; Erickson D Point of Care Technologies for Sepsis Diagnosis and Treatment. Lab Chip 2019, 19, 728–737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Reddy B; Hassan U; Seymour C; Angus DC; Isbell TS; White K; Weir W; Yeh L; Vincent A; Bashir R Point-of-Care Sensors for the Management of Sepsis. Nat. Biomed. Eng. 2018, 2, 640–648. [DOI] [PubMed] [Google Scholar]
- (20).Gaieski DF; Drumheller BC; Goyal M; Fuchs BD; Shofer FS; Zogby K Accuracy of Handheld Point-of-Care Fingertip Lactate Measurement in the Emergency Department. West J. Emerg. Med. 2013, 14, 58–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Molinero-Fernández Á; López MÁ; Escarpa A An On-Chip Microfluidic-Based Electrochemical Magneto-Immunoassay for the Determination of Procalcitonin in Plasma Obtained from Sepsis Diagnosed Preterm Neonates. Analyst 2020, 145, 5004–5010. [DOI] [PubMed] [Google Scholar]
- (22).Rascher D; Geerlof A; Kremmer E; Krämer P; Michael S; Hartmann A; Rieger M Total Internal Reflection (TIRF)-Based Quantification of Procalcitonin for Sepsis Diagnosis – a Point-of-Care Testing Application. Biosens. Bioelectron. 2014, 59, 251–258. [DOI] [PubMed] [Google Scholar]
- (23).Okamura Y; Yokoi H Development of a Point-of-Care Assay System for Measurement of Presepsin (Scd14-St). Clin. Chim. Acta 2011, 412, 2157–2161. [DOI] [PubMed] [Google Scholar]
- (24).Valera E; Berger J; Hassan U; Ghonge T; Liu J; Rappleye M; Winter J; Abboud D; Haidry Z; Healey R; Hung NT; Leung N; Mansury N; Hasnain A; Lannon C; Price Z; White K; Bashir R A Microfluidic Biochip Platform for Electrical Quantification of Proteins. Lab Chip 2018, 18, 1461–1470. [DOI] [PubMed] [Google Scholar]
- (25).Min J; Nothing M; Coble B; Zheng H; Park J; Im H; Weber GF; Castro CM; Swirski FK; Weissleder R; Lee H Integrated Biosensor for Rapid and Point-of-Care Sepsis Diagnosis. ACS Nano 2018, 12, 3378–3384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Buchegger P; Sauer U; Toth-Székély H; Preininger C Miniaturized Protein Microarray with Internal Calibration as Point-of-Care Device for Diagnosis of Neonatal Sepsis. Sensors (Basel) 2012, 12, 1494–1508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Sauer U; Domnanich P; Preininger C Protein Chip for the Parallel Quantification of High and Low Abundant Biomarkers for Sepsis. Anal. Biochem. 2011, 419, 46–52. [DOI] [PubMed] [Google Scholar]
- (28).Grazia Revello M; Percivalle E; Zannino M; Rossi V; Gerna G Development and Evaluation of a Capture ELISA for IgM Antibody to the Human Cytomegalovirus Major DNA Binding Protein. J. Virol. Methods 1991, 35, 315–329. [DOI] [PubMed] [Google Scholar]
- (29).Pohanka M; Zakova J A Smartphone Camera Colorimetric Assay of Acetylcholinesterase and Butyrylcholinesterase Activity. Sensors (Basel) 2021, 21, 1796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Kemmler M; Sauer U; Schleicher E; Preininger C; Brandenburg A Biochip Point-of-Care Device for Sepsis Diagnostics. Sens. Actuators B Chem. 2014, 192, 205–215. [Google Scholar]
- (31).Zhang B; Price J; Hong G; Tabakman SM; Wang H; Jarrell JA; Feng J; Utz PJ; Dai H Multiplexed Cytokine Detection on Plasmonic Gold Substrates with Enhanced Near-Infrared Fluorescence. Nano Research 2013, 6, 113–120. [Google Scholar]
- (32).Momenbeitollahi N; Cloet T; Li H Pushing the Detection Limits: Strategies Towards Highly Sensitive Optical-Based Protein Detection. Anal. Bioanal. Chem. 2021, 413, 5995–6011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Yang KS; Ciprani D; O’Shea A; Liss AS; Yang R; Fletcher-Mercaldo S; Mino-Kenudson M; Fernández-Del Castillo C; Weissleder R Extracellular Vesicle Analysis Allows for Identification of Invasive IPMN. Gastroenterology 2021, 160, 1345–1358.e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Park S; Xiao X; Min J; Mun C; Jung HS; Giannini V; Weissleder R; Maier SA; Im H; Kim D Self-Assembly of Nanoparticle-Spiked Pillar Arrays for Plasmonic Biosensing. Adv. Funct. Mater. 2019, 29, 1904257. [Google Scholar]
- (35).Yang J-Y; Park S-G; Jung S; Byeon E-Y; Kim D-G; Jung HS; Kim HJ; Lee S SERS Substrates Based on Self-Organized Dimple Nanostructures on Polyethylene Naphthalate Films Produced via Oxygen Ion Beam Sputtering. Appl. Surf. Sci. 2022, 572, 151452. [Google Scholar]
- (36).Itagaki N; Nakamura Y; Narishige R; Takeda K; Kamataki K; Koga K; Hori M; Shiratani M Growth of Single Crystalline Films on Lattice-Mismatched Substrates through 3D to 2D Mode Transition. Sci. Rep. 2020, 10, 4669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Mao P; Liu C; Favraud G; Chen Q; Han M; Fratalocchi A; Zhang S Broadband Single Molecule SERS Detection Designed By Warped Optical Spaces. Nat. Commun. 2018, 9, 5428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Chin LK; Son T; Hong JS; Liu AQ; Skog J; Castro CM; Weissleder R; Lee H; Im H Plasmonic Sensors for Extracellular Vesicle Analysis: From Scientific Development to Translational Research. ACS Nano 2020, 14, 14528–14548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).Costela-Ruiz VJ; Illescas-Montes R; Puerta-Puerta JM; Ruiz C; Melguizo-Rodríguez L SARS-CoV-2 Infection: The Role of Cytokines in Covid-19 Disease. Cytokine Growth Factor Rev. 2020, 54, 62–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Simon D; Tascilar K; Krönke G; Kleyer A; Zaiss MM; Heppt F; Meder C; Atreya R; Klenske E; Dietrich P; Abdullah A; Kliem T; Corte G; Morf H; Leppkes M; Kremer AE; Ramming A; Pachowsky M; Schuch F; Ronneberger M et al. Patients with Immune-Mediated Inflammatory Diseases Receiving Cytokine Inhibitors Have Low Prevalence of SARS-CoV-2 Seroconversion. Nat. Commun. 2020, 11, 3774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (41).Song P; Li W; Xie J; Hou Y; You C Cytokine Storm Induced by SARS-CoV-2. Clin. Chim. Acta 2020, 509, 280–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42).Cohen S; Fishman P Targeting the a3 Adenosine Receptor to Treat Cytokine Release Syndrome in Cancer Immunotherapy. Drug Des. Devel. Ther. 2019, 13, 491–497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (43).Filippou PS; Karagiannis GS Cytokine Storm During Chemotherapy: A New Companion Diagnostic Emerges. Oncotarget 2020, 11, 213–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (44).Mirsoian A; Murphy WJ Obesity and Cancer Immunotherapy Toxicity. Immunotherapy 2015, 7, 319–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (45).Abdel-Latif M; Abdel-Moneim AA; El-Hefnawy MH; Khalil RG Comparative and Correlative Assessments of Cytokine, Complement and Antibody Patterns in Paediatric Type 1 Diabetes. Clin. Exp. Immunol. 2017, 190, 110–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (46).Gomes CP; Torloni MR; Gueuvoghlanian-Silva BY; Alexandre SM; Mattar R; Daher S Cytokine Levels in Gestational Diabetes Mellitus: A Systematic Review of the Literature. Am. J. Reprod. Immunol. 2013, 69, 545–557. [DOI] [PubMed] [Google Scholar]
- (47).Min J; Chin LK; Oh J; Landeros C; Vinegoni C; Lee J; Lee SJ; Park JY; Liu AQ; Castro CM; Lee H; Im H; Weissleder R Cytopan-Portable Cellular Analyses for Rapid Point-of-care Cancer Diagnosis. Sci. Transl. Med. 2020, 12, eaaz9746. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Materials are available upon request by contacting the corresponding authors.


