Abstract
CRISPR-Cas9 gene editing is emerging as a prospective therapy for genomic mutations. However, current editing approaches are directed primarily toward relatively small cohorts of patients with specific mutations. Here, we describe a cardioprotective strategy potentially applicable to a broad range of patients with heart disease. We used base editing to ablate the oxidative activation sites of CaMKIIδ, a primary driver of cardiac disease. We show in cardiomyocytes derived from human induced pluripotent stem cells that editing the CaMKIIδ gene to eliminate oxidation-sensitive methionine residues confers protection from ischemia/reperfusion (IR) injury. Moreover, CaMKIIδ editing in mice at the time of IR enables the heart to recover function from otherwise severe damage. CaMKIIδ gene editing may thus represent a permanent and advanced strategy for heart disease therapy.
CRISPR-Cas9 gene editing is being developed as a therapeutic approach to correct monogenic mutations causing hereditary diseases (1–6). However, most CRISPR-Cas9 editing strategies are focused on correction of specific genetic mutations that occur in a small subset of patients, limiting broader applications of the approach. We sought to design a CRISPR-Cas9 gene editing therapy potentially applicable to a broad range of adult patients with cardiovascular disease, the leading cause of worldwide morbidity and mortality (7). Ca2+/calmodulin–dependent protein kinase IIδ (CaMKIIδ) is a central regulator of cardiac signaling and function (8). However, chronic overactivation of CaMKIIδ causes several cardiac diseases in humans and mice, including ischemia/reperfusion (IR) injury, heart failure, hypertrophy, and arrhythmias (9–16). Mechanistically, CaMKIIδ overactivation in the heart has been linked to disturbances in Ca2+ homeostasis, inflammation, apoptosis, and fibrosis, leading to cardiac dysfunction (9–13, 17). Oxidation of two methionine residues, Met281 and Met282, located in the regulatory domain of CaMKIIδ, promotes hyperactivation of the kinase by preventing association of the catalytic domain with the autoinhibitory region (18). Modification of these methionine residues to other amino acids prevents oxidation and overactivation of CaMKIIδ, thereby conferring cardioprotection as shown in knock-in mice, where both methionine residues were replaced with valines in the germline (14, 18, 19). This genetic modification did not cause adverse effects. Both methionines are encoded by exon 11, which is not subject to alternative splicing, so targeting the oxidative activation sites would affect all CaMKIIδ splicing variants (for example CaMKIIδB, δC, and δ9 as the major cardiac variants) (20).
Results
Design of a gene editing strategy to ablate the oxidative activation sites of CaMKIIδ
CRISPR-Cas9 adenine base editing (ABE) allows the precise conversion of adenine to guanine nucleotides without introducing double-stranded DNA breaks (1, 2, 21–23). We reasoned that ABE could potentially render CaMKIIδ insensitive to oxidative activation by converting ATG to GTG codons and thereby replacing the oxidation-sensitive methionines with valines (Fig. 1A). Instead of using CRISPR-Cas9 technology to correct genetic mutations, we used the technology to disrupt a pathological signaling pathway, offering a potential therapeutic approach for cardiac disease.
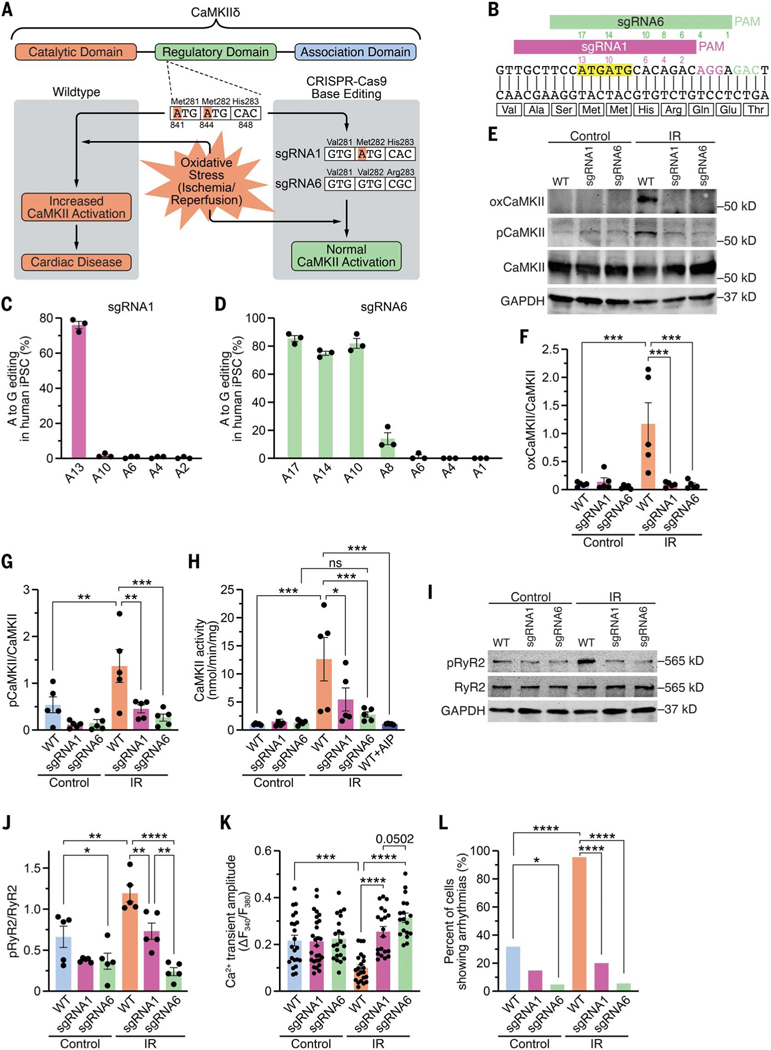
Fig. 1. Genomic editing of CaMKIIδ in human iPSC-cardiomyocytes.
(A) Schematic of CaMKIIδ and its three domains. Both critical methionines (Met281 and Met282) are located in the regulatory domain. Upon oxidative stress, these methionines are oxidized, resulting in increased CaMKII activity and cardiac disease. Using CRISPR-Cas9 adenine base editing, we identified sgRNA1, which edited only c.A841G (p.M281V; sgRNA1), and sgRNA6, which edited c.A841G (p.M281V), c.A844G (p.M282V), and c.A848G (p.H283R; sgRNA6), thereby preventing CaMKII activation upon oxidative stress. (B) Sequence of a segment of exon 11 of CaMKIIδ genomic DNA encoding part of the regulatory domain of CaMKIIδ. Alignment of sgRNA1 and sgRNA6 with CaMKIIδ. PAM sequences for sgRNA1 and sgRNA6 are in purple and green, respectively. Both ATGs encoding methionines are highlighted in yellow. Adenines within the sequences of sgRNA1 (purple) and sgRNA6 (green) are numbered (starting from the PAM). (C) Percentage of adenine (A) to guanine (G) editing in human iPSCs for each adenine in sgRNA1 after base editing with ABE8e and sgRNA1, as determined by Sanger sequencing. (D) Percentage of adenine (A) to guanine (G) editing in human iPSCs for each adenine in sgRNA6 after base editing with ABE8e and sgRNA6, as determined by deep amplicon sequencing. (E) Western blot analysis of oxidized CaMKII (specific antibody), autophosphorylated CaMKII (specific antibody), total CaMKII, and GAPDH in human WT, sgRNA1, and sgRNA6 iPSC-CMs for control group and after simulated ischemia/reperfusion (IR). (F) Mean densitometric analysis for oxidized CaMKII normalized to total CaMKII in control and post-IR iPSC-CMs (n = 5 independent iPSC-CM differentiations). (G) Mean densitometric analysis for autophosphorylated CaMKII normalized to total CaMKII in control and post-IR iPSC-CMs (n = 5 independent iPSC-CM differentiations). (H) Scatter bar plot showing mean CaMKII activity in control and post-IR iPSC-CMs and in lysates of WT post-IR iPSC-CMs in presence of the CaMKII inhibitor AIP (n = 5 independent iPSC-CM differentiations). (I) Western blot analysis of ryanodine receptor type 2 (RyR2) phosphorylation at the CaMKII site (serine 2814), total RyR2, and GAPDH in control and post-IR iPSC-CMs (n = 5 independent iPSC-CM differentiations). (J) Mean densitometric analysis for phosphorylated RyR2 normalized to total RyR2 in control and post-IR iPSC-CMs (n = 5 independent iPSC-CM differentiations). (K) Mean Ca2+ transient amplitude for each group (based on the number of cardiomyocytes). (L) Percentage of iPSC-CMs showing arrhythmias, as measured by epifluorescence microscopy. Statistical comparisons are based on one-way analysis of variance (ANOVA) post-hoc corrected by Holm-Sidak [(F) to (H)] and [(J) and (K)] and on Fisher’s exact test (L). Data are presented either as individual data points with means ± SEM or as percent of cells (L).
To identify optimal CRISPR-Cas9 base editing components, we used HEK293 cells to screen six different single guide RNAs (sgRNAs; table S1) that covered the genomic region encoding methionines 281 (adenine at nucleotide position c.841) and 282 (adenine at nucleotide position c.844). The sgRNAs were tested with two adenine base editors, ABEmax and ABE8e, in which the engineered deaminases were fused to either SpCas9 nickase or its variant SpRY nickase (3, 23, 24). Sanger sequencing revealed that sgRNA1 combined with ABE8e-SpCas9 had the highest efficiency for editing c.A841G (p.M281V), without editing c.A844 (p.M282). sgRNA6 combined with ABE8e-SpRY showed a broader editing window, editing c.A841G (p.M281V), c.A844G (p.M282V), and c.A848G (p.H283R; fig. S1 and table S2). We validated the sgRNA1 + ABE8e-SpCas9 and sgRNA6 + ABE8e-SpRY editing strategies in human induced pluripotent stem cells (iPSCs) using nucleofection and observed the same editing pattern seen in HEK293 cells (Fig. 1, B to D). We picked several iPSC clones to test whether exposure to sgRNA6 + ABE8e-SpRY resulted in a heterozygous or homozygous genotype. Sanger sequencing revealed that 75, 17, and 8% of the clones were homozygous, heterozygous, and wildtype, respectively (fig. S2A).
Analysis of potential off-target editing in human iPSCs
The two oxidation-sensitive methionines of human CaMKIIδ are encoded by exon 11 of the CaMKIIδ gene, which shares 79% nucleotide homology with CaMKIIα and 76% nucleotide homology with CaMKIIγ. The sgRNA6 sequence shared 85% homology with CaMKIIα and CaMKIIγ (fig. S2B). We used deep amplicon sequencing to validate the specificity of ABE genomic editing of CaMKIIδ with sgRNA6 + ABE8e-SpRY. In human iPSCs, we observed no genomic changes of the CaMKIIα or CaMKIIγ genes (fig. S2, C and D, and table S3). However, sgRNA6 has sequence identity with the human CaMKIIβ gene, and sequencing analysis showed that the human CaMKIIβ gene was edited by sgRNA6. Fortunately, CaMKIIβ is not expressed in human cardiomyocytes, so genomic editing of the CaMKIIβ gene in the heart would be inconsequential (fig. S2, E and F) (12). To assess off-target editing, we used CRISPOR to identify the top eight potential off-target sites (table S4) (25). Sequencing analysis showed adenine to guanine editing only in the DAZL gene at the adenine base 13 nucleotides upstream from the protospacer adjacent motif (PAM; fig. S2G and tables S4 and S5). This edited site is located in an intronic region that is not expressed and therefore should not have deleterious consequences (25). All other adenines of the predicted top eight potential off-target sites showed adenine to guanine editing of <0.2%, which is considered unspecific background (26).
Functional analyses of CaMKIIδ-edited human iPSC-derived cardiomyocytes
To investigate the physiological consequences of both editing patterns, we generated three independent human homozygous iPSC lines with sgRNA1, sgRNA6, or no sgRNA (wildtype) and differentiated them into cardiomyocytes (iPSC-CMs) that were subjected to simulated IR injury using a hypoxia chamber. There was no difference in the amounts of CaMKII protein in wild-type (WT) or edited iPSC-CMs (fig. S3). After IR, WT iPSC-CMs showed an increase in CaMKII oxidation, as measured by Western blot with an antibody that specifically recognizes oxidized CaMKII, whereas CaMKII oxidation was strongly reduced in sgRNA1 iPSC-CMs and sgRNA6 iPSC-CMs (Fig. 1, E and F, and fig. S3A). WT iPSC-CMs showed substantial increases in CaMKII autophosphorylation and activity post-IR, which were both reduced in sgRNA1- and sgRNA6-edited iPSC-CMs (Fig. 1, E, G, and H). In accordance with the changes in CaMKII activity, we observed increased CaMKII-dependent phosphorylation of ryanodine receptor type 2 (RyR2) at serine 2814 in WT but not in sgRNA1 or sgRNA6 iPSC-CMs post-IR (Fig. 1, I and J, and fig. S3). Function of iPSC-CMs was assessed by measuring cellular Ca2+ transients using epifluorescence microscopy. After IR, WT iPSC-CMs showed an increase in diastolic Ca2+ levels, a decrease in Ca2+ transient amplitude, and arrhythmias (Fig. 1, K and L, and fig. S4). By contrast, iPSC-CMs edited with sgRNA1 and sgRNA6 were protected from deleterious Ca2+ alterations post-IR.
CaMKIIδ editing in mice subjected to IR injury
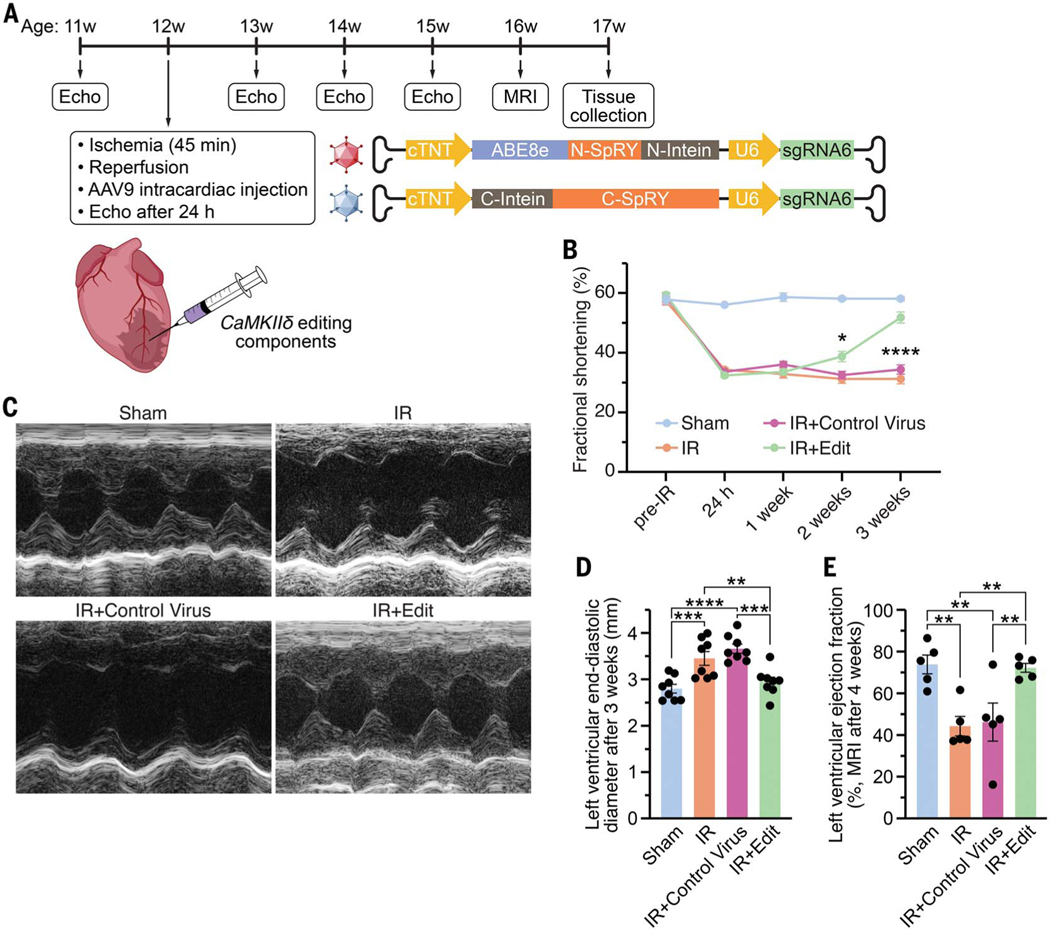
Since editing with sgRNA6 conferred greater protection to iPSC-CMs than with sgRNA1, we used mouse-sgRNA6 (with 95% homology to human-sgRNA6) and ABE8e-SpRY for base editing to ablate the CaMKIIδ oxidative activation sites in vivo in 12-week-old male C57Bl6 mice (Fig. 2A and fig. S5). We packaged the ABE components in adeno-associated virus serotype-9 (AAV9) using a split-intein trans-splicing system to accommodate the large size of ABE8e and sgRNA6. AAV9 was chosen as the delivery system because it effectively infects the hearts of mice and large mammals (2, 4). To ensure cardiac specificity, we used the cardiac troponin T (cTnT) promoter to drive ABE8e expression. After cardiac IR, AAV9 expressing sgRNA6 and ABE8e-SpRY (AAV-ABE-sgRNA6) was injected [7.5 × 1011 viral genomes (vg) per kg bodyweight of each component] directly into the area of cardiac injury (Fig. 2A). Control mice were subjected to IR with either an injection of control AAV9 or no injection. Sham-treated mice were also subjected to 45-min open chest surgery. Before IR, all mice exhibited normal cardiac function and similar fractional shortening between groups, as measured by echocardiography (Fig. 2B and fig. S6). As expected, cardiac function decreased 24 hours after IR surgery to a similar extent in all groups (fig. S7). While cardiac function remained stable for the first week (fig. S8), after 2 weeks mice that had been administered AAV-ABE-sgRNA6 began to functionally recover, as assessed by echocardiography (Fig. 2B and fig. S9). This recovery time is consistent with a previous study showing that genomic editing begins within 1 week after AAV9 delivery of CRISPR-Cas9 components in vivo (5). AAV-ABE-sgRNA6–edited mice showed further cardiac improvement 3 weeks post-IR and attained a level of fractional shortening comparable to that of the sham-treated mice (Fig. 2, B and C, and fig. S10). In addition, left ventricular end-diastolic dilation, a hallmark feature of heart failure, was observed after IR in control mice but was not seen post-IR in mice injected with AAV-ABE-sgRNA6 (Fig. 2D). Furthermore, cardiac magnetic resonance imaging, performed in a subgroup of mice at 4 weeks post-IR, showed impaired cardiac function in control mice and rescue of cardiac function in mice receiving AAV-ABE-sgRNA6 editing components, similar to the echocardiography findings (Fig. 2E, fig. S11, and movies S1 to S4).
Fig. 2. CaMKIIδ base editing improves cardiac function in vivo post-IR.
(A) Experimental design for subjecting mice to IR, injecting AAV-ABE-sgRNA6 for CaMKIIδ editing in vivo and monitoring heart function by echocardiography and cardiac magnetic resonance imaging (MRI). The AAV9 delivery system carrying the CRISPR-Cas9 base editing components with a split-intein trans-splicing system is shown. (B) Time course of fractional shortening for each group before IR as well as 24 hours, 1 week, 2 weeks, and 3 weeks post-IR (n =8 mice for each group; x axis not shown to scale). (C) Representative M-mode recordings of hearts of a sham-treated mouse, a mouse subjected to IR, a mouse subjected to IR with intracardiac injection of a control virus, and a mouse subjected to IR with intracardiac injection of AAV-ABE-sgRNA6 (IR+Edit) at 3 weeks post-IR. (D) Mean left ventricular end-diastolic diameter 3 weeks post-IR (n =8 mice for each group). (E) Mean left ventricular ejection fraction determined by cardiac MRI 4 weeks post-IR (n = 5 mice for each group). All replicates are individual mice. Statistical comparisons are based on two-way (B) and one-way [(D) and (E)] ANOVA post-hoc corrected by Holm-Sidak. Data are presented as means ± SEM.
Analysis of editing efficiency and potential off-target editing after in vivo ABE
Molecular analyses of heart tissue were performed at 5 weeks post-IR. Deep amplicon sequencing of DNA revealed an adenine to guanine editing efficiency of 7.6 ± 0.2% (c.A841G, p.M281V), 7.5 ± 0.2% (c.A844G, p.M282V), and 8.4 ± 0.2% (c.A848G, p.H283R) of the genomic DNA, and 46.1 ± 1.1% (c.A841G, p.M281V), 46.1 ± 1.1% (c.A844G, p.M282V), and 46.6 ± 1.0% (c.A848G, p.H283R) at the cDNA level (Fig. 3A and table S3). This difference can be explained because most cardiac CaMKIIδ is expressed in cardiomyocytes (27), which is the only cell type targeted by a troponin T-driven editing system. Notably, we detected a much higher editing efficiency at the anterior wall with 82.7 ± 1.2% (c.A841G, p.M281V), 85.7 ± 0.7% (c.A844G, p.M282V), and 85.8 ± 1.2% (c.A848G, p.H283R) at the cDNA level (Fig. 3B and C). This indicates that both critical methionines were ablated in almost all cardiomyocytes in the injured area of the heart. No off-target editing of the other CaMKII isoforms (α, β, γ) was seen in the hearts of mice injected with AAV-ABE-sgRNA6, as determined by deep amplicon sequencing (fig. S12, A and B, and table S3). As expected, mouse hearts injected with control AAV9 showed no genomic editing in the CaMKIIδ gene. Since CaMKII is expressed in many different tissues (27), editing CaMKII in organs other than the heart may potentially cause severe adverse effects. Assessment of CaMKII editing in other tissues did not reveal genomic editing of any CaMKII isoforms in the brain, the tibialis anterior muscle, or the liver, validating the cardiac specificity of the cTnT promoter used in our AAV9 editing system (fig. S12). We also detected no increase in transcriptome-wide adenine to inosine editing in post-IR mice injected with AAV-ABE-sgRNA6 (Fig. 3D).
Fig. 3. Analysis of mouse hearts after CaMKIIδ in vivo genomic editing by administration of AAV-ABE-sgRNA6.
(A) Percentage of adenine (A) to guanine (G) editing of DNA and cDNA for each adenine along sgRNA6 in the myocardium of mice treated with AAV-ABE-sgRNA6, as determined by deep amplicon sequencing. (B) Spatial analysis of adenine (A) to guanine (G) editing efficiency at the cDNA level for each adenine along sgRNA6 in the anterior and the inferior cardiac wall of mice injected with AAV-ABE-sgRNA6 in the anterior cardiac wall, as determined by Sanger sequencing. (C) Sequencing of a TOPO-TA clone shows in vivo editing of the CaMKIIδ gene at the cDNA level. (D) Percentage of transcriptome-wide adenine (A) to inosine (I) editing in the myocardium of sham-treated, IR, IR treated with a control virus, and IR-edited mice. (E) Western blot analysis of oxidized CaMKII, autophosphorylated CaMKII, total CaMKII, and GAPDH for all groups. (F) Mean densitometric analyses for oxidized CaMKII normalized to total CaMKII for sham-treated, IR, IR treated with a control virus, and IR-edited mice. (G) Mean densitometric analyses for autophosphorylated CaMKII normalized to total CaMKII for all groups. (H) Mean CaMKII activity for all groups and for lysates of IR and IR+Control Virus mice, both in presence of the CaMKII inhibitor AIP. (I) Western blot analysis of ryanodine receptor type 2 (RyR2) phosphorylation at the CaMKII site (serine 2814), total RyR2, and GAPDH for all groups. (J) Mean densitometric analysis for phosphorylated RyR2 normalized to total RyR2 for all groups. (K) Heat map of 209 differentially expressed genes between mice subjected to IR and injected with either a control AAV9 (n = 3) or AAV-ABE-sgRNA6 for editing of the CaMKIIδ gene (n = 4). (L) Gene ontology terms associated with the 101 genes up-regulated in mouse hearts injected with AAV-ABE-sgRNA6 compared to mice receiving control AAV9. (M) Gene ontology terms associated with the 108 genes down-regulated in mouse hearts injected with AAV-ABE-sgRNA6 compared to mice injected with control AAV9. All replicates are individual mice. Statistical comparisons are based on one-way ANOVA post-hoc corrected by Holm-Sidak. Data are presented as individual data points with means ± SEM.
Protein analysis showed a 4.4-fold increase in the amount of oxidized CaMKII in control mice post-IR. The level of oxidized CaMKII post-IR was normalized in the hearts of mice injected with AAV-ABE-sgRNA6 (Fig. 3, E and F, and fig. S13). The residual signal of oxidized CaMKII in post-IR mice injected with AAV-ABE-sgRNA6 may either be unedited and oxidized CaMKIIδ, oxidized CaMKIIγ, or unspecific background. Consistent with the amount of oxidized CaMKII, CaMKII autophosphorylation and activity were increased in control mice post-IR but not in post-IR mice injected with AAV-ABE-sgRNA6 (Fig. 3, E, G, and H, and fig. S13). Moreover, we found CaMKII-dependent phosphorylation of RyR2 to be increased in control mice post-IR but not in CaMKIIδ-edited mice post-IR (Fig. 3, I and J, and fig. S13).
Mechanisms of cardioprotection and long-term effects conferred by CaMKIIδ editing in vivo
RNA sequencing of control and AAV-ABE-sgRNA6–edited hearts revealed three different types of transcriptomes by principal component analysis (fig. S14). Although the transcriptome changed after IR, no differences were detected between the two different control groups post-IR. However, hearts subjected to IR- and CaMKIIδ-editing had a transcriptome different from that of control WT mouse hearts and formed a third cluster. In total, we identified 211 genes that were differentially expressed in mice subjected to IR with injection of control AAV9 compared with sham-treated mice (fig. S14B). Gene ontology analysis of the 163 genes up-regulated in IR (with control AAV9) revealed pathways related to cardiac disease whereas pathways associated with the 48 down-regulated genes were mainly related to cardiac function (fig. S14, C and D). Compared with mice treated with the control virus, we found 101 up-regulated and 108 down-regulated genes in mouse hearts with edited CaMKIIδ (Fig. 3K). Analysis of gene ontology terms revealed that pathways related to cardiac performance and disease—which were dysregulated in control AAV9 mice post-IR—were rescued in CaMKIIδ-edited mice post-IR (Fig. 3, L and M).
We found a substantial increase in the percentage of apoptotic cells in TUNEL-stained heart sections of mice post-IR compared with heart sections of sham-treated mice (Fig. 4, A and B). In contrast to the hearts of control mice post-IR, the percentage of apoptotic cells in CaMKIIδ-edited mice was comparable to the hearts of sham-treated mice. We also found a 2.7-fold increase in the area of fibrotic tissue in mice post-IR whereas the CaMKIIδ-edited mice were protected against fibrosis post-IR (Fig. 4, C and D, and figs. S15 and S16). We observed an increase in myocardial infiltration of inflammatory cells in mice post-IR, but this was not seen in hearts of CaMKIIδ-edited mice (fig. S15).
Fig. 4. Genomic editing of CaMKIIδ prevents cardiac cell death and fibrosis after IR.

(A) Immunohistochemistry of TUNEL (green, arrows), Hoechst 33342 (blue, for all nuclei), and cardiac troponin (recolored in purple for colorblind accessibility) in representative heart sections of mice subjected to sham surgery, IR, IR treated with a control AAV9, and IR treated with AAV-ABE-sgRNA6 (IR+Edit; scale bar 20 μm). (B) Mean percentage of TUNEL-positive cells in each group. (C) Whole transverse cross-sections of trichrome stained hearts for each group (scale bar 500 μm). (D) Mean percentage of fibrotic cardiac tissue in each group. Replicates are individual mice. Statistical comparisons are based on one-way ANOVA post-hoc corrected by Holm-Sidak. Data are presented as individual data points with means ± SEM.
To test for potential long-term adverse effects of CaMKIIδ editing, we analyzed mice 260 days after intraperitoneal injection of AAV-ABE-sgRNA6 at P5. Compared with noninjected littermates, we detected no difference in bodyweight (fig. S17). Since oxidized CaMKII has previously been linked to exercise performance (especially in skeletal muscle), we challenged these mice with a treadmill exhaustion test (28). We observed no difference in exercise performance, and immediate echocardiography after exhaustion revealed normal cardiac function in mice subjected to cardiac-specific CaMKIIδ editing 260 days earlier (fig. S17).
Discussion
CaMKIIδ inhibition has previously been proposed as a therapy for cardiac disease, and several CaMKII inhibitors have been tested in preclinical studies (15–17). However, these CaMKII inhibitors face several challenges (15), as some were reported to inhibit other ion channels (for example potassium channels) (29) and others showed few therapeutic benefits (15–17). Specific CaMKIIδ inhibitors that are ATP-competitive inhibitors also inhibit other kinases with potential deleterious effects, and other inhibitors are not bio-available or cell permeable (15, 16). Another challenge in using CaMKIIδ inhibitors is that CaMKIIδ is ubiquitously expressed and its global inhibition can have adverse effects in tissues other than the heart (27). Another clinical limitation of using CaMKIIδ inhibitors is the requirement for daily administration. Using the CRISPR-Cas9 ABE system to edit the genome provides a permanent change to the CaMKIIδ gene and overrides many of the above limitations. Incorporation of the cTnT promoter to restrict expression of the ABE components exclusively to the heart also prevents possible adverse consequences of CaMKIIδ inhibition in other tissues. Moreover, CRISPR-Cas9 gene editing is permanent, representing a “one and done” therapy (30).
In patients, administration of CaMKIIδ editing components after a myocardial infarction could be achieved in conjunction with the standard of care in response to a heart attack. The first therapeutic step after a myocardial infarction is coronary angiography and revascularization of the infarct artery, which requires a catheter that could also be used to deliver CaMKIIδ editing components to the infarct artery or to the infarct area (31). Before starting initial human clinical trials, future studies are needed including more analyses of the pharmacological profile, optimization of the viral dosage, more studies regarding the toxicity and safety of the treatment (such as potential immunogenicity of the base editor), and assessment of animal long-term survival.
We analyzed the top eight predicted sites in the human genome for potential off-target editing, but a deeper analysis (32), especially after extended exposure to the base editor, will be required for formal regulatory review. It will also be necessary to analyze the interaction with other drugs and treatments as well as the effectiveness of CaMKIIδ editing compared to currently available heart failure medication. Further studies showing a benefit in larger animals such as pigs and nonhuman primates would also be an important step toward clinical advancement of this approach.
CRISPR-Cas9 gene editing technology is typically used to correct specific genetic mutations before disease onset (1–6). Since the total number of patients carrying one specific mutation is usually low, the offered treatment affects only a limited group of patients. CRISPR-Cas9 gene editing has already been used to knock down WT PCSK9 gene in the liver as a strategy for hereditary familial hypercholesterolemia (32). Our strategy is designed to ablate a detrimental pathway in the adult heart and thereby provide therapeutic benefits for already-established heart disease. The concept of using CRISPR-Cas9 to block activation of deleterious pathways is also translatable to other signaling cascades in other human diseases.
Supplementary Material
ACKNOWLEDGMENTS
We thank A. Chai, Y. Zhang, and all other members of the Olson laboratory for helpful discussions; D. Alzhanov for technical assistance; J. Cabrera for graphics; J. Xu and Y. J. Kim from the CRI for Illumina NextSeq sequencing; C. Rodriguez-Caycedo for help with the iPSCs; J. Wansapura from the UTSW Advanced Imaging Research Center for mouse MRIs; Y. Zhang and the Boston Children’s Hospital Viral Core for virus production; J. Shelton from the Molecular Histopathology Core for helping with histology; the UTSW McDermott Center Sanger Sequencing Core; the UTSW McDermott Center Next Generation Sequencing Core; and the UTSW Flow Cytometry Core. Funding was provided by the following: National Institutes of Health grant R01HL130253 (to E.N.O. and R.B.-D.); National Institutes of Health grant R01HL157281 (to E.N.O. and R.B.-D.); National Institutes of Health grant P50HD087351 (to E.N.O. and R.B.-D.); Fondation Leducq Transatlantic Network of Excellence (to E.N.O.); Robert A. Welch Foundation 1-0025 (to E.N.O.); German Research Foundation (DFG) LE 5009/1-1 (to S.L.); German Cardiac Society (to S.L.); Cancer Prevention and Research Institute of Texas RP210099 (UTSW Advanced Imaging Research Center).
Footnotes
Competing interests: E.N.O. is a consultant for Vertex Pharmaceuticals, Tenaya Therapeutics, and Cardurion Pharmaceuticals. S.L., R.B.-D., and E.N.O. are inventors on patent application (63/352,804) submitted by UT Southwestern Medical Center that covers “Stress Editing of CaMKIIδ”. The other authors declare that they have no competing interests.
Data and materials availability: RNA-seq data have been deposited at Gene Expression Omnibus (https://www.ncbi.nlm.nih.gov/geo/) under accession number GSE210494. All other data are available in the main text or the supplementary materials.
REFERENCES AND NOTES
- 1.Liu N, Olson EN, Circ. Res 130, 1827–1850 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chemello F. et al. , Sci. Adv 7, eabg4910 (2021). [Google Scholar]
- 3.Richter MF et al. , Nat. Biotechnol 38, 883–891 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Amoasii L. et al. , Science 362, 86–91 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Amoasii L. et al. , Nat. Commun 10, 4537 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Koblan LW et al. , Nature 589, 608–614 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Virani SS et al. , Circulation 143, e254–e743 (2021). [DOI] [PubMed] [Google Scholar]
- 8.Beckendorf J, van den Hoogenhof MMG, Backs J, Basic Res. Cardiol 113, 29 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Neef S. et al. , Circ. Res 106, 1134–1144 (2010). [DOI] [PubMed] [Google Scholar]
- 10.Backs J. et al. , Proc. Natl. Acad. Sci. U.S.A 106, 2342–2347 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lebek S. et al. , Circ. Res 126, 603–615 (2020). [DOI] [PubMed] [Google Scholar]
- 12.Zhang T, Brown JH, Cardiovasc. Res 63, 476–486 (2004). [DOI] [PubMed] [Google Scholar]
- 13.Ling H. et al. , Circ. Res 112, 935–944 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Luo M. et al. , J. Clin. Invest 123, 1262–1274 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nassal D, Gratz D, Hund TJ, Front. Pharmacol 11, 35 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pellicena P, Schulman H, Front. Pharmacol 5, 21 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lebek S. et al. , J. Mol. Cell. Cardiol 118, 159–168 (2018). [DOI] [PubMed] [Google Scholar]
- 18.Erickson JR et al. , Cell 133, 462–474 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Purohit A. et al. , Circulation 128, 1748–1757 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang M. et al. , Nat. Cell Biol 21, 1152–1163 (2019). [DOI] [PubMed] [Google Scholar]
- 21.Gaudelli NM et al. , Nature 551, 464–471 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Komor AC, Kim YB, Packer MS, Zuris JA, Liu DR, Nature 533, 420–424 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Koblan LW et al. , Nat. Biotechnol 36, 843–846 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Walton RT, Christie KA, Whittaker MN, Kleinstiver BP, Science 368, 290–296 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Concordet JP, Haeussler M, Nucleic Acids Res. 46 (W1), W242–W245 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Clement K. et al. , Nat. Biotechnol 37, 224–226 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Uhlén M. et al. , Science 347, 1260419 (2015). [DOI] [PubMed] [Google Scholar]
- 28.Wang Q. et al. , Nat. Commun 12, 3175 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hegyi B. et al. , J. Mol. Cell. Cardiol 89 (Pt B), 173–176 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Karri DR et al. , Mol. Ther. Nucleic Acids 28, 154–167 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lawton JS et al. , Circulation 145, e18–e114 (2022). [DOI] [PubMed] [Google Scholar]
- 32.Musunuru K. et al. , Nature 593, 429–434 (2021). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.