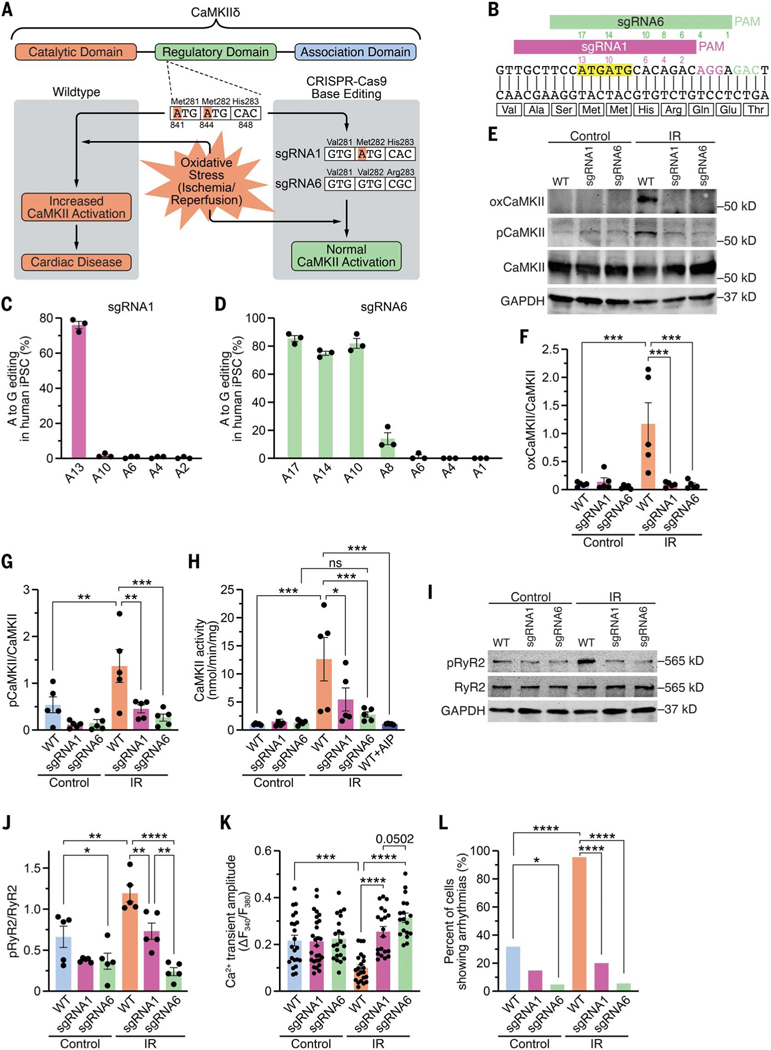
Fig. 1. Genomic editing of CaMKIIδ in human iPSC-cardiomyocytes.
(A) Schematic of CaMKIIδ and its three domains. Both critical methionines (Met281 and Met282) are located in the regulatory domain. Upon oxidative stress, these methionines are oxidized, resulting in increased CaMKII activity and cardiac disease. Using CRISPR-Cas9 adenine base editing, we identified sgRNA1, which edited only c.A841G (p.M281V; sgRNA1), and sgRNA6, which edited c.A841G (p.M281V), c.A844G (p.M282V), and c.A848G (p.H283R; sgRNA6), thereby preventing CaMKII activation upon oxidative stress. (B) Sequence of a segment of exon 11 of CaMKIIδ genomic DNA encoding part of the regulatory domain of CaMKIIδ. Alignment of sgRNA1 and sgRNA6 with CaMKIIδ. PAM sequences for sgRNA1 and sgRNA6 are in purple and green, respectively. Both ATGs encoding methionines are highlighted in yellow. Adenines within the sequences of sgRNA1 (purple) and sgRNA6 (green) are numbered (starting from the PAM). (C) Percentage of adenine (A) to guanine (G) editing in human iPSCs for each adenine in sgRNA1 after base editing with ABE8e and sgRNA1, as determined by Sanger sequencing. (D) Percentage of adenine (A) to guanine (G) editing in human iPSCs for each adenine in sgRNA6 after base editing with ABE8e and sgRNA6, as determined by deep amplicon sequencing. (E) Western blot analysis of oxidized CaMKII (specific antibody), autophosphorylated CaMKII (specific antibody), total CaMKII, and GAPDH in human WT, sgRNA1, and sgRNA6 iPSC-CMs for control group and after simulated ischemia/reperfusion (IR). (F) Mean densitometric analysis for oxidized CaMKII normalized to total CaMKII in control and post-IR iPSC-CMs (n = 5 independent iPSC-CM differentiations). (G) Mean densitometric analysis for autophosphorylated CaMKII normalized to total CaMKII in control and post-IR iPSC-CMs (n = 5 independent iPSC-CM differentiations). (H) Scatter bar plot showing mean CaMKII activity in control and post-IR iPSC-CMs and in lysates of WT post-IR iPSC-CMs in presence of the CaMKII inhibitor AIP (n = 5 independent iPSC-CM differentiations). (I) Western blot analysis of ryanodine receptor type 2 (RyR2) phosphorylation at the CaMKII site (serine 2814), total RyR2, and GAPDH in control and post-IR iPSC-CMs (n = 5 independent iPSC-CM differentiations). (J) Mean densitometric analysis for phosphorylated RyR2 normalized to total RyR2 in control and post-IR iPSC-CMs (n = 5 independent iPSC-CM differentiations). (K) Mean Ca2+ transient amplitude for each group (based on the number of cardiomyocytes). (L) Percentage of iPSC-CMs showing arrhythmias, as measured by epifluorescence microscopy. Statistical comparisons are based on one-way analysis of variance (ANOVA) post-hoc corrected by Holm-Sidak [(F) to (H)] and [(J) and (K)] and on Fisher’s exact test (L). Data are presented either as individual data points with means ± SEM or as percent of cells (L).