Abstract
Cathelicidins are a family of antimicrobial peptides prominent in the host defense mechanisms of several mammalian species. In addition to their antimicrobial activities, these peptides have been implicated in wound healing, angiogenesis, and other innate immune mechanisms. To investigate the regulatory mechanisms of cathelicidin gene expression, we conducted in vitro experiments evaluating the bone marrow cell expression of two porcine cathelicidins, PR-39 and protegrin, and cloned and evaluated the promoter sequence of PR-39. In addition, we evaluated in vivo kinetics of cathelicidin gene expression in pigs during an infection with Salmonella enterica serovar Typhimurium. Lipopolysaccharide (LPS) increased PR-39 and protegrin mRNA expression, which was ameliorated by polymyxin B. Concentrations of PR-39 in supernatants from bone marrow cell cultures were increased 10-fold after LPS stimulation. Similarly, interleukin-6 (IL-6) and all-trans retinoic acid (RA) markedly induced cathelicidin gene expression. To verify the transcriptional activation of the PR-39 gene by these agents, we made a PR-39 promoter-luciferase construct containing the full-length PR-39 promoter driving luciferase gene expression and transiently transfected PK-15 epithelial cells. RA and IL-6 increased luciferase activity in PK-15 cells transfected with the PR-39 promoter-luciferase reporter. Similarly, Salmonella-challenged pigs showed increased expression of PR-39 and protegrin mRNA in bone marrow cells at 6 and 24 h postchallenge. Taken together, these findings show that bacterial products (LPS), IL-6, RA, and Salmonella infection enhance the expression of the cathelicidins, PR-39 and protegrin, in bone marrow progenitor cells, and we suggest that extrinsic modulation of this innate host defense mechanism may be possible.
Antimicrobial peptides are an important first line of defense against microbial invasion and play a prominent role in host defense mechanisms of innate immunity (5, 6, 12, 13, 23, 29). Among these natural antibiotics are a family of antimicrobial peptides called cathelicidins (40). Cathelicidins are the largest family of antimicrobial peptides in pigs and include PR-39, a proline-arginine-rich 39-amino-acid residue antimicrobial peptide (2, 16, 37, 43, 46); protegrins 1 to 5 (20, 45, 47); prophenins 1 and 2 (17, 46); and porcine myeloid antimicrobial peptides 23, 36, and 37 (36). Cathelicidins isolated from humans include the antimicrobial peptide LL-37 and human cationic antimicrobial protein 18 (1, 9, 10, 15, 21). In addition, cathelicidins have been found in cattle, sheep, goats, horses, mice, and guinea pigs either by cDNA cloning or by direct purification from mature neutrophils (11, 14, 24, 27, 32, 33).
Most cathelicidins are constitutively expressed in myeloid progenitor cells and stored as pro-peptides in mature neutrophil granules, where few or no transcripts are expressed (26, 28, 41). However, some cathelicidins are expressed in other tissues and are inducible, such as human LL-37 in the testis (1) and in keratinocytes (9) and PR-39 and protegrin in lymphoid tissues in young pigs (39). Several genes of the cathelicidin family have been characterized, and all encode a conserved prepro region, which is similar to that of cathelin, and a variable C-terminal antimicrobial domain (1, 9, 10, 15–17, 20, 21, 36, 40, 45–47). Porcine cathelicidin genes are all organized in the same manner, compromised of four exons and three introns; exons 1 to 3 encode the prepro sequence, and exon 4 encodes several residues of the pro sequence and the mature peptide sequence (16, 17, 36, 41, 43, 45–47). The 5′-promoter region of the cathelicidin genes contains clusters of potential transcription regulatory elements, such as nuclear factor (NF)-κB, NF interleukin-6 (NF–IL-6), and IL-6 response elements (IL-6 RE) (15, 16, 45, 46), suggesting that cathelicidin gene expression may be actively regulated during infection and inflammation.
Although in vitro and in vivo induction of antimicrobial peptides has been reported previously for some antimicrobial peptides, such as LL-37 (9) and defensins (7, 8, 18, 19, 30, 31, 35, 38), information on the modulation of cathelicidin gene expression is sparse. The presence of potential transcription factor binding sites in the promoter region of cathelicidin genes, coupled with data indicating that some antimicrobial peptides are inducible, suggests that it is likely that cathelicidin genes are induced during bacterial infection. We report here on the regulatory mechanisms involved in gene expression and synthesis of porcine cathelicidins in response to lipopolysaccharide (LPS), inflammatory mediators, and bacterial infection. We show that all of these agents increase the expression of the porcine cathelicidins PR-39 and protegrin. In addition, promoter analysis of the PR-39 gene further supports the finding that cathelicidins are inducible and are not dependent on cell differentiation.
MATERIALS AND METHODS
Animals and reagents.
Healthy 6- to 8-week old crossbred pigs were obtained from the Kansas State University Swine Research Unit and housed in an environmentally controlled isolation facility. Pigs were determined to be free of both clinical signs of salmonellosis and detectable Salmonella organisms in fecal cultures 1 day before use in our experiments. Media (RPMI 1640, Dulbecco modified Eagle medium, and Hanks balanced salt solution, antibiotics, and TRIzol reagent all were purchased from Life Technologies (Rockville, Md.). Fetal bovine serum (FBS; low endotoxin) was from HyClone Laboratories (Logan, Utah). LPS from Salmonella enterica serovar Typhimurium, polymyxin B and all-trans retinoic acid (RA) were purchased from Sigma (St. Louis, Mo.). Recombinant porcine IL-6 was obtained from Endogen, Inc. (Woburn, Mass.). Oligonucleotides were synthesized by Integrated DNA Technologies, Inc. (Coralville, Iowa). Reagents for Northern blot analysis were purchased from Ambion, Inc. (Austin, Tex.). Reagents for reverse transcription-PCR (RT-PCR) were from Perkin-Elmer (Branchburg, N.J.). Synthetic PR-39 was synthesized by the Kansas State University Biotechnology Core Facility (Manhattan, Kans.) as previously described (34).
Cell isolation, culture, and stimulation.
Bone marrow cells were obtained from the femurs of healthy pigs and separated into light-density mononuclear cells and granulocytic-lineage cells by density gradient centrifugation as previously described (39). Briefly, granulocytic-lineage cell pellets were collected, washed with serum-free medium, filtered through sterile gauze, and centrifuged to pellet the cells. Erythrocytes were removed by hypotonic lysis, and cells were resuspended in RPMI 1640 medium supplemented with 10% FBS.
For stimulation experiments, bone marrow cells were seeded at 5 × 106 cells/ml in six-well plates and incubated with inducers for indicated times. The concentrations of inducers were as follows: LPS (0.1 or 1 μg/ml), RA (1 or 10 μM), IL-6 (1 or 10 ng/ml), or combined treatment with LPS (0.1 μg/ml) and polymyxin B (1 or 10 μg/ml).
Porcine kidney (PK)-15 epithelial cells were obtained from the American Type Culture Collection (Rockville, Md.) and grown at 37°C and 5% CO2 in minimal essential medium supplemented with 10% heat-inactivated FBS. PK-15 cells were used for transient-transfection assays and stimulated with LPS, RA, or IL-6.
Northern blot analyses.
Northern analyses were performed as previously described with slight modifications (39). Total RNA (10 μg) was extracted from bone marrow cells using TRIzol reagent, heat denatured, fractionated on 1% agarose-formaldehyde gels, and blotted onto positively charged nylon membranes (Boehringer Mannheim). Membranes were baked at 80°C for 15 min, prehybridized for 40 min at 60°C in ExpressHyb hybridization solution (Clontech, Palo Alto, Calif.), and hybridized for 2 h under the same conditions with 32P-labeled cDNA probes for PR-39, protegrin, or glyceraldehyde 3-phosphate dehydrogenase (GAPDH). The probes for PR-39, protegrin, and GAPDH were cDNA fragments, which were generated by RT-PCR and cloned into the pGEM-T Easy vector (Promega, Madison, Wis.) as previously described (39). Positive clones were selected and sequenced, and correct cDNA fragments were digested from positive clones, randomly labeled with [α-32P]dCTP using Ready-To-Go DNA Labeling Beads (Pharmacia Biotech), and used as probes. Posthybridization washes were done twice for 10 min each time at 42°C with 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate), followed by two washes of 20 min at 50°C with 0.1× SSC–0.5% sodium dodecyl sulfate. Blots were exposed to Kodak X-Omat films (Eastman Kodak, Rochester, N.Y.) with intensifying screens at −70°C. Intensity of the hybridization signals was quantified from autoradiographs using UN-SCAN-IT Gel Automated Digitizing System (Silk Scientific Corp., Orem, Utah), and the pixel values of each specific band were recorded from digitized images. The expression levels of PR-39 or protegrin were normalized to the expression of the housekeeping gene GAPDH by calculating the ratios of pixel values of PR-39 or protegrin over those of the housekeeping gene.
PR-39 ELISA.
To measure concentrations of PR-39 in supernatants from bone marrow cells incubated with LPS, a sandwich enzyme-linked immunosorbent assay (ELISA) was used as previously described (44). Briefly, 96-well microplates were coated overnight at 4°C with an anti-PR-39 monoclonal antibody. After washing twice, plates were blocked for 2 h with 1% bovine serum albumin in phosphate-buffered saline. Synthetic PR-39 standards and test samples were added to wells and incubated at 22°C for 2 h with shaking. After four washes, biotinylated goat polyclonal antibodies to PR-39 were added to each well, and plates were incubated for 2 h at 22°C while shaking. Plates were then incubated with peroxidase-labeled streptavidin, followed by color development with substrate. The absorbance was measured at 405 nm using a microplate reader.
Construction of luciferase fusion plasmids and transient-transfection assays.
An 836-bp nucleotide fragment of the 5′-flanking region of the PR-39 gene upstream from the start codon (GenBank accession nos. X87236 and X89201) was cloned from porcine genomic DNA by PCR amplification. The following primers were used: sense primer, 5′-CCC TCG AGG GTT CTT GGG ATG TAA GGG CG-3′ (from nucleotide −823 with a XhoI restriction site [underlined]); antisense primer, 5′-CCA AGC TTG GTG CCC AGG TGA GCC TCC TC-3′ (from nucleotide +13 with a HindIII restriction site [underlined]). The PCR products were digested with XhoI and HindIII and cloned into the multiple cloning sites of the pGL3 Luciferase Reporter Vector (Promega). The recombinant constructs were designated pGL3−836/+13, which contains the PR-39 promoter region between nucleotides −823 and +13, and pGL3-Basic (promoterless). The authenticity of the plasmid constructs was conformed by DNA sequencing using the T7 Sequenase version 2.0 DNA kit (Amersham Life Science, Cleveland, Ohio). The plasmids were purified with Wizard Plus Medipreps DNA Purification System (Promega) and used for transient-transfection assays.
For transfection assays, PK-15 cells were seeded into 24-well tissue culture plates 18 to 24 h before transfection and grown to 60 to 80% confluence. Plasmid DNA (1 μg in 100 μl of serum-free medium) was mixed with an equal volume of serum-free medium containing 2 μl of TransFast Transfection Reagent (Promega), incubated at room temperature for 15 min, and added to the cells. The cells were returned to the incubator for 1 h for transfection, and then 1 ml of medium containing 10% FBS was added. After 24 h, transfected cells were stimulated with or without LPS (0.1 or 1 μg/ml), IL-6 (1 or 10 ng/ml), or RA (1 or 10 μM). After 24 h of incubation with inducers, cells were lysed and assayed for luciferase activity using the Luciferase Assay System (Promega) as previously described (42).
S. enterica serovar Typhimurium infection.
Eighteen healthy 4-week-old pigs were used for Salmonella challenge experiments. All experimental procedures were performed in accordance with guidelines of the Kansas State University Institutional Animal Care and Use Committee and the Biosafety Committee. Pigs were housed in an environmentally controlled isolation facility and challenged orally with 9.5 × 109 CFU of S. enterica serovar Typhimurium suspended in growth medium. The bacterial strain was a primary isolate from a clinical case of salmonellosis in pigs and was confirmed to be a pure culture of S. enterica serovar Typhimurium by the U.S. Department of Agriculture National Veterinary Services Laboratory (Ames, Iowa). Three pigs from challenged or control groups were euthanized at 6, 12, and 24 h postinfection. Femurs were dissected from each pig and bone marrow cells were collected as described above, and used for the extraction of total RNA. The expression of PR-39 and protegrin mRNA was evaluated using semiquantitative RT-PCR.
Semiquantitative RT-PCR.
Total RNA (0.1 μg) was used as the template for RT-PCR. RT was performed at 42°C for 30 min in a volume of 50 μl of reaction mixture and then heated at 94°C for 5 min. The PCR profile for β-actin included denaturation at 94°C for 2 min, followed by 20 cycles of denaturation at 94°C for 1 min, annealing at 55°C, and extension at 72°C for 1 min, and a final extension at 72°C for 7 min. The PCR profile for PR-39 and protegrin was similar to that of β-actin, except that annealing was done at 50°C. Primers for PR-39, protegrin, and β-actin used for RT-PCR have been described previously (39, 42). Each PCR reaction included a negative control (no template). A 20-μl portion of each PCR product was subjected to electrophoresis on a 1.5% agarose gel with ethidium bromide. Negative films were used to determine the band intensity. PCR products were normalized according to the amount of β-actin detected in the same cDNA sample, and PR-39/β-actin or protegrin/β-actin ratios were calculated.
Statistical evaluation.
Data were analyzed by using the Student's t test. Data with a P value of ≤0.05 were considered significant.
RESULTS
LPS increases cathelicidin gene and protein expression.
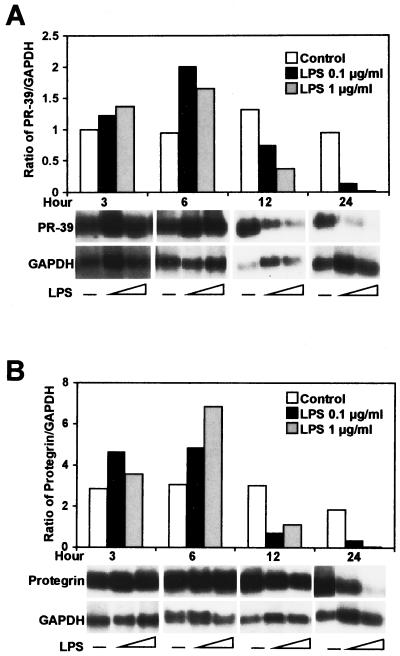
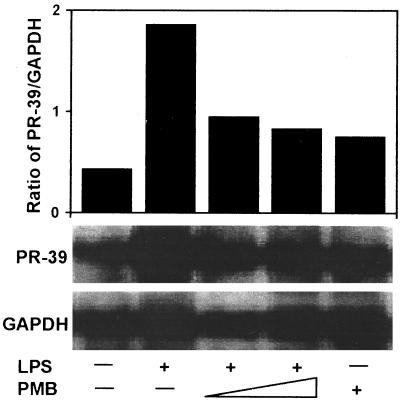
To investigate cathelicidin gene expression, we first evaluated the ability of a major constituent of the gram-negative bacterial membrane, LPS, to induce PR-39 and protegrin expression in primary-culture, porcine bone marrow cells. LPS at 0.1 or 1 μg/ml increased PR-39 (Fig. 1A) and protegrin (Fig. 1B) mRNA expression at 3 h, with peak expression occurring at 6 h. Expression of PR-39 and protegrin mRNA was decreased at 12 h after incubation with LPS. No difference in PR-39 and protegrin mRNA expression was found between the two concentrations of LPS used. To confirm the specificity of the LPS-induced increase in cathelicidin gene expression, bone marrow cells were treated with LPS (0.1 μg/ml) alone or a combination of LPS and polymyxin B (1 and 10 μg/ml). As shown in Fig. 2, Northern analysis revealed that LPS-induced PR-39 gene expression was blocked by polymyxin B. These results demonstrate that cathelicidin gene expression is inducible in bone marrow cells in response to LPS stimulation.
FIG. 1.
Northern analysis of PR-39(A) and protegrin (B) in porcine bone marrow cells in response to LPS. Bone marrow cells were treated with LPS (0.1 or 1 μg/ml) for 3 to 24 h, and total RNA (10 μg) was extracted, resolved by electrophoresis, and transferred to nylon membranes. Blots were hybridized with PR-39, protegrin, or GAPDH probes in duplicate. Densitometric analysis of PR-39 and protegrin were normalized to GAPDH and are shown as PR-39/GAPDH (A) and protegrin/GAPDH (B) ratios. The results are representative of three independent experiments using three pigs.
FIG. 2.
Effect of polymyxin B on LPS induction of PR-39 mRNA expression. Total RNA was isolated from bone marrow cells treated with LPS (0.1 μg/ml) alone or in the presence of polymyxin B (PMB, 1 or 10 μg/ml) for 6 h. Northern blot analysis was as described in Fig. 1. The results are representative of three independent experiments using three pigs.
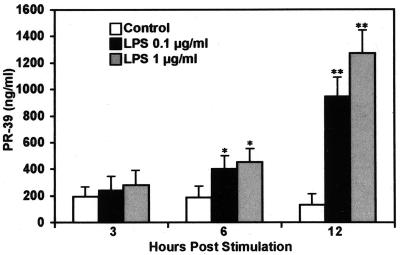
To confirm translation of the PR-39 gene, a PR-39 specific ELISA was used to measure concentrations of PR-39 in supernatants from bone marrow cells stimulated with LPS. As shown in Fig. 3, the concentrations of PR-39 in cell supernatants doubled after 3 h and peaked at 12 h. The concentrations of PR-39 were more than 10-fold greater after 12 h of incubation with LPS than those of unstimulated bone marrow cells.
FIG. 3.
Concentrations of PR-39 in supernatants of bone marrow cells stimulated with LPS (0.1 or 1 μg/ml). Supernatants of bone marrow cells were collected at the indicated times and assayed for concentrations of PR-39 using a sandwich ELISA. The values are means ± the standard deviation (SD) from three independent experiments. ∗, P < 0.05; ∗∗, P < 0.01.
IL-6 and RA upregulate cathelicidin gene expression.
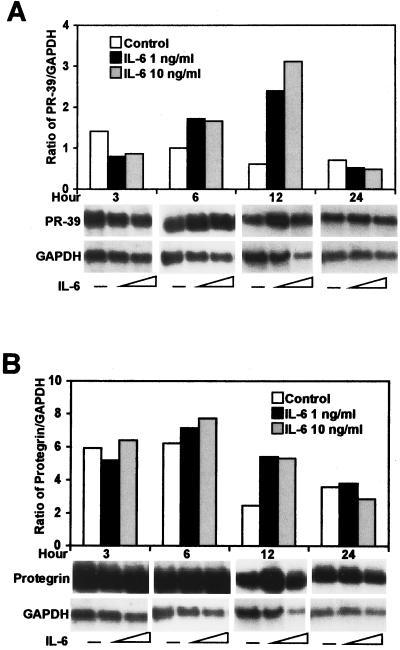
To investigate whether the inflammatory cytokine IL-6 affects cathelicidin expression, the recombinant cytokine was incubated with bone marrow cells, and PR-39 and protegrin transcription was evaluated by Northern blot analysis. Similar to the LPS findings, IL-6 (at 1 or 10 ng/ml) increased the expression of PR-39 (Fig. 4A) and protegrin (Fig. 4B) mRNA at 6 h poststimulation, with a fivefold maximal increase in PR-39 mRNA expression at 12 h poststimulation.
FIG. 4.
IL-6 induced the expression of PR-39 (A) and protegrin (B) mRNA in bone marrow cells. Porcine recombinant IL-6 was used at 1 or 10 ng/ml. Northern blot analysis was done as described in Fig. 1. The results are representative of three independent experiments using three pigs.
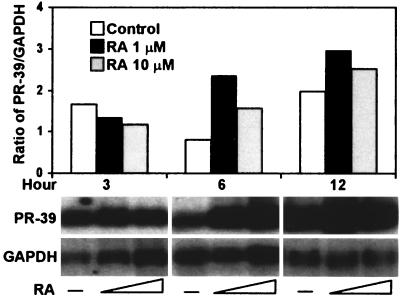
Because RA has been shown to be a strong inducer of defensins (18) and induces granulocytic cell differentiation (22), we analyzed PR-39 gene transcription in bone marrow cells exposed to RA using Northern blot analysis. PR-39 gene expression was markedly increased in bone marrow cells treated with RA at 6 or 12 h poststimulation (Fig. 5).
FIG. 5.
Northern blot analysis of PR-39 mRNA levels in bone marrow cells in response to RA. Bone marrow cells were incubated with RA at 1 or 10 μM for 3 to 12 h. Northern blot analysis was done as described in Fig. 1. The results are representative of three independent experiments using three pigs.
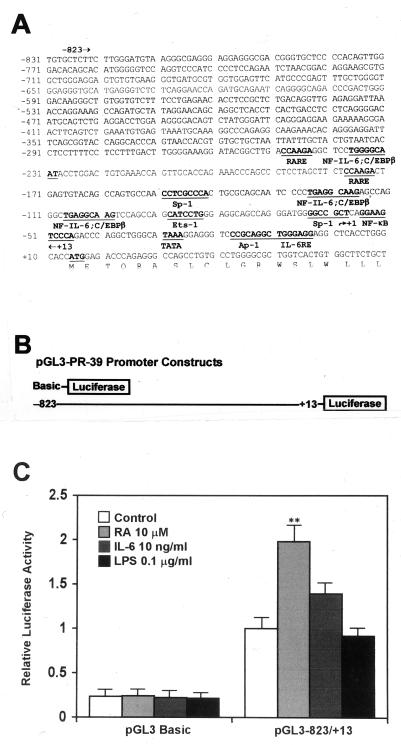
IL-6 and RA activate the PR-39 promoter.
The 3′ region of the PR-39 promoter contains clusters of potential transcription factor binding sites for NF-κB, NF-IL-6, IL-6 RE, and retinoic acid response element (RARE) (Fig. 6A). In addition, sequence analysis of this region of the promoter revealed other potential binding motifs for transcription factors Ets-1, Sp-1, activator protein-1 (Ap-1) and CCAAT/enhancer-binding protein (C/EBP). To determine whether the induction of PR-39 mRNA in primary cell cultures was due to changes in transcription of the gene or cell differentiation, we constructed reporter plasmids containing the PR-39 promoter driving luciferase gene expression and conducted transient-transfection assays (Fig. 6B). PK-15 cells were transfected with plasmids pGL3−836/+13 or pGL3-Basic and then incubated with RA (1 or 10 μM), IL-6 (1 or 10 ng/ml), or LPS (0.1 or 1 μg/ml). Consistent with Northern blot analysis, activation of the PR-39 promoter in PK-15 cells was increased by RA and IL-6 (Fig. 6C). RA caused a doubling of PR-39 promoter activity, and IL-6 induced a 40% increase of luciferase expression in the cells transfected with the full-length PR-39 promoter. However, LPS did not induce an increase in luciferase activity in PK-15 cells transfected with the full-length PR-39 promoter. The promoterless plasmid, pGL3-Basic, showed low basal level reporter gene expression and, as expected, did not respond to RA, IL-6, or LPS stimulation (Fig. 6C).
FIG. 6.
Nucleotide sequence of the PR-39 promoter (A), diagram of PR-39 promoter-luciferase constructs (B), and PR-39 promoter-luciferase construct activity in response to RA, IL-6, and LPS (C). The PR-39 promoter contains potential transcription factor binding sites (underlined) for RARE, NF–IL-6 plus C/EBPβ, Sp-1, Ets-1, NF-κB, Ap-1, and IL-6 RE. PR-39 promoter-luciferase constructs containing the whole PR-39 promoter (pGL3−836/+13) and a promoterless construct (pGL3-Basic) were transfected into PK-15 cells. Cells were transfected with plasmids for 24 h and then incubated with RA (1 or 10 μM), IL-6 (1 or 10 ng/ml), or LPS (0.1 or 1 μg/ml) for 24 h. The same trend was observed for both concentrations of inducers used; therefore, only data for one concentration are shown. The data were normalized against unstimulated cells transfected with plasmid pGL3−836/+13 and are shown as relative luciferase activity values. Values are means ± the SD from three independent experiments. ∗∗, P < 0.01.
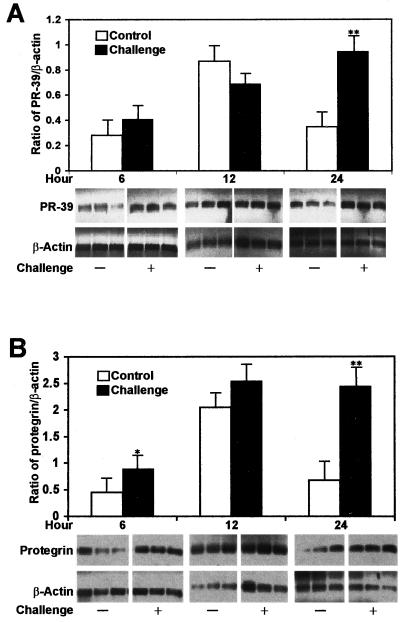
Cathelicidin gene expression is increased during infection with S. enterica serovar Typhimurium.
To evaluate cathelicidin gene expression in vivo, pigs were challenged orally with 9.5 × 109 CFU of S. enterica serovar Typhimurium, and semiquantitative RT-PCR was used to evaluate PR-39 and protegrin mRNA expression. Expression of both cathelicidins, PR-39 and protegrin, increased after infection; significant increases were found at 24 h postchallenge for PR-39 and at 6 and 24 h postchallenge for protegrin (Fig. 7).
FIG. 7.
Salmonella infection increased PR-39 (A) and protegrin (B) mRNA expression in bone marrow cells. Semiquantitative RT-PCR was used to quantify mRNA. Total RNA (0.1 μg) was used as templates using 28 cycles for PR-39 or protegrin and 20 cycles for β-actin. PR-39/β-actin or protegrin/β-actin ratios were determined. Values are means ± the SD from three challenged and control pigs at each time point. ∗, P < 0.05; ∗∗, P < 0.01.
DISCUSSION
Cathelicidin antimicrobial peptides generally are expressed only in granulocytic leukocytes or their precursors (16, 26–28, 37); however, some exceptions exist. Recently, we reported that PR-39 and protegrin mRNA were expressed in several lymphoid tissues in young pigs (39) and the human cathelicidin, LL-37, has been found in the testis (1) and in keratinocytes (9). In bone marrow, cathelicidin peptides are synthesized during the maturation and differentiation of neutrophil precursors (26, 28, 41). In this study, we evaluated cathelicidin gene expression by analyzing PR-39 and protegrin mRNA transcription in bone marrow progenitor cells, with transfection studies using the PR-39 promoter, and in Salmonella-challenged animals.
Our results show that LPS and IL-6 increase PR-39 and protegrin mRNA expression in porcine bone marrow cells. Cathelicidin transcription increased rapidly in bone marrow cells stimulated with LPS as early as 3 h post-LPS treatment and peaked at 6 h after stimulation. Similarly, increased PR-39 and protegrin mRNA expression was observed after 6 to 12 h of incubation with IL-6. The mechanism(s) of LPS- and IL-6-induced increases in PR-39 and protegrin gene expression has not been completely described. Sequence analysis of the PR-39 promoter revealed the existence of several potential transcriptional binding sites for NF–IL-6, IL-6 RE, and NF-κB (1, 16, 46, 47). The expression of NF-IL-6 is markedly increased in all tissues after stimulation with LPS, IL-1, or IL-6 (3). This transcription factor binds to regulatory regions of several acute-phase protein and cytokine genes, implying that NF–IL-6 has a role in the regulation of not only the IL-6 gene but also other genes involved in acute-phase reactions, inflammation, and hemopoiesis (3). The acute-phase response factor is a transcription factor that binds to the IL-6 RE in promoters of various genes and is translocated into the nucleus in response to IL-6 (4). Therefore, we reasoned that LPS and IL-6 might sequentially activate the NF–IL-6 and acute-phase response factor genes and then initiate cathelicidin gene transcription. Our data clearly demonstrate that LPS and IL-6 upregulate PR-39 and protegrin mRNA expression in bone marrow cells, suggesting that transcription factors, such as NF-κB, NF–IL-6, acute-phase response factor, and others may be involved in the transcriptional activation of PR-39 and protegrin genes.
PR-39 and protegrin mRNA transcription began to decrease at 12 h after stimulation with LPS. The mechanism by which this gene expression is downregulated is not known. Diamond et al. (7, 8) showed that LPS induced an increase in β-defensin mRNA transcription by 6 h with a further increase at 16 h; longer incubations failed to induce any further increase in mRNA levels. Although lower PR-39 mRNA levels were not observed at longer incubation times when cells were stimulated with IL-6 or RA, it is possible that LPS induces changes that alter cathelicidin mRNA production and/or stability. Alternatively, cathelicidin gene expression may increase with stimulation and then decrease in bone marrow progenitor cells as they mature and differentiate. To test this hypothesis, we evaluated the influence of RA, which induces the granulocytic differentiation of hematopoietic cells (18), on PR-39 gene expression. In contrast to LPS, RA stimulation caused robust increases in PR-39 mRNA expression in bone marrow cells after 12 h of incubation. In addition, our preliminary data suggest that stimulus-induced repression is a characteristic of the PR-39 promoter (H. Wu, G. Zhang, C. R. Ross, and F. Blecha, Abstr. 80th Conf. Res. Workers Anim. Dis., abstr. 136, 1999), which also may account for the LPS effect that was observed.
To limit the multiple interpretations that occur when differentiating bone marrow cells are used to study cathelicidin gene expression, we performed transient-transfection assays with PR-39 promoter-luciferase reporters in the porcine epithelial cell line, PK-15. Luciferase activity increased in cells transfected with the PR-39 promoter construct and incubated with RA and IL-6. RA binds its cognate nuclear receptor, interacts directly with a specific DNA sequence termed the RARE, and modulates target gene expression (22, 25). This activation process by RA is defined as an immediate-early gene regulation event, and cooperative response genes require secondary proteins as transcriptional factors to activate target gene expression. Likely candidates for the secondary transcription factors are members of the C/EBP family (22). Sequence analysis of the PR-39 promoter revealed consensus RARE half-sites and C/EBP potential binding motifs, indicating that RA may directly modulate PR-39 gene expression via these sites. Interestingly, LPS did not influence the activity of the PR-39 promoter in transfected PK-15 cells. It is possible that LPS influences cathelicidin expression indirectly and that secondary regulators, such as IL-6, may not be present in this system.
The expression of antimicrobial peptide genes, such as LL-37, lingual antimicrobial peptide, and enteric β-defensin is increased during infection (9, 19, 31, 35, 38). We also have reported that Salmonella infection increases serum concentrations of PR-39 (44). In this study, we extend those findings and show that PR-39 and protegrin gene expression increases after infection with S. enterica serovar Typhimurium. Whether these increased transcription levels result in antimicrobial peptide translation in vivo is not known; however, given the results of our previous study (44) and since our present findings show increased PR-39 peptide in the supernatants of LPS-stimulated cells, this is likely. Taken together, our findings support the hypothesis that increased expression of cathelicidin genes and the release of antimicrobial peptides by proinflammatory mediators or bacterial infection may contribute to the induction of enhanced host responses to infection. Further studies will explore the feasibility of extrinsic modulation of this important innate immune mechanism.
ACKNOWLEDGMENTS
We thank Danielle Goodband and Elena Donskaya for their excellent technical assistance.
This work was supported in part by USDA NRI Competitive Grants 95-37204-2141 and 98-35204-6397.
REFERENCES
- 1.Agerberth B, Gunne H, Odeberg J, Kogner P, Boman H G, Gudmundsson G H. FALL-39, a putative human peptide antibiotic, is cysteine-free and expressed in bone marrow and testis. Proc Natl Acad Sci USA. 1995;92:195–199. doi: 10.1073/pnas.92.1.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Agerberth B, Lee J, Bergman T, Carlquist M, Boman H G, Mutt V, Jörnvall H. Amino acid sequence of PR-39: isolation from pig intestine of a new member of the family of proline-arginine-rich antibacterial peptides. Eur J Biochem. 1991;202:849–854. doi: 10.1111/j.1432-1033.1991.tb16442.x. [DOI] [PubMed] [Google Scholar]
- 3.Akira S, Isshiki H, Sugita T, Tanabe O, Kinoshita S, Nishio Y, Nakajima T, Hirano T, Kishimoto T. A nuclear factor for IL-6 expression (NF-IL6) is a member of a C/EBP family. EMBO J. 1990;9:1897–1906. doi: 10.1002/j.1460-2075.1990.tb08316.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Akira S, Nishio Y, Inoue M, Wang X J, Wei S, Matsusaka T, Yoshida K, Sudo T, Naruto M, Kishimoto T. Molecular cloning APRF, a novel IFN-stimulated gene factor 3 p91-related transcription factor involved in the gp130-mediated signaling pathway. Cell. 1994;77:63–71. doi: 10.1016/0092-8674(94)90235-6. [DOI] [PubMed] [Google Scholar]
- 5.Boman H G. Peptide antibiotics and their role in innate immunity. Annu Rev Immunol. 1995;13:61–92. doi: 10.1146/annurev.iy.13.040195.000425. [DOI] [PubMed] [Google Scholar]
- 6.Boman H G. Gene-encoded peptide antibiotics and the concept of innate immunity: an update review. Scand J Immunol. 1998;48:15–25. doi: 10.1046/j.1365-3083.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- 7.Diamond G, Kaiser V, Rhodes J, Russell J P, Bevins C L. Transcriptional regulation of beta-defensin gene expression in tracheal epithelial cells. Infect Immun. 2000;68:113–119. doi: 10.1128/iai.68.1.113-119.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Diamond G, Russell J P, Bevins C L. Inducible expression of an antibiotic peptide gene in lipopolysaccharide-challenged tracheal epithelial cells. Proc Natl Acad Sci USA. 1996;93:5156–5160. doi: 10.1073/pnas.93.10.5156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Frohm M, Agerberth B, Ahangari G, Ståhle-Bäckdahl J, Lidén S, Wigzell H, Gudmundsson G H. The expression of the gene coding for the antibacterial peptide LL-37 is induced in human keratinocytes during inflammatory disorders. J Biol Chem. 1997;272:15258–15263. doi: 10.1074/jbc.272.24.15258. [DOI] [PubMed] [Google Scholar]
- 10.Frohm Nilsson M, Sandstedt B, Sorensen O, Weber G, Borregaard N, Stahle, Backdahl M. The human cationic antimicrobial protein (hCAP18), a peptide antibiotic, is widely expressed in human squamous epithelia and colocalizes with interleukin-6. Infect Immun. 1999;67:561–566. doi: 10.1128/iai.67.5.2561-2566.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gallo R L, Kim K J, Bernfield M, Kozak C A, Zanetti M, Merluzzi L, Gennaro R. Identification of CRAMP, a cathelin-related antimicrobial peptide expressed in the embryonic and adult mouse, J. Biol Chem. 1997;272:13088–13093. doi: 10.1074/jbc.272.20.13088. [DOI] [PubMed] [Google Scholar]
- 12.Ganz T, Lehrer R I. Antimicrobial peptides of leukocytes. Curr Opin Hematol. 1997;4:53–58. doi: 10.1097/00062752-199704010-00009. [DOI] [PubMed] [Google Scholar]
- 13.Ganz T, Lehrer R I. Antimicrobial peptides of vertebrates. Curr Opin Immunol. 1998;10:41–44. doi: 10.1016/s0952-7915(98)80029-0. [DOI] [PubMed] [Google Scholar]
- 14.Gennaro R, Skerlavaj B, Romeo D. Purification, composition, and activity of two bactenecins, antibacterial peptides of bovine neutrophils. Infect Immun. 1989;57:3142–3146. doi: 10.1128/iai.57.10.3142-3146.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gudmundsson G H, Agerberth B, Odeberg J, Bergman T, Olsson B, Salcedo R. The human gene FALL-39 and processing of the cathelin precursor to the antibacterial peptide LL-37 in granulocytes. Eur J Biochem. 1996;238:325–332. doi: 10.1111/j.1432-1033.1996.0325z.x. [DOI] [PubMed] [Google Scholar]
- 16.Gudmundsson G H, Magnusson K P, Chowdhary B P, Johansson M, Andersson L, Boman H G. Structure of the gene for porcine peptide antibiotic PR-39, a cathelin gene family member: comparative mapping of the locus for the human peptide antibiotic FALL-39. Proc Natl Acad Sci USA. 1995;92:7085–7089. doi: 10.1073/pnas.92.15.7085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Harwig S S, Kokryakov V N, Swiderek K M, Aleshina G M, Zhao C, Lehrer R I. Prophenin-1, an exceptionally proline-rich antimicrobial peptide from porcine leukocytes. FEBS Lett. 1995;362:65–69. doi: 10.1016/0014-5793(95)00210-z. [DOI] [PubMed] [Google Scholar]
- 18.Herwig S, Su Q, Zhang W, Ma Y, Tempst P. Distinct temporal patterns of defensin mRNA regulation during drug-induced differentiation of human myeloid leukemia cells. Blood. 1996;87:350–364. [PubMed] [Google Scholar]
- 19.Hiratsuka T, Nakazato M, Date Y, Ashitani J, Minematsu T, Chino N, Matsukura S. Identification of human beta-defensin-2 in respiratory tract and plasma and its increase in bacterial pneumonia. Biochem Biophys Res Commun. 1998;249:943–947. doi: 10.1006/bbrc.1998.9239. [DOI] [PubMed] [Google Scholar]
- 20.Kokryakov V N, Harwig S S, Panyutich E A, Shevchenko A A, Aleshina G M, Shamova O V, Korneva H A, Lehrer R I. Protegrins: leukocyte antimicrobial peptides that combine features of corticostatic defensins and tachyplesins. FEBS Lett. 1993;327:231–236. doi: 10.1016/0014-5793(93)80175-t. [DOI] [PubMed] [Google Scholar]
- 21.Larick J W, Hirata M, Balint R F, Lee J, Zhong J, Wright S C. Human CAP18: a novel antimicrobial lipopolysaccharide-binding protein. Infect Immun. 1995;63:1291–1297. doi: 10.1128/iai.63.4.1291-1297.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lawson N D, Berliner N. Neutrophil maturation and the role of retinoic acid. Exp Hematol. 1999;27:1355–1367. doi: 10.1016/s0301-472x(99)00085-5. [DOI] [PubMed] [Google Scholar]
- 23.Lehrer R I, Ganz T. Antimicrobial peptides in mammalian and insect host defense. Curr Opin Immunol. 1999;11:23–27. doi: 10.1016/s0952-7915(99)80005-3. [DOI] [PubMed] [Google Scholar]
- 24.Mahoney M M, Lee A Y, Brezinski-Caliguri D J, Huttner K M. Molecular analysis of the sheep cathelin family reveals a novel antimicrobial peptide. FEBS Lett. 1995;377:519–522. doi: 10.1016/0014-5793(95)01390-3. [DOI] [PubMed] [Google Scholar]
- 25.Medh R D, Schmidt T J. Trans-retinoic acid and glucocorticoids synergistically induce transcription from the mouse mammary tumor virus promoter in human embryonic kidney cells. J Steroid Biochem Mol Biol. 1997;62:129–142. doi: 10.1016/s0960-0760(97)00033-2. [DOI] [PubMed] [Google Scholar]
- 26.Nagaoka I, Hirata M, Sugimoto K, Tsutsumi-Ishii Y, Someya A, Saionli K, Igari J. Evaluation of the expression of human CAP18 gene during neutrophil maturation in the bone marrow. J Leukoc Biol. 1998;64:845–852. doi: 10.1002/jlb.64.6.845. [DOI] [PubMed] [Google Scholar]
- 27.Nagaoka I, Ishihara N, Yamashita T. Characterization of the promoters of the guinea pig neutrophil cationic peptide-1 and -2 genes. FEBS Lett. 1994;356:33–38. doi: 10.1016/0014-5793(94)01229-6. [DOI] [PubMed] [Google Scholar]
- 28.Nagaoka I, Tsutsumi-Ishii Y, Yomogida S, Yamashita T. Isolation of cDNA encoding guinea pig neutrophil cationic antibacterial polypeptide of 11 kDA (CAP11) and evaluation of CAP11 mRNA expression during neutrophil maturation. J Biol Chem. 1997;272:22742–22750. doi: 10.1074/jbc.272.36.22742. [DOI] [PubMed] [Google Scholar]
- 29.Nicholas P, Mor A. Peptides as weapons against microorganisms in the chemical defense system of vertebrates. Annu Rev Microbiol. 1995;49:277–304. doi: 10.1146/annurev.mi.49.100195.001425. [DOI] [PubMed] [Google Scholar]
- 30.Russell J P, Diamond G, Tarver A P, Scanlin T F, Bevins C L. Coordinate induction of two antibiotic genes in tracheal epithelial cells exposed to the inflammatory mediators lipopolysaccharide and tumor necrosis factor alpha. Infect Immun. 1996;64:1565–1568. doi: 10.1128/iai.64.5.1565-1568.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schonwetter B S, Stolzenberg E D, Zasloff M A. Epithelial antibiotics induced at sites of inflammation. Science. 1995;267:1645–1648. doi: 10.1126/science.7886453. [DOI] [PubMed] [Google Scholar]
- 32.Scocchi M, Bontempo D, Boscolo S, Tomasinsig L, Giulotto E, Zanetti M. Novel cathelicidins in horse leukocytes 1. FEBS Lett. 1999;457:459–464. doi: 10.1016/s0014-5793(99)01097-2. [DOI] [PubMed] [Google Scholar]
- 33.Shamova O, Brogden K A, Zhao C, Nguyen T, Kokryakov V N, Lehrer R I. Purification and properties of proline-rich antimicrobial peptides from sheep and goat leukocytes. Infect Immun. 1999;67:4106–4111. doi: 10.1128/iai.67.8.4106-4111.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shi J, Ross C R, Leto T L, Blecha F. PR-39, a proline-rich antibacterial peptide that inhibits phagocyte NADPH oxidase activity by binding to Src homology 3 domains of p47phox. Proc Natl Acad Sci USA. 1996;93:6014–6018. doi: 10.1073/pnas.93.12.6014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stolzenberg E D, Anderson G M, Ackermann M R, Whitlock R H, Zasloff M. Epithelial antibiotic induced in states of disease. Proc Natl AcadSci USA. 1997;94:8686–8690. doi: 10.1073/pnas.94.16.8686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Storici P, Scocchi M, Tossi A, Gennaro R, Zanetti M. Chemical synthesis and biological activity of a novel antibacterial peptide deduced from a pig myeloid cDNA. FEBS Lett. 1994;337:303–307. doi: 10.1016/0014-5793(94)80214-9. [DOI] [PubMed] [Google Scholar]
- 37.Storici P, Zanetti M. A cDNA derived from pig bone marrow cells predicts a sequence identical to the intestinal antibacterial peptide PR-39. Biochem Biophys Res Commun. 1993;196:1058–1056. doi: 10.1006/bbrc.1993.2358. [DOI] [PubMed] [Google Scholar]
- 38.Tarver A, P, D, Clark P, Diamond G, Russell J P, Erdjument-Bromage H, Tempest P, Cohen K S, Jones D E, Sweeney R W, Wines M, Hwang S, Bevins C L. Enteric β-defensin: molecular cloning and characterization of a gene with inducible intestinal epithelial cell expression associated with Cryptosp.ridium parvum infection. Infect Immun. 1998;66:1045–1056. doi: 10.1128/iai.66.3.1045-1056.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wu H, Zhang G, Ross C R, Blecha F. Cathelicidin gene expression in porcine tissues: roles in ontogeny and tissue specificity. Infect Immun. 1999;67:439–442. doi: 10.1128/iai.67.1.439-442.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zanetti M, Gennaro R, Romeo D. Cathelicidins: a novel protein family with a common proregion and a variable C-terminal antimicrobial domain. FEBS Lett. 1995;374:1–5. doi: 10.1016/0014-5793(95)01050-o. [DOI] [PubMed] [Google Scholar]
- 41.Zanetti M, Litteri L, Gennaro R, Horstmann H, Romeo D. Bactenecins, defense polypeptides of bovine neutrophils, are generated from precursor molecules stored in the large granules. J Cell Biol. 1990;111:1363–1371. doi: 10.1083/jcb.111.4.1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang G, Hiraiwa H, Yasue H, Wu H, Ross C R, Troyer D, Blecha F. Cloning and characterization of the gene for a new epithelial beta-defensin. Genomic structure, chromosomal localization, and evidence for its constitutive expression. J Biol Chem. 1999;274:24031–24037. doi: 10.1074/jbc.274.34.24031. [DOI] [PubMed] [Google Scholar]
- 43.Zhang G, Ross C R, Blecha F. Porcine antimicrobial peptides: new prospects for ancient molecules of host defense. Vet Res. 2000;31:277–296. doi: 10.1051/vetres:2000121. [DOI] [PubMed] [Google Scholar]
- 44.Zhang G, Ross C R, Dritz S S, Nietfeld J D, Blecha F. Salmonella infection increases porcine antibacterial peptide concentration in serum. Clin Diagn Lab Immunol. 1997;4:774–777. doi: 10.1128/cdli.4.6.774-777.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhao C, Ganz T, Lehrer R I. The structure of porcine protegrin genes. FEBS Lett. 1995;368:197–202. doi: 10.1016/0014-5793(95)00633-k. [DOI] [PubMed] [Google Scholar]
- 46.Zhao C, Ganz T, Lehrer R I. Structure of genes for two cathelin-associated antimicrobial peptides: prophenin-2 and PR-39. FEBS Lett. 1995;376:130–134. doi: 10.1016/0014-5793(95)01237-3. [DOI] [PubMed] [Google Scholar]
- 47.Zhao C, Liu L, Lehrer R I. Identification of a new member of the protegrin family by cDNA cloning. FEBS Lett. 1994;346:285–288. doi: 10.1016/0014-5793(94)00493-5. [DOI] [PubMed] [Google Scholar]