Abstract
Li-Fraumeni syndrome (LFS) is an autosomal dominant cancer-predisposition disorder. Approximately 70% of individuals who fit the clinical definition of LFS harbor a pathogenic germline variant in the TP53 tumor suppressor gene. However, the remaining 30% of patients lack a TP53 variant and even among variant TP53 carriers, approximately 20% remain cancer-free. Understanding the variable cancer penetrance and phenotypic variability in LFS is critical to developing rational approaches to accurate, early tumor detection and risk-reduction strategies. We leveraged family-based whole-genome sequencing and DNA methylation to evaluate the germline genomes of a large, multi-institutional cohort of patients with LFS (n = 396) with variant (n = 374) or wildtype TP53 (n = 22). We identified alternative cancer-associated genetic aberrations in 8/14 wildtype TP53 carriers who developed cancer. Among variant TP53 carriers, 19/49 who developed cancer harbored a pathogenic variant in another cancer gene. Modifier variants in the WNT signaling pathway were associated with decreased cancer incidence. Furthermore, we leveraged the noncoding genome and methylome to identify inherited epimutations in genes including ASXL1, ETV6, and LEF1 that confer increased cancer risk. Using these epimutations, we built a machine learning model that can predict cancer risk in patients with LFS with an area under the receiver operator characteristic curve (AUROC) of 0.725 (0.633–0.810).
Significance:
Our study clarifies the genomic basis for the phenotypic variability in LFS and highlights the immense benefits of expanding genetic and epigenetic testing of patients with LFS beyond TP53. More broadly, it necessitates the dissociation of hereditary cancer syndromes as single gene disorders and emphasizes the importance of understanding these diseases in a holistic manner as opposed to through the lens of a single gene.
Introduction
Li-Fraumeni syndrome (LFS; OMIM #151623) is a highly penetrant cancer-predisposition disorder associated with pathogenic germline variants in the TP53 tumor suppressor gene (1). Families affected with LFS are susceptible to a diverse spectrum of neoplasms including, but not limited to, bone and soft-tissue sarcomas, premenopausal breast cancers, carcinomas of the adrenal cortex, leukemias, and various brain tumors (2, 3).
The classical LFS criteria define a proband as having developed a sarcoma before the age of 45 years, with at least one first-degree relative with any cancer before 45 years and an additional first- or second-degree relative with any cancer before 45 years, or a sarcoma at any age (3). The identification of pathogenic germline TP53 variants in individuals who did not fulfill the strict definition of LFS led to the revised “Chompret” criteria for germline TP53 testing, which accounts for a broader spectrum of clinical heterogeneity (4). A recent study by Ceyhan-Bersoy and colleagues reinforces the need to consider a molecular definition of the diverse spectrum of LFS phenotypes (5) to improve cancer risk prediction and define the role of other genes in LFS families. Up to 75% of families with classical LFS harbor germline pathogenic/likely pathogenic (P/LP) TP53 variants. Among P/LP TP53-variant carriers, the cumulative lifetime risk of developing cancer is approximately 68% in males and 93% in females (6). However, molecular events that modify cancer risk in TP53-variant carriers remain poorly understood (7–10). Furthermore, the identification of genetic alterations that explain the increased cancer risk in TP53-wildtype LFS families is critical in guiding the prospective management of these patients.
The incomplete penetrance, despite the presence of pathogenic variants of TP53 in LFS suggests: (i) the presence of additional genetic and/or epigenetic driver events that contribute to cancer risk in certain individuals; and (ii) resilience mechanisms that explain the absence of cancer in some TP53-variant carriers, possibly initiated through alternative variants in compensatory pathways (11). Candidate gene approaches have been used to consider other relevant genetic causes in LFS families (10, 12–20). However, whole-genome DNA sequencing and/or epigenetic analyses have, to date, only been utilized in isolated case reports to explain these gaps in understanding of cancer occurrence (21–24). In this work, we leveraged family-based whole-genome sequencing (WGS) and methylation of DNA procured from peripheral blood leukocytes (PBL) to evaluate the germline genetic and epigenetic landscapes of a large cohort of patients that fit the clinical definitions of LFS (n = 396) and who harbor either pathogenic variant (n = 374) or wildtype TP53 (n = 22). Overall, our study identifies diagnostic biomarkers, prognostic signatures, actionable therapeutic targets and correlates of clinical heterogeneity that will allow for improved clinical management of patients with LFS.
Materials and Methods
WGS Patient Cohort
We performed WGS (RRID:SCR_016385) on 84 LFS family members from 47 families: 22 with wildtype TP53 and 62 with variant TP53 (Fig. 1A). Pedigrees for these families were established using information available from clinical notes of probands (Supplementary Figs. S5 and S7). Inclusion criteria for wildtype and variant TP53 families were determined by resemblance to characteristic patterns of cancer phenotype well documented in literature to be associated with LFS (i) patterns of inheritance (LFS criteria; refs. 1, 4), (ii) cancer type spectrum (adrenal, brain, leukaemia, bone, soft tissue, breast; refs. 6, 25, 26), and (iii) early age of onset that occurs in bimodal cancer-type specific manner (6, 25, 26).
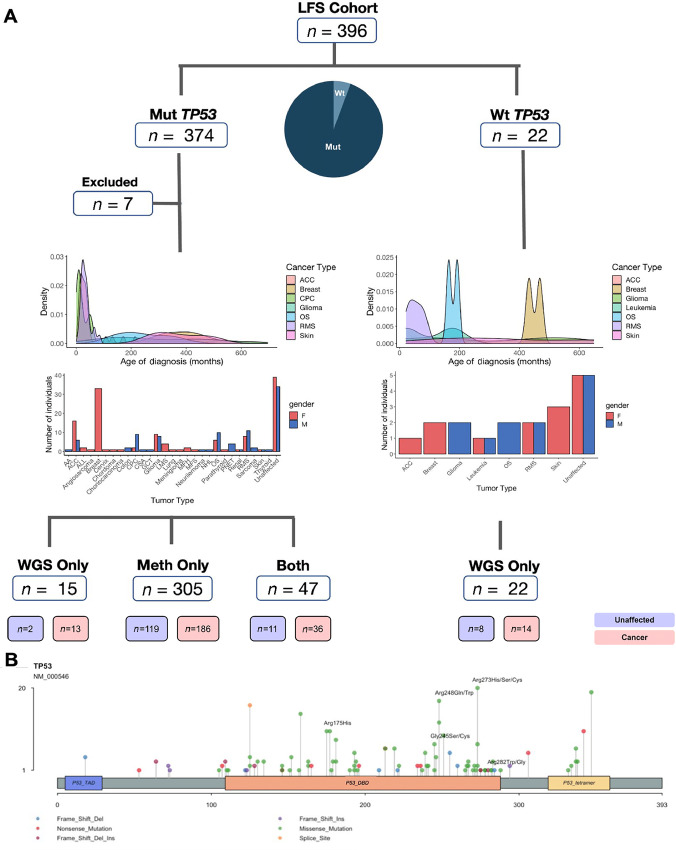
FIGURE 1.
The LFS cohort. A, Patient cohort and characteristics (age of first cancer onset, tumor type) by TP53 status and whether profiling was performed using WGS, methylation or both. AA = adrenal adenoma; ACC = adrenocortical carcinoma; ALL = acute lymphoid leukemia; CPC = choroid plexus carcinoma; CSA = chondrosarcoma; GCT = germ cell tumor; LMS = leiomyosarcoma; MFH = malignant fibrous histiocytoma; MFS = myxofibrosarcoma; NHL = non–Hodgkin lymphoma; OS = osteosarcoma; RMS = rhabdomyosarcoma. B, Lollipop plot displaying the location and number of SNVs and indels on the canonical TP53 transcript: NM_000546, which consists of a TAD, DBD, and tetramerization domain. TP53 hotspot variants present in our cohort (175, 245, 248, 273, 282) are highlighted.
Patterns of inheritance were determined through family history of cancer and characterization of LFS criteria (Supplementary Figs. S5 and S7). The variant TP53 cohort consists of 49 individuals who developed cancer and 13 individuals who remain cancer-free; 34 were from 13 families with 2–4 individuals sequenced within a given family and the remaining 28 had no family members sequenced. Among the variant TP53 cohort, two of the families fit the classical LFS criteria, 10 fit the Chompret criteria, and one family fit the incomplete LFS criteria. The wildtype cohort consists of 14 individuals who developed cancer and 8 individuals who are cancer-free, from six families (Fig. 1B). Four of the wildtype TP53 families fit the Chompret criteria while the remaining two fit the classical LFS criteria. Two to 6 individuals within each TP53 wildtype family had WGS performed. Moreover, patients in both the variant and wildtype TP53 cohort develop a similar spectrum of characteristic LFS-associated cancers (e.g., adrenal, brain, soft tissue, bone, blood) and age of cancer onset (Fig. 1A).
WGS and Processing
A total of 500 ng to 1 μg of genomic DNA was submitted to The Centre for Applied Genomics (TCAG) at The Hospital for Sick Children for genomic library preparation and WGS. DNA samples were quantified using the Qubit High Sensitivity Assay and purity was assessed using the Nanodrop OD 260/280 ratio. Approximately 500–700 ng of DNA was used as input material for library preparation using the Illumina TruSeq PCR-free DNA Library Prep Kit following the manufacturer's recommended protocol. In brief, DNA was fragmented to 400 bp on average using sonication on a Covaris LE220 instrument. Fragmented DNA was end-repaired, A-tailed, and indexed TruSeq Illumina adapters with overhang-T were added to the DNA. Libraries were then validated on a Bioanalyzer DNA High Sensitivity chip to check for size and absence of primer dimers and quantified by qPCR using Kapa Library Quantification Illumina/ABI Prism Kit protocol (KAPA Biosystems). Validated libraries were pooled in equimolar quantities and paired-end sequenced on an Illumina HiSeq X platform following Illumina's recommended protocol to generate paired-end reads of 150 bases in length and an average depth of 40X. FASTQ files were aligned to the hg19 reference genome using BWA-mem v0.78 (ref. 27; RRID:SCR_017619). PCR duplicates were marked with Picard MarkDuplicates v1.1.08 (ref. 28; RRID:SCR_006525), and base recalibration and realignment was performed using GATK v2.8.1 (RRID:SCR_001876; ref. 29).
Single-nucleotide Variant and Indels
Single-nucleotide variant (SNV) and indel detection was performed using GATK v.4.0.2.1 (RRID:SCR_001876; ref. 29) according to best practices for germline cohort data. All detected variants were further filtered to remove false positives using a set of filters designed for short read sequences as follows: read position 10–90, strandedness 1%–99%, distance to 3′ > 20, homopolymer <5, map quality difference <30, read length difference <25, and MMQS difference<100 (30). Variants with less than five alternate reads detected using bam-readcount were removed.
Identification of Pathogenic SNVs and Indels
Population filters were applied to remove common variants found in non-cancer “normal” populations: gnomAD, ExAC and 1kGP, at an allele frequency >0.01 (31). InterVar, an implementation of the ACMG-AMP guidelines (32), was used to classify variants into five categories: pathogenic (P), likely pathogenic (LP), variant of uncertain significance (VUS), likely benign (LB), and benign (B). In addition, all null variants (nonsense, frameshift, canonical ±2 splice sites, initiation codon, single or multiexon deletion) in a gene where loss of function is a known mechanism of disease, that met population filters, were annotated as LP. If the variant was found in all affected members of a family, it was coded as 1 for PP1 and as 0 for BS4. If the variant was not found in any affected members of a family, it was coded as 0 for PP1 and as 1 for BS4. Variants identified in singleton samples were coded as 0 for both PP1 and BS4 (33).
Structural Variants
Structural Variant Discovery
Germline structural variant (SV) discovery was performed using DELLY v0.7.7 (RRID:SCR_004473; ref. 34) and Manta v1.0.3 (35) to discover deletions, duplications, and inversions.
Filtering and Annotation of Deletions, Duplications, and Inversions
An ensemble approach was taken, retaining only the deletions, duplications, and inversions called by both DELLY and Manta with a maximum allowed distance of 1 kb between breakpoints. Manta relies on read-pair, split-read, and local-assembly support to call variants, while Delly uses read-pair and split-read support. The following size-specific reciprocal overlap constraints were used for breakpoint consensus between the two tools: breakpoints within 100 bp for SVs ≤10 kbp, breakpoints within 1 kbp for SVs ≥10 kbp and ≤50 kbp and breakpoints within 10 kbp for SVs ≥50 kbp. A high-quality SV set was obtained by applying additional filtering criteria. A panel of SVs occurring as a result of sequencing artefacts was created to remove technical false positives using 72 control germline genomes sequenced on the same HiSeqX platform (150 bp paired end sequencing, minimum of 30X depth coverage). SVs present in ≥3 samples in the panel of normals were removed. Annotation was performed with AnnotSV (36) and subsequently filtered to remove SVs with an AnnotSV score <3 and present in publically available non-cancer “normal” databases: 1000 Genomes Project (RRID:SCR_008801; ref. 37) and gnomAD (RRID:SCR_014964; ref. 38).
Copy-number Variation
Copy-number variation (CNV) was identified using read depth based CNV detection algorithms: CNVnator (RRID:SCR_010821; ref. 39) and ERDS (40). The intersection of the CNVnator and ERDS calls were subsequently filtered to remove repetitive and low-complexity regions using a predefined list formed by Trost and colleagues (41) This was followed by annotation using ANNOVAR (42).
Prioritizing Variants Using Tiered Cancer Gene Lists
Although a whole-genome approach was used to detect cancer-associated germline events, four gene lists were compiled to allow for the prioritization of our findings. From highest known cancer predisposition potential to lowest:
Tier 1: Autosomal dominant cancer predisposition genes (n = 60): 60 autosomal dominant CPG [from Zhang and colleagues (43); Supplementary Table S2].
Tier 2: Autosomal recessive cancer predisposition genes (n = 29): 29 autosomal recessive CPG [from Zhang and colleagues (43); Supplementary Table S2].
Tier 3: Cancer genes (n = 489): 58 tumor suppressors, 24 tyrosine kinases and 407 other cancer genes frequently mutated in the somatic context. This list was adapted from Zhang and colleagues (43) with the addition of nine genes that have been added since the publication in 2015, to the COSMIC Cancer Gene Census (Supplementary Table S2; ref. 44).
Tier 4: All genes (n = 21,380): This list encompasses all the other mapped genes in the genome, not present in the previous tiers.
Cancer Variant Classification Schema
The modern classification of variants for cancer makes it difficult to distinguish between pathogenic (P)/likely pathogenic (LP), and likely benign (LB)/benign (B) variants because (i) the relationship between a given gene and the cancer developed is not fully understood, and (ii) the functional consequence of a variant in a gene is difficult to comprehend without functional experimentation. As a result, we used a five-tier classification scheme for pathogenicity as follows:
Class 1: P/LP genetic variant in a known autosomal dominant, CPG (Tier 1).
Class 2: P/LP genetic variant in a known autosomal recessive, CPG (Tier 2).
Class 3: P/LP genetic variant in a known cancer gene frequently mutated in the somatic context (Tier 3).
Class 4: P/LP genetic variant in novel, candidate cancer gene (Tier 4) included if supported by sufficient evidence in the literature.
Class 5: Cancer-segregating VUS in a known cancer gene (Tier 1–3)
Analysis of WGS Data from 1000 Genomes Project (1kGP)
Genotype data for 2,504 individuals, generated by the 1kGP project in Variant Call Format from version 5 data release were downloaded by ftp (ftp://ftptrace.ncbi.nih.gov/1000genomes/ftp/release/20130502/supporting/vcf_with_sample_level_annotation/; ref. 37). All SNVs, indels, and structural variants were subject to pathogenicity classification as outlined above.
Analysis of WGS Data from SickKids Cancer Sequencing Program
The SickKids Cancer Sequencing (KiCS) Program is a prospective study of a demographically diverse population of children, adolescents and young adults with refractory, metastatic, relapsed or rare cancers, as well as children with unresolved suspicion for cancer predisposition (45). Whole-genome data from 185 KiCS patients were utilized as a “control” cohort of individuals who developed cancer who lack a germline TP53 variant. We excluded all patients with a previous diagnosis of a cancer predisposition disorder, more specifically individuals diagnosed with hereditary paraganglioma syndrome and Lynch syndrome. All KiCS FASTQ files were processed through the same pipeline as the LFS FASTQ files. SNVs, indels, and structural variants were also identified and subject to pathogenicity classification using the same methodology as the variants identified in the LFS cohort, as described above.
Pathway Analysis
The hallmark gene sets from the MSigDB collections were utilized to perform a pathway analysis. Fisher exact test was implemented for each of the 50 pathways, followed by FDR correction (46).
Survival Analysis Based on P/LP Variants in Cancer Genes
Survival data for 49 patients with WGS were right censored at death or last follow-up (Supplementary Table S1A). Each patient was assigned to a group based on having: (i) at least one class 1–3 variant (n = 15), (ii) P/LP variant in the WNT signaling pathway (n = 4), and (iii) no class 1–3 or P/LP WNT signaling variant (n = 31). Each patient fit the criteria for a single group, with the exception of one patient (Supplementary Fig. S6L – Patient 1355), that harbored a P/LP variant in AXIN1 (ii) and XRCC1 (i). Kaplan–Meier survival curves were created using survminer (v0.4.9) and survival (v3.3–1) with P values calculated using a log-rank test.
Methylation Sample Cohort
We profiled DNA methylation of PBLs procured from patients with LFS with a germline TP53 variant (n = 359) from 287 families. A subset of the patients in this cohort overlapped with the WGS cohort (n = 47). The number of individuals profiled for methylation per family ranged from 1 to 7 with a median of 1. Our discovery cohort (n = 89) consisted of samples collected from previous work (9) composed of individuals with cancer (n = 63) and individuals who did not have cancer at the time of analysis (n = 26). Methylation analysis for these samples was performed using the Illumina HumanMethylation 450k BeadChip array. Our internal validation cohort (n = 124) consisted primarily of patients with a pathogenic germline TP53 variant from the Hospital for Sick Children who developed cancer (n = 78) and those that were cancer-free (n = 46). Our external validation cohort (n = 146) consisted exclusively of individuals with pathogenic germline TP53 variants from the NCI who developed cancer (n = 88) and those that were unaffected (n = 58) (47). Both validation cohorts were profiled using the Illumina EPIC (850k) array. By limiting the samples processed on different array types exclusively to one cohort, we mitigated any biases that may have arisen due to array type.
Preprocessing and Addressing Confounders in the Methylation Data
Preprocessing and bias correction were performed on the discovery and each validation cohort separately. Raw beta values were corrected for dye-bias using ssNoob (Supplementary Fig. S11; ref. 48). The normalized beta values were then corrected for the batch effects between 96-well plates using ComBat (49). We used Probabilistic Estimation of Expression Residuals (PEER; RRID:SCR_009326)—a factor analysis method that infers hidden determinants and their effects on molecular profiles—to remove broad variance from known confounders (array type, batch, systemic treatment at draw, age of sample collection, gender) as well as 100 hidden (latent) factors (50). Following PCA transformation of each cohort (Supplementary Fig. S12), samples within 3 SDs of the mean of PC1 (μPC1 ± 3σ) and PC2 (μPC2 ± 3σ) were retained. This resulted in the removal of 1, 2, and 4 outliers from the discovery, internal validation, and external validation cohort, respectively, and a modest decrease to the size of our discovery cohort (n = 88), internal validation cohort (n = 122), and external validation cohort (n = 142).
Discovery of Cancer-associated Secondary Constitutional Epimutation in LFS Patients with a TP53 Variant
Using the preprocessed methylation data, an Epigenome-Wide Association Study (EWAS) was performed by testing each of the 452,497 methylation probes for their association with cancer status as follows:
EWAS of the discovery cohort using the Aziz test (ref. 51; methylation ∼ cancer status). P values were adjusted using FDR and probes with FDRdiscovery < 0.1 were considered significant.
EWAS of the internal and external validation cohorts using the Aziz Test (methylation ∼ cancer status). P values were adjusted using FDR and probes with FDRvalidation < 0.1 in both validation cohorts were considered significant.
The methylation probes found in the discovery cohort with FDRdiscovery <0.1 that were validated in both the internal and external validation cohorts with FDRvalidation < 0.1 resulted in 931 significant EWAS probes.
The patients with both WGS and methylation (n = 47) were used to identify cancer-associated secondary constitutional epimutations (CSCE). The premise behind this analysis is that a CSCE at a given probe will reveal underlying genetic cancer predisposition facilitated by epigenetic regulation (52, 53). We focused our analysis on cis-CSCE, as we were underpowered for the discovery of trans-CSCE due the size of our cohort. A cis-CSCE was defined as a EWAS probe significantly associated with a SNP within 10 kb upstream or downstream (cis-SNP). This association between a significant EWAS probe and a cis-SNP was determined using Spearman correlation. Each SNP was coded additively as 1 (homozygous reference), 3 (heterozygous), or 4 (homozygous alternative). For each EWAS probe, the strongest cis-CSCE was retained to represent that probe. Subsequently, FDR correction was performed on all the cis-CSCE. Using a FDR cutoff of 0.05 resulted in the discovery of 259 probes with cancer-associated cis-CSCEs. We tested all 259 cis-CSCE for their association with cancer type using an ANOVA.
Epimutation Clustering and Survival Analysis
We used Uniform Manifold Approximation and Projection (UMAP) for dimensionality reduction of the 259 cis-CSCE with the following parameters: n_neighbors = 15, n_components = 2, metric = euclidean. K-means clustering was used to cluster the UMAP projection; this revealed three distinct clusters of TP53 variant carriers. Available survival data (n = 328) were right censored at death or last follow-up (Supplementary Table S1A). Survival differences were evaluated between the three clusters using Kaplan–Meier survival curve with survminer (v0.4.9) and survival (v3.3-1).
LFS Cancer Risk Scores
We used methylation at the 259 cis-CSCE loci to build a random forest model to predict cancer risk in LFS (caret v6.0.91). To avoid overfitting, the external validation cohort (n = 142) was used as the training data, the internal validation cohort (n = 122) as the validation data and the discovery cohort (n = 88) as the test data. We trained a random forest with repeated 5-fold cross-validation, tuning the number of features considered at each split point (mtry) using grid search. The prediction probabilities from our model can be interpreted as an LFS cancer risk score—an indicator of elevated cancer risk in TP53 variant carriers. We then used our cancer risk model to predict risk scores on our validation and test sets. The optimal specificity threshold was determined as 0.667 in the discovery cohort using Youden index.
EWAS Exploring the Effect of Systemic Treatment on Methylation
Systemic treatment information is available for the majority of our internal and external validation cohorts (n = 254), consisting of 59 individuals that received systemic treatment prior to sample collection and 195 that received systemic treatment after sample collection or not at all. A total of 40% (78/195) of the individuals that did not receive systemic treatment developed cancer and 60% (117/195) remain cancer-free (Supplementary Table S1A). In addition to correcting for broad variation due to systemic treatment using PEER, we performed an EWAS to identify methylation probes significantly associated with systemic treatment. The Aziz test was used to test the association between each methylation probe and systemic treatment status in the internal and external validation cohorts. P values were adjusted using FDR and probes with FDRvalidation < 0.1 in both validation cohorts were considered significant.
Feature Enrichment Analysis of Cancer-associated cis-CSCE
We performed a feature enrichment analysis of the 259 significant cis-CSCE using 10 genomic properties, which included proximity to histone marks, open chromatin and expression levels from blood-derived samples. ChromImpute P-value signal tracks (bigwig files) were downloaded from the Roadmap Epigenomics Consortium (https://egg2.wustl.edu/roadmap/data/byFileType/signal/consolidatedImputed/) for the following genomic properties: H3K4me1, H3K4me3, H3K27me3, H3K9me3, H3K27ac, H3K36me3, DNase, H2A.Z, H3K79me2, and RNA-sequencing, in 23 blood-derived samples (E062, E034, E045, E033, E044, E043, E039, E041, E042, E040, E037, E048, E038, E047, E029, E031, E035, E051, E050, E036, E032, E046, E030). A total of 100,000 methylation probes were sampled with replacement to calculate the baseline enrichment level. Average signal values were calculated at the genomic loci of the 100,000 probes for all features in each sample. Average signal values were calculated at the genomic loci of the 259 significant cis-CSCE probes for all features in each sample. A paired Wilcoxon test was used to calculate a P value between the signal value of the null probes and the cis-CSCE probes for each feature. Similarly, effect size was calculated using Cohen distance (d).
Methylation of LEF1 in Plasma cell-free DNA of Patients with LFS Compared with Healthy Blood Controls
A total of 173 individuals with a germline TP53 variant and 28 healthy blood controls (HBC) with wildtype TP53 were profiled using cell-free methylated DNA immunoprecipitation (cfMeDIP-seq). cfMeDIP-seq samples were processed, aligned, and quantified into nonoverlapping 300 bp bins, as described in detail in another study by our group (54). Using the 300 bp bin at position chr4:109056901-109057200, which overlapped the probe cg03041109, we compared LEF1 methylation between HBC and patients with LFS.
Methylation of LEF1 in Choroid Plexus Tumors
Genome-wide DNAm profiles were generated using the Illumina HumanMethylation450 BeadChip array for 34 primary samples of choroid plexus tumor (CPT), consisting of 16 choroid plexus carcinomas (CPC), eight with wildtype TP53 and eight with a germline TP53 variant, and 20 choroid plexus papillomas, 19 with wildtype TP53 and one with variant TP53. All samples were processed as described in detail in our previous work (55, 56). We compared methylation of LEF1 (cg03041109) from individuals that developed CPTs, with and without a germline TP53 variant.
Methylation of LEF1 in Wilms Tumors
DNA methylation was collected and processed for 95 individuals from the United Kingdom and 81 from Toronto, Canada, as previously described by our collaborators (57). The UK cohort consisted of 36 normal kidney samples, 22 nephrogenic rest (NR), 32 Wilms tumor (WT) with favorable histology, and five with anaplasia (58). The Toronto cohort consisted of 18 blood (control), 25 kidney samples, 30 WT with favorable histology, and eight with anaplasia. To avoid batch effects, methylation of LEF1 (cg03041109) was compared between groups, within each cohort.
Consent
This study was conducted in accordance with the ethical principles of the Declaration of Helsinki and approved by an Institutional Review Board. All LFS (#1000051699) and KiCS (#0019910602) patients were approved for molecular profiling by the SickKids Research Ethics Board. The NCI LFS cohort study was approved by the NCI Institutional Review Board (ClinicalTrials.gov identifier NCT01443468). Written informed consent was signed by all participants or their legal guardian prior to sample collection.
Data Availability
All WGS and methylation data files are available in the European Genome-Phenome Archive (EGA) under the accession numbers EGAS00001007075 and EGAS00001007061. Analysis scripts can be found at doi:10.5281/zenodo.5563379.
Results
Patient Cohort Characteristics
Our international, multi-institutional (n = 30) LFS cohort consisted of 396 individuals, 374 with a pathogenic TP53 variant from 298 families (Supplementary Table S1A), and 22 with wildtype TP53 from six families (Fig. 1A; Supplementary Table S1B). Overall, 18 tumor types are observed among the patients with ages of onset ranging from 0–70.4 years and a mean age at diagnosis of 22.5 years [95% confidence interval: (20.4–24.5); Fig. 1A]. A total of 43% of the family members with cancer developed a tumor by the age of 18. The most prevalent tumor types were breast cancer (n = 69), central nervous system tumors (n = 40), adrenocortical carcinoma (ACC, n = 28), rhabdomyosarcoma (RMS, n = 28), and osteosarcoma (n = 24). In addition, we profiled 140 cancer-free family members of probands with clinical follow-up information, 8 from wildtype TP53 families and 132 from variant TP53 families (Fig. 1A). We performed WGS and/or methylation of DNA from PBL. In total, WGS was performed on 84 patients with wildtype (n = 22) and pathogenic TP53 variants (n = 62), and methylation of PBL was performed on 359 patients with LFS with a pathogenic TP53 variant. Given limited sample availability, only a subset (n = 47) of the LFS cohort had both WGS and methylation profiling.
Germline TP53 Variants
Pathogenic/likely pathogenic (P/LP) germline TP53 variants in the LFS cohort consisted of SNVs (n = 316), deletions (n = 41), insertions (n = 7), and insertion-deletions (n = 11; Fig. 1B). Of the 250 missense variants, 86.4% (n = 216) interrupted the DNA-binding domain (DBD), and the remaining 13.6% (n = 34) affected the tetramerization domain (TAD; Fig. 1B). Five hotspot residues made up 20.8% of all the variants, consisting of R175 (n = 10), R245 (n = 8), R248 (n = 29), R273 (n = 29), and R282 (n = 2). The majority of the TP53 missense variants were cytosine-to-thymine (C>T) transitions (n = 126). One patient diagnosed with an anaplastic embryonal rhabdomyosarcoma (ERMS) at age 5 years, harbored a compound heterozygous genotype with two pathogenic TP53 variants in trans: the hotspot R248Q variant (MAF = 0.29) and a pathogenic splicing variant in intron 5 (MAF = 0.22). Another notable patient, harbored an extremely rare, homozygous truncating TP53 variant and developed two aggressive synchronous primary malignancies (59).
Pathogenic Germline Genetic Variants in LFS
WGS of 84 individuals, 62 with variant TP53 and 22 with wildtype TP53, revealed 595,975,507 SNVs/indels and 19,241 SVs. On average, the LFS genomes had 7,261,182 variants per individual (Supplementary Table S1C). All genetic variants were classified into five classes using the Cancer Variant Classification Schema as described in the Materials and Methods (Fig. 2A; Supplementary Table S2). Two control cohorts with wildtype TP53 were used for comparison: (i) whole-exome data from the 1000 Genomes Project (1kGP) consisting of cancer-free individuals (n = 2,504) and (ii) whole-genome data from the KiCS Project consisting of individuals with no known cancer predisposition disorder, who developed cancer (n = 185). For the subsequent analyses, TP53 variants were omitted from the LFS cohort as they present an inherent bias to any statistical test comparing variants between cohorts. We compared the frequency of class 1–3 variants (P/LP variants in known cancer genes) between patients with LFS cancer, KiCS (n = 185) and 1kGP (n = 2,504). The prevalence of class 1–3 variants was 9.42% in 1kGP (Supplementary Table S3) and 19.05% in KiCS (Supplementary Table S4), which was significantly lower than the 39.3% and 57.14% prevalence observed in the cancer-affected TP53-variant (Fisher exact test, 1kGP: P = 4.98 × 10−9; KiCS: P = 8.90 × 10−4) and TP53-wildtype (Fisher exact test, 1kGP: P = 1.29 × 10−5; KiCS: P = 1.56 × 10−3) groups (Supplementary Table S5), respectively (Fig. 2B). This suggests that an enrichment of P/LP variants in additional cancer genes among patients with LFS may be contributing to their high rate of malignant transformation. However, the presence of a P/LP variant in a cancer gene (class 1–3) does not appear to contribute to the earlier age of cancer onset that is characteristic of patients with LFS (Supplementary Fig. S1). Furthermore, patients with LFS with at least one class 1–3 variant displayed lower overall survival than individuals lacking a class 1–3 variant (Fig. 2C).
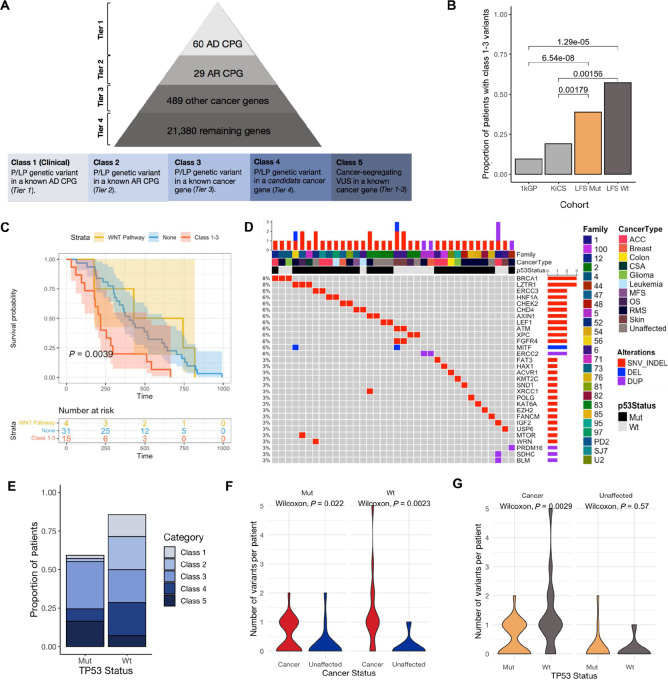
FIGURE 2.
Germline genetic variants in LFS. A, Classification scheme for genetic variants identified from WGS. The pyramid (top) shows how genes were prioritized into four tiers from highest known cancer predisposition potential to lowest. All variants were then classified using a Cancer Variant Classification Schema (Materials and Methods) into five classes (bottom) based on pathogenicity and whether the gene fell into Tier 1–4. AD: autosomal dominant; AR: autosomal recessive; CPG: cancer predisposition gene. B, Frequency of P/LP variants in known cancer genes (class 1–3) in LFS, KiCS and 1kGP. Fisher exact test P values for comparisons between the LFS cohorts and KiCS/1kGP shown. C, Kaplan–Meier survival curve comparing patients with P/LP variants in the WNT signaling pathway (yellow), patients with at least one class 1–3 variant (red) and patients with neither WNT signaling or class 1–3 variants (blue). D, Landscape of genetic alterations in cancer genes (class 1–3) in LFS. Annotated with family, cancer type and TP53 variant status. Genetic variants are indicated as a deletion (blue), SNV or indel (red) or duplication (purple). E, The proportion of individuals in the wildtype and variant TP53 cohorts with classes 1–5 variants (Materials and Methods). F, The number of P/LP variants in cancer genes (class 1–3) in individuals that developed cancer compared with unaffected individuals, stratified by TP53 status. G, The number of P/LP variants (class 1–3) in wildtype versus variant TP53 carriers, stratified by cancer status. On the basis of the premise that variants in the WNT signaling pathway are associated with decreased cancer incidence in LFS, these variants were removed from the comparisons made in F and G.
BRCA1 was the only autosomal dominant CPG (class 1; Table 1) mutated in our cohort. Eight autosomal recessive CPG (class 2; Table 1) and 12 well-characterized cancer genes (class 3) were found to be mutated in the LFS cohort (Fig. 2D). Recurrent P/LP variants were identified in individuals diagnosed with cancer from distinct families, in the following cancer genes: BRCA1 (n = 3), LZTR1 (n = 3), HNF1A (n = 2), CHEK2 (n = 2), MITF (n = 2), and CHD4 (n = 2; Fig. 2D). In addition, 10% (8/84) of patients harbored therapeutically targetable variants, as previously reported in the non-LFS germline context (60), like PARP inhibitors, mTOR inhibitors, EZH2 inhibitors, or tyrosine kinase inhibitors (Supplementary Fig. S2). Pathway analysis of the P/LP SNVs, indels, and SVs across all genes revealed that the p53 pathway (freq = 13%; FDR = 0.19) and the epithelial-to-mesenchymal transition pathway (freq = 13%; FDR = 0.10) were the most frequently mutated pathways across patients with LFS with cancer (Supplementary Fig. S3).
TABLE 1.
Variants in cancer predisposition genes (class 1–2) in the wildtype and variant TP53 cohorts, annotated with cancer type, sex, LFS criteria, inheritance status, segregation, variant location, variant function, and zygosity
| Patient ID | Family ID | TP53 status | Cancer type (age at diagnosis, months) | Sex | Gene | Class | LFS criteria | Inheritance status | Segregation | Variant | Variant function | Zygosity |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 3298A | MUT12 | Mut | Adrenocortical carcinoma (21) | F | BRCA1 | 1 | Unknown | Unknown | Unknown | NM_007294:exon2: c.68_69del:p.E23fs | frameshift deletion | Het |
| 2766 | WT4 | Wt | Embryonal rhabdomyosarcoma (24) | M | BRCA1 | 1 | Classic | Inherited | Cancer segregating | NM_007294:exon10:c. 5266dup:p.Gln1756fs | frameshift deletion | Het |
| 2768 | WT4 | Wt | Low grade glioma (516) | M | BRCA1 | 1 | Classic | Unknown | Cancer segregating | NM_007294:exon10:c. 5266dup:p.Gln1756fs | nonsynonymous SNV | Het |
| 1478 | MUT52 | Mut | Unaffected | M | ERCC3 | 2 | Chompret | Unknown | Unaffected segregating | NM_000122:exon3:c. 325C>T:p.Arg109Ter | nonsynonymous SNV | Het |
| 1478 | MUT52 | Mut | Unaffected | M | WRN | 2 | Chompret | Unknown | Unaffected segregating | NM_000553:exon24:c. 2856_2857del:p.S952fs | nonsynonymous SNV | Het |
| 3273 | MUT76 | Mut | Unaffected | M | FANCM | 2 | Classic | Not inherited from Mom | Unaffected segregating | NM_020937:exon11:c. 5791C>T:p.Arg1931Ter | stopgain | Het |
| SJACT007 | MUTSJ7 | Mut | Adrenocortical carcinoma | M | ERCC3 | 2 | Unknown | Unknown | Unknown | NM_000122:exon8: c.1120_1121insAGCAGT: p.W374delinsX | frameshift deletion | Het |
| 1970 | WT2 | Wt | Osteosarcoma (164) | M | XPC | 2 | Birch | Inherited | Shared | NM_004628:exon1: c.C55T:p.Q19X | stopgain | Het |
| 1972 | WT2 | Wt | Unaffected | F | XPC | 2 | Birch | Unknown | Shared | NM_004628:exon1: c.C55T:p.Q19X | stopgain | Het |
| 3021 | WT6 | Wt | Basal cell carcinoma (648) | F | ATM | 2 | Classic | Unknown | Cancer segregating | NM_000051:exon49: c.T7271G:p.V2424G | splicing | Het |
| 4093 | WT6 | Wt | Anorectal melanoma (319) | F | ATM | 2 | Classic | Inherited | Cancer segregating | NM_000051:exon49: c.T7271G:p.V2424G | stopgain | Het |
Cancer-associated Germline Genetic Modifiers in LFS Patients with Variant TP53
We observed that patients with LFS with a germline TP53 P/LP variant harbor an additional germline P/LP variant in another cancer gene that may prime them for cancer development (61). Among the patients with variant TP53 LFS who developed cancer (n = 49), 38.8% (19/49) of the patients had a second germline variant in a known cancer predisposition or cancer-associated gene (class 1–3; Fig. 2E; Supplementary Table S5A–S5D). Upon the inclusion of P/LP variants in novel, candidate cancer genes (class 4), we identified a secondary germline variant in 8.2% (4/49) of the patients who developed tumors (Fig. 2E; Supplementary Table S5E and S5F). These additional germline hits were scarce in the cancer-free individuals (Fig. 2F), with the exception of variants in the WNT signaling pathway, which we later discuss as being associated with decreased cancer incidence. With the exclusion of variants in the WNT signaling pathway, variant TP53 carriers that developed cancer were significantly enriched for class 1–3 variants (Wilcoxon rank-sum test, P = 0.022) and class 4 (Wilcoxon rank-sum test, P = 0.022) variants in comparison with the cancer-free variant TP53 carriers (Supplementary Fig. S4). For a subset of the variant TP53 carriers (n = 34), samples from cancer-free family members were available for WGS (Supplementary Fig. S5). For these patients, we performed family-based sequencing analysis, which allowed the identification of disease-segregating variants. Including disease-segregating VUS in known cancer genes (class 5), 12.2% (6/49) of the patients with cancer were found to harbor a secondary germline hit (Supplementary Table S5G).
Molecular Differences Between LFS Patients with Variant TP53 Compared to Those Harboring Wildtype TP53
Among the patients with LFS who developed cancer (n = 63), the wildtype TP53 patients (n = 14) had a greater burden of P/LP variants in CPG (class 1–2; Wilcoxon rank-sum test, P = 3.9 × 10−4; Supplementary Fig. S4) and cancer genes (class 1–3; Wilcoxon rank-sum test, P = 2.9 × 10−3) compared with the variant TP53 cohort (n = 49; Fig. 2G). This suggests that in the absence of a strong driver like a pathogenic TP53 variant, patients with LFS with wildtype TP53 require a greater burden of P/LP variants in other cancer-associated genes to be primed for cancer development. Compared with the variant TP53 patients who developed cancer, the wildtype TP53 patients who developed cancer were enriched for P/LP variants in genes found to be upregulated by reactive oxygen species (ROS; freq = 9%; FDR = 0.11; Supplementary Fig. S6) and genes important for mitotic spindle assembly (freq = 14%; FDR = 0.11; Supplementary Fig. S6). Although no one pathway was significantly enriched in the variant TP53 cohort over the wildtype cohort, KRAS signaling downregulation approached significance (freq = 15%; FDR = 0.09; Supplementary Fig. S6) as the most frequently mutated pathway in the variant TP53 cohort, with no P/LP variants identified in the wildtype cohort.
Identification of Alternative Susceptibility Genes in LFS Patients with Wildtype TP53
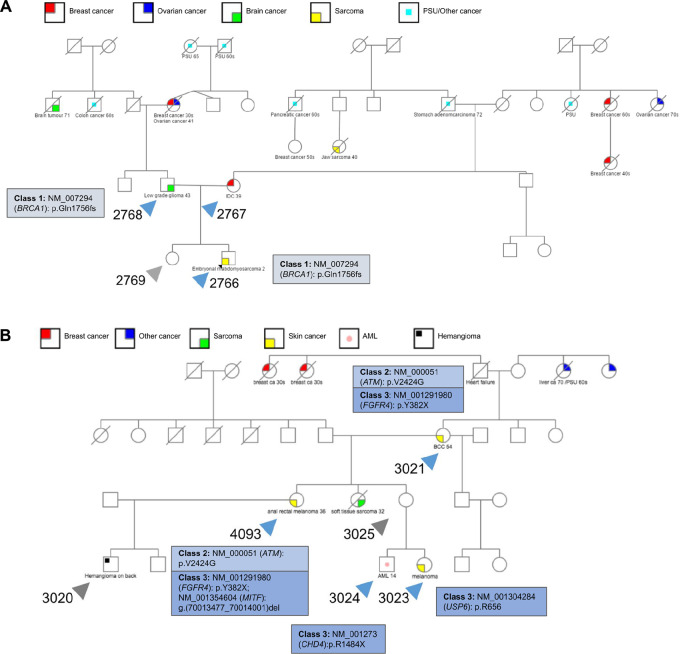
Among the patients with LFS with wildtype TP53 who developed cancer (n = 14), we sought a genetic explanation for their cancer in the absence of a pathogenic variant TP53 allele. A total of 14% (2/14) of the wildtype TP53 patients harbored a P/LP variant in an autosomal dominant CPG (class 1), both of which are a p.Gln1756fs frameshift variant in BRCA1 present within a single LFS family (Supplementary Table S5A). The BRCA1 variant was initially present in the father who developed a low-grade glioma (LGG) and inherited by his son who developed an ERMS (Fig. 3A). A total of 21% (3/14) harbored a P/LP variant in an autosomal recessive CPG (class 2; Supplementary Table S5B). One such example is an ATM p.V242G variant that was passed down from a mother in one family (Fig. 3B) with basal cell carcinoma to her daughter with anorectal melanoma. A total of 21% (3/14) of the patients with wildtype LFS with cancer had a P/LP variant in a known cancer gene (class 3; Supplementary Table S5C and S5D). There are five such variants in the WT6 family, a premature stop codon in FGFR4 (p.Y382X), CHD4 (p.R148X), and USP6 (p.R656X) and a deletion in MITF. Interestingly, the patient who harbored a deletion in MITF—a gene that is well characterized in the context of familial melanoma as having an essential role in melanin synthesis in melanocytes—also developed an extremely rare and aggressive form of melanoma known as anorectal melanoma (Fig. 3B; refs. 62, 63).
FIGURE 3.
Pedigrees and class 1–3 variants for two wildtype TP53 families that fit the classic LFS criteria. A, Pedigree of wildtype TP53 family (WT4) that fits the classic LFS criteria with four family members sequenced, two that developed cancer and two that are cancer-free. B, Pedigree of wildtype TP53 family (WT6) that fits the classic LFS criteria with six family members sequenced: five that developed cancer and one that is cancer-free.
Among the identified class 1–3 variants, all but one—a pathogenic variant in XPC—segregated with individuals that developed cancer in a given wildtype TP53 family (Supplementary Fig. S7B – Patients 1970 and 1972). We also noticed a higher burden of P/LP variants across known cancer predisposition (class 1–2; P = 3.9 × 10−4) and cancer-associated genes (class 1–3) was associated with cancer status in wildtype TP53 carriers (P = 2.3 × 10−3; Supplementary Fig. S4). P/LP variants in novel, candidate cancer genes (class 4) including SF3B4 (Fig. 3B – Patient 4093), FMN2 (Fig. 3B – Patient 4093), and MMP13 (Supplementary Fig. S7C – Patient 1774) are found in 21% (3/14) of patients with cancer (Supplementary Table S5E and S5F). Although the clinical significance of these variants is difficult to ascertain based on current knowledge, repeated documented cases are key in identifying novel cancer drivers and redundancies in critical signaling pathways. Finally, disease-segregating VUS in cancer genes (class 5) provide insights on an additional 7% (1/14) of the TP53 wildtype patients (Supplementary Table S5G), in particular a patient who developed an invasive ductal carcinoma with VUSs in PRF1 and BRD4 (Supplementary Fig. S7D – Patient 2767).
Leveraging Epimutations to Personalize Cancer Surveillance in TP53 Variant Carriers
To identify modifiers of cancer incidence among patients with LFS with variant TP53 in the noncoding genome, we evaluated epimutations. Using methylation data from DNA extracted from PBL of patients with LFS (n = 359), we identified 931 probes significantly associated with cancer incidence in our discovery cohort and validated them in an internal and external cohort (Fig. 4A); 4.2% (39/931) of the probes lie in cancer genes (Supplementary Table S6). A total of 27.8% (259/931) of the probes were cis methylation quantitative trait loci (meQTL; FDR < 0.05; Fig. 4B). These 259 loci represent cis cancer-associated secondary constitutional epimutations (cis-CSCE)—inherited SNPs in cis with methylation marks associated with cancer (52, 53).
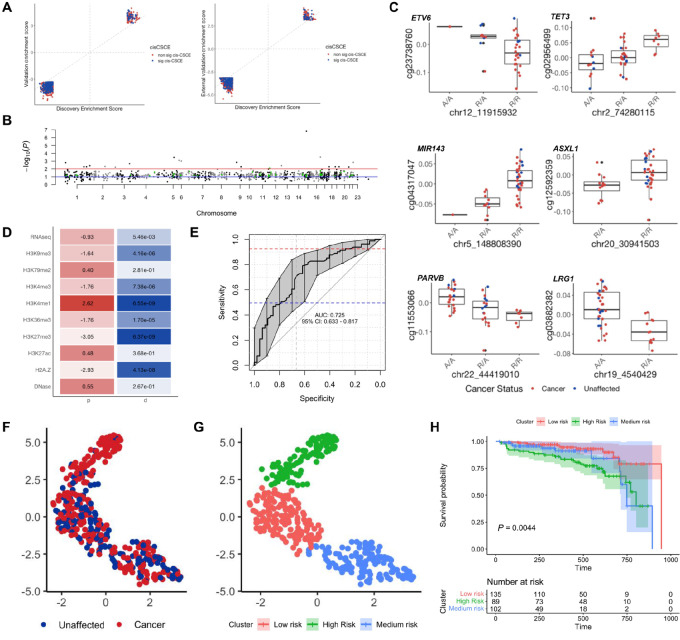
FIGURE 4.
Secondary constitutional epimutations in LFS. A, Concordance between enrichment scores of 931 EWAS probes found significantly associated with cancer status in the discovery cohort and the validation cohort, for both the internal (left) and external (right) validation cohort. Blue = associated with cancer status and a cis-CSCE; Red = associated with cancer status but not a cis-CSCE. B, Manhattan plot of adjusted P values from meQTL association tests between the 931 EWAS probes and their most correlated rsSNP. Red line: FDR = 0.1; Blue line: FDR = 0.01; Green points: probes in cancer genes with FDR < 0.1. C, Top cis-CSCE in ETV6, TET3, MIR143, ASXL1, PARVB, and LRG1.D, Feature enrichment heatmap of the 259 significant cis-CSCE loci showing adjusted P value (p) and effect size (d). E, ROC curve of the epimutation cancer risk model on the test set. F, UMAP projection of methylation at 259 cis-CSCE for all patients, colored by cancer status. G, UMAP projection of methylation at 259 cis-CSCE for all patients, colored by clusters associated with risk. H, Kaplan–Meier survival curve demonstrating significant differences in survival probability between the three epimutation clusters.
Similar to the pathway analysis performed on the genetic variants, pathway analysis of the genes that the 259 significant cis-CSCE reside in identified the p53 pathway as being the most significantly enriched pathway (FDR = 0.04). Eight of the cis-CSCE lie in cancer genes on our Tier 3 list, including ASXL1 (qCSCE = 1.04 × 10−2;rhoCSCE = −0.371) and ETV6 (qCSCE = 2.41 × 10−2;rhoCSCE = 0.386;Fig. 4C). A total of 50.6% (131/259) were associated with increased methylation with more copies of the alternate allele. 86.5% (224/259) of cis-CSCE were associated with an increased cancer risk upon hypomethylation. The remaining 13.5% (35/259) of cis-CSCE were positively associated with cancer risk. The majority of the cis-CSCE lie within the open sea in intergenic regions or within the body of a gene (Supplementary Fig. S8). Feature enrichment analysis at the genomic loci of the significant cis-CSCE showed enrichment of H3K4me1 marks, which are associated with the presence of enhancers (Cohen d = 2.62, P = 6.55 × 10−9; Fig. 4D).
We then leveraged the 259 cis-CSCE to train a random forest model to predict cancer risk in patients with LFS. Our model achieved an AUROC of 0.725 (0.633–0.810) and area under the precision recall curve (AUPRC) of 0.783 (0.775–0.792) on the test set (Fig. 4E). The optimal specificity of our model was determined as 42.0% (Materials and Methods), and corresponds to a sensitivity and positive predictive value of 85.1% and 46.0%, respectively, on the test set. Interestingly, our model was able to correctly classify an additional 22.4% (11/49) of the germline TP53 carriers that developed cancer for whom we were unable to identify a class 1–3 variant. However, it also misclassified 13 patients with a class 1–3 variant as cancer-free, suggesting that in the presence of a strong driver, epimutations may have a dampened role in contributing to elevated cancer risk. K-means clustering of the UMAP projection of the 259 cis-CSCE revealed three distinct clusters, whereby cluster membership was significantly associated with cancer status (Fig. 4F; χ2 test, P = 4.4 × 10−7) and survival differences (Fig. 4G and H; P = 0.0044). There was no clear relationship between the clusters and age of cancer onset or tissue type (Supplementary Fig. S9).
LEF1 and the WNT Signaling Pathway as Modifiers Associated with Decreased Cancer Risk in TP53 Variant Carriers
The pathway enriched for genetic variants in the cancer-free LFS family members was the WNT β-catenin signaling pathway (FDR = 0.04; Supplementary Fig. S3). It has been previously recognized that the loss of function of p53 results in the upregulation of downstream WNT target genes (TCF/LEF target genes, CD44, MMP7, MYCN), suggesting that p53 represses canonical WNT signaling (64). Among the patients with LFS with variant TP53, the WNT β-catenin signaling pathway was significantly mutated in the germline of 23% (3/13) of individuals who were cancer-free compared with 2% (1/49) of patients that developed cancer (Wilcoxon rank-sum test, FDR = 0.08). Likewise, individuals with a modifier in the WNT signaling pathway had better overall survival than those without a WNT signaling modifier (Fig. 2C). In one family (Supplementary Fig. S5B), the two unaffected family members (Patients 2815 and 2565) harbored a pathogenic variant in LEF1 that was absent in their cancer-affected relatives, suggesting its potential protective role. LEF1 is a transcription factor that activates oncogenes such as cyclin D1 and MYC to mediate cellular transformation and it has been shown that in the presence of a pathogenic variant in TP53, there are elevated levels of LEF1 that drive aberrant WNT signaling (64–66).
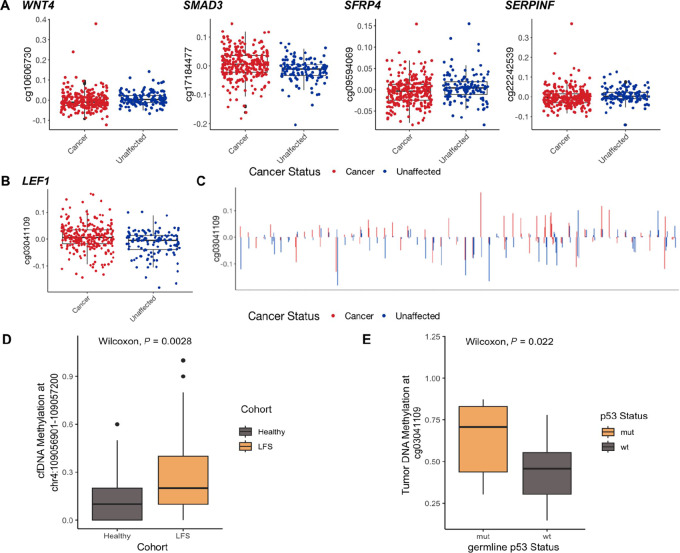
Among the 931 methylation probes found significantly associated with cancer status, several were located within genes involved in the WNT signaling pathway (Fig. 5A). This included the probe cg03041109 in the body of LEF1, which was significantly associated with cancer status (FDR = 0.02; Fig. 5B). We found probands almost universally exhibited higher LEF1 methylation than their unaffected family members (Fig. 5C). This further suggests the presence of molecular differences in the WNT signaling pathway between patients with LFS with and without cancer.
FIGURE 5.
Differential regulation of WNT signaling and LEF1 in LFS. A, Probes in the WNT signaling pathway (WNT4, SMAD3, SFRP4, SERPINF) significantly associated with cancer status. B, Probe in the body of LEF1 (cg03041109) associated with cancer status (FDR = 0.07). C, Methylation of probe in body of LEF1 (cg03041109) across members within each LFS family. Probands almost unanimously have higher LEF1 methylation than their unaffected family members. D, Plasma cfDNA methylation of the body of LEF1 (chr4:109056901-109057200), between patients with LFS and HBCs. E, Tumor methylation of the body of LEF1 (cg03041109) in CPTs, with and without a germline variant in TP53.
To further explore this, we evaluated methylation at the genomic location of cg03041109 in tumors and plasma cell-free DNA (cfDNA) of individuals with and without a germline variant in TP53. We found cfDNA of individuals with LFS were hypermethylated at LEF1 compared with HBCs with wildtype TP53 (Wilcoxon rank-sum test, P = 2.8 × 10−3;Fig. 5D). We also compared LEF1 methylation across CPTs—a characteristic LFS tumor—with or without a germline TP53 variant. Similar to PBLs and cfDNA of cancer-affected patients with LFS, we found increased LEF1 methylation in CPT with a germline TP53 variant compared with those with wildtype TP53 (Wilcoxon rank-sum test, P = 0.022;Fig. 5E).
To assess whether LEF1 methylation was associated with prognosis across paediatric cancers, we compared methylation of LEF1 in blood (control), normal kidney, NRs, WT with favorable histology, and WT with anaplastic histology across two cohorts, one from Toronto, Canada (57) and the other from the United Kingdom (58). Notably, anaplasia in WT is characteristic of tumors with a somatic TP53 variant and is associated with poor prognosis (67). We found WT with anaplasia were significantly hypomethylated at LEF1 compared with blood (pToronto = 6.4 × 10−7), normal kidney (pToronto = 7.2 × 10−8, pUK = 5.3 × 10−6), NR (pUK = 2.4 × 10−3) and WT with favorable outcomes (pToronto = 4.5 × 10−2, pUK = 5.1 × 10−3; Supplementary Fig. S10A). To evaluate whether this pattern was reflected in adult cancers lacking a germline TP53 variant, we compared LEF1 methylation in tumors with or without a somatic TP53 variant in LFS-associated cancers from The Cancer Genome Atlas. We found variable patterns of LEF1 methylation across tumors, with or without a somatic TP53 variant (Supplementary Fig. S10B). For instance, tumors with a somatic TP53 variant had increased methylation in LGG, but decreased methylation in sarcomas and breast cancers, compared with tumors with wildtype TP53.
Discussion
Hereditary cancers comprise 10% of all cancer incidence worldwide (68), of which it is estimated that up to 16% of childhood cancers are associated with pathogenic germline variants in CPG (43, 69, 70); in actuality these figures are likely much greater. Although in recent years, the discovery of common variants with small effect sizes has resulted in a paradigm shift of our understanding of cancer as a monogenic disease to a multifactorial disease, hereditary cancer syndromes, including LFS, have historically been considered monogenic diseases in which a single gene has been causally implicated. As a result, other contributing factors are frequently overlooked, even with variable cancer penetrance and phenotypes.
In this study, we present combined germline WGS and methylation data on a large multi-institutional cohort of individuals who fit the clinical definitions of LFS. We have demonstrated the novel utility of genetic and epigenetic data for personalized stratification of patients with LFS by two important clinical outcomes: cancer risk and overall survival. In individuals that lack a germline TP53 variant we identified alternative cancer-causing aberrations in 57.1% (8/14) of individuals with wildtype TP53 who developed cancer; this will enable the diagnosis of LFS in patients that would have been otherwise missed through standard screening procedures. Among variant TP53 carriers, we found 38.8% (19/49) who developed cancer harbored an additional P/LP variant in another cancer gene. Of note, modifier variants in the WNT signaling pathway were associated with decreased cancer incidence and improved survival outcomes in variant TP53 carriers. Furthermore, we evaluated the noncoding genome and methylome and identified inherited epimutations in genes including ASXL1, ETV6, and LEF1 that confer increased cancer risk. Leveraging these epimutations, we built a machine learning model that can predict cancer risk in patients with LFS with an AUROC of 0.725 (0.633–0.810). Taken together, this will enable the personalized stratification of patients with LFS based on the (i) the presence of a P/LP variant in a known cancer gene, (ii) predicted probability of developing cancer using our epimutation cancer risk model, (iii) epimutation cluster analysis, (iv) presence of a P/LP variant in the WNT signaling pathway, and (v) LEF1 methylation (Table 2). It clarifies the genomic basis for the phenotypic variability in LFS and highlights the immense benefits of expanding genetic and epigenetic testing of patients with LFS beyond TP53.
TABLE 2.
Genomics informed risk stratification guidelines for patients with LFS with a germline TP53 variant
| High-risk variant TP53 carrier | Low-risk variant TP53 carrier |
|---|---|
| P/LP variant in a cancer gene | P/LP variants in the WNT signaling pathway |
| High probability of developing cancer using our epimutation cancer risk model | Low probability of developing cancer using our epimutation cancer risk model |
| High-risk epimutation cluster | Low-risk epimutation cluster |
| LEF1 hypermethylation | LEF1 hypomethylation |
Approximately 30% of individuals who fit the clinical criteria used to diagnose LFS lack a pathogenic germline variant in TP53 (71). This suggests the involvement of other genetic and/or epigenetic mechanisms that contribute to cancer development in LFS. Candidate gene approaches have been taken to consider other relevant genetic causes in individual cases of LFS, proposing the role of alternative genes including CHEK2 (13–15), TP63 (15), BRCA1 (16, 17), BRCA2 (18), and CDKN1A (12). One study performed whole-exome sequencing of two index cases with cardiac angiosarcoma from a TP53-negative LFS family and identified an inherited pathogenic variant in POT1 (19). However, these studies fail to provide a comprehensive view of the germline genome in patients with LFS lacking a pathogenic variant in TP53. In addition, the majority of studies to-date have evaluated a single gene or utilized a limited panel of clinically recognized CPG (i.e., class 1) to screen for inherited cancer susceptibilities. In this study, we performed a genome-wide analysis of patients with LFS lacking a pathogenic germline variant in TP53 and adopted a tiered approach to evaluate cancer risk. In doing so, we identified alternative cancer-associated genetic aberrations (class 1–3) in 57% (8/14) of cancer-affected individuals diagnosed with LFS but lacking a pathogenic germline variant in TP53. This suggests that it is crucial that genetic testing in patients who fit the clinical definition of LFS be expanded beyond TP53 to permit the prospective management of these patients and their families. In addition to known cancer-associated variants, we identified P/LP variants in the novel candidate cancer genes SF3B4, FMN2, and MMP13 (class 4). Although promising, the P/LP variants we identified in novel candidate cancer genes (class 4) and disease-segregating VUSs in cancer-associated genes (class 5) require experimental validation to functionally characterize these alterations and determine their oncogenic capability.
In the 1980s, Vogelstein and Kinzler proposed a model for colorectal tumorigenesis that required four sequential alterations involving the activation of an oncogene coupled with the loss of three tumor suppressors (61). More recently, in a pan-cancer analysis, cancer genomes were found to contain driver variants affecting an average of four to five genes (70). Previous studies of pediatric cancer cases have also demonstrated that pathogenic germline variants in different genes can synergistically drive tumorigenesis (72). In line with these findings, we observed that patients with LFS harboring a P/LP TP53 variant with a co-occurring germline hit in another cancer gene (class 1–3) were associated with increased cancer incidence and decreased cancer survival compared with variant TP53 carriers lacking any class 1–3 variants. We also found that BRCA1 was the only autosomal dominant CPG (i.e., class 1), besides TP53, that harbored a P/LP variant. However, several other relevant cancer genes harbored P/LP variants that would have been otherwise overlooked, some of which are therapeutically actionable.
Furthermore, it is estimated that approximately 20% of patients with LFS with a pathogenic germline variant in TP53 never develop cancer. Understanding the reason for this apparent resiliency and the incomplete penetrance in LFS is crucial for risk stratification and ultimately informing patient management. We found variants in the WNT signaling pathway were associated with reduced cancer incidence and improved cancer survival compared with variant TP53 carriers lacking variants in the WNT signaling pathway, and even more so compared with those with at least one class 1–3 variant. Moreover, LEF1 hypermethylation was associated with increased cancer risk in patients with LFS with a germline TP53 variant. However, in WT and adult tumors with a somatic TP53 variant, hypomethylation of LEF1 was associated with poor prognosis, suggesting the relationship between LEF1 and patient outcomes is dependent on the timing of the TP53 variant (i.e., early germline event vs. later somatic event). Previously published work supports a model whereby aberrant WNT signaling abrogates p53 dysfunction (64), and future work should investigate this mechanism in the context of germline events.
Epigenetics has often been proposed as a mechanism to explain the “missing” causality and heritability of cancer (73–75). DNA methylation of PBL—as a proxy for the germline—is implicated in the transcriptional modulation of several cancers at regulatory regions of tumor suppressors and oncogenes (76–79). Susceptibility loci are enriched in these regulatory regions and can modulate the epigenome (80–82). Inherited methylation marks linked to genetic variants are referred to as secondary constitutional epimutations—formed by an initial genetic variant, which can then be inherited by offspring through genetic transmission of the initial variant or epigenetic transmission of the subsequent epimutation (83, 84). Previous research suggests patients with LFS with germline TP53 variants possess distinct PBL methylation signatures in comparison to their sporadic counterparts, suggesting a role for epimutations in LFS tumorigenesis (9). In our study, we leveraged the noncoding genome and methylation of patients with LFS and identified 931 methylation probes that are associated with increased cancer risk, of which 259 were associated with a cis-SNP. These epimutations suggest another mechanism by which the phenotype of patients with LFS is altered to favor a malignant state. However, it is difficult to discern the complementarity of the P/LP variants and epimutations, in an unbiased manner, without an additional validation cohort for which both WGS and methylation data are available.
We note, in our identification of cis-CSCE, the majority of PBL samples used for methylation profiling were collected from individuals that developed cancer, at or following diagnosis. While little is understood about the effects of chemotherapeutic drugs on DNA methylation of PBL, it remains an important consideration. To address this, we first removed broad, cancer- and treatment-associated signals from the methylation data. In previous work, Lemire and colleagues identified the absence of a global or local effect of chemotherapy on methylation in individuals with colorectal cancer (85). In contrast, Yao and colleagues found 4.2% of the CpG sites measured using the Illumina 450K methylation array underwent significant changes in patients with breast cancer after chemotherapy (86). Only 1.2% (7/568) of the probes associated with chemotherapy in Yao and colleagues overlapped with the 931 EWAS probes significantly associated with cancer status in variant TP53 carriers. Furthermore, we found no methylation probes significantly associated with systemic treatment status among variant TP53 carriers. Although this suggests chemotherapy is not a confounder in our analysis, reverse causation remains a potential limitation and further prospective studies are required.
Our study captures a snapshot of the clinical heterogeneity in LFS. To gain a comprehensive understanding of the germline landscape of LFS, further collaborative, prospective genetic and epigenetic profiling of similarly large cohorts of patients is crucial. Overall, our findings highlight the potential of expanding genetic and epigenetic testing of LFS patients beyond TP53, as TP53 alone does not explain the vast clinical heterogeneity in LFS. It also further necessitates the dissociation of hereditary cancer syndromes as single gene disorders given the existence of other factors that contribute to a cancer phenotype. Furthermore, these diseases need to be studied in a holistic manner as opposed to through the lens of a single gene. Future surveillance and treatment of hereditary cancer syndromes, like LFS, should incorporate genome-wide germline molecular profiling in order to provide personalized patient management.
Supplementary Material
Supplementary Figures
LFS Cohort
Tier 1-3 Gene Lists
1000G Variants
KiCS Variants
LFS Variants
cisCSCE
Acknowledgments
This study was funded with support from the Terry Fox Research Institute New Frontiers Program Project (no. 1081, D. Malkin) and Canadian Institutes for Health Research Foundation Scheme Grant (no. 143234, D. Malkin). We thank TCAG NGS facility for their sequencing services. The SickKids Cancer Sequencing (KiCS) program is supported by the Garron Family Cancer Centre with funds from the SickKids Foundation. D.M. is supported in part by the CIBC Children's Foundation Chair in Child Health Research. V. Subasri is supported by Ontario Graduate Scholarship and a Vector institute grant. Samples and data from J.R. Hansford were supplied by the Children's Cancer Centre Tissue Bank at the Murdoch Childrens Research Institute and The Royal Children's Hospital (RCH; www.mcri.edu.au/childrenscancercentretissuebank). Establishment and running of the Children's Cancer Centre Tissue Bank is made possible through generous support by CIKA (Cancer In Kids at RCH; http://www.cika.org.au), Leukaemia Auxiliary at RCH (LARCH), the Murdoch Childrens Research Institute and RCH. J.R. Hansford is supported by grants from the McClurg Foundation, Hospital Research Foundation, Robert Connor Dawes Foundation and My Room Children's Cancer Charity. The work of K.C. de Andrade, P.P. Khincha, and S.A. Savage was supported by the Intramural Research Program of the Division of Cancer Epidemiology and Genetics, NCI. We would also like to thank the late Ana Novokmet for her critical contributions to this study.
Footnotes
Note: Supplementary data for this article are available at Cancer Research Communications Online (https://aacrjournals.org/cancerrescommun/).
Authors’ Disclosures
J.R. Hansford reports personal fees from Bayer Pharmaceuticals, Alexion Pharmaceuticals, and Boxer Capital outside the submitted work. J.D. Schiffman reports other from Peel Therapeutics, Inc. outside the submitted work. T.J. Pugh reports personal fees from AstraZeneca, Canadian Pension Plan Investment Board, Chrysalis Biomedical Advisors, Illumina, Merck, PACT Pharma, SAGA Diagnostics, and grants from Roche/Genentech outside the submitted work. No disclosures were reported by the other authors.
Authors’ Contributions
V. Subasri: Conceptualization, data curation, software, formal analysis, validation, investigation, visualization, methodology, writing-original draft, project administration, writing-review and editing. N. Light: Resources, data curation, investigation, writing-review and editing. N. Kanwar: Investigation, writing-review and editing. J. Brzezinski: Resources, data curation, writing-review and editing. P. Luo: Resources, data curation. J.R. Hansford: Resources, writing-review and editing. E. Cairney: Resources, writing-review and editing. C. Portwine: Resources, writing-review and editing. C. Elser: Resources, writing-review and editing. J.L. Finlay: Resources, writing-review and editing. K.E. Nicholas: Resources, data curation, writing-review and editing. N. Alon: Data curation, project administration, writing-review and editing. L. Brunga: Resources, data curation, project administration, writing-review and editing. J. Anson: Resources, data curation, writing-review and editing. W. Kohlmann: Resources, data curation, writing-review and editing. K.C. de Andrade: Resources, data curation, writing-review and editing. P.P. Khincha: Resources, writing-review and editing. S.A. Savage: Resources, data curation, writing-review and editing. J.D. Schiffman: Resources, data curation, supervision, funding acquisition, writing-review and editing. R. Weksberg: Conceptualization, resources, data curation, supervision, funding acquisition, writing-original draft, writing-review and editing. T.J. Pugh: Conceptualization, resources, data curation, supervision, funding acquisition, writing-original draft, writing-review and editing. A. Villani: Conceptualization, resources, data curation, supervision, funding acquisition, writing-original draft, writing-review and editing. A. Shlien: Conceptualization, resources, data curation, supervision, funding acquisition, writing-original draft, writing-review and editing. A. Goldenberg: Conceptualization, resources, data curation, supervision, funding acquisition, writing-original draft, writing-review and editing. D. Malkin: Conceptualization, resources, data curation, supervision, funding acquisition, writing-original draft, writing-review and editing.
References
- 1. Malkin D, Li F, Strong L, Fraumeni J, Nelson C, Kim D, et al. Germ line p53 mutations in a familial syndrome of breast cancer, sarcomas, and other neoplasms. Science 1990;250:1233–8. [DOI] [PubMed] [Google Scholar]
- 2. Li FP. Soft-tissue sarcomas, breast cancer, and other neoplasms: a familial syndrome? Ann Intern Med 1969;71:747–52. [DOI] [PubMed] [Google Scholar]
- 3. Malkin D, Jolly KW, Barbier N, Look AT, Friend SH, Gebhardt MC, et al. Germline mutations of the p53 tumor-suppressor gene in children and young adults with second malignant neoplasms. N Engl J Med 1992;326:1309–15. [DOI] [PubMed] [Google Scholar]
- 4. Kratz CP, Achatz MI, Brugières L, Frebourg T, Garber JE, Greer M-LC, et al. Cancer screening recommendations for individuals with li-fraumeni syndrome. Clin Cancer Res 2017;23:e38–45. [DOI] [PubMed] [Google Scholar]
- 5. Ceyhan-Birsoy O, Selenica P, Chui MH, Jayakumaran G, Ptashkin R, Misyura M, et al. Paired tumor-normal sequencing provides insights into TP53-related cancer spectrum in li-fraumeni patients. J Natl Cancer Inst 2021;113:1751–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Malkin D. Li-fraumeni syndrome. Genes Cancer 2011;2:475–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Said BI, Malkin D. A functional variant in miR-605 modifies the age of onset in Li-Fraumeni syndrome. Cancer Genet 2015;208:47–51. [DOI] [PubMed] [Google Scholar]
- 8. Tabori U, Nanda S, Druker H, Lees J, Malkin D. Younger age of cancer initiation is associated with shorter telomere length in Li-Fraumeni syndrome. Cancer Res 2007;67:1415–8. [DOI] [PubMed] [Google Scholar]
- 9. Samuel N, Wilson G, Lemire M, Said BI, Lou Y, Li W, et al. Genome-wide DNA methylation analysis reveals epigenetic dysregulation of microRNA-34A in TP53 -associated cancer susceptibility. J Clin Oncol 2016;34:3697–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Marcel V, Palmero EI, Falagan-Lotsch P, Martel-Planche G, Ashton-Prolla P, Olivier M, et al. TP53 PIN3 and MDM2 SNP309 polymorphisms as genetic modifiers in the Li-Fraumeni syndrome: impact on age at first diagnosis. J Med Genet 2009;46:766–72. [DOI] [PubMed] [Google Scholar]
- 11. Camplejohn RS, Sodha N, Gilchrist R, Lomax ME, Duddy PM, Miner C, et al. The value of rapid functional assays of germline p53 status in LFS and LFL families. Br J Cancer 2000;82:1145–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Andrade RC, dos Santos ACE, de Aguirre Neto JC, Nevado J, Lapunzina P, Vargas FR. TP53 and CDKN1A mutation analysis in families with Li–Fraumeni and Li–Fraumeni like syndromes. Fam Cancer 2017;16:243–8. [DOI] [PubMed] [Google Scholar]
- 13. Bell DW. Heterozygous germ line hCHK2 mutations in Li-Fraumeni syndrome. Science 1999;286:2528–31. [DOI] [PubMed] [Google Scholar]
- 14. Zhuang X, Li Y, Cao H, Wang T, Chen J, Liu J, et al. Case report of a Li–Fraumeni syndrome-like phenotype with a de novo mutation in CHEK2. Medicine 2016;95:e4251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Bougeard G. Detection of 11 germline inactivating TP53 mutations and absence of TP63 and HCHK2 mutations in 17 French families with Li-Fraumeni or Li-Fraumeni-like syndrome. J Med Genet 2001;38:253–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Silva AG, Ewald IP, Sapienza M, Pinheiro M, Peixoto A, de Nóbrega AF, et al. Li-Fraumeni-like syndrome associated with a large BRCA1 intragenic deletion. BMC Cancer 2012;12:237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Cho Y, Kim J, Kim Y, Jeong J, Lee K-A. A case of late-onset li-fraumeni-like syndrome with unilateral breast cancer. Ann Lab Med 2013;33:212–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Evans DG, Wu CL, Birch JM. BRCA2: a cause of Li-Fraumeni-like syndrome. J Med Genet 2008;45:62–3. [DOI] [PubMed] [Google Scholar]
- 19. Calvete O, Martinez P, Garcia-Pavia P, Benitez-Buelga C, Paumard-Hernández B, Fernandez V, et al. A mutation in the POT1 gene is responsible for cardiac angiosarcoma in TP53-negative Li–Fraumeni-like families. Nat Commun 2015;6:8383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Bougeard G. Impact of the MDM2 SNP309 and p53 Arg72Pro polymorphism on age of tumour onset in Li-Fraumeni syndrome. J Med Genet 2006;43:531–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ariffin H, Hainaut P, Puzio-Kuter A, Choong SS, Chan ASL, Tolkunov D, et al. Whole-genome sequencing analysis of phenotypic heterogeneity and anticipation in Li-Fraumeni cancer predisposition syndrome. Proc Natl Acad Sci U S A 2014;111:15497–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Franceschi S, Spugnesi L, Aretini P, Lessi F, Scarpitta R, Galli A, et al. Whole-exome analysis of a Li–Fraumeni family trio with a novel TP53 PRD mutation and anticipation profile. Carcinogenesis 2017;38:938–43. [DOI] [PubMed] [Google Scholar]
- 23. Hu H, Liu J, Liao X, Zhang S, Li H, Lu R, et al. Genetic and functional analysis of a Li Fraumeni syndrome family in China. Sci Rep 2016;6:20221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Penkert J, Schmidt G, Hofmann W, Schubert S, Schieck M, Auber B, et al. Breast cancer patients suggestive of Li-Fraumeni syndrome: mutational spectrum, candidate genes, and unexplained heredity. Breast Cancer Res 2018;20:87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Amadou A, Waddington Achatz MI, Hainaut P. Revisiting tumor patterns and penetrance in germline TP53 mutation carriers: temporal phases of Li–Fraumeni syndrome. Curr Opin Oncol 2018;30:23–9. [DOI] [PubMed] [Google Scholar]
- 26. de Andrade KC, Khincha PP, Hatton JN, Frone MN, Wegman-Ostrosky T, Mai PL, et al. Cancer incidence, patterns, and genotype–phenotype associations in individuals with pathogenic or likely pathogenic germline TP53 variants: an observational cohort study. Lancet Oncol 2021;22:1787–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Li H. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv; 2013. Available from: http://arxiv.org/abs/1303.3997.
- 28. Broad Institute. Picard tools; 2018. Available from: http://broadinstitute.github.io/picard/.
- 29. McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, et al. The genome analysis toolkit: a mapreduce framework for analyzing next-generation DNA sequencing data. Genome Res 2010;20:1297–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Koboldt DC, Zhang Q, Larson DE, Shen D, McLellan MD, Lin L, et al. VarScan 2: somatic mutation and copy number alteration discovery in cancer by exome sequencing. Genome Res 2012;22:568–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Dick KJ, Nelson CP, Tsaprouni L, Sandling JK, Aïssi D, Wahl S, et al. DNA methylation and body-mass index: a genome-wide analysis. Lancet North Am Ed 2014;383:1990–8. [DOI] [PubMed] [Google Scholar]
- 32. Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med 2015;17:405–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Maxwell KN, Hart SN, Vijai J, Schrader KA, Slavin TP, Thomas T, et al. Evaluation of ACMG-guideline-based variant classification of cancer susceptibility and non-cancer-associated genes in families affected by breast cancer. Am J Hum Genet 2016;98:801–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Rausch T, Zichner T, Schlattl A, Stutz AM, Benes V, Korbel JO. DELLY: structural variant discovery by integrated paired-end and split-read analysis. Bioinformatics 2012;28:i333–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Chen X, Schulz-Trieglaff O, Shaw R, Barnes B, Schlesinger F, Källberg M, et al. Manta: rapid detection of structural variants and indels for germline and cancer sequencing applications. Bioinformatics 2016;32:1220–2. [DOI] [PubMed] [Google Scholar]
- 36. Geoffroy V, Herenger Y, Kress A, Stoetzel C, Piton A, Dollfus H, et al. AnnotSV: an integrated tool for structural variations annotation. Bioinformatics 2018;34:3572–4. [DOI] [PubMed] [Google Scholar]
- 37. The 1000 Genomes Project Consortium; Auton A, Brooks LD, Durbin RM, Garrison EP, Kang HM, et al. A global reference for human genetic variation. Nature 2015;526:68–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Karczewski KJ, Francioli LC, Tiao G, Cummings BB, Alföldi J, Wang Q, et al. The mutational constraint spectrum quantified from variation in 141,456 humans. Nature 2020;581:434–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Abyzov A, Urban AE, Snyder M, Gerstein M. CNVnator: An approach to discover, genotype, and characterize typical and atypical CNVs from family and population genome sequencing. Genome Res 2011;21:974–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Zhu M, Need AC, Han Y, Ge D, Maia JM, Zhu Q, et al. Using ERDS to infer copy-number variants in high-coverage genomes. Am J Hum Genet 2012;91:408–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Trost B, Walker S, Wang Z, Thiruvahindrapuram B, MacDonald JR, Sung WWL, et al. A comprehensive workflow for read depth-based identification of copy-number variation from whole-genome sequence data. Am J Hum Genet 2018;102:142–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res 2010;38:e164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Zhang J, Walsh MF, Wu G, Edmonson MN, Gruber TA, Easton J, et al. Germline mutations in predisposition genes in pediatric cancer. N Engl J Med 2015;373:2336–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Forbes SA, Beare D, Gunasekaran P, Leung K, Bindal N, Boutselakis H, et al. COSMIC: exploring the world's knowledge of somatic mutations in human cancer. Nucleic Acids Res 2015;43:D805–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Villani A, Davidson S, Kanwar N, Lo WW, Li Y, Cohen-Gogo S, et al. The clinical utility of integrative genomics in childhood cancer extends beyond targetable mutations. Nat Cancer 2023;4:203–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A 2005;102:15545–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Mai PL, Best AF, Peters JA, DeCastro RM, Khincha PP, Loud JT, et al. Risks of first and subsequent cancers among TP53 mutation carriers in the National Cancer Institute Li-Fraumeni syndrome cohort. Cancer 2016;122:3673–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Fortin J-P, Triche TJ, Hansen KD. Preprocessing, normalization and integration of the Illumina HumanMethylationEPIC array with minfi. Bioinformatics 2017;33:558–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Zhang Y, Jenkins DF, Manimaran S, Johnson WE. Alternative empirical Bayes models for adjusting for batch effects in genomic studies. BMC Bioinformatics 2018;19:262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Stegle O, Parts L, Piipari M, Winn J, Durbin R. Using probabilistic estimation of expression residuals (PEER) to obtain increased power and interpretability of gene expression analyses. Nat Protoc 2012;7:500–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Mezlini AM, Das S, Goldenberg A. Finding associations in a heterogeneous setting: statistical test for aberration enrichment. Genome Med 2021;13:68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Huan T, Joehanes R, Song C, Peng F, Guo Y, Mendelson M, et al. Genome-wide identification of DNA methylation QTLs in whole blood highlights pathways for cardiovascular disease. Nat Commun 2019;10:4267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Houlahan KE, Shiah Y-J, Gusev A, Yuan J, Ahmed M, Shetty A, et al. Genome-wide germline correlates of the epigenetic landscape of prostate cancer. Nat Med 2019;25:1615–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Wong D, Luo P, Oldfield L, Gong H, Brunga L, Rabinowicz R, et al. Integrated analysis of cell-free DNA for the early detection of cancer in people with Li-Fraumeni syndrome. Genetic and Genomic Medicine; 2022. Available from: http://medrxiv.org/lookup/doi/10.1101/2022.10.07.22280848.
- 55. Merino DM, Shlien A, Villani A, Pienkowska M, Mack S, Ramaswamy V, et al. Molecular characterization of choroid plexus tumors reveals novel clinically relevant subgroups. Clin Cancer Res 2015;21:184–92. [DOI] [PubMed] [Google Scholar]
- 56. Pienkowska M, Choufani S, Turinsky AL, Guha T, Merino DM, Novokmet A, et al. DNA methylation signature is prognostic of choroid plexus tumor aggressiveness. Clin Epigenetics 2019;11:117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Brzezinski J, Choufani S, Romao R, Shuman C, Chen H, Cunanan J, et al. Clinically and biologically relevant subgroups of Wilms tumour defined by genomic and epigenomic analyses. Br J Cancer 2021;124:437–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Charlton J, Williams RD, Weeks M, Sebire NJ, Popov S, Vujanic G, et al. Methylome analysis identifies a Wilms tumor epigenetic biomarker detectable in blood. Genome Biol 2014;15:434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Brown NJ, Bhatia K, Teague J, White SM, Lo P, Challis J, et al. Report of a bi-allelic truncating germline mutation in TP53. Fam Cancer 2019;18:101–4. [DOI] [PubMed] [Google Scholar]
- 60. Thavaneswaran S, Rath E, Tucker K, Joshua AM, Hess D, Pinese M, et al. Therapeutic implications of germline genetic findings in cancer. Nat Rev Clin Oncol 2019;16:386–96. [DOI] [PubMed] [Google Scholar]
- 61. Vogelstein B, Fearon ER, Hamilton SR, Kern SE, Preisinger AC, Leppert M, et al. Genetic alterations during colorectal-tumor development. N Engl J Med 1988;319:525–32. [DOI] [PubMed] [Google Scholar]
- 62. Seberg HE, Van Otterloo E, Cornell RA. Beyond MITF: Multiple transcription factors directly regulate the cellular phenotype in melanocytes and melanoma. Pigment Cell Melanoma Res 2017;30:454–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Soura E, Eliades PJ, Shannon K, Stratigos AJ, Tsao H. Hereditary melanoma: update on syndromes and management. J Am Acad Dermatol 2016;74:411–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Hao Y-H, Lafita-Navarro MC, Zacharias L, Borenstein-Auerbach N, Kim M, Barnes S, et al. Induction of LEF1 by MYC activates the WNT pathway and maintains cell proliferation. Cell Commun Signal 2019;17:129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Gutierrez A, Tschumper RC, Wu X, Shanafelt TD, Eckel-Passow J, Huddleston PM, et al. LEF-1 is a prosurvival factor in chronic lymphocytic leukemia and is expressed in the preleukemic state of monoclonal B-cell lymphocytosis. Blood 2010;116:2975–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Zhao Y, Zhu J, Shi B, Wang X, Lu Q, Li C, et al. The transcription factor LEF1 promotes tumorigenicity and activates the TGF-β signaling pathway in esophageal squamous cell carcinoma. J Exp Clin Cancer Res 2019;38:304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Bardeesy N, Falkoff D, Petruzzi M-J, Nowak N, Zabel B, Adam M, et al. Anaplastic Wilms’ tumour, a subtype displaying poor prognosis, harbours p53 gene mutations. Nat Genet 1994;7:91–7. [DOI] [PubMed] [Google Scholar]
- 68. Nagy R, Sweet K, Eng C. Highly penetrant hereditary cancer syndromes. Oncogene 2004;23:6445–70. [DOI] [PubMed] [Google Scholar]
- 69. Wong M, Mayoh C, Lau LMS, Khuong-Quang D-A, Pinese M, Kumar A, et al. Whole genome, transcriptome and methylome profiling enhances actionable target discovery in high-risk pediatric cancer. Nat Med 2020;26:1742–53. [DOI] [PubMed] [Google Scholar]
- 70. ICGC/TCGA Pan-Cancer Analysis of Whole Genomes Consortium. Pan-cancer analysis of whole genomes. Nature 2020;578:82–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Varley J, Evans D, Birch J. Li-Fraumeni syndrome – a molecular and clinical review. Br J Cancer 1997;76:1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Kuhlen M, Taeubner J, Brozou T, Wieczorek D, Siebert R, Borkhardt A. Family-based germline sequencing in children with cancer. Oncogene 2019;38:1367–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Eichler EE, Flint J, Gibson G, Kong A, Leal SM, Moore JH, et al. Missing heritability and strategies for finding the underlying causes of complex disease. Nat Rev Genet 2010;11:446–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Maher B. Personal genomes: the case of the missing heritability. Nature 2008;456:18–21. [DOI] [PubMed] [Google Scholar]
- 75. Petronis A. Epigenetics as a unifying principle in the aetiology of complex traits and diseases. Nature 2010;465:721–7. [DOI] [PubMed] [Google Scholar]
- 76. Robertson KD. DNA methylation and human disease. Nat Rev Genet 2005;6:597–610. [DOI] [PubMed] [Google Scholar]
- 77. Joo JE, Dowty JG, Milne RL, Wong EM, Dugué P-A, English D, et al. Heritable DNA methylation marks associated with susceptibility to breast cancer. Nat Commun 2018;9:867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Luo X, Huang R, Sun H, Liu Y, Bi H, Li J, et al. Methylation of a panel of genes in peripheral blood leukocytes is associated with colorectal cancer. Sci Rep 2016;6:29922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. McCarthy MI, Hirschhorn JN. Genome-wide association studies: potential next steps on a genetic journey. Hum Mol Genet 2008;17:R156–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Zhang Y, Schöttker B, Ordóñez-Mena J, Holleczek B, Yang R, Burwinkel B, et al. F2RL3 methylation, lung cancer incidence and mortality: F2RL3 methylation, lung cancer incidence and mortality. Int J Cancer 2015;137:1739–48. [DOI] [PubMed] [Google Scholar]
- 81. Whitington T, Gao P, Song W, Ross-Adams H, Lamb AD, Yang Y, et al. Gene regulatory mechanisms underpinning prostate cancer susceptibility. Nat Genet 2016;48:387–97. [DOI] [PubMed] [Google Scholar]
- 82. Cowper-Sal·lari R, Zhang X, Wright JB, Bailey SD, Cole MD, Eeckhoute J, et al. Breast cancer risk–associated SNPs modulate the affinity of chromatin for FOXA1 and alter gene expression. Nat Genet 2012;44:1191–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Heyn H, Sayols S, Moutinho C, Vidal E, Sanchez-Mut JV, Stefansson OA, et al. Linkage of DNA methylation quantitative trait loci to human cancer risk. Cell Rep 2014;7:331–8. [DOI] [PubMed] [Google Scholar]
- 84. Hitchins MP. Constitutional epimutation as a mechanism for cancer causality and heritability? Nat Rev Cancer 2015;15:625–34. [DOI] [PubMed] [Google Scholar]
- 85. Lemire M, Zaidi SHE, Zanke BW, Gallinger S, Hudson TJ, Cleary SP. The effect of 5-fluorouracil/leucovorin chemotherapy on CpG methylation, or the confounding role of leukocyte heterogeneity: an illustration. Genomics 2015;106:340–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Yao S, Hu Q, Kerns S, Yan L, Onitilo AA, Misleh J, et al. Impact of chemotherapy for breast cancer on leukocyte DNA methylation landscape and cognitive function: a prospective study. Clin Epigenetics 2019;11:45. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figures
LFS Cohort
Tier 1-3 Gene Lists
1000G Variants
KiCS Variants
LFS Variants
cisCSCE
Data Availability Statement
All WGS and methylation data files are available in the European Genome-Phenome Archive (EGA) under the accession numbers EGAS00001007075 and EGAS00001007061. Analysis scripts can be found at doi:10.5281/zenodo.5563379.