Summary
Prairie voles are among a small group of mammals that display long-term social attachment between mating partners. Many pharmacological studies show that signaling via the oxytocin receptor (Oxtr) is critical for the display of social monogamy in these animals. We used CRISPR-mutagenesis to generate three different Oxtr null mutant prairie vole lines. Oxtr mutants displayed social attachment such that males and females showed a behavioral preference for their mating partners over a stranger of the opposite sex, even when assayed using different experimental setups. Mothers lacking Oxtr delivered viable pups, and parents displayed care of their young and raised them to the weanling stage. Together, our studies unexpectedly reveal that social attachment, parturition, and parental behavior can occur in the absence of Oxtr signaling in prairie voles.
eTOC Blurb:
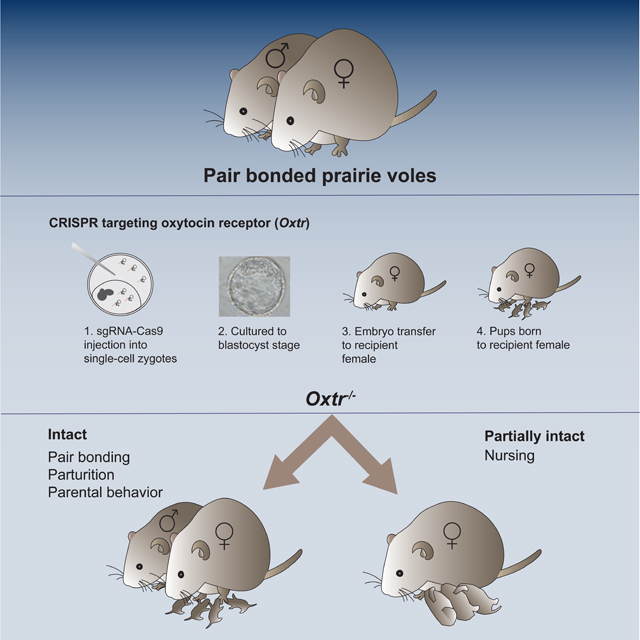
Berendzen et al. report the surprising finding that prairie voles lacking the oxytocin receptor (Oxtr) display pair bonding and parental behaviors, including nursing. Despite many pharmacological studies suggesting a requirement for Oxtr, these findings indicate that Oxtr is genetically dispensable for pair bond formation and parental behaviors in voles.
Graphical Abstract
Introduction
Strong, specific, and sustained relationships between mates and kin are displayed by a fascinating, but limited, subset of species across the animal kingdom1–3. Such attachments, which form the basis of diverse and complex social systems, are observed in species that have evolved the capacity to form lasting bonds between individuals, suggesting that they are innate with a strong underlying genetic component4–7. Progress in understanding the molecular or neural networks that promote social attachment has been hindered because traditional genetic model organisms such as C. elegans, D. melanogaster, D. rerio, and M. musculus do not display enduring attachments as adults. Here we report the use of CRISPR-based targeting in prairie voles (Microtus ochrogaster) to probe signaling pathways implicated in social attachment.
Adult prairie voles exhibit social attachment behavior such that mating partners form an enduring bond with each other. This attachment behavior, commonly referred to as pair bonding, has been observed in ethological studies in the wild as well as in the laboratory setting8,9. Pair bonded voles spend time together in close proximity (huddling behavior) and show a social preference for each other over a potential new mating partner. Neuropeptide signaling has long been known to control the display of social behaviors across diverse species10–14. Differences in neural populations regulated by these pathways correlate with interspecies variations in social structure15–17. Intriguingly, the evolution of varied complex social systems and affiliative behaviors, including social monogamy, has repeatedly converged upon the nonapeptide hormones oxytocin (Oxt) and arginine vasopressin (Avp) and their orthologs across phylogenies18–20.
Ethological studies, using live-trapping of wild prairie voles, reported that mating pairs are more likely to be trapped together than is expected by chance9. Similar studies of closely related species such as meadow voles (M. pennsylvanicus), that do not pair bond, showed that live-traps contained single animals9. Subsequent studies demonstrated that pair bonding behavior can also be observed in a laboratory setting21,22. Pair bonding is displayed as a suite of behavioral traits, the most commonly measured of which is a preference for the familiar partner over a novel stranger. Pair bonded animals prefer to huddle with their partners compared to exploring unfamiliar conspecifics of the opposite sex23. Such partner preference is also accompanied by aggression toward unfamiliar opposite sex conspecifics, indicative of active rejection of potential new mates24.
Comparative studies between socially monogamous and non-monogamous vole species revealed striking differences in Oxtr expression in brain regions thought to be important for social attachment, and implicated natural variation within species in specific aspects of pair bonding and attachment behaviors15,16,25–28. Pharmacological studies from multiple groups have shown that Oxt is sufficient to induce pair bonding behavior in otherwise naive voles and the administration of Oxtr-antagonists induces loss of these behaviors12,17,21,29. Viral manipulations of Oxtr expression in specific brain regions of prairie voles also recapitulate findings from such pharmacological studies30,31. Taken together, these findings suggest a critical role for Oxt signaling via its cognate receptor, Oxtr, in driving pair bonding behaviors in this species.
Prairie voles, like many other animals that display pair bonding behavior, exhibit biparental care of their young, and oxytocin signaling is thought to control these behaviors as well31,32. Oxytocin is additionally critical for milk let-down, the reflexive release of milk triggered by sensory stimuli associated with suckling33,34. All pups born to female mice null for Oxt or Oxtr die shortly after birth because of complete failure of milk let-down35,36. In addition to this role in lactation, stimulation of Oxt-expressing neurons in virgin female mice induces pup retrieval behaviors typical of lactating females37. Thus, decades of research implicates both Oxt and its cognate receptor Oxtr in a large repertoire of parenting behaviors. typical of lactating females37. Thus, decades of research implicates both Oxt and its cognate receptor Oxtr in a large repertoire of parenting behaviors.
To test the genetic requirement of Oxtr in pair bonding and parental behaviors, we employed a CRISPR-based approach to generate mutant prairie voles null for this receptor. Surprisingly, male and female prairie voles homozygous for each of the three distinct loss-of-function Oxtr alleles displayed pair bonding. Moreover, we observed that Oxtr null females were capable of raising pups to weaning. We therefore conclude that, contrary to previous assumptions, pair bonding and parental behaviors in prairie voles do not require Oxtr function.
Results
CRISPR-targeting can reliably generate multiple null alleles of Oxtr in prairie voles
To perform CRISPR-based gene targeting in prairie voles, we developed a protocol to obtain single cell embryos for injection of Cas9 ribonucleoprotein complexes. We were unable to achieve successful superovulation using hormonal supplementation protocols including those previously reported38, and therefore we implemented a timed-mating strategy to harvest embryos synchronized at specific developmental time points. We harvested 0.5 day single cell embryos using this protocol, an approach that yielded 3.7±1.2 embryos/female.
Because we maintain prairie voles in an outbred background, we were concerned about natural variation in Oxtr coding sequence that could potentially reduce targeting efficiency by specific small guide RNAs (sgRNAs). Indeed, we observed 3 synonymous substitutions in exon 1 of the Oxtr locus from 4 voles in our colony (Figure S1A). We designed 8 protospacer adjacent motif (PAM) site-anchored sgRNAs based on conserved sequences in exon 1. We initially tested whether these sgRNAs could generate mutations in exon 1 in vitro, injecting them as Cas9 ribonucleoprotein complexes individually into single cell embryos and genotyping 4 days later at the blastocyst stage (Figures S1B-D). Two sgRNAs consistently yielded mutations in exon 1, and sequencing revealed multiple mutations in individual blastocysts, suggesting that CRISPR-targeting had also occurred after the single cell stage (Figures S1E-F).
We proceeded to co-inject these two sgRNAs into single cell embryos, cultured them in vitro to the blastocyst stage, and then transferred these healthy blastocysts into recipient pseudopregnant prairie voles (Figure 1A). Each recipient female gave birth to 1–2 pups (~10% of embryos transferred) ~16 days following embryo transfer. Given the significant chimerism we observed following genotyping of blastocysts injected with our chosen sgRNAs (Figures S1E-F) and the low yield of liveborn pups following embryo transfer, we mated each founder (G0) to a wildtype (WT) partner even when genotyping tail samples from these founders revealed no mutations, as germline chimerism may not be reflected in the small, peripheral tissue samples used in genotyping39. This strategy yielded 3 distinct Oxtr alleles that transmitted via the germline of founders with WT tail DNA (Figures 1B,C). Oxtr1 has a 1 base pair (bp) insertion that is predicted to yield an 84 amino acid (aa) peptide, Oxtr4 contains 2 small, multi-bp deletions that are predicted to yield an 81 aa peptide, and Oxtr5 contains a large deletion spanning and extending 1.5 kbp beyond the sequence targeted by the two sgRNAs. There is currently no completely assembled and annotated prairie vole genome to aid in sequencing-based analysis of off-target events. Therefore, to isolate Oxtr alleles and minimize carry-over of potential off-target mutations in our lines, we independently outcrossed all lines of each of the three mutant alleles to WT voles in our colonies.
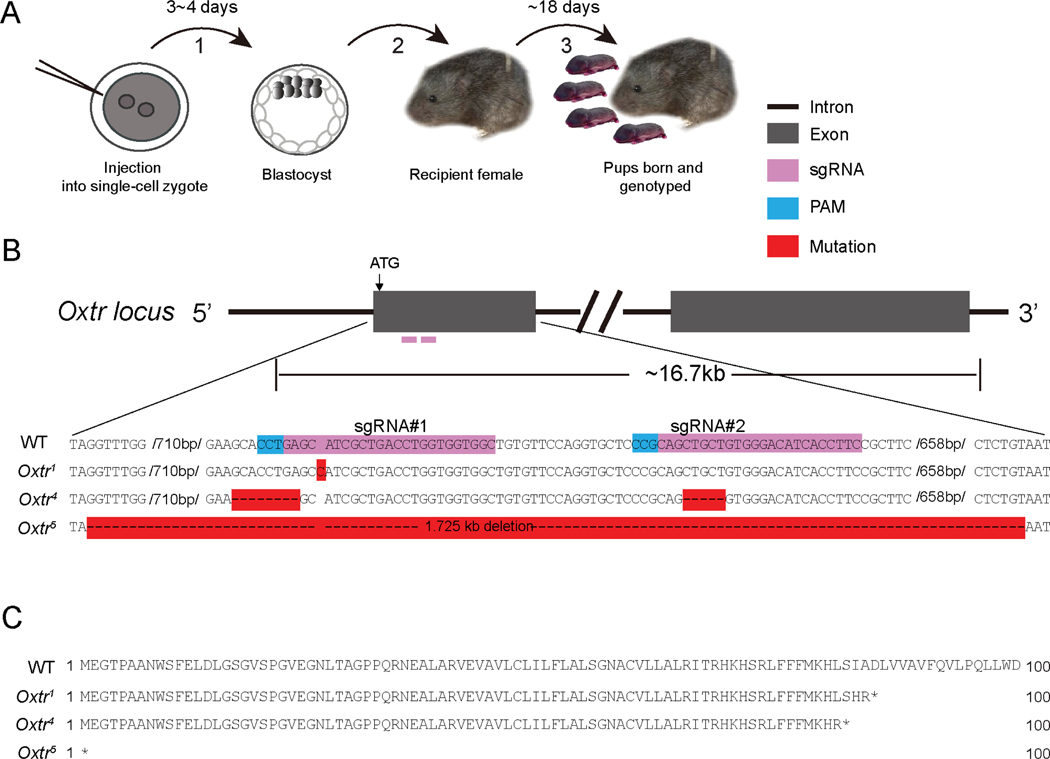
Figure 1. CRISPR mutagenesis yields multiple null alleles of Oxtr in prairie voles.
A. Schematic of CRISPR-based targeting to generate Oxtr mutant prairie voles. Single cell embryos injected with Cas9-sgRNA ribonucleoprotein were cultured (1) to the blastocyst stage and transferred (2) to pseudopregnant recipient females who carried the embryos to term (3).
B. Schematic (top) of Oxtr locus encompassing first two exons. DNA sequence of WT and targeted Oxtr alleles. Dash/missing nucleotide represents a deletion (Oxtr4, Oxtr5) and red highlighted “C” (Oxtr1) is an insertion. PAM, protospacer adjacent motif.
C. Predicted amino acid sequence of WT Oxtr, Oxtr1, Oxtr4, and Oxtr5 (only first 100 amino acids shown).
See also Figure S1.
CRISPR-generated mutations in Oxtr produce loss of function alleles
Each of the three Oxtr alleles we generated is predicted to generate non-functional Oxtr (Figure 1C). We tested this directly by performing a ligand binding assay in situ, using a radiolabeled small molecule competitive agonist for Oxtr. These studies showed a complete lack of binding in homozygous null mutants of both sexes of Oxtr1, Oxtr4, and Oxtr5 (Figures 2A-E, S2A-H), demonstrating absence of all ligand-binding Oxtr in vivo. Loss of Oxtr signaling may lead to changes in binding of the functionally related neuropeptide vasopressin (Avp) to its receptor, Avpr1a, or in expression of the Avp or Oxt peptides themselves. We could discern no differences in Avpr1a binding in a small number of Oxtr null mutants that we tested (data not shown). Furthermore, we found no increase in expression of Oxt or AVP peptide in Oxtr nulls compared with WT (Figure S2I-L). Consistent with the lack of requirement for Oxtr for survival, we observed Mendelian ratios of WT, heterozygous, and homozygous pups born to heterozygous parents (Oxtr1 29:60:24, Oxtr4 37:62:32 and Oxtr5 25:55:26).
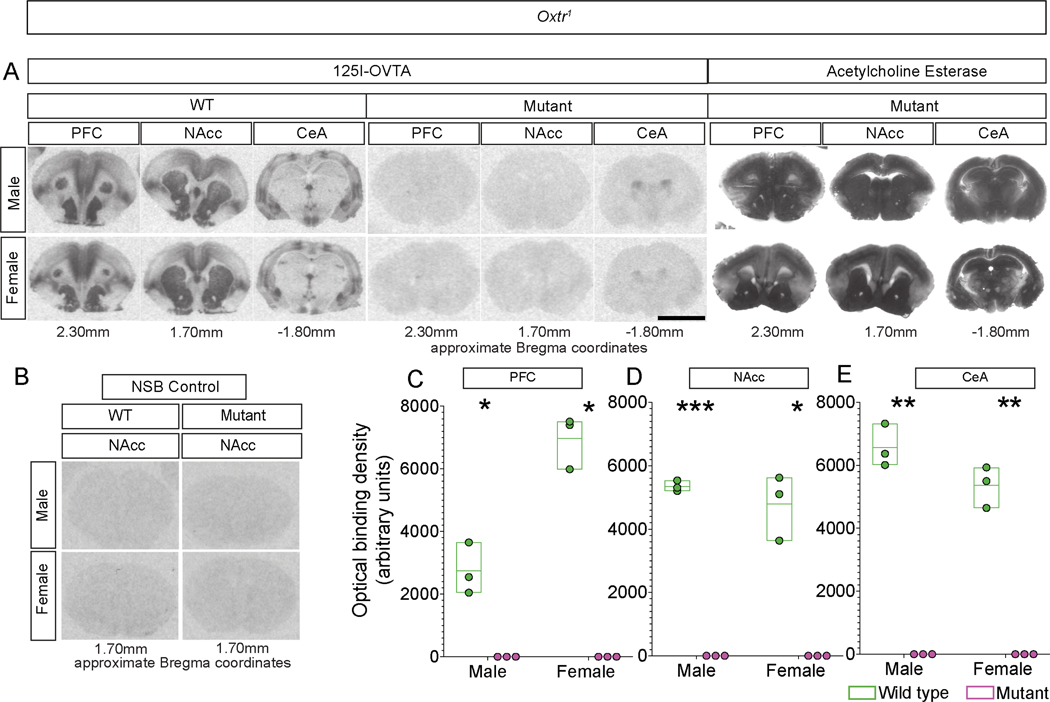
Figure 2. Oxtr mutant voles lack functional, ligand-binding Oxtr.
A. Loss of binding with the competitive agonist 125I-OVTA visualized in coronal sections through the rostral telencephalon: Left: Labeling in PFC (prefrontal cortex), NAcc (nucleus accumbens), and CeA (central amygdala) of WT sibling controls; Middle: Labeling in equivalent sections through PFC, Nacc, and CeA from Oxtr1 homozygous mutant voles; Right: The same mutant sections as in the middle panels, stained for Acetylcholine esterase to demonstrate equivalence of sections chosen for WT and mutants.
B. Non-specific binding (NSB) control shows no off-target binding in WT or Oxtr1 mutant sections.
C-E. Optical density-based quantification of binding to 125I-OVTA shows that binding is essentially undetectable in mutants null for Oxtr1 in PFC (C), NAcc (D), or CeA (E). Scale bar =5 mm; boxplot depicts max-minScale bar =5 mm; boxplot depicts max-min, midline denotes mean; n=3 for WT and mutant males and females (C-E). Scale bar =5 mm; boxplot depicts max-minSee also Figure S2.
Female and male prairie voles lacking Oxtr exhibit pair bonding
Prairie voles are induced ovulators, and following a short period of cohabitation between opposite sex animals, co-housed pairs will mate and subsequently display pair bonding23,40. To test whether our Oxtr null mutants display deficits in pair bonding, we co-housed sexually naive males and females for 7 days, a period previously shown to be sufficient to induce pair bonding23,40. WT or homozygous mutants were paired with an unfamiliar, unrelated WT animal of the opposite sex. We used two different setups commonly used to assay partner preference and behaviors between the experimental subject and a cohoused partner or novel unfamiliar conspecific of the opposite sex21,41. In the branched chamber apparatus, the experimental animal, its pair bonded partner, and an unfamiliar, opposite-sex conspecific are housed in separate interconnected chambers such that only the experimental subject has free access to all chambers (Figure 3A)21. We also used a linear, partially-divided chamber in which the cohoused partner and unfamiliar conspecific are tethered at opposite ends from each other, and only the experimental subject has access to all chambers (Figure 3B). In both experimental setups, the experimental animal can exhibit affiliative or other behaviors toward either tethered animal41,42.
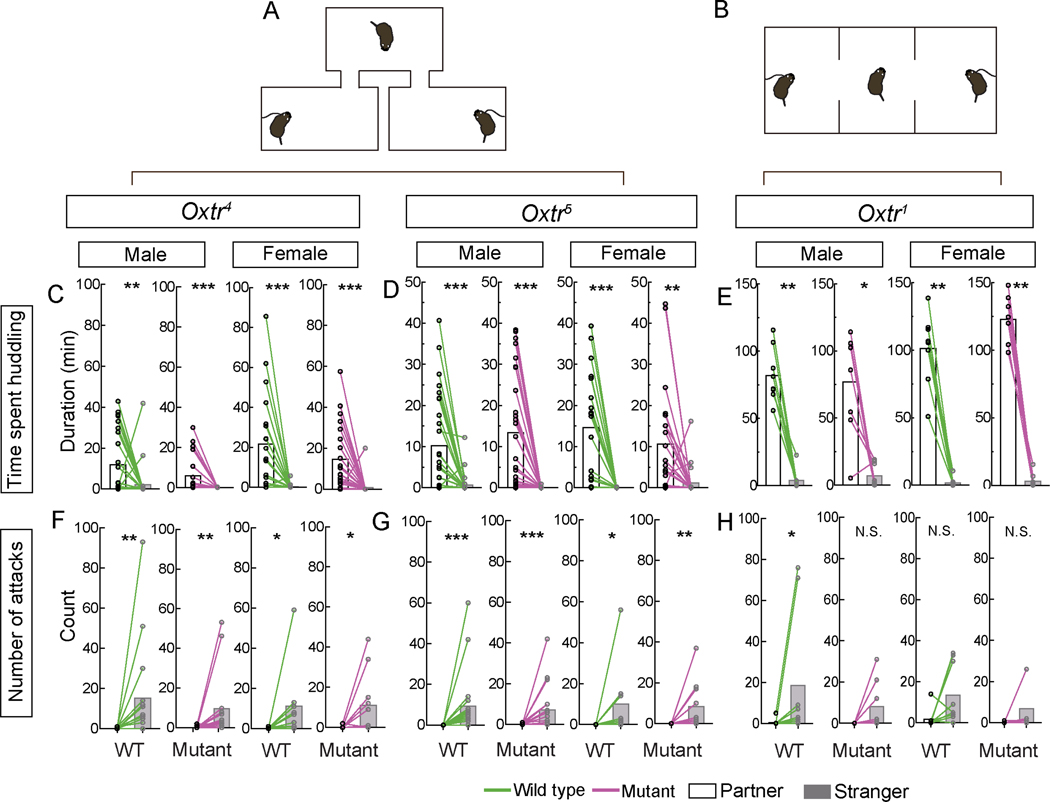
Figure 3. Female and male prairie voles lacking Oxtr exhibit pair bonding Scale bar =5 mm; boxplot depicts max-min.
A-B. Schematic of partner preference test performed in a branched (A) or linear chamber with two partitions (B). Experimental vole is free to move between chambers (A) or partitioned space (B) whereas stimulus voles (opposite sex partner and stranger) are restrained by tethers. Scale bar =5 mm; boxplot depicts max-min
C-E. WT, Oxtr4−/− (C), Oxtr5−/− (D), and Oxtr1−/− (E) voles spent more time with their partner than a stranger of the opposite sex.
F-H. WT, Oxtr4−/− (F), and Oxtr5−/− (G) voles attacked the stranger more frequently than their partner. WT Oxtr1 males attacked the stranger more frequently (H).
Mean ± SEM; n = 25 WT and 21 mutant males, 19 WT and 21 mutant females (C); 27 WT and mutant males each, 19 WT and 21 mutant females (D); 8 WT and 8 mutant males, 8 WT and 8 mutant females each (E); 15 WT and 14 mutant males, 9 WT and 11 mutant females (F); 18 WT and mutant males each, 9 WT and 12 mutant females 15 (G); 8 WT and mutant males and females each (H); *p<0.05, **p<0.01, ***p<0.001; N.S., not significant.
See also Figure S3.
We tested pair bonding of Oxtr1 mutants and their control group in the linear chamber design and that of Oxtr4 and Oxtr5 and their control groups in the branched chamber apparatus. Surprisingly, we observed that males and females homozygous for each of the three, null Oxtr alleles all spent more time huddling with their partners. They also displayed aggression towards the unfamiliar conspecific of the opposite sex, indicative of rejection of other potential mates. These displays of huddling with partners and rejection of unfamiliar potential partners were comparable to those of WT animals in both experimental paradigms we deployed (Figures 3, S3). However, Oxtr1 mutant males showed no significant difference in aggression toward partner and stranger females, in contrast to WT controls who showed variable but significant aggression towards stranger females, (Figure 3H). In any event, we find that prairie voles demonstrate pair bonding behaviors in the absence of functional Oxtr.
Prairie voles lacking Oxtr bear viable pups and display bi-parental care similar to wild types
Given the concordance in pair bonding displays by animals homozygous for each of the three mutant Oxtr alleles, we tested a subset of these for performance in parenting. Oxtr4 or Oxtr5 homozygous mutants and their WT siblings were paired with WT animals of the opposite sex until parturition and then tested for parental care of their progeny. Both male and female prairie vole parents interact intensively with their pups, spending prolonged periods huddling, licking, and grooming them. Experimental interference during the pre-weaning period can disrupt both subsequent displays of parental care as well as the repertoire of adult social behaviors exhibited by the pups43,44. Accordingly, the standard assay is to observe the duration of parental interactions with their pups in the first few days after parturition and document pup survival and health at weaning45.
We observed that Oxtr null parents interacted equivalently with their pups compared to their WT counterparts (Figures 4A-F). Both WT and mutant parents spent the majority of their time in the nest, in direct contact with their litters, and, in the case of mothers, nursing pups. Unlike mice lacking Oxtr36, we never observed cages in which pups were scattered across the cage floor, indicative of effective retrieval to the nesting area of pups who had wandered away.
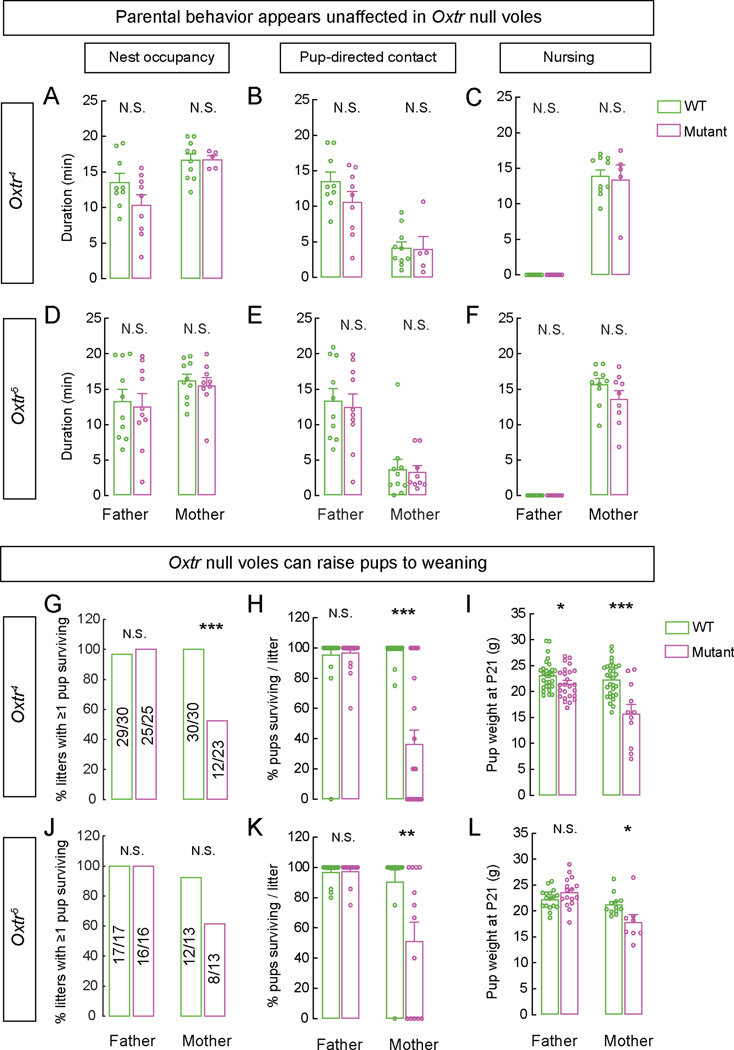
Figure 4. Prairie voles lacking Oxtr display bi-parental care and can raise pups to weaning.
A-F. WT, Oxtr4–/–, and Oxtr5–/– mothers and fathers exhibit equivalent nest occupancy (A, D), pup-directed contacts (B, E), and nursing behavior (C, F) with their pups.
G, J. Oxtr4–/– mothers wean fewer litters compared to WT mothers.
H, K. Litters from Oxtr4−/− and Oxtr5−/− mothers have fewer pups surviving to weaning compared to WT mothers
I, L. Oxtr4−/− mothers and fathers and Oxtr5−/− mothers weaned pups with significantly lower body weight compared to WT parents. Only litters with ≥1 pup surviving to weaning were analyzed.
Mean ± SEM; n = 9 WT and mutant males each, 10 WT and 5 mutant females (A-C); 10 WT and mutant males each, 10 WT and 9 mutant females (D-F); 30 WT and 25 mutant males, 30 WT and 23 mutant females (G,H); 29 WT and 25 mutant males, 30 WT and 11 mutant females (I); 17 WT and 16 mutant males, 13 WT and mutant females each (J,K); 17 WT and 16 mutant males, 12 WT and 8 mutant females (L);*p<0.05, **p<0.01, ***p<0.001; N.S., not significant.
See also Figure S4.
Surprisingly, we noticed that pups born of mothers homozygous for either of two Oxtr alleles survived past the first few hours of birth, with some weaning successfully, suggesting that mutant females could nurse some of their young (Figures 4G,H,J,K). Oxtr4 null mothers had significantly fewer surviving litters at weaning compared to WT mothers (Figure 4G). Oxtr5 null mothers also had fewer surviving litters at weaning, although this did not reach statistical significance (Figure 4J). Of the litters that survived to weaning to experimental females, Oxtr4 and Oxtr5 null mothers had fewer pups per litter (Figures 4H,K). Pups born to all mutant mothers weighed significantly less at weaning than pups born to WT mothers, suggesting a defect in milk let-down or subtle deficits in nursing behavior (Figures 4I,L, S4A). Consistent with a deficit in milk let-down, gross examination of the nipple area revealed less engorgement in females mutant for Oxtr compared to WT mothers (not shown).
Both WT and mutant fathers successfully raised their litters to weaning (Figures 4G-L, S4). Pups raised by Oxtr4, but not Oxtr1 or Oxtr5, null fathers weighed less when compared to those raised by WT fathers, suggesting a variably penetrant role for Oxtr in fathers in their ability to raise thriving pups. Weights at birth and time of weaning were decreased from Oxtr null mothers compared with WT (Figure 4I, J, S4). Together, our findings show that, in prairie voles, biparental care and some milk let-down occur in the absence of Oxtr signaling36.
Discussion
CRISPR-mutagenesis has been successfully used to manipulate a large variety of species of animals, allowing scientists the freedom to establish new model organisms for biological phenomena that are not observed in worms, fruit flies, zebrafish, or mice. We have utilized CRISPR-based targeting to test the role of Oxtr in prairie voles in modulating attachment and social behaviors, a suite of behaviors not displayed by standard genetic model organisms46. We find that pair bonding behaviors, nursing, and weaning of pups can all occur in the absence of Oxtr-mediated signaling. Our findings, consistent across multiple paradigms, three labs, and three null alleles of Oxtr, are in contrast to prior studies that highlight the importance of Oxtr signaling in pair bonding and parental behaviors in prairie voles13,21,27,30,31,40,47.
A key difference between our work and preceding studies is that the latter were conducted in adult animals, using pharmacological or viral misexpression strategies to determine a role for Oxtr function in behavior. Despite efforts to demonstrate the specificity of pharmacologic approaches to manipulate Oxtr activity, it remains possible that the compounds employed in these studies alter yet-to-be-identified pathways that contribute to pair bonding. In such a model, exogenous ligands that activate Oxtr bind to and stimulate additional receptors, while compounds that antagonize Oxt binding to Oxtr could also inhibit either its binding to additional receptors or other ligand-receptor interactions48,49. It is possible that compensatory pathways activated due to constitutive loss of function of Oxtr in our studies obscure a functional role for this receptor in pair bonding and parenting50,51. We did not find an increase in Oxt or Avp expression or discernible changes in Avpr1a expression indicative of such compensation, though we cannot exclude more subtle changes in the expression of these neuropeptides or other pathways that substitute for the loss of Oxtr signaling. Beyond Avp, additional neuroendocrine or neuromodulator pathways may be engaged following the initial social interactions that promote bonding52–54. Irrespective of these pathways, which now must be determined, the neural circuits mediating pair bonding and parental behaviors in prairie voles can facilitate the display of these behaviors independent of Oxtr signaling.
Future studies with targeted loss of Oxtr in specific brain regions of adult voles will reveal whether it is important for these behaviors. Alternatively, Oxtr and Avpr1a signaling might redundantly regulate pair bonding and parenting; Oxt can also signal via Avpr1a and it may therefore modulate these behaviors entirely through Avpr1a rather than Oxtr55,56. These possibilities can be disambiguated by testing the behavioral performance of voles null for Avpr1a or Avpr1a and Oxtr. Determining identity of the non-Oxtr targets of the pharmacological agents that have been used to modulate pair bonding might also lead to insights into the underlying neural circuits and molecular signaling pathways. In any event, our studies rigorously demonstrate that, in three different loss of function mutations generated and tested independently, Oxtr signaling is not genetically required in male and female prairie voles for either pair bonding or multiple aspects of parenting.
Female mice null for Oxtr show a complete failure of milk let-down and nursing behavior such that none of their pups survives beyond the day of birth34,36,57. In contrast, many pups born to prairie vole mothers lacking Oxtr survive to weaning, albeit with reduced weight. Furthermore, both female and male Oxtr null voles display intact parental behavior, consistent with recent findings indicating preserved alloparenting behavior in male prairie voles lacking Oxtr38. It is possible that the phenotypic difference between mice and voles mutant for Oxtr is a consequence of rapid evolution of pathways that regulate nursing and parenting in mammals6,46,58. Alternatively, this difference in phenotypes could reflect the fact that the Oxtr mutant mice were maintained on an inbred background whereas our colony is consistently outbred to wild voles59,60. It will be interesting to determine whether Oxtr null mice on an outbred, wild background can also exhibit parental behaviors. If true, this would suggest that inbreeding has led to reliance on Oxtr signaling for parenting in mice rather than a fundamental difference in species-specific roles for Oxtr in parental displays.
Oxt signaling has also been implicated in affiliative displays in humans suggesting a conserved role for this neuropeptide hormone in these social behaviors across ~90 million years of evolution28. Based on observations in prairie voles and other mammals, including humans, clinical trials have used exogenous oxytocin or small molecule ligands to Oxtr to ameliorate the deficits in social attachment and cognition seen in multiple psychiatric conditions; these studies however have yielded mixed results25,61–65. Together with these clinical studies, our observation that Oxtr signaling is not required genetically for pair bond formation or parenting in prairie voles suggests that we require a more refined understanding of the molecular pathways underlying social attachment behaviors. New genetic models such as the Oxtr prairie vole mutants we have generated may better allow the rigorous dissection of the molecular and circuit mechanisms mediating attachment behavior and its disruption in disease. Whole-animal mutants better represent what may occur in patients with mutations associated with neuropsychiatric disorders, and molecular genetic approaches in prairie voles now permit us to test directly the impact of such genetic disruptions in the context of complex social and attachment behaviors.
Star Methods
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Devanand Manoli (devanand.manoli@ucsf.edu).
Materials availability
All unique/stable reagents generated in this study are available from the Lead Contact with a completed Materials Transfer Agreement.
Data and Code Availability
All data reported in this paper will be shared by the lead contact upon request.
Code
This paper does not report original code.
Experimental model and subject details
Animals
Subjects were laboratory-bred prairie voles (Microtus ochrogaster) which originated through systematic outbreeding of a wild stock captured near Champaign, Illinois. Prairie voles were bred, maintained, and tested at three distinct sites: University of California San Francisco, University of California Davis, and Stanford University. Sexually naïve male and female animals were group weaned at 21 ± 1 days and separated to pair-housing with either a same-sex sibling or an age-matched same-sex non-sibling (about half to each type of pairing). Voles were maintained under a 14:10 h light-dark cycle in clear plastic cages (45 × 25 × 15 cm) with bedding, nesting material (nestlet), and a PVC hiding tube. Rooms were maintained at approximately 20°C, and food and water were available ad libitum.
Breeding pairs were established between two heterozygotes or a homozygous Oxtr mutant male and heterozygous female partner from a breeding line maintained by the respective labs. Pups from Oxtr1 breeding pairs were weighed at weaning, and those from Oxtr4 and Oxtr5 pairs (mutant line and WT partner) were weighed at birth and at weaning date. Percent of pups surviving is calculated based on the number of pups alive at weaning at 21 days out of the total number of pups born in a litter, with two litters assessed per pair. Voles were assigned into experimental or control groups based on genotype when they reached 7–9 weeks of age at the start of testing. This study was carried out in accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals published by the National Research Council. The protocol was approved by the Institutional Animal Care and Use Committee at each respective institution.
Method details
Isolation of prairie vole embryos for gene targeting
Unlike mice, ovulation in prairie voles is behaviorally induced. All attempts to artificially super-ovulate prairie voles yielded poor results in our hands, differing from findings of a prior study38. We therefore developed a timed mating protocol where we placed 6–8 week old females with adult males for 12 hours and then placed a physical barrier between them and housed them for another 24 hours. We empirically determined that this induced estrus in all the females we paired, and the females all yield fertilized embryos approximately 18 hours after the removal of the barrier.
Mutagenesis and embryo manipulation for gene targeting
The current assembly of the prairie vole genome, which includes just over 70% annotation, reveals only one region for Oxtr that currently maps to scaffold sequence. Additionally, we screened a BAC library (CHORI BACPAC) that covers the entire vole genome for clones containing sequences similar to the prairie vole Oxtr coding sequence66. Of the 10 clones we obtained, all contain a single locus identical to the region scaffold that aligns to Oxtr in the current version of the genome, suggesting that prairie voles contain only one copy of Oxtr. We used these sequences and obtained the genomic sequence of exon 1 of Oxtr from 4 randomly chosen animals from our colony and identified polymorphisms that correspond to 3 synonymous substitutions. Based on these sequences, we designed 8 sgRNAs targeting exon 1, and adopted microinjection protocols commonly used to manipulate embryos from mice to test varying concentrations of the guides along with either 20 ng/ul or 100ng/ul of Cas9 mRNA based on studies in mice, rats, and other rodent species67,68. We cultured embryos to the blastocyst stage, with ~45% of embryos surviving following injection. We then harvested genomic DNA from individual blastocysts, and assayed for mutagenesis of Oxtr by Surveyor PCR, and sequencing of genomic PCR products69. These analyses revealed significant mosaicism in blastocysts, suggesting that mutagenesis in vole embryos occurred significantly later than the first division. Based on these studies, we identified 2 sgRNAs (sgRNA1: GAGCATCGCTGACCTGGTGGTGGC, sgRNA2: CAGCTGCTGTGGGACATCACCTTC) that reliably yielded detectable mutations in embryos and selected these for further use. We performed pronuclear injections of the 2 sgRNA guides, 2μl Cas9 protein (PNA BIO, INC CP02, 5μg/μl) and 2μl cas9 mRNA (TRILINK BIOTECHNOLOGIES, INC L-7206, 1μg/μl) and cultured the manipulated embryos to the blastocyst stage. We generated pseudopregnant recipient females using our timed mating protocol with vasectomized males and surgically implanted the manipulated embryos into their oviduct68. Using such an approach, we developed a protocol whereby ~10 embryos are transferred to each uterine horn to obtain 3–5 live born pups per female.
Isolation and Outcrossing of Oxtr Alleles
Tail samples were taken from the founders (G0s) and screened for mutations in the Oxtr locus. Based on the chimerism we observed in our embryo experiments, it was possible that the G0 carried a mutation in their germ cells but not their tails. We therefore paired all G0s with wild types and screened F1 tails to look for germ-line mutations. We were able to isolate F1s with Oxtr mutations and design PCR genotyping schemes to identify the mutation in subsequent generations, hence verifying germ-line transmission. Every F1 was outcrossed (paired with a wild type of the opposite sex, Oxtr4 and Oxtr5, >3 generations; Oxtr1, 7 generations) to remove off-target effects of CRISPR mutagenesis. As there is currently no completely assembled and annotated prairie vole genome to aid in sequencing-based analysis of off-target events, we established an outcrossing protocol based on standard genetic practices in mice and flies to isolate specific mutations. Assuming that the fraction of the original background that is unlinked is ½^N for N generations, we outcrossed Oxtr1 for 7 generations to the wild type strain.
Autoradiography
Wildtype and homozygous mutant siblings were sacrificed using CO2 and their brains were dissected and rapidly frozen on powdered dry ice before storing at −80ºC. 20μm sections were thaw mounted onto super mount frost plus slides. Slide mounted sections were thawed until dry and fixed for two minutes in 0.1% paraformaldehyde (0.1% paraformaldehyde in 0.1M PBS). Slides were rinsed 2×10 minutes in 50mM Tris pH 7.4, then incubated for 90 minutes at room temperature in a solution (50 mM Tris, 10 mM MgCl2, 0.1% BSA, 0.05% bacitracin, 50 pM radioligand) containing the radioactively labeled 125I-ornithine vasotocin analog vasotocin, d(CH2)5[Tyr(Me)2,Thr4,Orn8,(125I)Tyr9-NH2] (125I-OVTA, PerkinElmer, Inc.). Slides processed for non-specific binding were incubated with an additional 50 uM non-radioactive ligand [Thr4Gly7]-oxytocin (Bachem). All slides were washed 3×5 min in chilled Tris-MgCl2 (50mM Tris, 10mM MgCl2, pH 7.4), followed by a 30-minute soak in Tris-MgCl2 on ice. Slides were quickly dipped in cold, distilled water and then air dried and exposed to Kodak BioMax MR film (Kodak, Rochester, NE, USA) for 3 days and subsequently developed. To quantify ligand binding, we first defined anatomical centers using the Allen Mouse Brain Atlas. Acetylcholine esterase was stained using the enzyme’s conversion of thiocholine ester.
The 125I-OVTA binding was quantified directly (measured as optical binding density, OBD) from the film using a light box, a top mounted camera, and the MCID Core Digital Densitometry system (Cambridge, UK) according to methods previously described70. Values for OBD from a set of 1251 autoradiography standards (American Radiolabeled Chemicals, Inc., St. Louis, MO, USA) were loaded into MCID in order to generate a standard curve, from which 5 OBD values for each ROI were extrapolated (Oxtr4 and Oxtr5). Average OBD values were calculated for each ROI within each individual specimen, as well as for one background area where no binding was detected, which served as a measure of non-specific binding. This average background/non-specific binding was subtracted from the ROI measurements to yield normalized OBDs across specimens and correct for individual variation in non-specific binding across individuals. Autoradiography performed on Oxtr1 was calculated with respect to the background before adjusting the background to zero. Non-standard binding controls performed on Oxtr1 showed that there was no difference between signal observed in the mutant and background. The signal intensities were compared using the student’s T test across males and females of both wild type and mutant siblings of a given mutant strain.
RT- quantitative PCR
Adult WT and Oxtr mutant voles were deeply anesthetized by IP injection with 2.5% Avertin and euthanized by decapitation. The brain was quickly dissected out and sectioned into 500 μm coronal slices using a brain matrix mold (BrainTree Scientific) chilled on ice. Slices were floated in ice-cold phosphate buffered saline (PBS) and the paraventricular nucleus (PVN) was identified using anatomical landmarks and dissected using a Zeiss microscope. For Oxtr4, tissue was dissected and RNA purified from individual animals. For Oxtr5, tissue from 2–3 animals was pooled prior to RNA extraction to reduce variability across samples. Tissue was flash-frozen on dry ice and stored at −80°C until further processing. Total RNA was extracted using TRIzol according to manufacturer’s instructions and quantified using NanoDrop (Thermo Scientific). We used 5 μl total RNA to program a 20 μl reverse transcription reaction using the ProtoScript cDNA synthesis kit with random hexamer priming. We used 2μl of the RT reaction to run real-time quantitative PCR using FastStart SYBR Green Master Mix and the StepOnePlus™ Real-Time PCR System. To quantify mRNA levels we used a relative quantitation method wherein we constructed a standard curve for each gene using six serial 1:10 dilutions of cDNA pooled from all of our samples. We ran two technical replicates for each standard curve and biological sample and normalized relative Avp and Oxt levels to the housekeeping gene Gapdh after verifying that Gapdh mRNA level did not vary by genotype. The following oligonucleotide primers were used: Avp, TGCGTGTTTCCTGAGCC (forward) and ATGTTGGGTCCGAAGCAG (reverse); Oxt CTGCTACATCCAGAACTGTCC (forward) and AAACAGCCCAGCTCATCG (reverse); Gapdh, GGTAAAGTCATCCCAGAGCTG (forward) and CCTGCTTCACCACCTTCTTG (reverse). Before the RT-qPCR reaction we sequenced the Oxt and Avp PCR products to confirm the specificity of our primers.
Partner preference assay
The partner preference test23 was used to assess pair bond formation71. Subjects were cohoused with an opposite-sex, wildtype animal (partner). Analysis of partner preference between wild type siblings from the Oxtr1 background and wild type animals from this colony showed no difference on the partner preference test. Pairing of Oxtr1 was followed by a timed mating protocol for synchronization of estrous in which paired animals are separated by a clear, plastic barrier 18 hours following pairing. The barrier was left in place for 24 hours and then removed allowing the subject free access to the partner animal thereafter. Regardless of which laboratory performed testing, all animals were allowed to cohabitate for one week prior to behavioral testing. As Oxtr1 was generated and maintained independently from Oxtr4 and Oxtr5, we used two different, well-established apparatuses for testing partner preference in the Oxtr1 line vs Oxtr4 and Oxtr5 based on the apparatus commonly used by the generating laboratory. In the branched design used to test Oxtr4 and Oxtr5, the subject, their partner, and an unfamiliar opposite-sex conspecific (stranger) were placed in an apparatus made of three small polycarbonate cages (27×16×16 cm) joined by clear Plexiglas tubes. Strangers were selected to be of similar age and size as the partner animal. Both the partner and the stranger were tethered in two separate, end cages, and the test subject was placed, untethered in a central cage. The test subject was free to access any of the three cages over a period of 3 hours. All behaviors were video recorded from an approach centered on the two end cages. In the apparatus design used to assay Oxtr1 animals, the subject, partner, and an opposite sex stranger are placed in a linear apparatus with open top and 10 × 32 in walls. The partner and stranger animals are tethered on either end of the three-chamber arena and the subject allowed free access, again over a period of 3 hours. Behaviors are recorded from a top view camera capturing the entire apparatus. Videos were scored post-test by validated scorers blind to condition. Observed behaviors included location (i.e., duration of time in partner, stranger, and neutral cages), duration of stationary huddling or >50% side-by-side contact between the partner and stranger animals, and frequency of aggressive behavior (i.e., lunges).
Parental behavior
A total of 80 individuals, distributed across sex (male or female), strain (Oxtr4 or Oxtr5) and genotype (wild-type or mutant) were paired with a wild type colony animal of the other sex. Following partner preference testing, pairs were left in large polycarbonate cages (44×22×16cm) with sterilized cotton for nesting material. All litters from 21 breeder pairs were observed on two separate days in the morning (08:00–11:00) and two separate days in the evening (15:00–18:00) for a total of four, 20-minute focal observations during the neonatal period (P1-P3). Parenting behavior was observed according to methods previously outlined45. Briefly, behaviors recorded included maternal and paternal huddling, non-huddling contact, licking/grooming, retrievals, hunching, nest building, and autogrooming. The time spent in behaviors that involved direct contact with pups (all those listed besides nest building) were combined for the total duration of pup-directed contact. In the mothers, lateral, active, and neutral nursing postures were scored. Active nursing is defined as pups attached while mother is locomoting around the cage, lateral nursing as mother laying on side with pups laying in front, and neutral nursing as standing over pups in a relaxed position without locomotion. The time in all of these nursing positions were combined for analysis of nursing duration. Eight individuals were not observed because of their death or the death of their partner (2 sires, 6 dams). Notably, of the two sires, one was wild type and one was mutant; and of the six dams, all six were mutant, split between Oxtr4 (n = 5) and Oxtr5 (n = 1). Increased rates of culling or natural death may have been due to reproductive complications in which oxytocin plays a significant role (pregnancy, birthing, and lactation). All observations were made in the home cage. Each parent was observed and left undisturbed during each 20-minute observation. The duration (seconds) of each behavior was recorded, and durations of all direct parenting behaviors (e.g. licking/grooming, huddling, etc.) were recorded within each observation period. All behaviors were live recorded using behavioral software (www.behaviortracker.com) or ScoreVideo (matlab).
Data analyses and statistics
The number of animals used was based on previous studies in the field by our group and others, combined with a power analysis. Assumptions of independence, homogeneity of variance, and normality were considered with multiple tests. The assumption of homogeneity of variance was considered through visual inspection of box plots and plots of residuals, and were further tested for homogeneity of variance using Bartlett’s test of homogeneity of variances. Partner preference behaviors in all three mutant backgrounds were considered with paired two-tailed T-test for parametric data or Wilcoxon signed-rank test for non-parametric. Differential time between partner and stranger was calculated as partner huddle duration – stranger huddle duration. The level of statistical significance for each test was set at p = 0.05. Outliers were detected using a combination of z-score (values +/− 3 or greater were considered outliers) and plots of original and log-transformed data. One WT Oxtr5 male (duration with stranger = 3287 seconds), one WT Oxtr1 male (duration with stranger = 6928 seconds) and one WT Oxtr1 female (duration with stranger = 7356 seconds) were excluded as outliers due to durations in partner preference assay. Analyses were completed in R (version 1.1.423) and GraphPad Prism (version 7.03).
Supplementary Material
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Chemicals, peptides, and recombinant proteins | ||
| Cas 9 protein | PNA BIO, Inc | CP02 |
| Cas9 mRNA | TRILINK BIOTECHNOLOGIES, INC | L-7206 |
| d(CH2)5[Tyr(Me)2,Thr4,Orn8,(125I)Tyr9-NH2] (125I-OVTA) | PerkinElmer, Inc. | NEX254010UC |
| [Thr4Gly7]-oxytocin | Bachem | 4013837.0005 |
| Experimental models: Organisms/strains | ||
| Prairie vole: Oxtr1−/− | This paper | |
| Prairie vole: Oxtr4−/− | This paper | |
| Prairie vole: Oxtr5−/− | This paper | |
| Oligonucleotides | ||
| sgRNA1: GAGCATCGCTGACCTGGTGGTGGC | This paper | |
| sgRNA2: CAGCTGCTGTGGGACATCACCTTC | This paper | |
| Avp forward: TGCGTGTTTCCTGAGCC | This paper | |
| Avp reverse: ATGTTGGGTCCGAAGCAG | This paper | |
| Oxt forward: CTGCTACATCCAGAACTGTCC | This paper | |
| Oxt reverse: AAACAGCCCAGCTCATCG | This paper | |
| Gapdh forward: GGTAAAGTCATCCCAGAGCTG | This paper | |
| Gapdh reverse: CCTGCTTCACCACCTTCTTG | This paper | |
| Software and algorithms | ||
| R (version 1.1.423) | R Core | https://www.R-project.org/ |
| GraphPad Prism (version 7.03) | GraphPad Software | https://graphpad.com/scientificsoftware/prism/ |
| BehaviorTracker (version 1.5) | BehaviorTracker | www.behaviortracker.com |
| MATLAB | MathWorks |
https://www.mathworks.com/products.html RRID: SCR_001622 |
Highlights:
Prairie voles lacking oxytocin receptor (Oxtr) generated with CRISPR-targeting.
Oxtr−/−voles form pair bonds or social attachments.
Oxtr−/− voles show parental behavior.
Oxtr−/− females nurse many of their pups to weaning.
Acknowledgments
The authors would like to thank members of the Bales lab for assistance and advice in animal husbandry and the Bales, Shah, and Manoli labs for helpful discussion and comments on the manuscript. In addition, we would like to thank Jake Freimer, Ron Parchem, and Archana Shenoy for advice regarding embryo harvesting and manipulation, and the Doudna, Jaenisch, and Zhang labs for advice regarding CRISPR mutagenesis.
Funding
National Institutes of Health grant R01MH123513 (DM)
National Science Foundation grant 1556974 (DM)
Burroughs Wellcome Fund 1015667 (DM)
Whitehall Foundation grant 2018-08-83 (DM)
National Institutes of Health grant R01MH108319 (KLB, NMS)
National Institutes of Health Director’s Pioneer award DP1MH099900 (NMS)
National Institutes of Health grant R25MH060482 (KMB)
AP Giannini Foundation Fellowship (KMB)
Larry L. Hillblom Foundation Fellowship (KMB)
Human Frontiers Science Program (SP)
Inclusion and Diversity
We worked to ensure sex balance in the selection of non-human subjects.
One or more of the authors of this paper self-identifies as an underrepresented ethnic minority in their field of research or within their geographical location. One or more of the authors of this paper self-identifies as a gender minority in their field of research. One or more of the authors of this paper self-identifies as a member of the LGBTQIA+ community. One or more of the authors of this paper self-identifies as living with a disability. One or more of the authors of this paper received support from a program designed to increase minority representation in their field of research. While citing references scientifically relevant for this work, we also actively worked to promote gender balance in our reference list.
Footnotes
Declaration of interests
Dr. Nirao Shah is a member of the advisory board for Neuron. The authors declare no other competing interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Kleiman DG (1977). Monogamy in Mammals. The Quarterly Review of Biology 52, 39–69. 10.1086/409721. [DOI] [PubMed] [Google Scholar]
- 2.Robinson KJ, Bosch OJ, Levkowitz G, Busch KE, Jarman AP, and Ludwig M. (2019). Social creatures: Model animal systems for studying the neuroendocrine mechanisms of social behaviour. J Neuroendocrinol 31. 10.1111/jne.12807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pfennig DW, and Sherman PW (1995). Kin Recognition. Sci Am 272, 98–103. 10.1038/scientificamerican0695-98. [DOI] [PubMed] [Google Scholar]
- 4.Insel T. (2000). Neuropeptides and the evolution of social behavior. Current Opinion in Neurobiology 10, 784–789. 10.1016/S0959-4388(00)00146-X. [DOI] [PubMed] [Google Scholar]
- 5.Maruska KP (2015). Social Transitions Cause Rapid Behavioral and Neuroendocrine Changes. Integr. Comp. Biol 55, 294–306. 10.1093/icb/icv057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Staes N, Guevara EE, Helsen P, Eens M, and Stevens JMG (2021). The Pan social brain: An evolutionary history of neurochemical receptor genes and their potential impact on sociocognitive differences. Journal of Human Evolution 152, 102949. 10.1016/j.jhevol.2021.102949. [DOI] [PubMed] [Google Scholar]
- 7.Wilson EO, and Hölldobler B. (2005). Eusociality: Origin and consequences. Proc. Natl. Acad. Sci. U.S.A 102, 13367–13371. 10.1073/pnas.0505858102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carter C, and Keverne E. (2002). The Neurobiology of Social Affiliation and Pair Bonding. In Hormones, Brain and Behavior (Elsevier), pp. 299–337. [Google Scholar]
- 9.Getz LL., Carter CS., and Gavish L. (1981). The mating system of the prairie vole, Microtus ochrogaster: Field and laboratory evidence for pair-bonding. Behav Ecol Sociobiol 8, 189–194. 10.1007/BF00299829. [DOI] [Google Scholar]
- 10.Donaldson ZR, and Young LJ (2008). Oxytocin, vasopressin, and the neurogenetics of sociality. Science 322, 900–904. 10.1126/science.1158668. [DOI] [PubMed] [Google Scholar]
- 11.Garrison JL, Macosko EZ, Bernstein S, Pokala N, Albrecht DR, and Bargmann CI (2012). Oxytocin/Vasopressin-Related Peptides Have an Ancient Role in Reproductive Behavior. Science 338, 540–543. 10.1126/science.1226201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Insel TR, Winslow JT, Wang ZX, Young L, and Hulihan TJ (1995). Oxytocin and the molecular basis of monogamy. Adv. Exp. Med. Biol 395, 227–234. [PubMed] [Google Scholar]
- 13.Insel TR, Winslow JT, Wang Z, and Young LJ (1998). Oxytocin, vasopressin, and the neuroendocrine basis of pair bond formation. Adv. Exp. Med. Biol 449, 215–224. [DOI] [PubMed] [Google Scholar]
- 14.Wagenaar DA, Hamilton MS, Huang T, Kristan WB, and French KA (2010). A Hormone-Activated Central Pattern Generator for Courtship. Current Biology 20, 487–495. 10.1016/j.cub.2010.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ophir AG, Gessel A, Zheng D-J, and Phelps SM (2012). Oxytocin receptor density is associated with male mating tactics and social monogamy. Hormones and behavior 61, 445–453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shapiro LE, and Insel TR (1992). Oxytocin receptor distribution reflects social organization in monogamous and polygamous voles. Ann. N. Y. Acad. Sci 652, 448–451. [DOI] [PubMed] [Google Scholar]
- 17.Winslow JT, Hastings N, Carter CS, Harbaugh CR, and Insel TR (1993). A role for central vasopressin in pair bonding in monogamous prairie voles. Nature 365, 545–548. 10.1038/365545a0. [DOI] [PubMed] [Google Scholar]
- 18.Klatt JD, and Goodson JL (2013). Oxytocin-like receptors mediate pair bonding in a socially monogamous songbird. Proc. R. Soc. B 280, 20122396. 10.1098/rspb.2012.2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nowicki JP, Pratchett MS, Walker SPW, Coker DJ, and O’Connell LA (2020). Gene expression correlates of social evolution in coral reef butterflyfishes. Proc. R. Soc. B 287, 20200239. 10.1098/rspb.2020.0239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.O’Connor CM., Marsh-Rollo SE., Aubin-Horth N., and Balshine S. (2016). Species-specific patterns of nonapeptide brain gene expression relative to pair-bonding behavior in grouping and non-grouping cichlids. Hormones and Behavior 80, 30–38. 10.1016/j.yhbeh.2015.10.015. [DOI] [PubMed] [Google Scholar]
- 21.Cho MM, DeVries AC, Williams JR, and Carter CS (1999). The effects of oxytocin and vasopressin on partner preferences in male and female prairie voles (Microtus ochrogaster). Behav. Neurosci 113, 1071–1079. [DOI] [PubMed] [Google Scholar]
- 22.Insel TR, and Hulihan TJ (1995). A gender-specific mechanism for pair bonding: oxytocin and partner preference formation in monogamous voles. Behav. Neurosci 109, 782–789. [DOI] [PubMed] [Google Scholar]
- 23.Williams JR, Catania KC, and Carter CS (1992). Development of partner preferences in female prairie voles (Microtus ochrogaster): the role of social and sexual experience. Hormones and behavior 26, 339–349. [DOI] [PubMed] [Google Scholar]
- 24.Wang Z, Hulihan TJ, and Insel TR (1997). Sexual and social experience is associated with different patterns of behavior and neural activation in male prairie voles. Brain Res 767, 321–332. [DOI] [PubMed] [Google Scholar]
- 25.Heinrichs M, von Dawans B, and Domes G. (2009). Oxytocin, vasopressin, and human social behavior. Front Neuroendocrinol 30, 548–557. 10.1016/j.yfrne.2009.05.005. [DOI] [PubMed] [Google Scholar]
- 26.Insel TR, Wang ZX, and Ferris CF (1994). Patterns of brain vasopressin receptor distribution associated with social organization in microtine rodents. J. Neurosci 14, 5381–5392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.King LB, Walum H, Inoue K, Eyrich NW, and Young LJ (2016). Variation in the oxytocin receptor gene predicts brain region–specific expression and social attachment. Biological psychiatry 80, 160–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Walum H, and Young LJ (2018). The neural mechanisms and circuitry of the pair bond. Nat Rev Neurosci 19, 643–654. 10.1038/s41583-018-0072-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Carter CS, Grippo AJ, Pournajafi-Nazarloo H, Ruscio MG, and Porges SW (2008). Oxytocin, vasopressin and sociality. Prog. Brain Res 170, 331–336. 10.1016/S0079-6123(08)00427-5. [DOI] [PubMed] [Google Scholar]
- 30.Keebaugh AC, and Young LJ (2011). Increasing oxytocin receptor expression in the nucleus accumbens of pre-pubertal female prairie voles enhances alloparental responsiveness and partner preference formation as adults. Hormones and Behavior 60, 498–504. 10.1016/j.yhbeh.2011.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Keebaugh AC, Barrett CE, Laprairie JL, Jenkins JJ, and Young LJ (2015). RNAi knockdown of oxytocin receptor in the nucleus accumbens inhibits social attachment and parental care in monogamous female prairie voles. Social neuroscience 10, 561–570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ophir AG., Sorochman G., Evans BL., and Prounis GS. (2013). Stability and Dynamics of Forebrain Vasopressin Receptor and Oxytocin Receptor During Pregnancy in Prairie Voles. J Neuroendocrinol 25, 719–728. 10.1111/jne.12049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lee H-J, Caldwell HK, Macbeth AH, Tolu SG, and Young WS (2008). A Conditional Knockout Mouse Line of the Oxytocin Receptor. Endocrinology 149, 3256–3263. 10.1210/en.2007-1710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Young III WS, Shepard E, Amico J, Hennighausen L, Wagner K-U, LaMarca ME, McKinney C, and Ginns EI (1996). Deficiency in Mouse Oxytocin Prevents Milk Ejection,but not Fertility or Parturition. Journal of Neuroendocrinology 8, 847–853. 10.1046/j.1365-2826.1996.05266.x. [DOI] [PubMed] [Google Scholar]
- 35.Nishimori K, Young LJ, Guo Q, Wang Z, Insel TR, and Matzuk MM (1996). Oxytocin is required for nursing but is not essential for parturition or reproductive behavior. Proc. Natl. Acad. Sci. U.S.A 93, 11699–11704. 10.1073/pnas.93.21.11699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Takayanagi Y, Yoshida M, Bielsky IF, Ross HE, Kawamata M, Onaka T, Yanagisawa T, Kimura T, Matzuk MM, Young LJ, et al. (2005). Pervasive social deficits, but normal parturition, in oxytocin receptor-deficient mice. PNAS 102, 16096–16101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Marlin BJ, Mitre M, D’amour JA, Chao MV, and Froemke RC (2015). Oxytocin enables maternal behaviour by balancing cortical inhibition. Nature 520, 499–504. 10.1038/nature14402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Horie K, Inoue K, Suzuki S, Adachi S, Yada S, Hirayama T, Hidema S, Young LJ, and Nishimori K. (2019). Oxytocin receptor knockout prairie voles generated by CRISPR/Cas9 editing show reduced preference for social novelty and exaggerated repetitive behaviors. Hormones and Behavior 111, 60–69. 10.1016/j.yhbeh.2018.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wilkie TM, Brinster RL, and Palmiter RD (1986). Germline and somatic mosaicism in transgenic mice. Developmental Biology 118, 9–18. 10.1016/0012-1606(86)90068-0. [DOI] [PubMed] [Google Scholar]
- 40.Insel TR, Preston S, and Winslow JT (1995). Mating in the monogamous male: Behavioral consequences. Physiology & Behavior 57, 615–627. 10.1016/0031-9384(94)00362-9. [DOI] [PubMed] [Google Scholar]
- 41.Beery AK, and Shambaugh KL (2021). Comparative Assessment of Familiarity/Novelty Preferences in Rodents. Front. Behav. Neurosci 15, 648830. 10.3389/fnbeh.2021.648830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Beery AK, Christensen JD, Lee NS, and Blandino KL (2018). Specificity in Sociality: Mice and Prairie Voles Exhibit Different Patterns of Peer Affiliation. Frontiers in Behavioral Neuroscience 12, 50. 10.3389/fnbeh.2018.00050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bales KL, Lewis-Reese AD, Pfeifer LA, Kramer KM, and Carter CS (2007). Early experience affects the traits of monogamy in a sexually dimorphic manner. Dev Psychobiol 49, 335–342. 10.1002/dev.20216. [DOI] [PubMed] [Google Scholar]
- 44.Danoff JS., Wroblewski KL., Graves AJ., Quinn GC., Perkeybile AM., Kenkel WM., Lillard TS., Parikh HI., Golino HF., Gregory SG., et al. (2021). Genetic, epigenetic, and environmental factors controlling oxytocin receptor gene expression. Clin Epigenet 13, 23. 10.1186/s13148-021-01017-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Perkeybile AM, Griffin LL, and Bales KL (2013). Natural variation in early parental care correlates with social behaviors in adolescent prairie voles (Microtus ochrogaster). Front. Behav. Neurosci 7. 10.3389/fnbeh.2013.00021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Feldman R, Monakhov M, Pratt M, and Ebstein RP (2016). Oxytocin Pathway Genes: Evolutionary Ancient System Impacting on Human Affiliation, Sociality, and Psychopathology. Biological Psychiatry 79, 174–184. 10.1016/j.biopsych.2015.08.008. [DOI] [PubMed] [Google Scholar]
- 47.Bales KL, Kim AJ, Lewis-Reese AD, and Sue Carter C. (2004). Both oxytocin and vasopressin may influence alloparental behavior in male prairie voles. Horm Behav 45, 354–361. 10.1016/j.yhbeh.2004.01.004. [DOI] [PubMed] [Google Scholar]
- 48.Bales KL, Plotsky PM, Young LJ, Lim MM, Grotte N, Ferrer E, and Carter CS (2007). Neonatal oxytocin manipulations have long-lasting, sexually dimorphic effects on vasopressin receptors. Neuroscience 144, 38–45. 10.1016/j.neuroscience.2006.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chini B, and Manning M. (2007). Agonist selectivity in the oxytocin/vasopressin receptor family: new insights and challenges. Biochemical Society Transactions 35, 737–741. 10.1042/BST0350737. [DOI] [PubMed] [Google Scholar]
- 50.El-Brolosy MA, and Stainier DYR (2017). Genetic compensation: A phenomenon in search of mechanisms. PLoS Genet 13, e1006780. 10.1371/journal.pgen.1006780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Teng X, Dayhoff-Brannigan M, Cheng W-C, Gilbert CE, Sing CN, Diny NL, Wheelan SJ, Dunham MJ, Boeke JD, Pineda FJ, et al. (2013). Genome-wide Consequences of Deleting Any Single Gene. Molecular Cell 52, 485–494. 10.1016/j.molcel.2013.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Taylor SA, Salo AL, and Dewsbury DA (1992). Estrus induction in four species of voles (Microtus). Journal of Comparative Psychology 106, 366–373. 10.1037/0735-7036.106.4.366. [DOI] [PubMed] [Google Scholar]
- 53.Gunnet JW, and Freeman ME (1983). The Mating-induced Release of Prolactin: A Unique Neuroendrocine Response*. Endocrine Reviews 4, 44–61. 10.1210/edrv-4-1-44. [DOI] [PubMed] [Google Scholar]
- 54.Bronson FH, and Desjardins C. (1982). Endocrine Responses to Sexual Arousal in Male Mice. Endocrinology 111, 1286–1291. 10.1210/endo-111-4-1286. [DOI] [PubMed] [Google Scholar]
- 55.Anacker AMJ, Christensen JD, LaFlamme EM, Grunberg DM, and Beery AK (2016). Septal oxytocin administration impairs peer affiliation via V1a receptors in female meadow voles. Psychoneuroendocrinology 68, 156–162. 10.1016/j.psyneuen.2016.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chini B, and Fanelli F. (2000). Molecular basis of ligand binding and receptor activation in the oxytocin and vasopressin receptor family. Experimental Physiology 85, 59s–66s. 10.1111/j.1469-445X.2000.tb00008.x. [DOI] [PubMed] [Google Scholar]
- 57.Wakerley JB, Dyball REJ, and Lincoln DW (1973). MILK EJECTION IN THE RAT: THE RESULT OF A SELECTIVE RELEASE OF OXYTOCIN. Journal of Endocrinology 57, 557–558. 10.1677/joe.0.0570557. [DOI] [PubMed] [Google Scholar]
- 58.Paré P., Paixão-Côrtes VR., Tovo-Rodrigues L., Vargas-Pinilla P., Viscardi LH., Salzano FM., Henkes LE., and Bortolini MC. (2016). Oxytocin and arginine vasopressin receptor evolution: implications for adaptive novelties in placental mammals. Genet. Mol. Biol 39, 646–657. 10.1590/1678-4685-gmb-2015-0323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Crawley JN, Belknap JK, Collins A, Crabbe JC, Frankel W, Henderson N, Hitzemann RJ, Maxson SC, Miner LL, Silva AJ, et al. (1997). Behavioral phenotypes of inbred mouse strains: implications and recommendations for molecular studies. Psychopharmacology 132, 107–124. 10.1007/s002130050327. [DOI] [PubMed] [Google Scholar]
- 60.Simpson EM, Linder CC, Sargent EE, Davisson MT, Mobraaten LE, and Sharp JJ (1997). Genetic variation among 129 substrains and its importance for targeted mutagenesis in mice. Nat Genet 16, 19–27. 10.1038/ng0597-19. [DOI] [PubMed] [Google Scholar]
- 61.Di Simplicio M, and Harmer CJ (2016). Oxytocin and emotion processing. J. Psychopharmacol. (Oxford) 30, 1156–1159. 10.1177/0269881116641872. [DOI] [PubMed] [Google Scholar]
- 62.Feifel D, Macdonald K, Nguyen A, Cobb P, Warlan H, Galangue B, Minassian A, Becker O, Cooper J, Perry W, et al. (2010). Adjunctive intranasal oxytocin reduces symptoms in schizophrenia patients. Biol. Psychiatry 68, 678–680. 10.1016/j.biopsych.2010.04.039. [DOI] [PubMed] [Google Scholar]
- 63.Green JJ, and Hollander E. (2010). Autism and oxytocin: new developments in translational approaches to therapeutics. Neurotherapeutics 7, 250–257. 10.1016/j.nurt.2010.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Rubin LH, Carter CS, Drogos L, Pournajafi-Nazarloo H, Sweeney JA, and Maki PM (2010). Peripheral oxytocin is associated with reduced symptom severity in schizophrenia. Schizophr. Res 124, 13–21. 10.1016/j.schres.2010.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sikich L, Kolevzon A, King BH, McDougle CJ, Sanders KB, Kim S-J, Spanos M, Chandrasekhar T, Trelles MDP, Rockhill CM, et al. (2021). Intranasal Oxytocin in Children and Adolescents with Autism Spectrum Disorder. N Engl J Med 385, 1462–1473. 10.1056/NEJMoa2103583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.McGraw LA, Davis JK, Lowman JJ, ten Hallers BFH, Koriabine M, Young LJ, de Jong PJ, Rudd MK, and Thomas JW (2010). Development of genomic resources for the prairie vole (Microtus ochrogaster): construction of a BAC library and vole-mouse comparative cytogenetic map. BMC Genomics 11, 70. 10.1186/1471-2164-11-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yang H., Wang H., Shivalila CS., Cheng AW., Shi L., and Jaenisch R. (2013). One-step generation of mice carrying reporter and conditional alleles by CRISPR/Cas-mediated genome engineering. Cell 154, 1370–1379. 10.1016/j.cell.2013.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Behringer R. (2014). Manipulating the mouse embryo: a laboratory manual Fourth edition. (Cold Spring Harbor Laboratory Press; ). [Google Scholar]
- 69.Ran FA, Hsu PD, Lin C-Y, Gootenberg JS, Konermann S, Trevino AE, Scott DA, Inoue A, Matoba S, Zhang Y, et al. (2013). Double nicking by RNA-guided CRISPR Cas9 for enhanced genome editing specificity. Cell 154, 1380–1389. 10.1016/j.cell.2013.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Rogers FD, Freeman SM, Anderson M, Palumbo MC, and Bales KL (2021). Compositional variation in early‐life parenting structures alters oxytocin and vasopressin 1a receptor development in prairie voles ( Microtus ochrogaster ). J Neuroendocrinol 33. 10.1111/jne.13001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ahern TH, and Young LJ (2009). The impact of early life family structure on adult social attachment, alloparental behavior, and the neuropeptide systems regulating affiliative behaviors in the monogamous prairie vole (Microtus ochrogaster). Frontiers in behavioral neuroscience 3, 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data reported in this paper will be shared by the lead contact upon request.