PURPOSE
Novel biomarkers are needed to differentiate outcomes in intermediate-risk rhabdomyosarcoma (IR RMS). We sought to evaluate strategies for identifying circulating tumor DNA (ctDNA) in IR RMS and to determine whether ctDNA detection before therapy is associated with outcome.
PATIENTS AND METHODS
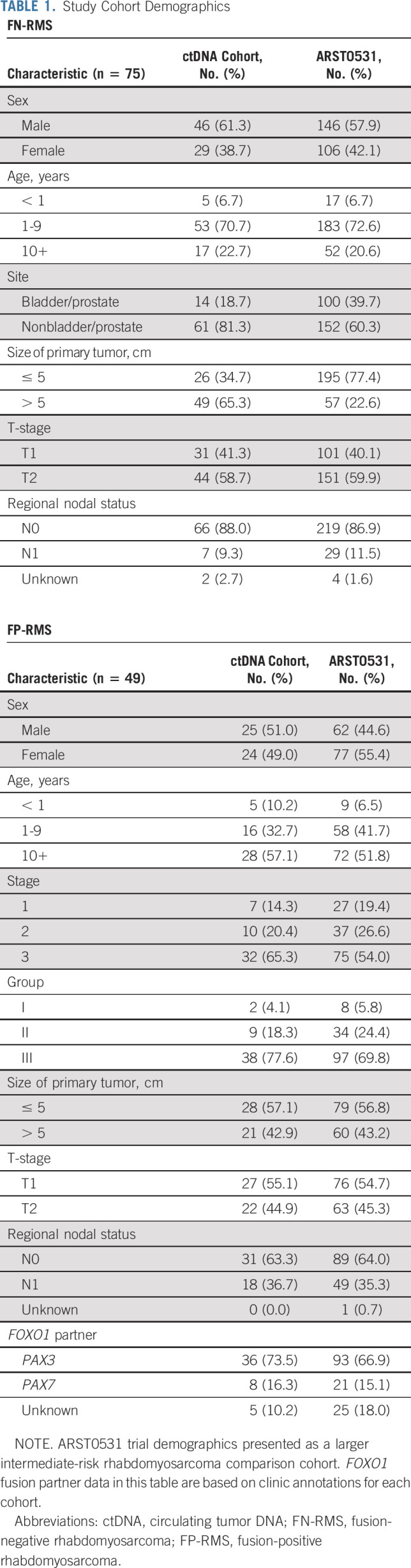
Pretreatment serum and tumor samples were available from 124 patients with newly diagnosed IR RMS from the Children's Oncology Group biorepository, including 75 patients with fusion-negative rhabdomyosarcoma (FN-RMS) and 49 with fusion-positive rhabdomyosarcoma (FP-RMS) disease. We used ultralow passage whole-genome sequencing to detect copy number alterations and a new custom sequencing assay, Rhabdo-Seq, to detect rearrangements and single-nucleotide variants.
RESULTS
We found that ultralow passage whole-genome sequencing was a method applicable to ctDNA detection in all patients with FN-RMS and that ctDNA was detectable in 13 of 75 serum samples (17%). However, the use of Rhabdo-Seq in FN-RMS samples also identified single-nucleotide variants, such as MYOD1L122R, previously associated with prognosis. Identification of pathognomonic translocations between PAX3 or PAX7 and FOXO1 by Rhabdo-Seq was the best method for measuring ctDNA in FP-RMS and detected ctDNA in 27 of 49 cases (55%). Patients with FN-RMS with detectable ctDNA at diagnosis had significantly worse outcomes than patients without detectable ctDNA (event-free survival, 33.3% v 68.9%; P = .0028; overall survival, 33.3% v 83.2%; P < .0001) as did patients with FP-RMS (event-free survival, 37% v 70%; P = .045; overall survival, 39.2% v 75%; P = .023). In multivariable analysis, ctDNA was independently associated with worse prognosis in FN-RMS but not in the smaller FP-RMS cohort.
CONCLUSION
Our study demonstrates that baseline ctDNA detection is feasible and is prognostic in IR RMS.
INTRODUCTION
Rhabdomyosarcoma (RMS) is the most common soft tissue sarcoma in children and adolescents with approximately 350 new cases per year in the United States.1 RMS is subdivided into two genetic subtypes. Fusion-positive RMS (FP-RMS) is characterized by a somatic translocation between the FOXO1 gene and either the PAX3 or PAX7 gene (PAX/FOXO1). Fusion-negative RMS (FN-RMS) is characterized by aneuploidy and recurrent single-nucleotide variants (SNVs) often in RAS pathway genes.2
CONTEXT
Key Objective
Is circulating tumor DNA (ctDNA) detectable in the blood of patients with intermediate-risk rhabdomyosarcoma (RMS) and is ctDNA detection associated with outcome?
Knowledge Generated
In this retrospective cohort, detection of ctDNA was feasible and relied on the identification of known somatic variants, such as aneuploidy and translocations, that define the genetic subtypes of RMS. Detection of ctDNA was significantly associated with a shorter event-free and overall survival, refining prognosis for patients with intermediate-risk RMS. Multivariable analysis demonstrated that ctDNA was independently associated with outcome in patients with fusion-negative RMS but did not reach significance in fusion-positive RMS.
Relevance (S. Bhatia)
ctDNA could serve as a prognostic tool and could inform future risk-stratified treatment in patients with intermediate risk RMS, when confirmed in prospective trials.*
*Relevance section written by JCO Associate Editor Smita Bhatia, MD, MPH, FASCO.
Clinical and histologic features are used for risk stratification and treatment selection in RMS. Intermediate-risk (IR) disease is associated with a 3-year event-free survival (EFS) ranging from 50% to 75% and accounts for the largest portion of newly diagnosed patients.3 Optimal treatment selection at diagnosis is important in RMS as survival after relapse is unusual. Additional biomarkers of prognosis would more precisely identify patients who will be cured with standard IR RMS treatment and patients who should enroll on clinical trials for high-risk disease.
Circulating tumor DNA (ctDNA) has recently emerged as a new prognostic biomarker in pediatric and adult cancers, including other sarcomas.4-6 Next-generation sequencing (NGS) strategies for the detection of ctDNA allows for quantification of ctDNA levels and identification of somatic variants that may provide additional prognostic information.7-9 Previous work from our group and others has shown that detection, quantification, and profiling of ctDNA in the blood of patients with RMS are feasible.10-12 In this study, we apply two NGS approaches to serum samples collected through a Children's Oncology Group (COG) banking study of 124 patients with IR RMS. We determine which somatic features are most useful for identifying ctDNA, whether molecular prognostic features can be identified by ctDNA profiling, and whether the presence of detectable ctDNA is prognostic in IR RMS.
PATIENTS AND METHODS
Patient Selection
All patients were enrolled on the COG biology study D9902, had pretreatment serum banked, and were coenrolled on a COG IR RMS trial (D9803 or ARST0531) or met criteria for IR RMS as defined in ARST0531. Matched germline samples and tumor tissue were obtained from a subset of cases. All patients signed written informed consent for D9902 at the time of enrollment. Separate approval for this retrospective use of patient samples and clinical data was obtained from the COG and the Dana-Farber/Harvard Cancer Center institutional review board. Additional details are provided in the Data Supplement (online only).
Design of an RMS-Specific Hybrid Capture Assay
We used two methods to profile tumor and serum samples. Copy number alterations (CNAs) were detected using ultralow passage whole-genome sequencing (ULP-WGS) as previously described (Data Supplement).13 To identify translocations and SNVs, we designed an RMS-specific hybrid capture assay. This Rhabdo-Seq panel targets intronic regions of FOXO1, PAX3, PAX7, VGLL2, CITED2, NCOA1, and NCOA2, which are translocated in RMS (Data Supplement).10 In the same hybrid capture panel, we targeted coding regions of 24 genes recurrently mutated in RMS (Data Supplement).14 Sequencing and analytical methods are presented in the Data Supplement.
RESULTS
Patients
Our study included pretreatment diagnostic serum samples from 124 patients with IR RMS, including 75 with FN-RMS and 49 with FP-RMS. The clinical characteristics of our analytical cohort were similar to those of patients treated on the prior IR RMS COG trial, ARST0531 (Table 1).15 Matched tumor tissue and germline DNA were available for 69 and 67 patients with FN-RMS, respectively. Matched tumor tissue was available for 35 patients with FP-RMS. Among the FN-RMS, histologies included nine botryoid, two spindle cell, 61 embryonal, and three for whom histology was not specified.
TABLE 1.
Study Cohort Demographics
Rhabdo-Seq for Tumor and ctDNA Profiling
To validate the performance of Rhabdo-Seq, we first sequenced cell lines and tumors from patients with FP-RMS. We identified the known complex PAX3/FOXO1 rearrangement in Rh4 and PAX7/FOXO1 in CW9019 cell lines. We also performed a serial dilution of DNA extracted from Rh4 into normal DNA to determine the limit of detection (LOD). We found that the fusion could be detected when Rh4 DNA comprised as little as 0.4% of the total DNA sample (Data Supplement). We detected the translocation and patient-specific breakpoints in 28 of 30 tumors with Rhabdo-Seq data. Two patients without an identifiable translocation were reported to have a FOXO1 fusion by fluorescence in situ hybridization, but we were unable to confirm these primary clinical data. We also analyzed Rhabdo-Seq data from 57 FN-RMS tumors and, when available, matched normal DNA. SNVs were detected in genes frequently mutated in RMS at rates similar to those reported in previous studies (Data Supplement).14
ctDNA Detection in FN-RMS
To determine which cases of FN-RMS had detectable CNAs and SNVs, we profiled available tumor tissue from 69 patients by both ULP-WGS and Rhabdo-Seq. Matched germline DNA was also profiled by Rhabdo-Seq. All 69 cases had detectable CNAs (Data Supplement), confirming that detection of ctDNA by ULP-WGS would be expected in all patients with ctDNA levels above the LOD (3% of total cell-free DNA; Fig 1A). There was sufficient sequencing library remaining for Rhabdo-Seq profiling of 57 tumors. We detected a somatic SNV in one or more targeted genes in only 40 of 57 tumor samples (70%), indicating that for a third of patients, we would not be able to detect ctDNA by Rhabdo-Seq regardless of ctDNA content (Fig 1B).
FIG 1.
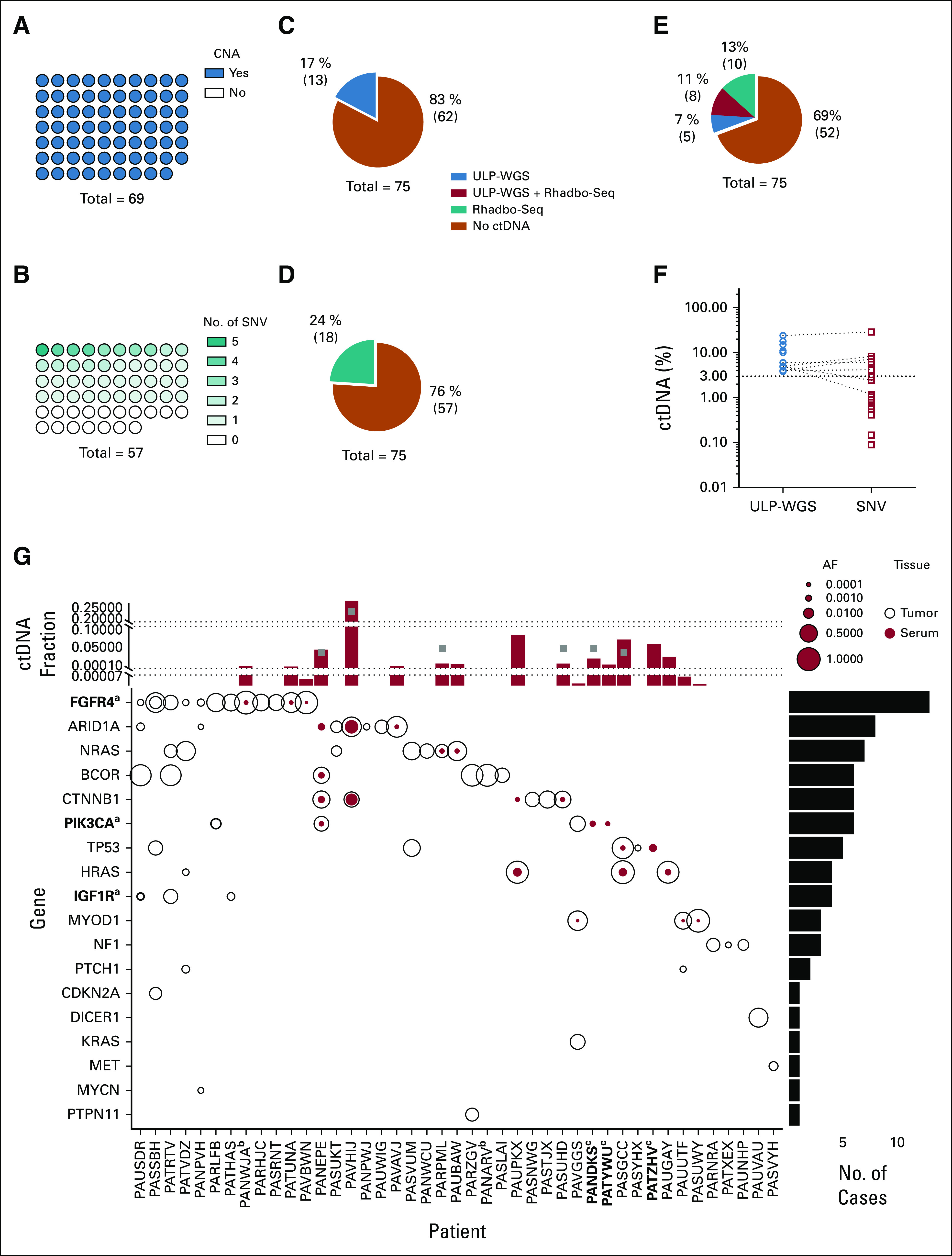
Characterization of FN-RMS genomics and ctDNA detection and quantification. (A) CNAs were identified in all 69 tumor samples by ULP-WGS. (B) SNVs were identified by the Rhabdo-Seq panel in 40 of 57 tumor samples with adequate sequencing coverage. ctDNA was detected in the serum in (C) 17% of the patients by ULP-WGS and (D) 24% of the patients by SNV identification with Rhabdo-Seq. (E) In total, 31% of patients had detectable ctDNA using a combination of ULP-WGS for CNA detection and Rhabdo-Seq panel for SNV detection. (F) Plotted are the percent ctDNA values for FN-RMS samples with detectable ctDNA by ULP-WGS (blue circles) and by Rhabdo-Seq (red circles). ctDNA content estimates are similar in cases where ctDNA is detected by both methods. However, the lack of SNVs in some cases results in ctDNA being unmeasurable by Rhabdo-Seq. In other cases, the lower limit of detection in Rhabdo-Seq allows for identification of very low content of ctDNA in some samples. (G) The plot depicts SNVs in genes (y-axis) targeted by the Rhabdo-Seq panel in patients with FN-RMS (x-axis) who have at least one variant found in either a tumor or serum sample. Open black circles represent variants found in the tumor, and solid red circles represent variants in serum. Circle size reflects allelic fraction of the identified variant. The histogram on top depicts ctDNA levels in serum samples from the patient (indicated on the x-axis) by ULP-WGS (gray squares) or SNV allelic fraction (red bar). When there are two SNVs, only the highest allelic fraction is represented by the red bar. The histogram is partitioned into three sections to help visualize values in cases with very low content of ctDNA. The histogram to the right represents the number of variants identified in the cohort. aGenes in bold indicate that a case had evidence of two mutations in that gene (FGFR4 in PASSBH, PIK3CA in PARLFB, and IGF1R in PAUSDR). bPatient IDs that are in bold indicate cases without available tumor data. cPatient IDs indicate cases without available germline DNA. The five cases with SNVs detected in the serum and not the tumor (PANEPE, PAUPKX, PANDKS, PATYWU, and PATZHV) were analyzed with a patient-matched normal DNA sample. AF, allelic fraction; CNA, copy number alteration; ctDNA, circulating tumor DNA; FN-RMS, fusion-negative rhabdomyosarcoma; SNV, single-nucleotide variant; ULP-WGS, ultralow passage whole-genome sequencing.
We applied both assays to serum samples from 75 patients with FN-RMS. ctDNA could be detected in 13 patients (17%) by ULP-WGS (Fig 1C and Data Supplement) and in 18 patients (24%) by Rhabdo-Seq (Fig 1D). In total, ctDNA was detected in 23 patients (31%) by either assay (Fig 1E). In 5 cases where ctDNA was detectable by ULP-WGS but negative by Rhabdo-Seq, no targeted SNVs had been detected in the tumor. In all cases for which ctDNA was detected by Rhabdo-Seq but not by ULP-WGS, ctDNA levels were below the LOD for ULP-WGS (Fig 1F). In cases with ctDNA detected by both methods, estimates of ctDNA content were concordant (Data Supplement)
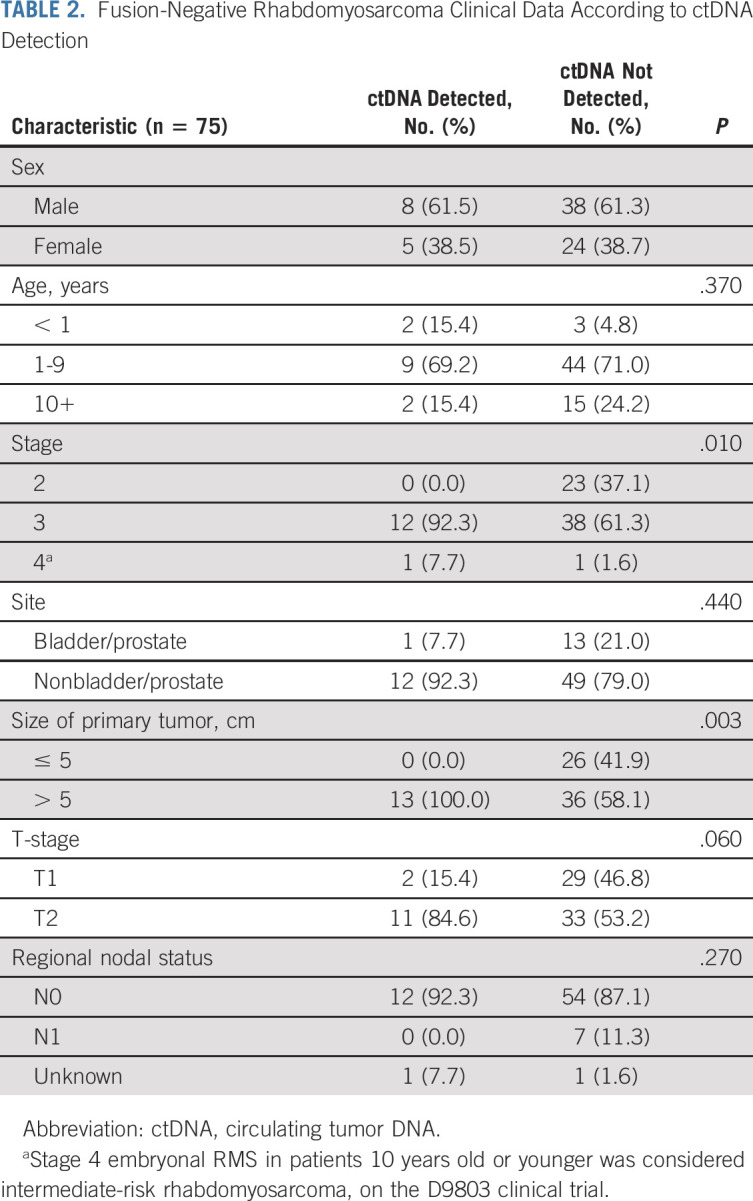
To determine whether ctDNA detection was associated with clinical features, we used the ULP-WGS data applicable to all FN-RMS cases. Detection of ctDNA was associated with disease stage and tumor size (Table 2). We also examined the specific SNVs detected in the tumor and serum samples by Rhabdo-Seq (Fig 1G). SNVs were always detected in serum in cases where SNVs were identified in tumor tissue and for which ctDNA levels were > 3% by ULP-WGS. In two patients, we detected a somatic SNV in the serum that was not detectable in the tumor (an ARID1A mutation in PANEPE and a CTNNB1 mutation in PAUPKX), suggesting that these variants were present in subclonal tumor cell populations. Moreover, we detected SNVs in three patients (PANDKS, PATYWU, and PATZHV) without available tumor data (Fig 1G). All five cases were analyzed with matched germline DNA to verify that these events were somatic. Three patients with MYOD1L122R mutations in the tumor had ctDNA in the serum detectable by identification of the MYOD1L122R variant by Rhabdo-Seq but below the LOD for ULP-WGS.
TABLE 2.
Fusion-Negative Rhabdomyosarcoma Clinical Data According to ctDNA Detection
ctDNA Detection in FP-RMS
FP-RMS tumors are characterized by PAX/FOXO1 translocations, whereas CNAs are present in only a portion of cases.2 Tumors from 35 FP-RMS cases were profiled with ULP-WGS and Rhabdo-Seq. ULP-WGS detected segmental CNAs in 25 of 35 tumors (71%; Fig 2A). There was sufficient sequencing library remaining for Rhabdo-Seq from 30 tumors, which detected a PAX/FOXO1 translocation in 28 samples (93%; Fig 2B). All translocations had a unique patient-specific genomic breakpoint (Data Supplement).10 Two samples did not have a detectable translocation despite adequate sequencing coverage and evidence of tumor content by ULP-WGS, making translocation detection impossible in liquid biopsies.
FIG 2.
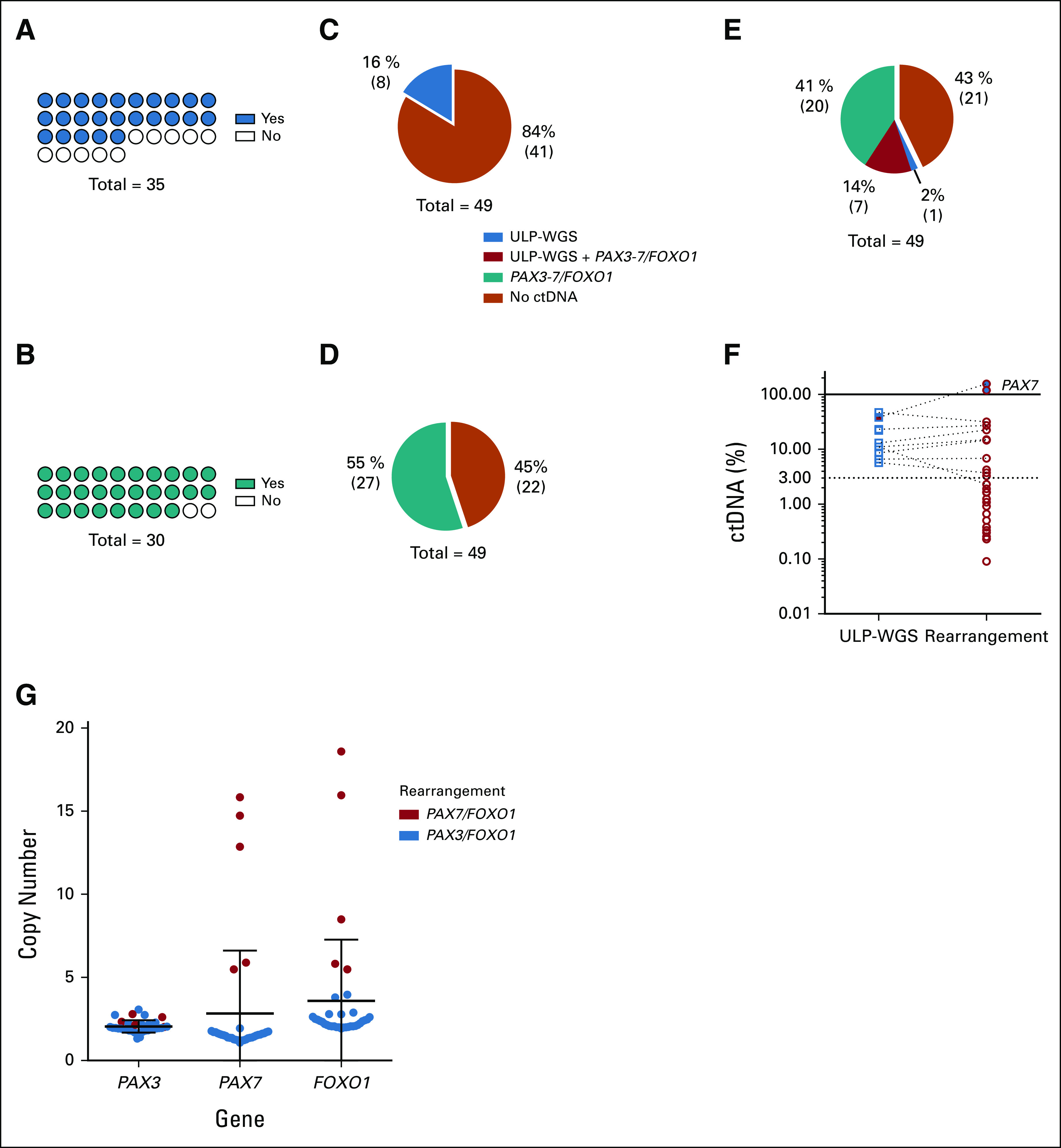
Characterization of fusion-positive rhabdomyosarcoma genomics and ctDNA detection and quantification. (A) CNAs were identified in 25 of 35 samples (71%) by ULP-WGS. (B) Translocations (rearrangement) were detected in 28 of 30 samples (93%) with sufficient sequencing library available for sequencing by Rhabdo-Seq. Of 35 tumors, five did not have sufficient DNA material quality for Rhabdo-Seq panel sequencing. (C) Sixteen percent of serum samples had detectable ctDNA by ULP-WGS. (D) ctDNA was detected by identifying the PAX/FOXO1 translocation in 55% of serum samples. (E) In total, 57% of patients had detectable ctDNA using both ULP-WGS for CNA detection and Rhabdo-Seq panel for gene rearrangement detection. (F) Plotted are the percent ctDNA values for fusion-negative rhabdomyosarcoma samples with detectable ctDNA by ULP-WGS (blue circles) and by Rhabdo-Seq (red circles). ctDNA content estimates are similar in most cases where ctDNA is detected by both methods. However, the lack of aneuploidy in some cases results in ctDNA being unmeasurable by ULP-WGS. (G) Absolute copy numbers of PAX3, PAX7, and FOXO1 locus were inferred from the gene locus using ULP-WGS data from tumor samples. In all the PAX7/FOXO1 tumors available, both partner genes were amplified from 5 to 18 times explaining the overestimate of ctDNA content in three cases with detectable ctDNA by PAX7/FOXO1 detection. CNA, copy number alteration; ctDNA, circulating tumor DNA; ULP-WGS, ultralow passage whole-genome sequencing.
We next profiled cell-free DNA with ULP-WGS and Rhabdo-Seq from 49 FP-RMS serum samples. We detected ctDNA with ULP-WGS in eight (16%) samples (Fig 2C) and with Rhabdo-Seq in 27 (55%; Fig 2D). The translocation breakpoint was identical in all patient-matched tumor and serum samples. In total, we identified ctDNA in 28 (57%) FP-RMS cases (Fig 2E). Rhabdo-Seq (with a lower LOD) identified a translocation in all cases where ctDNA was detectable by ULP-WGS, except where no rearrangement was found in the tumor. ULP-WGS detected the presence of ctDNA only in cases where ctDNA was estimated to be 3% or greater by Rhabdo-Seq and for which the tumor sample demonstrated CNAs (Fig 2F). ctDNA detection by Rhabdo-Seq was associated with the disease group and local T2 disease (Table 3).
TABLE 3.
Fusion-Positive Rhabdomyosarcoma Clinical Data According to ctDNA Detection
Serum samples for these studies were collected before the development of optimized liquid biopsy collection procedures. Evidence of cellular DNA contamination was found in 22 of 49 FP-RMS serum samples (Data Supplement), preventing accurate estimation of ctDNA content. However, in two cases (PASFIN and PARITH), the ctDNA content was estimated to be > 100%, which is impossible. Trans-Seq analysis of these samples yielded the same results (Data Supplement).10 Both cases involved PAX7/FOXO1 translocations, known to be amplified in RMS.16,17 ULP-WGS data from PAX7/FOXO1 cases in our cohort demonstrated copy gains in both PAX7 and FOXO1 loci (Fig 2G), accounting for the overestimate of ctDNA content. For case PASFIN, the ctDNA content was recalculated to 14.5% when adjusting for the measured amplification from tumor data (tumor data from PARITH were not available). For another PAX7/FOXO1 case, PATGUL, ctDNA content estimates were adjusted from 0.7% to 0.1%. No other cases with PAX7/FOXO1 translocations had detectable ctDNA.
ctDNA Detection Before Therapy Is Prognostic in IR RMS
Previous studies in pediatric and adult sarcomas have shown that detection of ctDNA, before the start of therapy, is associated with a worse outcome.18 In IR RMS, we found that detection of ctDNA (using ULP-WGS for FN-RMS and translocation for FP-RMS) was associated with a significantly lower 5-year EFS (35.9%; 95% CI, 21.4 to 50.7) and overall survival (OS, 37.3%; 95% CI, 22.3 to 52.3) compared with patients without detectable ctDNA by ULP-WGS (EFS, 69.3%; 95% CI, 58 to 78.1%; P = .0001; OS, 81.2%; 95% CI, 70.7 to 88.2; P = .0001; Figs 3A and 3B). MYOD1L122R mutations are associated with extremely poor outcomes in FN-RMS, and patients found to have these mutations are receiving intensified therapy on the current COG ARST2032 trial.14 We expected this mutation to be a stronger biomarker of outcome than detection of ctDNA. In fact, although the three patients found to have MYOD1L122R mutations did not have detectable levels of ctDNA by ULP-WGS, outcomes were significantly worse than for the rest of the patients with FN-RMS (Data Supplement). After excluding these three cases a priori, detection of pretreatment ctDNA was associated with a 5-year EFS of 35.9% (95% CI, 21.4 to 50.7) compared with 83.2% (95% CI, 72.8 to 89.9) for patients without detectable ctDNA (P = .0001) and a 5-year OS of 33.3% (95% CI, 10.3 to 58.8) compared with 86% (95% CI, 73.9 to 2.7; P = .0001; Figs 3C and 3D). Univariable analysis also showed that local T2 disease (invasion) was associated with outcome, although with a smaller hazard ratio (HR) than ctDNA (Data Supplement). Multivariable analysis of the whole cohort, including patients with a MYOD1L122R mutation, showed that ctDNA was significantly associated with EFS (HR, 2.8; 95% CI, 1.6 to 5.1; P = .0005) and OS (HR, 3.9; 95% CI, 2.1 to 7.5; P < .0001) as was local T2 disease (EFS HR, 2.0; 95% CI, 1.1 to 3.8; P = .02; OS HR, 2.5; 95% CI, 1.2 to 5; P = .01) and MYOD1L122R mutation (EFS HR, 9.2; 95% CI, 2.6 to 32.3; P = .0005; OS HR, 5.2; 95% CI, 1.1 to 23.5; P = .03). These associations were confirmed using additional methods for multivariable modeling (Data Supplement).
FIG 3.
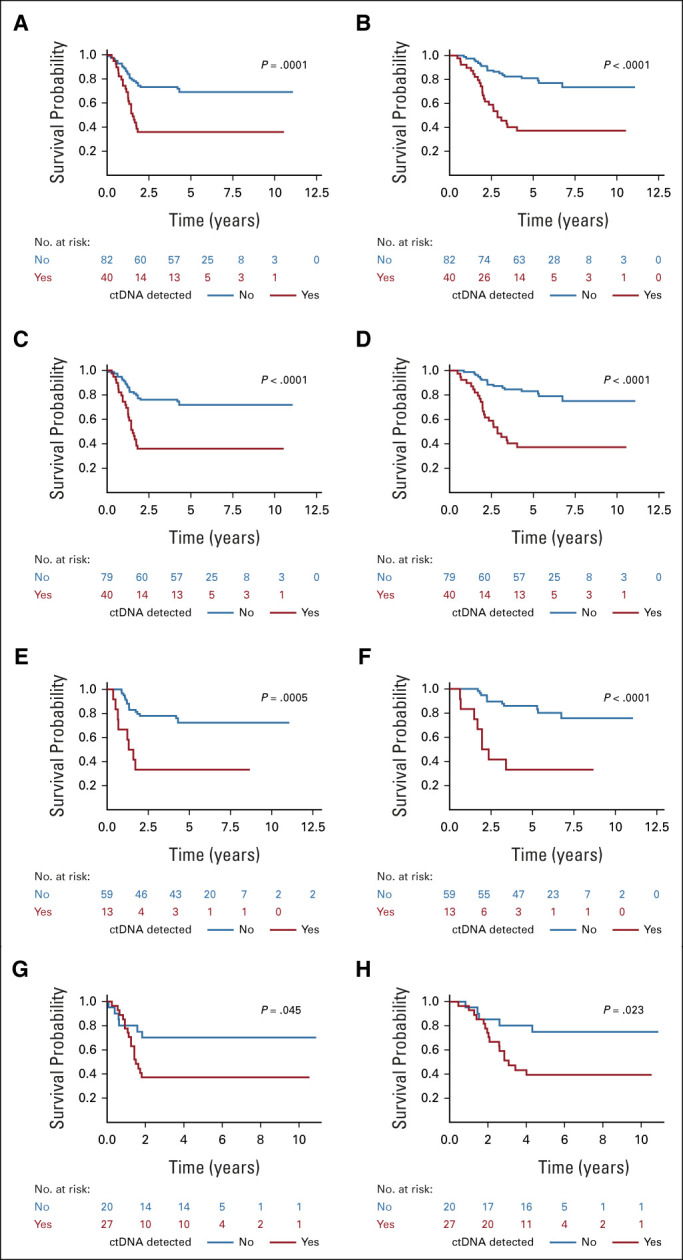
(A) EFS and (B) OS by ctDNA detection status, by ULP-WGS for FN-RMS, and translocation detection for FP-RMS of the entire cohort. (C) EFS and (D) OS by ctDNA detection status, determined by ULP-WGS for FN-RMS and translocation detection for FP-RMS, of the entire cohort excluding cases with MYOD1L122R mutations. (E) EFS and (F) OS by ctDNA status, determined by ULP-WGS, in patients with FN-RMS after excluding cases with MYOD1L122R mutations. (G) EFS and (H) OS by ctDNA status, determined by translocation detection, in patients with FP-RMS. P value from the log-rank test. ctDNA, circulating tumor DNA; EFS, event-free survival; FN-RMS, fusion-negative rhabdomyosarcoma; FP-RMS, fusion-positive rhabdomyosarcoma; OS, overall survival; RMS, rhabdomyosarcoma; ULP-WGS, ultralow passage whole-genome sequencing.
To exclude the possibility that differences in the LOD between ULP-WGS and Rhabdo-Seq contributed to our findings, we studied the association between ctDNA detection and outcome using each method. When using only ULP-WGS, detection of ctDNA was associated with a significantly lower 5-year EFS (35.1%; 95% CI, 15.7 to 55.3) and OS (34.3%; 95% CI, 14.9 to 54.8) compared with patients without detectable ctDNA (EFS, 69.5%; 95% CI, 57.7 to 78.6; P = .0007; OS, 79.8%; 95% CI, 68.7 to 87.3; P = .0001; Data Supplement). Using only Rhabdo-Seq, detection of ctDNA was again associated with a significantly lower 5-year EFS (38.7%; 95% CI, 24.5 to 52.6) and OS (45.1%; 95% CI, 29.6 to 59.4) than for patients without detectable ctDNA (EFS, 71.0%; 95% CI, 55.4 to 82.0; P = .001; OS, 71%; 95% CI, 55.4 to 82; P = .001; Data Supplement).
We also examined the association between ctDNA and outcome for each of the genomic subtypes of RMS. For FN-RMS, we used ULP-WGS to detect ctDNA (Rhabdo-Seq analysis would be limited to only cases with an SNV in the tumor). Detection of ctDNA was associated with a worse EFS and OS for FN-RMS (Data Supplement). After excluding patients with MYOD1L122R mutations, detection of ctDNA was associated with a 5-year EFS of 33.3% (95% CI, 10.3 to 58.8) compared with 72.4% (95% CI, 59 to 82.1) for patients without detectable ctDNA (P = .0005) and a 5-year OS of 33.3% (95% CI, 10.3 to 58.8) compared with 86% (95% CI, 73.9 to 92.7), respectively (P < .0001; Figs 3E and 3F). Univariable analysis showed that local T2 disease was also associated with outcome (Data Supplement). Stepwise multivariable analysis, including patients with MYOD1L122R, showed that ctDNA was significantly associated with EFS (HR, 4; 95% CI, 1.7 to 9.4; P = .0016) and OS (HR, 6.5; 95% CI, 2.5 to 16.6; P < .0001) as was MYOD1L122R (EFS HR, 14.2; 95% CI, 4.0 to 51.3; P < .0001; OS HR, 9.3; 95% CI, 2.0 to 44.4; P = .005). This was confirmed by additional multivariable models (Data Supplement). Although limited to a subset of cases in FN-RMS (n = 41), ctDNA detection by Rhabdo-Seq was also associated with a worse EFS and OS (with a 5-year EFS of 33.5% [95% CI, 12.2 to 56.6]) compared with 75.8% (95% CI, 53.8 to 88.3) for patients without detectable ctDNA (P = .0018) and a 5-year OS of 49.1% (95% CI, 21.4 to 72) compared with 83.6% (95% CI, 62.4 to 93.6), respectively (P = .0086; Data Supplement).
In FP-RMS, ctDNA detection by translocation (applicable to the largest portion of cases) had a 5-year EFS of 37% (95% CI, 19.6 to 54.6) compared with 70% (95% CI, 45.1 to 85.3) for patients without detectable ctDNA (P = .045) and a 5-year OS of 39.2% (95% CI, 21 to 57) and 75% (95% CI, 50 to 88.7), respectively (P = .023; Figs 3G and 3H). Univariable analysis also showed that T2 was associated with outcome (Data Supplement). Stepwise multivariable analysis showed that ctDNA was significantly associated with EFS (HR, 2.5; 95% CI, 1 to 6.4; P = .05) and OS (HR, 3.1; 95% CI, 1.1 to 8.4; P = .03), whereas other multivariable models, including the full model with all variables, showed that T2 stage was associated with outcome (Data Supplement). Finally, in the small cohort of FP-RMS confirmed to have CNAs in their tumors (n = 25), ctDNA detection by ULP-WGS was not significantly associated with outcome (Data Supplement).
DISCUSSION
The mainstay of therapy for RMS in North America includes chemotherapy cycles of vincristine, dactinomycin, and cyclophosphamide, often alternating with cycles of vincristine and irinotecan.15 Local control includes surgery, radiation, or both. Patients with low-risk RMS do well with less intense chemotherapy, whereas few patients with high-risk disease are cured.19-21 Patients with IR RMS are defined by a combination of biologic and clinical features, but as many as 25%-50% will relapse and die.15,22 More precise risk stratification for patients with IR RMS could better identify patients who will be cured with vincristine, dactinomycin, and cyclophosphamide/vincristine and irinotecan and those who will not but could be made eligible for high-risk trials.
Studies in other pediatric sarcomas demonstrate that detection of pretreatment ctDNA is associated with a worse outcome.18,23 In this study, we hypothesized that detection of ctDNA in patients with IR RMS would be associated with a worse outcome and therefore useful for further risk stratification. However, identification of ctDNA requires the detection of somatic variants that define the patient's cancer, differentiating ctDNA from cell-free DNA originating from normal cells.4 FP-RMS and FN-RMS are defined by distinct classes of somatic variants.2 FP-RMS is defined by a PAX/FOXO1 fusion and less frequent CNAs. FN-RMS is defined by aneuploidy in nearly every tumor, and SNVs in a small number of genes in 60% of cases.2,17 To identify ctDNA in the serum of patients with both genetic subtypes, we applied ULP-WGS that identifies aneuploidy in cell-free DNA and Rhabdo-Seq, designed to detect PAX/FOXO1, VGLL2/CITED2, VGLL2/NCOA2, and other translocations and SNVs in 22 RMS-relevant genes.
ULP-WGS analysis was applicable for ctDNA analysis in all cases of FN-RMS, classifying ctDNA as detectable in 17%. Although Rhabdo-Seq was unsuitable for ctDNA analysis in 30% of FN-RMS cases, this assay provided additional information, including the detection of prognostic MYOD1L122R variants and SNVs in potentially targetable genes such as FGFR4 and the RAS pathway.14,24-27 In FP-RMS, identification of PAX/FOXO1 rearrangements was applicable for the majority of cases and detected ctDNA in 55%. Although ULP-WGS was unsuitable for ctDNA detection in 29% of copy-neutral FP-RMS cases, this assay identified PAX7/FOXO1 amplifications (allowing for correction of ctDNA content estimates) and prognostic CNAs such as MYCN and CDK4.14
The detection of ctDNA before the start of therapy in RMS was associated with aggressive disease features such as tumor size, stage, and invasiveness, which may reflect mechanisms of ctDNA shed. Detection of ctDNA was also associated with a worse outcome in IR RMS with an estimated 5-year EFS and OS consistent with high-risk RMS.28,29 Multivariable analyses, using a model that includes all clinical variables, demonstrated that ctDNA is an independent prognostic biomarker of outcome in the full IR RMS cohort and the FN-RMS group but was not independently prognostic in the smaller FP-RMS cohort. Local disease invasion (local T2 disease) and MYOD1L122R were also associated with outcome in multivariable analyses. Although these results indicate that ctDNA analysis is a new prognostic factor in IR RMS, they also indicate that more sophisticated risk stratification approaches, such as those using recursive partitioning, may be able to best refine prognosis by integrating ctDNA detection, clinical features, and specific genomic events.
Our study has some limitations. The use of serum samples, banked before the development of ctDNA-specific blood collection protocols, prevented ctDNA quantification and might have decreased the proportion of positive samples. Another limitation was that serial samples were not available. Numerous studies now demonstrate the prognostic value of tracking changes in ctDNA levels over time.7,8,10,18,30-34 One recent report in Ewing sarcoma, another translocation-driven sarcoma, demonstrated that detectable levels of ctDNA present after two cycles of therapy were associated with relapse.35 Future studies will be needed to determine whether changes in ctDNA during therapy are prognostic in RMS or whether measurements of ctDNA after completion of initial chemotherapy could help guide the use and duration of maintenance therapy.36,37 Furthermore, additional studies of FP-RMS are warranted to demonstrate the prognostic value of ctDNA in multivariable analyses compared with clinical factors. Finally, this was a retrospective study and patients were not treated as uniformly as they would be on a prospective clinical trial.
Future efforts, such as the ongoing study of ctDNA in patients enrolled on the COG ARST1431 trial for IR RMS, will be needed to build a comprehensive prognostic score that includes ctDNA together with other prognostic variables. We plan to apply both NGS approaches presented in this study, which will allow for subtype-specific detection of ctDNA and detection of additional clinically relevant CNAs and SNVs.
In summary, the findings presented here strongly suggest that ctDNA will be an important tool for future prognostication and risk-stratified treatment strategies in RMS. Collection of high-quality pretreatment, serial on-treatment, and surveillance liquid biopsy samples on large prospective trials will be needed to refine these results.
ACKNOWLEDGMENT
The authors thank the Children's Oncology Group (COG) protocol coordinators, research coordinators, Clinical Research Assistants, and other health professionals who contributed to acquiring samples used in this study. The authors would like to thank the extended D9803 and ARST0531 teams who supported the clinical trial and biobanking data and samples used in this study. Finally, we are very grateful to the patients and their families for consenting to sample deposition through D9902.
Donald A. Barkauskas
Employment: Genentech (I)
Patents, Royalties, Other Intellectual Property: U.S. patent based on PhD research in glioblastoma (I)
Mark D. Krailo
Consulting or Advisory Role: Merck Sharp & Dohme
Travel, Accommodations, Expenses: Merck Sharp & Dohme
Steven G. DuBois
Consulting or Advisory Role: Bayer, Amgen, Jazz Pharmaceuticals
Research Funding: Merck (Inst), Roche/Genentech (Inst), Lilly (Inst), Curis (Inst), Loxo (Inst), BMS (Inst), Eisai (Inst), Pfizer (Inst), Turning Point Therapeutics (Inst), Bayer (Inst), Salarius Pharmaceuticals (Inst)
Travel, Accommodations, Expenses: Roche/Genentech, Salarius Pharmaceuticals
Uncompensated Relationships: Y-mAbs Therapeutics Inc
Douglas S. Hawkins
Research Funding: Bayer (Inst), Lilly (Inst), Incyte (Inst), Jazz Pharmaceuticals (Inst), Pfizer (Inst)
Brian D. Crompton
Employment: Acceleron Pharma (I), Generate Biomedicines (I)
Stock and Other Ownership Interests: Acceleron Pharma (I)
Consulting or Advisory Role: PetDx, Animal Cancer Foundation, Osteosarcoma Institute (Foundation)
Research Funding: Gradalis
No other potential conflicts of interest were reported.
DISCLAIMER
The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
PRIOR PRESENTATION
Presented in part at the The Connective Tissue Oncology Society meeting, Rome, Italy, November 14-17, 2018, and American Association for Cancer Research Advances in Liquid Biopsies, Miami, FL, January 13-16, 2020.
SUPPORT
Supported by a Children's Oncology Group Translational Pilot Studies Program for Solid Malignancies (B.D.C.), Hyundai Hope on Wheels (B.D.C.), the QuadW Foundation (B.D.C.), and Nuovo-Soldati Foundation for Cancer Research (S.A.). The project was also supported by the COG Chair's Grant (U10CA098543), the NCTN Network Group Operations Center Grant (U10CA180886; D.S.H.), the Statistics & Data Center Grant (U10CA098413), the NCTN Statistics & Data Center (U10CA180899), Human Specimen Banking in NCI-Sponsored Clinical Trials (U24CA114766), Human Specimen Banking in NCI-Sponsored Clinical Trials (1U24-CA196173), and St Baldrick's Foundation (D.S.H.).
S.A. and K.K. equally participated to this work.
DATA SHARING STATEMENT
Sequencing data are available through dbGAP Accession phs002866.v1.p1.
AUTHOR CONTRIBUTIONS
Conception and design: Samuel Abbou, Kelly Klega, Jack F. Shern, Brian D. Crompton
Financial support: Brian D. Crompton
Administrative support: Samuel Pollock, Jack F. Shern
Provision of study materials or patients: Jack F. Shern
Collection and assembly of data: Samuel Abbou, Kelly Klega, David Hall, Carrie Cibulskis, Aaron R. Thorner, Samuel Pollock, Brian D. Crompton
Data analysis and interpretation: Samuel Abbou, Kelly Klega, Junko Tsuji, Mohammad Tanhaemami, David Hall, Donald A. Barkauskas, Mark D. Krailo, Carrie Cibulskis, Anwesha Nag, Alma Imamovic-Tuco, Steven G. DuBois, Rajkumar Venkatramani, Douglas S. Hawkins, Brian D. Crompton
Manuscript writing: All authors
Final approval of manuscript: All authors
Accountable for all aspects of the work: All authors
AUTHORS' DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
Circulating Tumor DNA Is Prognostic in Intermediate-Risk Rhabdomyosarcoma: A Report From The Children's Oncology Group
The following represents disclosure information provided by authors of this manuscript. All relationships are considered compensated unless otherwise noted. Relationships are self-held unless noted. I = Immediate Family Member, Inst = My Institution. Relationships may not relate to the subject matter of this manuscript. For more information about ASCO's conflict of interest policy, please refer to www.asco.org/rwc or ascopubs.org/jco/authors/author-center.
Open Payments is a public database containing information reported by companies about payments made to US-licensed physicians (Open Payments).
Donald A. Barkauskas
Employment: Genentech (I)
Patents, Royalties, Other Intellectual Property: U.S. patent based on PhD research in glioblastoma (I)
Mark D. Krailo
Consulting or Advisory Role: Merck Sharp & Dohme
Travel, Accommodations, Expenses: Merck Sharp & Dohme
Steven G. DuBois
Consulting or Advisory Role: Bayer, Amgen, Jazz Pharmaceuticals
Research Funding: Merck (Inst), Roche/Genentech (Inst), Lilly (Inst), Curis (Inst), Loxo (Inst), BMS (Inst), Eisai (Inst), Pfizer (Inst), Turning Point Therapeutics (Inst), Bayer (Inst), Salarius Pharmaceuticals (Inst)
Travel, Accommodations, Expenses: Roche/Genentech, Salarius Pharmaceuticals
Uncompensated Relationships: Y-mAbs Therapeutics Inc
Douglas S. Hawkins
Research Funding: Bayer (Inst), Lilly (Inst), Incyte (Inst), Jazz Pharmaceuticals (Inst), Pfizer (Inst)
Brian D. Crompton
Employment: Acceleron Pharma (I), Generate Biomedicines (I)
Stock and Other Ownership Interests: Acceleron Pharma (I)
Consulting or Advisory Role: PetDx, Animal Cancer Foundation, Osteosarcoma Institute (Foundation)
Research Funding: Gradalis
No other potential conflicts of interest were reported.
REFERENCES
- 1.US Department of Health and Human Services; National Cancer Institute : Cancer Incidence and Survival Among Children and Adolescents: United States SEER Program 1975-1995. 1999. http://www.crossref.org/deleted_DOI.html [Google Scholar]
- 2.Shern JF, Chen L, Chmielecki J, et al. : Comprehensive genomic analysis of rhabdomyosarcoma reveals a landscape of alterations affecting a common genetic axis in fusion-positive and fusion-negative tumors. Cancer Discov 4:216-231, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arndt CAS, Stoner JA, Hawkins DS, et al. : Vincristine, actinomycin, and cyclophosphamide compared with vincristine, actinomycin, and cyclophosphamide alternating with vincristine, topotecan, and cyclophosphamide for intermediate-risk rhabdomyosarcoma: Children's Oncology Group Study D9803. J Clin Oncol 27:5182-5188, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Abbou SD, Shulman DS, DuBois SG, et al. : Assessment of circulating tumor DNA in pediatric solid tumors: The promise of liquid biopsies. Pediatr Blood Cancer 66:e27595, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cescon DW, Bratman SV, Chan SM, et al. : Circulating tumor DNA and liquid biopsy in oncology. Nat Cancer 1:276-290, 2020 [DOI] [PubMed] [Google Scholar]
- 6.Alix-Panabières C, Pantel K: Liquid biopsy: From discovery to clinical application. Cancer Discov 11:858-873, 2021 [DOI] [PubMed] [Google Scholar]
- 7.Oxnard GR, Thress KS, Alden RS, et al. : Association between plasma genotyping and outcomes of treatment with osimertinib (AZD9291) in advanced non-small-cell lung cancer. J Clin Oncol 34:3375-3382, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chabon JJ, Simmons AD, Lovejoy AF, et al. : Circulating tumour DNA profiling reveals heterogeneity of EGFR inhibitor resistance mechanisms in lung cancer patients. Nat Commun 7:11815, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kurtz DM, Scherer F, Jin MC, et al. : Circulating tumor DNA measurements as early outcome predictors in diffuse large B-cell lymphoma. J Clin Oncol 36:2845-2853, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Klega K, Imamovic-Tuco A, Ha G, et al. : Detection of somatic structural variants enables quantification and characterization of circulating tumor DNA in children with solid tumors. JCO Precis Oncol 10.1200/PO.17.00285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mc Connell L, Gazdova J, Beck K, et al. : Detection of structural variants in circulating cell-free DNA from sarcoma patients using next generation sequencing. Cancers (Basel) 12:E3627, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Peneder P, Stütz AM, Surdez D, et al. : Multimodal analysis of cell-free DNA whole-genome sequencing for pediatric cancers with low mutational burden. Nat Commun 12:3230, 2021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Adalsteinsson VA, Ha G, Freeman SS, et al. : Scalable whole-exome sequencing of cell-free DNA reveals high concordance with metastatic tumors. Nat Commun 8:1324, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shern JF, Selfe J, Izquierdo E, et al. : Genomic classification and clinical outcome in rhabdomyosarcoma: A report from an International Consortium. J Clin Oncol 39:2859-2871, 2021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hawkins DS, Chi Y-Y, Anderson JR, et al. : Addition of vincristine and irinotecan to vincristine, dactinomycin, and cyclophosphamide does not improve outcome for intermediate-risk rhabdomyosarcoma: A report from the Children's Oncology Group. J Clin Oncol 36:2770-2777, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Frascella E, Lenzini E, Schafer BW, et al. : Concomitant amplification and expression of PAX7-FKHR and MYCN in a human rhabdomyosarcoma cell line carrying a cryptic t(1;13)(p36;q14). Cancer Genet Cytogenet 121:139-145, 2000 [DOI] [PubMed] [Google Scholar]
- 17.Barr FG, Nauta LE, Davis RJ, et al. : In vivo amplification of the PAX3-FKHR and PAX7-FKHR fusion genes in alveolar rhabdomyosarcoma. Hum Mol Genet 5:15-21, 1996 [DOI] [PubMed] [Google Scholar]
- 18.Shulman DS, Klega K, Imamovic-Tuco A, et al. : Detection of circulating tumour DNA is associated with inferior outcomes in Ewing sarcoma and osteosarcoma: A report from the Children's Oncology Group. Br J Cancer 119:615-621, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Oberlin O, Rey A, Sanchez de Toledo J, et al. : Randomized comparison of intensified six-drug versus standard three-drug chemotherapy for high-risk nonmetastatic rhabdomyosarcoma and other chemotherapy-sensitive childhood soft tissue sarcomas: Long-term results from the International Society of pediatric Oncology MMT95 study. J Clin Oncol 30:2457-2465, 2012 [DOI] [PubMed] [Google Scholar]
- 20.Weigel BJ, Lyden E, Anderson JR, et al. : Intensive multiagent therapy, including dose-compressed cycles of ifosfamide/etoposide and vincristine/doxorubicin/cyclophosphamide, irinotecan, and radiation, in patients with high-risk rhabdomyosarcoma: A report from the Children's Oncology Group. J Clin Oncol 34:117-122, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Beverly Raney R, Walterhouse DO, Meza JL, et al. : Results of the intergroup rhabdomyosarcoma study group D9602 protocol, using vincristine and dactinomycin with or without cyclophosphamide and radiation therapy, for newly diagnosed patients with low-risk embryonal rhabdomyosarcoma: A report from the soft tissue sarcoma Committee of the Children's Oncology Group. J Clin Oncol 29:1312-1318, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pappo AS, Anderson JR, Crist WM, et al. : Survival after relapse in children and adolescents with rhabdomyosarcoma: A report from the intergroup Rhabdomyosarcoma Study Group. J Clin Oncol 17:3487-3493, 1999 [DOI] [PubMed] [Google Scholar]
- 23.Lak NSM, Voormanns TL, Zappeij-Kannegieter L, et al. : Improving risk stratification for pediatric patients with rhabdomyosarcoma by molecular detection of disseminated disease. Clin Cancer Res 27:5576-5585, 2021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li SQ, Cheuk AT, Shern JF, et al. : Targeting wild-type and mutationally activated FGFR4 in rhabdomyosarcoma with the Inhibitor ponatinib (AP24534). PLoS One 8:e76551, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Taylor JG VI, Cheuk AT, Tsang PS, et al. : Identification of FGFR4-activating mutations in human rhabdomyosarcomas that promote metastasis in xenotransplanted models. J Clin Invest 119:3395-3407, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ney GM, McKay L, Koschmann C, et al. : The emerging role of Ras pathway signaling in pediatric cancer. Cancer Res 80:5155-5163, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nakagawa N, Kikuchi K, Yagyu S, et al. : Mutations in the RAS pathway as potential precision medicine targets in treatment of rhabdomyosarcoma. Biochem Biophys Res Commun 512:524-530, 2019 [DOI] [PubMed] [Google Scholar]
- 28.Rudzinski ER, Anderson JR, Chi Y-Y, et al. : Histology, fusion status, and outcome in metastatic rhabdomyosarcoma: A report from the Children's Oncology Group. Pediatr Blood Cancer 64:e26645, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Malempati S, Weigel BJ, Chi Y-Y, et al. : The addition of cixutumumab or temozolomide to intensive multiagent chemotherapy is feasible but does not improve outcome for patients with metastatic rhabdomyosarcoma: A report from the Children's Oncology Group. Cancer 125:290-297, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bettegowda C, Sausen M, Leary RJ, et al. : Circulating tumor cells bad news in breast cancer. Sci Transl Med 4:144ec132, 2012 [Google Scholar]
- 31.Dawson S-J, Tsui DWY, Murtaza M, et al. : Analysis of circulating tumor DNA to monitor metastatic breast cancer. N Engl J Med 368:1199-1209, 2013 [DOI] [PubMed] [Google Scholar]
- 32.Garcia-Murillas I, Schiavon G, Weigelt B, et al. : Mutation tracking in circulating tumor DNA predicts relapse in early breast cancer. Sci Translational Med 7:302ra133, 2015 [DOI] [PubMed] [Google Scholar]
- 33.Chicard M, Colmet-Daage L, Clement N, et al. : Whole-exome sequencing of cell-free DNA reveals temporo-spatial heterogeneity and identifies treatment-resistant clones in reuroblastoma. Clin Cancer Res 24:939-949, 2018 [DOI] [PubMed] [Google Scholar]
- 34.Stankunaite R, George SL, Gallagher L, et al. : Circulating tumour DNA sequencing to determine therapeutic response and identify tumour heterogeneity in patients with paediatric solid tumours. Eur J Cancer 162:209-220, 2022 [DOI] [PubMed] [Google Scholar]
- 35.Krumbholz M, Eiblwieser J, Ranft A, et al. : Quantification of translocation-specific ctDNA provides an integrating parameter for early assessment of treatment response and risk stratification in Ewing sarcoma. Clin Cancer Res 27:5922-5930, 2021 [DOI] [PubMed] [Google Scholar]
- 36.Bisogno G, De Salvo GL, Bergeron C, et al. : Vinorelbine and continuous low-dose cyclophosphamide as maintenance chemotherapy in patients with high-risk rhabdomyosarcoma (RMS 2005): A multicentre, open-label, randomised, phase 3 trial. Lancet Oncol 20:1566-1575, 2019 [DOI] [PubMed] [Google Scholar]
- 37.Bisogno G, Hawkins DS: An unresolved issue in rhabdomyosarcoma treatment: The duration of chemotherapy. Pediatr Blood Cancer 67:e28174, 2020 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Sequencing data are available through dbGAP Accession phs002866.v1.p1.


