Abstract
While the majority of BRAF-mutant melanomas respond to BRAF/MEK inhibitors, these agents are not typically curative. Moreover, they are largely ineffective in NRAS- and NF1-mutant tumors. Here we report that genetic and chemical suppression of HDAC3 potently cooperates with MAPK pathway inhibitors in all three Ras pathway-driven tumors. Specifically, we show that entinostat dramatically enhances tumor regression when combined with BRAF/MEK inhibitors, both in models that are sensitive or relatively resistant to these agents. Interestingly, MGMT expression predicts responsiveness and marks tumors with latent defects in DNA repair. BRAF/MEK inhibitors enhance these defects by suppressing homologous recombination genes, inducing a BRCA-like state; however, entinostat addition triggers the concomitant suppression of NHEJ genes, resulting in a chemical synthetic lethality caused by excessive DNA damage. Together these studies identify melanomas with latent DNA repair defects, describe a promising drug combination that capitalizes on these defects, and reveal a tractable therapeutic biomarker.
Introduction
Melanomas can be classified into four genomic subtypes based on the presence or absence of mutations in Ras pathway genes: BRAF, NRAS, NF1 and Triple-wild-type (1). Fortunately, selective BRAF and MEK inhibitors, and more recently BRAF/MEK inhibitor combinations, have improved prognosis and overall survival in patients with metastatic BRAF-mutant disease (2). Nevertheless, all individuals ultimately relapse, and do so on average in 11 months (3, 4). A fraction of NRAS-mutant tumors (15%) exhibit partial responses to MEK inhibitors, albeit with shorter durations (5). The MEK pathway is also hyperactivated in NF1-mutant melanomas, however the clinical activity of MEK inhibitors in this subtype is not known. Regardless, these observations suggest that while RAF/MEK pathway inhibition will remain an important cornerstone of melanoma treatment, improved combinations and/or sequential therapies are needed. Accordingly, additional meaningful targets must be identified.
Agents that target epigenetic enzymes are increasingly being developed as potential cancer therapies (6). HDAC inhibitors are one such class of compounds and various drugs have been approved for use in hematopoietic malignancies (7). While single agent efficacy in solid tumors has not been observed, HDAC inhibitors are currently being evaluated in combination with other targeted agents in several diseases (8). However, the key to discovering successful combinations, if they exist, will likely lie in 1) identifying the most selective/potent agent for the specified target to minimize potential toxicities; 2) elucidating the mechanism of action of the combination identified; and 3) using this insight to prospectively identify patients that are most likely to respond. While these criteria are important for developing any successful combination therapy, they may be critical for developing combinations with HDAC inhibitors, many of which inhibit numerous HDAC isoforms, and therefore have major effects on chromatin.
Here we identify an epigenetic-based combination therapy for BRAF-, NRAS-, and NF1-mutant melanomas. Specifically, we show that HDAC3 is an important therapeutic target in these tumors and that the selective Class I inhibitor, entinostat, not only dramatically enhances the in vivo efficacy of BRAF/MEK inhibitors in BRAF-mutant malignancies with varying sensitivities to these agents, but also cooperates with MEK inhibitors in NRAS and NF1-mutant tumors. We further demonstrate that these agents function by coordinately suppressing the transcription of homologous recombination (HR) and nonhomologous end joining (NHEJ) genes, thereby triggering excessive DNA damage in sensitive tumors. Finally, we identify a tractable biomarker that marks melanomas with broad defects in DNA repair genes and predicts efficacy. Together these studies have identified a promising mechanism-based combinatorial strategy for treating melanomas with several RAS/RAF pathway defects and have outlined a path for clinical translation.
Results
HDAC inhibitors dramatically potentiate the effects of BRAF/MEK inhibitors in BRAF-, NRAS- and NF1-mutant melanomas
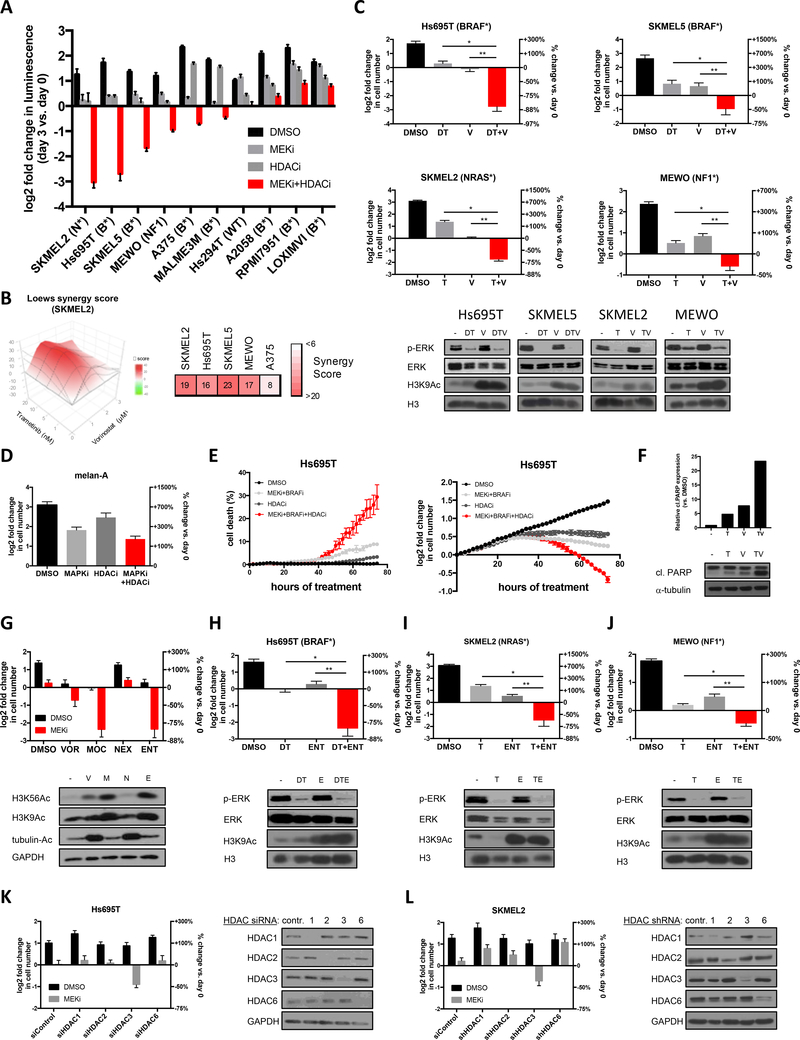
MITF is a lineage-specific survival gene that is amplified in a subset of melanomas and has been shown to confer resistance to MAPK pathway inhibition (9). Because HDAC inhibitors have been reported to suppress MITF expression (10), we reasoned that these agents might potentiate the therapeutic effects of BRAF and/or MEK inhibitors. A panel of melanoma cell lines harboring mutations in BRAF, NRAS or NF1 were examined in a CellTiter-Glo based screen. The MEK inhibitor, trametinib, was used to broadly suppress the MAPK pathway in all cell lines. Notably, the pan-HDAC inhibitor, vorinostat, dramatically potentiated the effects of trametinib in 6 out of 10 lines, including BRAF-, NRAS- and NF1-mutant cells (Fig. 1A). These effects were determined to be synergistic using the Loewe excess additivity model (Fig. 1B).
Figure 1: HDAC inhibitors potentiate the effects of BRAF/MEK pathway inhibitors in BRAF-, NRAS- and NF1-mutant melanoma.
(A) Melanoma cell lines with specified genotypes (BRAF-mutant (B*), NRAS-mutant (N*), NF1-mutant (NF1) or wild-type for BRAF, NRAS and NF1 (WT)) were treated with DMSO, 10nM trametinib (MEK inhibitor), 2μM vorinostat (HDAC inhibitor) or both agents. Graph depicts log2 transformation of the fold change in luminescence using a CellTiter-Glo assay in cells after 3 days of treatment versus day 0 (left axis) +/− SD. (B) Synergy scores (to characterize the combination effects in excess of Loewe additivity) for Vorinostat combined with Trametinib for SKMEL2 (3D synergy map, left) and additional sensitive cell lines from Fig. 1A (right). Strong synergy is indicated in red. (C) BRAF-, NRAS- and NF1-mutant melanoma cell lines were treated with DMSO, 100nM dabrafenib (D) and/or 10nM trametinib (T), 2μM vorinostat (V) or the specified agents combined, as indicated. Cells were manually counted prior to the addition of compounds and 3 days after treatment. Graphs represent log2 transformation of the fold change in cell number at day 3 versus day 0. Right axis shows percent change in cell number relative to day 0. Negative values represent a decrease in cell number over time and positive values represent an increase in cell number. Immunoblots show levels of phosphorylated ERK (p-ERK), total ERK, histone H3 acetylation at lysine 9 (H3K9Ac) and total H3 after 48 hours of indicated treatment. (Hs695T) *p=0.001896, **p=0.002332; (SKMEL5) *p=0.000011, **p=0.000077; (SKMEL2) *p=0.000007, **p=0.000074; (MEWO) *p=0.003025, **p=0.001545 (D) Normal melanocytes (melan-A) were treated with DMSO, 100nM dabrafenib and 10nM trametinib (MAPKi), 1μM HDAC inhibitor (HDACi) and the combination of MEK/BRAF and HDAC inhibitors. Cells were manually counted prior to the addition of compounds and 3 days after treatment. Graphs represent log2 transformation of the fold change in cell number at day 3 versus day 0. (E) Real-time quantification of cell death (left panel) and cell numbers (right panel) using the live cell imager IncuCyte Zoom. Red (nuclear restricted NucLight Red fluorescent protein for quantifiying live cells) and green (Cytotox Green reagent for quantifying dead cells) fluorescent objects were monitored in the Incucyte ZOOM acquiring images every 2 hours (for 72 hours) following treatment with vehicle (DMSO) or drugs (MEK inhibitor, BRAF inhibitor, HDAC inhibitor) and then quantified with the IncuCyte integrated analysis software. (F) Western blot depicting cleaved PARP protein levels after 48hrs of treatment with vehicle (−), MEK inhibitor (trametinib, T) and/or HDAC inhibitor (vorinostat, V). Alpha-tubulin serves as a loading control. Bar graphs represent quantification of cleaved PARP for different treatment arms, relative to untreated control. (G) Hs695T cells were treated with vorinostat (VOR, 1μM), mocetinostat (MOC, 1μM), nexturastat (NEX, 2μM) or entinostat (ENT, 2μM), alone (black) and in combination with 10nM trametinib (MEKi, red). Graph depicts the mean log2 fold change of cell number after 72 hours, relative to day 0. Right axis shows percent change in cell number relative to day 0. Immunoblot shows acetylated H3 at lysine 9 (H3K9Ac), acetylated H3 at lysine 56 (H3K56Ac) and acetylated tubulin (tubulin-Ac, surrogate marker for HDAC6 inhibition) after 48 hours of indicated treatment. GAPDH serves as a loading control. (H-J) Melanoma cell lines were treated with DMSO, 100nM dabrafenib (D) and/or 10nM trametinib (T), 1μM entinostat (ENT) or the specified drug combinations. Graphs represent log2 transformation of the fold change in cell number at day 3 versus day 0. Right axis shows percent change in cell number relative to day 0. Negative values represent cell death and a decrease in cell number. Immunoblots show levels of phosphorylated ERK (p-ERK), total ERK, histone H3 acetylation at lysine 9 (H3K9Ac) and total H3 after 48 hours of indicated treatment. (Hs695T) *p=0.005997, **p=0.005364; (SKMEL2) *p=0.008834, **p=0.017467; (MEWO) *p=0.003176, **p=0.000183 (K) Hs695T cells were transfected with pooled siRNAs targeting HDAC1, HDAC2, HDAC3, HDAC6 or control non-targeting siRNAs and treated with DMSO (black) or 10nM trametinib (MEKi, grey). Graph depicts the mean log2 fold change of cell number after 72 hours, relative to day 0. Right axis shows percent change in cell number relative to day 0. Immunoblot below depicts knockdown of the respective siRNAs. GAPDH serves as a loading control. (L) SKMEL2 cells were infected with lentiviral constructs targeting HDAC1, HDAC2, HDAC3, HDAC6 or control and treated with DMSO (black) or 10nM trametinib (MEKi, grey). Graph depicts the mean log2 fold change of cell number after 72 hours, relative to day 0. Right axis shows percent change in cell number relative to day 0. Immunoblot below depicts knockdown of the respective shRNAs. GAPDH serves as a loading control.
Cytotoxicity was confirmed by cell counting assays and live cell imaging in the presence of clinically relevant drug combinations. Combined BRAF and MEK inhibitors (dabrafenib/trametinib) were used to suppress the MAPK pathway in BRAF-mutant cells, as this represents the standard of care (2), whereas trametinib alone was used in NRAS and NF1-mutant cell lines. In both settings, combined MAPK/HDAC suppression resulted in a dramatic loss of viable cells in just 72 hours in sensitive lines representing all three genotypes (Fig. 1C). Dabrafenib/trametinib and vorinostat effectively suppressed their respective targets in all cell lines and the combination did not further reduce ERK phosphorylation in sensitive cells nor attenuate phospho-ERK suppression in resistant cells (Fig. 1C and Supplementary Fig. 1A,B). These agents were not generally toxic, as they did not affect the viability of normal melanocytes, unresponsive melanoma cell lines, and a variety of other unrelated cell types (Fig. 1D and Supplementary Fig. 1A–D).
Live-cell incucyte imaging was used to concomitantly measure cell death and changes in cell number over time, using a fluorescent assay that detects live and dead cells. BRAF/MEK inhibitors alone killed 9% of cells, consistent with previous reports (Fig. 1E, left panel) (11). However, while the HDAC inhibitor had little effect on its own, it dramatically potentiated cell death triggered by dabrafenib/trametinib, resulting in 29% cell death within just 72 hours (Fig. 1E, left panel). Cell death was mediated by apoptosis, as confirmed by PARP cleavage and a fluorescent caspase reporter, which similarly revealed that one-third of the cell population was undergoing apoptosis within 72 hours (Fig. 1F and Supplementary Fig. 1E). Importantly, the onset of cell death corresponded to a concomitant reduction in cell number (Fig. 1E, right panel). Consistent with previous observations, BRAF/MEK inhibitors alone appeared to exert delayed cytostatic effects in vitro (11); however, this apparent cytostasis was likely due to the net effects of concomitant proliferation and lower levels of cell death (Fig. 1E). By contrast, BRAF/MEK and HDAC inhibitors together synergized to cause much more cell death, thereby eliminating more resistant tumor cells (Fig. 1A–C,E).
Therapeutic responses are unrelated to MITF status or expression changes
Melanomas can either be categorized as MITFhigh or MITFlow (12). Based on our original hypothesis we expected that MITFhigh cells might be more sensitive to these agents, and if so MITF suppression by HDAC inhibitors would correspond to sensitivity. Surprisingly however, sensitivity was unrelated to MITF status, as both MITFhigh and MITFlow cells responded to this combination (Supplementary Fig. 1F). In addition, MITF suppression did not correlate with sensitivity (Supplementary Fig. 1G). Taken together these observations suggest that HDAC inhibitors broadly potentiate the therapeutic effects of BRAF/MEK pathway inhibitors in a high percentage of melanomas harboring mutations in one of several genes affecting the RAS/RAF pathway, but that these effects are unrelated to MITF status or expression changes.
Suppression of HDAC3 is sufficient to kill melanomas when combined with MAPK pathway inhibitors
Before embarking on mechanistic studies we sought to identify the most clinically tractable agent(s). Vorinostat is a pan-HDAC inhibitor, however more selective inhibitors have been developed (7). Importantly, the use of more selective compounds might minimize potential toxicities in humans, especially in the context of drug combinations. The effects of vorinostat were compared to mocetinostat (which inhibits Class I HDACs 1–3 and Class IV HDAC 11) (13), nexturastat (a selective HDAC6 inhibitor that primarily affects acetylation of proteins other than histones) (14), and entinostat (which inhibits Class I HDACs 1–3) (7). Nexturastat did not kill cells when combined with MAPK pathway inhibitors, indicating that HDAC6 inhibition is not sufficient for these effects (Fig. 1G). However, mocetinostat and entinostat, both cooperated with trametinib and were even more potent than vorinostat, demonstrating that the suppression of Class I HDAC proteins is sufficient for a maximal therapeutic response (Fig. 1G).
We were particularly enthusiastic about entinostat because it is the most selective agent in this panel that exerted therapeutic effects, and it has shown promising responses in Phase 2 drug combination studies in breast cancer (15). It is also currently being evaluated in a variety of solid tumors, underscoring the potential translatability of entinostat-based combinations. Importantly, entinostat also cooperated with MAPK pathway inhibitors in BRAF-, NRAS- and NF1-mutant cell lines (Fig. 1H–J).
Entinostat suppresses the activity of HDAC1, 2 and 3, and does not affect protein expression, as previously reported (Supplementary Fig. 1H) (16). To determine whether the inhibition of a specific HDAC gene was mediating the therapeutic effects, individual HDACs were genetically suppressed using pooled siRNAs. Interestingly, only HDAC3 suppression was sufficient to kill melanomas when combined with trametinib, which was confirmed using an unrelated panel of shRNA sequences in a second sensitive cell line (Fig. 1K,L). Because entinostat is currently the most selective, clinically available agent that suppresses HDAC3 and is well tolerated even when combined with other agents, we continued our analysis using entinostat. In addition, it should be noted that while pan-HDAC inhibitors also exert activity in this setting, toxicities associated with broad HDAC inhibition have limited their clinical utility in the context of some drug combinations (17), further supporting the selection of entinostat.
Entinostat potently cooperates with dabrafinib/trametinib in vivo in models with differing sensitivities to BRAF/MEK inhibitors
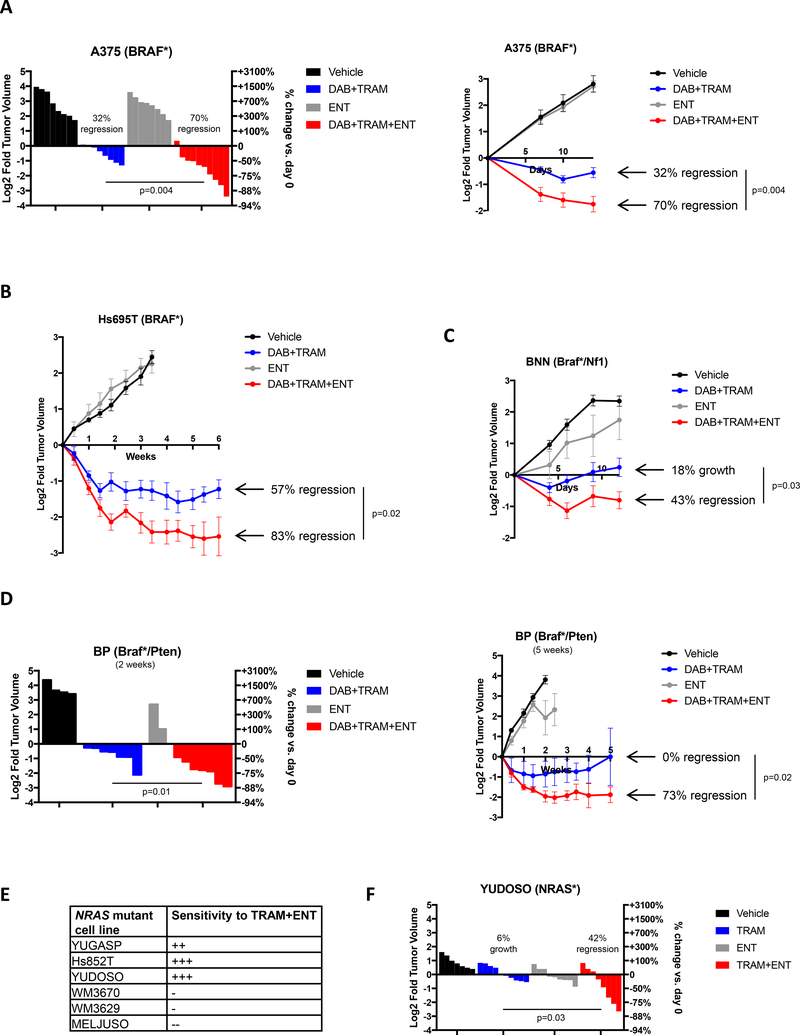
Next, we investigated whether these agents could exert similar cooperative effects in vivo, using drug concentrations that mimic human exposures (18). In clinical trials the BRAF inhibitor dabrafenib exhibits activity as a single agent in BRAF-mutant melanomas, however when combined with trametinib, efficacy is enhanced and toxicities are reduced, due to the suppression of feedback pathways (3, 4). As such, combined BRAF/MEK inhibitors are now the standard of care in BRAF-mutant melanoma. Nevertheless, a range of therapeutic responses are observed in patients. Therefore, we examined several BRAF-mutant melanoma models with differing sensitivities to these agents. The A375 xenograft model was moderately sensitive to the standard of care; average tumor regression in response to dabrafenib/trametinib was 32% (Fig. 2A). However, the inclusion of entinostat at 1mg/kg per week, a dose that is comparable to the human dose of 5mg once weekly being used in other clinical combination studies, dramatically enhanced efficacy (p=0.004). While entinostat on its own had no effect on tumor growth, when combined with dabrafenib/trametinib, tumors regressed by 70% on average (Fig. 2A).
Figure 2: Entinostat improves the efficacy of BRAF/MEK pathway inhibitors in vivo.
(A) Waterfall plot (left panel) depicting change in tumor volume in a BRAF-mutant melanoma xenograft model (A375 cells) after 2 weeks of treatment with single and combined agents as indicated. Each bar represents an individual tumor (Vehicle n=9, DAB+TRAM n=8, ENT n=8, DAB+TRAM+ENT n=10). Left axis indicates the log2 of fold change in tumor volume, and right axis indicates the percentage change in tumor volume relative to day 0. Dabrafenib (DAB, BRAF inhibitor), trametinib (TRAM, MEK inhibitor), entinostat (ENT, Class I HDAC inhibitor). Growth curve for the entire duration of the study is shown on the right. Graphs represent mean values +/− standard error of the mean. (B) Growth curve of an additional human BRAF-mutant melanoma xenograft model (Hs695T) treated with single and combined agents as indicated (Vehicle n=6, DAB+TRAM n=8, ENT n=6, DAB+TRAM+ENT n=8). Left axis indicates the log2 of fold change in tumor volume relative to day 0. Graphs represent mean values +/− standard error of the mean. (C) Growth curve depicting change in tumor volume of GEMM-derived Braf/Nf1-mutant melanoma allografts after treatment with single and combined agents as indicated. (Braf/Nf1: Vehicle n=7, DAB+TRAM n=6, ENT n=6, DAB+TRAM+ENT n=5). Graphs represent mean values +/− standard error of the mean. (D) Waterfall plot (left panel) depicting change in tumor volume in a Braf/Pten-mutant melanoma allograft model after 2 weeks of treatment with single and combined agents as indicated. Each bar represents an individual tumor (Vehicle n=4, DAB+TRAM n=7, ENT n=2, DAB+TRAM+ENT n=7). Growth curve for the entire duration of the study is shown on the right. Graphs represent mean values +/− standard error of the mean. (E) Table summarizing in vitro sensitivity of 6 human NRAS-mutant cell lines against combined trametinib and entinostat treatment. (F) Waterfall plot depicting change in tumor volume of NRAS-mutant melanoma xenografts (YUDOSO cells) treated with single and combined agents as indicated (Vehicle n=7, TRAM n=9, ENT n=9, TRAM+ENT n=8). Each bar represents an individual tumor.
Next, we examined the effects of entinostat in a human xenograft model that is more sensitive to dabrafenib/trametinib in vivo (Hs695T) (Fig. 2B). In this study we were also interested in examining the durability of the response and therefore continued treatment for 6 weeks. Strikingly, even in this model entinostat substantially enhanced the therapeutic response to dabrafenib/trametinib, promoting deeper regressions (83% versus 57%, p=0.02), demonstrating that this combination is able to kill more residual disease. Moreover, regressions remained durable throughout the entire study.
Finally, the effects of these agents were evaluated in two different GEMM allograft models, with cooperating mutations in genes associated with resistance to BRAF inhibitors (19, 20). Importantly, the immune system is also intact in each of these models. The first model harbored mutations in Braf and Nf1 (19), as NF1 mutations have been shown to functionally confer resistance to MAPK pathway inhibitors (19, 21). As predicted, MEK/BRAF inhibition did not cause any durable regressions in this model, and instead, tumors grew 18% on average; however, combined MEK/BRAF/HDAC suppression triggered tumor regression in every animal, and tumors shrunk by 43% (Fig. 2C, p=0.03). PTEN mutations have been proposed to function by augmenting survival in response to MAPK pathway inhibition and are associated with smaller and less durable clinical responses in patients (22, 23). Consistent with clinical observations, the effect of dabrafenib/trametinib on Braf/Pten-mutant GEMM allografts was also relatively modest in most tumors (Fig. 2D, left), and these lesions ultimately became resistant (Fig. 2D, right). Nevertheless, the inclusion of entinostat caused tumors to shrink by more than 73% and responses in all tumors were stable (Fig. 2D; p=0.02). Altogether, preclinical studies in four distinct BRAF-mutant models, representing tumors with distinct genetic alterations and different sensitivities to BRAF/MEK inhibitors (sensitive, moderately sensitive, and resistant), suggest that entinostat can substantially enhance the therapeutic effects of BRAF/MEK pathway inhibitors in vivo in all of these settings. Fig. 2C also demonstrates that this combination is effective in vivo in tumors that harbor NF1 mutations.
Entinostat sensitizes NRAS-mutant melanomas to trametinib
We also expanded our analysis of NRAS-mutant melanomas. Using a panel of six additional NRAS-mutant lines we found that three were sensitive to combined MEK and HDAC inhibitors while three were not, consistent with the frequency of sensitivity to this combination overall (Fig. 2E). Importantly, these agents also cooperatively enhanced the regression of NRAS-mutant xenografts (Fig. 2F). Whereas trametinib alone exerted largely cytostatic effects (on average tumors grew by 6%), trametinib and entinostat together induced a 42% tumor regression (p=0.03). This observation is particularly important given that there are currently no effective treatments for NRAS-mutant tumors.
MGMT is a biomarker that predicts sensitivity to combined MAPK/HDAC inhibitors
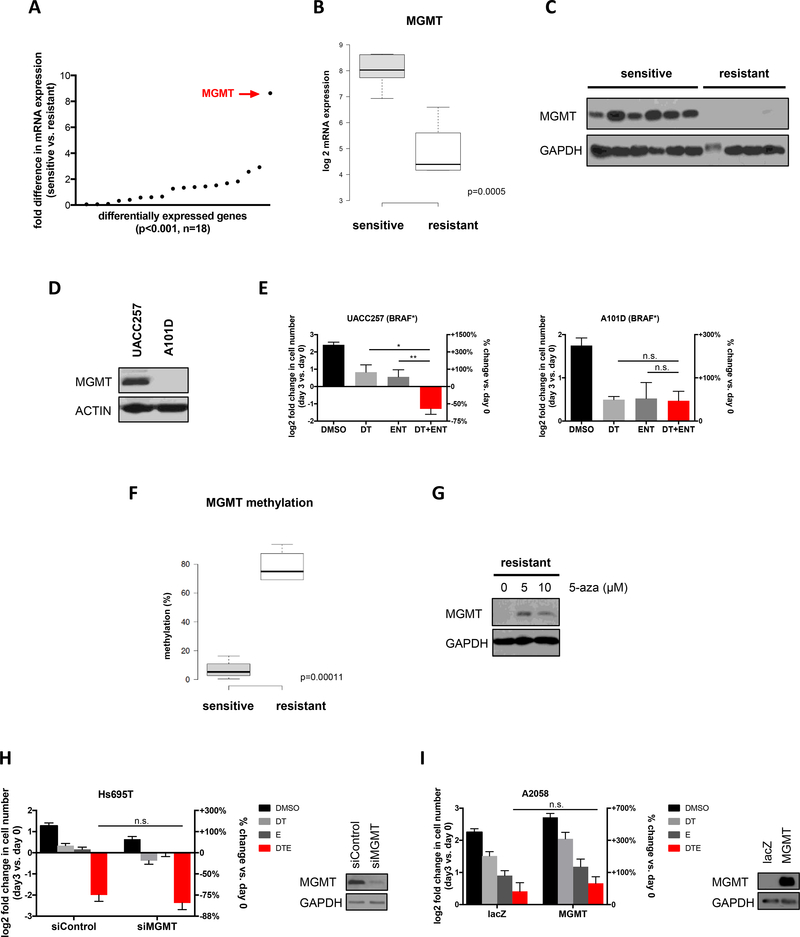
While several genetically distinct melanoma cell lines and tumor models were sensitive to combined HDAC and MAPK pathway inhibitors, a subset were resistant to this combination (Fig. 1A). Therefore, we hypothesized that these differential responses could be exploited to identify potential biomarkers of sensitivity or resistance. Extensive genomic analysis did not reveal any recurrent mutations or copy number alterations that distinguished sensitive or resistant cells. We therefore performed a two-class comparison of transcriptional profiles from the sensitive (n=6) and resistant (n=4) cell lines shown in Fig. 1A. Eighteen genes that were differentially expressed (p<0.001) between these two populations were identified (Fig. 3A, Supplementary Table 1); however dramatic differences in expression were observed for only one of these genes, O6-methylguanine DNA methyltransferase (MGMT), which was elevated in sensitive cells by almost 9-fold (Fig. 3A and B, p=0.0005). Western blot analysis further revealed that MGMT protein was readily detected in sensitive cell lines whereas it was undetectable or minimally expressed in resistant cells (Fig. 3C). Based on these observations two additional BRAF-mutant cell lines, predicted by the CCLE to have either high or low levels of MGMT mRNA, were selected for analysis. Immunoblots confirmed the expected differences in MGMT protein levels (Fig. 3D). Importantly, the MGMT expressing cells were sensitive to dabrafenib/trametinib/entinostat, whereas the cell line that lacked MGMT expression was insensitive to this combination (Fig. 3E). Similarly, the NRAS-mutant melanomas that were sensitive to these agents expressed MGMT, whereas insensitive cells did not (Supplementary Fig. 2). Altogether, analysis of 18 different sensitive and resistant cell lines indicates that MGMT expression is a strong predictive biomarker of sensitivity to this drug combination.
Figure 3: MGMT expression predicts sensitivity to BRAF/MEK and HDAC inhibition but does not play a functional role.
(A) Graph depicts fold difference in mRNA expression of 18 differentially expressed genes in cell lines sensitive (n=6) or resistant (n=4) to combined MAPK and HDAC inhibition (p<0.001). MGMT is upregulated by 8.6-fold in sensitive cells (p=0.0005). (B) Boxplot illustrating differential mRNA expression of MGMT in cell lines sensitive (n=6) or resistant (n=4) to combined MAPK and HDAC inhibition (p=0.0005). (C) Immunoblot depicting MGMT protein levels in sensitive and resistant cell lines. Order of cell lines is the same as in Fig. 1A. (D) Two additional melanoma cell lines characterized by the CCLE to have either high (UACC257) or low (A101D) MGMT mRNA levels were analyzed for MGMT protein levels by Western blot. Actin serves as a loading control. (E) Both cell lines were treated with either DMSO, 100nM dabrafenib and 10nM trametinib (DT), 1μM entinostat (ENT) or DT+ENT. Cells were manually counted prior to the addition of compounds and 3 days after treatment. Graphs represent log2 transformation of the fold change in cell number at day 3 versus day 0. Right axis shows percent change in cell number relative to day 0. Negative values represent cell death and a decrease in cell number. (UACC257) *p=0.002090, **p=0.002767; (A101D) n.s., not significant (F) Boxplot illustrating differential MGMT DNA methylation in cell lines sensitive (n=4) or resistant (n=3) to combined MAPK and HDAC inhibition (p=0.00011). (G) Western blot depicting MGMT protein levels before and after treatment with 5-azacitidine (5-aza) for 5 days in a cell line resistant to combined MEK and HDAC inhibition. (H) Hs695T cells were transfected with pooled siRNAs targeting MGMT or control non-targeting siRNAs and treated with drugs as indicated. Graphs represent log2 transformation of the fold change in cell number at day 3 versus day 0. Right axis shows percent change in cell number relative to day 0. Immunoblot depicts knockdown of MGMT. GAPDH serves as a loading control. n.s., not significant (I) A2058 cells were infected with a lentiviral construct expressing MGMT or lacZ control. Stable cell lines were treated with drugs as indicated. Graphs represent log2 transformation of the fold change in cell number at day 3 versus day 0. Right axis shows percent change in cell number relative to day 0. Immunoblot depicts ectopic expression of MGMT. GAPDH serves as a loading control. n.s., not significant
MGMT directly reverses the mutagenic DNA lesion O6-methylguanine, which is caused by alkylating agents (24). Accordingly, in glioblastomas, high MGMT levels are associated with resistance to alkylating chemotherapies such as temozolomide (25, 26). In a subset of glioblastomas, the MGMT promoter is epigenetically silenced by methylation, which is thought to be responsible for conferring sensitivity to temozolomide. Therefore, MGMT promoter methylation testing is routinely used in clinical practice as a predictive biomarker to guide patient management in glioblastoma. Consistent with this mechanism of regulation, we found that the MGMT promoter was differentially methylated in MGMT+ versus MGMT- melanoma cell lines (Fig. 3F, p=0.00011) and that treatment with the DNA demethylating agent 5-azacitidine restored MGMT expression in MGMT- cells (Fig. 3G). However, it is important to note that in this setting MGMT expression correlates with sensitivity, rather than resistance, to BRAF/MEK/HDAC inhibitors. Regardless, this pre-existing clinical test could be readily implemented to select patients for clinical trials. Importantly, two separate studies have shown that the MGMT promoter is methylated in only 21.5–26.0% of human metastatic melanomas using this clinical assay (27, 28). Therefore, these observations suggest that up to 79% of patients with RAS/RAF pathway mutations could benefit from this combination.
To determine whether MGMT was a functional or passive biomarker we genetically ablated it in sensitive cells and ectopically expressed it in resistant cells. Genetic ablation of MGMT did not make sensitive cells become resistant to these agents (Fig. 3H) and ectopic MGMT expression did not confer sensitivity (Fig. 3I). Therefore, we conclude that MGMT does not play a functional role in mediating sensitivity or resistance to this drug combination, but rather marks a distinct subset of melanomas. Regardless, the fact that a clinical test already exists makes this a clinically useful biomarker.
MGMT expressing melanomas exhibit broader defects in DNA repair genes
The strong association between MGMT expression and drug sensitivity prompted us to determine whether we could use a larger set of (primary) tumors to identify a specific genetic defect, responsible for sensitivity or resistance, that might co-segregate with high or low MGMT expression. To maximize potential differences we compared the top 10% of tumors in the TCGA database expressing the highest levels of MGMT mRNA (MGMT+) with the bottom 10% of tumors, expressing the lowest levels of MGMT mRNA (MGMT−). Similar to cell line studies, comparative analysis of mutations and copy number changes did not identify any recurrent genetic alterations that were enriched in MGMT+ or MGMT− tumors. However, these tumor cohorts exhibited distinct transcriptional profiles.
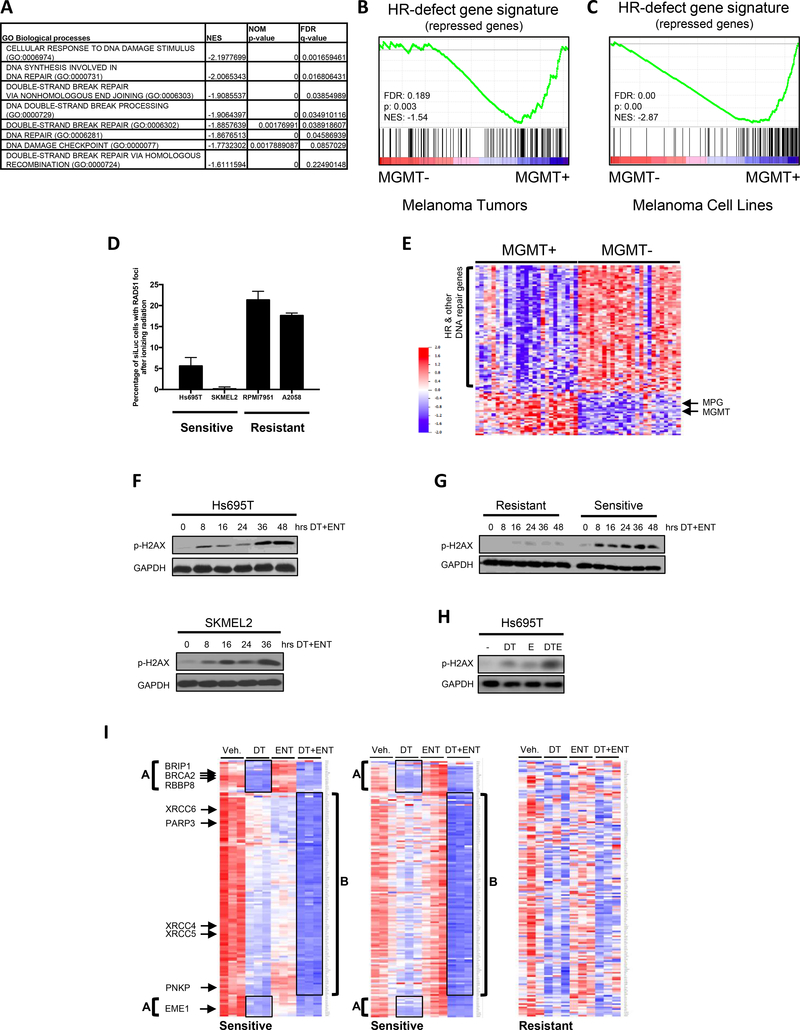
To identify potentially defective pathways and/or vulnerabilities in MGMT+ melanomas, we examined transcriptional signatures that were suppressed in these tumors using the GO:Biological Processes database. Interestingly, among the top 35 suppressed gene sets, numerous signatures associated with DNA repair, in particular double-strand break repair, were identified (Fig. 4A and Supplementary Table 2), suggesting that these tumors might possess inherent defects in DNA repair processes.
Figure 4: Baseline and drug induced suppression of DNA repair genes and consequential DNA damage in MGMT+ tumors/cells.
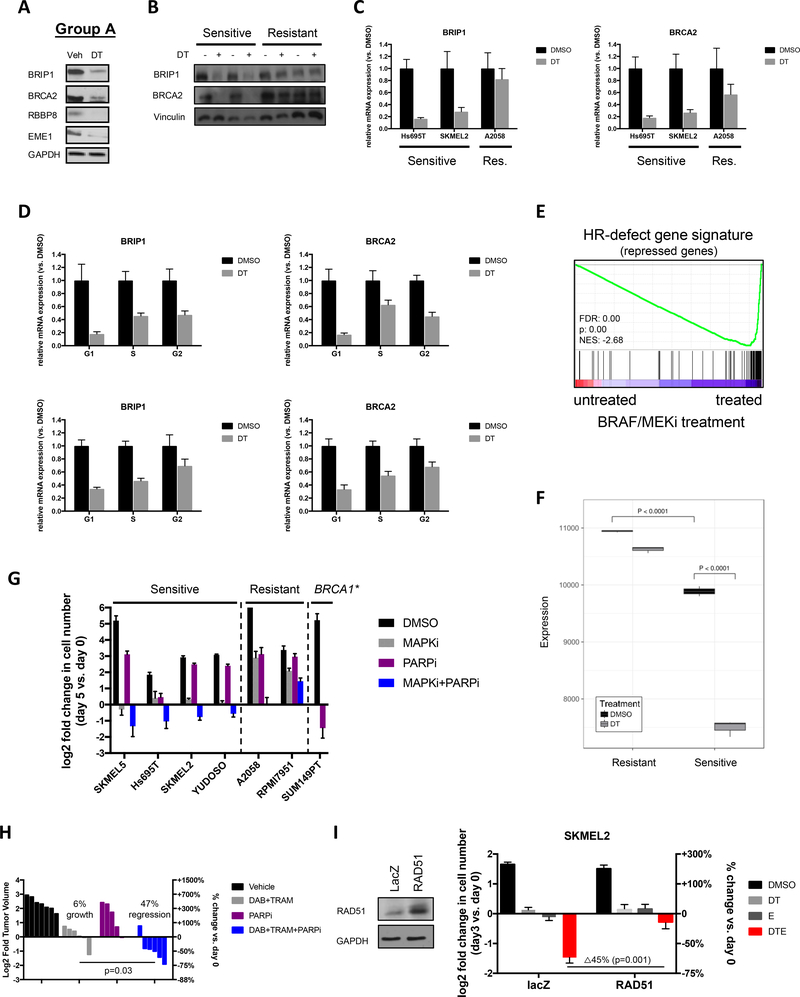
(A) Gene sets from the GO:Biological Processes database involved in DNA repair and HR are significantly suppressed in MGMT “high” versus MGMT “low” samples. “High” expression was defined as the top 10% of tumors in the TCGA SKCM provisional dataset stratified by MGMT mRNA expression, whereas “low” was defined as the bottom 10%. Normalized Enrichment Scores (NES), p-values and False Discovery Rates (FDRs) calculated by Gene Set Enrichment Analysis (GSEA) software from the Broad Institute. The entire list of the top 35 suppressed gene sets are shown in Supplementary Table 2. (B) Gene Set Enrichment Analysis (GSEA) was used to develop a plot depicting the enrichment of the HR-defect (HRD) gene signature (29) in primary tumors with high versus low MGMT expression. “High” expression (MGMT+) was defined as the top 10% of tumors in the TCGA SKCM provisional dataset stratified by MGMT mRNA expression, whereas “low” (MGMT-) was defined as the bottom 10%. Note: The HRD gene signature is a compilation of genes that are significantly suppressed after single-gene ablation of HR components (29). MGMT+ tumors show a reduction of these same genes. FDR: false discovery rate, NES: normalized enrichment score. (C) Gene Set Enrichment Analysis (GSEA) was used to develop a plot depicting the enrichment of the HR-defect (HRD) gene signature (29) in cell lines with high versus low MGMT expression. “High” expression was defined as the top 10% of melanoma cell lines in the CCLE stratified by MGMT mRNA expression, whereas “low” was defined as the bottom 10%. Note: The HRD gene signature is a compilation of genes that are significantly suppressed after single-gene ablation of HR components (29). MGMT+ cells show a reduction of these same genes. FDR: false discovery rate, NES: normalized enrichment score. (D) Melanoma cells, transfected with control (siLuc) or pooled siRNAs targeting BRCA2 were irradiated with 10Gy. Cells were fixed 5 hours later and analyzed by immunofluorescence for RAD51 and gamma H2AX. The bar graph depicts the percentage of siLuc cells with RAD51 foci. Images are shown in Supplementary Fig. 4A. (E) Heatmap of differentially expressed DNA repair genes (significance level p=0.001) in (MGMT+) versus (MGMT-) melanomas. (F) Western blot depicting phospho-gamma H2AX levels in two sensitive cell lines in response to dabrafenib/trametinib (DT) plus entinostat (ENT) over time. GAPDH serves as a loading control. (G) Western blot comparing phospho-gamma H2AX levels in sensitive and resistant cell lines after treatment with DT+ENT. GAPDH serves as a loading control. (H) Western blot depicting phospho-gamma H2AX levels in sensitive cell line after treatment with either DMSO, 100nM dabrafenib and 10nM trametinib (DT), 1μM entinostat (E) or DT+E. GAPDH serves as a loading control. (I) Microarray analysis of sensitive and resistant cell lines after 24 hours of treatment with vehicle, 100nM dabrafenib/10nM trametinib (DT), 1μM entinostat (ENT), or DT+ENT. Heatmap (triplicate samples per treatment arm) depicts the upregulated (red) or downregulated (blue) non-cell cycle regulated DNA repair genes, reaching a significance level of p=0.001. Group A defines a subset of potently suppressed genes that cluster together in two groups in the DT treatment arm. A subset of these genes are listed. Group B defines an additional large set of DNA repair genes suppressed in DT+ENT arm. A subset of these genes are also listed.
Concomitantly, we examined transcriptional profiles of sensitive cells exposed to MAPK/HDAC inhibitors, prior to the onset of cell death. In drug-treated cells, the Hallmark_DNA_Repair gene set was identified as one of the top significantly suppressed pathways in response to dabrafenib/trametinib/entinostat, suggesting that DNA repair processes were also being inhibited by this combination (Supplementary Table 3). These observations raised the intriguing possibility that preexisting defects in DNA repair, compounded by the chemical suppression of these pathways, might be mediating cell death by causing excessive DNA damage. Of note, an oxidative phosphorylation signature was also observed, but we found that reactive oxygen species were not consistently elevated by these agents, and were neither necessary nor sufficient for this therapeutic response (Supplementary Fig. 3A–E).
To investigate a potential role for DNA repair defects in this response, we further characterized MGMT+ tumors and cell lines. The transcriptional signatures identified in Fig. 4A suggested that MGMT+ tumors might harbor defects in homologous recombination (HR). Using a more specific transcriptional dataset we confirmed that MGMT+ tumors were, in fact, enriched for a common signature that is induced by RNAi-mediated ablation of many HR genes, termed the HR-defect gene signature (Fig. 4B) (29). The HR-defect signature was also enriched in MGMT+ melanoma cell lines as compared to MGMT- cells (Fig. 4C). However, to confirm that sensitive cells harbor functional defects in HR, we performed a Rad51 redistribution assay. In response to ionizing radiation, Rad51 normally accumulates in numerous distinct foci at sites of DNA damage (30). However the appearance of these foci is prevented or reduced in cells with defects in various HR proteins (30). Indeed, while ionizing radiation induced a dramatic increase in Rad51 foci that co-localized with phospho-gamma H2AX in the two resistant cell lines, the two sensitive cell lines were substantially impaired in their ability to form Rad51 foci (Fig. 4D, Supplementary Fig. 4A). These observations demonstrate that sensitive cell lines harbor a preexisting impairment in HR.
MGMT+ melanomas exhibit a global reduction of DNA repair genes
In breast and ovarian cancers, BRCA1/2 mutations are known to underlie defects in HR (31). However, more recently castration-resistant prostate cancers have been shown to harbor alterations in any one of five different HR genes (32). Therefore we reanalyzed genomic data to determine whether we might have missed a similar heterogeneous mutational pattern of HR genes but did not find this to be the case.
Because there were no recurrent genetic alterations in HR genes, the relative expression levels of all genes implicated in DNA repair were examined in MGMT+ versus MGMT- tumors (list obtained from (33)). Strikingly, the majority of DNA repair genes were downregulated in MGMT+ tumors as compared to MGMT− tumors (Fig. 4E). Genes known to be involved in HR as well as other DNA repair pathways were suppressed in MGMT+ tumors. Importantly, this observation held true even when known cell cycle-regulated genes were removed from the analysis, reinforcing that this is a true DNA repair effect, and not a by-product of cell cycle differences between treatments (Supplementary Fig. 4B) (33). A subset of repair genes, which included MGMT and MPG, both involved in the repair of alkylated bases, along with genes that repair hydrolyzed and oxidized bases, clustered together and exhibited the inverse expression pattern as compared to the broader group of genes: these genes were expressed at higher levels in MGMT+ tumors and were lower in MGMT- tumors (Fig. 4E). Importantly, this distinct expression pattern of DNA repair genes was not associated with prior exposure to therapy, suggesting that these genes were not selectively repressed or induced as a consequence of treatment. Therefore, there appears to be two potentially distinct populations of melanomas: DNA repair low(MGMT+) and DNA repair intact(MGMT−), which we hypothesize are differentially sensitive to BRAF/MEK/HDAC inhibitors. While the mechanisms that underlie the inverse expression pattern of these gene clusters could not be ascertained, it is well known that genes with similar functions are often co-regulated. We believe that this may be occurring in melanoma, especially given the related function of the co-regulated genes.
HDAC and MAPK pathway inhibitors cooperatively induce DNA damage
To determine whether the HDAC/MAPKi combination might be capitalizing on this potential defect and killing cells by triggering unresolvable DNA damage, we first compared phospho-gamma H2AX expression levels, a marker of double strand DNA breaks, in sensitive and resistant cells. In sensitive cell lines two waves of phosphor-gamma H2AX induction were observed. Phospho-gamma H2AX initially increased within 8 hours of treatment, increased more dramatically between 24–36 hours, and remained elevated (Fig. 4F). Importantly, gamma H2AX phosphorylation occurred prior to cell death, which begins at 40 hours (Fig. 1E and Supplementary 1E), suggesting that this event precedes cell death and is not merely a consequence of DNA damage in dying cells. In contrast, phospho-gamma H2AX levels remained low in resistant cells (Fig. 4G). Notably, both agents induced low levels of gamma H2AX phosphorylation as single agents; however, the drug combination was required to induce maximal DNA damage, demonstrating that these agents cooperatively induce DNA damage (Fig. 4H).
BRAF/MEK and HDAC inhibitors coordinately suppress the expression of DNA repair genes
To investigate the molecular mechanism by which this combination was functioning, transcriptional profiles were examined in cells treated with vehicle, dabrafenib/trametinib, entinostat or all three agents at 24 hours, prior to the commencement of cell death. Striking changes in the expression of DNA repair genes in response to these agents were observed. As depicted in the heatmaps shown, dabrafenib/trametinib alone suppressed the expression of many DNA repair genes in sensitive cell lines, which clustered together in two groups, herein referred to as Group A genes (Fig. 4I, left and middle panels). Interestingly, Group A genes included several HR pathway genes such as BRCA2, BRIP1, EME1 and RBBP8 (33, 34). These four genes were suppressed between 60–84%. Entinostat had no effect on the expression of group A genes but in both sensitive cell lines, dabrafenib/trametinib/entinostat together not only inhibited Group A genes but also suppressed an additional large set of DNA repair genes (Group B) (Fig. 4I, left and middle panels). Notably, the most potently suppressed genes in Group B (suppressed 65–80%) included genes that function in the NHEJ pathway (e.g. XRCC4, XRCC5, XRCC6, PNKP and PARP3). This striking pattern of transcriptional repression was not observed in resistant cells (Fig. 4I, right panel).
It should be noted that HDAC inhibitors have previously been reported to induce DNA damage in cells through a variety of direct and indirect mechanisms (35–37). Nevertheless, in this setting we observed a potent and broad suppression of DNA repair genes that only occurred in the presence of MAPK pathway inhibitors, which was quite unexpected. Therefore, while HDAC inhibitors may contribute to DNA damage via additional mechanisms, the deep suppression of numerous DNA repair genes that occurs in response to this combination is likely to play a major role in this response. The observation that DNA repair gene networks are already impaired in sensitive cell lines further supports this model.
MAPK pathway inhibitors potently suppress HR pathway genes in sensitive but not resistant melanomas
As noted above, Group A genes included several HR pathway genes such as BRCA2, BRIP1, EME1 and RBBP8 (Fig. 4I) (33, 34). Western blot analysis confirmed that dabrafenib/trametinib potently suppressed BRIP1, BRCA2, RBBP8 and EME1 protein expression in sensitive cells (Fig. 5A). BRIP1 and BRCA2 expression were further evaluated in two sensitive and resistant cell lines. Dabrafenib/trametinib nearly depleted the expression of BRIP1 and BRCA2 proteins in sensitive cell lines, but did not do so in resistant cells (Fig. 5B). Quantitative PCR further confirmed that BRAF/MEK inhibitors suppressed the transcription of these HR genes, and did so in sensitive but not resistant cells (Fig. 5C).
Figure 5: MAPK pathway inhibitors potently reduce the expression of HR pathway genes in sensitive melanomas.
(A) Western blot depicting BRIP1, BRCA2, RBBP8 and EME1 protein levels in Hs695T cells after 48hrs of treatment with vehicle (Veh) or 100nM dabrafenib and 10nM trametinib (DT). GAPDH serves as a loading control. (B) Western blot depicting BRIP1 and BRCA2 protein levels in sensitive (Hs695T and SKMEL2) and resistant (A2058 and RPMI7951) cells after 48hrs of treatment with vehicle (Veh) or 100nM dabrafenib and 10nM trametinib (DT). GAPDH serves as a loading control. (C) mRNA levels of BRIP1 and BRCA2 in sensitive (Hs695T and SKMEL2) and resistant (A2058) cells after 24 hours of treatment with vehicle (Veh) or 100nM dabrafenib and 10nM trametinib (DT) as determined by quantitative PCR. (D) Cells from 2 sensitive cell lines (top and bottom panels, respectively) were separated in different phases of the cell cycle for subsequent gene expression analysis. Bar graphs show mRNA expression of BRCA2 and BRIP1 in specific cell subpopulations after 24 hours of treatment with Vehicle (Veh) or 100nM dabrafenib and 10nM trametinib (DT). (E) Plot of Gene Set Enrichment Analysis (GSEA) showing suppression of HRD associated genes (29) in melanoma cells in response to trametinib (10nM) and dabrafenib (100nM). Note: The HRD gene signature is a compilation of genes that are significantly suppressed after single-gene ablation of HR components (29). Cells treated with BRAF/MEKi show a reduction of these same genes. FDR: false discovery rate, NES: normalized enrichment score. (F) Single Sample Gene Set Enrichment Analysis (ssGSEA) depicting the relative expression of HRD signature genes in response to dabrafenib/trametinib (DT) treatment in sensitive and resistant cell lines. Note: The HRD gene signature is a compilation of genes that are significantly suppressed after single-gene ablation of HR components (29). (G) Sensitive cells (SKMEL5, Hs695T, SKMEL2 and YUDOSO) and resistant cells (A2058 and RPMI7951) were treated with DMSO, 100nM dabrafenib/10nM trametinib (DT), 5μM olaparib (PARP inhibitor) or the combination of DT + PARP inhibitor. As a positive control the BRCA1-mutant breast cancer cell line SUM149PT was treated with the PARP inhibitor. Cells were manually counted prior to the addition of compounds and 5 days after treatment. Graphs represent log2 transformation of the fold change in cell number at day 5 versus day 0. Negative values represent a net loss of cells. (H) Waterfall plot depicting change in tumor volume in a Braf/Nf1-mutant melanoma allograft model after 9 days of treatment with single and combined agents as indicated. Each bar represents an individual tumor (Vehicle n=7, DAB+TRAM n=6, PARPi n=5, DAB+TRAM+PARPi n=6). Left axis indicates the log2 of fold change in tumor volume, and right axis indicates the percentage change in tumor volume relative to day 0. Dabrafenib (DAB, BRAF inhibitor), trametinib (TRAM, MEK inhibitor), olaparib (PARPi, PARP inhibitor). (I) Overexpression of RAD51 suppresses death in response to MAPK/HDAC inhibition. Western blot confirms overexpression.
To exclude the possibility that differences in expression were a secondary consequence of effects on the cell cycle, sensitive cells were treated with vehicle or dabrafenib/trametinib and cells in G1, S and G2/M were separated by FACS. In both sensitive cell lines, BRIP1 and BRCA2 mRNA levels were suppressed in all phases of the cell cycle (Fig. 5D), demonstrating that the suppression of these genes is not merely due to changes in cell cycle distribution.
MAPK pathway inhibitors induce a BRCAness phenotype by further suppressing the transcription of HR pathway genes
Figures 4A–E demonstrate that sensitive melanomas already have broad defects in the expression of DNA repair genes that result in HR defects, as inferred by transcriptional profiles and validated by Rad51 redistribution assays. However, Fig. 5A–C demonstrate that dabrafenib/trametinib causes a nearly complete suppression of a subset of these HR genes, which would presumably enhance these defects. Indeed, we found that dabrafenib/trametinib caused a potent enrichment of the HR-defect signature in sensitive cells (Fig. 5E). In order to visualize the progressive suppression of this pathway, we performed ssGSEA analysis on sensitive and resistant cell lines. Figure 5F demonstrates that genes that are suppressed in response to HR defects are lower in (untreated) sensitive compared to resistant cells (sample 3 versus sample 1, p<0.0001). Moreover, dabrafenib/trametinib causes a further potent reduction of these genes in sensitive cells (sample 4 versus 3, p<0.0001). Taken together these observations suggest that BRAF/MEK inhibitors are potentiating inherent defects in the HR pathway by suppressing multiple HR pathway genes.
The Rad51 redistribution experiment confirmed that there are substantial defects in HR in untreated sensitive cells, however the dynamic range of this assay precluded us from determining whether BRAF/MEK inhibitors could further potentiate these defects. HR impairment can also be measured by a I-Sce-I-based double strand break repair assay, however this requires reporter integration into a single genomic site, specifically in sensitive cells, which was unachievable after extensive effort. However, it is well established that genetic defects in HR genes, such as BRCA1 and BRCA2, confer sensitivity to PARP inhibitors, which has become a hallmark functional assay for measuring HR defects (31). Given the potent suppression of HR gene expression, in particular BRCA2, we hypothesized that BRAF/MEK inhibitors would sensitize (or further sensitize) responsive cell lines to PARP inhibitors.
Interestingly, we found that the PARP inhibitor, olaparib, did not exert potent cytotoxic effects as a single agent in sensitive melanomas, in contrast to BRCA1-deficient breast cancer cells (Fig. 5G). These observations suggest that the DNA repair defects present in these melanomas are less pronounced than those in BRCA1-mutant tumor cells, consistent with the observation that these cells still express HR genes (Fig. 5A). However, dabrafenib/trametinib potently sensitized melanomas to olaparib and together dabrafenib/trametinib/olaparib killed multiple sensitive cell lines and did not kill non-responsive cell lines (Fig. 5G). Strikingly, these observations were also recapitulated in vivo. While the PARP inhibitor olaparib alone was unable to cause tumor regression, when combined with dabrafenib/trametinib, tumors regressed by 47% on average (p=0.03, Fig. 5H). Taken together these results suggest that while baseline defects in DNA repair gene expression prime sensitive cells, MAPK pathway inhibition is required to functionally impair the HR pathway in these melanomas to the extent observed in BRCA1-mutant breast cancers.
Finally, to prove that defects in HR functionally contribute to the therapeutic response to combined MAPK/HDAC inhibitors, we examined the effects of Rad51 overexpression. A key step in HR is the recruitment of the Rad51 recombinase to double strand breaks, which normally requires HR proteins such as BRCA2 (34). However it is well known that overexpression of Rad51 can override this regulatory step in cells with various upstream HR defects (34). Therefore we reasoned that Rad51 would be the one component that might rescue effects caused by the concomitant suppression of multiple HR proteins. Importantly, Rad51 overexpression suppressed death in response to MAPK/HDAC inhibition, confirming that defects in HR and consequently excessive DNA damage play critical functional roles in mediating the therapeutic response to these agents (Fig. 5I). It should be noted that Rad51 did not rescue the limited response to BRAF/MEK inhibitors alone, indicating that these agents mediate their canonical effects through other pathways, as would be expected.
MAPK and HDAC inhibitors cooperatively suppress NHEJ genes in sensitive melanomas
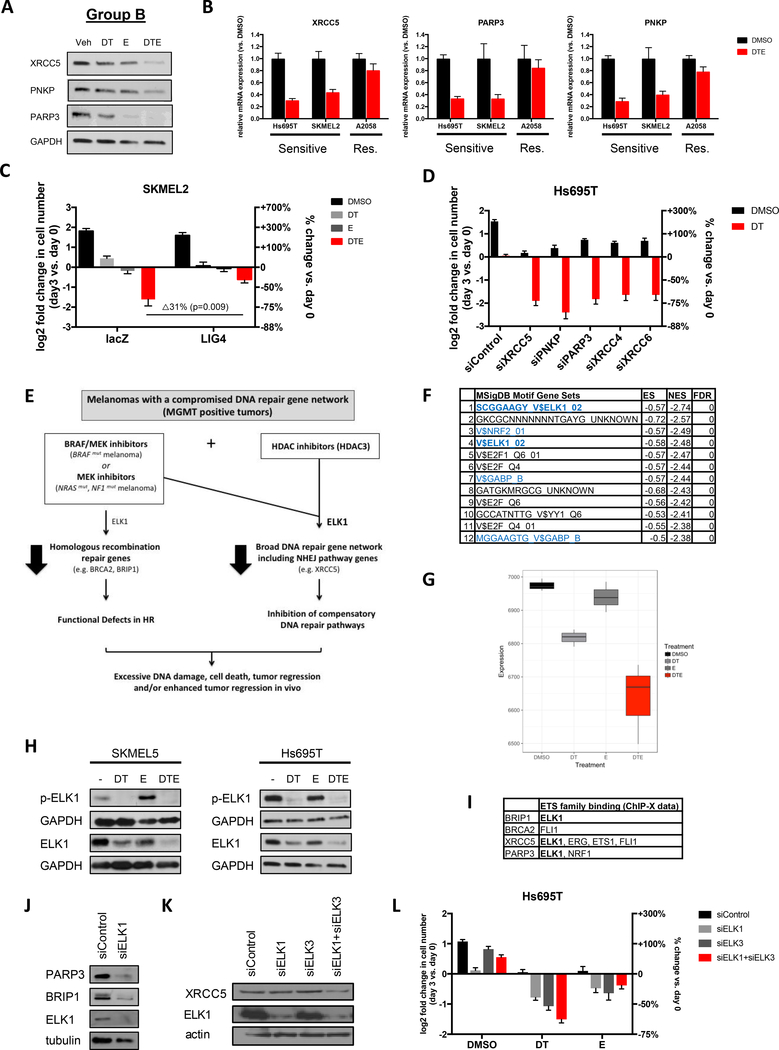
We next sought to identify critical genes in Group B that might be responsible for the cooperativity between dabrafenib/trametinib and entinostat. Interestingly, the most potently suppressed Group B genes that could be ascribed to a specific DNA repair pathway are known to function in the NHEJ pathway (a subset listed in Fig. 4I). Importantly, it is well established that when HR is impaired, NHEJ can compensate (38). Therefore we examined a subset of genes in the NHEJ pathway that were the most transcriptionally suppressed, specifically XRCC5 (33, 39), PNKP (33, 39), and PARP3 (33, 40), and confirmed that protein expression was also potently inhibited by dabrafenib/trametinib/entinostat (Fig. 6A). We also showed that suppression occurred at the level of transcription, occurred in different sensitive cell lines, and that these genes were not suppressed in insensitive cells (Fig. 6B). Together, these results demonstrate that combined HDAC and MAPK inhibitors potently suppress both HR and NHEJ genes.
Figure 6: Entinostat enhances dabrafenib/trametinib activity by cooperatively triggering the broad suppression of other DNA repair genes including NHEJ genes.
(A) Western blot depicting XRCC5, PNKP and PARP3 protein levels in Hs695T cells after treatment with vehicle (Veh), 100nM dabrafenib/10nM trametinib (DT), 1μM entinostat (E) or DT+E. GAPDH serves as a loading control. (B) mRNA levels of XRCC5, PARP3 and PNKP in sensitive (Hs695T and SKMEL2) and resistant (A2058) cells after 24 hours of treatment with vehicle (Veh) or 100nM dabrafenib, 10nM trametinib and 1μM entinostat (DTE) as determined by quantitative PCR. (C) Overexpression of LIG4 suppresses cell death in response to MAPK/HDAC inhibition. (D) Hs695T cells were transfected with pooled siRNAs targeting XRCC5, PNKP, PARP3, XRCC4, XRCC6 or control non-targeting siRNAs and treated with DMSO (black) or 100nM dabrafenib and 10nM trametinib (DT, red). Graph depicts the mean log2 fold change of cell number after 72 hours, relative to day 0. Right axis shows percent change in cell number relative to day 0. Immunoblots in Supplementary Fig. 5A confirm siRNA target suppression. (E) Model depicting the mechanism by which BRAF/MEK pathway and HDAC inhibitors promote excessive DNA damage, cell death and tumor regression of melanomas with a compromised DNA repair gene network (MGMT+ tumors). (F) Table depicting top scoring transcription factor binding sites enriched in genes suppressed by BRAF/MEK/HDACi, using the MSigDB Transcription Factor Motif database. (blue = ETS family transcription factor binding sites; bold: ELK1 binding sites) (G) Single Sample Gene Set Enrichment Analysis (ssGSEA) projection scores for ELK1 target genes following drug treatment. ssGSEA quantifies ELK1 target gene activation for each treatment group, showing a decrease in ELK1-driven transcription in single agent treatment (MAPKi (DT) and HDACi (E)) followed by further suppression of ELK1 targets by combination therapy (DTE). (H) Western blots depicting phosphor-ELK1 and total ELK1 protein levels in 2 sensitive cell lines treated with 100nM dabrafenib (D), 10nM trametinib (T) and/or 1μM entinostat (E). (I) Experimentally identified direct ETS family binding sites (ChIP-X data) in BRIP1, BRCA2, XRCC5 and PARP3. (J,K) Western blots depicting PARP3, BRIP1, XRCC5 and ELK1 protein levels after knockdown of ELK1 and/or ELK3 using siRNA smartpools. (L) RNAi-mediated suppression of ELK1 and/or ELK3 cooperates with either MAPK (Dabrafenib/Trametinib, DT) or HDAC (Entinostat, E) inhibitors.
The cooperative effects of MAPK/HDAC inhibition are due to the coordinate suppression of HR and NHEJ pathways
To functionally determine whether the suppression of NHEJ pathways was required for cell death in response to this combination, we ectopically expressed Lig 4. Lig 4 directly mediates the NHEJ ligation step and its overexpression, analogous to Rad51 overexpression, is the one distal gene that can largely override deficiencies in (multiple) upstream components (39). Importantly, Lig 4 substantially inhibited the cytotoxic effects of MAPK/HDAC suppression (Fig. 6C; p=0.009). Taken together with the Rad51 overexpression experiment in Fig. 5I, these studies demonstrate that the impairment of both HR and NHEJ are required for cell death in response to combined MAPK/HDAC inhibition.
The converse experiment was also performed. We reasoned that if cell death was occurring because 1) MAPK pathway inhibitors suppress HR and 2) MAPK/HDAC inhibitors together suppress NHEJ, then ablation of NHEJ genes should kill cells in the presence of MAPK pathway inhibitors. Individual NHEJ genes were suppressed by pooled siRNA sequences and cells were treated with dabrafenib/trametinib. Suppression of any one of the NHEJ genes killed cells but only in the presence of dabrafenib/trametinib (Fig. 6D and Supplementary Fig. 5A), suggesting that NHEJ pathway suppression was able to recapitulate the effects of entinostat in this context.
Taken altogether these observations suggest that BRAF/MEK inhibitors suppress DNA repair genes function in the HR pathway, resulting in a state that functionally resembles tumors with BRCA pathway defects (model shown in Fig. 6E) (38). However, when combined with entinostat, these agents now cooperatively and potently suppress a second broader set of DNA repair genes, including NHEJ pathway genes. Importantly, this broad transcriptional repression of these major DNA repair genes is lethal in tumors with a preexisting impairment in a DNA repair gene expression.
HDAC and MAPK pathway inhibitors cooperatively suppress ELK, which regulates the expression of key DNA repair genes
To investigate the molecular mechanism(s) by which MAPK and HDAC inhibitors might be cooperatively suppressing DNA repair genes, we first identified transcription factor binding sites that were enriched in genes suppressed by this combination, using the MSigDB Transcription factor targets database. ELK1 binding sites were the most significantly enriched in the suppressed genes and several other ETS family members binding sites were also identified (Fig. 6F). ssGSEA analysis was then used to examine the pattern of ELK1 regulated transcriptional signatures in response to single and combined agents. MAPK suppression partially inhibited these signatures, as expected, given that ERK phosphorylation contributes to the activation of ELK1 (41) (Fig. 6G). HDAC inhibitors slightly suppressed ELK1 signatures, however these signatures were maximally suppressed in the presence of combined MAPK/HDAC inhibitors (Fig. 6G). We found that ELK1 phosphorylation was inhibited in response to BRAF/MEK inhibitors as would be predicted (Fig. 6H). However, unexpectedly both agents individually and cooperatively suppressed total ELK1 protein expression. Microarray data indicated that this was occurring at the level of transcription, which was confirmed by Q-PCR (Supplementary Fig. 5B). The related gene, ELK3, showed a similar pattern of suppression (Supplementary Fig. 5B).
Twenty percent of the DNA repair genes suppressed by MAPK/HDAC inhibitors have been experimentally determined to be direct ELK1 targets (32/160 genes; (42)) and many more of these genes are targets of other ETS family proteins. For practical reasons we focused on determining whether ELK was controlling the expression of any of the most prominently suppressed group A and B genes (BRIP1, BRCA2, XRCC5 and PARP3). Experimental ChIP data indicate that ETS family proteins directly bind sites in the promoters of all four genes, although only BRIP1, XRCC5 and PARP3 contain the ELK1 target sequence (Fig. 6I). Because ELK1 and 3 bind the same DNA sequences and therefore may be redundant, we investigated the consequences of genetically ablating ELK1 and/or ELK3. Notably, ELK1 ablation was sufficient to potently suppress the expression of PARP3 and BRIP1 (Fig. 6J), whereas XRCC5 expression was inhibited by the loss of both ELK1 and 3 (Fig. 6K). The less dramatic effect on XRCC5 expression, as compared to PARP3 and BRIP1, may be due to the presence of several ETS family binding sites in its promoter (Fig. 6I). Nevertheless, these observations demonstrate that ELK1 critically regulates the expression of these important HR and NHEJ proteins.
More importantly however, we found that RNAi-mediated suppression of ELK genes cooperate with either MAPK or HDAC inhibitors to kill these melanomas (Fig. 6L), consistent with the observation that ELK regulates both types of genes (e.g. HR and NHEJ) (Fig. 6l). The effects of ELK suppression were more pronounced when combined with MAPK inhibitors, versus HDAC inhibitors, as might be expected, since it is likely that other MAPK pathway targets are important in BRAF-mutant melanomas. Growth arrest triggered by ELK1 overexpression precluded rescue experiments. However the observation that 1) ELK activity and expression is potently suppressed in drug treated cells, 2) BRIP1, PARP3, XRCC5 (and many other DNA repair genes) are direct ELK1 targets, 3) ELK suppression substantially inhibits the expression of these genes in melanomas and 4) ELK ablation can partially recapitulate the effects of both drugs, all support the conclusion that ELK critically regulates this DNA repair network in response to HDAC/MAPK inhibitors. Other ETS family members may play a cooperative role in this process as well. Taken together, these findings provide a mechanistic basis for understanding the cooperativity of MAPK and HDAC inhibitors on DNA repair pathways and cell death.
Discussion
Using multiple animal models of BRAF-mutant melanoma we have shown that the Class I HDAC inhibitor, entinostat, dramatically enhances the efficacy of BRAF/MEK inhibitors, both in tumors that are sensitive and relatively resistant to these agents. In addition we have uncovered a strategy for treating NRAS- and NF1-mutant tumors, for which there are currently no approved targeted therapies. Finally, we have elucidated the mechanism by which these agents function and have discovered that a clinically available biomarker can be used to select patients that are most likely to benefit from MAPK/HDAC inhibitor combinations. Together these findings provide a promising and readily translatable strategy for improving treatments for these RAS/RAF pathway-driven melanomas.
Interestingly, in the course of this work we also discovered a population of melanomas that exhibit unconventional defects in DNA repair. Mutations in BRCA1 or BRCA2 represent examples of classical cancer-associated DNA repair defects, and confer sensitivity to agents that stall replication forks, such as PARP inhibitors (31, 43). However, additional alterations have been proposed to phenocopy these mutations, resulting in a state sometimes referred to as ‘BRCAness’ (discussed in (38)). Examples of this include mutations in other HR genes, hypermethylation of BRCA genes, amplification of genes that disrupt HR, and mutations in the transcriptional regulator CDK12. Here we observe a related, but more complex paradigm. “Sensitive” melanomas exhibit measureable defects in HR genes due to the transcriptional suppression of a broad group of DNA repair genes (29). Nevertheless, these defects are not potent enough to reach a functional threshold that confers sensitivity to PARP inhibitors alone. Intriguingly, BRAF/MEK pathway inhibitors unmask these defects, by further reducing the expression of several HR/BRCA pathway genes, thereby triggering a BRCA-like state (38). Accordingly, these studies reveal a new type of ‘priming’ defect in melanomas. Moreover, they demonstrate that in this setting BRAF/MEK inhibitors can broadly suppress the expression of DNA repair genes, most notably in the HR/BRCA pathways, thereby creating a new therapeutic vulnerability.
Importantly, we show that combined suppression of BRAF/MEK and HDAC3 capitalizes on this vulnerability. While BRAF/MEK pathway inhibitors suppress one set of DNA repair genes, most notably HR pathway genes, the addition of entinostat induces a potent suppression of a broader set of genes including essential NHEJ genes. It should be noted that these effects are not due to an additive effect of MAPK and HDAC inhibition, but rather these agents cooperate by potently suppressing a broad transcriptional network of DNA repair genes. We have shown that key nodes of this network are regulated by ELK1, which is cooperatively suppressed by combined MAPK/HDAC suppression, via effects on phosphorylation and expression. Notably, ELK1 has been shown to directly bind and regulate 20% of the DNA repair genes that are suppressed by these agents, suggesting a broader role for ELK1 in maintaining this network. Other ETS family members may also contribute to this response.
It is well established that the NHEJ pathway can compensate for defects in HR. As such, synthetic lethal interactions between HR genes and NHEJ genes have been observed (38, 44). Here we have discovered a chemical synthetic lethality between BRAF/MEK inhibitors and Class I HDAC inhibitors, which when combined coordinately suppress these pathways. Importantly, we have experimentally demonstrated that suppression of both HR and NHEJ pathways are required for cell death.
While the molecular basis for the priming defect in melanomas is not known, it could be related to an epigenetic event or state, or alternatively caused by specific defects in transcriptional regulatory proteins. Fortunately however, this defect consistently segregates with MGMT expression, which has already proven to be a tractable clinical biomarker in other settings (25, 26). In this respect it is interesting to note that there are two clusters of DNA repair genes that show inverse expression patterns; a broad set of genes that are suppressed in MGMT+ tumors, and a smaller set of genes, several of which are also involved in directly repairing base modifications, that are high in MGMT+ tumors (and vice versa). This observation further supports the notion that these gene clusters are co-regulated in melanomas. Regardless, the identification of this potent therapeutic combination, coupled with the elucidation of its mechanism of action and a tractable predictive biomarker of efficacy, will enable new mechanism-based combination trials that have the potential to improve treatments for these three major melanoma subtypes.
Methods
Cell lines and reagents
All cell lines were purchased from ATCC, except for MALME3M (obtained from Levi Garraway, Dana-Farber Cancer Institute), Meljuso (obtained from William Hahn, Dana-Farber Cancer Institute), SUM149PT (obtained from Frank McCormick, University of California, San Francisco), Yugasp and Yudoso (obtained from Yale Dermatology Center) and WM3670 and WM3629 (obtained from Rockland Immunochemicals). No further authentication of these cell lines was performed. All of the cell lines were determined to be negative for mycoplasma using the MycoAlert Mycoplasma Detection Kit (Lonza, LT07–318). Cells were used for experiments within 15 to 20 passages from thawing. Antibodies were obtained from the following sources: Cell Signaling Technologies: p-ERK (4370), ERK (9102), GAPDH (2118), BRIP1 (4578), XRCC5 (2753), H3K56-Ac (4243), HDAC1 (5356), HDAC2 (5113), HDAC3 (3949), HDAC6 (7612), RAD51 (8875), p-ELK1 (9181), HA (3724), cleaved PARP (9541); Sigma Aldrich: Actin (A2066); EMD Millipore: H3K9-Ac (06–942), BRCA2 (OP95), p-H2AX (05–636); Santa Cruz Biotechnology: XRCC4 (365055), XRCC6 (17789), PARP3 (390771), EME1 (53275); Bethyl Laboratories: RBBP8 (A300–488A-M); Novus Biologicals: PNKP (NBP1–87257); BD Pharmingen: MGMT (557045); Abcam: ELK1 (32106), LIG4 (26039). Trametinib, dabrafenib and olaparib were purchased from LC labs. Vorinostat, mocetinostat, nexturastat and entinostat were purchased from Selleck Chemicals. 5-azacytidine, NAC, Vitamin C and BSO were purchased from Sigma-Aldrich. Carboxy-H2DCFDA was purchased from Life Technologies (#C400).
RNAi
Non-targeting, HDAC1, HDAC2, HDAC3, HDAC6, XRCC4, XRCC5, XRCC6, PARP3, PNKP, BRCA2, MGMT, ELK1 and ELK3 siRNA pools were purchased from GE Healthcare/Dharmacon (D-001810–10, L-003493–00, L-003495–02, L-003496–00, L-003499–00, L-004494–00, L-010491–00, L-005084–00, L-009297–00, L-006783–00, L-003462–00, L-008856–01, L-003885–00, L-010320–00, respectively). SiRNAs were transfected into cells using RNAiMax lipofectamine from Invitrogen. Cells were transfected approximately 24 hours before starting proliferation experiments. Control, HDAC1, HDAC2, HDAC3, and HDAC6 shRNAs were purchased from Sigma-Aldrich (SHC016, SHCLNG-NM_004964, SHCLNG-NM_008229, SHCLNG-NM_010411 and SHCLNG-NM_006044, respectively).
Expression constructs
cDNA clones were obtained from Sino Biologicals (MGMT) and the PlasmID Repository at Harvard Medical School (LIG4 and RAD51), sequence verified and subsequently cloned into the pHAGE-FLAG-HA mammalian lentiviral expression vector.
Cell growth assays
For the initial MAPK/HDACi screen and the dose-response matrix assay, cells were plated in 96-well plates. 5 replicates were done for each condition. At 24 hours, one plate of cells was frozen (−80°C) representing the time 0 plate. At this time compounds were added to the remaining plates. After 72 hours each of the plates was frozen. After freezing, the plates (day 0 and 3) were thawed simultaneously and cells were quantified using CellTiter-Glo (Promega) as per manufacturer’s instructions. SynergyFinder (45) was used to analyze drug combination dose-response matrix data. To determine the combination effects in excess of Loewe additivity, a Synergy Score was calculated to characterize the strength of synergistic interaction. For all other proliferation experiments, cells were seeded in triplicate into 6-well plates. Approximately 24 hours after plating, day 0 counts were taken using a hemocytometer. For inhibitor experiments, drug treatments were started at this time. Final cell counts were taken 72 hours after day 0 to determine changes in cell number versus day 0. For western blots to determine drug efficacy or efficient knockdown, lysates were collected 48 hours following the addition of inhibitors.
Live cell imaging
The live cell imager IncuCyte ZOOM (Essen Biosciences, Ann Arbor, MI, USA) was used for multiplexed measurements of cell proliferation alongside cell death in a single well. The mKate2 red fluorescent protein (Essen Biosciences) was transfected into the Hs695T melanoma cell line and selected for RFP-infected cells using puromycin. Successful insertion of the nuclear-restricted RFP was confirmed via direct visualization of transfected cells on the IncuCyte ZOOM fluorescent microscope. For proliferation experiments, stably transfected cells were plated in 96-well plates and allowed to settle overnight at 37°C. The green Incucyte Cytotox or Caspase-3/7 Apoptosis reagents were then added to the tissue culture growth media to assess real-time quantification of cell death. Red and green fluorescent objects were monitored in the Incucyte ZOOM acquiring images every 2 hours (for 72 hours) following treatment with vehicle or drugs and then quantified with the IncuCyte integrated analysis software. Relative cell death was defined as the ratio between cells with overlapping red and green cells (dead cells) and cells with red nuclei only (live cells).
Animal Studies and Treatments
Animal procedures were approved by the Center for Animal and Comparative Medicine in Harvard Medical School in accordance with the NIH Guide for the Care and Use of Laboratory Animals and the Animal Welfare Act. For cancer cell xenograft experiments immunodeficient Nu/Nu (A375 and Hs695T) or NSG (YUDOSO) mice were inoculated subcutaneously with 3×106 human BRAF- or NRAS-mutant melanoma cells. For allograft experiments, immunocompetent C56/Bl6 mice were inoculated subcutaneously with 1.5×105 murine Braf/Nf1-mutant melanoma cells or 4×105 murine Braf/Pten-mutant melanoma cells. Tumor volumes were calculated by measuring length and width of the lesions and with the formula [(length) x (width)2 × 0.52]. When tumors reached approximately 200–300mm3, mice were randomly divided into different treatment groups that were administered either the MEK inhibitor trametinib (0.6mg/kg QD, oral gavage (OG)), the BRAF inhibitor dabrafenib (30mg/kg QD, OG), the HDAC inhibitor entinostat (1mg/kg QW, intraperitoneal injection (IP)), the PARP inhibitor olaparib (10mg/kg, QD IP) or their combinations as indicated. To track changes in tumor volume, tumor size was measured at day 0 and subsequently every 3–5 days by Vernier calipers. Unpaired 2-tailed t-tests with unequal variance were used to compare data sets where indicated and p-values are shown.
MGMT methylation assay
MGMT methylation analysis was performed using the OneStep qMethyl kit (Zymo Research, D5310). Briefly, DNA from 7 different melanoma cell lines was digested with methylation sensitive restriction enzymes in the test reaction while DNA in the reference reaction was not. Following digestion, DNA from both samples was amplified using real-time PCR in the presence of fluorescent dye and quantified. Investigator was blinded to group allocation and when assessing outcome. MGMT primer sequences used: Forward: 5’-GCGCTCTCTTGCTTTTCTCA-3’, Reverse: 5’-CTGCAGGACCACTCGAGG-3’.
RAD51 foci assay
Melanoma cells transfected with control (siLuc) or siRNA pools against BRCA2 were irradiated with 10Gy. Cells were fixed 5 hours later and analyzed by immunofluorescence for RAD51 foci and gamma H2AX.
DCFDA staining
Cells were treated as indicated and then stained with H2DCFDA at 10μM and analyzed by flow cytometry.
Cell cycle phase separation
Live cells were separated in different phase of the cell cycle for subsequent gene expression analysis. Cells treated with vehicle or drugs were incubated with 10μg/ml Hoechst33342 for 1 hour at 37°C in the dark. After trypsinization, cells were resuspended at a concentration of 10×106 cells per ml for sorting. Cells were sorted based on the amount of DNA by defining two regions for sorting: one for G1 and the other for G2/M. The separated cells were collected in RNAse free conditions and further processed for RNA extraction.
Microarray and gene set enrichment analysis
RNA was isolated from Hs695T, SKMEL2 and A2058 cells 24 hours after treatment with indicated drugs. Prior to RNA extraction ERCC spike-in control mix was added to Trizol and RNA isolation was performed following the manufacturer’s protocol. RNA clean-up was performed using the Qiagen RNeasy kit (74201). The Molecular Biology Core Facility at Dana-Farber Cancer Institute hybridized RNA to the Affymetrix Human 2.0 ST array chip. Analyses used BRB-ArrayTools developed by R. Simon and the BRB-ArrayTools development team. Thresholds were set at p=0.001. Microarray data can be accessed from the Gene Expression Omnibus (GEO) database (accession number: GSE125565). Gene set enrichment analysis (GSEA) and ssGSEA was performed using the Broad Institute interface (http://software.broadinstitute.org/gsea/index.jsp). The signature of Homologous Recombination-associated genes (PENG_HRD_SIGNATURE) was developed from genome-wide transcriptome profiling of Homologous Recombination DNA Repair (29) and added to the KEGG signature file from MSIGDB for subsequent analysis (http://software.broadinstitute.org/gsea/msigdb/genesets.jsp?collection=CP:KEGG).
Tumor sample transcriptional profiles were obtained from the TCGA SKCM provisional dataset (http://firebrowse.org/?cohort=SKCM) and stratified based on median-centered z-scores of MGMT mRNA expression. The top 10% of MGMT expressors comprised the “MGMT+” cohort, whereas the bottom 10% of MGMT expressors comprised the “MGMT−” cohort used in GSEA analyses. Melanoma cell line transcriptional profiles were obtained from CCLE (https://portals.broadinstitute.org/ccle/home) and stratified based on gene-centric RMA-normalized MGMT mRNA expression. The top 10% of MGMT expressors comprised the “MGMT+” cohort, whereas the bottom 10% of MGMT expressors comprised the “MGMT-” cohort used in GSEA analyses. Experimental ChIP data were obtained from the ChIP-X database (http://amp.pharm.mssm.edu/Harmonizome/).
Statistical analysis for in vitro experiments
For quantitative measurements, graphs represent mean values +/− standard deviation. Where indicated the data is presented as log2 fold (left axis) and percent change (right axis) over initial measurements. Unpaired 2-tailed t-tests with unequal variance were used to compare data sets where indicated and p-values are shown. Data were graphed and analyzed using GraphPad Prism v.6.
Statement of replication
All proliferation studies in Figures 1, 3, 4, 5 and Supplementary Figures 1 and 3 were performed ≧ 3 times. Synergy studies were performed in 5 cell lines at different times with qualitatively similar findings. The large preclinical animal studies were performed one time, however qualitatively similar results were obtained in 5 different animal models and responses mimicked effects observed numerous times in vitro. The gamma H2AX experiment (Fig. 4D) is shown in 3 cell lines and has been repeated in a separate set of cell lines. The microarray experiment was performed once (3 technical replicates) in 3 different cell lines and key targets were confirmed by qPCR and western blot (n>3) as shown throughout Figures 5 and 6.
Supplementary Material
Statement of significance.
BRAF/MEK inhibitors are not typically curative in BRAF-mutant melanomas and are ineffective in NRAS- and NF1-mutant tumors. We show that HDAC inhibitors dramatically enhance the efficacy of BRAF/MEK inhibitors in sensitive and insensitive Ras pathway-driven melanomas, by coordinately suppressing two DNA repair pathways, and identify a clinical biomarker that predicts responsiveness.
Acknowledgements
This work was supported by grants from the NCI (R01CA111754, KC), the Ludwig Center at Harvard (KC, SE) and the NF Research Initiative at Boston Children’s Hospital made possible by an anonymous gift (OM).
Footnotes
Conflict of interest statement
K. Cichowski is a consultant for Genentech. S.J. Elledge is a consultant/advisory board member for CRISPR Therapeutics, Homology Medicines, Maze Therapeutics, X-CHEM, MPM Capital and Tenaya Therapeutics; and is a founder of Miramus and Maze Therapeutics. K. Flaherty serves on the Board of Directors of Loxo Oncology, Clovis Oncology, Strata Oncology and Vivid Biosciences; on the Corporate Advisory Boards of X4 Pharmaceuticals and PIC Therapeutics; on the scientific advisory boards of Sanofi, Amgen, Asana, Adaptimmune, Fount, Aeglea, Array BioPharma, Shattuck Labs, Arch Oncology, Tolero, Apricity, Oncoceutics, Fog Pharma, Neon Therapeutics and Tvardi; and as a consultant to Novartis, Genentech, BMS, Merck, Takeda, Verastem, Checkmate, Boston Biomedical, Pierre Fabre, Cell Medica and Debiopharm. L.A. Garraway is Sr. Vice President at Eli Lilly and Company and has ownership interest (including stock, patents, etc.) in Tango Therapeutic and Foundation Medicine. Based on subject matter, there is no conflict with the current manuscript. No potential conflicts of interest were disclosed by the other authors.
References
- 1.Cancer Genome Atlas N Genomic Classification of Cutaneous Melanoma. Cell 2015;161:1681–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Luke JJ, Flaherty KT, Ribas A, Long GV. Targeted agents and immunotherapies: optimizing outcomes in melanoma. Nature reviews Clinical oncology 2017;14:463–82 [DOI] [PubMed] [Google Scholar]
- 3.Long GV, Stroyakovskiy D, Gogas H, Levchenko E, de Braud F, Larkin J, et al. Dabrafenib and trametinib versus dabrafenib and placebo for Val600 BRAF-mutant melanoma: a multicentre, double-blind, phase 3 randomised controlled trial. Lancet 2015;386:444–51 [DOI] [PubMed] [Google Scholar]
- 4.Robert C, Karaszewska B, Schachter J, Rutkowski P, Mackiewicz A, Stroiakovski D, et al. Improved overall survival in melanoma with combined dabrafenib and trametinib. The New England journal of medicine 2015;372:30–9 [DOI] [PubMed] [Google Scholar]
- 5.Dummer R, Schadendorf D, Ascierto PA, Arance A, Dutriaux C, Di Giacomo AM, et al. Binimetinib versus dacarbazine in patients with advanced NRAS-mutant melanoma (NEMO): a multicentre, open-label, randomised, phase 3 trial. The Lancet Oncology 2017;18:435–45 [DOI] [PubMed] [Google Scholar]
- 6.Jones PA, Issa JP, Baylin S. Targeting the cancer epigenome for therapy. Nature reviews Genetics 2016;17:630–41 [DOI] [PubMed] [Google Scholar]
- 7.West AC, Johnstone RW. New and emerging HDAC inhibitors for cancer treatment. The Journal of clinical investigation 2014;124:30–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Falkenberg KJ, Johnstone RW. Histone deacetylases and their inhibitors in cancer, neurological diseases and immune disorders. Nature reviews Drug discovery 2014;13:673–91 [DOI] [PubMed] [Google Scholar]
- 9.Johannessen CM, Johnson LA, Piccioni F, Townes A, Frederick DT, Donahue MK, et al. A melanocyte lineage program confers resistance to MAP kinase pathway inhibition. Nature 2013;504:138–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yokoyama S, Feige E, Poling LL, Levy C, Widlund HR, Khaled M, et al. Pharmacologic suppression of MITF expression via HDAC inhibitors in the melanocyte lineage. Pigment cell & melanoma research 2008;21:457–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tsai J, Lee JT, Wang W, Zhang J, Cho H, Mamo S, et al. Discovery of a selective inhibitor of oncogenic B-Raf kinase with potent antimelanoma activity. Proceedings of the National Academy of Sciences of the United States of America 2008;105:3041–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Konieczkowski DJ, Johannessen CM, Abudayyeh O, Kim JW, Cooper ZA, Piris A, et al. A melanoma cell state distinction influences sensitivity to MAPK pathway inhibitors. Cancer discovery 2014;4:816–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fournel M, Bonfils C, Hou Y, Yan PT, Trachy-Bourget MC, Kalita A, et al. MGCD0103, a novel isotype-selective histone deacetylase inhibitor, has broad spectrum antitumor activity in vitro and in vivo. Molecular cancer therapeutics 2008;7:759–68 [DOI] [PubMed] [Google Scholar]
- 14.Miyake Y, Keusch JJ, Wang L, Saito M, Hess D, Wang X, et al. Structural insights into HDAC6 tubulin deacetylation and its selective inhibition. Nature chemical biology 2016;12:748–54 [DOI] [PubMed] [Google Scholar]
- 15.Connolly RM, Li H, Jankowitz RC, Zhang Z, Rudek MA, Jeter SC, et al. Combination Epigenetic Therapy in Advanced Breast Cancer with 5-Azacitidine and Entinostat: A Phase II National Cancer Institute/Stand Up to Cancer Study. Clinical cancer research 2017;23:2691–701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhu S, Denman CJ, Cobanoglu ZS, Kiany S, Lau CC, Gottschalk SM, et al. The narrow-spectrum HDAC inhibitor entinostat enhances NKG2D expression without NK cell toxicity, leading to enhanced recognition of cancer cells. Pharm Res 2015;32:779–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Oki Y, Buglio D, Fanale M, Fayad L, Copeland A, Romaguera J, et al. Phase I study of panobinostat plus everolimus in patients with relapsed or refractory lymphoma. Clinical cancer research : an official journal of the American Association for Cancer Research 2013;19:6882–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.King AJ, Arnone MR, Bleam MR, Moss KG, Yang J, Fedorowicz KE, et al. Dabrafenib; preclinical characterization, increased efficacy when combined with trametinib, while BRAF/MEK tool combination reduced skin lesions. PloS one 2013;8:e67583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Maertens O, Johnson B, Hollstein P, Frederick DT, Cooper ZA, Messiaen L, et al. Elucidating distinct roles for NF1 in melanomagenesis. Cancer discovery 2013;3:338–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Paraiso KH, Xiang Y, Rebecca VW, Abel EV, Chen YA, Munko AC, et al. PTEN loss confers BRAF inhibitor resistance to melanoma cells through the suppression of BIM expression. Cancer Res 2011;71:2750–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Whittaker SR, Theurillat JP, Van Allen E, Wagle N, Hsiao J, Cowley GS, et al. A genome-scale RNA interference screen implicates NF1 loss in resistance to RAF inhibition. Cancer discovery 2013;3:350–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Catalanotti F, Cheng DT, Shoushtari AN, Johnson DB, Panageas KS, Momtaz P, et al. PTEN Loss-of-Function Alterations Are Associated With Intrinsic Resistance to BRAF Inhibitors in Metastatic Melanoma. JCO Precision Oncology 2017;1:1–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Van Allen EM, Wagle N, Sucker A, Treacy DJ, Johannessen CM, Goetz EM, et al. The genetic landscape of clinical resistance to RAF inhibition in metastatic melanoma. Cancer discovery 2014;4:94–109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fu D, Calvo JA, Samson LD. Balancing repair and tolerance of DNA damage caused by alkylating agents. Nature reviews Cancer 2012;12:104–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hegi ME, Diserens AC, Gorlia T, Hamou MF, de Tribolet N, Weller M, et al. MGMT gene silencing and benefit from temozolomide in glioblastoma. The New England journal of medicine 2005;352:997–1003 [DOI] [PubMed] [Google Scholar]
- 26.Weller M, Felsberg J, Hartmann C, Berger H, Steinbach JP, Schramm J, et al. Molecular predictors of progression-free and overall survival in patients with newly diagnosed glioblastoma: a prospective translational study of the German Glioma Network. Journal of clinical oncology : official journal of the American Society of Clinical Oncology 2009;27:5743–50 [DOI] [PubMed] [Google Scholar]
- 27.Schraml P, von Teichman A, Mihic-Probst D, Simcock M, Ochsenbein A, Dummer R, et al. Predictive value of the MGMT promoter methylation status in metastatic melanoma patients receiving first-line temozolomide plus bevacizumab in the trial SAKK 50/07. Oncology reports 2012;28:654–8 [DOI] [PubMed] [Google Scholar]
- 28.Tuominen R, Jewell R, van den Oord JJ, Wolter P, Stierner U, Lindholm C, et al. MGMT promoter methylation is associated with temozolomide response and prolonged progression-free survival in disseminated cutaneous melanoma. International journal of cancer 2015;136:2844–53 [DOI] [PubMed] [Google Scholar]
- 29.Peng G, Chun-Jen Lin C, Mo W, Dai H, Park YY, Kim SM, et al. Genome-wide transcriptome profiling of homologous recombination DNA repair. Nature communications 2014;5:3361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gildemeister OS, Sage JM, Knight KL. Cellular redistribution of Rad51 in response to DNA damage: novel role for Rad51C. J Biol Chem 2009;284:31945–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Farmer H, McCabe N, Lord CJ, Tutt AN, Johnson DA, Richardson TB, et al. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature 2005;434:917–21 [DOI] [PubMed] [Google Scholar]
- 32.Dhawan M, Ryan CJ, Ashworth A. DNA Repair Deficiency Is Common in Advanced Prostate Cancer: New Therapeutic Opportunities. Oncologist 2016;21:940–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mjelle R, Hegre SA, Aas PA, Slupphaug G, Drablos F, Saetrom P, et al. Cell cycle regulation of human DNA repair and chromatin remodeling genes. DNA repair 2015;30:53–67 [DOI] [PubMed] [Google Scholar]
- 34.Moynahan ME, Jasin M. Mitotic homologous recombination maintains genomic stability and suppresses tumorigenesis. Nature reviews Molecular cell biology 2010;11:196–207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Robert C, Rassool FV. HDAC inhibitors: roles of DNA damage and repair. Adv Cancer Res 2012;116:87–129 [DOI] [PubMed] [Google Scholar]
- 36.Thurn KT, Thomas S, Raha P, Qureshi I, Munster PN. Histone deacetylase regulation of ATM-mediated DNA damage signaling. Molecular cancer therapeutics 2013;12:2078–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Miller KM, Tjeertes JV, Coates J, Legube G, Polo SE, Britton S, et al. Human HDAC1 and HDAC2 function in the DNA-damage response to promote DNA nonhomologous end-joining. Nat Struct Mol Biol 2010;17:1144–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lord CJ, Ashworth A. BRCAness revisited. Nature reviews Cancer 2016;16:110–20 [DOI] [PubMed] [Google Scholar]
- 39.Lieber MR. The mechanism of double-strand DNA break repair by the nonhomologous DNA end-joining pathway. Annual review of biochemistry 2010;79:181–211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Beck C, Boehler C, Guirouilh Barbat J, Bonnet ME, Illuzzi G, Ronde P, et al. PARP3 affects the relative contribution of homologous recombination and nonhomologous end-joining pathways. Nucleic acids research 2014;42:5616–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shaw PE, Saxton J. Ternary complex factors: prime nuclear targets for mitogen-activated protein kinases. Int J Biochem Cell Biol 2003;35:1210–26 [DOI] [PubMed] [Google Scholar]
- 42.Lachmann A, Xu H, Krishnan J, Berger SI, Mazloom AR, Ma’ayan A. ChEA: transcription factor regulation inferred from integrating genome-wide ChIP-X experiments. Bioinformatics 2010;26:2438–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lord CJ, Ashworth A. The DNA damage response and cancer therapy. Nature 2012;481:287–94 [DOI] [PubMed] [Google Scholar]
- 44.Hemann MT. From breaking bad to worse: exploiting homologous DNA repair deficiency in cancer. Cancer discovery 2014;4:516–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ianevski A, He L, Aittokallio T, Tang J. SynergyFinder: a web application for analyzing drug combination dose-response matrix data. Bioinformatics 2017;33:2413–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.