Abstract
Genetic perturbation in different genetic backgrounds can cause a range of phenotypes within a species. These phenotypic differences can be the result of the interaction between the genetic background and the perturbation. Previously, we reported that perturbation of gld-1, an important player in the developmental control of Caenorhabditis elegans, released cryptic genetic variation (CGV) affecting fitness in different genetic backgrounds. Here, we investigated the change in transcriptional architecture. We found 414 genes with a cis-expression quantitative trait locus (eQTL) and 991 genes with a trans-eQTL that were specifically found in the gld-1 RNAi treatment. In total, we detected 16 eQTL hotspots, of which 7 were only found in the gld-1 RNAi treatment. Enrichment analysis of those 7 hotspots showed that the regulated genes were associated with neurons and the pharynx. Furthermore, we found evidence of accelerated transcriptional aging in the gld-1 RNAi–treated nematodes. Overall, our results illustrate that studying CGV leads to the discovery of hidden polymorphic regulators.
Keywords: C. elegans, cryptic genetic variation, eQTL, genetic perturbation
Introduction
Cryptic genetic variation (CGV) is genetic variation that generates little or no phenotypic variation in a population under normal conditions. The effect of this hidden genetic variation can be unlocked by environmental or genetic perturbation (Gibson and Dworkin 2004). A classic example of CGV due to mutational perturbation was reported for the fruit fly Drosophila melanogaster. The effect of mutations in genes involved in wing development on wing shape was tested in different fly genetic backgrounds. It appeared that the effect of the mutation differed strongly across the different genetic backgrounds (Dworkin and Gibson 2006). Hidden genetic variation is widespread across species and affects numerous complex traits including disease and disorder (Merlo and Boyle 2003; Chen et al. 2016; Riordan and Nadeau 2017; Niepoth and Bendesky 2020). Investigating the interaction between a gene perturbation in different genetic backgrounds facilitates the detection of hidden polymorphic regulators of complex traits.
The model nematode species Caenorhabditis elegans allows for studying CGV due to its genetic tractability and the possibility to combine forward and reverse genetic perturbation tools with populations of recombinant inbred lines (RILs) (Li et al. 2006; Gutteling et al. 2007; Viñuela et al. 2010). Studies that focus on CGV in C. elegans have used quantitative trait locus (QTL) analysis, with the aim of detecting polymorphic loci associated with trait variance between RILs (Evans et al. 2021). Many QTL studies in C. elegans have focused on CGV due to environmental perturbations (Rodriguez et al. 2012; Balla et al. 2015; Zdraljevic et al. 2017; Evans et al. 2018; Zamanian et al. 2018; Snoek et al. 2019). But quantitative genetics studies investigating the phenotypic response due to genetic perturbation are less well studied. These studies have the potential to reveal “modifier loci” affecting the mutant phenotype (Milloz et al. 2008; Duveau and Felix 2012). Genetic perturbation in QTL studies can be accomplished by introgressing a mutation in a RIL population, inserting a mutation via CRISPR/Cas9, or by exposing the RILs to an RNAi treatment to create a loss or reduction of function phenotype. Elvin et al. (2011) used RNAi to knock down specific genes in an N2xCB4856 RIL population and subsequently measured the fitness effects. QTL close to ppw-1 (a PAZ/PIWI protein) were found for several different genetic perturbations. The gene ppw-1 is known to be involved in germline sensitivity to RNAi (Tijsterman et al. 2002; Elvin et al. 2011); however, although ppw-1 plays a role, it is not the only genetic factor determining the phenotypic response (Pollard and Rockman 2013). Moreover, the penetrance of the RNAi treatment has been shown to be a complex interplay between target gene and genetic background (Paaby et al. 2015; Vu et al. 2015).
Gene expression QTL (eQTL) reflect the genetic architecture of complex traits (Cookson et al. 2009). eQTL are polymorphic loci associated with gene expression variation and can be mapped either locally (in cis) or distantly (in trans). Cis-eQTL are eQTL where DNA variation within the affected gene causes expression variation, whereas genes of trans-eQTL are distantly regulated by, for example, a transcription factor. Within this paper, we will refer to cis-eQTL as locally mapped eQTL and to trans-eQTL as distantly mapped eQTL. Many trans-eQTL can map to the same genomic region forming clusters called trans-bands (eQTL hotspots). It is these trans-eQTL and trans-bands that are affected the most by environmental and genetic perturbations (Viñuela et al. 2012; Sterken et al. 2017). In the context of cryptic genetic variation, eQTL studies aim to reveal the hidden transcriptional network. Environmental and genetic perturbations have revealed eQTL patterns in C. elegans (Viñuela et al. 2010; Snoek et al. 2017; Sterken et al. 2017). Viñuela et al. (2010) found that variation in gene expression increases with age, and that trans-acting eQTL are relatively more abundant than cis-acting eQTL in older nematodes compared to younger nematodes. Furthermore, genetic perturbations by introgression of the gain-of-function mutation of let-60 in an N2xCB4856 RIL population showed mostly effects on trans-eQTL (Sterken et al. 2017). Taken together, these studies paint a picture of the transitory and reactive nature of trans-eQTL to perturbation.
Here, we studied CGV that is released upon perturbation of gld-1. The gene gld-1 encodes for an RNA-binding protein involved in the regulation of meiotic entry and oocyte development in C. elegans and is an important target of the glp-1/NOTCH pathway. This signaling pathway is highly conserved across species and has been implicated with key cell fate decisions such as proliferation, differentiation, and cellular reprogramming (Artavanis-Tsakonas et al. 1999; Jarriault et al. 2008; Liu et al. 2010; Greenwald and Kovall 2013). Differences in the genetic background can influence an animal's physiological response to gld-1 perturbation, as exposing N2xCB4856 RILs to gld-1 RNAi releases CGV that affects fitness traits (Elvin et al. 2011; Vu et al. 2015).
We extended the study of Elvin et al. (2011) by investigating the transcriptional architecture after gld-1 perturbation by exposing an N2xCB4856 RIL population to gld-1 RNAi. The penetration of the gld-1 RNAi treatment can differ per strain, as germline sensitivity for RNAi was found to be different across genotypes and the responsible underlying loci are segregated in this RIL population (Tijsterman et al. 2002; Elvin et al. 2011; Pollard and Rockman 2013; Paaby et al. 2015). Therefore, the gene expression changes and the mapped eQTL will not only gain insight into the molecular mechanisms, pathways, and modifiers associated with gld-1, it will also reveal CGV associated with gld-1 RNAi. We found that gld-1 perturbation had a major impact on global gene expression and that transcriptional aging was accelerated in gld-1 RNAi–treated animals compared to animals treated with an empty vector. eQTL analysis showed distinct trans-eQTL patterns between the gld-1 RNAi–treated and the empty vector–treated animals.
Materials and methods
Strains used and culturing
The C. elegans strains used in this study were the wild-type strains Bristol N2 and CB4856 and 48 RILs derived from a cross between N2 and CB4856 (genotypes are described in Li et al. 2006). The RILs used were genetically characterized by SNP markers and low-coverage sequencing (Li et al. 2006; Thompson et al. 2015). The strains were kept on nematode growth medium (NGM) with E. coli OP50 as a food source at 12°C (Sulston and Hodgkin 1988).
RNAi exposure
The RNAi bacteria clones were kindly provided by Julie Ahringer. The control (empty vector) used in this experiment was L4440. Part of the gld-1 sequence (T23G11.3, from WormBase), excluding both UTRs, was PCR amplified and inserted in the L4440 backbone, hereby obtaining the gld-1 vector (Kamath et al. 2003).
To ensure consistency between the current and previous experiments, RNAi exposure was performed following the earlier described method and exposed in one single batch (Fraser et al. 2000; Kamath et al. 2001, 2003; Elvin et al. 2011). In short, gravid adults were bleached, and eggs were allowed to hatch in M9 buffer on a rocking platform at 20 RPM resulting in synchronized L1 larvae. Approximately 200 L1 larvae were deposited on 9-cm RNAi plates seeded with 600 μl of an overnight culture of empty vector (control) or gld-1 RNAi bacteria and left to grow at 20°C for 72 h (Elvin et al. 2011). Gravid adults were harvested in M9 and bleached. Embryos were washed in M9, placed onto freshly seeded RNAi plates, and incubated at 20°C for 47 h. L4 stage nematodes were harvested with M9 buffer, centrifuged, and stored at −80°C until further use. The parental strains were treated with the gld-1 RNAi and empty vector twice, 39 RILs were exposed to the empty vector, and 46 RILs were exposed to the gld-1 RNAi treatment (Supplementary Table 1).
RNA isolation, cDNA synthesis, and labeling
The mRNA was isolated with the RNEasy Micro kit from Qiagen (Hilden, Germany) according to the manufacturer's protocol (purification of total RNA from animal and human tissues) with a modified lysing procedure according to Volkers et al. (2013). Labeling of samples, hybridization of the arrays, and feature extraction were done following the manufacturer's instructions in the “Two-Color Microarray-Based Gene Expression Analysis—Low Input Quick Amp Labeling” manual (Agilent Technologies, Santa Clara, CA, USA).
Data analysis
Data was analyzed using R (version 3.6.1 Windows x64) (Team 2019), and for processing the data, the ggplot2, dplyr, and limma packages were used (Ritchie et al., 2015; Wickham, 2016; Wickham et al., 2020). Scripts that were used for this project are available online (https://git.wur.nl/published_papers/vanwijk_etal_2020_).
Microarray hybridization and normalization
The input of total RNA was 200 ng for each sample on C. elegans (V2) Gene Expression Microarray 4X44K slides. The microarrays were scanned using an Agilent High Resolution C Scanner using the settings as recommended in the abovementioned manual. Data was extracted with the Agilent Feature Extraction Software version 10.5, following manufacturer's guidelines. For normalization, the limma package for the “R” environment (version 2.13.1 x64) was used (Ritchie et al. 2015). No background correction of the RNA array data was performed as recommended (Zahurak et al. 2007). For within-array normalization of the RNA array data, the Loess method was used, and for between-array normalization, the quantile method was used (Smyth and Speed 2003). The obtained log2-normalized intensities (single channel data) were used for further analysis. Data is accessible at ArrayExpress under E-MTAB-9742.
Treatment response
The transcriptional responses to the RNAi treatments were determined by explaining the gene expression over the treatments with a linear model,
where y is the log2-normalized intensity as measured by the microarray spot i (i = 1, 2, …, 45,220) and T is treatment (either empty vector or gld-1 RNAi treatment). Genotype was not taken into account in this analysis. The Bonferroni method in Padj (P < 0.05) was applied over the significances to correct for multiple testing (Benjamini and Yekutieli 2001).
The gene expression data of the RILs was used in a principal component analysis (PCA). Therefore, the data was transformed to a log2 ratio with the mean of the 2 treatments using the formula:
where R is the log2 relative expression of spot i (i = 1, 2, …, 45,220) in strain j (RIL) and y is the intensity of spot i in strain j. The function prcomp with scale = TRUE, which normalizes the input data, was used to perform the PCA.
Developmental variation between the RILs
The relative age of the samples was estimated by their transcription profiles. A genetic ruler consisting of 2,195 spots with a significant [log10(P) > 6 and effect size > 0.1 per hour] linearly up- or downregulation during the time period from 41.5 to 72 h was used, based on a previously published method and data (Snoek et al. 2014). The raw data used for this analysis can be obtained from E-MTAB-7019. The expression data from our samples, the same spots that were used to generate the genetic ruler, were compared to the genetic ruler to estimate the transcriptional age of our samples. The relative age was calculated by transforming the predicted ages (in hours and derived from the genetic ruler) into the standard normal distribution.
eQTL mapping and threshold determination
Expression quantitative trait loci mapping was done using a linear model on the log2-normalized intensities. For the empty vector and gld-1 RNAi treatment, a single-marker model was used,
where y is the log2-normalized intensity as measured by the microarray of spot i (i = 1, 2, …, 45,220) of RIL j. This is explained over the genotype (either CB4856 or N2) on marker location x (x = 1, 2, …, 729) of RIL j.
Genome-wide thresholds were determined by permutations per spot per treatment. In this permutation, the log2-normalized intensities were randomly distributed over the strains. The randomized data set was then used in eQTL mapping to determine the false discovery rate (FDR) (Benjamini and Yekutieli 2001). This was repeated for 10 randomized data sets. To calculate the genome-wide thresholds, the following formula was used:
where FDS (false discoveries) was the outcome of the permutations and RDS (real discoveries) the outcome of the eQTL mapping at q = 0.05. The value of m (number of hypotheses tested) was set at 45,220, representing the number of spots on the microarray. The m0 (number of true hypotheses tested) was 45,220 RDS. In this way, the genome-wide thresholds of −log10(P) > 3.9 and −log10(P) > 3.7 were determined for the empty vector and the gld-1 treatments, respectively. The threshold of −log10(P) > 3.9 was used for further analysis for both treatments.
Statistical power analysis
The statistical power for eQTL mapping in the RIL populations was determined in a power analysis (Sterken et al. 2017). Per marker, random noise was introduced based on a standard normal distribution with sigma = 1 and mu = 0. For each marker, eQTL were simulated that explained 20, 25, 30, …, 80% of the variance on top of the random introduced noise. Based on the permutation, the threshold of −log10(P) > 3.9 was used to determine the number of correctly detected QTL, the number of false positives, and the number of undetected QTL. This power analysis also included the precision of the QTL location and the effect size estimation.
eQTL analysis
After eQTL mapping, distinction was made between cis- and trans-eQTL based on the position of the gene and the eQTL peak. An eQTL was assigned as cis if the peak of the eQTL was within 1 Mb of the gene; other eQTL were assigned trans (Snoek et al. 2017).
The amount of variance that each eQTL explained was calculated by ANOVA, by analysis of the gene expression over the marker underlying the eQTL peak. This analysis was conducted per peak if spots had multiple eQTL. The eQTL were compared between the empty vector and gld-1 RNAi treatment by filtering the spots with a similar eQTL type (cis or trans). Trans-eQTL were overlapping when eQTL had overlapping confidence intervals.
To define the trans-bands in this experiment, the genome was divided in 203 bins of 0.5 Mb, and trans-eQTL were counted per bin. Based on the Poisson distribution (Rockman et al. 2010; Snoek et al. 2017), it was calculated how many trans-eQTL a bin should contain in order to represent an overrepresentation (P < 0.0001) and thus contain a trans-band.
All eQTL data and results are available on WormQTL2 (Snoek et al. 2020).
Functional enrichment analysis
Gene group enrichment analysis was performed by filtering genes of interest through several databases provided by WormBase.org: the WS220 gene class annotations, the WS256 GO-annotation, anatomy terms, phenotypes, RNAi phenotypes, developmental stage expression, and disease-related genes (Harris et al. 2014). Modencode.org provided the MODENCODE release 32 transcription factor binding sites that were mapped to transcription start site (Gerstein et al. 2010; Niu et al. 2011; Tepper et al. 2013). Genome.jp/keg/ provided the KEGG pathway release 65.0 (Ogata et al. 1999). Enrichments were selected if the size of category was n > 3 and the overlap between genes of interest and enrichment group was n > 2. Selected enrichments were tested using a hypergeometric test. Here, we corrected the P-values for multiple testing using the Bonferroni correction.
Comparing eQTL datasets
For determining if there were similar trans-bands between this study and Viñuela et al. (2010), trans-bands were called with the aforementioned method (Viñuela et al. 2010; Snoek et al. 2017). First, overlapping trans-bands were determined by filtering for common trans-bands between the 2 data sets. Secondly, it was investigated if the two overlapping trans-bands contained identical genes with a trans-eQTL. Finally, a hypergeometric test was conducted (phyper) to determine if the overlap of identical genes within a trans-band or the overlap of cis- and/or trans-eQTL was greater than we could expect by chance.
Results
gld-1 RNAi has a major impact on gene expression and transcriptional aging
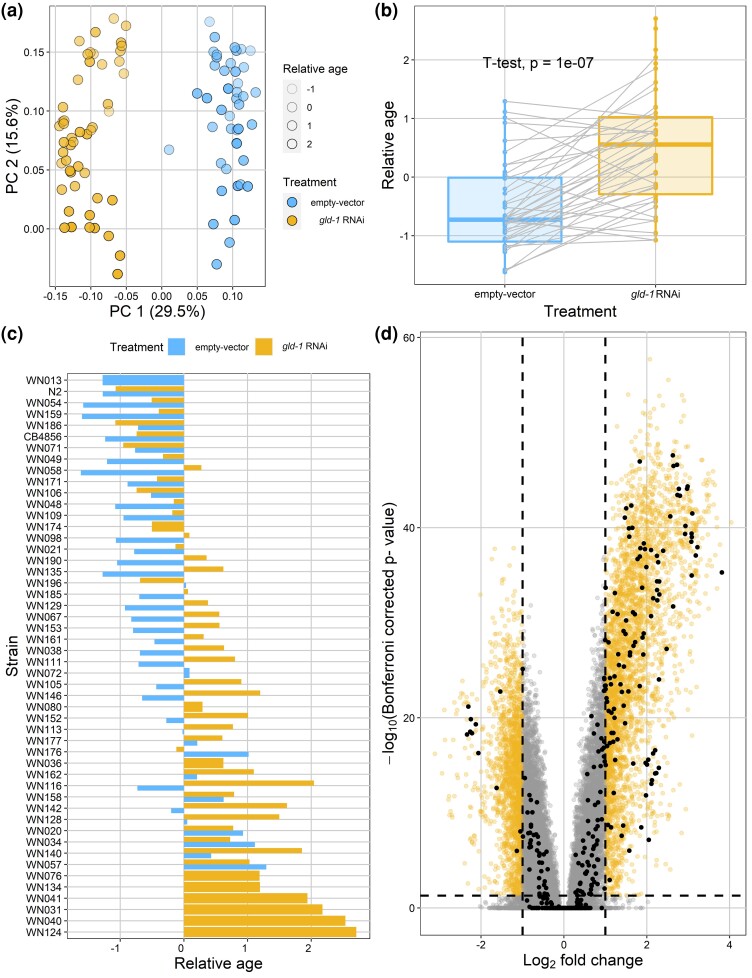
We measured gene expression in a set of RILs derived from N2 and CB4856 that were exposed to 2 treatments (empty vector and gld-1 RNAi). To determine how gene expression variance could be explained by the 2 treatments, we conducted a principal component analysis (PCA). The first axis of the PCA captured the variance that separated the 2 treatments and accounted for 29.5% of the variance in this experiment, indicating that the gld-1 RNAi treatment caused significant changes in global gene expression when compared to the empty vector treatment (Fig. 1a). The second axis captured 15.6% of the variance. From previous experiments, we knew that RILs have differences in developmental timing (Snoek et al. 2019) which can be reflected by changes in gene expression; therefore, it is likely that PC2 was related to differences in transcriptional age (Viñuela et al. 2012; Snoek et al. 2014) (Fig. 1a).
Fig. 1.
gld-1 RNAi has a major effect on global gene expression. a) Most of the of the variance in gene expression of the complete data set can be explained by differences in RNAi treatment (29.5%) as measured by principal component analysis (PCA). PC2 is linked to transcriptional age differences. Every dot represents a strain that had been subjected to either the gld-1 RNAi or the empty vector treatment. b) Strains that underwent the gld-1 RNAi treatment are significantly transcriptionally older compared to animals that had been exposed to the empty vector (1-tailed t-test P = 1 × 10−7). c) The relative age is influenced by genotype and treatment. d) Higher expression of 1,927 genes and lower expression of 1,056 genes in the gld-1 RNAi treatment compared to the empty vector treatment (Bonferroni, Padj < 0.05 and log2 fold change > 2). Also, 58 out of 131 genes with a known interaction with gld-1/GLD-1 were differentially expressed (Bonferroni, Padj < 0.05 and log2 fold change > 2) which is more than we can expect by chance (hypergeometric test, P < 3.2 × 10–15). Every dot represents a microarray spot, and some genes are represented by multiple spots. Yellow spots represent the significantly regulated spots. Black spots represent the spots representing genes that have a known interaction with gld-1/GLD-1.
To further investigate the potential transcriptional age differences, we used previously published data on gene expression over the L4 stage to estimate the relative transcriptional age of the RILs (Snoek et al. 2014). Here, we used genes that are linearly regulated over the L4 stage and used them to infer the relative age of the nematodes in our RIL population using gene expression data. As expected, PC2 was indeed linked to differences in transcriptional age, although all animals were harvested 47 h after bleaching and subsequent RNAi exposure (Fig. 1a). Strains that were exposed to the gld-1 RNAi treatment were, on average, transcriptionally older than empty vector–treated strains (1-tailed t-test, P < 1 × 10−6), indicating that accelerated aging is induced by gld-1 RNAi treatment (Fig. 1b).
Comparing the strains, treatments, and relative age showed the complex interplay between genotype and RNAi treatment on transcriptional age (Fig. 1c). It is notable that the differences in transcriptional age between the empty vector and the gld-1 RNAi treatment are relatively small between the parents N2 and CB4856, whereas much larger differences in transcriptional age can be observed in the RIL progeny. If this increased transcriptional age was solely caused by natural variation in ppw-1, an important gene in germline RNAi sensitivity, then, we would expect much larger differences in relative transcriptional age between the empty vector and gld-1 RNAi treatments between N2 and CB4856. Furthermore, RILs showed a pattern of transgressive behavior for relative transcriptional age in either the empty vector or the gld-1 RNAi treatment and for the difference between relative transcriptional age between the empty vector and gld-1 RNAi treatment. This indicates that multiple alleles are segregating in this population that influence transcriptional aging after gld-1 RNAi exposure (Fig. 1c).
Perturbation effects are primarily due to gld-1 RNAi, not RNAi itself
Observations on the level of gene expression are in line with the observations on population growth (Elvin et al. 2011). Namely, gld-1 RNAi exposed populations grew faster compared to populations that were fed with an empty vector (Supplementary Fig. 1a). Population growth was determined by adding C. elegans to a 96-well plate with E. coli, containing the appropriate RNAi vector, and to subsequently measure the OD600 at different time points. Here, a high relative OD600 meant less food had been consumed and translated to a small nematode population, whereas a low relative OD600 meant that more food had been consumed and translated to a larger nematode population. Moreover, if the differences in fitness between the RILs were solely the result of RNAi penetration variation instead of gld-1 perturbation, we would expect all target genes to behave the same in a particular RIL. However, we clearly see that each RNAi construct has a genotype-dependent effect on fitness. This means that natural variation in fitness after gld-1 RNAi perturbation is primarily caused by gld-1 perturbation (Supplementary Fig. 1a).
The transcriptional age is correlated with population growth effects. Because there were 39 identical RILs used between our study and the study of Elvin et al. (2011), we reanalyzed their data and compared the inferred transcriptional age of our samples to the OD600 of their samples. We observed that the largest differences in OD600 could be observed between 96 and 144 h (Supplementary Fig. 1a). Next, we correlated the relative age of the samples from our study with the normalized OD600 and found that the relative transcriptional age negatively correlated with the OD600 (Pearson correlation 96 h: P < 0.005, 120 h: P < 0.01, 144 h: P < 0.05) (Supplementary Fig. 1b). Therefore, we conclude that the transcriptionally older RILs in our study were also growing faster in the study of Elvin et al. (2011).
Next, we wanted to explore the extent and variety of differentially expressed genes between gld-1 RNAi and empty vector–treated animals. We found that 1,927 genes were higher expressed in the gld-1 RNAi treatment compared to the empty vector and 1,056 genes were lower expressed in the gld-1 treatment compared to the empty vector (Bonferroni, Padj < 0.05 and log2 fold change > 2) (Fig. 1d) (Supplementary Table 2). Enrichment of genes that were higher expressed in the empty vector condition were mainly involved in neuronal anatomy and oxidation processes, whereas genes that were higher expressed in the gld-1 RNAi–treated animals were mainly involved in embryo development and reproduction (Supplementary Table 3). To see whether the RNAi pathway was differentially regulated between the 2 treatments, we obtained a list of genes known to be involved in the RNAi pathway (e.g. alg-1, dcr-1, and eri-1) from WormBase. We found that 5 out of 27 genes contained transcripts that were significantly higher expressed in the gld-1 RNAi treatment compared to the empty vector treatment (Bonferroni, Padj < 0.05 and log2 fold change > 2) (Supplementary Table 4). Therefore, we must conclude that gene expression differences between the 2 treatments were the result of gld-1 perturbation and the effect of RNAi itself.
Because gld-1 is known to be an important translational regulator, we investigated for the transcriptional response of known gld-1 interaction targets. We previously observed a positive correlation between mRNA and protein abundance in N2 and CB4856 (Kamkina et al. 2016). Therefore, we hypothesized that if there would be a change in GLD-1 abundance, there would be a change in protein concentration of translational GLD-1 targets which would influence GLD-1 target gene mRNAs. We obtained a list of genes that have a genetic, physical, regulatory, and/or predicted interaction with gld-1 from WormBase. Of these 209 known interactions, we were able to detect the transcription levels of 131 unique genes. In total, we found 58 genes that were differentially expressed as a result of the gld-1 RNAi treatment, which is more than we can expect by chance alone (Fig. 1d) (Bonferroni, Padj < 0.05 and log2 fold change > 2 and hypergeometric test, P < 3.2 × 10−15) (Supplementary Table 5).
All in all, the clear separation between the gld-1 RNAi–treated animals and the empty vector animals in the PCA (Fig. 1a), the difference in transcriptional age between the 2 treatments (Fig. 1b and c), the significant correlation between the transcriptional ages of the samples in this study and the population fitness of the samples in Elvin et al. (2011) (Supplementary Fig. 1, a and b), the large number of differentially expressed genes between the 2 treatments (Fig. 1d), and the overrepresentation of differentially expressed genes that are known to be interacting with gld-1/GLD1 provide evidence of gld-1 perturbation.
Differences in eQTL architecture between the empty vector and gld-1 RNAi treatment
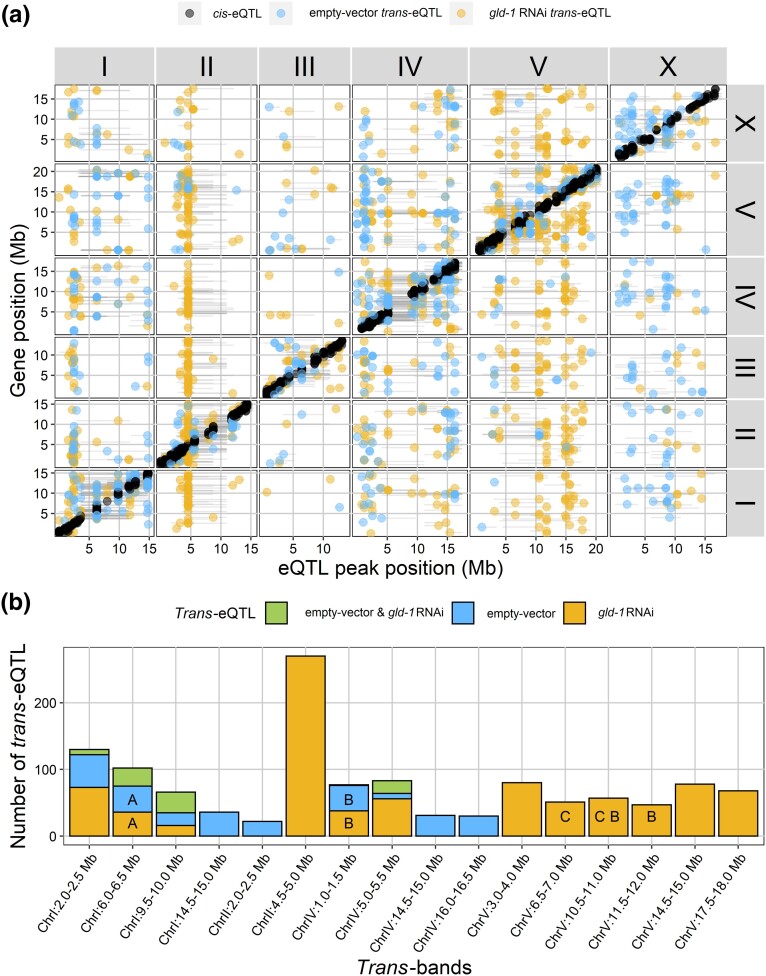
To investigate the effect of the genetic variation on gene expression after gld-1 perturbation, we mapped eQTL. We used a single-marker model for the eQTL mapping and compared the identified eQTL (FDR ≤ 0.05) between the empty vector and gld-1 RNAi treatment. Statistical power analyses showed that the populations represented in the empty vector and the gld-1 RNAi treatment had the power to detect 64.7 and 78.9% of the eQTL that explain at least 35% of the variance, respectively (Supplementary Table 6). In total, we found 1,358 genes with an eQTL in the empty vector treatment and 1,949 genes with an eQTL in the gld-1 RNAi treatment (Fig. 2a) (Supplementary Table 7). This difference in the number of eQTL can partly be explained by the difference in statistical power between the data sets. Trans-eQTL often explain less variance compared to cis-eQTL and are therefore more likely to be missed when calling eQTL.
Fig. 2.
eQTL mapped in this experiment. a) The black dots represent the spots with a cis-eQTL and the blue/gold dots represent the spots with a trans-eQTL detected in the empty vector or gld-1 RNAi treatment, respectively. The y-axis represents the genomic location of the differentially expressed spots; the x-axis represents the genomic location of the mapped eQTL. The gray horizontal lines represent the confidence interval of the eQTL (based on a 1.5 drop in −log10(P). b) Trans-bands that are found in this study can be specific for the empty vector treatment or the gld-1 RNAi treatment or can colocate and share some eQTL (Poisson distribution, P < 0.0001). Numbers of trans-eQTL are represented in spots, not genes. Trans-bands that were found in Viñuela et al. (2010) that share more eQTL with eQTL in our trans-bands at that specific trans-band interval than can be expected by chance are marked with a letter (A, juvenile; B, adult; C, old) (hypergeometric test, P < 0.05).
To compare eQTL between the 2 treatments, we defined shared cis-eQTL as genes with an eQTL and classified as cis-eQTL (eQTL that map within 1.0 Mb of the physical location of the gene), whereas shared trans-eQTL were genes with eQTL that were classified as trans-eQTL (eQTL that do not map within 1.0 Mb of the physical location of the gene) and had overlapping confidence intervals. We found 531 genes with a cis-eQTL that overlapped between the empty vector and the gld-1 RNAi treatment and 240 and 414 genes with a cis-eQTL that were only observed in the empty vector and gld-1 RNAi treatment, respectively (Table 1; Fig. 2a). Enrichment of cis-eQTL of each treatment individually yielded gene classes, bath, math, and pals, which were also found in previous eQTL studies in C. elegans (Volkers et al. 2013; Snoek et al. 2017; Sterken et al. 2017). Furthermore, we found 76 genes with a trans-eQTL that overlapped between the empty vector and the gld-1 RNAi treatment; 566 and 991 genes with a trans-eQTL were only found in the empty vector or gld-1 RNAi treatment, respectively (Table 1; Fig. 2a).
Table 1.
Number of genes with an eQTL per RNAi treatment. Numbers of spots on the microarray with an eQTL that represent the number of genes with an eQTL are depicted in bold.
| gld-1 | Empty vector | |
|---|---|---|
| Specific cis-eQTL | 414 genes, 513 spots | 240 genes, 298 spots |
| Overlap cis-eQTL | 531 genes, 779 spots | |
| Total cis-eQTL | 900 genes, 1,292 spots | 742 genes, 1,077 spots |
| Specific trans-eQTL | 991 genes, 1,533 spots | 556 genes, 891 spots |
| Overlap trans-eQTL | 76 genes, 234 spots | |
| Total trans-eQTL | 1,049 genes, 1,767 spots | 616 genes, 1,125 spots |
| Total eQTL | 1,949 genes, 3,059 spots | 1,358 genes, 2,202 spots |
Because trans-eQTL typically form hotspots of loci that regulate the transcription of many genes, we determined the location of hotspots in our data. Trans-eQTL hotspots were identified by comparing the number of trans-eQTL in a specific bin to the average number of eQTL mapped in the experiment (Poisson distribution, P < 0.0001). In total, we found 9 trans-bands for the empty vector treatment and 12 trans-bands for the gld-1 RNAi treatment. Most trans-bands harbored trans-eQTL that were specific for either the gld-1 RNAi treatment or the empty vector condition. However, trans-eQTL hotspots on chromosomes I and IV contained trans-eQTL that were treatment specific but also included trans-eQTL that were shared between treatments (Fig. 2b). These shared eQTL are most likely to be an underrepresentation because of differences in statistical power between the data sets. However, this might also suggest that regulatory loci underlying those trans-eQTL hotspots change transcription specificity after gld-1 perturbation.
To elucidate the underlying biology of the different trans-bands, we performed an enrichment analysis on the genes that mapped to each trans-band. The genes that mapped to the trans-eQTL hotspots in the gld-1 trans-bands revealed enrichment for protein (de)phosphorylation (ChrII:4.5–5.0 Mb); transcription factors (ChrIV:1.0–1.5 Mb); endopeptidase activity (ChrV:6.5–7.0 Mb); nerve system, hormone activity, and phenotypes associated with movement (ChrV:14.5–15.0 Mb); and unfolded protein response (ChrV:17.5–18.0 Mb).
Perturbation using gld-1 RNAi not only revealed the genetic architecture of gld-1 perturbation-specific effects; it also may have revealed RNAi-specific processes since RNAi penetration is genotype dependent (Elvin et al. 2011; Paaby et al. 2015; Vu et al. 2015). Elvin et al. (2011) discovered that natural variation in ppw-1 explained the part of the RNAi penetration variation between N2 and CB4856. Here, we did not find an eQTL for ppw-1 or a trans-band that mapped to the ppw-1 genomic location (ChrI:4.2 Mb). Furthermore, we did not find differences in RNAi pathway–specific eQTL between the 2 treatments, nor did we observe trans-bands that mapped to the genomic location of important RNAi genes that contain predicted high impact variation between N2 and CB4856 (Supplementary Table 4). Therefore, we can conclude that RNAi penetration differences between genetic backgrounds were unlikely to have a major impact on the eQTL mapping.
The eQTL architecture upon gld-1 RNAi indicates increased aging
Because we had found an accelerated transcriptional aging effect upon gld-1 RNAi perturbation, we were curious to see if we could find age-related eQTL patterns in our data. Therefore, we used the eQTL dataset of Viñuela et al. (2010) where eQTL patterns in C. elegans at 3 different ages were investigated: juveniles (40 h), adults (96 h), and old nematodes (214 h) (Viñuela et al. 2010). When we compared our eQTL to the juvenile eQTL data, we found that one trans-band (ChrI:6.0–6.5 Mb) had more overlapping eQTL between the empty vector and juveniles and the gld-1 RNAi and juveniles than could be expected by chance (hypergeometric test, P = 1.5 × 10−11 and 3.8 × 10−8, respectively) (Fig. 2b). This indicates that the trans-band ChrI:6.0–6.5 Mb between the 3 conditions is the same and might be regulated by the same regulator. When we compared our empty vector trans-bands to trans-bands from the adult and old nematode life stages, we only found 1 trans-band (ChrIV:1.0–1.5 Mb) with more overlapping eQTL than we could expect by chance (hypergeometric test, P < 7.0 × 10−5). Strikingly, when we compared the gld-1 RNAi treatment trans-bands to the trans-bands from the adult and old nematode life stages, we found 5 trans-bands (adult, ChrIV:1.0–1.5 Mb, ChrV:10.5–11.0 Mb, and ChrV:11.5–12.0 Mb; old nematodes, ChrV:6.5–7.0 Mb and ChrV:10.5–11.0 Mb) with more overlapping eQTL than we could expect by chance (hypergeometric test, P = 2.7 × 10−3, 2.7 × 10−2, 1.8 × 10−11, 4.3 × 10−3, and 4.6 × 10−4, respectively). This relatively large number of overlapping trans-bands between the gld-1 RNAi eQTL and the adult and old nematode eQTL of Viñuela et al. (2010) points towards increased aging processes upon gld-1 perturbation.
Discussion
Genes linked to gld-1 are affected by CGV
In our study, we observed a clear effect in gene expression caused by the gld-1 RNAi treatment (in the PCA, in the DEG analysis, transcriptional age). Elvin et al. (2011) and Vu et al. (2015) observed a clear phenotypic effect when animals were exposed to gld-1 RNAi compared to control animals (Elvin et al. 2011; Vu et al. 2015). We expected to find lower levels of gld-1 in the gld-1 RNAi–treated animals compared to the empty vector–treated animals. However, we observed a transcriptional increase of gld-1, indicating a feedback mechanism for obtaining adequate GLD-1, like in Brenner and Schedl (2016). There they found that gld-1[gld-1(q485)/+] animals showed similar phenotypic features compared to full gld-1 knockouts, indicating that a lower gene dose of gld-1 has an effect on the animals. However, GLD-1 levels in adult gld-1[gld-1(q485)/+] nematodes were normal, suggesting a feedback mechanism between gld-1 and GLD-1 (Brenner and Schedl 2016). Investigating the precise interaction between gld-1 and GLD-1 is beyond the scope of this paper but may be an interesting topic for future research.
The genetic perturbation caused by gld-1 RNAi will have varied between genotypes since germline sensitivity depends on the genetic background (Tijsterman et al. 2002; Pollard and Rockman 2013; Paaby et al. 2015; Vu et al. 2015). Much of the phenotypic variance between N2 and CB4856 after RNAi treatments can be attributed to allelic differences in ppw-1; CB4856 harbors a natural null allele, making CB4856 more resistant to the effects of germline active RNAi (Tijsterman et al. 2002; Elvin et al. 2011; Pollard and Rockman 2013; Vu et al. 2015). However, both Vu et al. (2015) and Elvin et al. (2011) showed that the phenotypic variation between N2 and CB4856 after gld-1 RNAi can only partially be explained by natural variation in ppw-1. Furthermore, we did not observe an eQTL for ppw-1 nor did we find a trans-band located at the ppw-1 locus, indicating that allelic variation in ppw-1 did not have a large effect on the perturbated transcriptional architecture. Taken together, we conclude that any phenotypic effects in this RIL panel were the result of a combination of gld-1 RNAi penetration and genetic background effects.
Next to an abundance of differentially regulated genes, we found many different trans-eQTL and trans-bands due to gld-1 RNAi, confirming that trans-eQTL are affected by genetic perturbations (Sterken et al. 2017). However, it should be noted that differences in eQTL observed in this study are influenced by multiple factors, i.e. statistical power differences between the 2 experimental conditions, developmental effects, natural variation in RNAi specificity, mishybridization on the microarray due to polymorphisms, and the gld-1 perturbation. Off-target effects of the used gld-1 RNAi construct could have altered gene expression of unintended target genes (Jackson et al. 2003). However, we found no differences in eQTL of genes that are involved in the RNAi pathway between the 2 treatments nor did we find trans-bands at genomic locations harboring polymorphic RNAi genes. This suggests that genetic variation did not strongly affect gene expression of RNAi, therefore indicating that the RNAi itself did not have a large influence on genetic architecture.
Perturbation with gld-1 RNAi affects transcriptional aging
Development plays an important role in the detection of eQTL. (e)QTL patterns can be influenced by developmental speed differences within a RIL population and have been observed before (Snoek et al. 2019; Snoek et al. 2021). Furthermore, eQTL patterns change with age (Viñuela et al. 2010). In this study, we found that gld-1 RNAi–treated animals were transcriptionally older than the empty vector–treated animals. Moreover, when we compared eQTL patterns between our data and Viñuela et al. (2010), we observed that gld-1 RNAi–treated animals shared more trans-bands with adult and old nematodes than the empty vector animals share with adult and old nematodes. This is interesting since the glp-1/NOTCH pathway has been implicated in the development of age-related diseases in human and thus might play an important role in aging in general (Balistreri et al. 2016). A possible explanation for the enhanced transcriptional age of the gld-1 RNAi–treated nematodes is the connection between the NOTCH pathway and insulin/IGF-1 signaling. The downregulation of gld-1 could influence glp-1 expression and translation, which in turn might act on insulin/IGF-1 signaling genes (Marin and Evans 2003; Schaffitzel and Hertweck 2006). Upregulation of the insulin/IGF-1 pathway promotes reproductive development, thereby aging the nematodes (Schaffitzel and Hertweck 2006). Further investigations into the interplay between genetic background, altered transcriptional age, and gld-1 perturbation might be an interesting research topic for the future.
Linking eQTL architecture to previously observed fitness responses
This study started as an extension of Elvin et al. (2011) where they exposed an N2xCB4856 RIL panel to gld-1 RNAi and mapped the fitness responses. Comparison of trans-bands mapped in this research with QTL peaks associated with fitness found by Elvin et al. (2011) resulted in the overlap of 2 regions: ChrI:6:0–6:5 Mb and ChrIV:16.0–16.5 Mb. The QTL on chromosome IV in Elvin et al. (2011) was mapped in the empty vector and in the gld-1 RNAi treatment, but in our study, we only found a trans-band at that location for the empty vector treatment. The overlapping trans-band on chromosome I in our study is present in the empty vector treatment and the gld-1 RNAi treatment, whereas Elvin et al. (2011) only mapped a QTL in the gld-1 RNAi treatment and attributed it to ppw-1. The gene ppw-1 has been implicated with germline sensitivity to RNAi and is located on chromosome I at 4.2 Mb. However, no trans-band was found in our research on that locus and neither did we find an eQTL for ppw-1. This suggests that natural variation in gene expression of ppw-1 does not explain differences in germline sensitivity to RNAi, which is in line with the natural variation segregated in the RIL panel, which includes the CB4856 null allele (Tijsterman et al. 2002; Elvin et al. 2011; Pollard and Rockman 2013; Vu et al. 2015). Furthermore, the ppw-1 null allele does not influence germline sensitivity to RNAi via the regulation of many transcripts.
Conclusion
Here, we present the transcriptional response and the contribution of CGV on eQTL between empty vector– and gld-1 RNAi–exposed animals. The mapped eQTL might explain part of the underlying genetics of the differences in fitness observed in Elvin et al. (2011) and could potentially help to elucidate differences in other phenotypic traits between N2 and CB4856 related to gld-1 perturbation. Furthermore, we provided evidence that gld-1 perturbation is causing accelerated transcriptional aging in C. elegans. In conclusion, we demonstrate that genetic perturbation causes genotype-dependent shifts in transcriptional architecture that most likely affect phenotypic differences. Therefore, we not only suggest to take natural variation into account for studying the complex genetic architecture of the glp-1/NOTCH pathway but also when studying the effects of other important genetic perturbations.
Supplementary Material
Acknowledgements
We would like to thank Emma Cuperus for her support in data analysis, Miriam Rodriguez for her experimental support, Harm Nijveen for adding the QTL results to WormQTL2, and WormBase and CeNDR for being important resources to the C. elegans research community.
Contributor Information
Marijke H van Wijk, Laboratory of Nematology, Wageningen University & Research, Droevendaalsesteeg 1, 6708 PB Wageningen, The Netherlands.
Joost A G Riksen, Laboratory of Nematology, Wageningen University & Research, Droevendaalsesteeg 1, 6708 PB Wageningen, The Netherlands.
Mark Elvin, Peak Proteins, Birchwood House, Larkwood Way, Macclesfield, Cheshire, SK10 2XR, UK.
Gino B Poulin, Division of Molecular and Cellular Function, School of Biological Sciences, Faculty of Biology, Medicine and Health, University of Manchester, Manchester Academic Health Science Centre, Manchester, Oxford Road, M13 9PL, UK.
Muhammad I Maulana, Laboratory of Nematology, Wageningen University & Research, Droevendaalsesteeg 1, 6708 PB Wageningen, The Netherlands.
Jan E Kammenga, Laboratory of Nematology, Wageningen University & Research, Droevendaalsesteeg 1, 6708 PB Wageningen, The Netherlands.
Basten L Snoek, Theoretical Biology and Bioinformatics, Utrecht University, 3584 CH Utrecht, The Netherlands.
Mark G Sterken, Laboratory of Nematology, Wageningen University & Research, Droevendaalsesteeg 1, 6708 PB Wageningen, The Netherlands.
Data availability
Scripts that were used for this project are available online (https://git.wur.nl/published_papers/vanwijk_etal_2020_). Transcriptome data were deposited at ArrayExpress under E-MTAB-9742. eQTL results are publicly accessible on WormQTL2.
Supplemental material available at G3 online.
Author contributions
The experiments were designed by JEK and GBP and were performed by ME, JAGR, and MHW. The data was analyzed by MHW, MGS, BLS, and MIM. The manuscript was written by MHW in close collaboration with MIM, MGS, and JEK, with comments from all coauthors.
Literature cited
- Artavanis-Tsakonas S, Rand MD, Lake RJ. Notch signaling: cell fate control and signal integration in development. Science. 1999;284(5415):770–776. doi: 10.1126/science.284.5415.770. [DOI] [PubMed] [Google Scholar]
- Balistreri CR, Madonna R, Melino G, Caruso C. The emerging role of Notch pathway in ageing: focus on the related mechanisms in age-related diseases. Ageing Res Rev. 2016;29:50–65. doi: 10.1016/j.arr.2016.06.004. [DOI] [PubMed] [Google Scholar]
- Balla KM, Andersen EC, Kruglyak L, Troemel ER. A wild C. elegans strain has enhanced epithelial immunity to a natural microsporidian parasite. PLoS Pathog. 2015;11(2):e1004583. doi: 10.1371/journal.ppat.1004583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, Yekutieli D. The control of the false discovery rate in multiple testing under dependency. Ann Stat. 2001;29(4):1165–1188. doi: 10.1214/aos/1013699998. [DOI] [Google Scholar]
- Brenner JL, Schedl T. Germline stem cell differentiation entails regional control of cell fate regulator GLD-1 in Caenorhabditis elegans. Genetics. 2016;202(3):1085–1103. doi: 10.1534/genetics.115.185678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen R, Shi L, Hakenberg J, Naughton B, Sklar P, Zhang J, Zhou H, Tian L, Prakash O, Lemire M, et al. Analysis of 589,306 genomes identifies individuals resilient to severe Mendelian childhood diseases. Nat Biotechnol. 2016;34(5):531–538. doi: 10.1038/nbt.3514. [DOI] [PubMed] [Google Scholar]
- Cookson W, Liang L, Abecasis G, Moffatt M, Lathrop M. Mapping complex disease traits with global gene expression. Nat Rev Genet. 2009;10(3):184–194. doi: 10.1038/nrg2537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duveau F, Felix MA. Role of pleiotropy in the evolution of a cryptic developmental variation in Caenorhabditis elegans. PLoS Biol. 2012;10(1):e1001230. doi: 10.1371/journal.pbio.1001230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dworkin I, Gibson G. Epidermal growth factor receptor and transforming growth factor-beta signaling contributes to variation for wing shape in Drosophila melanogaster. Genetics. 2006;173(3):1417–1431. doi: 10.1534/genetics.105.053868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elvin M, Snoek LB, Frejno M, Klemstein U, Kammenga JE, Poulin GB. A fitness assay for comparing RNAi effects across multiple C. elegans genotypes. BMC Genomics. 2011;12(1):510. doi: 10.1186/1471-2164-12-510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans KS, Brady SC, Bloom JS, Tanny RE, Cook DE, Giuliani SE, Hippleheuser SW, Zamanian M, Andersen EC. Shared genomic regions underlie natural variation in diverse toxin responses. Genetics. 2018;210(4):1509–1525. doi: 10.1534/genetics.118.301311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans KS, van Wijk MH, McGrath PT, Andersen EC, Sterken MG. From QTL to gene: C. elegans facilitates discoveries of the genetic mechanisms underlying natural variation. Trends Genet. 2021;37(10):933–947. doi: 10.1016/j.tig.2021.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser AG, Kamath RS, Zipperlen P, Martinez-Campos M, Sohrmann M, Ahringer J. Functional genomic analysis of C. elegans chromosome I by systematic RNA interference. Nature. 2000;408(6810):325–330. doi: 10.1038/35042517. [DOI] [PubMed] [Google Scholar]
- Gerstein MB, Lu ZJ, Van Nostrand EL, Cheng C, Arshinoff BI, Liu T, Yip KY, Robilotto R, Rechtsteiner A, Ikegami K, et al. Integrative analysis of the Caenorhabditis elegans genome by the modENCODE project. Science. 2010;330(6012):1775–1787. doi: 10.1126/science.1196914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson G, Dworkin I. Uncovering cryptic genetic variation. Nat Rev Genet. 2004;5(9):681–690. doi: 10.1038/nrg1426. [DOI] [PubMed] [Google Scholar]
- Greenwald I, Kovall R. Notch signaling: genetics and structure (January 17, 2013), WormBook, ed. The C. elegans Research Community, WormBook. doi: 10.1895/wormbook.1.10.2, http://www.wormbook.org. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutteling EW, Riksen JA, Bakker J, Kammenga JE. Mapping phenotypic plasticity and genotype-environment interactions affecting life-history traits in Caenorhabditis elegans. Heredity (Edinb). 2007;98(1):28–37. doi: 10.1038/sj.hdy.6800894. [DOI] [PubMed] [Google Scholar]
- Harris TW, Baran J, Bieri T, Cabunoc A, Chan J, et al. WormBase 2014: new views of curated biology. Nucleic Acids Res. 2014;42(D1):D789–D793. doi: 10.1093/nar/gkt1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson AL, Bartz SR, Schelter J, Kobayashi SV, Burchard J, Mao M, Li B, Cavet G, Linsley PS. Expression profiling reveals off-target gene regulation by RNAi. Nat Biotechnol. 2003;21(6):635–637. doi: 10.1038/nbt831. [DOI] [PubMed] [Google Scholar]
- Jarriault S, Schwab Y, Greenwald I. A Caenorhabditis elegans model for epithelial–neuronal transdifferentiation. Proc Natl Acad Sci U S A. 2008;105(10):3790–3795. doi: 10.1073/pnas.0712159105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamath RS, Fraser AG, Dong Y, Poulin G, Durbin R, Gotta M, Kanapin A, Le Bot N, Moreno S, Sohrmann M, et al. Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature. 2003;421(6920):231–237. doi: 10.1038/nature01278. [DOI] [PubMed] [Google Scholar]
- Kamath RS, Martinez-Campos M, Zipperlen P, Fraser AG, Ahringer J. Effectiveness of specific RNA-mediated interference through ingested double-stranded RNA in Caenorhabditis elegans. Genome Biol. 2001;2(1):RESEARCH0002. doi: 10.1186/gb-2000-2-1-research0002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamkina P, Snoek LB, Grossmann J, Volkers RJ, Sterken MG, Daube M, Roschitzki B, Fortes C, Schlapbach R, Roth A, et al. Natural genetic variation differentially affects the proteome and transcriptome in Caenorhabditis elegans. Mol Cell Proteomics. 2016;15(5):1670–1680. doi: 10.1074/mcp.M115.052548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Alvarez OA, Gutteling EW, Tijsterman M, Fu J, Riksen JAG, Hazendonk E, Prins P, Plasterk RHA, Jansen RC, et al. Mapping determinants of gene expression plasticity by genetical genomics in C. elegans. PLoS Genet. 2006;2(12):e222. doi: 10.1371/journal.pgen.0020222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Sato C, Cerletti M, Wagers A. Notch signaling in the regulation of stem cell self-renewal and differentiation. Curr Top Dev Biol. 2010;92:367–409. doi: 10.1016/S0070-2153(10)92012-7. [DOI] [PubMed] [Google Scholar]
- Marin VA, Evans TC. Translational repression of a C. elegans Notch mRNA by the STAR/KH domain protein GLD-1. Development. 2003;130(12):2623–2632. doi: 10.1242/dev.00486. [DOI] [PubMed] [Google Scholar]
- Merlo CA, Boyle MP. Modifier genes in cystic fibrosis lung disease. J Lab Clin Med. 2003;141(4):237–241. doi: 10.1067/mlc.2003.29. [DOI] [PubMed] [Google Scholar]
- Milloz J, Duveau F, Nuez I, Felix MA. Intraspecific evolution of the intercellular signaling network underlying a robust developmental system. Genes Dev. 2008;22(21):3064–3075. doi: 10.1101/gad.495308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niepoth N, Bendesky A. How natural genetic variation shapes behavior. Annu Rev Genomics Hum Genet. 2020;21(1):437–463. doi: 10.1146/annurev-genom-111219-080427 [DOI] [PubMed] [Google Scholar]
- Niu W, Lu ZJ, Zhong M, Sarov M, Murray JI, Brdlik CM, Janette J, Chen C, Alves P, Preston E, et al. Diverse transcription factor binding features revealed by genome-wide ChIP-seq in C. elegans. Genome Res. 2011;21(2):245–254. doi: 10.1101/gr.114587.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogata H, Goto S, Sato K, Fujibuchi W, Bono H, Kanehisa M. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 1999;27(1):29–34. doi: 10.1093/nar/27.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paaby AB, White AG, Riccardi DD, Gunsalus KC, Piano F, Rockman Matthew V. Wild worm embryogenesis harbors ubiquitous polygenic modifier variation. Elife. 2015;4:e09178. doi: 10.7554/eLife.09178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard DA, Rockman MV. Resistance to germline RNA interference in a Caenorhabditis elegans wild isolate exhibits complexity and nonadditivity. G3 (Bethesda). 2013;3(6):941–947. doi: 10.1534/g3.113.005785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riordan JD, Nadeau JH. From peas to disease: modifier genes, network resilience, and the genetics of health. Am J Hum Genet. 2017;101(2):177–191. doi: 10.1016/j.ajhg.2017.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, Smyth GK. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43(7):e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rockman MV, Skrovanek SS, Kruglyak L. Selection at linked sites shapes heritable phenotypic variation in C. elegans. Science. 2010;330(6002):372–376. doi: 10.1126/science.1194208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez M, Snoek LB, Riksen JAG, Bevers RP, Kammenga JE. Genetic variation for stress-response hormesis in C. elegans lifespan. Exp Gerontol. 2012;47(8):581–587. doi: 10.1016/j.exger.2012.05.005. [DOI] [PubMed] [Google Scholar]
- Schaffitzel E, Hertweck M. Recent aging research in Caenorhabditis elegans. Exp Gerontol. 2006;41(6):557–563. doi: 10.1016/j.exger.2006.02.008. [DOI] [PubMed] [Google Scholar]
- Smyth GK, Speed T. Normalization of cDNA microarray data. Methods. 2003;31(4):265–273. doi: 10.1016/S1046-2023(03)00155-5. [DOI] [PubMed] [Google Scholar]
- Snoek BL, Sterken MG, Bevers RPJ, Volkers RJM, Van't Hof A, Brenchley R, Riksen JAG, Cossins A, Kammenga JE. Contribution of trans regulatory eQTL to cryptic genetic variation in C. elegans. BMC Genomics. 2017;18(1):500. doi: 10.1186/s12864-017-3899-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snoek BL, Sterken MG, Hartanto M, van Zuilichem AJ, Kammenga JE, de Ridder D, Nijveen H. WormQTL2: an interactive platform for systems genetics in Caenorhabditis elegans. Database (Oxford). 2020;2020:baz149. doi: 10.1093/database/baz149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snoek BL, Sterken MG, Nijveen H, Volkers RJM, Riksen J, Rosenstiel PC, Schulenburg H, Kammenga JE. The genetics of gene expression in a Caenorhabditis elegans multiparental recombinant inbred line population. G3 (Bethesda). 2021;11(10):jkab258. doi: 10.1093/g3journal/jkab258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snoek LB, Sterken MG, Volkers RJ, Klatter M, Bosman KJ, Bevers RPJ, Riksen JAG, Smant G, Cossins AR, Kammenga JE. A rapid and massive gene expression shift marking adolescent transition in C. elegans. Sci Rep. 2014;4(1):3912. doi: 10.1038/srep03912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snoek BL, Volkers RJM, Nijveen H, Petersen C, Dirksen P, Sterken MG, Nakad R, Riksen JAG, Rosenstiel Philip, Stastna JJ, et al. A multi-parent recombinant inbred line population of C. elegans allows identification of novel QTLs for complex life history traits. BMC Biol. 2019;17(1):24. doi: 10.1186/s12915-019-0642-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sterken MG, van Bemmelen van der Plaat L, Riksen JAG, Rodriguez M, Schmid T, Hajnal A, Kammenga JE, Snoek BL. Ras/MAPK modifier loci revealed by eQTL in Caenorhabditis elegans. G3 (Bethesda). 2017;7(9):3185–3193. doi: 10.1534/g3.117.1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulston JE, Hodgkin J. The nematode Caenorhabditis elegans; 1988.
- Team RC. R: a language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing; 2019. [Google Scholar]
- Tepper RG, Ashraf J, Kaletsky R, Kleemann G, Murphy CT, Bussemaker HJ. PQM-1 complements DAF-16 as a key transcriptional regulator of DAF-2-mediated development and longevity. Cell. 2013;154(3):676–690. doi: 10.1016/j.cell.2013.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson OA, Snoek LB, Nijveen H, Sterken MG, Volkers RJM, Brenchley R, van’t Hof A, Bevers RPJ, Cossins AR, Yanai I, et al. Remarkably divergent regions punctuate the genome assembly of the Caenorhabditis elegans Hawaiian strain CB4856. Genetics. 2015;200(3):975–989. doi: 10.1534/genetics.115.175950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tijsterman M, Okihara KL, Thijssen K, Plasterk RHA. PPW-1, a PAZ/PIWI protein required for efficient germline RNAi, is defective in a natural isolate of C. elegans. Curr Biol. 2002;12(17):1535–1540. doi: 10.1016/S0960-9822(02)01110-7. [DOI] [PubMed] [Google Scholar]
- Viñuela A, Snoek LB, Riksen JA, Kammenga JE. Genome-wide gene expression regulation as a function of genotype and age in C. elegans. Genome Res. 2010;20(7):929–937. doi: 10.1101/gr.102160.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viñuela A, Snoek LB, Riksen JA, Kammenga JE. Aging uncouples heritability and expression-QTL in Caenorhabditis elegans. G3 (Bethesda). 2012;2(5):597–605. doi: 10.1534/g3.112.002212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volkers RJ, Snoek LB, Hubar CJ, Coopman R, Chen W, Yang W, Sterken MG, Schulenburg H, Braeckman BP, Kammenga JE. Gene-environment and protein-degradation signatures characterize genomic and phenotypic diversity in wild Caenorhabditis elegans populations. BMC Biol. 2013;11(1):93. doi: 10.1186/1741-7007-11-93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vu V, Verster AJ, Schertzberg M, Chuluunbaatar T, Spensley M, Pajkic D, Hart GT, Moffat J, Fraser AG. Natural variation in gene expression modulates the severity of mutant phenotypes. Cell. 2015;162(2):391–402. doi: 10.1016/j.cell.2015.06.037. [DOI] [PubMed] [Google Scholar]
- Wickham H. ggplot2: Elegant Graphics for Data Analysis. New York: Springer-Verlag; 2016. [Google Scholar]
- Wickham H, François R, Müller K. dplyr: a grammar of data manipulation; 2020.
- Zahurak M, Parmigiani G, Yu W, Scharpf RB, Berman D, Schaeffer E, Shabbeer S, Cope L. Pre-processing Agilent microarray data. BMC Bioinformatics. 2007;8(1):142. doi: 10.1186/1471-2105-8-142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamanian M, Cook DE, Zdraljevic S, Brady SC, Lee D, Lee J, Andersen EC. Discovery of genomic intervals that underlie nematode responses to benzimidazoles. PLoS Negl Trop Dis. 2018;12(3):e0006368. doi: 10.1371/journal.pntd.0006368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zdraljevic S, Strand C, Seidel HS, Cook DE, Doench JG, Andersen EC. Natural variation in a single amino acid substitution underlies physiological responses to topoisomerase II poisons. PLoS Genet. 2017;13(7):e1006891. doi: 10.1371/journal.pgen.1006891. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Scripts that were used for this project are available online (https://git.wur.nl/published_papers/vanwijk_etal_2020_). Transcriptome data were deposited at ArrayExpress under E-MTAB-9742. eQTL results are publicly accessible on WormQTL2.
Supplemental material available at G3 online.