Abstract
Prostate cancer (PCa) is one of the two solid malignancies in which a higher T cell infiltration in the tumor microenvironment (TME) corresponds with a worse prognosis for the tumor. The inability of T cells to eliminate tumor cells despite an increase in their number reinforces the possibility of impaired antigen presentation. In this study, we investigated the TME at single-cell resolution to understand the molecular function and communication of dendritic cells (DCs) (as professional antigen-presenting cells). According to our data, tumor cells stimulate the migration of immature DCs to the tumor site by inducing inflammatory chemokines. Many signaling pathways such as TNF-α/NF-κB, IL2/STAT5, and E2F up-regulated after DCs enter the tumor location. In addition, some molecules such as GPR34 and SLCO2B1 decreased on the surface of DCs. The analysis of molecular and signaling alterations in DCs revealed some suppression mechanisms of tumors, such as removing mature DCs, reducing the DC's survival, inducing anergy or exhaustion in the effector T cells, and enhancing the differentiation of T cells to Th2 and Tregs. In addition, we investigated the cellular and molecular communication between DCs and macrophages in the tumor site and found three molecular pairs including CCR5/CCL5, CD52/SIGLEC10, and HLA-DPB1/TNFSF13B. These molecular pairs are involved in the migration of immature DCs to the TME and disrupt the antigen-presenting function of DCs. Furthermore, we presented new therapeutic targets by the construction of a gene co-expression network. These data increase our knowledge of the heterogeneity and the role of DCs in PCa TME.
Keywords: Prostate cancer, Tumor microenvironment, Dendritic cells, Immunotherapy, Single-cell RNA sequencing
1. Introduction
Prostate cancer (PCa) is the second most common malignant neoplasm in men globally [1] and also the fifth leading cause of cancer-related death [2]. The prognosis for patients with metastasis is poor; their 5-year relative survival rate is only 30.2% [1,3]. Age, genetics, environmental toxins, chemical agents, and radiation are a few risk factors that can contribute to the development of PCa pathogenesis [4]. Surgical and non-surgical treatments such as androgen deprivation therapy (ADT), radiation therapy (RT), ablative therapies, and chemotherapy are among the conventional therapeutic approaches [5]. Depending on the clinical situation, these methods can be applied singly or in combination [5]. Compared to individuals who choose radiation therapy, those who undergo radical prostatectomy are more likely to have urine incontinence and get erection maintenance [6]. Most patients experience erectile dysfunction, low libido, obesity, and loss of bone mass even after curative resection [[7], [8], [9]].
In light of the negative side effects of conventional treatments, current cancer research has focused on novel treatments such as immunotherapies, new delivery technologies, and precise targeting of the tumor microenvironment (TME) for PCa treatment [10]. One of the most significant difficulties in cancer research is the tumor-suppressive microenvironment. This complex microenvironment, which can be different among various tumors, plays crucial roles in tumor initiation, progression, and metastasis [11]. In this microenvironment, immune cells are suppressed, and numerous cell-based immunotherapies fail to work.
A deeper understanding of the TME and immune system suppression mechanisms facilitated the development of new immunotherapies, some of which have had remarkable success. For example, after identifying the role of some immune checkpoints (ICs) such as programmed cell death 1 (PD1) and cytotoxic-T-lymphocyte-associated protein 4 (CTLA4) in suppressing immune system cells, the inhibitors of these molecules were used as a therapeutic strategy in the treatment of many cancers, including melanoma, renal cell carcinoma (RCC), non-small cell lung cancer (NSCLC), bladder cancers, head and neck squamous cell carcinoma (HNSCC), gastric cancer, ovarian cancer, Hodgkin's lymphoma, and tumors with mismatch repair (MMR) deficiency [12,13]. Although these immune checkpoint blockades (ICB) are also utilized in PCa patients [[14], [15], [16]], the outcomes are inconsistent, which may be related to the heterogeneity of tumor cells or variations in the PCa microenvironment [[17], [18], [19], [20]]. Additionally, tumor cells attempt to evade being detected by CD8+T cells by downregulating the expression of MHC-I and co-receptors. CART cells are a developing cell-based technology designed to overcome the evasion of tumor cells from presenting antigens to the immune system [21]. These modified cells can recognize tumor-related antigens in a way that is not HLA complex-restricted [21]. Several promising studies are now being conducted employing CART cells to target antigens expressed in PCa (NCT02744287), (NCT01140373), (NCT03013712), and (NCT03159585) [22]. Therefore, a better understanding of the suppressive mechanisms employed by cancer cells as well as a focus on the modifications that immune cells experience when exposed to the TME can help in the creation of novel, efficient treatments.
Antigen-presenting cells (APCs) are one of the main targets of tumor immunotherapy. APCs play a critical role in the activation of anti-tumor responses by capturing, processing, and presenting tumor antigens to T cells. Similar to other cells, these cells experience alterations in the tumor suppressor microenvironment that impair antigen presentation. The TME recruits immature and functionally deficient dendritic cells (DCs), which causes defects in tumor antigen presentation [23]. Among other abnormalities observed in tumor-infiltrating dendritic cells (TIDCs) is a decrease in the expression of co-stimulatory molecules, a reduction in IL12 production, and an inhibition of the antigen processing processes. These cells are not able to activate CD4+ and CD8+ T lymphocytes [23]. Blockage of DC functioning caused by a tumor is a distinct method of tumor evasion. Furthermore, new research has shown the role of TIDCs in cancer treatments such as radiation therapy, chemotherapy, adoptive cell therapy, and ICs blockage [24].
Single-cell RNA sequencing (scRNA-seq) techniques that have recently undergone rapid development have made it possible to identify rare and novel cell types, characterize numerous different cell states simultaneously [25], and study cell-cell communication via ligand-receptor signaling in the TME [26]. Recently, limited studies based on this method have been conducted on the PCa microenvironment. Although these reports have revealed important aspects of this complexity, there is still a long way to go in delineating the immunological landscape of the TME.
Considering the importance of DCs in inducing anti-tumor immune responses and the loss of this function in a tumor suppressor microenvironment, we investigated the TME at single-cell resolution to better understand the molecular and cellular communication of these cells. We also hope that our findings help in the improvement of existing treatment strategies and the development of novel therapeutic approaches.
2. Materials and methods
2.1. Data acquisition
The experiment by Carriero R et al., with accession number GSE153892, is where the raw data came from (https://www.ncbi.nlm.nih.gov/geo/) [27]. They employed scRNA-seq with a total of six samples from two conditions (normal and tumor) and three donors of PCa tissue (Table 1). The GEO expression matrix was identified with gene symbols using information from the GPL18573 Illumina NextSeq 500. The data analysis process followed a standard Scanpy pipeline workflow (Fig. 1) [28].
Table 1.
The overall design of GSE153892. This dataset includes a total of six samples; two conditions (normal and tumor) and three patients.
| Donor | Number of Cells (Total) | Number of Cells (Tumor) | Number of Cells (Normal) |
|---|---|---|---|
| Donor 1 | 6416 | 3818 | 2598 |
| Donor 2 | 5387 | 4165 | 1222 |
| Donor 3 | 6194 | 3634 | 2560 |
| Total | 17997 | 11617 | 6380 |
Fig. 1.
The flowchart of the study. Preprocessing of PCa scRNA-seq data analysis, identification of TIDCs, molecular function and intercellular communication analysis of TIDCs, and construction of a gene co-expression network for potential therapeutic target identification were the main steps in this study.
2.2. Quality control, dimensionality reduction, clustering, and visualization
Quality control was used to exclude cells with poor quality, and we only included cells that complied with the following standards: more than 450 genes, fewer than 32000 counts, and less than 20% of readings matched mitochondrial genes. Normalized expression was performed to reduce bias in cell counts and improve intracellular replication of cell expression levels by using the Scanpy normalized-total function or by analyzing size factors for each cell. The combat algorithm corrected the batch effects from the six loaded samples [29]. To enable unsupervised grouping, the top 4000 highly variable genes (HVGs) had their variable count reduced using principal component analysis (PCA). The next step involved inserting a k-nearest-neighbor graph with a resolution of 1.0 for the Leiden community finding. The clusters were identified by the expression levels of specific marker genes [30]. The TIDCs were annotated with specific markers such as MRC1, CD1E, CD207, and CLEC10A. After that, we constructed UMAP embeddings with a minimum distance of 0.5 and a spread of 1.0 to display the most closely similar neighbor graph [31].
2.3. Differently expressed genes and cell cycle analysis of TIDCs
To determine the disparities between TIDCs in healthy and cancerous tissues, a comparative analysis of differently expressed genes (DEGs) was conducted. This analysis involved the utilization of the sc.tl.rank_genes_groups function, which is a feature selection and clustering tool used in the field of scRNA-seq analysis. It allows for the identification and grouping of genes that show significant differences in expression levels between different groups of cells. The sc.tl.rank_genes_groups function utilizes a t-test method for efficient evaluation of genes. The genes were selected based on certain criteria, such as an adjusted p-value of less than <0.05 and log fold change greater than |2|. The p-value was also corrected through the Benjamini-Hochberg method to ensure the validity and reliability of the results. These chosen genes were then set for further examination and interpretation.
After identifying DEGs in TIDC subpopulations, gene ontology analysis was used to determine the molecular function and KEGG Pathway (2021) of the identified DEGs. Gene Set Enrichment Analysis (GSEA; http://software.broadinstitute.org/gsea) is a computer program that allows researchers to determine whether a set of genes has statistically significant levels of expression in a specific cell type, biological process, cellular component, molecular function, or biological pathway. The GSEA uses MSigDB, the Molecular Signature Database, to provide gene sets for the gene set enrichment analysis. In this study, the WEB-based GEne SeT AnaLysis Toolkit (webgestalt; http://www.webgestalt.org/option.php) was used to annotate the DEGs' probable signaling pathways. Also, Enrichr (https://maayanlab.cloud/Enrichr/) was applied for Gene Ontology (GO) analysis.
The Scanpy function (scanpy.tl.score_genes_cell_cycle) was used to score S and G2M-specific genes to forecast the cell cycle stage. Each individual cell is given an S- and G2M-score, which are determined by the class with the greatest score. The cells are designated as being in the G1 phase if neither the S-score nor the G2M-score surpasses 0.5. The article by Kowalczyk et al. contains the cell cycle phase reference genes used for scoring [32].
2.4. Pseudotime analysis
The pseudotime analysis was performed with scFates v0.8.1 (https://pypi.org/project/scFates/), which is fully compatible with Scanpy and has GPU-accelerated features for faster and more scalable inference [33]. Early and late branch-specific features were identified using differential expression analysis and pseudotime values. For each bifurcation point of interest, a set of cells associated with the clusters involved in the bifurcation were selected.
2.5. ICs expression pattern in TIDCs
To find the expression behavior of ICs in TIDCs, we conducted differential expression analysis between tumor and healthy samples for the 13 most common inhibitory ICs, including TIGIT, LY9, PDCD1, LAG3, CTLA4, CD276, NT5E, PDCD1LG2, CD274, IDO1, VSIR, HAVCR2, and ENTPD. Following that, IC expression was visualized using pseudo-time, and the cluster specificity of their expression was determined using UMAP. After that, we checked to see whether the relevant ICs' expression patterns matched those of their counterparts in the TCGA PCa dataset by utilizing the GEPIA database.
2.6. Cell-cell interaction analysis
SquidPy [34], which gives analytic methods for depositing, manipulating, and interactively clarifying single-cell RNA sequencing data, was used to study cell-cell interaction. It makes use of an efficient re-implementation of the CellPhoneDB method [35], which can handle a large number of interacting pairs (100k+) and cluster combinations (100+). In this study, using the technique with 1000 permutations, only interactions whose ligand and receptor genes were expressed in at least 10% of the cells were taken into consideration.
2.7. Identification of transcription factors, miRNAs and regulatory drugs
We overlapped the biologically relevant genes to DrugBank and DGIdb to find potential therapeutic candidates for our important ligands and receptors (data released on December 25, 2022). DGIdb (www.dgidb.org) is a database that provides a vast collection of druggable targets from scientific papers, databases, and other online sources in addition to bioinformatics and chemoinformatics information on pharmaceuticals [36,37]. The candidates for the targeted genes were evaluated using a number of criteria, such as pharmacologically active medications, human efficacy, authorized annotations, or clinical studies. The position of the selected medications within the current treatment strategies was also looked into using the Clinical Trials website (accessed on December 25, 2022; https://clinicaltrials.gov/).
Then, CyTargetLinker (https://cytargetlinker.github.io/) was used to identify the transcription factors (TFs) engaged in the expression of these six genes. The miRTarbase database (http://mirtarbase.mbc.nctu.edu.tw/php/index.php) was utilized to find upstream miRNAs for significant genes. The data was then imported into Cytoscape (version: 3.8.2) (https://cytoscape.org/) for display after the screening process.
3. Results
3.1. PCa TME cell fractions
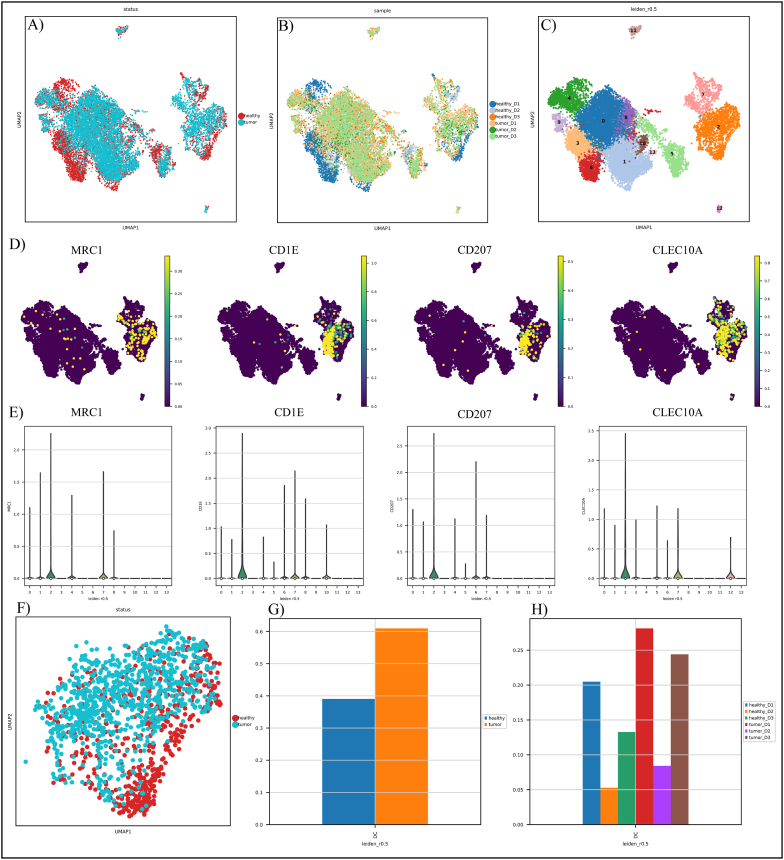
We included 13638 cells from 6 tissue samples of PCa donors for our scRNA-seq analysis after eliminating low-quality cells. Cells were classified based on their origin (tumor (n = 7744; 56.7%) or healthy (n = 5894; 43.3%)) (Table 2) (Fig. 2A–B). In the next step, cells were divided into 14 groups based on the expression of canonical gene markers in the TME of PCa, using UMAP and unsupervised graph-based clustering (Fig. 2C).
Table 2.
Quality control process.
| QC level | Number of Cells/Genes before QC | Criteria | Number of Filtered out Cells/Genes | Number of Cells/Genes after QC |
|---|---|---|---|---|
| Cells | 17997 | With less than 1000 counts | 4266 | 13638 |
| With more than 32000 counts | 5 | |||
| With Mitochondrial genes fraction more than 20% | 4 | |||
| With less than 450 expressed genes | 57 | |||
| With Ribosomal genes fraction more than 5% | 27 | |||
| Genes | 33538 | Detected in less than 20 cells | 18769 | 14769 |
Fig. 2.
Characterization of the PCa tumor microenvironment. A) Status-based UMAP visualization of cells; B) Sample-based UMAP visualization of cells; C) Leiden-based UMAP visualization of cells; D) UMAP visualization of TIDC marker genes; E) Violin displays the expression pattern of TIDC gene markers as well as other clusters; and F) Status-based UMAP visualization of TIDCs. The healthy cells are presented as red and turquoise cells, which demonstrate the tumor cells. G) TIDC cell fraction in healthy and tumor samples; H) TIDC cell fraction in different donors.
Cluster 2 was classified as DC according to marker gene expression profiles like MRC1, CD1E, CD207, and CLEC10A (Fig. 2D–E) [38,39]. Cluster 2 includes a total of 6655 cells, out of which 3623 are derived from the tumor. Tumor-derived DCs were distributed differently from their healthy counterparts (Fig. 2F). After that, we compared the TIDC fractions in each status and donor and observed that the abundance of TIDCs varies across states and donors, suggesting considerable tumor heterogeneity (Fig. 2G–H).
3.2. DEG and cell cycle analysis of the PCa TME derived TIDCs
After comparing gene expression of TIDCs in normal and tumor PCa samples, pseudogenes, mitochondrial encoded, and ribosomal genes were excluded. 1862 DEGs, including 1089 up-regulated genes and 773 down-regulated genes, were found (Fig. 3A) (p.adj 0.05). DEG analysis showed a significant increase in the expression of CDC45, CDCA8, VSIG10L, CD200, IGLC7, and GMEB2 genes in tumor samples compared to healthy samples (Fig. 3A). Wan et al. confirmed that CDCA8 plays a role in promoting malignant tumor progression. However, the exact function of CDCA8 in the development and progression of PCa remains unclear. CDCA8 was significantly overexpressed in PCa cells compared with normal prostate cells. High CDCA8 expression predicts poor prognosis in PCa patients, and CDCA8 expression was higher in high-grade PCa [40]. On the other hand, the expression of SEMG1, TIGD4, SEMG2, GOLGA8Q, and RMRP genes in the tumor sample has decreased compared to the healthy sample (Fig. 3A). The results revealed that DEGs were mainly involved in the cytokine-mediated signaling pathways, cellular response to cytokine stimulus, neutrophil activation involved in immune response, and positive regulation of cytokine production in the BP category. According to MSigDB, DEGs are mostly involved in TNF-α signaling via NF-κB, inflammatory response, allograft rejection, apoptosis, IL2/STAT5 signaling, hypoxia, complement, mTORC1 signaling, and interferon alpha response (Fig. 3B). Also, previous study has found that there is a robust expression of TNF-α in localized PCa [41].
Fig. 3.
DEG and cell cycle analysis of TIDCs in PCa TME. A) TIDCs DEGs volcano plot. Genes with an adjusted p-value <0.05 were considered significant and were highlighted in red; B) TIDCs DEGs biological process; C) TIDC cell cycle in healthy samples; C) TIDC cell cycle in tumor samples; E) The area plot shows the cell cycle phase of TIDCs in healthy and tumor samples. Although TIDCs that were in the G1 phase decreased from healthy to tumor samples, TIDCs that were in the G2M phase increased from healthy to tumor samples. The S-phase cell didn't show a significant alteration; F) GSEA analysis of TIDCs DEGs based on MSigDB.
3.3. Pseudo-time trajectory revealed TIDCs dynamic changes in PCa TME
We did a pseudo-temporal reconstruction using the program scFates, which infers global lineage structures and pseudo-time variables for each cell along a lineage. Eight distinct nodes were identified that corresponded to unique transcriptional changes in our clusters (Fig. 4A), of which node 4 was considered as TIDCs. Gene expression was plotted as a function of pseudo-time to track changes across different trajectories. As shown in Fig. 4A, MS4A6A, MS4A7, C1QA, C1QB, C1QC, FCGR2A, CSF1R, A2M, SLCO2B1, and GPR34 were the top 10 upregulated genes in the TIDCs node. These genes were compared with the DEG list and similarly selected genes for evaluation of expression behavior alongside pseudo-time trajectory (Fig. 4B). The pseudo-time expression pattern showed that the eight similar genes between the two lists, including MS4A6A, CSF1R, C1QA, C1QB, C1QC, A2M, SLCO2B1, and GPR34, were expressed uniquely in the TIDCs cluster (Fig. 4C). To verify these findings, the expression of each gene was also visualized through UMAP presentation. Additionally, as shown in Figure A1A, the pattern of pseudo-temporal gene expression was confirmed. Future studies could use this pattern as a disease-and cluster-specific biomarker. According to MSigDB, these genes are mostly involved in complement pathways, IL6/JAK/STAT3 signaling, and coagulation (Figure A1B). The enrichment results also demonstrated that similar genes were involved in cell junction disassembly, synapse pruning, complement activation through the classical pathway, humoral immune response mediated by circulating immunoglobulin, and regulation of complement activation in the BP term list (Figure A1C).
Fig. 4.
The pseudo-time analysis of PCa TME. A) The top ten upregulated genes in the TIDCs node. B) Comparison between the top ten upregulated genes in the TIDCs with the DEG list, and it indicates that eight genes are common between the two lists. C) Exhibition of the pseudo-time analysis of the eight genes, and all eight genes are specific to the TIDCs cluster.
3.4. Inhibitory ICs expression pattern in the TIDCs of the PCa TME
The possibility of variations in the expression of 13 common inhibitory ICs in TIDCs of tumor and healthy samples were then examined. Our results indicated considerable heterogeneity in the expression of ICs in the TIDCs of PCa samples (Fig. 5). According to our analysis, only three genes matched with the DEGs list based on a p.adj 0.05 cutoffs (Fig. 5A). The expression level of VSIR, HAVCR2, and ENTPD1 genes significantly decreased in the TIDCs of tumor samples compared to healthy samples (Fig. 5B–C). The expression changes of other ICs, including TIGIT, LY9, PDCD1, LAG3, CTLA4, CD276, NT5E, PDCD1LG2, CD274, and IDO1, were not significant between TIDCs of two statuses. The three targeted genes were then tested for their cluster-specificity using pseudo-time and UMAP, and their expression in the TIDC cluster was verified (Fig. 5D–E). Additionally, to validate the ICs expression results, we used TCGA PCa patient specimens. The TCGA cohort's results showed the expression of the VSIR gene has been considerably down regulated in PCa tumor samples. No significant changes have been observed in the HAVCR2 and ENTPD1 gene expression.
Fig. 5.
Inhibitory IC characterization in PCa TIDCs. A) A comparison of the 13 common inhibitory ICs found in TIDCs with the DEG list; B) dot plot analysis of the 13 common inhibitory ICs found in PCa TIDCs. As shown, only VSIR, HAVCR2, and ENTPD1 had significant changes from healthy to tumor samples; C) log2 fold change and adjusted p value of inhibitory ICs in TIDCs; D) and E) The pseudo-time and UMAP analysis of 3 significant Inhibitory ICs. As shown, all three genes are specific for the TIDCs cluster.
3.5. The cell-cell interaction of the TIDCs in the TME of PCa
Then, Squidpy was used to examine the cross-talk between TIDCs and other cells in the PCa TME. Overall, among TME cell populations, the degree of interaction between cluster 7 and TIDCs was the highest based on Squidpy score which was determined by calculating the total ligand and receptor scores between various clusters (Fig. 6A). Due to the expression of MARCO, SAT1, TYROBP, CD163, and FTL in cluster 7, this cluster was determined to be tumor-associated macrophages (TAMs) (Figure A2). As a result, we focused on molecular and cellular interactions in these two cells. Themost critically involved communications pairs between TIDCs (as source) and TAMs (as target) were the BMP8B/PLAUR, ANXA1/FPR1, LGALS9/CD44, HLA-E/KLRK1, HLA-E/KLRC2, and HLA-E/KLRC2 pairs (Fig. 6B). When we considered TAMs as the source and TIDCs as the target, the communications that were most seriously involved were MIF/CD74, COPA/CD74, CXCL8/CXCR1, CXCL8/CXCR2, CCL3/CCR1, CCL3/CCR5 and NRG1/HLA-DBP1 pairs (Fig. 6C).
Fig. 6.
Quantification of cell-cell interactions occurring in the PCa tumor microenvironment. A) Cell-cell communication scores of TIDCs and other clusters. As shown, TIDCs interacted the most with cluster 7, which was annotated as TAMs; B) Cell-cell communication in the PCa TME between TIDCs as receivers and TAMs as sources; C) Cell-cell communication in the PCa TME between TIDCs as sources and TAMs as receivers; D) and E) MSigDB and biological processes of significant ligands and receptors of TIDCs and TAMs in the PCa TME; F) pair-wise gene correlation analysis of CCR5-CLL5, SIGLEC10-CD52, and TNFSF13B-HLA-DPB1 genes in the TCGA dataset; G) UMAP visualization of significant correlated ligands and receptors of TIDCs and TAMs in the PCa TME.
These ligands and receptors were all found to be involved in TNF-α signaling, allograft rejection, inflammatory response, epithelial-mesenchymal transition, apoptosis, hypoxia, KRAS signaling up, interferon-gamma response, and IL6/JAK/STAT3 signaling, according to MSigDB study (Fig. 6D). Additionally, the GO biological process revealed that these genes control the inflammatory response to injury, chemokine signaling pathway, positive regulation of CD4+, positive regulation of prostaglandin biosynthesis, DC migration, and the release of sequestered calcium ion into the cytosol by the sarcoplasmic reticulum, and granulocytes chemotaxis (Fig. 6E).
Then the correlation between the mRNA level of ligands and receptors was investigated using the GEPIA web tool. Our findings showed that in TCGA PCa patient specimens, CCL5, CD52, and TNFSF13B ligands were positively correlated with the expression levels of CCR5, SIGLEC10, and HLA-DPB1 receptors (Pearson's product-moment correlation coefficient r 0.6, p0.01) (Fig. 6F). Fig. 6G showed the expression levels of these correlated ligand and receptor pairs. TNFSF13B was mostly expressed in the TAMs cluster, while SIGLEC10 and HLA-DPB1 showed a TIDCs cluster-specific expression pattern. UMAP visualization, on the other hand, showed that CD52, CCR5, and CCL5 were expressed in various clusters.
3.6. Construction of correlated ligand-receptor genes co-expression network
To further explore the six prescreened correlated ligands and receptors, a gene co-expression network was constructed using the GeneMANIA online database without the addition of related genes and attributes. The six correlated genes were then subjected to analyses by CyTargetLinker to predict associated TFs. To determine the pathways and biological processes that the TFs were responsible for inducing, we also entered the TFs into the MSigDB and GO databases (Figure A3). The miRTarbase database was also employed to predict the upstream miRNAs of crucial genes. In addition, by using the DGIdb and Drugbank databases, related drugs that are in different stages of clinical trials were identified. Finally, in a single illustration, the entire set of detected interactions was illustrated (Fig. 7).
Fig. 7.
Co-expression network construction and finding related drugs, miRNAs, and TFs.
4. Discussion
DCs as the most professional antigen-presenting cells, play a crucial role in inducing T cell activation in various diseases, including cancer. However, these cells become functionally defective after being placed in the TME and even participate in promoting the tumor suppressor microenvironment [42]. Therefore, we examined the cellular and functional analysis of TIDCs in the PCa microenvironment using scRNA-seq.
Our findings indicated that the number of TIDCs was higher in tumor tissue than in healthy tissue (Fig. 2G and H). Tumor cell-associated inflammation may be the reason for the higher frequency of DCs in tumor tissue compared to healthy tissue. Based on the studies, chronic inflammation is necessary for the survival and growth of the majority of solid malignancies [43]. Tumor cell-associated inflammation increases the inflammatory chemokines and mediators in the site and promotes the migration of innate and adaptive immune cells to the tumor site [43]. Finally, cancer cells employ these immune cells to further their objectives.
Examining the biological function of DCs in both healthy and tumor tissues showed that some signaling pathways have increased in the TIDCs (Fig. 3B and C). The TNF-α/NF-κB signaling pathway was one of these, which significantly increased in TIDCs compared to DCs in healthy samples. Activation of this pathway can lead to the release of inflammatory mediators. Wang et al. showed that TIDCs are more capable of producing chemokines like CCL2, CCL3, CCL4, CCL5, CCL8, CXCL8, cytokines such as IL6, IL16, IL23A, TNF-α, and growth factors such as VEGFA, PDGFA, FGF11, and NRG1 than DCs in healthy tissue [44]. Consistent with Wang's results, we also confirmed increased inflammatory responses in TIDCs. Contrary to earlier beliefs, it is now well established that some inflammatory cytokines, like TNF-α and IL6, play a crucial role in the maintenance, growth, and even metastasis of cancer cells [45,46]. Some chemokines, including CCL2 and CCL4, have also been implicated in the spread of cancer cells or the migration of suppressor cells to the tumor site [47]. Since the activity of the TNF-α/NF-κB signaling pathway is enhanced in TIDCs, the expression of P50 will probably also increase. It is important to note that an environment rich with IL10 and TGF-β, like a TME, causes the nuclear accumulation of P50 [48]. In this study has shown that the accumulation of P50 in the nucleus of DCs causes an increase in the expression of IDO and a decrease in the production of IL1, IL18, and IFN-β by DCs [48]. Additionally, it has been demonstrated that the expression of P50 by DCs enhances their capacity to induce Th2 and Treg cells [49].
Our investigations also indicated increased induction of angiogenesis by TIDCs, which can be related to the high ability of these cells to secrete growth factors such as VEGF and CCL5 [44,50].
According to Balachander et al., DCs temporarily produce IL2 after being stimulated by PAMPs [51]. Autocrine IL2 signaling in DCs activates the JAK/STAT5 pathway and induces apoptosis in PAMP-matured DCs to maintain immunological tolerance after inflammation [51]. Our results confirmed that this signaling pathway is significantly activated in TIDCs, which can indicate the tumor's attempt to eliminate the mature DCs and prevent the development of anti-tumor responses. One of the most significant pathways examined in this study was the E2F pathway, which was elevated in TIDCs compared to DCs in healthy tissue. E2F has been identified as a critical regulator of DC maturation, and multiple examinations have demonstrated that E2F overexpression prevents LPS-induced DC maturation [52].
According to our data analysis, TIDCs expressed eight DC-specific genes lower than DCs from healthy tissue (MS4A6A, C1QA, C1QB, C1QC, CSF1R, A2M, SLCO2B1 and GPR34). These genes are involved in a number of processes, such as synapse pruning, IL6/JAK/STAT3 signaling, complement pathways, cell junction disintegration, and coagulation (Fig. 4 and Figure A1). Among these eight genes, the decrease in expression of SLCO2B1 and GPR34 has been considerable compared to other genes. Jäger and colleagues believe that the activation of NF-κB and MAPK signaling pathways down regulate the GPR34 mRNA expression in DCs [53]. As a result of decreased GPR34 expression, DCs have less of this receptor on their surface and increased caspase 3 and 7 activity, which lowers cell survival and increases the induction of apoptosis [53]. This process can be one of the induction strategies of the TME for reducing the survival of TIDCs and presenting antigens. The influx transporters of the SLCO family are involved in the uptake of medicines, hormones, leukotrienes, prostaglandins, and metabolites of arachidonic acid. DCs only expressed SLCO2B1, SLCO3A1, and SLCO4A1 genes [54]. It has been established that SLCO2B1 is crucial for the absorption of many drugs; however, the function of SLCO2B1 in DCs is still unclear. It is possible that the decrease of SLCO2B1 in TIDCs represents a mechanism for the development of drug resistance.
The limited treatment options for PCa patients and the favorable results of treatment with ICIs in some cancers have caused the use of ICIs for the treatment of PCa patients to receive more attention [55]. As a result, we investigated the state and existence of ICs on the surface of DCs. ICs panel examination showed that only three immunological checkpoints (VSIR, HAVCR2, and ENTPD1) changed in TIDCs compared to DCs (Fig. 5C–D). Although the expression of these genes has decreased in TIDCs at single-cell resolution but the analysis of gene expression in bulk level only confirms the decrease in expression in VSIR (Fig. 5F). It can be concluded that the development of PCa is not related to the increased expression of any of the ICs. This also seems to explain the poor efficacy of anti-immunization checkpoint monotherapies for PCa treatment. In addition, the lack of a correlation between the expression of VISTA and that of PDCD1LG2, TIGIT, TIM3, and LAG3 in PCa showed that several IC proteins cooperate to inhibit anti-tumor immunity and that co-targeting these molecules may have synergistic effects.
Macrophages (MQ) make up a significant portion of the immune cells in the TME. MQs are typically divided into two subsets (M1 and M2). M1 is induced by inflammatory mediators like interferon-gamma (IFN-γ) and lipopolysaccharide (LPS), while M2 is induced by anti-inflammatory mediators like interleukin IL4 and IL13. It is evident that the tumor suppressor microenvironment induces M2s, and because of their capacity to suppress immune responses, they are called tumor-associated macrophages (TAMs). These cells support angiogenesis and provide a tumor-suppressive microenvironment by secreting cytokines and other mediators. The rise in the number of TAMs in the TME, their significant contribution to the advancement of tumor goals, and the high score these cells received following the Squidpy analysis in connection with DCs drew our attention to the relationship between TIDCs and TAMs. The cell-cell interaction analysis of TIDCs and TAMs identified the most important ligand-receptor pairs (Fig. 6B–C). These ligand-receptor interactions were related to different biological processes and pathways (Fig. 6D–E). We selected CCR5/CCL5, CD52/SIGLEC10, and HLA-DPB1/TNFSF13B as hub pairs by confirming their co-expression correlation in the TCGA PCa dataset (Fig. 6F).
The analysis of the data revealed that CCR5 and CCL5 expression have increased in both TIDCs and TAMs. Monocytes are one of the CCR5-expressing cells that are attracted to the CCL5-rich TME. Monocytes develop into M2 (TAMs) after entering the TME, which promotes the generation of a tumor-suppressive environment [56,57]. TAMs with high expression of PDL-1 and CD206 and increased secretion of IL10 and TGF-β play an important role in the induction of Tregs [58]. CCR5 is a crucial molecular player in directing DCs to the site of inflammation. Immature DCs migrate to the site of inflammation because they have high expression of CCR5, whereas mature DCs down regulate CCR5 and up regulate CCR7 [59]. Immature DCs migrate to the tumor site as a result of CCL5, which is secreted by tumor cells or cells in the TME, including TAMs. The uptake of tumor antigens causes anergy when co-stimulatory molecule expressions and DC maturation are absent (because of overexpression of E2F). Additionally, immature DCs play a role in the migration of other cells into the tumor site by producing CCL5 [59]. TIDCs and TAMs are both sources of CCL5 production in the tumor environment. The CCL5/CCR5 axis is involved in tumor growth, the proliferation of cancer stem cells, metastasis, angiogenesis, and drug resistance [50].
The immunoglobulin-like type I transmembrane protein is known as Siglecs, which binds sialic acid, and has immune receptor tyrosine inhibitory motifs (ITIM) in the cytoplasm. The structure containing sialic acid is recognized by siglecs. SIGLEC10, as an inhibitory receptor, is expressed on the surface of immune cells, including DCs, B cells, monocyte, and a small population of NK and T cells [60]. The primary ligand for this inhibitory receptor is soluble CD52; however, studies have reported that SIGLEC10 can also bind to CD24 and vascular adhesion protein 1 (VAP-1) [61]. The connection of CD52 to SIGLEC10 can cause inhibitory signal transmission and disruption of the maturation and function of DCs.
According to the results of Chen et al., TAMs are one of the most important producers of TNFSF13B in the TME [62]. TNFSF13B has already been shown to be involved in immunosuppression and malignant cell invasion [62]. However, there is no data on the relation between HLA-DPB1and TNFSF13B.
Drug repositioning, often known as old drugs for new purposes, is a highly efficient, low-cost, and risk-free approach to discovering new indications for existing pharmaceuticals. We, therefore, presented chemicals and immunotherapy medication candidates that influence the interactions of TIDCs and TAMs with each other and with other cells in this study with the use of the DGIdb, Drugbank, and clinical trial databases (Fig. 7). Belimumab which inhibits TNFSF13B protein could prevent cancer invasion and immune suppression in the PCa microenvironment. Belimumab was approved by the FDA on March 9, 2011, and is the first targeted biological drug specifically designed to treat Systemic lupus erythematosus (SLE) [63]. Alemtuzumab by covering soluble CD52 prevents its binding to SIGLEC10 and the transmission of the inhibitory signal to the DCs. This drug can stop antigen-presenting dysfunction and anergy induction by DCs. The first formulation (Mabcampath® 1033 mg administered over 12 weeks) was used to treat CD52+ T and B cell malignancies, particularly chronic lymphocytic leukemias, and other lymphocyte-mediated diseases [64]. Maraviroc, which is used in the treatment of HIV, by targeting CCR5 can block angiogenesis and even improve the results of treatment of PCa with DC vaccines [65,66]. In addition, a set of miRNAs that target the selected molecules is listed in Fig. 7. The transcription factors that play a role in the expression of these molecules have also been identified and characterized. Each of these molecules can be a therapeutic target that enables the restoration of the function of immature DCs in the TME.
After analyzing the data, we can ultimately provide an overview of how the TME affects DCs. Tumor cells stimulate the migration of immune cells, including immature DCs, to the tumor site by secreting inflammatory cytokines and chemokines, such as CCL5. Signaling pathways change as a result of DCs entering the tumor location. Tumor cells attempt to eliminate mature DCs and reduce the survival of immature DCs to decrease the possibility of tumor antigens presenting. In addition, tumor cells prevent the maturation of immature DCs in the TME, so that in the absence of co-stimulatory molecules, antigen-presenting to T cells induces anergy or exhaustion in the effector T cells. The increased TNF-α/NF-κB pathway in DCs makes them a source of the tumor-needed cytokines and chemokines and enhances their capacity to differentiate and induce Th2 and Tregs. Immature DCs can influence other cells and promote the microenvironment in favor of the tumor.
Each of these signaling pathways or molecules that change after DCs are placed in the TME can be a target for immunotherapy. Targeting these pathways and molecules could restore the maturation and function of DCs. Considering the complexity of tumor mechanisms to escape from the immune system, it can be seen that monotherapy cannot be a suitable solution to overcome the complicated TME. Advanced technologies such as scRNA sequencing can provide an immunological overview of the interactions and cellular changes in the TME. This immunological landscape can improve existing treatment strategies and develop novel therapeutic approaches (Fig. 8).
Fig. 8.
Schematic representation of ligand-receptors and potential related drugs.
5. Conclusions
In summary, our study investigated the molecular function and communication of TIDCs in the PCa TME. We found that DCs undergo molecular and signaling alterations in the TME, leading to the suppression of anti-tumor immunity. We also identified molecular pairs involved in the migration of TIDCs to the TME and disruption of TIDC antigen-presenting function. Finally, our study provides potential therapeutic targets for PCa by constructing a gene co-expression network. Our findings contribute to a better understanding of the heterogeneity and function of TIDCs in the PCa TME, which may lead to the development of novel therapeutic strategies. Last but not least, it should be highlighted that although the results of bioinformatics analysis are crucial, they need to be supported by cohort studies and experiments for greater validity.
Author contribution statement
Adib Miraki Feriz, Arezou Khosrojerdi, Neusha Shamsaki, Mohammad Fereidouni: Performed the experiments; Wrote the paper.
Mohammad Lotfollahi: Analyzed and interpreted the data; Wrote the paper.
Mohammad GhasemiGol, Edris HosseiniGol, Mohammad Hossein Rohban: Analyzed and interpreted the data; Wrote the paper.
Ahmad Reza Sebzari, Samira Saghafi, Patrizia Leone, Nicola Silvestris: Contributed reagents, materials, analysis tools or data; Wrote the paper.
Hossein Safarpour, Vito Racanelli: Conceived and designed the experiments; Wrote the paper.
Funding statement
This research received no external funding.
Data availability statement
Data sharing is not applicable to this article as no datasets were generated during the current study.
Declaration of interest's statement
The authors declare no conflict of interest.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.heliyon.2023.e15694.
Contributor Information
Hossein Safarpour, Email: safarpour701@yahoo.com.
Vito Racanelli, Email: vito.racanelli1@uniba.it.
Appendix A. Supplementary data
The following are the Supplementary data to this article.
fig. s1.
fig. s2.
fig. s3.
References
- 1.Khazaei Z., et al. Global cancer statistics 2018: globocan estimates of incidence and mortality worldwide prostate cancers and their relationship with the human development index. Advances in Human Biology. 2019;9(3):245. [Google Scholar]
- 2.Rawla P. Epidemiology of prostate cancer. World J. Oncol. 2019;10(2):63. doi: 10.14740/wjon1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Steele C.B., et al. Prostate cancer survival in the United States by race and stage (2001‐2009): findings from the CONCORD‐2 study. Cancer. 2017;123:5160–5177. doi: 10.1002/cncr.31026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fujita K., et al. Obesity, inflammation, and prostate cancer. 2019;8(2):201. doi: 10.3390/jcm8020201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Evans A.J.J.M.P. Treatment effects in prostate cancer. 2018;31(1):110–121. doi: 10.1038/modpathol.2017.158. [DOI] [PubMed] [Google Scholar]
- 6.Brawley S., Mohan R., Nein C. Localized prostate cancer: treatment options. 2018;97(12):798–805. [PubMed] [Google Scholar]
- 7.Oun R., Moussa Y.E., Wheate N.J.J.D.t. The side effects of platinum-based chemotherapy drugs: a review for chemists. 2018;47(19):6645–6653. doi: 10.1039/c8dt00838h. [DOI] [PubMed] [Google Scholar]
- 8.Kishan A.U., et al. Long-term outcomes of stereotactic body radiotherapy for low-risk and intermediate-risk prostate cancer. 2019;2(2) doi: 10.1001/jamanetworkopen.2018.8006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dobbs R.W., et al. Estrogens and prostate cancer. 2019;22(2):185–194. doi: 10.1038/s41391-018-0081-6. [DOI] [PubMed] [Google Scholar]
- 10.Chyuan I.-T., Chu C.-L., Hsu P.-N. Targeting the tumor microenvironment for improving therapeutic effectiveness in cancer immunotherapy: focusing on immune checkpoint inhibitors and combination therapies. Cancers. 2021;13(6):1188. doi: 10.3390/cancers13061188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang M., et al. Role of tumor microenvironment in tumorigenesis. J. Cancer. 2017;8(5):761. doi: 10.7150/jca.17648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Thompson J.A. New NCCN guidelines: recognition and management of immunotherapy-related toxicity. J. Natl. Compr. Cancer Netw. 2018;16(5S):594–596. doi: 10.6004/jnccn.2018.0047. [DOI] [PubMed] [Google Scholar]
- 13.Das S., Johnson D.B. Immune-related adverse events and anti-tumor efficacy of immune checkpoint inhibitors. Journal for immunotherapy of cancer. 2019;7(1):1–11. doi: 10.1186/s40425-019-0805-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Beer T.M., et al. Randomized, double-blind, phase III trial of ipilimumab versus placebo in asymptomatic or minimally symptomatic patients with metastatic chemotherapy-naive castration-resistant prostate cancer. J. Clin. Oncol. 2017;35(1):40–47. doi: 10.1200/JCO.2016.69.1584. [DOI] [PubMed] [Google Scholar]
- 15.Hansen A., et al. Pembrolizumab for advanced prostate adenocarcinoma: findings of the KEYNOTE-028 study. Ann. Oncol. 2018;29(8):1807–1813. doi: 10.1093/annonc/mdy232. [DOI] [PubMed] [Google Scholar]
- 16.Antonarakis E.S., et al. Pembrolizumab for treatment-refractory metastatic castration-resistant prostate cancer: multicohort, open-label phase II KEYNOTE-199 study. J. Clin. Oncol. 2020;38(5):395. doi: 10.1200/JCO.19.01638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vitkin N., et al. The tumor immune contexture of prostate cancer. Front. Immunol. 2019;10:603. doi: 10.3389/fimmu.2019.00603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.De Velasco M.A., Uemura H. Prostate cancer immunotherapy: where are we and where are we going? Curr. Opin. Urol. 2018;28(1):15–24. doi: 10.1097/MOU.0000000000000462. [DOI] [PubMed] [Google Scholar]
- 19.Yang Y., et al. High intratumoral CD8+ T‐cell infiltration is associated with improved survival in prostate cancer patients undergoing radical prostatectomy. Prostate. 2021;81(1):20–28. doi: 10.1002/pros.24068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Handa S., et al. Immunotherapy in prostate cancer: current state and future perspectives. Therapeutic advances in urology. 2020;12 doi: 10.1177/1756287220951404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Marofi F., et al. CAR T cells in solid tumors: challenges and opportunities. Stem Cell Res. Ther. 2021;12(1):1–16. doi: 10.1186/s13287-020-02128-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wolf P., et al. The potential of CAR T cell therapy for prostate cancer. Nat. Rev. Urol. 2021;18(9):556–571. doi: 10.1038/s41585-021-00488-8. [DOI] [PubMed] [Google Scholar]
- 23.Ma Y., et al. Dendritic cells in the cancer microenvironment. J. Cancer. 2013;4(1):36. doi: 10.7150/jca.5046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim C.W., Kim K.-D., Lee H.K. The role of dendritic cells in tumor microenvironments and their uses as therapeutic targets. BMB reports. 2021;54(1):31. doi: 10.5483/BMBRep.2021.54.1.224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Derakhshani A., et al. The expression pattern of VISTA in the PBMCs of relapsing-remitting multiple sclerosis patients: a single-cell RNA sequencing-based study. Biomed. Pharmacother. 2022;148 doi: 10.1016/j.biopha.2022.112725. [DOI] [PubMed] [Google Scholar]
- 26.Tirosh I., et al. Dissecting the multicellular ecosystem of metastatic melanoma by single-cell RNA-seq. Science. 2016;352(6282):189–196. doi: 10.1126/science.aad0501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Masetti M., et al. Lipid-loaded tumor-associated macrophages sustain tumor growth and invasiveness in prostate cancer. J. Exp. Med. 2021;219(2):e20210564. doi: 10.1084/jem.20210564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wolf F.A., Angerer P., Theis F.J. SCANPY: large-scale single-cell gene expression data analysis. Genome Biol. 2018;19(1):1–5. doi: 10.1186/s13059-017-1382-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang Y., Parmigiani G., Johnson W.E. ComBat-seq: batch effect adjustment for RNA-seq count data. NAR genomics and bioinformatics. 2020;2(3):lqaa078. doi: 10.1093/nargab/lqaa078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Soneson C., Robinson M.D. Bias, robustness and scalability in single-cell differential expression analysis. Nat. Methods. 2018;15(4):255–261. doi: 10.1038/nmeth.4612. [DOI] [PubMed] [Google Scholar]
- 31.Becht E., et al. Dimensionality reduction for visualizing single-cell data using UMAP. Nat. Biotechnol. 2019;37(1):38–44. doi: 10.1038/nbt.4314. [DOI] [PubMed] [Google Scholar]
- 32.Kowalczyk M.S., et al. Single-cell RNA-seq reveals changes in cell cycle and differentiation programs upon aging of hematopoietic stem cells. Genome Res. 2015;25(12):1860–1872. doi: 10.1101/gr.192237.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Faure L., et al. scFates: a scalable python package for advanced pseudotime and bifurcation analysis from single-cell data. 2023;39(1):btac746. doi: 10.1093/bioinformatics/btac746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Palla G., et al. Squidpy: a scalable framework for spatial omics analysis. Nat. Methods. 2022;19(2):171–178. doi: 10.1038/s41592-021-01358-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Efremova M., et al. CellPhoneDB: inferring cell–cell communication from combined expression of multi-subunit ligand–receptor complexes. Nat. Protoc. 2020;15(4):1484–1506. doi: 10.1038/s41596-020-0292-x. [DOI] [PubMed] [Google Scholar]
- 36.Wishart D.S., et al. DrugBank 5.0: a major update to the DrugBank database for 2018. Nucleic Acids Res. 2018;46(D1):D1074–D1082. doi: 10.1093/nar/gkx1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Freshour S.L., et al. Integration of the drug–gene interaction database (DGIdb 4.0) with open crowdsource efforts. Nucleic Acids Res. 2021;49(D1):D1144–D1151. doi: 10.1093/nar/gkaa1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Villani A.C., et al. Single-cell RNA-seq reveals new types of human blood dendritic cells, monocytes, and progenitors. Science. 2017;356(6335) doi: 10.1126/science.aah4573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen B., et al. Unraveling the heterogeneity and ontogeny of dendritic cells using single-cell RNA sequencing. 2021;12 doi: 10.3389/fimmu.2021.711329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wan S., et al. Overexpression of CDCA8 predicts poor prognosis and promotes tumor cell growth in prostate cancer. Front. Oncol. 2022;12:1228. doi: 10.3389/fonc.2022.784183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Deichaite I., et al. Differential regulation of TNFα and IL-6 expression contributes to immune evasion in prostate cancer. J. Transl. Med. 2022;20(1):527. doi: 10.1186/s12967-022-03731-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Veglia F., Gabrilovich D.I. Dendritic cells in cancer: the role revisited. Curr. Opin. Immunol. 2017;45:43–51. doi: 10.1016/j.coi.2017.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Grivennikov S.I., Greten F.R., Karin M. Immunity, inflammation, and cancer. Cell. 2010;140(6):883–899. doi: 10.1016/j.cell.2010.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang Z., et al. Dendritic cells in tumor microenvironment promoted the neuropathic pain via paracrine inflammatory and growth factors. Bioengineered. 2020;11(1):661–678. doi: 10.1080/21655979.2020.1771068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kumari N., et al. Role of interleukin-6 in cancer progression and therapeutic resistance. Tumor Biol. 2016;37(9):11553–11572. doi: 10.1007/s13277-016-5098-7. [DOI] [PubMed] [Google Scholar]
- 46.Balkwill F. TNF-α in promotion and progression of cancer. Cancer Metastasis Rev. 2006;25(3):409–416. doi: 10.1007/s10555-006-9005-3. [DOI] [PubMed] [Google Scholar]
- 47.Qin R., et al. Role of chemokines in the crosstalk between tumor and tumor-associated macrophages. Clin. Exp. Med. 2022:1–15. doi: 10.1007/s10238-022-00888-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Larghi P., et al. The p50 subunit of NF-κB orchestrates dendritic cell lifespan and activation of adaptive immunity. PLoS One. 2012;7(9) doi: 10.1371/journal.pone.0045279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Artis D., et al. Dendritic cell-intrinsic expression of NF-κB1 is required to promote optimal Th2 cell differentiation. J. Immunol. 2005;174(11):7154–7159. doi: 10.4049/jimmunol.174.11.7154. [DOI] [PubMed] [Google Scholar]
- 50.Zeng Z., et al. Genes & diseases; 2021. CCL5/CCR5 axis in Human Diseases and Related Treatments. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Balachander A., et al. Dendritic cell derived IL‐2 inhibits survival of terminally mature cells via an autocrine signaling pathway. Eur. J. Immunol. 2015;45(5):1494–1499. doi: 10.1002/eji.201445050. [DOI] [PubMed] [Google Scholar]
- 52.Fang F., et al. Transcription factor E2F1 suppresses dendritic cell maturation. J. Immunol. 2010;184(11):6084–6091. doi: 10.4049/jimmunol.0902561. [DOI] [PubMed] [Google Scholar]
- 53.Jäger E., et al. Dendritic cells regulate GPR34 through Mitogenic signals and undergo apoptosis in its absence. J. Immunol. 2016;196(6):2504–2513. doi: 10.4049/jimmunol.1501326. [DOI] [PubMed] [Google Scholar]
- 54.Skazik C., et al. Differential expression of influx and efflux transport proteins in human antigen presenting cells. Exp. Dermatol. 2008;17(9):739–747. doi: 10.1111/j.1600-0625.2008.00745.x. [DOI] [PubMed] [Google Scholar]
- 55.Venkatachalam S., et al. Immune checkpoint inhibitors in prostate cancer. Cancers. 2021;13(9):2187. doi: 10.3390/cancers13092187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Oppermann M. Chemokine receptor CCR5: insights into structure, function, and regulation. Cell. Signal. 2004;16(11):1201–1210. doi: 10.1016/j.cellsig.2004.04.007. [DOI] [PubMed] [Google Scholar]
- 57.Aldinucci D., Borghese C., Casagrande N. The CCL5/CCR5 axis in cancer progression. Cancers. 2020;12(7):1765. doi: 10.3390/cancers12071765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Aldinucci D., Borghese C., Casagrande N. Formation of the immunosuppressive microenvironment of classic Hodgkin lymphoma and therapeutic approaches to counter it. Int. J. Mol. Sci. 2019;20(10):2416. doi: 10.3390/ijms20102416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cravens P.D., Lipsky P.E. Dendritic cells, chemokine receptors and autoimmune inflammatory diseases. Immunol. Cell Biol. 2002;80(5):497–505. doi: 10.1046/j.1440-1711.2002.01118.x. [DOI] [PubMed] [Google Scholar]
- 60.Wang J., et al. Siglec receptors modulate dendritic cell activation and antigen presentation to T cells in cancer. Front. Cell Dev. Biol. 2022;10 doi: 10.3389/fcell.2022.828916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Schanin J., et al. BMJ Specialist Journals; 2022. Antibody Blockade of the Immunoinhibitory Receptor Siglec 10 Polarizes Tumor Associated Myeloid Cells and Promotes Anti-tumor Immunity. [Google Scholar]
- 62.Chen R., et al. TNFSF13 is a novel onco-inflammatory marker and correlates with immune infiltration in gliomas. Front. Immunol. 2021;12 doi: 10.3389/fimmu.2021.713757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Dubey A.K., et al. Belimumab: first targeted biological treatment for systemic lupus erythematosus. J. Pharmacol. Pharmacother. 2011;2(4):317–319. doi: 10.4103/0976-500X.85930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Baker D., et al. The irony of humanization: alemtuzumab, the first, but one of the most immunogenic, humanized monoclonal antibodies. Front. Immunol. 2020;11:124. doi: 10.3389/fimmu.2020.00124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Woollard S.M., Kanmogne G.D. Maraviroc: a review of its use in HIV infection and beyond. Drug Des. Dev. Ther. 2015;9:5447. doi: 10.2147/DDDT.S90580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ng-Cashin J., et al. Host absence of CCR5 potentiates dendritic cell vaccination. J. Immunol. 2003;170(8):4201–4208. doi: 10.4049/jimmunol.170.8.4201. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data sharing is not applicable to this article as no datasets were generated during the current study.