Abstract
AIMS:
Activating somatic mutations or gene amplification of KIT result in constitutive activation of its receptor tyrosine kinase, which is targetable in various solid tumors. Here, we sought to investigate the presence of KIT genetic alterations in breast cancer (BC) and characterize the histologic and genomic features of these tumors.
METHODS:
A retrospective analysis of 5,575 BCs previously subjected to targeted sequencing using the FDA-authorized MSK-IMPACT assay was performed to identify BCs with KIT alterations. A histologic assessment of KIT-altered BCs was conducted, and their repertoire of genetic alterations was compared to that of BCs lacking KIT genetic alterations, matched for age, histologic type, estrogen receptor (ER)/HER2 status and sample type.
RESULTS:
We identified 18 BCs (0.32%), including 9 primary and 9 metastatic BCs, with oncogenic/likely oncogenic genetic alterations affecting KIT, including activating somatic mutations (n=4) or gene amplification (n=14). All KIT-altered BCs were of high histologic grade, although no distinctive histologic features were observed. When compared to BCs lacking KIT genetic alterations, no distinctive genetic features were identified. In two metastatic KIT-altered BCs in which the matched primary BC had also been analyzed by MSK-IMPACT, the KIT mutations were found to be restricted to the metastatic samples, suggesting that they were late events in the evolution of these cancers.
CONCLUSIONS:
KIT genetic alterations are vanishingly rare in BC. KIT-altered BCs are of high grade but lack distinctive histological features. Genetic alterations in KIT might be late events in the evolution and/or progression of BC.
Keywords: Breast cancer, KIT, Receptor tyrosine kinase, Massive parallel sequencing, MSK-IMPACT
INTRODUCTION
KIT maps to 4q11–q12 and encodes c-KIT (CD117), a type III transmembrane receptor tyrosine kinase [1]. Constitutive activation of KIT by hotspot activating mutations targeting its cytoplasmic juxta-membrane domain, extracellular region and tyrosine kinase domains have been reported in gastrointestinal stromal tumors (GIST), acral and mucosal melanomas, renal cell carcinomas, dysgerminomas and malignant gliomas [2–9], whereas KIT gene amplification has been described in GISTs [10] and melanomas [7,11]. Importantly, KIT genetic alterations are well established key therapeutic targets, as highlighted by the effectiveness of imatinib and ripretinib in GISTs [12,13]. Early preclinical studies suggested a potential efficacy of KIT inhibition in breast cancer (BC) models [14–16]. Nonetheless, clinical trials evaluating KIT inhibition in BC patients, either alone or in combination with endocrine therapy and/or chemotherapy, showed no evidence of clinical efficacy [17–20]. These disappointing results led the discontinuation of anti-KIT drug development in BC.
Previous studies focused on BCs displaying c-KIT overexpression rather than KIT genetic alterations. Increased c-KIT protein expression has been documented in a small subset (1–13%) of BCs [21,22], and to be rather prevalent in adenoid cystic carcinomas (AdCCs) of the breast [23]. The role of KIT oncogenic alterations in BC remains to be investigated. Hence, here, we sought to determine the frequency of KIT activating somatic genetic alterations in primary and metastatic BC and to describe the clinicopathologic and genomic features of these tumors.
MATERIALS AND METHODS
Cases
This study was approved by the Institutional Review Board of Memorial Sloan Kettering Cancer Center (MSK). We retrospectively investigated the presence of oncogenic/likely oncogenic somatic mutations affecting KIT in targeted sequencing data from 5,575 BCs previously subjected to the FDA-authorized MSK Integrated Mutation Profiling of Actionable Targets (MSK-IMPACT) assay [24] in the clinical setting.
Targeted sequencing analysis
Non-synonymous somatic mutations, amplifications, and homozygous deletions for the cases included in our cohort were retrieved from cBioPortal [25]. The fraction of genome altered (FGA; i.e. the number of copy number segments which are not copy neutral divided by the total number of copy number segments [26]), and the non-synonymous tumor mutation burden (TMB) (i.e. the number of non-synonymous mutations divided by the total genomic region assessed by MSK-IMPACT, per megabase), were retrieved from cBioPortal. Mutational signatures were inferred using SigMA [27], using all synonymous and non-synonymous somatic mutations in BCs with at least five single nucleotide variants (SNVs), as previously described [28,29]. In addition, we retrieved the raw MSK-IMPACT sequencing data (i.e., FASTQ files) and reprocessed them using our validated bioinformatics pipeline [30,31] for two cases with paired primary and metastatic samples to infer the copy number alterations and cancer cell fraction (CCF) using ABSOLUTE [32].
Histopathologic assessment
The histopathologic review and classification of BCs harboring KIT oncogenic/likely oncogenic alterations was conducted by four pathologists (MV, FD, JSR-F, FP) following the criteria put forward by the World Health Organization (WHO) [33]. Tumors were graded according to the Nottingham grading system [33,34]. Estrogen receptor (ER) and HER2 status, determined according to the American Society of Clinical Oncology/College of American Pathologists guidelines [35,36], were retrieved from the pathology reports.
Comparison with BCs lacking KIT genetic alterations
We compared the frequency of non-synonymous somatic mutations, amplifications and homozygous deletions, non-synonymous TMB, FGA and mutational signatures of the KIT-altered BCs (n=18) to those of BCs lacking genetic alterations affecting KIT from the study by Razavi et al [37], matched by age, menopausal status, sample type, histologic type and ER/HER2 status to the KIT-altered cases at a 3:1 ratio (n=54).
Immunohistochemistry
c-KIT expression was assessed by immunohistochemistry (IHC) using a Benchmark ULTRA system (Ventana, Oro Valley, AZ). Following heat-based antigen retrieval with the CC1 buffer for 32 minutes, tissue sections were incubated with the anti-CD-117 polyclonal antibody (catalog number: A4502) from DAKO (Glostrup, Denmark) at a 1:2000 dilution for 20 minutes. Subsequently, the primary antibody was detected with a polymer-based secondary kit. Positive and negative controls were included in each slide run. Only c-KIT membranous expression was considered.
Statistical analysis
Statistical analyses were conducted using R (v3.1.2). Comparisons of categorical and continuous variables were performed by using Fisher’s exact and Mann-Whitney U test, respectively. Multiple testing correction using the Benjamini-Hochberg method was applied to control for the false discovery rate whenever appropriate. P < 0.05 was considered as statistically significant. All tests used were two-tailed.
RESULTS
Following a retrospective query of 5,575 BCs previously subjected to targeted sequencing using the FDA-authorized MSK-IMPACT [24], we identified 18/5,575 (0.3%) cases harboring V560_Y578del in-frame deletion (case KIT2; Fig. 1A–1B, Table 1). In addition, to validate the frequency of KIT genetic alterations observed, we interrogated the whole-exome sequencing data of 1,108 BCs from The Cancer Gene Atlas (TCGA) [12] for the presence of oncogenic/likely oncogenic alterations in this gene and identified 9/1,108 (0.8%) BCs harboring KIT gene amplification (Supplementary Table 1). No KIT oncogenic/likely oncogenic mutations were identified in the Breast TCGA cohort.
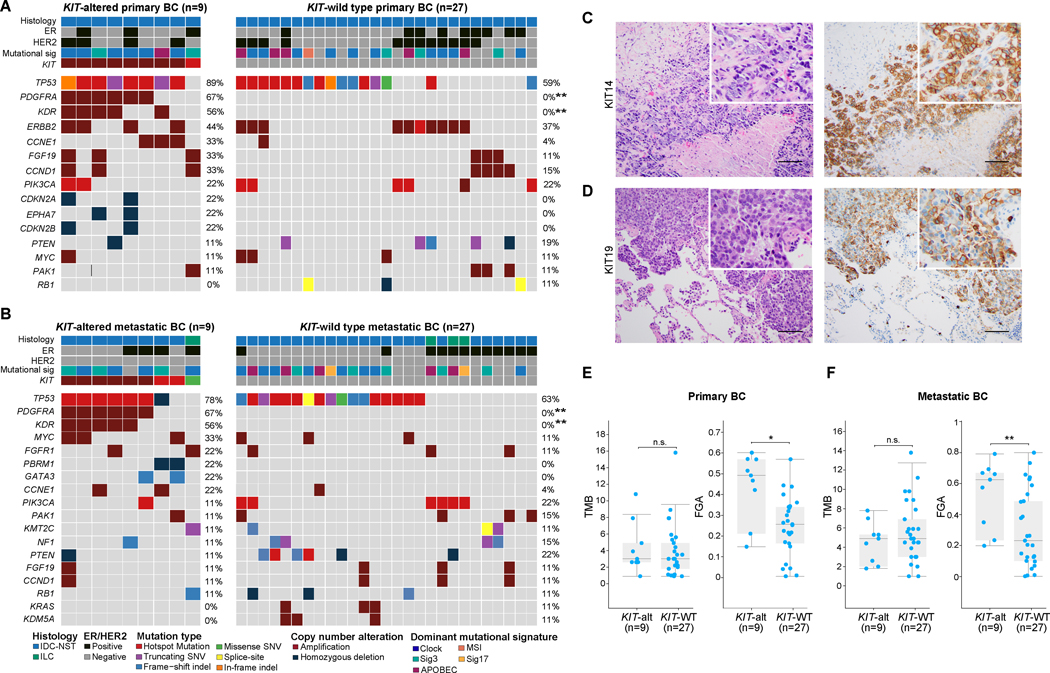
Figure 1. Repertoire of genetic alterations in breast cancers with KIT genetic alterations.
(A-B) Heatmaps depicting the non-synonymous somatic mutations, amplifications and homozygous deletions in (A) primary (n=9) and (B) metastatic (n=9) KIT-altered breast cancers (BC) compared to BCs lacking KIT genetic alterations matched by clinicopathologic features at a 3:1 ratio (n=27, primary and metastatic BC, each). Recurrently affected (≥2 cases) genes in KIT-altered BCs and the most frequently altered (≥3 cases) genes in KIT-wild type (WT) BCs are shown. Fisher’s exact test, FDR adjusted P value, **, <0.01. (C-D) Representative hematoxylin and eosin (H&E) micrographs (left) and corresponding KIT protein expression micrographs (right) assessed by immunohistochemistry for the (C) primary BC case KIT14, and (D) metastatic BC case KIT19. Scale bar, 50 microns. (E-F) Box-plots displaying the non- synonymous tumor mutation burden (TMB; left) and fraction of genome altered (FGA; right) in (E) primary and (F) metastatic BCs harboring KIT genetic alterations (n=9, each) compared to KIT-WT BCs matched by clinicopathologic features at a 3:1 ratio (n=27, each). Mann-Whitney U test, *, P<0.05; **, P<0.01; n.s., non-significant.
Table 1.
Clinicopathologic characteristics of breast cancers harboring oncogenic/likely oncogenic alterations affecting KIT from the MSK-IMPACT cohort
| Sample ID | Sample type | Histology | Grade/ Differentiation | ER | HER2 | KIT genetic alteration | Hotspot mutation (KIT) | KIT expression (IHC) |
|---|---|---|---|---|---|---|---|---|
| KIT1 | Metastasis | IDC-NST | Poorly differentiated | Negative | Negative | M552_Y570del | Yes | NP |
| KIT2 | Metastasis | IDC-NST | Poorly differentiated | Positive | Negative | V560_Y578del | Yes | Positive |
| KIT3 | Primary | IDC-NST | Grade 3 | Positive | Negative | V559G | Yes | NP |
| KIT4 | Metastasis | Pleomorphic ILC | Poorly differentiated | Positive | Negative | R634Q | No | NP |
| KIT5 | Metastasis | IDC-NST | Poorly differentiated | Positive | Negative | Amplification | NA | NP |
| KIT6 | Primary | IDC-NST | Grade 3 | Positive | Positive | Amplification | NA | NP |
| KIT7 | Metastasis | IDC-NST | Poorly differentiated | Positive | Negative | Amplification | NA | NP |
| KIT9 | Primary | IDC-NST | Grade 3 | Negative | Positive | Amplification | NA | NP |
| KIT10 | Primary | IDC-NST | Grade 3 | Negative | Negative | Amplification | NA | NP |
| KIT11 | Primary | IDC-NST | Grade 3 | Positive | Positive | Amplification | NA | NP |
| KIT12 | Metastasis | IDC-NST | Poorly differentiated | Negative | Negative | Amplification | NA | NP |
| KIT13 | Primary | IDC-NST | Grade 3 | Negative | Positive | Amplification | NA | NP |
| KIT14 | Primary | IDC-NST | Grade 3 | Negative | Negative | Amplification | NA | Positive |
| KIT16 | Primary | IDC-NST | Grade 3 | Negative | Negative | Amplification | NA | NP |
| KIT17 | Metastasis | IDC-NST | Poorly differentiated | Negative | Negative | Amplification | NA | Positive |
| KIT18 | Metastasis | IDC-NST | Poorly differentiated | Negative | Negative | Amplification | NA | NP |
| KIT19 | Metastasis | IDC-NST | Poorly differentiated | Negative | Negative | Amplification | NA | Positive |
| KIT20 | Primary | IDC-NST | Grade 3 | Negative | Negative | Amplification | NA | NP |
ER, estrogen receptor; IDC-NST, invasive ductal carcinoma of no special type; IHC, immunohistochemistry; ILC, invasive lobular carcinoma; NA, not applicable; NP, not performed.
Clinicopathologic characteristics
We sought to determine whether BCs harboring KIT oncogenic genetic alterations would display distinctive histologic features. All primary (n=9) and most (8/9, 89%) metastatic KIT-altered BCs from the MSK-IMPACT cohort were invasive ductal carcinomas of no special type (IDC-NSTs). One metastatic BC harboring a KIT R634Q missense mutation was an invasive lobular carcinoma. Notably, all primary and metastatic KIT-altered BCs identified were of histologic grade 3/ poorly differentiated (Figure 1C–1D, Table 1). Most primary KIT-altered BCs were either ER-negative/HER2-negative or HER2-positive (4/9, 44%, each), whereas most metastatic KIT-altered BCs were ER-negative/HER2-negative (5/9, 56%; Table 1). Likewise, all KIT-altered primary BCs from TCGA (n=11) were IDC-NSTs and were of histologic grade 3. In agreement with our observations in the MSK-IMPACT cohort, most KIT-altered primary BCs from TCGA were ER-negative/HER2-negative or HER2-positive (3/9, 33%, each; Supplementary Table 1). Immunohistochemical analysis of c-KIT expression in four KIT-altered BCs (primary, n=1; metastatic, n=3) with available material revealed moderate to strong membranous expression in all cases interrogated (Figure 1C–1D, Table 1). Taken together, our findings indicate that BCs harboring KIT oncogenic genetic alterations display aggressive histologic features, but no distinctive histologic features.
Repertoire of somatic genetic alterations in KIT-altered BCs
We next sought to determine whether KIT-altered BCs would genetically differ from cases lacking alterations affecting this gene. We compared the repertoire of non-synonymous somatic genetic alterations in primary and metastatic KIT-altered BCs (n=9, each) with that of primary and metastatic KIT-wild type (WT) BCs (n=27, each) from the study by Razavi et al [37], matched for age, menopausal status, histologic type and ER/HER2 status at a 3:1 ratio, respectively. TP53 was the gene found to be most frequently mutated in primary (8/9; 89%) and metastatic (7/9; 78%) KIT-altered BCs. Compared to KIT-WT BCs, no gene was found to be affected in a statistically significantly different frequency in the KIT-altered BCs (Fig. 1A–1B), besides PDGFRA and KDR that map to the same amplicon as KIT and showed frequent co-amplification with this gene in both primary and metastatic BCs (PDGFRA, 67% vs 0%, P<0.01; KDR, 56% vs 0%, P<0.01; Fig. 1A–1B). Whilst no differences were detected in the non- synonymous TMB between the different groups (Fig. 1E–1F), the FGA of primary (median, 0.49; range, 0.15–0.6) and metastatic (median, 0.63; range, 0.2–0.8) KIT-altered BCs was significantly higher than that of primary (median, 0.26; range, 0.01–0.57; P<0.05) and metastatic (median, 0.23; range, 0–0.8; P<0.01) BCs lacking alterations in this gene matched by clinicopathologic characteristics, respectively (Fig. 1E–1F). Due to the limited sample size, however, type II or β errors cannot be entirely ruled out. Akin to their matched controls, most primary (6/9; 67%) and metastatic (4/7; 57%) KIT-altered BCs displayed a dominant aging (clock-like) mutational signature (Fig. 1A–1B).
Comparative analysis of paired primary and metastatic samples of KIT-altered BC
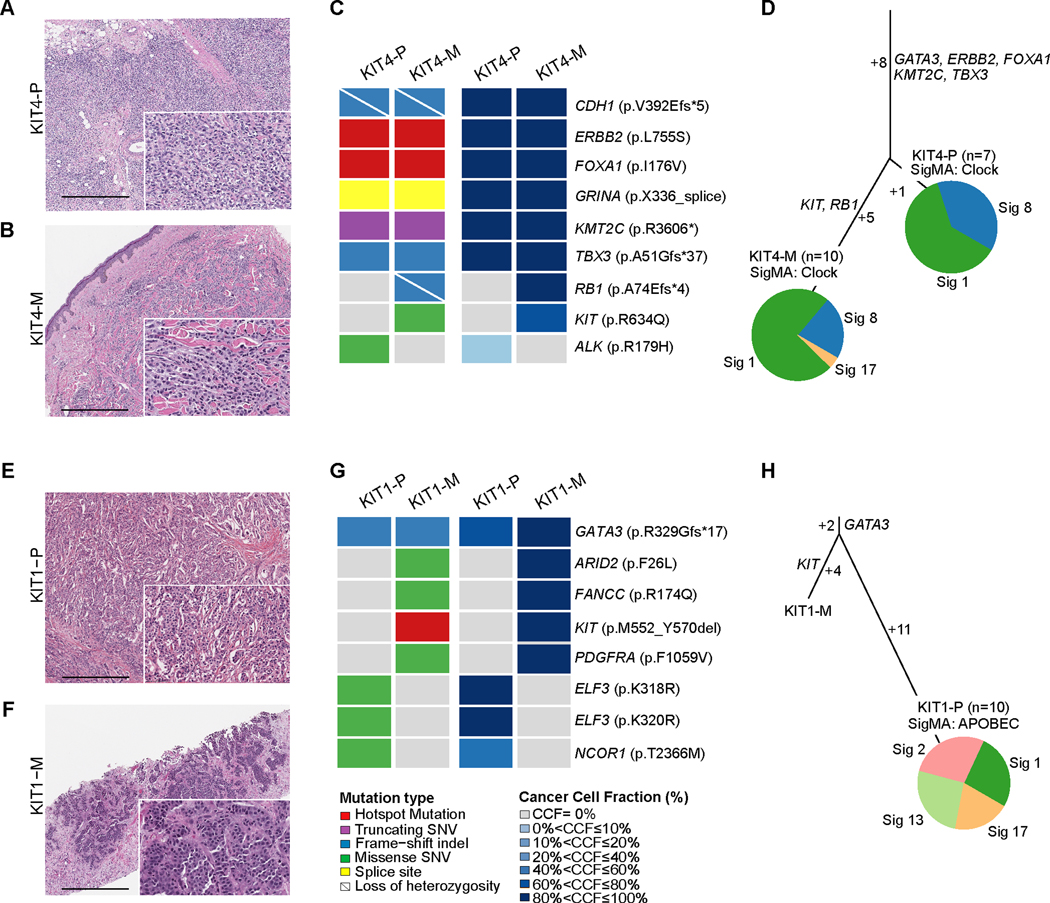
To investigate the role of KIT genetic alterations in BC progression, we analyzed two KIT-altered metastatic BCs from our cohort for which paired primary BC samples had also been subjected to targeted sequencing using MSK-IMPACT. Case KIT-4 corresponded to a woman in her late 70s who presented with an ER-positive/HER2-negative pleomorphic invasive lobular carcinoma (Fig. 2A). Following mastectomy, despite receiving two lines of therapy including everolimus in combination with an aromatase inhibitor (4 months) and palbociclib plus tamoxifen (12 months), sixteen months later, the patient progressed with a metastatic outgrowth in the skin (Fig. 2B). Our analysis of the paired primary (KIT4-P) and metastatic (KIT4-M) BC samples reveals a clonal CDH1 frameshift mutation associated with loss-of-heterozygosity (LOH) of the wild-type allele and truncal mutations affecting other genes classically enriched in lobular carcinomas [38,39], including TBX3 and KMT2C loss-of-function mutations, as well as FOXA1 (I176V) and ERBB2 (L755S) hotspot mutations (Fig. 2C-2D). We observed a clonal RB1 frameshift mutation associated with LOH and a likely oncogenic KIT R634Q missense mutation restricted to the metastatic sample (Fig. 2C). RB1 mutations have been shown to be enriched in ER-positive metastatic BC compared to early BC and to be associated with resistance to CDK4/6 inhibitors [40,41]. Nonetheless, it is possible that the p.R634Q KIT mutation identified in the metastatic sample of this case may have contributed, at least in part, to BC progression in this case. Both primary and metastatic BC samples displayed a dominant aging mutational signature (Fig. 2D).
Figure 2. Clonal decomposition and mutational signatures of paired primary and metastatic KIT-altered breast cancers.
(A,B,E,F) Representative hematoxylin and eosin (H&E) micrographs of the paired primary (P) and metastatic (M) breast cancer (BC) samples of case KIT4 including (A) KIT4-P and (B) KIT4-M, and of case KIT1, including (E) KIT1-P and (F) KIT1-M. Scale bar, 500 microns. (C,G) Heatmaps depicting the non-synonymous somatic mutations (left) and cancer cell fraction (right) of the paired primary and metastatic BC samples of cases (C) KIT4 and (E) KIT1. (D,H) Mutation based phylogenetic trees depicting the clonal evolution of the paired primary and metastatic BC samples of (D) case KIT4 and (H) case KIT1. The length of the trunk and branches of the trees is proportional to the number of shared and private mutations in the primary and metastatic BC samples. Mutational signatures identified in the primary and metastatic BCs with ≥5 SNVs as inferred by SigMA are depicted in pie charts.
Case KIT-1 corresponded to a woman in her mid 40s who presented with an ER-positive/HER2- negative IDC-NST (Fig. 2E). Following surgical excision of the primary tumor, the patient was treated with adjuvant chemotherapy and endocrine therapy. Five years later, the patient relapsed with metastatic BC involving liver, which lacked expression of ER and HER2 (Fig. 2F). Our analysis of the paired primary and metastatic BC samples revealed a truncal GATA3 frameshift mutation as well as a KIT hotspot M552_Y570 inframe deletion, absent in the primary BC sample (Fig. 2G-2H). Taken together, these findings demonstrate that KIT genetic alterations may occur as relative late events in BC evolution and suggest a potential role for KIT in disease progression and/or acquired treatment resistance in a small subset of BCs.
DISCUSSION
Through the reanalysis of targeted sequencing data of a large cohort of primary and metastatic BCs, we demonstrated that oncogenic alterations affecting KIT are vanishingly rare in BC, in contrast to other cancer types such as GIST, melanoma, seminoma, ovarian dysgerminoma and gliomas [26,42–44]. Approximately 75–80% of GISTs harbor gain-of-function mutations in KIT, whereas only <3% of GISTs have a KIT gene amplification [10,45]. In contrast, in our cohort of KIT-altered BCs only a small subset of cases were found to harbor gain-of-function KIT mutations, whereas most cases displayed KIT gene amplification, akin to what has been reported for melanoma [11,46], dysgerminoma[2], medulloblastomas and primitive neuroectodermal tumors (PNET) [47].
Although KIT genetic alterations have been successfully targeted in other tumor types, as exemplified by the success of imatinib in GISTs [12], clinical trials investigating imatinib monotherapy or combined with chemotherapy and endocrine therapy in BC have yielded disappointing results [17–20]. Although KIT gene amplification is considered potentially targetable similarly to KIT activating mutations, the efficacy of imatinib in KIT-amplified tumors remains contentious [11,48,49]. Given that the selection of cases in previous studies in BC were conducted based on overexpression of c-KIT, rather than on KIT genetic alterations, it is possible that the limited efficacy of pharmacologic KIT inhibition observed in BC might be due to the fact that KIT-altered BCs are mainly KIT-amplified and only minority harbor activating mutations. Three of the BCs studied here harbored mutations targeting the exon 11 of KIT, encoding for the juxtamembrane domain, that confer sensitivity to imatinib in GISTs [50], while one of them harbored an exon 13 mutation, frequently associated to resistance to this drug [51]. Whether the rare BCs harboring KIT oncogenic mutations would respond to Imatinib remains to be determined.
AdCCs express c-KIT, which is used as ancillary diagnostic tool for this entity [23]. None of the KIT-altered cases we identified here were AdCCs, they were all IDC-NSTs instead. These data are in agreement with our previous findings indicating that AdCCs, which are underpinned by MYB-NFIB fusion gene, MYBL1 rearrangements or MYB gene amplifications [52], do not harbor KIT genetic alterations [23,53]. The mechanism by which c-KIT is upregulated in AdCC is unknown.
Our cohort included two BC in which paired primary and metastatic samples were analyzed, and in which oncogenic/likely oncogenic mutations in KIT were restricted to the metastasis, suggesting that, at least in a subset of cases, genetic alterations in this gene might constitute a late event in the evolution and/or progression of BC. Further studies aimed at evaluating the role of KIT alterations in progression and in determining resistance to standard treatments in BC, such as endocrine therapy, are warranted.
Our study has important limitations. The small size of the cohort given the rarity of KIT genetic alterations in BC did not allow for the comparison of clinical and genomic features with adequate statistical power. Moreover, we were not able to assess the expression of c-KIT in all cases systematically due to unavailability of material. Despite these limitations, our findings indicate that genetic alterations affecting KIT are exceedingly rare in BC, but detectable in a subset of cases. Although KIT-altered BCs were found to be uniformly of high histologic grade, they do not display a distinctive histologic phenotype. In at least a subset of cases, genetic alterations targeting KIT might represent a late event in BC evolution or progression and may even might play roles in the acquisition of resistance to standard BC treatments.
Supplementary Material
KEY MESSAGES.
What is already known on this topic
KIT activating mutations or gene amplification, which result in tyrosine kinase activation, are well known therapeutic targets in various tumors.
What this study adds
Breast cancers harboring oncogenic alterations affecting KIT are rare and display aggressive histologic features, but not a distinctive phenotype
How this study might affect research, practice or policy
KIT oncogenic alterations might represent late events in breast cancer progression in a subset of cases
FUNDING
This study was partially funded by the Breast Cancer Research Foundation. Research reported in this article was supported in part by a Cancer Center Support Grant of the National Institutes of Health/ National Cancer Institute (grant no. P30CA008748). BW, JSRF and FP are funded in part by a National Institutes of Health/ National Cancer Institute P50 CA247749 01 grant. JSR-F is a Komen Scholar and a recipient of a Susan G Komen Scholarship grant. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. BW is funded in part by a Cycle for Survival grant.
Footnotes
COMPETING INTERESTS
JSR-F reports receiving personal/consultancy fees from Goldman Sachs, REPARE Therapeutics and Paige.AI, membership of the scientific advisory boards of VolitionRx, REPARE Therapeutics, Personalis, Bain Capital and Paige.AI, membership of the Board of Directors of Grupo Oncoclinicas, and ad hoc membership of the scientific advisory boards of Roche Tissue Diagnostics, Ventana Medical Systems, Merck, Daiichi Sankyo and Astrazeneca, outside the scope of this study. BW reports ad hoc membership of the scientific advisory board of REPARE Therapeutics, outside the scope of the submitted work. All other authors declare no conflicts of interest.
ETHICS APPROVAL STATEMENT
This study was approved by the Institutional Review Board of Memorial Sloan Kettering Cancer Center.
CONTRIBUTORSHIP STATEMENT
JSR-F and FP conceived and planned the study. MV, FD, JRS-F and FP reviewed the cases. MV, FD, ADCP, HD, AM, AMG, DNB, PS, DR, PR, HZ, BW, HYW, EB, JSR-F and FP analyzed and interpreted the data. MV, FD and FP wrote the first manuscript, which was reviewed by all coauthors. MV, FD and ADCP contributed equally to this study.
REFERENCES
- 1.Du Z, Lovly CM. Mechanisms of receptor tyrosine kinase activation in cancer. Mol Cancer 2018;17(1):58 doi: 10.1186/s12943-018-0782-4[published Online First: Epub Date]|. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cheng L, Roth LM, Zhang S, et al. KIT gene mutation and amplification in dysgerminoma of the ovary. Cancer 2011;117(10):2096–103 doi: 10.1002/cncr.25794[published Online First: Epub Date]|. [DOI] [PubMed] [Google Scholar]
- 3.Lennartsson J, Ronnstrand L. Stem cell factor receptor/c-Kit: from basic science to clinical implications. Physiol Rev 2012;92(4):1619–49 doi: 10.1152/physrev.00046.2011[published Online First: Epub Date]|. [DOI] [PubMed] [Google Scholar]
- 4.Ohshima K, Fujiya K, Nagashima T, et al. Driver gene alterations and activated signaling pathways toward malignant progression of gastrointestinal stromal tumors. Cancer Sci 2019;110(12):3821–33 doi: 10.1111/cas.14202[published Online First: Epub Date]|. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jones C, Rodriguez-Pinilla M, Lambros M, et al. c-KIT overexpression, without gene amplification and mutation, in paediatric renal tumours. J Clin Pathol 2007;60(11):1226–31 doi: 10.1136/jcp.2007.046441[published Online First: Epub Date]|. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Joensuu H, Puputti M, Sihto H, Tynninen O, Nupponen NN. Amplification of genes encoding KIT, PDGFRalpha and VEGFR2 receptor tyrosine kinases is frequent in glioblastoma multiforme. J Pathol 2005;207(2):224–31 doi: 10.1002/path.1823[published Online First: Epub Date]|. [DOI] [PubMed] [Google Scholar]
- 7.Meng D, Carvajal RD. KIT as an Oncogenic Driver in Melanoma: An Update on Clinical Development. Am J Clin Dermatol 2019;20(3):315–23 doi: 10.1007/s40257-018-0414-1[published Online First: Epub Date]|. [DOI] [PubMed] [Google Scholar]
- 8.Jiang X, Gurel O, Mendiaz EA, et al. Structure of the active core of human stem cell factor and analysis of binding to its receptor kit. EMBO J 2000;19(13):3192–203 doi: 10.1093/emboj/19.13.3192[published Online First: Epub Date]|. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Butti R, Das S, Gunasekaran VP, Yadav AS, Kumar D, Kundu GC. Receptor tyrosine kinases (RTKs) in breast cancer: signaling, therapeutic implications and challenges. Mol Cancer 2018;17(1):34 doi: 10.1186/s12943-018-0797-x[published Online First: Epub Date]|. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tabone S, Theou N, Wozniak A, et al. KIT overexpression and amplification in gastrointestinal stromal tumors (GISTs). Biochim Biophys Acta 2005;1741(1–2):165–72 doi: 10.1016/j.bbadis.2005.03.011[published Online First: Epub Date]|. [DOI] [PubMed] [Google Scholar]
- 11.Carvajal RD, Antonescu CR, Wolchok JD, et al. KIT as a therapeutic target in metastatic melanoma. JAMA 2011;305(22):2327–34 doi: 10.1001/jama.2011.746[published Online First: Epub Date]|. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Berger AC, Korkut A, Kanchi RS, et al. A Comprehensive Pan-Cancer Molecular Study of Gynecologic and Breast Cancers. Cancer Cell 2018;33(4):690–705 e9 doi: 10.1016/j.ccell.2018.03.014[published Online First: Epub Date]|. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Blay JY, Serrano C, Heinrich MC, et al. Ripretinib in patients with advanced gastrointestinal stromal tumours (INVICTUS): a double-blind, randomised, placebo-controlled, phase 3 trial. Lancet Oncol 2020;21(7):923–34 doi: 10.1016/S1470-2045(20)30168-6[published Online First: Epub Date]|. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.DiPaola RS, Kuczynski WI, Onodera K, et al. Evidence for a functional kit receptor in melanoma, breast, and lung carcinoma cells. Cancer Gene Ther 1997;4(3):176–82 [PubMed] [Google Scholar]
- 15.Hines SJ, Organ C, Kornstein MJ, Krystal GW. Coexpression of the c-kit and stem cell factor genes in breast carcinomas. Cell Growth Differ 1995;6(6):769–79 [PubMed] [Google Scholar]
- 16.Roussidis AE, Mitropoulou TN, Theocharis AD, et al. STI571 as a potent inhibitor of growth and invasiveness of human epithelial breast cancer cells. Anticancer Res 2004;24(3a):1445–7 [PubMed] [Google Scholar]
- 17.Modi S, Seidman AD, Dickler M, et al. A phase II trial of imatinib mesylate monotherapy in patients with metastatic breast cancer. Breast Cancer Res Treat 2005;90(2):157–63 doi: 10.1007/s10549-004-3974-0[published Online First: Epub Date]|. [DOI] [PubMed] [Google Scholar]
- 18.Cristofanilli M, Morandi P, Krishnamurthy S, et al. Imatinib mesylate (Gleevec) in advanced breast cancer-expressing C-Kit or PDGFR-beta: clinical activity and biological correlations. Ann Oncol 2008;19(10):1713–9 doi: 10.1093/annonc/mdn352[published Online First: Epub Date]|. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chew HK, Barlow WE, Albain K, et al. A phase II study of imatinib mesylate and capecitabine in metastatic breast cancer: Southwest Oncology Group Study 0338. Clin Breast Cancer 2008;8(6):511–5 doi: 10.3816/CBC.2008.n.062[published Online First: Epub Date]|. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yam C, Murthy RK, Rauch GM, et al. A phase II study of imatinib mesylate and letrozole in patients with hormone receptor-positive metastatic breast cancer expressing c-kit or PDGFR-beta. Invest New Drugs 2018;36(6):1103–09 doi: 10.1007/s10637-018-0672-z[published Online First: Epub Date]|. [DOI] [PubMed] [Google Scholar]
- 21.Simon R, Panussis S, Maurer R, et al. KIT (CD117)-positive breast cancers are infrequent and lack KIT gene mutations. Clin Cancer Res 2004;10(1 Pt 1):178–83 doi: 10.1158/1078-0432.ccr-0597-3[published Online First: Epub Date]|. [DOI] [PubMed] [Google Scholar]
- 22.Tsuura Y, Suzuki T, Honma K, Sano M. Expression of c-kit protein in proliferative lesions of human breast: sexual difference and close association with phosphotyrosine status. J Cancer Res Clin Oncol 2002;128(5):239–46 doi: 10.1007/s00432-002-0329-2[published Online First: Epub Date]|. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Marchio C, Weigelt B, Reis-Filho JS. Adenoid cystic carcinomas of the breast and salivary glands (or ‘The strange case of Dr Jekyll and Mr Hyde’ of exocrine gland carcinomas). J Clin Pathol 2010;63(3):220–8 doi: 10.1136/jcp.2009.073908[published Online First: Epub Date]|. [DOI] [PubMed] [Google Scholar]
- 24.Cheng DT, Mitchell TN, Zehir A, et al. Memorial Sloan Kettering-Integrated Mutation Profiling of Actionable Cancer Targets (MSK-IMPACT): A Hybridization Capture-Based Next-Generation Sequencing Clinical Assay for Solid Tumor Molecular Oncology. J Mol Diagn 2015;17(3):251–64 doi: 10.1016/j.jmoldx.2014.12.006[published Online First: Epub Date]|. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gao J, Aksoy BA, Dogrusoz U, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal 2013;6(269):pl1 doi: 10.1126/scisignal.2004088[published Online First: Epub Date]|. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zehir A, Benayed R, Shah RH, et al. Mutational landscape of metastatic cancer revealed from prospective clinical sequencing of 10,000 patients. Nat Med 2017;23(6):703–13 doi: 10.1038/nm.4333[published Online First: Epub Date]|. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gulhan DC, Lee JJ, Melloni GEM, Cortes-Ciriano I, Park PJ. Detecting the mutational signature of homologous recombination deficiency in clinical samples. Nat Genet 2019;51(5):912–19 doi: 10.1038/s41588-019-0390-2[published Online First: Epub Date]|. [DOI] [PubMed] [Google Scholar]
- 28.Pareja F, Ferrando L, Lee SSK, et al. The genomic landscape of metastatic histologic special types of invasive breast cancer. NPJ Breast Cancer 2020;6:53 doi: 10.1038/s41523-020-00195-4[published Online First: Epub Date]|. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Selenica P, Marra A, Choudhury NJ, et al. APOBEC mutagenesis, kataegis, chromothripsis in EGFR-mutant osimertinib-resistant lung adenocarcinomas. Ann Oncol 2022. doi: 10.1016/j.annonc.2022.09.151[published Online First: Epub Date]|. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pareja F, Brown DN, Lee JY, et al. Whole-Exome Sequencing Analysis of the Progression from Non-Low-Grade Ductal Carcinoma In Situ to Invasive Ductal Carcinoma. Clin Cancer Res 2020;26(14):3682–93 doi: 10.1158/1078-0432.CCR-19-2563[published Online First: Epub Date]|. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pareja F, Lee JY, Brown DN, et al. The Genomic Landscape of Mucinous Breast Cancer. J Natl Cancer Inst 2019;111(7):737–41 doi: 10.1093/jnci/djy216[published Online First: Epub Date]|. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Carter SL, Cibulskis K, Helman E, et al. Absolute quantification of somatic DNA alterations in human cancer. Nat Biotechnol 2012;30(5):413–21 doi: 10.1038/nbt.2203[published Online First: Epub Date]|. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.WHO Classification of Tumors Editorial Board. Breast tumours. WHO Classification of Tumors. 5th Edition. 5th Edition ed. IARC: Lyon, 2019. [Google Scholar]
- 34.Elston CW, Ellis IO. Pathological prognostic factors in breast cancer. I. The value of histological grade in breast cancer: experience from a large study with long-term follow-up. Histopathology 2002;41(3A):154–61 [PubMed] [Google Scholar]
- 35.Allison KH, Hammond MEH, Dowsett M, et al. Estrogen and Progesterone Receptor Testing in Breast Cancer: ASCO/CAP Guideline Update. J Clin Oncol 2020;38(12):1346–66 doi: 10.1200/JCO.19.02309[published Online First: Epub Date]|. [DOI] [PubMed] [Google Scholar]
- 36.Wolff AC, Hammond MEH, Allison KH, et al. Human Epidermal Growth Factor Receptor 2 Testing in Breast Cancer: American Society of Clinical Oncology/College of American Pathologists Clinical Practice Guideline Focused Update. J Clin Oncol 2018;36(20):2105–22 doi: 10.1200/JCO.2018.77.8738[published Online First: Epub Date]|. [DOI] [PubMed] [Google Scholar]
- 37.Razavi P, Chang MT, Xu G, et al. The Genomic Landscape of Endocrine-Resistant Advanced Breast Cancers. Cancer Cell 2018;34(3):427–38 e6 doi: 10.1016/j.ccell.2018.08.008[published Online First: Epub Date]|. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ciriello G, Gatza ML, Beck AH, et al. Comprehensive Molecular Portraits of Invasive Lobular Breast Cancer. Cell 2015;163(2):506–19 doi: 10.1016/j.cell.2015.09.033[published Online First: Epub Date]|. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Desmedt C, Zoppoli G, Sotiriou C, Salgado R. Transcriptomic and genomic features of invasive lobular breast cancer. Semin Cancer Biol 2017;44:98–105 doi: 10.1016/j.semcancer.2017.03.007[published Online First: Epub Date]|. [DOI] [PubMed] [Google Scholar]
- 40.Bertucci F, Ng CKY, Patsouris A, et al. Genomic characterization of metastatic breast cancers. Nature 2019;569(7757):560–64 doi: 10.1038/s41586-019-1056-z[published Online First: Epub Date]|. [DOI] [PubMed] [Google Scholar]
- 41.Li Z, Razavi P, Li Q, et al. Loss of the FAT1 Tumor Suppressor Promotes Resistance to CDK4/6 Inhibitors via the Hippo Pathway. Cancer Cell 2018;34(6):893–905 e8 doi: 10.1016/j.ccell.2018.11.006[published Online First: Epub Date]|. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Roskoski R Jr. The role of small molecule Kit protein-tyrosine kinase inhibitors in the treatment of neoplastic disorders. Pharmacol Res 2018;133:35–52 doi: 10.1016/j.phrs.2018.04.020[published Online First: Epub Date]|. [DOI] [PubMed] [Google Scholar]
- 43.De Miguel MP, Cheng L, Holland EC, Federspiel MJ, Donovan PJ. Dissection of the c-Kit signaling pathway in mouse primordial germ cells by retroviral-mediated gene transfer. Proc Natl Acad Sci U S A 2002;99(16):10458–63 doi: 10.1073/pnas.122249399[published Online First: Epub Date]|. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.McIntyre A, Summersgill B, Grygalewicz B, et al. Amplification and overexpression of the KIT gene is associated with progression in the seminoma subtype of testicular germ cell tumors of adolescents and adults. Cancer Res 2005;65(18):8085–9 doi: 10.1158/0008-5472.CAN-05-0471[published Online First: Epub Date]|. [DOI] [PubMed] [Google Scholar]
- 45.Kang G, Yun H, Sun CH, et al. Integrated genomic analyses identify frequent gene fusion events and VHL inactivation in gastrointestinal stromal tumors. Oncotarget 2016;7(6):6538–51 doi: 10.18632/oncotarget.3731[published Online First: Epub Date]|. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Siroy AE, Boland GM, Milton DR, et al. Beyond BRAFV600: Clinical Mutation Panel Testing by Next-Generation Sequencing in Advanced Melanoma. Journal of Investigative Dermatology 2015;135(2):508–15 doi: 10.1038/jid.2014.366[published Online First: Epub Date]|. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Blom T, Roselli A, Hayry V, et al. Amplification and overexpression of KIT, PDGFRA, and VEGFR2 in medulloblastomas and primitive neuroectodermal tumors. J Neurooncol 2010;97(2):217–24 doi: 10.1007/s11060-009-0014-2[published Online First: Epub Date]|. [DOI] [PubMed] [Google Scholar]
- 48.Guo J, Si L, Kong Y, et al. Phase II, open-label, single-arm trial of imatinib mesylate in patients with metastatic melanoma harboring c-Kit mutation or amplification. J Clin Oncol 2011;29(21):2904–9 doi: 10.1200/JCO.2010.33.9275[published Online First: Epub Date]|. [DOI] [PubMed] [Google Scholar]
- 49.Hodi FS, Corless CL, Giobbie-Hurder A, et al. Imatinib for melanomas harboring mutationally activated or amplified KIT arising on mucosal, acral, and chronically sun-damaged skin. J Clin Oncol 2013;31(26):3182–90 doi: 10.1200/JCO.2012.47.7836[published Online First: Epub Date]|. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rausch JL, Ali AA, Lee DM, et al. Differential antitumor activity of compounds targeting the ubiquitin-proteasome machinery in gastrointestinal stromal tumor (GIST) cells. Scientific Reports 2020;10(1):5178 doi: 10.1038/s41598-020-62088-7[published Online First: Epub Date]|. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Heinrich MC, Corless CL, Blanke CD, et al. Molecular correlates of imatinib resistance in gastrointestinal stromal tumors. J Clin Oncol 2006;24(29):4764–74 doi: 10.1200/JCO.2006.06.2265[published Online First: Epub Date]|. [DOI] [PubMed] [Google Scholar]
- 52.Pareja F, Weigelt B, Reis-Filho JS. Problematic breast tumors reassessed in light of novel molecular data. Mod Pathol 2021;34(Suppl 1):38–47 doi: 10.1038/s41379-020-00693-7[published Online First: Epub Date]|. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wetterskog D, Wilkerson PM, Rodrigues DN, et al. Mutation profiling of adenoid cystic carcinomas from multiple anatomical sites identifies mutations in the RAS pathway, but no KIT mutations. Histopathology 2013;62(4):543–50 doi: 10.1111/his.12050[published Online First: Epub Date]|. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.