Significance
We demonstrate the highest-resolution MR images ever obtained of the mouse brain. The diffusion tensor images (DTI) @ 15 μm spatial resolution are 1,000 times the resolution of most preclinical rodent DTI/MRI. Superresolution track density images are 27,000 times that of typical preclinical DTI/MRI. High angular resolution yielded the most detailed MR connectivity maps ever generated. High-performance computing pipelines merged the DTI with light sheet microscopy of the same specimen, providing a comprehensive picture of cells and circuits. The methods have been used to demonstrate how strain differences result in differential changes in connectivity with age. We believe the methods will have broad applicability in the study of neurodegenerative diseases.
Keywords: magnetic resonance histology, light sheet microscopy, connectome, mouse brain
Abstract
We have developed workflows to align 3D magnetic resonance histology (MRH) of the mouse brain with light sheet microscopy (LSM) and 3D delineations of the same specimen. We start with MRH of the brain in the skull with gradient echo and diffusion tensor imaging (DTI) at 15 μm isotropic resolution which is ~ 1,000 times higher than that of most preclinical MRI. Connectomes are generated with superresolution tract density images of ~5 μm. Brains are cleared, stained for selected proteins, and imaged by LSM at 1.8 μm/pixel. LSM data are registered into the reference MRH space with labels derived from the ABA common coordinate framework. The result is a high-dimensional integrated volume with registration (HiDiver) with alignment precision better than 50 µm. Throughput is sufficiently high that HiDiver is being used in quantitative studies of the impact of gene variants and aging on mouse brain cytoarchitecture and connectomics.
MRI of the human brain has become a mainstay of both clinical care and basic neuroscience research since Lauterbur’s catalytic studies in the 1970s (1). Resolution, contrast, and imaging speed have improved at a rapid rate. Resolution is limited in humans by movement artifacts, sensitivity, and short scan durations. Voxels are typically 1 mm on edge. Consequently, structural MRI is unable to resolve fine-grained anatomical structure at the scale of tissue cytoarchitecture. But, these studies have been enormously rewarding because throughput is high, and sample sizes can include thousands of subjects.
As a result, there is now a burgeoning new field that combines MRI with genetics and genome-wide association studies (GWAS) (2, 3). These joint MRI and genetic studies are now sufficiently well powered to detect DNA variants and haplotypes that are causally linked to both normal differences in brain architecture and differential susceptibility and resilience to neurological diseases. Examples include ENIGMA (4), the UK Biobank (5), and the Alzheimer’s Disease Neuroimaging Initiative (ADNI) (6, 7). In comparison, some MRI studies using small mammals—rats and mice—have taken an orthogonal approach in which the major focus has been to achieve ever higher spatial resolution rather than higher throughput. This has involved high-field MRI acquisition strategies from postmortem specimens, i.e., magnetic resonance histology (MRH) (8–11). We can now image the whole mouse brain at spatial resolution as high as 25 µm (12). These murine MRH datasets can be merged with whole-brain LSM immunohistochemistry (IHC) to provide up to two orders of magnitude higher spatial resolution.
Rodent brain MRH and LSM studies typically have modest sample sizes—often less than 20 cases per study and often include only one or two genotypes, ages, or treatment groups. The bottleneck is long acquisition times for both MRH and LSM. There is an equally serious constraint in processing, aligning, and merging terabyte 3D images (13). These problems have thwarted even moderate-throughput rodent neurogenetics of the types heralded in humans by ENIGMA (4), ADNI (7), and UK Biobank (5).
We have developed atlases and methods to accurately register MRH and LSM with 3D volumetric labeling of 180 regions of interest (ROIs) in each hemisphere. We demonstrate that high-dimensional integrated volume with registration (HiDiver) generalizes to different sexes, ages, and genotypes with a precision better than 50 μm. Imaging and computational workflows have a throughput of hundreds of specimens per year. This enables systematic global analyses and genetic dissection of variation in CNS architecture as a function of genotype, age, sex, sensorimotor experience, environmental exposure, behavioral phenotype, and interventional treatment at both regional and cellular levels.
There are many alternative methods for postmortem analysis of the mouse brain. Expansion microscopy of cleared tissues has been used to image whole brains at submicron resolution (14). Knife-edge scanning microscopy has been used to provide connectivity measure using Golgi stains at micron resolution (15). Optical coherence microscopy has been used to generate whole-brain images at ~ 5 µm resolution (16). These methods have been used thus far to provide reference atlases. Typical acquisition/processing times are weeks per specimen. And several investigators have merged high-resolution MR with optical methods (17–19).
The work we present here differs from the previous work in four ways: 1) The resolution index (20) of the MR data presented here is nearly 10 times that of any previous MRH acquisition, enabling far more accurate reconstruction of whole-brain connectomes; 2) the MRH is acquired in the skull, allowing accurate measures of regional volumes; 3) the light sheet images are registered to this in skull geometry, making cell density measures much more accurate; and 4) the entire process can be accomplished with sufficient throughput to allow systematic studies (N > 100) of neurogenetics, linking genome to regional brain morphometry, cell density, and whole-brain connectivity.
1. Results
We addressed three technical challenges of imaging and registering MRH with LSM for the mouse brain: 1) acquisition of very large (250GB) MRH at microscopic resolution with adequate throughput; 2) accurate registration of very large geometrically distorted LSM data with MRH data; and 3) workflows to map these data into a common reference space. We have previously demonstrated the utility of MRH in neurogenetics at moderate throughput—12 strains with two replicates each (21). For that work, we registered MRH data into the INCF Waxholm reference space (10) and generated volumetric data for 35 ROIs. In the last decade, we have increased both the spatial and contrast resolution using diffusion tensor imaging (12, 22).
SI Appendix, Table S1 summarizes four experiments used to develop and validate HiDiver:
-
1.
C57BL/6 mice studied at 90 d of age—an age close to that of most data generated by GENSAT (23), the ABA, and the BRAIN initiative (24, 25). Specimens were scanned to create new 3D HiDiver reference atlases at 24 X the spatial resolution of any previous DTI atlas. (SI Appendix, Table S1, specimens 200302 and 200316).
-
2.
A second set of mice was used to explore the impact of spatial and angular resolution on scalar DTI metrics to improve throughput and test LSM registration accuracy. For this purpose, we generated track density images (TDI) at 5 μm superresolution, in a C57BL/6J transgenic line (B6.Cg-Tg(Thy1-YFP)HJrs/J (26, 27) that expresses yellow fluorescent protein intensely in subsets of large neurons and their axons, particularly pyramidal neurons in neocortical layer 5 and pyramidal and granule neurons in hippocampus.
-
3.
The third experiment tested the robustness of HiDiver registration in the face of significant variation in genotype and age of specimens. For this work, we used BXD89 mice, an inbred strain generated by crossing of C57BL/6J to DBA/2J (28). Its genome has been fully sequenced (29) and differs from the reference genome at ~2.87 million loci—87% single-nucleotide polymorphisms (SNPs) and 13% other variant classes (DG Ashbrook, personal communication). One of the BXD89s was a young adult at 111 d; the other was an old adult at 687 d. The mean life expectancy of this strain is ~700 d (30).
-
4.
The fourth experiment applied the methods to determine which strains and regions of the brain were most affected by aging in 13 diverse strains of the BXD mouse. We employed a higher-throughput, lower-spatial/-angular resolution (45 μm/46 angle) that could be executed in under 12 h, allowing us to collect data on 111 different specimens.
1.1. Reference Framework Creation.
1.1.1. Registered scalar MRH volumes.
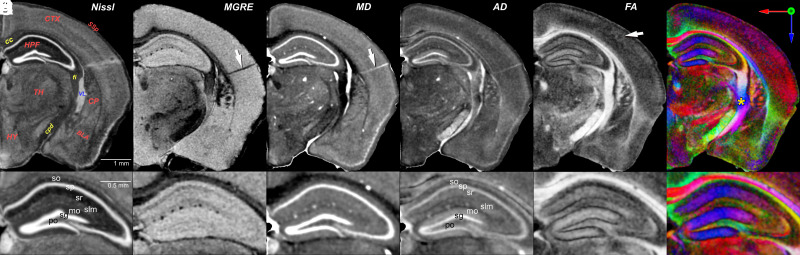
Fig. 1 provides an overview of the differing anatomical detail evident in MGRE and scalar images from the DTI acquisitions. A Nissl section from the ABA is included for reference (Fig. 1A). The average MGRE (Fig. 1B) combines four echoes of the 4D MGRE volume. Scalar diffusion images were derived from the DTI (31) and GQI algorithms (32). These scalar images yield unique contrast that is related to the ultrastructure of cerebral tissue (e.g., axonal densities, myelin content, and configuration of intra-/extra-cellular compartments). These MRH base datasets provide 12 complementary views of cyto- and myeloarchitecture, five of which are shown in Fig. 1. SI Appendix, Table S2 summarizes all the 12 scalar volumes and the algorithms used to generate the contrast. Blood vessels are prominent in the MGRE and AD images (Fig. 1 B and C), whereas mean diffusivity highlights hippocampal layers particularly well (MD, Fig. 1C). In marked contrast, the fractional anisotropy (FA, Fig. 1E) accentuates white matter tracts and those regions in which dendritic arbors have a common orientation, such as stratum radiatum in the CA fields of the hippocampus and the molecular layer of the dentate gyrus.
Fig. 1.
Overview of MRH contrast (SI Appendix, Table S1, specimen 200302-1:1) at a plane equivalent to level 74 of the ABA coronal series (https://tinyurl.com/CCFv3-Level74). (A) Reference image from the ABA of an inverted contrast Nissl-stained section. Several regions and tracts are labeled: cortex (CTX), corpus callosum (cc), primary somatosensory area (SSp), hippocampus (HPF), fimbria (fi), thalamus (TH), hypothalamus (HY), basolateral amygdala (BLA), and lateral ventricle (VL, in blue). (B) Average MGRE with dark blood vessels (arrow – perforating artery) and darker heavily myelinated tracts (e.g., cc, cpd, and fi in A and B). (C) Mean diffusivity (MD) with light regions defining both cell-dense regions such as the granule cell layer (D, sg) dentate gyrus, and blood vessels (arrow) and ventricle. (D) Axial diffusivity (AD) is similar to C, but differentiates additional hippocampal layers. (E) Fractional anisotropy (FA) defines finer details of neurite orientation—white in the heavily myelinated corpus callosum and corticofugal projections and the margins of barrels in the somatosensory cortex (arrow). The barrel field cortex is revealed in an enface tangential plane in SI Appendix, Fig. S2. (F) Color FA provides additional differentiation of fiber tracts based on their orientation (Top Right Inset). The yellow asterisk marks a complex of intersecting tracts around the internal capsule (SI Appendix, Fig. S1). The corpus callosum (red) is distinct from both the cingulum (green above) and the dorsal hippocampal commissure (green below). Lower rows (a–f) are corresponding 2× magnifications of the hippocampus that illustrate MRH resolution of layers. Abbreviations—so: stratum oriens; sp: pyramidal cell layer; sr: stratum radiatum; slm: stratum lacunosum-moleculare; mo molecular layer of the dentate gyrus; sg: granule cell layer of the dentate gyrus; po: polymorphic layer of the dentate gyrus. Video available @ https://bit.ly/MouseBrain15micron
The utility of color FA is emphasized by the complex intersection of tracts around the internal capsule—asterisk in Fig. 1F. The intensity represents the magnitude of diffusion anisotropy, whereas color encodes the primary axis of diffusion (Fig. 1F, f). The intersecting tracts—the fimbria, the stria terminalis, the cerebral peduncle, the internal capsule, and the optic tract—are difficult to resolve by conventional histology. These tracts can be dissected unambiguously in the color FA. A comparative view with the ABA tract demarcations is given in SI Appendix, Fig. S1. By combining multiple MRH scalar images, one can resolve structures that are hard to detect using standard light microscopy.
1.1.2. Definition of ROIs.
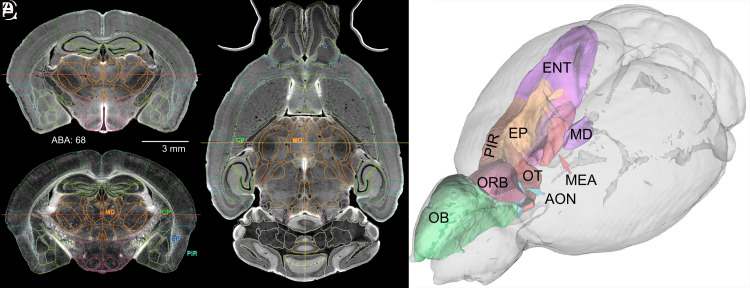
The ABA common coordinate framework (CCFv3) defines regions of interest (ROIs) for 461 structures (33). We registered these into each hemispheres of the C57BL/6J in Fig. 2 using ANTs (34). More than half of all ROIs are so small that they cannot be reliably registered even within the same strain. Registering across genotypes is even more challenging, which compromises the accuracy with which we were able to estimate volumes, cell densities, and cell counts. Almost all ROI volumes with a left–right coefficient of error (CE) greater than 0.05 are generated by poor technical accuracy rather than fluctuating asymmetry in brain structure (see refs. 35 and 36). We opted to generate a reduced subset of ROIs that combines smaller adjacent subvolumes. We refer to this subset as the first reduced version of CCFv3 or r1CCFv3. It consists of 180 ROIs/hemisphere and provides an isotropic parcellation of the whole brain (SI Appendix, Table S3). This is comparable to the human connectome project in which there are 180 ROIs in each hemisphere. It represents a significant improvement over previous MRI atlases, e.g., Waxholm space with 35 ROIs/hemisphere and the Calabrese connectome atlas with 27 ROIs/hemisphere.
Fig. 2.
Full-brain volumetric rendering of r1CCFv3. All lines that define ROIs use the ABA color conventions. (A) The DWI in a coronal plane with ROI demarcations. (B) The FA image at the same level with four ROIs: the mediodorsal nucleus of the thalamus (MD), the caudoputamen (CP), the endopiriform nucleus (EP), and the piriform area (PIR). (C) Axial DWI of a horizontal section with corresponding borders and two ROIs labeled in common with B. The red lines in A and B and the yellow line in C define orthogonal images. (D) 3D delineations of major components of the olfactory system displayed with DSI Studio. Abbreviations—OB: olfactory bulb; AON: anterior olfactory nucleus; ENT: entorhinal area; MEA: medial amygdaloid nucleus; OT: olfactory tubule (specimen 200316-1:1)
The parcellation of r1CCFv3 was refined by two neuroanatomists (LEW, RWW). The product was a set of ROIs that can be embedded reliably in all MRH data, regardless of sex, age, and genotype, and with a low mean CE (<0.05) without extensive manual curation.
1.1.3. Light sheet and MRH registration.
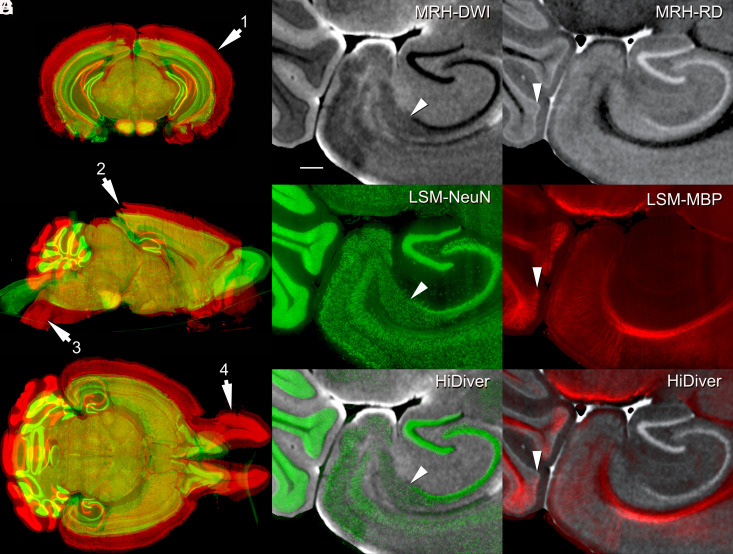
MRH provides excellent geometric fidelity; multiple registered contrasts (Fig. 1); and the practicality of conducting whole-brain morphometry of in situ volumes, ROI histology, and tractography. The benefits of LSM are high spatial resolution and the application of numerous labeling and staining protocols. The main drawbacks are poor geometric fidelity and problematic penetration of reagents. HiDiver combines the strength of both methods. Fig. 3 shows an original LSM dataset before (red) and after (green) registration to the MRH data. Swelling using SHIELD is over 60% relative to the MRH reference volumes (SI Appendix, Table S1), and this expansion is not uniform. For example, swelling in the olfactory bulb is nearly 100%.
Fig. 3.
HiDiver integration of MRH and LSM. Distortion of LSM samples was corrected by registering to the MRH reference volume of the same specimen. Panel (A) flags four types of registration problems in the LSM data—before (red) and after (green) correction with specimen 200316-1:1. Arrow 1 highlights variable expansion during LSM processing that can range up to 100% for the olfactory bulbs, but averages about ~60% globally in this case. Arrow 2 flags tears in the visual cortex introduced during dissection. Arrow 3 marks an exaggerated flexure of the brainstem, while arrow 4 indicates spread of the olfactory bulbs. All the three LSM channels were brought into alignment with MRH by diffeomorphic registration. Panels (B–D) (specimen 191209-1:1) provide an overview of HiDiver alignment where (B) is DWI, (C) is NeuN, and (D) is the HiDiver product. The arrowheads in these overlays point to the CA1, subiculum boundary. (E–G) show the RD with MBP and a similar trio that points to a layer of Purkinje cells and Bergmann glia in the cerebellar cortex. The MBP label extends into and just above this layer. (Scale bar in B is 200 µm.)
Fig. 3 highlights two key elements relevant to resolution and alignment of HiDiver. First, the resolution of MRH is at the threshold of resolving cell layers and fascicles of axons. Trios of oblique arrowheads in Fig. 3 B–C point out the tight correspondence of the CA1–subiculum boundary. A similar trio of images (Fig. 3 E–G) demonstrates the melding of MRH-RD and LSM-MBP labeling of the Purkinje cell layer. The availability of multiple scalars from the MRH acquisition provides a range of anatomic features to drive registration of many different immunohistochemistry markers (37).
The precision of registration was measured by visualizing coregistered volumes in Imaris. Small (<50 μm diameter) vessels were identified in the axial plane of the NeuN image (specimen 200316-1:1) in which the vessels are dark (37). Cross hairs were placed in the center of the vessel. The plane of section was incremented interactively to locate an intersection of a branch of the same vessel. The three-dimensional coordinates were recorded. The process was repeated in the RD image in which the vessels are bright. Eleven different measurements were made across the volume with an average alignment of 22 ± 14 μm (37).
1.2. Spatial Resolution and Tractography.
1.2.1. Spatial resolution: in vivo vs. MRH.
The voxel volume in the new reference atlas is 3.4 × 10−6 mm3. SI Appendix, Fig. S3 shows a comparison of the FA for an in vivo mouse DTI @ 0.15 mm resolution (voxel volume of 3.4 × 10−3 mm3) to MRH DTI in this work (0.015 mm), i.e., a decrease in voxel volume of 1000 X.
1.2.2. Trade-Off in spatial and angular resolution.
Experiment 2 compared trade-offs between spatial and angular resolution (SI Appendix, Table S1). The TR, TE, and b values were fixed for all the three scans. SI Appendix, Fig. S4 shows the FA images which have been scaled for comparison. The first two scans at 15 μm (SI Appendix, Fig. S4A) vs. 25 μm (SI Appendix, Fig. S4B) were acquired with approximately the same total scan time, the slight variation coming from the desire to choose angular invariant sampling strategies in which the b vectors were equally spaced on the unit sphere. As one would expect, the 15-μm data are noisier than the 25-µm data. The definition of cortical layers and layers in the hippocampus is superior in the lower-resolution (25 μm) data. Noise in the FA image is dependent on the noise in the constituent images and the propagation of error through the multiple pipelines. As a surrogate for noise, we include the histogram of a central region of white matter (corpus callosum) which we believe is relatively uniform. The impact of angular sampling, i.e., comparison of SI Appendix, Fig. S4B (at 108 angles) with SI Appendix, Fig. S4C (at 61 angles), is not as obvious, yet it results in a reduction of scan time by ~1/2. Recent work has demonstrated an inflection point in reduction of false negatives in the connectomes at ~61 angles when comparing connectome metrics vs. angular sampling (figure 8h in ref. 38). Finally, the denoising step addresses some of the loss in SNR in moving to reduced angular sampling. SI Appendix, Fig. S5 shows the impact of denoising on the 15 μm/108 angle volume. Reduction of the spatial resolution (15 to 25 μm) and angular resolution (108 to 61) with the addition of the denoising and implementation of a fast spin echo variant has yielded a protocol (25 μm, 61 angles) that can be executed in 20 h (SI Appendix, Fig. S6).
1.2.3. LSM alignment to superresolution MRH track density images.
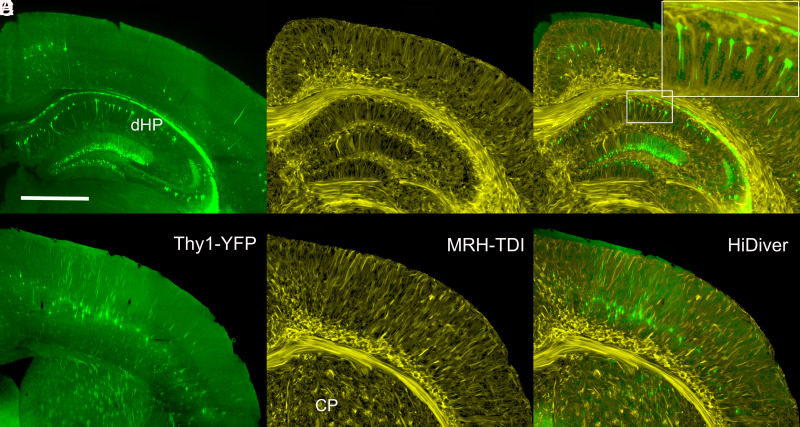
Track density images (TDI) were generated using the method developed by Calamante (39, 40) with nominal superresolution of 5 μm. The TDI have been registered to their LSM in which a subset of large neurons is labeled with yellow fluorescent protein (Fig. 4). The TDI are in yellow and the LSM in green. Despite the nonuniform distortions highlighted in Fig. 3, we can achieve alignment that is usually at the level of cytoarchitectonic boundaries and cell layers (Fig. 4 C and F).
Fig. 4.
Joint LSM and superresolution TDI at two levels through cortex, dorsal hippocampus, and caudoputamen. (A) and (D) are Thy1 fluorescence images (green) from a 90-d-old B6.Cg-Tg(Thy1-YFG)HJrs/J (sample 190415-2:1). (B and E) are TDI at the same levels at a superresolution of 5 µm. (C and F) are merged HiDiver images that highlight the alignment of TDI and Thy1-positive pyramidal cells, dentate gyrus granule cells, and axon fascicles penetrating the caudoputamen. Inset in (C) is a 3× magnification of a radial section of CA1. (D–F) are LSM, TDI, and HiDiver at the level of the primary motor cortex (MOp), respectively. All images have been rendered at the same effective slice thickness of 14.4 µm. (Scale bar in A is 1 mm.) Video available @ https://bit.ly/MRHLightSheet.
The joint LSM and TDI facilitate interpretation of the fine structure in Fig. 4 B and E compared to the previous state-of-the-art work (e.g., figure 5C from ref. 40). The fine strands in the corpus callosum are consistent with the mean orientation of fascicles of myelinated axons and contrast with darker interfascicular callosal oligodendrocytes (41, 42) (Fig. 4 B and E). These TDI strands are an order of magnitude larger than the diameter of the largest myelinated fiber—about 3 µm (43). A striking feature of the strands is their radial uniformity in fortuitous planes through the cortex and hippocampus (Fig. 4E). As TDI strands reach superficial layers of the cortex, they spread and form an arcade as expected of distal dendrites of pyramidal cells and ascending axonal projections and collaterals (Fig. 4B).
1.2.4. Embedding 3D volumetric labels from MRH into LSM.
The transfer of labels from MRH to LSM is good but still imperfect. The correspondence of the mirror-imaged horizontal planes—DWI (SI Appendix, Fig. S7, Right) and NeuN (Left)—is sufficiently accurate as judged by the overall concordance of independently defined boundaries, blood vessels, and cytoarchitectonic boundaries. Vessels provide useful landmarks for comparison (37).
1.3. Application in Neurogenetics: Impact of Age and Strain Variation on HiDiver.
To test HiDiver across genotype and age, we transferred the r1CCFv3 ROIs into two BXD89 mice that differ from C57BL/6J at about 1 in 1,000 base pairs of DNA—the typical level of genetic variation seen among any two humans. The r1CCFv3 labels were transferred from the male reference (specimen 200302-1:1) to the new strains using our dedicated pipeline for rodents (44) (SI Appendix, Fig. S7). The HiDiver transfer of volumetric labels from r1CCFv3 from C57BL/6J to both BXD89 cases is visually accurate as judged by the boundaries imposed on the axial DWI and the corresponding LSM channels (Figs. 4 and 5).
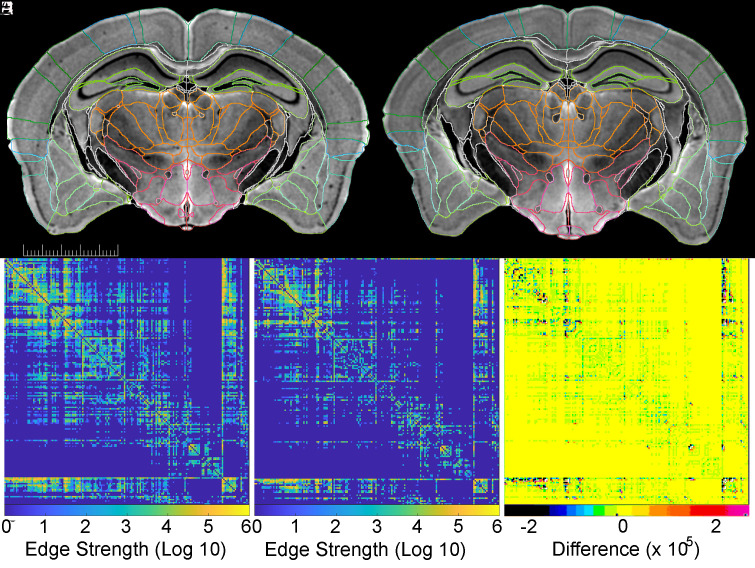
Fig. 5.
Quantitative tractography applied to BXD89 males. (A) (111 d) and (B) (687 d) are matched DWIs of specimens that differ by 19 mo. The scale bar is 5 mm. (C and D) are matched connectomes of the same young and old specimens that provide a measure (log10 scale) of the connection strength of each of 179 of 180 ROIs in the right hemisphere with all other right-hemisphere ROIs (one ROI is the whole hemisphere). (E) Difference of (C and D) scaled to allow the reader to appreciate where there is difference.
We generated whole-brain connectomes by seeding 5 million points distributed across all ROIs (Fig. 5 C and D). For simplicity, only the connectomes of the right hemisphere are shown for young and old animals. Maier-Hein et al. have highlighted the complications of false positive connections in human studies (45) where voxels range from 1.25 to 2 mm3 (46). In contrast, connectomes in Fig. 5 C and D have voxels that are up to 2.5 million times smaller with commensurate reduction in the number of crossing or merging fibers in any voxel (47). Fig. 5E shows the linear difference scaled to allow the reader to appreciate where there are differences. The bands along the bottom and right sides are the fiber tracts.
1.4. High Throughput.
Experiment 4 demonstrates the utility of HiDiver in a study of over 100 cases in 13 strains. We acquired whole-brain connectivity with a higher-throughput protocol (11.8 h/specimen) at 45 μm/46 angles across both sexes and two age cohorts as part of an ongoing study of aging and Alzheimer’s disease. Forty-seven cases were young, between the ages of 72 and 176 d. Sixty-four cases matched by strain were postreproductive and ranged in age between 341 and 687 d. Using life expectancy estimates of all these BXD family members (30), we rescaled chronological age to the fraction of expected lifespan as shown in SI Appendix, Fig. S8.
Statistical methods described in refs. 36 and 48 were used to find the global differences in connectivity as a function of strain and age. A minimum deformation template (MDT) was generated in our SAMBA pipeline for all the specimens in a given strain. A visual quality assurance check was executed in Slicer which allows multicontrast multiplanar visualization. Our canonical atlas (with labels) was mapped to the MDT. The individual specimens (now with labels) were mapped back to their original laboratory space by inverting the transform to the MDT. The connectome pipeline was used to generate connectivity between each ROI. A dimensional reduction was applied to map the vectors of each of the 179 ROIs into a reduced space to enable ANOVA comparisons in young and old animals across all the strains. Multidimensional scaling was used to embed the results into two dimensions to visualize clustering (48). This enables rank ordering of the nodes in which there are differences within a strain and between ages (36). Our previous work has demonstrated that more than 150 vertices of the whole-brain connectome (generated with 179 ROIs in both hemispheres) had significant dependence on the strain (genome) (36). SI Appendix, Fig. S9A emphasizes this by plotting the MDS of the BXD65b and BXD34 in the same space. For each strain, clusters in a different part of the plot highlight the genetic dependence of the connectome. Perhaps more interesting is the fact that the two strains sequester differently with age. The largest global connectivity change during aging (BXD65b) and the smallest connectivity change (BXD34) are calculated from the Euclidean distances between the means of their respective clusters. The young BXD65b specimens (green) were tightly clustered, while the old specimens (purple) were clearly separated from the young and were not clustered tightly. The separation in the dimensionless MDS plot is 322. The dotted lines indicate one SD of the cluster centroid. The BXD34 data on the right show a much smaller separation of 38. The SD of the centroid is smaller for the young animals than it is for the old, i.e., as the animals age, there is divergence in their connectomes.
A comparable analysis was performed for all regions of interest and the results rank ordered to highlight which nodes and strains underwent the largest change in connectivity with age. Over 100 (of 179) regions changed with age in the omni MANOVA comparisons. The subiculum, one of the regions in which the connectivity changed with age in all of the strains, is shown in SI Appendix, Fig. S9 B and C. The young specimens are again clustered more tightly. The difference between the centroids for the BXD29 (75) is more than three times the difference for the BXD34 24. SI Appendix, Fig. S9 D and E provide a demonstration of the consequence of the large change in subiculum connectivity for the BXD29 for the two specimens identified by the arrows in SI Appendix, Fig. S9B.
SI Appendix, Fig. S10 shows comparison of two of the old BXD65b animals used in the analysis described in SI Appendix, Fig. S9. As animals age, there is a greater chance of error in the labeling pipeline because of age-related atrophy. But, as SI Appendix, Fig. S10 demonstrates, the labels are transferred with good fidelity.
SI Appendix, Fig. S11 shows the integrated method applied in our ongoing studies of strain-related resilience in Alzheimer’s disease. A total of 40 different BXD strains with and without the 5XFAD transgene are being imaged at 6 and 14 mo. More than 150 specimens have been imaged thus far using the 25-μm high-throughput protocol described in SI Appendix, Fig. S6. Brains have been removed from the skull and transferred to LifeCanvas Technologies (Cambridge, MA) where they were stained for NeuN and a-β and scanned using structured plane illumination. The LSM volumes were returned to Duke where they were aligned with the MRH volume obtained in the skull. The 14-mo-old female 5XFADBXD77 mice shown in SI Appendix, Fig. S11 were (group) analyzed using Omni MANOVA which identified the subiculum as an area in which there was significant reduction in connectivity in the 5XFAD animal. SI Appendix, Fig. S11A shows the FA image. SI Appendix, Fig. S11B shows the a-β LSM that has been aligned to the FA. Heavy plaque deposits in the subiculum are consistent with the Omni MANOVA analysis. SI Appendix, Fig. 11C (control nTgBXD77) and SI Appendix, Fig. 11D (5XFADBXD77) show the tractograms obtained by seeding the right subiculum. There is substantial reduction in the connectivity in the 5XFADBXD77.
2. Discussion
2.1. Synopsis of HiDiver.
The methods described here rely on a fundamental redesign of infrastructure that enables the collection and registration of structural data for complete individual brains. This allows us to probe the gene–structure relations far more systematically than has been possible to date. A quantitative analysis of regional volumes, cell types (49, 50), and brain wide connectomes (51) is now practical across hundreds of unique genomes. We believe this approach will offer ways to understand genetic variation and environmental perturbations in cellular complement and distribution, circuit architecture, and behavioral phenotype as a function of stage, age, life experience, and/or interventional treatment.
Our methods differ from those of previous work (17, 52, 53) in five specific ways: 1) the spatial resolution of MRH is improved by more than two orders of magnitude; 2) the contrast resolution is higher, and there are multiple scalar images from the DTI/GQI acquisition that provide varied anatomical definition; 3) MRH data are acquired in the skull with limited distortion, enabling LSM to be spatially remediated with excellent geometric accuracy with respect to in vivo morphometry; 4) we labeled multimodal packages for each specimen within a common reference space; and finally, 5) the throughput is sufficiently high that the methods can be targeted at neurogenetic and genome-wide mapping studies which require large numbers of specimens (21, 54).
Our most immediate target is neurogenetics of Alzheimer’s disease. DTI has generated considerable interest as a tool for diagnosing and staging Alzheimer’s disease (55, 56). While we and others have demonstrated the ability to detect atrophy in animal models of Alzheimer’s disease (57), the methods we have described here provide a much richer set of measures. Volume measures are possible with much higher precision because of the increased spatial resolution and improved segmentation. Scalar measures of axial diffusivity provide potential measure of axonal loss. Radial diffusivity provides insight into myelination. Mean diffusivity may be an early measure of change in cortical microstructure (58). Recent work by Stone et al has suggested that measuring diffusion properties in specific tracts may provide one of the most sensitive and specific methods for staging Alzheimer’s disease (59). Translating these methods to the mouse, as outlined above, is challenging because of the need for much higher spatial resolution than is routine in the clinical domain. The work we have outlined here creates a path for translation between mouse models and clinical application.
2.2. Limitations of HiDiver.
While throughput still remains a bottleneck, we have made significant progress from 45 μm resolution in 10 d (47). The acceleration in SI Appendix, Fig. S6 has reduced the acquisition time for a 25 μm/61 angle volume to 20 h. A second promising method exploits the structural redundancy in q space, i.e., joint compression in k and q space (60). We have examined the trade-offs in angular sampling and spatial resolution from which it has become clear that as the spatial resolution is increased, the angular resolution can be decreased. Figure 8 in the study by Crater et al. (38) demonstrates that with spatial resolution at 25 μm, reducing the angular resolution from 60 to 40 angles has negligible effects on the false positive ratio with a small change (0.17 to 0.20) in the false negative ratio.
2.3. HiDiver and Genetic and Environmental Dissection of the Murine CNS.
The conundrum now is how to connect these very large volumes of neuroimaging data in ways that can help answer major questions related to brain function, aging, and disease. Multiscalar data registration is just a key first step. What is also needed is a computational framework to systematically extract or test the causality of putative genome–phenome relations. In many cases that means large and highly diverse samples such as are common in human neuroimaging. As is typical in our reductionist age, most experimental preclinical work in rodents still relies on single canonical genotypes. The archetypal mouse is a 56-d-old C57BL/6J male (61). This n-of-1 design is not computationally robust. It is not possible to use a single genome to derive structure–function relations that generalize to other members of the same species, let alone to other species and of course, to humans. As we improve throughput, we anticipate that experimental studies will shift to robust population models that incorporate levels of genetic variation comparable to human populations.
3. Materials and Methods
SI Appendix, Fig. S12 provides an overview of the workflow. There are two main processing streams. The first focuses on the generation of scalar volumes derived from the diffusion tensor images (DTI) and multigradient echo images (MGRE) with the brain in the cranial vault. The second focuses on whole-brain 3D LSM images. The two streams are merged into a common space where the morphology of the LSM is restored by registration to the MRH which defines a common coordinate reference frame (CCF3v3) (33) to allow generation of image-derived phenotypes of cells and circuits covering the whole brain. The methods are facilitated through an integrated high-performance computing structure and specialized processing pipelines described in more detail in SI Appendix.
Supplementary Material
Appendix 01 (PDF)
Acknowledgments
Our special thanks to Dr. Glenn D. Rosen for invaluable help in developing the r1CCFv3 used in this publication. Thanks to Drs. Lu and Suheeta Roy for access to BXD mice and aged animals from the -University of Tennessee Health Science Center Aging Colony. We are grateful to Lucy Upchurch for technical assistance. We thank Tatiana Johnson for special care in manuscript preparation and submission. This work was supported by National Institute on Aging (R01AG070913), National Institute of Neurological Disorders and Stroke (R01NS096729), National Institute of Biomedical Engineering (P41EB015897), and NIH (S10OD010683).
Author contributions
G.A.J. designed research; G.A.J., G.P.C., K.H., N.W., and R.W.W. performed research; G.A.J., D.G.A., J.J.C., J.C.G., A.H., F.-C.Y., and N.W. contributed new reagents/analytic tools; G.A.J., Y.T., Y.Q., and L.E.W. analyzed data; Y.Q. maintained database; and G.A.J., L.E.W., and R.W.W. wrote the paper.
Competing interests
The authors declare no competing interest.
Footnotes
This article is a PNAS Direct Submission.
Data, Materials, and Software Availability
Software is available at https://github.com/YuqiTianCIVM/MRH_LSM_registration (62). We have made the data for experiments 1-3 available under creative commons CC by NC-SA at https://civmimagespace.civm.duhs.duke.edu/login.php/client/4 (63). The data is stored in H5 format to enable interactive examination using Neuroglancer (https://github.com/google/neuroglancer; https://zenodo.org/record/5573294#.ZCrkIezMIvo) (64, 65).
Supporting Information
References
- 1.Lauterbur P., Image formation by induced local interactionsL examples employing nuclear magnetic resonance. Nature 242, 190–191 (1973). [PubMed] [Google Scholar]
- 2.Van Essen D. C., et al. , The WU-Minn human connectome project: An overview. Neuroimage 80, 62–79 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Toga A. W., et al. , Mapping the human connectome. Neurosurgery 71, 1–5 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Thompson P. M., et al. , The ENIGMA Consortium: Large-scale collaborative analyses of neuroimaging and genetic data. Brain Imaging Behav. 8, 153–182 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Elliott L. T., et al. , Genome-wide association studies of brain imaging phenotypes in UK Biobank. Nature 562, 1–17 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Saykin A. J., et al. , Genetic studies of quantitative MCI and AD phenotypes in ADNI: Progress, opportunities, and plans. Alzheimers Dement. 11, 792–814 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dumitrescu L., et al. , Genetic variants and functional pathways associated with resilience to Alzheimer’s disease. Brain 143, 2561–2575 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Johnson G. A., et al. , Histology by magnetic resonance microscopy. Magn. Reson. Q. 9, 1–30 (1993). [PubMed] [Google Scholar]
- 9.Johnson G. A., et al. , Morphologic phenotyping with magnetic resonance microscopy: The visible mouse. Radiology 222, 789–793 (2002). [DOI] [PubMed] [Google Scholar]
- 10.Johnson G. A., et al. , Waxholm Space: An image-based reference for coordinating mouse brain research. Neuroimage 53, 365–372 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang N., et al. , Whole mouse brain structural connectomics using magnetic resonance histology. Brain Struct. Funct. 223, 4323–4335 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang N., et al. , Cytoarchitecture of the mouse brain by high resolution diffusion magnetic resonance imaging. Neuroimage 216, 116876 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vogelstein J. T., et al. , A community-developed open-source computational ecosystem for big neuro data. Nat. Methods 15, 846–847 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Murakami T. C., et al. , A three-dimensional single-cell-resolution whole-brain atlas using CUBIC-X expansion microscopy and tissue clearing. Nat. Neurosci. 21, 625–637 (2018). [DOI] [PubMed] [Google Scholar]
- 15.Chung J. R., et al. , Multiscale exploration of mouse brain microstructures using the knife-edge scanning microscope brain atlas. Front. Neuroinform. 5, 29 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lefebvre J., et al. , Whole mouse brain imaging using optical coherence tomography: Reconstruction, normalization, segmentation, and comparison with diffusion MRI. Neurophotonics 4, 041501 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Goubran M., et al. , Multimodal image registration and connectivity analysis for integration of connectomic data from microscopy to MRI. Nat. Commun. 10, 5504 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liang Z., et al. , Virtual mouse brain histology from multi-contrast MRI via deep learning. Elife 11, e72331 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Qu L., et al. , Cross-modal coherent registration of whole mouse brains. Nat. Methods 19, 111–118 (2022). [DOI] [PubMed] [Google Scholar]
- 20.Johnson G. A., et al. , Whole mouse brain connectomics. J. Comp. Neurol. 12, 1–12 (2018). [Google Scholar]
- 21.Badea A., Johnson G. A., Williams R. W., Genetic dissection of the mouse brain using high-field magnetic resonance microscopy. Neuroimage 45, 1067–1079 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jiang Y., Johnson G. A., Microscopic diffusion tensor atlas of the mouse brain. Neuroimage 56, 1235–1243 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gerfen C. R., Paletzki R., Heintz N., GENSAT BAC cre-recombinase driver lines to study the functional organization of cerebral cortical and basal ganglia circuits. Neuron 80, 1368–1383 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lein E. S., et al. , Genome-wide atlas of gene expression in the adult mouse brain. Nature 445, 168–176 (2007). [DOI] [PubMed] [Google Scholar]
- 25.BRAIN Initiative Cell Census Network (BICCN), A multimodal cell census and atlas of the mammalian primary motor cortex. Nature 598, 86–102 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Feng G., et al. , Imaging neuronal subsets in transgenic mice expressing multiple spectral variants of GFP. Neuron 28, 41–51 (2000). [DOI] [PubMed] [Google Scholar]
- 27.Porrero C., et al. , Mapping of fluorescent protein-expressing neurons and axon pathways in adult and developing Thy1-eYFP-H transgenic mice. Brain Res. 1345, 59–72 (2010). [DOI] [PubMed] [Google Scholar]
- 28.Ashbrook D. G., et al. , A platform for experimental precision medicine: The extended BXD mouse family. Cell Syst. 12, 235–247.e9 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sasani T. A., et al. , A natural mutator allele shapes mutation spectrum variation in mice. Nature 605, 497–502 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Roy S., et al. , Gene-by-environment modulation of lifespan and weight gain in the murine BXD family. Nat. Metab. 3, 1217–1227 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Basser P. J., Mattiello J., LeBihan D., MR diffusion tensor spectroscopy and imaging. Biophys. J. 66, 259–67 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yeh F.-C., Wedeen V. J., Tseng W.-Y.I., Generalized q-sampling imaging. IEEE Trans. Med. Imaging 29, 1626–1635 (2010) [DOI] [PubMed] [Google Scholar]
- 33.Wang Q., et al. , The allen mouse brain common coordinate framework: A 3D reference atlas. Cell 181, 936–953.e20 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Avants B. B., Tustison N. J., Wu J., Cook P. A., Gee J. C., An open source multivariate framework for n-tissue segmentation with evaluation on public data. Neuroinformatics 9, 381–400 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Williams R. W., et al. , Genetic and environmental control of variation in retinal ganglion cell number in mice. J. Neurosci. 16, 7193–205 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang N., et al. , Variability and heritability of mouse brain structure: Microscopic MRI atlases and connectomes for diverse strains. NeuroImage 222, 117274 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tian Y., James C. J., Johnson G. A., Restoring morphology of light sheet microscopy data based on magnetic resonance histology. Front. Neurosci. 16, 1011895 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Crater S., et al. , Resolution and b value dependent structural connectome in ex vivo mouse brain. Neuroimage 255, 119199 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Calamante F., Tournier J. D., Jackson G. D., Connelly A., Track-density imaging (TDI): Super-resolution white matter imaging using whole-brain track-density mapping. NeuroImage 53, 1233–1243 (2010). [DOI] [PubMed] [Google Scholar]
- 40.Calamante F., et al. , Super-resolution track-density imaging studies of mouse brain: Comparison to histology. NeuroImage 59, 286–296 (2012). [DOI] [PubMed] [Google Scholar]
- 41.Lee H. H., et al. , Along-axon diameter variation and axonal orientation dispersion revealed with 3D electron microscopy: Implications for quantifying brain white matter microstructure with histology and diffusion MRI. Brain Struct. Funct. 224, 1469–1488 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tanaka T., et al. , Large-scale electron microscopic volume imaging of interfascicular oligodendrocytes in the mouse corpus callosum. Glia 69, 2488–2502 (2021). [DOI] [PubMed] [Google Scholar]
- 43.West K. L., et al. , Quantitative analysis of mouse corpus callosum from electron microscopy images. Data Brief 5, 124–128 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Anderson R. J., et al. , Small animal multivariate brain analysis (SAMBA)—A high throughput pipeline with a validation framework. Neuroinformatics 17, 451–472 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Maier-Hein K. H., et al. , The challenge of mapping the human connectome based on diffusion tractography. Nat. Commun. 8, 1349 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ugurbil K., et al. , Pushing spatial and temporal resolution for functional and diffusion MRI in the Human Connectome Project. NeuroImage 80, 80–104 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Calabrese E., et al. , A diffusion MRI tractography connectome of the mouse brain and comparison with neuronal tracer data. Cereb. Cortex 25, 4628–4637 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Levin K. A. A., Tan M., Lyzinski V., Park Y., Priebe C. E., A central limit theorem for an omnibus embedding of multiple random graphs and implications for multiscale network inference. arXiv [Preprint] (2017). https://arxiv.org/abs/1705.09355.
- 49.Zhu F., et al. , Architecture of the mouse brain synaptome. Neuron 99, 781–799.e10 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cizeron M., et al. , A brainwide atlas of synapses across the mouse life span. Science 369, 270–275 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chen X., et al. , High-throughput mapping of long-range neuronal projection using in situ sequencing. Cell 179, 772–786.e19 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Renier N., et al. , Mapping of brain activity by automated volume analysis of immediate early genes. Cell 165, 1789–1802 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chandrashekhar V., et al. , CloudReg: Automatic terabyte-scale cross-modal brain volume registration. Nat. Methods 18, 845–846 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Badea A., Johnson G. A., Williams R. W., Genetic dissection of the mouse CNS using magnetic resonance microscopy. Curr. Opin. Neurol. 22, 379–386 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fornito A., Zalesky A., Breakspear M., The connectomics of brain disorders. Nat. Rev. Neurosci. 16, 159–172 (2015). [DOI] [PubMed] [Google Scholar]
- 56.Mayo C. D., et al. , Relationship between DTI metrics and cognitive function in Alzheimer’s disease. Front. Aging. Neurosci. 10, 436 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Badea A., Johnson G. A., Jankowsky J. L., Remote sites of structural atrophy predict later amyloid formation in a mouse model of Alzheimer’s disease. Neuroimage 50, 416–427 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Weston P. S. J., et al. , Measuring cortical mean diffusivity to assess early microstructural cortical change in presymptomatic familial Alzheimer’s disease. Alzheimers. Res. Ther. 12, 112 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Stone D. B., et al. , Specific white matter tracts and diffusion properties predict conversion from mild cognitive impairment to Alzheimer’s disease. Front. Aging. Neurosci. 13, 711579 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sun J., Entezari A., Vemuri B. C., Exploiting structural redundancy in q-space for improved EAP reconstruction from highly undersampled (k, q)-space in DMRI. Med. Image Anal. 54, 122–137 (2019). [DOI] [PubMed] [Google Scholar]
- 61.Morris J. A., et al. , Divergent and nonuniform gene expression patterns in mouse brain. Proc. Natl. Acad. Sci. U.S.A. 107, 19049–19054 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tian Y., Cook J. C., Johnson G A., Restoring morphology of light sheet microscopy data based on magnetic resonance histology. Github. https://github.com/YuqiTianCIVM/MRH_LSM_registration. Deposited 17 November 2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Crifield S., Cook J. C., Mansour H., Upchurch L., Johnson G. A., Merged magnetic resonance and light sheet microscopy of the whole mouse brain. Duke CIVM Imagespace. https://civmimagespace.civm.duhs.duke.edu/login.php/client/4. Deposited 11 October 2022. [Google Scholar]
- 64.Maitin-Shepard J., et al. , google/neuroglancer. Github. https://github.com/google/neuroglancer. Deposited 29 March 2023.
- 65.Maitin-Shepard J., et al. , google/neuroglancer. Zenodo. https://zenodo.org/record/5573294#.ZCtNtXtBw2x. Deposited 16 October 2021.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix 01 (PDF)
Data Availability Statement
Software is available at https://github.com/YuqiTianCIVM/MRH_LSM_registration (62). We have made the data for experiments 1-3 available under creative commons CC by NC-SA at https://civmimagespace.civm.duhs.duke.edu/login.php/client/4 (63). The data is stored in H5 format to enable interactive examination using Neuroglancer (https://github.com/google/neuroglancer; https://zenodo.org/record/5573294#.ZCrkIezMIvo) (64, 65).