Abstract
Introduction
The continued emergence and spread of multidrug-resistant (MDR) bacterial pathogens require a new strategy to improve the efficacy of existing antibiotics. Proline-rich antimicrobial peptides (PrAMPs) could also be used as antibacterial synergists due to their unique mechanism of action.
Methods
Utilizing a series of experiments on membrane permeability, In vitro protein synthesis, In vitro transcription and mRNA translation, to further elucidate the synergistic mechanism of OM19r combined with gentamicin.
Results
A proline-rich antimicrobial peptide OM19r was identified in this study and its efficacy against Escherichia coli B2 (E. coli B2) was evaluated on multiple aspects. OM19r increased antibacterial activity of gentamicin against multidrug-resistance E. coli B2 by 64 folds, when used in combination with aminoglycoside antibiotics. Mechanistically, OM19r induced change of inner membrane permeability and inhibited translational elongation of protein synthesis by entering to E. coli B2 via intimal transporter SbmA. OM19r also facilitated the accumulation of intracellular reactive oxygen species (ROS). In animal models, OM19r significantly improved the efficacy of gentamicin against E. coli B2.
Discussion
Our study reveals that OM19r combined with GEN had a strong synergistic inhibitory effect against multi-drug resistant E. coli B2. OM19r and GEN inhibited translation elongation and initiation, respectively, and ultimately affected the normal protein synthesis of bacteria. These findings provide a potential therapeutic option against multidrug-resistant E. coli.
Keywords: multidrug-resistant bacteria, SbmA, transcription, antimicrobial peptide, PrAMPs
1. Introduction
The emergence of multi-drug resistant (MDR) bacterial pathogens is a major threat to global public health (Aslam et al., 2018). At present, due to the extensive use of antibiotics, the family Enterobacteriaceae associated with animals have developed multidrug resistance (Van Boeckel et al., 2019). Escherichia coli (E. coli) is one of the most common pathogens of this family leading to infections in animals and humans with pathogenicity of 10–15% and mortality of 3.8–72%. Usually, aminoglycosides have been used for therapy against E. coli infections; however, recent studies have shown that a large number of E. coli strains are resistant to aminoglycosides. The resistance rate of aminoglycosides resistant genes and its related phenotypes in E. coli from lactating dairy cows have been reported for about 20% in the US (Jeamsripong et al., 2021). Likewise, aminoglycosides resistant bacterial strains of Enterobacteriaceae family isolated from different sources in Ireland have also been reported. The bacteria develop different frequencies of aminoglycosides resistance based on their habitat and geographical distribution. The highly exposed areas to antibiotics receives the highest frequency of resistance (Hooban et al., 2022). With the continuous evolution of pathogenic bacteria under the pressure of antimicrobial drugs, the existing antimicrobial drugs are no longer effective in the clinical treatment of bacterial infections; therefore, alternatives to traditional antibiotics are required on priority basis.
Using antibiotic synergists is a promising strategy as it could extend the half-life of existing antibiotics, alleviate the current shortage of effective antibiotics, and reduce the lengthy drug development time and thereby the economic risk. Currently, novel β-lactam antibacterial synergists (Si et al., 2020; Durand-Reville et al., 2021), tetracycline antibacterial synergists (Zheng et al., 2017; Liu Y. et al., 2020), polymyxin antibacterial synergists (Stokes et al., 2017; Song et al., 2021) and macrolides antibacterial synergists (Sturge et al., 2019) have been identified. Furthermore, bee venom enhances the bacteriostatic effect of gentamicin (GEN) and vancomycin (VAN) against methicillin-resistant Staphylococcus aureus (MRSA) (Peng et al., 2021). In addition, thiosemicarbazide combined with gentamicin (GEN) prevent Gram-positive bacterial infection by inhibiting DNA gyrase activity by approximately 50% (Wang et al., 2019). However, data on antimicrobial peptides as synergists to restore gentamicin (GEN) sensitivity of Gram-negative bacteria, especially drug-resistant E. coli, are limited.
Cationic antimicrobial peptides (AMPs) have also been used as synergists to enhance the antibacterial activity of antibiotics owing to their unique membrane-breaking mechanism (Randhawa et al., 2016; Si et al., 2020; Song et al., 2020; Bai et al., 2021; Zhu et al., 2021). Certain proline-rich antimicrobial peptides (PrAMPs) can directly cross the bacterial cell membrane and exert their antibacterial effects (Roy et al., 2015; Böttger et al., 2016). However, there have been no studies on the use of PrAMPs as adjuvants to improve the antibacterial effect of existing antibiotics. Proline-rich antimicrobial peptide OM19r (VDKPPYLPRPRPIRrPGGr-NH2) is a sequence-derived peptide from the existing AMP sequences. The new peptide OM19r of this study was designed by substitution of D-type amino acid at positions 15 and 19 of OM19R (Cui et al., 2021), while OM19R(VDKPPYLPRPRPIRRPGGR-NH2) was a hybrid obtained from Oncocin and MDAP-2 fragments (Liu L. et al., 2020). OM19r shown the effective restoration the gentamicin (GEN) sensitivity of multidrug resistant E. coli. The effect of OM19r combined with gentamicin (GEN) on the cell membrane and DNA of E. coli B2 (mcr-1) was determined via a series of fluorescence permeability and DNA binding assays. Meanwhile, an in vitro transcription-translation system (TX-TL) was constructed to investigate the effects of OM19r against multidrug resistant E. coli B2. Our findings suggest the potential use of OM19r as an aminoglycosides antibacterial synergist to improve the antibacterial activity of aminoglycosides.
2. Materials and methods
2.1. Bacterial strains and cell culture
Mueller-Hinton (MH) broth and Mueller-Hinton (MH) agar culture medium were purchased from Haibo Biotechnology Co., Ltd. (Qingdao, China) and the antibiotics were purchased from the Institute of China Food and Drug Administration. The bacterial strains were obtained from the Key Laboratory of New Veterinary Drug Research and Development of Jilin Province. The E. coli ATCC25922 strain served a quality control. Multidrug resistant E. coli B2 (mcr-1) strain was provided by the China Agricultural University while E. coli ATCC25922 (ΔSbmA) strain was provided by the Key Laboratory of Animal Production, Product Quality and Security, Ministry of Education. HeLa CellS and mouse macrophage cells (RAW264.7) were cultured in Dulbecco’s Modified Eagle Medium (Gibco) supplemented with 1% (w:v) penicillin–streptomycin and 10% (w:v) heat-inactivated FBS (Sigma-Aldrich) at 37°C in a 5% CO2 atmospheric environment.
2.2. Experimental animals
BALB/c mice (4 weeks old, weighing 20–22 g) were obtained from the laboratory of Jilin Agricultural University Changchun, China. All the animals were housed in an environment with a temperature of 22 ± 1°C, a relative humidity of 50 ± 1%, and a light/dark cycle of 12/12 h. The laboratory animal usage license (SYXK-2018-0023) was issued by the Laboratory Animal Center of Jilin Agricultural University Changchun, China. All animal studies (including the mice euthanasia procedure) were performed in compliance with the regulations and guidelines of Institutional Animal Care of Jilin Agricultural University Changchun, China.
2.3. Antimicrobial activity
The minimum inhibitory concentration (MIC) of antimicrobials was determined by the broth microdilution method (Miao et al., 2020; Zhong et al., 2021). Briefly, the strains to be tested were inoculated into 5 ml of MH broth medium and cultured to logarithmic growth phase (OD600 = 0.5), and diluted with fresh MH broth to obtain 1× 106 CFU/ml. The diluted bacterial solution (100 μl) was added to 96-well plates containing different concentrations of antibiotics or different concentrations of antimicrobial peptides. The final concentrations of OM19r strain were 1–128 μg/ml (0.45–57.5 μM). A 100 μl bacterial solution +100 μl MH broth medium served as positive control while 200 μl MH broth medium was kept as negative control. The 96-well plate was incubated at 37°C for 18–20 h, and the MICs were measured at 600 nm. The experiment was conducted in triplicate.
2.4. Checkerboard assays
The synergistic antibacterial activity was measured by the checkerboard method (Zhong et al., 2019). Briefly, a 50 μl of diluted OM19r (0–8 μg/ml) and 50 μl of diluted antibiotics (0–512 μg/ml) were put into a 96-well plate, then added a 100 μl of the diluted bacterial solution (1 × 106 CFU/ml) and incubated at 37°C for 16–18 h, and measured the FIC at 600 nm. The experiment was performed in triplicate. The FIC index was calculated as:FIC = MIC (group A combined)/MIC (group A used alone) + MIC (group B combined)/MIC (group B used alone). Where the following were considered as, FIC ≤ 0.5 for synergistic effect, 0.5 < FIC ≤ 1 for additive effect, 1 < FIC ≤ 4 for no effect, and FIC > 4 for antagonistic effect.
2.5. Bacterial growth curve and time-kill curve analysis
Growth curves were measured according to the previously described method at OD600 nm wavelength at 37°C for 24 h at every 1 h interval (Song et al., 2020). Briefly, different concentrations of OM19r (2 μg/ml), GEN (32 μg/ml), or OM19r combined with GEN (2 + 32 μg/ml) were added to 96-well microplates with an equal amounts of bacterial dilutions (1 × 106 CFU/ml). The time-kill curve was drawn with time as the abscissa and the Log value of the number of colonies as the ordinate (Falciani et al., 2020). Different concentrations of OM19r (8 μg/ml), GEN (128 μg/ml), OM19r combined with GEN (4 + 2 μg/ml) or OM19r combined with GEN (4 + 4 μg/ml) were diluted to contain indicator bacteria (OD = 0.5) in a 1.5 ml centrifuge tube. PBS was kept as a negative control group. After every half an hour, a 100 μl of the diluted bacterial solution was taken out and spread on MH agar medium, and counted after culturing at 37°C for 16–18 h. The procedures were conducted triplicate.
2.6. Hemolytic activity
The fresh sheep blood (1–2 ml) was collected at 4°C and centrifuged at 1238 × g for 10 min according to a previously described method (Zhong et al., 2020). The collected red blood cells were washed three times with PBS (10 mM, PH = 7.4), and then resuspended in a 9 ml PBS with approximate cell concentration of 109 cells/mL. Erythrocyte suspension (100 μl) was successively added to 96-well plates that were added with antimicrobial peptides (ranging from1 μg/mL to 64 μg/ml) and PBS. After incubation at 37°C for 2 h, the samples were centrifuged at 8800 × g for 10 min. The supernatants (180 μl) were transferred to a new 96-well plate, and the absorbance was measured at 570 nm. The absorbance of PBS (100 μl) treated-erythrocyte supernatants (100 μl) served as negative control while the absorbance of 0.1% Triton X-100 (100 μl) treated-erythrocyte supernatants (100 μl) as positive control. Hemolysis was calculated using the following formula:
Hemolysis (%) = [(A 420, peptide – A420, PBS) / (A420, 0.2% Triton X-100 – A420, PBS)] * 100.
2.7. Cytotoxicity assays
The cytotoxicity of OM19r was determined using 3-(4,5-dimethylthiozol-2-yl)-2,5 diphenyltetrazolium bromide (MTT, United Kingdom) dye reduction test according to a previously described method (Mwangi et al., 2019). Briefly, mouse mononuclear macrophage RAW264.7 and HeLa cells (2.0 × 104 cells/well) were added into 96-well plates at 37°C overnight. Then OM19r was added to the cell cultures to reach a final concentration of 1–64 μg/ml and cultured under a 5% CO2 atmospheric environment at 37°C for 24 h. After incubation with MTT (50 ml, 0.5 mg/ml) at 37°C for 2 h, the cell cultures without supernatants were collected by centrifuging at 413 × g for 5 min. Subsequently, a 150 μl of dimethyl sulfoxide (DMSO) was added to dissolve the formazan crystals that were created during experiemnt. Absorbance (490 nm) was measured using a microplate reader. Cytotoxicity was calculated using the following formula:
Cytotoxicity (%) = 100 – [(A570 of peptide treated cells / A570 of control) * 100].
2.8. Membrane permeability assays
The outer membrane changes were measured using 1-N-phenylnaphthylamine (NPN) dye (Ma et al., 2020). NPN is a hydrophobic fluorescent agent normally used for emission of weak fluorescence in aqueous solution but it emits strong fluorescence upon entering to a hydrophobic medium. We added a10 μL of 1 mM NPN solution to each 990 μl of bacterial suspension (1 × 106 CFU/ml) and incubated it at room temperature for 30 min in the dark environment. The groups were as follow: experimental group (Fbos) holding a 100 μl of NPN-containing bacterial suspension +100 μl of antimicrobial peptide mixture, control group (F0) holding a 100 μl of NPN bacterial suspension +100 μl of PBS and control group (F100) holding a 100 μl of NPN-containing bacterial suspension +100 μl of polymyxin B. Fluorescence intensity was detected after every 10 min for 150 min. The mixture was placed in a 96-well dark plate, the excitation wavelength and emission wavelength was adjusted to 350 nm and 420 nm, respectively, and the fluorescence intensity was detected. Each experiment was set up with 3 replicates. Absorption was measured as,
NPN absorption rate = (Fbos-F0)/(F100-F0) × 100%.
The membrane changes were measured using propidium iodide (PI) dye (Mattiuzzo et al., 2010; Mwangi et al., 2019; Ma et al., 2020). Propidium iodide (PI) is a popular red-fluorescent nuclear and chromosome counterstain. Since propidium iodide is not permeant to live cells, it is commonly used to detect dead cells in a population. PI binds to DNA by intercalating between the bases with little or no sequence preference. The bacterial suspension was adjusted to 1 × 106 CFU/ml by the same method mentioned above. First PI dye was added to the bacterial suspension to reach a final concentration of 10 μM. Then PI-containing suspension was mixed at 1:1 with the antimicrobial peptide mixture in a total volume of 200 μl and added to a 96-well dark plate. The excitation wavelength and emission wavelength was set to 535 nm and 615 nm, respectively. Fluorescence intensity was detected after every 10 min for 150 min.
2.9. Proton motive force assays
The PMF consists of a pH gradient (ΔpH) and a potential gradient (Δφ), which together constitute an electrochemical gradient (Corbalan et al., 2013). Membrane depolarization was measured using 3,3-dipropylthiodicarbocyanine iodide DiSC3(5) fluorescent dye (Xu et al., 2020). DiSC3(5) was added to the bacterial suspension (1 × 106 CFU/ml) to a final concentration of 0.5 μM, and then the bacterial suspension containing DiSC3(5) was mixed at 1:1 with different concentrations of peptide mixture in a total volume of 200 μl. It was placed on a 96-well light-shielding plate, and the excitation wavelength and emission wavelength was set at 670 nm and 622 nm, respectively. Polymyxin B and PBS served as the positive and negative control, respectively. Membrane depolarization was measured by using BCECF-AM fluorescent dye (Ding et al., 2020). BCECF-AM was added to the bacterial suspension (1 × 106 CFU/ml) to a final concentration of 20 μM, then mixed with the different concentrations of peptide mixture at 1:1 to a total volume of 200 μl, and then added to a 96-well plate protected in dark environment. The excitation wavelength and emission wavelength was set to 522 nm and 488 nm, respectively. The positive control was glucose, and the negative control was PBS, and each group had three replicates.
2.10. Efflux pump assays
Fluorescent probe Ethidium bromide (EtBr) was used to detect the effect of drug on non-specific bacterial pumping system and to monitor EtBr efflux from the cell. The bacterial solution was re-suspended in PBS buffer until OD600 = 0.5, then a 2 μg/ml EtBr was added, mixed evenly, and incubated in the dark for 30 min. The suspensions containing EtBr in the two experiments were then mixed at 1:1 with antimicrobial peptides to make a total volume of 200 μl. It was placed on a 96-well dark plate, and the excitation wavelength and emission wavelength was set at 600 nm and 530 nm, respectively. The fluorescence intensity was detected after every 10 min for 150 min. The positive control was the efflux pump inhibitor CCCP, and the negative control was PBS, and each group had three replicates (Wang et al., 2015).
2.11. Real-time PCR
The qPCR was performed according to a previously described method (Bustin et al., 2009). The bacterial suspension was adjusted to 1 × 106 CFU/ml by the same method as stated. RNA was extracted using RNAiso Plus and the values at 260 nm/280 nm were measured on spectrophotometer. The primers used were SbmA primers (SbmA-F: GAACCTCGAGCTGATCTTCG; SbmA-R: CTGAGCTCCGATTCGAAGG) and internal reference gene primers (rsmC-F: GAAATTCTGGGCGAATACA; rsmC-R: CTTTCACCTCGGAAAAGACG). The reaction system was 10 μl of SYBR Green I Master (2×), 8 μl of ddH2O, 1 μl of cDNA upstream and downstream primers, and 0.5 μl of upstream and downstream primers. Reaction program was as follow: pre-denaturation at 95°C for 5 min, denaturation at 95°C for 10 s, annealing at 60°C for 15 s, extension at 72°C for 20 s and a total of 40 cycles. Each experiment was repeated three times. And the fold change of gene expression was calculated by the 2-ΔΔCt method.
2.12. Recombinant expression of SbmA
The pET28a- SbmA-6His recombinant expression vector was constructed and transformed to E. coli BL21 competent cell for the overexpression SbmA-6His protein. The SbmA protein was purified using Ni-NTA 6FF column. [The process was as follows: Lysed E. coli cell with Buffer A (20 mM Tris–HCl pH 7.4, 5% glycerol, 500 mM NaCl. That 6 mM β-Mercaptoethenol, 0.1 mM Benzamidine and 0.1 mM PMSF added immediately before use). Non-affinity protein was washed off by Buffer A added 20 mM imidazole. The target protein was finally eluted by Buffer A added 200 mM imidazole.] The effect of SbmA on the bacteriostatic activity of OM19r against E. coli was determined by in vitro competitive inhibition assay. The secondary structure of OM19r was constructed by https://zhanggroup.org/I-TASSER/. The structure of SbmA protein was constructed by using the online server Swiss-Model.1
2.13. Molecular docking
The HDOCK online software was used for molecular docking. The resulting conformations were set to 100 and the scoring function was used to select the conformation with the negative energy. The docking results were visualized by using pymol software.
2.14. Confocal laser scanning microscopy
Confocal laser scanning microscopy (CLSM) was performed according to the previously described method (Si et al., 2020). E. coli B2 precipitates in logarithmic growth phase (OD600 value 0.5) were collected at 2000 × g for 5 min and washed three times with sterile PBS. C-terminal FITC-labeled antimicrobial peptides were added to a final concentration of 0.5 × MIC and1 × MIC, and incubated at 37°C for 30 min. The site of FITC-labeled antimicrobial peptide OM19r entering E. coli B2 was observed by confocal laser scanning microscope (FV1000).
2.15. Transmission electron microscopy
The intracellular changes in E. coli B2 cells induced by OM19r were determined by transmission electron microscope (TEM) according a previously described method (Türkez et al., 2019). The E. coli B2 treated with OM19r and Melittin (0.5 × MIC,1 × MIC) were streaked on slides and washed with PBS 3 times, fixed with 2.5% glutaraldehyde for 12 h and 1% osmium for 4 h, then gradient elution was performed with different concentrations (30, 50, 70, 90 and 100%) of acetone, embedded in epoxy resin polymer. Finally, the sections were coated and stained with 2% bisoxyl acetate and lead citrate, and the bacterial samples were observed by using E-1010 TEM (HITACHI JEOL, Tokyo, Japan).
2.16. Measurement of total reactive oxygen species
The accumulation of intracellular ROS was measured using DCFH-DA fluorescent dye (Maisuria et al., 2015). DCFH-DA was added to bacterial suspension (1 × 106 CFU/ml) with a final concentration of 10 μM and incubated at 37°C for 30 min. Then centrifuged at 1000 × g for 10 min, resuspended in PBS, and removed the fluorescent probe that did not enter the cells. The bacterial suspension containing DCFH-DA was mixed at 1:1 with the different concentrations of peptide mixture in a total volume of 200 μl and added to a 96-well dark plate. The fluorescence emission was measured by excitation and emission wavelength of 535 nm and of 485 nm, respectively. Hydrogen peroxide and PBS served as positive and negative control, respectively and three replicates were set for each group.
2.17. DNA binding assay
The genome of E. coli B2 was extracted using genome extraction kit according to manufacturer’s instructions (Shanghai Shangon Biotech Co., LTD., China), and the concentration was adjusted to 100 ng/μL. A 20 μl of peptides and antibiotics with different concentrations were combined with the genome were set in the reaction system. LRPG was used as positive control (Jia et al., 2020). After incubation at 37°C for 1 h, a 2 μl 10 × loading buffer was added and the effect of OM19r on DNA migration was analyzed by 1% agarose electrophoresis.
2.18. In vitro protein synthesis
In vitro protein synthesis was performed according to a previously described method (Garamella et al., 2016). myTXTL® Sigma 70 Master Mix Kit (Daicel Arbor Biosciences, United States) was used to determine the effect of GEN combined with OM19r on protein synthesis in vitro. The concentration of OM19r was 0.5 μg/ml and GEN was 0.05, 0.25, 0.5, 1, 2, 5 and 50 μg/ml.
2.19. In vitro transcription
In vitro transcription assays were performed according to a previously described method (Marras et al., 2004). Molecular beacons were purchased from ComateBio, which sequence was: 5′-6-FAM-CCGCGCATCTCGGTTGATTTCTTTTCCTCGGGC GCGG-Dabcyl-3′. The 5′ end was linked to a fluorescent dye (6-FAM), while the 3′–end was coupled to a quencher molecule (Dabcyl). The region which base-pairs with the target rRNA was indicated under the black solid line. Both sides of the end contained repeated GC bases, which were considered the stem of the Molecular beacon, made it quench normally. After mating with the target rRNA sequence, the fluorescence was emitted. OM19r was competitively bound to the target sequence, the molecular beacon configuration did not change and the fluorescence was quenched. The Components of experimental group were as follow: 5 × Buffer (80 μl), NTPs (8 μl), DNA template (4 μl), RNA polymerase (50 μl), RNA inhibitor (1 μl), different concentrations of OM19r (18 μl) and H2O replenishment to 200 μl. The Components of negative control group were as follow: 5 × Buffer(80 μl), NTPs (8 μl), RNA inhibitor (1 μl), different concentrations of OM19r (18 μl) and H2O and H2O replenishment to 200 μl. The Components of positive control group were as follow: 5 × Buffer(80 μl), NTPs (8 μl), 23SrRNA (2 μl), RNA polymerase (50 μl), RNA inhibitor (1 μl), different concentrations of OM19r (18 μl) and H2O and H2O replenishment to 200 μl. Each group was incubated at 37°C for 20 min. Subsequently, 100% ethanol and 3 M NaAc(pH-5.2) were added, followed by an ice bath for 30 min, and the precipitate was removed by centrifugation at 13523 × g for 10 min at 4°C. After addition of 80% ethanol, the precipitate was removed by centrifugation at 13523 × g for 10 min at 4°C. After vacuum drying for 30 min, a 2 μl molecular beacon was added. Subsequently, a 100 μl of Tris–HCL (pH = 8.8) was added to each well, followed by incubation at 95°C for 2 min and 45°C for 10 min. It was transferred to a 96-well dark plate, and the excitation and emission wavelength was 485 nm and 520 nm, respectively.
2.20. mRNA translation
E. coli S30 extract translation system(Brandi et al., 2007): The reaction mixture to test standard mRNA translation in bacterial extracts contains, in 30 μl of 10 mM Tris–HCl (pH 7.7): 7 mM Mg acetate, 100 mM NH4Cl, 2 mM DTT, 2 mM ATP, 0.4 mM GTP, 10 mM PEP, 0.025 mg/ml PK, 0.12 mM 10-formyl-terahydrofolate, 3 μg/μL tRNA (Escherichia coli BL21), an amino acid mixture containing 0.2 mM of all amino acids (with the exception of phenylalanine); 9 μM [3H] phenylalanine and 36 μM non-radioactive phenylalanine, an optimized amount of the S30 cell extract (generally 2–6 μl/reaction mixture) and 1–3 μM mRNA (pre-heated 5 min at 65°C). After 30–60 min incubation at 37°C, 20 μl aliquots from each reaction mixture are spotted on 3 mm paper disks which are dropped into 10% ice-cold TCA and processed according to the hot TCA procedure (Mans and Novelli, 1961).
Poly(U) hybrid translation system (Kaminishi et al., 2015): Translation was carried out in Poly(U) hybrid translation system. A 30 μl of buffer containing 3 mM phosphoenolpyruvate (PEP), 0.05 μg/ml pyruvate kinase (PK), 1 mM GTP, 0.15 μg/μL poly(U), 10 μM [3H]-Phe-tRNA and 0.2 μM of Escherichia coli 30S and 50S were added to the mix after a 5 min incubation at room temperature in the presence of increasing concentrations of OM19r. After 30 min at 37°C the level of poly(U) translation was quantified from the amount of acid-insoluble [3H] Phe-tRNA incorporated.
2.21. Animal infection model
Mouse peritonitis model: The sample size was selected according to the preliminary infection test (n = 8 per group). Some 6-week-old female mice were intraperitoneally injected with 0.5 ml of E. coli B2 (3 × 108 CFU/ml) to establish a mouse peritonitis model. Then after 1 h, different doses of OM19r (8 mg/kg), GEN (8 mg/kg) and OM19r combined with GEN (4 + 2 mg/kg) were injected intraperitoneally. The blank control mice were injected with the same dose of normal saline. Survival of mice treated and colony counts in each organ were recorded over 7 days.
G. mellonella infection model: G. mellonella larvae were randomly divided into 6 groups (n = 8 per group) and infected with 10 μl of E. coli B2 (2 × 106 CFU/ml) in the right hind ventral foot. Then after 1 h, different doses of OM19r (8 mg/kg), GEN (8 mg/kg) and OM19r combined with GEN (4 + 2 mg/kg) were injected intraperitoneally. The blank control mice were injected with the same dose of normal saline. Survival in response to treatment was recorded over 5 days.
2.22. Data analysis
All the experimental data were processed by SPSS software using an unpaired two-tailed Student’s t-test. Quantitative data were expressed as mean ± standard deviation, p-values was considered statistically significant as * p < 0.05, ** p < 0.01, *** p < 0.001, and ns represented non-significant.
3. Results
3.1. Antimicrobial activity and checkerboard assays
A total of Seven AMPs with good antibacterial activity E. coli were synthesized from the existing AMP sequences and evaluated for their efficacy on multiple aspects. OM19r in combination with gentamicin revealed the lowest FIC (FIC = 0.156; Supplementary Table S2) among seven experimental antibacterial peptides. Therefore, a proline-rich antimicrobial peptide OM19r was derived and synthesized (Supplementary Figure S1; Supplementary Table S3). The results of OM19r combined with other antibiotics acting on different sites showed that when combined with gentamicin, the synergistic antibacterial activity was the strongest, and it could restore the sensitivity of multi-drug resistant E. coli to gentamicin (Supplementary Table S4). The MIC of OM19r against some Gram-negative bacteria was 1-8 μg/ml (Table 1).
Table 1.
Minimum inhibitory concentrations (MICs) of OM19r against different bacteria.
| Bacterial Species | OM19r (μg/mL) |
|---|---|
| Gram-negative | |
| E. coli ATCC25922 | 1 |
| E. coli ATCC25922 (△SbmA) | >128 |
| E. coli k88 | 2 |
| S. enterica ATCC 14028 | 2 |
| S. typhimurium CMCC50115 | 2 |
| S. flexneri ATCC12022 | 2 |
| S. dysenteriae CMCC51252 | 2 |
| K. pneumoniae CMCC46117 | 4 |
| Gram-negative (Clinical isolate strains) | |
| E. coli SN5 | 1 |
| E. coli SN3 | 2 |
| E. coli Q12 | 4 |
| E. coli Q14 | 4 |
| E. coli Q45 | 4 |
| E. coli W23 | 4 |
| E. coli QY | 4 |
| E. coli B2(mcr-1) | 4 |
| S. typhimurium 1A | 4 |
| A. baumannii JS1 | 8 |
| S. dysenteriae A3 | 4 |
| S. flexneri RH | 8 |
| K. Pneumoniae JP20 | 4 |
| Gram-positive | |
| S. aureus ATCC25923 | >512 |
| S. aureus (MRSA) | >512 |
| S. faecalis ATCC29212 | >512 |
| B. subtilis ATCC63501 | >512 |
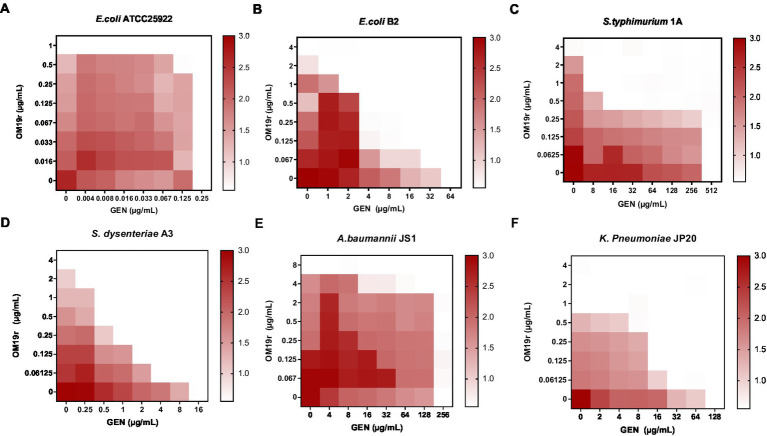
The antibacterial effect of OM19r combined with GEN against drug-resistant bacteria including E. coli B2, S. typhimurium 1A, A. baumannii JS1, S. dysenteriae A3, and K. pneumoniae JP20 (Supplementary Table S5), was determined using the microbroth dilution method and checkerboard assay. The results showed that OM19r combined with GEN had a synergistic antibacterial effect (FIC ≤ 0.5) against some Gram-negative bacteria (Figures 1A–F). When OM19r was used at a sub-minimum inhibitory concentration (2 μg/ml), the MIC of GEN against resistant E. coli B2 decreased from 64 to 1 μg/ml. The FIC of OM19r combined with kanamycin, streptomycin, tobramycin and spectinomycin against drug-resistant E. coli B2 was ≤0.5. These results indicate that OM19r combined with aminoglycoside produced synergistic antibacterial effect against drug-resistant E. coli B2 (Supplementary Table S6).
Figure 1.
OM19r enhances the antibacterial activity of GEN against some Gram-negative bacteria. (A–F) Checkerboard test results of OM19r combined with GEN against E. coli ATCC25922, E. coli B2, S. typhimurium 1A, A. baumannii JS1, S. dysenteriae A3, and K. pneumoniae JP20. The X-axis represents gentamicin 0-MIC concentration, and the Y-axis represents OM19r 0-MIC concentration.
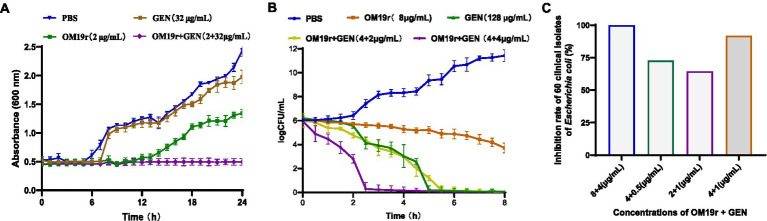
3.2. Growth curves and time-kill curves of bacteria
Growth curve analysis showed that OM19r combined with GEN (2 + 32 μg/ml) inhibited the growth of E. coli B2, indicating no antagonistic effect between OM19r combined with GEN against E. coli B2 (Figure 2A). The time-kill curve of OM19r combined with GEN (4 + 4 μg/ml) showed that E. coli B2 could be eliminated within 3 h in a dose dependent manner (Figure 2B). The combined antibacterial activity of OM19r and GEN against 60 other clinical isolates of E. coli was also determined. The result showed that OM19r at a low dose (2 μg/ml) combined with GEN (1 μg/ml) inhibited growth of 36 E. coli strains (60%) (Figure 2C), while OM19r at a high dose (4 μg/ml) combined with GEN (0.5 μg/ml) inhibited growth of 54 E. coli strains (90%) (Figure 2C).
Figure 2.
Antibacterial effect of OM19r combined with GEN against E. coli. (A) 24 h growth curves of OM19r (2 μg/ml) combined with GEN (32 μg/ml) against E. coli B2. (B) 8 h Time-kill curves of OM19r (8 μg/ml), GEN (128 egg/ml) and OM19r combined with GEN (4 + 2 μg/ml and 4 + 4 μg/ml) against E. coli B2. (C) Synergistic antibacterial activity of OM19r combined with GEN against 60 clinically isolated E. coli. Graphs show means from at least three biological replicates, and error bars indicate standard deviations.
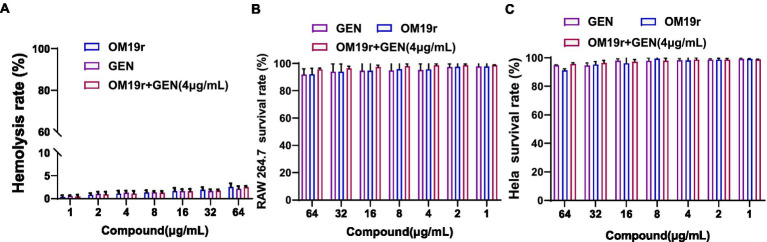
3.3. Hemolytic and cytotoxicity assays
The hemolysis rate of sheep red blood cells was <5% following treatment with different concentrations of antimicrobial peptide OM19r combined with GEN (Figure 3A). Alternatively, cytotoxicity assays showed that the survival rate of mouse macrophages (RAW264.7) and human cervical cancer cells (HeLa) exceeded 90% after treatment with different concentrations of OM19r combined with GEN (Figures 3B,C).
Figure 3.
Safety evaluation of OM19r combined with GEN at cell level. (A), Hemolysis of sheep red blood cells treated with OM19r combined with GEN. (B) Cytotoxicity of RAW264.7 cells treated with OM19r, GEN and OM19r combined with GEN. (C) Cytotoxicity of HeLa cells treated with OM19r, GEN and OM19r combined with GEN. Graphs show mean of three biological replicates, p-values were determined using an unpaired two-tailed Student’s t-test.
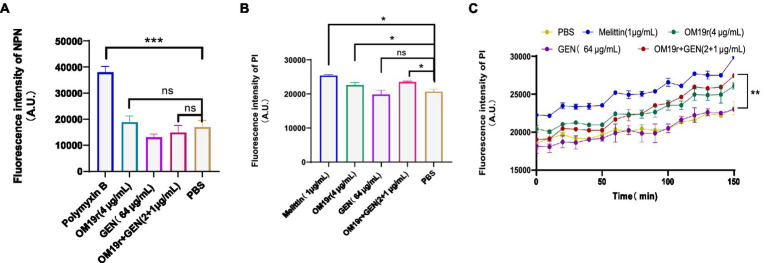
3.4. Membrane permeability
The results of outer membrane permeability assays showed that OM19r combined with GEN had no effect on the outer membrane permeability of E. coli B2 (Figure 4A; Supplementary Figure S2). Meanwhile, inner membrane permeability assays showed that OM19r combined with GEN (2 + 1 μg/ml) increased the inner membrane permeability of E. coli B2. There was no significant difference in membrane permeability between OM19r combined with GEN (2 + 1 μg/ml) group and OM19r(4 μg/ml) group (Figures 4B,C). These results indicate that OM19r affected the inner membrane integrity of E. coli B2, while GEN had no effect on the cell membrane.
Figure 4.
Effect of OM19r combined with GEN on membrane permeability of E. coli B2. (A) Outer membrane permeability of E. coli B2 was measured by NPN probe at 30 min. (B), Inner membrane permeability of E. coli B2 was measured by PI probe at 30 min. (C) Inner membrane permeability of E. coli B2 was measured by PI probe, fluorescence intensity was detected after every 10 min for 150 min. Graphs show mean of three biological replicates, p-values were determined using an unpaired two-tailed Student’s t-test.
3.5. Proton motive force assays
PMF assays results showed no significant differences in △φ and △pH of E. coli B2 in OM19r combined with GEN group compared with the negative control group (Supplementary Figures S3A,B).
3.6. Efflux pump assays
The fluorescent probe EtBr was used to detect the effects of drugs on the non-specific bacterial pumping system. OM19r combined with GEN did not inhibit the bacterial efflux pump (Supplementary Figures S4A,B).
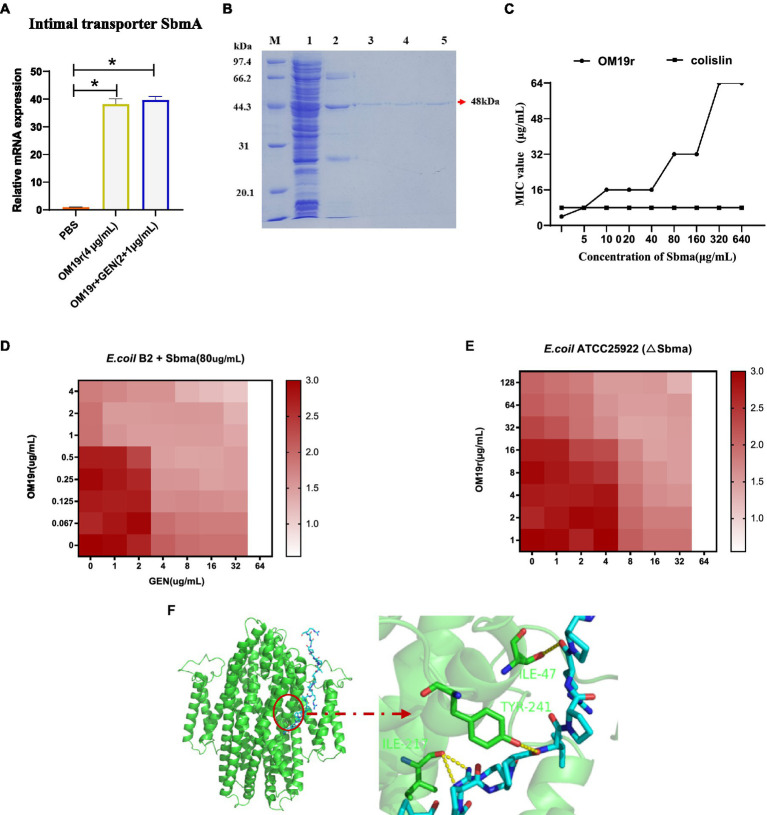
3.7. Real-time PCR and in vitro expression of SbmA
SbmA is a member of the peptide uptake permease family (PUP) and involved in transporting antimicrobial peptides (Armas et al., 2021). The results of qPCR revealed that OM19r combined with GEN upregulated (p < 0.05) the mRNA expression level of SbmA in E. coli B2 significantly (Figure 5A). But there was no significant difference in SbmA expression between OM19r combined with GEN (2 + 1 μg/ml) group and OM19r(4 μg/ml) group and indicated that SbmA plays a vital role in the entry OM19r to E. coli B2. Therefore, SbmA protein (48 kDa) was expressed and purified using E. coli expression system, Ni+ column and SDS-PAGE (Figure 5B). The effect of SbmA on the antibacterial activity of OM19r was determined in vitro. The results showed that the MIC of OM19r against E. coli B2 was increased as SbmA protein concentration increased (Figure 5C). In addition, SbmA protein also affected the synergistic antibacterial effect of OM19r combined with GEN against E. coli B2 (Figure 5D). The SbmA gene deletion in E. coli ATTCC25922 resulted in the loss of the bacteriostatic activity of OM19r(MIC>128 μg/ml) (Figure 5E). Molecular docking showed that OM19r had a good binding affinity with SbmA (binding energy was −262.92 kcal/mol), the binding sites of OM19r and SbmA transporter were ILE-47, ILE-217 and TYR-241 (Figure 5F).
Figure 5.
OM19r transported to cytoplasm by the SbmA protein on bacterial inner membrane. (A) The mRNA expression of SbmA gene increased after OM19r and OM19r combined with GEN against E. coli B2. (B) SDS-PAGE results of SbmA protein. M: Premixed Protein Marker(low), 1: flow through 2: 50 mM imidazole wash, 3: 100 mM imidazole wash, 4: 300 mM imidazole first wash, 5: 300 mM imidazole second wash. (C) Effect of SbmA on the antibacterial activity of OM19r and colistin against E. coli B2 was determined by adding SbmA protein in vitro. (D) Effect of SbmA on the FIC of OM19r combined with GEN against E. coli B2 was determined by adding SbmA protein in vitro. (E) FIC of OM19r combined with GEN against E.coli ATCC25922 (△SbmA) was determined in vitro. (F) Molecular docking results of both OM19r and SbmA protein binding.
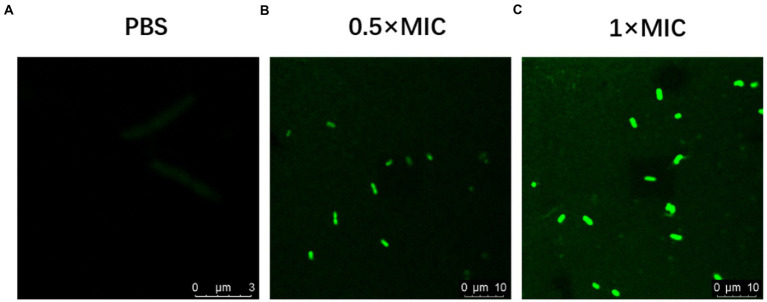
3.8. Confocal laser scanning microscopy
FITC-OM19r in the 0.5 × MIC treatment group displayed a small amount of green fluorescence in the cytoplasm of E. coli B2 after 30 min., On the other hand, FITC-OM19r in the 1 × MIC treatment group showed a large amount of green fluorescence in the cytoplasm of E. coli B2 (Figures 6A–C). Indicating the binding ability of the antimicrobial peptide OM19r to the cell membrane or accumulation in the cytoplasm.
Figure 6.
Confocal laser scanning micrograph of E. coli B2 treated with OM19r and FITC-OM19r. (A-C) Images obtained by fluorescence microscope.
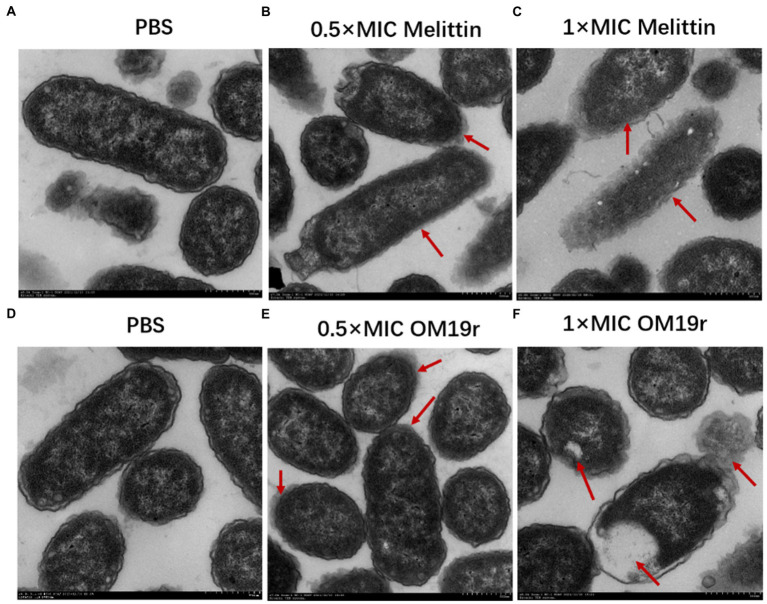
3.9. Transmission electron microscopy
The TEM results showed that the cell membrane of E. coli B2 was smooth, while the cell contents were full prior to OM19r treatment (Figures 7A,D). The membrane of E. coli B2 cells treated with Melittin was not clear and the contents were separated (Figures 7B,C). After one hour of OM19r treatment with Melittin at 0.5 × MIC, some contents were leaked and the edge of E. coli B2 membrane (Figure 7E). While E coli B2 treated for one hour with OM19r at 1 × MIC shown a complete leakage of the contents (Figure 7F). This suggests that OM19r entered bacteria and exerted its antibacterial effect.
Figure 7.
Transmission electron microscopy results of E. coli B2 treated with OM19r. (A) Transmission electron microscopic images of E. coli B2. (B) Transmission electron microscopic images of E coli B2 treated with Melittin (0.5 × MIC) for 1 h. (C) Transmission electron microscopic images of E coli B2 treated with Melittin (1 × MIC) for 1 h. (D) Transmission electron microscopic images of E. coli B2. E,Transmission electron microscopic images of E coli B2 treated with OM19r (0.5 × MIC) for 1 h. D, Transmission electron microscopic images of E coli B2 treated with OM19r(1 × MIC) for 1 h. Red arrows in B, C, and E points to the cell membrane;The red arrow in F points to intracellular vacuolization.
3.10. Measurement of total reactive oxygen species
DCFH-DA fluorescent probe was used to determine the intracellular ROS accumulation in E. coli B2 following OM19r combined with GEN treatment. The intracellular ROS accumulation of E. coli B2 was significantly increased (p < 0.01) in OM19r and OM19r combined with GEN treated groups compared with the control group (Figure 8). This suggests that OM19r treatment accumulated intracellular ROS, but gentamicin alone had no effect.
Figure 8.
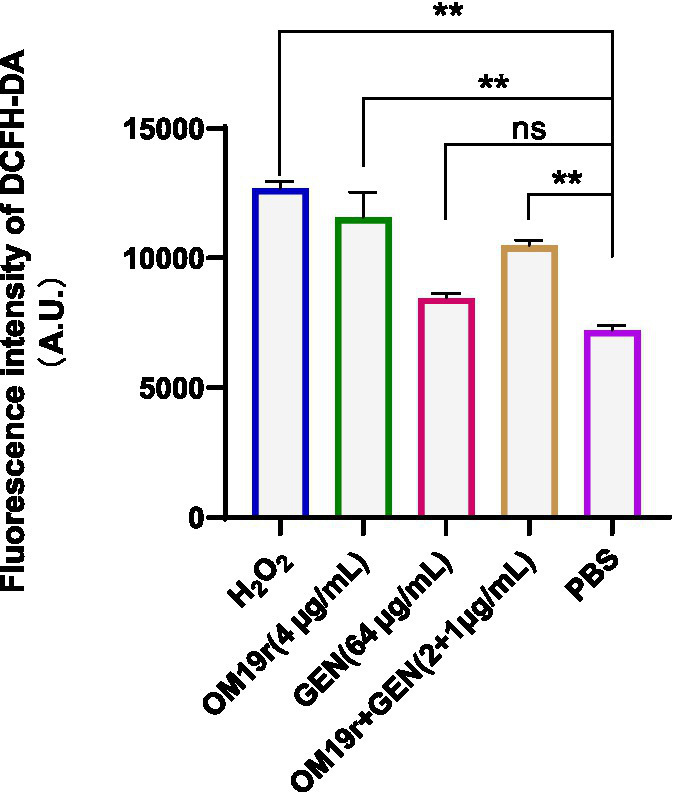
ROS accumulation in E. coli B2 cells determined by DCFH-DA fluorescent probe. Graphs show mean of three biological replicates, p-values were determined using an unpaired two-tailed Student’s t-test.
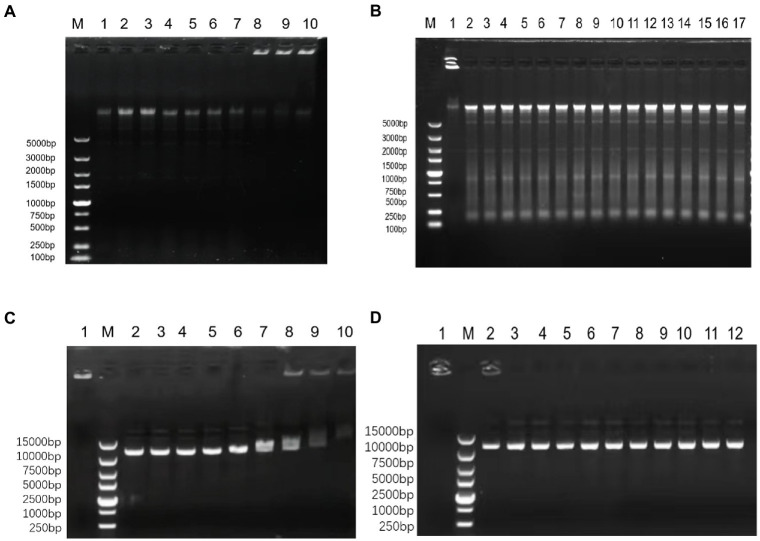
3.11. DNA binding assay
OM19r (≥128 μg/ml) bound to E. coli B2 genomic DNA (Figures 9A–B), and OM19r (≥64 μg/ml) attached to the plasmid DNA (Figures 9C,D). However, OM19r combined with GEN (8 + 4 μg/ml) had no binding effect on E. coli B2 genomic DNA and plasmid DNA. These results indicated that the bacterial genomic DNA was not the target for OM19r combined with GEN.
Figure 9.
The effect of OM19r combined with GEN on the DNA of E. coli determined by DNA binding assay. (A) The effect of different concentrations of OM19r on the genomic DNA of E. coli B2, M: DL5000Marker; 1: E. coli B2 genome; 2: OM19r (2 μg/ml); 3: OM19r (4 μg/ml); 4: OM19r (8 μg/ml); 5: OM19r (16 μg/ml); 6: OM19r (32 μg/ml); 7: OM19r (64 μg/ml); 8: OM19r (128 μg/ml); 9:OM19r (256 μg/ml); 10: Positive control. (B) Effects of different concentrations of GEN and OM19r combined with GEN on the genomic DNA of E. coli B2. M: DL5000Marker; 1: Positive control 2: GEN (512 μg/ml); 3: GEN (256 μg/ml); 4: GEN (128 μg/ml); 5: GEN (64 μg/ml); 6: GEN (32 μg/ml); 7: GEN (16 μg/ml); 8: GEN (8 μg/ml); 9: GEN (4 μg/ml); 10: GEN (2 μg/ml); 11: GEN (1 μg/ml); 12: OM19r combined with GEN (2 + 1 μg/ml); 13: OM19r combined with GEN (2 + 2 μg/ml); 14: OM19r combined with GEN (4 + 2 μg/ml); 15: OM19r combined with GEN (4 + 4 μg/ml);16: OM19r combined with GEN (8 + 4 μg/ml);17:E. coli B2 genome. (C) The effects of different concentrations of OM19r on plasmid, 1: Positive control; M: 15000 DNA Marker; 2: plasmid; 3: OM19r (2 μg/ml); 4: OM19r (4 μg/ml); 5: OM19r (8 μg/ml); 6: OM19r (16 μg/ml); 7: OM19r (32 μg/ml);8: OM19r (64 μg/ml); 9: OM19r (128 μg/ml); 10: OM19r (256 μg/ml); (D) Effects of different concentrations of GEN and OM19r combined with GEN on plasmid. 1: Positive control; M: 15000 DNA Marker; 2: GEN (512 μg/ml); 3: GEN (256 μg/ml);4: GEN (128 μg/ml); 5: GEN (64 μg/ml); 6: GEN (32 μg/ml); 7: OM19r combined with GEN (2 + 1 μg/ml); 8: OM19r combined with GEN (2 + 2 μg/ml); 9: OM19r combined with GEN (4 + 2 μg/ml); 10: OM19r combined with GEN (4 + 4 μg/ml);11: OM19r combined with GEN (8 + 4 μg/ml); 12: plasmid.
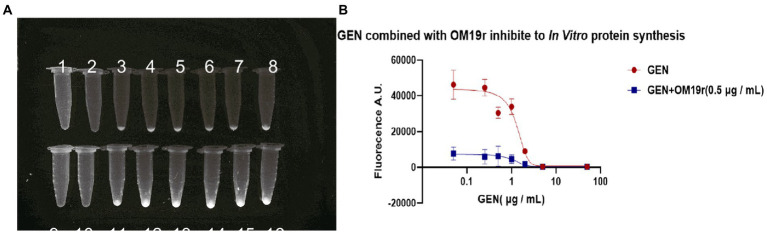
3.12. In vitro protein synthesis
EGFP green fluorescence was observed under UV light after the reaction of GEN and OM19r with the mixture at 29°C for 16 h. The results showed that the fluorescence intensity of EGFP decreased with the increase in GEN concentration (Figure 10A). These results suggested that GEN can inhibit EGFP expression in vitro. However, when OM19r (0.5 μg/ml) was added to each group, the fluorescence value of EGFP decreased significantly more than that GEN alone (Figure 10B). The above results indicated that OM19r alone could affect the synthesis of EGFP fluorescent protein.
Figure 10.
The effect of OM19r combined with GEN on in vitro protein synthesis. (A) EGFP green fluorescence observed under UV light by different concentrations of GEN and OM19r(0.5 μg/ml) combined with GEN for 16 h. 1: OM19r combined with GEN (50 μg/ml), 2: OM19r combined with GEN (5 μg/ml), 3: OM19r combined with GEN (2 μg/ml), 4: OM19r combined with GEN (1 μg/ml), 5: OM19r combined with GEN (0.5 μg/ml), 6: OM19r combined with GEN (0.25 μg/ml), 7: OM19r combined with GEN(0.05 μg/ml), 8: OM19r (0.5 μg/ml), 9: GEN (50 μg/ml), 10: GEN (5 μg/ml), 11: GEN (2 μg/ml), 12: GEN (1 μg/ml), 13: GEN (0.5 μg/ml), 14: GEN (0,25 μg/ml), 15: GEN (0.05 μg/ml), 16: negative control. (B) The fluorescence value of EGFP induced by the GEN and OM19r (0.5 μg/ml). Graphs show mean of three biological replicates.
3.13. In vitro transcription
The effect of OM19r on bacterial transcription was detected in vitro. The molecular beacon fluorescence signal of E. coli transcribed in vitro remained unchanged following treatment with OM19r, suggesting that OM19r had no effect on the transcription of E. coli in vitro (Figure 11).
Figure 11.
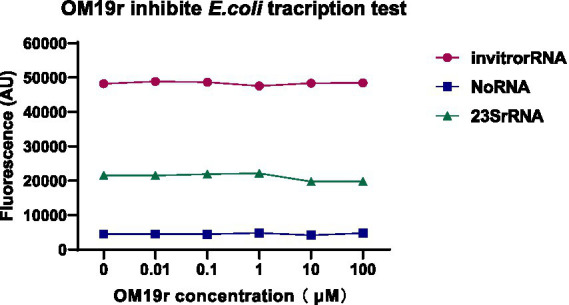
The effect of OM19r on the transcription process of E. coli determined by constructing the transcription system in vitro. Graphs show mean of three biological replicates.
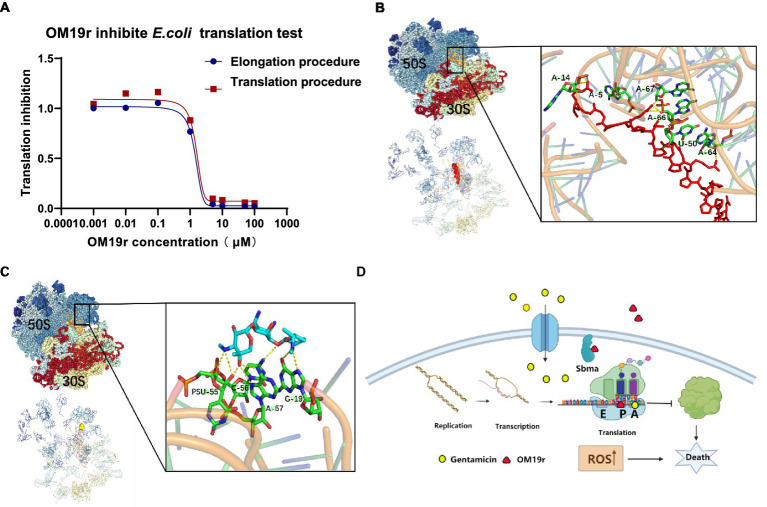
3.14. mRNA translation
The dose–response curves for OM19r showed the inhibition of mRNA translation and Poly (U) formation overlap (Figure 12A), suggesting that inactivation of peptidyl transferase center (PTC) was caused by OM19r blocking mRNA translation. OM19r appeared to be an inhibitor of PTC activity, effectively blocking PTC specifically from producing the “first peptide bond” and mRNA translation. This may explain why the dose–response curves for inhibition of mRNA translation and Poly (U) formation were essentially overlapped. The molecular docking revealed that OM19r and GEN bind to ribosomes at different sites. The binding energy of OM19r to the ribosome was −115.33 kcal/mol (Supplementary Table S7). OM19r was predicted to bind to the ribosome’s A-5, A-14, A-64, A-66, A-67 and U-50 sites (Figure 12B). The binding energy of GEN to the ribosome was −140.8 kcal/mol (Supplementary Table S7). GEN was predicted to bind to the ribosome’s C-56, A-57, and G-19 sites (Figure 12C), proposing that OM19r entered the bacteria via SbmA and influenced the translation elongation process and ROS accumulation (Figure 12D).
Figure 12.
Translation elongation process of OM19r-targeted mRNA. (A) Dose–response of OM19r inhibition mRNA translation (red) and fMet-Phe dipeptide formation (blue) in vitro. (B) Molecular docking results of OM19r and ribosome. (C) Molecular docking results of GEN and ribosome. (D) Schematic diagram of synergistic antibacterial action mechanism of OM19r combined with GEN.
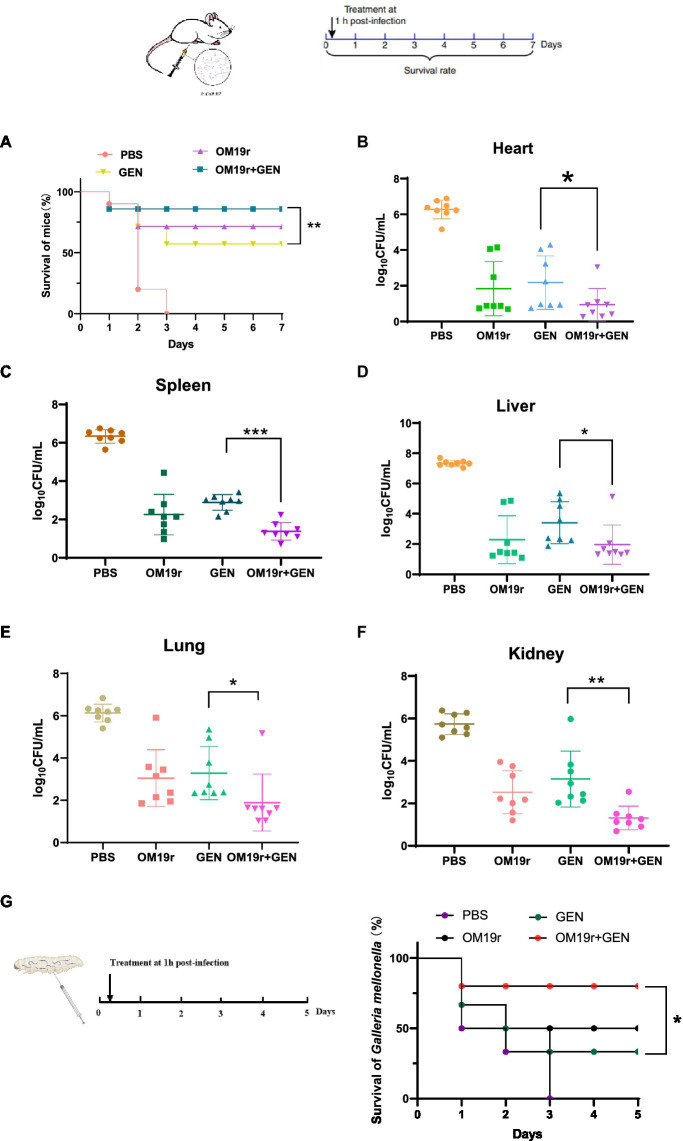
3.15. Animal infection model
OM19r restored susceptibility of drug-resistant strains to GEN, which was assessed in animal models infected with E. coli B2 (Figure 13). In mouse model of peritonitis, 80% of mice survived within 7 days of treatment with OM19r combined with GEN (4 + 2 mg/kg), significantly higher(p < 0.01) than the GEN (8 mg/kg) treated group (Figure 13A). The bacterial load test results showed that OM19r combined with GEN (4 + 2 mg/kg) significantly reduced the bacterial content in mice. The bacterial content in the liver, spleen, lung, and kidney was reduced by five-, four-, four-, and five-fold, respectively (Figures 13B–F). In Galleria mellonella (G. mellonella) larval model, 80% of G. mellonella survived within 7 days of treatment with OM19r combined with GEN (4 + 2 mg/kg), significantly higher (p < 0.01) than the GEN (8 mg/kg, 25% survival) treated group (Figure 13G). These findings suggested that LRGG is an antibiotic potentiator to enhance the antibacterial activity of GEN against E. coil B2 in vivo.
Figure 13.
OM19r rescues GEN activity in two animal models of infection. (A) Survival curve of mice infected with non-lethal dose of E. coli B2 (3 × 108 cfu/ml) for 7 days. p-values determined using a two-sided log[rank] (Mantel-Cox) test. (B–F) Bacterial load in all organs (heart, spleen, liver, lung and kidney) of mice measured after OM19r combined with GEN treatment. G, Survival curve of G. mellonella larvae infected with a non-lethal dose of E. coli B2 (1× 105 cfu/ml) for 5 days. p-values determined using a two-sided log[rank] (Mantel-Cox) test.
4. Discussion
Antibacterial synergists when used in combination with antibiotics could restore the sensitivity of drug-resistant pathogens up to some extent and prolong the effective life of antibiotics by reducing the resistance. Although there have been reports that AMPs used as synergists can enhance the antibacterial activity of antibiotics (Randhawa et al., 2016; Zhong et al., 2020), studies on antimicrobial peptides combined with gentamicin to reverse drug resistance in E. coli are lacking. Most AMPs contain cationic and amphiphilic molecules, which may be drawn to negatively charged cell membranes and damage the lipid bilayer (Shai, 2002). Therefore, AMPs usually exhibit low selectivity and broad-spectrum antibacterial activity against pathogenic bacteria (Matsuzaki, 2009). However, the antibacterial targets of some proline-rich antimicrobial peptides (PrAMPs) are often located inside the bacterial cells and do not cause obvious membrane damage (Nicolas, 2009; Scocchi et al., 2011, 2016; Graf et al., 2017). An antimicrobial peptide OM19r containing both proline and arginine was derived from the available peptide sequences in our laboratory. Compared with the broad-spectrum antimicrobial peptides, OM19r had a narrow antibacterial spectrum and antibacterial activity only against some Gram-negative bacteria. Meanwhile, OM19r had no combined antibacterial effect with other antibiotics at different action sites (FIC > 0.5), and OM19r reduced the MIC of GEN against multidrug-resistant E. coli B2 from 64 to 1 μg/ml. Some disadvantages of AMPs, such as hemolysis, cytotoxicity, and instability reported by other scientists hinder their clinical application (Kumar et al., 2017). In this study, the cytotoxicity and hemolytic activity of OM19r was measured in vitro. The results of safety assessment experiments at the cellular level indicated that OM19r had no hemolytic activity and cytotoxicity at OM19r ≤ 64 μg/ml. The results of animal infection model showed that OM19r restored GEN sensitivity of E. coli B2 in vivo. In conclusion, OM19r acted as a potential aminoglycoside antibiotic synergist.
The antibacterial mechanism of antimicrobial peptides involves membranous and non-membranous targeting. The antibacterial mechanism of OM19r on bacterial cell membranes was investigated via membrane permeability assay. The inner membrane permeability assays results showed that OM19r increased the inner membrane permeability of E. coli B2. SbmA as a member of the peptide uptake permease family (PUP) was located in some Gram-negative bacteria and played an important role in antibiotic transport (Paulsen et al., 2016). In previous studies, Oncocin was shown to enter the cell by the SbmA transporter. OM19r is an antimicrobial peptide heterozygous by Oncocin with the MDAP-2 fragment (Liu L. et al., 2020). So, the effect of OM19r on the bacterial inner membrane transporter SbmA protein was further determined by qPCR assays, SmbA protein was added in vitro and gene deletion strains were constrcuted. The qPCR results showed that mRNAs expression of SbmA was up-regulated following treatment with OM19r or OM19r combined with GEN. And the MIC of OM19r against E. coli B2 increased by adding purified SbmA protein in vitro. Meanwhile, SbmA gene deletion in E. coli ATTCC25922 resulted in the loss of the bacteriostatic activity of OM19r. It was further demonstrated that OM19r could enter the bacteria through membrane transporter SbmA. Molecular docking results showed that OM19r could bound to the ILE-47, ILE-217 and TYR-241 sites of SbmA. Finally, the CLSM and TEM results showed that OM19r entered E. coli B2 resulted in the release of the bacterial contents. In summary, OM19r was transported into E. coli B2 via SbmA to exert the bacteriostatic mechanism.
For non-membrane targeted antimicrobial peptides, current research on the mechanism has mainly focused on the effect of antimicrobial peptides on bacterial DNA (Bai et al., 2021) and protein levels (Roy et al., 2015). Therefore, the antibacterial mechanism of OM19r combined with GEN was studied in this study using DNA binding assay and cell-free expression assay. DNA binding test showed that OM19r could bind to E. coli genome at 128 μg/ml and E. coli plasmid at 64 μg/ml. GEN can bind to E. coli plasmid at concentration of 512 μg/ml. This indicate that high concentrations of OM19r and GEN (higher than the MIC) have binding effect on DNA. Therefore, OM19r combined with GEN mainly exerted antibacterial effects via mechanisms other than bacterial DNA. The effects of GEN and OM19r combined with GEN on protein synthesis were measured by fluorescence intensity of EGFP protein. GEN alone or OM19r combined with GEN affected the EGFP protein expression in vitro. Moreover, OM19r combined with GEN had a greater effect on the expression of EGFP protein than GEN alone. GEN influences the initial phase of bacterial translation, leading to mRNA misreading, and might explain the effect of GEN on EGFP protein expression in vitro. In summary, these results suggest that OM19r had no effect on DNA, but can affect protein synthesis.
Transcription and translation are important stages of protein synthesis in cells (Marras et al., 2004). The stage at which a drug acts on bacterial transcription and translation can be measured by constructing transcriptional translation systems in vitro (Marras et al., 2004; Brandi et al., 2007). Therefore, a transcription and translation systems were established to determine the specific inhibitory mechanism of OM19r on protein synthesis in vitro. Firstly, we proved by in vitro transcription experiments that the peptide has no effect on the transcription process. Secondly, since the in vitro protein synthesis could be inhibited by OM19r, but having no effect on RNA synthesis. Therefore, we focused on the protein translation process in vitro. A series of experiments was conducted and the results showed that OM19r had inhibition activity on the translation elongation process. Finally, molecular docking results further demonstrated that OM19r and GEN binds to different sites in the ribosome, with OM19r inhibiting the extension phase of protein translation and GEN inhibiting the initiation phase of translation.
Meanwhile, ROS has been reported to play a key role in bacterial resistance and the bactericidal activity of antibiotics (Van Acker and Coenye, 2017; Hong et al., 2019). Non-lethal concentrations of antibiotics cause ROS produced by bacteria, which can promote the SOS-DNA damage repair system, activate the stress protection mechanism, and form drug-resistant bacteria. Conversely, ROS produced by bacteria with lethal concentration of antibiotics can further accelerate the death of pathogens (Kohanski et al., 2010). The OM19r combined with GEN caused ROS accumulation in E. coli B2. In conclusion, OM19r can enter the cell through the intimal transporter SbmA, causing ROS accumulation in bacteria and inhibiting the translation extension stage of the protein, and ultimately causing death of the pathogen.
5. Conclusion
This study showed that OM19r combined with GEN had a strong synergistic inhibitory effect (FIC = 0.156) against multi-drug resistant E. coli B2. The combined inhibitory mechanism of OM19r and GEN shown that OM19r entered bacterial cells through SbmA transporter on cell membranes, thus causing intracellular ROS accumulation. Simultaneously, OM19r and GEN inhibited translation elongation and initiation, respectively, and ultimately affected the normal protein synthesis of bacteria. Two animal models shown that OM19r can also restore the sensitivity of multidrug-resistant E. coli B2 to GEN in animals. These findings offer a potential treatment option for the infections caused globally by multidrug-resistant E. coli.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary material, further inquiries can be directed to the corresponding authors.
Ethics statement
The animal study was reviewed and approved by Jilin Agricultural University guidelines.
Author contributions
H-XM, L-CK, and C-GH conceived and designed research. Q-JX, H-DY, YL, AG, AF, and X-YJ conducted experiments. H-PZ and P-HL contributed new reagents or analytical tools. Q-JX analyzed data. QC wrote the manuscript. All authors read and approved the manuscript.
Funding
This study was funded by National Natural Science Foundation of China (31872519) and Jilin Scientific and Technological Development Program (20210202033NC, 20220508049RC, YDZJ202203CGZH050, 20230402037GH).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We are grateful to China Agricultural University for multi-drug resistant E. coli B2.
Footnotes
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2023.1144946/full#supplementary-material
References
- Armas F., di Stasi A., Mardirossian M., Romani A. A., Benincasa M., Scocchi M. (2021). Effects of Lipidation on a proline-rich antibacterial peptide. Int. J. Mol. Sci. 22:7959. doi: 10.3390/ijms22157959, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aslam B., Wang W., Arshad M. I., Khurshid M., Muzammil S., Rasool M. H., et al. (2018). Antibiotic resistance: a rundown of a global crisis. Infect. Drug Resist. 11, 1645–1658. doi: 10.2147/IDR.S173867, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai S., Wang J., Yang K., Zhou C., Xu Y., Song J., et al. (2021). A polymeric approach toward resistance-resistant antimicrobial agent with dual-selective mechanisms of action. Sci. Adv. 7:eabc9917. doi: 10.1126/sciadv.abc9917, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Böttger R., Knappe D., Hoffmann R. (2016). Readily adaptable release kinetics of prodrugs using protease-dependent reversible PEGylation. J. Control. Release 230, 88–94. doi: 10.1016/j.jconrel.2016.04.010 [DOI] [PubMed] [Google Scholar]
- Brandi L., Fabbretti A., Milon P., Carotti M., Pon C. L., Gualerzi C. O. (2007). Methods for identifying compounds that specifically target. Methods Enzymol. 431, 229–267. doi: 10.1016/S0076-6879(07)31012-4 [DOI] [PubMed] [Google Scholar]
- Bustin S. A., Benes V., Garson J. A., Hellemans J., Huggett J., Kubista M., et al. (2009). The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 55, 611–622. doi: 10.1373/clinchem.2008.112797 [DOI] [PubMed] [Google Scholar]
- Corbalan N., Runti G., Adler C., Covaceuszach S., Ford R. C., Lamba D., et al. (2013). Functional and structural study of the dimeric inner membrane protein SbmA. J. Bacteriol. 195, 5352–5361. doi: 10.1128/JB.00824-13, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui Q., Qi-Jun X., Lei L., Li-Li G., Xiu-Yun J., Muhammad I., et al. (2021). Preparation, characterization and pharmacokinetic study of N-terminal PEGylated D-form antimicrobial peptide OM19r-8. J. Pharm. Sci. 110, 1111–1119. doi: 10.1016/j.xphs.2020.10.048, PMID: [DOI] [PubMed] [Google Scholar]
- Ding X., Yang C., Moreira W., Yuan P., Periaswamy B., Sessions P. F., et al. (2020). A macromolecule reversing antibiotic resistance phenotype and repurposing drugs as potent antibiotics. Adv. Sci. 7:2001374. doi: 10.1002/advs.202001374, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durand-Reville T. F., Miller A. A., O'Donnell J. P., Wu X., Sylvester M. A., Guler S., et al. (2021). Rational design of a new antibiotic class for drug-resistant infections. Nature 597, 698–702. doi: 10.1038/s41586-021-03899-0, PMID: [DOI] [PubMed] [Google Scholar]
- Falciani C., Zevolini F., Brunetti J., Riolo G., Gracia R., Marradi M., et al. (2020). Antimicrobial peptide-loaded nanoparticles as inhalation therapy for Pseudomonas aeruginosa infections. Int. J. Nanomedicine 15, 1117–1128. doi: 10.2147/IJN.S218966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garamella J., Marshall R., Rustad M., Noireaux V. (2016). The all E. coli TX-TL toolbox 2.0: a platform for cell-free synthetic biology. ACS Synth. Biol. 5, 344–355. doi: 10.1021/acssynbio.5b00296, PMID: [DOI] [PubMed] [Google Scholar]
- Graf M., Mardirossian M., Nguyen F., Seefeldt A. C., Guichard G., Scocchi M., et al. (2017). Proline-rich antimicrobial peptides targeting protein synthesis. Nat. Prod. Rep. 34, 702–711. doi: 10.1039/c7np00020k, PMID: [DOI] [PubMed] [Google Scholar]
- Hong Y., Zeng J., Wang X., Drlica K., Zhao X. (2019). Post-stress bacterial cell death mediated by reactive oxygen species. Proc. Natl. Acad. Sci. U. S. A. 116, 10064–10071. doi: 10.1073/pnas.1901730116, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooban B., Fitzhenry K., O'Connor L., Miliotis G., Joyce A., Chueiri A., et al. (2022). A longitudinal survey of antibiotic-resistant Enterobacterales in the Irish environment, 2019-2020. Sci. Total Environ. 828:154488. doi: 10.1016/j.scitotenv.2022.154488, PMID: [DOI] [PubMed] [Google Scholar]
- Jeamsripong S., Li X., Aly S. S., Su Z., Pereira R. V., Atwill E. R. (2021). Antibiotic resistance genes and associated phenotypes in Escherichia coli and Enterococcus from cattle at different production stages on a dairy farm in Central California. Antibiotics (Basel) 10:1042. doi: 10.3390/antibiotics10091042, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia B. Y., Wang Y. M., Zhang Y., Wang Z., Wang X., Muhammad I., et al. (2020). High cell selectivity and bactericidal mechanism of symmetric peptides centered on d-pro-Gly pairs. Int. J. Mol. Sci. 21:1140. doi: 10.3390/ijms21031140, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaminishi T., Schedlbauer A., Fabbretti A., Brandi L., Ochoa-Lizarralde B., He C. G., et al. (2015). Crystal lographic characterization of the ribosomal binding site and molecular mechanism of action of Hygromycin a. Nucleic Acids Res. 43, 10015–10025. doi: 10.1093/nar/gkv975, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohanski M., DePristo M. A., Collins J. J. (2010). Sublethal antibiotic treatment leads to multidrug resistance via radical-induced mutagenesis. Mol. Cell 37, 311–320. doi: 10.1016/j.molcel.2010.01.003, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar A., Mahajan M., Awasthi B., Tandon A., Harioudh M. K., Shree S., et al. (2017). Piscidin-1-analogs with double L- and D-lysine residues exhibited different conformations in lipopolysaccharide but comparable anti-endotoxin activities. Sci. Rep. 7:39925. doi: 10.1038/srep39925, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Jia Y., Yang K., Li R., Xiao X., Zhu K., et al. (2020). Metformin restores Tetracyclines susceptibility against multidrug resistant bacteria. Adv. Sci. (Weinh) 7:1902227. doi: 10.1002/advs.201902227, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L., Liu J., Cui Q., Jia B. Y., Ma H. X. (2020). Design and characterization of a novel hybrid antimicrobial peptide om19r based on oncocin and mdap-2. Int. J. Pept. Res. Ther. 26, 1839–1846. doi: 10.1007/s10989-019-09984-3 [DOI] [Google Scholar]
- Ma L., Xie X., Liu H., Huang Y., Wu H., Jiang M., et al. (2020). Potent antibacterial activity of MSI-1 derived from the magainin 2 peptide against drug-resistant bacteria. Theranostics 10, 1373–1390. doi: 10.7150/thno.39157, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mans R. J., Novelli G. D. (1961). Measurement of the incorporation of radioactive amino acids into protein by a filter-paper disk method. Archives of Biochemistry and Biophysics. 94, 48–53. doi: 10.1016/0003-9861(61)90009-1 [DOI] [Google Scholar]
- Maisuria V. B., Hosseinidoust Z., Tufenkji N. (2015). Polyphenolic extract from maple syrup potentiates antibiotic susceptibility and reduces biofilm formation of pathogenic bacteria. Appl. Environ. Microbiol. 81, 3782–3792. doi: 10.1128/AEM.00239-15, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marras S. A., Gold B., Kramer F. R., Smith I., Tyagi S. (2004). Real-time measurement of in vitro transcription. Nucleic Acids Res. 32:e72. doi: 10.1093/nar/gnh068, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuzaki K. (2009). Control of cell selectivity of antimicrobial peptides. Biochim. Biophys. Acta 1788, 1687–1692. doi: 10.1016/j.bbamem.2008.09.013 [DOI] [PubMed] [Google Scholar]
- Mattiuzzo M., Bandiera A., Gennaro R., Benincasa M., Pacor S., Antcheva N., et al. (2010). Role of the Escherichia coli SbmA in the antimicrobial activity of proline-rich peptides. Mol. Microbiol. 66, 151–163. doi: 10.1111/j.1365-2958.2007.05903.x, PMID: [DOI] [PubMed] [Google Scholar]
- Miao X., Zhou T., Zhang J., Xu J., Guo X., Hu H., et al. (2020). Enhanced cell selectivity of hybrid peptides with potential antimicrobial activity and immunomodulatory effect. Biochim. Biophys. Acta Gen. Subj. 1864:129532. doi: 10.1016/j.bbagen.2020.129532, PMID: [DOI] [PubMed] [Google Scholar]
- Mwangi J., Yin Y., Wang G., Yang M., Li Y., Zhang Z., et al. (2019). The antimicrobial peptide ZY4 combats multidrug-resistant Pseudomonas aeruginosa and Acinetobacter baumannii infection. Proc. Natl. Acad. Sci. U. S. A. 116, 26516–26522. doi: 10.1073/pnas.1909585117, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicolas P. (2009). Multifunctional host defense peptides: intracellular-targeting antimicrobial peptides. FEBS J. 276, 6483–6496. doi: 10.1111/j.1742-4658.2009.07359.x, PMID: [DOI] [PubMed] [Google Scholar]
- Paulsen V. S., Mardirossian M., Blencke H. M., Benincasa M., Runti G., Nepa M., et al. (2016). Inner membrane proteins YgdD and SbmA are required for the complete susceptibility of E. coli to the proline-rich antimicrobial peptide arasin 1(1-25). Microbiology 162, 601–609. doi: 10.1099/mic.0.000249 [DOI] [PubMed] [Google Scholar]
- Peng T., Huiyang F., Xi M. (2021). Design, optimization, and nanotechnology of antimicrobial peptides: from exploration to applications. Nano Today 39:101229. doi: 10.1016/j.nantod.2021.101229 [DOI] [Google Scholar]
- Randhawa H. K., Gautam A., Sharma M., Bhatia R., Varshney G. C., Raghava G. P., et al. (2016). Cell-penetrating peptide and antibiotic combination therapy: a potential alternative to combat drug resistance in methicillin-resistant Staphylococcus aureus. Appl. Microbiol. Biotechnol. 100, 4073–4083. doi: 10.1007/s00253-016-7329-7, PMID: [DOI] [PubMed] [Google Scholar]
- Roy R. N., Lomakin I. B., Gagnon M. G., Steitz T. A. (2015). The mechanism of inhibition of protein synthesis by the proline-rich peptide oncocin. Nat. Struct. Mol. Biol. 22, 466–469. doi: 10.1038/nsmb.3031, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scocchi M., Mardirossian M., Runti G., Benincasa M. (2016). Non-membrane Permeabilizing modes of action of antimicrobial peptides on bacteria. Curr. Top. Med. Chem. 16, 76–88. doi: 10.2174/1568026615666150703121009, PMID: [DOI] [PubMed] [Google Scholar]
- Scocchi M., Tossi A., Gennaro R. (2011). Proline-rich antimicrobial peptides: converging to a non-lytic mechanism of action. Cell. Mol. Life Sci. 68, 2317–2330. doi: 10.1007/s00018-011-0721-7, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shai Y. (2002). Mode of action of membrane active antimicrobial peptides. Biopolymers 66, 236–248. doi: 10.1002/bip.10260 [DOI] [PubMed] [Google Scholar]
- Si Z., Lim H. W., Tay M. Y. F., Du Y., Ruan L., Qiu H., et al. (2020). A glycosylated cationic block poly(β-peptide) reverses intrinsic antibiotic resistance in all ESKAPE gram-negative bacteria. Angew. Chem. Int. Ed. Engl. 59, 6819–6826. doi: 10.1002/anie.201914304, PMID: [DOI] [PubMed] [Google Scholar]
- Song M., Liu Y., Huang X., Ding S., Wang Y., Shen J., et al. (2020). A broad-spectrum antibiotic adjuvant reverses multidrug-resistant gram-negative pathogens. Nat. Microbiol. 5, 1040–1050. doi: 10.1038/s41564-020-0723-z [DOI] [PubMed] [Google Scholar]
- Song M., Liu Y., Li T., Liu X., Hao Z., Ding S., et al. (2021). Plant natural flavonoids against multidrug resistant pathogens. Adv. Sci. 8:e2100749: 2100749. doi: 10.1002/advs.202100749, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stokes J. M., MacNair C. R., Ilyas B., French S., Côté J. P., Bouwman C., et al. (2017). Pentamidine sensitizes gram-negative pathogens to antibiotics and overcomes acquired colistin resistance. Nat. Microbiol. 2:17028. doi: 10.1038/nmicrobiol.2017.28, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturge C. R., Felder-Scott C. F., Pifer R., Pybus C., Jain R., Geller B. L., et al. (2019). AcrAB-TolC inhibition by peptide-conjugated Phosphorodiamidate Morpholino oligomers restores antibiotic activity in vitro and in vivo. ACS Infect. Dis. 5, 1446–1455. doi: 10.1021/acsinfecdis.9b00123, PMID: [DOI] [PubMed] [Google Scholar]
- Türkez H., Enes A. M., Erdal S., Fatime G., Metin A., Abdulgani T. (2019). Microarray assisted toxicological investigations of boron carbide nanoparticles on human primary alveolar epithelial cells. Chem. Biol. Interact. 300, 131–137. doi: 10.1016/j.cbi.2019.01.021, PMID: [DOI] [PubMed] [Google Scholar]
- Van Acker H., Coenye T. (2017). The role of reactive oxygen species in antibiotic-mediated killing of bacteria. Trends Microbiol. 25, 456–466. doi: 10.1016/j.tim.2016.12.008, PMID: [DOI] [PubMed] [Google Scholar]
- Van Boeckel T. P., Pires J., Silvester R., Zhao C., Song J., Criscuolo N. G., et al. (2019). Global trends in antimicrobial resistance in animals in low- and middle-income countries. Science 365:eaaw1944: 365. doi: 10.1126/science.aaw1944, PMID: [DOI] [PubMed] [Google Scholar]
- Wang J., Chou S., Xu L., Zhu X., Dong N., Shan A., et al. (2015). High specific selectivity and membrane-active mechanism of the synthetic centrosymmetric α-helical peptides with Gly-Gly pairs. Sci. Rep. 5:15963. doi: 10.1038/srep15963, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Dou X., Song J., Lyu Y., Zhu X., Xu L., et al. (2019). Antimicrobial peptides: promising alternatives in the post feeding antibiotic era. Med. Res. Rev. 39, 831–859. doi: 10.1002/med.21542, PMID: [DOI] [PubMed] [Google Scholar]
- Xu L., Shao C., Li G., Shan A., Chou S., Wang J., et al. (2020). Conversion of broad-Spectrum antimicrobial peptides into species-specific antimicrobials capable of precisely targeting pathogenic bacteria. Sci. Rep. 10:944. doi: 10.1038/s41598-020-58014-6, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Z., Tharmalingam N., Liu Q., Jayamani E., Kim W., Fuchs B. B., et al. (2017). Synergistic efficacy of Aedes aegypti antimicrobial peptide Cecropin A2 and tetracycline against Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 61, e00686–e00617. doi: 10.1128/AAC.00686-17, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong C., Gou S., Liu T., Zhu Y., Zhu N., Liu H., et al. (2019). Study on the effects of different dimerization positions on biological activity of partial d-amino acid substitution analogues of Anoplin. Microb. Pathog. 139:103871. doi: 10.1016/j.micpath.2019.103871, PMID: [DOI] [PubMed] [Google Scholar]
- Zhong C., Zhang F., Zhu N., Zhu Y., Yao J., Gou S., et al. (2021). Ultra-short lipopeptides against gram-positive bacteria while alleviating antimicrobial resistance. Eur. J. Med. Chem. 212:113138. doi: 10.1016/j.ejmech.2020.113138, PMID: [DOI] [PubMed] [Google Scholar]
- Zhong C., Zhu N., Zhu Y., Liu T., Gou S., Xie J., et al. (2020). Antimicrobial peptides conjugated with fatty acids on the side chain of D-amino acid promises antimicrobial potency against multidrug-resistant bacteria. Eur. J. Pharm. Sci. 141:105123. doi: 10.1016/j.ejps.2019.105123, PMID: [DOI] [PubMed] [Google Scholar]
- Zhu N., Zhong C., Liu T., Zhu Y., Gou S., Bao H., et al. (2021). Newly designed antimicrobial peptides with potent bioactivity and enhanced cell selectivity prevent and reverse rifampin resistance in gram-negative bacteria. Eur. J. Pharm. Sci. 158:105665. doi: 10.1016/j.ejps.2020.105665, PMID: [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary material, further inquiries can be directed to the corresponding authors.