Significance
We describe the use of a DNA hydrogel that promotes bone repair in defects that would otherwise not heal. This approach could overcome some of the inherent disadvantages of the currently used bone grafts. DNA hydrogels demonstrate considerable promise as bone-promoting scaffolds because of their biocompatibility, ease of synthesis, and biodegradation. Importantly, these findings support the potential utility of DNA hydrogels as scalable biomaterials for use in bone repair.
Keywords: DNA nanotechnology, DNA hydrogels, biomineralization, bone regeneration
Abstract
DNA-based biomaterials have been proposed for tissue engineering approaches due to their predictable assembly into complex morphologies and ease of functionalization. For bone tissue regeneration, the ability to bind Ca2+ and promote hydroxyapatite (HAP) growth along the DNA backbone combined with their degradation and release of extracellular phosphate, a known promoter of osteogenic differentiation, make DNA-based biomaterials unlike other currently used materials. However, their use as biodegradable scaffolds for bone repair remains scarce. Here, we describe the design and synthesis of DNA hydrogels, gels composed of DNA that swell in water, their interactions in vitro with the osteogenic cell lines MC3T3-E1 and mouse calvarial osteoblast, and their promotion of new bone formation in rat calvarial wounds. We found that DNA hydrogels can be readily synthesized at room temperature, and they promote HAP growth in vitro, as characterized by Fourier transform infrared spectroscopy, X-ray diffraction, scanning electron microscopy, atomic force microscopy, and transmission electron microscopy. Osteogenic cells remain viable when seeded on DNA hydrogels in vitro, as characterized by fluorescence microscopy. In vivo, DNA hydrogels promote the formation of new bone in rat calvarial critical size defects, as characterized by micro-computed tomography and histology. This study uses DNA hydrogels as a potential therapeutic biomaterial for regenerating lost bone.
DNA nanotechnology is a field where synthetic DNA strands are designed to fold into predetermined designer shapes in two-dimensional (2D) and three-dimensional (3D) assemblies, including extended networks and cross-linked hydrogels (1–4). Self-assembled DNA scaffolds have controllable structural, mechanical, and biochemical properties with envisioned biomedical applications in extracellular matrix (ECM) mimetics for tissue regeneration, biosensing, targeted drug delivery, and cancer therapy (5–8). The use of DNA-based biomaterials for bone tissue regeneration aims to overcome inherent disadvantages of currently used bone grafts that rely on autogenous (bone from the same patient receiving the graft) or allogeneic (bone from another human donor) bone transplantation (9, 10). The application of autogenous bone grafts is often restricted due to limitations imposed by graft harvesting, donor site morbidity, and availability, while transmission of diseases and other pathogenic organisms restrict the utility of allogeneic bone grafts (10).
The feasibility of using DNA-based materials in dynamically remodeling tissues has been established for encapsulation and release of small molecules, nanoparticles, and cells in biologically relevant environments (11). In the case of bone repair, recent reports described self-assembled 2D DNA scaffolds as promoters of osteogenic differentiation of preosteoblasts (12), while other studies have reported the effect of DNA tetrahedron nanostructures in promoting osteogenic differentiation of mesenchymal stem cells (13–15). Self-assembled DNA nanostructures have been shown to template hydroxyapatite (HAP) mineralization in vitro (16–19). These reports suggest a potentially osteoconductive nature for DNA-based materials. Furthermore, DNA degradation releases phosphate ions (20), which exhibit a strong affinity for Ca2+ and that produce calcium phosphate (CaP), an important constituent of bone matrix. Notably, extracellular phosphate can promote osteogenic differentiation and calcification of preosteoblasts and mesenchymal stem cells (21–23). Extracellular adenine, another side product of DNA degradation, has also been shown to be a promoter of osteogenic differentiation (24–28).
DNA hydrogels are 3D DNA networks assembled using DNA nanotechnology principles. Similar to other DNA-based materials, DNA hydrogels are expected to have osteogenesis-promoting properties, possibly due to their phosphate-rich backbone and their inclusion of adenine. DNA hydrogels have the ability to polymerize and conform to crevices in situ. Furthermore, compared to discrete DNA nanostructures, DNA hydrogels can maintain a more localized effect due to being polymerized within and confined to the defect area. DNA hydrogels have also been used as a cell delivery material, with successful incorporation of cells prior to their in situ injection (29).
Herein, we investigate whether DNA hydrogels promote bone repair in defects that would otherwise not heal. Specifically, we describe the design and synthesis of a DNA hydrogel, its ability to promote HAP mineral growth in vitro, the behavior of osteogenic cell lines MC3T3-E1 and mouse calvarial osteoblast (MCO) when seeded on the DNA hydrogels, and the efficacy of the DNA hydrogel as a scaffold for bone repair in a rat calvarial model when compared to autogenous bone as the gold standard.
Results
Design and Characterization of DNA Hydrogels.
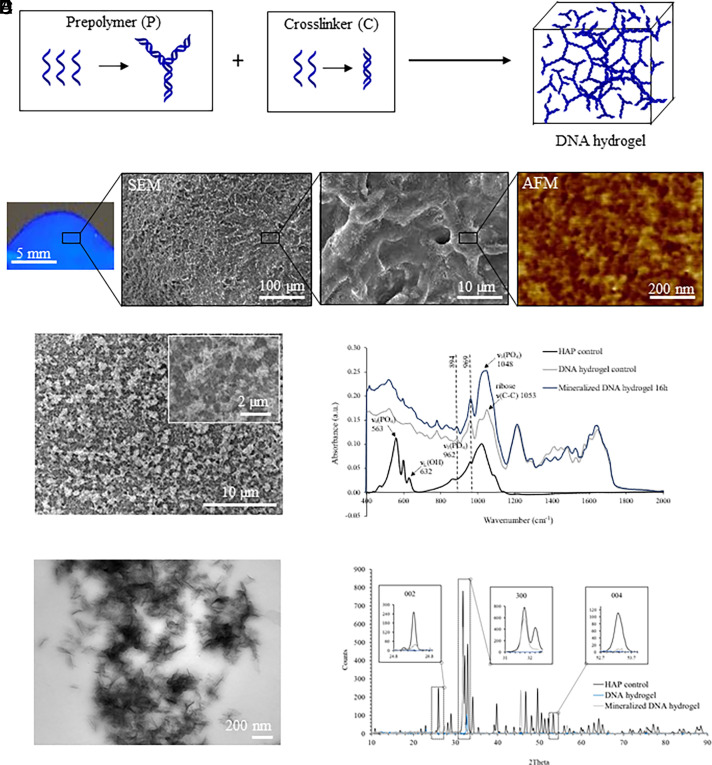
DNA hydrogels were assembled from five DNA strands with complementary sequences, following a modified previously published protocol (30). The DNA hydrogel is assembled from two DNA constructs, DNA prepolymer and DNA cross-linker, that hybridize to one another in a 1:3 ratio through a 12-base pair sticky-end cohesion (Tm: 46.2 °C). The DNA prepolymer is composed of three (Y1, Y2, and Y3) strands, while the DNA cross-linker is composed of two (L1 and L1′) strands (Fig. 1A and SI Appendix, Table S1). To confirm DNA strand purity and proper assembly, denaturing and native polyacrylamide gel electrophoresis (PAGE) were performed, respectively. Hybridization efficiency was detected by the decrease in mobility of the DNA bands in native PAGE experiments (SI Appendix, Fig. S1). Hydrogel polymerization occurred within seconds of mixing DNA prepolymer and cross-linker in a 1:3 ratio and total DNA content of 2.7% (w/w) at room temperature, as shown in the UV-illuminated photograph (Fig. 1B). The nanostructure of DNA hydrogels was characterized by scanning electron microscopy and atomic force microscopy (SEM and AFM), where detailed information of their internal structure was observed (Fig. 1B). SEM results demonstrated that these hydrogels were composed of interconnected fibers that cross-linked into a network. AFM characterization revealed nanopores of approximately 25 nm in diameter throughout the DNA, in agreement with the theoretical calculations (73 bases, 24.8 nm, between the center-to-center distance of the Y cross-linkers in the assembled gels). Pore dimensions influence ion diffusion processes critical to biomaterial function and define the microenvironment in which HAP crystals will ultimately form. As our data suggest, the DNA hydrogel platform offers local control over pore dimensions that would be challenging to achieve using traditional covalent or physically cross-linked polymer networks. The mechanical properties of the DNA hydrogel were analyzed by rheological testing using oscillatory time sweeps for 3 min. The G′/G″ ratio at 1 Hz frequency and 1.5% strain was 26, which is greater than previously reported values for a similar DNA hydrogel (G′/G″ of 21 at 1 Hz frequency and 1% strain) (31). Furthermore, our DNA hydrogel had a G′ value of 953 Pa, which is comparable to the 1,100 Pa value reported by the same group for their DNA hydrogel (32) and also comparable to a 5 mg/mL collagen gel [test done at 1 Hz frequency and 0.8% strain (33)].
Fig. 1.
DNA hydrogel synthesis and characterization. (A) Schematic representation of DNA hydrogel formation after mixing DNA prepolymer (P) with the DNA cross-linker (C). (B) Picture of the DNA hydrogel under UV irradiation (Left), and SEM along with AFM imaging showing a fibrous porous morphology at microscale and nanoscale, respectively. (C) SEM imaging of DNA hydrogels after 16 h of mineralization. (D) Infrared spectra of mineralized DNA hydrogels confirming that the observed mineral phase is HAP. (E) TEM imaging of DNA hydrogels after 16 h of mineralization. (F) XRD spectra of DNA hydrogels after 16 h mineralization confirming that the observed mineral phase is HAP.
In Vitro Calcium Phosphate Mineralization on DNA Hydrogels.
Although natural bone healing processes are cell mediated, effective regenerative scaffolds typically exhibit some intrinsic ability to support HAP mineral growth in the absence of cells, such as bioceramics, polymers, nanoparticles, composites, and metals (34). To assess the HAP mineralization ability of DNA hydrogels, hydrogels assembled on glass coverslips or Transmission Electron Microscopy (TEM) grids were immersed for up to 16 h in a mineralizing solution (35). After mineralization, surfaces were rinsed briefly with distilled H2O, air-dried, and characterized by AFM, SEM, and TEM. AFM characterization of initial mineralization steps of DNA hydrogel disclosed regions of spherical particles distributed throughout the hydrogel surface after 3 min of mineralization and packed plate-like nanocrystals giving rise to aggregates of approximately 400 nm after 16 h of mineralization (SI Appendix, Fig. S2). SEM imaging showed crystalline aggregates covering the entire surface of the hydrogel after 16 h of mineralization throughout the hydrogel scaffolds (Fig. 1C). Fourier transform infrared spectroscopy (FTIR) analysis of hydrogel incubated in mineralizing conditions for 16 h verified that the mineral phase observed on DNA hydrogels is HAP (Fig. 1D). In particular, bands located in the 1,750 to 1,600 cm−1 region are common to C=O, C=N, and C=C stretching and exocyclic –NH2 bending vibrations in the DNA bases (36, 37). Furthermore, sensitive bands including 1,053 cm−1 [stretching of ribose ν(C–C)] of the phosphodiester–deoxyribose backbone and 1,226 cm−1 [asymmetric stretching mode of PO2− groups/νas(PO2−)] are evident and can provide further information on the interaction between the DNA backbone and the newly formed mineral (37). Absorbance peaks at 969 and 894 cm−1 are attributed to the B form of DNA (37). Characteristic peaks for HAP were observed at 1,048 cm−1 v3(PO4), 962 cm−1 v1(PO4), and 632 cm−1 due to OH− ions in apatitic environments, and a broad shoulder was observed on the low-wavenumber region (563 cm−1) of the v4(PO4) band (38). In addition, TEM characterization revealed needle-like crystals embedded within the mineralized DNA hydrogel (Fig. 1E). The presence of HAP within the mineralized DNA hydrogels was also verified by X-ray diffraction (XRD), where the relative peak intensities at 26°, 33°, and 53° were detected correlating to the (002), (300), and (004) HAP planes, respectively (Fig. 1F).
Cell Seeding on DNA Hydrogels.
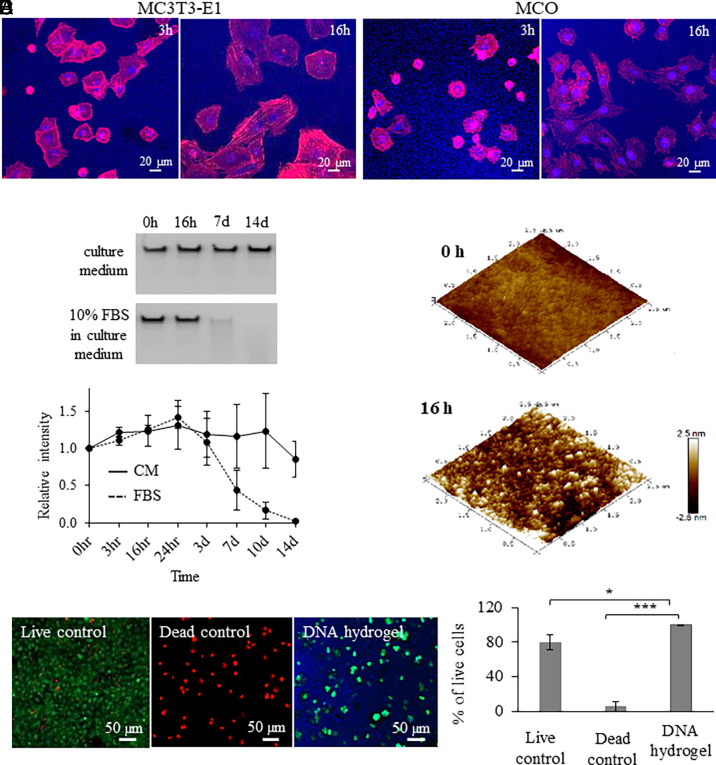
The attachment and differentiation of anchorage-dependent osteogenic cells to form a bone-conducive matrix is a prerequisite for endogenous bone formation. Biomaterials may promote these processes during bone regeneration; however, as the lesion volume increases, cell exclusion from the bulk of the applied biomaterial would ultimately limit this therapeutic effect. To evaluate osteogenic cell interaction with the DNA hydrogels, mouse MC3T3-E1 and MCO (39) cell lines were seeded on DNA hydrogels. Fluorescence microscopy imaging revealed that MC3T3-E1 and MCO cells were distributed at discrete planes throughout the thickness of the hydrogel (Fig. 2 A and B), which is an interesting finding that points to the potential ability of DNA hydrogels to support some degree of cell adherence and subsequent migration through it. While this finding was surprising given our measurement of pore dimensions that are orders of magnitude smaller than the diameter of the cells, a general capacity for cell diffusion through nanoporous DNA hydrogels is further supported by a previous study (8).
Fig. 2.
DNA hydrogel interaction and degradation with osteogenic cells. Fluorescence microscopy images of (A) MC3T3-E1 and (B) MCO cell interaction on DNA hydrogels up to 16 h. (C) (Top), Denaturing gel electrophoresis of DNA hydrogel incubated in cell culture medium or in cell culture medium with 10% FBS for up to 14 d. (Bottom), Degradation kinetics of DNA hydrogel in the presence of cell culture medium with and without 10% FBS. (D) AFM images of DNA hydrogels after 0- and 16-h incubation in cell culture medium with 10% FBS. (E) MCO cell viability assay in the presence of DNA hydrogel. Significant difference is indicated by brackets (*P < 0.05 and ***P < 0.001). Values were compared by the repeated measures ANOVA test.
DNA Hydrogel Degradation.
DNA degradation in serum-containing environments will lead to DNA hydrogel dissolution, a process that limits the duration of hydrogel-mediated biological responses while being essential for the ultimate clearance of the applied hydrogel. To evaluate the degradation rates of our DNA hydrogels, we incubated the hydrogels in serum-containing in vitro conditions [10% fetal bovine serum (FBS)] to remotely simulate in vivo conditions and thereby provide a better insight about the rate by which the DNA hydrogel would degrade when implanted in vivo. The hydrogel samples were incubated in serum-containing culture medium (10% FBS) for up to 2 wk; aliquots were collected at specific time points (0, 3, and 16 h and 1, 3, 7, 10, and 14 d), and DNA fragment generation from degraded hydrogel was monitored by denaturing PAGE. As controls, DNA hydrogels were incubated in serum-free culture medium, and aliquots were collected following the same time points. DNA in serum-containing culture medium started to degrade within 3 d, with complete degradation occurring by day 14 and an estimated half-life of 6.6 d; whereas samples incubated in serum-free medium did not show any appreciable degradation by day 14, and their half-life was not yet reached by the end of the 2-wk incubation period (Fig. 2C). Interestingly, AFM images of the DNA hydrogels incubated in cell culture medium showed signs of DNA hydrogel degradation in as little as 16 h of incubation, as observed by the 2.9× increase in measured mean surface roughness (Ra of 0.498 nm), as calculated through AFM image analysis (Fig. 2D).
We also measured the amount of adenine in degraded DNA hydrogel solutions. Briefly, DNA hydrogel solutions were incubated for 7 d in cell culture medium containing 10% FBS. SI Appendix, Fig. S3 shows that the degraded DNA hydrogel resulted in an adenine concentration of 15.5 picomoles per well. This value corresponds to a concentration of 3.1 mM of adenine in the culture medium. Previously, it was reported that adenine up-regulates osteogenic genes using exogenous adenine supplementation of 100 µM for 24 h (28).
Biocompatibility of DNA Hydrogels.
To evaluate DNA hydrogel biocompatibility in vitro, a cell viability assay was performed for MCO cells embedded within the DNA hydrogels. Results after 16 h of incubation showed that cells remained viable within the DNA hydrogels. A statistically significant difference in cell viability was observed when comparing the hydrogel group with the positive (live cells) and negative (dead cells) controls, showing respective cell viability levels of 99.6%, 79.8%, and 5.4% after 16 h of incubation (Fig. 2E).
We also assessed whether the DNA hydrogel promoted inflammation in vivo by implantation of the DNA hydrogels in the dorsal subcutaneous tissue of rats. At 24 and 72 h after surgery, subcutaneous tissue samples with the DNA hydrogels were collected and stained with hematoxylin and eosin. The peri-implant connective tissue in the group that received the DNA hydrogel (SI Appendix, Fig. S4) showed similar morphological characteristics to those observed in the control/buffer animals. This included few inflammatory cells, in accordance with the chronology of the repair process, along with collagen fibers.
Calvarial Bone Regeneration in an In Vivo Rat Model.
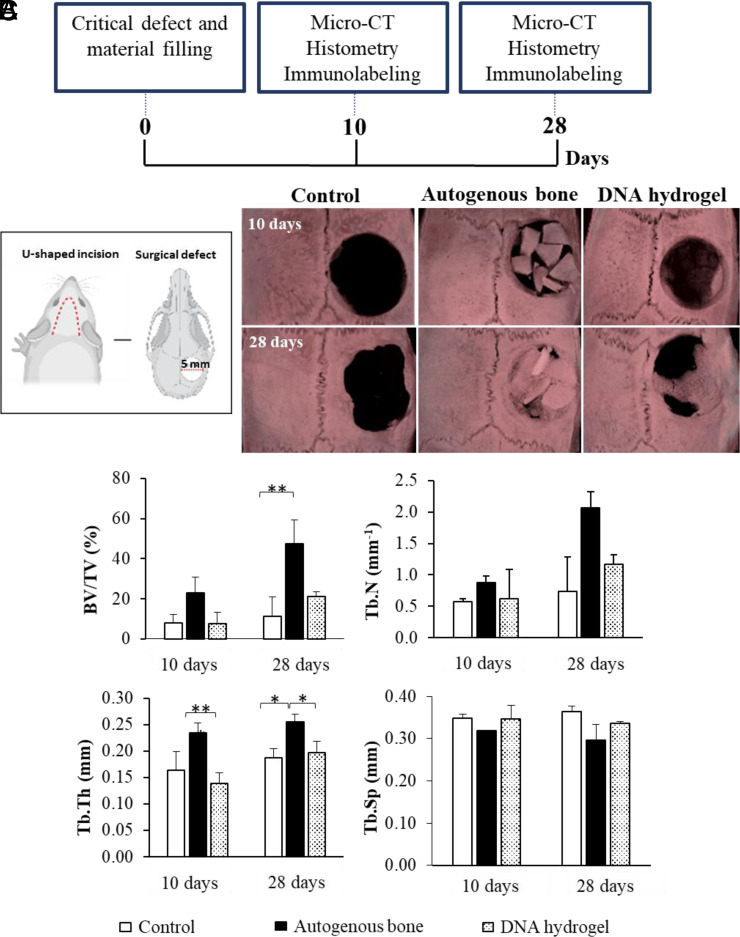
The rat calvarial model has been used extensively as an in vivo model that does not heal spontaneously depending on the size of the defect and period of evaluation (40–42). Furthermore, the tissue does not require fixation for skeleton stabilization. To evaluate bone regeneration properties of DNA hydrogels in vivo, DNA hydrogels were injected into 5-mm rat calvarial critical size bone defects. The quantity and quality of the formed bone in the defect area were evaluated by micro-CT, histology, and immunohistochemistry at 10 and 28 d after injection (Fig. 3A). For comparison, rats were treated with buffer only (negative control) and with autogenous bone (positive control), the current gold standard treatment for bone repair. Importantly, no signs of inflammation were detected in any of the groups 28 d after the rats were killed (SI Appendix, Fig. S4).
Fig. 3.
In vivo rat calvarial regeneration using DNA hydrogels. (A) Timeline of the performed analysis. (B) Experimental design according to the procedures performed in the study and micro-CT characterization of the studied groups. (C) Measured parameters by micro-CT including BV/TV (%), trabecular number (Tb.N; mm−1), trabecular thickness (Tb.Th; mm), and trabecular separation (Tb. Sp; mm). Significant difference is indicated by brackets (*P < 0.05 and **P < 0.01). Values were compared by a one-way ANOVA test.
Surgical procedure and micro-CT images of all samples at 10 and 28 d after surgery are shown in Fig. 3B and SI Appendix, Fig.S5. As anticipated, autogenous bone, as the positive control, promoted extensive bone formation with almost complete defect closure by day 28. The negative control group (buffer) promoted little-to-no new bone formation, even at 28 d after surgery. In the defects injected with DNA hydrogel, there was partial filling in of the defect, with bone forming along the edges by day 10, and larger amounts of new bone being observed at day 28, including in the center of the defect. The DNA hydrogel was able to promote new bone formation, even though it degraded relatively early on (as judged by the in vitro experiments). This supports the hypothesis that DNA exerts a rather early osteogenic inductive effect, which can potentially be heightened as DNA degrades and releases extracellular phosphate ions from its phosphate-rich backbone and likely adenine, which have been shown to induce migration and up-regulate osteogenic differentiation (21–28). The ratio of bone volume to tissue volume (BV/TV) in the region of interest was measured for each sample to characterize the amount of trabecular bone formed in the calvarial defect. To characterize the quality of the new bone, trabecular thickness (Tb.Th) and trabecular number (Tb.N) were measured. The measured parameters, BV/TV, Tb.Th, and Tb.N, have been previously described to characterize the quality of newly formed bone (43). In all of our experiments, the performance trend followed: positive control > DNA hydrogels > negative control (Fig. 3C). Trabecular separation (Tb.Sp) is a measurement of the distance between bone particles, where higher values are inversely correlated with bone quality. In our experiments, the trend followed: positive control < DNA hydrogel < negative control (Fig. 3C), although it was not statistically significant.
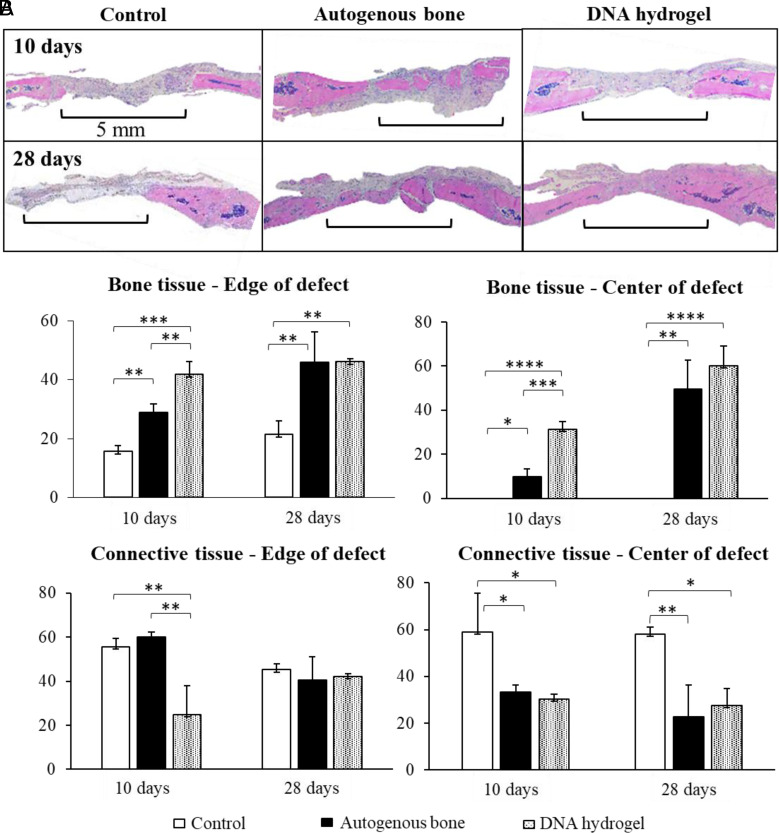
Histological analysis was performed on the calvarial defects of each group at 10 and 28 d after surgery (Fig. 4 and SI Appendix, Fig. S6). Panoramic images of sagittal sections of the defect within the rat skull are shown in Fig. 5A. At 10 d after surgery, the negative control sample was filled with collagen fibers, with no new bone detected. In the samples treated with autogenous bone, fragments of autogenous bone could be observed at the center of the defect, with newly formed bone connecting the autogenous bone fragments. The defect treated with DNA hydrogel was filled with collagen fibers at the center, whereas newly deposited bone (woven bone) was detected at the edge of the defect along with aggregates of what appeared to be osteoblast cells lining the newly formed bone. At 28 d, a period where the bone repair process is more advanced, differences among the groups were heightened. In the negative control group, only collagen fibers and connective tissue were observed filling the center of the defect, while the beginning of new bone formation was detected along the edges of the defect. As expected, the negative control showcased the greatest delay in the repair process. In the autogenous bone group, bone formation filled the majority of the defect, with a large quantity of autogenous bone interconnected with new bone. The DNA hydrogel group also presented a large quantity of bone, starting at the edges of the defect, and filling in toward its center. At 28 d, the amount of bone filling the center of the defect was greatly increased for the DNA hydrogel group, presenting a significant improvement in the repair process compared to the negative control. Interestingly, a nonsignificant difference was calculated when compared to the gold standard autogenous bone. SI Appendix, Fig. S7 describes a qualitative analysis of the tissue composition for the three studied groups.
Fig. 4.
Histological analysis of rat calvarial regeneration using DNA hydrogels. (A) Histological images of treated and untreated rat calvarial defects at 10 and 28 d after surgery. (B) Bone and connective tissue measurements among the studied groups. Significant difference is indicated by brackets (*P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001). Values were compared by a one-way ANOVA test.
Fig. 5.
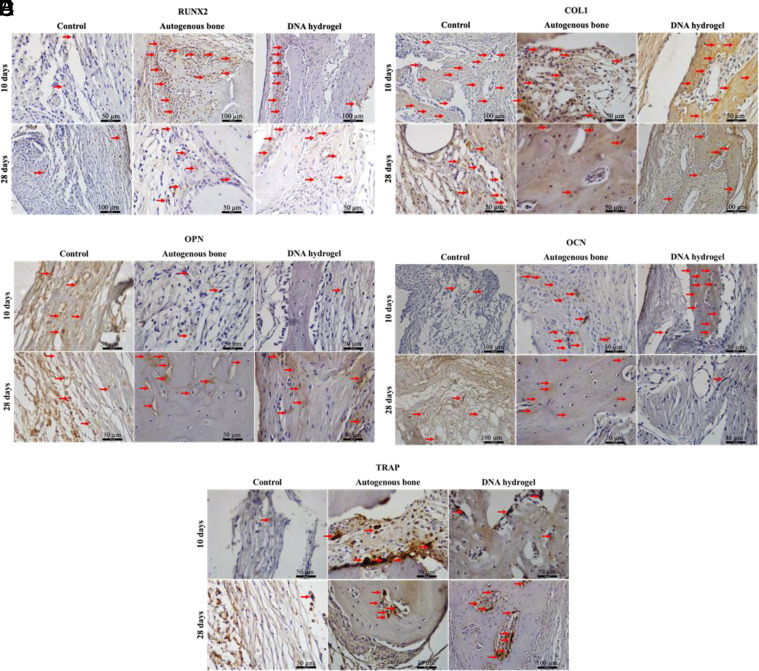
Immunolabeling analysis during DNA hydrogel calvarial healing. Immunohistochemical detection of osteogenic differentiation markers. (A) Runt-related transcription factor 2 (RUNX2), (B) collagen type I (Col1), (C) osteopontin (OPN), (D) osteocalcin (OCN), and (E) tartrate-resistant acid phosphatase (TRAP) among the studied groups at 10 and 28 d after surgery.
Immunolabeling of Bone Formation Markers.
Immunolabeling analysis of the defect area was performed in all groups at 10 and 28 d after surgery (Fig. 5 and SI Appendix, Fig. S8). The proteins stained were 1) runt-related transcription factor 2 (RUNX2), a transcription factor that labels preosteoblastic cells; 2) collagen type I (Col1), the main component of bone ECM; 3) osteopontin (OPN), a protein present at the start of the mineralization process; 4) osteocalcin (OCN), a protein present during the mineralization process; and 5) tartrate-resistant acid phosphatase (TRAP), a marker for osteoclastic activity. The analysis was performed in the region of interest that presented positive labeling (highlighted with red arrows) for each protein. Scores (discrete, moderate, or intense) were given to describe the distribution area of the markers in all groups.
At day 10, the negative control group was mostly composed of parallel collagen fibers, with no new bone detected. Tissue labeling was discrete for RUNX2, moderate for Col1, and discrete for OPN (on cells and ECM), OCN, and TRAP. In the positive control, areas of autogenous bone and new bone were observed, with aggregates of cells close to the bone trabeculae showing intense labeling for RUNX2. Tissue labeling was moderate for Col1, discrete for OPN, and moderate for OCN and TRAP. In the DNA hydrogel group, areas of new bone formation and parallel collagen fibers were observed. The tissue staining was intense for RUNX2 along individual cells. The staining was intense for Col1, discrete for OPN, intense for OCN, and discrete for TRAP, which indicates ongoing mineralization within the newly formed bone regions and reduced osteoclastic activity relative to the positive control.
At day 28, the negative control was still mostly composed of collagen fibers at the center of the defect, with incipient bone present along the edges of the defect. The tissue staining was discrete for RUNX2, Col1, and OPN, moderate for OCN (especially along the nonmineralized ECM and cells), and discrete for TRAP along the edges of the defect. In the positive control group, the center and edges of the defect were filled with bone containing osteocytes. The tissue staining was moderate for RUNX2, discrete for Col1, and moderate for OPN (including along reversal lines), OCN (including osteocytes present within the newly formed bone), and TRAP. The DNA hydrogel group showcased new bone formation along the center and edges of the defect, albeit to a lesser extent than the positive control. The tissue staining was moderate for RUNX2, Col1, and OPN (along reversal lines), discrete for OCN (especially in osteocytes located within the newly formed bone), and moderate for TRAP, which suggests that cell activity associated with healing is less pronounced than at the earlier time point, yet still ongoing by day 28.
Discussion
Herein, we describe synthetic DNA hydrogels self-assembled through complementary DNA hybridization as promising scaffolds for bone regeneration. DNA gelation occurred at room temperature immediately upon mixing DNA prepolymer and cross-linker solutions in Ca2+-containing buffers. The porosity of DNA hydrogels allows for exchange of ions and water molecules within. Unlike other hydrogel systems, DNA hydrogels are also capable of sustaining CaP mineral growth, recalling features described in biomimetic mineralization studies on DNA nanostructure scaffolds (16–19, 44).
In recent years, bioactive hydrogel scaffolds have been introduced as an alternative biomaterial for bone tissue regeneration. Hydrogels contain both free and bound water in their structure, thereby mimicking biological structures (45). For example, core–shell fibrous hydrogel scaffolds composed of cobalt and bone morphogenetic protein-2 (BMP-2) have successfully induced osteogenesis and angiogenesis for synergistic bone regeneration process in a rat calvarial defect (46). Furthermore, gelatin- and hydroxybenzoic acid–cross-linked fibrous hydrogels allowed appropriate cell dispersion and viability for various applications in tissue engineering (47). Hydrophilic 3D hydrogels have been beneficial for cell survival and new bone growth. They have been used in cell culture, cell encapsulation, and cell delivery, revealing fundamental regulations on cell differentiation of various cell types (8, 29, 48).
Previous studies show that 2D DNA networks enhance cell adhesion, leading to an increase in the cell area as well as greater uptake of endocytic membrane cargos (49). Cell adhesion determines their physiological function within tissues by stimulating signals for cell differentiation and migration. 2D DNA nanoporous scaffolds have been used previously to simulate a bone microenvironment, providing a biocompatible support for MC3T3-E1 preosteoblasts which favored their osteogenic differentiation (12). In this work, we show that DNA hydrogels can be used to generate a scaffold that supports the viability of osteogenic MC3T3-E1 and MCO cells in vitro, as well as supporting osteogenic differentiation in vivo. As the engagement of specific adhesion receptors on the plasma membrane of osteogenic cells is critical for osteodifferentiation (50), membrane-associated cell attachment molecules could possibly be mediating cellular adhesion to DNA, although the nature of such molecules has not been elucidated. As such, DNA hydrogels can provide a scaffold that is favorable for osteogenic cell viability and growth and therefore has promise for possible future translation as a tissue-engineering scaffold.
As previously shown (51), DNA scaffolds made from salmon sperm DNA underwent complete degradation within 5 d of subcutaneous implantation. However, the current literature does not offer information about the degradation profiles of DNA hydrogels, which, unlike naturally occurring salmon sperm DNA strands, form a 3D network of DNA with predesigned complementary base pairs. As demonstrated by our in vitro experiments, DNA hydrogels also undergo degradation in serum-containing environments, likely due to the effect of nucleases present in the serum (52, 53). Likewise, in vivo degradation would be anticipated, although the degradation profile may vary due to different DNAse concentrations as well as the presence of other factors such as DNAse inhibitors in body fluids.
In vivo results show that DNA hydrogels can be implanted successfully in rat calvarial defects after surgical resection with no inflammation even at 28 d after surgery. The DNA hydrogels supported the formation of trabecular bone, as indicated by micro-CT (bone microarchitecture) and histology (cellular responses) experiments. As a gold standard graft material, autogenous bone (positive control) still provided the best results due to its desired properties of osteoinduction, osteoconduction, and osteogenesis. However, DNA hydrogels showed better responses in most aspects when compared with the negative control. Considering the very early stage of the material development as a tissue scaffolding material, our results highlight that DNA hydrogels show promise as biomaterials capable of activating cellular responses related to bone repair, and with design optimization, they could potentially be developed into powerful bone regeneration therapeutics.
Cellular activation leading to bone formation was observed as early as 10 d postimplantation, as characterized by immunohistochemistry. This response was confirmed at 28 d after surgery, where a large portion of the calvarial defect was filled with new bone and bone ECM in the DNA hydrogel group. In immunolabeling analysis, the DNA hydrogel group had a tissue architecture that is characteristic of bone, staining positive for Col1, showing an active process of bone matrix deposition. Furthermore, positive RUNX2 staining confirmed the presence of osteoblasts in the defect area. Corroborating these results, OCN labeling was positive throughout the majority of the defect, confirming that the mineralization process was underway. As bone was formed in the calvarial defect, osteoclastic activity was observed through TRAP-positive immunolabeling. While all these markers were positive in the DNA hydrogel group, the mineralization process was delayed when compared to the positive control but significantly faster when compared to the negative control.
The search for ideal biomaterials capable of playing a key role as scaffolds for bone repair is ongoing. DNA hydrogels emerge as unique materials capable of up-regulating osteoblastic activity and collagen synthesis, the pillars of bone organic matrix. Furthermore, DNA degradation is another important property that allows for the remodeling process to occur, although the rate by which it occurs requires further optimization to more closely match the rate of bone ECM deposition. Our study identifies DNA hydrogels as scaffolds for bone repair and poses important questions about their mineralization and osteogenic cell interactions and how these processes can be used in favor of promoting bone regeneration. Future studies will include incorporating biomolecules that may potentiate the repair process, thereby activating cellular responses that can improve the regenerative outcome. Discrete nanostructured DNA assemblies have been functionalized with HAP-promoting peptides (18). Thus, the mineralization capability of DNA hydrogels could potentially be further improved through the incorporation of mineral-promoting peptide conjugates within the DNA hydrogels.
Materials and Methods
Synthesis of DNA Hydrogel.
DNA hydrogels were assembled in a two-step reaction, as described previously (30). Briefly, single-stranded DNA sequences complementary to each other were purchased from GeneLink Inc. (SI Appendix, Fig. S1). DNA prepolymer (composed of Y1, Y2, and Y3) and DNA cross-linker (L1 and L1′) solutions were prepared in 1× Tris(hydroxymethyl)aminomethane (Tris)–Ca2+ buffer (pH 7.4) in separate tubes. The DNA prepolymer and DNA cross-linker solutions were mixed in a ratio of 1:3, respectively, with the DNA hydrogel (2.25 mM) forming within seconds. The samples were characterized by denaturing experiments, which were run at 15 mA and 300 V for 1.5 h, and by native gel experiments, which were run at 15 mA and 80 V for 16 h, after which the gel was stained with GelRed® (Biotium). Gel imaging was performed using a Gene Genius Bioimaging System (Syngene). Quantification of the gel panels was performed using ImageJ software (version 1.53e).
Rheology.
Rheological testing for the DNA hydrogels was performed using the Discovery HR-3 Rheometer at The Hospital for Sick Children’s Structural & Biophysical Core Facility (Toronto, ON, Canada). Oscillatory time sweep tests were done at 25 °C, at 1 Hz frequency and 1.5% fixed strain, for 3 min.
In Vitro Mineralization of DNA Hydrogels.
To examine calcium phosphate mineralization on DNA hydrogels in vitro, hydrogels were self-assembled on glass coverslips and were immersed in a mineralizing solution for up to 16 h at room temperature. The mineralizing solution consisted of two parts of Tris-buffered saline (TBS) (125 mM NaCl and 50 mM Tris, pH 7.4), one part of calcium chloride solution (6.8 mM CaCl2 in TBS), and one part of disodium phosphate solution (38 mM Na2HPO4 in TBS). The samples were characterized by AFM, SEM, TEM, XRD, and FTIR.
AFM.
DNA hydrogel with or without mineralization was air-dried and imaged in tapping mode under ambient conditions using a MultiMode AFM with a Nanoscope III controller (Digital Instruments, Santa Barbara) and OTESPA-R3 cantilevers (Bruker, California). Image analysis was performed using NanoScope Analysis (Bruker) where the images were flattened to remove curvature and slope, and the RMS roughness was calculated.
SEM.
DNA hydrogel with or without mineralization was characterized by SEM. Samples were frozen in a vacuum freeze dryer overnight and subsequently mounted on aluminum stubs and coated with a 4-nm platinum layer using a Fisons Polaron SC515 sputter coater. The hydrogels and mineralization were imaged with a Hitachi FlexSEM 1000 scanning electron microscope at 10 kV.
TEM.
DNA hydrogel was placed on glow-discharged carbon-coated 400 mesh copper grids and exposed to a mineralizing solution (12.5 mM Tris–Ca2+ and 0.1 M phosphate buffer at 1:1.67 Ca:P ratio) in a wet chamber at room temperature for 16 h. At the end of the incubation period, excess liquid was removed with a filter paper, and the sample was air-dried prior to imaging. Samples were imaged within 24 h of sample preparation using a Hitachi HT7800 TEM (120 kV) at the SickKids Nanoscale Biomedical Imaging Facility (Toronto, ON, Canada).
FTIR.
DNA hydrogel with or without mineralization was characterized by FTIR. Spectra were collected in absorbance mode using a FTIR spectrometer (Thermo Scientific FTIR Nicolet iS20) equipped with a germanium crystal at 8 cm−1 nominal resolution. The spectra were obtained by averaging 32 scans and processed by OMNIC software (version 9.11.697).
XRD.
DNA hydrogel was placed on a coverslip and exposed to a mineralizing solution (12.5 mM Tris–Ca2+ and 0.1 M phosphate buffer at 1:1.67 Ca:P ratio) in a wet chamber at room temperature for 16 h. Samples were then removed from solution, dried, and mounted onto a lightly greased glass rod (broad amorphous scattering from the grease, and glass was observed in the data at 11° and 21°). 2D diffraction data were collected in the MAX Diffraction Facility at the McMaster University (Hamilton, ON, Canada) using a Bruker D8 DISCOVER diffractometer.
DNA Hydrogel Degradation in Serum.
Degradation of DNA hydrogels was examined under two conditions: serum-containing culture medium (10% FBS and 1% antibiotics in α-MEM) and serum-free culture medium (1% antibiotics in α-MEM) for the controls. Twenty microliter of the culture medium was added to 2 µL polymerized hydrogel. Degradation was assessed by denaturing PAGE after 0 h (control time point), 3 h, 16 h, 1 d, 3 d, 7 d, 10 d, and 14 d of incubation at 37 °C and 5% CO2. At the lapse of the specified time points, the samples were frozen and stored at −80 °C. For the gel run, the samples were thawed at room temperature, followed by addition of 20 µL formamide and thoroughly mixing via pipetting and vortexing. Then, 0.5 µL of this mix was then added to 7.25 µL distilled water and 7.25 µL formamide, making a total volume of 15 µL to be loaded in each of the gel lanes. The denaturing gel was run at 15 mA and 300 V for 1.5 h, after which the gel was stained with GelRed® (Biotium). Gel imaging was performed using a Gene Genius Bioimaging System (Syngene). Each experiment was repeated in triplicates. Quantification of the gel panels was performed using ImageJ software (version 1.53e).
Adenine Quantification.
In 20 µL serum-containing culture medium, 2 µL of the DNA hydrogel was incubated. After 7 d, adenine concentration in the incubation mixture was measured using an adenine quantification kit (BioVision, Abcam) and compared to a standard curve, following the protocol provided with the kit. The samples were diluted to 1:1,000 to fit in the standard curve. Fluorescence was measured using an Ex/Em of 535/587 using the BioTek’s Cytation 3 Imaging Reader. Adenine concentration and standard curve measurements were repeated in triplicates.
Cell Culture.
Mouse calvaria osteoblasts (MCO) and MC3T3-E1 (clone 14) cells were obtained from WT and CFKO mice and immortalized, as previously described (39). MCOs and MC3T3-E1 cells were cultured in AMEM supplemented with 10% FBS and 1% penicillin/streptomycin.
Confocal Microscopy.
Cells seeded on the DNA hydrogels were imaged using a Zeiss LSM 880 confocal fluorescent microscope with a 20× objective. The samples were placed on 8-well cell culture plates and incubated at 37 °C and 5% CO2 for 3 and 16 h. They were then washed with phosphate-buffered saline (PBS/1X, pH 7.4), fixed with 4% paraformaldehyde, and stained with DAPI for DNA and rhodamine phalloidin for actin. Samples were imaged under excitation at 406 nm for DAPI and at 561 nm for rhodamine phalloidin. The number of attached cells and cell spreading were measured using Image J software (version 1.8).
Cell Viability.
Cell viability testing was done using a LIVE/DEAD® Viability/Cytotoxicity Kit for mammalian cells (Invitrogen, L3224). For the experimental group, a cell pellet containing 5 × 105 cells was resuspended in 10 μL of the crosslinker solution and dispensed onto a glass-bottom dish. This was followed by adding 10 μL of the prepolymer solution to generate a cell-laden 20 μL DNA hydrogel. Culture medium (10% FBS and 1% antibiotics in α-MEM) was then added to the dish. For the controls, the cells were added directly to the glass-bottom dish, without the hydrogel. The dishes were incubated for 16 h at 37 °C and 5% CO2. After the end of the incubation period, the samples were washed with 1× PBS and stained with 2 µM calcein AM and 4 µM EthD-1 solution (in 1× PBS) and DAPI for the hydrogel group to stain the DNA hydrogel. For the dead control, an additional step prior to staining was done, where 70% ethyl alcohol was added to the cells and incubated for 30 min. This was followed by washing five times with 1× PBS and then adding the staining kit and proceeding to imaging using a Zeiss LSM 880 confocal microscope. Six microscopic fields were used per sample (at 10× magnification), and the cells were counted and quantified using ImageJ software (version 1.53e).
Surgical Proceedings.
All procedures for the in vivo rat bone regeneration using DNA hydrogels were approved by the Ethics Committee of Animal Use from Araçatuba Dental School—UNESP (protocol number 2021/549). Forty-two male adult (6-mo-old) rats (Rattus norvegicus albinus, Wistar), weighing approximately 350 g each, were submitted to the surgical proceeding. Thirty-six animals (12 in each group, 6 measured at 10 d and 6 measured at 28 d after surgery). There were six additional animals submitted for subcutaneous analysis (three for 24 h and three for 72 h postsubcutaneous implantation). The animals were anesthetized with xylazine (Coopazine; Coopers, Ltda; Brazil) and intravenous ketamine hydrochloride (Vetaset; Fort Dodge Saúde Animal, Ltda; Brazil), and the trichotomy and antisepsis of the calvarial region were performed. Local anesthesia was performed with lidocaine (without a vasoconstrictor) diluted to 0.5% in a saline solution, 1 to 3 mg/kg. For each animal, a U-shaped incision was made in the occipitofrontal direction, and the soft tissue was detached. Using a 5-mm-diameter inner drill bit (3i Implant Innovations, Inc.), a 5-mm-diameter surgical defect was made in the right parietal bone, maintaining the integrity of the dura mater. The materials that were used to fill the defects were determined for each group from the present study. For the negative control group, the defect was filled with the same buffer used to prepare the DNA hydrogel samples. For the positive control group, the bone collected with the drill was broken into small pieces with a bone grinder and was used to fill the area of the defect. For the DNA hydrogel group, DNA hydrogel was introduced with micropipette to the area of the defect. For autogenous bone, DNA hydrogel and control groups, the amount of material used was standardized across all animals to fill the defect area. After the filling procedure, the soft tissue was carefully repositioned and sutured with nylon thread. No membrane was used to cover the material positioned in the defect. In the immediate postoperative period, each animal received a single intramuscular dose of 0.2 mL penicillin G benzathine (Pentabiótico Veterinário Pequeno Porte; Fort Dodge Saúde Animal Ltda; Brazil). The animals were killed at 10 or 28 d after the surgery through an excessive dose of anesthetic (sodium thiopental—dose 100 mg/kg intraperitoneally) to collect samples for the study analyses.
Software was used to randomly select the samples that were destined for micro-CT, immunolabeling, and histometric analyses. Samples containing the calvaria of the animals were fixed in 10% formaldehyde for 48 h; samples were then washed under running water for 24 h. After these procedures, the samples were divided into undecalcified group, which was maintained in 70% ethanol for microtomography analysis, and decalcified group, which was stored in 10% EDTA for the demineralization process. After conclusion of this step, samples in both groups were embedded in paraffin and sectioned for histometric and immunohistochemical analysis.
Histometric Analysis.
After demineralization of the collected calvaria, gradual dehydration followed by xylol and paraffin embedding was performed. Serial sections with a thickness of 5 μm were cut using a microtome (Leica Biosystems), then mounted on slides, and stained with hematoxylin and eosin. Two regions (center and edge of the defect) of each sample were digitally captured at ×20 magnification (Leica DMLB) and blindly analyzed by an expert observer. An ImageJ software tool was used to perform the morphometry, which contained 391 points on an 8,000-pixel model. The points corresponding to the neo-formed bone tissue, the remaining biomaterial, and the adjacent connective tissue were summed, and the total of each variable was obtained.
Immunolabeling Analysis.
The slices separated for immunolabeling analysis were proceeded to paraffin removal and after that gone through sequence baths of decreasing alcohol concentrations until complete rehydration of the sections. Then, hydrogen peroxide was added to block endogenous peroxidases, followed by antigen retrieval using phosphate buffer and citric acid to expose the antigenicity sites for the proteins that will be labeled. Polyclonal primary antibodies produced in goat (Santa Cruz Biotechnology) and against RUNX2, Col1, OPN, OCN, and TRAP (Santa Cruz Biotechnology, Inc., Dallas, Texas, USA) were selected to label events important for bone formation such as preosteoblast differentiation, bone matrix labeling, mineralization process, and bone resorption. Anti-goat secondary antibody functionalized with biotin was produced in rabbits (Pierce Biotechnology, Thermo Fisher Scientific Inc.), while signal amplification was achieved using the ABC system (Vector Laboratories, Inc.) using diaminobenzidine (Dako North America Inc.) as chromogen. At the end of the reaction, Meyer's hematoxylin was used to label the cytoarchitecture of the interest area. Microscopical analysis was performed to evaluate the size of area with positive labeling from each protein, through the attribution of scores, using ordinal qualitative analysis. The images were captured with Digital Camera (Leica Microsystems) coupled to an Optical Microscope (Leica Microsystems) using objectives with different magnifications.
Microtomographic Analysis.
Using the SkyScan microtomograph (SkyScan 1272, Bruker Micro-CT, Belgium, 2003), the samples were scanned in 11-µm slices with a 0.5-mm Al filter and a 0.6-mm rotation step. The scanning was done at 180° with a frame of 2, a resolution of 2016 × 1344, and an acquisition time of approximately 50 min. The images, obtained by X-ray, were stored and reconstructed by determining the area of interest with NRecon software (SkyScan, version 1.6.6.0, 2011) using a smoothing of 2, an artifact ring correction of 5, a beam-hardening correction of 20%, and an image conversion range varying from 0.007 to 0.061. The images were reconstructed in Data Viewer (SkyScan, version 1.4.4, 64 bit) and observed on three planes (transversal, longitudinal, and sagittal). Then, using CT analyzer software (CTAn, 2003-11, and SkyScan, 2012; Bruker Micro-CT; version 1.12.4.0), the region of the created defect (40 slices for each sample) was evaluated using a histogram of 110 to 205 to remove the denser material. The parameters evaluated were bone volume percentage (BV/TV), trabecular bone thickness (Tb.Th), and separation and number of trabeculae (Tb.Sp and Tb.N).
Statistical Analysis.
The quantitative results obtained from the histometric and computed microtomography analyses were submitted to statistical analysis. The statistical analysis was performed using GraphPad Prism software (GraphPad Software, Inc.; 2017). An analysis of homoscedasticity was performed using the Shapiro–Wilk test to distinguish parametric and nonparametric data. For the analysis of the micro-CT data, the one-way ANOVA test and Tukey posttest were used. A significance level of P < 0.05 was adopted for all tests.
Supplementary Material
Appendix 01 (PDF)
Acknowledgments
We thank the CAMiLoD Microscopy Center for access to fluorescence and scanning electron microscopes and imaging analysis software, Dr. Laurent Bozec for access to the Fourier transform infrared (FTIR) spectrometer, MAS Diffraction Facility for X-ray diffraction (XRD) characterization, The Hospital for Sick Children’s Structural & Biophysical Core Facility for access to the Discovery HR-3 Rheometer, Nanoscale Biomedical Imaging Facility for access to the transmission electron microscope (TEM), and Mr. Alexander Danesi and Ms. Isabel Ding for performing pilot experiments. This study was supported by the Natural Sciences and Engineering Research Council of Canada (RGPIN-2017-06885) to K.M.M.C. and the Brazilian Federal Agency for Support and Evaluation of Graduate Education Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES) in the scope of the Program CAPES-Print process number 88887.310463/2018-00.
Author contributions
C.A.M., R.O., and K.M.M.C. designed research; D.A., N.M., N.G.M., A.C.E.-S., R.L.C., R.O., and K.M.M.C. performed research; C.A.M., R.O., and K.M.M.C. contributed new reagents/analytic tools; D.A., N.M., N.G.M., A.C.E.-S., R.L.C, C.A.M., R.O., and K.M.M.C. analyzed data; K.M.M.C. provided funding; and D.A., N.M., R.O., and K.M.M.C. wrote the paper.
Competing interests
The authors declare no competing interest.
Footnotes
This article is a PNAS Direct Submission.
Data, Materials, and Software Availability
All study data are included in the article and/or SI Appendix.
Supporting Information
References
- 1.Seeman N. C., Sleiman H. F., DNA nanotechnology. Nat. Rev. Mater. 3, 17068 (2018). [Google Scholar]
- 2.Hong F., Zhang F., Liu Y., Yan H., Origami D. N. A., Scaffolds for creating higher order structures. Chem. Rev. 117, 12584–12640 (2017). [DOI] [PubMed] [Google Scholar]
- 3.Wang D., Liu P., Luo D., Putting DNA to work as generic polymeric materials. Angew. Chem. Int. Ed. 61, e202110666 (2022). [DOI] [PubMed] [Google Scholar]
- 4.Carneiro K. M. M., Avakyan N., Sleiman H. F., Long-range assembly of DNA into nanofibers and highly ordered networks. Wires Nanomed. Nanobi. 5, 266–285 (2013). [DOI] [PubMed] [Google Scholar]
- 5.Sethi S., Sugiyama H., Endo M., Biomimetic DNA nanotechnology to understand and control cellular responses. ChemBioChem 23, e202100446 (2022). [DOI] [PubMed] [Google Scholar]
- 6.Bush J., Hu C. H., Veneziano R., Mechanical properties of DNA hydrogels: Towards highly programmable biomaterials. Appl. Sci. 11 (2021). [Google Scholar]
- 7.Shahbazi M.-A., Bauleth-Ramos T., Santos H. A., DNA hydrogel assemblies: Bridging synthesis principles to biomedical applications. Adv. Ther. 1, 1800042 (2018). [Google Scholar]
- 8.Wang Y., et al. , Constructing tissue-like complex structures using cell-laden DNA hydrogel bricks. ACS Appl. Mater. Interf. 9, 12311–12315 (2017). [DOI] [PubMed] [Google Scholar]
- 9.Hernigou J., Vertongen P., Rasschaert J., Hernigou P., Role of scaffolds, subchondral, intra-articular injections of fresh autologous bone marrow concentrate regenerative cells in treating human knee cartilage lesions: Different approaches and different results. Int. J. Mol. Sci. 22, 3844 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang W., Yeung K. W. K., Bone grafts and biomaterials substitutes for bone defect repair: A review. Bioact. Mater. 2, 224–247 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ma W., et al. , The biological applications of DNA nanomaterials: Current challenges and future directions. Signal Transduct. Target Ther. 6, 351 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Baig M., Dissanayaka W. L., Zhang C., 2D DNA nanoporous scaffold promotes osteogenic differentiation of pre-osteoblasts. Int. J. Biol. Macromol. 188, 657–669 (2021). [DOI] [PubMed] [Google Scholar]
- 13.Shao X. R., et al. , Effect of tetrahedral DNA nanostructures on osteogenic differentiation of mesenchymal stem cells via activation of the Wnt/beta-catenin signaling pathway. Nanomed. Nanotechnol. Biol. Med 13, 1809–1819 (2017). [DOI] [PubMed] [Google Scholar]
- 14.Zhou M., et al. , Effect of tetrahedral DNA nanostructures on proliferation and osteo/odontogenic differentiation of dental pulp stem cells via activation of the notch signaling pathway. Nanomed. Nanotechnol. Biol. Med. 14, 1227–1236 (2018). [DOI] [PubMed] [Google Scholar]
- 15.Zhou M., et al. , Effect of tetrahedral DNA nanostructures on proliferation and osteogenic differentiation of human periodontal ligament stem cells. Cell Prolif. 52, e12566 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu X. G., et al. , DNA framework-encoded mineralization of calcium phosphate. Chem 6, 472–485 (2020). [Google Scholar]
- 17.Athanasiadou D., Carneiro K. M. M., DNA nanostructures as templates for biomineralization. Nat. Rev. Chem. 5, 93–108 (2021). [DOI] [PubMed] [Google Scholar]
- 18.Danesi A. L., et al. , Peptide-decorated DNA nanostructures promote site-specific hydroxyapatite growth. Acs Appl. Mater. Inter. 14, 1692–1698 (2022). [DOI] [PubMed] [Google Scholar]
- 19.Kim F., et al. , Functionalized DNA nanostructures as scaffolds for guided mineralization. Chem. Sci. 10, 10537–10542 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Katsumata Y., Kajiya H., Okabe K., Fukushima T., Ikebe T., A salmon DNA scaffold promotes osteogenesis through activation of sodium-dependent phosphate cotransporters. Biochem. Biophys. Res. Commun. 468, 622–628 (2015). [DOI] [PubMed] [Google Scholar]
- 21.Beck G. R., Zerler B., Moran E., Phosphate is a specific signal for induction of osteopontin gene expression. Proc. Natl. Acad. Sci. U.S.A. 97, 8352–8357 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chai Y. C., et al. , Current views on calcium phosphate osteogenicity and the translation into effective bone regeneration strategies. Acta Biomater. 8, 3876–3887 (2012). [DOI] [PubMed] [Google Scholar]
- 23.Lin H., et al. , Effect of inorganic phosphate on migration and osteogenic differentiation of bone marrow mesenchymal stem cells. BMC Dev. Biol. 21, 1 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Evans B., Ham J., An emerging role for adenosine and its receptors in bone homeostasis. Front. Endocrinol. 3, 1–8 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Evans B. A., et al. , Human osteoblast precursors produce extracellular adenosine, which modulates their secretion of IL-6 and osteoprotegerin. J. Bone Miner. Res. 21, 228–236 (2006). [DOI] [PubMed] [Google Scholar]
- 26.D’Alimonte I., et al. , Adenosine A1 receptor stimulation enhances osteogenic differentiation of human dental pulp-derived mesenchymal stem cells via WNT signaling. Stem Cell Res. 11, 611–624 (2013). [DOI] [PubMed] [Google Scholar]
- 27.Shih Y.-R.V., et al. , Calcium phosphate-bearing matrices induce osteogenic differentiation of stem cells through adenosine signaling. Proc. Natl. Acad. Sci. U.S.A. 111, 990–995 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen Y.-P., et al. , Anti-inflammatory effects of adenine enhance osteogenesis in the osteoblast-like MG-63 cells. Life 10, 116 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yan X., et al. , Anti-friction MSCs delivery system improves the therapy for severe osteoarthritis. Adv. Mater. 33, e2104758 (2021). [DOI] [PubMed] [Google Scholar]
- 30.Um S. H., et al. , Enzyme-catalysed assembly of DNA hydrogel. Nat. Mater. 5, 797–801 (2006). [DOI] [PubMed] [Google Scholar]
- 31.Xing Y., et al. , Self-assembled DNA hydrogels with designable thermal and enzymatic responsiveness. Adv. Mater. 23, 1117–1121 (2011). [DOI] [PubMed] [Google Scholar]
- 32.Cheng E., et al. , A pH-triggered, fast-responding DNA hydrogel. Angew. Chem. Int. Ed. 48, 7660–7663 (2009). [DOI] [PubMed] [Google Scholar]
- 33.Yang Y.-L., Leone L. M., Kaufman L. J., Elastic moduli of collagen gels can be predicted from two-dimensional confocal microscopy. Biophys. J. 97, 2051–2060 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Koons G. L., Diba M., Mikos A. G., Materials design for bone-tissue engineering. Nat. Rev. Mater. 5, 584–603 (2020). [Google Scholar]
- 35.Deshpande A. S., Beniash E., Bioinspired synthesis of mineralized collagen fibrils. Cryst. Growth Des. 8, 3084–3090 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Brewer S. H., Anthireya S. J., Lappi S. E., Drapcho D. L., Franzen S., Detection of DNA hybridization on gold surfaces by polarization modulation infrared reflection absorption spectroscopy. Langmuir 18, 4460–4464 (2002). [Google Scholar]
- 37.Taki A., John B., Arakawa S., Okamoto M., Structure and rheology of nanocomposite hydrogels composed of DNA and clay. Eur. Polym. J. 49, 923–931 (2013). [Google Scholar]
- 38.Drouet C., Apatite formation: Why it may not work as planned, and how to conclusively identify apatite compounds. Biomed. Res. Int. 2013, 1–12 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rajshankar D., Wang Y., McCulloch C. A., Osteogenesis requires FAK-dependent collagen synthesis by fibroblasts and osteoblasts. FASEB J. 31, 937–953 (2017). [DOI] [PubMed] [Google Scholar]
- 40.Spicer P. P., et al. , Evaluation of bone regeneration using the rat critical size calvarial defect. Nat. Protoc. 7, 1918–1929 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Luvizuto E. R., et al. , The effect of BMP-2 on the osteoconductive properties of β-tricalcium phosphate in rat calvaria defects. Biomaterials 32, 3855–3861 (2011). [DOI] [PubMed] [Google Scholar]
- 42.Gomes-Ferreira P. H. S., et al. , Sonochemical time standardization for bioactive materials used in periimplantar defects filling. Ultrason. Sonochem. 56, 437–446 (2019). [DOI] [PubMed] [Google Scholar]
- 43.Bouxsein M. L., et al. , Guidelines for assessment of bone microstructure in rodents using micro–computed tomography. J. Bone Miner. Res. 25, 1468–1486 (2010). [DOI] [PubMed] [Google Scholar]
- 44.Gkioni K., Leeuwenburgh S. C. G., Douglas T. E. L., Mikos A. G., Jansen J. A., Mineralization of hydrogels for bone regeneration. Tissue Eng. Part B Rev. 16, 577–585 (2010). [DOI] [PubMed] [Google Scholar]
- 45.Allen M. E., Hindley J. W., Baxani D. K., Ces O., Elan Y., Hydrogels as functional components in artificial cell systems. Nat. Rev. Chem. 6, 562–578 (2022), 10.1038/s41570-022-00404-7. [DOI] [PubMed] [Google Scholar]
- 46.Perez R. A., Kim J. H., Buitrago J. O., Wall I. B., Kim H. W., Novel therapeutic core-shell hydrogel scaffolds with sequential delivery of cobalt and bone morphogenetic protein-2 for synergistic bone regeneration. Acta Biomater. 23, 295–308 (2015). [DOI] [PubMed] [Google Scholar]
- 47.Hu M., et al. , Cell immobilization in gelatin-hydroxyphenylpropionic acid hydrogel fibers. Biomaterials 30, 3523–3531 (2009). [DOI] [PubMed] [Google Scholar]
- 48.Yao C., et al. , Double rolling circle amplification generates physically cross-linked DNA network for stem cell fishing. J. Am. Chem. Soc. 142, 3422–3429 (2020). [DOI] [PubMed] [Google Scholar]
- 49.Walia S., et al. , Designer DNA hydrogels stimulate 3D cell invasion by enhanced receptor expression and membrane endocytosis. ACS Biomater. Sci. Eng. 7, 5933–5942 (2021). [DOI] [PubMed] [Google Scholar]
- 50.Tang Y., et al. , Laminin alpha 4 promotes bone regeneration by facilitating cell adhesion and vascularization. Acta Biomater. 126, 183–198 (2021). [DOI] [PubMed] [Google Scholar]
- 51.Matsumoto A., et al. , Degradation rate of DNA scaffolds and bone regeneration. J. Biomed. Mater. Res. Part B Appl. Biomater. 107, 122–128 (2019). [DOI] [PubMed] [Google Scholar]
- 52.von Kockritz-Blickwede M., Chow O. A., Nizet V., Fetal calf serum contains heat-stable nucleases that degrade neutrophil extracellular traps. Blood 114, 5245–5246 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Napirei M., Ludwig S., Mezrhab J., Klockl T., Mannherz H. G., Murine serum nucleases–contrasting effects of plasmin and heparin on the activities of DNase1 and DNase1-like 3 (DNase1l3). FEBS J. 276, 1059–1073 (2009). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix 01 (PDF)
Data Availability Statement
All study data are included in the article and/or SI Appendix.