Significance
Proteins function through their ability to form specific three-dimensional structures or interactions, but only a small proportion of these features within a proteome have been experimentally determined. Protein structural modelers have now generated predictions for millions of proteins, including for complexes, but the accuracy of these predictions lacks systematic experimental assessment. We generated a significant resource of protein cross-links within the human cell, experimentally mapping spatially constrained pairs of amino acids within proteins or between interaction interfaces. Critically, these capture proteins with native sequences, posttranslational modifications, subcellular niches, and cofactors. We demonstrate how our resource—and large-scale cross-linking mass spectrometry in general—can be used for the mapping, assessment, and contextualization of the recently expanded structural proteome.
Keywords: cross-linking mass spectrometry, protein–protein interactions, protein structure prediction, AlphaFold2, structural proteomics
Abstract
Significant recent advances in structural biology, particularly in the field of cryoelectron microscopy, have dramatically expanded our ability to create structural models of proteins and protein complexes. However, many proteins remain refractory to these approaches because of their low abundance, low stability, or—in the case of complexes—simply not having yet been analyzed. Here, we demonstrate the power of using cross-linking mass spectrometry (XL-MS) for the high-throughput experimental assessment of the structures of proteins and protein complexes. This included those produced by high-resolution but in vitro experimental data, as well as in silico predictions based on amino acid sequence alone. We present the largest XL-MS dataset to date, describing 28,910 unique residue pairs captured across 4,084 unique human proteins and 2,110 unique protein–protein interactions. We show that models of proteins and their complexes predicted by AlphaFold2, and inspired and corroborated by the XL-MS data, offer opportunities to deeply mine the structural proteome and interactome and reveal mechanisms underlying protein structure and function.
Proteins are the primary effectors in biology. Their function is determined in large part by their three-dimensional structure and by the protein–protein interactions (PPIs) that they form. A system-wide understanding of protein structure and interactions has thus been a long-standing goal.
To this end, the protein–protein interactome has been systemically catalogued using several approaches, including yeast two-hybrid (Y2H), affinity-purification mass spectrometry (AP-MS) and by the Bio-ID proximity assay and variants (reviewed in ref. 1). These methods have been very successful in identifying both direct and indirect protein interactions and at least one interactor has been curated for almost 50% of the human proteome (2). However, these data do not provide structural or mechanistic information on interactions and do not necessarily analyze proteins interacting in their native state. These significant shortcomings call for additional strategies to better define the interactome.
Despite intensive efforts in the fields of X-ray crystallography, NMR, and cryoelectron microscopy (cryo-EM), only 35% of human proteins have any representation in the Protein Data Bank (PDB) (3, 4). Many are only partially resolved: Only 17% of residues in human proteins are present in the PDB, and experimental structures exist for only 6% of known human PPIs (5). Furthermore, the heterologous overexpression and purification of proteins are often necessary to produce enough material for these techniques, with only 5% of the human proteins in the PDB produced from native sources (3, 4). This situation raises possible concerns about the integrity of structures and complexes that are generated using such approaches.
In lieu of experimental structures, machine learning-based structural modelers such as AlphaFold2 (6) have been shown to be highly accurate under controlled tests (7) and have greatly expanded the coverage of structural proteomes across a range of species (8, 9). However, because the training dataset (the PDB) has a very low representation of truly native structures, the accuracy of these models and the question of how to assess them remain to be determined.
Cross-linking mass spectrometry (XL-MS) provides a means to assay native protein structure and PPIs in a parallel fashion (reviewed in refs. 10 and 11). Chemical cross-linkers containing at least two reactive groups are introduced into a protein sample to covalently link amino acids within spatially constrained reactions. The identification of cross-linked peptides via mass spectrometry enables the definition of the linked residues, providing conformational constraints for a protein chain or PPI interface. These constraints, despite having low effective resolution, can be used to validate experimental structures, help inform modeling of proteins with unknown structures, and guide docking studies of PPIs (reviewed in ref. 10). Taking into consideration the relatively low protein sample requirements, XL-MS can provide a first view of the structures and interactions of many poorly studied or less accessible proteins (12).
Recent technical developments in XL-MS have established its use at scale and in native contexts such as in intact human cells (13, 14) and even mammalian tissues (15). However, the complexity and dynamic range of the proteome and interactome leads to under-sampling. One means to achieve better proteome coverage while preserving in vivo or near-in vivo proteoform states is to isolate and then cross-link intact organelles (16, 17). Furthermore, the use of diverse cross-linker reactivities can increase the density of structural constraints by sampling a larger protein sequence space (18, 19).
To establish the utility of large-scale XL-MS for the characterization of the structural proteome and interactome, we have generated a high-coverage and high-density cross-link dataset for a cultured human cell line. Fractionated organelles were cross-linked with three different chemistries [DHSO (20), disuccinimidyl sulfoxide (DSSO) (21), and 4-(4,6-dimethoxy-1,3,5-triazin-2-yl)-4- methylmorpholinium (DMTMM) (22)], and a multistep analytical pipeline used to extensively fractionate and identify cross-linked peptides. We identified 91,709 cross-link spectral matches (CSMs) representing 28,910 unique residue pairs (URP) across 4,084 proteins and 2,110 PPIs. This resource is the largest reported to date for any species.
Benchmarking against the protein structure and PPI literature demonstrates the integrity and utility of our dataset, which provides orthogonal validation for many high-resolution (but in vitro) experimental structures. In parallel, our cross-links also identify PPIs and confirm many PPIs that were previously only identified using systems-level approaches. Importantly, our data also provide structural information for a broad range of proteins and PPIs, including proteins that lack any experimental characterization. We demonstrate how cross-links can be utilized to evaluate, validate, and support structure modeling platforms such as AlphaFold2. Overall, we conclude that XL-MS can provide corroborative data for structural modeling pipelines to extend our understanding of the structural proteome, including in the context of the interactome.
Results
Orthogonal Cross-Linkers, Sample Enrichment and Improved Database Search Strategies Generate the Largest Cross-Linked Proteome To Date.
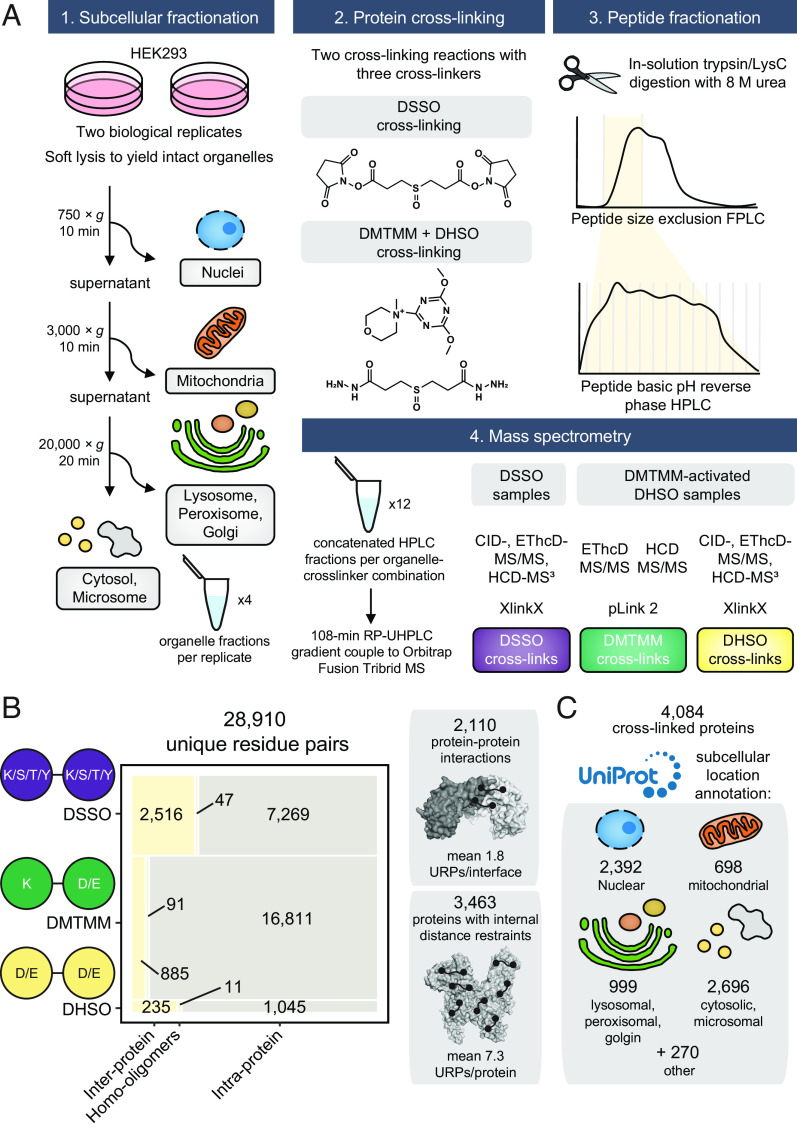
To provide broad coverage of the human structural proteome, we performed crude subcellular fractionation of HEK293 cells and cross-linked these fractions in two reactions with three orthogonal cross-linkers: DHSO (20), DSSO (21), and DMTMM (22). We reasoned that by prefractionating the cells, we would capture more (and different) proteins across the broad dynamic range of the human proteome. Importantly, membrane-bound organelles (e.g., nucleus, mitochondria, lysosomes, and Golgi) remain compartmentalized before cross-linking, and should retain their proteins in a state that is more native than in a lysate (although not to the same extent as in vivo XL-MS). Following protein digestion, we fractionated the resulting peptides using sequential offline size exclusion chromatography and high pH reversed-phase liquid chromatography (Fig. 1A).
Fig. 1.
Generation of the most comprehensive human cross-linking mass spectrometry dataset. (A) Experimental overview. (B) Breakdown and summary statistics of the cross-linked unique residue pairs (URP) and the underlying captured unique protein–protein interactions or internally linked proteins. Interprotein refers to cross-linking between peptides mapping to two distinct UniProt accessions, intraprotein to the same accession, and homooligomer to same accession but with overlapping peptide sequences (and hence must derive from two separate molecules). (C) The size and breakdown of annotated subcellular localizations detected in the cross-linked proteome. Note that redundancies are caused by proteins with multiple annotated subcellular assignments.
From mass spectrometry, we identified 4,084 unique proteins linked by 28,910 unique cross-linked residue pairs (which we define as “unique residue pairs”, or URPs) from 91,709 CSMs (Fig. 1 B and C and Dataset S1). These URPs can be further classified into 3,785 and 25,125 interprotein (including homooligomers) and intraprotein URPs, respectively.
Our URP list was generated using stringent quality control measures (Methods, SI Appendix, Fig. S1, and ref. 16). These enabled the control of the false discovery rate (FDR) to ≤1% at the URP level for both intraprotein and interprotein links, and 1.9% at the PPI level for interprotein links. All three cross-linkers produced interprotein to intraprotein URP ratios in line with theoretical expectations for their maximum distance constraints (23), indicating an appropriately controlled FDR.
To our knowledge, this is the largest XL-MS dataset to date, containing more than twice the number of URPs of the next largest studies (14, 24, 25). Protein abundance data from the PaxDB database (26) revealed that we sampled the proteome at a significantly deeper level than previous work. We identified 1.6-fold more proteins than the previous benchmark (14), and the median abundance of a cross-linked protein in our dataset was 1.8-fold lower (SI Appendix, Fig. S2A). Furthermore, the density of cross-links defining each PPI and intralinked protein was higher than observed in the previous benchmark study (14), including for low-abundance proteins (SI Appendix, Fig. S2B). We note that the improvements in cross-linking density from the use of orthogonal chemistries was more pronounced for intraprotein cross-links, almost doubling the densities reported in other similar studies that use only one cross-linker (7.3 in our study vs. ~4 intraprotein URPs per intralinked protein in refs. 1–3, Dataset S2). On the other hand, our density of interprotein URPs per PPI was more similar to previous studies [1.8 vs. 1.4 to 2.3, Dataset S2, (14, 24, 25)], likely reflecting the composition of our dataset in which most interprotein URPs were captured by a single cross-linker (DSSO, which is longer and lysine-reactive) (Fig. 1B). This observation might also explain the lower number of PPIs described by our dataset when compared to Wheat et al. (14) (Dataset S2), which employed a single but longer (+4 Å relative to DSSO) enrichable and lysine-reactive Alkyne-A-DSBSO cross-linker. Overall, our dataset is substantial and of high quality.
Most proteome-wide XLMS studies to date have relied on N-hydroxysuccinimide-based (NHS) cross-linkers, which primarily target the ε-amino side chain of lysine (K) residues (21). Although reactions with the hydroxyl side chains of serine (S), threonine (T) and tyrosine (Y) are also possible (27, 28), they are often not considered due to the significant (and in most cases, prohibitive) increase in computation time during peptide database searches. In contrast to most other comparable studies, we included possible cross-links to S/T/Y during in our analysis of DSSO spectra. This strategy yielded 1.3-fold more DSSO URPs compared to K-K cross-link searching alone (9,832 vs. 7,829 URPs). Linkages involving S/T/Y residues made up 24% of all DSSO URPs (SI Appendix, Fig. S3A), consistent with previous smaller scale studies (27, 28). Many S/T/Y-linked residues were initially localized to a nearby K residue within the same peptide (SI Appendix, Fig. S3B). For many of these peptides, their identification scores significantly improved when S/T/Y reactivity were considered (SI Appendix, Fig. S3C), indicating an improved accuracy in the localization of cross-linking sites. It also enabled us to identify ~20% more PPIs than a K-K-only search strategy (1,464 rather than 1,243 PPIs).
Our data indicate that different cross-linker chemistries might be better suited to certain subcellular niches. The proportion of URPs arising from each cross-linker varied significantly between organellar fractions (nucleus, cytoplasm, mitochondria, and Golgi). DSSO was the most effective at cross-linking the nuclear fraction, whereas DHSO was most effective for the mitochondrial and Golgi fractions (SI Appendix, Fig. S4A). These differences might reflect variation in amino acid composition, local pH, or even the solubility or permeability of a cross-linker in a given compartment. We also noted that the nuclear fraction produced a significantly higher proportion of interprotein URPs than any other fraction, suggesting denser protein packing or better preservation of protein complexes (SI Appendix, Fig. S4B).
Benchmarking against Experimental Structures Demonstrates the High Quality of the Dataset.
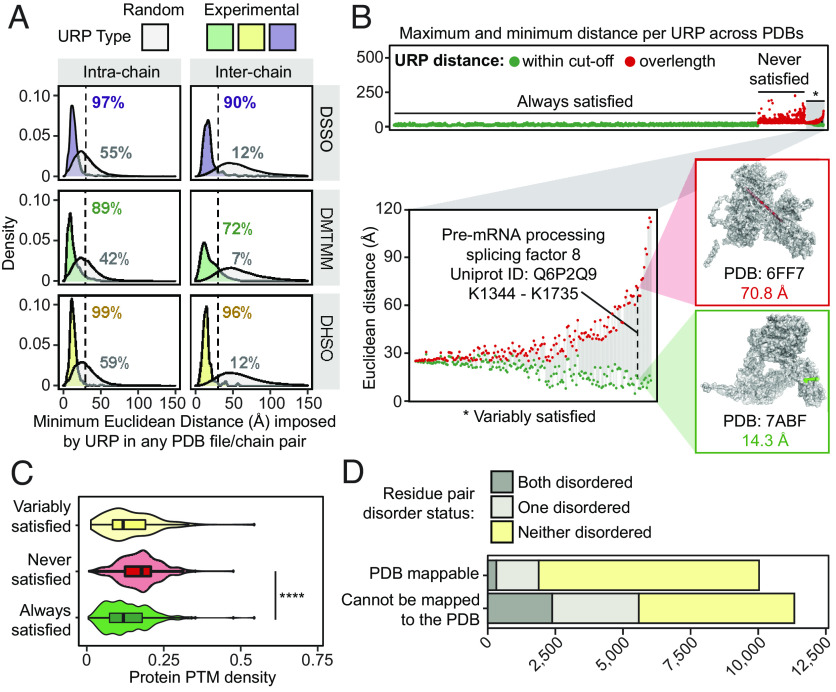
We next used structures from the PDB (4) to assess the quality of our data. Forty-three percent (9,152) of our 21,367 unambiguous URPs (i.e., URPs from cross-linked peptides for which each peptide sequence could be uniquely mapped to a single UniProt accession code) could be mapped onto 10,332 unique experimental structures (Dataset S3). Euclidean distances (Cα-Cα) were calculated using Xwalk (29) and URPs considered satisfied if their minimum mapped distance on any PDB entry was within the maximum theoretical distance of 30 Å for DHSO (20) and DSSO (21), and within 25 Å for DMTMM (22, 30).
Considering only intrachain URPs, 7,860 URPs mapped onto 10,110 structures of 1,406 proteins, of which 99% (DHSO), 97% (DSSO), and 89% (DMTMM) were satisfied (Fig. 2A and Dataset S3). In contrast, randomly sampled residue pairs within each structure met the distance cutoffs in only ~40 to 60% of cases (Fig. 2A). For interchain linkages (including homodimeric URPs from overlapped peptide sequences), 1,292 URPs describing 519 unique PPIs were mapped onto 2,274 distinct PDB entries. We observed slightly lower distance satisfaction rates of 90% for DSSO, 72% for DMTMM, and 96% for DHSO [consistent with previous work (31)], whereas randomly sampled interchain URPs had very low satisfaction rates (~7 to 12%).
Fig. 2.
Cross-link distance constraints are validated by, and uniquely contextualize the variability within, in vitro experimental structures curated in the Protein Data Bank. (A) The distribution of minimum Euclidean distances (Å, Cα-Cα) imposed by unique residue pairs (URPs) across redundant possible chain-pairs and/or Protein Data Bank (PDB) entries. Only “unambiguous” URPs are considered in these analyses, which are those with underlying cross-linked peptide sequences that were uniquely mapped to a single protein sequence in the canonical human proteome. Random residue pairs with appropriate sidechain reactivities were simulated for each individual PDB structure. The percentage of URPs falling within the cross-linker specific distance cutoffs (dotted lines, 25 Å for DMTMM or 30 Å for DHSO/DSSO) are also indicated. (B) The range of Euclidean distances imposed by URPs mapped across multiple unique PDB entries, considering only PDB entries with one possible chain-pair configuration for the URP. The minimum and maximum distances observed for each URP are plotted in-line as circles joined by a gray line. Green fill denotes a structure for which the distance in question is falls within the relevant cutoff, and red fill denotes a structure in which the distance violates the cutoff. The Top panel shows the range distribution for all unambiguous URPs, stratified by their global satisfaction rate and ordered by increasing difference in distances within these categories. The distribution of the URP subset with variable satisfaction across PDB entries (*), is shown on the Lower Left. Inset structures to the Right show an example of a variably satisfied URP from the pre-mRNA-processing-splicing factor 8 protein (Q6P2Q9), indicated by the dashed line. (C) The density of PhosphoSitePlus-annotated posttranslational modifications (number of distinct annotated modification sites/length of protein) of cross-linked protein(s) (with the maximum value for the two proteins used for interprotein links) for each URP mapping stratification type (by variability of URP satisfaction as described above). **** is P = 2.2 × 10−16, from a one-tailed Wilcoxon rank sum tests with continuity corrections. (D) The distribution of unambiguous URPs involving residues from regions predicted to be disordered, stratified by whether the URPs are resolved in PDB structures.
It is also notable that our cross-linking was carried out under significantly more native-like conditions than those typically used for the determination of protein structures. Structures are generally determined from heterologously expressed polypeptides that often comprise only a fraction of the full protein sequence and are determined in the absence of interactions with partners. The remarkably high overall satisfaction rates we observe therefore provide large-scale experimental corroboration of thousands of in vitro experimental structures.
Cross-Links Capture and Confirm Alternative Structural Conformers.
We noted many URPs that could map to more than one PDB structure for the same protein and therefore asked whether our data could provide insights into possible conformational plasticity of these proteins. We examined 4,857 URPs that could be mapped onto multiple PDB structures and compared the minimum and maximum Euclidean distances for these URPs across all of their PDB entries (Fig. 2B and Dataset S3). Most URPs (85.4%, “Always satisfied” in Fig. 2 B, Top) were always mapped within the cross-linker cutoff distances regardless of PDB structure.
In contrast, although 529 URPs were not satisfied in any available structure, we observed 178 URPs that were satisfied in some but not all structures (“never satisfied” or “variably satisfied”, respectively, Fig. 2B). For example, the URP K1344-K1735 in the pre-mRNA splicing factor 8 (Uniprot: Q6P2Q9) differed dramatically in distance depending on which specific precursor subcomplex of the spliceosome it was mapped onto. The two residues show a distance of 71 Å in the structure of a spliceosome core variant that contains the U2/U6 catalytic RNA network [PDB: 6FF7 (32)], whereas the same residues are 14 Å apart in a structure lacking these RNAs [PDB: 7ABF, (33), Fig. 2 B, Inset]. In another example, the URP E673-K890 in the DNA replication licensing factor MCM2 (UniProt: P49736) shows a distance of 10 Å when the complex that it is a part of–the CDC45-MCM-GINS helicase–is not engaged in the replisome [PDB: 6XTX (34)] and 39 Å when it is engaged in the replisome [PDB: 7PFO (35)] (SI Appendix, Fig. S5). These observations suggest that URPs that are not satisfied in currently available protein structures might in some cases flag the existence of alternative conformations or architectures that are yet to be experimentally characterized.
Alternative structural conformations can arise from the formation of distinct complexes, as above, or directly from allosteric changes induced by the deposition of post-translational modifications (PTMs). We observe that the never satisfied URPs tended to fall in proteins that are more densely posttranslationally modified according to the PhosphoSitePlus database (36) compared to those satisfied in all PDB structures (Fig. 2C; P < 2.2 × 10−16). This finding underscores the fact we are probing endogenous proteins in a near-cellular environment, and therefore probably capturing protein conformations and complex architectures that more closely reflect the in vivo state than do experimental high-resolution structures of isolated polypeptides expressed from heterologous systems (e.g., Escherichia coli) that do not possess relevant human PTMs.
As noted above, we were unable to map more than half (~53%) of our unambiguous URPs due to the absence of cognate experimental structures. Because intrinsically disordered regions are resistant to structure determination, we investigated whether our unmappable URPs resided in such regions. Surprisingly, more than half of the URPs without PDB resolution did not lie in a disordered region, as annotated by a consensus of predictions curated in MobiDB (37) (Fig. 2D). These URPs therefore likely define experimental distance restraints for ordered regions within structures that are either difficult to resolve (because of conformational dynamics, recalcitrance to purification efforts, or highly contextual conformations) or simply involve unstudied proteins.
Cross-Links Enable the High-Throughput Experimental Assessment of Protein Structure Predictions.
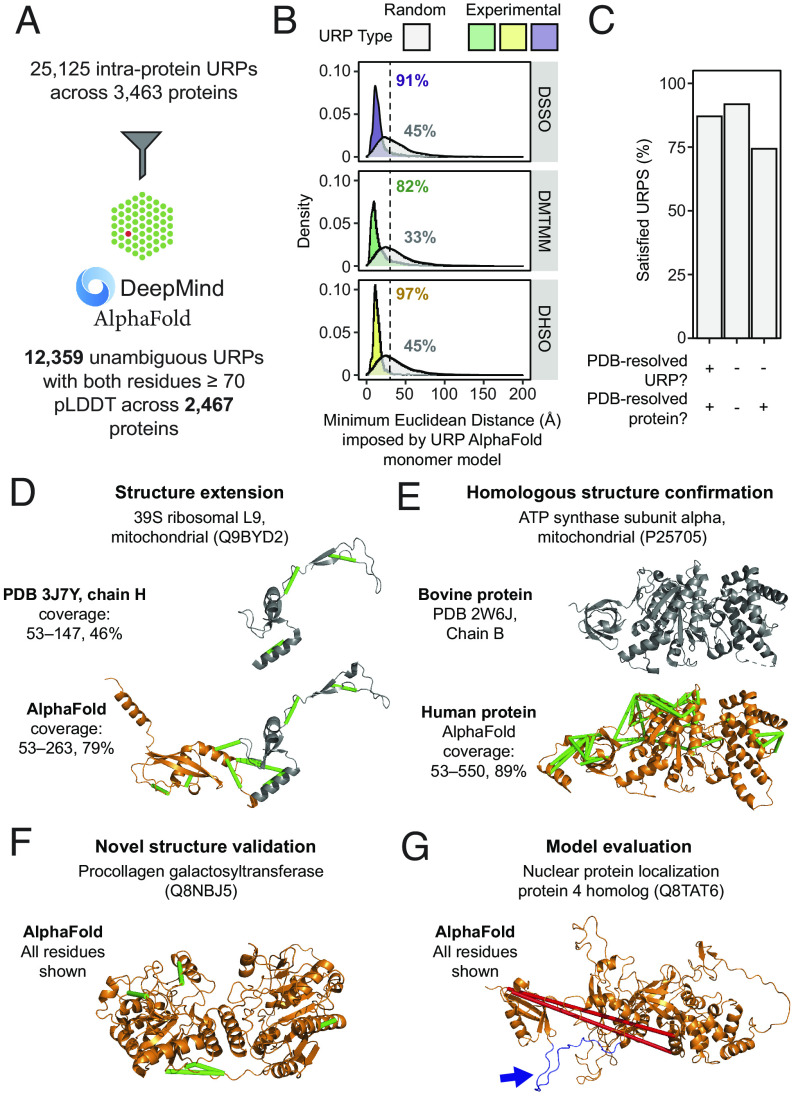
Next-generation structural modeling programs such as AlphaFold2 (6) have generated enormous recent interest because of their performance in controlled environments such as CASP14 (7). However, the accuracy of such structural predictions for proteins in their native cellular context is less well established. The near-native structural constraints provided by our cross-link resource provides a unique opportunity to address this question, and we therefore asked how well our unambiguous intraprotein URPs map onto the recent AlphaFold2 (AF2) predictions of the human proteome (9). We only considered URPs that mapped to “high-confidence” residues—defined by a value of ≥70 for the AlphaFold2 pLDDT quality metric. Despite this restriction, the AF2 dataset significantly increased the number of unambiguous URPs that we could map, with 12,359 URPs able to be resolved across 2,467 unique proteins (Fig. 3A and Dataset S4). This represents a 1.4-fold increase in resolved URPs compared to our PDB mapping above, and an increase of 1.8-fold in the number of proteins to which URPs could be mapped.
Fig. 3.
The high-throughput experimental assessment of AlphaFold2 monomeric protein structure predictions using cross-link distance constraints. (A) Statistics for the mapping of unique URPs onto AlphaFold2 (AF2) monomer models. Only high-confidence URPs (both residues within pLDDT ≥70) were used for further analyses. (B) Fulfillment of URPs on AF2 models. For each AF2 monomer model, five random URPs were generated for each cross-linker with appropriate side-chain reactivities. The distribution of Euclidean distances (Å, Cα-Cα) determined for experimental and random URPs is shown, stratified by cross-linker. Annotations show the percentage of URPs fulfilled for each subset. The cutoff for each cross-linker is indicated by a dotted line (25 Å for DMTMM or 30 Å for DHSO/DSSO). (C) Overall URP satisfaction rate across URPs with different degrees of PDB resolution. (D–G) Examples of the use of URPs for model validation and evaluation. Protein names are official UniProt entry names. The range of residues shown are indicated to the left of each structure. Green URPs are satisfied, red URPs are overlength. (D) The model for Q9BYD2 shows an example of a PDB-resolved human protein that has benefitted from increased structural coverage (orange) and is now corroborated by nine experimental URPs. Gray regions within structures represent those derived from experimental PDB structures. (E and F) show proteins without any PDB entry for the human proteins (orange). Gray regions within structures represent those derived from experimental PDB structures. (E) The AF2 model of the mitochondrial ATP synthase subunit alpha with a known bovine structural homologue, 44 URPs support this model. (F) The AF2 model of the procollagen galactosyltransferase (Q8NBJ5) that does not have any PDB entries for itself or of homologous proteins. Five URPs support this model. (G) An example where the two identified cross-links do not fit the AF2 model. However, while the cross-linked residues are within well-modeled domains, the two domains are separated by a low-confidence, disordered loop (colored in blue and highlighted with a blue arrow).
The distance distribution and satisfaction rates of these high-confidence URPs on the AF2 models (Fig. 3B and SI Appendix, Fig. S6A) were comparable to the values observed for intraprotein URPs mapped to PDB structures (Fig. 2A), spanning 82 to 97% satisfaction across cross-linkers, compared to ~33 to 45% for randomly sampled URPs. This observation confirms that the predictions made by AF2 are of very high quality.
Given that AF2 is trained on the PDB, we next asked whether URP satisfaction rates on AF2 models varied depending on whether or not an experimental structure also existed for that protein (Dataset S4). We first stratified the high-confidence URPs into three categories based on their PDB status at both the URP level and the overall protein level (Fig. 3C). URPs for which both residues could be observed in a PDB structure had an 87% satisfaction rate on their corresponding AF2 model. Unexpectedly, URPs in proteins for which no experimental structure is currently available had even better satisfaction rates (92%). A significantly lower satisfaction rate of 74% was observed for URPs from proteins that have at least one PDB structure available, but that describe a residue or residues without any experimental resolution in these structures. These URPs might capture protein regions that display significant conformational dynamics, highlighting the fact that each AF2 model captures only a single snapshot of a protein’s conformational landscape.
Our data demonstrate the potential for integrated systems-wide approaches (such as the combination of AF2 and XL-MS) to address known biases of structural proteome coverage in the PDB—for example, toward well-behaved and highly ordered domains/proteins.
Cross-Links Experimentally Corroborate Hundreds of AlphaFold2 Models of Proteins with Unknown Structures.
As we mapped our URPs to the AF2 models, we noted that these data could generate structural insights for the regions within proteins that are not able to be resolved by other methods. For example, the L9 subunit of the mitochondrial 39S ribosome (Uniprot: Q9BYD2) is partially resolved in several PDB entries; for example, in PDB: 3J7Y (38) residues 53 to 147 are observable, and we can map three URPs to this region, all of which are satisfied (Fig. 3 D, Top). However, the AF2 model resolved an additional 116 ordered C-terminal residues compared to the experimental structure, all of which have pLDDT scores of ≥70. An additional six URPs mapped to this region (including three that bridge the PDB-resolved and AF2-only regions) and all were satisfied in the AF2 model (Fig. 3 D, Bottom). This agreement provides strong corroboration for the predicted model and in this context, we reiterate that the cross-linking data were obtained in a native-like context, giving additional confidence that this is the relevant structure in the environment of an intact ribosome in vivo.
Of the set of 737 proteins that were absent from the PDB but for which cross-links were observed, 624 had all of their high-confidence URPs satisfied in the corresponding AF2 model (SI Appendix, Fig. S6B and Dataset S4). Our dataset provides experimental validation for these models in one of several ways. First, there were 268 instances where the structure of the human protein is unknown, but a structure exists for a homologue. This included the alpha subunit of mitochondrial ATP synthase (Uniprot: P25705). The structure of the human protein has not been reported, but the structure of the bovine protein (Uniprot: P19483) (97% sequence identity) is resolved in PDB: 2W6J (Fig. 3E). Not surprisingly, the AF2 model for the human protein closely resembles the bovine structure, and our set of 44 experimental URPs verifies the conserved fold.
Second, there were 356 proteins for which we observed cross-links that were both absent from the PDB and also displayed a very low degree of structural precedent. For example, the procollagen galactosyltransferase 1 enzyme (Uniprot: Q8NBJ5) has neither an existing PDB entry nor a PDB structure for a homologous protein [using protein Basic Local Alignment Search Tool (BLAST) against the PDB database (39)]. AF2, however, produced a model with over 85% of residues having pLDDT ≥70 (Fig. 3F). Importantly, all five high-confidence URPs were satisfied within this model, compared to only 8% in the random URP control. Similarly, the very-long-chain 3-oxoacyl-CoA reductase (Uniprot: Q53GQ0) had little structural precedent but AF2 produced a model with >95% of residues having pLDDT ≥70 and for which all nine URPs were satisfied (SI Appendix, Fig. S6C).
Third, cross-links that do not corroborate AF2 models are also of interest as they indicate incongruence between experimental data and modeled structures. In total, there were 66 proteins with structure predictions for which all cross-links were violated in the AF2 model. For example, despite >90% of the sequence the nuclear protein localization protein 4 (NPL4; Uniprot: Q8TAT6) being well-modeled by AF2, neither of the two mappable URPs were satisfied on the model (Fig. 3G). Examination of the model revealed that a ~20-residue linker with low pLDDT scores separates an N-terminal domain from the bulk of the protein and that both cross-links bridge these two domains. Repositioning of the N-terminal domain to satisfy the cross-links would involve a straightforward rigid-body rearrangement. Proteome-wide XL-MS data thus provide a resource to aid interpretation of—and potentially improve upon—structural models, making use of either manual “structural sculpting” [e.g., as implemented in XMAS (40)] or integrative modeling pipelines (reviewed in ref. 41) that can be driven by cross-linking restraints.
Cross-Links Reveal and Corroborate Thousands of PPIs.
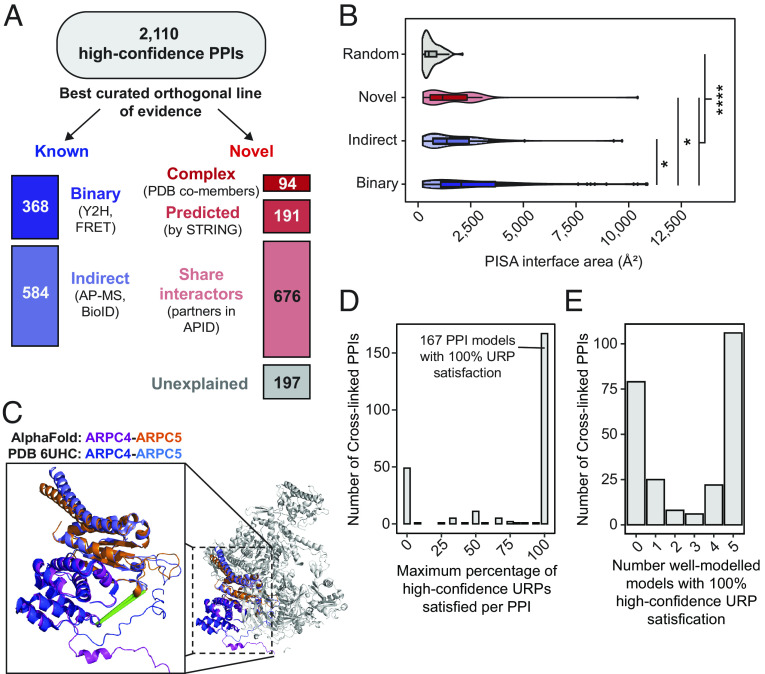
Our cross-link resource identifies and provides insights into the interfaces of thousands of PPIs. A total of 3,785 interprotein URPs describe 2,110 PPIs, including 84 unambiguous homooligomers (Fig. 4A).
Fig. 4.
Discovery and AlphaFold Multimer-v2 modeling of protein–protein interactions. (A) Stratification of protein–protein interactions by their best existing curated evidence. Categories for “known” PPIs include those annotated with a binary, or if not, an indirect interaction mapping technique in the APID database. Categories for “novel” PPIs include proteins cocurated in the same CORUM complex, or PDB entry, or those predicted by STRING (combined scores at least 0.4), or those that share local interactome partners. (B) The largest interface surface area per PPI calculated by PISA for well-modeled AlphaFold Multimer-v2 models (clashscores ≤ 100, average interface residue pLDDT ≥70, interface areas > 200 Å2) generated for interactions with at least 2 URPs (stratified by best evidence—Binary, Indirect, or Novel) and for random protein pairs. (C) The predicted AlphaFold Multimer-v2 model of the ARPC4-ARPC5 overlays very well (RMSD = 0.9 Å) with the known PDB: 6UHC structure of the full 7-subunit Arp2/3 complex. (D and E) Only high-confidence URPs (both cross-linked residues with pLDDT ≥ 70) were used for assessment. (D) The distribution of maximum percentage satisfaction rates of high-confidence URPs per well-modeled PPI. (E) The number of well-modeled AlphaFold Multimer-v2 models with fully satisfied URPs per PPI.
Comparison of our dataset to the APID PPI metadatabase (2) showed that 55% (1,158) of these PPIs had not been previously described (Dataset S5). We therefore assessed the degree of orthogonal evidence available for each protein pair by sorting the PPIs according to their highest level of supporting evidence. Strikingly, 584 (more than 60%) of our 952 known interactions had only been described by indirect interaction mapping methods, such as affinity purification mass spectrometry and proximity ligation assays.
Of the novel interactions, most could be explained by leveraging existing systems-level annotations (Fig. 4A). First, a small but significant number (94) appeared to be previously characterized interactions that had simply escaped annotation in APID (or by systematic interactome screens), based on the fact that the cross-linked proteins reside in the same CORUM-annotated complex (42) or even the same PDB entry. Second, 191 of the remaining novel PPIs were predicted with at least medium confidence by the STRING database (combined score of at least 0.4) (43), which integrates information across multiple lines of evidence including known interactions curated for homologous protein pairs, gene and protein coexpression, and literature text-mining. Third, 676 of the remaining novel PPIs shared APID-annotated interaction partners and hence local interactomes (at one degree of separation).
Last, using the resources above, 197 PPIs of the novel PPIs remain “unexplained”. Of these, ~16% (32) involved the heat-shock protein Hspa1b [Uniprot: P0DMV9, (44)]. Because the function of this protein involves promiscuous binding to many protein substrates, our cross-links might well be capturing biologically relevant interactions. In contrast, the CCT complex subinteractome, which is known to be a more specific chaperone system, had many previously undetected PPIs but none that were categorized in the above scheme as unexplained.
AlphaFold Multimer-v2 Predicts Structural Interfaces for Hundreds of Cross-Linked PPIs.
One advantage of XL-MS over other systematic interactome mapping approaches is its ability to localize interaction interfaces rather than simply identify interactions. Because AF2 has recently been extended to predict the structures of protein complexes (45), we used our data to inform and assess AF2-generated models of complexes. We chose the subset of 590 PPIs from our dataset that were captured by at least two URPs and subjected 530 of these (some interactions were too large to model our high-performance computing hardware setup) to AlphaFold Multimer-v2 modeling (Dataset S6). As a control, 400 randomly sampled protein pairs from the pool of proteins identified in our study were also modeled.
We assessed the overall quality of predicted structures of complexes by examining several structural measures. First, we examined the number of clashes within the models [identified by MolProbity (46)], which was reported to be unusually high (47) in the initial release of AlphaFold Multimer. However, most models had acceptably low levels of clashes (clashscore ≤ 100; i.e., less than 10% of atoms clashing) (SI Appendix, Fig. S7A). We then used PISA (48) to identify structural interfaces in our models and defined “well-modeled” interfaces as those that had an average pLDDT score of ≥70 for PISA-identified interface residues (SI Appendix, Fig. S7B), a PISA-defined interface size of >200 Å2 (SI Appendix, Fig. S7C) and a clashscore ≤100. Using these criteria, AlphaFold Multimer-v2 generated at least one well-modeled structure (from the five generated per run) for a total of 343 of 530 (69%) cross-linked PPIs compared to 78 of the 400 (19%) random protein pairs (Dataset S6).
Well-modeled AlphaFold Multimer-v2 models for PPIs defined by cross-links had significantly larger interaction surface areas than randomly sampled protein pairs (P < 2.2 × 10−16), corroborating the novel interactions defined by our data. Interestingly, the size of interaction interfaces also differed for PPIs that had previously been detected by different interactome mapping approaches. For example, PPIs described by binary interactome mapping approaches had, by comparing median values, 1.5-fold larger interface areas than those described only by indirect methods (AP-MS, BioID) (P = 0.003) (Fig. 4B). Binary PPI interfaces were also significantly larger than novel PPI interfaces (>1.5-fold larger, P = 0.007). On the other hand, novel and known (but only) indirect PPIs did not significantly differ in interface size (P = 0.23). This observation perhaps flags differences in the nature of PPIs that can be detected by binary assays carried out in a heterologous system (e.g., Y2H) vs. approaches (such as XL-MS or AP-MS) that retain native PTMs and additional complex partners. Hits in the former assays will be restricted to PPIs that are more robust and less dependent on biological context.
We were also able to use the experimental structures in the PDB to assess the AlphaFold Multimer-v2 dimer predictions. Of the 343 well-modeled complexes defined above, 151 comprised pairs of proteins that could be found in the same PDB entry. Furthermore, an additional 81 PPIs had PDB entries curated for homologous protein pairs (at least 40% sequence similarity). We superimposed the AlphaFold Mulitmer-v2-derived structures onto their corresponding PDB structures for these 232 proteins and found that 154 pairs (66%) aligned well, with a median RMSD value of 1.6 Å (Dataset S6).
Limitations in the Use of AlphaFold Multimer-v2 Modeling for the Identification of Protein Interactions.
In our dataset, AlphaFold Multimer-v2 Performed most poorly in situations where the two proteins are part of the same complex but do not make direct contacts in the experimental structure. For example the Arp2/3 complex, which is involved in the formation of branched actin networks (49), comprises seven subunits and has had its structure determined by cryo-EM (PDB: 6UHC (50), Fig. 4C). Based on the existence of cross-links, we predicted models for three subcomplexes: ARPC4-ARPC5 (Uniprot: P59998 and O15511), ARP2-ARPC2 (Uniprot IDs: P61160 and O15144), and ARP2-ARPC3 (Uniprot: P61160 and O15145). The prediction for ARPC4-ARPC5, which featured an interface of 830 Å2, closely matched the experimental structure (RMSD = 0.9 Å) and is supported by five URPs (Fig. 4C). However, although the other two subunit pairs do not make direct contact in the experimental structure, AlphaFold Multimer-v2 predicted spurious complexes that had interfaces of 830 and 430 Å2, respectively. All three URPs (two for ARP2-ARPC3, one for ARP2-ARPC2) did not fit the AlphaFold Multimer-v2 models for ARP2-ARPC2 (SI Appendix, Fig. S8A) and ARP2-ARPC3 (SI Appendix, Fig. S8B). In contrast, all three URPs were fulfilled in the experimental structure of the seven-subunit complex.
Similarly, we observed cross-links between pairs of histones such as histone H2A and histone H4. Although these subunits do make contact in the native nucleosome structure, they form more intimate dimers with H2B and H3 (PDB: 1AOI), respectively. However, because all four of these core histones have the same fold, AlphaFold Multimer-v2 incorrectly predicts a structure for the H2A-H4 complex that reflects the H3-H4 (or H2A-H2B) complex instead (SI Appendix, Fig. S8C). It is noteworthy that misaligned histones alone account for 32 out of the 78 (41%) models that poorly match their experimental counterparts.
In summary, we find that although AlphaFold Multimer-v2 is largely correct in its model predictions, we would caution its use as a “discovery” tool to identify PPIs. Such models should be corroborated with additional orthogonal experimental data such as XL-MS data.
Cross-Links Provide Insight into AlphaFold Multimer-v2 Complex Models.
We asked whether our URP distance restraints could experimentally corroborate the 343 well-modeled dimer interfaces calculated above. These dimers each feature between two and 39 URPs, although many URPs connect residues that were not confidently modeled by AlphaFold Multimer-v2. We therefore used only URPs involving high-confidence residues (i.e., pLDDT ≥70 for both cross-linked residues) for model assessment. As a result, 97 models did not have any URPs that met this criterion, leaving 246 models that were described by 1 to 32 URPs (Dataset S6). For comparison, we also simulated random URPs for each model. We found that the Euclidean distances measured for our experimental interprotein URPs were considerably shorter than the randomly sampled URPs, and also shorter than the randomly sampled URPs in the 400 randomly sampled protein pairs (SI Appendix, Fig. S9, P < 2.2 × 10−16 for both comparisons). Furthermore, even a single high-confidence URP was useful for corroborating the location of a predicted interface, performing better than a single randomly sampled but still high-confidence URP (76% of cross-linked PPIs assessed using an experimental URP were satisfied compared to just 14% when using a random URP). Thus, our experimental cross-links are enriched at the interface of the predicted models, consistent with AlphaFold Multimer-v2 generating predictions that reflect the true structures of these complexes.
When assessing the overall degree of cross-link satisfaction in our well-modeled PPIs, we found that 167 out of 246 PPIs (68%) had a 100% URP satisfaction rate in at least one of the five AlphaFold Multimer-v2 models (Fig. 4D), and 106 of those 167 had 100% URP satisfaction for all five models (Fig. 4E). Interestingly, the distribution of cross-link satisfaction rates for dimer predictions was clearly bimodal, which we did not observe for monomeric predictions (SI Appendix, Fig. S6A). This suggests that AlphaFold Multimer-v2 was either getting the models largely right or largely wrong. It is possible that “wrong” models arise from AlphaFold Multimer-v2 not having all the necessary information (e.g., additional interaction partners, PTMs). In summary, it further highlights the need for experimental data, such as XL-MS, to validate AlphaFold Multimer-v2 generated models.
Of the 246 PPIs with at least 1 high-confidence URP, 171 had either a preexisting PDB structure or a PDB structure involving homologous protein pairs. As described above, superimposition of AlphaFold Multimer-v2 predictions and these experimental structures revealed that there were 129 PPI models that aligned well and 42 that did not align well. Strikingly, 83% (107 of 129) of the well-aligned models had 100% URP satisfaction rates, compared to just 60% (25 of 42) for the poorly aligned models. This difference between the two groups is increased further to 81% (well-aligned models) vs. 46% (poorly aligned models) when histones were excluded (on the basis that histones are small proteins, meaning that cross-links are much more likely to be satisfied regardless of how the proteins were arranged relative to each other). This suggests that high-confidence URPs could be used to discriminate between a correct well-modeled AlphaFold Multimer-v2 model and an incorrect but well-modeled model.
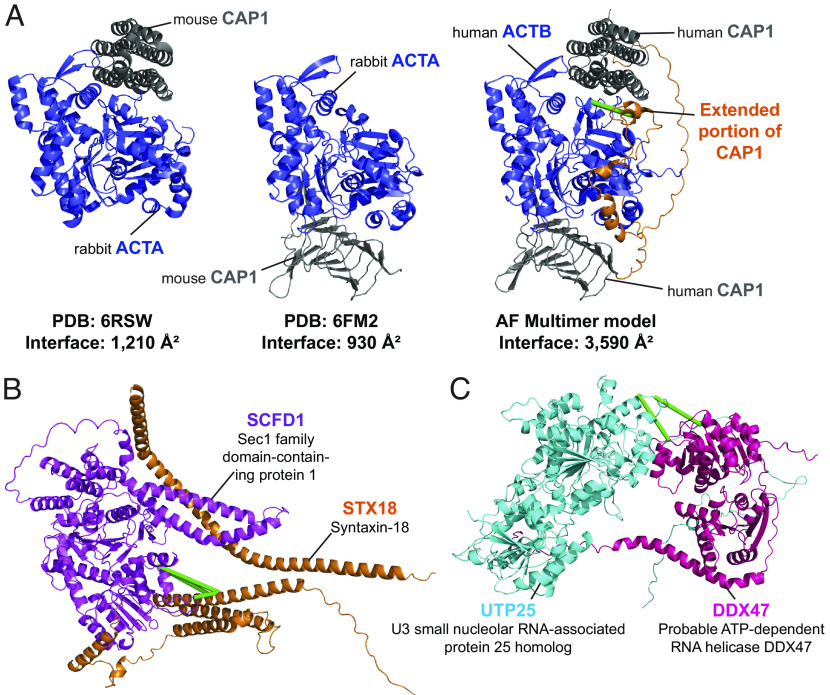
Similar to the situation for monomeric AF2 models (Fig. 3 D and E), some dimer models confirmed and extended existing experimental structures of human PPIs. For example, although no PDB structure exists for the human beta-actin ACTB (Uniprot: G5E9R0) in complex with the adenyl cyclase-associated protein 1 CAP1 (Uniprot: Q01518), there are two PDB entries [6FM2 and 6RSW (51, 52)] comprising the close homologues alpha-actin ACTA from rabbit (Uniprot: P68135) and CAP1 from mouse (Uniprot: P40124). Our AlphaFold Multimer-v2 model was both able to confirm conservation of the overall structure in the human ACTB-CAP1 complex and extend coverage of the interface itself via two corroborating interprotein URPs (Fig. 5A). This previously unresolved and cross-link-validated interface indicates that the interaction between ACTB and the CAP1 protein is more intimate than previously appreciated, featuring 30 hydrogen bonds, 20 salt bridges, and an interface area of 3,590 Å2 in our AlphaFold Multimer-v2 model vs. a combined 19 hydrogen bonds, 17 salt bridges and an interface area 2,140 Å2 in the experimental X-ray crystal structures.
Fig. 5.
Cross-links enable assessment of computational predictions of structural interfaces mediating protein–protein interactions. (A) AlphaFold Mulitmer-v2 model of human beta actin ACTB in complex with the adenyl cyclase-associated protein 1 CAP1 (Middle) vs. the PDB structures 6FM2 (Left) and 6RSW (Right) of homologous proteins alpha actin ACTA from rabbit in complex with the mouse CAP1 protein. The AlphaFold multimer-v2 model largely recapitulates the regions resolved in the PDB structures and extends the interaction surface further (orange). (B and C) Examples of AlphaFold Multimer-v2 models of structurally undefined PPIs that have satisfied all URPs. Green lines denote satisfied URPs. (B) The model of the interaction between STX18 (Q9P2W9; orange) and SCFD1 (Q8WVM8; magenta). Four interprotein URPs support this model. (C) The model of the interaction between UTP25 (Q68CQ4; cyan) and DDX47 (Q9H0S4; pink). Two interprotein URPs support this model.
From our analyses, we also identified 75 PPIs with well-modeled AlphaFold Multimer-v2 interfaces that were structurally undefined (i.e., have no shared or homologous PDB entries available). Many of these PPIs had been previously characterized by binary and direct interaction mapping approaches. Of these 75 PPIs, 35 models had 100% URP satisfaction; these 35 models thus have a high likelihood of being correct. It also appears that our cross-links were better satisfied in predicted models when homologous structures were available (38/59, 64% with 100% URP satisfaction). This result might reflect a combination of i) a bias of the AlphaFold-Multimer-v2 software toward its training dataset (the PDB) and ii) the poorer evolutionary information available (through lower sequence coverage in the multiple sequence alignments) for proteins in entirely novel dimer structures (53). An example of a corroborated dimer interface without any structural precedent in the PDB is the interaction of STX18 (Uniprot: Q9P2W9) and SCFD1 (Uniprot: Q8WVM8) was previously described by four studies (54–57). Our four interprotein URPs were satisfied in all five models, and the top model featured an interface area of 3,688 Å2 with 21 hydrogen bonds and 15 salt bridges (Fig. 5B). The formation of this PPI is implicated in the regulation of skeletal development (58) and, interestingly, there is evidence for an amyotrophic lateral sclerosis disease-causing missense mutation involving amino acid I70 in SCFD1 (I70T, ClinVar accession RCV001260556.1), a residue that is near cross-linked residues K63 and K61 at the interface. The combination of our data with AlphaFold Multimer-v2 thus provides precise and accurate structural context for biologically relevant PPIs.
Our data can also be used to both define PPIs and experimentally corroborate their predicted interaction interface. For example, the top ranked model for the proposed interaction between UTP25 (Uniprot: Q68CQ4) and DDX47 (Uniprot: Q9H0S4) displays a 1,030 Å2 interface area and features two satisfied URPs (one of high confidence) (Fig. 5C). The interface area and number of hydrogen bonds and salt bridges contributing to the interface (two for both categories) are significantly smaller than those seen in the previous two modeled PPIs described above. However, the two proteins were confidently predicted to interact in the STRING database (combined score of 0.98 out of a maximum of 1) and appear to have functional overlap. DDX47 is a DEAD-box RNA helicase known to associate with pre-RNAs (59) and UTP25 is implicated in preribosomal rRNA processing (60, 61). Notably, both URPs defining this PPI were detected in the nuclear fraction, consistent with a shared role in ribosome biogenesis. It is possible that the presence of RNA cofactors or other protein complex partners could be required to stabilize this PPI. This again highlights the ability of XL-MS to capture weaker interactions than some other interactome screening technologies and the value of the native context provided in our XL-MS approach.
Cross-Links Define the Binary Interactions That Underlie the Assembly of Larger Protein Complexes.
Because our URPs can also be derived from higher order assemblies, we mapped our data onto the CORUM database of manually curated protein complexes (42). Our URPs could be mapped onto 366 unique CORUM-documented complexes (Dataset S7). Of these 366 complexes, 165 displayed XLs for at least two distinct pairs of subunits. We considered the most densely cross-linked CORUM complexes, where ≥70% of proteins annotated in the complex were involved in at least one cross-linked PPI. In total, there were 51 such complexes. Some, such as the CCT complex (Dataset S7), are found in the PDB, with all cross-linked PPIs sharing PDB entries and 88% of the mappable URPs satisfied. However, almost half (21) of the densely cross-linked complexes were missing structural information for at least one PPI for which we found cross-linking evidence. These complexes could benefit from integrative structural modeling approaches, such as the cross-link guided modeling pipelines used in Assembline (62) and IMProv (63).
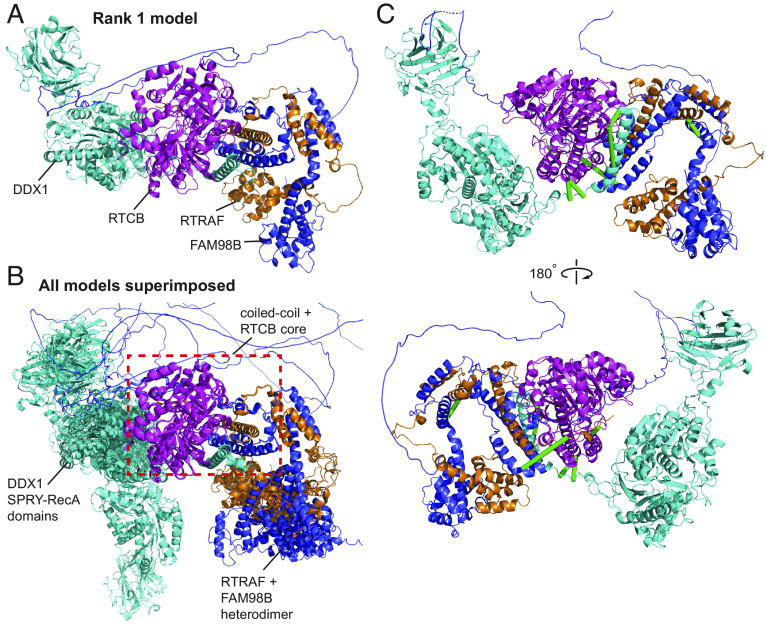
Additionally, we asked whether AlphaFold Multimer-v2 could be used to generate structures for higher order complexes and whether our cross-links could corroborate the models. The pentameric tRNA ligase complex comprising DDX1, FAM98B, RTRAF, RTCB, and ASHWIN (Uniprot: Q92499, Q52LJ0, Q9Y224, Q9Y3I0 and Q9BVC5; CORUM #6301) plays an essential role in tRNA splicing (64). While there are structures (including homologous structures) of some individual subunits and domains [PDB: 7P3B (65); PDB: 7P3A (65); PDB: 4XW3 (66); PDB: 6O5F (67)], no structures of subcomplexes or the complete complex are available. Although AlphaFold Multimer-v2 failed to produce a model of the full complex, it successfully predicted a model for a core four-membered complex, based on existing biochemical data that showed that only DDX1, FAM98B, RTRAF, and RTCB are essential for complex formation (65) (Fig. 6A).
Fig. 6.
Cross-links support the AlphaFold Multimer-v2 model of the tRNA ligase core complex. DDX1 (Q92499) is in cyan, RTCB (Q9Y3I0) is in magenta, RTRAF (Q9Y224) is in orange and FAM98B (Q52LJ0) is in blue. (A) Rank 1 AF Multimer-v2 model of the tRNA ligase core complex. (B) All five AlphaFold Multimer-v2 models superimposed onto the Rank 1 model using the coiled-coiled core as the reference point. The coiled-coil core comprising of the C-terminal helices of DDX1, RTRAF, and FAM98B together with RTCB is consistent across all five models (red dashed box). (C) Twelve URPs mapped onto the Rank 1 model of the tRNA ligase core complex. Eight URPs are fulfilled (green lines) The remaining four overlength URPs (not shown) were cross-linked to mobile regions of the DDX1 SPRY-RecA domains and the disordered C-terminal tail of RTRAF.
All five AlphaFold Multimer-v2 models featured a central helical bundle comprising the C-terminal helices of DDX1, FAM98B and RTRAF; this bundle packs against the RTCB ligase. In addition, the N-terminal domains of FAM98B and RTRAF formed an intimate heterodimer. Importantly, both observations are supported by available biochemical data (65). While the helical core of the complex was conserved in all models (superimposed RMSDs < 0.5 Å), the orientations of the SPRY-RecA domains of DDX1 and the FAM98B-RTRAF heterodimer showed considerable variability relative to the core (Fig. 6B). Of the 12 URPs we measured, eight were fulfilled in all models (Fig. 6C). Of the remaining four cross-links, two were to the disordered C-terminal tail of RTRAF and two were to the “mobile” SPRY-RecA domains of DDX1. We had URPs between all members of the complex except to ASHWIN. Therefore, our cross-links support the overall arrangement of the core of the complex that AlphaFold Multimer-v2 has predicted.
Our cross-links were also able to assist in the construction of models for other known multimeric protein complexes, for example the MICOS complex where cross-links clearly supported predicted structures generated using specific subunit stoichiometries (Supplementary Results, SI Appendix, Figs. S10 and S11).
In summary, we show that the combination of XL-MS data, AlphaFold Multimer-v2, and preexisting biochemical data creates a powerful strategy for integrative modeling of higher order native protein complexes without the requirement for recombinant expression and purification.
Discussion
We have generated a cross-link resource reporting on more than 4,000 human proteins in near-native states with native sequences, abundances and contexts (e.g., the presence of native PTMs, relevant partner proteins as well as other macromolecules, cofactors and metabolites), which is a significant advantage over many other methods used to map protein structure and interactions. We have demonstrated that our cross-links are both of high quality and utility, and that they significantly extend and annotate the human structural proteome and interactome with very high throughput and biological relevance.
Moving Beyond the Lysine Cross-Linked Proteome.
Our study highlights the value of moving beyond the lysine (K) to lysine (K) cross-linked proteome. Alternative cross-linker chemistries, such as bismaleimide (BMSO) (68) (homobifunctional, cysteines), can provide further, orthogonal information but have yet to be used routinely in proteome-wide studies. Photoreactive diazirine-based cross-linkers such as sulfosuccinimidyl 4,4′-azipentanoate (sulfo-SDA) (69), which allow cross-linking at any residue to be explored, represent a further extension. Although the greatest limitation for non-MS-cleavable cross-linkers of this type is the computing time required during peptide identification, the recent development of the MS-cleavable succinimidyl diazirine sulfoxide cross-linker (70) may aid in the feasibility and scale of such endeavors.
A recent community-wide, multilab study recommended that the search space for NHS-ester-based cross-linking should be expanded to include S/T/Y residues (27). However, due to its prohibitive computational cost, searches including S/T/Y linkages are not routinely performed, especially in large-scale cross-linking studies. In our dataset and in line with previous conclusions (27, 28), a significant portion (~24%) of DSSO URPs involved S/T/Y residues. However, we note that only ~1% had S/T/Y to S/T/Y linkages, with most being K to S/T/Y linkages. This highlights that future studies should define NHS-ester reactivity as K to K/S/T/Y to balance computation time with localization improvements and cross-link gains. This hybrid search strategy is currently configurable in several cross-link search engines such as pLink 2 (71) and MeroX (72). With a small compromise (reducing the size of the sequence database), we demonstrate that including S/T/Y linkages is possible on a large-scale basis.
Finally, although we have employed a diversified strategy to improve on the coverage of the cross-linked proteome using several cross-linkers combined with enrichment and fractionation strategies, many further possible enhancements exist. For example, the use of alternative proteases is relatively easy to implement but has seen limited use thus far (73, 74). Promising enzymes that will likely provide orthogonal information include pepsin (cuts at Y/F/W), AspN (cuts at D), and ProAlanase (cuts at P/A) (75).
A Substantial Cross-Linked Proteome Resource That Complements AI-Based Structure Predictions.
We have demonstrated how our dataset can be leveraged to gain a deeper understanding of the biochemistry and biology of a higher eukaryote. Our structurally near-native cross-links corroborate experimentally determined protein structures—and with the advantage of being relatively fast, having modest sample requirements and providing data for proteins that are otherwise difficult to handle in isolation. We also identified regions of potential conformational variability and noted that PTMs might contribute to these variabilities. Furthermore, we show that XL-MS data can provide powerful corroboration of AI-based structure predictors like AlphaFold2, which have democratized access to protein structure modeling for nonspecialist groups and provide access to experimentally intractable proteins. With the recent release of more than 200 million predicted structures by AlphaFold2 (76) and 600 million in the ESM Metagenomic Atlas (77), it is now more important than ever to have orthogonal data to corroborate these models. However, it is important to note that, for large proteins (>2,700 amino acids), the AlphaFold2 database (78) combines multiple predictions of overlapping amino acid sequences that cover the full sequence). Furthermore, the hardware limitations of our supercomputer GPU prevented the modeling of larger multimeric complexes (>2,000 amino acids). Therefore, development of more computationally efficient structural prediction software [e.g., OpenFold (79)], will be required to enable the prediction of missing subsets of the structural proteome and interactome.
Perhaps most importantly, our data define 2,110 PPIs, a considerable extension in the coverage and diversity of the structural interactome. The URPs that define these PPIs can be used in combination with AI-based tools to generate and assess models of protein complexes, and such models can be subjected to experimental verification using biochemical and cellular approaches. Furthermore, regardless of AlphaFold2 modeling status, all 2,110 PPIs defined here contain cross-linking constraints that can help localize their interfaces at the sequence level without any reference to structure. Many interactions are known to occur exclusively in disordered regions, mediated by sequence motifs and domains (reviewed in ref. 80). Our cross-linking resource can therefore also be used to assist the annotation of domain–domain interactions and linear interaction motifs [as, for example, in Pfam (81)]—efforts that help to infer function for the thousands of understudied human proteins (12).
The wide accessibility of different cross-linkers, alongside critical efforts to standardize XL-MS search engines and analyses (82, 83), will enable high-volume and high-confidence cross-link constraint sets to be generated for proteins from diverse conditions and organisms. We note that the datasets provided by such large-scale studies are best used to corroborate structural predictions and to define interactions. For the purposes of detailed structural modeling, follow-up XL-MS studies on purified proteins and complexes will provide superior cross-link densities that will be valuable in integrative modeling pipelines (62, 84). One could envisage a (near) future in which archived XL-MS data is harvested automatically by AlphaFold2 or similar systems and optionally displayed on predicted structures. Likewise, interprotein cross-links could be displayed to alert the user that they could benefit from incorporating additional proteins into their prediction.
Methods
Detailed descriptions on how the subcellular fractionation, protein cross-linking, peptide preparation for mass spectrometry, mass spectrometry data acquisition, data analysis, statistical analysis, implementation of AlphaFold Multimer-v2 model predictions and associated analyses can be found in SI Appendix.
Supplementary Material
Appendix 01 (PDF)
Dataset S01 (XLSX)
Dataset S02 (XLSX)
Dataset S03 (XLSX)
Dataset S04 (XLSX)
Dataset S05 (XLSX)
Dataset S06 (XLSX)
Dataset S07 (XLSX)
Acknowledgments
We acknowledge the Sydney Mass Spectrometry Core Research Facility at the University of Sydney for providing access to mass spectrometers and thank the technical staff for the maintenance of the instruments. This project was undertaken with the assistance of resources and services from the National Computational Infrastructure, which is supported by the Australian Government. T.K.B. acknowledges the support of an Australian Government Research Training Program scholarship. M.R.W. acknowledges support from the Australian Research Council, Australia, and J.P.M acknowledges support from the National Health and Medical Research Council, Australia. M.R.W. and X.V.-C. acknowledge support from the New South Wales State Government Research Attraction and Acceleration Program (RAAP) scheme and the University of New South Wales Research Infrastructure Scheme (RIS). J.P.M. acknowledges support from the Australian National Health and Medical Research Council.
Author contributions
T.K.B., M.R.W., J.P.M., and J.K.K.L. designed research; T.K.B., X.V.-C., C.L., and J.K.K.L. performed research; X.V.-C., A.N., and R.J.P. contributed new reagents/analytic tools; T.K.B., X.V.-C., C.L., M.J., J.P.M., and J.K.K.L. analyzed data; and T.K.B. and J.K.K.L. wrote the paper.
Competing interests
The authors declare no competing interest.
Footnotes
This article is a PNAS Direct Submission.
Data, Materials, and Software Availability
Key data processing scripts (including for the filtering and consolidation of cross-linked spectral matches across software suites) have been uploaded to FigShare and are available at https://doi.org/10.6084/m9.figshare.21561645.v1 (85). All mass spectrometry data and XLMS search results have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository (86) with the dataset identifier PXD035844. The dimeric models are available in ModelArchive (modelarchive.org) with the accession codes ma-low-csi and at https://modelarchive.org/doi/10.5452/ma-low-csi (87).
Supporting Information
References
- 1.Iacobucci I., Monaco V., Cozzolino F., Monti M., From classical to new generation approaches: An excursus of -omics methods for investigation of protein-protein interaction networks. J. Proteomics 230, 103990 (2021). [DOI] [PubMed] [Google Scholar]
- 2.Alonso-Lopez D., et al. , APID database: Redefining protein-protein interaction experimental evidences and binary interactomes. Database (Oxford) 2019, baz005 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dana J. M., et al. , SIFTS: Updated structure integration with function, taxonomy and sequences resource allows 40-fold increase in coverage of structure-based annotations for proteins. Nucleic Acids Res. 47, D482–D489 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Berman H. M., et al. , The protein data bank. Nucleic Acids Res. 28, 235–242 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mosca R., Ceol A., Aloy P., Interactome3D: Adding structural details to protein networks. Nat. Methods 10, 47–53 (2013). [DOI] [PubMed] [Google Scholar]
- 6.Jumper J., et al. , Highly accurate protein structure prediction with AlphaFold. Nature 596, 583–589 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pereira J., et al. , High-accuracy protein structure prediction in CASP14. Proteins 89, 1687–1699 (2021). [DOI] [PubMed] [Google Scholar]
- 8.Porta-Pardo E., Ruiz-Serra V., Valentini S., Valencia A., The structural coverage of the human proteome before and after AlphaFold. PLoS Comput. Biol. 18, e1009818 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tunyasuvunakool K., et al. , Highly accurate protein structure prediction for the human proteome. Nature 596, 590–596 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Graziadei A., Rappsilber J., Leveraging crosslinking mass spectrometry in structural and cell biology. Structure 30, 37–54 (2022). [DOI] [PubMed] [Google Scholar]
- 11.Matzinger M., Mechtler K., Cleavable cross-linkers and mass spectrometry for the ultimate task of profiling protein-protein interaction networks in vivo. J. Proteome Res. 20, 78–93 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kustatscher G., et al. , An open invitation to the Understudied Proteins Initiative. Nat. Biotechnol. 40, 815–817 (2022). [DOI] [PubMed] [Google Scholar]
- 13.Jiang P. L., et al. , A membrane-permeable and immobilized metal affinity chromatography (IMAC) enrichable cross-linking reagent to advance in vivo cross-linking mass spectrometry. Angew. Chem. Int. Ed. Engl. 61, e202113937 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wheat A., et al. , Protein interaction landscapes revealed by advanced in vivo cross-linking-mass spectrometry. Proc. Natl. Acad. Sci. U.S.A. 118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chavez J. D., et al. , Chemical crosslinking mass spectrometry analysis of protein conformations and supercomplexes in heart tissue. Cell Syst. 6, 136–141.e5 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bartolec T. K., et al. , Cross-linking mass spectrometry analysis of the yeast nucleus reveals extensive protein-protein interactions not detected by systematic two-hybrid or affinity purification-mass spectrometry. Anal. Chem. 92, 1874–1882 (2020). [DOI] [PubMed] [Google Scholar]
- 17.Liu F., Lossl P., Rabbitts B. M., Balaban R. S., Heck A. J. R., The interactome of intact mitochondria by cross-linking mass spectrometry provides evidence for coexisting respiratory supercomplexes. Mol. Cell Proteomics 17, 216–232 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Belsom A., Rappsilber J., Anatomy of a crosslinker. Curr. Opin. Chem. Biol. 60, 39–46 (2021). [DOI] [PubMed] [Google Scholar]
- 19.Mintseris J., Gygi S. P., High-density chemical cross-linking for modeling protein interactions. Proc. Natl. Acad. Sci. U.S.A. 117, 93–102 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gutierrez C. B., et al. , Developing an acidic residue reactive and sulfoxide-containing MS-cleavable homobifunctional cross-linker for probing protein-protein interactions. Anal. Chem. 88, 8315–8322 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kao A., et al. , Development of a novel cross-linking strategy for fast and accurate identification of cross-linked peptides of protein complexes. Mol. Cell Proteomics 10, M110 002212 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Leitner A., et al. , Chemical cross-linking/mass spectrometry targeting acidic residues in proteins and protein complexes. Proc. Natl. Acad. Sci. U.S.A. 111, 9455–9460 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Keller A., Chavez J. D., Felt K. C., Bruce J. E., Prediction of an upper limit for the fraction of interprotein cross-links in large-scale in vivo cross-linking studies. J. Proteome Res. 18, 3077–3085 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jiao F., et al. , Exploring an alternative cysteine-reactive chemistry to enable proteome-wide PPI analysis by cross-linking mass spectrometry. Anal. Chem. 95, 2532–2539 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gao H., et al. , In-depth in vivo crosslinking in minutes by a compact, membrane-permeable, and alkynyl-enrichable crosslinker. Anal. Chem. 94, 7551–7558 (2022). [DOI] [PubMed] [Google Scholar]
- 26.Wang M., Herrmann C. J., Simonovic M., Szklarczyk D., von Mering C., Version 4.0 of PaxDb: Protein abundance data, integrated across model organisms, tissues, and cell-lines. Proteomics 15, 3163–3168 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Iacobucci C., et al. , First community-wide, comparative cross-linking mass spectrometry study. Anal. Chem. 91, 6953–6961 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Smith D. L., Gotze M., Bartolec T. K., Hart-Smith G., Wilkins M. R., Characterization of the interaction between arginine methyltransferase Hmt1 and its substrate Npl3: Use of multiple cross-linkers, mass spectrometric approaches, and software platforms. Anal. Chem. 90, 9101–9108 (2018). [DOI] [PubMed] [Google Scholar]
- 29.Kahraman A., Malmstrom L., Aebersold R., Xwalk: Computing and visualizing distances in cross-linking experiments. Bioinformatics 27, 2163–2164 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cammarata M. B., Macias L. A., Rosenberg J., Bolufer A., Brodbelt J. S., Expanding the scope of cross-link identifications by incorporating collisional activated dissociation and ultraviolet photodissociation methods. Anal. Chem. 90, 6385–6389 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tuting C., Iacobucci C., Ihling C. H., Kastritis P. L., Sinz A., Structural analysis of 70S ribosomes by cross-linking/mass spectrometry reveals conformational plasticity. Sci. Rep. 10, 12618 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Haselbach D., et al. , Structure and conformational dynamics of the human spliceosomal B(act) complex. Cell 172, 454–464.e11 (2018). [DOI] [PubMed] [Google Scholar]
- 33.Townsend C., et al. , Mechanism of protein-guided folding of the active site U2/U6 RNA during spliceosome activation. Science 370, eabc3753 (2020). [DOI] [PubMed] [Google Scholar]
- 34.Rzechorzek N. J., Hardwick S. W., Jatikusumo V. A., Chirgadze D. Y., Pellegrini L., CryoEM structures of human CMG-ATPgammaS-DNA and CMG-AND-1 complexes. Nucleic Acids Res. 48, 6980–6995 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jones M. L., Baris Y., Taylor M. R. G., Yeeles J. T. P., Structure of a human replisome shows the organisation and interactions of a DNA replication machine. EMBO J. 40, e108819 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hornbeck P. V., et al. , PhosphoSitePlus, 2014: Mutations, PTMs and recalibrations. Nucleic Acids Res. 43, D512–D520 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Piovesan D., et al. , MobiDB: Intrinsically disordered proteins in 2021. Nucleic Acids Res. 49, D361–D367 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Brown A., et al. , Structure of the large ribosomal subunit from human mitochondria. Science 346, 718–722 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Altschul S. F., et al. , Protein database searches using compositionally adjusted substitution matrices. FEBS J. 272, 5101–5109 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lagerwaard I. M., Albanese P., Jankevics A., Scheltema R. A., Xlink mapping and analysis (XMAS)–Smooth integrative modeling in ChimeraX. bioRxiv [Preprint] (2022). 10.1101/2022.04.21.489026 (Deposited 1 July 2022). [DOI]
- 41.Rout M. P., Sali A., Principles for integrative structural biology studies. Cell 177, 1384–1403 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Giurgiu M., et al. , CORUM: The comprehensive resource of mammalian protein complexes-2019. Nucleic Acids Res. 47, D559–D563 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Szklarczyk D., et al. , The STRING database in 2021: Customizable protein-protein networks, and functional characterization of user-uploaded gene/measurement sets. Nucleic Acids Res. 49, D605–D612 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mayer M. P., Hsp70 chaperone dynamics and molecular mechanism. Trends Biochem. Sci. 38, 507–514 (2013). [DOI] [PubMed] [Google Scholar]
- 45.Evans R., et al. , Protein complex prediction with AlphaFold-Multimer. bioRxiv [Preprint] (2022). 10.1101/2021.10.04.463034 (Deposited 1 April 2022). [DOI]
- 46.Chen V. B., et al. , MolProbity: All-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 66, 12–21 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gao M., Nakajima An D., Parks J. M., Skolnick J., AF2Complex predicts direct physical interactions in multimeric proteins with deep learning. Nat. Commun. 13, 1744 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Krissinel E., Henrick K., Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 372, 774–797 (2007). [DOI] [PubMed] [Google Scholar]
- 49.Molinie N., Gautreau A., The Arp2/3 regulatory system and its deregulation in cancer. Physiol. Rev. 98, 215–238 (2018). [DOI] [PubMed] [Google Scholar]
- 50.Zimmet A., et al. , Cryo-EM structure of NPF-bound human Arp2/3 complex and activation mechanism. Sci. Adv. 6, eaaz7651 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kotila T., et al. , Structural basis of actin monomer re-charging by cyclase-associated protein. Nat. Commun. 9, 1892 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kotila T., et al. , Mechanism of synergistic actin filament pointed end depolymerization by cyclase-associated protein and cofilin. Nat. Commun. 10, 5320 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Orlando G., Raimondi D., Vranken W. F., Observation selection bias in contact prediction and its implications for structural bioinformatics. Sci. Rep. 6, 36679 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Huttlin E. L., et al. , The bioplex network: A systematic exploration of the human interactome. Cell 162, 425–440 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Huttlin E. L., et al. , Architecture of the human interactome defines protein communities and disease networks. Nature 545, 505–509 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hirose H., et al. , Implication of ZW10 in membrane trafficking between the endoplasmic reticulum and Golgi. EMBO J. 23, 1267–1278 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Aoki T., et al. , Identification of the neuroblastoma-amplified gene product as a component of the syntaxin 18 complex implicated in Golgi-to-endoplasmic reticulum retrograde transport. Mol. Biol. Cell 20, 2639–2649 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hou N., Yang Y., Scott I. C., Lou X., The Sec domain protein Scfd1 facilitates trafficking of ECM components during chondrogenesis. Dev. Biol. 421, 8–15 (2017). [DOI] [PubMed] [Google Scholar]
- 59.Sekiguchi T., Hayano T., Yanagida M., Takahashi N., Nishimoto T., NOP132 is required for proper nucleolus localization of DEAD-box RNA helicase DDX47. Nucleic Acids Res. 34, 4593–4608 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tao T., et al. , The pre-rRNA processing factor DEF is rate limiting for the pathogenesis of MYCN-driven neuroblastoma. Oncogene 36, 3852–3867 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Charette J. M., Baserga S. J., The DEAD-box RNA helicase-like Utp25 is an SSU processome component. RNA 16, 2156–2169 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rantos V., Karius K., Kosinski J., Integrative structural modeling of macromolecular complexes using assembline. Nat. Protoc. 17, 152–176 (2022). [DOI] [PubMed] [Google Scholar]
- 63.Ziemianowicz D. S., et al. , IMProv: A resource for cross-link-driven structure modeling that accommodates protein dynamics. Mol. Cell Proteomics 20, 100139 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Popow J., et al. , HSPC117 is the essential subunit of a human tRNA splicing ligase complex. Science 331, 760–764 (2011). [DOI] [PubMed] [Google Scholar]
- 65.Kroupova A., et al. , Molecular architecture of the human tRNA ligase complex. Elife 10, e71656 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kellner J. N., Meinhart A., Structure of the SPRY domain of the human RNA helicase DDX1, a putative interaction platform within a DEAD-box protein. Acta Crystallogr. F Struct. Biol. Commun. 71, 1176–1188 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Song H., Ji X., The mechanism of RNA duplex recognition and unwinding by DEAD-box helicase DDX3X. Nat. Commun. 10, 3085 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Gutierrez C. B., et al. , Development of a novel sulfoxide-containing MS-cleavable homobifunctional cysteine-reactive cross-linker for studying protein-protein interactions. Anal. Chem. 90, 7600–7607 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Belsom A., Schneider M., Fischer L., Brock O., Rappsilber J., Serum albumin domain structures in human blood serum by mass spectrometry and computational biology. Mol. Cell Proteomics 15, 1105–1116 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Gutierrez C., et al. , Enabling photoactivated cross-linking mass spectrometric analysis of protein complexes by novel MS-cleavable cross-linkers. Mol. Cell Proteomics 20, 100084 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Chen Z. L., et al. , A high-speed search engine pLink 2 with systematic evaluation for proteome-scale identification of cross-linked peptides. Nat. Commun. 10, 3404 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Iacobucci C., et al. , A cross-linking/mass spectrometry workflow based on MS-cleavable cross-linkers and the MeroX software for studying protein structures and protein-protein interactions. Nat. Protoc. 13, 2864–2889 (2018). [DOI] [PubMed] [Google Scholar]
- 73.Dau T., Gupta K., Berger I., Rappsilber J., Sequential digestion with trypsin and elastase in cross-linking mass spectrometry. Anal. Chem. 91, 4472–4478 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Leitner A., et al. , Expanding the chemical cross-linking toolbox by the use of multiple proteases and enrichment by size exclusion chromatography. Mol. Cell Proteomics 11, M111 014126 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Samodova D., et al. , Proalanase is an effective alternative to trypsin for proteomics applications and disulfide bond mapping. Mol. Cell Proteomics 19, 2139–2157 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Callaway E., “The entire protein universe”: AI predicts shape of nearly every known protein. Nature 608, 15–16 (2022). [DOI] [PubMed] [Google Scholar]
- 77.Lin Z., et al. , Evolutionary-scale prediction of atomic level protein structure with a language model. bioRxiv [Preprint] (2022). 10.1101/2022.07.20.500902 (Deposited 1 September 2022). [DOI] [PubMed]
- 78.Varadi M., et al. , AlphaFold protein structure database: Massively expanding the structural coverage of protein-sequence space with high-accuracy models. Nucleic Acids Res. 50, D439–D444 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ahdritz G., et al. , OpenFold: Retraining AlphaFold2 yields new insights into its learning mechanisms and capacity for generalization. bioRxiv [Preprint] (2022). 10.1101/2022.11.20.517210 (Deposited 1 December 2022). [DOI] [PMC free article] [PubMed]
- 80.Ivarsson Y., Jemth P., Affinity and specificity of motif-based protein-protein interactions. Curr. Opin. Struct. Biol. 54, 26–33 (2019). [DOI] [PubMed] [Google Scholar]
- 81.Mistry J., et al. , Pfam: The protein families database in 2021. Nucleic Acids Res. 49, D412–D419 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Matzinger M., et al. , Mimicked synthetic ribosomal protein complex for benchmarking crosslinking mass spectrometry workflows. Nat. Commun. 13, 3975 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Leitner A., et al. , Toward increased reliability, transparency, and accessibility in cross-linking mass spectrometry. Structure 28, 1259–1268 (2020). [DOI] [PubMed] [Google Scholar]
- 84.van Zundert G. C. P., et al. , The HADDOCK2.2 web server: User-friendly integrative modeling of biomolecular complexes. J. Mol. Biol. 428, 720–725 (2016). [DOI] [PubMed] [Google Scholar]
- 85.Bartolec T., Human XL-MS scripts. Figshare. 10.6084/m9.figshare.21561645.v1. Deposited 30 March 2023. [DOI]
- 86.Perez-Riverol Y., et al. , The PRIDE database resources in 2022: A hub for mass spectrometry-based proteomics evidences. Nucleic Acids Res. 50, D543–D552 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Bartolec T. K., et al. , Structural interfaces predicted for dimer human protein-protein interactions. ModelArchive. https://modelarchive.org/doi/10.5452/ma-low-csi. Deposited 12 October 2022.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix 01 (PDF)
Dataset S01 (XLSX)
Dataset S02 (XLSX)
Dataset S03 (XLSX)
Dataset S04 (XLSX)
Dataset S05 (XLSX)
Dataset S06 (XLSX)
Dataset S07 (XLSX)
Data Availability Statement
Key data processing scripts (including for the filtering and consolidation of cross-linked spectral matches across software suites) have been uploaded to FigShare and are available at https://doi.org/10.6084/m9.figshare.21561645.v1 (85). All mass spectrometry data and XLMS search results have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository (86) with the dataset identifier PXD035844. The dimeric models are available in ModelArchive (modelarchive.org) with the accession codes ma-low-csi and at https://modelarchive.org/doi/10.5452/ma-low-csi (87).