Comprehensive profiling of the enhancer landscape and 3D genome structure in liposarcoma identifies extensive enhancer-oncogene coamplification and enhancer hijacking events, deepening the understanding of how oncogenes are regulated in cancer.
Abstract
Liposarcoma (LPS) is the most common soft-tissue sarcoma in adults with two major subtypes, well differentiated and dedifferentiated. Both subtypes are characterized with the pathognomonic giant ring or marker chromosomes that harbor high copy numbers of known oncogenes. Here, we reported a comprehensive molecular characterization of both tumor and normal tissues from the same patients with LPS, including whole-genome sequencing (WGS), transcriptome, enhancer landscape, and genome-wide three-dimensional (3D) genome structure by Hi-C. Tumor-specific transcripts and regulatory elements were identified, and enhancer coamplification and hijacking events were discovered as novel mechanisms upregulating oncogenes such as MDM2, CDK4, and HMGA2. Combining Hi-C, optical mapping, nanopore long reads, and WGS data partially resolved complex structural variations and reconstructed the local genome and the giant chromosome. Overall, this study provides a comprehensive resource for LPS research and offers insights into how altered enhancers and the 3D genome contribute to gene dysregulation in cancer.
Significance:
Comprehensive profiling of the enhancer landscape and 3D genome structure in liposarcoma identifies extensive enhancer-oncogene coamplification and enhancer hijacking events, deepening the understanding of how oncogenes are regulated in cancer.
Introduction
Liposarcoma (LPS) is the most common type of soft tissue sarcoma, with reports of increased incidence in recent years (1). Among these tumors, well-differentiated LPS (WD-LPS) and dedifferentiated LPS (DD-LPS) are the most frequent subtypes, and account for about half of all cases (2). WD-LPS does not metastasize, and histologically exhibits adipocyte-like morphology. In contrast, DD-LPS is a high-grade non-lipogenic sarcoma and is associated with high recurrence and poor prognosis (2). Up to one-half of patients die within 5 years, and surgery remains the sole therapeutic approach in these potentially devastating tumors (3–5). Additional molecular insights are essential to spur development of novel treatment targets with high efficacy.
WD-LPS and DD-LPS harbor similar genomic alterations, including chromosomal amplification of 12q13–15 and neo-chromosomes, which occur in a giant marker chromosome or a ring chromosome (2). DD-LPS is further associated with additional abnormalities such as copy-number alterations (CNA). The amplification of chromosome 12q13–15 includes many cancer-related genes, such as the oncogene MDM2, cell-cycling regulator CDK4, and chromatin structure regulator HMGA2 (4, 6). Furthermore, supernumerary ring or giant chromosomes in both WD-LPS and DD-LPS, contain almost all the amplified genomic regions from different chromosomes (7). In addition to these common alterations, recurrent amplifications of 1p32 and 6q23, which lead to overexpression of the adipocyte dedifferentiation associated genes JUN and ASK1, have been reported in DD-LPS (8). Further, deletions in chromosome 1p, 11q, 13q, 15q, 17p, and amplifications at chromosome 1q, 5p, 6q, 8q, 11p, 12q, 14q, and 15q were also previously reported in DD-LPS (2), although the clinical significance of these events is poorly understood.
In addition to genomic alterations, aberrant epigenomic features such as DNA methylation and histone modifications have been implicated in DD-LPS tumorigenesis. For example, promoter hypermethylation was found to silence expression of the pro-adipogenic transcription factors (TF), CEBPA and KLF4, and downregulate pro-apoptotic regulator FASLG (4). DNA methylation status has been used as a predictor of patient survival among the previously reported The Cancer Genome Atlas (TCGA) dataset (4). Increased H3K9me3 signal may also be responsible for driving a dedifferentiated phenotype, as it is previously shown to reduce expression of the tumor suppressor KLF6 (9). Studies have also investigated the super enhancer landscape of these tumors and identified a BET protein-dependent core transcriptional regulatory circuit that consists of FOSL2, MYC, and RUNX1 (10). Indeed, genetic depletion of the BET genes, core transcription factors, or their target gene SNAI2 mitigates the LPS malignancy (10).
Despite these initial observations, it remains unclear how genomic alterations are related to aberrant epigenomic and transcriptional regulation in WD/DD-LPS. More importantly, the role of three-dimensional (3D) genome organization in liposarcoma has not been investigated yet. Therefore, in this study, we generated Hi-C, chromatin immunoprecipitation sequencing (ChIP-seq) for H3K27ac, RNA sequencing (RNA-seq), and whole-genome sequencing (WGS) in four pairs of LPS patient samples and one LPS cell line (Lipo863B). By taking advantage of our recently published software NeoLoopFinder (11), we resolved complex structural variants (SV) with the Hi-C data, and also identified SV-induced chromatin loops. We demonstrated that the coamplification of local regulatory elements and hijacking of distal regulatory elements contributed to the regulation of key oncogenes, such as MDM2, CDK4, and HMGA2. Furthermore, by integrating WGS, Hi-C with additional Bionano optical mapping and Nanopore long read sequencing in Lipo863B, one of the most widely used cell lines in LPS studies, we reported the general composition and potential reconstructions of the giant chromosome in LPS. Collectively, our study characterizes the chromatin regulatory landscape of LPS, identifies oncogenic enhancer coamplification and enhancer hijacking events, and offers a comprehensive view of how genomic structure variation contribute to oncogenic regulation.
Materials and Methods
Cell culture
Liposarcoma cell line (Lipo863B) were cultured in DMEM supplemented with 15% FBS and additional 1 mmol/L GlutaMax supplement (Thermo Fisher Scientific, 35050061).
Nuclei isolation from patient samples
Patient liposarcoma samples were ground in liquid nitrogen, lysed in cold lysis buffer (20 mmol/L Tris-HCl, pH 8.0, 15 mmol/L NaCl, 60 mmol/L KCl, 320 mmol/L sucrose, 1 mmol/L DTT, 0.1% triton X-100, 0.1 mmol/L PMSF, 1× proteinase inhibitor cocktail), and dounced by homogenizer around 20 times using pestle B on the ice. Then the supernatant containing the nuclei was transferred to a 15 mL tube. Nuclei were pelleted by spinning for 6 minutes at 600 × g in a swing-bucket centrifuge and were resuspended in lysis buffer. The same volume of 1.4 M sucrose buffer (20 mmol/L Tris-HCl, pH 8.0, 15 mmol/L NaCl, 60 mmol/L KCl, 1.4 M sucrose, 1 mmol/L DTT, 0.1 mmol/L PMSF, 1× proteinase inhibitor cocktail) was carefully added to the bottom of 15 mL tube underneath of nuclei solution. Then, the nuclei were purified by spinning for 30 minutes at 2,500 × g in swing-bucket centrifuge and resuspended in 200 μL of 320 mmol/L sucrose buffer (20 mmol/L Tris-HCl, pH 8.0, 15 mmol/L NaCl, 60 mmol/L KCl, 320 mmol/L sucrose, 1 mmol/L DTT, 0.1 mmol/L PMSF, 1× proteinase inhibitor cocktail). A small portion of nuclei was taken out to a new 1.5 mL tube and added trypan blue to count the number under the microscope. Tissue specimens were obtained in accordance with institutionally approved protocols at the Ohio State University (IRB: #2014-C0028).
RNA-seq
Twenty micrograms of ground patient samples or 1.5 million liposarcoma cells were used for RNA-seq. The total RNA was extracted from TRizol according to the manufacturer's protocol (Invitrogen). PolyA RNA was purified from 1,000 ng of total RNA using oligo (dT) beads (Invitrogen). Extracted RNA was first fragmented, then followed by reverse transcription, end-repair, adenylation, adaptor ligation, and subsequent PCR amplification. The final product was checked by size distribution and concentration using a BioAnalyzer High Sensitivity DNA Kit (Agilent) and Kapa Library Quantification Kit (Kapa Biosystems). The libraries were sequenced on Hi-seq 2500 or NovaSeq platform with 2 × 100 bp setting.
ChIP-seq
ChIP-seq experiments were performed as described previously (12) with some modifications shown as follows. About 30,000 of nuclei or cells were used for one ChIP. Native nuclei or cells were taken out from −80°C freezer and resuspended in cold assay for transposase-accessible chromatin with sequencing (ATAC-seq) lysis buffer (10 mmol/L Tris-HCl, pH 7.5, 10 mmol/L NaCl, 3 mmol/L MgCl2, 0.1% IGEPAL CA-630, 1× proteinase inhibitor cocktail). 1 ug of H3K27ac antibody (Active Motif, 39133) was used per ChIP. After PCR amplification, libraries were size-selected by 0.6×/1.0× KAPA pure beads. The final libraries were sequenced on Hi-seq 2500 or NovaSeq platform with 2 × 100 bp setting.
Hi-C
Hi-C was performed using 0.1 to 0.5 million isolated nuclei or 1 million liposarcoma cells as described previously (13), but with some modifications. Briefly, 2% formaldehyde cross-linked nuclei were digested by DpnII and then underwent biotin incorporation and proximity ligation. Next, the reverse crosslinked DNA was purified by 0.5× KAPA pure beads and was fragmented by sonication using Covaris E220 with the following parameters: 140 W, duty factor 10, 200 per burst, 50 seconds. Then, size selection was performed by 0.55×/0.85× KAPA pure beads and DNA was dissolved in 1× TE buffer. Streptavidin beads (Thermo Fisher Scientific, 65601) was used to pull down biotin-labeled DNA by rotation at room temperature for 15 minutes and then washed twice with 600 μL of wash buffer [5 mmol/L Tris-HCl (pH 7.5), 0.5 mmol/L EDTA, 1 mmol/L NaCl, 0.05% Tween 20] and one time with 600 μL of 10 mmol/L Tris-HCl (pH 8.0). The DNA bound on the beads was prepared for sequencing using KAPA HyperPrep Kit (Roche) according to the manufacturer's protocol and amplified eight cycles by PCR. After purification by 0.85× KAPA pure beads, the libraries were sequenced on Hi-seq 2500 or NovaSeq platform with 2 × 100 bp setting.
Whole-genome sequencing
The genomic DNA from ground patient tissues or liposarcoma cell lines was extracted using DNeasy Blood & Tissue Kit (Qiagen, 69504). Then, 500 ng of genomic DNA was used for DNA library preparation with the Illumina DNA PCR-Free Prep Kit according to manufacturer's protocol.
BioNano optical mapping
The genomic DNA from Lipo863B was used for BioNano optical mapping. gDNA was extracted using BioNano Blood and Cell Culture DNA Isolation Kit (80004). Next, the homogenized gDNA was directly labeled using DLE enzyme (BioNano, 80005) according to the BioNano Prep Direct Label and Stain (DLS)-30206 protocol. The labeled and stained genomic DNA was then loaded onto a Saphyr chip for optical data collection.
Nanopore sequencing
The genomic DNA from Lipo863B was used for Nanopore sequencing. gDNA was isolated using BioNano Blood and Cell Culture DNA Isolation Kit (80004). Then, the homogenized gDNA was sheared by pipetting and nanopore library was made using NEBNext Companion Module for Oxford Nanopore Technologies Ligation Sequencing Kit (E7180S) and Nanopore Ligation Sequencing Kit (SQK-LSK110). One microgram of library was loaded per GridION Flow Cell (R9.4.1).
FISH experiment
Preparation and hybridization of FISH probes
FISH was performed using Agilent SureFISH probes CDK4 localizing to 12q14.1 (chr12:57968669–58276609) and MDM2 localizing to 12q15 (chr12:69137889–69317809). The probe/hybridization buffer mixture was applied to the sample and co-denatured using a Thermobrite (Abbott Molecular) for 5 minutes at 78°, followed by a hybridization for 24 hours at 37° in a humidified chamber.
FISH detection, visualization, and image capture
After hybridization, the FISH slides were washed in a 0.4×SSC solution at 72° for 1 minute followed by 2×SSC solution at 37° for 15 seconds, then counterstained with DAPI (Abbot Molecular). Fluorescent signals were visualized on an Olympus BX51 microscope workstation (Applied Spectral Imaging) with DAPI, TRITC and FITC filter sets. FISH images were captured using a digital U-TV0.5XC-3 camera (ASI) and FISHView ASI software.
RNA-seq data analysis
The raw reads were first trimmed for adapter and then mapped against hg38 human reference genome using STAR (RRID:SCR_004463; ref. 14). Gene-level abundance was quantitated using RSEM program. The raw counts were then normalized using variance stabilizing transformation (VST) function from DEseq2 (RRID:SCR_015687) package. Top 500 variable genes from the 19 pairs of patient samples were used to perform t-SNE analysis. We then used DEseq2 to identify the differential expressed genes between paired tumor and peritumor samples. Pathway enrichment analysis for up/downregulated genes was performed using clusterProfiler.
For all the samples, we used the Arriba (version 2.2.1; ref. 15) to detect fusion gene events.
ChIP-seq/ATAC-seq data analysis
For ChIP-seq and ATAC-seq data analysis, we used the standard ENCODE ChIP-seq/ATAC-seq analysis pipeline (https://github.com/ENCODE-DCC/chip-seq-pipeline2). Detected peaks for patient samples were filtered with cutoff q-value < 1e−8. Motif enrichment analysis were performed with HOMER using the findMotifsGenome program. Heatmaps were generated using deeptools by centering on identified peaks center. Super enhancers were identified using the ROSE algorithm (https://bitbucket.org/young_computation/rose).
Hi-C data analysis
We first trimmed the adapters of the Hi-C raw reads and then mapped the trimmed files against hg38 human reference genome using runHiC python package (https://pypi.org/project/runHiC/). The copy-number variants (CNV) profile in each sample was estimated from the Hi-C matrices using a generalized additive model. The breakpoints of large SVs, as input to the NeoLoopFinder (https://github.com/XiaoTaoWang/NeoLoopFinder), were identified from Hi-C using Hi-C breakfinder (https://github.com/dixonlab/hic_breakfinder). Local genome structure and neoloop were constructed and identified using the NeoLoopFinder.
WGS data analysis
The raw reads were first trimmed for adapter and then mapped against hg38 human reference genome using BWA-MEM algorithm (0.7.17-r1188). Duplicate reads were removed using Picard Tools (2.6.0-SNAPSHOT). For SV detection, we used three independent softwares including lumpy (v 0.2.13), Delly (Version: 0.8.3), and SvABA (Version: 1.1.0) with default parameters. For patients, peritumor normal samples were used as control input to identify somatic structure variations. SVs were filtered by the following criteria: length greater than 50bp, with at least 10 supporting reads, identified by at least two of the methods, blacklist regions including telomeric, centromeric, and 12 heterochromatic regions provided by the Delly software were masked for the SV detections in all the methods. We used SURVIVOR (16) for the SV filtering and merging (https://github.com/fritzsedlazeck/SURVIVOR). Svviz2 (17) were used for single SV event visualization.
To identify SV events identified from both Hi-C and WGS, we also use SURVIVOR merge function and allowing maximum 1Mb distance between the breakpoints.
Bionano optical mapping data analysis
DNA molecules were captured from the Saphyr instrument and converted to BNX digital text file by Bionano Access (v1.6.1), which contained the label position of each molecule. We required that the molecules to be at least 150 kb in length and with more than nine labels per molecule. Cell line consensus genomic maps (cmap) were generated through de novo assembly of DNA optical reads using Bionano Solve 3.6.1 pipeline.
Specifically, A total of 1,741 Gb data were generated and 1,268 Gb data were left after filtering. The filtered data are corresponding to 353× coverage of the hg38 human genome. These filtered molecules were aligned using RefAligner with default parameters. It produced 1057 genome maps with an N50 of 29.45 Mbp.
SV detection was performed after de novo assembly using the Bionano built-in SVs caller module by comparing the assembled genomic maps (cmap) to the hg38 reference genome with default parameters.
Nanopore sequencing data analysis
In total, we generated 119.8 Gb Nanopore sequencing data, which were corresponding to 34× converge of the human genome with N50 57.5 kb. The raw data were mapped against the hg38 reference genome using the minimap2 (18). The structure variations were identified using Sniffles (19) and filtered using the same criteria as from WGS. The example structure variations were visualized using Ribbon (20).
We tried nextDenovo (https://github.com/Nextomics/NextDenovo) for de novo assembly, which resulted 304 contigs with N50 49.5 Mb and the largest contig 124.8 Mb. We also tried Flye (21), which resulted 2670 contigs with N50 4.2 Mb.
Identification of CNV regions, CNV genes, and gene enhancer coamplification event
CNV profiles and segments were generated by Control-FREEC. The ploidy parameter was set as two in all the samples. In total, we identified 2,396 CNV regions from all the patient tumor samples. To obtain the recurrent somatic CNV regions, we first subtracted the CNV regions if 80% of them had overlap with CNV regions identified from the corresponding peri-tumor samples. To count for the recurrencies of the CNV regions, all the filtered CNV regions (1,876 regions) were intersected by multiIntersectBed and 38 of the regions were recurrent from all the 4 patient samples.
To calculate the number of enhancers located in the CNV regions, we counted the H3K27ac peaks located 2.5kb away from gene promoters that entirely located within the CNV regions (CNV gain: ≥5 copies, CNV loss: <2 copies) within each patient. To study how CNV affect gene expressions, we specifically focused on the CNV gain regions, and required 50% gene body to be overlap with CNV regions. CNV region with both entire gene bodies and enhancers would be considered as one gene enhancer coamplification event.
Data availability
Processed RNA-seq, WGS, Hi-C, and H3K27ac ChIP-seq data generated from primary patient samples have been deposited at the Gene Expression Omnibus under accession GSE201056. Raw and processed RNA-seq, WGS, Hi-C, H3K27ac ChIP-seq, Nanpore sequencing data in Lipo863B cell line have been deposited at the Gene Expression Omnibus under accession GSE201056.
Results
Comprehensive genomic and epigenomic profiling in liposarcoma
To identify and understand the aberrant CNAs and alterations in epigenomic regulation and 3D genome organization, we studied five pairs of WD-LPS and 14 pairs of DD-LPS patient samples, and a commonly used DD-LPS cell line (Fig. 1A). Each pair of patient samples contains one tumor tissue and one matched peritumoral tissue as normal control. To profile the transcribed regions and to reveal the differential transcriptomes between tumors and peritumoral tissues, we performed mRNA sequencing (mRNA-seq) in all patient samples and cell lines (Supplementary Table S1). In four pairs of DD-LPS patient samples (P241, P298, P69, and P209) and one DD-LPS cell line (Lipo863B), we performed (i) chromatin immunoprecipitation followed by next-generation sequencing (ChIP-seq) for histone modifications H3 lysine 27 acetylation (H3K27ac; Supplementary Table S2) to chart active enhancer regions; (ii) Hi-C experiments to study high-order chromatin structure and link distal enhancers to their target genes (Supplementary Table S3); and (iii) WGS to profile CNVs and dissect genome rearrangements (Supplementary Table S4). To better reveal the sequence details of genomic rearrangements, we performed Nanopore long-read sequencing and BioNano optical mapping in Lipo863B cells to help assemble the rearranged regions (Supplementary Table S5). In total, we generated 73 genomic datasets, including 62 for the patient tissue samples and 11 for the cell line Lipo863B. To date, this is the most comprehensive analysis of epigenomic regulation and 3D genome organization in liposarcoma.
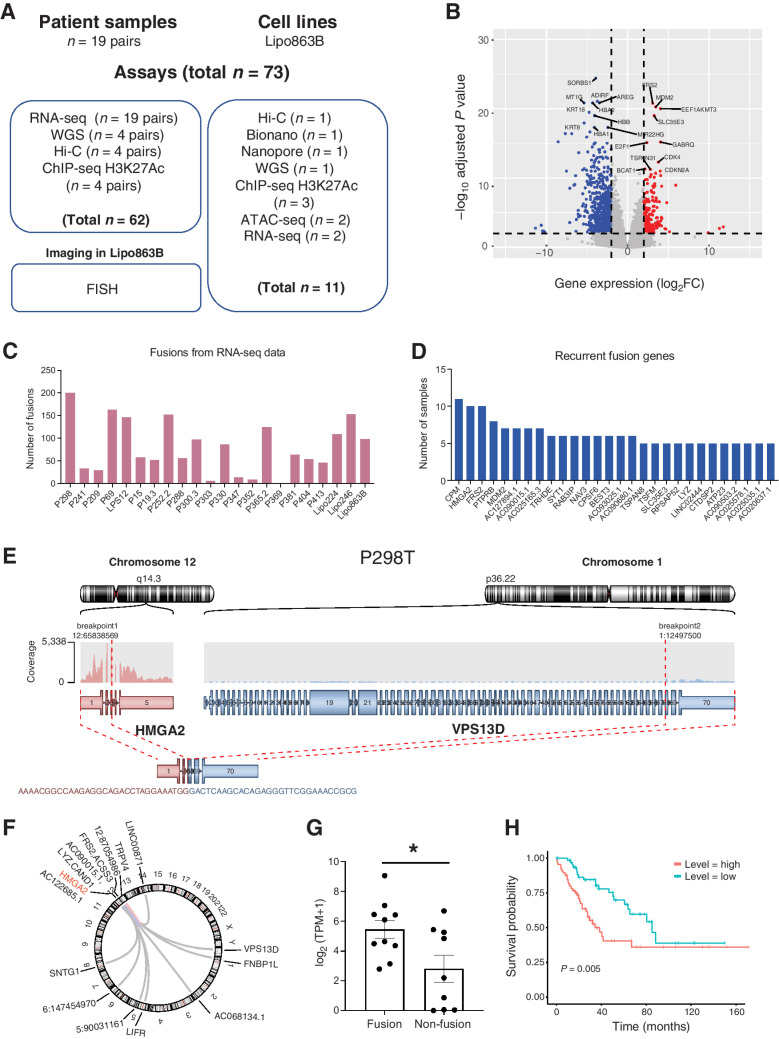
Figure 1.
Gene expression and fusion events in liposarcoma patient samples. A, A summary of the LPS patient samples and genomic profiling assays performed in this study. B, The volcano plot for 19 pairs of patients RNA-seq data. C, The number of gene fusion events detected from patients and cell line samples using RNA-seq data. D, List of recurrent fusion gene partners (fusion events occurred in at least four samples for the gene). E, Example of HMGA2-VPS13D fusion in P298T sample. F, The HMGA2-related fusion events in all the patient tumor samples. G, Expression level of HMGA2 in HMGA2 fusion-detected samples and non-fusion samples (one tail t test P value = 0.0126). H, Kaplan-Meier survival analysis from TCGA sarcoma patient samples stratified by the expression level of HMGA2. The high-level expression group consists of patients with top 25% HMGA2 expression and low-level expression group has patients with bottom 25% HMGA2 expression.
Comparison of transcriptomes between LPS tumors and peritumoral tissues
First, we performed clustering analysis for the 19 paired tumor versus normal LPS samples with the RNA-seq data by t-SNE plot (Supplementary Fig. S1A). The sample information and the statistics of the sequencing data mapping were listed in Supplementary Table S1. In general, tumor samples were clearly separated from the peritumor normal tissues (Supplementary Fig. S1A). Surprisingly, the DD-LPS and WD-LPS samples were not well separated, indicating that genome-wide RNA-seq data alone might not be able to fully capture the features that differs these two subtypes of tumors. Then, we performed the differential gene expression analysis to determine the tumor-specific gene expression patterns using the same 19 pairs of patient samples. We identified 1,186 genes with elevated expression and 1,027 downregulated genes in tumor (Fig. 1B). FRS2, MDM2, CDK4, as well as E2F1 and CDKN2A genes were among the most upregulated ones, consistent with current literature (4). SORBS1, KRT8, and MT1G were among the top downregulated genes (Fig. 1B). Interestingly, MT1G was previously reported to be a tumor suppressor and was silenced by DNA methylation in hepatocellular carcinoma (22, 23). Also, low-level expression of SORBS1 was associated with poor clinical outcomes in breast cancer and the inhibition of SORBS1 promotes the epithelial-to-mesenchymal transition (EMT) process (24). Gene Ontology term analysis showed that upregulated genes were more enriched in cell proliferation pathways, whereas downregulated genes were more involved in metabolic process, which may suggest a transition from adipogenesis to proliferative pathways associated with transformation (Supplementary Fig. S1B).
Recurrent fusion genes identified from liposarcoma patient samples
Gene fusion caused by genomic rearrangements plays a causal role in tumorigenesis across various types of cancer, including BCR-ABL1 in chronic myelogenous leukemia (CML), EML4-ALK in non–small cell lung cancer (NSCLC), and so on (25, 26). To identify recurrent fusion genes in liposarcoma, we exploited the 19 pairs of RNA-seq data and identified 1,391 fusion gene events for the 19 patient tumor samples (Fig. 1C; Supplementary Fig. S2A). By further investigating the fusion genes across all the patient samples, we identified genes that recurrently (at least in 4 patient tumor samples) fused with other genes, including CPM, HMGA2, FRS2, PTPRB, and MDM2, and so on (Fig. 1D).
HMGA2 gene fusion has been identified in various mesenchymal tumors (27–29). In our liposarcoma patient samples, about 50% were identified with HMGA2 gene fusion events (Fig. 1D). Here, we demonstrated an example of the HMGA2-VPS13D gene fusion identified from P298 patient tumor sample (Fig. 1E). In this example, the first three exons of HMGA2 were fused with the last three exons from VPS13D. The loss of HMGA2 3′ UTR and let-7 binding sites in these chimeric fusion events would probably lead to the activation of HMGA2 pathway according to previous studies (27). We also noticed that the HMGA2 was able to fuse with many different genes (Fig. 1F), indicating that it was probably the truncation of the HMGA2 instead of the fusion partners that play a role in tumorigenesis. Further investigation demonstrated that HMGA2 expression was significantly higher (one tail t-test P-value = 0.0126) in samples with HMGA2 fusion events comparing with samples without fusion (Fig. 1G). The HMGA2 expression level was, to some extent, an indicator of patient survival as shown from TCGA datasets that HMGA2 expression was significantly anti-correlated with sarcoma patient survival (Fig. 1H).
WD/DD-LPS-specific enhancer landscape in patient samples
To study how the LPS genes were regulated, we performed ChIP-seq for H3K27ac to identify the potential enhancers in tumor and normal tissues from patients P241, P298, P69, and P209 (Supplementary Table S2). We also improved the ChIP-seq protocol so that it could work with only ∼30,000 cells. We observed strikingly tumor-specific enhancers near the well-known LPS oncogenes such as FRS2 and MDM2 (Fig. 2A). In total, we identified 5,510 tumor-specific enhancers and 1,487 peritumor-specific enhancers (Fig. 2B). We next performed correlation analysis of the H3K27ac ChIP-seq signals at the identified specific distal enhancer regions for our datasets and the public available datasets. The hierarchical clustering of the correlations demonstrated three major clusters: the normal samples, the cell lines, and the tumor samples (Supplementary Fig. S3A). These results indicate that tumor/peritumor-specific distal enhancers we identified here were able to separate the tumor, normal, and cell line samples from external datasets. We also predicted super-enhancers as described previously (Supplementary Fig. S3B; ref. 30), and observed that FRS2, HMGA2, and SDK2 genes are recurrently located in the super-enhancer regions.
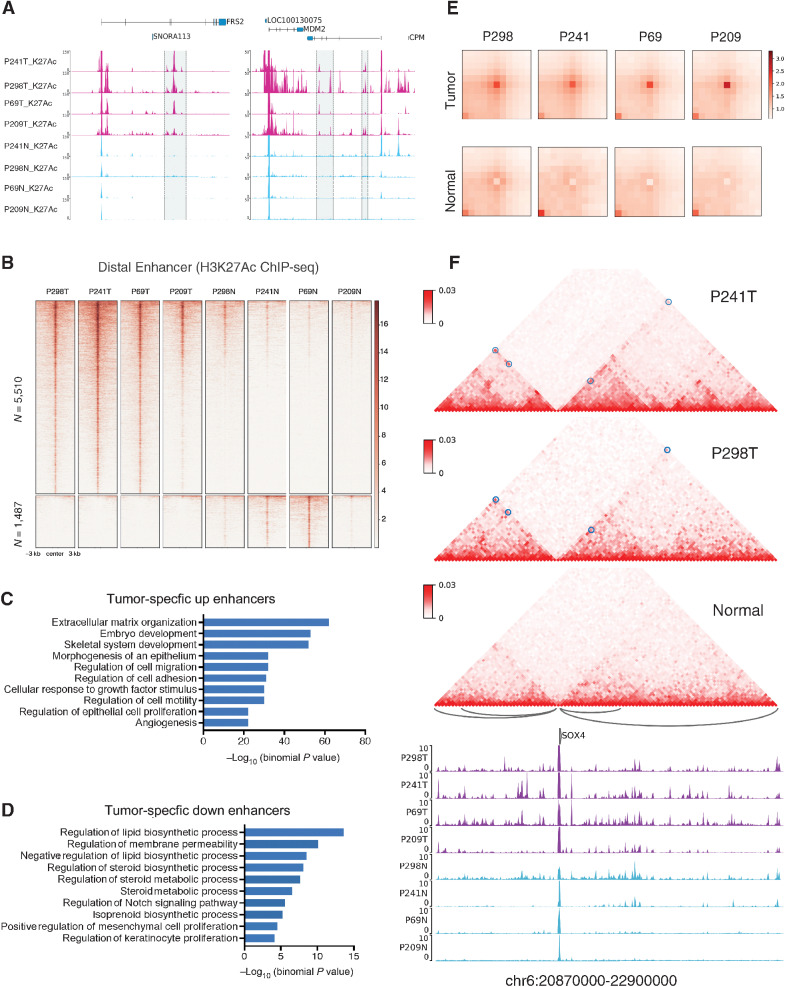
Figure 2.
Enhancer and chromatin interaction landscape in liposarcoma patient samples. A, Example regions around chr12: 69,445,000–69,585,000 and chr12: 68,800,000–68,965,000 corresponding to genes FRS2 and MDM2 of tumor-specific enhancers. B, Heatmap of genome-wide tumor-specific distal enhancers at CNV free regions. C and D, The GREAT analysis for the pathway enrichment of the tumor-specific enhancers. E, APA for cancer-specific chromatin interactions in P298, P241, P69, and P209 tumor and normal samples (n = 1,823). F, Cancer-specific chromatin interactions involving SOX4 genomic region. The blue circles on the Hi-C map highlights the positions of the cancer-specific interactions as well as the arcs below the Hi-C maps. The top two panels of the Hi-C maps are from the P241 and P298 patient tumor samples and the bottom panel shows the merged Hi-C matrix from all the normal tissue samples. The tracks below are the H3K27Ac ChIP-seq profiles for the patient samples from both tumor (purple) and normal (blue) tissues.
We further performed GREAT analysis for the pathway enrichment of the tumor-specific enhancer regions identified in Fig. 2B. Tumor-specific enhancers were more enriched in cell migration and proliferation pathways whereas the peritumor-specific enhancers were more enriched in lipid biosynthetic process or metabolic process, which is consistent with the observations from the gene expression results (Fig. 2C and D). Next, to identify which transcription regulators potentially modulate these transcriptional enhancers between tumor and normal samples, we performed motif enrichment analysis. Transcription factors from Jun family, E2F1, ATF, TWIST1, ARID3A, and NFIX were highly enriched in the tumor-specific enhancers (Supplementary Fig. S3C).
WD/DD-LPS-specific chromatin interaction landscape in patient samples
Spatial chromatin interaction plays an important role in the regulation of gene expression, including long-range control by distant enhancers and repressors (31). Here, to address whether and how the 3D chromatin interactions are different between cancer and normal samples from the same patient samples, we performed Hi-C in tumor and normal tissues from patients P241, P298, P69, and P209. By genome-wide search of chromatin interactions using Peakachu (32), we identified a merged loop set of 46,987 in all samples. By comparing the normalized signal at these loop regions, we identified 1,823 cancer-specific chromatin interactions and generated aggregated peak analysis (APA) plot in both tumor and normal samples (Fig. 2E). As expected, we observed significant aggregation signal in tumor samples but not in normal samples.
We further investigated how these cancer-specific chromatin interactions affect gene expression. We observed genomic regions with cancer-specific chromatin interactions involving significant upregulated genes in tumor samples, for example, TGFB2 and SOX4 (Fig. 2F; Supplementary Fig. S3D). We observed strong long-distance interaction for TGFB2 and four interactions for SOX4 in tumor samples. In addition, for the leftmost interaction of SOX4, we also observed tumor-specific enhancers in all the four tumor samples. To evaluate how gene expression were affected in general, we intersected the interaction regions with gene promoter regions and obtained cancer-specific chromatin interaction-related genes. We observed an elevating trend of gene expression level in the four patient tumor samples comparing to the corresponding normal samples (Supplementary Fig. S3C). These results demonstrated that the 3D chromatin interactions would contribute to the upregulation of genes potentially by facilitating the regulation from distal enhancers.
CNA and enhancer-gene coamplification
Liposarcoma is well known to have extensive CNVs and structure variations (SV). To capture the comprehensive list of variations in the patients, we first performed WGS at ∼30-fold of coverage for the four pairs of primary tumors versus peritumoral tissues (Supplementary Table S4) and a widely used liposarcoma cell line Lip863B (Fig. 3A; Supplementary Fig. S4A), at ∼30 coverage in each sample. Overall, we identified 2,396 CNV regions, with 38 regions recurrent in all the 4 patient samples. As we and others have shown that Hi-C can also be used to estimate CNVs (11, 33), we then computed the genome-wide CNV profiles with Hi-C data, and they were highly similar to those computed from WGS data (Supplementary Fig. S4B).
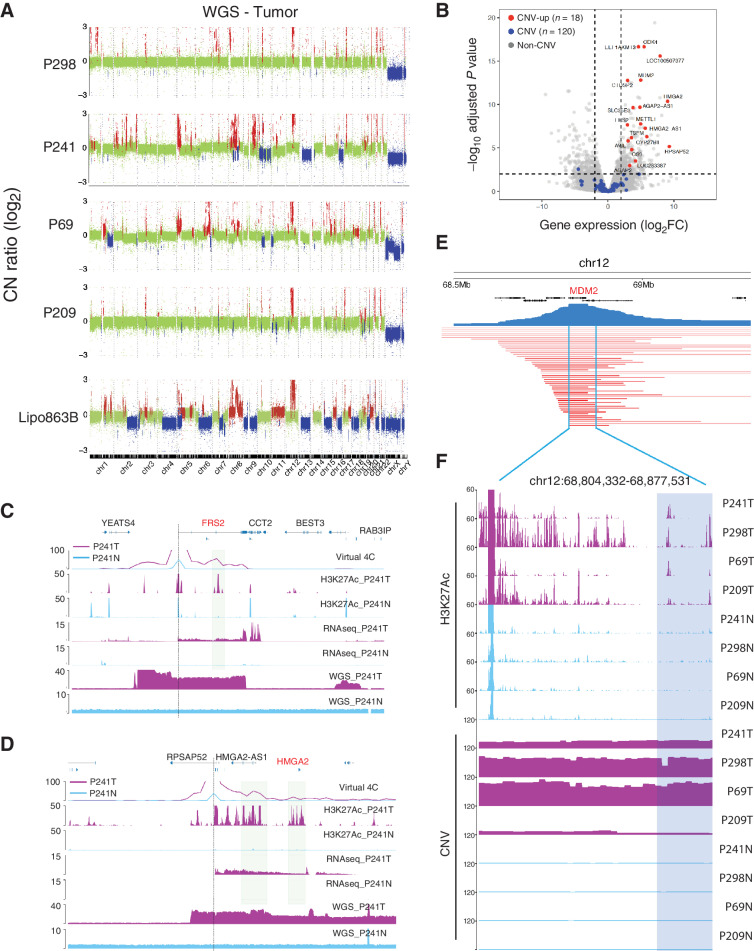
Figure 3.
The identification of the enhancer coamplification events. A, The CNV profiles for Lipo863B, P298, P241, P69, and P209 identified from the WGS data. B, Volcano plot for the four pairs of patient samples. Red, copy number gained genes with significant differential expression; blue: copy number gained genes with no significant differential expression. C and D, Examples of enhancer coamplification for FRS2 (chr12:69,300,000–69,800,000) and HMGA2 (chr12:65,600,000–66,100,000) gene in P241. Gene transcription start site is marked with a dash line. The coamplified enhancer is highlighted in sierra blue rectangles. E, The CNV segments for the TCGA LPS patient samples covering the MDM2 gene. F, The H3K27Ac ChIP-seq track and CNV profiles for P298, P241, P69, and P209 samples at the recurrent CNV regions from TCGA.
As expected, we observed amplification of chr12q13–15 in all LPS samples, which contains MDM2 and CDK4 genes, a signature of liposarcoma (Fig. 3A; ref. 2). The Hi-C map and WGS track at this signature amplification region showed high consistency from all patient and cell line samples (Supplementary Figs. S4C–S4F). We also observed other genes with extensive and recurrent copy-number gains, such as METTL1, TSFM, AVIL, CTDSP2 (Supplementary Fig. S5A). As these genes were also significantly upregulated in the patient tumor samples, we suspected that CNVs, especially copy-number gains, could possibly contribute to the altered gene expression. Therefore, we systematically investigated the relationship between CNVs and gene expression. For this analysis, we used the four pairs of tumors versus peritumor samples, in which we have both the WGS and the RNA-seq data in tumor and peritumor tissues. We identified 138 genes that were located inside an CNV gained regions in at least three tumor samples. The log2FC of these 138 genes were significantly higher than the rest of the genes (Supplementary Figs. S5B and S5C). Surprisingly, only 18 of these genes were significantly upregulated (Fig. 3B; Supplementary Fig. S5D), suggesting that CNV gain itself only contributed a small portion of the upregulated genes.
As enhancers are critical for proper gene expression regulation, we next examined how they were affected by the CNVs. On average, we identified 3,316 enhancers located in CNV regions, with 1,925 gain of copies and 1,191 in loss of copy regions. Strikingly, we observed several oncogenes, and their linked enhancers were amplified together. For examples, there was a ∼178 kb region containing the FRS2 oncogene that were amplified by ∼25 times on average in different LPS samples (Fig. 3C). Nearly 60 kb downstream from the FRS2 gene promoter, there was an enhancer (marked by H3K27ac) in the same amplified region. We examined the virtual 4C data for this region and the results showed there were strong chromatin interactions between the FRS2 gene promoter and this enhancer. We made similar observation for another oncogene HMGA2 (Fig. 3D), whose promoter region was linked with multiple enhancer elements downstream. For both genes, neither the enhancers nor the chromatin loops between the enhancers and promoters were present in the normal tissues.
Genome-widely, we observed 264 such gene enhancer coamplification cases. All the protein coding genes we identified in Fig. 3B with both significant elevated expression and CNV gains demonstrated enhancer coamplification in at least two patient samples, including OS9, MDM2, CDK4, CTDSP2, FRS2, METTL1, TSFM, and so on. We further investigated the TCGA liposarcoma samples, and strikingly, 87% of the patient samples (45 of 52) have amplified regions that contain the MDM2 gene and the coamplified enhancers that we identified in this study (Fig. 3E and F). In addition, we identified 58 oncogenes, including DDIT3, TWIST2, SNAI2, YEATS4, and so on, with enhancer coamplification in at least one patient sample. Their expression patterns are highly consistent with the coamplification events (Supplementary Fig. S5B). These results indicated that enhancer coamplification would potentially contribute to the oncogene dysregulations, and it would be interesting in future studies to investigate how such scenarios directly contribute to oncogenesis.
Extensive structural variants and enhancer hijacking in LPS
We systematically identified structural variants such as translocations and inversions in the patient genomes. First, we identified SVs using the WGS data with three different SV callers including Lumpy, Delly, and SvABA (34–36), and kept the SVs identified by at least two methods for further analysis (Materials and Methods). An example of interchromosomal translocations was shown in Supplementary Fig. S6A. This translocation was between chr1:93.47Mb and chr12:65.77Mb and was identified by all three methods. On average, we detected several hundred inversions and translocations in each tumor (Fig. 4A).
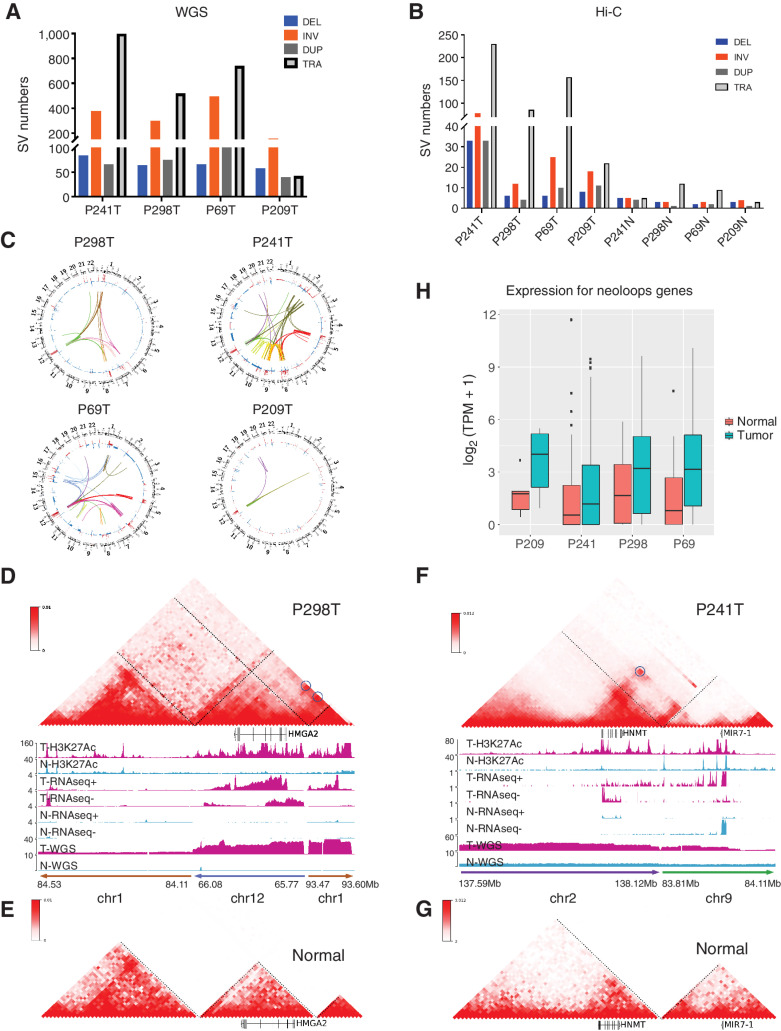
Figure 4.
Enhancer hijacking events contribute to the oncogene misregulation in liposarcoma patient samples. A and B, The stratified number of SVs identified from WGS (A) and Hi-C (B). C, The circos plot showing SVs and CNVs identified in P241, P298, P69, P209 patient samples detected from both Hi-C and WGS data. D and E, The reconstructed Hi-C map and genomic ChIP-seq, RNA-seq, and WGS tracks for translocation events chr1:84.11Mb-84.53Mb, chr12:65.77Mb-66.08Mb, and chr1:93.47–93.60Mb from P298T (D) and the Hi-C map interactions from the same regions in merged normal samples (E). F and G, The reconstructed Hi-C map and genomic ChIP-seq, RNA-seq, and WGS tracks for translocation events chr2:137.59Mb-138.12Mb, chr9:83.81Mb-84.11Mb from P241T (F) and the Hi-C map interactions from the same regions in merged normal samples (G). H, Boxplot for expression levels of genes located within neo-loops anchor regions in patient samples.
Previously, we and other groups have shown that Hi-C can be used to detect a full range of SVs (33, 37), and it provides a unique list of SVs that are missed by WGS. Here, we utilized HiCBreakFinder (33) to obtain another list of SVs in these tumor samples (Fig. 4B). We then compared the SVs predicted by Hi-C and WGS (Supplementary Fig. S6B) and defined a list of high-confident SVs detected by both methods. We noted that Hi-C tends to detect large-scale SVs compared with WGS. To provide genome-wide overview of the SV landscape, we drew the circos plot (38) with the high-confident SVs and the CNV profiles (Fig. 4C). We observed that SVs could be very specific in different patient samples, but we also noticed several SVs occurred at the chr12 amplified region across all the samples.
Recent work has shown that SVs could lead to the “enhancer hijacking” events by re-positioning the enhancers to the proximity of oncogenes and elevating their expression (39, 40). Enhancer hijacking events have been demonstrated in contributing to tumorigenesis in different types of cancer (41–44). Using a method that our group recently developed (NeoLoopFinder; ref. 11), we systematically detected enhancer-hijacking events in all the LPS samples. We showed such an example in Fig. 4D, where there were interchromosomal translocations between chr1 and chr12. We reconstructed the local chromatin interaction map surrounding the breakpoints with NeoLoopFinder. Because of the interchromosomal translocation, the promoter of the HMGA2 gene (which was on chr12) was connected to a cluster of enhancers on chr1 (Fig. 4D). There were no interactions between the HMGA2 gene and this enhancer cluster in the corresponding normal sample (Fig. 4E), because they were located in two different chromosomes and no such translocation occurred. A similar enhancer-hijacking event involving the HNMT1 gene was also observed in tumor but not in the normal sample (Fig. 4F and G). We demonstrated several more examples of enhancer-hijacking events involving the TERT, TGFBR1, and TWIST2 oncogenes (Supplementary Figs. S7A–S7F). In total, we detected 1,906 neo-loops from all the patient samples. APA on one of the patients P241 (1,440 neo-loops) demonstrated strong enrichment comparing to the merged control normal samples (Supplementary Fig. S7G).
Next, to evaluate the effects of neo-loops on gene expression level genome-widely, we identified 805 genes located within the neo-loop anchor (±20kb) regions as neo-loop involved genes from all the four patient tumor samples. Significantly, these genes demonstrated higher gene expression levels in the tumor samples comparing to the corresponding normal samples in all patients (Fig. 4H). In addition, we also noticed that the average H3K27Ac levels within the neo-loop anchors demonstrated higher signals in tumors comparing with normal samples (Supplementary Fig. S7H), indicating that the neo-loops involved genes upregulation were potentially through the enhancer-hijacking events in general.
Interestingly, we noticed that Hi-C data were able to identify gene fusion events in patient samples. The HMGA2-VPS13D and HMGA2-AC068134.1 fusion events (Fig. 1E and F) were also identified using Hi-C data in P298 and P241 tumor samples correspondingly (Supplementary Fig. S8). Combining the H3K27Ac ChIP-seq and the Hi-C data, we observed the enhancer hijacking events for the fusion gene (as highlighted in Supplementary Figs. S8A and S8B), which could be another contributor to the activation of HMGA2.
Complex SVs and local genome assembly for the lipo863b cell line
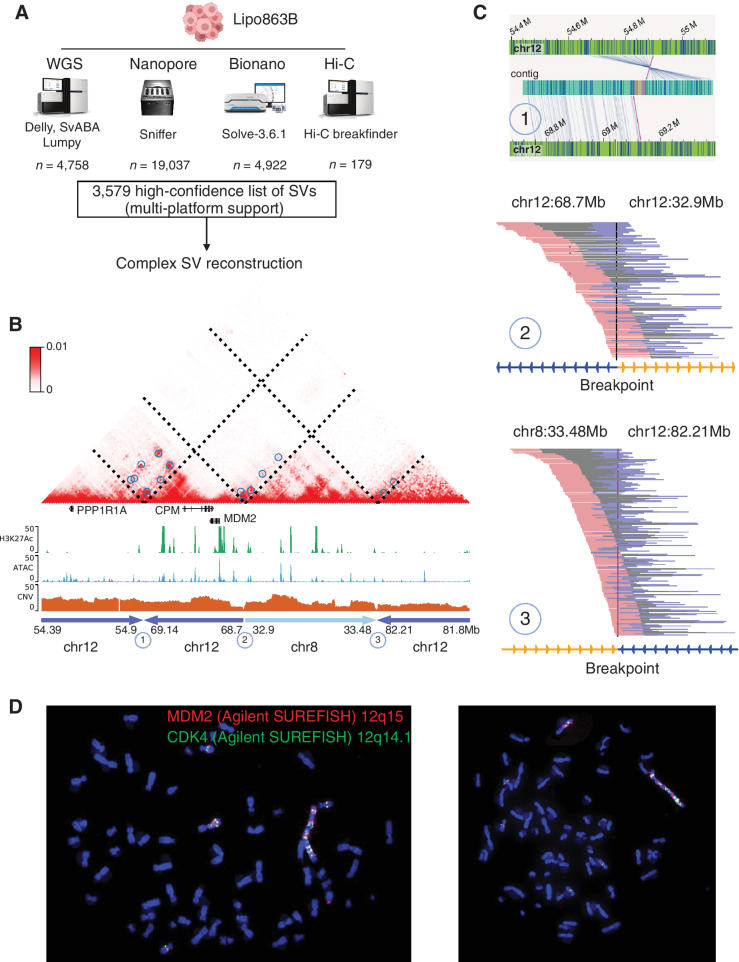
The human Lipo863B cell line was established from surgically resected retroperitoneal DDLPS in 2011 (45), and has been widely used for the studies of LPS tumors. To obtain a comprehensive view of the SVs and study their role in altered gene expression in LPS, we performed WGS, Hi-C, and two long-read sequencing: Bionano optical mapping and Nanpore sequencing (Supplementary Table S5). We observed a large variation of the numbers of SVs from each platform (Fig. 5A, details in Materials and Methods). Among them, Nanopore identified the most SVs (19,037), whereas Hi-C reported the least (179). For the WGS data, we only considered SVs supported by at least two SV caller methods to mitigate the false positives (Supplementary Figs. S9A and S9B), which predicted 4,758 SVs in total. As reported previously (46), Nanopore uniquely captured a large number of insertions compared with WGS (Supplementary Fig. S9C). Bionano also identified several thousand deletions and insertion. In general, there are 1,810 SVs detected by both of the two long-molecule sequencing platforms, with 1,519 insertions at kilobase in length. WGS and Nanopore also support each other for 1,327 SVs mostly for short deletions (102–103 bp; Supplementary Figs. S9D–S9F). Finally, we reported 3,579 high-confident SVs, which were defined as SVs predicted by at least two of all the technologies. One such example was shown in Supplementary Figs. S10A to S10D, where an inversion event, supported by all the four platforms, was identified between chr1:153.61Mb and 195.96Mb.
Figure 5.
Constructing the genome, including giant chromosome, of Lipo863B cells. A, A multi-platform SV detection. The SVs were first identified independently from each technology. A high-confidence list of SVs (supported by at least two platforms) was generated based on the merged SV calls. These high-confidence list of SVs was further employed in the complex SV reconstruction. B, The Hi-C map demonstrated a reconstruction of the complex local structure of a derivative chromosome in Lipo863b by the integration of optical mapping, WGS, Hi-C, and nanopore long reads. The rearranged region consists of four regions: chr12: 54.39–54.9Mb, chr12: 68.7–69.14Mb, chr8: 32.9–33.48Mb, chr12: 81.8–82.21Mb, and the junctions of the Hi-C map are marked as 1, 2, and 3. C, Supporting evidence from different technologies for each junction point: 1, optical mapping; 2 and 3, WGS and Nanopore (Supplementary Figs. S10E and S10F). D, FISH experiments for the two oncogenes MDM2 and CDK2 in Lipo863B.
Next, we tried to perform local genome assembly, especially for the regions that contain the known oncogenes for LPS, using the long-read sequencing data from BioNano and Nanopore. An example is shown in Fig. 5B, where we were able to assemble fragments from four genomic loci together and the LPS signature gene MDM2 was in this region. The three breakpoints were individually supported by different technologies (Fig. 5C). For example, breakpoint 1 was supported by Bionano and Hi-C, breakpoint 2 was supported by WGS and Nanopore and breakpoint 3 by WGS, HiC, and Nanopore (Supplementary Figs. S10E and S10F). We further studied the enhancer hijacking events induced by SVs. As shown in Supplementary Fig. S10A, we provided the supporting evidence from Hi-C by the reconstructed Hi-C map and noticing that this event also induced neo-loop formation, which could potentially lead to the dysregulation of the S100A13 gene expression. This event was further supported by WGS, Bionano, and Nanopore (Supplementary Figs. S10B–S10D). More examples of such reconstructed Hi-C maps were shown in Supplementary Figs. S11A to S11F. Also, we observed multiple Bionano contigs mapped to the chr12: ∼55–102Mb regions, indicating heavy rearrangement (Supplementary Fig. S12A). We then also performed the genome reconstruction using the AmpliconReconstrutor and achieved so far, the longest cancer genome amplicon reconstruction (31.34Mb; Supplementary Figs. S12B–S12D).
To further investigate how prevalent the MDM2 and CDK4 genes are in LPS genomes, we performed FISH experiments. There are three interesting observations: (i) consistent with the current literature in LPS, there were tens of copies of each oncogene, as shown in the FISH images. This is also consistent with the CNV profiles computed from the WGS data; (ii) there was a giant chromosome in essentially all the cells, and many copies of the MDM2 and CDK4 were on the giant chromosomes; and (iii) the number of chromosomes in the Lipo863B cells increased extensively, and it varied from cell to cell (∼60 chromosomes in each cell; Fig. 5D). Therefore, it is imperative for future genomic and epigenomic studies in LPS to adopt single cell-based approaches.
Discussion
In this study, we have presented a systematic integration of genome topology, epigenome landscape, genomic abnormalities, and transcriptome in liposarcoma. In particular, our data and analysis revealed how genomic aberrations contribute to the dysregulation of oncogenes. First, the transcriptome datasets from 19 pairs of liposarcoma patient samples charted the differentially expressed genes between tumor and peritumor tissues. Analysis of the H3K27ac ChIP-seq from four pairs of the patient samples revealed a library of tumor specific enhancers. Further, we identified genome-wide enhancer coamplification and enhancer hijacking events as novel mechanisms for oncogenic expression in liposarcoma patients, for example, MDM2, CDK4, HMGA2, by integration of the genomic alteration information, 3d genomic interaction, transcriptome, and the enhancer landscape from patient tumor and the matched normal samples. In addition, we screened gene fusion events genome-widely from all the 19 patient samples and identified thousands of fusion events with high recurrency for the highly amplified oncogenes such as MDM2, CDK4, FRS2, and HMGA2. These CNV gains, enhancer coamplification and hijacking, as well as gene fusions could play redundant roles in the activations of oncogenes at chr12q13–15. Further studies are needed to dissect the exact roles of these potential mechanisms. Finally, we fully charted the genomic landscape using multi-platform datasets including Nanopore long read sequencing, Bionano optical mapping, WGS and Hi-C as well as epigenomic landscape from H3K27ac ChIP-seq and ATAC-seq for a widely used liposarcoma cell line Lipo863B.
The WDLPS and DDLPS were not well separated based on RNA-seq data, which is likely due to that the WDLPS and DDPLS share many common features. Clinically, WDLPS and DDLPS can have similar microscopic appearances, although WDLPS typically has more lipoblastic-appearing cells, whereas DDLPS typically contains more prominent spindle shaped cells. Genetically, WDLPS and DDLPS both have amplifications on chr12q13–15 and have markedly overexpressed MDM2 and a set of other genes located in this amplified region. More importantly, both subtypes can recur as multifocal lesions and it is likely that WDLPS and DDLPS could exist in the same sample at different proportions.
An increasing number of studies have demonstrated that disruption of distal enhancers is implicated as an important cancer-driven mechanism in various tumors (40, 47–53). Super enhancers have been observed and BET protein inhibitor, an enhancer inhibitor, was also proved antitumor efficacy in liposarcoma (10). However, the mechanisms of how enhancers contribute to liposarcoma tumorigenesis remains unclear. In this study, we demonstrated the existence of both enhancer coamplification and enhancer hijacking events with oncogenes in liposarcoma patient samples. Such tumorigenesis mechanisms were in line with previous examples of enhancer amplifications (47, 48, 54–58) and enhancer hijacking (42, 44, 49–51, 53, 59), activating oncogenes in various tumors. Among these studies, Andrew and colleagues demonstrated that of 4,577 tumor samples across 9 different solid cancer, 743 tumors had high-level focal enhancer oncogene coamplifications (56). Zhang. and colleagues found expression of hundreds of genes was altered within 100kb of a structure variation breakpoint in over 1,200 cancer genomes, which could be attributed to enhancer hijacking (60). These indicate that the roles of both enhancer coamplification and enhancer hijacking in cancer were currently under-appreciated (61). In liposarcoma, we observed the coexistence of enhancer coamplification and enhancer hijacking in the same patient involving the same oncogenes such as MDM2 and CDK4. Further study is required to investigate whether and how these aberrant enhancers cooperatively contribute to the dysregulation of the oncogenes.
In this study, we have shown that the MDM2 gene upregulation, fusion, coamplification, and enhancer hijacking events can occur in the same patient (Supplementary Figs. S13A–S13C) for P298 and P69. This is potentially due to the importance of MDM2 in liposarcoma. MDM2 amplification is the hallmark of both well-dedifferentiated liposarcomas (WD-LPS) and dedifferentiated liposarcomas (DD-LPS). Its amplification has been used as a diagnostic tool to distinguish WD-LPS and DD-LPS from benign lipomatous neoplasms and other high-grade sarcomas in clinic (62). In addition, the large genomic region (12q13–15) that contains MDM2 gene is usually amplified and possesses frequent structural alterations, such as fusions and inversions. Such genomic structural alterations usually lead to the massive genome arrangement and the formation of extra ring/giant chromosome, as evidenced by the FISH experiments shown in Fig. 5D. As shown in this work, the MDM2 gene expression was dramatically increased through a combination of copy gain, enhancer coamplification, gene fusion, and enhancer hijacking, and its upregulation has been shown to lead to tumor initiation and progression (63, 64).
More generally, we demonstrated that multiple copies of MDM2 and CDK4 exist on the liposarcoma giant marker chromosome. The giant chromosome represents a specific kind of amplification structure different from both ecDNA and HSR (65). Recent studies have revealed the multifaceted features of the extra-chromosomal circular DNA in cancer (56, 66–68). However, little is known about the role of the giant chromosome in cancer. In this work, we performed the FISH experiments for CDK4 and MDM2, two of the most amplified genes in liposarcoma. As shown Fig. 5D, we didn't observe any CDK4 or MDM2-containing ecDNA. Instead, we observed many copies of these two oncogenes on the giant chromosome, in line with previous studies (7, 65). However, we cannot completely exclude the existence of ecDNA (that might contain other oncogenes) in liposarcoma and it is possible that ecDNA and giant chromosomes coexist in liposarcoma.
In addition, we characterized a high confidence list of structure variations from the Lipo863B cell line, which provided a more comprehensive view of the liposarcoma cancer genome. Based on this, we were able to further reconstruct the amplified segments containing oncogenes. The reconstructions were re-affirmed by multi-platform support. Genomic and epigenomic architectures were resolved at these reconstructed genome segments. These greatly improved our understanding about the transcriptional regulations of the oncogenes in the local context. It would greatly deepen our understanding about the giant chromosome in cancers by resolving the whole genomic architectures in this cell line.
Supplementary Material
Patient Sample Information
H3K27Ac ChIP-seq Patient Sample Statistics
Patient Sample Hi-C data Statistics
Patient Sample WGS data statistics
Sequence Data Information in Lipo863B
All Supplementary Figures
Acknowledgments
F. Yue was supported by NIH grants R35GM124820, 1R01HG009906, R01HG011207, and U24HG012070. R. Pollock was supported by Translational Team Science Award from Department of Defense (CA210874) and NCI P30CA016058 OSU Cancer Center Support Grant. The authors thank all the members of Yue lab for helpful discussions.
The publication costs of this article were defrayed in part by the payment of publication fees. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Footnotes
Note: Supplementary data for this article are available at Cancer Research Online (http://cancerres.aacrjournals.org/).
Authors' Disclosures
F. Yue reports other support from Sariant Therapeutics Inc. outside the submitted work. No disclosures were reported by the other authors.
Authors' Contributions
T. Liu: Conceptualization, writing-review and editing, performed most of the experiments. J. Wang: Data curation, formal analysis, visualization, writing–original draft, writing–review and editing. H. Yang: Conceptualization. Q. Jin: Validation. X. Wang: Formal analysis. Y. Fu: Formal analysis. Y. Luan: Formal analysis. Q. Wang: Formal analysis, writing–review and editing. M.W. Youngblood: Writing–review and editing. X. Lu: Validation. L. Casadei: Resources. R. Pollock: Conceptualization, resources. F. Yue: Conceptualization, supervision, funding acquisition, writing–original draft, writing–review and editing.
References
- 1. Nassif NA, Tseng W, Borges C, Chen P, Eisenberg B. Recent advances in the management of liposarcoma. F1000Res 2016;5:2907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Keung EZSN. Overview of liposarcomas and their genomic landscape. J Transl Genet Genom 2019;3:8. [Google Scholar]
- 3. Kraybill WG, Harris J, Spiro IJ, Ettinger DS, DeLaney TF, Blum RH, et al. Long-term results of a phase 2 study of neoadjuvant chemotherapy and radiotherapy in the management of high-risk, high-grade, soft tissue sarcomas of the extremities and body wall: radiation therapy oncology group trial 9514. Cancer 2010;116:4613–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Cancer Genome Atlas Research Network. Electronic address edsc, Cancer Genome Atlas Research N: Comprehensive and integrated genomic characterization of adult soft tissue sarcomas. Cell 2017;171:950–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Haddox CL, Riedel RF. Recent advances in the understanding and management of liposarcoma. Fac Rev 2021;10:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Saada-Bouzid E, Burel-Vandenbos F, Ranchere-Vince D, Birtwisle-Peyrottes I, Chetaille B, Bouvier C, et al. Prognostic value of HMGA2, CDK4, and JUN amplification in well-differentiated and dedifferentiated liposarcomas. Mod Pathol 2015;28:1404–14. [DOI] [PubMed] [Google Scholar]
- 7. Garsed DW, Marshall OJ, Corbin VD, Hsu A, Di Stefano L, Schroder J, et al. The architecture and evolution of cancer neochromosomes. Cancer Cell 2014;26:653–67. [DOI] [PubMed] [Google Scholar]
- 8. Mariani O, Brennetot C, Coindre JM, Gruel N, Ganem C, Delattre O, et al. JUN oncogene amplification and overexpression block adipocytic differentiation in highly aggressive sarcomas. Cancer Cell 2007;11:361–74. [DOI] [PubMed] [Google Scholar]
- 9. Keung EZ, Akdemir KC, Al Sannaa GA, Garnett J, Lev D, Torres KE, et al. Increased H3K9me3 drives dedifferentiated phenotype via KLF6 repression in liposarcoma. J Clin Invest 2015;125:2965–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Chen Y, Xu L, Mayakonda A, Huang ML, Kanojia D, Tan TZ, et al. Bromodomain and extraterminal proteins foster the core transcriptional regulatory programs and confer vulnerability in liposarcoma. Nat Commun 2019;10:1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Wang X, Xu J, Zhang B, Hou Y, Song F, Lyu H, et al. Genome-wide detection of enhancer-hijacking events from chromatin interaction data in rearranged genomes. Nat Methods 2021;18:661–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Brind'Amour J, Liu S, Hudson M, Chen C, Karimi MM, Lorincz MC. An ultra-low-input native ChIP-seq protocol for genome-wide profiling of rare cell populations. Nat Commun 2015;6:6033. [DOI] [PubMed] [Google Scholar]
- 13. Rao SS, Huntley MH, Durand NC, Stamenova EK, Bochkov ID, Robinson JT, et al. A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell 2014;159:1665–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 2013;29:15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Uhrig S, Ellermann J, Walther T, Burkhardt P, Frohlich M, Hutter B, et al. Accurate and efficient detection of gene fusions from RNA sequencing data. Genome Res 2021;31:448–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Jeffares DC, Jolly C, Hoti M, Speed D, Shaw L, Rallis C, et al. Transient structural variations have strong effects on quantitative traits and reproductive isolation in fission yeast. Nat Commun 2017;8:14061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Spies N, Zook JM, Salit M, Sidow A. svviz: a read viewer for validating structural variants. Bioinformatics 2015;31:3994–6. [DOI] [PubMed] [Google Scholar]
- 18. Li H. Minimap2: pairwise alignment for nucleotide sequences. Bioinformatics 2018;34:3094–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Sedlazeck FJ, Rescheneder P, Smolka M, Fang H, Nattestad M, von Haeseler A, et al. Accurate detection of complex structural variations using single-molecule sequencing. Nat Methods 2018;15:461–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Nattestad M, Aboukhalil R, Chin CS, Schatz MC. Ribbon: intuitive visualization for complex genomic variation. Bioinformatics 2021;37:413–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kolmogorov M, Yuan J, Lin Y, Pevzner PA. Assembly of long, error-prone reads using repeat graphs. Nat Biotechnol 2019;37:540–6. [DOI] [PubMed] [Google Scholar]
- 22. Wang Y, Wang G, Tan X, Ke K, Zhao B, Cheng N, et al. MT1G serves as a tumor suppressor in hepatocellular carcinoma by interacting with p53. Oncogenesis 2019;8:67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Zeng JD, Zhang N, Zhao GJ, Xu LX, Yang Y, Xu XY, et al. MT1G is silenced by DNA methylation and contributes to the pathogenesis of hepatocellular carcinoma. J Cancer 2018;9:2807–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Song L, Chang R, Dai C, Wu Y, Guo J, Qi M, et al. SORBS1 suppresses tumor metastasis and improves the sensitivity of cancer to chemotherapy drug. Oncotarget 2017;8:9108–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Groffen J, Stephenson JR, Heisterkamp N, de Klein A, Bartram CR, Grosveld G. Philadelphia chromosomal breakpoints are clustered within a limited region, bcr, on chromosome 22. Cell 1984;36:93–99. [DOI] [PubMed] [Google Scholar]
- 26. Soda M, Choi YL, Enomoto M, Takada S, Yamashita Y, Ishikawa S, et al. Identification of the transforming EML4-ALK fusion gene in non-small-cell lung cancer. Nature 2007;448:561–6. [DOI] [PubMed] [Google Scholar]
- 27. Unachukwu U, Chada K, D'Armiento J. High mobility Group AT-Hook 2 (HMGA2) oncogenicity in mesenchymal and epithelial neoplasia. Int J Mol Sci 2020;21:3151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ouchi K, Miyachi M, Yagyu S, Kikuchi K, Kuwahara Y, Tsuchiya K, et al. Oncogenic role of HMGA2 in fusion-negative rhabdomyosarcoma cells. Cancer Cell Int 2020;20:192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Dahlen A, Mertens F, Rydholm A, Brosjo O, Wejde J, Mandahl N, et al. Fusion, disruption, and expression of HMGA2 in bone and soft tissue chondromas. Mod Pathol 2003;16:1132–40. [DOI] [PubMed] [Google Scholar]
- 30. Whyte WA, Orlando DA, Hnisz D, Abraham BJ, Lin CY, Kagey MH, et al. Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell 2013;153:307–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Dekker J. Gene regulation in the third dimension. Science 2008;319:1793–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Salameh TJ, Wang X, Song F, Zhang B, Wright SM, Khunsriraksakul C, et al. A supervised learning framework for chromatin loop detection in genome-wide contact maps. Nat Commun 2020;11:3428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Dixon JR, Xu J, Dileep V, Zhan Y, Song F, Le VT, et al. Integrative detection and analysis of structural variation in cancer genomes. Nat Genet 2018;50:1388–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Layer RM, Chiang C, Quinlan AR, Hall IM. LUMPY: a probabilistic framework for structural variant discovery. Genome Biol 2014;15:R84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Rausch T, Zichner T, Schlattl A, Stutz AM, Benes V, Korbel JO. DELLY: structural variant discovery by integrated paired-end and split-read analysis. Bioinformatics 2012;28:i333–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Wala JA, Bandopadhayay P, Greenwald NF, O'Rourke R, Sharpe T, Stewart C, et al. SvABA: genome-wide detection of structural variants and indels by local assembly. Genome Res 2018;28:581–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Wang S, Lee S, Chu C, Jain D, Kerpedjiev P, Nelson GM, et al. HiNT: a computational method for detecting copy number variations and translocations from Hi-C data. Genome Biol 2020;21:73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Krzywinski M, Schein J, Birol I, Connors J, Gascoyne R, Horsman D, et al. Circos: an information aesthetic for comparative genomics. Genome Res 2009;19:1639–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Spielmann M, Lupianez DG, Mundlos S. Structural variation in the 3D genome. Nat Rev Genet 2018;19:453–67. [DOI] [PubMed] [Google Scholar]
- 40. Weischenfeldt J, Dubash T, Drainas AP, Mardin BR, Chen Y, Stutz AM, et al. Pan-cancer analysis of somatic copy-number alterations implicates IRS4 and IGF2 in enhancer hijacking. Nat Genet 2017;49:65–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Drier Y, Cotton MJ, Williamson KE, Gillespie SM, Ryan RJ, Kluk MJ, et al. An oncogenic MYB feedback loop drives alternate cell fates in adenoid cystic carcinoma. Nat Genet 2016;48:265–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Groschel S, Sanders MA, Hoogenboezem R, de Wit E, Bouwman BAM, Erpelinck C, et al. A single oncogenic enhancer rearrangement causes concomitant EVI1 and GATA2 deregulation in leukemia. Cell 2014;157:369–81. [DOI] [PubMed] [Google Scholar]
- 43. Yang M, Safavi S, Woodward EL, Duployez N, Olsson-Arvidsson L, Ungerback J, et al. 13q12.2 deletions in acute lymphoblastic leukemia lead to upregulation of FLT3 through enhancer hijacking. Blood 2020;136:946–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Ooi WF, Nargund AM, Lim KJ, Zhang S, Xing M, Mandoli A, et al. Integrated paired-end enhancer profiling and whole-genome sequencing reveals recurrent CCNE1 and IGF2 enhancer hijacking in primary gastric adenocarcinoma. Gut 2020;69:1039–52. [DOI] [PubMed] [Google Scholar]
- 45. Peng T, Zhang P, Liu J, Nguyen T, Bolshakov S, Belousov R, et al. An experimental model for the study of well-differentiated and dedifferentiated liposarcoma; deregulation of targetable tyrosine kinase receptors. Lab Invest 2011;91:392–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Aganezov S, Goodwin S, Sherman RM, Sedlazeck FJ, Arun G, Bhatia S, et al. Comprehensive analysis of structural variants in breast cancer genomes using single-molecule sequencing. Genome Res 2020;30:1258–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Shi J, Whyte WA, Zepeda-Mendoza CJ, Milazzo JP, Shen C, Roe JS. et al. Role of SWI/SNF in acute leukemia maintenance and enhancer-mediated Myc regulation. Genes Dev 2013;27:2648–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Zhang X, Choi PS, Francis JM, Imielinski M, Watanabe H, Cherniack AD, et al. Identification of focally amplified lineage-specific super-enhancers in human epithelial cancers. Nat Genet 2016;48:176–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Northcott PA, Lee C, Zichner T, Stutz AM, Erkek S, Kawauchi D, et al. Enhancer hijacking activates GFI1 family oncogenes in medulloblastoma. Nature 2014;511:428–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Haller F, Bieg M, Will R, Korner C, Weichenhan D, Bott A, et al. Enhancer hijacking activates oncogenic transcription factor NR4A3 in acinic cell carcinomas of the salivary glands. Nat Commun 2019;10:368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Helmsauer K, Valieva ME, Ali S, Chamorro Gonzalez R, Schopflin R, Roefzaad C, et al. Enhancer hijacking determines extrachromosomal circular MYCN amplicon architecture in neuroblastoma. Nat Commun 2020;11:5823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Smeenk L, Ottema S, Mulet-Lazaro R, Ebert A, Havermans M, Varea AA, et al. Selective requirement of MYB for oncogenic hyperactivation of a translocated enhancer in leukemia. Cancer Discov 2021;11:2868–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Montefiori LE, Bendig S, Gu Z, Chen X, Polonen P, Ma X, et al. Enhancer hijacking drives oncogenic BCL11B expression in lineage-ambiguous stem cell leukemia. Cancer Discov 2021;11:2846–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Takeda DY, Spisak S, Seo JH, Bell C, O'Connor E, Korthauer K, Ribli D, et al. A somatically acquired enhancer of the androgen receptor is a noncoding driver in advanced prostate cancer. Cell 2018;174:422–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Viswanathan SR, Ha G, Hoff AM, Wala JA, Carrot-Zhang J, Whelan CW, et al. Structural alterations driving castration-resistant prostate cancer revealed by linked-read genome sequencing. Cell 2018;174:433–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Morton AR, Dogan-Artun N, Faber ZJ, MacLeod G, Bartels CF, Piazza MS, et al. Functional enhancers shape extrachromosomal oncogene amplifications. Cell 2019;179:1330–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Wang J, Huang TY, Hou Y, Bartom E, Lu X, Shilatifard A, et al. Epigenomic landscape and 3D genome structure in pediatric high-grade glioma. Sci Adv 2021;7:eabg4126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Liu Y, Wu Z, Zhou J, Ramadurai DKA, Mortenson KL, Aguilera-Jimenez E, et al. A predominant enhancer co-amplified with the SOX2 oncogene is necessary and sufficient for its expression in squamous cancer. Nat Commun 2021;12:7139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Yang L, Chen F, Zhu H, Chen Y, Dong B, Shi M, et al. 3D genome alterations associated with dysregulated HOXA13 expression in high-risk T-lineage acute lymphoblastic leukemia. Nat Commun 2021;12:3708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Zhang Y, Chen F, Fonseca NA, He Y, Fujita M, Nakagawa H, et al. High-coverage whole-genome analysis of 1220 cancers reveals hundreds of genes deregulated by rearrangement-mediated cis-regulatory alterations. Nat Commun 2020;11:736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Claringbould A, Zaugg JB. Enhancers in disease: molecular basis and emerging treatment strategies. Trends Mol Med 2021;27:1060–73. [DOI] [PubMed] [Google Scholar]
- 62. Ware PL, Snow AN, Gvalani M, Pettenati MJ, Qasem SA. MDM2 copy numbers in well-differentiated and dedifferentiated liposarcoma: characterizing progression to high-grade tumors. Am J Clin Pathol 2014;141:334–41. [DOI] [PubMed] [Google Scholar]
- 63. Hauck PM, Wolf ER, Olivos DJ 3rd, Batuello CN, McElyea KC, McAtarsney CP, et al. Early-stage metastasis requires Mdm2 and not p53 gain of function. Mol Cancer Res 2017;15:1598–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Jones SN, Hancock AR, Vogel H, Donehower LA, Bradley A. Overexpression of Mdm2 in mice reveals a p53-independent role for Mdm2 in tumorigenesis. Proc Natl Acad Sci U S A 1998;95:15608–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Pedeutour F, Forus A, Coindre JM, Berner JM, Nicolo G, Michiels JF, et al. Structure of the supernumerary ring and giant rod chromosomes in adipose tissue tumors. Genes Chromosomes Cancer 1999;24:30–41. [PubMed] [Google Scholar]
- 66. Wu S, Turner KM, Nguyen N, Raviram R, Erb M, Santini J, et al. Circular ecDNA promotes accessible chromatin and high oncogene expression. Nature 2019;575:699–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Zhu Y, Gujar AD, Wong CH, Tjong H, Ngan CY, Gong L, et al. Oncogenic extrachromosomal DNA functions as mobile enhancers to globally amplify chromosomal transcription. Cancer Cell 2021;39:694–707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. van Leen E, Bruckner L, Henssen AG. The genomic and spatial mobility of extrachromosomal DNA and its implications for cancer therapy. Nat Genet 2022;54:107–14. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Patient Sample Information
H3K27Ac ChIP-seq Patient Sample Statistics
Patient Sample Hi-C data Statistics
Patient Sample WGS data statistics
Sequence Data Information in Lipo863B
All Supplementary Figures
Data Availability Statement
Processed RNA-seq, WGS, Hi-C, and H3K27ac ChIP-seq data generated from primary patient samples have been deposited at the Gene Expression Omnibus under accession GSE201056. Raw and processed RNA-seq, WGS, Hi-C, H3K27ac ChIP-seq, Nanpore sequencing data in Lipo863B cell line have been deposited at the Gene Expression Omnibus under accession GSE201056.