Dual functions of circFNDC3B in enhancing the metastatic ability of cancer cells and promoting vasculature formation through regulation of multiple pro-oncogenic signaling pathways drive lymph node metastasis of OSCC.
Abstract
Emerging evidence has demonstrated that circular RNAs (circRNA) are involved in cancer metastasis. Further elucidation of the role of circRNAs in oral squamous cell carcinoma (OSCC) could provide insights into mechanisms driving metastasis and potential therapeutic targets. Here, we identify a circRNA, circFNDC3B, that is significantly upregulated in OSCC and is positively associated with lymph node (LN) metastasis. In vitro and in vivo functional assays showed that circFNDC3B accelerated the migration and invasion of OSCC cells and the tube-forming capacity of human umbilical vein endothelial cells and human lymphatic endothelial cells. Mechanistically, circFNDC3B regulated ubiquitylation of the RNA-binding protein FUS and the deubiquitylation of HIF1A through the E3 ligase MDM2 to promote VEGFA transcription, thereby enhancing angiogenesis. Meanwhile, circFNDC3B sequestered miR-181c-5p to upregulate SERPINE1 and PROX1, which drove epithelial–mesenchymal transition (EMT) or partial-EMT (p-EMT) in OSCC cells and promoted lymphangiogenesis to accelerate LN metastasis. Overall, these findings uncovered the mechanistic role of circFNDC3B in orchestrating cancer cell metastatic properties and vasculature formation, suggesting circFNDC3B could be a potential target to reduce OSCC metastasis.
Significance:
Dual functions of circFNDC3B in enhancing the metastatic ability of cancer cells and promoting vasculature formation through regulation of multiple pro-oncogenic signaling pathways drive lymph node metastasis of OSCC.
Introduction
Oral cancer, predominantly oral squamous cell carcinoma (OSCC), is the eighth most common cancer type, with 170,000 deaths and over 370,000 newly added cases in 2020 alone worldwide (1, 2). Despite recent remarkable advances in the diagnosis and treatment of OSCC, the 5-year survival rate of patients remains under 50% owing to high rates of tumor metastasis, and the presence of cervical lymph node (LN) metastasis is considered the most important prognostic determinant of OSCC (3, 4). However, the mechanisms that lead to OSCC spread via the lymphatic vessels remain largely unknown. Therefore, it is urgently needed to investigate the underlying mechanisms for identifying potential biomarkers and therapeutic targets for improving the survival of affected patients.
LN metastasis, a major metastatic pattern of OSCC, is a multistep process involving epithelial–mesenchymal transition (EMT) and the intermediate state of EMT (p-EMT) associated with migration, invasion, and orchestrating the metastatic tumor microenvironment (TME) such as angiogenesis and lymphangiogenesis (5, 6). Of note is that lymphangiogenesis is a key determinant of metastatic progression and a promising predictor of patient outcomes (7). The prospero homeobox 1 (PROX1) and sex-determining region Y-box 18 (SOX18) transcription factors regulate the lymphangiogenic process by controlling the initiation of endothelial branching and spreading (8). Highly metastatic tumor cells can also form structures similar to lymphatic vessels, fusing with native lymphatic structures and thus facilitating metastatic dissemination (9). Furthermore, the density of lymphatic vessels can predict OSCC patient metastasis-free survival (10). Recently, emerging evidence highlights the roles of circular RNAs (circRNA) in lymphangiogenesis, metastasis, and related oncogenic processes through their transcriptional and posttranscriptional regulatory activities (11, 12), whereas the physiologic functions of circRNAs in OSCC have not been completely established.
circRNAs are noncoding RNAs with a covalent closed-loop structure (13). Bioinformatics technologies have led to the characterization of tissue-specific expression profiles for thousands of different circRNAs (14). Although the reverse splicing of precursor RNAs necessary to generate circRNAs is less efficient than linear splicing such that circRNAs transcription levels are decreased relative to those of parental genes, circRNAs are extremely stable and persist within cells owing to their inherent exonuclease-resistant properties (15, 16). This stability has led to growing clinical interest in leveraging circRNAs as specific biomarkers of varying diseases (17, 18). Mechanistically, circRNAs can sequester complementary microRNAs (miRNA) and thus alter gene expression, as in the case of circUHRF1, which drives OSCC development via sequestering miR-526b-5p and promoting c-Myc upregulation (19). circRNAs can also interact with RNA-binding proteins (RBP) to alter target gene transcription and stability or to aid enzyme colocalization (20). For example, circFoxo3 promotes the MDM2-mediated ubiquitylation and degradation of p53 in breast cancer (21). In certain settings, circRNA-derived peptides can also regulate certain diseases (22). circ-LINC-PINT-derived FBXW-185aa can suppress the transcriptional elongation of several oncogenes by interacting with PAF1c, in turn inhibiting glioblastoma development (23). Therefore, exploring specific circRNAs functions in OSCC might provide improved diagnosis and treatment for OSCC patients.
The fibronectin type III domain containing 3B (FNDC3B) gene encodes a protein primarily found at the endoplasmic reticulum (ER) membrane that is linked with cellular migration and adhesion (24, 25). FNDC3B dysregulation has been observed in cervical, nasopharyngeal, and oral cancers (26–28). Here, a circRNA derived from FNDC3B (exons 2–3; hsa_circ_0001361, circFNDC3B) significantly upregulated in OSCC and linked with LN metastasis. Mechanistically, circFNDC3B promotes the ubiquitin-mediated degradation of the FUS and then regulates HIF1A and VEGFA expression, in turn, enhanced angiogenesis in TME. Moreover, circFNDC3B sequesters miR-181c-5p to promote upregulation of the lymphatic endothelial cell-fate regulator PROX1 and the partial-EMT marker Serpine1, thus driving metastatic tumor activity and lymphangiogenesis in TME. To our knowledge, this is the first evidence linking metastatic cancer cells to vasculature formation in TME through circFNDC3B, contributing to LN metastasis of OSCC.
Materials and Methods
Patient samples
In total, 104 paired tumor and paracancerous tissue samples from OSCC patients (74 males and 30 females, aged from 26 to 88 years old) were harvested in the Department of Oral and Maxillofacial Surgery, Affiliated Stomatological Hospital of Nanjing Medical University. Histologic and pathologic diagnoses of tissue samples were independently confirmed by 2 experienced histopathologists. Patient demographic and clinical characteristics are summarized in Supplementary Table S1. Blood samples (2 mL) of OSCC patients and healthy controls were collected and centrifuged at 3,500 rpm for 10 minutes at room temperature. The upper serum was frozen at −80°C until use. The experimental content and protocol of this study were evaluated and reviewed by the Ethics Committee of Affiliated Stomatological Hospital of Nanjing Medical University. All patients mentioned above have agreed to participate and signed written informed consent.
Cell culture
Head and neck squamous cell carcinoma (HNSCC) cell lines (SCC4, HN4, HN6, and FADU (RRID: CVCL_7751, RRID: CVCL_8127, RRID: CVCL_8129, and RRID: CVCL_1218)) and human oral keratinocytes (HOK) cell lines were obtained from the Chinese Academy of Sciences, HSC3 (RRID: CVCL_1288), CAL27 (RRID: CVCL_1107), and SSC9 (RRID: CVCL_1685) cells were obtained from ATCC. SCC9 cell was cultured in DMEM/F-12 (Gibco) medium containing 10% FBS (Biological Industries) and 1% penicillin/streptomycin (Invitrogen). Other cell lines were appropriately cultured in high-glucose DMEM (Gibco) containing 10% FBS and 1% penicillin/streptomycin. Human umbilical vein endothelial cells (HUVEC) were gifts from Ning Chen of Nanjing Medical University (Nanjing, China) and were cultured in Endothelial Cell Medium (ECM, Gibco) containing 5% FBS. Human lymphatic endothelial cells (HLEC) were obtained from Genio Biotechnology and were maintained in ECM containing 15% FBS. All cells were grown in a humidified 37°C 5% CO2 incubator. All cell lines are qualified after identification and Mycoplasma testing.
Fluorescent in situ hybridization
Fluorescent in situ hybridization (FISH) assays and circFNDC3B probes synthesized were performed with a Ribo FISH Kit (RiboBio). Cells were seeded on coverslips in 24-well plates (1×104 cells/well). After a 2-day incubation, cells were then fixed using 4% paraformaldehyde at room temperature for 30 minutes and treated with Triton X-100 for 15 minutes. Next, after incubating in prehybridization solution at 37°C for 30 minutes, coverslips were incubated with circFNDC3B or internal reference FISH probes with hybridization solution at 37°C overnight. The following day, the excess probe and hybridization solution were removed by washing with 4×, 2× and 1× Saline Sodium Citrate Buffer sequentially, and 4′, 6-diamidino-2-phenylindole (DAPI) was used for nuclear counterstaining. Cells were then imaged via laser scanning confocal microscope (#LSM710, Zeiss).
Zebrafish metastasis model
To evaluate the impact of circFNDC3B on OSCC cell metastasis ability, zebrafish were used to explore important data regarding metastatic events. Wild-type zebrafish with better developmental status were selected for culture. On the fourth day, each embryo was microinjected with 250 green fluorescent protein (GFP)-labeled tumor cells. Zebrafish were quickly transferred to a 28°C incubator for 1 hour, then transferred to a 31°C incubator for further incubation. On the sixth day, zebrafish were fixed, and images of tumor cell migration in caudal hematopoietic tissue were captured using the stereoscopic fluorescence microscope (Leica).
RNA pulldown assays
Pierce Magnetic RNA-protein Pulldown Kit (#20164, Thermo) was used to perform circFNDC3B pulldown. Biotin-labeled circFNDC3B and oligonucleotide probes were designed and synthesized by RiboBio. 1×107 HSC3 or CAL27 cells were lysed with 100 μL IP lysis buffer (Thermo) and collected into enzyme-free EP tubes. After centrifugation at 4°C for 10 minutes, the supernatant was obtained and then added with 1 μL RNase inhibitor. Next, circFNDC3B sense and scramble probes were incubated with cell lysates at 37°C for 4 hours. Meanwhile, 50 μL streptavidin magnetic beads mixed with 50 μL 1× RNA capture and 50 μL 1× RNA-protein binding buffer were incubated with cell lysates containing probes overnight at 4°C. The second day, magnetic beads were purified with 20 mmol/L Tris (PH = 7.5) 3 times and treated with 50 μL elution buffer at 37°C for 30 minutes with oscillation. For protein detection, 50 μL elution buffer with magnetic beads was added with 5× SDS loading buffer (12.5 μL) and then boiled for 10 minutes. For enriched miRNAs detection, 50 μL elution buffer should be added with 550 μL TRIzol reagent to extract RNA. Detailed information on probes sequences was listed in Supplementary Table S2.
Western blotting analysis
Radio immunoprecipitation assay buffer (Beyotime) containing phenylmethanesulfonyl fluoride (PMSF; Beyotime) and protease inhibitor (PI; Dalian Meilun Biotechnology) was used to lyse samples and followed by centrifugation at 12,000 rpm for 10 minutes at 4°C. The supernatant with 5× SDS loading buffer was boiled for 10 minutes. Subsequently, proteins were separated by 8%, 10%, or 12% SDS-polyacrylamide gel electrophoresis and transferred to polyvinylidene difluoride membranes (PVDF; Millipore). Blots were blocked using 5% skim milk, probed with primary antibodies (Supplementary Table S3) overnight at 4°C. The second day, the PVDF membranes were immersed with the secondary antibodies for 1 hour and imaged using Tanon High-sig ECL Western Blotting Substrate (180-5001; Tanon). RRIDs of all antibodies are provided in Supplementary Table S3.
Bioinformatics analysis of circRNAs in Gene Expression Omnibus
The keywords "OSCC" and "circRNA" were searched through the Gene Expression Omnibus (GEO) data sets. GSE131182 and GSE197138 were obtained for analysis. DESeq2, fold change (FC) ≥ 2, and P < 0.05 were used to analyze differentially expressed circRNAs. The common highly expressed circRNAs were screened out by Venn analysis. The demographic and clinical details of these data sets were provided in Supplementary Table S4.
In vivo orthotopic xenograft model
In total, 20 female BALB/c nude mice were randomized into 4 groups. The tongue mucosa of each mouse was then injected with GFP-control– or GFP-sh-circFNDC3B–transfected HSC3 cells, or with GFP-control– or GFP-LV-circFNDC3B–transfected CAL27 cells, with each mouse being implanted with 5×106 cells in 50 μL of DMEM and Matrigel. The weight of the mice was measured every 3 days. After 14 days, in vivo imaging was used to confirm tongue cancer model establishment, and mice were euthanized, after which, tongue tumors and cervical LNs were collected for size measurement, histopathologic, immunofluorescence, and IHC staining to examine tumor cells proliferation, micrometastases, and cancer-associated vasculature formation. Tumor and LNs volume (mm3) = width (mm)2 × length (mm)/2. The area ratio of LN micrometastases and tongue tumors and the number of neovascularization were measured by ImageJ software.
In vivo translational experiment
Fourty female BALB/c nude mice were randomized into 8 groups. After nude mice adapted to the environment, the tongue mucosa of each mouse in group 1, groups 2 to 4, group 5, and groups 6 to 8 nude mice were injected with 5×106 HSC3-sh-NC, HSC3-sh-circFNDC3B, CAL27-LV-NC, and CAL27-LV-cicFNDC3B cells, respectively. On day 7 after implantation, bevacizumab (0.1 mg per mouse, HY-P9906, MedChemExpress) was intratumorally injected into group 3 and 7 mice every 4 days, with 4 total injections. Similarly, 5 nmol/L of miR-181c-5p antagomir (RiboBio) was intratumorally injected into groups 4 and 8. And other nude mice were injected with the same amount of PBS. On day 21, mice were euthanized and then tongue tumors and cervical LNs were collected for analysis of neovascularization and metastasis.
Statistical analyses
All statistical calculations were performed using GraphPad Prism 8 software. Student t test (two groups), analysis of variance (multiple groups), and Χ2 test (clinical parameters) were used to determine statistical significance. Experiments were repeated at least three times. *, P < 0.05 was the significance threshold.
Further applied methods
Additional methodological details and database URLs were provided in the Supplementary Materials.
Ethics approval and consent to participate
Written informed consent was obtained from all patients, and the study was approved by the Ethics Committee of Affiliated Stomatological Hospital of Nanjing Medical University (Approval no. PJ2019-109-001). Animal experimental procedures were approved by the Laboratory Animal Care and Use Committee at Nanjing Medical University (Approval no. IACUC-2101038).
Data availability
The accession numbers of data for analyzing differentially expressed circRNAs reported in this paper are GEO: GSE131182 and GSE197138. The data generated in this study are also available upon request from the corresponding author.
Results
Hsa_circ_0001361 is upregulated and correlated with LN metastasis in OSCC patients
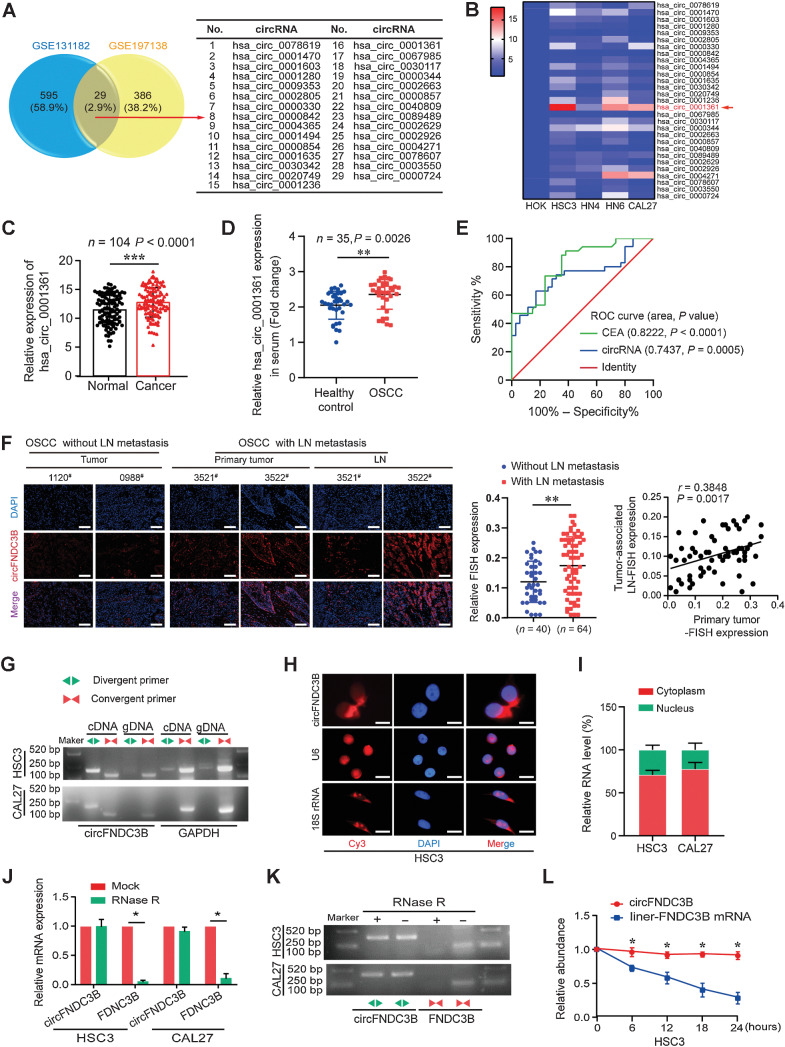
The GSE131182 and GSE197138 data sets were used to analyze circRNAs expression in paired OSCC samples, 624 and 415 overexpressed circRNAs were identified, respectively. Among them, 29 co-upregulated circRNAs were detected including hsa_circ_0001361, hsa_circ_0002805, hsa_circ_0089489 (Fig. 1A). Then, qRT-PCR was used to detect the level of these circRNAs in four HNSCC cell lines, revealing pronounced hsa_circ_0001361 overexpression, particularly in tongue squamous cell carcinoma cells (CAL27, HSC3, and HN6; Fig. 1B). Consistently, hsa_circ_0001361 levels were increased in OSCC tissues relative to paracancerous tissues, and overexpression was positively associated with poor recurrence-free survival (RFS, P = 0.0279; Fig. 1C; Supplementary Fig. S1A). Additionally, to assess the value of hsa_circ_0001361 expression as a biomarker for distinguishing healthy people from OSCC patients, the expression of hsa_circ_0001361 in the serum of 35 OSCC patients and 35 healthy controls was detected and ROC curve analysis was performed. The results showed that hsa_circ_0001361 in the serum of OSCC patients was significantly higher than that of healthy individuals, and the area under the ROC curve (AUC) of hsa_circ_0001361 was 0.7437, the best cutoff value was 2.395 with a sensitivity of 62.86% and a specificity of 82.86%. Meanwhile, carcino-embryonic antigen (CEA), a well-known biomarker, was considered as an internal control to identify the effectiveness of hsa_circ_0001361. ROC curve analysis of CEA was also detected, and AUC was 0.8222 with a cutoff value of 1.515 (Fig. 1D and E). These results suggested that hsa_circ_0001361 expression had a promising value of OSCC diagnosis. Subsequently, we detected the expression of hsa_circ_0001361 in different HNSCC cells and human oral mucosa keratinocytes (HOK) cell line, and focused on HSC3 and CAL27 cell lines as research objects (Supplementary Fig. S1B). In addition, we investigated whether hsa_circ_0001361 expression was correlated with clinicopathologic features in the 104 OSCC patients and found hsa_circ_0001361 upregulation significantly associated with LN metastasis (Supplementary Table S1). FISH staining also showed that hsa_circ_0001361 was significantly upregulated in primary tumor with LN metastasis and positively correlated with hsa_circ_0001361 expression in metastatic LNs (Fig. 1F). Together, our data indicate that hsa_circ_0001361 is highly expressed in OSCC tissues and cells and correlated with LN metastasis.
Figure 1.
The identification and characterization of hsa_circ_0001361 in OSCC. A, Downloaded GEO data sets for comprehensive analysis of differentially expressed circRNAs. Twenty-nine upregulated expressed circRNAs were listed. B, The relative expression levels of 29 circular RNAs were detected in four groups of HNSCC cell lines and HOK by qRT-PCR. Hsa_circ_0001361 has the highest relative expression. C, qRT-PCR analysis of hsa_circ_0001361 expression in 104 paired OSCC tissues and normal tissues. D, Serum hsa_circ_0001361 expression in healthy controls and OSCC patients were determined by qRT-PCR (n = 35, P = 0.0026). E, ROC curves were used to evaluate the diagnostic value of serum circFNDC3B in OSCC, compared with CEA. F, FISH staining assay showed that circFNDC3B expression in OSCC tissues with LN (**, P = 0.0030) metastasis was higher than that without LNs, and higher circFNDC3B expression in OSCC primary tumor was positively correlated with its expression in paired LNs. G, Combining PCR with an electrophoresis assay indicated the presence of circFNDC3B using divergent and convergent primers from cDNA or genomic DNA in HSC3 and CAL27 cells. H, Representative FISH images showed the cellular localization of circFNDC3B. The circFNDC3B probe was labeled with Cy3 (red), and nuclei were stained with DAPI (blue). The images were photographed at ×400 magnification. I, The location of circFNDC3B was confirmed using a subcellular fractionation assay and qRT-PCR data indicated that circFNDC3B is mainly located in the cytoplasm, and a minor part is in the nucleus. J and K, qRT-PCR and electrophoresis analysis for the resistance of circFNDC3B and linear FNDC3B to RNase R in HSC3 and CAL27 cells. Two-tailed t test was used. L, Actinomycin D assay to evaluate the stability of circFNDC3B and FNDC3B mRNA in HSC3 cells. Two-tailed t test was used. *, P < 0.05. #, Case numbers of patients. Scale bars, 100 μm (F) and 10 μm (H).
Characterization of circFNDC3B (hsa_circ_0001361) in OSCC cells
The structure of hsa_circ_0001361 was analyzed by the circBase database, revealing 215 nucleotide circRNA to be encoded at chr3:171830241–171851336 and to be the result of the back-splicing of exons 2 and 3 of FNDC3B (Supplementary Fig. S1C). We also found that the other 6 circRNAs originated from FNDC3B were all downregulated in HNSCC cells through qRT-PCR assays (Supplementary Fig. S1D). The circularization of hsa_circ_0001361 (designated circFNDC3B) was confirmed by producing convergent and divergent primers for the respective amplification of FNDC3B mRNA and circFNDC3B. Sanger sequencing indicated that divergent primer amplicons contained the predicted circFNDC3B head-to-tail splicing site predicted by the circBase database, and the secondary structure of circFNDC3B was predicted by RegRNA2.0 (Fig. 1G; Supplementary Fig. S1E). Subsequently, the existence of circFNDC3B was confirmed by qRT-PCR using different reverse transcription primers (Supplementary Fig. S1F). Furthermore, we investigated the cellular localization of circFNDC3B in OSCC cells. Subcellular fractionation and FISH analyses of circFNDC3B revealed it primarily localizing in the cytosol, with limited levels of nuclear distribution (Fig. 1H and I; Supplementary Fig. S1G). The circularization of circFNDC3B was further confirmed through its resistance to RNase R digestion, in contrast to linear FNDC3B mRNA (Fig. 1J and K). circFNDC3B stability was detected through actinomycin D treating OSCC cells and confirmed by an increased half-life of circFNDC3B transcript relative to that of linear FNDC3B (Fig. 1L; Supplementary Fig. S1H). These results suggest that circFNDC3B is a highly stable circRNA in OSCC.
circFNDC3B promotes the migration, invasion, EMT, and metastasis of OSCC cells
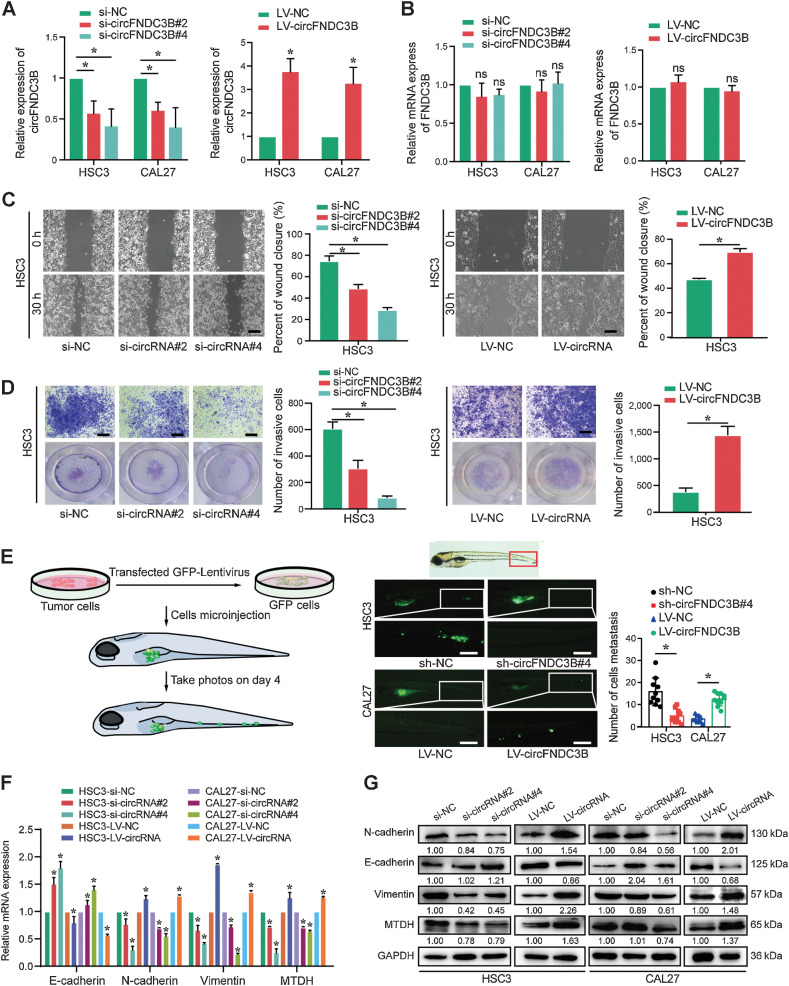
Initial functional characterization of circFNDC3B in OSCC was performed using small interfering RNAs (siRNA) specific for the circFNDC3B back-splicing junction, and the results showed that siRNAs of circFNDC3B effectively knocked down its expression in HSC3 and CAL27 without any impact on FNDC3B levels, meanwhile, circFNDC3B overexpression (cells transfected with LV-circFNDC3B) increased its level without impacting FNDC3B (Fig. 2A and B; Supplementary Fig. S2A). The proliferation of OSCC cells was unaffected by circFNDC3B overexpression or knockdown (Supplementary Fig. S2B and S2C). Considering a positive correlation between circFNDC3B expression and LN metastasis in OSCC patients, we assessed the roles of circFNDC3B in regulating migration and invasion of OSCC cells, and found that circFNDC3B knockdown significantly suppressed OSCC cells migration and invasion, whereas circFNDC3B overexpression had the opposite effect (Fig. 2C and D; Supplementary Fig. S2D–S2F). Currently, zebrafish is emerging as an attractive animal model in cancer research, which provided important data regarding metastatic events in vivo (9). Accordingly, the perivitelline space of zebrafish was microinjected using GFP-labeled HSC3 cells transfected with sh-circFNDC3B/sh-NC, or CAL27 cells with LV-circFNDC3B/LV-NC. Significantly reduced tail metastasis was observed in the sh-circFNDC3B group compared with the sh-NC group, whereas the opposite result was observed for the LV-circFNDC3B group (Fig. 2E). EMT enables epithelial cells to acquire migratory and invasive capabilities; thereby, we further assayed the effects of circFNDC3B on EMT- and metastasis-associated indexes via qRT-PCR and Western blot, and found that N-cadherin, vimentin, and MTDH in si-circFNDC3B group decreased, but E-cadherin increased. However, the opposite results were observed in the LV-circFNDC3B group (Fig. 2F and G). Collectively, our results indicate that circFNDC3B promotes the migration, invasion, EMT, and metastasis of OSCC cells.
Figure 2.
circFNDC3B accelerates the migration, invasion, EMT, and metastasis of OSCC cells. A and B, circFNDC3B and FNDC3B mRNA levels were assessed by qRT-PCR in HSC3 and CAL27 cells treated with si-circFNDC3Bs and LV-circFNDC3B, as indicated. C, The migration capability was suppressed in HSC3 cells treated with si-circFNDC3B#2 and si-circFNDC3B#4, whereas migration was promoted in HSC3 cells transfected with the LV-circFNDC3B, as determined using the wound-healing assay. D, The cell invasion ability was measured using transwell Matrigel invasion assays after transfection with si-circFNDC3B or circFNDC3B overexpression in HSC3 cells. E, Zebrafish xenografts revealed that the HSC3-sh-NC group had more tumor metastasis to tail areas of the fish compared with the HSC3-sh-circFNDC3B group, whereas the CAL27-LV-circFNDC3B group had more tumor metastasis cancer cells than the CAL27-LV-NC group. F and G, mRNA and protein expression levels of EMT and metastasis markers were detected via qRT-PCR and Western blot analysis in HSC3 and CAL27 cells. Data are presented as the mean of >3 independent experiments ± SD. *, P < 0.05. ns, no statistical significance. Scale bars, 100 μm (C–E).
circFNDC3B enhances angiogenesis and lymphangiogenesis
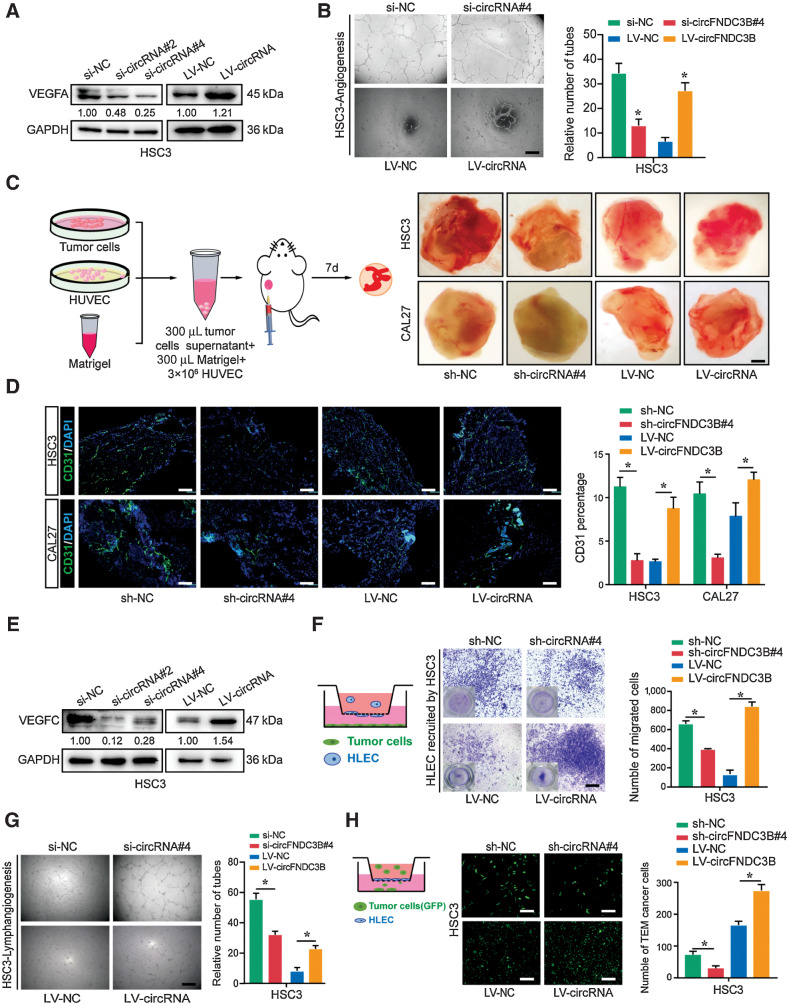
Given that angiogenesis and lymphangiogenesis play critical roles in the metastasis of carcinoma, we further explored whether circFNDC3B promoted angiogenesis and lymphangiogenesis. First, the expression of VEGFA and CD31 in 104 OSCC samples was evaluated using IHC. Combined with circFNDC3B expression and IHC score of VEGFA/CD31, we found a positive correlation between circFNDC3B and VEGFA/CD31 (Supplementary Fig. S3A). Furthermore, circFNDC3B knockdown or overexpression in CAL27 and HSC3 evidently inhibited or increased the expression of VEGFA (Fig. 3A; Supplementary Fig. S3B). In vitro assays showed that the conditioned medium from circFNDC3B-silencing or -overexpressing cells markedly suppressed or promoted the tube formation of HUVEC (Fig. 3B; Supplementary Fig. S3C). To further identify the proangiogenic function of circFNDC3B in vivo, we used a Matrigel-implanted nude mouse model acknowledged as a standard method for research on morphologic and functional angiogenesis (29). Consistently, enhanced neovascularization was demonstrated in the circFNDC3B overexpression group, which was further revealed by immunofluorescent staining (Fig. 3C and D). Considering the critical role of lymphangiogenesis in LN metastasis of OSCC, we also explored whether circFNDC3B promoted lymphangiogenesis. Similarly, IHC of VEGFC and LYVE1 was performed and the positive relationship of circFNDC3B and VEGFC/LYVE1 was discovered (Supplementary Fig. S3D). In cells, circFNDC3B knockdown or overexpression, respectively, suppressed or enhanced VEGFC expression (Fig. 3E; Supplementary Fig. S3E). Moreover, the migration and tube formation of HLEC were dramatically inhibited or promoted while cocultured with circFNDC3B-silencing or -overexpressing cells (Fig. 3F and G; Supplementary Fig. S3F and S3G). Endothelial adhesion is critical to transendothelial migration (TEM) of tumor cells, which is the initiation of distant metastasis of tumor cells (30). In this study, we found that circFNDC3B overexpression or knockdown, respectively, enhanced or inhibited the TEM of CAL27 and HSC3 cells (Fig. 3H; Supplementary Fig. S3H). Taken together, these results indicate that circFNDC3B in OSCC markedly enhances angiogenesis and lymphangiogenesis.
Figure 3.
circFNDC3B enhances angiogenesis and lymphangiogenesis. A, Western blot assays showed that si-circFNDC3B downregulated VEGFA expression, and LV-circFNDC3B promoted its expression in HSC3. B, The conditioned medium from si-circFNDC3B HSC3 cells dramatically inhibited the tube formation of HUVECs in vitro, whereas HSC3-LV-circFNDC3B had the opposite results. C, Pattern diagram of Matrigel plug assay for in vivo evaluation of angiogenesis and representative images of resected Matrigel plug. Intensity of the red color was used as a parameter for qualitative assessment. D, Representative image of Matrigel plug cryosection. Endothelial cells were stained for CD31 (green), and nuclei were stained with DAPI (blue). E, Western blot assays showed that si-circFNDC3B downregulated VEGFC expression in HSC3, whereas LV-circFNDC3B upregulated its expression. F and G, Transwell assays (F) and tube formation (G) of HLECs treated with conditioned medium from circFNDC3B-silencing or overexpressing HSC3 cells. H, The indicated tumor cells were seeded on HLEC monolayers reaching confluence in 24-well transwell inserts. After incubation for 24 hours, the transmigrated HSC3 cells were imaged. These experiments were replicated three times. Data are presented as the mean ± SD. *, P < 0.05. Scale bars, 100 μm (B, D, and F–H); 0.25 cm (C).
circFNDC3B binds the RNA-binding protein FUS in OSCC cells
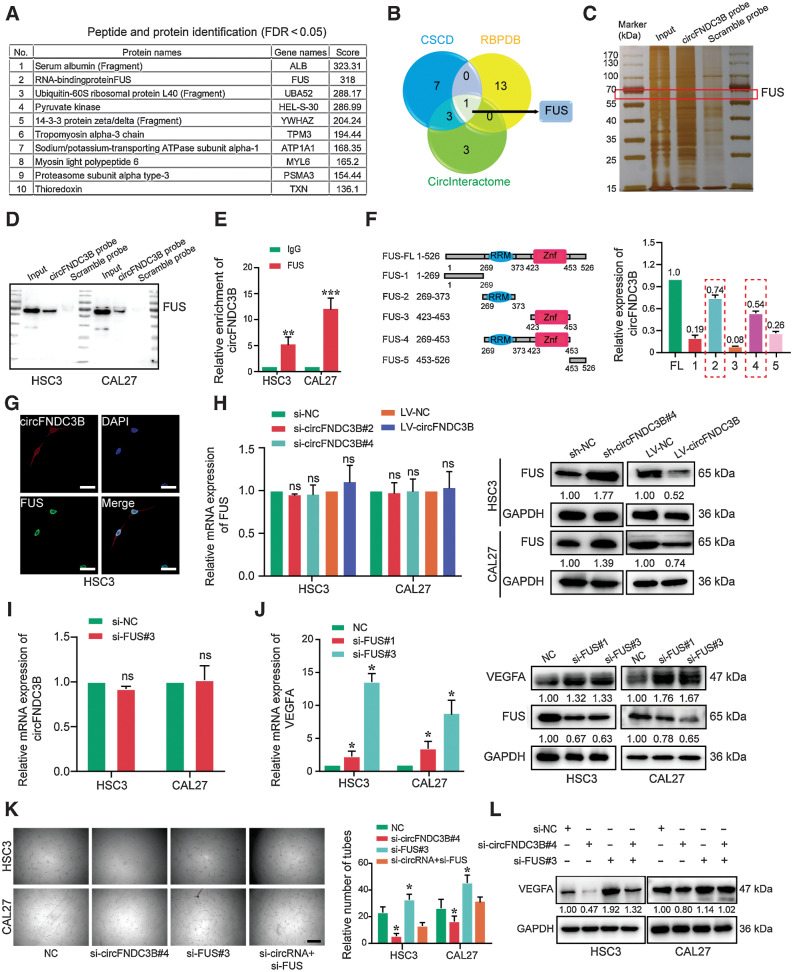
To investigate the mechanisms of circFNDC3B in promoting OSCC processes, biotinylated circFNDC3B-specific probes were used for an RNA pulldown assay. Enriched proteins by circFNDC3B-mediated pulldown were identified, and top-ranked proteins were listed in Fig. 4A and Supplementary Table S5. CSCD, RBPDB, and CircInteractome databases were used to predict the interaction of circFNDC3B with FUS (Fig. 4B). Consistent with the mass spectrometry results, FUS was a ranked forward protein enriched by circFNDC3B pulldown. After silver staining, the sense-specific band at about 65 kDa (red box) was excised (Fig. 4C). The interaction between circFNDC3B and FUS was further confirmed in both HSC3 and CAL27 cell lysates by RNA pulldown and RNA-binding protein immunoprecipitation (RIP) assays (Fig. 4D). Meanwhile, anti-FUS antibodies also pulled down circFNDC3B (Fig. 4E). Based on the predicted binding sites of circFNDC3B with FUS (Supplementary Fig. S4A), vectors encoding FLAG-tagged FUS deletion mutant constructs were generated, and circFNDC3B interacting with the RNA recognition motif domains of FUS was identified by anti-FLAG-mediated RIP (Fig. 4F). Immunofluorescent and FISH staining also showed that FUS and circFNDC3B colocalized in OSCC cell nuclei (Fig. 4G). Functionally, circFNDC3B overexpression decreased FUS protein levels without affecting FUS mRNA expression (Fig. 4H). We also found that the circFNDC3B expression was negatively correlated with the IHC score of FUS in 104 OSCC samples (Supplementary Fig. S4B). Accordingly, we found that circFNDC3B expression was unaffected by FUS knockdown (Fig. 4I; Supplementary Fig. S4D–S4E), indicating that FUS was the downstream of circFNDC3B. Next, the cBioPortal database and Metascape assays demonstrated that FUS-related genes were enriched in the VEGFA–VEGFR2 signaling pathway (Supplementary Fig. S4C). Moreover, the expression of VEGFA in FUS-silencing cells was upregulated, and a negative correlation of FUS and VEGFA IHC score was shown in OSCC samples (Fig. 4J; Supplementary Fig. S4F). We further showed that FUS-silencing functionally rescued circFNDC3B knockdown-mediated changes of VEGFA expression and angiogenesis (Fig. 4K and L). Collectively, these findings suggest that circFNDC3B promotes angiogenesis by destabilizing FUS protein.
Figure 4.
circFNDC3B promotes angiogenesis by binding FUS in OSCC. A, List of the top 10 differentially expressed proteins identified by mass spectrometry, FDR < 0.05. B, Venn diagram demonstrating the overlapping of the interacting RBPs of circFNDC3B predicted by CSCD, RBPDB, and CircInteractome. C, Silver staining of circFNDC3B pulldown in OSCC cells. The red box shows different expressions of FUS (65 kDa) between the sense and antisense lanes. D, Western blot showed circFNDC3B pulldown of the FUS. E, Expression of circFNDC3B was detected by qRT-PCR after RIP for FUS in HSC3 and CAL27. **, P = 0.0093; ***, P = 0.0009. F, Truncated versions of FLAG-FUS were produced according to the FUS domain. mRNAs isolated from the RIP assays with anti-FLAG tag antibody were identified by qRT-PCR analysis using circFNDC3B primers in 293T cells. Three independent experiments were performed. G, The colocalization of FUS and circFNDC3B was detected by immunofluorescence staining in HSC3 cells. H, qRT-PCR and Western blot were used to detect the expression of FUS after transfection with si-circFNDC3B and LV-circFNDC3B. I, Transcript level of circFNDC3B was not regulated by FUS. This means that FUS was downstream of circFNDC3B. J, The transcriptional levels and protein expression of VEGFA were upregulated by FUS silencing. K and L, Vessel formation (K) and Western blot (L) analysis showed that si-FUS functionally rescued angiogenesis and the expression of VEGFA upon circFNDC3B silencing. *, P < 0.05. ns, no statistical significance. Scale bars, 10 μm (G) and 100 μm (K).
FUS suppresses the transcription of VEGFA via modulating MDM2–HIF1A interactions
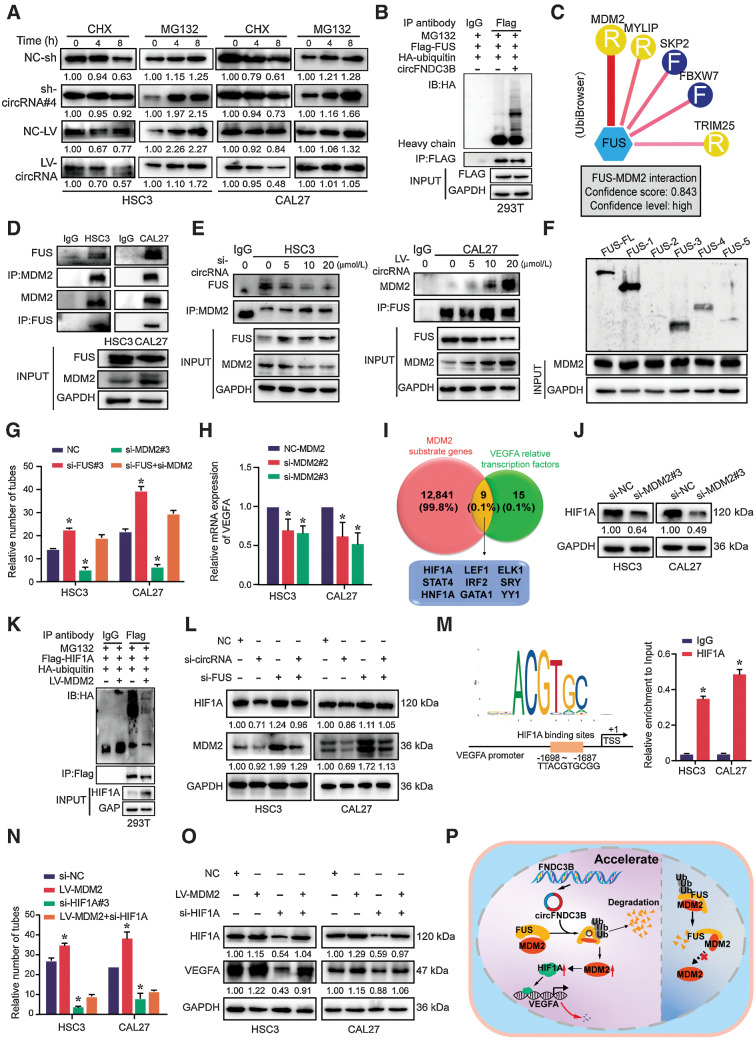
Given the stable FUS mRNA levels following circFNDC3B knockdown (Fig. 4H), cycloheximide and MG132 assays were used to further evaluate FUS stability in the context of circFNDC3B-overexpressing or -silencing (Supplementary Fig. S5A). Strikingly, circFNDC3B overexpression or knockdown, respectively, decreased or increased the half-life of FUS in HSC3 and CAL27 cells (Fig. 5A; Supplementary Fig. S5B). Analyses of 293T cells further indicated that FUS ubiquitylation was enhanced upon circFNDC3B overexpression (Fig. 5B), suggesting that circFNDC3B drove proteasome-dependent FUS degradation. UbiBrowser was used to identify FUS-interacting E3 ubiquitin ligases and showed that murine double minute 2 (MDM2) was predicted to interact most strongly (Fig. 5C), and the interaction of FUS-MDM2 was identified by coimmunoprecipitation (Fig. 5D). With circFNDC3B downregulating, the FUS–MDM2 interaction was disrupted, whereas the interaction was enhanced with circFNDC3B upregulation (Fig. 5E). Then, we performed 293T cells expressing a series of FUS deletion mutants and found the 1–269 and ZNF domains of FUS specifically coimmunoprecipitated with MDM2 (Fig. 5F). Furthermore, we also showed that FUS-silencing promoted MDM2 upregulation (Supplementary Fig. S5C). Together, these results suggest that circFNDC3B facilitates FUS–MDM2 interaction, thus promoting MDM2-induced FUS ubiquitylation and degradation.
Figure 5.
FUS suppresses the transcription of VEGFA via modulating MDM2–HIF1A interactions. A, After treatment with cycloheximide (CHX; 10 μg/mL) and MG132(10 μg/mL) for indicated times, protein levels of FUS were determined by Western blot analyses of OSCC cells transfected with sh-/LV-circFNDC3B. B, Western blot analysis of ubiquitin immunoprecipitated with anti-HA tag antibodies in 293T cells. C, The UbiBrowser tool identified the E3 ligase that interacts with FUS. D, IP analysis revealed the FUS/MDM2 interaction in HSC3 and CAL27 cells. E, The FUS–MDM2 interaction was decreased in circFNDC3B downregulated HSC3 cells, whereas overexpression of circFNDC3B strengthened the interaction in CAL27 cells. F, Immunoprecipitation analysis determined that FUS 1-269 and Znf domains specifically immunoprecipitated with MDM2. G, Quantification of tube formation presented that si-FUS could partially rescue the number of tubes through si-MDM2. Data are shown as mean ± SD of n > 3/group. H, Transcript level of VEGFA was detected after transfecting si-MDM2. I, Venn diagram demonstrating the overlapping of MDM2 substrate genes and VEGFA relative transcription factors. J, The protein expression of HIF1A was detected in OSCC cells transfected with the si-MDM2. K, Overexpression of MDM2 inhibited HIF1A ubiquitination in 293T cells. L, Western blot analysis showed that the si-FUS could functionally rescue the expression of HIF1A and MDM2 upon circFNDC3B silencing. M, The JASPAR database found a binding site within the HIF1A and VEGFA promoter region. Chromatin immunoprecipitation demonstrated that the HIF1A antibody could be effectively precipitated with the region (−1698 to −1687) of the VEGFA promoter. N and O, Quantification of tube formation (N) and Western blot (O) presented that si-HIF1A could partially rescue the number of tubes and VEGFA expression through LV-MDM2. Data are shown as mean ± SD of n > 3/group. P, The pattern diagram of circFNDC3B in the nucleus might mediate FUS/MDM2 ubiquitination and attenuate MDM2/HIF1A/VEGFA axis in OSCC angiogenesis. *, P < 0.05.
Next, we determined whether FUS regulated angiogenesis via MDM2 and found that FUS knockdown partially reversed the decreased tube formation of HUVEC in MDM2-silencing (Fig. 5G; Supplementary Fig. S5D and S5E). Moreover, MDM2 knockdown also reduced the mRNA and protein expression of VEGFA (Fig. 5H; Supplementary Fig. S5F). However, considering E3 ligases could not directly alter transcriptional activity, we speculated that MDM2 might ubiquitylate specific transcription factors to influence VEGFA transcription. Accordingly, UbiBrowser was used to identify MDM2 substrates, and VEGFA-regulating transcription factors were identified through the PROMO and UCSC databases. Of the 9 transcription factors intersecting between the two lists, HIF1A exhibited the highest score (Fig. 5I; Supplementary Fig. S5G). Meanwhile, we showed that HIF1A expression was evidently reduced in OSCC cells with MDM2-silencing (Fig. 5J). Consistently, MDM2 overexpression in 293T cells reduced HIF1A ubiquitylation (Fig. 5K). FUS knockdown reversed the expression change of HIF1A and MDM2 in the context of circFNDC3B-silencing (Fig. 5L). Notably, the JASPAR database revealed the HIF1A binding site in the VEGFA promoter, and our chromatin immunoprecipitation assays further confirmed the precipitation of HIF1A-specific antibodies with this VEGFA promoter (Fig. 5M). In addition, the GEPIA database showed a positive correlation of VEGFA and HIF1A expression in HNSCC patients. Intriguingly, we verified that VEGFA protein levels decreased in OSCC cells with HIF1A-silencing (Supplementary Fig. S5H–S5J), and HIF1A-silencing partially reversed the increased tube formation of HUVEC and VEGFA expression of OSCC cells under MDM2-overexpressing (Fig. 5N and O; Supplementary Fig. S5K). Taken together, these results highlight that FUS suppresses VEGFA expression via regulating interactions between MDM2 and HIF1A (Fig. 5P). Above all, we conclude that circFNDC3B accelerates FUS ubiquitin degradation and then promotes VEGFA expression and angiogenesis.
circFNDC3B functions as a miR-181c-5p sponge to promote p-EMT in OSCC cells
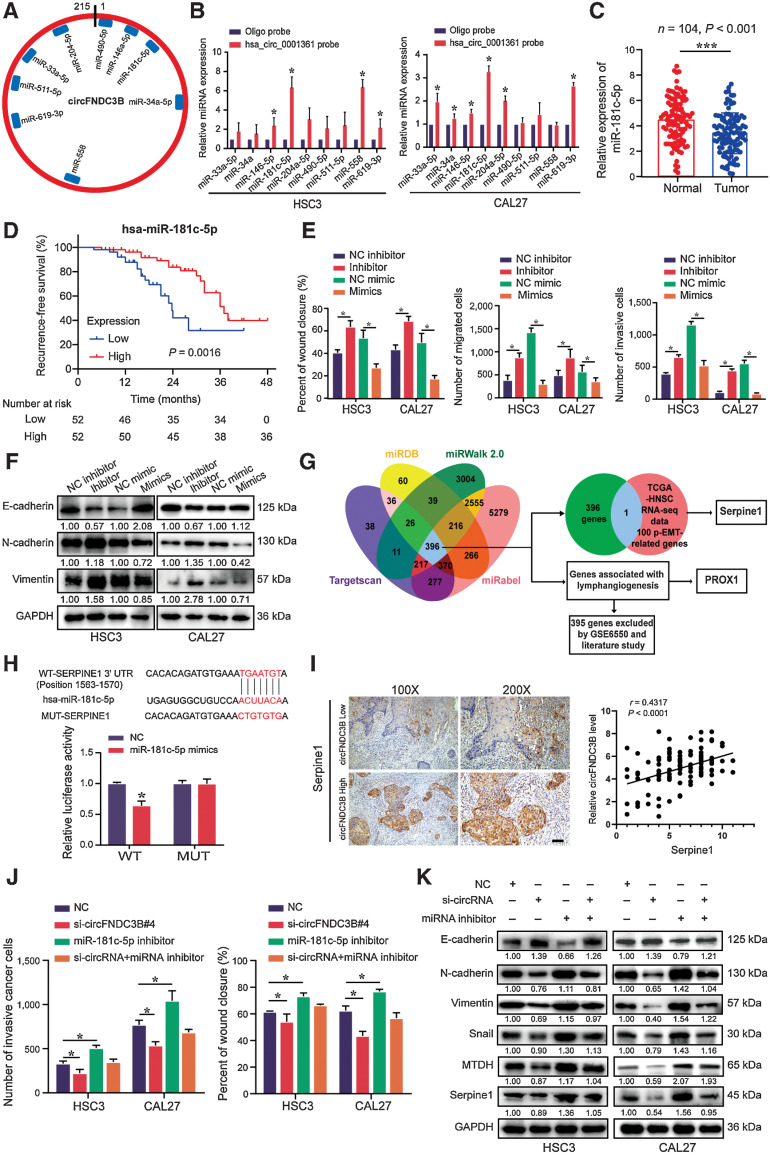
Given circRNAs often act as a molecular “sponge” for miRNAs, we first confirmed that circFNDC3B could interact with AGO2 by RNA pulldown (Supplementary Fig. S6A), and found 9 miRNAs predicted to bind circFNDC3B by CircInteractome and circMIR (Fig. 6A). Of these 9 transcripts, only miR-181c-5p was found to be enriched by circFNDC3B in pulldown assays (Fig. 6B). Therefore, we further verified that miR-181c-5p was significantly downregulated in tissues and cells (Fig. 6C; Supplementary Fig. S6B). Importantly, we confirmed that lower miR-181c-5p was associated with worse RFS in our cohort (Fig. 6D). Moreover, in vitro data showed that miR-181c-5p overexpression suppressed the migration, invasion, and EMT markers expression of CAL27 and HSC3, whereas miR-181c-5p inhibition showed the opposite outcome (Fig. 6E and F; Supplementary Fig. S6C and S6D). These findings indicate that circFNDC3B functions as a miR-181c-5p sponge, and miR-181c-5p inhibits the migration, invasion, and EMT of OSCC.
Figure 6.
circFNDC3B promotes OSCC metastasis through the miR-181c-5p/Serpine1 pathway. A, The potential target miRNAs of circFNDC3B were predicted using CircInteractome and circMIR. B, RNA pulldown assays revealed that miRNAs directly interact with circFNDC3B in HSC3 and CAL27 cells. C, Expression of miR-181c-5p was detected by qRT-PCR in 104 paired OSCC tissues and normal tissues. D, The survival analysis revealed that lower miR-181c-5p levels were significantly associated with worse RFS. E, Statistics of wound healing, transwell migration, and invasion assays were performed to identify the cell motility upon miR-181c-5p inhibitor or mimics in HSC3 and CAL27 cells. F, Western blot assays detected the expression of EMT markers in HSC3 and CAL27 cells transfected miR-181c-5p inhibitor or mimics. G, Schematic illustration showing potential target genes of miR-181c-5p as predicted by four miRNA databases, TCGA-HNSC RNA sequencing (RNA-seq) data, and GSE6550. H, Schematic illustration showed the alignment of miR-181c-5p with Serpine1, and the red portion indicated the mutagenesis nucleotides. Dual luciferase reporter assays of miR-181c-5p with Serpine1 were performed. I, The correlation between circFNDC3B expression and IHC score of serpine1 was determined. Scale bar, 100 μm. J and K, Quantification of wound healing, transwell invasion assays (J), and Western blot (K) presented that miR-181c-5p inhibitor could partially rescue the cell motility and the expression of EMT markers, MTDH and Serpine1 through si-circFNDC3B, respectively. Data were shown mean ± SD of n > 3/group. *, P < 0.05.
Considering p-EMT as an independent predictor in HNSCC patients (31), p-EMT-related genes by The Cancer Genome Atlas RNA sequencing data were compared with 396 putative miR-181c-5p target genes predicted by miRDB, miRWalk 2.0, Targetscan, and miRabel. We identified Serpine1 as a potential p-EMT-related miR-181c-5p target (Fig. 6G), and confirmed the physiologic function of miR-181c-5p by luciferase reporter assay (Fig. 6H). Consistently, miR-181c-5p inhibitor and mimics, respectively, enhanced and suppressed Serpine1 expression (Supplementary Fig. S6E and S6F). Moreover, through evaluating the relative expression of circFNDC3B and IHC score of Serpine1 in 104 OSCC samples, we showed a positive correlation between circFNDC3B and Serpine1 (Fig. 6I). Meanwhile, we further demonstrated that higher Serpine1 expression was associated with poorer patient prognosis, and a positive correlation of Serpine1 expression in primary tumor and metastatic LNs (Supplementary Fig. S6G and S6H).
To investigate whether miR-181c-5p inhibition impacts the effects of circFNDC3B knockdown, si-circFNDC3B and miR-181c-5p inhibitors were cotransfected in HSC3 and CAL27. The results showed that miR-181c-5p inhibition partially reversed the decreased migration and invasion of OSCC cells in the context of circFNDC3B-silencing (Fig. 6J; Supplementary Fig. S6I and S6J). miR-181c-5p inhibition not only rescued the effect of circFNDC3B knockdown on p-EMT but also reversed corresponding changes of metastasis-associated markers and EMT markers (Fig. 6K). Notably, key transcription factors for EMT, ZEB1, and SNAI2 were also identified as miR-181c-5p targets (Supplementary Fig. S6K and S6L). These data suggest that circFNDC3B promotes the metastatic ability of OSCC cells through sequestrating miR-181c-5p and then releasing key factors for p-EMT and EMT.
circFNDC3B promotes lymphangiogenesis via miR-181c-5p/PROX1
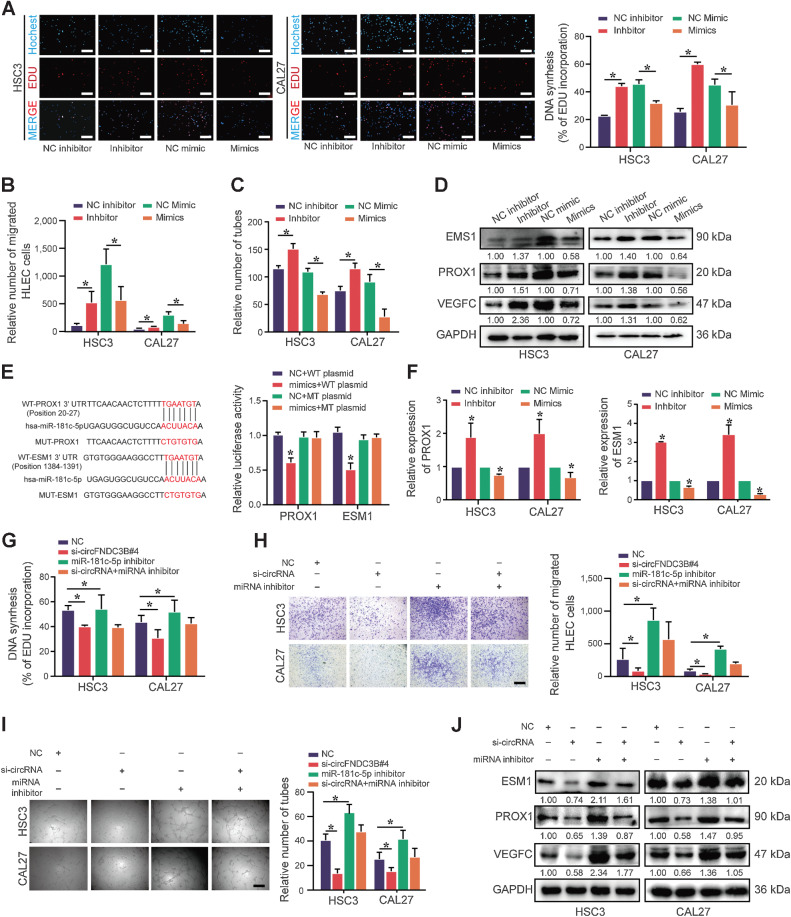
We further investigated whether circFNDC3B promoted lymphangiogenesis via sponging miR-181c-5p. Our findings revealed that the conditioned medium derived from miR-181c-5p inhibitor-transfected OSCC cells enhanced the tube formation, proliferation, and migration of HLEC, whereas miR-181c-5p mimics suppressed this promalignant activity (Fig. 7A–C; Supplementary Fig. S7A and S7B). Moreover, miR-181c-5p knockdown significantly enhanced the expression of VEGFC, which is a characteristic factor of lymphangiogenesis (Fig. 7D).
Figure 7.
circFNDC3B promotes lymphangiogenesis via the miR-181c-5p/PROX1 axis in OSCC. A–C, The conditioned medium from miR-181c-5p inhibitor OSCC cells was dramatically beneficial to the proliferation (A), migration (B), and tube formation (C) of HLECs, whereas the miR-181c-5p mimics group had the opposite results. D, The protein expression of VEGFC, PROX1, and ESM1 in HSC3 and CAL27 cells with miR-181c-5p inhibitor or mimics transfection was detected by Western blot. E, Dual luciferase reporter assays showed the luciferase activity of WT or MUT PROX1 and ESM1 following cotransfection with miR-181c-5p mimics or NC. Relative firefly luciferase expression was normalized to that of Renilla luciferase. F, The mRNA expression of PROX1 and ESM1 in HSC3 and CAL27 cells with miR-181c-5p inhibitor or mimics transfection was detected by qRT-PCR. G–I, EdU (G), transwell migration assays (H), and tube formation (I) presented that the conditioned medium from miR-181c-5p inhibitor could partially rescue proliferation, migration, and tube formation of HLEC cocultured with the conditioned medium from si-circFNDC3B, respectively. J, Western blot analysis of ESM1, PROX1, and VEGFC. Data are presented as the mean ± SEM from three independent experiments. *, P < 0.05. Scale bars, 100 μm (A, H, and I).
PROX1 acts as a master transcriptional regulator of lymphangiogenesis and lymphatic endothelial cell growth, whereas ESM1 mediates the lymphangiogenic process (32, 33). We identified PROX1 and ESM1 as targets of miR-181c-5p associated with lymphangiogenesis through the GSE6550 data set (Fig. 6G) and the Targetscan database (Supplementary Fig. S7C), and confirmed the ability of miR-181c-5p regulating 3′-UTR sequences of PROX1 and ESM1 (Fig. 7E). IHC score of PROX1 in primary tumor and LNs showed a positive correlation, which indicated that PROX1 was beneficial to LN metastasis (Supplementary Fig. S7D). Moreover, miR-181c-5p inhibitor significantly increased the expression of PROX1 and ESM1, whereas miR-181c-5p mimics showed the opposite expression (Fig. 7D and F). Notably, miR-181c-5p inhibitor promoted the proliferation, migration, and tube formation of HLEC, whereas circFNDC3B-silencing reversed these effects (Fig. 7G–I; Supplementary Fig. S7E). Downregulated ESM1, PROX1, and VEGFC in the context of circFNDC3B-silencing were also partially reversed by an miR-181c-5p inhibitor (Fig. 7J). Together, these data suggest that circFNDC3B promotes lymphangiogenesis via the miR-181c-5p/PROX1 axis.
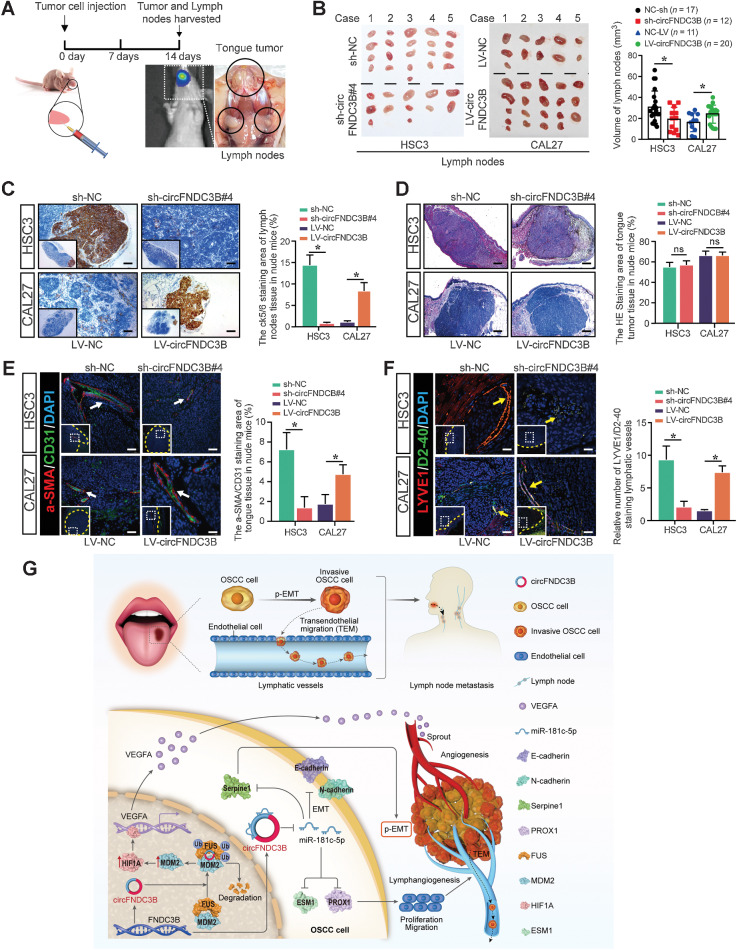
circFNDC3B enhances metastasis of OSCC in vivo
Lastly, 4 groups of nude mice were used to establish a tongue cancer model via injecting 5×106 HSC3-sh-NC/HSC3-sh-circFNDC3B or CAL27-LV-NC/CAL27-LV-circFNDC3B, respectively (Fig. 8A). Fourteen days later, both LNs and tumors were harvested for analysis. Results showed that circFNDC3B knockdown was associated with a lower LN volume, whereas its overexpression increased LN micrometastasis compared with control cell implantation (Fig. 8B and C). However, primary tumor volume was unaffected by circFNDC3B (Fig. 8D; Supplementary Fig. S8A). Subsequent a-SMA/CD31 and LYVE1/D2-40 immunofluorescent staining demonstrated that the markers of lymphangiogenesis and angiogenesis were downregulated in the sh-circFNDC3B group, whereas upregulated expressions were showed in the LV-circFNDC3B group (Fig. 8E and F). In addition, we further showed that the sh-circFNDC3B group exhibited decreased markers expression of EMT and lymphangiogenesis compared with the control group, whereas these markers upregulated in the circFNDC3B overexpression group (Supplementary Fig. S8B).
Figure 8.
circFNDC3B enhances OSCC metastasis and associated neovascularization in vivo. A, Schematic diagram of an animal model to study LN metastasis of OSCC. B, Representative images of cervical LNs. C, IHC staining of LN micrometastases. D, Hematoxylin and eosin staining of tongue tumor. E and F, Immunofluorescence staining of angiogenesis (white arrow; E) and lymphangiogenesis (yellow arrow; F) in the tongue tumor microenvironment. Yellow dashed lines demarcate the boundary of tumor. G, Schematic illustration showing the suggested mechanism by which circFNDC3B functions as an oncogene for OSCC angiogenesis, metastasis, and lymphangiogenesis through the FUS/MDM2/HIF1A/VEGFA axis and contributes as a miR-181c-5p sponge to upregulate Serpine1 and PROX1, respectively. *, P, 0.05. ns, no statistical significance. Scale bars, 100 μm (C and D) and 50 μm (E and F).
To further explore the roles of VEGFA and miR-181c-5p in circFNDC3B-mediated OSCC metastasis, bevacizumab (BEV; specifically binding to VEGFA isoforms with high affinity) and miR-181c-5p antagomir were used in a tongue cancer model (Supplementary Fig. S9A). At day 21, primary tumors and LNs were harvested and measured (Supplementary Fig. S9B and S9C). IHC and IF staining showed that BEV cooperated with sh-circFNDC3B to inhibit angiogenesis in TME and metastatic LN, whereas miR-181c-5p antagomir aggravated the effects of LV-circFNDC3B on promoting tumor-related lymphangiogenesis and LNs metastasis (Supplementary Fig. S9D and S9E). Together, these in vivo results indicate that circFNDC3B enhances metastasis of OSCC via promoting angiogenesis/lymphangiogenesis in metastatic TME.
Discussion
Metastasis is the leading cause of cancer-related mortality, and OSCC patients exhibit poor 5-year survival rates (34). As such, the identification of novel diagnostic and therapeutic targets in OSCC patients is urgently needed. The expression and functional roles of circRNAs in a range of cancer have been studied in recent years (11, 19, 35). However, the underlying mechanism of circRNAs in the metastasis and neovascularization of OSCC remains puzzling. In this study, we found multiple circRNAs derived from the FNDC3B gene, and only hsa_circ_0001361 was overexpressed in OSCC, which might be due to the different biogenesis of these circRNAs. Several studies indicated that circFNDC3B (hsa_circ_0006156) could dramatically inhibit tumor progression including bladder cancer and colorectal cancer (36, 37). Furthermore, Yang and colleagues also found that high circFNDC3B (hsa_circ_0001361) expression in OSCC could repress cell ferroptosis (38). In this study, higher hsa_circ_0001361 (circFNDC3B) expression was associated with LN metastasis and neovascularization in OSCC patients. Meanwhile, to our knowledge, this is the first evidence linking metastatic cancer cells to vasculature formation in the TME through circRNAs and indicates that circFNDC3B may be a potential therapeutic target for OSCC.
Angiogenesis is a critical mechanism that provides nutrients and energy to support tumor cells. In this study, circFNDC3B was found to promote the angiogenic activity of OSCC. Given that multiple functions of circRNAs, including sponging microRNAs, interacting with RBPs, and translating peptides, have been reported in a growing number of studies (39), we further explored the possible role of circFNDC3B in regulating angiogenesis in OSCC. FUS is a multifunctional RBP in various diseases, including inhibition of cardiovascular repair (40). In this report, circFNDC3B was found to bind nuclear FUS and posttranscriptionally reduce FUS protein levels without impacting FUS mRNA expression. Recent studies have identified ADAMTS9-AS2 as a FUS scaffold capable of facilitating its MDM2-mediated ubiquitylation, and FUS protein binding to ncRNA could cause its closed-to-open conformational change to inhibit downstream molecule expression, such as CCND1 (41, 42). As an E3 ubiquitin ligase, MDM2 ubiquitylates a range of target proteins (43). Our findings also supported that circFNDC3B promoted MDM2-medicated FUS ubiquitination and degradation. Given that FUS and circFNDC3B/MDM2 exhibit distinct binding sites, we speculate that circFNDC3B promotes a conformational change in FUS facilitating its ubiquitylation. Subsequently, our data verified that MDM2 stabilized HIF1A and thereby enhanced VEGFA transcription. Overall, these data suggest that nuclear circFNDC3B may promote FUS/MDM2 ubiquitylation and modulate MDM2/HIF1A/VEGFA signaling in the context of OSCC-promoting angiogenesis.
Cancer metastasis often means poor prognosis and lymphangiogenesis plays a critical role in metastatic progression (44, 45). Here, circFNDC3B was found to promote OSCC metastasis by driving lymphangiogenesis. However, no studies to date have focused on the regulation of circRNAs and lymphangiogenesis in oral cancer. Extensive studies have shown that circRNAs located in the cytoplasm could bind to miRNAs, thereby terminating the regulation of miRNAs on their target genes. circFNDC3B is primarily localized in the cytosol and predicted to bind miR-181c-5p as confirmed in pulldown assays. Notably, miR-181c-5p has been identified as a putative tumor suppressor in OSCC, cervical cancer, and breast cancer (46–48). Consistent with previous studies, miR-181c-5p was here found to suppress lymphangiogenesis and metastasis in OSCC.
Our work indicated that circFNDC3B functioned in OSCC as a miR-181c-5p sponge, and PROX1 and Serpine1 were identified as miR-181c-5p target genes associated with two malignant processes. Serpine1 is associated with partial EMT induction, which is linked to greater metastatic risk relative to full EMT induction (31). Importantly, PROX1, which regulates HLEC differentiation, proliferation, and lymphatic vessel elongation by regulating the expression of HLEC-specific proteins such as podoplanin, VEGFR3, and LYVE-1, is considered a master transcriptional switch involved in lymphatic endothelial cell-fate and lymphangiogenesis (49). Meanwhile, we also further identified the lymphangiogenic mediator ESM1 and the EMT markers Snail and ZEB1 as miR-181c-5p targets. Critically, miR-181c-5p inhibitor treatment significantly reversed circFNDC3B knockdown-related changes in target gene expression. Therefore, circFNDC3B regulates these key malignant processes by sequestering miR-181c-5p.
Conclusion
In summary, our study demonstrates the pleiotropic function of circFNDC3B as an oncogene in OSCC, which on the one hand enhances the invasive and metastatic capacity of OSCC cells and on the other hand promotes angiogenesis and lymphangiogenesis in the tumor microenvironment. circFNDC3B regulates the FUS/MDM2/HIF1A/VEGFA axis in the nucleus and miR-181c-5p/Serpine1/PROX1 axis in the cytoplasm, respectively, via interacting with RBPs and sponging microRNAs (Fig. 8G). Our study broadens the knowledge of circRNAs in promoting cancer metastasis and proposes new insights that circFNDC3B can be a potential target for the diagnosis and treatment of OSCC.
Supplementary Material
Supplementary Materials and Methods.
Supplementary Figures and Legends.
Supplementary Tables.
Full original pictures.
Acknowledgments
The authors thank the patients and all investigators who participated in this study. This work was supported by the Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD, 2018-80).
The publication costs of this article were defrayed in part by the payment of publication fees. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Footnotes
Note: Supplementary data for this article are available at Cancer Research Online (http://cancerres.aacrjournals.org/).
Authors' Disclosures
No disclosures were reported.
Authors' Contributions
X. Li: Conceptualization, resources, data curation, software, formal analysis, validation, investigation, methodology, writing–original draft, writing–review and editing. C. Wang: Resources, data curation, investigation, methodology, writing–review and editing. H. Zhang: Investigation, methodology. Y. Li: Resources, investigation, methodology. D. Hou: Resources, funding acquisition, methodology. D. Liu: Data curation, methodology. R. Xu: Supervision, validation, methodology, writing–review and editing. J. Cheng: Formal analysis, visualization. L. Liu: Resources, data curation. Y. Fu: Conceptualization, formal analysis, supervision, methodology. J. Ye: Resources, data curation, formal analysis, supervision, visualization, writing–review and editing. H. Jiang: Conceptualization, formal analysis, supervision, funding acquisition, methodology, project administration, writing–review and editing.
References
- 1. Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2021;71:209–49. [DOI] [PubMed] [Google Scholar]
- 2. Liu J, Jiang X, Zou A, Mai Z, Huang Z, Sun L, et al. circIGHG-induced epithelial-to-mesenchymal transition promotes oral squamous cell carcinoma progression via miR-142–5p/IGF2BP3 signaling. Cancer Res 2021;81:344–55. [DOI] [PubMed] [Google Scholar]
- 3. Miyauchi S, Kim SS, Pang J, Gold KA, Gutkind JS, Califano JA, et al. Immune modulation of head and neck squamous cell carcinoma and the tumor microenvironment by conventional therapeutics. Clin Cancer Res 2019;25:4211–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. SH J, Hysi D. Methods and risk of bias in molecular marker prognosis studies in oral squamous cell carcinoma. Oral Dis 2018;24:115–19. [DOI] [PubMed] [Google Scholar]
- 5. Dieterich LC, Tacconi C, Ducoli L, Detmar M. Lymphatic vessels in cancer. Physiol Rev 2022;102:1837–79. [DOI] [PubMed] [Google Scholar]
- 6. Raj S, Kesari KK, Kumar A, Rathi B, Sharma A, Gupta PK, et al. Molecular mechanism(s) of regulation(s) of c-MET/HGF signaling in head and neck cancer. Mol Cancer 2022;21:31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hu C, Huang Q, Sun Q. The regulation of lymph node pre-metastatic niche formation in head and neck squamous cell carcinoma. Front Oncol 2022;12:852611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Gramolelli S, Elbasani E, Tuohinto K, Nurminen V, Gunther T, Kallinen RE, et al. Oncogenic Herpesvirus engages endothelial transcription factors SOX18 and PROX1 to increase viral genome copies and virus production. Cancer Res 2020;80:3116–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Karinen S, Juurikka K, Hujanen R, Wahbi W, Hadler-Olsen E, Svineng G, et al. Tumour cells express functional lymphatic endothelium-specific hyaluronan receptor in vitro and in vivo: lymphatic mimicry promotes oral oncogenesis? Oncogenesis 2021;10:23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Jardim JF, Galvis MM, Fabelo IR, Soares FA, Pinto CAL, Kowalski LP. Intratumoral lymphatic vascular density is an independent factor for disease-free and overall survival in advanced stage oral squamous cell carcinoma. Oral Surg Oral Med Oral Pathol Oral Radiol 2021;132:580–88. [DOI] [PubMed] [Google Scholar]
- 11. Yang H, Li X, Meng Q, Sun H, Wu S, Hu W, et al. CircPTK2 (hsa_circ_0005273) as a novel therapeutic target for metastatic colorectal cancer. Mol Cancer 2020;19:13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Liu F, Zhang H, Xie F, Tao D, Xiao X, Huang C, et al. Hsa_circ_0001361 promotes bladder cancer invasion and metastasis through miR-491-5p/MMP9 axis. Oncogene 2020;39:1696–709. [DOI] [PubMed] [Google Scholar]
- 13. Li J, Sun D, Pu W, Wang J, Peng Y. Circular RNAs in cancer: biogenesis, function, and clinical significance. Trends Cancer 2020;6:319–36. [DOI] [PubMed] [Google Scholar]
- 14. Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK, et al. Natural RNA circles function as efficient microRNA sponges. Nature 2013;495:384–8. [DOI] [PubMed] [Google Scholar]
- 15. Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 2013;495:333–8. [DOI] [PubMed] [Google Scholar]
- 16. Jeck WR, Sorrentino JA, Wang K, Slevin MK, Burd CE, Liu J, et al. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA 2013;19:141–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Meng S, Zhou H, Feng Z, Xu Z, Tang Y, Li P, et al. CircRNA: functions and properties of a novel potential biomarker for cancer. Mol Cancer 2017;16:94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wang S, Zhang K, Tan S, Xin J, Yuan Q, Xu H, et al. Circular RNAs in body fluids as cancer biomarkers: the new frontier of liquid biopsies. Mol Cancer 2021;20:13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Zhao W, Cui Y, Liu L, Qi X, Liu J, Ma S, et al. Splicing factor derived circular RNA circUHRF1 accelerates oral squamous cell carcinoma tumorigenesis via feedback loop. Cell Death Differ 2020;27:919–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kristensen LS, Andersen MS, Stagsted LVW, Ebbesen KK, Hansen TB, Kjems J. The biogenesis, biology and characterization of circular RNAs. Nat Rev Genet 2019;20:675–91. [DOI] [PubMed] [Google Scholar]
- 21. Du WW, Fang L, Yang W, Wu N, Awan FM, Yang Z, et al. Induction of tumor apoptosis through a circular RNA enhancing Foxo3 activity. Cell Death Differ 2017;24:357–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Shi Y, Jia X, Xu J. The new function of circRNA: translation. Clin Transl Oncol 2020;22:2162–69. [DOI] [PubMed] [Google Scholar]
- 23. Zhang M, Zhao K, Xu X, Yang Y, Yan S, Wei P, et al. A peptide encoded by circular form of LINC-PINT suppresses oncogenic transcriptional elongation in glioblastoma. Nat Commun 2018;9:4475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Nishizuka M, Kishimoto K, Kato A, Ikawa M, Okabe M, Sato R, et al. Disruption of the novel gene fad104 causes rapid postnatal death and attenuation of cell proliferation, adhesion, spreading and migration. Exp Cell Res 2009;315:809–19. [DOI] [PubMed] [Google Scholar]
- 25. Fucci C, Resnati M, Riva E, Perini T, Ruggieri E, Orfanelli U, et al. The interaction of the tumor suppressor FAM46C with p62 and FNDC3 proteins integrates protein and secretory homeostasis. Cell Rep 2020;32:108162. [DOI] [PubMed] [Google Scholar]
- 26. Han B, Wang H, Zhang J, Tian J. FNDC3B is associated with ER stress and poor prognosis in cervical cancer. Oncol Lett 2020;19:406–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Li YQ, Chen Y, Xu YF, He QM, Yang XJ, Li YQ, et al. FNDC3B 3'-UTR shortening escapes from microRNA-mediated gene repression and promotes nasopharyngeal carcinoma progression. Cancer Sci 2020;111:1991–2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Zhong Z, Zhang H, Hong M, Sun C, Xu Y, Chen X, et al. FNDC3B promotes epithelial-mesenchymal transition in tongue squamous cell carcinoma cells in a hypoxic microenvironment. Oncol Rep 2018;39:1853–59. [DOI] [PubMed] [Google Scholar]
- 29. Kastana P, Zahra FT, Ntenekou D, Katraki-Pavlou S, Beis D, Lionakis MS, et al. Matrigel plug assay for in vivo evaluation of angiogenesis. Methods Mol Biol 2019;1952:219–32. [DOI] [PubMed] [Google Scholar]
- 30. Reymond N, d'Agua BB, Ridley AJ. Crossing the endothelial barrier during metastasis. Nat Rev Cancer 2013;13:858–70. [DOI] [PubMed] [Google Scholar]
- 31. Kisoda S, Shao W, Fujiwara N, Mouri Y, Tsunematsu T, Jin S, et al. Prognostic value of partial EMT-related genes in head and neck squamous cell carcinoma by a bioinformatic analysis. Oral Dis 2020. doi: 10.1111/odi.13351. [DOI] [PubMed] [Google Scholar]
- 32. Bui K, Hong YK. Ras pathways on prox1 and lymphangiogenesis: insights for therapeutics. Front Cardiovasc Med 2020;7:597374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Shin JW, Huggenberger R, Detmar M. Transcriptional profiling of VEGF-A and VEGF-C target genes in lymphatic endothelium reveals endothelial-specific molecule-1 as a novel mediator of lymphangiogenesis. Blood 2008;112:2318–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kanojia D, Sawant SS, Borges AM, Ingle AD, Vaidya MM. Alterations in keratins and associated proteins during 4-Nitroquinoline-1-oxide induced rat oral carcinogenesis. J Carcinog 2012;11:14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Huang G, Liang M, Liu H, Huang J, Li P, Wang C, et al. CircRNA hsa_circRNA_104348 promotes hepatocellular carcinoma progression through modulating miR-187–3p/RTKN2 axis and activating Wnt/β-catenin pathway. Cell Death Dis 2020;11:1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Zeng W, Zhu JF, Guo J, Huang GJ, Ai LS, Zeng Y, et al. m(6)A-modified circFNDC3B inhibits colorectal cancer stemness and metastasis via RNF41-dependent ASB6 degradation. Cell Death Dis 2022;13:1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Liu H, Bi J, Dong W, Yang M, Shi J, Jiang N, et al. Invasion-related circular RNA circFNDC3B inhibits bladder cancer progression through the miR-1178–3p/G3BP2/SRC/FAK axis. Mol Cancer 2018;17:161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Yang J, Cao XH, Luan KF, Huang YD. Circular RNA FNDC3B protects oral squamous cell carcinoma cells from ferroptosis and contributes to the malignant progression by regulating miR-520d-5p/SLC7A11 axis. Front Oncol 2021;11:672724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Huang A, Zheng H, Wu Z, Chen M, Huang Y. Circular RNA-protein interactions: functions, mechanisms, and identification. Theranostics 2020;10:3503–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Garikipati VNS, Verma SK, Cheng Z, Liang D, Truongcao MM, Cimini M, et al. Circular RNA CircFndc3b modulates cardiac repair after myocardial infarction via FUS/VEGF-A axis. Nat Commun 2019;10:4317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Yan Y, Xu Z, Chen X, Wang X, Zeng S, Zhao Z, et al. Novel function of lncRNA ADAMTS9-AS2 in promoting temozolomide resistance in glioblastoma via upregulating the FUS/MDM2 ubiquitination axis. Front Cell Dev Biol 2019;7:217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Hamad N, Mashima T, Yamaoki Y, Kondo K, Yoneda R, Oyoshi T, et al. RNA sequence and length contribute to RNA-induced conformational change of TLS/FUS. Sci Rep 2020;10:2629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Zhang X, Min X, Wang S, Sun N, Kim KM. Mdm2-mediated ubiquitination of β-arrestin2 in the nucleus occurs in a Gβγ- and clathrin-dependent manner. Biochem Pharmacol 2020;178:114049. [DOI] [PubMed] [Google Scholar]
- 44. Patel SG, Amit M, Yen TC, Liao CT, Chaturvedi P, Agarwal JP, et al. Lymph node density in oral cavity cancer: results of the international consortium for outcomes research. Br J Cancer 2013;109:2087–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Zheng W, Aspelund A, Alitalo K. Lymphangiogenic factors, mechanisms, and applications. J Clin Invest 2014;124:878–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Xie D, Li S, Wu T, Wang X, Fang L. MiR-181c suppresses triple-negative breast cancer tumorigenesis by targeting MAP4K4. Pathol Res Pract 2022;230:153763. [DOI] [PubMed] [Google Scholar]
- 47. Li N, Cheng C, Wang T. MiR-181c-5p mitigates tumorigenesis in cervical squamous cell carcinoma via targeting glycogen synthase kinase 3β interaction protein (GSKIP). Onco Targets Ther 2020;13:4495–505. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 48. Shigeishi H, Yokoyama S, Murodumi H, Sakuma M, Fukada S, Okuda S, et al. Melatonin enhances cisplatin-induced cell death through inhibition of DERL1 in mesenchymal-like CD44(high) OSCC cells. J Oral Pathol Med 2022;51:281–89. [DOI] [PubMed] [Google Scholar]
- 49. Chen JM, Luo B, Ma R, Luo XX, Chen YS, Li Y. Lymphatic endothelial markers and tumor lymphangiogenesis assessment in human breast cancer. Diagnostics 2021;12:4. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Materials and Methods.
Supplementary Figures and Legends.
Supplementary Tables.
Full original pictures.
Data Availability Statement
The accession numbers of data for analyzing differentially expressed circRNAs reported in this paper are GEO: GSE131182 and GSE197138. The data generated in this study are also available upon request from the corresponding author.