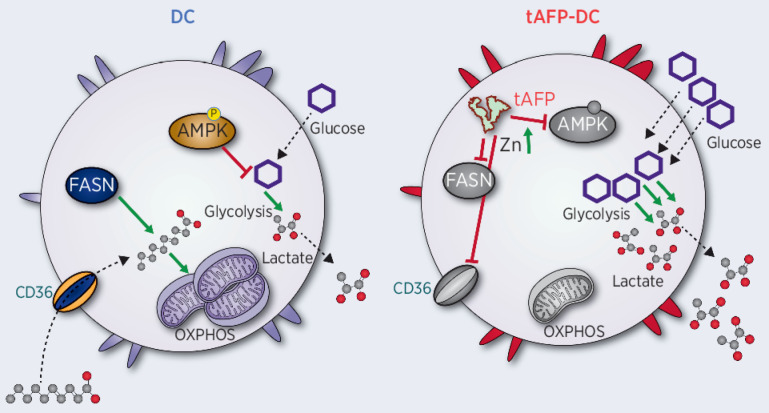
α-Fetoprotein (AFP) is a secreted tumor protein and biomarker with impact on immunity. Fatty acid–bound AFP promotes immune suppression by skewing human dendritic cell metabolism toward glycolysis and reduced immune stimulation.
Abstract
α-Fetoprotein (AFP) is expressed by stem-like and poor outcome hepatocellular cancer tumors and is a clinical tumor biomarker. AFP has been demonstrated to inhibit dendritic cell (DC) differentiation and maturation and to block oxidative phosphorylation. To identify the critical metabolic pathways leading to human DC functional suppression, here, we used two recently described single-cell profiling methods, scMEP (single-cell metabolic profiling) and SCENITH (single-cell energetic metabolism by profiling translation inhibition). Glycolytic capacity and glucose dependence of DCs were significantly increased by tumor-derived, but not normal cord blood–derived, AFP, leading to increased glucose uptake and lactate secretion. Key molecules in the electron transport chain in particular were regulated by tumor-derived AFP. These metabolic changes occurred at mRNA and protein levels, with negative impact on DC stimulatory capacity. Tumor-derived AFP bound significantly more polyunsaturated fatty acids (PUFA) than cord blood–derived AFP. PUFAs bound to AFP increased metabolic skewing and promoted DC functional suppression. PUFAs inhibited DC differentiation in vitro, and ω-6 PUFAs conferred potent immunoregulation when bound to tumor-derived AFP. Together, these findings provide mechanistic insights into how AFP antagonizes the innate immune response to limit antitumor immunity.
Significance:
α-Fetoprotein (AFP) is a secreted tumor protein and biomarker with impact on immunity. Fatty acid–bound AFP promotes immune suppression by skewing human dendritic cell metabolism toward glycolysis and reduced immune stimulation.
Graphical Abstract
Introduction
Liver cancer accounts for 8.3% of cancer-related deaths worldwide, making it the third leading cause of cancer-related mortality (1). Hepatocellular carcinoma (HCC) represents 70% to 85% of primary liver cancers (2). Important drivers of HCC rates include chronic hepatitis B (HBV) and C (HCV) infections and control of these infections has decreased HCC rates in East Asia and Southern Europe (3). Unfortunately, downward trends in HBV and HCV infections are offset by increases in other HCC-risk factors, including alcohol consumption, smoking, and obesity. Obesity can lead to fatty infiltration into the liver causing nonalcoholic fatty liver disease (NAFLD), leading to nonalcoholic steatohepatitis (NASH; ref. 4). In the United States, more than 1 in 3 people have some form of NAFLD, and 6 million people have NASH (4). Given HCC's lethality, coupled with the concerning rise in HCC risk factors, new therapies are urgently needed.
Treatments for patients with early stages of HCC include surgery, ablative therapies, embolization approaches, or liver transplantation (5) can be effective. For the majority of patients with more advanced stages of disease, systemic therapy options have expanded in recent years to include small-molecule multikinase inhibitors, monoclonal antibodies targeting VEGF or its receptors, and most recently, immune checkpoint inhibition (6–12)
The combination of bevacizumab and atezolizumab, targeting VEGF and PD-L1, respectively, has emerged as a new global standard for first-line therapy based upon substantial improvement in outcomes compared with the multikinase inhibitor, sorafenib, with median overall survival (OS) of 19.2 months for the combination versus 13.4 months for sorafenib (HR, 0.66; P = 0.0009; refs. 9, 13) Objective responses occurred in 30% of patients treated with the combination, including 8% with complete responses, with median duration of response not reached. Other immunotherapy combinations have also shown striking improvements in rates of objective radiographic response compared with historical controls (10, 14, 15), and the combination of the PD-L1 inhibitor, durvalumab, with the CTLA4 inhibitor, tremelimumab, improved OS compared with sorafenib in a randomized, phase III trial (8, 16). These studies demonstrate the potential for robust and durable immune responses in a subset of patients with HCC and underscore the urgent necessity to identify and address mechanisms of resistance in the majority of patients who do not achieve prolonged responses. Although immunotherapies blocking exhaustion markers (PD-1, PD-L1, and CTLA4) and/or VEGF are encouraging, there are additional barriers in vivo that limit the potency of antitumor immunity.
α-Fetoprotein (AFP) is an oncofetal glycoprotein, similar to albumin, which is expressed by the majority of HCC tumors (tAFP) and can be detected in serum as well as the tumor microenvironment. Elevated serum concentration of AFP is associated with poor prognosis across stages of HCC, and tumors with high AFP expression may represent a distinct biologic subtype associated with activation of proliferative pathways and VEGF signaling (17–19). Like albumin, tAFP is a secreted protein that can bind multiple metabolites and enter activated lymphocytes, hepatocytes, natural killer (NK) cells, and monocytes. Because its initial discovery in a patient with HCC in the 1960s (20), interest in tAFP has focused on its prognostic (21) and diagnostic potential in HCC (22), as a cancer vaccine antigen target (23–25), and its immunoregulatory properties on NK cells (26), macrophages (27, 28), monocytes, and dendritic cells (DC; refs. 29, 30). AFP is also being targeted in TCR-engineered adoptive cell transfer studies (31, 32). Our group demonstrated that tAFP has more potent immunoregulatory properties than cord blood-derived “normal” AFP (nAFP; ref. 29). The molecular features of AFP that are immunoregulatory have been attributed to differences in glycosylation patterns (33, 34), isoforms (35, 36) or isoelectric points (37), and the presence of specific ligands (29, 38–40). In addition, our group has determined that tAFP-mediated suppression of DCs’ function depends on a low molecular mass (LMM; ref. 29) molecule that is neither protein nor glycan.
Here, using novel single-cell methods and lipid profiling in both in vitro models and in vivo human HCC patient blood samples, we have determined that tAFP uptake by DC causes reduced fatty acid uptake and metabolism and a switch to glycolysis accompanied by increased glucose uptake and lactate secretion. This metabolic skewing is accompanied by a shift in immune phenotype, with reduced costimulatory molecule expression and increased DC CD14 and PD-L1 expression. For the first time, we identify differences in the ligand composition between nAFP and tAFP and show that these fatty acids are essential for the immunoregulatory features of tAFP. These findings have important implications for understanding how AFP+ HCC limits innate immune responses, identifying strategies to improve DC function in vivo, and development of more potent DC vaccines.
Materials and Methods
Patient samples
HCC patient blood (with written informed consent; Table 1) and healthy donor (HD) blood [purchased (Trima Residuals RE202, Vitalant)] was collected in BD Vacutainer heparin tubes (Cat. #02–689–6), and in some cases, BD Vacutainer serum tubes (Cat. #B-D367820Z) were collected. Heparinized blood was centrifuged to separate the blood and plasma components. Plasma was stored at −80°C. The remaining cellular fraction was overlaid over Ficoll (Cytiva, Cat. #45–001–749) in Leucosep tubes (Greiner, Cat. #07–000–983) and centrifuged to isolate peripheral blood mononuclear cells (PBMC). PBMCs were washed with PBS, and viable cells were quantified via trypan blue (Gibco, Cat. #15–250–061) on a Nexcelom Cellometer Spectrum. If cell pellets had substantial red blood cells, they were briefly lysed using ACK lysing buffer (Thermo Fisher Scientific, Cat. #A1049201). Cells were resuspended in freezing media (80% CellGenix + 20% DMSO (MP Biomedicals, Cat. #ICN19141880), stored at −80°C overnight, and stored in gas-phase LN2.
Table 1.
Characteristics of patients with HCC.
| Characteristic (N = 8 patients) | Number or median (range) | (%) |
|---|---|---|
| Gender (n) | ||
| Male | 6 | 75% |
| Female | 2 | 25% |
| Age, y | ||
| Median (range) | 72 (61–83) | |
| Race | ||
| African-American | 2 | 25% |
| Asian | 2 | 25% |
| Caucasian | 4 | 50% |
| Ethnicity | ||
| Non-Hispanic/Latino | 8 | 100% |
| Hispanic/Latino | 0 | 0% |
| Liver Disease Etiology | ||
| Hepatitis C (cAb+) | 4 | 50% |
| Hepatitis B (sAg+) | 2 | 25% |
| Child Pugh Score at Enrollment | ||
| Child Pugh A | 7 | 87% |
| Child Pugh B | 1 | 13% |
| Serum AFP (μg/L) | ||
| Median (range) | 229 (<2.0–7287.9) | |
| Disease stage | ||
| Stage IIIB | 1 | 13% |
| Stage IVA | 3 | 38% |
| Stage IVB | 4 | 50% |
| Histologic grade | ||
| Moderately differentiated | 4 | 50% |
| Poorly differentiated | 2 | 25% |
| Unknown | 2 | 25% |
In vitro DC differentiation
DCs were differentiated in vitro similarly as previously described (29). In brief, cryopreserved PBMCs were thawed and CD14+ monocytes were magnetically labeled using CD14 MicroBeads (Miltenyi, Cat. #130–050–201) and isolated by LS columns (Miltenyi, Cat. #130–042–401) per the manufacturer's instructions. Viable eluted cells were enumerated using trypan blue on a Nexcelom Cellometer Spectrum. To generate immature DCs (iDC), monocytes were stimulated for 5 days in the presence of 800 IU/mL of rGM-CSF (Miltenyi, Cat. #130–093–862) and 500 IU/mL of rIL-4 (Miltenyi, Cat. #130–095–373) as well as OVA, nAFP, or tAFP in CellGenix GMP DC media (Cat. #20801–0500) 37°C at 5% CO2. Highly purified grade tAFP was obtained from Bio-Rad, and the AFP-L3 is approximately 70% as compared with 10% in human cord serum, by PAGE analysis. The nAFP was obtained from Cell Sciences (Cat. #CSI0379), with a purity of >99% by SDS-PAGE analysis and sterile filtered. The chicken ovalbumin was obtained by Sigma (Cat. #A5503–1G) with a purity of >98% by agarose gel electrophoresis. All proteins were aliquoted to prevent multiple freeze-thaw cycles and stored at −80C. Finally, an additional 24-hour stimulation with 1,000 IU/mL of rIFNγ (Peprotech, Cat. #300–02) and 250 ng/mL of LPS (Sigma-Aldrich, Cat. #L2630–10MG) to produce monocytic DCs (mDC). To harvest cells, DCs were detached using TrypLE Select (Gibco, Cat. #12563011) for 15 minutes at 37°C and then washed several times with cold PBS.
SCENITH
SCENITH was performed as described in ref. 41. The SCENITH reagents kit (inhibitors, puromycin and antibodies) was obtained from www.scenith.com/try-it and used according to the provided protocol for in vitro–derived myeloid cells. Briefly, control and tol-moDC cultures at desired timepoints, were treated for 18 minutes with Control (DMSO), 2-deoxy-glucose (2DG; 100 mmol/L), oligomycin (O; 1 μmol/L), a combination of 2DG and oligomycin (DGO) or Harringtonine (H; 2 μg/mL). Following metabolic inhibitors, Puromycin (final concentration 10 μg/mL) was added to cultures for 17 minutes. After puromycin treatment, cells were detached from wells using TrypLE Select (Thermo Fisher Scientific, 505914419), washed in cold PBS and stained with a combination of Human TruStain FcX (BioLegend, 422301) and fluorescent cell viability dye (BioLegend, 423105) for 10 minutes 4°C in PBS. Following PBS wash step, primary antibodies against surface markers were incubated for 25 minutes at 4°C in Brilliant Stain Buffer (BD Biosciences, 563794). Next, cells were fixed and permeabilized using True-Nuclear Transcription Factor Buffer Set (BioLegend, 424401) as per the manufacturer's instructions. Intracellular staining of puromycin and protein targets was performed for 1 hour in diluted (10x) permeabilization buffer at 4°C. Finally, data acquisition was performed using the Cytek Aurora flow cytometer. Primary conjugated antibody information used in SCENITH panel is listed in Supplementary Table S1. All antibodies were titrated to reduce spillover and increase resolution using single-stained moDC (generated as described above) samples. Unstained cell controls used for autofluorescence extraction were generated for each time point, culture conditions (OVA, nAFP, and tAFP) and metabolic inhibitor treatments (C, 2DG, O, DGO). Samples were unmixed using reference controls generated in combination with stained Ultracomp beads (Thermo Fisher Scientific, 01–2222–41) and stained cells using the SpectroFlo Software v2.2.0.1. The unmixed FCS files were used for data processing and analysis using FlowJo (BD Biosciences, version 10.7.1). Manually gated CD14−HLA-DR+CD86+ cells were used for downstream analysis. gMFI expression values were imported into R environment for correlation and heatmap analysis.
Glucose and lactate measurements
Glucose and lactate were measured by applying approximately 5 μL of supernatant to Clarity BG1000 Blood Glucose strips (Cat. #75840–798) and meter (Cat. #75840–800) system or the Lactate Plus strips (Nova Biomedical, Cat. #40813) and meter version 2 (Nova Biomedical, Cat. #62624) system. Each meter was quality checked with control glucose and lactate solutions and CellGenix media before each experiment.
CyTOF phenotypic profiling
scMEP (single-cell metabolic profiling) analysis was performed as recently described in ref. 42. In short, antibodies targeting metabolic features were conjugated in-house using an optimized conjugation protocol (Hartmann and colleagues, ref. 42) and validated on multiple sample types. Cells were prepared for scMEP analysis by incubation with small molecules to be able to assess biosynthesis rates of DNA, RNA and protein, cisplatin-based live/dead staining, PFA-based cell fixation and cryopreservation (dx.doi.org/10.17504/protocols.io.bkwkkxcw). Next, cells were stained with metabolic antibodies in a procedure that includes surface staining for 30 minutes at room temperature, PFA-fixation for 10 minutes at room temperature, MeOH-based permeabilization for 10 minutes on ice, intracellular staining for 1 hour at room temperature, and DNA intercalation (dx.doi.org/10.17504/protocols.io.bntnmeme). Finally, cells were acquired on a CyTOF2 mass cytometer (Fluidigm). Protein targets and antibody information used in scMEP are listed in Supplementary Table S2.Raw mass spectrometry data were preprocessed, de-barcoded and imported into R environment using the flowCore package (version 2.0.1; ref. 43). Values were arcsinh transformed (cofactor 5) and normalized (42) for downstream analyses based on previously reported workflow (44).
Microarray and gProfiler
OVA, nAFP, and tAFP-treated DC were lysed, and total mRNA was obtained for microarray (Affymetrix HG-U133A). DE genes were uploaded in g:Profiler in R Studio for pathway analysis and visualization (45).
Zinc measurement
Intracellular zinc (Zn) was quantified by flow cytometry using the Zinc Assay Kit (Cell-based; Abcam, Cat. #ab241014). Monocytes were differentiated to iDCs as described above in the presence of OVA, nAFP, tAFP, or ZnSO4. Zn staining was performed per the manufacturer's suggested protocol with positive (Zn) and negative (Zn + chelator) controls as well as a Zn FMO included in each experiment. Cells were stained with LD Aqua for 10 minutes at room temperature. Cells were washed in 1X Assay Buffer, then stained in 100 μL of Assay Buffer + 0.2 μL of Zn Probe for 30 minutes at 37°C. Cells were then washed twice with 1X Assay buffer then stained with HLA-DR-APC-H7 (BD, Clone: GF6–6, Cat. #561358, Lot #0023290, 0339025), CD86-BV785 (BioLegend, Clone: IT2.2, Cat. #305441, Lot #B277560), CD206 PE-Cy7 (BioLegend, Clone: 15–2, Cat. #321123, Lot #B331254), and CD14-BUV805 (BD Biosciences, Clone: M5E2, Cat. #612903, Lot #0297714), in Brilliant Stain Buffer (BD Horizon, Cat. #566349, Lot #0121427) for 20 minutes at 4°C. Cells were washed twice in FACS Buffer and fixed in 1% paraformaldehyde (Thermo Fisher Scientific, Cat. #J19943-K2, Lot #195273, diluted in PBS) for at least 30 minutes before acquisition on a BD LSRFortessa X-50. As a negative control, we briefly treated cells with Zn but did not stain for Zn as a fluorescence-minus-one (FMO) control [mean fluorescence intensity (MFI) = 421] or stained with a Zn probe as a positive control (MFI = 25,850). Zn-treated cells were treated with a Zn chelator included in the kit before staining, and this resulted in a marked approximately 97% reduction in Zn MFI compared with the positive control.
Lipid analysis by mass spectrometry or gas chromatography
Commercially available OVA (N = 3; Sigma-Aldrich, Cat. #A5503–1G, Lot #SLCB8249), nAFP (N = 3; Cell Sciences, Cat. #CSI10379, Lot #4111714), and tAFP (N = 3; Bio-Rad, Cat. #13752600, Lot #64110896) were submitted diluted in PBS (Gibco, Cat. #20–012–050) at 1,000 μg/mL on dry ice. CellGenix GMP DC Medium (N = 1; CellGenix, Cat. #20801–0500) media and supernatants of mDCs from an HD (N = 1) differentiated in the presence of 5 μg per mL of OVA, nAFP, or tAFP were tested. Lipid analysis (Supplementary Table S1) was performed at the UCSD Lipidomics Core (46).
Fatty acid screen
Fatty acids (Supplementary Table S3) were acquired from Cayman Chemical, including 16:0 (palmitic acid, Item # 10006627, Batch #0523612–48), 18:1 (oleic acid, Item #90260, Batch #0540276–62), 20:3 N6 (dihomo-γ-linolenic acid, Item #90230, Batch #0532009–37), 20:3 N9 (5,8,11-eicosatrienoic acid, Item #90190, Batch #0564724–7), 20:4 (arachidonic acid, Item #90010, Batch #0570304–50), 22:4 (adrenic acid, Item #90300, Batch #0537603–20), 20:5 (eicosapentaenoic acid, Item #26415, Batch #0583627), 22:5 N3 (docosapentaenoic acid, Item #90165, Batch #0569492–11), 22:5 N6 (docosapentaenoic acid, Item #10008335, Batch #0462864–36), and 22:6 (docosahexaenoic acid, Item #90310, Batch #0593448–15).
Fatty acids were resuspended in ethanol and stored at −20°C at 100 μmol/L. High molar mass (HMM) fractions of OVA, nAFP, and tAFP were obtained by removing the LMM contents with the Amicon Ultra – 0.5 mL Centrifugal Filters Ultracel—3K (Millipore, Cat. #UFC500324, Lot #R9HA51100) per the manufacturer's suggested protocol and stored at −80°C. Both the native preparations and HMM fractions contained similar amounts of protein (∼0.5 mg/mL), whereas protein was undetectable (<0 mg/mL) in the LMM fraction. In addition, we determined the A260/A280 ratio as a measure of purity and found the LMM fraction had an approximately 3-fold increase in the A260/A280 ratio, indicating a large proportion of non-protein compounds in the LMM fraction, as expected. Fats (± HMM) were added to pre-warmed media and incubated for 1 hour, mixing at 37°C before adding to cells (47). Fats were combined with HMM at a 3:1 molar ratio, as previously described (47).
Statistical analysis and visualization
Statistical comparisons between groups were performed using paired-sample t tests unless otherwise stated using R (version 4.0.2) and R Studio (version 1.3.1093) or Prism (version 9.0.2). P values are represented as *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001; ****, P ≤ 0.0001. P values of <0.05 were considered statistically significant. Numerical labels indicate near significant values). Figure graphs were generated using the R package ggplot2 (version 3.3.3) or in Prism.
Study approval
Blood collection from patients with HCC was approved by the UCSF Hepatobiliary Tissue Bank and Registry Oversight Committee (CC#124512). The UCSF Cancer Immunotherapeutics Tissue Use Committee approved samples from HDs at UCSF (CC#16983).
Data availability statement
The data generated in this study as well as data from prior publications are available upon request from the corresponding author. The array data discussed in this publication were previously deposited in NCBI's Gene Expression Omnibus and are accessible through GEO Series accession number GSE62005 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE62005).
Results
tAFP induces immunometabolic dysregulation of DCs
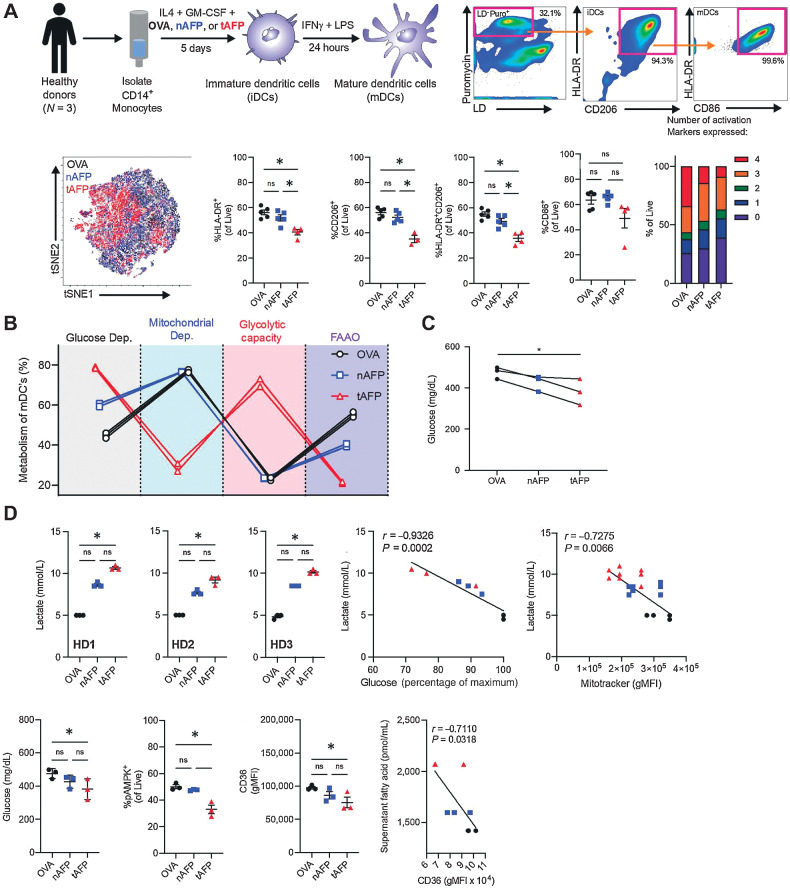
To determine the mechanism of immune suppression induced by AFP, we performed immune and metabolic profiling human DC. Previously, we demonstrated using population-based assays that tAFP decreases the differentiation mDCs and reduces their T-cell stimulatory potential (29). We demonstrated that tAFP limited DC HLA-DR and CD206 expression with a trend for reduced CD86. Furthermore, the Boolean analysis revealed a decreased coexpression of multiple activation markers (HLA-DR, CD206, CD86, and ICOSLG/CD275) among tAFP-treated DC.
To understand the immunometabolic impact of tAFP on DCs at the single-cell level, we used the recently described single-cell energetic metabolism by profiling translation inhibition (SCENITH) assay (41). Ovalbumin (OVA; negative control), nAFP, and tAFP-treated DCs were generated in vitro (Fig. 1A). Viable cells (LD−) actively translating RNA into protein (Puro+) were analyzed (Supplementary Fig. S1) that expressed cell surface molecules associated with mDCs (HLA-DR+, CD206+, CD86+; previously shown to be representative of many common DC phenotypic markers; ref. 30). To assess the broad immunometabolic state of the cells, a tSNE analysis was performed on all parameters that indicated tAFP-treated cells tended to cluster separately from nAFP or OVA-treated mDCs. The calculated metabolic profiles are shown for glucose dependency, mitochondrial dependency, glycolytic capacity, and fatty acid and glutaminolysis (FAAO; Fig. 1B). Even among mDC with strong expression of activation markers (HLA-DR+, CD206+, and CD86+), there was a dramatic increase in glycolysis and a reduction in mitochondrial dependency and FAAO in tAFP-treated DCs. Consistent with a greater reliance on glycolysis, tAFP-treated cells had significantly less glucose in culture supernatants at day 6 (Fig. 1C). OVA and nAFP-treated DCs had relatively higher frequencies of pAMPK+ DCs, which is consistent with their increased mitochondrial dependency and mitochondrial mass as opposed to tAFP-treated DCs. This result is consistent with our previous study, in which increased pAMPK signaling as opposed to mTOR activation upregulated mitochondrial metabolism and FAAO in DCs (48). In conjunction with a decrease in FAAO, a decline in expression of the fatty acid transporter CD36 was detected (Fig. 1D). Similarly, free fatty acids in the culture supernatants at day 6 were inversely correlated (r = −0.7110, P = 0.0318) with the expression of CD36. Taken together, these data indicate that tAFP-treated DCs rely on glycolysis and have a decreased ability to take up and oxidize fatty acids.
Figure 1.
tAFP exposure skews dendritic cell metabolism to glucose dependency. CD14+ monocytes were isolated from healthy donors and treated with IL4 + GM-CSF in the presence of OVA, nAFP, or tAFP to produce iDCs and in some experiments treated with IFNγ and LPS to produce mDCs. A, A representative gating scheme is shown to identify live cells containing puromycin, as well as expressing markers consistent with iDCs (HLA-DR and CD206) and mDCs (CD86). DCs treated with OVA (black), nAFP (blue), and tAFP (red) were clustered on the basis of their immune parameters by tSNE, as well as the percentage of live cells expressing HLA-DR, CD206, and CD86. The proportion of cells expressing HLA-DR, CD206, CD86, and ICOSLG is shown from 3 to 5 technical replicates from a single donor. B, Shown are three technical replicates of one healthy donor of a SCENITH assay to quantify the glucose dependency (black), mitochondrial dependency (blue), glycolytic capacity (red), and FAAO (purple) with mDCs treated with OVA (black), nAFP (blue), or tAFP (red). Glucose and concentrations of cellular supernatants and intracellular glucose are shown with three technical replicates from a single donor, as well as the intracellular glucose uptake as determined by the influx of the fluorescent glucose analog 2NDBG from a single replicate from a single donor. C, Shown are cellular supernatants of in vitro–generated DCs from three healthy donors performed in technical replicates, as well as correlations with glucose in the supernatant and mitochondrial size. D, The glucose supernatant concentrations, %pAMPK, and CD36 gMFI are shown from healthy donors (N = 3) treated with OVA, nAFP, or tAFP. The correlation between supernatant fatty acids and CD36 levels are shown. ns, nonsignificant; *, P ≤ 0.05.
In agreement with decreased mitochondrial capacity by SCENITH (Fig. 1B), we previously confirmed decreased mitochondrial mass in tAFP-treated DCs (30). With decreased mitochondrial activity and the DC reliance on glycolysis, we investigated the potential release of lactate. Given the immunoregulatory functions of lactate, we determined whether tAFP could promote lactate secretion by DC. Lactate was measured in the media of OVA, nAFP, and tAFP-treated DCs (Fig. 1D). In all HDs, tAFP-treated DC secreted the most lactate at approximately twice the concentration of OVA-treated DCs, which may in part explain tAFP-treated DCs diminished capacity to stimulate T cells (29). The increased concentrations of lactate inversely correlated with glucose in the supernatant (r = −0.9326, P = 0.0002), suggesting this build-up of lactate results from increased reliance on glycolysis, as opposed to oxidative phosphorylation or FAAO, for the production of ATP.
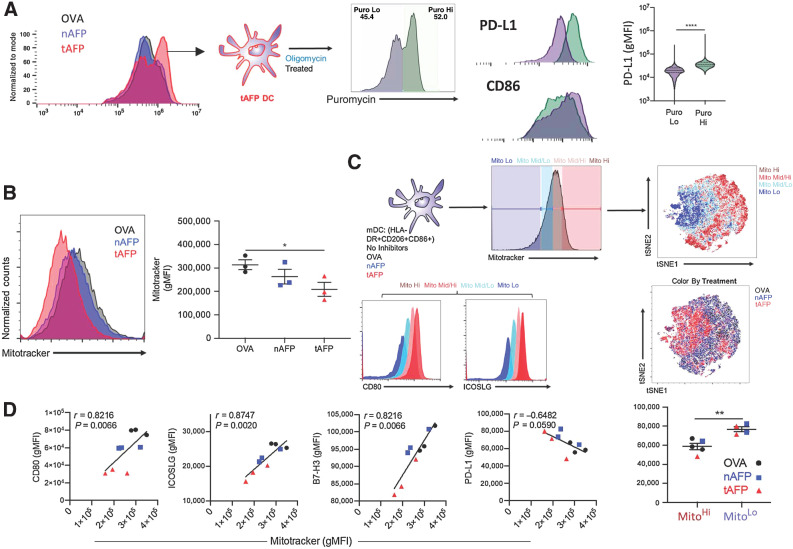
Given that tAFP induced both immune and metabolic changes, we examined correlations between costimulatory markers and metabolic state. Cells were gated on the basis relative mitochondrial mass (Fig. 2). As mitochondrial size decreased, the cells coalesced around a single cluster. To determine the impact of altered mitochondrial load on the expression of key costimulatory molecules, we determined the relative expression of activation markers (i.e., CD80 and ICOSLG) based on mitochondrial size. We observed strong positive correlations between mitochondrial size and the expression of CD80 (r = 0.8216, P = 0.0066), ICOSLG (r = 0.8747, P = 0.0020), and B7-H3/CD276 (r = 0.8216, P = 0.0066; Fig. 2C). Although there was a trend toward a negative correlation between mitochondrial size and PD-L1/CD274 (r = −0.6482, P = 0.0590), this reached statistical significance when directly comparing PD-L1 levels between MitoHi versus MitoLo cells (Fig. 2D). These findings indicate that decreased mitochondrial mass skews the cells toward glycolysis and is also associated with weaker expression of activating ligands (CD80, ICOSLG, B7-H3) and increased expression of inhibitory ligands (PD-L1).
Figure 2.
Mitochondria expression correlates with costimulatory molecule expression. A, Shown are puromycin histograms (a measurement of translation and a surrogate for ATP production) for OVA (black), nAFP (blue), or tAFP (red)-treated DCs. tAFP-treated DCs treated with oligomycin were separated into puromycin low (purple) and high (green). The expression levels of PD-L1 and CD86 are shown in the puromycin low and high DCs. B, Mitochondrial size, as measured by MitoTracker was determined in OVA, nAFP, and tAFP-treated DCs. C, mDCs treated with OVA, nAFP, or tAFP were characterized by mitochondrial size with high (dark red), mid/high (pink), mid/low (teal), and low (dark blue), and clustered on the basis of immune parameters by tSNE colored by mitochondrial size or treatment condition. Shown are the expression levels of CD80 and ICOSLG based on mitochondrial size. D, Correlations between MitoTracker and CD80, ICOSLG, B7-H3, and PD-L1 are shown. Differences in PD-L1 expression levels are shown between Mito hi and Mito lo cells color coded by treatment condition. All data are from one donor performed in 3 technical replicates. *, P ≤ 0.05; **, P ≤ 0.01; ****, P ≤ 0.0001.
Single-cell phenotypic profiling
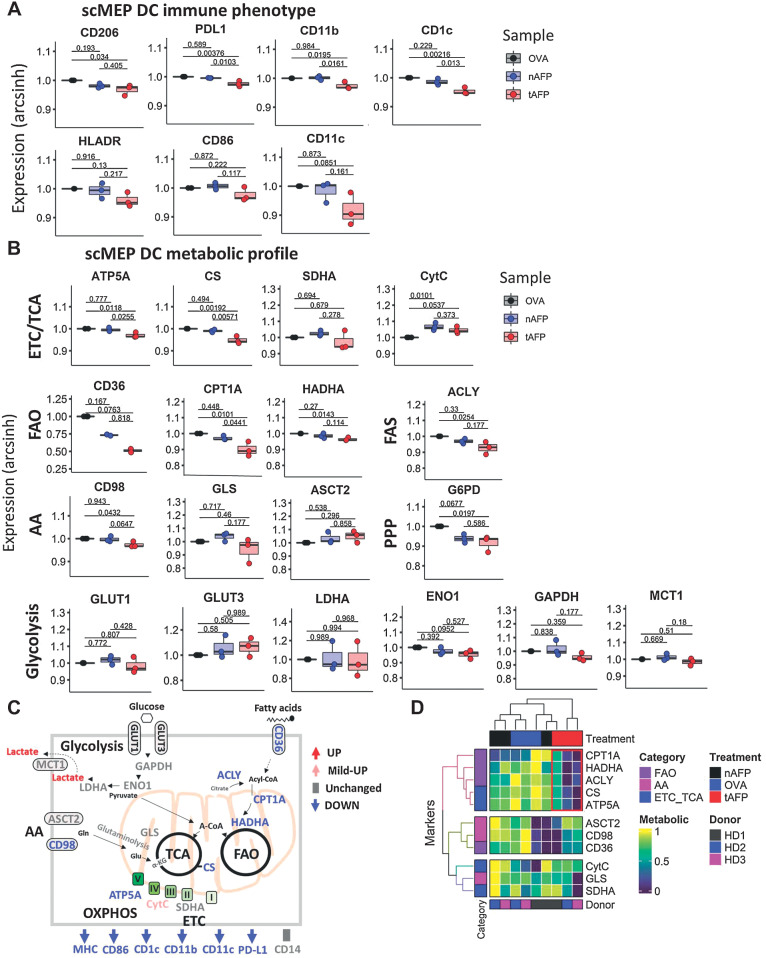
To further dissect the metabolic molecular pathways affected by AFP, we used the recently described single-cell metabolic regulome profiling (scMEP; ref. 42) on OVA, nAFP, and tAFP-treated cells. Consistent with a less differentiated phenotype and with our previous studies21,22, tAFP DC had reduced expression of DC markers, including CD206, PD-L1, CD11b, CD1c, HLA-DR, CD86, and CD11c (Fig. 3A). Multiple metabolic parameters related to the electron transport chain (ETC)/TCA, including ATP5A, CS and SDHA, were lower in tAFP-treated cells (Fig. 3B). However, levels of cytochrome C (CytC) were elevated in both nAFP and tAFP-treated DCs (Fig. 3B). Fatty acid oxidation (FAO) associated or fatty acid synthase (FAS) proteins CD36, CDPT1A, HADHA, and ACLY were decreased in tAFP-treated mDCs. Proteins involved in amino acid (AA) pathways had modest changes, CD98 and G6PD were significantly decreased and GLS tended to be lower, whereas ASCT2 was unchanged (Fig. 3B). Proteins involved in glycolysis (GLUT1, GLUT3, LDHA, EN01, GAPDH, and MTC1) displayed modest differences that were not statistically significant. A schematic summarizing the proteins that were upregulated, unchanged or downregulated is shown (Fig. 3C). A heatmap with hierarchical clustering on the basis of metabolic markers (FAO, AA, and ETC/TCA) and treatment condition (Fig. 3D). The tAFP-treated mDC cluster separately from the nAFP or OVA-treated mDCs. This clustering was due in large part to FAO and ETC/TCA proteins CPT1A, HADHA, ACLY, CS, and ATP5A.
Figure 3.
Immune and metabolic profile by scMEP. A and B, mDC were generated in vitro in the presence of OVA, nAFP, and tAFP and analysis of the immune-metabolic profile was determined by scMEP using CyTOF. Shown are the arcsinh-transformed values for immune response–related molecules (A) and metabolic pathway proteins (B). C, A schematic is shown to summarize the tAFP-induced immune-metabolic changes. D, A heatmap of various metabolic markers was generated and hierarchical clustering was performed on the basis of marker expression and treatment condition. The red box indicates the unique clustering of the tAFP-treated cells. Data are representative of three separate healthy donors, each performed in a single replicate.
tAFP induces transcriptional changes consistent with polyunsaturated fatty acid exposure
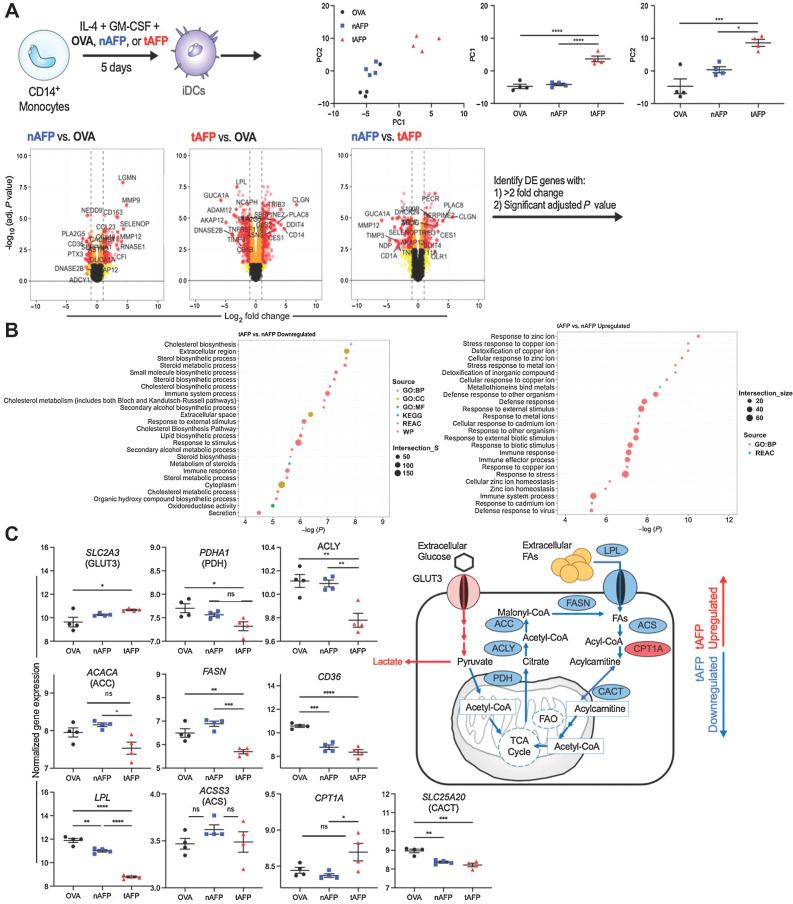
Given the AFP-induced protein level changes in multiple transcription factors, we determined the tAFP-induced transcriptional changes in DCs by microarray (Fig. 4). Principle component (PC) analysis revealed tAFP clustered separately from OVA and nAFP, and PC1 and PC2 were independently significant (Fig. 4A). We developed volcano plots for the three possible comparisons to determine differentially expressed genes (DEG). Consistent with the metabolic data, analysis revealed that pathways associated with lipid metabolism were significantly downregulated in tAFP compared with nAFP. The upregulated gene pathways demonstrated a stress response to metal ions, particularly zinc (Fig. 4B).
Figure 4.
DC gene expression profiles. A, Monocytes from healthy donors (N = 4) were differentiated into iDCs in the presence of OVA, nAFP, and tAFP and gene expression profiles were determined by microarray. Principle components were determined and color coded by treatment condition. Volcano plots were generated to identify genes that were differentially enriched (>2-fold change, significant adjusted P value) and that list of genes was used in a functional enrichment analysis by g:Profiler. B, Shown are predicted upregulated and downregulated pathways between tAFP and nAFP. C, Shown are differences in genes involved in glycolysis and fatty acid metabolism pathways, and a schematic summary of tAFP upregulated (red) and downregulated (blue) genes. Data are from four healthy donors, each performed once. ns, nonsignificant; *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001; ****, P ≤ 0.0001.
To dissect these classifications, we analyzed individual genes that could be important in promoting glycolysis and downregulated FA uptake or oxidation. Consistent with tAFP decreasing free glucose in the media and increasing lactate concentrations, we observed an increase in SLC2A3 (GLUT3) gene expression (Fig. 4C). In contrast, we observed a consistent tAFP-induced downregulation with genes involved in fatty acid metabolism, including genes encoding PDH, ACLY, ACC, FASN, LPL, CD36, and CACT (Fig. 4C). Interestingly, the ACS gene did not reach statistical significance, and tAFP increased the gene expression of CPT1A. Overall, these data provide strong orthogonal evidence in agreement with the SCENITH and scMEP experiments that tAFP alters fatty acid metabolism at the transcript level.
tAFP ligands are enriched for Zn and polyunsaturated fatty acids
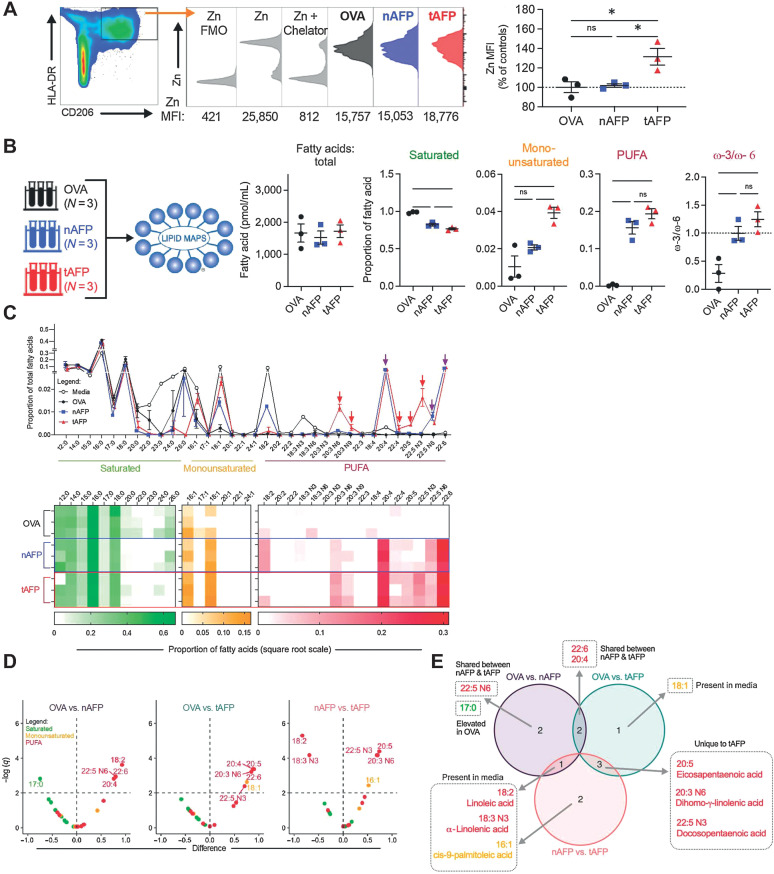
To further define the mechanism by which tAFP alters metabolism and function of DC (more so than nAFP), we examined the AFP-bound LMM ligands (29) that we previously demonstrated altered immunoregulatory properties of tAFP22. nAFP has been previously shown to bind more polyunsaturated fatty acids (PUFA; ref. 49) and Zn (39, 40) than albumin. Zn can induce tolerogenic DC (50), and PUFAs are known inhibitors of DC differentiation (51) and have previously been shown to limit lipid metabolism in hepatocytes, in particular through the direct downregulation of the FASN gene. These findings, taken together, suggest that the tAFP may bind more Zn and PUFAs compared with nAFP, and therefore result in more potent immunometabolic changes.
On the basis of the g:Profiler Zn gene signature (Fig. 4B), we quantified the amount of intracellular Zn in OVA, nAFP, and tAFP-treated iDCs. Monocytes were differentiated to iDCs in the presence of OVA, nAFP, or tAFP. The tAFP-treated DC had a statistically significant approximately 30% increase in Zn MFI compared with nAFP (P = 0.0293) or OVA-treated (P = 0.0228) iDCs (Fig. 5A). These findings are consistent the transcriptional data (Fig. 4), and demonstrate that tAFP is more efficient at increasing intracellular Zn concentrations in iDCs, when compared with OVA or nAFP.
Figure 5.
tAFP bound lipids are enriched for PUFAs. A, Levels of zinc were quantified in iDCs treated with OVA (black), nAFP (blue) or tAFP (red) in three technical replicates from a single donor and are shown as histograms as well as Zn MFIs (percentage of controls). B, Shown are the total mass of all bound fatty acids and the proportion that are saturated, monounsaturated, and polyunsaturated (PUFA) fatty acids. C, Displayed are the proportions of individual fatty acids present in CellGenix DC media, OVA, nAFP, or tAFP. Shown is a heatmap of the proportion of individual fatty acids bound to each protein. D, Volcano plots were generated on the basis of each protein compared with each other protein and color-coded on the basis of the class of fatty acid: saturated (green), monounsaturated (yellow), and PUFA (red). The horizontal dashed line indicates the significance threshold based on an FDR of 1%. E, A Euler diagram demonstrates the various combinations of differentially bound fatty acids (from C), indicating that they are present in the media and are unique or shared amongst the proteins. Error bars are based on mean ± SD. Statistical differences in the mass or proportion of saturated, monounsaturated, or polyunsaturated fatty acids were determined on the basis of a one-way ANOVA with Tukey multiple comparison test. Volcano plots were generated based upon unpaired t tests using a single pooled variance; multiple comparisons were accounted for using an FDR of 1% via a two-stage step-up. ns, nonsignificant; *, P ≤ 0.05.
OVA, nAFP, and tAFP LMM ligands were quantified by mass spectrometry and gas chromatography. The total quantity of bound fatty acids was similar among all proteins, with a mean concentration of approximately 1,500 pmol/mL. In contrast, tAFP bound less saturated fatty acids (SUFA; mean = 77%) compared with nAFP (82%, P = 0.0003) or OVA (99%, P < 0.0001). Although the amount of monounsaturated fatty acids (MUFA)was low among all proteins, tAFP bound 2- and 4-fold more MUFAs compared with OVA (P = 0.0042) and nAFP (P = 0.0318), respectively (Fig. 5B). PUFAs were greater on tAFP (P < 0.0001) and nAFP (P = 0.0003) when compared with OVA. On the basis the terminal double-bond location, PUFAs can be further divided into ω-3 and ω-6. Both tAFP (P = 0.0067) and nAFP (P = 0.0264) had greater ω-3:ω-6 ratios than OVA. Next, we examined each protein's ligand composition based upon the carbon length and the number of double bonds of each FA. Although we did not observe a bias based on FA length, nAFP and tAFP tended to bind FAs with 4 or more double bonds. Next, we analyzed the proportion of individual FAs from each protein (Fig. 5C). We hypothesized that FAs present in high quantities in the media would be unlikely to mediate tAFP's immunoregulatory properties, and FAs unique to tAFP would be compelling candidates. To identify FAs statistically unique to tAFP, we generated volcano plots for all three possible comparisons (Fig. 5D). Only a single fatty acid, 17:0, was increased on OVA. As expected, both nAFP and tAFP bound several PUFAs at a greater concentration relative to OVA. When comparing nAFP and tAFP, PUFAs were enriched on nAFP (18:2, 18:3 N3) and tAFP (16:1, 20:3 N6, 22:5 N3, 20:5). To determine which were shared or unique based on each comparison, a Euler diagram of all the differentially bound fatty acids was generated (Fig. 5E). Several of these differentially bound FAs were present in the media (18:2, 18:3 N3, 16:1, 18:1) or attached equally to nAFP and tAFP (20:4, 22:6, 22:5N6). However, the three fatty acids 20:5, 20:3 N6, 22:5N3 were statistically increased on tAFP and not present in the other comparisons.
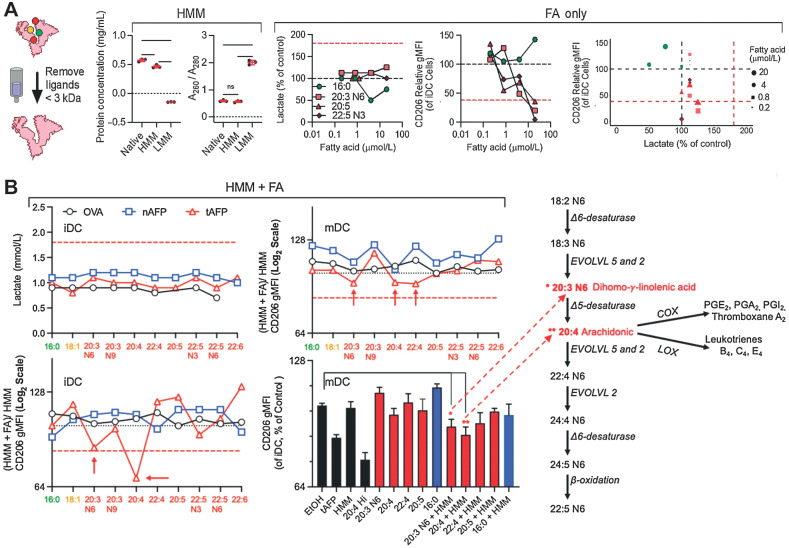
PUFA restore tAFP's suppression of DCs
We developed an in vitro assay to screen-specific FAs that are necessary for tAFP-mediated suppression of DC formation (Fig. 6A; Supplementary Fig. S2). Many of the FAs screened have known roles in promoting (52, 53) or limiting DC differentiation (51). Therefore, to determine their necessity for tAFP-mediated DC suppression, we titrated several FAs unique to tAFP (20:3 N6, 20:5, and 22:5 N3), and other PUFAs to determine the concentration they lost their inherent ability to suppress DC differentiation in the absence of OVA, nAFP, or tAFP (Fig. 6A). None of the FAs, at any concentration, tested induced production of lactate to levels caused by tAFP, indicating that lactate secretion and reduced costimulatory molecule expressions are separable immune suppression effects. Treatment with high concentrations (5–20 μmol/L) of 16:0 (palmitic acid) tended to decrease the production of lactate relative to control cells (black-dashed line), suggesting a less glycolytic phenotype (Fig. 6B). All three PUFAs at high concentrations inhibited CD206 expression on DCs (51), at levels equivalent to or greater than tAFP treatment. In contrast, the SUFA 16:0 (palmitic acid) tended to promote DC differentiation (52). All PUFAs lost immunoregulatory activity at the 0.2 μmol/L concentration (Fig. 6B).
Figure 6.
Low molar mass–binding partner screening. A, Low molar mass ligands were removed from OVA, nAFP, and tAFP. Fatty acids (FA) were titered onto iDCs and supernatant lactate was measured. Control levels are indicated by a black dashed line and native tAFP lactate induction indicated by a red dashed line. Levels of CD206 were also measured, with black and red dashed lines indicating control and native tAFP-treated cells, respectively. B, Individual fatty acids were added back to HMM-purified OVA, nAFP, and tAFP proteins and supernatant lactate, and CD206 levels were measured at the iDC and mDC stage. CD206 gMFI of the HMM + FA were normalized to the HMM only control. The red dashed line indicates the level of suppression seen with native tAFP. The red asterisks indicate the fatty acids most significantly associated with a decrease in CD206. Also shown is a schematic of their role in fatty acid metabolism. Data were performed with one to three technical replicates from one healthy donor. ns, nonsignificant.
Although all of the PUFAs could inhibit CD206 expression of DC, they did not robustly increase lactate production under these conditions; in contrast, the saturated FA palmitic acid was unique in its ability to increase CD206 expression and decrease lactate secretion (Fig. 6A). None of the FAs combined with either OVA, nAFP or tAFP induced lactate secretion comparable with native tAFP. When measuring CD206 expression at the iDC stage, we observed immunoregulatory activity with FAs 20:3 N6 and 20:4 when combined with tAFP—but not with OVA or nAFP (Fig. 6A). For mDC, we observed inhibition (∼15%) of CD206 expression with 20:3 N6, 20:4, and 22:4 when combined with tAFP (Fig. 6B). The more modest reduction at the matured DC (matDC) timepoint suggests that treatment with rIFNγ and LPS can partially, but not entirely reverse, the effects of HMM tAFP + PUFAs. When multiple FA + HMM tAFP were compared with controls, only 20:3N6 and 20:4 showed significantly reduced CD206 expression (Fig. 6B). Importantly, in the metabolism of ω-6 FAs, 20:3 N6 is converted to 20:4, which can then be further converted into a variety of molecules by COX and LOX enzymes (Fig. 6B). Taken together, these data suggest that PUFAs are necessary for tAFP inhibition of DC differentiation in vitro. Furthermore, these effects may result from exposure to increased 20:3 N6 and/or 20:4, COX/LOX enzymatic derivates of 20:4.
HCC patient monocyte and DC metabolic profiling
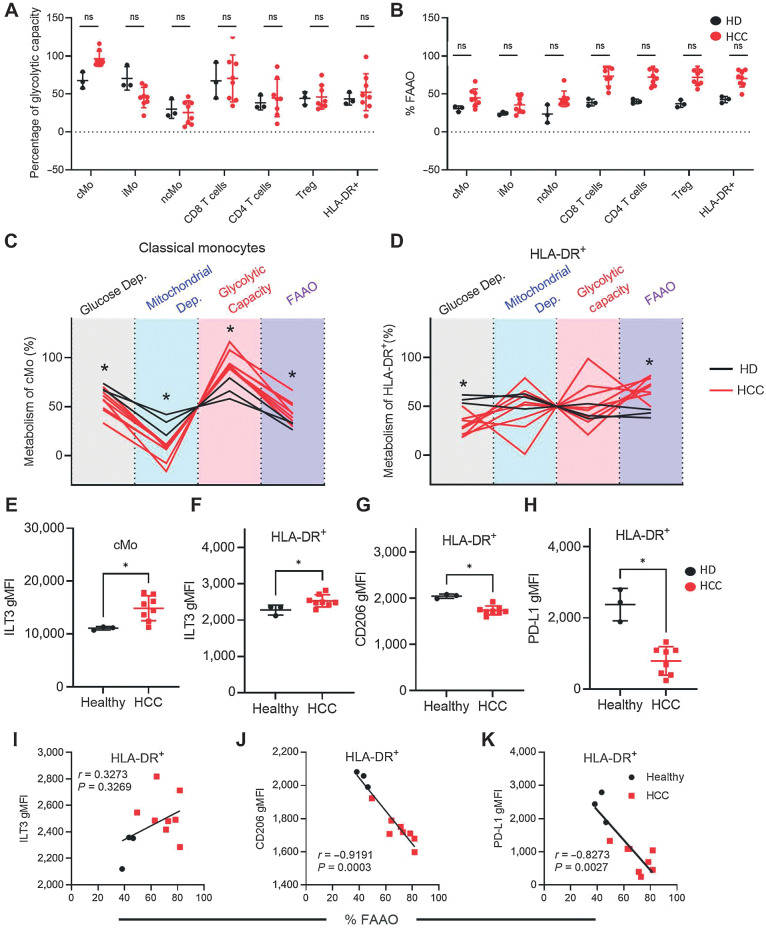
To better understand how these in vitro results extend to in vivo circulating patient myeloid cells, we measured the immunometabolic profile of patient with HCC and HD PBMCs. SCENITH was used to determine the percentage of glycolytic capacity and FAAO (Fig. 7A and B) across cell types. Monocytes were classified as classical (CD14+CD16−, cMo), intermediate (CD14+CD16+, iMo) or non-classical (CD14−CD16+, ncMo). Among the monocyte subsets, metabolic differences between HCC and HD were most consistent among the cMo (Fig. 7A and B). cMo from patients with HCC had decreased glucose and mitochondrial dependence and increased glycolytic capacity and FAAO compared with HD (Fig. 7C). Patient-derived classical monocytes resembled in vitro-differentiated DCs treated with AFP (Fig. 1B), showing decreased mitochondrial dependency and elevated glycolytic capacity. In contrast with the in vitro–generated DCs, patients with HCC had decreased glucose dependence and increased FAAO (Fig. 7D). These in vivo findings are in partial contrast with the prior in vitro data that suggested AFP-treated DCs were more glucose-dependent and had decreased FAAO (Fig. 1B). Regardless of the source of DCs, both in vitro treatment with tAFP or ex vivo derived from patients with HCC were associated with DCs retaining a more monocyte-like metabolic phenotype, consistent with tAFP impairing the immunometabolic reprogramming required for generating immunostimulatory DCs.
Figure 7.
HCC patient monocytes and DCs have a dysregulated immunometabolism. A and B, PBMCs were isolated from patients with HCC for SCENITH analysis. Shown are the percentages of glycolytic capacity (A) and percentage of FAAO (B) in multiple immune cell subsets. C and D, The total SCENITH metabolic profiles are shown for classic monocytes (C) and HLA-DR+ cells (D), with healthy donors (HD; black) and HCC (red). Statistically significant differences are indicated with an asterisk. E and F, Levels of ILT3 are indicated on classical monocytes (cMo; E) and HLA-DR-positive cells (F). G and H, In addition, shown are CD206 and PD-L1 expression levels on HLA-DR+ cells. I–K, Correlations between ILT3 (I), CD206 (J), and PD-L1 (K) are shown with percentage of FAAO in healthy donors (black circles) and patients with HCC (red squares). Data are presentative from three healthy donors and 8 patients with HCC. ns, nonsignificant; *, P ≤ 0.05.
Given the correlation between metabolic state and suppression of key stimulatory molecules (Fig. 2), we measured immune markers on cMo and DCs. The immunoglobulin-like transcript 3 (ILT3) is an important inhibitory receptor expressed on multiple myeloid cells, including monocytes and DCs (54–56). Consistent with a more immunoregulatory phenotype, HCC cMo (Fig. 7E) and DCs (Fig. 7F) expressed more ILT3 compared with HDs. Consistent with the in vitro data, CD206 was decreased in HCC patient DCs. Interestingly, and in contrast with the in vitro data, HCC DCs expressed less PD-L1 (Fig. 7H). Although this may suggest a less immunoregulatory phenotype because iDCs tend to express less PD-L1 than matDCs, it could also result from a blockade in full DC differentiation (57). To understand the connection between these immune markers and their metabolic profiles, we performed correlations between these immune markers on DCs and the %FAAO. Although patients with HCC DCs tended to have greater %FAAO and ILT3 expression, these variables were not significantly correlated (P = 0.3269, r = 0.3273, Fig. 7I).
The serum concentration of AFP in patient blood (Table 1) did not show a direct correlation with circulating myeloid cell phenotypes or metabolic profiles (not shown). This may indicate that the type or amount of tumor-associated ligands binding tAFP in vivo vary between patients and tumors. This was not entirely unexpected, as our previous clinical trial analyses of DC vaccination, and of NK, CD8+, and CD4+ T-cell activity in AFP+ and AFP− patients with HCC showed significant skewing and dysfunction that was not directly correlated to in vivo serum AFP concentrations (23, 24, 25, 58, 59, 60). Similarly, the tumor microenvironment concentrations of AFP are likely different from circulating concentrations.
Discussion
Here, using scMEP, we have identified key metabolic pathways and both transcriptional and protein-level regulatory molecules used by tAFP to suppress DC function. nAFP modestly increases glucose uptake and glucose-dependent metabolism and similarly reduces FAO. tAFP has a much more potent impact on DC metabolism, promoting a complete dysregulation of all measured metabolic pathways. tAFP-exposed DCs take up more glucose and secrete high levels of lactate, which is a well-recognized immune suppressive mediator (61, 62, 63). We recently showed that monocytes cultured in vitamin D3 to become functionally tolerogenic also have increased reliance on glycolysis and secrete high levels of lactate (48). Lactate blockade reversed the immune-suppressive phenotype of the tolerogenic DC. Here, we show that tAFP has a similar effect on DC.
We have also identified specific FA-binding partners of AFP that mediate some of these effects. These findings are consistent with groups that have shown that PUFAs inhibit DCs, some of these have been previously described (DPA, AA, and EPA) whereas others are newly described here (Dihomo-gamma). We also found consistent data that palmitic acid can promote DC differentiation and are the first to observe that palmitic acid can promote OxPhos. In our examination of the transcriptional pathways associated with Zn, our findings are consistent with groups who have shown Zn can induce tolerogenic DC. We now show that tAFP delivers more Zn intracellularly than nAFP and induces a glycolytic phenotype in DCs. Although AFP has been known to bind Zn, here, we report that the Zn bound to tAFP is important for the observed glycolytic switch.
It is important to consider is that metabolism of in vitro cultured DC may not fully reflect cellular metabolism in circulating cells in vivo, given the high concentration of glucose commonly present in culture media. Our comparative SCENITH analysis of HD and HCC PBMC revealed DCs that more closely resembled the metabolic profile of monocytes than of DCs. Both in vitro-generated DCs treated with tAFP and ex vivo DCs from patients with HCC had decreased mitochondrial dependency and increased glycolytic capacity when compared with controls. However, though in vitro generated DCs had increased glucose dependency and decreased FAAO compared with controls, we saw the opposite pattern in HCC-derived DCs. Despite the similarities and differences between the in vitro and ex vivo DCs, in both instances, DCs treated with tAFP or derived from patients with HCC more closely resembled their respective monocyte metabolic profiles. These findings are consistent with tAFP limiting the immune-metabolic reprogramming during monocyte differentiation yielding DCs retaining monocyte profiles. Of note, there are several drugs (including TPST1120; ref. 64) being studied in HCC to inhibit PPARa and FAO. Such an approach could negatively impact the myeloid compartment and immune reactivity while targeting metabolic dysfunction in tumor cells, which could be investigated.
These data provide mechanistic insights on how AFP antagonizes the innate immune response to limit antitumor immunity in vivo. Understanding the impact of tAFP on the tumor immune microenvironment may inform the development of future immune checkpoint inhibition combination strategies in HCC overall and in the subset of patients with high tumor AFP expression. Furthermore, these data suggest novel strategies to generate more potent DC vaccines for patients with HCC, including supplementing culture media with SUFAs and inclusion of Zn chelators.
Supplementary Material
Supplementary Figures, Table, Supplementary Tables
Acknowledgments
The authors wish to acknowledge the Hepatobiliary Tissue Bank Registry (HBTBR), which receives funding from the Bili Project Foundation, Inc. and the Cholangiocarcinoma Foundation; Dr. O. Quehenberger and Milda Simonaitis of the UCSD Lipidomics Core for the quantification of fatty acids; and Vinh Nguyen and Ashley Carlos of the UCSF Flow Cytometry Core for their consultation on flow cytometry assays (Ashley Carlos). This work was generously supported by the Parker Institute for Cancer Immunotherapy (PICI).
The publication costs of this article were defrayed in part by the payment of publication fees. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Footnotes
Note: Supplementary data for this article are available at Cancer Research Online (http://cancerres.aacrjournals.org/).
Authors' Disclosures
A.J. Combes reports grants from ImmunoX during the conduct of the study and grants from Genentech, Eli Lilly, and Corbus outside the submitted work. R.K. Kelley reports grants from Agios, Astra Zeneca, Bayer, BMS, Eli Lilly, EMD Serono, Exelixis, as well as other support from Ipsen, and grants from Genentech/Roche, Merck, Novartis, Partner Therapeutics, QED, Relay Therapeutics, Surface Oncology, and personal fees from Kinnate and from Compass Therapeutics outside the submitted work. L.H. Butterfield reports grants from Parker Institute for Cancer Immunotherapy during the conduct of the study. No disclosures were reported by the other authors.
Authors' Contributions
P.V. Munson: Conceptualization, data curation, investigation, writing–original draft. J. Adamik: Conceptualization, data curation, formal analysis, investigation, writing–original draft, writing–review and editing. F.J. Hartmann: Investigation. P.M.B. Favaro: Investigation. D. Ho: Investigation. S.C. Bendall: Resources, supervision, writing–review and editing. A.J. Combes: Resources, investigation, writing–review and editing. M.F. Krummel: Resources, supervision, investigation. K. Zhang: Resources, writing–review and editing. R.K. Kelley: Resources, supervision, writing–original draft, writing–review and editing. L.H. Butterfield: Conceptualization, resources, supervision, funding acquisition, writing–original draft, project administration, writing–review and editing.
References
- 1. Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2021;71:209–49. [DOI] [PubMed] [Google Scholar]
- 2. Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin 2011;61:69–90. [DOI] [PubMed] [Google Scholar]
- 3. Bertuccio P, Turati F, Carioli G, Rodriguez T, La Vecchia C, Malvezzi M, et al. Global trends and predictions in hepatocellular carcinoma mortality. J Hepatol 2017;67:302–9. [DOI] [PubMed] [Google Scholar]
- 4. R Team, LaBrecque DR, Abbas Z, Anania F, Ferenci P, Khan AG, et al. World Gastroenterology Organisation global guidelines: nonalcoholic fatty liver disease and nonalcoholic steatohepatitis. J Clin Gastroenterol 2014;48:467–73. [DOI] [PubMed] [Google Scholar]
- 5. Llovet JM, Kelley RK, Villanueva A, Singal AG, Pikarsky E, Roayaie S, et al. Hepatocellular carcinoma. Nat Rev Dis Primer 2021;7:6. [DOI] [PubMed] [Google Scholar]
- 6. Bruix J, Qin S, Merle P, Granito A, Huang Y-H, Bodoky G, et al. Regorafenib for patients with hepatocellular carcinoma who progressed on sorafenib treatment (RESORCE): a randomised, double-blind, placebo-controlled, phase 3 trial. Lancet Lond Engl 2017;389:56–66. [DOI] [PubMed] [Google Scholar]
- 7. Kudo M, Finn RS, Qin S, Han K-H, Ikeda K, Piscaglia F, et al. Lenvatinib versus sorafenib in first-line treatment of patients with unresectable hepatocellular carcinoma: a randomised phase 3 non-inferiority trial. Lancet Lond Engl 2018;391:1163–73. [DOI] [PubMed] [Google Scholar]
- 8. Finn RS, Qin S, Ikeda M, Galle PR, Ducreux M, Kim T-Y, et al. Atezolizumab plus bevacizumab in unresectable hepatocellular carcinoma. N Engl J Med 2020;382:1894–905. [DOI] [PubMed] [Google Scholar]
- 9. Finn RS, Cheng A-L. Atezolizumab and bevacizumab in hepatocellular carcinoma. reply. N Engl J Med 2020;383:695. [DOI] [PubMed] [Google Scholar]
- 10. Yau T, Kang Y-K, Kim T-Y, El-Khoueiry AB, Santoro A, Sangro B, et al. Efficacy and safety of nivolumab plus ipilimumab in patients with advanced hepatocellular carcinoma previously treated with sorafenib: the CheckMate 040 randomized clinical trial. JAMA Oncol 2020;6:e204564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Zhu AX, Kang Y-K, Yen C-J, Finn RS, Galle PR, Llovet JM, et al. Ramucirumab after sorafenib in patients with advanced hepatocellular carcinoma and increased α-fetoprotein concentrations (REACH-2): a randomised, double-blind, placebo-controlled, phase 3 trial. Lancet Oncol 2019;20:282–96. [DOI] [PubMed] [Google Scholar]
- 12. Llovet JM, Ricci S, Mazzaferro V, Hilgard P, Gane E, Blanc J-F, et al. Sorafenib in advanced hepatocellular carcinoma. N Engl J Med 2008;359:378–90. [DOI] [PubMed] [Google Scholar]
- 13. Finn RS, Qin S, Ikeda M, Galle PR, Ducreux M, Kim T-Y, et al. IMbrave150: updated overall survival (OS) data from a global, randomized, open-label phase III study of atezolizumab (atezo) + bevacizumab (bev) versus sorafenib (sor) in patients (pts) with unresectable hepatocellular carcinoma (HCC). J Clin Oncol 2021;39:267–. [Google Scholar]
- 14. Finn RS, Ikeda M, Zhu AX, Sung MW, Baron AD, Kudo M, et al. Phase Ib study of lenvatinib plus pembrolizumab in patients with unresectable hepatocellular carcinoma. J Clin Oncol Off J Am Soc Clin Oncol 2020;38:2960–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kelley RK, Sangro B, Harris W, Ikeda M, Okusaka T, Kang Y-K, et al. Safety, efficacy, and pharmacodynamics of tremelimumab plus durvalumab for patients with unresectable hepatocellular carcinoma: randomized expansion of a phase I/II study. J Clin Oncol Off J Am Soc Clin Oncol 2021;39:2991–3001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Abou-Alfa GK, Lau G, Kudo M, Chan SL, Kelley RK, Furuse J, et al. Tremelimumab plus durvalumab in unresectable hepatocellular carcinoma. NEJM Evid 2022;1. Available from: https://evidence.nejm.org/doi/10.1056/EVIDoa2100070. [DOI] [PubMed] [Google Scholar]
- 17. Bai D-S, Zhang C, Chen P, Jin S-J, Jiang G-Q. The prognostic correlation of AFP level at diagnosis with pathological grade, progression, and survival of patients with hepatocellular carcinoma. Sci Rep 2017;7:12870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hoshida Y, Moeini A, Alsinet C, Kojima K, Villanueva A. Gene signatures in the management of hepatocellular carcinoma. Semin Oncol 2012;39:473–85. [DOI] [PubMed] [Google Scholar]
- 19. Montal R, Andreu-Oller C, Bassaganyas L, Esteban-Fabró R, Moran S, Montironi C, et al. Molecular portrait of high alpha-fetoprotein in hepatocellular carcinoma: implications for biomarker-driven clinical trials. Br J Cancer 2019;121:340–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Tatarinov IS. [Detection of embryo-specific alpha-globulin in the blood serum of a patient with primary liver cancer]. Vopr Med Khim 1964;10:90–1. [PubMed] [Google Scholar]
- 21. Kelley RK, Meyer T, Rimassa L, Merle P, Park J-W, Yau T, et al. Serum alpha-fetoprotein levels and clinical outcomes in the phase III CELESTIAL study of cabozantinib versus placebo in patients with advanced hepatocellular carcinoma. Clin Cancer Res Off J Am Assoc Cancer Res 2020;26:4795–804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Farinati F, Marino D, De Giorgio M, Baldan A, Cantarini M, Cursaro C, et al. Diagnostic and prognostic role of alpha-fetoprotein in hepatocellular carcinoma: both or neither? Am J Gastroenterol 2006;101:524–32. [DOI] [PubMed] [Google Scholar]
- 23. Butterfield LH, Koh A, Meng W, Vollmer CM, Ribas A, Dissette V, et al. Generation of human T-cell responses to an HLA-A2.1–restricted peptide epitope derived from alpha-fetoprotein. Cancer Res 1999;59:3134–42. [PubMed] [Google Scholar]
- 24. Butterfield LH, Ribas A, Meng WS, Dissette VB, Amarnani S, Vu HT, et al. T-cell responses to HLA-A*0201 immunodominant peptides derived from α-fetoprotein in patients with hepatocellular cancer. Clin Cancer Res 2003;9:5902–8. [PubMed] [Google Scholar]
- 25. Butterfield LH, Ribas A, Dissette VB, Lee Y, Yang JQ, De la Rocha P, et al. A phase I/II trial testing immunization of hepatocellular carcinoma patients with dendritic cells pulsed with four alpha-fetoprotein peptides. Clin Cancer Res 2006;12:2817–25. [DOI] [PubMed] [Google Scholar]
- 26. Vujanovic L, Stahl EC, Pardee AD, Geller DA, Tsung A, Watkins SC, et al. Tumor-derived α-fetoprotein directly drives human natural killer–cell activation and subsequent cell death. Cancer Immunol Res 2017;5:493–502. [DOI] [PubMed] [Google Scholar]
- 27. Lu CY, Changelian PS, Unanue ER. Alpha-fetoprotein inhibits macrophage expression of Ia antigens. J Immunol 1984;132:1722–7. [PubMed] [Google Scholar]
- 28. Aussel C, Fehlmann M. α-Fetoprotein stimulates leukotriene synthesis in P388D1 macrophages. Cell Immunol 1986;101:415–24. [DOI] [PubMed] [Google Scholar]
- 29. Pardee AD, Shi J, Butterfield LH. Tumor-derived α-fetoprotein impairs the differentiation and T-cell stimulatory activity of human dendritic cells. J Immunol 2014;193:5723–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Santos PM, Menk AV, Shi J, Tsung A, Delgoffe GM, Butterfield LH. Tumor-derived α-fetoprotein suppresses fatty acid metabolism and oxidative phosphorylation in dendritic cells. Cancer Immunol Res 2019;7:1001–12. [DOI] [PubMed] [Google Scholar]
- 31. Docta RY, Ferronha T, Sanderson JP, Weissensteiner T, Pope GR, Bennett AD, et al. Tuning T-cell receptor affinity to optimize clinical risk-benefit when targeting alpha-fetoprotein–positive liver cancer. Hepatology 2019;69:2061–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Zhu W, Peng Y, Wang L, Hong Y, Jiang X, Li Q, et al. Identification of α-fetoprotein–specific T-cell receptors for hepatocellular carcinoma immunotherapy. Hepatology 2018;68:574–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Vessella RL, Santrach MA, Bronson D, Smith CJP, Klicka MJ, Lange PH. Evaluation of AFP glycosylation heterogeneity in cancer patients with AFP-producing tumors. Int J Cancer 1984;34:309–14. [DOI] [PubMed] [Google Scholar]
- 34. Aoyagi Y, Suzuki Y, Isemura M, Nomoto M, Sekine C, Igarashi K, et al. The fucosylation index of alpha-fetoprotein and its usefulness in the early diagnosis of hepatocellular carcinoma. Cancer 1988;61:769–74. [DOI] [PubMed] [Google Scholar]
- 35. Lamerz R. AFP isoforms and their clinical significance (overview). Anticancer Res 1997;17:2927–30. [PubMed] [Google Scholar]
- 36. Mizejewski GJ. Alpha-fetoprotein structure and function: relevance to isoforms, epitopes, and conformational variants. Exp Biol Med Maywood NJ 2001;226:377–408. [DOI] [PubMed] [Google Scholar]
- 37. Burditt LJ, Johnson MM, Johnson PJ, Williams R. Detection of hepatocellular carcinoma- specific alpha-fetoprotein by isoelectric focusing. Cancer 1994;74:25–9. [DOI] [PubMed] [Google Scholar]
- 38. Benassayag C, Vallette G, Delorme J, Savu L, Nunez EA. High affinity of nonesterified polyunsaturated fatty acids for rat alpha-fetoprotein (AFP). Oncodev Biol Med 1980;1:27–36. [PubMed] [Google Scholar]
- 39. Wu JT, Monir-Vaghefi SM, Clayton F. Human alpha-fetoprotein and albumin: differences in zinc binding. Clin Physiol Biochem 1987;5:85–94. [PubMed] [Google Scholar]
- 40. Permyakov SE, Oberg KA, Cherskaya AM, Shavlovsky MM, Permyakov EA, Uversky VN. Human alpha-fetoprotein as a Zn(2+)-binding protein. Tight cation binding is not accompanied by global changes in protein structure and stability. Biochim Biophys Acta 2002;1586:1–10. [DOI] [PubMed] [Google Scholar]
- 41. Argüello RJ, Combes AJ, Char R, Gigan J-P, Baaziz AI, Bousiquot E, et al. SCENITH: a flow cytometry-based method to functionally profile energy metabolism with single-cell resolution. Cell Metab 2020;32:1063–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Hartmann FJ, Mrdjen D, McCaffrey E, Glass DR, Greenwald NF, Bharadwaj A, et al. Single-cell metabolic profiling of human cytotoxic T cells. Nat Biotechnol 2021;39:186–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Hahne F, LeMeur N, Brinkman RR, Ellis B, Haaland P, Sarkar D, et al. flowCore: a Bioconductor package for high throughput flow cytometry. BMC Bioinformatics. 2009;10:106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Nowicka M, Krieg C, Crowell HL, Weber LM, Hartmann FJ, Guglietta S, et al. CyTOF workflow: differential discovery in high-throughput high-dimensional cytometry datasets. F1000Research. 2017;6:748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Raudvere U, Kolberg L, Kuzmin I, Arak T, Adler P, Peterson H, et al. g: Profiler: a web server for functional enrichment analysis and conversions of gene lists (2019 update). Nucleic Acids Res 2019;47:W191–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Quehenberger O, Armando AM, Brown AH, Milne SB, Myers DS, Merrill AH, et al. Lipidomics reveals a remarkable diversity of lipids in human plasma. J Lipid Res 2010;51:3299–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Alsabeeh N, Chausse B, Kakimoto PA, Kowaltowski AJ, Shirihai O. Cell culture models of fatty acid overload: problems and solutions. Biochim Biophys Acta 2018;1863:143–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Adamik J, Munson PV, Hartmann FJ, Combes AJ, Pierre P, Krummel MF, et al. Distinct metabolic states guide maturation of inflammatory and tolerogenic dendritic cells. Nat Commun 2022;13:5184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Parmelee DC, Evenson MA, Deutsch HF. The presence of fatty acids in human alpha-fetoprotein. J Biol Chem 1978;253:2114–9. [PubMed] [Google Scholar]
- 50. George MM, Subramanian Vignesh K, Landero Figueroa JA, Caruso JA, Deepe GS. Zinc induces dendritic cell tolerogenic phenotype and skews regulatory T-cell-Th17 balance. J Immunol Baltim Md 1950 2016;197:1864–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Zeyda M, Säemann MD, Stuhlmeier KM, Mascher DG, Nowotny PN, Zlabinger GJ, et al. Polyunsaturated fatty acids block dendritic cell activation and function independently of NF-κB activation. J Biol Chem 2005;280:14293–301. [DOI] [PubMed] [Google Scholar]
- 52. Weatherill AR, Lee JY, Zhao L, Lemay DG, Youn HS, Hwang DH. Saturated and polyunsaturated fatty acids reciprocally modulate dendritic cell functions mediated through TLR4. J Immunol 2005;174:5390–7. [DOI] [PubMed] [Google Scholar]
- 53. Lancaster GI, Langley KG, Berglund NA, Kammoun HL, Reibe S, Estevez E, et al. Evidence that TLR4 is not a receptor for saturated fatty acids but mediates lipid-induced inflammation by reprogramming macrophage metabolism. Cell Metab 2018;27:1096–110.e5. [DOI] [PubMed] [Google Scholar]
- 54. Cella M, Döhring C, Samaridis J, Dessing M, Brockhaus M, Lanzavecchia A, et al. A novel inhibitory receptor (ILT3) expressed on monocytes, macrophages, and dendritic cells involved in antigen processing. J Exp Med 1997;185:1743–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Manavalan JS, Rossi PC, Vlad G, Piazza F, Yarilina A, Cortesini R, et al. High expression of ILT3 and ILT4 is a general feature of tolerogenic dendritic cells. Transpl Immunol 2003;11:245–58. [DOI] [PubMed] [Google Scholar]
- 56. Chang CC, Ciubotariu R, Manavalan JS, Yuan J, Colovai AI, Piazza F, et al. Tolerization of dendritic cells by T(S) cells: the crucial role of inhibitory receptors ILT3 and ILT4. Nat Immunol 2002;3:237–43. [DOI] [PubMed] [Google Scholar]
- 57. Pen JJ, Keersmaecker BD, Heirman C, Corthals J, Liechtenstein T, Escors D, et al. Interference with PD-L1/PD-1 co-stimulation during antigen presentation enhances the multifunctionality of antigen-specific T cells. Gene Ther 2014;21:262–71. [DOI] [PubMed] [Google Scholar]
- 58. Liu Y, Daley S, Evdokimova VN, Zdobinski DD, Potter DM, Butterfield LH. Hierarchy of alpha fetoprotein (AFP)-specific T-cell responses in subjects with AFP-positive hepatocellular cancer. J Immunol Baltim Md 1950 2006;177:712–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Evdokimova VN, Butterfield LH. Alpha-fetoprotein and other tumour-associated antigens for immunotherapy of hepatocellular cancer. Expert Opin Biol Ther 2008;8:325–36. [DOI] [PubMed] [Google Scholar]
- 60. Evdokimova VN, Liu Y, Potter DM, Butterfield LH. AFP-specific CD4+ helper T-cell responses in healthy donors and HCC patients. J Immunother Hagerstown Md 1997 2007;30:425–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Certo M, Tsai C-H, Pucino V, Ho P-C, Mauro C. Lactate modulation of immune responses in inflammatory versus tumour microenvironments. Nat Rev Immunol 2021;21:151–61. [DOI] [PubMed] [Google Scholar]
- 62. de la Cruz-López KG, Castro-Muñoz LJ, DO R-H, García-Carrancá A, Manzo-Merino J. Lactate in the regulation of tumor microenvironment and therapeutic approaches. Front Oncol 2019;9:1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Hirschhaeuser F, Sattler UGA, Mueller-Klieser W. Lactate: a metabolic key player in cancer. Cancer Res 2011;71:6921–5. [DOI] [PubMed] [Google Scholar]
- 64. Yarchoan M, Powderly JD, Bastos BR, Karasic TB, Crysler OV, Munster PN, et al. A phase 1 study of TPST-1120 as a single agent and in combination with nivolumab in subjects with advanced solid tumors. J Clin Oncol 2022;40:3005–. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figures, Table, Supplementary Tables
Data Availability Statement
The data generated in this study as well as data from prior publications are available upon request from the corresponding author. The array data discussed in this publication were previously deposited in NCBI's Gene Expression Omnibus and are accessible through GEO Series accession number GSE62005 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE62005).